Abstract
Pseudomonas aeruginosa is an opportunistic pathogen responsible for chronic, drug-resistant lung infections in individuals with cystic fibrosis (CF). Although extensive heterogeneity in antimicrobial resistance (AMR) phenotypes of P. aeruginosa CF lung populations has been previously described, there has yet to be a thorough investigation on how genomic diversification drives the evolution of AMR diversity within a population. In this study, we harnessed sequencing from a collection of 300 clinical isolates of P. aeruginosa to unravel the evolution of resistance diversity in four individuals with CF. We found that genomic diversity was not always a reliable predictor of phenotypic AMR diversity within a population, and notably, the least genetically diverse population in this cohort displayed AMR diversity comparable to that of populations with up to two orders of magnitude more SNPs. Hypermutator strains often displayed increased sensitivity to antimicrobials, even when there was a history of use of antimicrobial in the treatment of the patient. Lastly, we sought to determine whether diversity in AMR could be explained by evolutionary trade-offs with other traits. Our results showed no strong evidence of collateral sensitivity between aminoglycoside, beta-lactam, or fluoroquinolone antibiotics within these populations. Additionally, there was no evidence of trade-offs between AMR and growth in a sputum-mimicking environment. Overall, our findings highlight that (i) genomic diversity within a population is not a necessary precursor to phenotypic diversity in AMR; (ii) hypermutator populations can evolve increased sensitivity to antimicrobials even under apparent antibiotic selection; and that (iii) resistance to a single antibiotic may not impose enough of a fitness cost to elicit trade-offs with fitness.
Keywords: population heterogeneity, antibiotic resistance, hypermutation, evolution
INTRODUCTION
Pseudomonas aeruginosa is a dominant bacterial pathogen in chronic infections of the airways of adults with cystic fibrosis (CF), a genetic disorder that results in thickened mucus, persistent lung infection, and progressive decline in lung function (1). P. aeruginosa has multiple intrinsic and acquired mechanisms of antimicrobial resistance (AMR), with clinical strains frequently displaying multi-drug resistance (MDR). While antibiotic treatment can be effective against early-stage, transient P. aeruginosa infections, antibiotic regimens ultimately fail to eradicate chronic infections of P. aeruginosa from the CF lung (2). This may be due to the high degree of phenotypic and genomic heterogeneity that naturally evolves in P. aeruginosa populations inhabiting CF airways (3, 4), allowing the population to exploit various pathways of resistance and for the emergence of rare clones that evade treatment and re-establish infection afterwards (5, 6). Most individuals with CF are initially infected by a single environmental or transmissible epidemic strain of P. aeruginosa, which then diversifies in the CF lung over the course of many years of infection (7). Mutations in DNA mismatch repair (MMR) mechanisms act as a catalyst for this diversification, potentially providing an evolutionary advantage in an environment that demands rapid adaptation for survival, though potentially at a fitness cost (8, 9).
Maintaining diversity in populations can be advantageous for bet-hedging in a complex infection environment where there are a multitude of external stressors such as competing microbiota, antibiotic exposure, and host immune responses. Heterogeneity in populations may develop as individual members of the population evolve specialized functions to occupy different ecological niches (10); however, adaptations to a particular niche may come at an expense to other energetically costly traits (i.e., fitness costs) (11, 12). The vast diversity of P. aeruginosa in CF lung infection suggests that individual isolates within the population could have different specializations resulting in trade-offs with other traits. Of particular interest is collateral sensitivity— increased sensitivity to one antimicrobial as a trade-off with increased resistance to another— as a potential avenue for targeting drug-resistant populations using combination therapy or antibiotic cycling.
Although P. aeruginosa population diversity in the CF lung is widely accepted, this diversity is often overlooked. Within-host adaptations of P. aeruginosa to the CF lung have previously been investigated and described, primarily via longitudinal single-isolate sampling (13–23). Longitudinal sampling of single or small subsets of isolates from a population only reflects a fraction of the total evolutionary pathways exhibited within a population and may result in significant underestimation of the diversity of antimicrobial susceptibility profiles. As population diversity may impact infection outcomes via heteroresistance (24), microbial social interactions (25, 26), or the ability of a population to survive evolutionary bottlenecks (2), this warrants a shift in our sampling and susceptibility testing of chronic microbial infections to reflect our understanding of them as complex, dynamic populations. A few studies have thoroughly investigated population diversity in this infection context, in which their analyses were focused on (i) phenotypic diversity (27–30), (ii) genetic analyses via pooled population sequencing (31, 32), or (iii) both extensive sequencing and phenotyping, but lacking analysis linking the two at the isolate-level (4). As a result, we still have an incomplete understanding of how genomic diversification drives AMR heterogeneity within a population, and what trade-offs are involved in these evolutionary processes.
Here, we investigated the evolution of AMR diversity for chronic P. aeruginosa lung populations in four unique individuals with CF. We sought to test whether genomic diversity is a strong predictor of phenotypic diversity in AMR within a population. We further explored the role that hypermutation plays in driving resistance, specific links between genotype and phenotype at the isolate-level, and enrichments in mutations and gene content changes relevant to AMR. Lastly, we searched for evidence that resistance to one antimicrobial may trade-off with susceptibility to other antimicrobials and fitness in a CF-like environment.
METHODS
Cohort selection and strain isolation.
We selected four adult individuals for this study from a cohort of CF patients at Emory University in Atlanta who were chronically infected with P. aeruginosa at the time of sampling. From each patient, we collected and processed a single expectorated sputum sample. We processed sputum by supplementing each sample with 5 ml synthetic cystic fibrosis medium (SCFM) (33) and autoclaved glass beads, homogenizing the mixture via vortexing for 2 mins, centrifuging the homogenized sputum mixture for 4 mins at 4200 rpm, removing the supernatant, and conducting a 10x serial dilution of cell pellet re-suspended in phosphate buffered saline to streak on Pseudomonas isolation agar (PIA) plates. These plates were incubated at 37°C overnight, then at room temperature for up to 72 h. From each expectorated sputum sample, we randomly isolated 75 P. aeruginosa colonies for a total of 300 isolates.
Whole genome sequencing.
To conduct sequencing, we first grew all 300 isolates overnight in 15 ml conical tubes in lysogeny broth (LB) at 37°C with shaking at 200 rpm. We extracted DNA from these cultures using the Promega Wizard Genomic DNA Purification Kit according to the manufacturer’s instructions. We prepared sequencing libraries using the Nextera XT DNA Library Preparation Kit and used the Illumina Novaseq platform to obtain 250 bp paired-end reads for a mean coverage of 70x. 28 samples either failed or did not meet the minimum sequencing coverage or quality requirements, so we re-sequenced these using the Illumina MiSeq platform for 250 bp paired-end reads and combined the reads from both sequencing runs to analyze these 28 samples. We randomly selected one isolate from each patient to serve as the reference strain for the other 74 isolates isolated from that patient. For these reference isolates, we additionally obtained Oxford Nanopore long read sequences through the Microbial Genome Sequencing Center (GridION Flow Cell chemistry type R9.4.1 with Guppy high accuracy base calling v4.2.2) at 35x coverage.
Multi-locus sequence typing.
Our multi-locus sequence typing was implemented in Bactopia v1.6.5 (34), which employs the PubMLST.org schema (35).
Constructing annotated reference assemblies.
We used Unicyler v0.5.0 (36) to create long-read assemblies for the four reference isolates. We then conducted one round of long-read polishing on these assemblies using Medaka v1.0.3 (37), which produced preliminary consensus sequences. We conducted quality control on all 300 Illumina reads using the Bactopia v1.6.5 (34) pipeline. We conducted two further short-read assembly polishing steps on the long-read assemblies by aligning the quality-adjusted short reads of each of the four reference isolates to its respective consensus sequence using Polypolish v0.5.0 (38) and Pilon v1.24 (39). We validated the final consensus sequences by mapping the Illumina reads of each reference to its respective assembly using Snippy v4.6.0 (40) and confirming that 0 variants were called. We used (i) Prokka v1.14.6 (41) and (ii) RATT v1.0.3 (42) to (i) annotate our reference strains using a P. aeruginosa pan-genome database collated by Bactopia, and to (ii) transfer gene annotations from PAO1 to their respective positions in each of the reference strains, respectively.
Variant calling.
We used Snippy v4.6.0 (39) to call variants from the other 296 isolates against their respective reference strain and create a core genome alignment. Using PhyML v3.3.20211231 (43), we created a maximum likelihood phylogeny. Then, using VCFtools v0.1.16 (44) and Disty McMatrixface v0.1.0 (45), we generated a pairwise SNP matrix for each patient. For Disty, we only considered alleles in the core genome and chose to ignore ambiguous bases pairwisely (-s 0). We then employed SnpEff and SnpSift v4.3t (46) to identify the affected genes and sort the variants by predicted effect. We identified hypermutators in these populations by the presence of non-synonymous mutations in mutL, mutS, and uvrD.
Antimicrobial susceptibility testing.
To assess antimicrobial susceptibility profiles, we followed the guidelines and standards provided by the Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing M100S, 30th edition. We first grew all isolates overnight in LB in 24-well microtiter plates at 37°C with shaking at 200 rpm. We diluted cultures to a Macfarland standard of 0.5 (OD600 ~0.06) and streaked a lawn on 100×15 mm Petri dishes with 20 ml Mueller-Hinton agar using pre-sterilized cotton swabs. We then stamped amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ) on each plate and incubated for 17 h at 37°C. We measured the zone of inhibition at 17 h and classified the values as resistant, intermediate, or susceptible per the established CLSI interpretive criteria. We used P. aeruginosa strain ATCC 27853 as a quality control. We tested all isolates in biological triplicates. We ran a Mann-Whitney U test to compare the means of antimicrobial susceptibilities between hypermutators and normomutators (non-hypermutators) and a linear regression to determine relationships between susceptibilities to different antimicrobials, both using α = .05.
Resistome genotyping.
We assessed genotypes relevant to resistance by uploading the de novo assemblies to the Resistance Gene Identifier (RGI) v6.1.0 web portal, which predicts resistomes using the Comprehensive Antibiotic Resistance Database (CARD) v3.2.6 (43). We excluded loose and nudge hits from this analysis.
Enrichment analysis.
We conducted an enrichment analysis to determine which functional categories of genes were differentially impacted by mutations than would be expected by random chance. We used an in-house Python script to retrieve the PseudoCAP functional group of each gene where a non-synonymous SNP or microindel was identified. We accounted for the varying lengths of genes in each functional category in our analysis, based off their lengths and prevalence in the PAO1 genome. We used a chi-squared goodness of fit test to conduct the enrichment analyses for Patients 1–3 to determine which functional categories were disproportionately impacted by non-synonymous variants. We used the R package XNomial v1.0.4 (44) to conduct an exact multinomial goodness of fit test using Monte-Carlo simulations for Patient 4 because the SNP frequencies of Patient 4 did not meet the assumptions for a chi-squared test. Given the formula for calculating the chi-squared statistic: if the value for a particular PseudoCAP functional category was in the top 30 percentile of all values (top 8 of 27 total categories) in the analyses of at least three patients, we noted this as an enrichment.
Predicting putative recombination events.
We input the core genome alignment from each patient to Gubbins v3.3.0 (45) to predict potential recombinant regions in each population.
Analyzing gene content variability.
We used the Bactopia v1.6.5 (34) pipeline to process all paired-end Illumina reads. Following quality control, Bactopia implements Shovill v1.1.0 (46) to prepare de novo assemblies and Prokka v1.14.6 (41) to annotate these assemblies. We then used Roary v3.13.0 (47) to conduct a pangenome-style analysis and create a core genome alignment from the annotated assemblies. Our parameters for Roary were as follows: 90% identity threshold for BlastP, 98% prevalence for consideration in the core genome, and paralog splitting turned off. We further filtered gene families from our output that had a median length of <200 bp. We then implemented IQ-TREE v2.1.2 (48) to create a maximum likelihood phylogeny of the core genome alignment.
Analyzing growth curves.
To assess growth, we cultured strains in for 24 h in 96-well microtiter plates (Corning) at 37°C static, in 200μL synthetic cystic fibrosis sputum medium (SCFM) (33), shaking for 4 s before reading optical density at 600 nm every 20 min. We tested all clinical isolates in biological triplicates. We used GrowthCurver (49) to analyze the resulting growth curves and calculate growth rate (r). We then assessed the relationship between growth rate and susceptibility profiles using a linear mixed model in brms (50).
Visualizations.
We conducted graphical analyses in R v4.3.0. We employed a modified Python script from PIRATE v1.0.5 (51) to visualize the genomes.
Data availability.
The sequences in this study will be made available in the NCBI SRA database upon publication.
RESULTS
Description of the four patient cohort selected for this study.
The four individuals selected for this study were aged 24–31 years and had been chronically infected with P. aeruginosa for 10–15 years at the time of sampling. All four individuals had at least one copy of the F508del CFTR mutation, but none were on CFTR modulator therapy. Patients 1, 2, and 4 were seeking outpatient treatment for an acute pulmonary exacerbation at the time of sampling, while Patient 3 was in stable medical condition. These individuals were in the early (%FEV1 > 70) to intermediate (%FEV1 ≤ 70, ≥ 40) stages of lung disease, with %FEV1 scores ranging from 60.30% to 74.92%. The antibiotic regimens for each patient at the time of sampling were as follows: Patient 1 was receiving inhaled tobramycin and oral azithromycin; Patient 2 was receiving inhaled tobramycin and oral trimethoprim/ sulfamethoxazole; Patient 3 was receiving inhaled tobramycin, oral azithromycin, and inhaled aztreonam; and Patient 4 was receiving inhaled tobramycin, oral trimethoprim/ sulfamethoxazole, and oral levofloxacin (Table 1).
Table 1.
Metadata on the four patients in our cohort: sex, cystic fibrosis transmembrane conductance regulator (CFTR) mutation status, length of P. aeruginosa infection, clinical status, forced expiratory volume (% FEV1), modulator therapy, antibiotic treatment, and dominant infection strain type.
| Patent 1 | Patent 2 | Patent 3 | Patent 4 | |
|---|---|---|---|---|
| Patient Sex | F | F | F | M |
| CFTR Mutation | F508del/R1162X | F508del/F508del | F508del/L467P | F508del/ 621+1G->T |
| Length of Pa infection | 15 years, 2 months | 12 years, 5 months | 10 years, 4 months | 13 years |
| Clinical status | APE Outpatient | APE Outpatient | Stable | APE Outpatient |
| FEV1 (%) | 67.96% | 74.92% | 67.83% | 60.30% |
| Modulator | None | None | None | None |
| Antibiotic Treatment | Inhaled tobramycin, oral azithromycin | Inhaled tobramycin, oral Trimethoprim / Sulfamethoxazole | Inhaled tobramycin, inhaled aztreonam, oral azithromycin | Inhaled tobramycin, oral Trimethoprim / Sulfamethoxazole, oral levofloxacin |
| Dominant ST | 870 | 2999 | 1197 | 274 |
P. aeruginosa populations display significant within-patient diversity in antimicrobial resistance profiles.
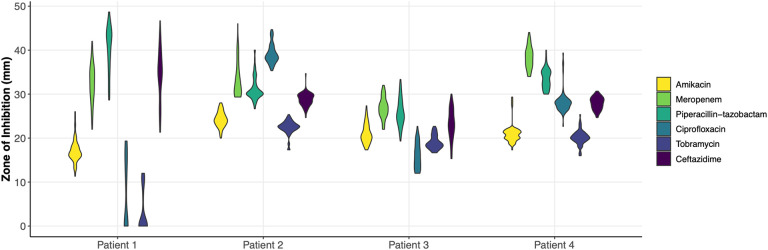
In order to first assess diversity in AMR, we selected 75 isolates from a single sputum sample of each of four individuals with CF for a total of 300 isolates. Using a standard disc diffusion assay, we assessed these 300 isolates for their susceptibilities to six antimicrobials commonly prescribed in CF treatment: amikacin, meropenem, piperacilin-tazobactam, ciprofloxacin, tobramycin, and ceftazidime (Tables S1–S4). The majority of isolates presented values well within the range of susceptibility for the tested antibiotics, despite ineffective clearing of infection in the clinic for these patients chronically infected with P. aeruginosa (Fig. 1; Fig. S1). Only two patients harbored isolates that tested in the range of clinical resistance to any antimicrobial: amikacin, ciprofloxacin, and tobramycin for Patient 1; and ciprofloxacin for Patient 3. Three of the four patients harbored populations that presented phenotypes spanning across the clinical thresholds for resistant, intermediate, and susceptible for at least one, if not multiple, antibiotics. Zone of inhibition values within a population for a given antibiotic displayed a statistical range (minimum subtracted from the maximum value of a population) between 6 and 25.3 mm, with an average of 12.75 mm. Standard deviations of these values ranged from 1.4 to 8.0 mm, with an average standard deviation of 3.0 mm.
Figure 1.
Violin plot of the antimicrobial susceptibility profiles of all four populations against amikacin, meropenem, piperacillin-tazobactam, ciprofloxacin, tobramycin, and ceftazidime as measured by zone of inhibition in a standard disc diffusion assay shows phenotypic diversity across all populations.
The four patients are chronically infected by a single P. aeruginosa strain, populations of which display a range of genomic diversity levels.
In order to quantify the level of within-patient genomic diversity for these populations, we sequenced the 75 isolates from each of the four individuals of this cohort. We prepared the sequences of all 300 isolates using de novo assembly and annotation. We assembled the genomes in 20 to 444 contigs (mean = 53 contigs; Table S5). Genomes in this dataset ranged in size from 5,888,197 to 6,746,489 nucleotides, with 5,209 to 5,970 genes (Table S5). The median genome sizes of isolates sourced from Patients 1–4, respectively, were 6,222,786, 6,331,110, 6,742,689, and 6,308,671 nucleotides, with 5,523, 5,571, 5,964, and 5,567 genes, respectively (Table S5). A phylogenetic tree of the core genome alignment revealed that the populations infecting Patients 1, 2, and 4 clustered closely with PAO1, while that of Patient 3 more closely resembled PA14 (Fig. S2). Strain typing of the isolates showed that there was a single, dominant strain type in each patient— ST870, ST2999, ST1197, and ST274 for Patients 1–4, respectively (Table 1). For the rest of the text, we will simply refer to each population by its respective patient number.
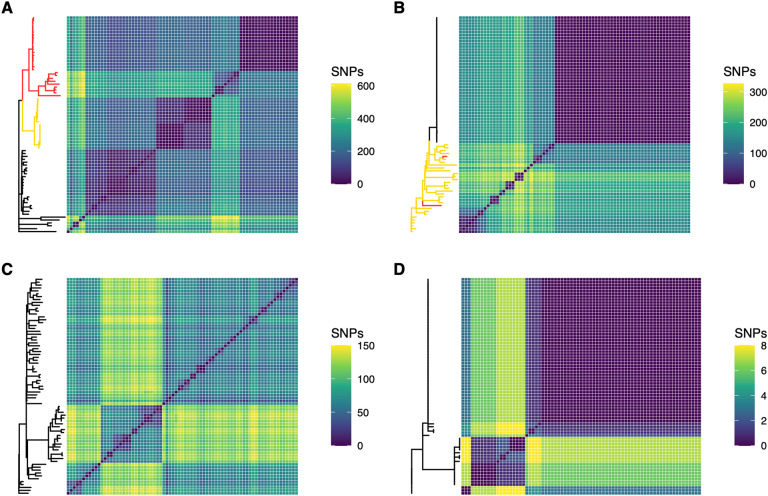
We assessed the genomic diversity in these populations according to the number of single nucleotide polymorphisms (SNPs) and microindels (insertions and deletions). We found that genomic diversity varied significantly between patients. The total number of unique SNPs discovered across 75 isolates for Patient 1 was 4,592 (maximum number of pairwise SNPs = 611, median number of pairwise SNPs = 199, mean = 208); for Patient 2 was 1,972 (max. = 326, median = 145, mean = 118); for Patient 3 was 1,638 (max. = 150, median = 76, mean = 87); and for Patient 4 was 31 (max. = 8, median = 1, mean = 3) (Fig. 2; Table 2). Across the population of Patient 1 we found 498 unique microindels, 307 for Patient 2, 330 for Patient 3, and 14 for Patient 4 (Table 2).
Figure 2. Genomic diversity as measured by core genome SNPs varies greatly from one population to another.
Populations are presented in order of decreasing genomic diversity: Patient 1 (A), Patient 2 (B), Patient 3 (C), and Patient 4 (D). Isolates with one DNA mismatch repair mutation are highlighted in yellow on phylogenies. Isolates with two DNA mismatch repair mutations are highlighted in red.
Table 2.
Genetic variations in each population: single nucleotide polymorphisms (SNPs), multiple nucleotide polymorphisms (MNPs), and insertions and deletions (indels).
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
|---|---|---|---|---|
| Total # unique SNPs/ MNPs | 4592 | 1972 | 1638 | 31 |
| # SNPs/ MNPs separating most divergent isolates | 611 | 326 | 150 | 8 |
| Non-synonymous SNPs/ MNPs | 2803 | 1294 | 1024 | 24 |
| Synonymous SNPs/ MNPs | 1248 | 484 | 425 | 5 |
| SNPs in non-coding regions | 541 | 194 | 189 | 2 |
| Total # indels | 498 | 307 | 330 | 14 |
| Indels in non-coding regions | 204 | 99 | 115 | 2 |
Genomic diversity may not be a consistent predictor of antimicrobial resistance diversity in a population.
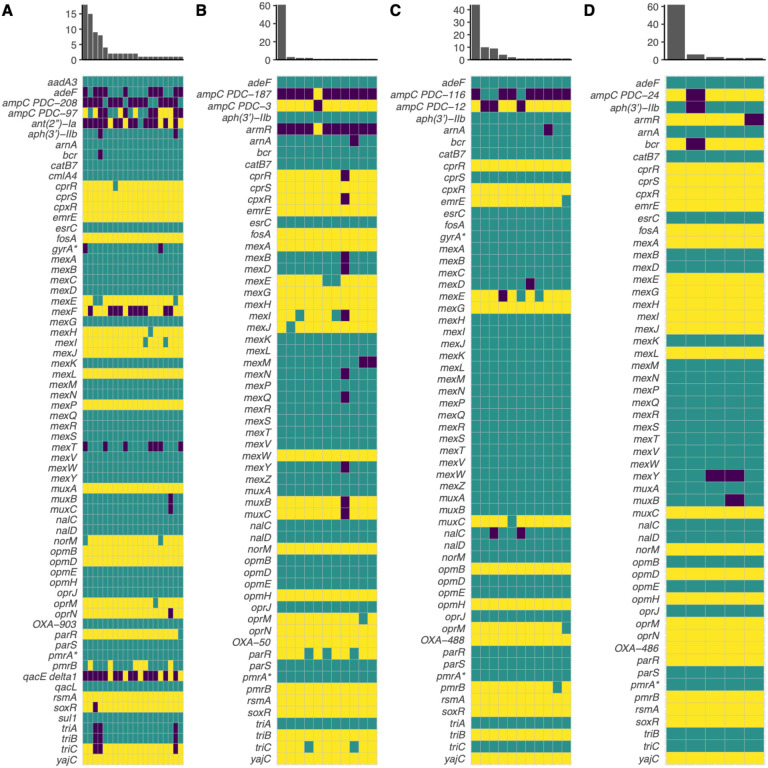
We next determined whether genomic diversity could serve as a predictor of diversity in AMR phenotypes in our cohort. We hypothesized that genetically diverse populations would also display more diversity in AMR. We chose to quantify genomic diversity in terms of SNPs. We quantified AMR diversity using the number of distinct AMR profiles (i.e., distinct zone of inhibition values) for a given antibiotic within a population. Total SNP count in a population was a strong indicator of AMR diversity for amikacin (R2 = .90, F(1, 2) = 18.94, p = .049), meropenem (R2 = .93, F(1, 2) = 25.3, p = .037), and piperacilin-tazobactam (R2 = .95, F(1, 2) = 39.86, p = .024). However, SNP count was a poor indicator of AMR diversity for ciprofloxacin (R2 = .12, F(1,2) = .27, p = .65) and ceftazidime (R2 = .71, F(1,2) = 4.78, p = .16), and was inversely related to AMR diversity for tobramycin (R2 = .97, F(1,2) = 66.61, p = .015). We next used the number of distinct CARD resistance genotype profiles within a population (Fig. 3) as a proxy for genomic diversity to eliminate bias from SNPs not relevant to AMR and to account for the epistatic effect that combinations of various alleles may have. This yielded similar results to the previous analysis (data not shown). When only taking into consideration the AMR genotypes relevant to the appropriate antimicrobial class, this still did not improve the strength of the associations for ciprofloxacin, tobramycin, and ceftazidime (data not shown). We next used the standard deviation of zone of inhibition values within a population as a proxy for AMR diversity to see if this would improve the strength of the association between genomic diversity and phenotypic diversity for these antimicrobials. We found that the number of distinct CARD profiles within a population was a better predictor of standard deviation for ciprofloxacin (R2 = .79, F(1,2) = 7.35, p = .11), tobramycin (R2 = .77, F(1,2) = 6.73, p = .12), and ceftazidime (R2 = .81, F(1,2) = 8.44, p = .10), though these associations were still not significant.
Figure 3. Resistomes of Patients 1 (A), 2 (B), 3 (C), and 4 (D) as predicted by the Comprehensive Antibiotic Resistance Database Resistance Gene Identifier (CARD RGI).
Yellow indicates a perfect hit to the database, teal indicates a strict hit, and purple indicates no hit (or loose hit in some cases). X-axis of the histogram indicates the number of unique resistome profiles in the population, and y-axis indicates the number of isolates in the population that share a unique resistome profile. An asterisk (*) indicates a gene with resistance conferred by a mutation (i.e. CARD RGI protein variant model).
P. aeruginosa diversity is primarily driven by de novo mutations, especially mutations in DNA mismatch repair.
We next wanted to further understand the processes by which P. aeruginosa diversified in our cohort. We first sought to predict putative recombination events. In Patients 1–4, 527 (11.5%), 19 (<1%), 86 (5.25%), and 0 SNPs were predicted to be in 31, 3, 17, and 0 recombinant regions, respectively. These data show that de novo mutation was a much more prominent driver of intra-specific diversity than recombination in our particular cohort. As expected, we found that the infections with the highest SNP diversity harbored strains with DNA MMR mutations. Patients 1 and 2 harbored DNA MMR mutants (hypermutators); however, we found no hypermutators in Patients 3 or 4 (Fig. 2). The phylogeny of Patient 1 indicates that a non-synonymous SNP in mutS (Ser31Gly) evolved first in the population, after which a frameshift deletion in mutS (Ser544fs) piggybacked. In total, mutS mutants comprise 61.3% of this population. In Patient 2, a non-synonymous SNP in mutL resulting in a pre-mature stop codon (Glu101*) evolved first, found in 41.3% of the population. Two of these mutL mutants further independently acquired a single non-synonymous mutation in mutS (Phe445Leu, Ala507Thr) (Fig. 2).
In Patient 1, there were two distinct branches of the phylogenetic tree, one with hypermutators and the other composed of normomutators (38.7%) (Fig. 2). Interestingly, there was a significant amount of genetic diversity within both the normomutators (mean SNP distance = 156.9 SNPs, median = 91 SNPs) and hypermutators (mean = 174.6 SNPs, median = 197 SNPs). There was a distinct small cluster of normomutator isolates that significantly diverged from the others. Of the hypermutators, these further diverged into those with one DNA MMR mutation (39.1%) and those with two MMR mutations (60.9%). In Patient 2, there was largely a lack of genetic diversity in the normomutators (mean = 0.36 SNPs, median = 0 SNPs), with one clone dominating 48% of the population (Fig. 2). The emergence of hypermutators appears to have been responsible for the large majority of all the genetic diversity in this population (mean = 211.2 SNPs, median = 224 SNPs). In Patient 3, there were three major lineages, comprising 58.7%, 26.7%, and 14.7% of the total population (mean = 61.9, 55.5, and 65.4 SNPs; median = 62, 61, and 64 SNPs, respectively; Fig. 2). In Patient 4, there was one dominant clone encompassing 66.6% of the population, with a small number of SNPs (mean = 4 SNPs, median = 3 SNPs) differentiating the other 33.3% of the population (Fig. 2).
Hypermutation can drive the evolution of increased susceptibility to antimicrobials, even under apparent selective pressure.
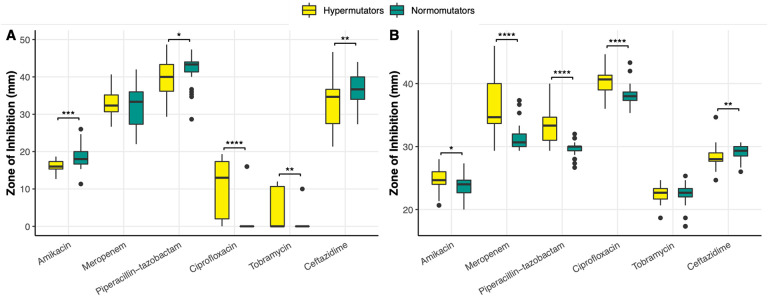
As our cohort had two populations with DNA MMR mutants, we used this opportunity to ascertain how hypermutation drives the evolution of AMR. In Patient 1, AMR genotypes cluster by DNA MMR genotype. Hypermutators were significantly more resistant to amikacin than normomutators (U = 315.5, p = .00013) (Fig. 4), although this difference could not be attributed to any hits in the CARD database. Hypermutators were also significantly more resistant to beta-lactams piperacilin-tazobactam (U = 457.5, p = .023) and ceftazidime (U = 428, p = .0095), although there was no significant difference in the resistance profiles of hyper- and normomutators in regards to the beta-lactam meropenem (U = 630, p =.69) (Fig. 4). Some normomutators in this population acquired a SNP in ampC (461 A > G, Asp154Gly) (Fig. 3), which increased susceptibility to piperacilin-tazobactam and ceftazidime. Of the isolates with one DNA MMR mutation, some lost ampC entirely, also leading to increased susceptibility to these antimicrobials. Of the isolates with both DNA MMR mutations, some had acquired a SNP in ampC (1066 G > A, Val356Ile), which increased their resistance to these antimicrobials (Fig. 3).
Figure 4. Comparative antimicrobial susceptibility profiles of hypermutators and normomutators in Patient 1 (A) and Patient 2 (B) as measured by zone of inhibition in a standard disc diffusion assay highlight increased sensitivities and resistance levels by hypermutators.
(A) In Patient 1, hypermutators were significantly more resistant to amikacin (U = 315.5, p = .00013), piperacilin-tazobactam (U = 457.5, p = .023), and ceftazidime (U = 428, p = .0095) than normomutators, although there was no significant difference in the resistance profiles of hyper- and normomutators in regards to meropenem (U = 630, p =.69). Hypermutator isolates in Patient 1 displayed zone of inhibition (ZOI) values that were on average 10 times larger for ciprofloxacin (U = 218, p < .00001) and >13 times larger for tobramycin (U = 379.5, p = .0018) than normomutators, and isolates with both DNA MMR mutations in this population additionally presented ZOI values that were 36 times larger than normomutators for tobramycin (U = 172.5, p < .00001), indicating increased sensitivity displayed by hypermutators. (B) In Patient 2, hypermutators displayed increased susceptibility to amikacin (U = 479, p = .029), meropenem (U = 194, p < .00001), piperacilin-tazobactam (U = 121.5, p < .00001), and ciprofloxacin (U = 213.5, p < .00001) relative to normomutators. Hypermutators in Patient 2 were more resistant to ceftazidime (U = 417.5, p = .0045). There was no statistically significant difference between the tobramycin susceptibility profiles of hyper- and normomutators in this population (U = 634.5, p = .61). (*) indicates p ≤ .05, (**) indicates p ≤ .01, (***) indicates p ≤ .001, and (****) indicates p < .0001 in a Mann-Whitney U test.
Interestingly, hypermutator isolates in this population displayed zone of inhibition (ZOI) values that were on average 10 times larger for ciprofloxacin (U = 218, p < .00001) and >13 times larger for tobramycin (U = 379.5, p = .0018) than normomutators, indicating increased sensitivity of hypermutators to these antimicrobials (Fig. 4). Isolates with both DNA MMR mutations in this population additionally presented ZOI values that were 36 times larger than normomutators for tobramycin (U = 172.5, p < .00001) (Fig. 4). The altered ciprofloxacin phenotype may be explained in part by SNPs in gyrA (248 T > C, Ile83Thr) or norM (61 G > A, Ala21Thr) (Fig. 3). However, there are isolates in this population whose phenotypes are not explained by either of these genotypes. The increased susceptibility to tobramycin is strongly linked to the aforementioned SNP in norM (Fig. 3). We saw evidence of one of these hypermutators reversing this increased susceptibility to tobramycin by acquisition of the aminoglycoside nucleotidyltransferase ant(2”)-Ia (Fig. 3). There was a normomutator isolate with an outlier tobramycin susceptibility phenotype. Interestingly, 12 isolates from Patient 1 had improved growth in the presence of tobramycin (determined by visual observation of denser growth in the region surrounding the antibiotic disc in a disc diffusion assay), a phenotype which could not be explained by any hits in the database. All of the normomutator isolates had a truncated mexF (Fig. 3), although this did not appear to impact any of the tested phenotypes.
In Patient 2, hypermutators displayed increased susceptibility to meropenem (U = 194, p < .00001), piperacilin-tazobactam (U = 121.5, p < .00001), and ciprofloxacin (U = 213.5, p < .00001) relative to normomutators (Fig. 4). This appears to be caused by a SNP in mexB (2257 T > C, Trp753Arg) (Fig. 4). However, there were outliers that could not be explained by this genotype. Hypermutators were also more susceptible to amikacin (U = 479, p = .029) and more resistant to ceftazidime (U = 417.5, p = .0045) (Fig. 4), although these strains harbored no apparent genes or SNPs associated with these phenotypes in the CARD database. There was no statistically significant difference between the tobramycin susceptibility profiles of hyper- and normomutators in this population (U = 634.5, p = .61) (Fig. 4). One hypermutator isolate in Patient 2 had an unusual density of truncated pseudogenes, 10 of which are involved in resistance mechanisms and 9 of which specifically play roles in resistance-nodulation-cell division efflux— mexY, mexQ, mexN, cpxR, muxB, muxC, mexI, mexB, mexD, and cprR (Fig. 3). Although RGI denoted these genes as missing due to truncation, this isolate was equally or more resistant to every antimicrobial tested relative to other DNA MMR mutants in the population, suggesting that many of these genes are still functional.
In the two normomutator populations, there was significantly decreased resistome diversity. In Patient 3, a SNP in ampC (716 T > C, Val239Ala) resulted in increased resistance to ceftazidime (Fig. 3). Some of the isolates with this SNP additionally were missing nalC (Fig. 3), and displayed increased susceptibility to meropenem and increased resistance to ciprofloxacin relative to other isolates within this group. In Patient 4, variability in susceptibilities to amikacin, ciprofloxacin, and tobramycin were attributable to a truncation in mexY (Fig. 3). Surprisingly, isolates that were missing a hit to aph(3’)-IIb were more resistant to aminoglycosides amikacin and tobramycin (Fig. 3). However, no hits in the CARD database explained the variations in beta-lactams meropenem, piperacilin-tazobactam, or ceftazidime. Seeing as some of these relationships are unexpected, it is likely that there are other genetic variations not cataloged in the CARD database that are influencing these phenotypes.
Protein export/ secretion systems and transcriptional regulators are hotspots for de novo mutations.
To determine whether these populations were enriched for mutations in genes with roles in resistance, we categorized non-synonymous SNPs and microindels that occurred within coding regions of genes according to the PseudoCAP functional categories and conducted an enrichment analysis. We did not find that AMR genes were enriched for such variants in this cohort (Fig. S3). However, we found that protein secretion and export apparatuses and transcriptional regulators were enriched for such mutations (Fig. S3). Additionally, two of the four genes impacted by non-synonymous mutations in all four patients in this study were related to protein secretion, fha1 and pscP (Table S6). We found that phage/transposon/plasmid genes were less likely to be impacted by such mutations (Fig. S3). Non-coding RNAs were also less likely to be impacted by mutations than other functional categories (Fig. S3; see Table S7 for all supporting statistical values), which is unsurprising given that small non-coding RNAs are known to hold important regulatory functions in bacteria (52). 57 genes were impacted by non-synonymous mutations in 3 of the 4 patients, which included genes with previously described functions in alginate biosynthesis, primary metabolism, antibiotic resistance and efflux, iron uptake, biofilm formation, stress response, amino acid biosynthesis, type IV pili, lipopolysaccharide, quorum sensing, and virulence (Table S8). A full list of all SNPs discovered in this dataset can be found in Tables S9–S12.
Mobile genetic elements and determinants of pathogenicity are commonly variable genes within a population.
We further investigated the potential pathways of adaptation employed by P. aeruginosa by identifying the genes that displayed variation within a population, especially those involved in AMR. Our analysis sought to identify variable genes, not whether genes had been specifically lost or acquired in the population. Within a given patient, we found that variations in gene content were limited, with the exception of one patient. In Patients 2, 3, and 4, 96.7%, 97.7%, and 97.0% of discovered genes were found in >98% of isolates within the population, respectively, whereas only 84.8% of genes were found in >98% of isolates in Patient 1, indicating a much larger proportion of variable genes in Patient 1 (Fig. 5). There were 934 variable genes in Patient 1. Many of these are involved in previously described pathogenic functions, such as type VI secretion, type IV pili, phenazine biosynthesis, quorum sensing, fimbrial biogenesis, flagellar motility, triclosan efflux, biofilm signaling regulators, chemotaxis, aerotaxis, and iron acquisition. Unsurprisingly, these also included integrative and conjugative elements, as well as prophages including integrases, recombinases, and tranposases. In Patient 2, there were 185 variable genes, the most notable being a large enrichment of truncated pseudogenes— 10 of which are involved in AMR as described earlier (Fig. 3)— in isolate 74. There were 171 variable genes in Patient 3, notably all 10 genes of the pil2 operon, 10 integrative and conjugative element genes, as well as dsbA2, katA, and other genes encoding hypothetical proteins. In Patient 4, there were 196 variable genes notable variable genes, including 6 genes in the hpa operon, ampC and ampR, bfmRS, ccoN, aph, fepBCDG, fvbA, acoABCRX, and mpaR. There were 13 genes that were variable in all 4 patients, and 33 genes that were variable in 3 of 4 patients. The phenazine biosynthetic pathway was the most frequently variable across multiple patients, with phzC1, phzD1, phzF2, phzE, and phzG1 all being variable in 3 or more patients.
Figure 5. Patient 1 has a larger proportion of variable genes than Patients 2, 3, and 4, as displayed by the aggregated genomes of the 300-isolate dataset.
Phylogeny is given on the left, and the aggregated genome matrix on the right. Each tile on the x-axis of the aggregated genome matrix indicates a gene family, where a blue tile indicates presence of that gene family at ≥90% BlastP sequence identity in a sample, and a white tile indicates absence.
Populations display poor evidence for evolutionary trade-offs to explain heterogeneity in resistance profiles.
We next wanted to ascertain if there was any evidence of evolutionary trade-offs involving AMR in these populations. Collateral sensitivity is sensitive to genetic background (53–56) and must be proven robust across a wide range of genetic backgrounds in order to be broadly applicable as a therapeutic strategy (57). Therefore, we searched for evidence of collateral sensitivity within our populations, and additionally for evidence of trade-offs between AMR and fitness (i.e., growth rate) in a CF sputum-like medium, SCFM (33). We found no evidence of collateral sensitivity across any of the six antimicrobials tested for any patient (Fig. S4). We analyzed growth curves for all 300 isolates (Tables S13–S16), and using a linear mixed model, determined that there was not a significant relationship between resistance and fitness for any of the tested antimicrobials (Fig S5; Table S17 for supporting code and statistical values).
DISCUSSION
The goal of this project was to better understand how genomic diversification in P. aeruginosa CF lung populations drives the evolution of AMR. For this study, we selected four distinct patients with varying levels of P. aeruginosa genomic population diversity, ranging from a few dozen to multiple thousands of SNPs within a given population. We found that (i) genomic diversity was not consistently a reliable predictor of AMR diversity for this cohort; (ii) hypermutators in one population evolved increased sensitivity to tobramycin, even when undergoing treatment by tobramycin; and that (iii) there was no evidence for collateral sensitivity or trade-offs between AMR and fitness in these populations.
Previous studies have reported both on genomic and phenotypic diversity of P. aeruginosa in CF airways (3, 4, 27–32); however, the clinical implications of genomic diversity within these populations on resistance diversity have not been assessed. Our results suggest that genomic diversity may not be a reliable predictor of phenotypic diversity for all antibiotics. However, there are a number of limitations to this finding: (i) our sample size for this analysis was small; (ii) we cannot account for diverse genotypes that result in converging phenotypes; and (iii) there are likely many genetic variants that act on AMR that have not been catalogued in CARD. Nonetheless, we highlight that Patient 4 displayed a number of distinct AMR profiles that was, in the case of ciprofloxacin, comparable to that of Patient 1, which had 148x more SNPs and 4x as many distinct CARD genotype profiles within the population. In the case of tobramycin, Patient 4 displayed more distinct AMR profiles and higher zone of inhibition standard deviation values compared to Patients 2 and 3, which both had 2x as many distinct CARD genotype profiles and over 53x more population SNPs compared to Patient 4. Ultimately, because of our limited ability at present to predict the phenotypic impact of novel genetic variants or the epistatic interactions of alleles in silico, it may prove challenging to ascertain the phenotypic heterogeneity of an infection in a parsimonious manner that could be translated to the clinic (58). In addition to improved in silico prediction capabilities, greater understanding of the social interactions that impact how co-infecting microbes with varying resistance levels collectively respond to antibiotic treatment and development of reliable methodology for assessing population-level resistance may be instrumental in future approaches for tackling chronic infections.
Our data further highlight that even our ability to assess resistance at the isolate-level is inadequate. All individuals in this cohort were being treated with inhaled tobramycin, yet only one population displayed clinical resistance to tobramycin, and an overwhelming number of isolates in our study were classified as clinically susceptible to the majority of antimicrobials we tested despite persisting within the lung for over a decade. We were particularly concerned to find isolates with increased growth in the presence of tobramycin, as inhaled tobramycin is one of the most commonly prescribed drugs for CF patients with P. aeruginosa infection. These findings are in accordance with the wide array of literature that has already called into question the utility of antimicrobial susceptibility testing in the clinic, where testing results correlate poorly with patient outcomes (59).
Combining single-isolate whole genome sequencing and phenotypic characterization approaches further allowed us to understand how the evolution of genotypes and combination of alleles impact AMR within a population. Although we were able to identify a number of candidate genotypes responsible for these phenotypic variations, there were a number of unexplained phenotypic outliers, highlighting the presence of novel genetic signatures of AMR that have yet to be discovered. Previous reports have primarily focused on the role that hypermutation plays in evolving increased AMR in clinical P. aeruginosa populations (60–66). We found ample evidence that hypermutation can also lead to increased susceptibility, such as the hypermutator isolates in Patient 1 that were significantly more sensitive to tobramycin, despite this patient undergoing treatment with inhaled tobramycin. This may be a function of antimicrobial treatment regimens exerting uneven selection pressure on the population. Or, it may be that the evolution of genetic resistance for these populations is inconsequential because antimicrobials are failing to penetrate phenotypic barriers, such as biofilms, persister cells, and other mechanisms of antibiotic tolerance (67–71). Although antimicrobial treatment leads to increased resistance in vitro (72–78), the development of resistance or sensitivity in vivo may, in some ways, be a result of stochastic processes or other evolutionary drivers if antibiotic treatment regimens are only exerting weak selective pressure.
It is often assumed that the evolution of AMR involves a fitness cost, although this has predominantly been tested in lab-evolved strains (78–82). We found no evidence for collateral sensitivity or trade-offs between resistance and fitness in a CF-like medium for these clinical populations. However, in interpreting these results, we must consider that in vitro susceptibility and growth testing may not accurately recapitulate the conditions of an in vivo lung. Therefore, trade-offs between these measures may be present in the lung but not detectable under laboratory conditions. Collateral sensitivity, although shown in evolutionary experiments, has yet to be demonstrated as widely prevalent in naturally occurring clinical strains. Further work is needed to show that collateral sensitivity is a viable approach for future therapeutic consideration. A recent report found evidence for trade-offs between fitness and multi-drug resistance in clinical P. aeruginosa populations (83). Taken together with our results, we hypothesize that resistance to a single antibiotic may not exert sufficient fitness cost to act as a driving force for trade-offs with growth rate, while resistance to multiple antibiotics perhaps does. Furthermore, this study found stronger evidence for trade-offs in mixed strain infections, whereas all of the individuals in our cohort were infected with a single strain of P. aeruginosa. Moreover, as the majority of our strains were technically clinically sensitive to the tested antimicrobials, we may not have had the power to detect trade-offs if they are only elicited at high resistance levels. If resistance does indeed trade-off with resistance, this suggests that slow-growing strains may prove to be the most resistant to treatment. The implication of this for the clinic is concerning, as slow-growing strains are the most likely to remain undetected during routine susceptibility testing in the clinic, where quick results are favored in order to expedite treatment.
Conducting deep sampling of clinical P. aeruginosa populations has allowed us to illuminate population structure, evolution, and population diversity in CF in a manner that single-isolate sampling or population-level sequencing cannot. These methods suffer in their ability to identify rare variants, accurately resolve population structure, and in the case of pooled deep-sequencing, link genotype to phenotype for individual isolates. A 2016 study claimed that single-isolate sampling of longitudinal isolates was sufficient to capture the evolutionary pathways of P. aeruginosa in CF lung infection; however, the authors conducted metagenome sequencing at a low depth of 10–31x and only sought to determine if SNPs within individual isolates could be re-discovered in the metagenomes, not whether the individual isolates captured the full diversity of the metagenome (84). However, we believe there is still incredible value in conducting longitudinal analyses. Building upon previous work (85), we propose that conducting deeper sampling of populations over long time scales will help illuminate the full evolutionary dynamics of P. aeruginosa populations in the CF lung and lead to insights that will assist in tackling chronic infections.
Supplementary Material
Supplemental Table 1. Antimicrobial susceptibility testing measurements for Patient 1 as measured by zone of inhibition (ZOI) in a standard disc diffusion assay for amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Data in the left columns represent raw measurements of zone of inhibition radii (mm units). Data in the right columns represent calculated zone of inhibition values as diameters (mm units).
Supplemental Table 2. Antimicrobial susceptibility testing measurements for Patient 2 as measured by zone of inhibition (ZOI) in a standard disc diffusion assay for amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Data in the left columns represent raw measurements of zone of inhibition radii (mm units). Data in the right columns represent calculated zone of inhibition values as diameters (mm units).
Supplemental Table 3. Antimicrobial susceptibility testing measurements for Patient 3 as measured by zone of inhibition (ZOI) in a standard disc diffusion assay for amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Data in the left columns represent raw measurements of zone of inhibition radii (mm units). Data in the right columns represent calculated zone of inhibition values as diameters (mm units).
Supplemental Table 4. Antimicrobial susceptibility testing measurements for Patient 4 as measured by zone of inhibition (ZOI) in a standard disc diffusion assay for amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Data in the left columns represent raw measurements of zone of inhibition radii (mm units). Data in the right columns represent calculated zone of inhibition values as diameters (mm units).
Supplemental Table 5. Genome size, average sequencing coverage, and number of contigs of each assembly.
Supplemental Table 6. Genes that were impacted by non-synonymous mutations in at least one isolate in all four populations.
Supplemental Table 7. Full details of the chi-squared goodness of fit and Monte Carlo simulation exact multinomial tests, with all associated chi-squared and p-values.
Supplemental Table 8. Genes that were impacted by non-synonymous mutations in at least one isolate in three out of four populations.
Supplemental Table 9. All annotated genetic variants discovered in Patient 1.
Supplemental Table 10. All annotated genetic variants discovered in Patient 2.
Supplemental Table 11. All annotated genetic variants discovered in Patient 3.
Supplemental Table 12. All annotated genetic variants discovered in Patient 4.
Supplemental Table 13. Raw OD600 reads for growth in SCFM used to create growth curves and analyze growth rate (r) for Patient 1. Time is given in hours, and all isolates were tested in biological triplicates.
Supplemental Table 14. Raw OD600 reads for growth in SCFM used to create growth curves and analyze growth rate (r) for Patient 2. Time is given in hours, and all isolates were tested in biological triplicates.
Supplemental Table 15. Raw OD600 reads for growth in SCFM used to create growth curves and analyze growth rate (r) for Patient 3. Time is given in hours, and all isolates were tested in biological triplicates.
Supplemental Table 16. Raw OD600 reads for growth in SCFM used to create growth curves and analyze growth rate (r) for Patient 4. Time is given in hours, and all isolates were tested in biological triplicates.
Supplemental Table 17. Supporting brms R code and statistical values for the linear mixed model run to assess the relationship between growth rate (r) and antimicrobial resistance. Results of the model, with growth rate in SCFM as a fixed effect and patient as a random effect, show that the 95% confidence interval of the fixed effect of growth rate spans 0 for all six antimicrobials. Therefore, the null hypothesis that the fixed effect of growth on antimicrobial susceptibility is 0 cannot be rejected, providing no evidence for trade-offs or any significant relationship between resistance and growth rate in these populations.
Supplemental Figure 1. Violin plot of the antimicrobial susceptibility profiles of all four populations against amikacin, meropenem, piperacillin-tazobactam, ciprofloxacin, tobramycin, and ceftazidime as measured by zone of inhibition in a standard disc diffusion assay. This plot displays the same data as Fig. 1, displayed in clusters by antibiotic rather than by population. Black horizontal bars indicate the cut-off values for susceptibility (top bar) and resistance (bottom bar) for each antibiotic as determined by the Clinical and Laboratory Standards Institute (CLSI).
Supplemental Figure 2. Phylogeny of Patients 1–4 with PAO1 and PA14. Patients 1, 2, and 4 cluster with PAO1, while Patient 3 clusters with PA14.
Supplemental Figure 3. Enrichment analysis of the frequency of functional categories in which non-synonymous SNPs and microindels are found in each of the four populations relative to the proportions of these functional categories in the PAO1 genome shows that protein secretion/export apparatuses and transcriptional regulators are enriched for such variants, while phage/transposon/ plasmid and non-coding RNA are less likely to be impacted by such variants. Donut plot of the relative frequencies of genes categorized within each of the 27 different PseudoCAP functional categories in the PAO1 genome (A). Donut plots of the relative frequencies of non-synonymous SNPs and indels located in each of the 27 different PseudoCAP functional categories in Patient 1 (B), 2 (C), 3 (D), and 4 (E).
Supplemental Figure 4. Lack of statistically significant negative correlations between any two antimicrobial susceptibility profiles provides no evidence for collateral sensitivity trade-offs. Pearson’s correlation coefficient (upper right quadrant), scatterplots (lower left quadrant), and density plots (diagonal) for pairwise comparisons of susceptibility profiles across all six tested antimicrobials: amikacin (AK), meropenem (MEM), piperacillin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ).
Supplemental Figure 5. Scatterplots of zone of inhibition (ZOI) versus growth rate (r) in SCFM for all six tested antibiotics: amikacin (AK), meropenem (MEM), piperacillin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Results of linear mixed model (Table S17), with growth rate in SCFM as a fixed effect and patient as a random effect, demonstrate that there is no significant effect of growth rate on resistance, and therefore, no evidence for trade-offs between growth rate and resistance in these four populations.
ACKNOWLEDGEMENTS
For funding, we thank the Georgia Institute of Technology, the Cystic Fibrosis Foundation for grants (DIGGLE18I0 and DIGGLE20G0) to S.P.D. and a fellowship to S.A. (AZIMI18F0), CF@LANTA for a fellowship to S.A. (3206AXB), the National Institutes of Health for a grant (R01AI153116) to S.P.D and T.R., and the National Science Foundation for a fellowship to J.V. (DGE-2039655). Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies. Access to the Cystic Fibrosis Biospecimen Registry at Emory Children’s Center for CF and Airways Disease Research was provided through Children’s Healthcare of Atlanta and the Emory University Pediatric CF Discovery Core. We thank Arlene Stecenko and Katy Clemmer for assistance in acquiring sputum samples.
Footnotes
Preprint servers: This manuscript has been submitted as a preprint to bioRxiv
Bibliography
- 1.C. F. Foundation (2021) Cystic Fibrosis Foundation Patient Registry 2020 Annual Data Report.
- 2.Jackson L., Waters V., Factors influencing the acquisition and eradication of early Pseudomonas aeruginosa infection in cystic fibrosis. J Cyst Fibros 20, 8–16 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Darch S. E. et al. , Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci Rep 5, 7649 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorth P. et al. , Regional Isolation Drives Bacterial Diversification within Cystic Fibrosis Lungs. Cell Host Microbe 18, 307–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark S. T., Guttman D. S., Hwang D. M., Diversification of Pseudomonas aeruginosa within the cystic fibrosis lung and its effects on antibiotic resistance. FEMS Microbiol Lett 365 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Bartell J. A. et al. , Omics-based tracking of Pseudomonas aeruginosa persistence in “eradicated” cystic fibrosis patients. Eur Respir J 57 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camus L., Vandenesch F., Moreau K., From genotype to phenotype: adaptations of Pseudomonas aeruginosa to the cystic fibrosis environment. Microb Genom 7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta H. H. et al. , The Essential Role of Hypermutation in Rapid Adaptation to Antibiotic Stress. Antimicrob Agents Chemother 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lujan A. M. et al. , Polymicrobial infections can select against Pseudomonas aeruginosa mutators because of quorum-sensing trade-offs. Nat Ecol Evol 6, 979–988 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Armbruster C. R. et al. , Heterogeneity in surface sensing suggests a division of labor in Pseudomonas aeruginosa populations. Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter S. S., Rice K. J., Trade-offs, spatial heterogeneity, and the maintenance of microbial diversity. Evolution 67, 599–608 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Ferenci T., Trade-off Mechanisms Shaping the Diversity of Bacteria. Trends Microbiol 24, 209–223 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Cramer N. et al. , Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ Microbiol 13, 1690–1704 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Marvig R. L., Johansen H. K., Molin S., Jelsbak L., Genome analysis of a transmissible lineage of pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet 9, e1003741 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feliziani S. et al. , Coexistence and within-host evolution of diversified lineages of hypermutable Pseudomonas aeruginosa in long-term cystic fibrosis infections. PLoS Genet 10, e1004651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markussen T. et al. , Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. mBio 5, e01592–01514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marvig R. L. et al. , Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients. BMC Microbiol 15, 218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marvig R. L., Sommer L. M., Molin S., Johansen H. K., Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 47, 57–64 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Bianconi I. et al. , Persistence and Microevolution of Pseudomonas aeruginosa in the Cystic Fibrosis Lung: A Single-Patient Longitudinal Genomic Study. Front Microbiol 9, 3242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klockgether J., Cramer N., Fischer S., Wiehlmann L., Tummler B., Long-Term Microevolution of Pseudomonas aeruginosa Differs between Mildly and Severely Affected Cystic Fibrosis Lungs. Am J Respir Cell Mol Biol 59, 246–256 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Gabrielaite M., Johansen H. K., Molin S., Nielsen F. C., Marvig R. L., Gene Loss and Acquisition in Lineages of Pseudomonas aeruginosa Evolving in Cystic Fibrosis Patient Airways. mBio 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datar R. et al. , Phenotypic and Genomic Variability of Serial Peri-Lung Transplantation Pseudomonas aeruginosa Isolates From Cystic Fibrosis Patients. Front Microbiol 12, 604555 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wardell S. J. T. et al. , Genome evolution drives transcriptomic and phenotypic adaptation in Pseudomonas aeruginosa during 20 years of infection. Microb Genom 7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson D. I., Nicoloff H., Hjort K., Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol 17, 479–496 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Reece E., Bettio P. H. A., Renwick J., Polymicrobial Interactions in the Cystic Fibrosis Airway Microbiome Impact the Antimicrobial Susceptibility of Pseudomonas aeruginosa. Antibiotics (Basel) 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galdino A. C. M., Vaillancourt M., Celedonio D., Jorth P., “Polymicrobial Biofilms in Cystic Fibrosis Lung Infections: Effects on Antimicrobial Susceptibility” in Multispecies Biofilms: Technologically Advanced Methods to Study Microbial Communities, Kaushik K. S., Darch S. E., Eds. (Springer International Publishing, Cham, 2023), 10.1007/978-3-031-15349-5, pp. 231–267. [DOI] [Google Scholar]
- 27.Mowat E. et al. , Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med 183, 1674–1679 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Ashish A. et al. , Extensive diversification is a common feature of Pseudomonas aeruginosa populations during respiratory infections in cystic fibrosis. J Cyst Fibros 12, 790–793 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workentine M. L. et al. , Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One 8, e60225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark S. T. et al. , Phenotypic diversity within a Pseudomonas aeruginosa population infecting an adult with cystic fibrosis. Sci Rep 5, 10932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams D. et al. , Divergent, coexisting Pseudomonas aeruginosa lineages in chronic cystic fibrosis lung infections. Am J Respir Crit Care Med 191, 775–785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams D. et al. , Transmission and lineage displacement drive rapid population genomic flux in cystic fibrosis airway infections of a Pseudomonas aeruginosa epidemic strain. Microb Genom 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer K. L., Aye L. M., Whiteley M., Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189, 8079–8087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petit R. A. 3rd, Read T. D., Bactopia: a Flexible Pipeline for Complete Analysis of Bacterial Genomes. mSystems 5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jolley K. A., Bray J. E., Maiden M. C. J., Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3, 124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wick R. R., Judd L. M., Gorrie C. L., Holt K. E., Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O. N. T. Research (2018) Medaka. (Oxford Nanopore Technologies; ). [Google Scholar]
- 38.Wick R. R., Holt K. E., Polypolish: Short-read polishing of long-read bacterial genome assemblies. PLoS Comput Biol 18, e1009802 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker B. J. et al. , Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seemann T. (2020) Snippy.
- 41.Seemann T., Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Otto T. D., Dillon G. P., Degrave W. S., Berriman M., RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res 39, e57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alcock B. P. et al. , CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res 51, D690–D699 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engels B. (2015) XNomial.
- 45.Croucher N. J. et al. , Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43, e15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seemann T., Shovill. (2020).
- 47.Page A. J. et al. , Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sprouffske K., Wagner A., Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17, 172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bürkner P., brms: An R Package for Bayesian Multilevel Models using Stan. Journal of Statistical Software 80, 1–28 (2017). [Google Scholar]
- 51.Bayliss S. C., Thorpe H. A., Coyle N. M., Sheppard S. K., Feil E. J., PIRATE: A fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. Gigascience 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toledo-Arana A., Repoila F., Cossart P., Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol 10, 182–188 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Hernando-Amado S., Laborda P., Valverde J. R., Martinez J. L., Mutational background influences P. aeruginosa ciprofloxacin resistance evolution but preserves collateral sensitivity robustness. Proc Natl Acad Sci U S A 119, e2109370119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hernando-Amado S., Laborda P., Valverde J. R., Martinez J. L., Rapid Decline of Ceftazidime Resistance in Antibiotic-Free and Sublethal Environments Is Contingent on Genetic Background. Mol Biol Evol 39 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Genova R., Laborda P., Cuesta T., Martinez J. L., Sanz-Garcia F., Collateral Sensitivity to Fosfomycin of Tobramycin-Resistant Mutants of Pseudomonas aeruginosa Is Contingent on Bacterial Genomic Background. Int J Mol Sci 24 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hernando-Amado S. et al. , Rapid Phenotypic Convergence towards Collateral Sensitivity in Clinical Isolates of Pseudomonas aeruginosa Presenting Different Genomic Backgrounds. Microbiol Spectr 11, e0227622 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roemhild R., Andersson D. I., Mechanisms and therapeutic potential of collateral sensitivity to antibiotics. PLoS Pathog 17, e1009172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su M., Satola S. W., Read T. D., Genome-Based Prediction of Bacterial Antibiotic Resistance. J Clin Microbiol 57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somayaji R. et al. , Antimicrobial susceptibility testing (AST) and associated clinical outcomes in individuals with cystic fibrosis: A systematic review. J Cyst Fibros 18, 236–243 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Macia M. D. et al. , Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother 49, 3382–3386 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cabot G. et al. , Evolution of Pseudomonas aeruginosa Antimicrobial Resistance and Fitness under Low and High Mutation Rates. Antimicrob Agents Chemother 60, 1767–1778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khil P. P. et al. , Dynamic Emergence of Mismatch Repair Deficiency Facilitates Rapid Evolution of Ceftazidime-Avibactam Resistance in Pseudomonas aeruginosa Acute Infection. mBio 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rees V. E. et al. , Characterization of Hypermutator Pseudomonas aeruginosa Isolates from Patients with Cystic Fibrosis in Australia. Antimicrob Agents Chemother 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Colque C. A. et al. , Hypermutator Pseudomonas aeruginosa Exploits Multiple Genetic Pathways To Develop Multidrug Resistance during Long-Term Infections in the Airways of Cystic Fibrosis Patients. Antimicrob Agents Chemother 64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colque C. A. et al. , Longitudinal Evolution of the Pseudomonas-Derived Cephalosporinase (PDC) Structure and Activity in a Cystic Fibrosis Patient Treated with beta-Lactams. mBio 13, e0166322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dulanto Chiang A. et al. , Hypermutator strains of Pseudomonas aeruginosa reveal novel pathways of resistance to combinations of cephalosporin antibiotics and beta-lactamase inhibitors. PLoS Biol 20, e3001878 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moriarty T. F., McElnay J. C., Elborn J. S., Tunney M. M., Sputum antibiotic concentrations: implications for treatment of cystic fibrosis lung infection. Pediatr Pulmonol 42, 1008–1017 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Ciofu O., Tolker-Nielsen T., Jensen P. O., Wang H., Hoiby N., Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv Drug Deliv Rev 85, 7–23 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Martin I., Waters V., Grasemann H., Approaches to Targeting Bacterial Biofilms in Cystic Fibrosis Airways. Int J Mol Sci 22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santi I., Manfredi P., Maffei E., Egli A., Jenal U., Evolution of Antibiotic Tolerance Shapes Resistance Development in Chronic Pseudomonas aeruginosa Infections. mBio 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Witzany C., Regoes R. R., Igler C., Assessing the relative importance of bacterial resistance, persistence and hyper-mutation for antibiotic treatment failure. Proc Biol Sci 289, 20221300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong A., Rodrigue N., Kassen R., Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet 8, e1002928 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jorgensen K. M. et al. , Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57, 4215–4221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jorth P. et al. , Evolved Aztreonam Resistance Is Multifactorial and Can Produce Hypervirulence in Pseudomonas aeruginosa. mBio 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahmed M. N., Porse A., Sommer M. O. A., Hoiby N., Ciofu O., Evolution of Antibiotic Resistance in Biofilm and Planktonic Pseudomonas aeruginosa Populations Exposed to Subinhibitory Levels of Ciprofloxacin. Antimicrob Agents Chemother 62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanz-Garcia F., Hernando-Amado S., Martinez J. L., Mutation-Driven Evolution of Pseudomonas aeruginosa in the Presence of either Ceftazidime or Ceftazidime-Avibactam. Antimicrob Agents Chemother 62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wardell S. J. T. et al. , A large-scale whole-genome comparison shows that experimental evolution in response to antibiotics predicts changes in naturally evolved clinical Pseudomonas aeruginosa. Antimicrob Agents Chemother 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahmed M. N. et al. , The evolutionary trajectories of P. aeruginosa in biofilm and planktonic growth modes exposed to ciprofloxacin: beyond selection of antibiotic resistance. NPJ Biofilms Microbiomes 6, 28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hernando-Amado S., Sanz-Garcia F., Blanco P., Martinez J. L., Fitness costs associated with the acquisition of antibiotic resistance. Essays Biochem 61, 37–48 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Barbosa C., Romhild R., Rosenstiel P., Schulenburg H., Evolutionary stability of collateral sensitivity to antibiotics in the model pathogen Pseudomonas aeruginosa. Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laborda P., Martinez J. L., Hernando-Amado S., Evolution of Habitat-Dependent Antibiotic Resistance in Pseudomonas aeruginosa. Microbiol Spectr 10, e0024722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jorth P. et al. , Evolved bacterial siderophore-mediated antibiotic cross-protection. Preprint available at 10.21203/rs.3.rs-2644953/v1 (2023). [DOI] [Google Scholar]
- 83.Diaz Caballero J. et al. , Mixed strain pathogen populations accelerate the evolution of antibiotic resistance in patients. Nat Commun 14, 4083 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sommer L. M. et al. , Is genotyping of single isolates sufficient for population structure analysis of Pseudomonas aeruginosa in cystic fibrosis airways? BMC Genomics 17, 589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Diaz Caballero J. et al. , Selective Sweeps and Parallel Pathoadaptation Drive Pseudomonas aeruginosa Evolution in the Cystic Fibrosis Lung. mBio 6, e00981–00915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Antimicrobial susceptibility testing measurements for Patient 1 as measured by zone of inhibition (ZOI) in a standard disc diffusion assay for amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Data in the left columns represent raw measurements of zone of inhibition radii (mm units). Data in the right columns represent calculated zone of inhibition values as diameters (mm units).
Supplemental Table 2. Antimicrobial susceptibility testing measurements for Patient 2 as measured by zone of inhibition (ZOI) in a standard disc diffusion assay for amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Data in the left columns represent raw measurements of zone of inhibition radii (mm units). Data in the right columns represent calculated zone of inhibition values as diameters (mm units).
Supplemental Table 3. Antimicrobial susceptibility testing measurements for Patient 3 as measured by zone of inhibition (ZOI) in a standard disc diffusion assay for amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Data in the left columns represent raw measurements of zone of inhibition radii (mm units). Data in the right columns represent calculated zone of inhibition values as diameters (mm units).
Supplemental Table 4. Antimicrobial susceptibility testing measurements for Patient 4 as measured by zone of inhibition (ZOI) in a standard disc diffusion assay for amikacin (AK), meropenem (MEM), piperacilin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Data in the left columns represent raw measurements of zone of inhibition radii (mm units). Data in the right columns represent calculated zone of inhibition values as diameters (mm units).
Supplemental Table 5. Genome size, average sequencing coverage, and number of contigs of each assembly.
Supplemental Table 6. Genes that were impacted by non-synonymous mutations in at least one isolate in all four populations.
Supplemental Table 7. Full details of the chi-squared goodness of fit and Monte Carlo simulation exact multinomial tests, with all associated chi-squared and p-values.
Supplemental Table 8. Genes that were impacted by non-synonymous mutations in at least one isolate in three out of four populations.
Supplemental Table 9. All annotated genetic variants discovered in Patient 1.
Supplemental Table 10. All annotated genetic variants discovered in Patient 2.
Supplemental Table 11. All annotated genetic variants discovered in Patient 3.
Supplemental Table 12. All annotated genetic variants discovered in Patient 4.
Supplemental Table 13. Raw OD600 reads for growth in SCFM used to create growth curves and analyze growth rate (r) for Patient 1. Time is given in hours, and all isolates were tested in biological triplicates.
Supplemental Table 14. Raw OD600 reads for growth in SCFM used to create growth curves and analyze growth rate (r) for Patient 2. Time is given in hours, and all isolates were tested in biological triplicates.
Supplemental Table 15. Raw OD600 reads for growth in SCFM used to create growth curves and analyze growth rate (r) for Patient 3. Time is given in hours, and all isolates were tested in biological triplicates.
Supplemental Table 16. Raw OD600 reads for growth in SCFM used to create growth curves and analyze growth rate (r) for Patient 4. Time is given in hours, and all isolates were tested in biological triplicates.
Supplemental Table 17. Supporting brms R code and statistical values for the linear mixed model run to assess the relationship between growth rate (r) and antimicrobial resistance. Results of the model, with growth rate in SCFM as a fixed effect and patient as a random effect, show that the 95% confidence interval of the fixed effect of growth rate spans 0 for all six antimicrobials. Therefore, the null hypothesis that the fixed effect of growth on antimicrobial susceptibility is 0 cannot be rejected, providing no evidence for trade-offs or any significant relationship between resistance and growth rate in these populations.
Supplemental Figure 1. Violin plot of the antimicrobial susceptibility profiles of all four populations against amikacin, meropenem, piperacillin-tazobactam, ciprofloxacin, tobramycin, and ceftazidime as measured by zone of inhibition in a standard disc diffusion assay. This plot displays the same data as Fig. 1, displayed in clusters by antibiotic rather than by population. Black horizontal bars indicate the cut-off values for susceptibility (top bar) and resistance (bottom bar) for each antibiotic as determined by the Clinical and Laboratory Standards Institute (CLSI).
Supplemental Figure 2. Phylogeny of Patients 1–4 with PAO1 and PA14. Patients 1, 2, and 4 cluster with PAO1, while Patient 3 clusters with PA14.
Supplemental Figure 3. Enrichment analysis of the frequency of functional categories in which non-synonymous SNPs and microindels are found in each of the four populations relative to the proportions of these functional categories in the PAO1 genome shows that protein secretion/export apparatuses and transcriptional regulators are enriched for such variants, while phage/transposon/ plasmid and non-coding RNA are less likely to be impacted by such variants. Donut plot of the relative frequencies of genes categorized within each of the 27 different PseudoCAP functional categories in the PAO1 genome (A). Donut plots of the relative frequencies of non-synonymous SNPs and indels located in each of the 27 different PseudoCAP functional categories in Patient 1 (B), 2 (C), 3 (D), and 4 (E).
Supplemental Figure 4. Lack of statistically significant negative correlations between any two antimicrobial susceptibility profiles provides no evidence for collateral sensitivity trade-offs. Pearson’s correlation coefficient (upper right quadrant), scatterplots (lower left quadrant), and density plots (diagonal) for pairwise comparisons of susceptibility profiles across all six tested antimicrobials: amikacin (AK), meropenem (MEM), piperacillin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ).
Supplemental Figure 5. Scatterplots of zone of inhibition (ZOI) versus growth rate (r) in SCFM for all six tested antibiotics: amikacin (AK), meropenem (MEM), piperacillin-tazobactam (TZP), ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ). Results of linear mixed model (Table S17), with growth rate in SCFM as a fixed effect and patient as a random effect, demonstrate that there is no significant effect of growth rate on resistance, and therefore, no evidence for trade-offs between growth rate and resistance in these four populations.
Data Availability Statement
The sequences in this study will be made available in the NCBI SRA database upon publication.