Abstract
Researchers and policymakers have proposed systems to detect novel pathogens early by monitoring samples from hospital patients, wastewater, and air travel, in order to mitigate future pandemics. How much benefit would such systems offer? We developed, empirically validated, and mathematically characterized a quantitative model that simulates disease spread and detection time for any given disease and detection system. We find that hospital monitoring could have detected COVID-19 in Wuhan 0.4 weeks earlier than it was actually discovered, at 2,300 cases compared to 3,400. Wastewater monitoring would not have accelerated COVID-19 detection in Wuhan, but provides benefit in smaller catchments and for asymptomatic or long-incubation diseases like polio or HIV/AIDS. Monitoring of air travel provides little benefit in most scenarios we evaluated. In sum, early detection systems can substantially mitigate some future pandemics, but would not have changed the course of COVID-19.
It has been widely debated which policies, if any, could have mitigated the initial stages of the COVID-19 pandemic in late 2019 and early 2020 as community transmission became established and widespread. Early studies compared non-pharmaceutical interventions (NPIs) such as mobility restrictions (1, 2), school closures (3, 4), voluntary home quarantine (5) and testing policies (6), and optimized NPI parameters like testing frequency (7), quarantine length (8), testing modality (9), test pooling (10) and intervention timing and ordering (11). While such NPIs undoubtedly slowed the early spread of COVID-19 (12) and previous outbreaks (13, 14), there has been little investigation of whether a separate strategy focused on earlier detection of COVID-19 would have enabled more successful mitigation. In theory, earlier detection enables a response when the outbreak is smaller: thus resource-intensive mitigation strategies like test-trace-isolate become less costly, and the earlier interventions are applied, the larger the number of infections and deaths that can be delayed until healthcare capacity is increased (15). However, the relevant question is not whether early detection helps, but quantitatively how much of a difference it would make. This question is especially urgent given current international and national policy proposals to invest billions of dollars in such systems (16, 17).
Researchers and policymakers have proposed immediate investments in systems to continuously monitor for novel pathogens in (i) patients with infectious symptoms in hospitals (18), (ii) community wastewater treatment plants (19, 20), and (iii) airplane sewage or bridge air on international flights (21–23), as well as other sites (24–28). These three sites have attracted interest because they have been frequent testing sites in COVID-19: hospitals since the pandemic’s beginning (29), and wastewater and air travel more recently (30, 31) because hospital cases can lag community cases (32). COVID-19 also spurred methodological innovation and characterization of sampling from these sites, particularly wastewater (33–35). Detecting novel pandemics at these sites has occasionally been piloted (20, 36) but has not been implemented at scale, in part because it is unclear if these proposed systems sufficiently expedite detection of outbreaks. The systems under consideration would use multiplex testing for conserved nucleic acid sequences of known pathogen families, exploiting the fact that many past emerging diseases belonged to such families, including SARS-CoV-2 (2019), Ebola (2013), MERS-CoV (2012), and pandemic flu (2009). Proposed technologies include multiplex PCR (37–40), CRISPR-based multiplex diagnostics (41), and metagenomic sequencing (42), possibly implemented with pooling (10).
To determine whether early detection of novel pathogens at these sites could be effective in changing the course of a pandemic, we first examined whether COVID-19 could have been detected earlier in Wuhan if systems had been in place in advance to monitor hospital, wastewater or air travel. To do this, we developed, empirically validated, and mathematically characterized a quantitative, branching process simulation-based model that predicts the number of cases at the time of detection given a detection system and a set of outbreak epidemiological parameters. We then used this model and COVID-19 epidemiological parameters (43) to estimate how early COVID-19 would initially have been detected in Wuhan by the three early detection systems, and compare this to the actual date of COVID-19 detection. Finally, we use our model to estimate detection times of infectious agents with different epidemiological properties, such as monkeypox and polio in recent outbreaks (44, 45), to inform pathogen-agnostic surveillance for future pandemics.
Model to estimate earliness of detection
Previous research (15) and our analysis (Supplementary text, figs. S1—5 and table S1) suggest that earlier COVID-19 lockdowns could have delayed cases and deaths. Thus it is critical to understand which early detection systems, if any, could have effectively enabled earlier response. To do this, we built a model that simulates outbreak spread and earliness of detection for a given outbreak and detection system (Materials and methods, Supplementary materials). This builds upon branching process models that have previously been used to model the spread of COVID-19 (46, 47) and other infectious diseases (48). A traditional branching process model starts from an index case and iteratively simulates each new generation of infections. Our model follows this pattern, but with each new infection we also simulate whether the infected person is detected by the detection system with some probability (Fig. 1A), and the simulation stops when the number of detected individuals equals the detection threshold and the detection delay has passed. Thus each detection system is characterized by these three parameters of detection probability, threshold, and delay (table S2). For example, in hospital monitoring, an infected individual’s detection probability is the probability they are sick enough to enter the hospital, which is the hospitalization rate (assuming testing has a negligible false negative rate). In systems that test individuals (hospital and air travel individual monitoring), the threshold is measured in an absolute number of cases. In systems that test wastewater (wastewater monitoring), the threshold is measured in terms of outbreak prevalence because wastewater monitoring can only sample a small percentage of sewage flows (49); thus a higher number of cases is required to trigger detection in a bigger community. We gathered literature estimates of detection system and outbreak parameters (tables S2 and S3) and validated wastewater monitoring sensitivity in independent data (fig. S6 and Materials and methods, Supplementary materials). We then empirically validated the model by testing its ability to predict the detection times for the first COVID-19 outbreaks in 50 US states in 2020. We gathered the dates of the first COVID-19 case reported by the public health department of each US state (table S4) as well as literature estimates of true (tested and untested) statewide COVID-19 case counts in early 2020 (50). Using our model, we were able to predict the number of weeks until travel-based detection in each US state to within a mean absolute error of 0.97 weeks (fig. S7 and S8).
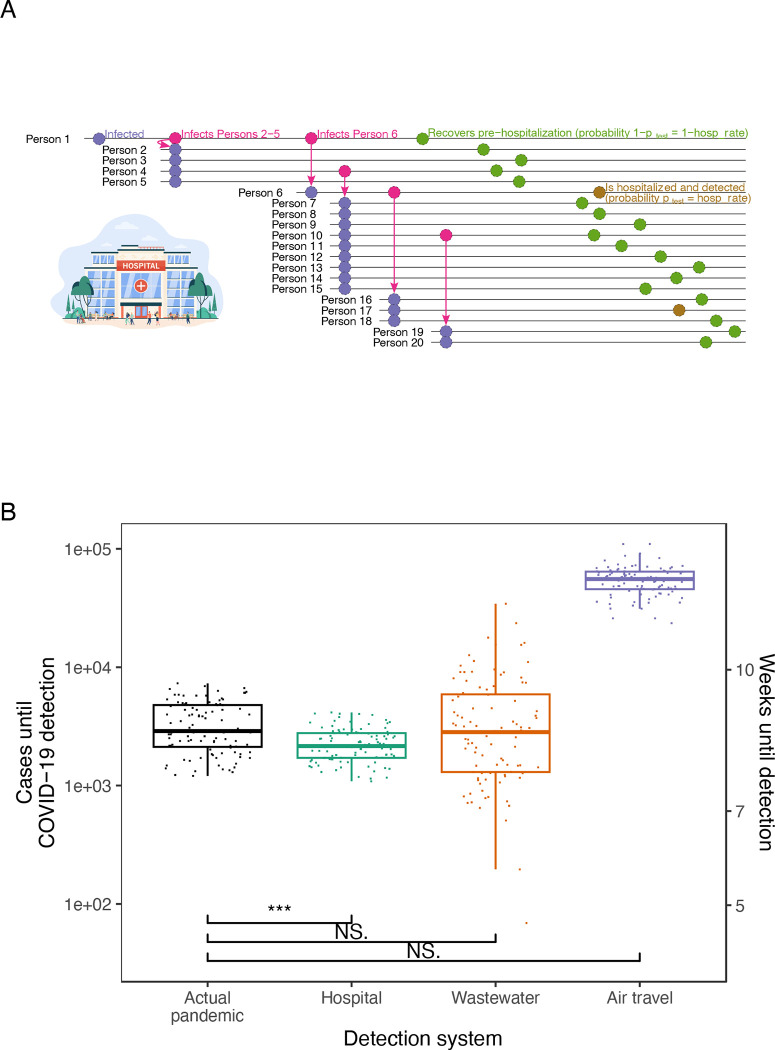
Fig. 1. Comparison of COVID-19 detection times in the actual pandemic versus with proposed early detection systems.
(A) Schematic of first 20 infections in a simulated run of the detection model. In this run, Person 1 seeds an outbreak in a community covered by a hospital detection system. Each person infects a number of individuals determined by a draw from a negative binomial distribution. Each person is then detected by the detection system with probability (gold) or goes undetected (olive); in the hospital system, equals the hospitalization rate. (B) Estimated cases until COVID-19 detection in the actual pandemic versus model-simulated cases until detection for proposed detection systems. Estimates for the actual pandemic are drawn from (51). Points for proposed detection systems are simulated case counts from the model (actual pandemic (black), hospital (teal), wastewater (orange) and air travel (purple)) assuming a Wuhan-sized catchment (650,000 people). Three, two, and one asterisk(s) signify statistically significant differences at the 0.001, 0.01, and 0.05 levels, respectively, in one-sided t-tests of between each detection system and the actual pandemic. NS. signifies not statistically significant at p=0.05. Equivalent weeks until detection are shown on the right y-axis.
Early detection’s impact on COVID-19 detection in Wuhan
Next we use our model to examine the detection systems’ ability to detect the first major COVID-19 outbreak in Wuhan (Fig. 1B and table S2). To estimate cases at detection in the actual pandemic, we used literature estimates of total (tested and untested) COVID-19 case counts in Wuhan in late 2019 and early 2020 (51). Our model shows that, on average, hospital monitoring could have detected COVID-19 after 2,292 cases. In reality, the pandemic was identified after 3,413 cases on average. Thus, hospital monitoring would have caught the outbreak 1,121 cases earlier (approximately 0.43 weeks earlier), a statistically significant difference with p = 1.9e-09 and t = −6.3 in one-sided Welch two-sample t-test. Wastewater monitoring would have lagged detection in the actual pandemic; it caught the outbreak after 4,575 cases, or 1,162 cases later, on average (p = 0.018 and t = 2.1). We tested this wastewater prediction empirically by calculating the cases until COVID-19 wastewater detection in Massachusetts in early 2020, using literature-estimated Massachusetts COVID-19 cases (50) and Massachusetts wastewater SARS-CoV-2 PCR data (52); our model prediction was consistent with this analysis (fig. S9). Because we model wastewater monitoring to detect later in larger communities (Materials and methods, Supplementary materials), the Wuhan result is in part due to Wuhan’s 650,000-person catchments. Wastewater monitoring would lead status quo detection of COVID-19 in catchments smaller than 480,000 people, well above the global mean catchment size of 25,000 people (53). Air travel monitoring did not provide any acceleration of detection because of the low probability of simultaneously traveling and being sick.
Early detection’s impact for other diseases: mathematical analysis and simulation
To make our model easily usable for outbreaks beyond COVID-19, we derived a compact formula that approximates the model’s simulations. We observed that, without accounting for the delay of generations between the threshold case’s infection and detection, the number of cases until detection, , is a random variable that follows a negative binomial distribution by definition: each infected case is a Bernoulli trial, “success” in that trial occurs when that case enters the detection system (with a probability we name ), and we count the number of cases until the number of successes equals the detection threshold . After accounting for , we derived a formula approximating the mean of when the outbreak starts in a community covered by the detection system (see Supplementary Text for full derivation):
| (1) |
We confirmed our formula approximates the simulation model closely by comparing the detection times predicted by both for all the detection systems for multiple diseases (fig. S10). Thus the formula allows us to interpret the model and the quantitative relationships between detection times and various variables: the formula shows that the number of cases until detection increases linearly with the detection threshold, increases polynomially with and exponentially with the delay as , and decreases as the fraction of cases being tested increases. This formula also makes the model easily usable for detection systems beyond the ones studied here.
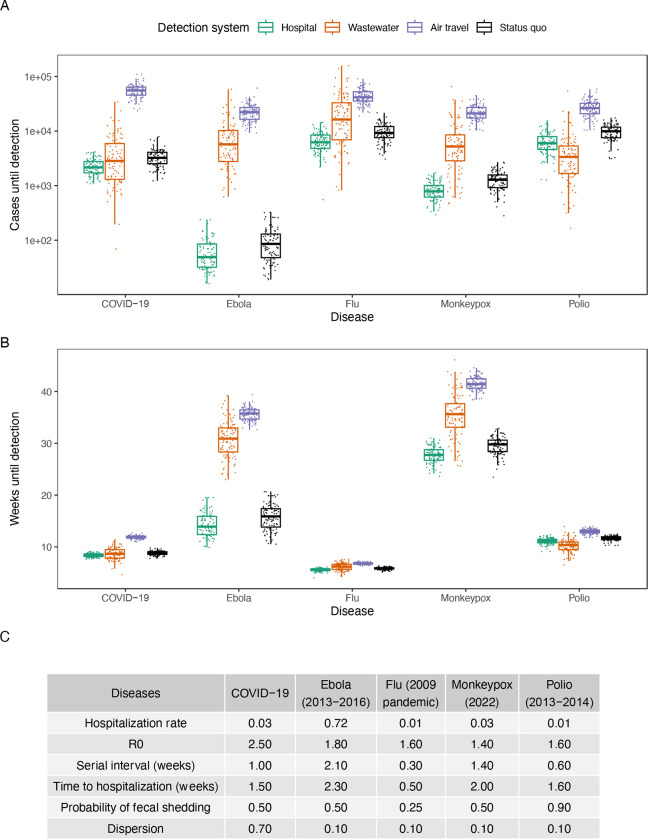
We applied our model to several outbreaks of recent interest–including COVID-19, monkeypox (2022), polio (2013–2014), Ebola (2013–2016) and flu (2009 pandemic)–and found that the detection systems vary in their success depending on the epidemiological parameters of the agent (Figs. 2, S11 and S12, and table S3). For example, in our model hospital monitoring tends to outperform wastewater monitoring when the hospitalization rate is high, as in the case of Ebola, but tends to underperform for diseases like polio, in which the hospitalization rate is low and when there is high asymptomatic spread in the delay from detection to hospitalization. This is consistent with Equation (1), as well as previous observations that Ebola was first detected in hospitals (54) and that wastewater monitoring has been more effective than hospital monitoring for detecting polio (55). We also modeled the status quo detection times for these outbreaks: the number of cases until these outbreaks were detected in the status quo, without the proposed detection systems in place. We found that early detection systems can catch outbreaks when they are up to 52% smaller (wastewater for polio) or 110 weeks earlier (hospital for HIV/AIDS) (figs. S13, S14, S15 and S16).
Fig. 2. Comparison of detection systems for different infectious diseases.
(A) Earliness of detection for detection systems in cases across infectious diseases (hospital (teal), wastewater (orange), air travel (purple) and status quo (black)) in a 650,000-person catchment. (B) Earliness of detection for detection systems in weeks across infectious diseases in a 650,000-person catchment. (C) Epidemiological parameters of the studied diseases.
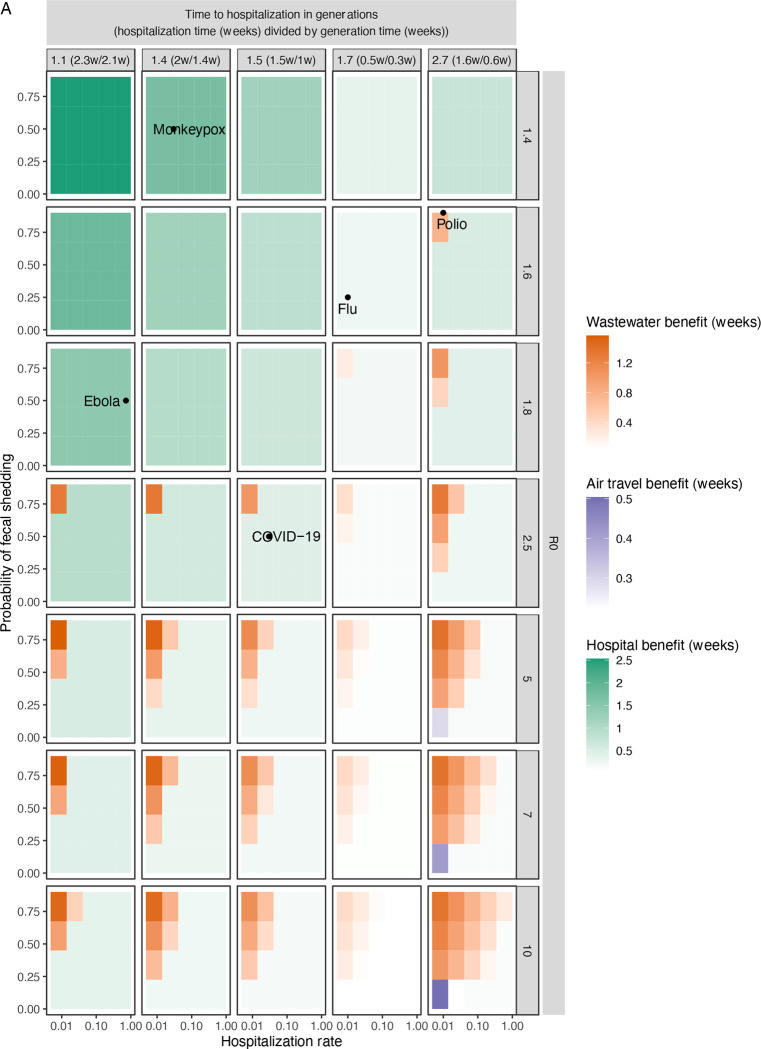
Because future infectious diseases are likely to have different epidemiological parameters, we generalized the previous analysis and calculated detection times for many possible diseases spanning the epidemiological parameter space (Figs. 3 and S17). As expected, hospital monitoring is the best system for diseases with higher hospitalization rates and lower times to hospitalization. For diseases with higher R0s and times to hospitalization, wastewater monitoring emerges as the best system more often, because hospital monitoring has a longer detection delay (mainly the time from infection to hospitalization) than wastewater (mainly the time from infection to fecal shedding), during which cases grow exponentially with R0. However, this holds mainly for diseases with high probability of fecal shedding and low hospitalization rate. Air travel monitoring, which did not perform well in the previous modeled diseases (Figs. 1 and 2), actually performed best for a few diseases for which fecal shedding is low (disadvantaging wastewater monitoring) and the time to hospitalization and R0 are too large (disadvantaging hospital monitoring).
Fig. 3. Comparison of detection systems across the space of possible diseases of varying epidemiological parameters.
(A) Average weeks gained over status quo detection by the proposed detection systems across the epidemiological space of possible diseases. Within each panel, each uniformly colored cell corresponds to a specific disease with the hospitalization rate and probability of fecal shedding indicated on the x- and y-axes, as well as the R0 and time to hospitalization (generations) indicated by the panel row and column. The cell has a hue corresponding to the detection system that detects the disease the earliest (hospital (teal), wastewater (orange) and air travel (purple)) and an intensity corresponding to the number of weeks gained by the earliest system over status quo detection. Times are calculated by the derived mathematical approximation in a 650,000-person catchment.
Our results show that the benefits of early detection systems vary from marginal (0.4 weeks earlier for COVID-19) to significant (110 weeks earlier for HIV/AIDS) (Figs. 1B, 2, and S16). Our detection time model (Fig. 1A) can be used for many diseases and detection systems, including other systems beyond this study (24, 25), by varying the fraction of the infected population being tested in each system. Two further points are worth emphasizing. First, early detection only aids mitigation if it leads to a coordinated early response. Many factors beyond detection affect the pace of response, including the economic and political feasibility of lockdowns, the availability of medicines and personal protective equipment, and whether there are pre-determined policies to be implemented upon detection. Second, when deciding to invest in these systems, one must consider factors such as cost-effectiveness and whether the system provides evidence of disease severity. Although wastewater monitoring gives earlier detection than hospital monitoring in multiple diseases (Fig. 3A), it does not discriminate between mild and severe disease. In contrast, hospital monitoring provides evidence that the detected pathogen produces symptoms that require hospital treatment.
These results can inform ongoing international and national policy debates about which policies are needed to mitigate future pandemics. In the wake of COVID-19, the World Health Organization Intergovernmental Negotiating Body is actively negotiating a new treaty on international pandemic preparedness which updates the International Health Regulations (2005). Drafts of this treaty highlight “early warning and alert systems” as key measures (16). Similarly, the presidential administration of the United States has proposed investing $5.3 billion over 7 to 10 years in early warning and real-time monitoring systems, including in hospitals and wastewater (17). In this study, we have assessed detection systems’ detection times and have developed a model to assess current and future detection system proposals. Along with additional cost-effectiveness analysis and technical pilots (20), these results can help inform which detection systems are most effective and thus worth funding in pandemic preparedness efforts.
Supplementary Material
Acknowledgments:
We gratefully acknowledge Mauricio Santillana, Nicholas B. Link, Fred S. Lu, and Andre T. Nguyen for sharing their estimates on COVID-19 incidence in the US states. We also acknowledge Jonathan Pekar, Joel Wertheim, and Michael Worobey for sharing their estimates on COVID-19 incidence in late 2019 and early 2020. We finally acknowledge Michael McLaren, Becky Ward, and Quincey Justman for feedback on the manuscript.
Funding:
Lynch Foundation Fellows Program in Systems Biology at Harvard Medical School (ABL) National Library of Medicine grant T15LM007092 (DL)
National Institutes of Health grant R01GM120122 (APJ)
CDC contract 200-2016-91779 (WPH)
National Institutes of Health grant R01GM120122 (MS)
Footnotes
Competing interests: WPH is a member of the scientific advisory board and has stock options in BioBot Analytics. MS is a cofounder of Rhinostics and consults for the diagnostic consulting company Vectis Solutions LLC. The other authors declare that they have no competing interests.
References and Notes:
- 1.Kraemer M. U. G., Yang C.-H., Gutierrez B., Wu C.-H., Klein B., Pigott D. M., OPEN COVID-19 DATA WORKING GROUP, du Plessis L., Faria N. R., Li R., Hanage W. P., Brownstein J. S., Layan M., Vespignani A., Tian H., Dye C., Pybus O. G., Scarpino S. V., The effect of human mobility and control measures on the COVID-19 epidemic in China. Science. 368, 493–497 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyerowitz-Katz G., Bhatt S., Ratmann O., Brauner J. M., Flaxman S., Mishra S., Sharma M., Mindermann S., Bradley V., Vollmer M., Merone L., Yamey G., Is the cure really worse than the disease? The health impacts of lockdowns during COVID-19. BMJ Global Health. 6, e006653 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamey G., Walensky R. P., Covid-19: Re-opening universities is high risk. BMJ. 370, m3365 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Brauner J. M., Mindermann S., Sharma M., Johnston D., Salvatier J., Gavenčiak T., Stephenson A. B., Leech G., Altman G., Mikulik V., Norman A. J., Monrad J. T., Besiroglu T., Ge H., Hartwick M. A., Teh Y. W., Chindelevitch L., Gal Y., Kulveit J., Inferring the effectiveness of government interventions against COVID-19. Science. 371, eabd9338 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson N. M., Laydon D., Nedjati-Gilani G., Imai N., Ainslie K., Baguelin M., Bhatia S., Boonyasiri A., Cucunub Z., Cuomo-Dannenburg G., Dighe A., Dorigatti I., Fu H., Gaythorpe K., Green W., Hamlet A., Hinsley W., Okell L. C., van Elsland S., Thompson H., Verity R., Volz E., Wang H., Wang Y., Walker P. G., Walters C., Winskill P., Whittaker C., Donnelly C. A., Riley S., Ghani A. C., “Report 9 - Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand” (2020), (available at https://www.imperial.ac.uk/medicine/departments/school-public-health/infectious-disease-epidemiology/mrc-global-infectious-disease-analysis/covid-19/report-9-impact-of-npis-on-covid-19/).
- 6.Levine-Tiefenbrun M., Yelin I., Uriel H., Kuint J., Schreiber L., Herzel E., Katz R., Ben-Tov A., Gazit S., Patalon T., Chodick G., Kishony R., SARS-CoV-2 RT-qPCR Test Detection Rates Are Associated with Patient Age, Sex, and Time since Diagnosis. The Journal of Molecular Diagnostics. 24, 112–119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larremore D. B., Wilder B., Lester E., Shehata S., Burke J. M., Hay J. A., Tambe M., Mina M. J., Parker R., Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Science Advances, eabd5393 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu A. B., Davidi D., Landsberg H. E., Francesconi M., Platt J. T., Nguyen G. T., Yune S., Deckard A., Puglin J., Haase S. B., Hamer D. H., Springer M., Association of COVID-19 Quarantine Duration and Postquarantine Transmission Risk in 4 University Cohorts. JAMA Network Open. 5, e220088 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyllie A. L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J. L., Geng B., Muenker M. C., Moore A. J., Vogels C. B. F., Petrone M. E., Ott I. M., Lu P., Venkataraman A., Lu-Culligan A., Klein J., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L. R., Valdez J., White E. B., Lapidus S., Kalinich C. C., Jiang X., Kim D. J., Kudo E., Linehan M., Mao T., Moriyama M., Oh J. E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.-E., Wong P., Yang Y., Bermejo S., Odio C. D., Omer S. B., Dela Cruz C. S., Farhadian S., Martinello R. A., Iwasaki A., Grubaugh N. D., Ko A. I., Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. New England Journal of Medicine. 383, 1283–1286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yelin I., Aharony N., Tamar E. S., Argoetti A., Messer E., Berenbaum D., Shafran E., Kuzli A., Gandali N., Shkedi O., Hashimshony T., Mandel-Gutfreund Y., Halberthal M., Geffen Y., Szwarcwort-Cohen M., Kishony R., Evaluation of COVID-19 RT-qPCR Test in Multi sample Pools. Clinical Infectious Diseases. 71, 2073–2078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karin O., Bar-On Y. M., Milo T., Katzir I., Mayo A., Korem Y., Dudovich B., Yashiv E., Zehavi A. J., Davidovitch N., Milo R., Alon U., Cyclic exit strategies to suppress COVID-19 and allow economic activity (2020), doi: 10.1101/2020.04.04.20053579. [DOI] [Google Scholar]
- 12.Flaxman S., Mishra S., Gandy A., Unwin H. J. T., Mellan T. A., Coupland H., Whittaker C., Zhu H., Berah T., Eaton J. W., Monod M., Ghani A. C., Donnelly C. A., Riley S., Vollmer M. A. C., Ferguson N. M., Okell L. C., Bhatt S., Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 584, 257–261 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Hatchett R. J., Mecher C. E., Lipsitch M., Public health interventions and epidemic intensity during the 1918 influenza pandemic. Proceedings of the National Academy of Sciences. 104, 7582–7587 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peak C. M., Childs L. M., Grad Y. H., Buckee C. O., Comparing nonpharmaceutical interventions for containing emerging epidemics. Proceedings of the National Academy of Sciences. 114, 4023–4028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei S., Kandula S., Shaman J., Differential effects of intervention timing on COVID-19 spread in the United States. Science Advances. 6, eabd6370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bureau of the Intergovernmental Negotiating Body, World Health Organization, “Conceptual zero draft for the consideration of the Intergovernmental Negotiating Body at its third meeting” (2022), p. 32. [Google Scholar]
- 17.Lander E., Sullivan J., American Pandemic Preparedness: Transforming Our Capabilities, 27 (2021). [Google Scholar]
- 18.Ecker D. J., How to Snuff Out the Next Pandemic. Scientific American Blog Network (2020), (available at https://blogs.scientificamerican.com/observations/how-to-snuff-out-the-next-pandemic/). [Google Scholar]
- 19.Peccia J., Zulli A., Brackney D. E., Grubaugh N. D., Kaplan E. H., Casanovas-Massana A., Ko A. I., Malik A. A., Wang D., Wang M., Warren J. L., Weinberger D. M., Arnold W., Omer S. B., Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nature Biotechnology. 38, 1164–1167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Nucleic Acid Observatory Consortium, A Global Nucleic Acid Observatory for Biodefense and Planetary Health. arXiv:2108.02678 [q-bio] (2021) (available at http://arxiv.org/abs/2108.02678). [Google Scholar]
- 21.Hjelmsø M. H., Mollerup S., Jensen R. H., Pietroni C., Lukjancenko O., Schultz A. C., Aarestrup F. M., Hansen A. J., Metagenomic analysis of viruses in toilet waste from long distance flights—A new procedure for global infectious disease surveillance. PLOS ONE. 14, e0210368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muntean J., Howard K., Atwood P., CDC has tested wastewater from aircraft amid concerns over Covid-19 surge in China. CNN; (2023), (available at https://www.cnn.com/2023/01/05/health/airplane-wastewater-covid-testing/index.html). [Google Scholar]
- 23.Nordahl Petersen T., Rasmussen S., Hasman H., Carøe C., Bælum J., Charlotte Schultz A., Bergmark L., Svendsen C. A., Lund O., Sicheritz-Pontén T., Aarestrup F. M., Metagenomic analysis of toilet waste from long distance flights; a step towards global surveillance of infectious diseases and antimicrobial resistance. Scientific Reports. 5, 11444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mina M. J., Metcalf C. J. E., McDermott A. B., Douek D. C., Farrar J., Grenfell B. T., A Global Immunological Observatory to meet a time of pandemics. eLife. 9, e58989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu H. Y., Englund J. A., Starita L. M., Famulare M., Brandstetter E., Nickerson D. A., Rieder M. J., Adler A., Lacombe K., Kim A. E., Graham C., Logue J., Wolf C. R., Heimonen J., McCulloch D. J., Han P. D., Sibley T. R., Lee J., Ilcisin M., Fay K., Burstein R., Martin B., Lockwood C. M., Thompson M., Lutz B., Jackson M., Hughes J. P., Boeckh M., Shendure J., Bedford T., Early Detection of Covid-19 through a Citywide Pandemic Surveillance Platform. New England Journal of Medicine. 383, 185–187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brownstein J. S., Freifeld C. C., Madoff L. C., Digital Disease Detection — Harnessing the Web for Public Health Surveillance. The New England journal of medicine. 360, 2153–2157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dugas A. F., Jalalpour M., Gel Y., Levin S., Torcaso F., Igusa T., Rothman R. E., Influenza Forecasting with Google Flu Trends. PLOS ONE. 8, e56176 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajema N., Beaver W., Parthemore C., “Toward a Global Pathogen Early Warning System: Building on the Landscape of Biosurveillance Today” (2021), (available at https://councilonstrategicrisks.org/wp-content/uploads/2021/07/Toward-A-Global-Pathogen-Early-Warning-System_2021_07_20-1.pdf).
- 29.Lee V. J., Chiew C. J., Khong W. X., Interrupting transmission of COVID-19: Lessons from containment efforts in Singapore. Journal of Travel Medicine. 27 (2020), doi: 10.1093/jtm/taaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boehm A. B., Hughes B., Wolfe M. K., White B. J., Duong D., Chan-Herur V., Regional Replacement of SARS-CoV-2 Variant Omicron BA.1 with BA.2 as Observed through Wastewater Surveillance. Environmental Science & Technology Letters. 9, 575–580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J. E., Lee J. H., Lee H., Moon S. J., Nam E. W., COVID-19 screening center models in South Korea. Journal of Public Health Policy. 42, 15–26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meakin S., Abbott S., Bosse N., Munday J., Gruson H., Hellewell J., Sherratt K., Chapman L. A. C., Prem K., Klepac P., Jombart T., Knight G. M., Jafari Y., Flasche S., Waites W., Jit M., Eggo R. M., Villabona-Arenas C. J., Russell T. W., Medley G., Edmunds W. J., Davies N. G., Liu Y., Hué S., Brady O., Pung R., Abbas K., Gimma A., Mee P., Endo A., Clifford S., Sun F. Y., McCarthy C. V., Quilty B. J., Rosello A., Sandmann F. G., Barnard R. C., Kucharski A. J., Procter S. R., Jarvis C. I., Gibbs H. P., Hodgson D., Lowe R., Atkins K. E., Koltai M., Pearson C. A. B., Finch E., Wong K. L. M., Quaife M., O’Reilly K., Tully D. C., Funk S., CMMID COVID-19 Working Group, Comparative assessment of methods for short-term forecasts of COVID-19 hospital admissions in England at the local level. BMC Medicine. 20, 86 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregory D. A., Wieberg C. G., Wenzel J., Lin C.-H., Johnson M. C., Monitoring SARS-CoV-2 Populations in Wastewater by Amplicon Sequencing and Using the Novel Program SAM Refiner. Viruses. 13, 1647 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C., Bayati M., Hsu S.-Y., Hsieh H.-Y., Lindsi W., Belenchia A., Zemmer S. A., Klutts J., Samuelson M., Reynolds M., Semkiw E., Johnson H.-Y., Foley T., Wieberg C. G., Wenzel J., Lyddon T. D., LePique M., Rushford C., Salcedo B., Young K., Graham M., Suarez R., Ford A., Antkiewicz D. S., Janssen K. H., Shafer M. M., Johnson M. C., Lin C.-H., Population Normalization in SARS-CoV-2 Wastewater-Based Epidemiology: Implications from Statewide Wastewater Monitoring in Missouri (2022), doi: 10.1101/2022.09.08.22279459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson C. A., Hsieh H.-Y., Hsu S.-Y., Wang Y., Salcedo B. T., Belenchia A., Klutts J., Zemmer S., Reynolds M., Semkiw E., Foley T., Wan X., Wieberg C. G., Wenzel J., Lin C.-H., Johnson M. C., Defining biological and biophysical properties of SARS-CoV-2 genetic material in wastewater. Science of The Total Environment. 807, 150786 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bibby K., Peccia J., Identification of Viral Pathogen Diversity in Sewage Sludge by Metagenome Analysis. Environmental Science & Technology. 47, 1945–1951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creager H. M., Cabrera B., Schnaubelt A., Cox J. L., Cushman-Vokoun A. M., Shakir S. M., Tardif K. D., Huang M.-L., Jerome K. R., Greninger A. L., Drobysheva D., Spaulding U., Rogatcheva M., Bourzac K. M., Hinrichs S. H., Broadhurst M. J., Fey P. D., Clinical evaluation of the BioFire® Respiratory Panel 2.1 and detection of SARS-CoV-2. Journal of Clinical Virology. 129, 104538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edin A., Eilers H., Allard A., Evaluation of the Biofire Filmarray Pneumonia panel plus for lower respiratory tract infections. Infectious Diseases. 52, 479–488 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Murphy C. N., Fowler R., Balada-Llasat J. M., Carroll A., Stone H., Akerele O., Buchan B., Windham S., Hopp A., Ronen S., Relich R. F., Buckner R., Warren D. A., Humphries R., Campeau S., Huse H., Chandrasekaran S., Leber A., Everhart K., Harrington A., Kwong C., Bonwit A., Dien Bard J., Naccache S., Zimmerman C., Jones B., Rindlisbacher C., Buccambuso M., Clark A., Rogatcheva M., Graue C., Bourzac K. M., Multicenter Evaluation of the BioFire FilmArray Pneumonia/Pneumonia Plus Panel for Detection and Quantification of Agents of Lower Respiratory Tract Infection. Journal of Clinical Microbiology. 58, e00128–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quick J., Grubaugh N. D., Pullan S. T., Claro I. M., Smith A. D., Gangavarapu K., Oliveira G., Robles-Sikisaka R., Rogers T. F., Beutler N. A., Burton D. R., Lewis-Ximenez L. L., de Jesus J. G., Giovanetti M., Hill S. C., Black A., Bedford T., Carroll M. W., Nunes M., Alcantara L. C., Sabino E. C., Baylis S. A., Faria N. R., Loose M., Simpson J. T., Pybus O. G., Andersen K. G., Loman N. J., Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature Protocols. 12, 1261–1276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackerman C. M., Myhrvold C., Thakku S. G., Freije C. A., Metsky H. C., Yang D. K., Ye S. H., Boehm C. K., Kosoko-Thoroddsen T.-S. F., Kehe J., Nguyen T. G., Carter A., Kulesa A., Barnes J. R., Dugan V. G., Hung D. T., Blainey P. C., Sabeti P. C., Massively multiplexed nucleic acid detection with Cas13. Nature. 582, 277–282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu C. Y., Miller S. A., Clinical metagenomics. Nature Reviews Genetics. 20, 341–355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bar-On Y. M., Sender R., Flamholz A. I., Phillips R., Milo R., A quantitative compendium of COVID-19 epidemiology. arXiv:2006.01283 [q-bio] (2020) (available at https://arxiv.org/abs/2006.01283). [Google Scholar]
- 44.Du Z., Shao Z., Bai Y., Wang L., Herrera-Diestra J. L., Fox S. J., Ertem Z., Lau E. H. Y., Cowling B. J., Reproduction number of monkeypox in the early stage of the 2022 multi-country outbreak. Journal of Travel Medicine, taac099 (2022). [DOI] [PubMed] [Google Scholar]
- 45.U.S. Centers for Disease Control and Prevention (CDC), United States confirmed as country with circulating vaccine-derived poliovirus. CDC; (2022), (available at https://www.cdc.gov/media/releases/2022/s0913-polio.html). [Google Scholar]
- 46.Bradshaw W. J., Alley E. C., Huggins J. H., Lloyd A. L., Esvelt K. M., Bidirectional contact tracing could dramatically improve COVID-19 control. Nature Communications. 12, 232 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hellewell J., Abbott S., Gimma A., Bosse N. I., Jarvis C. I., Russell T. W., Munday J. D., Kucharski A. J., Edmunds W. J., Sun F., Flasche S., Quilty B. J., Davies N., Liu Y., Clifford S., Klepac P., Jit M., Diamond C., Gibbs H., van Zandvoort K., Funk S., Eggo R. M., Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. The Lancet Global Health. 8, e488–e496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lloyd-Smith J. O., Schreiber S. J., Kopp P. E., Getz W. M., Superspreading and the effect of individual variation on disease emergence. Nature. 438, 355–359 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bushman M., Chai P. R., Duvallet C., Erickson T. B., Foppe K., Ghaeli N., Gu X., Hanage W. P., Huang K. H., Lee W. L., McElroy K. A., Rhode S. F., Matus M., Wuertz S., Thompson J., Alm E. J., Wastewater surveillance of SARS-CoV-2 across 40 U.S. States from February to June 2020. Water Research. 202, 117400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu F. S., Nguyen A. T., Link N. B., Molina M., Davis J. T., Chinazzi M., Xiong X., Vespignani A., Lipsitch M., Santillana M., Estimating the cumulative incidence of COVID-19 in the United States using influenza surveillance, virologic testing, and mortality data: Four complementary approaches. PLOS Computational Biology. 17, e1008994 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pekar J. E., Magee A., Parker E., Moshiri N., Izhikevich K., Havens J. L., Gangavarapu K., Malpica Serrano L. M., Crits-Christoph A., Matteson N. L., Zeller M., Levy J. I., Wang J. C., Hughes S., Lee J., Park H., Park M.-S., Ching Zi Yan K., Lin R. T. P., Mat Isa M. N., Noor Y. M., Vasylyeva T. I., Garry R. F., Holmes E. C., Rambaut A., Suchard M. A., Andersen K. G., Worobey M., Wertheim J. O., The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2. Science. 377, 960–966 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massachusetts Water Resources Authority, MWRA - Wastewater COVID-19 Tracking (2022), (available at https://www.mwra.com/biobot/biobotdata.htm).
- 53.Adhikari S., Halden R. U., Opportunities and limits of wastewater-based epidemiology for tracking global health and attainment of UN sustainable development goals. Environment International. 163, 107217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sack K., Fink S., Belluck P., Nossiter A., How Ebola Roared Back. The New York Times; (2014) (available at https://www.nytimes.com/2014/12/30/health/how-ebola-roared-back.html). [Google Scholar]
- 55.Brouwer A. F., Eisenberg J. N. S., Pomeroy C. D., Shulman L. M., Hindiyeh M., Manor Y., Grotto I., Koopman J. S., Eisenberg M. C., Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proceedings of the National Academy of Sciences. 115, E10625–E10633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M. A., Bushman M., Chai P. R., Duvallet C., Erickson T. B., Foppe K., Ghaeli N., Gu X., Hanage W. P., Huang K. H., Lee W. L., Matus M., McElroy K. A., Nagler J., Rhode S. F., Santillana M., Tucker J. A., Wuertz S., Zhao S., Thompson J., Alm E. J., SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Science of The Total Environment. 805, 150121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soller J., Jennings W., Schoen M., Boehm A., Wigginton K., Gonzalez R., Graham K. E., McBride G., Kirby A., Mattioli M., Modeling infection from SARS-CoV-2 wastewater concentrations: Promise, limitations, and future directions. Journal of Water and Health. 20, 1197–1211 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones D. L., Baluja M. Q., Graham D. W., Corbishley A., McDonald J. E., Malham S. K., Hillary L. S., Connor T. R., Gaze W. H., Moura I. B., Wilcox M. H., Farkas K., Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. The Science of the Total Environment. 749, 141364 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Analytics Biobot, “The Effect of Septic Systems on Wastewater-Based Epidemiology” (2022), (available at http://biobot.io/wp-content/uploads/2022/09/BIOBOT_WHITEPAPER_EFFECT_OF_SEPTIC_V01-1.pdf).
- 60.Mallela A., Neumann J., Miller E. F., Chen Y., Posner R. G., Lin Y. T., Hlavacek W. S., Bayesian Inference of State-Level COVID-19 Basic Reproduction Numbers across the United States. Viruses. 14, 157 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldberg C., Mass. Public Health Lab Can Now Test For New Coronavirus, Speeding Results (2020), (available at https://www.wbur.org/news/2020/02/28/mass-public-health-coronavirus-testing).
- 62.Pekar J., Worobey M., Moshiri N., Scheffler K., Wertheim J. O., Timing the SARS-CoV-2 index case in Hubei province. Science. 372, 412–417 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pekar J. E., Magee A., Parker E., Moshiri N., Izhikevich K., Havens J. L., Gangavarapu K., Serrano L. M. M., Crits-Christoph A., Matteson N. L., Zeller M., Levy J. I., Wang J. C., Hughes S., Lee J., Park H., Park M.-S., Yan K. C. Z., Lin R. T. P., Isa M. N. M., Noor Y. M., Vasylyeva T. I., Garry R. F., Holmes E. C., Rambaut A., Suchard M. A., Andersen K. G., Worobey M., Wertheim J. O., Sars-cov-2-origins / multi-introduction. GitHub; (2022), (available at https://github.com/sars-cov-2-origins/multi-introduction). [Google Scholar]
- 64.Yuan Yao, Yujie Ma, Jialu Zhou, Wenkun Hou, Xinhua Headlines: Chinese doctor recalls first encounter with mysterious virus - Xinhua English.news.cn. Xinhua (2020), (available at https://web.archive.org/web/20200423140325/https://www.xinhuanet.com/english/2020-04/16/c_138982435.htm). [Google Scholar]
- 65.The 2019-nCoV Outbreak Joint Field Epidemiology Investigation Team, Li Q, An Outbreak of NCIP (2019-nCoV) Infection in China — Wuhan, Hubei Province, 2019–2020. China CDC Weekly. 2, 79–80 (2020). [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenthal E., THE SARS EPIDEMIC: THE PATH; From China’s Provinces, a Crafty Germ Breaks Out. The New York Times; (2003) (available at https://www.nytimes.com/2003/04/27/world/the-sars-epidemic-the-path-from-china-s-provinces-a-crafty-germ-breaks-out.html). [Google Scholar]
- 67.Garrett L., The Coming Plague (1994).
- 68.Center for Systems Science and Engineering (CSSE) at Johns Hopkins University, COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (2022), (available at https://github.com/CSSEGISandData/COVID-19).
- 69.Mathieu E., Ritchie H., Rodés-Guirao L., Appel C., Giattino C., Hasell J., Macdonald B., Dattani S., Beltekian D., Ortiz-Ospina E., Roser M., Total COVID-19 tests per 1,000 people. Our World in Data (2020) (available at https://ourworldindata.org/coronavirus-testing#world-map-total-tests-performed-relative-to-the-size-of-population). [Google Scholar]
- 70.Harris T. E., “The theory of branching processes” (1964), (available at https://www.rand.org/content/dam/rand/pubs/reports/2009/R381.pdf).
- 71.Yoo S. J., Moon S. J., Kuak E.-Y., Yoo H. M., Kim C. K., Chey M.-J., Shin B.-M., Frequent Detection of Pandemic (H1N1) 2009 Virus in Stools of Hospitalized Patients. Journal of Clinical Microbiology. 48, 2314–2315 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vetter P., Fischer II W. A., Schibler M., Jacobs M., Bausch D. G., Kaiser L., Ebola Virus Shedding and Transmission: Review of Current Evidence. The Journal of Infectious Diseases. 214, S177–S184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Independent Evaluation Department, Asia Development Bank, “People’s Republic of China: Wuhan Wastewater and Stormwater Management Project” (2016), (available at https://www.adb.org/sites/default/files/evaluation-document/188852/files/pvr-447.pdf).
- 74.Faes C., Abrams S., Van Beckhoven D., Meyfroidt G., Vlieghe E., Hens N., Time between Symptom Onset, Hospitalisation and Recovery or Death: Statistical Analysis of Belgian COVID-19 Patients. International Journal of Environmental Research and Public Health. 17, 7560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He D., Zhao S., Xu X., Lin Q., Zhuang Z., Cao P., Wang M. H., Lou Y., Xiao L., Wu Y., Yang L., Low dispersion in the infectiousness of COVID-19 cases implies difficulty in control. BMC Public Health. 20, 1558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang S., Zhang F., Yuan Z., Xu M., Wang Z., Gao C., Guo R., Du Z., Serial intervals and incubation periods of the monkeypox virus clades. Journal of Travel Medicine, taac105 (2022). [DOI] [PubMed] [Google Scholar]
- 77.DeWitt M. E., Polk C., Williamson J., Shetty A. K., Passaretti C. L., McNeil C. J., Fairman R. T., Sampson M. M., Dalton C., Sanders J. W., Global monkeypox case hospitalisation rates: A rapid systematic review and meta-analysis. eClinicalMedicine. 54, 101710 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reda A., Hemmeda L., Brakat A. M., Sah R., El-Qushayri A. E., The clinical manifestations and severity of the 2022 monkeypox outbreak among 4080 patients. Travel Medicine and Infectious Disease. 50, 102456 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryan E. T., Hill D. R., Solomon T., Aronson N., Endy T. P., Hunter’s Tropical Medicine and Emerging Infectious Diseases E-Book (Elsevier Health Sciences, 2019). [Google Scholar]
- 80.Mailhe M., Beaumont A.-L., Thy M., Le Pluart D., Perrineau S., Houhou-Fidouh N., Deconinck L., Bertin C., Ferré V. M., Cortier M., De La Porte Des Vaux C., Phung B.-C., Mollo B., Cresta M., Bouscarat F., Choquet C., Descamps D., Ghosn J., Lescure F.-X., Yazdanpanah Y., Joly V., Peiffer-Smadja N., Clinical characteristics of ambulatory and hospitalized patients with monkeypox virus infection: An observational cohort study. Clinical Microbiology and Infection (2022), doi: 10.1016/j.cmi.2022.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tiwari A., Adhikari S., Kaya D., Islam Md. A., Malla B., Sherchan S. P., Al-Mustapha A. I., Kumar M., Aggarwal S., Bhattacharya P., Bibby K., Halden R. U., Bivins A., Haramoto E., Oikarinen S., Heikinheimo A., Pitkänen T., Monkeypox outbreak: Wastewater and environmental surveillance perspective. Science of The Total Environment. 856, 159166 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolfe M. K., Duong D., Hughes B., Chan-Herur V., White B. J., Boehm A. B., Detection of monkeypox viral DNA in a routine wastewater monitoring program (2022), doi: 10.1101/2022.07.25.22278043. [DOI] [Google Scholar]
- 83.U.S. Centers for Disease Control and Prevention (CDC) National Center for Health Statistics, Immunization (2022), (available at https://www.cdc.gov/nchs/fastats/immunize.htm).
- 84.World Health Organization, Circulating vaccine-derived poliovirus type 3 – Israel (2022), (available at https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON366).
- 85.Estivariz Concepcion F., Link-Gelles Ruth, Shimabukuro Tom, Pinkbook: Poliomyelitis CDC (2022), (available at https://www.cdc.gov/vaccines/pubs/pinkbook/polio.html).
- 86.Nelson K. E., Williams C. M., “Determinants of Epidemic Growth. In Chapter 6: Infectious Disease Dynamics” in Infectious Disease Epidemiology: Theory and Practice (Jones & Bartlett Learning, Burlington, Mass, 3rd edition., 2013), p. 135. [Google Scholar]
- 87.Chung P. W., Huang Y. C., Chang L. Y., Lin T. Y., Ning H. C., Duration of enterovirus shedding in stool. Journal of microbiology, immunology, and infection. 34, 167–170 (2001). [PubMed] [Google Scholar]
- 88.Onorato I. M., Modlin J. F., McBean A. M., Thoms M. L., Losonsky G. A., Bernier R. H., Mucosal Immunity Induced by Enhanced-Potency Inactivated and Oral Polio Vaccines. The Journal of Infectious Diseases. 163, 1–6 (1991). [DOI] [PubMed] [Google Scholar]
- 89.Wong Z. S. Y., Bui C. M., Chughtai A. A., Macintyre C. R., A systematic review of early modelling studies of Ebola virus disease in West Africa. Epidemiology & Infection. 145, 1069–1094 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Drake J. M., Kaul R. B., Alexander L. W., O’Regan S. M., Kramer A. M., Pulliam J. T., Ferrari M. J., Park A. W., Ebola Cases and Health System Demand in Liberia. PLOS Biology. 13, e1002056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Kerkhove M. D., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A., A review of epidemiological parameters from Ebola outbreaks to inform early public health decision-making. Scientific Data. 2, 150019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qureshi A. I., Chughtai M., Bah E. I., Barry M., Béavogui K., Loua T. O., Malik A. A., High Survival Rates and Associated Factors Among Ebola Virus Disease Patients Hospitalized at Donka National Hospital, Conakry, Guinea. Journal of Vascular and Interventional Neurology. 8, S4–S11 (2015). [PMC free article] [PubMed] [Google Scholar]
- 93.Fraser C., Donnelly C. A., Cauchemez S., Hanage W. P., Van Kerkhove M. D., Hollingsworth T. D., Griffin J., Baggaley R. F., Jenkins H. E., Lyons E. J., Jombart T., Hinsley W. R., Grassly N. C., Balloux F., Ghani A. C., Ferguson N. M., Rambaut A., Pybus O. G., Lopez-Gatell H., Alpuche-Aranda C. M., Chapela I. B., Zavala E. P., Ma D.. Guevara E., Checchi F., Garcia E., Hugonnet S., Roth C., Pandemic Potential of a Strain of Influenza A (H1N1): Early Findings. Science (New York, N.Y.). 324, 1557–1561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heijnen L., Medema G., Surveillance of Influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in the Netherlands. Journal of Water and Health. 9, 434–442 (2011). [DOI] [PubMed] [Google Scholar]
- 95.Presanis A. M., Angelis D. D., T. N. Y. C. S. F. I. Team3¶, Hagy A., Reed C., Riley S., Cooper B. S., Finelli L., Biedrzycki P., Lipsitch M., The Severity of Pandemic H1N1 Influenza in the United States, from April to July 2009: A Bayesian Analysis. PLOS Medicine. 6, e1000207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Furuya-Kanamori L., Cox M., Milinovich G. J., Magalhaes R. J. S., Mackay I. M., Yakob L., Heterogeneous and Dynamic Prevalence of Asymptomatic Influenza Virus Infections. Emerging Infectious Diseases. 22, 1052–1056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lessler J., Reich N. G., Cummings D. A. T., Outbreak of 2009 Pandemic Influenza A (H1N1) at a New York City School. New England Journal of Medicine. 361, 2628–2636 (2009). [DOI] [PubMed] [Google Scholar]
- 98.Louie J. K., Acosta M., Winter K., Jean C., Gavali S., Schechter R., Vugia D., Harriman K., Matyas B., Glaser C. A., Samuel M. C., Rosenberg J., Talarico J., Hatch D., for the California Pandemic (H1N1) Working Group, Factors Associated With Death or Hospitalization Due to Pandemic 2009 Influenza A(H1N1) Infection in California. JAMA. 302, 1896–1902 (2009). [DOI] [PubMed] [Google Scholar]
- 99.Vynnycky E., White R., An Introduction to Infectious Disease Modelling (Oxford University Press, New York, Illustrated edition., 2010). [Google Scholar]
- 100.Morgan D., Mahe C., Mayanja B., Okongo J. M., Lubega R., Whitworth J. A. G., HIV-1 infection in rural Africa: Is there a difference in median time to AIDS and survival compared with that in industrialized countries? AIDS (London, England). 16, 597–603 (2002). [DOI] [PubMed] [Google Scholar]
- 101.Todd J., Glynn J. R., Marston M., Lutalo T., Biraro S., Mwita W., Suriyanon V., Rangsin R., Nelson K. E., Sonnenberg P., Fitzgerald D., Karita E., Żaba B., Time from HIV seroconversion to death: A collaborative analysis of eight studies in six low and middle-income countries before highly active antiretroviral therapy. AIDS (London, England). 21, S55 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.CASCADE, Time from HIV-1 seroconversion to AIDS and death before widespread use of highly-active antiretroviral therapy: A collaborative re-analysis. Collaborative Group on AIDS Incubation and HIV Survival including the CASCADE EU Concerted Action. Concerted Action on SeroConversion to AIDS and Death in Europe. Lancet (London, England). 355, 1131–1137 (2000). [PubMed] [Google Scholar]
- 103.Crum-Cianflone N. F., HIV and the Gastrointestinal Tract. Infectious diseases in clinical practice (Baltimore, Md.). 18, 283–285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yolken R. H., Li S., Perman J., Viscidi R., Persistent Diarrhea and Fecal Shedding of Retroviral Nucleic Acids in Children Infected with Human Immunodeficiency Virus. The Journal of Infectious Diseases. 164, 61–66 (1991). [DOI] [PubMed] [Google Scholar]
- 105.World Health Organization, Tuberculosis (TB) (2022), (available at https://www.who.int/news-room/fact-sheets/detail/tuberculosis).
- 106.Behr M. A., Edelstein P. H., Ramakrishnan L., Revisiting the timetable of tuberculosis. The BMJ. 362, k2738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma Y., Horsburgh C. R., White L. F., Jenkins H. E., Quantifying TB transmission: A systematic review of reproduction number and serial interval estimates for tuberculosis. Epidemiology and Infection. 146, 1478–1494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.