Abstract
Human genetic studies indicate that suicidal ideation and behavior are both heritable. Most studies have examined associations between aberrant gene expression and suicide behavior, but behavior risk is linked to severity of suicidal ideation. Through a gene network approach, this study investigates how gene co-expression patterns are associated with suicidal ideation and severity using RNA-seq data in peripheral blood from 46 live participants with elevated suicidal ideation and 46 with no ideation. Associations with presence and severity of suicidal ideation were found within 18 and 3 co-expressed modules respectively (p < 0.05), not explained by severity of depression. Suicidal ideation presence and severity-related gene modules with enrichment of genes involved in defense against microbial infection, inflammation, and adaptive immune response were identified, and tested using RNA-seq data from postmortem brain that revealed gene expression differences in suicide decedents vs. non-suicides in white matter, but not gray matter. Findings support a role of brain and peripheral blood inflammation in suicide risk, showing that suicidal ideation presence and severity is associated with an inflammatory signature detectable in blood and brain, indicating a biological continuity between ideation and suicidal behavior that may underlie a common heritability.
1. INTRODUCTION
Between 2010 and 2020, suicide was the 12th most common cause of death in the US 1 . Suicidal behaviors contribute to significant health and economic burden, exceeding $70 billion per year in the United States alone (CDC, 2020). Although genetics, together with environmental and other individual factors, contribute to suicidal risk 2 , family and twin studies have been foundational in establishing the genetic contribution to suicidality, showing higher frequency of suicide attempt and death in monozygotic twins compared to dizygotic twins among suicide twin survivors but not non-suicide twin survivors 3 . Family studies suggest a significant genetic contribution with heritability ranging from 30 to 55% for suicidal ideation (SI), behavior, and death 4 5 , with additive genetic effect on the continuum of the phenotype spanning suicidal ideation, behavior, and death 6 .
Clinical studies link more severe SI, like that characterized by a plan and intent, to imminent risk of suicide attempts. Most studies have examined associations between aberrant gene expression and suicide behavior, but consideration of how such transcriptional profiles align with SI and evidence for a suicidal behavior-ideation phenotype continuum is lacking. Transcriptional studies that capture the spectrum of suicide risks using peripheral blood and brain samples can capture common elements across the molecular pathologies between ideation and suicidal behavior that may underlie a common phenotypic heritability5, 6. Such research may provide a genomic explanation for suicidal behavior as a risk factor for future suicidal acts7. To intervene effectively and efficiently, new methods are needed to identify not just who is at risk, but also to identify, using dynamic markers, times when they are at highest risk. This remains challenging, as individual vulnerability to suicide appears to be complex and multidimensional, with many contributing sociodemographic, genetic and environmental factors 8–10.
In recent years, there has been increasing interest in harnessing genomic technologies and large-scale genomic data to improve understanding of the underlying biology of suicide. One approach used to study the biology of suicide is to examine gene transcript expression patterns. Gene transcription are a functional output of genes expressed in a molecular pathway or process. Biological signals inducing gene transcript expression can vary in timescales from milliseconds, to seconds, hours, days, or even decades with varying transcriptional kinetics 11. Like the observed temporal dynamics in transcriptional activity, SI is also an inherently dynamic construct12, 13. As such, expression activity of genes associated with SI in comparison to expression patterns in brain of suicide decedents can provide an important snapshot of the underlying biology contributing to the intensity of suicidal state. One of the early seminal findings that resulted from genome-scale transcriptional regulatory studies in peripheral blood identified the gene SKA2 (Spindle and Kinetochore Associated Complex Subunit 2) as a potential biomarker of suicide risk and ideation14 that prospectively predicted suicidal ideation and behavior in psychiatric patients15, 16. Subsequently, using whole genome transcript profiling via RNA-Seq, altered gene expression for spermidine/spermine N1- Acetyltransferase1 (SAT1) in blood also was shown to be a potential peripheral suicide risk biomarker 14.
However, complex phenotypes of suicide may involve interactions of multiple intertwined genes, transcription factors and gene co-expression clusters (i.e. genes with correlated levels of expression) in cells with modular but diverse functional patterns 17. Co-expression gene clusters represent coherent functional pathways in both normal conditions18 as well as in disease states17. Gene network approaches for identification of clusters of co-expressed genes have been applied towards discovery of genes involved in common molecular processes associated with suicidal behavior, as well as psychiatric conditions19. Gene network approaches capture gene-gene interactions by regarding genes as nodes and interactions as links, in which the association between each gene pair is measured by a similarity score or statistical significance of the association, whereby co-expressed clusters are then identified. Commonly used methods to generate gene co-expression networks include the Weighted Gene Co-expression Network Analysis (WGCNA)20 and the Multiscale Embedded Gene Expression Network Analysis (MEGENA)21 used in the present study. Such approaches have been widely applied in systems biology and brain research to study gene co-expression across such psychiatric and neurodegenerative diseases as depression 22, bipolar disorder 23, schizophrenia 24, and Alzheimer’s disease 25, 26.
Although gene network approaches have been applied in transcriptome studies of suicide in human postmortem brain tissue from suicide decedents27, to our knowledge, no study has investigated transcriptome profiles of SI in clinical samples using gene co-expression network methods to uncover biological processes and pathways underlying SI. As SI and gene transcripts are both highly dynamic, identification of gene networks associated with SI or its severity could be of utility in clinical translational studies for development of blood biomarker profiles that can be used to identify individuals at risk for suicide. To this end, for the first aim of the present study, MEGENA21 was used to identify co-expressed gene modules associated with SI and severity in the two weeks before the blood sample was taken. The second aim of the present study was to determine whether these SI associated co-expressed modules were also differentially expressed in gray and white matter postmortem human brain specimens of suicide decedent cases and non-suicide controls, to determine which elements of peripheral blood gene expression changes related to SI are also found in brain gene expression related to suicide death. This common pathophysiology may reflect common heritable elements between ideation and suicide or perhaps the intense suicidal ideation at the time of suicide.
2. METHODS
2.1. Samples & Subjects
AH studies were approved by the Institutional Review Board of the New York State Psychiatric Institute, IRB number for live subjects is #4815 and postmortem studies is 7351R.
2.2. Live subject sample recruitment and evaluation
Study participants were adults 20–65 years old and were recruited in the New York metropolitan area by advertising, via the Columbia University Medical Center Portal for research subject volunteers and through patient referrals from clinics and mental health professionals. Participants were evaluated at New York Psychiatric Institute. Study participants comprised three groups: 1. Healthy controls (HC, n = 27); 2. participants with a major depressive disorder (MDD) diagnosis with no history of suicide attempt (MDD/NS, n = 50); and 3. participants with MDD diagnosis with history of suicide attempt (MDD/SA, n = 23) (Table 1). All participants were evaluated using the Structured Clinical Interview for DSM-IV (SCID I) 28 by trained clinicians, with raters being at least Master’s level psychologists or psychiatric nurses. Axis I and II disorders were assessed using Structured Clinical Interviews for DSM-IV and Structured Clinical Interviews for DSM-IV Axis II Disorders 29. Healthy volunteers had no personal history of Axis I disorders, cluster B personality disorder, substance use disorder and lifetime history of suicide attempt, and had no first-degree relatives with a history of mood disorders, psychotic disorders or suicidal behavior.
Table 1.
Demographic and clinical characteristics for 100 subjects for MEGENA network building.
| Total N = 100 |
MDD/SA N = 23 |
MDD/NS N = 50 |
HC N = 27 |
P-value | Sig. pair | |
|---|---|---|---|---|---|---|
| Age, Mean ± sd | 36.4 ± 11.4 | 36.3 ± 11.1 | 37.5 ± 11.2 | 34.7 ± 12.3 | 0.5906 | NA |
| Sex, n(%) | ||||||
| Male | 40 (40%) | 8 (35%) | 21 (42%) | 11 (41%) | 0.8393 | NA |
| Female | 60 (60%) | 15 (65%) | 29(58%) | 16(59%) | ||
| Ethnicity, n(%) | ||||||
| Hispanic | 17 (17%) | 3 (13%) | 8 (16%) | 6 (22%) | 0.7342# | NA |
| Non-Hispanic | 83 (83%) | 20 (87%) | 42 (84%) | 21 (78%) | ||
| Race, n(%) | ||||||
| White | 59 (59%) | 15 (65%) | 31 (62%) | 13 (48%) | 0.2138# | NA |
| Black or African American | 23 (23%) | 5 (22%) | 8 (16%) | 10 (37%) | ||
| Asian | 11 (11%) | 3 (13%) | 7 (14%) | 1 (4%) | ||
| Other | 7 (7%) | 0 (0%) | 4 (8%) | 3 (11%) | ||
| Beck Scale for Suicidal ideation (SSI), Mean ± sd | 6.7 ± 8.6 | 12.6 ± 9.0 | 7.6 ± 8.4 | 0±0 | < 0.0001# | MDD/NS > HC, MDD/SA > HC |
| Beck Depression Inventory (BDI), Mean ± sd | 17.1 ± 14.8 | 27.2 ± 12.0 | 22.1 ± 12.6 | 0.9 ± 2.2 | < 0.0001# | MDD/NS > HC, MDD/SA > HC |
| Hamilton Depression Rating Scales (HAM17), Mean ± sd | 13.6 ± 10.2 | 20.9 ± 5.8 | 19.9 ± 5.2 | 1.3 ± 1.5 | < 0.0001# | MDD/NS > HC, MDD/SA > HC |
| Lethality of most lethal actual suicide attempt median [IQR] | 1 [0–3] | |||||
| Suicidal ideation group, n* | ||||||
| SSI = 0 (No-SI) | 46 | 4 | 15 | 27 | ||
| SSI ≥ 5 (High-SI) | 46 | 19 | 27 | 0 | ||
| 0 < SSI < 5 (Low-SI)* | 8 | 0 | 8 | 0 | ||
A total of 92 subjects had No or High ideation group assignment.
non-parametric tests: Fisher’s exact for count, Kruskal-Wallis test for three group comparison with post-hoc Wilcoxon rank sum test using Bonferroni correction, and Wilcoxon rank sum test for two group comparison.
Low-SI subjects were excluded for SI group comparison.
Depressive symptoms were assessed by a research clinician using the 17-item Hamilton Depression Rating Scale (HAM17)30. Patients’ subjective perception of depression severity was assessed by means of the Beck Depression Inventory (BDI)31. The number, method and degree of medical damage of past suicide attempts were recorded on the Columbia Suicide History Form32. Suicidal ideation was measured by the Beck Scale for Suicidal ideation (SSI)33. Moderate to elevated SI was defined as SSI ≥ 5 corresponding to reported suicidal ideation in the last two weeks prior to the assessment, and no ideation was defined by SSI = 0.
Suicide attempt was defined as a self-injurious act with some degree of intent to end one’s life. Severity of suicide attempts was characterized by using the Suicide Intent Scale34, which assessed the patient’s expectation regarding the outcome of the suicidal behavior, and the Lethality Rating Scale34, which assessed the degree of medical injury resulting from the attempt. Psychiatric diagnoses and suicide attempts were verified in a consensus conference with research psychologists and psychiatrists. All subjects underwent physical examinations, and venous blood samples were collected in RNA Paxgene vacutainer tubes.
2.3. Postmortem brain samples
Brain samples were obtained from the Macedonian/New York State Psychiatric Institute Brain Collection in the Molecular Imaging and Neuropathology at the New York State Psychiatric Institute. Cause of death, excluding suicide, was determined by the medical examiner. All cases and controls were psychiatrically characterized by psychological autopsies, using a validated psychological autopsy interview method with at least one significant other 35. Diagnoses of major psychiatric disorder were determined using the SCID I 36 and suicide was determined using the Columbia Classification Algorithm for Suicide Assessment 37.
All suicide deaths and non-suicide deaths were sudden, without prolonged agonal state, absence of psychotropic or illegal drugs on history in the past three months confirmed by toxicological screens. Suicide decedents had a lifetime diagnosis of MDD, while sudden death controls did not have an Axis I psychiatric disorder. In total, the postmortem human brain samples included: 1. white matter cases with 9 controls i.e., non-psychiatric non-suicides who died of accidental causes and 15 suicide decedents and 2. grey matter cases with 29 controls and 21 suicide decedents (RNA-seq data for gray matter samples were downloaded from GEO database accession# GSE101521). Details on subject demographics and postmortem interval-PMI and brain pH are provided in supplemental Table S5. For tissue dissection, prefrontal cortical regions, specifically the dorsal gray matter (BA 9) and ventral white matter (BA 47) were used from frozen brain sections.
2.4. RNA Sequencing
For live samples, PAXgene blood tubes were thawed quickly and brought to room temperature prior to total RNA isolation according to the manufacturer’s instruction, and Globin mRNA was removed using GLOBINclear (Invitrogen/Ambion). For postmortem human brain specimens, total RNA was isolated from ~ 30–50 mg of gray or white matter tissue using RNeasy Lipid Tissue Mini Kit (Qiagen, #74804) according to the manufacturer’s instruction. All RNA samples were subject to Ribo-zero depletion, and libraries prepared using the Illumina Truseq library preparation kit and sequenced on the Illumina HiSeq 2500 (2 × 50), generating a mean of 48 million reads per sample. Reads were aligned to hg19 using STAR aligner and annotated to transcripts using Gencode v18 annotation.
2.5. Statistical and network analysis.
RNA-seq data for the live and postmortem samples were filtered separately, based on expression level and sample variability. Lowly-expressed genes with 0.5 count per million (CPM) in fewer than 2% of samples or with average log2CPM < 4 were excluded from all analyses. Variably expressed genes with log2CPM standard deviation ≥ 0.35 after adjustment for age and sex (via limma38), were included in the network analysis performed using MEGENA on the live samples only. Recommended default software parameters were used, i.e., minimum module size set to 10 and using Pearson correlation for calculation of correlation in gene co-expression in building Planar Filtered Networks (PFN)39. MEGENA multiscale clustering analysis was performed in the manner of hierarchical division to dissect the PFNs into coherent modules with nested clusters at various scales of resolution. Each module was then tested for association with presence of SI (SSI ≥ 5 vs. SSI = 0) and separately for ideation severity within the ideator group. Note, sample size for the low ideation group (i.e., SSI range 1-to-4, N = 8, Table 1) was limited and thus excluded from the comparison, yet still included in the MEGENA analysis.
For each module, gene expression patterns were represented by an eigengene, i.e., the first principal component of the expression levels computed using prcomp function in R. Differences in gene expression patterns between the group with moderate to elevated SI with SSI ≥ 5 (denoted as the high-SI group, n = 46) vs. the non-ideator group (SSI = 0, n = 46) was tested using the Wilcoxon rank sum test. Within the high-SI group, Spearman correlation coefficient with the eigengene expression was computed to assess association of suicidal ideation severity with gene expression patterns for each module. For modules found significantly associated with severity of SI, post-hoc analyses were also performed to test if observed associations with suicidal ideation severity’ persisted after adjusting for participants depression severity, measured by BDI and HAM17 with suicide related items within these instruments removed. These analyses were run in separate models using robust regression via lmrob function from robustbase40 in R, where the eigengene was set as the response, ideation severity set as the predictor and depression severity set as the covariate. In the postmortem brain RNA-seq data, age and sex adjusted log2CPM gene expression data were used to compute the eigengene using eigenvector coefficients obtained from MEGENA network analysis from the live samples. For comparison of module gene expression patterns between suicide decedent and non-psychiatric non-suicide cases, robust Cohen’s d (via d.robust from pysch41) effect sizes were computed and reported, instead of performing significance testing, due to sample size limitations.
2.6. Ingenuity pathway and upstream regulator analysis
All unnested modules significantly associated with suicidal ideation and severity (p < 0.05) were included in Ingenuity Pathway Analysis (IPA, QIAGEN Inc.,)42, using following thresholds: pathways that were significantly (Benjamini Hochberg adjusted BH p<0.1) upregulated (z-score>2) or downregulated (z-score < −2). Result of clusters with significant pathway were reported as circos plots using GOChord function from GOplot43 package in R. For upstream analysis and thresholds and filtering used as follows: p < 0.05, |z-score| > 2 and upstream regulators were filtered for genes, RNAs, and proteins. All p-values for IPA were computed using Fisher’s exact tests.
3. RESULTS
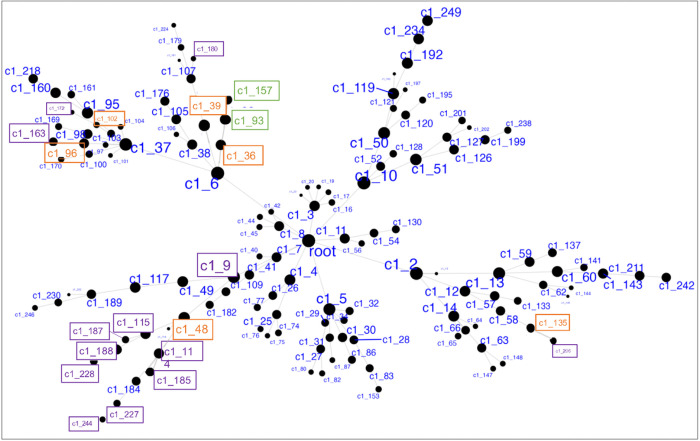
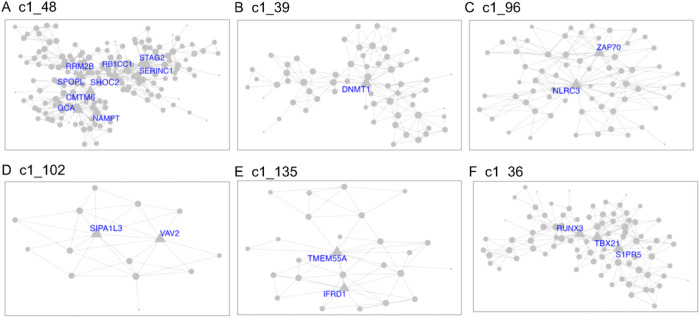
Demographics and relevant clinical measurements for the N = 100 live subjects are shown in Table 1, by recruitment group. Groups did not differ by age, sex, race or ethnicity, but as expected, differed on depression and suicidal ideation scales. Of these, 92 were included in gene network MEGENA analyses: 46 participants who reported elevated suicidal ideation (denoted as high-SI with SSI ≥ 5), and 46 who endorsed no ideation (denoted as no-SI, SSI = 0), irrespective of diagnostic status. To identify gene expression signatures associated with SI, peripheral blood gene co-expression analysis was performed in live samples. Using 2740 variably expressed genes from the live samples, the MEGENA analysis produced 135 modules ranging from 10 to 607 genes per module (Fig. 1). Eigengenes representing the overall gene expression pattern of each module were computed and compared between the high-SI vs. the no-SI groups using the Wilcoxon rank sum test, resulting in 18-modules associated with SI (Table S2). Of these, 5 modules with no overlapping genes (denoted as “unnested-modules”) were identified (c1_48, c1_39, c1_96, c1_102 and c1_135, p< 0.05, see Fig. 2A–E and Table S2). Although the c1_9 module consisting of 278 genes associated with SI, this parent cluster contained two child clusters i.e, c1_48 and c1_149 modules in which the c1_49 module with 113 genes showed no association with SI (p = 0.1586); MEGENA identified the SI associated module c1_48 with superior specificity, which was then used for downstream functional analyses. To determine whether these modules’ associations with SI could be explained by diagnostic group or depression severity-related differences, post-hoc pairwise analyses were performed by comparing each module’s eigengene across the three groups (HC vs. MDD/NS vs. MDD/SA) using the Kruskal Wallis test. Two modules with significant group differences (c1_39, p = 0.0493; and its child cluster c1_180, p = 0.0458) showed significant medium sized difference between MDD/SA vs. HC (p = 0.0421, d = 0.52, Table S2 and Fig. 3) for c1_39 and trend-wise medium sized difference between MDD/SA vs. HC (p = 0.0556, d = 0.60, Table S2 and Fig. 3) for c1_180. No significant differences were observed for the remaining two comparisons for the modules c1_39 and c1_180 (p > 0.05, Table S2). Amongst the high-SI participants, 3 nested modules (Table S3) were associated with ideation severity, corresponding to modules c1_36 (rs = −0.32, p = 0.0310, Fig. 2F) and corresponding child modules c1_93 (rs = −0.32, p = 0.0278), and c1_157 (rs= 0.34, p = 0.0205). The associations between ideation severity and gene expression patterns for these three modules remained significant after adjusting for depression severity (Table S3).
Figure 1.
Hierarchical network structure of clusters via MEGENA. Highlighted in purple are 5 un-nested clusters (and their child clusters, total 18 clusters) showing significant differences in expression between High-SI and No-SI. In green are 2 nested clusters showing significant association with suicidal ideation severity among High SI. In orange are the 6 parent clusters.
Figure 2.
MEGENA modules identified from suicide ideation group comparisons high-SI vs. no-SI (A-E) and ideation severity analysis within the high-SI group (F), with hub genes annotated by corresponding gene symbols.
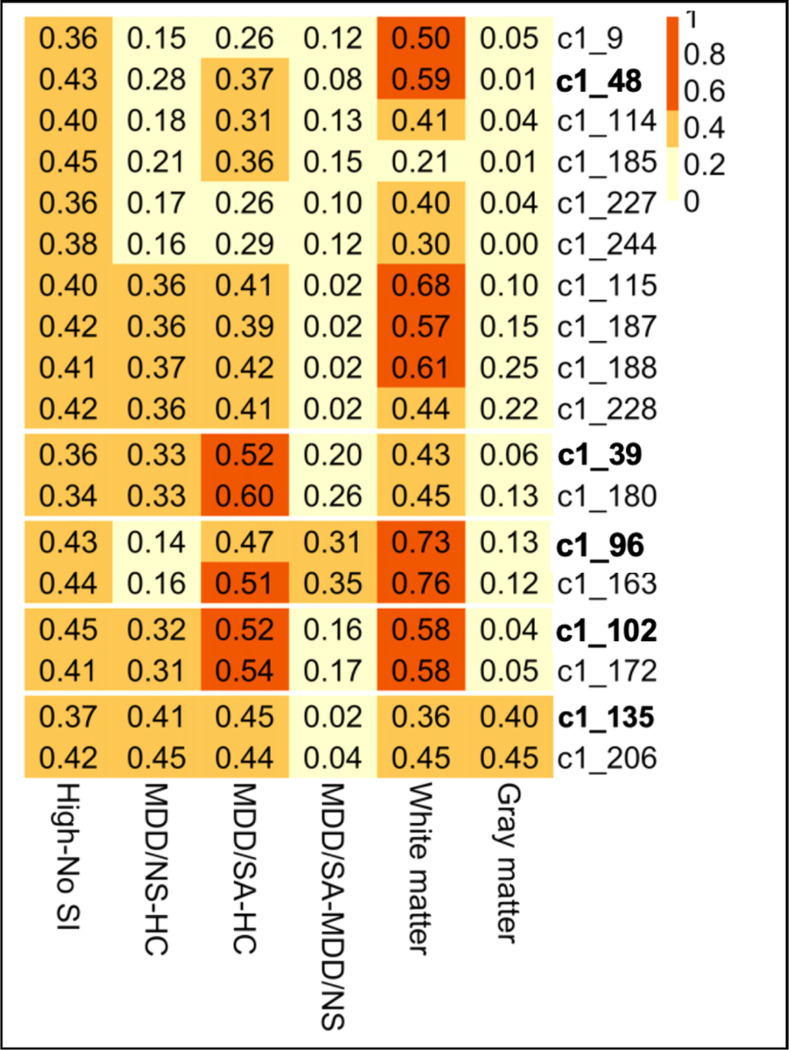
Figure 3.
Heatmap showing effect sizes (measured as absolute values of Cohen’s D) from group comparisons of clusters’ eigengene expression, with column labels depicting various group comparisons (last two columns show white/gray matter data contrasting samples of suicide death vs. non-psychiatric non-suicides death).
Gene co-expression modules associated with suicidal ideation and severity in peripheral blood were evaluated in both gray and white matter tissue from suicide decedents and non-psychiatric non-suicide controls using whole genome transcriptome data. It should be noted that for the postmortem cases, no group difference was observed for PMI (Wilcoxon p = 0.7649), pH (Wilcoxon p = 0.9523) or RIN (t test p = 0.7916) for the white matter, and gray matter tissues (PMI: p = 0.1182, pH: Wilcoxon p = 0.3255, RIN: Wilcoxon p = 0.3515 ). Thus, following analyses were not adjusted for PMI, pH, and RIN due to limited number of postmortem cases. Also given the limited sample size of the postmortem cases, high SI associated modules from blood were evaluated using effect sizes, namely robust Cohen’s d statistic. In white matter cases, medium effect sizes were detected for 9 high-SI associated modules (Fig. 3). Specifically, amongst the five unnested-modules in white matter tissue, the effect size ranged from minimum of 0.36 for c1_135 to a maximum effect size of 0.73 for c1_96 (Fig. 3). In grey matter tissue, effect sizes ranging from negligible (d = 0.01 for c1_48) to small (d = 0.4 for c1_135) were observed for the unnested modules (Fig. 3).
Gene ontology analyses were performed to delineate the functional importance of the genes within the modules associated with high SI and SI severity, using significant threshold Benjamini Hochberg adjusted BH p< 0.1 chosen a priori. Pathway analysis, performed via IPA using genes from the unnested modules (Fig. 2), showed consistent gene expression differences in both blood and brain (specifically, only white matter). Notably, the high SI and high suicide related modules c1_48 and c1_96 showed enrichment of genes involved in immune and inflammatory pathways. Inflammatory pathways, including pyroptosis signaling (BH p = 0.0015, Z = 2.65), TREM1 signaling (BH p = 0.0016, Z = 2.45), and neuroinflammation pathways (trend-wise significant, BH p = 0.125, Z = 2.83), were all upregulated in the high SI associated module c1_48 (Fig. 4A, Table S4). Additionally, the high SI-and-suicide associated module c1_96 showed enrichment of genes in the NF-κB (BH p = 0.0593, Z=−2.24), T cell receptor (BH p = 0.0593, Z=−2.24), and B cell signaling immune pathways (BH p = 0.0678, Z=−2.24) (Fig. 4B, Table S4). Module c1_36, associated with SI severity showed enrichment of genes involved in cell cycle regulation i.e., CREB signaling (BH p < 0.0001, Z=−3.61), FAK signaling (BH p < 0.0001, Z=−3.87) and Stathmin regulation (BH p = 0.0001, Z=−3.46) (Fig. 4C, Table S4). No significant findings were detected in pathway analyses of genes within the c1_39 (associated with suicide attempt), and c1_102 and c1_135 modules.
Figure 4.
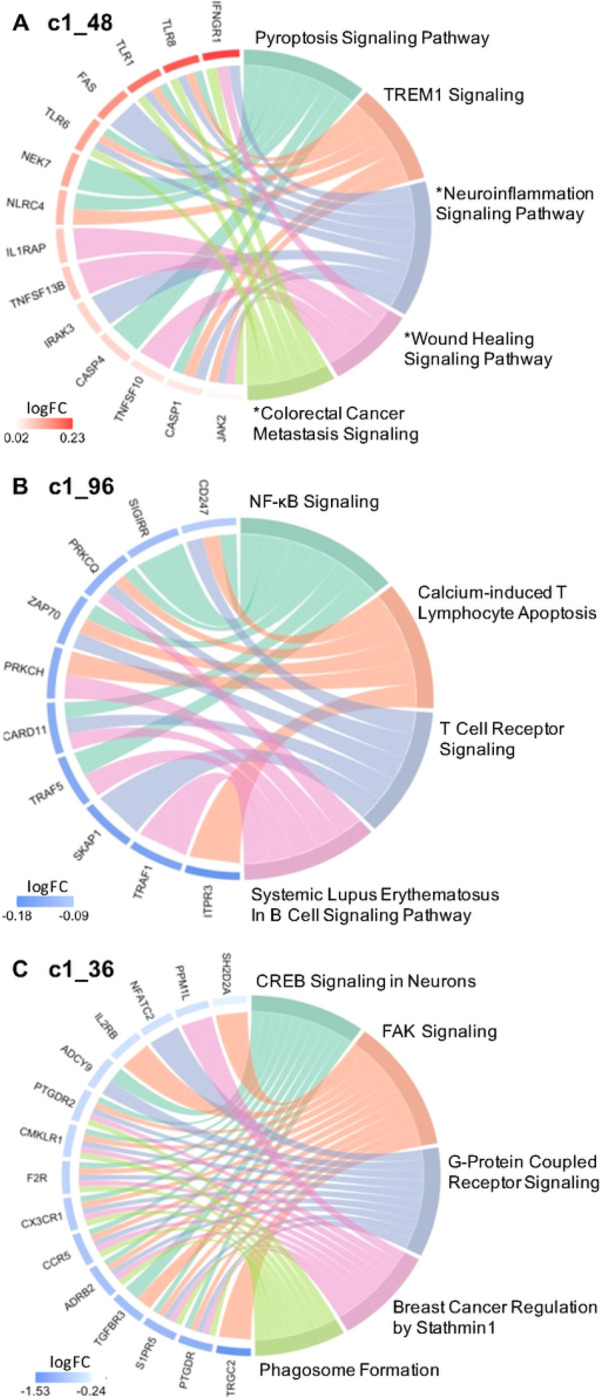
Circos plots show relationships between genes (left side) and significant IPA canonical pathways (right side). Significant IPA canonical pathways are identified for 2 SI group associated clusters (Panel A and B) and 1 suicide severity associated cluster (Panel C), based on |Z| > 2 and BH p < 0.1. Pathways marked with an asterisk (*) indicate a p-value less than uncorrected p < 0.05 but BH p >0.1. log FC = log 2 fold change expression.
4. DISCUSSION
Using whole genome transcriptional data from both live and postmortem samples, in the present study we investigated the associations between coordinated gene expression clusters and suicide ideation. Given the labile nature of gene expression and SI dynamics, we chose to focus on identifying transcript profiles that are associated with suicidal ideation presence and severity. Another cluster, C1_39, showed significant group difference in blood transcriptome between MDD/SA and HC groups. In addition to transcriptome data from peripheral blood from live participants, whole genome transcriptome data from gray and white matter postmortem brains of suicide decedents and controls were used for cross comparison of ideation with suicide death. Blood gene co-expression network analysis revealed a total of 18 SI associated modules transdiagnostically, and of these, 9 modules were detected in white matter with moderate effect sizes but none in gray matter.
Of the 16 hub genes (highly interconnected, Table 2) identified in these modules, 13 are novel in terms of association with suicidal ideation. Thus, they can be targeted for future studies including for consideration as potential predictors or pharmacological targets in preclinical and clinical suicide research studies. Notably among the hub genes identified, DNMT1, RRM2B and GCA have previously been shown to be associated with suicide or depression. In particular, the gene DNMT1, a hub gene in cluster c1_39, encodes the protein for DNA-methyltransferase 1 (DNMT1) 44, 45. Decreased DNMT1 expression has been reported in the brain of suicide decedents 46with altered expression also detected in the limbic system and brainstem of suicide decedents as compared with controls. The second hub gene RRM2B codes for one of two versions of the R2 subunit of ribonucleotide reductase, which generates nucleotide precursors required for DNA replication by reducing ribonucleoside diphosphates to deoxyribonucleoside diphosphates47, 48. Mutation in RRM2B is reported to cause Autosomal-Dominant Progressive External Ophthalmoplegia with variable symptoms including depression. The RRM2B-related mitochondrial disease also leads to distinct clinical and molecular characteristics including depression49. The third hub gene in cluster c1_48, GCA (grancalcin)50–52 is a calcium-binding protein abundant in neutrophils and macrophages50, previously linked to treatment response in depression 53potentially through mechanisms involving innate immune processes.
Table 2.
Hub genes of clusters with differential expression pattern between high-SI and no-SI groups.
| Cluster | Hub Genes (degree of intra-module connectivity) | Role of Hub Genes |
|---|---|---|
| C1_39 | DNMT1 (17) | Gene expression regulation (DNA methylation) |
| C1_48 | SHOC2(30),CMTM6(23),RRM2B(22),SPOPL(21),GCA(20),STAG2(19),SERINC1(18),RB1CC1(16),NAMPT(16) | Metabolism of lipid, protein, DNA and nicotinamide adenine dinucleotide (NAD); Innate immunity regulation; Cell cycle regulation; DNA repair |
| C1_96 | NLRC3(33),ZAP70(21) | Immunity regulation (both innate and adaptive responses): cytosolic regulator of innate immunity; Motility, adhesion and cytokine expression of mature T-cells |
| C1_102 | SIPA1L3 (11), VAV2 (9) | Cell proliferation; Endothelial cell migration Angiogenesis |
| C1_135 | TMEM55A (15), IRFD1(11) | Metabolism of lipids; Cell proliferation |
Across the Sl-associated modules, gene ontology analyses demonstrated enrichment of genes involved in immune and inflammatory processes. Our findings indicate that in peripheral blood moncytes, ideators show enhanced inflammatory signatures of molecules related not only to adaptive but also to innate immune responses, such as recognition of pathogen or damage-associated molecular patterns (PAMPs/DAMPs) and activation of proinflammatory signals. A salient finding in the present study is the peripheral blood monocyte gene co-expression related to inflammatory processes in high-SI participants, with enrichment of genes in Pyroptosis Signaling, TREM1 Signaling, NF-κB signaling, T Cell Receptor Signaling, Systemic Lupus Erythematosus In B Cell Signaling, Calcium-induced T Lymphocyte Apoptosis, and trend wise enrichment of genes in neuroinflammation pathways in the high SI vs. no SI groups. This is reflected in modules c1_48 and c1_96, with a number of genes overlapping across these pathways shown in Fig. 4A and 4B. Notably in the upregulated pathways from module c1_48 (Fig. 4A), the Toll-like receptor (TLR) genes shared across these pathways in the high-SI compared to the no-SI group is consistent with prior findings, in which mRNA and protein expression of the TLR-1 and TLR-6 loci was higher in serum and prefrontal cortex of depressed suicide decedents compared with non-suicide decedents, respectively. Innate immune receptors such as TLRs participate in initiation of the immune activation cascade, leading to the production of cytokines in the brain. Growing evidence supports the role of these receptors in facilitating the brain to mount immune responses during systemic infection and neuronal injury54 as well as in mood disorders55. Given the role that TLRs play in cytokine production, findings that the mRNA and protein expression of TLRs are elevated in dorsolateral prefrontal cortex (dlPFC) in suicide decedents can inform understanding of upstream mechanisms in neuroinflammation that lead to abnormalities in cytokine expression in suicidal patients across the continuum of suicide risk. Although none of the individual genes identified in this study showed robust fold changes in terms of magnitude of gene expression differences between the high-SI vs. no-SI groups, they did show coordinated gene expression differences associated with suicidal ideation, in pathways previously implicated in neurodegenerative and psychiatric disorders evidenced by both animal models and human studies56–67.
Across the 18 Sl-associated modules found in studying peripheral blood monocytes, 9 modules were also altered in white matter with moderate effect sizes, specifically in ventral PFC white matter postmortem in suicide decedents. The gene ontology analyses demonstrated enrichment of genes involved in both innate and adaptive immune responses shown in modules c1_48 and c1_96 as discussed above. Since normal white matter is essential for the brain`s executive function, a greater focus on white matter integrity appears indicated for suicide research. Intact white matter is critical for functioning of prefrontal cortical areas related to attention, self-control, planning, decision-making, mood regulation and other higher cognitive abilities68. Loss of myelin that affects disruption of the connectivity between the PFC and the limbic system, can lead to perceptual misrepresentations, misjudgment, impulsivity, and a distorted cognitive appraisal of reality. In vivo studies of frontal lobe in depression and suicidality report reduced metabolic response69, 70 and disrupted brain connectivity in grey and white matter of those who attempt or die by suicide71. Early studies using structural magnetic resonance imaging(SMRI) also support white matter alterations in patients with history of suicide attempt by showing an increase in white matter hyperintensities72–74. Diffusion tensor imaging (DTI) studies have shown diminished structural integrity of white matter that provides fronto-limbic connections in suicide attempters. Decreased fractional anisotropy (FA) has been reported in ventral frontal white matter in suicide attempters, including within the region of the uncinate fasciculus that carries major ventral PFC-amygdala connections, with an association to impulsivity75 76. Our findings of gene modules related to suicidal ideation detected in white matter of suicide decedents may also relate to suicidal behavior, as 41% of participants with suicidal ideation in the present study had prior history of suicide attempt, as compared to 9% in the no ideation group.
Neuropathological studies also implicate white matter in suicide, with reports of increased densities of activated microglia in PFC ventral white matter of suicide postmortem cases77, as well as increased microglial priming and macrophage recruitment in white matter of depressed suicides78. Microglial activation can be stimulated through transmission of inflammatory signals from the periphery to the brain via sensory afferent projections or humoral transmigration through areas with a potentially impaired blood-brain barrier79, 80. In an activated state, microglia adopt different phenotypes and, in response to PAMPs or DAMPs81 that bind to TLR and activate NF-κB, secrete numerous proinflammatory cytokines and chemokines82. This observed dynamic cross-talk between peripheral inflammation and microglial-induced neuroinflammation is consistent with our findings of increased expression of inflammatory genes both in blood and in white matter, as observed in modules c1_48 and c1_96. Although prior studies employing gene network methods have primarily investigated co-expression of genes in human postmortem gray matter, with findings in line with the present study 83–84.
This study also identified a cluster of genes associated with suicidal ideation severity, showing coordinated expression in module c1_36 with enrichment of genes involved in cell cycle regulation including CREB signaling, FAK signaling, and Stathmin regulation (Fig. 4C and Table S4). Notably, altered cell cycle regulation previously has been linked to suicidal ideation and behavior in both peripheral blood and postmortem brain studies85–87. Specifically, downregulation of the gene SKA2 (spindle and KT associated 2)88 has been associated with depression and suicidal ideation. Expression of SKA2 is regulated by transcription factors including CREB, a nuclear transcription factor that also regulates transcription activity of neuronal survival and expression of different growth factors 89. CREB expression is also downregulated in multiple major psychiatric disorders, including bipolar disorder, schizophrenia, and major depressive disorder90–92, and decreased protein and mRNA expression of CREB is observed in postmortem brain of depressed suicide decedents 92. These findings are consistent with the observed downregulation of the CREB signaling pathway associated with SI severity in the present study. Further, downregulation of Stathmin-1 signaling associated with suicidal ideation severity in the present study is also implicated in fear and anxiety behaviors in animal studies. Stathmin-1 signaling is a cell-cycle regulating pathway that regulates fear and anxiety both in rodents 93 and humans 94. In one study using a social defeat mouse model of stress, knockout of Stathmin-1 gene induced anxious hyperactivity, impaired object recognition, and decreased levels of neutral and social investigative behaviors compared to wild-type controls 95. Genes in the Stathmin-1 signaling pathway have additionally been implicated in psychiatric disorders 96–99 comorbid with suicidality.
Several strengths of the study design add confidence to the findings. The design allows comparison of genomic biology in peripheral blood related to suicidal ideation in brain to suicide death. The ideation severity findings were not attributable to depression severity. This study also has several limitations. The samples (although well-characterized through psychological assessments for live participants and psychological autopsy for postmortem cases) are small in size. Also, as is often the case in human postmortem brain studies, the measure of RNA quality RIN (RNA integrity number) can be highly variable. In this study the RIN values ranged from 2.9–9.0 for gray matter and 4.0–9.1 for white matter, with all RNAseq data passing quality control, indicating that RNA of low RIN can result in reliable RNA-seq data. This finding aligns with previous reports showing that RIN was not a sensitive measure of RNA quality for postmortem human brains100, since RIN is not a sensitive measure of RNA quality for substantially degraded samples, because first, the RIN score relies heavily on the amount of 18S and 28S ribosome RNAs but fails to measure the mRNA integrity directly, and second, the RIN is an overall assessment of RNA quality, and cannot serve as a specific criterion to adjust for differential RNA degradation among transcripts in downstream gene expression analyses. This limits its application in both pre-sequencing RNA sample screening and post-sequencing RNA-seq data analysis. Further, as is often the case in studies of suicidality, comorbid medical and mental health conditions are a potential confound both in study participants with SI and in the postmortem brain cases of suicide decedents and non-suicide controls used for validation of gene co-expression modules. As such, findings from this study may not generalize to other datasets with samples having varying comorbid conditions and life experiences as childhood or military trauma, etc. that may contribute to potential lack of reproducibility. Across the SI associated modules, individual genes within each module did not show significant expression differences in the SI vs. no-SI groups. This is not unexpected, since gene network approaches by design identify group(s) of genes that are changing in the same direction and magnitude, even if these changes are small. Modules of co-expressed genes thus identified in the present study, will likely be co-regulated or may belong to the same functional pathway, which need to be validated in future studies with additional experiments in vivo using animal models or in vitro using cell lines to confirm their effects on genes within relevant modules.
In conclusion, findings from this study suggest that gene expression networks are a valuable tool for identification of common pathways between suicide and psychiatric diseases, as well as factors unique to suicidality in the continuum of risk from suicidal ideation, suicidal behavior, and suicide. The SI associated co-expressed gene modules implicated with immune processes suggest potentially significant signals driving biological and molecular changes both in the periphery and in the CNS (especially in the white matter), which can potentially distinguish patients with elevated SI that may be at imminent risk for suicide. Identification of such gene expression networks occurring in both peripheral and CNS tissue as demonstrated in the present study can be integrated with other clinical measures in translational and biomarker studies of SI to improve models for suicide risk prediction.
Acknowledgements
Fatemeh Haghighi, PhD is a recipient of the VA CSR&D Research Career Scientist Award; CX002074, and her laboratory and work is supported by CX001728, CX001395, BX003794, RX003818 & RX001705 at the James J. Peters VA Medical Center. J. John Mann, PhD is supported by Conte Center for Suicide Prevention P50MH090964
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
Contributor Information
Shengnan Sun, Columbia University.
Qingkun Liu, Columbia University.
Zhaoyu Wang, Columbia University.
Yung-yu Huang, Columbia University.
M. Sublette, Columbia University
Andrew Dwork, Icahn School of Medicine at Mount Sinai.
Gorazd Rosoklija, Icahn School of Medicine at Mount Sinai.
Yongchao Ge, Icahn School of Medicine at Mount Sinai.
Hanga Galfalvy, Icahn School of Medicine at Mount Sinai.
J. John Mann, Icahn School of Medicine at Mount Sinai.
Fatemeh Haghighi, Icahn School of Medicine at Mount Sinai.
Data Availability:
The data that supports the findings of this study will be uploaded to GEO upon publication of this work.
References
- 1.Hedegaard H, Curtin SC, Warner M. Suicide Mortality in the United States, 1999–2017. NCHS Data Brief 2018; (330): 1–8. [PubMed] [Google Scholar]
- 2.Strawbridge RJ, Ward J, Ferguson A, Graham N, Shaw RJ, Cullen B et al. Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. EBioMedicine 2019; 41: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li QS, Shabalin AA, DiBlasi E, Gopal S, Canuso CM, Palotie A et al. Genome-wide association study meta-analysis of suicide death and suicidal behavior. Molecular psychiatry 2023; 28(2): 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Statham DJ, Heath AC, Madden PA, Bucholz KK, Bierut L, Dinwiddie SH et al. Suicidal behaviour: an epidemiological and genetic study. Psychological medicine 1998; 28(4): 839–855. [DOI] [PubMed] [Google Scholar]
- 5.Voracek M, Loibl LM. Genetics of suicide: a systematic review of twin studies. Wiener Klinische Wochenschrift 2007; 119. [DOI] [PubMed] [Google Scholar]
- 6.Mann JJ, Arango VA, Avenevoli S, Brent DA, Champagne FA, Clayton P et al. Candidate endophenotypes for genetic studies of suicidal behavior. Biological psychiatry 2009; 65(7): 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klonsky ED, May AM, Saffer BY. Suicide, suicide attempts, and suicidal ideation. Annual review of clinical psychology 2016; 12: 307–330. [DOI] [PubMed] [Google Scholar]
- 8.Fazel S, Runeson B. Suicide. N Engl J Med 2020; 382(3): 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mann JJ, Rizk MM. A brain-centric model of suicidal behavior. American journal of psychiatry 2020; 177(10): 902–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turecki G, Brent DA, Gunnell D, O’Connor RC, Oquendo MA, Pirkis J et al. Suicide and suicide risk. Nature reviews Disease primers 2019; 5(1): 1–22. [DOI] [PubMed] [Google Scholar]
- 11.Lenstra TL, Rodriguez J, Chen H, Larson DR. Transcription dynamics in living cells. Annual review of biophysics 2016; 45: 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butner JE, Bryan CJ, Tabares JV, Brown LA, Young-McCaughan S, Hale WJ et al. Temporal-dimensional examination of the Scale for Suicidal Ideation in a cohort of service members in treatment for PTSD. Psychological trauma: theory, research, practice, and policy 2021; 13(7): 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryan CJ, Rozek DC, Butner J, Rudd MD. Patterns of change in suicide ideation signal the recurrence of suicide attempts among high-risk psychiatric outpatients. Behaviour research and therapy 2019; 120: 103392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulsen DV, Stigsdotter UK, Refshage AD. Whatever happened to the soldiers? Nature-assisted therapies for veterans diagnosed with post-traumatic stress disorder: A literature review. Urban Forestry & Urban Greening 2015; 14(2): 438–445. [Google Scholar]
- 15.Kaminsky Z, Wilcox H, Eaton WW, Van Eck K, Kilaru V, Jovanovic T et al. Epigenetic and genetic variation at SKA2 predict suicidal behavior and post-traumatic stress disorder. Translational psychiatry 2015; 5(8): e627–e627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadeh N, Wolf EJ, Logue MW, Hayes JP Stone A, Griffin LM et al. Epigenetic variation at SKA2 predicts suicide phenotypes and internalizing psychopathology. Depression and anxiety 2016; 33(4): 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter SL, Brechbühler CM, Griffin M, Bond AT. Gene co-expression network topology provides a framework for molecular characterization of cellular state. Bioinformatics 2004; 20(14): 2242–2250. [DOI] [PubMed] [Google Scholar]
- 18.Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of” guilt-by-association” within gene coexpression networks. BMC bioinformatics 2005; 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo Y, Wei D, Zhu C, Naveed O, Hong W, Yang X. Unveiling the pathogenesis of psychiatric disorders using network models. Genes 2021; 12(7): 1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langfelder P Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 2008; 9(1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song W-M, Zhang B. Multiscale embedded gene co-expression network analysis. PLoS computational biology 2015; 11(11): e1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciobanu LG, Sachdev PS, Trollor JN, Reppermund S, Thalamuthu A, Mather KA et al. Co-expression network analysis of peripheral blood transcriptome identifies dysregulated protein processing in endoplasmic reticulum and immune response in recurrent MDD in older adults. Journal of psychiatric research 2018; 107: 19–27. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Gu H-Y, Zhu J, Niu Y-M, Zhang C, Guo G-L. Identification of hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Frontiers in physiology 2019; 10: 1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Cheng L, Grennan K, Fibiri F, Zhang C, Badner JA et al. Two gene co-expression modules differentiate psychotics and controls. Molecular psychiatry 2013; 18(12): 1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. Journal of neuroscience 2008; 28(6): 1410–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Ma C, Gearing M, Wang FG, Chin L-S, Li L. Integrated proteomics and network analysis identifies protein hubs and network alterations in Alzheimer’s disease. Acta neuropathologica communications 2018; 6: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng D, He S, Ma C, Wen Y, Song W, Xu Q et al. Network-based approach to identify molecular signatures in the brains of depressed suicides. Psychiatry Res 2020; 294: 113513. [DOI] [PubMed] [Google Scholar]
- 28.Spitzer R, Williams J, Gibbon M, First M. Structured Clinical Interview for DSM-NI-R-Fatient Version (SCID-F, 9/1/89 Version). New York, NY: Biometrics Research Department, New York State Psychiatric Institute; 1989. [Google Scholar]
- 29.First MB, Spitzer RL, Gibbon M, Williams JB. Structured clinical interview for DSM-IV axis I disorders-patient edition (SCID-I/F, Version 2.0). New York: Biometrics Research Department, New York State Psychiatric Institute 1995; 722. [Google Scholar]
- 30.Hamilton M. A rating scale for depression. Journal of neurology, neurosurgery, and psychiatry 1960; 23(1): 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Archives of general psychiatry 1961; 4(6): 561–571. [DOI] [PubMed] [Google Scholar]
- 32.Oquendo MA, Halberstam B, Mann JJ. Risk factors for suicidal behavior. Standardized evaluation in clinical practice 2003; 22: 103–129. [Google Scholar]
- 33.Beck AT, Kovacs M, Weissman A. Assessment of suicidal intention: the Scale for Suicide Ideation. Journal of consulting and clinical psychology 1979; 47(2): 343. [DOI] [PubMed] [Google Scholar]
- 34.Beck AT, Beck R, Kovacs M. Classification of suicidal behaviors: I. Quantifying intent and medical lethality. The American journal of psychiatry 1975. [DOI] [PubMed] [Google Scholar]
- 35.Kelly TM, Mann J. Validity of DSM-IM-R diagnosis by psychological autopsy: a comparison with clinician ante-mortem diagnosis. Acta Psychiatrica Scandinavica 1996; 94(5): 337–343. [DOI] [PubMed] [Google Scholar]
- 36.Spitzer RL, Williams JB, Gibbon M, First MB. User’s guide for the structured clinical interview for DSM-iii-R: SCID. American Psychiatric Association 1990. [Google Scholar]
- 37.Posner K, Oquendo MA, Gould M, Stanley B, Davies M. Columbia Classification Algorithm of Suicide Assessment (C-CASA): classification of suicidal events in the FDA’s pediatric suicidal risk analysis of antidepressants. American journal of psychiatry 2007; 164(7): 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 2015; 43(7): e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tumminello M, Aste T, Di Matteo T, Mantegna RN. A tool for filtering information in complex systems. Proceedings of the National Academy of Sciences 2005; 102(30): 10421–10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maechler M, Rousseeuw P Croux C, Todorov V, Ruckstuhl A, Salibian-Barrera M et al. robustbase: Basic Robust Statistics. R package version 0. 2021. [Google Scholar]
- 41.Revelle W. psych: procedures for personality and psychological research. Northwestern University, Evanston. 2018. [Google Scholar]
- 42.QIAGEN Ingenuity Pathway Analysis (QIAGEN IPA). https://digitalinsights.qiagen.com/IPA, 2022, Accessed Date Accessed 2022 Accessed.
- 43.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015; 31(17): 2912–2914. [DOI] [PubMed] [Google Scholar]
- 44.Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. Journal of molecular biology 2001; 309(5): 1189–1199. [DOI] [PubMed] [Google Scholar]
- 45.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer research 2007; 67(3): 946–950. [DOI] [PubMed] [Google Scholar]
- 46.Poulter MO, Du L, Weaver ICG, Palkovits M, Faludi G, Merali Z et al. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry 2008; 64(8): 645–652. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 2000; 404(6773): 42–49. [DOI] [PubMed] [Google Scholar]
- 48.Cho E-C, Kuo M-L, Cheng J-h Cheng Y-C, Hsieh Y-C, Liu Y-R et al. RRM2B-mediated regulation of mitochondrial activity and inflammation under oxidative stress. Mediators of Inflammation 2015; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pitceathly RD, Smith C, Fratter C, Alston CL, He L, Craig K et al. Adults with RRM2B-related mitochondrial disease have distinct clinical and molecular characteristics. Brain 2012; 135(11): 3392–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyhan A, Casimir CM, French JK, Teahan CG, Segal AW. Molecular cloning and characterization of grancalcin, a novel EF-hand calcium-binding protein abundant in neutrophils and monocytes. Journal of Biological Chemistry 1992; 267(5): 2928–2933. [PubMed] [Google Scholar]
- 51.Teahan CG, Totty NF, Segal AW. Isolation and characterization of grancalcin, a novel 28 kDa EF-hand calcium-binding protein from human neutrophils. Biochemical Journal 1992; 286(2): 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen C, Tarabykina S, La Cour JM, Lollike K, Berchtold MW. The PEF family proteins sorcin and grancalcin interact in vivo and in vitro. FEBS letters 2003; 545(2–3): 151–154. [DOI] [PubMed] [Google Scholar]
- 53.N0hr AK, Lindow M, Forsingdal A, Demharter S, Nielsen T, Buller R et al. A large-scale genome-wide gene expression analysis in peripheral blood identifies very few differentially expressed genes related to antidepressant treatment and response in patients with major depressive disorder. Neuropsychopharmacology 2021; 46(7): 1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crack PJ, Bray PJ. Toll-like receptors in the brain and their potential roles in neuropathology. Immunology and cell biology 2007; 85(6): 476–480. [DOI] [PubMed] [Google Scholar]
- 55.Fleshner M, Frank M, Maier SF. Danger signals and inflammasomes: stress-evoked sterile inflammation in mood disorders. Neuropsychopharmacology 2017; 42(1): 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian D-D, Wang M, Liu A, Gao M-R, Qiu C, Yu W et al. Antidepressant effect of paeoniflorin is through inhibiting pyroptosis CASP-11/GSDMD pathway. Molecular neurobiology 2021; 58: 761–776. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Song W, Tong Y, Zhang X, Zhao J, Gao X et al. Isoliquiritin ameliorates depression by suppressing NLRP3-mediated pyroptosis via miRNA-27a/SYK/NF-κB axis. Journal of Neuroinflammation 2021; 18: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon MS, Schiweck C, Arteaga-Henriquez G, Poletti S, Haarman BC, Dik WA et al. Monocyte mitochondrial dysfunction, inflammaging, and inflammatory pyroptosis in major depression. Progress in Neuro-Psychopharmacology and Biological Psychiatry 2021; 111: 110391. [DOI] [PubMed] [Google Scholar]
- 59.Weigelt K, Carvalho LA, Drexhage RC, Wijkhuijs A, de Wit H, van Beveren NJ et al. TREM-1 and DAP12 expression in monocytes of patients with severe psychiatric disorders. EGR3, ATF3 and PU. 1 as important transcription factors. Brain, behavior, and immunity 2011; 25(6): 1162–1169. [DOI] [PubMed] [Google Scholar]
- 60.Jiang T, Gong P-Y, Tan M-S, Xue X, Huang S, Zhou J-S et al. Soluble TREM1 concentrations are increased and positively correlated with total tau levels in the plasma of patients with Alzheimer’s disease. Aging clinical and experimental research 2019; 31: 1801–1805. [DOI] [PubMed] [Google Scholar]
- 61.Koo JW, Russo SJ, Ferguson D, Nestler EJ, Duman RS. Nuclear factor-KB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proceedings of the National Academy of Sciences 2010; 107(6): 2669–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patas K, Willing A, Demiralay C, Engler JB, Lupu A, Ramien C et al. T cell phenotype and T cell receptor repertoire in patients with major depressive disorder. Frontiers in immunology 2018; 9: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eilat E, Mendlovic S, Doron A, Zakuth V, Spirer Z. Increased apoptosis in patients with major depression: a preliminary study. The Journal of Immunology 1999; 163(1): 533–534. [PubMed] [Google Scholar]
- 64.Ivanova S, Semke VY, Vetlugina T, Rakitina N, Kudyakova T, Simutkin G. Signs of apoptosis of immunocompetent cells in patients with depression. Neuroscience and behavioral physiology 2007; 37: 527–530. [DOI] [PubMed] [Google Scholar]
- 65.Szuster-Ciesielska A, Słotwińska M, Stachura A, Marmurowska-Michafowska H, Dubas-Ślemp H, Bojarska-Junak A et al. Accelerated apoptosis of blood leukocytes and oxidative stress in blood of patients with major depression. Progress in Neuro-Psychopharmacology and Biological Psychiatry 2008; 32(3): 686–694. [DOI] [PubMed] [Google Scholar]
- 66.Fabbri C, Marsano A, Albani D, Chierchia A, Calati R, Drago A et al. PPP3CC gene: a putative modulator of antidepressant response through the B-cell receptor signaling pathway. The pharmacogenomics journal 2014; 14(5): 463–472. [DOI] [PubMed] [Google Scholar]
- 67.Holmes SE, Hinz R, Conen S, Gregory CJ, Matthews JC, Anton-Rodriguez JM et al. Elevated translocator protein in anterior cingulate in major depression and a role for inflammation in suicidal thinking: a positron emission tomography study. Biological psychiatry 2018; 83(1): 61–69. [DOI] [PubMed] [Google Scholar]
- 68.Bechara A, Damasio AR, Damasio H, Anderson SW . Insensitivity to future consequences following damage to human prefrontal cortex. Cognition 1994; 50(1–3): 7–15. [DOI] [PubMed] [Google Scholar]
- 69.Kegeles LS, Malone KM, Slifstein M, Ellis SP Xanthopoulos E, Keilp JG et al. Response of cortical metabolic deficits to serotonergic challenge in familial mood disorders. American Journal of Psychiatry 2003; 160(1): 76–82. [DOI] [PubMed] [Google Scholar]
- 70.Oquendo MA, Placidi GP Malone KM, Campbell C, Keilp J, Brodsky B et al. Positron emission tomography of regional brain metabolic responses to a serotonergic challenge and lethality of suicide attempts in major depression. Archives of general psychiatry 2003; 60(1): 14–22. [DOI] [PubMed] [Google Scholar]
- 71.Willeumier K, Taylor DV, Amen DG. Decreased cerebral blood flow in the limbic and prefrontal cortex using SPECT imaging in a cohort of completed suicides. Translational psychiatry 2011; 1(8): e28–e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ehrlich S, Breeze JL, Hesdorffer DC, Noam GG, Hong X, Alban RL et al. White matter hyperintensities and their association with suicidality in depressed young adults. J Affect Disord 2005; 86(2–3): 281–287. [DOI] [PubMed] [Google Scholar]
- 73.Pompili M, Innamorati M, Mann JJ, Oquendo MA, Lester D, Del Casale A et al. Periventricular white matter hyperintensities as predictors of suicide attempts in bipolar disorders and unipolar depression. Prog Neuropsychopharmacol Biol Psychiatry 2008; 32(6): 1501–1507. [DOI] [PubMed] [Google Scholar]
- 74.Serafini G, Pompili M, Innamorati M, Fusar-Poli P Akiskal HS, Rihmer Z et al. Affective temperamental profiles are associated with white matter hyperintensity and suicidal risk in patients with mood disorders. Journal of affective disorders 2011; 129(1–3): 47–55. [DOI] [PubMed] [Google Scholar]
- 75.Carballedo A, Amico F, Ugwu I, Fagan A, Fahey C, Morris D et al. Reduced fractional anisotropy in the uncinate fasciculus in patients with major depression carrying the met-allele of the Val66Met brain-derived neurotrophic factor genotype. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 2012; 159(5): 537–548. [DOI] [PubMed] [Google Scholar]
- 76.Bechara A, Damasio H, Damasio AR. Emotion, decision making and the orbitofrontal cortex. Cerebral cortex 2000; 10(3): 295–307. [DOI] [PubMed] [Google Scholar]
- 77.Schnieder TP Trencevska I, Rosoklija G, Stankov A, Mann JJ, Smiley J et al. Microglia of prefrontal white matter in suicide. J Neuropathol Exp Neurol 2014; 73(9): 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Torres-Platas SG, Cruceanu C, Chen GG, Turecki G, Mechawar N. Evidence for increased microglial priming and macrophage recruitment in the dorsal anterior cingulate white matter of depressed suicides. Brain Behav Immun 2014; 42: 50–59. [DOI] [PubMed] [Google Scholar]
- 79.Cytokine Dantzer R., sickness behavior, and depression. Immunology and Allergy Clinics 2009; 29(2): 247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Serna-Rodríguez MF, Bernal-Vega S, Camacho-Morales A, Pérez-Maya AA. The role of damage associated molecular pattern molecules (DAMPs) and permeability of the blood-brain barrier in depression and neuroinflammation. Journal of Neuroimmunology 2022: 577951. [DOI] [PubMed] [Google Scholar]
- 81.Mondelli V, Vernon AC, Turkheimer F, Dazzan P Pariante CM. Brain microglia in psychiatric disorders. Lancet Psychiatry 2017; 4(7): 563–572. [DOI] [PubMed] [Google Scholar]
- 82.Enache D, Pariante CM, Mondelli V. Markers of central inflammation in major depressive disorder: A systematic review and meta-analysis of studies examining cerebrospinal fluid, positron emission tomography and post-mortem brain tissue. Brain Behav Immun 2019; 81: 24–40. [DOI] [PubMed] [Google Scholar]
- 83.Zeng D, He S, Ma C, Wen Y, Song W, Xu Q et al. Network-based approach to identify molecular signatures in the brains of depressed suicides. Psychiatry research 2020; 294: 113513. [DOI] [PubMed] [Google Scholar]
- 84.Jabbi M, Arasappan D, Eickhoff SB, Strakowski SM, Nemeroff CB, Hofmann HA. Neuro-transcriptomic signatures for mood disorder morbidity and suicide mortality. Journal of psychiatric research 2020; 127: 62–74. [DOI] [PubMed] [Google Scholar]
- 85.Guintivano J, Brown T, Newcomer A, Jones M, Cox O, Maher BS et al. Identification and replication of a combined epigenetic and genetic biomarker predicting suicide and suicidal behaviors. American journal of psychiatry 2014; 171(12): 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Melhem NM, Munroe S, Marsland A, Gray K, Brent D, Porta G et al. Blunted HPA axis activity prior to suicide attempt and increased inflammation in attempters. Psychoneuroendocrinology 2017; 77: 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pandey GN, Rizavi HS, Zhang H, Bhaumik R, Ren X. The expression of the suicide-associated gene SKA2 is decreased in the prefrontal cortex of suicide victims but not of nonsuicidal patients. International Journal of Neuro-psychopharmacology 2016; 19(8): pyw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xie M, Bu Y. SKA2/FAM33A: A novel gene implicated in cell cycle, tumorigenesis, and psychiatric disorders. Genes & Diseases 2019; 6(1): 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang H, Xu J, Lazarovici P Quirion R, Zheng W. cAMP response element-binding protein (CREB): a possible signaling molecule link in the pathophysiology of schizophrenia. Frontiers in molecular neuroscience 2018; 11: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ren X, Rizavi HS, Khan MA, Bhaumik R, Dwivedi Y, Pandey GN. Alteration of cyclic-AMP response element binding protein in the postmortem brain of subjects with bipolar disorder and schizophrenia. Journal of affective disorders 2014; 152: 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sulser F. The role of CREB and other transcription factors in the pharmacotherapy and etiology of depression. Annals of medicine 2002; 34(5): 348–356. [DOI] [PubMed] [Google Scholar]
- 92.Ren X, Dwivedi Y, Mondal AC, Pandey GN. Cyclic-AMP response element binding protein (CREB) in the neutrophils of depressed patients. Psychiatry research 2011; 185(1–2): 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shumyatsky GP Malleret G, Shin R-M, Takizawa S, Tully K, Tsvetkov E et al. Stathmin, a gene enriched in the amygdala, controls both learned and innate fear. Cell 2005; 123(4): 697–709. [DOI] [PubMed] [Google Scholar]
- 94.Brocke B, Lesch KP Armbruster D, Moser DA, Müller A, Strobel A et al. Stathmin, a gene regulating neural plasticity, affects fear and anxiety processing in humans. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 2010; 153(1): 243–251. [DOI] [PubMed] [Google Scholar]
- 95.Nguyen TB, Prabhu VV, Piao YH, Oh YE, Zahra RF, Chung Y-C. Effects of stathmin 1 gene knockout on behaviors and dopaminergic markers in mice exposed to social defeat stress. Brain sciences 2019; 9(9): 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Katayama T, Hattori T, Yamada K, Matsuzaki S, Tohyama M. Role of the PACAP-PAC1-DISC1 and PACAP–PAC1–stathmin1 systems in schizophrenia and bipolar disorder: novel treatment mechanisms? Ph armacogenomics 2009; 10(12): 1967–1978. [DOI] [PubMed] [Google Scholar]
- 97.Paulson L, Martin P Persson A, Nilsson CL, Ljung E, Westman-Brinkmalm A et al. Comparative genome- and proteome analysis of cerebral cortex from MK-801-treated rats. Journal of Neuroscience Research 2003; 71(4): 526–533. [DOI] [PubMed] [Google Scholar]
- 98.Brocke B, Lesch K-P, Armbruster D, Moser DA, Müller A, Strobel A et al. Stathmin, a gene regulating neural plasticity, affects fear and anxiety processing in humans. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 2010; 153B(1): 243–251. [DOI] [PubMed] [Google Scholar]
- 99.Teyssier JR, Chauvet-Gelinier JC, Ragot S, Bonin B. Up-regulation of leucocytes genes implicated in telomere dysfunction and cellular senescence correlates with depression and anxiety severity scores. PLoS One 2012; 7(11): e49677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sonntag KC, Tejada G, Subburaju S, Berretta S, Benes FM, Woo TUW. Limited predictability of postmortem human brain tissue quality by RNA integrity numbers. Journal of neurochemistry 2016; 138(1): 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that supports the findings of this study will be uploaded to GEO upon publication of this work.