Abstract
Pontine nuclei (PN) neurons mediate the communication between the cerebral cortex andthe cerebellum to refine skilled motor functions. Prior studies showed that PN neurons fall into two subtypes based on their anatomic location and region-specific connectivity, but the extent of their heterogeneity and its molecular drivers remain unknown. Atoh1 encodes a transcription factor that is expressed in the PN precursors. We previously showed that partial loss of Atoh1 function in mice results in delayed PN development and impaired motor learning. In this study, we performed single-cell RNA sequencing to elucidate the cell state–specific functions of Atoh1 during PN development and found that Atoh1 regulates cell cycle exit, differentiation, migration, and survival of PN neurons. Our data revealed six previously not known PN subtypes that are molecularly and spatially distinct. We found that the PN subtypes exhibit differential vulnerability to partial loss of Atoh1 function, providing insights into the prominence of PN phenotypes in patients with ATOH1 missense mutations.
ScRNA-seq reveals that the Atoh1-dependent pontine nuclei, a hub for skilled motor function, are made up of six distinct subtypes.
INTRODUCTION
Motor skills such as picking up a cup of coffee or hitting a speedy baseball with a bat require communication between the cerebral cortex and the cerebellum (1). These connections are mediated through the pontine nuclei (PN) that are located in the ventral pons and composed of mostly glutamatergic neurons (2–7). Given their pivotal role in motor functions, several efforts have been made to understand how the PN develop in mammals. PN neurons originate from a group of proliferating neuroepithelial cells residing in the rhombic lip (RL) located in the developing hindbrain. Specifically, Wnt1- and Atoh1-expressing cells within the caudal RL (cRL) give rise to glutamatergic PN neurons (8–10). PN neurons are born during mouse embryonic day 12.5 (E12.5) to E18.5 and migrate tangentially along the anterior extramural stream (AES) until they reach the ventral pons (11, 12).
A previous study of PN in rabbit and cat has suggested that PN can be categorized into subpopulations on the basis of their anatomical location and connectivity (13). Anatomically, the PN are divided into the basal pontine nucleus (BPN) and the reticulotegmental nucleus (RtTg) at the anterior-ventral and posterior-dorsal part of the PN, respectively. Subpopulations of PN neurons have been proposed on the basis of their positions along the rostro-caudal axis, which inherit the expression pattern of Hox2-Hox5 from their progenitors at the cRL (14). Moreover, tracing experiments demonstrated that the corticopontine connectivity was established in a partially region-specific manner (15, 16), suggesting that PN neurons are functionally diverse on the basis of their regional cortical inputs. However, the extent of PN heterogeneity remains unclear, and the molecular determinants of PN heterogeneity are not known.
Another outstanding question is how a pool of seemingly homogenous Atoh1+ progenitors give rise to diverse progenies in the PN. Atoh1, or Atonal homolog 1, encodes a basic helix-loop-helix transcription factor ATOH1 that is required for the development of a variety of neurons in the hindbrain (9) and the dorsal spinal cord (17), hair cells in the inner ear (18), Merkel cells in the skin (19), and secretory cells in the gut (20). In the hindbrain, Atoh1-lineage neurons contribute to many key components of the proprioceptive pathway, including the PN (9, 21). A recent report implicated a homozygous missense variant in ATOH1 in two human patients with global developmental delay, motor function deficits, pontocerebellar hypoplasia, and hearing loss (22). It is thus of great interest and clinical relevance to understand how Atoh1 shapes proper PN development.
Loss of both copies of Atoh1 in mice results in complete absence of the PN, whereas loss of one copy of Atoh1 in mice has no observable change in the PN (23), making it difficult to study the function of Atoh1 during PN development using either the Atoh1 knockout or the heterozygous mouse model. We previously found that substituting the serine at position 193 with an alanine (S193A) resulted in an Atoh1 hypomorphic allele (24). Mice carrying Atoh1S193A over an Atoh1 null allele (Atoh1S193A/−) showed delayed development in the PN neurons at postnatal day 0 (P0) and impaired motor learning as adults. Given the heterogeneity of the PN neurons, it is unclear whether this hypomorphic mutation affects the development of all PN subtypes equally. In other contexts, we learn that different cell types have differential vulnerability to perturbation in genes implicated in neurodevelopmental disorders (25, 26). For example, despite being expressed in both neurogenic niches, increased level of the transcription factor Foxg1 compromises the neurogenesis of excitatory neurons but not inhibitory neurons (26). We therefore hypothesized that PN subtypes are molecularly distinct and have differential vulnerability to partial loss of function of Atoh1.
In this study, we profiled the single-cell transcriptomes of the PN neurons and characterized the molecular and cellular phenotypes of the PN in Atoh1S193A/− mice at the single-cell resolution. We found cell state–specific roles of Atoh1 during PN development including regulating cell cycle exit, differentiation, migration, survival, and cellular heterogeneity of the PN neurons. In addition, our single-cell RNA sequencing (scRNA-seq) data revealed six unreported subtypes of the PN neurons at P5 and identified subtype-specific markers. We found that the PN subtypes have differential vulnerabilities to partial loss of function of Atoh1. Our study provides comprehensive evidence of how a transcription factor plays multiple roles during neuronal development and demonstrates that neuronal subtypes could have differential sensitivity to perturbation of a gene that is expressed in the precursors of all subtypes.
RESULTS
Phospho-mutation of Atoh1 at serine-193 leads to PN hypoplasia in mice
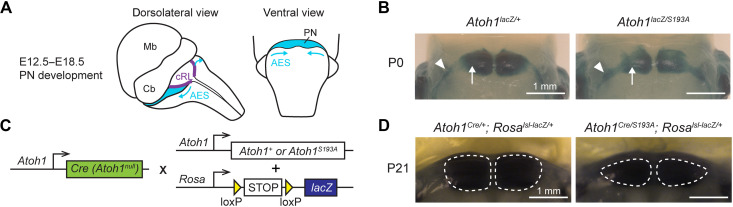
To test whether partial loss of function of Atoh1 results in PN abnormalities postnatally, we compared the morphology of the PN in mice with hypomorphic mutation to those in control mice at P0 and P21. We crossed Atoh1S193A/+ mice to Atoh1lacZ/+, whereby the lacZ allele replaced the coding region of Atoh1, creating a null allele. Following the tangential migration along the AES (Fig. 1A), most of the PN neurons finished migrating at P0 in control mice (Fig. 1B, left, arrow), with little lacZ signal in the AES (arrowhead). We observed a decreased intensity of the lacZ staining in the PN in Atoh1S193A/lacZ mice (Fig. 1B, right, arrow), indicating potential neuronal loss at P0 in Atoh1S193A/lacZ mice. In addition, consistent with our previous study (24), we found that there were more lacZ+ neurons retained at the AES in Atoh1S193A/lacZ mice (Fig. 1B, right, arrowhead), suggesting a developmental delay or a migration deficit in some PN neurons upon partial loss of Atoh1 function.
Fig. 1. Phospho-mutation of Atoh1 at serine-193 leads to PN hypoplasia in mice.
(A) Schematic representation of the PN development in mice. PN neurons migrate from cRL to ventral pons through AES during E12.5 to E18.5. Mb, midbrain; Cb, cerebellum. (B) Whole-mount X-galactosidase (X-gal) staining on mouse hindbrain at P0 (ventral view). The arrows and arrowheads denote the neurons at the PN and in the AES, respectively. Scale bars, 1 mm. (C) Strategy to label the Atoh1-lineage neurons. Atoh1Cre/+ knock-in mouse was crossed with mouse carrying either wild-type or hypomorphic Atoh1 (Atoh1S193A) and a Cre-dependent lacZ allele. (D) Whole-mount X-gal staining on mouse hindbrain at P21 (ventral view). The dashed lines outline the area of the PN. Scale bars, 1 mm.
To test whether those neurons retained at the AES ever reach their destination, we examined the PN morphology at a juvenile age, P21. Given that Atoh1 is turned off postnatally in the PN, we permanently labeled the Atoh1-lineage neurons using a Cre-dependent lacZ reporter (27) in combination with an Atoh1Cre/+ knock-in mouse in which one copy of Atoh1 is functionally a null allele because the Cre replaced the coding region of Atoh1 (28). This allows us to visualize the PN postnatally (Fig. 1C). We found that the size of the PN was reduced in Atoh1Cre/S193A; Rosalsl-lacZ/+ animals compared to Atoh1Cre/+; Rosalsl-lacZ/+ animals (Fig. 1D). We further confirmed this phenotype quantitatively by performing immunofluorescence staining with osteopontin antibody, a pan-PN neuronal marker (fig. S1). These data suggested that partial loss of function of Atoh1 not only leads to delayed development of PN but also leads to PN hypoplasia in mice reminiscent of the malformation of the PN in patients with ATOH1 missense variant (22). Together, these data reinforced the importance of Atoh1 in PN development and demonstrated that Atoh1 hypomorphic allele could provide a useful mouse model to dissect the functions of Atoh1 during PN development.
Progression of the PN development is impaired in Atoh1S193A/− animals
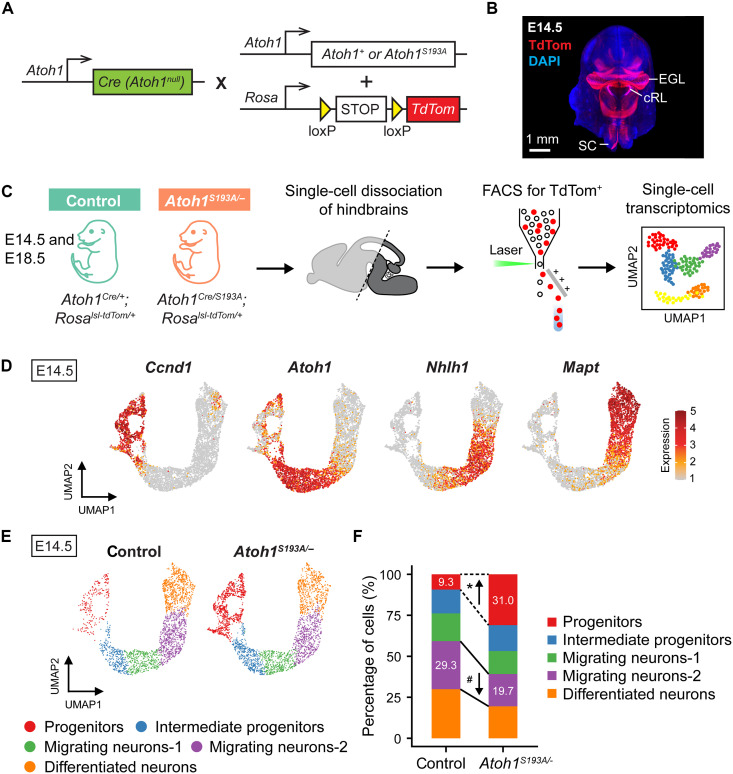
To investigate the mechanisms by which Atoh1 regulates normal PN development, we performed scRNA-seq in developing hindbrains from control mice and mice with Atoh1 hypomorphic mutation. We used a Cre-dependent fluorescent reporter (Rosalsl-TdTomato) to permanently label Atoh1-lineage neurons with Atoh1Cre/+ mice (Fig. 2, A and B). During development, PN progenitors and migrating PN neurons are spatially dispersed between the cRL to ventral pons. Therefore, to ensure that we capture all PN progenitors and migrating neurons, we collected whole hindbrains from Atoh1Cre/+; Rosalsl-tdTomato/+ (hereafter referred to as control) and Atoh1Cre/S193A; Rosalsl-tdTomato/+ (hereafter referred to as Atoh1S193A/−) embryos. After single-cell dissociation, Atoh1-lineage (TdTom+) neurons were isolated by fluorescence- activated cell sorting (FACS), followed by scRNA-seq with 10X Genomics Chromium platform (Fig. 2C). We collected samples at two time points, E14.5 and E18.5, to examine the phenotypes of Atoh1S193A/− mice in the middle and at the end of neurogenesis of the PN neurons, respectively.
Fig. 2. Atoh1S193A/− animals exhibited an impaired progression of PN development.
(A) Strategy to label the Atoh1-lineage neurons. Atoh1Cre/+ knock-in mouse was crossed with mouse carrying either wild-type or hypomorphic Atoh1 (Atoh1S193A) and a Cre-dependent TdTomato (TdTom) reporter. (B) An image of three-dimensional rendering of E14.5 mouse head using lightsheet microscopy (back view). The TdTom represents the Atoh1-lineage neurons in developing hindbrain. EGL, external granule layer; SC, spinal cord. (C) Workflow for scRNA-seq of Atoh1-lineage neurons from E14.5 and E18.5 mouse hindbrain. Hindbrains were collected from E14.5 and E18.5 control and Atoh1S193A/− embryos (n = 3 per genotype for each time point). After enrichment by sorting, single-cell transcription profiles were captured by 10X Genomics Chromium platform. (D) Expression levels of the selective markers visualized on uniform manifold approximation and projection (UMAP). Ccnd1, Atoh1, Nhlh1, and Mapt are known markers for progenitors, intermediate progenitors, migrating neurons, and differentiated neurons, respectively. (E) UMAP of the five cell states identified in developing PN at E14.5 by scRNA-seq. (F) Proportion of the cells in each cell state. The arrows indicate the direction of the change in Atoh1S193A/− animals. *False discovery rate (FDR) < 0.01 and #FDR < 0.05.
We first focused on the E14.5 time point to explore the role of Atoh1 during PN development. After filtering out low-quality cells (see Materials and Methods), a total of 22,191 cells were retained from control hindbrains, and 22,206 cells were retained from Atoh1S193A/− hindbrains (n = 3 animals per genotype). Among the cells in the hindbrain, we identified the PN cells based on the clusters that expressed the established markers of the AES and PN (fig. S2, A to D). This resulted in 2451 control PN cells and 3124 Atoh1S193A/− PN cells for downstream analyses. After unbiased clustering of the PN cells, we annotated each cluster by the expression of the known markers (Fig. 2D, fig. S2E, and data S1). Cells at different cell states during PN development were fully captured in our dataset, including proliferating progenitors (Ccnd1+), postmitotic intermediate progenitors (Atoh1high), migrating neurons (Nhlh1+), and differentiated neurons (Mapt+) in both control and Atoh1S193A/− samples (Fig. 2E). The migrating neurons constituted the largest population and were divided into two clusters. We therefore annotated the two migrating populations as migrating neurons-1 and migrating neurons-2 based on their maturity. The migrating neurons-1 are neurons starting to differentiate and migrate away from the cRL (Atoh1lowNhlh1high), whereas the migrating neurons-2 are neurons expressing not only high level of the marker for migration but also low level of the differentiated neuronal marker (Nhlh1highMapt low) (Fig. 2, D and E).
Next, we calculated the percentage of the cells in each cell state in control and Atoh1S193A/− animals and performed differential abundance test using propeller with speckle R package (29). We found that the proportion of the progenitors increased markedly from 9.3 to 31% with a significant decrease in the percentage of the migrating neurons-2 from 29.3 to 19.7% in Atoh1S193A/− animals (Fig. 2F and fig. S3A). The shift in the proportion of the cells in each state indicates that the progression of the PN development was impaired in Atoh1S193A/− mice. Trajectory analysis with Slingshot (30, 31) showed that in control embryos, more cells progressed further in pseudo-time than in Atoh1S193A/− embryos (fig. S3, B and C). Collectively, these data demonstrate that partial loss of Atoh1 function leads to accumulation of the PN progenitors at the expense of the differentiating PN neurons.
Partial loss of function of Atoh1 leads to decreased cell cycle exit in PN progenitors and deficits in differentiation and migration
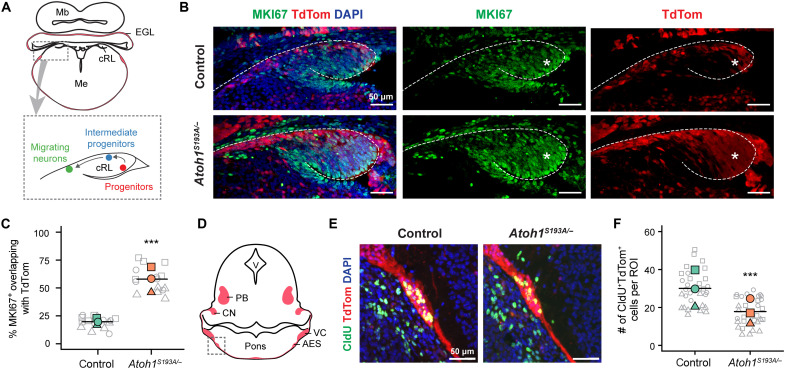
We sought to investigate the mechanisms underlying the altered proportions of the cells in each state in Atoh1S193A/− animals. While we observed an increased proportion of the proliferating progenitors in Atoh1S193A/− mice, the proportion of its downstream cell state (i.e., the intermediate progenitors) was not altered (Fig. 2F). Instead, the fraction of the migrating neurons-2 was significantly decreased. Therefore, we proposed two mechanisms underlying the observed phenotypes: Robust function of Atoh1 is required to (i) drive cell cycle exit and (ii) promote differentiation and migration (fig. S3D). To validate our scRNA-seq data, we performed immunofluorescence staining and histological analyses on the embryonic tissues. First, we performed immunofluorescence staining for MKI67, a marker for proliferating cells, on E14.5 hindbrains of control and Atoh1S193A/− animals. On the basis of the staining of ATOH1 and MKI67 at the cRL in control animals (fig. S4), the MKI67+ proliferating progenitors and ATOH1+ intermediate progenitors reside at the ventromedial and dorsolateral cRL, respectively (Fig. 3A). In control animals, there were only few TdTom+ cells at the cRL (Fig. 3B, asterisks), suggesting that the proliferating PN progenitors become postmitotic and leave the cRL upon the onset of Atoh1 expression. In contrast, there was an increased percentage of the MKI67+ cells that overlapped with TdTom at the cRL in Atoh1S193A/− mice (Fig. 3, B and C), indicating an accumulation of proliferating progenitors at the cRL. The data suggest that robust Atoh1 function is important for the cycling progenitors to become postmitotic.
Fig. 3. Partial loss of function of Atoh1 leads to decreased cell cycle exit in PN progenitors and deficits in differentiation and migration.
(A) Schematic illustration of mouse coronal section at E14.5. The area of cRL was enlarged on the bottom with locations of three cell states being labeled. Me, medulla. (B) Immunofluorescence staining of MKI67 on E14.5 mouse brain. The cropped view of cRL in control (top) and Atoh1S193A/− (bottom) mice. Atoh1-lineage neurons were labeled with TdTom, and the nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI). The asterisk denotes the progenitors at cRL. Scale bars, 50 μm. (C) Quantification of the percentage of MKI67 fluorescence overlapping with TdTom in control and Atoh1S193A/− mice. The gray symbols represent the five regions of interest quantified from each animal (n = 3 per genotype). The average percentage for each animal was shown in colored shape. The crossbar denotes the mean per genotype. ***Χ2(1) = 53.95, P = 2.06 × 10−13 by mixed-model analysis of variance (ANOVA). (D) Schematic illustration of coronal section at E14.5. The dashed gray box indicates the region of interest shown in (E). V, ventricle; PB, parabrachial nuclei; CN, cerebellar nuclei; VC, ventral cochlear nucleus. (E) Immunofluorescence staining of CldU on E14.5 mouse brain. The areas of AES for control (left) and Atoh1S193A/− (right) animals were shown. Atoh1-lineage neurons were labeled with TdTom, and the nuclei were labeled with DAPI. Scale bars, 50 μm. (F) The average number of the CldU+TdTom+ cells per region of interest (ROI) in control and Atoh1S193A/− mice. The gray symbols represent the 10 regions of interest quantified from each animal (n = 3 per genotype). The average number for each animal was shown in colored shape. The crossbar denotes the mean per genotype. ***Χ2(1) = 39.81, P = 2.80 × 10−10 by mixed-model ANOVA.
Next, to test whether the differentiating and migrating neurons are reduced in Atoh1S193A/− mice, we used the thymidine analog, 5-chloro-2′-deoxyuridine (CldU) to label and quantify the number of PN neurons born within the 24-hour window between E13.5 and E14.5. We injected the pregnant dams with CldU at E13.5 and harvested the brains from E14.5 embryos. It has been shown that it takes 1 to 2 days for the PN neurons to migrate from cRL to ventral pons (12). Consistent with the literature, we found that most of the CldU-labeled PN neurons (CldU+TdTom+) were located at the AES 24 hours after injection in control animals (Fig. 3, D and E). The number of the CldU+TdTom+ cells at the AES was significantly reduced in Atoh1S193A/− mice (Fig. 3F), suggesting that fewer neurons differentiated and migrated away from the cRL in Atoh1S193A/− animals. These data validate our scRNA-seq data in which we found that migrating population was decreased in Atoh1S193A/− mice. Together, our data demonstrate that Atoh1 governs multiple biological processes in addition to specifying PN neurons. Atoh1 is important for both cell cycle exit of the PN progenitors and the differentiation and migration of the intermediate progenitors.
The cell state–specific dysregulated pathways in Atoh1S193A/− mice
To understand how Atoh1 mediates different biological processes during PN development, we sought to identify the genes that were dysregulated in each cell state in Atoh1S193A/− mice compared to control mice. Using differential gene expression analysis, we identified 362 differentially expressed genes (DEGs) [log2 fold change (log2FC) > 0.25 and false discovery rate (FDR) < 0.05] (data S2). We found that intermediate progenitors had the highest number of DEGs, followed by migrating neurons-1 and progenitors (Fig. 4A). We verified that the different number of DEGs per cell state was not simply a result of differences in total cell number in each cell state (fig. S5A). The cell states with the greatest transcriptional dysregulation (i.e., progenitors, intermediate progenitors, and migrating neurons-1) also expressed high levels of Atoh1 (Figs. 4A and 2D), which led us to test whether these DEGs were directly regulated by Atoh1. We calculated the percentage of the DEGs that had ATOH1 binding peak(s) identified by ATOH1 chromatin immunoprecipitation sequencing (32) and performed enrichment analysis for each cell state. We found a significant enrichment of DEGs with ATOH1-binding in progenitors (47%), intermediate progenitors (48%), and migrating neurons-1 (42%) (Fig. 4B), highlighting the direct impact of Atoh1 in these cell states. We found several genes that were direct targets of Atoh1 and have been reported to play a role in PN development (Fig. 4C, shaded box). For instance, Atoh1, which is known to regulate itself (33), was down-regulated in the progenitors. Barhl1, important for migration and survival of the PN neurons (34), was down-regulated in both progenitors and intermediate progenitors upon partial loss of Atoh1 function. Last, Nhlh1, essential for migration of the PN neurons (35), was decreased in the intermediate progenitors and the migrating neurons. Moreover, we also identified other DEGs such as Pcp4 and Rab15, whose roles have not been characterized in PN development but have robust changes in expression levels in multiple cell states (Fig. 4C). Pcp4 has been identified as an Atoh1 target in cochlear hair cells (36). Rab15 was also reported as an Atoh1 target in different Atoh1-lineage cells including developing cerebellar granule neurons (32), Merkel cells (37), cochlear hair cells (36), and neurons in dorsal neural tube (38).
Fig. 4. Partial loss of Atoh1 function leads to multiple dysregulated pathways.
(A) The number of the up-regulated (left) and down-regulated (right) genes at each cell state in Atoh1S193A/− compared to control. The DEGs were filtered by log2FC > 0.25 and FDR < 0.05. (B) The enrichment of the DEGs with ATOH1-binding in each cell state. The percentage of the DEGs with ATOH1-binding peak was shown in number. The odds ratios were presented with 95% confidence interval performed by Fisher’s exact test. P value was adjusted by Bonferroni. (C) Volcano plot of the DEGs with ATOH1-binding peak grouped by cell state. The shaded box indicates the known Atoh1 targets that have been reported in PN development. (D) Gene ontology (GO) enrichment analysis of the DEGs. The representative biological processes are shown with −log10(FDR). (E) TUNEL staining on cRL in control (top) and Atoh1S193A/− (bottom) at E14.5. The Atoh1-lineage cells were labeled with TdTom, and the nuclei were stained with DAPI. (F) The average number of the TUNEL+ cells per region of interest in control and Atoh1S193A/− mice. The gray symbols represent the five ROIs quantified from each animal (n = 3 per genotype). The average number for each animal was shown in colored shape. The crossbar denotes the mean per genotype. ***Χ2(1) = 82.41, P = 2.20 × 10−16 by mixed-model ANOVA.
Next, we focused on the three most affected cell states and performed gene ontology (GO) enrichment analysis to identify the pathways that were dysregulated in Atoh1S193A/− mice beyond the known Atoh1 targets (Fig. 4D and data S3). In line with the finding of increased proliferating cells in Atoh1S193A/− mice, cell proliferation and cell cycle regulation including Notch signaling were among the top enriched GO terms for the up-regulated genes in the progenitor state (Fig. 4D, left). Consistent with the trajectory analysis that showed an impaired differentiation process in Atoh1S193A/− mice, down-regulated genes were enriched for neuron differentiation and cytoskeleton organization ontologies across all three cell states (Fig. 4D). These data supported our earlier findings that Atoh1 plays roles in both cell cycle regulation, neuronal differentiation, and migration. We found that up-regulated genes in both intermediate progenitors and migrating neurons-1 were enriched for regulators of apoptosis (Fig. 4D, middle and right). For instance, the average level of Hrk, a member of proapoptotic Bcl-2 family, was significantly increased in both intermediate progenitors and migrating neurons-1 in Atoh1S193A/− mice (fig. S5B). In addition, other genes involved in programmed cell death such as Pak3 and Dap were also up-regulated in the intermediate progenitor state (fig. S5, C and D). These data indicate that Atoh1 hypomorphic mutation might lead to increased cell death. To test this hypothesis, we performed terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay and found that the number of TUNEL-positive cells was significantly increased in Atoh1S193A/− mice at the lateral cRL (Fig. 4, E and F), where the intermediate progenitors (Atoh1high) are located (Fig. 3A and fig. S4). The increased cell apoptosis in Atoh1S193A/− animals may count for the reduced size of PN at older age and suggest that Atoh1 is important for the survival of the intermediate progenitors. In summary, these data highlight the important roles of Atoh1 in progenitors, intermediate progenitors, and migrating neurons by regulating cell cycle, differentiation, migration, and survival of the developing PN neurons.
PN neurons are molecularly heterogeneous
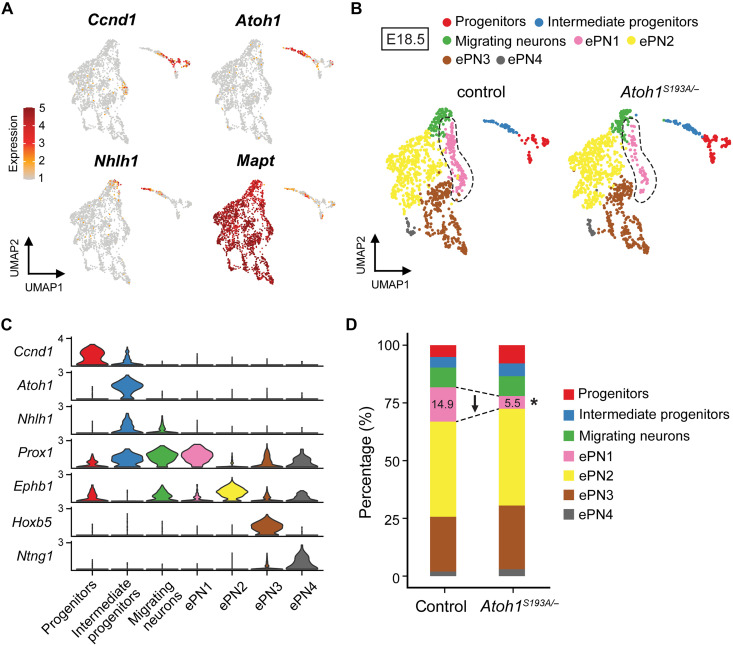
To determine whether the cellular identities of the differentiated PN neurons are altered in Atoh1S193A/− mice, we performed scRNA-seq in control and Atoh1S193A/− hindbrains at E18.5, when most of the PN neurons have been born and migrated to the ventral pons. We analyzed 1196 control and 1176 Atoh1S193A/− PN cells (n = 3 animals per genotype) (fig. S6, A to D). After unbiased clustering, we annotated the clusters based on the known marker genes (Fig. 5, A and B). As expected, most of the cells at E18.5 are differentiated neurons marked by Mapt expression (Fig. 5A). The differentiated neurons were classified into four subtypes (Fig. 5B), suggesting that they are molecularly heterogeneous. We characterized the marker genes for each subtype based on differential gene expression analysis and named the differentiated PN neuron subtypes as embryonic PN1(ePN1) to ePN4 (Fig. 5C, fig. S6E, and data S1). Similar to the E14.5 data, we found a slightly increased proportion of the progenitors in Atoh1S193A/− mice at E18.5 (Fig. 5D). In addition, the percentage of ePN1 cells was significantly reduced from 14.9 to 5.5% (Fig. 5D and fig. S6F), while there was no significant change in other ePN subtypes. These data indicate that ePN subtypes have differential vulnerability to the Atoh1 hypomorphic mutation.
Fig. 5. PN neurons at E18.5 are heterogeneous and exhibit differential vulnerability to Atoh1 hypomorphic mutation.
(A) Expression levels of the selective markers visualized on UMAP. Ccnd1, Atoh1, Nhlh1, and Mapt are known markers for progenitors, intermediate progenitors, migrating neurons, and differentiated neurons, respectively. (B) UMAP of the major cell states and ePN subtypes at E18.5. The dashed line denotes the subtype that was significantly reduced in Atoh1S193A/− mice. (C) Violin plot showing the expression levels of the marker genes. Prox1, Ephb1, Hoxb5, and Ntng1 are the marker genes for ePN1, ePN2, ePN3, and ePN4, respectively. (D) Proportion of the cells in each cell state and PN subtype. The arrow indicates the direction of the change in Atoh1S193A/− animals. *FDR < 0.01.
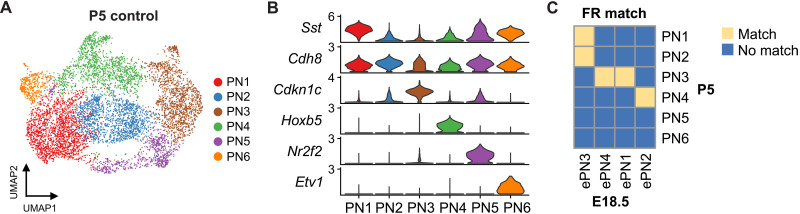
To test whether the differential vulnerability of the PN subtypes in Atoh1S193A/− embryos proceeds to the postnatal stage, we first determined whether the PN neurons maintained their molecular heterogeneity at P5. We enriched the PN neurons by dissecting and pooling the PN from control mice at P5 (n = 11 to 13 per replicate) and performed scRNA-seq for TdTom+ cells. Six PN subtypes (PN1 to PN6) were uncovered using unbiased clustering (Fig. 6A), suggesting that PN neurons maintained molecularly distinct at P5. In addition, we identified the marker genes for each subtype by differential gene expression analysis (Fig. 6B, fig. S7A, and data S1). Notably, several PN subtypes at P5 share similar markers with those at E18.5 (figs. S6E and S7A), indicating that PN subtypes might be conserved between these two time points. Thus, we performed Friedman-Rafsky (FR) test to match the cell type in E18.5 and P5 datasets (39). We found concordant signatures between the two time points (Fig. 6C), suggesting that the PN neurons have partially acquired their molecular signatures by E18.5. Notably, we did not find a matched cell type for PN5 and PN6 in the E18.5 data (Fig. 6C). Given that PN5 and PN6 are two of the smallest populations among all subtypes, those two subtypes could be underrepresented in the E18.5 data due to the low total number of the PN neurons being sampled. Together, these data confirm that differentiated PN neurons are molecularly heterogeneous at both E18.5 and P5. Moreover, the preferential loss of ePN1 subtype at E18.5 in Atoh1S193A/− embryos spurred our interest in testing whether the six PN subtypes at P5 are affected by Atoh1 hypomorphic mutation equally.
Fig. 6. scRNA-seq reveals six PN subtypes in mice at P5.
(A) UMAP of the major subtypes of the PN neurons at P5. (B) Violin plot showing the expression levels of the marker genes for each PN subtype. (C) Matching the cell type between E18.5 ePN subtypes and P5 PN subtypes by FR test.
The six molecularly defined PN subtypes are spatially segregated
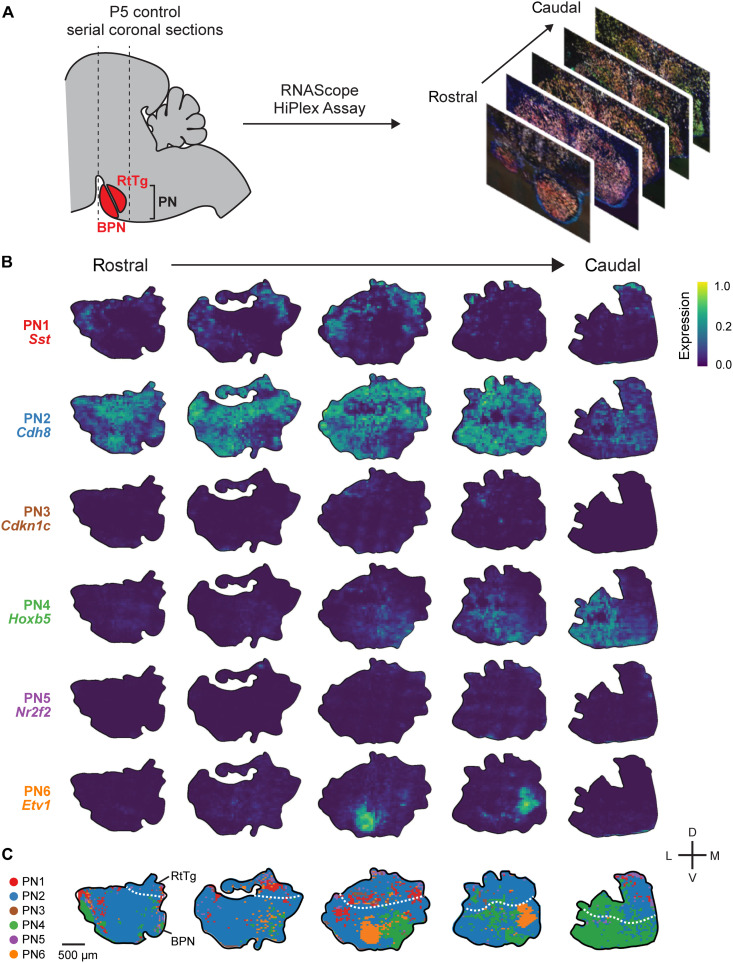
We ultimately wanted to test whether the six PN subtypes were differentially compromised in the Atoh1S193A/− mice. However, examining the cellular phenotypes in PN subtypes is challenging without knowing where those subtypes are located. Unlike the well-characterized laminated structure in the cerebral cortex, cerebellum, and retina, the cellular organization of the PN has not been delineated. Thus, we first characterized the anatomic location of the six PN subtypes in control animals and then used the spatial information to identify the cellular phenotypes in Atoh1S193A/− animals. We performed fluorescence RNA in situ hybridization (ISH) with RNAScope HiPlex assay on serial coronal sections from control mouse brain at P5 (Fig. 7A). We used TdTom probes to label Atoh1-lineage neurons and define the region of interest (i.e., PN). By binning the images, we calculated the average expression of the marker genes within each bin across the PN (Fig. 7B). We found that all the markers exhibit spatial specificity except Cdh8. For example, Hoxb5 is expressed in the caudal part of the PN, which is consistent with the previous study (14). Another marker, Etv1, is highly expressed in a restricted part of the medioventral PN. In contrast, Cdh8 is globally expressed across PN, which is expected based on our P5 scRNA-seq data (Fig. 6B). Notably, we found that Somatostatin (Sst), a neuropeptide that is typically expressed in inhibitory neurons, is expressed in a subset of the PN neurons that exhibit a shell-like pattern at the rostral PN (Fig. 7B). To date, there are only few studies showing that Sst is coexpressed in subsets of excitatory neurons in pre-Bötzinger complex, lateral hypothalamus, and dorsolateral periaqueductal gray matter (40–42). Here, we demonstrated that Sst is coexpressed in a subset of Slc17a6+ [Vesicular Glutamate Transporter 2 (VGLUT2+)] PN neurons (fig. S7, B and C).
Fig. 7. The spatial map of the PN subtypes in mice at P5.
(A) Workflow of the RNAScope HiPlex Assay on P5 control mouse. (B) Expression patterns of the selective markers for each PN subtype across the rostral to caudal axis of the PN. (C) Schematic summary of the distribution of the PN subtypes across the PN. The dashed line denotes the border between RtTg and BPN.
To find the corresponding PN subtypes between ISH and scRNA-seq data, we implemented K-nearest neighbor classification method to annotate the six PN subtypes on the representative regions of interest (Fig. 7C). PN neurons have been categorized into two nuclei, RtTg and BPN, based on their anatomic location. However, there has been no evidence showing that they are molecularly different. We found that PN6 is largely restricted to BPN, while PN3 and PN5 mostly reside at RtTg. In contrast, PN1, PN2, and PN4 are distributed across both nuclei. These findings suggest that RtTg and BPN have both shared and distinct neuronal subtypes. Together, our ISH data validate the expression of the marker genes identified by scRNA-seq and establish the spatial map of the PN subtypes that provides a higher resolution of the cytoarchitecture in the PN.
PN subtypes have differential vulnerability to Atoh1 hypomorphic mutation
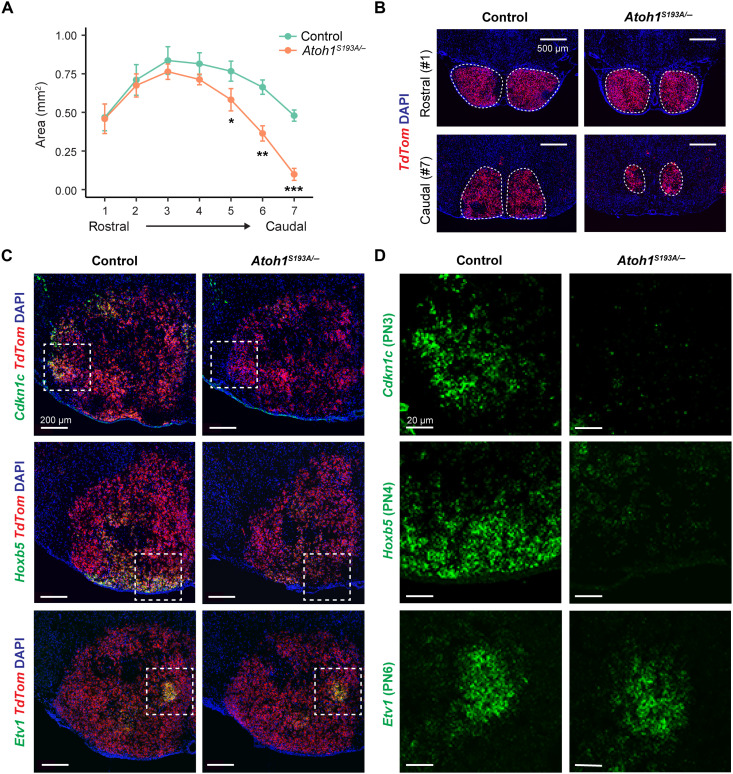
With the spatial map of the PN subtypes being established, we next performed fluorescence ISH to characterize PN subtypes in control and Atoh1S193A/− animals at P5. First, we used TdTom probes to identify PN across serial coronal sections. On the basis of the anatomic landmarks other than PN, we aligned the sections from different animals to proximity and quantified the size of PN in control and Atoh1S193A/− animals. Consistent with the whole-mount lacZ staining at P21 (Fig. 1D), we observed a reduced size of PN in Atoh1S193A/− animals at P5 (Fig. 8A). Moreover, we found that the caudal PN were most affected with 80% reduction at the most caudal section, while there is no significant difference at the rostral PN (Fig. 8, A and B).
Fig. 8. PN subtypes have different vulnerabilities to Atoh1 hypomorphic mutation.
(A) The size of the PN was determined by the areas across seven coronal sections (n = 3 per genotype). Data are presented as means ± SD. *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed unpaired t test. (B) Representative RNA ISH images for the most rostral (top) and the most caudal (bottom) sections from control (left) and Atoh1S193A/− (right) mice at P5. The PN neurons were labeled with TdTom probes. The nuclei were stained with DAPI. The dashed line encloses the area of the PN. Scale bars, 500 μm. (C) RNA ISH on control (left) and Atoh1S193A/− (right) mice at P5 using Cdkn1c (top), Hoxb5 (middle), and Etv1 (bottom) probes. PN neurons were labeled with TdTom probes. The nuclei were stained with DAPI. The box denotes the area shown in (D). Scale bars, 200 μm. (D) The zoom-in view of PN from the boxes in (C). The expression of Cdkn1c (top), Hoxb5 (middle), and Etv1 (bottom) were shown in green. Scale bars, 20 μm.
Given that different PN subtypes exhibit spatial specificity along the rostral-caudal axis (Fig. 7C), we hypothesized that the PN subtypes are differentially affected in Atoh1S193A/− mice. We thus examined three PN subtypes (PN3, PN4, and PN6) in control and Atoh1S193A/− at P5 by performing dual ISH using TdTom probes and marker genes Cdkn1c, Hoxb5, and Etv1, respectively. Cdkn1c is the marker gene for PN3 subtype, which matches ePN1 subtype at E18.5 (Fig. 6C) and is predicted to be reduced in Atoh1S193A/− animals according to our E18.5 scRNA-seq data (Fig. 5D). Moreover, on the basis of the spatial map of the PN (Fig. 7C) and the selective loss of the caudal PN in Atoh1S193A/− mice (Fig. 8, A and B), we predicted that Hoxb5+ (PN4) was compromised by partial loss of function of Atoh1. Last, to test whether there is one subtype that is not sensitive to Atoh1 hypomorphic mutation, we chose Etv1+ subtype (PN6). Given its specific location within the PN (Fig. 7C), changes in PN, if any, should be easily detected. We examined serial sections across the whole PN to exclude the possibility that certain subtype might be mislocated. We found fewer Cdkn1c+TdTom+ PN3 neurons in Atoh1S193A/− mice, accompanied by reduced size in the area where the Cdkn1c+ PN3 neurons are normally located in controls (Fig. 8, C and D, top). Hoxb5+TdTom+ PN4 neurons were also reduced in Atoh1S193A/− animals at the caudal sections (Fig. 8, C and D, middle). In contrast, Etv1+TdTom+ PN6 neurons were not affected in Atoh1S193A/− animals (Fig. 8, C and D, bottom). Together, these data demonstrated that although all PN subtypes were derived from Atoh1+ progenitors, certain PN subtypes such as PN3 and PN4 neurons were more vulnerable to partial loss of function of Atoh1, suggesting that the robustness of Atoh1 function is critical for PN subtype cell fate decisions.
DISCUSSION
In this study, we used a mouse model carrying an Atoh1 hypomorphic mutation and implemented scRNA-seq technology to elucidate the roles of Atoh1 in different cell states during PN development. We demonstrate that Atoh1 is involved in multiple biological processes including regulating the cell cycle, differentiation, cell survival, migration, and the heterogeneity of the PN neurons. Moreover, we show that Atoh1-lineage PN neurons are classified as six subtypes based on their molecular signatures and provide a list of marker genes for future studies. PN subtypes display differential vulnerability to Atoh1 hypomorphic mutation, which opens up questions such as how cell fate decisions are made and how perturbation of a transcription factor results in subtype-specific phenotypes.
The interplay between Atoh1 and Notch signaling
One interesting phenotype that we observed in Atoh1S193A/− mice was the marked increase in proliferating progenitors (Figs. 2F and 3B), suggesting that Atoh1 might promote cell cycle exit. In addition, differential gene expression analysis revealed several dysregulated pathways including up-regulated Notch signaling in the proliferating progenitors (Fig. 4D). The interaction between Atoh1 and Notch signaling has been demonstrated in the mammalian intestine (43, 44). In the epithelium lining the crypts in the small intestine, the stem cells differentiate into secretory cells when they escape Notch activation by up-regulation of Atoh1. In contrast, the stem cells in which Notch is activated remain as progenitors at the crypts where Wnt is high. Here, we hypothesize that in cRL, partial loss of function of Atoh1 leads to failure to escape Notch activation, which maintains the cells at the proliferating state. In line with this hypothesis, we observed up-regulation of the Notch receptor Notch1 and its ligand Dll1 as well as genes downstream from active Notch signaling including Hes1 and Hes5 in the proliferating progenitors of Atoh1S193A/− mice (data S2). Moreover, a recent study in zebrafish showed that inhibition of Notch activity at lower RL led to increased atoh1b+ postmitotic precursors at the expense of atoh1a+ proliferating progenitors (45). This study also supports our hypothesis that Atoh1 and Notch antagonize each other at the cRL in mammals.
The molecular heterogeneity of the PN neurons
One of the most puzzling questions in neurobiology is how to define a neuronal subtype. In the case of the PN, they have been considered as a group of heterogeneous neurons based on their anatomical location, origins at the cRL, and functions based on the cortico-ponto-cerebellar connectivity (13–16, 46). However, whether the PN neurons can be further defined by their molecular signature has not been shown. scRNA-seq provides a powerful approach to classify the cell type unbiasedly based on transcription profiles. Here, we uncovered six PN subtypes in P5 mice (Fig. 6, A and B). These six PN subtypes were spatially segregated (Fig. 7), which raises an interesting question regarding whether this spatial map reflects the topographic connectivity between the cerebral cortex and the PN. For example, Sst+ PN subtype reside at the rostral dorsal part of the PN (Fig. 7B). This location coincides with where PN receive inputs from brain regions involved in visual pathway including visual cortex, superior colliculus, inferior colliculus, and pretectum (47). Whether there is a subtype-specific connectivity that also reflects the functionality needs further investigation. However, those studies cannot be done without genetic tools to target different PN subtypes. Our study identified and validated the marker genes for the six PN subtypes. In future studies, one can test whether different PN subtypes preferentially connect with specific groups of neurons in the cerebral cortex and/or cerebellum by intersectional labeling and viral tracing approaches with the molecular markers. In addition to the connectivity, the molecular markers that we identified can be used to manipulate PN neurons in a subtype-specific manner for functional characterization.
From a pool of Atoh1+ progenitors to diverse PN subtypes
How does a pool of seemingly homogeneous progenitors give rise to a diverse population of neurons? In the case of Atoh1-lineage neurons in the hindbrain, Atoh1+ progenitors contribute to neurons in the cerebellum and dozens of brainstem nuclei depending on the rostrocaudal origins at RL and the timing of leaving from RL (9, 10, 48). PN, for example, were derived from rhombomere 6 to 8 where Hox2 to Hox5 are expressed (10). It has not been addressed, however, whether Atoh1 contributes to the diversity of the subpopulation within one lineage (i.e., PN in this study). To this end, we tested whether PN subtypes display differential vulnerability to Atoh1 hypomorphic mutation. On the basis of our scRNA-seq data at E18.5 and the histological analysis at P5, partial loss of function of Atoh1 not only reduced the size of the PN but also reduced the diversity of the PN neurons by preferentially affecting PN3 and PN4 subtypes (Fig. 8, C and D). These data suggest that Atoh1 may contribute to the acquisition or maintenance of the heterogeneity of the PN neurons.
In summary, this study dissected the functions of Atoh1 during PN differentiation by characterizing the phenotypes in Atoh1 hypomorphic mutant at single-cell resolution. Our data demonstrate that Atoh1 regulates cell cycle exit, differentiation, migration, and survival during PN development and contributes to the diversity of the PN subtypes. In addition, this study also uncovers the molecular heterogeneity of the PN, which opens new doors for understanding the neural fate decisions, connectivity, and functionality of the PN neurons.
MATERIALS AND METHODS
Mice
The following mouse lines were used in this study: Atoh1lacZ/+ (23), Atoh1S193A/+ (24), Atoh1Cre/+ (28), Rosalsl-lacZ/+ (JAX:02429), and Rosalsl-TdTomato/+ (JAX:007914). All mice were housed in a level 3, American Association for Laboratory Animal Science (AALAS)–certificated facility on a 14-hour light cycle. Husbandry, housing, euthanasia, and experimental guidelines were approved by the Institutional Animal Care & Use Committee (IACUC) at Baylor College of Medicine.
Whole-mount X-galactosidase staining
The brains of P0 pups and P21 mice were dissected out in ice-cold phosphate-buffered saline (PBS). The samples were fixed in 4% paraformaldehyde (PFA) at 4°C for 1 hour (P0) or 2 hours (P21). After brief PBS wash at room temperature (RT), the samples were incubated in equilibration buffer [2 mM MgCl2, 0.05% sodium deoxycholate, 0.02% NP-40, and 0.1 M sodium phosphate (pH 7.3)] at 4°C for 15 min, followed by 2-hour incubation at 37°C in X-galactosidase (X-gal) reaction solution [X-gal (1 mg/ml), 5 mM potassium ferrocyanide, and 5 mM potassium ferricyanide in equilibration buffer]. After staining, the samples were washed with PBS three times at RT and fixed again in 4% PFA at 4°C for 1 hour (P0) or 4 hours (P21) before imaging.
Sample preparation for scRNA-seq
The brains of embryos and P5 pups were dissected out in ice-cold HEBG medium (0.8× B27 and 0.25× GlutaMAX in hibernate E medium). For embryonic studies, hindbrains were collected from Atoh1S193A/− and its littermate control (one embryo per genotype, three independent replicates). For P5 study, PN were microdissected out and pooled from 11 to 13 pups (two independent replicates). Tissues were cut into small pieces and transferred to a 1.5-ml microcentrifuge tube using wide-bore pipette tips. The single-cell dissociation protocol was modified from the previous study (49). Briefly, the tissues were incubated with Worthington Papain solution at 37°C for 30 min at 800 rpm. At the end of incubation, the samples were transferred to a 15-ml falcon tube, followed by gentle trituration with serologic pipette. The cell pellets were collected by centrifuge at 200 rcf for 3 min at 4°C, washed, and resuspended in ice-cold sorting buffer (PBS with 0.05% fetal bovine serum). The single-cell resuspension was loaded to 30-μm cell drainer (CellTrics, SYSMEX 04-004-2326) to remove debris, followed by 4′,6-diamidino-2-phenylindole (DAPI) staining at RT for 5 min. TdTom+DAPI− cells (120,000 to 150,000 cells per sample) were sorted into bovine serum albumin–coated 15-ml falcon tube by Sony SH800S cell sorter. The cells were pelleted by centrifuge at 200 rcf for 5 min at 4°C and resuspended in sorting buffer to make the final concentration of 1000 cells/μl.
Library construction and sequencing
The cDNA libraries were constructed by 10X Genomics 3′ v3.1 kit following the user guide. Briefly, ~16,500 cells from one sample were mixed with reversed transcription master mix before loaded into Chromium Chip G. Droplets containing cells, reversed transcription reagents, and barcoded gel beads were generated by Chromium Controller. The first strand cDNA was amplified, fragmentated, and ligated with sequencing adaptors and sample indices. The cDNA libraries were sequenced by Illumina NovaSeq 6000.
RNAScope HiPlex assay
The brain of the P5 pup was flash-frozen and embedded in optimal cutting temperature (OCT) compound. Coronal sections were made by cryostat (Leica) at 20 μm and store in −80°C until use. The RNAScope HiPlex assay (ACD, catalog no. 324419) was performed according to the manufacturer’s instructions. Briefly, sections were fixed in 4% PFA for 1 hour at RT, followed by dehydration with 50, 70, and 100% ethanol. The protease treatment was done by incubating the sections with protease III at RT for 30 min. Sections were hybridized with the 12 probes (catalog nos. 487941-T1, 845141-T2, 480301-T3, 557891-T4, 319171-T5, 458331-T6, 485461-T7, 503461-T8, 404631-T9, 503481-T10, 317041-T11, and 433411-T12) at 40°C for 2 hours, followed by three rounds of amplification. The TdTom reporter was helpful during sectioning to ensure that we covered the whole PN. However, it also introduced hazy background in HiPlex assay. Thus, we quenched the TdTom signal by incubating the sections with 5% formalin-fixed paraffin-embedded (FFPE) reagent (included in the kit) at RT for 30 min before developing the first round of the targets (T1 to T3). The nuclei were stained with DAPI. The 12 targets were detected in four rounds of imaging with three targets per round. Between each round, the fluorophores were cleaved and washed away before the next round of signal development. After image acquisition of all targets, the fluorophores were cleaved, and a blank image of each section was taken to serve as background images.
Immunofluorescence staining
The heads of the E14.5 embryos were dissected in ice-cold PBS. After brief wash, the samples were fixed in 4% PFA at 4°C for 4 hours, followed by PBS wash and incubation in 30% sucrose in PBS at 4°C for 14 to 16 hours. The samples were cryopreserved and stored at −80°C until use. The coronal sections were collected on slides by cryostat (Leica) with 20 or 25 μm in thickness. The slides were rinsed with PBS to remove OCT, followed by permeabilization with 0.3% Triton X-100 in PBS for 15 min at RT. Antigen retrieval was performed by heating in antigen retrieval buffer [10 mM sodium citrate and 0.05% Tween-20 (pH 6.0)] at 85°C for 10 min [MKI67/red fluorescent protein (RFP) staining] or 30 min (CldU labeling). After blocking with blocking buffer (5% normal goat serum with 0.3% Triton X-100 in PBS) for 2 hours at RT, the sections were incubated with primary antibodies in blocking buffer at 4°C for 24 hours, followed by PBS wash three times. The sections were incubated with secondary antibodies in blocking buffer at RT for 2 hours. The counterstain was performed by DAPI staining at RT for 10 min. The slides were mounted with ProLong Gold mounting media (Invitrogen). The following antibodies were used in this study with the indicated dilutions: rat anti–5-bromo-2′-deoxyuridine (BrdU)/CldU (1:250; Abcam, ab6326), mouse anti-Ki67 (1:100; R&D Systems, AF7649), rabbit anti-RFP (1:2000; Rockland, 600-401-379), goat anti-rat Alexa Fluor 488 (1:500; Invitrogen, A-11006), goat anti-rabbit Alexa Fluor 555 (1:500; Invitrogen, A-21428), goat anti-mouse Alexa Fluor 647 (1:500; Invitrogen, A-21236).
CldU labeling
CldU was prepared in sterile saline at 4.82 mg/ml and injected into pregnant dams intraperitoneally at E13.5 (85 mg/kg). Twenty-four hours later, the embryos were collected at E14.5, followed by immunofluorescence staining using rat anti-BrdU/CldU antibody (1:250; Abcam, ab2326).
TUNEL assay
The sample preparation follows the same protocol that we used for immunofluorescence staining. Coronal sections were made by cryostat (Leica) at 25 μm. TUNEL assay was performed using DeadEnd Fluorometric TUNEL system (Promega) according to the user guide. Briefly, sections were fixed in 4% PFA at RT for 15 min, followed by proteinase K (20 μg/ml) treatment at RT for 12 min and a second fixation with 4% PFA for 5 min. The sections were equilibrated and incubated with fluorescein-labeled nucleotides and terminal deoxynucleotidyl transferase reaction mix at 37°C for 1 hour. After stopping the reaction by 2× saline-sodium citrate (SSC), the nuclei were stained with DAPI.
Dual-color fluorescence RNA ISH
The brains from P5 pups were embedded in OCT, frozen on dry ice, and stored in −80°C until use. The sections were cut by cryostat (Leica) at 20 μm. We generated a digoxigenin (DIG)–labeled mRNA antisense probes against Slc17a6 and TdTom and fluorescein isothiocyanate (FITC)–labeled mRNA against Sst, Etv1, and Hoxb5 using reverse-transcribed mouse cDNA as template and DIG or FITC RNA labeling kits from Roche (Sigma-Aldrich). Primer sequences for Sst, Slc17a6, and TdTom probes are available in Allen Brain Atlas (www.brain-map.org). The following primers were used: 5′-ttcagaactcgggtctgctt-3′ and 5′-gaatcatgcaaaaggtggct-3′ for Etv1 probe and 5′-gatggatctcagcgtcaacc-3′ and 5′-tatgagtctggctacagccg-3′ for Hoxb5 probe. ISH was performed by the RNA In Situ Hybridization Core at Baylor College of Medicine using an automated robotic platform as previously described (50) with modifications of the protocol for double ISH. Briefly, two probes were hybridized to the tissue simultaneously. After the wash and blocking steps, the DIG-labeled probes were visualized by incubating with tyramide-Cy3 Plus (1:75; PerkinElmer) for 15 min. After washing in TNT (0.05% Tween-20 in 150mM NaCl and 100mM Tris-HCl, pH7.5), the remaining horseradish peroxidase (HRP) activity was quenched by a 10-min incubation in 0.2 M HCl. The sections were then washed in TNT and blocked in TNB for 15 min, followed by incubation with HRP-labeled sheep anti-FITC antibody (1:500 in TNB; Roche) at RT for 30 min. After washes in TNT, the FITC-labeled probes were visualized by incubating with tyramide-FITC Plus (1:75; PerkinElmer) for 15 min. Following washes in TNT, the slides were stained with DAPI (Invitrogen), washed again, removed from the machine, and mounted with ProLong Diamond (Invitrogen).
Image acquisition
Images of the whole-mount tissues were obtained by Zeiss Axio Zoom.V16. All fluorescence images were obtained by Nikon Ti2E Inverted Motorized Microscope equipped with CSU-W1 Dual Camera-Dual Disk System with 405/488/561/640-nm lasers with 10×, 20×, or 40× objectives.
Data preprocessing
The reads were aligned to customized genome that composed of mouse mm10 reference genome and woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) sequence to ensure capturing the TdTom transcripts. The alignment and quantification of unique molecular identifier (UMI) were performed on 10× cloud analysis platform by CellRanger pipeline v5.0.1 with default parameters. Individual expression matrix of each sample was filtered by Seurat v4 package (51), followed by doublet removal using DoubletFinder v2 package (52). Briefly, cells with at least 1000 genes expressed and less than 1% of total UMIs that were mitochondrial genes were retained (see the code for detailed criteria depending on the age of the samples.) Doublets with high confidence score identified by DoubletFinder with default parameters were removed.
Integration and clustering
The data integration and clustering were performed by Seurat package. The filtered datasets of the samples from the same age were combined by integration pipeline. Briefly, the filtered matrices were normalized and scaled by regressing out the percentage of mitochondrial genes with SCTransform (53). For E14.5 and E18.5 samples in which different genotypes were compared, a reference-based integration method was used by setting the first replicate as reference to cut down the computational power and time needed. For P5 study, the default integration method was used. Following the integration, principal components analysis (PCA) was performed. We selected the top 30 PCAs to generate the K-nearest neighbor graph, which was used to perform clustering with variated resolutions depending on the complexity of the dataset. To annotate the clusters, the top 10 genes that were highly expressed in each cluster were identified by FindAllMarkers function with default parameters.
Trajectory analysis
The RNA assay of the Seurat object was extracted and transformed into SingleCellExperiment object (sce) by as.SingleCellExperiment function. The sce was used as input to infer the trajectory by slingshot (30). Kolmogorov-Smirnov test was used to assess whether the distribution of pseudo-time is identical between the genotypes.
Differential expression analysis
We performed differential expression analysis between control and Atoh1S193A/− samples for each cell state by FindMarkers function from Seurat package. Only genes that were expressed in at least 10% of the cells were included. MAST (54) was used as test method. The P value was corrected by Benjamini-Hochberg FDR method.
GO analysis
GO analysis was performed using a web-based tool called g:Profiler (55) with custom statistic domain scope. The up- and down-regulated DEGs (log2FC > 0.25 and FDR < 0.05) within progenitors, intermediate progenitors, and migrating neurons-1 were used as inputs independently. GO biological process was selected for the analysis.
Image processing for RNAScope HiPlex assay
Five confocal z-stacks of images were collected at 100- and 200-μm intervals along the rostral-caudal axis of the pontine nucleus for each of the nine transcripts. In addition, DAPI images were also collected to serve as reference images. Individual z-stacks consisted of 10 images separated by 0.9 μm. The z-stacks were first processed by taking the max intensity projection and cropping the resulting transcript image such that the midline of the pontine aligned with the right-hand border of each image.
Visualizing transcript expression
Overlaying the transcript images onto the DAPI reference images (56) revealed that some transcripts were not consistently colocalized to the nucleus. To address this ambiguity in our analysis, we did not associate transcript expression with individual cells. Rather, we binned each image into a set of tiles measuring 34 μm × 31 μm and measured the transcript expression in each tile (57). Since transcripts may be expressed at very different levels, we normalized the expression for each transcript by the max expression recorded across the five image locations in the pontine. Thus, in Fig. 7B, the expression for each transcript varies from 0 to 1 across the five rostral to caudal sections. To identify the boundaries of the pontine, the max intensity projection across all the transcripts at a given imaging location was smoothed with a Gaussian filter with an SD of 35 μm. This filtered image was converted to a binary image by mean thresholding, and any remaining holes were morphologically closed using a 5-μm × 5-μm structuring element. Last, the pontine boundary was taken to be the edge of the largest single region in the closed binary image.
Quantification of the immunostaining and statistics
The cell number of the CldU+TdTom+ (Fig. 3F) and TUNEL+ (Fig. 4F) was determined by manual counting on imageJ. The percentage of MKI67 signal overlapping with TdTom (Fig. 3C) was calculated based on Manders’ coefficients using JACoP (58). For the statistical test, we used mixed-model analysis of variance (ANOVA) to compare genotype using lme4 package on R (59) to count for variations among technical and biological replicates. The person who performed the quantification was blinded to the genotypes of the samples.
Classification of RNA ISH transcripts
The transcript expression levels for each tile in the ISH images can be represented as a nine-dimensional vector, one component for each transcript. To classify these transcript expression vectors as one of the six classes determined from the single-cell RNA-seq analysis, we built a K-nearest neighbor classifier (60). Specifically, for each cell (n = 7029) from the RNA-seq clustering results, we extracted the same transcripts as measured in the ISH and the cluster identity. Each of these nine-component vectors was normalized (60) and, along with their respective cluster IDs, was used to train and validate our classifier. The K-nearest neighbor classifier measures the distance between each nine-component vector and a preset number of neighbor vectors to predict class membership. To determine the number of neighbors, we performed sixfold cross-validation on the training dataset and found that 25 neighbors provided an average test accuracy of 80%. We then used this 25 nearest neighbor model to predict the cluster identity of each tile in the ISH images. The border of the RtTg and BPN in Fig. 7C was defined using the reference atlas of P6 mouse brain (61).
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute Of Child Health and Human Development or the National Institutes of Health. We thank the Baylor College of Medicine Center for Comparative Medicine for mouse colony management, D. Yu for assistance with microscopy, C. Ljungberg for assistance with RNA ISH, and C.-W. Logan Hsu for assistance with the tissue clearing and lightsheet microscopy. We thank the members of the Zoghbi lab and M. E. Van Der Heijden for discussions and comments on the manuscript.
Funding: This project was supported by Howard Hughes Medical Institute and funding from a Shared Instrumentation grant from the NIH (S10 OD016167) and the NIH IDDRC Grant P50 HD103555 from the Eunice Kennedy Shriver National Institute Of Child Health and Human Development for use of the RNA In Situ Hybridization Core and Neurovisualization Core. J.C.B. was supported by NIH F32 (NS117723). R.S.D. was supported by NIH NINDS F32 (NS127854). This work was supported by Howard Hughes Medical Institute (to H.Y.Z.), National Institutes of Health grant S10 OD016167, National Institutes of Health IDDRC P50 HD103555, National Institutes of Health grant NS117723 (to J.C.B.), and National Institutes of Health grant NS127854 (to R.S.D.).
Author contributions: Conceptualization: H.Y.Z., S.-R.W., J.C.B., and M.A.D. Methodology: S.-R.W., J.C.B., and M.A.D. Investigation: S.-R.W., J.C.B., and J.-P.R. Software: S.-R.W., M.S.C., R.S.D., and M.A.D. Visualization: S.-R.W. and M.S.C. Writing—original draft: S.-R.W. Writing—review and editing: H.Y.Z., S.-R.W., J.C.B., M.S.C., and R.S.D. Funding acquisition: H.Y.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data are available in the main text and/or the Supplementary Materials. The raw FASTQ files of the scRNA-seq and the processed files (output from CellRanger) are accessible through GEO (accession number: GSE224031). The code for data analyses is available on figshare (doi: 10.6084/m9.figshare.22490956) and via the link: https://figshare.com/s/ca568d8f44d020cb6389.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
Legends for data S1 to S3
References
Other Supplementary Material for this manuscript includes the following:
Data S1 to S3
REFERENCES AND NOTES
- 1.E. V. Evarts, W. T. Thach, Motor mechanisms of the CNS: Cerebrocerebellar interrelations. Annu. Rev. Physiol. 31, 451–498 (1969). [DOI] [PubMed] [Google Scholar]
- 2.C. R. Legg, B. Mercier, M. Glickstein, Corticopontine projection in the rat: The distribution of labelled cortical cells after large injections of horseradish peroxidase in the pontine nuclei. J. Comp. Neurol. 286, 427–441 (1989). [DOI] [PubMed] [Google Scholar]
- 3.P. Brodal, J. G. Bjaalie, Organization of the pontine nuclei. Neurosci. Res. 13, 83–118 (1992). [DOI] [PubMed] [Google Scholar]
- 4.C. Schwarz, P. Thier, Binding of signals relevant for action: Towards a hypothesis of the functional role of the pontine nuclei. Trends Neurosci. 22, 443–451 (1999). [DOI] [PubMed] [Google Scholar]
- 5.J. D. Schmahmann, R. Ko, J. MacMore, The human basis pontis: Motor syndromes and topographic organization. Brain 127, 1269–1291 (2004). [DOI] [PubMed] [Google Scholar]
- 6.K. Tziridis, P. W. Dicke, P. Thier, Pontine reference frames for the sensory guidance of movement. Cereb. Cortex 22, 345–362 (2012). [DOI] [PubMed] [Google Scholar]
- 7.G.-Y. Wu, S.-L. Liu, J. Yao, L. Sun, B. Wu, Y. Yang, X. Li, Q.-Q. Sun, H. Feng, J.-F. Sui, Medial prefrontal cortex-pontine nuclei projections modulate suboptimal cue-induced associative motor learning. Cereb. Cortex 28, 880–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.C. I. Rodriguez, S. M. Dymecki, Origin of the precerebellar system. Neuron 27, 475–486 (2000). [DOI] [PubMed] [Google Scholar]
- 9.V. Y. Wang, M. F. Rose, H. Y. Zoghbi, Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron 48, 31–43 (2005). [DOI] [PubMed] [Google Scholar]
- 10.A. F. Farago, R. B. Awatramani, S. M. Dymecki, Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron 50, 205–218 (2006). [DOI] [PubMed] [Google Scholar]
- 11.J. Altman, S. A. Bayer, Development of the precerebellar nuclei in the rat: IV. The anterior precerebellar extramural migratory stream and the nucleus reticularis tegmenti pontis and the basal pontine gray. J. Comp. Neurol. 257, 529–552 (1987). [DOI] [PubMed] [Google Scholar]
- 12.T. Okada, K. Keino-Masu, M. Masu, Migration and nucleogenesis of mouse precerebellar neurons visualized by in utero electroporation of a green fluorescent protein gene. Neurosci. Res. 57, 40–49 (2007). [DOI] [PubMed] [Google Scholar]
- 13.A. Brodal, J. Jansen, The ponto-cerebellar projection in the rabbit and cat; experimental investigations. J. Comp. Neurol. 84, 31–118 (1946). [DOI] [PubMed] [Google Scholar]
- 14.T. D. Meglio, C. F. Kratochwil, N. Vilain, A. Loche, A. Vitobello, K. Yonehara, S. M. Hrycaj, B. Roska, A. H. F. M. Peters, A. Eichmann, D. Wellik, S. Ducret, F. M. Rijli, Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science 339, 204–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.T. B. Leergaard, K. A. Lyngstad, J. H. Thompson, S. Taeymans, B. P. Vos, E. de Schutter, J. M. Bower, J. G. Bjaalie, Rat somatosensory cerebropontocerebellar pathways: Spatial relationships of the somatotopic map of the primary somatosensory cortex are preserved in a three-dimensional clustered pontine map. J. Comp. Neurol. 422, 246–266 (2000). [DOI] [PubMed] [Google Scholar]
- 16.J. U. Henschke, J. M. Pakan, Disynaptic cerebrocerebellar pathways originating from multiple functionally distinct cortical areas. eLife 9, e59148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.N. A. Bermingham, B. A. Hassan, V. Y. Wang, M. Fernandez, S. Banfi, H. J. Bellen, B. Fritzsch, H. Y. Zoghbi, Proprioceptor pathway development is dependent on Math1. Neuron 30, 411–422 (2001). [DOI] [PubMed] [Google Scholar]
- 18.N. A. Bermingham, B. A. Hassan, S. D. Price, M. A. Vollrath, N. Ben-Arie, R. A. Eatock, H. J. Bellen, A. Lysakowski, H. Y. Zoghbi, Math1: An essential gene for the generation of inner ear hair cells. Science 284, 1837–1841 (1999). [DOI] [PubMed] [Google Scholar]
- 19.S. M. Maricich, S. A. Wellnitz, A. M. Nelson, D. R. Lesniak, G. J. Gerling, E. A. Lumpkin, H. Y. Zoghbi, Merkel cells are essential for light-touch responses. Science 324, 1580–1582 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.N. F. Shroyer, M. A. Helmrath, V. Y.-C. Wang, B. Antalffy, S. J. Henning, H. Y. Zoghbi, Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478–2488 (2007). [DOI] [PubMed] [Google Scholar]
- 21.M. F. Rose, K. A. Ahmad, C. Thaller, H. Y. Zoghbi, Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc. Natl. Acad. Sci. U.S.A. 106, 22462–22467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.T. Višnjar, A. Maver, K. Writzl, O. Maloku, G. Bergant, H. Jaklǐ, D. Neubauer, F. Fogolari, N. P. Meglǐ, B. Peterlin, Biallelic ATOH1 gene variant in siblings with pontocerebellar hypoplasia, developmental delay, and hearing loss. Neurol. Genet. 8, e677 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N. Ben-Arie, B. A. Hassan, N. A. Bermingham, D. M. Malicki, D. Armstrong, M. Matzuk, H. J. Bellen, H. Y. Zoghbi, Functional conservation of atonal and Math1in the CNS and PNS. Development 127, 1039–1048 (2000). [DOI] [PubMed] [Google Scholar]
- 24.W. R. Xie, H.-I. Jen, M. L. Seymour, S.-Y. Yeh, F. A. Pereira, A. K. Groves, T. J. Klisch, H. Y. Zoghbi, An Atoh1-S193A phospho-mutant allele causes hearing deficits and motor impairment. J. Neurosci. 37, 8583–8594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.X. Jin, S. K. Simmons, A. Guo, A. S. Shetty, M. Ko, L. Nguyen, V. Jokhi, E. Robinson, P. Oyler, N. Curry, G. Deangeli, S. Lodato, J. Z. Levin, A. Regev, F. Zhang, P. Arlotta, In vivo perturb-seq reveals neuronal and glial abnormalities associated with autism risk genes. Science 370, eaaz6063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.I. Schaffner, M.-T. Wittmann, T. Vogel, D. C. Lie, Differential vulnerability of adult neurogenic niches to dosage of the neurodevelopmental-disorder linked gene Foxg1. Mol. Psychiatry 28, 497–514 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.P. Soriano, Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999). [DOI] [PubMed] [Google Scholar]
- 28.H. Yang, X. Xie, M. Deng, X. Chen, L. Gan, Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis 48, 407–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.B. Phipson, C. B. Sim, E. R. Porrello, A. W. Hewitt, J. Powell, A. Oshlack, Propeller: Testing for differences in cell type proportions in single cell data. Bioinformatics 38, 4720–4726 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.K. Street, D. Risso, R. B. Fletcher, D. Das, J. Ngai, N. Yosef, E. Purdom, S. Dudoit, Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.H. R. de Bézieux, K. Van den Berge, K. Street, S. Dudoit, Trajectory inference across multiple conditions with condiments: Differential topology, progression, differentiation, and expression. bioRxiv 10.1101/2021.03.09.433671 [Preprint]. 10 March 2021. 10.1101/2021.03.09.433671. [DOI]
- 32.T. J. Klisch, Y. Xi, A. Flora, L. Wang, W. Li, H. Y. Zoghbi, In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc. Natl. Acad. Sci. U.S.A. 108, 3288–3293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A. W. Helms, A. L. Abney, N. Ben-Arie, H. Y. Zoghbi, J. E. Johnson, Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127, 1185–1196 (2000). [DOI] [PubMed] [Google Scholar]
- 34.S. Li, F. Qiu, A. Xu, S. M. Price, M. Xiang, Barhl1 regulates migration and survival of cerebellar granule cells by controlling expression of the neurotrophin-3 gene. J. Neurosci. 24, 3104–3114 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.T. Schmid, M. Kruger, T. Braun, NSCL-1 and -2 control the formation of precerebellar nuclei by orchestrating the migration of neuronal precursor cells. J. Neurochem. 102, 2061–2072 (2007). [DOI] [PubMed] [Google Scholar]
- 36.T. Cai, H.-I. Jen, H. Kang, T. J. Klisch, H. Y. Zoghbi, A. K. Groves, Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 35, 5870–5883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. V. Yu, L. Tao, J. Llamas, X. Wang, J. D. Nguyen, T. Trecek, N. Segil, POU4F3 pioneer activity enables ATOH1 to drive diverse mechanoreceptor differentiation through a feed-forward epigenetic mechanism. Proc. Natl. Acad. Sci. U.S.A. 118, e2105137118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.H. C. Lai, T. J. Klisch, R. Roberts, H. Y. Zoghbi, J. E. Johnson, In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J. Neurosci. 31, 10859–10871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Y. Zhang, B. Aevermann, R. Gala, R. H. Scheuermann, Cell type matching in single-cell RNA-sequencing data using FR-match. Sci. Rep. 12, 9996 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R. L. Stornetta, D. L. Rosin, H. Wang, C. P. Sevigny, M. C. Weston, P. G. Guyenet, A group of glutamatergic interneurons expressing high levels of both neurokinin-1 receptors and somatostatin identifies the region of the pre-Bötzinger complex. J. Comp. Neurol. 455, 499–512 (2003). [DOI] [PubMed] [Google Scholar]
- 41.L. E. Mickelsen, M. Bolisetty, B. R. Chimileski, A. Fujita, E. J. Beltrami, J. T. Costanzo, J. R. Naparstek, P. Robson, A. C. Jackson, Single-cell transcriptomic analysis of the lateral hypothalamic area reveals molecularly distinct populations of inhibitory and excitatory neurons. Nat. Neurosci. 22, 642–656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.N. Winke, F. Aby, D. Jercog, G. Zoé, D. Girard, M. Landry, L. Castell, E. Valjent, S. Valerio, P. Fossat, C. Herry, Brainstem somatostatin-expressing cells control the emotional regulation of pain behavior. bioRxiv 10.1101/2022.01.20.476899 [Preprint]. 22 January 2022. 10.1101/2022.01.20.476899. [DOI]
- 43.Q. Yang, N. A. Bermingham, M. J. Finegold, H. Y. Zoghbi, Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294, 2155–2158 (2001). [DOI] [PubMed] [Google Scholar]
- 44.R. Sancho, C. A. Cremona, A. Behrens, Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 16, 571–581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.I. Belzunce, C. Belmonte-Mateos, C. Pujades, The interplay of atoh1 genes in the lower rhombic lip during hindbrain morphogenesis. PLOS ONE 15, e0228225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R. V. Sillitoe, Y. Fu, C. Watson, in The Mouse Nervous System, C. Watson, G. Paxinos, L. Puelles, Eds. (Academic Press, San Diego, 2012), pp. 260–397.
- 47.C. F. Kratochwil, U. Maheshwari, F. M. Rijli, The long journey of pontine nuclei neurons: From rhombic lip to cortico-ponto-cerebellar circuitry. Front. Neural Circuits 11, 33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R. Machold, G. Fishell, Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48, 17–24 (2005). [DOI] [PubMed] [Google Scholar]
- 49.R. Chen, X. Wu, L. Jiang, Y. Zhang, Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 18, 3227–3241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.M. B. Yaylaoglu, A. Titmus, A. Visel, G. Alvarez-Bolado, C. Thaller, G. Eichele, Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Dev. Dyn. 234, 371–386 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck III, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.C. S. McGinnis, L. M. Murrow, Z. J. Gartner, DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.C. Hafemeister, R. Satija, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.G. Finak, A. McDavid, M. Yajima, J. Deng, V. Gersuk, A. K. Shalek, C. K. Slichter, H. W. Miller, M. J. McElrath, M. Prlic, P. S. Linsley, R. Gottardo, MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.U. Raudvere, L. Kolberg, I. Kuzmin, T. Arak, P. Adler, H. Peterson, J. Vilo, G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. D. Hunter, Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007). [Google Scholar]
- 57.S. van der Walt, J. L. Schönberger, J. Nunez-Iglesias, F. Boulogne, J. D. Warner, N. Yager, E. Gouillart, T. Yu; scikit-image contributors , Scikit-image: Image processing in Python. PeerJ 2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. Bolte, F. P. Cordelières, A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 59.D. Bates, M. Mächler, B. Bolker, S. Walker, Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 60.C. R. Harris, K. J. Millman, S. J. van der Walt, R. Gommers, P. Virtanen, D. Cournapeau, E. Wieser, J. Taylor, S. Berg, N. J. Smith, R. Kern, M. Picus, S. Hoyer, M. H. van Kerkwijk, M. Brett, A. Haldane, J. F. Del Río, M. Wiebe, P. Peterson, P. Gérard-Marchant, K. Sheppard, T. Reddy, W. Weckesser, H. Abbasi, C. Gohlke, T. E. Oliphant, Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.G. Paxinos, Atlas of the Developing Mouse Brain at E17.5, P0 and P6 (Elsevier, Amsterdam; Boston, ed. 1st, 2007), pp. xi, 353 p. [Google Scholar]
- 62.M. F. Rose, J. Ren, K. A. Ahmad, H.-T. Chao, T. J. Klisch, A. Flora, J. J. Greer, H. Y. Zoghbi, Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron 64, 341–354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1
Legends for data S1 to S3
References
Data S1 to S3