Abstract
Summary
Biological data repositories are an invaluable source of publicly available research evidence. Unfortunately, the lack of convergence of the scientific community on a common metadata annotation strategy has resulted in large amounts of data with low FAIRness (Findable, Accessible, Interoperable and Reusable). The possibility of generating high-quality insights from their integration relies on data curation, which is typically an error-prone process while also being expensive in terms of time and human labour. Here, we present ESPERANTO, an innovative framework that enables a standardized semi-supervised harmonization and integration of toxicogenomics metadata and increases their FAIRness in a Good Laboratory Practice-compliant fashion. The harmonization across metadata is guaranteed with the definition of an ad hoc vocabulary. The tool interface is designed to support the user in metadata harmonization in a user-friendly manner, regardless of the background and the type of expertise.
Availability and implementation
ESPERANTO and its user manual are freely available for academic purposes at https://github.com/fhaive/esperanto. The input and the results showcased in Supplementary File S1 are available at the same link.
1 Introduction
Public repositories store increasing volumes of toxicogenomics (TGx) data, representing a powerful and heterogenous source of prior knowledge, valuable for chemical risk assessment. In the past, however, data were generated without thinking about its potential long-term usage, resulting in poor standardization and low-quality metadata. FAIR principles guide researchers to generate reusable data, aiding science efforts to shift from traditional animal testing to alternative approaches (Saarimäki et al. 2021). Despite the well-defined standards for omics data representation, a large proportion of such data remains non-compliant with the FAIR principles (Wilkinson et al. 2016), limiting their integration and exploitation (Saarimäki et al. 2022). Rigorous data curation is needed to meet this challenge, harmonizing poorly standardized data to allow their integration and turn them into valuable high-quality insights (Odell et al. 2017). Several solutions have been proposed to automate the process through text mining and AI techniques (Müller et al. 2018, Arnaboldi et al. 2020, Courtot et al. 2022), but despite the claimed accuracy, a final validation by an experienced curator is still needed. Here, we propose ESPERANTO, a R/Shiny (Chang et al. 2022) application performing a streamlined semi-supervised curation of TGx metadata in compliance with Good Laboratory Practice (GLP) standards. The user is actively involved in data harmonization in a consistent framework and in the enhancement of data FAIRness, merging the advantages of both automated and manual curation approaches. Regardless of the expertise, the interactive graphical interface guides the user intuitively through the whole data curation and integration pipeline and requires almost no informatics knowledge. ESPERANTO fosters GLP principles (OECD 2021), by supplying formal guidance to reconstruct the whole pipeline behind the curation of reliable, reproducible, and high-quality data.
Although designed to run on TGx metadata, the tool can curate any health data science datasets, provided a different reference vocabulary.
2 Comparison with other curation tools
To the best of our knowledge, ESPERANTO is the first free available tool to allow dataset standardization and harmonization. Its closest commercial competitor, Genestack ODM (Genestack ODM | Genestack, n.d.), accepts customized vocabularies and it is pre-loaded with several validated ontologies for dataset curation, but it is not designed to adhere to GLP principles.
On the other hand, other solutions have been developed to automate data harmonization through AI techniques, but they are designed to solve slightly different tasks. In fact, most of tools focus on parsing biomedical literature by applying natural language processing techniques to automatically identify entities (i.e. signalling pathways, proteins) of interest. The results are submitted to the user for validation only through the end of the curation. Instead, with a standardized pipeline and reference vocabulary, ESPERANTO offers the user flexible and punctual curation throughout the whole pipeline. Furthermore, our method is applicable for harmonization and standardization of datasets, even in case no literature is available. A detailed comparison among the different curation tools is shown in Table 1.
Table 1.
Comparison between ESPERANTO and other existing data curation tools.
| Tool | Free license | Customizable vocabulary | Graphical user interface | Unrestricted to literature | GLP compliance | Reference |
|---|---|---|---|---|---|---|
| ESPERANTO |

|

|

|

|

|
|
| Genestack ODM |

|

|

|

|

|
Genestack ODM | Genestack (2023 ) |
| Textpresso Central |

|

|

|

|

|
Müller et al. (2018) |
| WormBase AFP |

|

|

|

|

|
Arnaboldi et al. (2020) |
| BioSamples |

|

|

|

|

|
Courtot et al. (2022) |
3 Methods and features
ESPERANTO ensures a streamlined and standardized TGx metadata harmonization and the construction of a customized vocabulary. Both metadata and vocabulary are essential inputs, or optionally, a previously saved curation session can be restored. An empty vocabulary is provided, but a pre-existing ontology can also be utilized as long as it is structured as described in Section 3.2. During the data curation, the software assists the decision-making of the user on each modification before implementing it on the data and recording the operation automatically. In GLP mode, the researcher is required to integrate the taken action with a justification, resulting in a meticulous GLP-compliant report. The purpose of the user guide available online at https://github.com/fhaive/esperanto is not only to instruct and support the user but also to advise on action protocols in relation to specific scenarios.
3.1 Curation and integration
Through word(s) matching, ESPERANTO harmonizes uncurated TGx metadata through the cross-comparison with a precompiled reference vocabulary. The tool, while originally intended for TGx metadata, has the capability to curate health data science datasets of any type as long as a distinct reference vocabulary is provided. Essentially, curation takes place in three stages: (i) consistent renaming of table columns; (ii) deletion of column duplicates; and (iii) potential editing of any remaining content. Any cross-comparison result is shown to be validated, or as an alternative, the user can edit all unique instances, storing the processed terms in a temporary vocabulary. Once multiple datasets have been curated, the tool assesses their integration, highlighting potential inconsistencies among them and/or the vocabulary through a traffic light colour system. The colour code supports the user to evaluate and mark discrepancies as ‘Issue’ to refine in a successive curation round.
The effectiveness of the pipeline in metadata harmonization and integration (Fig. 1) is showcased on TGx Idiopathic Pulmonary Fibrosis datasets publicly available on Gene Expression Omnibus repository (Supplementary File S1). ESPERANTO is a powerful resource to improve FAIRness of metadata upstream of omics data preprocessing and analysis pipelines that can be easily interoperable with the tools in the Nextcast suite (Serra et al. 2022) (more details in Supplementary File S2). The entire design of the application was aimed to make any prerequisite knowledge unnecessary to the user for running the tool.
Figure 1.
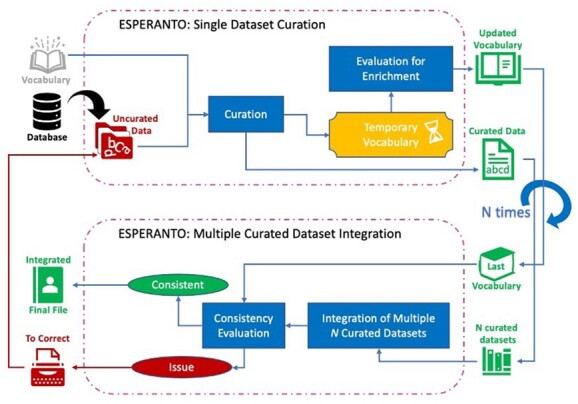
Pipeline of ESPERANTO in single and multiple dataset modes. Once the datasets of interest have been curated, they are used as input for the multiple mode. Red boxes indicate datasets or entries that need curation; dark blue represents the different processes inside ESPERANTO leading to a consistent curated dataset/entry (green). Once integration is completed and evaluated, the final output can be used for further analysis.
3.2 Vocabulary
The vocabulary is characterized by two linked pairs of key: synonym(s) dictionary type objects (Lai 2022). Each feature in metadata ties to all potential instances of that feature; both feature and instances are associated to their synonyms (i.e. ‘ethnicity’, ‘race’, ‘Black, White, Inuit’, ‘“B, African-American”, “Caucasian, C”’). Each round of curation offers the possibility to customize the vocabulary with new features and instances. Their evaluation follows the colour-coded mechanism established in the integration of multiple datasets.
3.3 GLP compliance
Being the curation influenced by the curator, it cannot be considered fully GLP compliant. With its standardized pipeline and its reference vocabulary, ESPERANTO minimizes the impact of subjectivity, at the same time, leaving the user active control over the task. Each action performed during the curation is automatically recorded (in the GLP mode together with its mandatory justification), resulting in a detailed report describing the entire process. The user cannot turn low-quality metadata into top-notch documentation; nevertheless, its FAIRness is improved by recording any data modification during curation. Reproducibility of data harmonization is ensured by following step by step the track described in the GLP-compliant report of ESPERANTO.
4 Conclusion
ESPERANTO supports metadata harmonization and integration in a flexible and user-friendly fashion, allowing tailored analytical procedures and detailed GLP-compliant reporting regardless of the type and level of expertise of the user. Supplementary File S1 shows the case study evaluation of publicly available TGx metadata.
Supplementary Material
Contributor Information
Emanuele Di Lieto, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland.
Angela Serra, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland; Tampere Institute for Advanced Study, Tampere, 33520, Finland.
Simo Iisakki Inkala, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland.
Laura Aliisa Saarimäki, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland.
Giusy del Giudice, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland.
Michele Fratello, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland.
Veera Hautanen, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland.
Maria Annala, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland.
Antonio Federico, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland; Tampere Institute for Advanced Study, Tampere, 33520, Finland.
Dario Greco, FHAIVE, Faculty of Medicine and Health Technology, Tampere University, Tampere 33520, Finland; Institute of Biotechnology, Helsinki Institute of Life Sciences (HiLife), University of Helsinki, Helsinki 00790, Finland; Division of Pharmaceutical Biosciences, Faculty of Pharmacy, University of Helsinki, Helsinki 00790, Finland.
Supplementary data
Supplementary data are available at Bioinformatics online.
Funding
This work was supported by the European Union Horizon 2020 Programme, NanoinformaTIX project (grant agreement no. 814426), the Academy of Finland (grant agreement no. 322761), and the European Research Council (ERC) Programme, Consolidator project ARCHIMEDES (grant agreement no. 101043848). A.S. and A.F. were supported by the Tampere Institute for Advanced Study.
Conflict of interest
The authors declare no conflicts of interest.
Data availability
The source code of ESPERANTO is publicly available at the following GitHub repository: https://github.com/fhaive/esperanto.
References
- Arnaboldi V, Raciti D, Van Auken K. et al. Text mining meets community curation: a newly designed curation platform to improve author experience and participation at WormBase. Database 2020;2020:baaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Cheng J, Allaire JJ. et al. Shiny: Web Application Framework for R, 2022. https://rstudio.github.io/shiny/authors.html.
- Courtot M, Gupta D, Liyanage I. et al. BioSamples database: FAIRer samples metadata to accelerate research data management. Nucleic Acids Res 2022;50:D1500–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genestack ODM | Genestack. Software Solutions for Life Science Data Management and Integration, 2023.
- Lai R. Collections: High Performance Container Data Types, 2022. https://randy3k.github.io/collections/authors.html#citation.
- Müller H-M, Van Auken KM, Li Y. et al. Textpresso Central: a customizable platform for searching, text mining, viewing, and curating biomedical literature. BMC Bioinformatics 2018;19:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell SG, Lazo GR, Woodhouse MR. et al. The art of curation at a biological database: principles and application. Current Plant Biology 2017;11-12:2–11. [Google Scholar]
- OECD. Number 22: Advisory Document of the Working Party on Good Laboratory Practice on GLP Data Integrity. Paris, France: Organization of Economic Cooperation and Development, 2021. https://www.oecd.org/chemicalsafety/testing/oecdseriesonprinciplesofgoodlaboratorypracticeglpandcompliancemonitoring.htmhttps://one.oecd.org/document/env/cbc/mono(2021)26/en/pdf. [Google Scholar]
- Saarimäki LA, Federico A, Lynch I. et al. Manually curated transcriptomics data collection for toxicogenomic assessment of engineered nanomaterials. Sci Data 2021;8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarimäki LA, Melagraki G, Afantitis A. et al. Prospects and challenges for FAIR toxicogenomics data. Nat Nanotechnol 2022;17:17–8. [DOI] [PubMed] [Google Scholar]
- Serra A, Saarimäki LA, Pavel A. et al. Nextcast: a software suite to analyse and model toxicogenomics data. Comput Struct Biotechnol J 2022;20:1413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg I. et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source code of ESPERANTO is publicly available at the following GitHub repository: https://github.com/fhaive/esperanto.


