Abstract
Modern neuroscience approaches including optogenetics, calcium imaging, and other genetic manipulations have facilitated our ability to dissect specific circuits in rodent models to study their role in neurological disease. These approaches regularly use viral vectors to deliver genetic cargo (e.g., opsins) to specific tissues and genetically-engineered rodents to achieve cell-type specificity. However, the translatability of these rodent models, cross-species validation of identified targets, and translational efficacy of potential therapeutics in larger animal models like nonhuman primates remains difficult due to the lack of efficient primate viral vectors. A refined understanding of the nonhuman primate nervous system promises to deliver insights that can guide the development of treatments for neurological and neurodegenerative diseases. Here, we outline recent advances in the development of adeno-associated viral vectors for optimized use in nonhuman primates. These tools promise to help open new avenues for study in translational neuroscience and further our understanding of the primate brain.
Keywords: Nonhuman primate, Adeno-associated viral vectors, Capsid variants, Enhancers, Brain, Gene delivery
Graphical abstract
Highlights
-
•
Nonhuman primates are a valuable model for study of human disease.
-
•
Technical hurdles have impeded translation of gene delivery from mice to primates.
-
•
Capsid engineering allows for systemic delivery to primate nervous system.
-
•
Identification of gene regulatory elements may further enable cell-type specificity.
1. Nonhuman primate as a valuable model for study of human diseases
A major goal of modern neuroscience is to inform our understanding of the human condition and brain-based disorders. However, this requires better comprehension of the distributed anatomical and molecularly diverse pathways, and functional circuits underlying disease. Recent advances in genetic technologies have made it possible to control and image neuronal circuits in living animals, through the delivery of various effectors, sensors, and reporters to the brain (Boyden et al., 2005; Fenno et al., 2011; Wang et al., 2019; Yang and Yuste, 2017). This breakthrough in technology has advanced our understanding of neural circuits, cell-types, molecules, neurotransmitters, and gene regulatory elements that work together to contribute to the progression of disease (e.g.(Coley et al., 2021; Cummings and Clem, 2020; Fadok et al., 2018; Pignatelli and Beyeler, 2019; Xu et al., 2019)). For example, research on anxiety-relevant circuits has leveraged optical control of specific cell-types (e.g. somatostatin and corticotrophin-releasing hormone expressing cells) and their projections to threat-relevant regions (e.g. central amygdala to periaqueductal gray interneurons) in order to elucidate multiple distinct mechanisms that underlie specific aspects of threat responding behavior (Ciocchi et al., 2010; Fadok et al., 2017; Holley and Fox, 2022; Tovote et al., 2016). This work has far-reaching implications for our understanding of anxiety disorders, by identifying multiple distinct mechanisms that likely contribute to differences in symptomatology. Similarly, in basic research studies of the mechanisms relevant to neurodegenerative diseases like Parkinson's, optical inhibition of cells in the subthalamic nucleus of parkinsonian rodents was sufficient to improve 6-hydroxydopamine-induced forelimb akinesia, opening the door to potential treatment avenues for patients with PD (Yoon et al., 2014). Unfortunately, these advances have not led to a large number of new and improved treatments for neurological diseases. This gap between rodent models of disease and translational outcomes is due, in part, to difficulty in validating the relevancy of these potential targets in human disease, as well as in understanding how these potential therapeutic compounds interact with their molecular targets in primates. Differences in evolutionary pressures have contributed to differences in brain structure and function across species, including the expansion of cortical regions during human evolution (Ahmed, 2018; Baker et al., 2020; Ménard et al., 2016; Pine et al., 2021; Preuss and Wise, 2022), potentially contributing to poor predictability of rodent disease models to the human condition. Nonhuman primates (NHPs) are well-suited to bridge this gap as their recent evolutionary divergence from a common ancestor have endowed them with many anatomical, physiological, and ethological similarities to humans (Kalin and Shelton, 2003; Öngür and Price, 2000; Petrides and Pandya, 1999, 2002; Phillips et al., 2014). This makes them well suited for anatomical and functional dissection in both the central nervous system (CNS) and peripheral nervous system (PNS), and as models for interrogation of potential therapeutic targets.
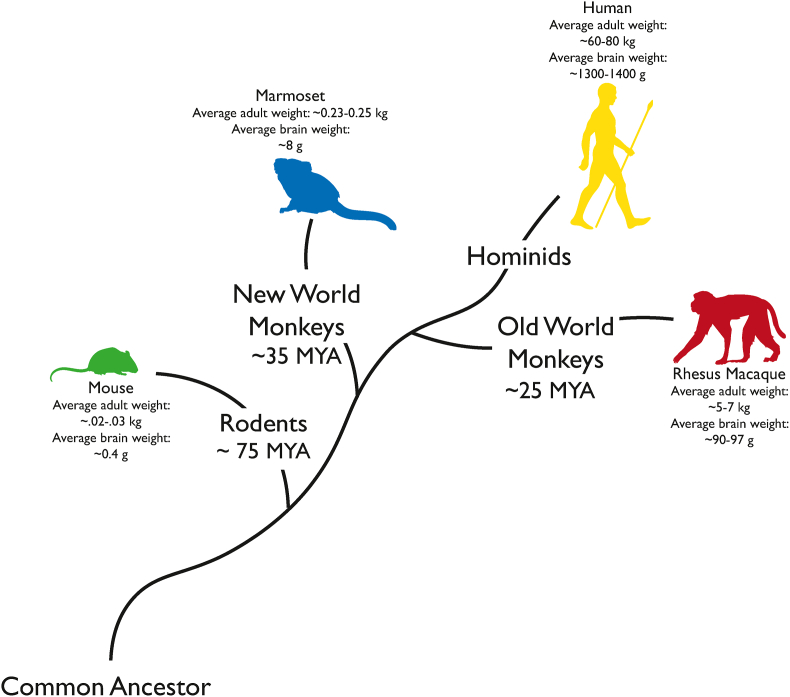
Perhaps the most notable distinction between human and rodent brains is the expansion of neocortex during human evolution (Kaas, 2012; Öngür and Price, 2000; Petrides and Pandya, 1999, 2002; Pine et al., 2021). This expansion is often thought to have contributed to the many high order abilities and social complexities related to human uniqueness (Kaas, 2012; Smaers et al., 2011). Many studies in humans have shed light on the neuronal circuits associated with these abilities, however, due to the limitations of the available tools for the study of the human brain, a more comprehensive understanding of the underlying biology is still needed (Craig, 2009; Cristofori et al., 2019; Gläscher et al., 2012; Horga et al., 2014). To this end, NHPs are of particular importance. To briefly review, NHPs can be roughly broken down into various simian species, which include monkeys and apes, and prosimians, such as lemurs. Monkeys can be further divided into Old World (Catarrhini) and New World (Platyrrhini) monkeys (Welker, 2017). Marmosets (Callithrix jacchus), which diverged from the human lineage approximately 35 million years ago (MYA), and rhesus macaques (Macaca mulatta), which diverged from humans even more recently, approximately ∼25 MYA (Fig. 1), are the two most common NHPs used in research. It is this recent evolutionary divergence from a common ancestor that has made NHPs a valuable model in neuroscience, as they possess a highly elaborated prefrontal cortex, including a well-developed internal granular layer (Bernardi and Salzman, 2019; Öngür and Price, 2000; Petrides and Pandya, 1999, 2002). Because they are our phylogenetic neighbors, NHPs share many behavioral and anatomical features with humans (Kalin and Shelton, 2003; Öngür and Price, 2000; Petrides and Pandya, 1999, 2002; Phillips et al., 2014). For example, the ability to navigate social complexities has been hypothesized to be enabled by the evolutionary expansion of the primate prefrontal cortex (Dunbar and Shultz, 2007; Pine et al., 2021). Indeed, unlike many animals, both NHPs and humans have developed complex social behaviors that have helped them navigate the complexities of living in large social groups (Chang and Platt, 2014). These include prosocial behaviors (Miller et al., 2016), social imitation (Subiaul et al., 2004), and in New World Monkeys like marmosets, monogamy and infant rearing (Miller et al., 2016; Saito, 2015). The shared social repertoires between monkeys and humans have been helpful in studying the underlying biology of social behaviors (Chang and Platt, 2014; Ziegler, 2018). In addition to social behaviors, the phylogenetic proximity of Old World monkeys, like rhesus macaques, to humans provides an avenue to study the primate brain which has a similar structure and cytoarchitecture to the human brain (Öngür and Price, 2000; Petrides and Pandya, 1999, 2002). For this reason, primates can contribute to understanding human-specific cognitive functions like higher-order cognition, attention, and working memory (Brady and Hampton, 2018; Deaner and Platt, 2003; Dezfouli et al., 2021; Rich and Wallis, 2016; Snyder et al., 2021; Xie et al., 2022).
Fig. 1.
Evolutionary tree depicting the phylogentic relationship of common research species. Among these species, Old World Monkeys, like the rhesus macaque, are approximately more similar to humans than New World Monkeys and rodents.
While the phylogenetic proximity of NHPs to humans has made them anatomically and behaviorally similar, it is also likely that throughout evolution, the composition and function of neuronal circuits have adapted based on the evolutionary pressures placed on the species (Katz and Harris-Warrick, 1999). That is to say, while rodents and humans may share basic organization of circuits, changes within these circuits can cause large and important changes in behaviors (Katz and Harris-Warrick, 1999). Indeed, recent studies have found that while there seems to be a strong evolutionary conservation of cell-types between rodents and primates, there are also a number of primate-specific cell-types distributed throughout the brain-in cortical and subcortical structures and in midbrain (Hodge et al., 2019; Kamath et al., 2022; Krienen et al., 2020; Schmitz et al., 2022). For example, von Economo neurons and fork neurons which were originally assumed to exist only in humans and great apes, have been identified in Old World Monkeys (Evrard, 2019). This discovery provides the opportunity to elucidate the function of these neurons, as they have been shown to have projections to regions often implicated in studies of psychological dysfunction in both humans and NHPs (Craig, 2009; Evrard, 2019; Fox et al., 2015). Similarly, primates have a larger diversity of interneurons (Fishell and Kepecs, 2020; Franjic et al., 2022; Krienen et al., 2020; Schmitz et al., 2022), which may impact circuit dynamics in ways that cannot be studied in rodent models. For example, researchers have identified a primate-specific striatal interneuron (i.e. GABAergic neurons that express TAC3), which is thought to have emerged through a developmental repurposing of dopaminergic periglomerular cells of the olfactory bulb (Krienen et al., 2020; Schmitz et al., 2022). Additionally, a novel excitatory cell type expressing neuropeptide Y (NPY) was recently discovered in the primate visual cortex (Wei et al., 2022). As we aim to further dissect and elucidate the distributed brain-body circuits related to human pathology, the ability to target and manipulate specific cells will become increasingly important as they may be key determinants in the emergence of brain based disorders in humans.
In brief, rodent models have been useful in understanding the molecular and cellular basis of diseases, due in large part to advances in molecular genetic technologies (Boyden et al., 2005; Fenno et al., 2011; Yang and Yuste, 2017). However, rodents and humans differ in physiology, anatomy, and social complexity. To this end, NHPs are of particular importance because of the recent evolutionary divergence between humans and primates that have made them similar to humans both biologically and socially. However, a critical component of translation includes the translation of the tools that have been commonplace in rodent neuroscience. In this review, we highlight the need for more efficient neurotropic AAVs that can be delivered systemically in NHPs, recently engineered capsid variants that can cross the blood brain barrier in NHPs, and advances made to target specific cell-types.
2. AAVs enable gene delivery and circuit interrogation
The ability to define, monitor, and manipulate a neural circuit requires precise delivery of reporters, sensors, and effectors to the individual circuit components (e.g. cell-types). Viral vectors such as herpes simplex virus (HSV), rabies, adenovirus, lentivirus (LV), and adeno-associated viruses (AAVs) have emerged as an effective tool for neuroscience in that they enable neuronal tracing and functional interrogation through the delivery of various transgenes (Davidson and Breakefield, 2003; Ghosh et al., 2020; Hui et al., 2022; Kristensson et al., 1982; Liu et al., 2022). Viral vectors are composed of: i) a capsid, an outer protein shell enclosing the genetic material and which determines the vector's tropism, or ability to infect different cell-types; ii) regulatory elements such as enhancers or promoters which restrict expression to specific cell or tissue types; and iii) a transgene (Bulcha et al., 2021). Transgenes include fluorescent proteins as genetic reporters for visualization, sensors for measuring neurotransmitter release (e.g., GCaMP, DLight, etc.), opsins and synthetic receptors for cellular manipulation (e.g., ChR2, DREADDs), and repair templates for CRISPR-Cas9 based gene editing and expression manipulation (e.g., using CRISPRa/i) (Boyden et al., 2005; Klapoetke et al., 2014; X. Li et al., 2005; Magnus et al., 2011; Patriarchi et al., 2018; Roth, 2016; Yim et al., 2020; F. Zhang et al., 2007). Such genetic tools have advanced our understanding of how various cell-types and specific circuits contribute to adaptive behaviors and emergent properties of the brain. Among the viral vectors, AAVs are considered to be the safest since they are non-pathogenic, and are naturally replication deficient i.e. they lack the genes necessary for replication and replicate only when co-infected with a helper virus (Buller et al., 1981; Rose and Koczot, 1972). In contrast, lentiviruses transduce cells with higher efficiency than AAVs but there is uncertainty surrounding their safety due to the possibility of random insertional mutagenesis (Zheng et al., 2018). This can affect the genetic code at the DNA insertion site, leading to adverse outcomes, including cancer (Zheng et al., 2018). Similarly, herpes viral vectors can cause strong inflammatory responses (Ghosh et al., 2020). These non-specific and adverse effects have precluded them from widespread use in NHPs (Tremblay et al., 2020). Additionally, AAVs are capable of transducing both dividing and post-mitotic cells such as neurons, and so are ideally suited for gene manipulation studies that require stable, long term transgene expression in cells that have already matured (Bartlett et al., 1998). For these reasons, recombinant AAVs (rAAVs) have become the viral vector of choice for in vivo gene therapy applications, with more than 285 registered clinical trials to date (Kuzmin et al., 2021; U.S. National Library of Medicine (www.clinicaltrials.gov)).
For the study and treatment of neurological and neurodegenerative diseases, widespread distribution of transgene expression could be transformative (Hadaczek et al., 2016; Muramatsu, 2010; Sun and Roy, 2021). For example, idiopathic Parkinson's disease is hypothesized to result from the aggregation of a protein called alpha-synuclein (α-Syn) first in the enteric nervous system, before it propagates up the vagus nerve to the basal forebrain, midbrain and ultimately the cerebral cortex (Braak et al., 2003). rAAVs can be used to deliver a pathogenic protein such as α-Syn to model PD in animals, and help tease apart the cell-types in the ENS and CNS that are susceptible to α-Syn pathology (Alam et al., 2022; Challis et al., 2020; Huntington and Srinivasan, 2021; Kirik and Björklund, 2003; Ulusoy et al., 2010). In such cases, widespread transgene expression is required. Conversely, the pathogenic protein can be silenced, or the disease phenotype may be reversed by delivering a therapeutic gene such as GBA1, which encodes the lysosomal enzyme Glucerebrosidase, and has been shown to reduce inflammation and aggregation of α-Syn in models of PD as well as Gaucher's disease-a lysosomal neurodegenerative disorder (Abeliovich et al., 2021; Sardi et al., 2011, 2013). The route of AAV administration, dose, age at the time of injection, and preexisting neutralizing antibodies against it in the host, are all key determinants of an AAV's safety, efficacy and tropism (Chan and Deverman, 2022). There are many advantages to direct in-brain injection. In fact, most studies to date have directly injected viruses into the brain to deliver genetic cargo to specific regions. This has been performed in animal models of PD to target the putamen or substantia nigra (Bartus et al., 2013; Christine et al., 2009; Kaplitt et al., 2007; Kells et al., 2012; Muramatsu, 2010). Despite its many advantages, which include relatively dense and robust expression surrounding the infusion site, targeting large, diffuse, or spatially distributed regions can require multiple injections. Thus, this route of administration is most suitable for localized targets (Wang, 2021). Covering entire brain regions remains a challenge due to the size of the primate brain and often requires multiple craniotomies. Systemic delivery of AAVs obviates the need for multiple direct injections and importantly reduces the health risks associated with extremely long and highly invasive surgeries. As a therapeutic approach, systemic administration via a single injection might be a safer alternative toward achieving brain wide gene transduction (Bourdenx et al., 2014; Kimura and Harashima, 2022). Ultimately, increased efficacy of BBB-crossing AAVs may be combined with other technologies to achieve localized expression. However, crossing the BBB, which acts as a gatekeeper by preventing toxins and pathogens in the systemic circulation from entering the CNS, is a major hurdle for efficient gene delivery.
To date, 13 distinct naturally-occurring or wildtype serotypes of AAVs, (AAV1-13) have been identified in humans and NHPs (Srivastava, 2016). Each of these serotypes differs in capsid structure and, therefore, tropism (Agbandje-McKenna and Kleinschmidt, 2012). The most commonly used serotypes in rodent research are AAV2, AAV5, AAV8, and AAV9, which transduce the CNS, although some transduce other organs as well (Aschauer et al., 2013). In NHPs, the most commonly used serotypes are AAV5 and AAV9 (Tremblay et al., 2020). AAV9 has been particularly widely studied because of its ability to cross the BBB and has been employed in several CNS-targeted gene therapies (Chan and Deverman, 2022; Chen et al., 2021; Foust et al., 2009; Song et al., 2022; Yang et al., 2014; Zhang et al., 2011). Efforts have also been made to characterize other serotypes that also have the capability to cross the BBB. For example, Gao et al. cloned and identified more than 100 novel rAAVs from human and NHP tissues (Gao et al., 2005; Gao et al., 2002). Among these, AAVrh8, AAVrh10 and AAVhu32 were found to cross the BBB with high efficiencies, similar to AAV9.
However, these AAVs, including AAV9, have limitations that have prevented their wider use. For instance, their cell-type tropism can vary across species. In neonatal mice and macaques, intravenously administered AAV9 transduces neurons preferentially, whereas in juvenile and adult mice and macaques, the tropism shifts toward astrocytes (Bevan et al., 2011; Dehay et al., 2012; Foust et al., 2009; Gray et al., 2011; Mattar et al., 2013; Samaranch et al., 2012). Moreover, AAV9 and the other BBB crossing serotypes mentioned above have a higher tropism for peripheral organs such as the liver than the brain (Gray et al., 2011; Zincarelli et al., 2008). This is especially concerning in large animals such as NHPs as they require large volumes of virus for systemic delivery and the high doses of AAV needed to achieve clinical relevance can lead to hepatotoxicity or sensory neuron toxicity (Hinderer et al., 2018). Additionally, humans as well as NHPs harbor neutralizing anti-AAV antibodies to certain wildtype AAV serotypes from pre-existing exposure or develop anti-AAV antibodies after therapeutic rAAV administration (Louis Jeune et al., 2013). This is a limiting factor for gene therapy applications where subsequent viral administration may be needed if the transgene expression wanes over time. Another important consideration is the ∼4.7 kb size limitation of the AAV vector genome, which is comparatively smaller than that of lentiviruses (∼9.7 kb) or herpes simplex virus (∼150 kb) (Kumar et al., 2001; Latchman, 2001; Sena-Esteves et al., 2000).
When designing a study, it is important to take these considerations into account. In studies of the primate brain, it is important to ensure that the target gene sequence is reliably expressed, to minimize off-target effects, and to ensure animal safety. This has led most studies to prefer AAVs. However, if the genetic cargo is larger than optimal for an AAV genome, researchers run the risk of lower transduction efficiency, affecting their ability to perform the desired manipulation (Wu et al., 2010). These cost-benefit calculations are study-specific and constantly changing. Development of cell-type specificity, as a function of the viral capsid or shortened enhancer and promoter cargo, as described below could mitigate off-target effect. In the following sections, we will discuss current efforts to develop novel systemic rAAV vectors with high transduction efficiency and optimized biodistribution, with minimal off-target delivery, and low immunogenicity for gene delivery to the NHP CNS (Challis et al., 2022; Chan and Deverman, 2022; Davidson et al., 2022).
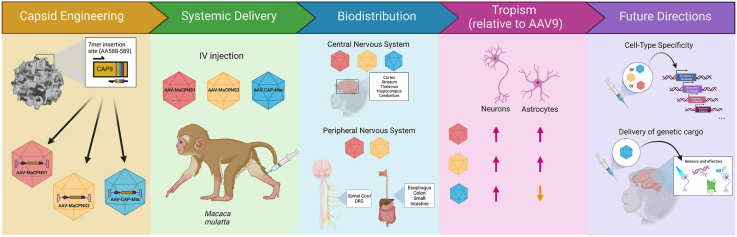
3. Capsid engineering for systemic delivery to achieve tissue specific biodistribution in NHPs
The capsid of an AAV is its primary point of interaction with receptors on the host cell surface which enable the virus to be internalized, and ultimately deliver their genetic cargo to the cell nucleus (Challis et al., 2022; Li and Samulski, 2020). Because of this, the capsid structure of AAVs have been widely researched in order to determine the protein domains responsible for cellular receptor binding, and consequently the virus' tropism and efficacy (Challis et al., 2022; Lee et al., 2018; Li and Samulski, 2020). Capsid modification or engineering is one route toward altering an AAV's tropism and efficacy as several permissive sites for rational and random amino acid substitutions and insertion have been identified (Challis et al., 2022). Through capsid engineering, we can enhance and refine AAV tropisms, as well as identify novel AAV serotypes with improved BBB crossing properties.
Capsid engineering can be carried out either through rational design or directed evolution. Rational design capitalizes on the knowledge of existing AAV serotypes to systematically predict and refine virus function (Lee et al., 2018). On the other hand, directed evolution is a high-throughput approach that involves using a selection process to generate variants with the desired properties such as antibody neutralization and/or tissue and cell-type tropism. By iteratively creating many non-naturally occurring viral serotypes and selecting those with the desired tropism for the next round of evaluation, researchers have been able to discover novel capsid variants that have a higher transduction efficiency at lower titers or concentration (Chan et al., 2017; Deverman et al., 2016; Körbelin et al., 2016; Kumar et al., 2020; Nonnenmacher et al., 2021). For example, our group has developed the Cre recombination-based AAV targeted evolution (CREATE) selection method (Deverman et al., 2016). In brief, the CREATE method enables the recovery of capsid sequences that transduce Cre-expressing cell populations in transgenic mice. Wherever Cre is present, a library fragment adjacent to the cap gene is inverted. PCR-based amplification can then detect the sequences that have successfully transduced the target population. After multiple rounds of evolution, top performing capsids can then be selected and characterized. This method led to the identification of AAV-PHP.B and AAV-PHP.eB, which target the mouse CNS (Chan et al., 2017; Deverman et al., 2016). Both AAV-PHP.B and AAV-PHP.eB are derived from AAV9, with AAV-PHP.eB showing >50% transduction of cells across brain regions of C57BL/6J mice (Chan et al., 2017). However, the ability of these engineered variants to transduce cells in the CNS of other mouse strains or NHPs is dependent on the administration route and whether it can cross the BBB (Mathiesen et al., 2020). Indeed, IV administration of AAV-PHP.B in marmosets showed poor transduction of neurons and astrocytes similar to AAV9 (Matsuzaki et al., 2018). Furthermore, Hordeaux et al. reported that a higher dose (7.9E13 gc/kg) of the AAV-PHP.B vector resulted in acute toxicity in an IV-injected macaque (Hordeaux et al., 2018). Subsequent studies revealed that the enhanced CNS tropism of AAV-PHP.B and AAV-PHP.eB in the C57BL/6J mice might be due to their interaction with the LY6A receptor, which is a GPI-anchored protein that is highly expressed by brain microvascular endothelial cells (Hordeaux et al., 2019; Mathiesen et al., 2020). Interestingly, there is no LY6A homolog in primates thus limiting the utility of these viruses in primates, and further highlighting the need for molecular target identification and validation studies in NHPs as a critical aspect of early research. We have begun to extend this directed evolution strategy to develop novel AAVs that can be used to target primate cells. Multiplexed-CREATE (M-CREATE) implements internal controls to reduce sequencing bias and increase the number of variants identified with enhanced CNS tropism (Kumar et al., 2020). This approach can be iterated across species, testing thousands of candidates in mice to identify top-performing capsids for primate testing. Recently, our lab used M-CREATE to identify systemic variants with enhanced CNS and PNS transduction in both Old World and New World monkeys. In marmosets, we found two variants evolved from AAV-PHP.eB, AAV-CAP-B10 and AAV-CAP-B22, to have enhanced CNS transduction after IV delivery in adult marmosets (Goertsen et al., 2022). AAV-CAP-B10 and AAV-CAP-B22 displayed four and two-fold increased neuronal transduction over AAV9, respectively. Furthermore, AAV-CAP-B10 showed decreased expression in the liver, where expression is typically associated with toxicity, as compared to AAV9. Importantly, broad and robust transgene expression was seen across cortical, subcortical, and cerebellar regions as well as in the dorsal root ganglia (DRG) and the spinal cord (SC).
More recently, we have developed AAV-MaCPNS1 and AAV-MaCPNS2 (Chen et al., 2022). Although these AAVs were designed to target the PNS in rodents, we found that they transduce PNS and CNS in both marmosets and rhesus macaques. Specifically, in adult marmosets, IV delivery of AAV-MaCPNS1/2 capsids carrying fluorescent reporter proteins (i.e., ssAAV:CAG-eGFP or ssAAV:CAG-tdTomato) targeted PNS and CNS more efficiently than AAV9. In the PNS, enhanced transduction was observed in DRG, the small intestine (SI), and the ascending fiber tracts in the dorsal column of the spinal cord (SC). Surprisingly, in the CNS, diffuse brain-wide transduction was seen in regions including the cortex, thalamus, globus pallidus, cerebellum, and brainstem. This tropism was recapitulated in infant rhesus macaques using IV delivery of AAV-MaCPNS1/2. In the PNS, enhanced transduction was seen in the SC, DRG, and gastrointestinal (GI) tract, including the esophagus, colon, and SI. Similar to what was observed in marmosets, in the rhesus monkey CNS, AAV-MaCPNS1/2 capsids mediated enhanced brain-wide transduction, including areas of the cortex, hippocampus, putamen, and brainstem. Additionally, AAV-MaCPNS1/2 displayed an increase in astrocyte transduction over AAV9 in the cortex and thalamus. However, of the two, MaCPNS2 had the greater fold increase in astrocytes. These results demonstrate the potential of these capsids in interrogating peripheral to brain circuitry.
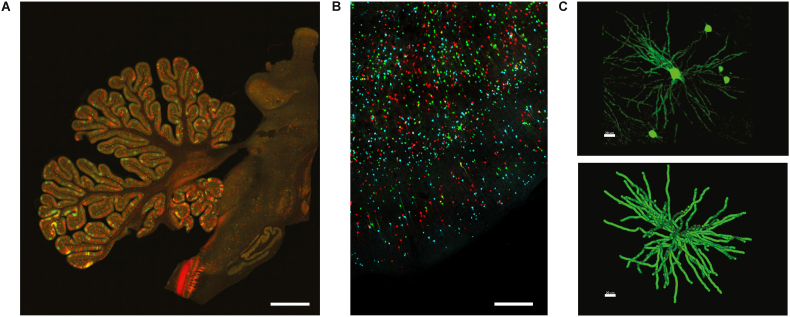
In addition to M-CREATE, a multi species screening and characterization strategy was used to identify another capsid variant, AAV-CAP-Mac, for systemic brain-wide delivery in Old World Monkeys, including rhesus macaques and green monkeys (Chlorocebus sabaeus) (Chuapoco et al., in press). To characterize the transduction properties of AAV-CAP-Mac, pools of AAV-capsids, including AAV9, AAV-PHP.eB, AAV.CAP-B10 and other previously engineered AAVs, were simultaneously injected intravenously into infant rhesus macaques. Each capsid variant contained a single stranded human FXN (frataxin) transgene fused to a hemagglutinin (HA) epitope tag under control of a CAG promoter (ssCAG-hFXN-HA). AAV-CAP-Mac was found to primarily transduce primate neurons in all lobes of the cortex, cerebellum, and multiple subcortical regions. IV injection of AAV-CAP-Mac outperformed its parent capsid AAV9 as well as other previously engineered AAVs, including AAV-PHP.eB. Interestingly, AAV-CAP-Mac also outperformed AAV-CAP-B10, suggesting genetic differences in the BBB even between Old World and New World monkeys. AAV-CAP-Mac, was also shown to have reduced tropism towards the liver across primate species. Further validating the utility of AAV-CAP-Mac for NHP research, delivering a mixture of fluorescent proteins achieved a sparse Golgi stain-like expression pattern that could facilitate large-scale studies of neuronal morphology. Again, in this experiment, AAV-CAP-Mac was seen to target neurons in the cortex and multiple subcortical regions including hippocampus, putamen, thalamus, and caudate, enabling reconstruction of both medium spiny neurons and cortical pyramidal cells (Fig. 2). Additional characterization was also performed in 8-month-old green monkeys. Injections of AAV-CAP-Mac (packaging ssCAG-eGFP) via IV delivery showed broad and strong neuronal expression in all lobes of the cortex and in various subcortical regions. Consistent with results from the rhesus experiments, AAV-CAP-Mac transduced a higher percentage of neurons than AAV9 in sampled cortex and subcortical regions. In summary, these novel AAV variants with tailored properties and tropism, offer greater CNS and/or PNS transduction than AAV9 in NHP.
Fig. 2.
(A) Representative image of MaCPNS1-mediated expression of eGFP and MaCPNS2-mediated expression of tdTomato in the cerebellum and brainstem of an infant rhesus macaque. (B) Multicolor labeling of neurons in cortex after IV administration of a cocktail of 3 fluorescent proteins (ssCAG-mNeonGreen, ssCAG-mRuby2, and ssCAG-mTurquoise2) packaged in AAV-CAP-Mac and (C) morphological reconstruction of a striatal medium spiny neuron in rhesus macaque (scale, 20 uM). Scale bars for A and B = 100 uM.
At present, in vivo characterization of AAV-CAP-Mac and AAV-MaCPNS1/2 has been limited to infant rhesus macaques (Chuapoco et al., in press). Higher vector doses could lead to increased immunogenicity due to preexisting neutralizing antibodies, thus, further characterization of these capsids in adult animals is necessary and currently underway. Encouragingly, though, these variants show significantly lower transduction of the liver compared to AAV9, potentially minimizing the risk of hepatotoxicity resulting from high systemic doses. Additionally, while these variants were engineered for systemic delivery, they may also be effective for local delivery. In vitro data from human-derived iPSCs supports the increased effectiveness of these AAVs over AAV9 (Chuapoco et al., in press), but further work is needed to test this, as well as the functional efficacy of the genetic cargo delivered by these novel AAVs.
In addition to our efforts, several other groups have also engineered AAVs with unique features that are relevant to NHPs. Using an in-vivo directed evolution strategy, Dalkara et al. engineered a novel AAV variant, 7m8, that specifically transduces photoreceptors in the NHP retina via intravitreal injections (Dalkara et al., 2013). Through iterative refinement, researchers have developed newly engineered variants that induce higher expression in foveal cones than 7m8 in cynomolgus macaques (Byrne et al., 2020). The in-vivo strategy DELIVER (directed evolution of AAV capsids leveraging in vivo expression of transgene RNA), has been used to identify muscle-tropic capsids that can be systemically administered to transduce primate muscles with high efficiency in cynomolgus macaques (Tabebordbar et al., 2021). Similarly, rAAV2-retro, which was developed for delivery of genetic cargo to retrogradely targeted cells, has also been tested for use in the primate brain (Bohlen et al., 2020) though see: (Cushnie et al., 2020)).
4. AAV-based approaches for targeting specific cell-types in NHPs
Developing tools to target specific cell-types to study their role in normal and disease circuitry remains a major challenge for primate neuroscience. Rodent models often rely on genetically-engineered Cre lines to achieve cell-type specificity. Unfortunately, primate gestational and maturational timelines preclude the widespread use of these genetic engineering approaches in primates (though see: (Drummer et al., 2021; Park et al., 2016; Sasaki et al., 2009; Tomioka et al., 2017)) so a different approach is needed. It is unlikely that capsid engineering alone will achieve the level of cell-type specificity required for NHP neuroscience research. This is due in large part because profiling AAV capsid variants generated by selection experiments is time-consuming and labor-intensive, and most remain uncharacterized (Zolotukhin and Vandenberghe, 2022). Although new molecular and computational tools, such as machine learning, might facilitate capsid profiling, these approaches also have limitations (Zolotukhin and Vandenberghe, 2022). Additionally, studies in NHPs suggest that novel transduction properties may not only arise from unique capsid binding properties, but also from uncharacterized capsid-promoter interactions (Bohlen et al., 2020). Therefore, it is likely that achieving cell-type specificity in NHPs will depend on a combination of BBB-crossing AAV capsid variants and regulatory elements.
Cis-acting regulatory DNA elements, such as promoters and enhancers, are sequences of DNA that proteins bind to in order to initiate and increase the likelihood of transcription respectively (Levine, 2010; Wittkopp and Kalay, 2012). Thus promoters and enhancers can determine the level of transgene expression and the cells they are expressed in. Ubiquitous promoters, such as cytomegalovirus (CMV), chicken β-actin (CBA), human elongation factor 1 alpha (EF1α) or combinations of these such as CMV early enhancer/chicken beta actin (CAG), drive high levels of transgene expression in most cell-types (Haery et al., 2019). However, high, widespread transgene expression is not always desired and can evoke immune responses to the transgene product (Perez et al., 2020; Samelson-Jones et al., 2020). Alternatively, cell-type specific promoters can be incorporated into the AAV cargo. These can be used, for instance, to target neurons (synapsin 1) or astrocytes (glial fibrillary acidic protein), or even more specifically dopaminergic neurons, cerebellar Purkinje cells, or parvalbumin (PVALB) neurons in the brain (El-Shamayleh et al., 2017; Hoshino et al., 2021; Matsuzaki et al., 2014; Nitta et al., 2017; Shinohara et al., 2016; Stauffer et al., 2016). Promoter sizes can range anywhere from ∼100 bp to 1000 bp (Domenger and Grimm, 2019). Due to the AAV's size limitation, ongoing efforts are focused on identifying shorter, and phylogenetically conserved, regulatory element sequences to direct cell-type specific transgene expression across species (de Leeuw et al., 2016; Domenger and Grimm, 2019; Matsuzaki et al., 2014; Nathanson, 2009).
Recently, chromatin profiling techniques coupled with next-generation sequencing led to the discovery of putative enhancers that are less than 600 bp and can drive cell-type specific activation of genes (Buenrostro et al., 2013; Cusanovich et al., 2015; Fang et al., 2021; Grandi et al., 2022; Graybuck et al., 2021; Hrvatin et al., 2019; Mich et al., 2021; Nair et al., 2020; Preissl et al., 2018; Rubin et al., 2020; Visel et al., 2013; Vormstein-Schneider et al., 2020). A distal-less homeobox (Dlx) gene enhancer sequence that targets GABAergic interneurons in the telencephalon of several vertebrate species including mouse and marmoset was identified (Dimidschstein et al., 2016; A. T. Lee et al., 2014; Zerucha et al., 2000). Additionally, the mouse ortholog of the Dlx5/6 enhancer (mDLX5/6), which is only ∼400 bp, packaged into either AAV1 or AAV9, showed similar specificity for GABAergic interneurons when locally injected into area V1 of the primary visual cortex of rhesus macaques (De et al., 2020). Mich et al. further optimized the human ortholog of the Dlx5/6 enhancer (hDLXI5/6i) by engineering a triple tandem of core elements taken from hDLXI5/6i and called it hDLX2.0 (Mich et al., 2021). AAV-PHP.eB containing hDLX2.0 upstream of a minimal beta-globin promoter and super yellow fluorescent protein-2 (SYFP2) reporter transduced GABAergic interneurons in ex vivo Macaca nemestrina cortical slice cultures and human neocortical slice cultures (Mich et al., 2021). Putative enhancers for targeting PVALB-expressing interneurons have similarly been identified, packaged in AAV-PHP.eB and tested in mice via retro-orbital injections, and in marmoset and macaque via local or intraparenchymal injections (Lawler et al., 2022; Mich et al., 2021; Vormstein-Schneider et al., 2020). These enhancers either targeted PVALB interneurons broadly or specific sub-classes of PVALB interneurons in the neocortex in both mouse and NHP. To identify regulatory elements that can drive faithful expression across species using AAV vectors, the selection method has largely focused on sequences that are conserved across species. However, Mich et al. reported that certain PVALB enhancer sequences present in the open chromatin analyses of the human neocortex but not in the mouse neocortex, were still able to drive selective expression in PVALB neurons in the mouse brain (Mich et al., 2021; Vormstein-Schneider et al., 2020). Thus, to minimize the number of experimental animals used for in vivo screening, it may be advantageous to develop machine-learning classifiers that can identify DNA sequence patterns important for driving species-agnostic cell-type specific activation (Lawler et al., 2022).
Single-cell and single-nucleus transcriptomics studies of the rodent and primate brain have revealed the molecular complexity and diversity of cell-types present based on their gene expression profiles (Hodge et al., 2019; Tasic et al., 2016, 2018; Zeisel et al., 2015). In the primary motor cortex alone, there are potentially 45 conserved cell-types among mouse, marmoset and human (Callaway et al., 2021). Only once a cell-type has been molecularly defined can researchers begin to identify DNA regulatory elements that are required for cell-type specific gene activation, and guide the development of tailored targeting strategies for treatment and functional interrogation. We have shown that broad CNS transduction in NHP is possible using the ubiquitous CAG promoter in our recently engineered vectors (AAV-CAP-Mac, AAV-MaCPNS1 and AAV-MaCPNS2) (Chen et al., 2022; Chuapoco et al., in press). These variants, which show comparatively higher neuronal transduction than AAV9 in NHPs, can be used to screen regulatory elements that specifically target neuronal subpopulations. One caveat of this approach is that injecting multiple AAVs with different enhancer and promoter elements in the same animal may cause interference between the regulatory elements, resulting in a loss of specificity compared to independent delivery (Mehta et al., 2019; Pouchelon et al., 2022). This may confound interpretation of pooled screens of putative regulatory elements.
Further cell- and tissue-type specificity can be achieved by incorporating microRNA (miRNA) target site sequences (∼22 nucleotides) into the 3’ untranslated coding region of AAVs. These bind to complementary miRNA sequences wherever they are expressed and inhibit mRNA expression. This approach has been used to detarget transgene expression from primary sensory neurons (miR183) and liver (miR122) to reduce dose-dependent toxicity in NHPs (Hordeaux et al., 2020; Kochunov et al., 2015). Recently, a database of miRNA expression across 196 primary cell-types was generated, enabling the potential testing of numerous combinations of miRNA-binding transgene cassettes (Patil et al., 2022). With spatial transcriptomics we can characterize the cell type-specific expression of various combinations of systemically-delivered AAV capsids and cargo, with the goal of expanding the gene delivery toolkit for NHPs (Jang et al., 2023).
5. Future directions
The effective delivery of sensors, effectors, and reporters for circuit tracing and manipulation, largely depends on the vector used. However, differences in brain size and immune function have hampered the widespread adoption of genetic technology in monkeys. Thus, the engineering of more efficient and specific viral vectors to target the CNS in NHPs addresses many of the challenges that have inhibited progress in translating rodent disease biology to better therapies and therapeutic approaches. Here, we review newly engineered systemic capsid variants that address these challenges. Specifically, using an adapted, cross-species directed evolution approach, our group has identified new variants that can transduce neuronal cells in CNS and PNS via peripheral injection in multiple NHP species commonly used in research.
In marmosets, AAV-CAP-B10 and AAV-CAP-B22, variants of AAV-PHP.eB, were identified to have enhanced CNS transduction compared to AAV9. In both marmosets and rhesus macaques, AAV-MaCPNS1 and AAV-MaCPNS2 variants transduced cells in both CNS and PNS. This may be particularly useful for studies where widespread transgene expression is desired as infusions in NHPs typically require multiple sites of injection to cover whole areas of tissue; however, this remains to be tested in relation to distributed brain function and/or behavior. In both rhesus macaques and green monkeys, AAV-CAP-Mac was found to transduce a higher percentage of neurons than AAV9. Importantly, these variants show significantly lower transduction in the liver compared to AAV9, thereby minimizing the risk of hepatotoxicity.
Still, while these novel AAVs may provide a new and necessary tool for delivering genetic cargo into the primate brain, many technical issues still remain. Below we briefly discuss these issues.
-
1.
Achieving cell-type specificity in NHPs. For decades, studies in NHPs have relied on lesions, reversible inactivation, and electrophysiology to elucidate the role of specific regions in a particular function (Balan et al., 2019; Dal Monte et al., 2015; Lak et al., 2014; Rudebeck et al., 2013). While this has provided invaluable insight into distributed circuits that underlie behavior, these studies are limited by their cell-type agnostic nature. Lesioning and reversible inactivation generally impact all cells in a particular region. In addition, lesion studies have resulted in conflicting reports on observed behaviors within the same region of the brain, often due to unintended damage to fibers of passage (Rudebeck et al., 2013). Similarly, electrophysiological recording techniques cannot differentiate molecular cell-types, and rely on electrophysiological-specific characterization (e.g. early-firing, late-firing, ramping, etc.). For example, in the ventral tegmental area, a minority of neurons share the same electrophysiological properties as dopamine neurons, leading to questions on whether some recordings have been misattributed to dopamine (Ungless and Grace, 2012). To address this issue, rodent studies have used TH-Cre mice to target dopamine-expressing VTA neurons for the expression of sensors and effectors (Bariselli et al., 2016; Lindeberg et al., 2004). However, in NHPs, Cre-lines do not yet exist or are not widely used. It is unlikely that capsid engineering alone will enable cell-type specificity. Instead, it is likely that a combination of capsid and DNA regulatory elements will achieve cell-type specificity. To this end, AAV-Cap-Mac and AAV-MaCPNS1/2 can be used to screen regulatory elements that specifically target neuronal subpopulations.
-
2.
Effective delivery and functional efficacy of genetic cargo. Advances in genetic cargo, like opsins and DREADDs, have been critical in dissecting circuits that are thought to contribute to disease -- in rodents. However, these techniques have not been widely adopted in NHPs because of difficulties in delivering genetic cargo efficiently into the primate brain. Currently, only a few published studies in NHPs have demonstrated successful delivery of opsins, with AAV5 and AAV9 being amongst the most commonly used vectors (Tremblay et al., 2020). Moreover, outcome measures-anatomy, physiology, and behavior in these studies-vary greatly (Bliss-Moreau et al., 2022). For example, AAV5 has been shown to preferentially target some brain regions but not others (Roseboom et al., 2021). Still, a major hurdle remains in determining the extent in which systemic delivery can express effector-cargo in a sufficient proportion of cells to affect behavior. Even so, infecting a small proportion of cells can still help us better understand the contributions of a small number of cells on behaviors as the functional efficacy of sensor-cargo does not necessitate a large proportion of cells. Currently, the effective delivery and functional relevance of sensors and effectors have not been tested using AAV-CAP-Mac or AAV-MaCPNS1/2. Further work is needed to show the functional efficacy of the genetic cargo delivered by these novel AAVs.
Understanding the emergence or origins of brain-based disease requires coordinated cross-species research. To this end, NHP models are particularly important because of their shared biology with humans. However, tools for interrogating anatomical pathways and functional circuits will need to be translated for widespread use in NHPs. While the vectors presented here address many common technical challenges seen in NHPs, there is a continued need for more efficient and specific AAVs. We hope that further optimization of these vectors can lead to more efficient delivery and ultimately, lead to new tools for the study of the primate brain and the development of new treatments for brain-based disorders.
CRediT authorship contribution statement
Lillian J. Campos: Writing – original draft, Writing – review & editing. Cynthia M. Arokiaraj: Writing – original draft, Writing – review & editing. Miguel R. Chuapoco: Writing – review & editing. Xinhong Chen: Writing – review & editing. Nick Goeden: Writing – review & editing. Viviana Gradinaru: Writing – original draft, Writing – review & editing. Andrew S. Fox: Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Viviana Gradinaru reports a relationship with Capsida Biotherapeutics that includes: board membership. Nick Goeden reports a relationship with Capsida Biotherapeutics that includes: employment.
Acknowledgments
Research in the Gradinaru and Fox labs is funded in part by Aligning Science Across Parkinson's [ASAP-020495] through the Michael J. Fox Foundation for Parkinson's Research (MJFF) (to V.G. and A.S.F.). For the purpose of open access, the author has applied a CC BY public copyright license to all Author Accepted Manuscripts arising from this submission. Additional support was provided by the California National Primate Research Center, NIH BRAIN Initiative Armamentarium grant UF1MH128336 (to V.G. and A.S.F.) and NIH P51OD011107 (to A.S.F). Figures were created using images from BioRender.com. We would like to thank Catherine Oikonomou and Rothem Kovner for help with manuscript editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crneur.2023.100086.
Contributor Information
Viviana Gradinaru, Email: viviana@caltech.edu.
Andrew S. Fox, Email: dfox@ucdavis.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- Abeliovich A., Hefti F., Sevigny J. Gene therapy for Parkinson's disease associated with GBA1 mutations. J. Parkinsons Dis. 2021;11(s2):S183–S188. doi: 10.3233/JPD-212739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbandje-McKenna M., Kleinschmidt J. In: Snyder R., Moullier P., editors. Vol. 807. Humana Press; 2012. AAV capsid structure and cell interactions. (Adeno-Associated Virus: Methods and Protocols). [DOI] [Google Scholar]
- Ahmed S.H. Individual decision-making in the causal pathway to addiction: contributions and limitations of rodent models. Pharmacol. Biochem. Behav. 2018;164:22–31. doi: 10.1016/j.pbb.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Alam M.M., Yang D., Li X.-Q., Liu J., Back T.C., Trivett A., Karim B., Barbut D., Zasloff M., Oppenheim J.J. Alpha synuclein, the culprit in Parkinson disease, is required for normal immune function. Cell Rep. 2022;38(2) doi: 10.1016/j.celrep.2021.110090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschauer D.F., Kreuz S., Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M., Hong S.-I., Kang S., Choi D.-S. Rodent models for psychiatric disorders: problems and promises. Laboratory Animal Research. 2020;36(1):9. doi: 10.1186/s42826-020-00039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan P.F., Gerits A., Zhu Q., Kolster H., Orban G.A., Wardak C., Vanduffel W. Fast compensatory functional Network changes caused by reversible inactivation of monkey parietal cortex. Cerebr. Cortex. 2019;29(6):2588–2606. doi: 10.1093/cercor/bhy128. [DOI] [PubMed] [Google Scholar]
- Bariselli S., Glangetas C., Tzanoulinou S., Bellone C. Ventral tegmental area subcircuits process rewarding and aversive experiences. J. Neurochem. 2016;139(6):1071–1080. doi: 10.1111/jnc.13779. [DOI] [PubMed] [Google Scholar]
- Bartlett J.S., Samulski R.J., McCown T.J. Selective and rapid uptake of adeno-associated virus type 2 in brain. Hum. Gene Ther. 1998;9(8):1181–1186. doi: 10.1089/hum.1998.9.8-1181. [DOI] [PubMed] [Google Scholar]
- Bartus R.T., Baumann T.L., Siffert J., Herzog C.D., Alterman R., Boulis N., Turner D.A., Stacy M., Lang A.E., Lozano A.M., Olanow C.W. Safety/feasibility of targeting the substantia nigra with AAV2-neurturin in Parkinson patients. Neurology. 2013;80(18):1698–1701. doi: 10.1212/WNL.0b013e3182904faa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi S., Salzman C.D. The contribution of nonhuman primate research to the understanding of emotion and cognition and its clinical relevance. Proc. Natl. Acad. Sci. USA. 2019;116(52):26305–26312. doi: 10.1073/pnas.1902293116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan A.K., Duque S., Foust K.D., Morales P.R., Braun L., Schmelzer L., Chan C.M., McCrate M., Chicoine L.G., Coley B.D., Porensky P.N., Kolb S.J., Mendell J.R., Burghes A.H., Kaspar B.K. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol. Ther. 2011;19(11) doi: 10.1038/mt.2011.157. Article 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss-Moreau E., Costa V.D., Baxter M.G. A pragmatic reevaluation of the efficacy of nonhuman primate optogenetics for psychiatry. Oxford Open Neurosci. 2022;1 doi: 10.1093/oons/kvac006. kvac006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen M.O., McCown T.J., Powell S.K., El-Nahal H.G., Daw T., Basso M.A., Sommer M.A., Samulski R.J. Adeno-associated virus capsid-promoter interactions in the brain translate from rat to the Nonhuman primate. Hum. Gene Ther. 2020;31(21–22):1155–1168. doi: 10.1089/hum.2020.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx M., Dutheil N., Bezard E., Dehay B. Systemic gene delivery to the central nervous system using Adeno-associated virus. Front. Mol. Neurosci. 2014;7 doi: 10.3389/fnmol.2014.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E.S., Zhang F., Bamberg E., Nagel G., Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8(9):1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Braak H., R☐b U., Gai W.P., Del Tredici K. Idiopathic Parkinson's disease: possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J. Neural. Transm. 2003;110(5):517–536. doi: 10.1007/s00702-002-0808-2. [DOI] [PubMed] [Google Scholar]
- Brady R.J., Hampton R.R. Post-encoding control of working memory enhances processing of relevant information in rhesus monkeys (Macaca mulatta) Cognition. 2018;175:26–35. doi: 10.1016/j.cognition.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulcha J.T., Wang Y., Ma H., Tai P.W.L., Gao G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Targeted Ther. 2021;6(1):53. doi: 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller R.M.L., Janik J.E., Sebring E.D., Rose J.A. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 1981;40(1):241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne L.C., Day T.P., Visel M., Strazzeri J.A., Fortuny C., Dalkara D., Merigan W.H., Schaffer D.V., Flannery J.G. In vivo–directed evolution of adeno-associated virus in the primate retina. JCI Insight. 2020;5(10) doi: 10.1172/jci.insight.135112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAIN Initiative Cell Census Network (BICCN), BRAIN Initiative Cell Census Network (BICCN) Corresponding authors. Callaway E.M., Dong H.-W., Ecker J.R., Hawrylycz M.J., Huang Z.J., Lein E.S., Ngai J., Osten P., Ren B., Tolias A.S., White O., Zeng H., Zhuang X., BICCN contributing principal investigators. Ascoli G.A., Behrens, Chun M.M., J, Sunkin S. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature. 2021;598(7879):86–102. doi: 10.1038/s41586-021-03950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis C., Hori A., Sampson T.R., Yoo B.B., Challis R.C., Hamilton A.M., Mazmanian S.K., Volpicelli-Daley L.A., Gradinaru V. Gut-seeded α-synuclein fibrils promote gut dysfunction and brain pathology specifically in aged mice. Nat. Neurosci. 2020;23(3):327–336. doi: 10.1038/s41593-020-0589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R.C., Ravindra Kumar S., Chen X., Goertsen D., Coughlin G.M., Hori A.M., Chuapoco M.R., Otis T.S., Miles T.F., Gradinaru V. Adeno-associated virus toolkit to target diverse brain cells. Annu. Rev. Neurosci. 2022;45(1):447–469. doi: 10.1146/annurev-neuro-111020-100834. [DOI] [PubMed] [Google Scholar]
- Chan Y.A., Deverman B.E. In: de Lange E.C.M., Hammarlund-Udenaes M., Thorne R.G., editors. Vol. 33. Springer International Publishing; 2022. Crossing the blood-brain barrier with AAVs: what's after SMA? pp. 629–654. (Drug Delivery to the Brain). [DOI] [Google Scholar]
- Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.-L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20(8):1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.W.C., Platt M.L. Oxytocin and social cognition in rhesus macaques: implications for understanding and treating human psychopathology. Brain Res. 2014;1580:57–68. doi: 10.1016/j.brainres.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Yao S., Wan J., Tian Y., Huang L., Wang S., Akter F., Wu Y., Yao Y., Zhang X. BBB-crossing adeno-associated virus vector: an excellent gene delivery tool for CNS disease treatment. J. Contr. Release. 2021;333:129–138. doi: 10.1016/j.jconrel.2021.03.029. [DOI] [PubMed] [Google Scholar]
- Chen X., Ravindra Kumar S., Adams C.D., Yang D., Wang T., Wolfe D.A., Arokiaraj C.M., Ngo V., Campos L.J., Griffiths J.A., Ichiki T., Mazmanian S.K., Osborne P.B., Keast J.R., Miller C.T., Fox A.S., Chiu I.M., Gradinaru V. Engineered AAVs for non-invasive gene delivery to rodent and non-human primate nervous systems. Neuron. 2022;110(14) doi: 10.1016/j.neuron.2022.05.003. Article 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christine C.W., Starr P.A., Larson P.S., Eberling J.L., Jagust W.J., Hawkins R.A., VanBrocklin H.F., Wright J.F., Bankiewicz K.S., Aminoff M.J. Safety and tolerability of putaminal AADC gene therapy for Parkinson disease. Neurology. 2009;73(20):1662–1669. doi: 10.1212/WNL.0b013e3181c29356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuapoco M.R., Flytzanis N.C., Goeden N., Octeau J.C., Roxas K.M., Chan K.Y., Scherrer J., Winchester J., Blackburn R.J., Campos L.J., Arokiaraj C.M., Miles T.F., Jang M.J., Vendemiatti J., Deverman B.E., Pickel J., Fox A.S., Gradinaru V. Intravenous gene transfer throughout the brain of infant old world primates using AAV. Nat. Nanotechnol. 2023 doi: 10.1038/s41565-023-01419-x. n.d (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciocchi S., Herry C., Grenier F., Wolff S.B.E., Letzkus J.J., Vlachos I., Ehrlich I., Sprengel R., Deisseroth K., Stadler M.B., Müller C., Lüthi A. Encoding of conditioned fear in central amygdala inhibitory circuits. Nature. 2010;468(7321):277–282. doi: 10.1038/nature09559. [DOI] [PubMed] [Google Scholar]
- Coley A.A., Padilla-Coreano N., Patel R., Tye K.M. Vol. 158. Elsevier; 2021. Valence processing in the PFC: reconciling circuit-level and systems-level views; pp. 171–212. (International Review of Neurobiology). [DOI] [PubMed] [Google Scholar]
- Craig A.D.B. How do you feel--now? The anterior insula and human awareness. Nat. Rev. Neurosci. 2009;10(1):59–70. doi: 10.1038/nrn2555. [DOI] [PubMed] [Google Scholar]
- Cristofori I., Cohen-Zimerman S., Grafman J. Vol. 163. Elsevier; 2019. Executive functions; pp. 197–219. (Handbook of Clinical Neurology). [DOI] [PubMed] [Google Scholar]
- Cummings K.A., Clem R.L. Prefrontal somatostatin interneurons encode fear memory. Nat. Neurosci. 2020;23(1):61–74. doi: 10.1038/s41593-019-0552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich D.A., Daza R., Adey A., Pliner H.A., Christiansen L., Gunderson K.L., Steemers F.J., Trapnell C., Shendure J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348(6237):910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushnie A.K., El-Nahal H.G., Bohlen M.O., May P.J., Basso M.A., Grimaldi P., Wang M.Z., de Velasco Ezequiel M.F., Sommer M.A., Heilbronner S.R. Using rAAV2-retro in rhesus macaques: promise and caveats for circuit manipulation. J. Neurosci. Methods. 2020;345 doi: 10.1016/j.jneumeth.2020.108859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Monte O., Costa V.D., Noble P.L., Murray E.A., Averbeck B.B. Amygdala lesions in rhesus macaques decrease attention to threat. Nat. Commun. 2015;6(1) doi: 10.1038/ncomms10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkara D., Byrne L.C., Klimczak R.R., Visel M., Yin L., Merigan W.H., Flannery J.G., Schaffer D.V. In vivo–directed evolution of a New adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med. 2013;5(189) doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- Davidson B.L., Breakefield X.O. Viral vectors for gene delivery to the nervous system. Nat. Rev. Neurosci. 2003;4(5):353–364. doi: 10.1038/nrn1104. [DOI] [PubMed] [Google Scholar]
- Davidson B.L., Gao G., Berry-Kravis E., Bradbury A.M., Bönnemann C., Buxbaum J.D., Corcoran G.R., Gray S.J., Gray-Edwards H., Kleiman R.J., Shaywitz A.J., Wang D., Zoghbi H.Y., Flotte T.R., Tauscher-Wisniewski S., Tifft C.J., Sahin M. Gene-based therapeutics for rare genetic neurodevelopmental psychiatric disorders. Mol. Ther. 2022;30(7):2416–2428. doi: 10.1016/j.ymthe.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A., El-Shamayleh Y., Horwitz G.D. Fast and reversible neural inactivation in macaque cortex by optogenetic stimulation of GABAergic neurons. Elife. 2020;9 doi: 10.7554/eLife.52658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw C.N., Korecki A.J., Berry G.E., Hickmott J.W., Lam S.L., Lengyell T.C., Bonaguro R.J., Borretta L.J., Chopra V., Chou A.Y., D'Souza C.A., Kaspieva O., Laprise S., McInerny S.C., Portales-Casamar E., Swanson-Newman M.I., Wong K., Yang G.S., Zhou M., et al. RAAV-compatible MiniPromoters for restricted expression in the brain and eye. Mol. Brain. 2016;9(1):52. doi: 10.1186/s13041-016-0232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaner R.O., Platt M.L. Reflexive social attention in monkeys and humans. Curr. Biol. 2003;13(18):1609–1613. doi: 10.1016/j.cub.2003.08.025. [DOI] [PubMed] [Google Scholar]
- Dehay B., Dalkara D., Dovero S., Li Q., Bezard E. Systemic scAAV9 variant mediates brain transduction in newborn rhesus macaques. Sci. Rep. 2012;2(1):253. doi: 10.1038/srep00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.-L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34(2) doi: 10.1038/nbt.3440. Article 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezfouli M.P., Zarei M., Constantinidis C., Daliri M.R. Task-specific modulation of PFC activity for matching-rule governed decision-making. Brain Struct. Funct. 2021;226(2):443–455. doi: 10.1007/s00429-020-02191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidschstein J., Chen Q., Tremblay R., Rogers S.L., Saldi G.-A., Guo L., Xu Q., Liu R., Lu C., Chu J., Grimley J.S., Krostag A.-R., Kaykas A., Avery M.C., Rashid M.S., Baek M., Jacob A.L., Smith G.B., Wilson D.E., et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 2016;19(12):1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenger C., Grimm D. Next-generation AAV vectors—do not judge a virus (only) by its cover. Hum. Mol. Genet. 2019;28(R1) doi: 10.1093/hmg/ddz148. R3–R14. [DOI] [PubMed] [Google Scholar]
- Drummer C., Vogt E.-J., Heistermann M., Roshani B., Becker T., Mätz-Rensing K., Kues W.A., Kügler S., Behr R. Generation and breeding of EGFP-transgenic marmoset monkeys: cell chimerism and implications for disease modeling. Cells. 2021;10(3):505. doi: 10.3390/cells10030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar R.I.M., Shultz S. Evolution in the social brain. Science. 2007;317(5843):1344–1347. doi: 10.1126/science.1145463. [DOI] [PubMed] [Google Scholar]
- El-Shamayleh Y., Kojima Y., Soetedjo R., Horwitz G.D. Selective optogenetic control of Purkinje cells in monkey cerebellum. Neuron. 2017;95(1) doi: 10.1016/j.neuron.2017.06.002. Article 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard H.C. The organization of the primate insular cortex. Front. Neuroanat. 2019;13:43. doi: 10.3389/fnana.2019.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok J.P., Krabbe S., Markovic M., Courtin J., Xu C., Massi L., Botta P., Bylund K., Müller C., Kovacevic A., Tovote P., Lüthi A. A competitive inhibitory circuit for selection of active and passive fear responses. Nature. 2017;542(7639):96–100. doi: 10.1038/nature21047. [DOI] [PubMed] [Google Scholar]
- Fadok J.P., Markovic M., Tovote P., Lüthi A. New perspectives on central amygdala function. Curr. Opin. Neurobiol. 2018;49:141–147. doi: 10.1016/j.conb.2018.02.009. [DOI] [PubMed] [Google Scholar]
- Fang R., Preissl S., Li Y., Hou X., Lucero J., Wang X., Motamedi A., Shiau A.K., Zhou X., Xie F., Mukamel E.A., Zhang K., Zhang Y., Behrens M.M., Ecker J.R., Ren B. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 2021;12(1):1337. doi: 10.1038/s41467-021-21583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L., Yizhar O., Deisseroth K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011;34(1):389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G., Kepecs A. Interneuron types as attractors and controllers. Annu. Rev. Neurosci. 2020;43(1):1–30. doi: 10.1146/annurev-neuro-070918-050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust K.D., Nurre E., Montgomery C.L., Hernandez A., Chan C.M., Kaspar B.K. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009;27(1):59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A.S., Oler J.A., Shackman A.J., Shelton S.E., Raveendran M., McKay D.R., Converse A.K., Alexander A., Davidson R.J., Blangero J., Rogers J., Kalin N.H. Intergenerational neural mediators of early-life anxious temperament. Proc. Natl. Acad. Sci. U. S. A. 2015;112(29):9118–9122. doi: 10.1073/pnas.1508593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franjic D., Skarica M., Ma S., Arellano J.I., Tebbenkamp A.T.N., Choi J., Xu C., Li Q., Morozov Y.M., Andrijevic D., Vrselja Z., Spajic A., Santpere G., Li M., Zhang S., Liu Y., Spurrier J., Zhang L., Gudelj I., et al. Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron. 2022;110(3):452–469.e14. doi: 10.1016/j.neuron.2021.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G.-P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99(18):11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Vandenberghe L., Wilson J. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 2005;5(3):285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Brown A.M., Jenkins C., Campbell K. Viral vector systems for gene therapy: a comprehensive literature review of progress and biosafety challenges. Appl. Biosafety. 2020;25(1):7–18. doi: 10.1177/1535676019899502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläscher J., Adolphs R., Damasio H., Bechara A., Rudrauf D., Calamia M., Paul L.K., Tranel D. Lesion mapping of cognitive control and value-based decision making in the prefrontal cortex. Proc. Natl. Acad. Sci. USA. 2012;109(36):14681–14686. doi: 10.1073/pnas.1206608109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertsen D., Flytzanis N.C., Goeden N., Chuapoco M.R., Cummins A., Chen Y., Fan Y., Zhang Q., Sharma J., Duan Y., Wang L., Feng G., Chen Y., Ip N.Y., Pickel J., Gradinaru V. AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat. Neurosci. 2022;25(1):106–115. doi: 10.1038/s41593-021-00969-4. [DOI] [PubMed] [Google Scholar]
- Grandi F.C., Modi H., Kampman L., Corces M.R. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 2022;17(6):1518–1552. doi: 10.1038/s41596-022-00692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S.J., Matagne V., Bachaboina L., Yadav S., Ojeda S.R., Samulski R.J. Preclinical differences of intravascular AAV9 delivery to Neurons and glia: a comparative study of adult mice and Nonhuman primates. Mol. Ther. 2011;19(6):1058–1069. doi: 10.1038/mt.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybuck L.T., Daigle T.L., Sedeño-Cortés A.E., Walker M., Kalmbach B., Lenz G.H., Morin E., Nguyen T.N., Garren E., Bendrick J.L., Kim T.K., Zhou T., Mortrud M., Yao S., Siverts L.A., Larsen R., Gore B.B., Szelenyi E.R., Trader C., et al. Enhancer viruses for combinatorial cell-subclass-specific labeling. Neuron. 2021;109(9):1449–1464.e13. doi: 10.1016/j.neuron.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadaczek P., Stanek L., Ciesielska A., Sudhakar V., Samaranch L., Pivirotto P., Bringas J., O'Riordan C., Mastis B., San Sebastian W., Forsayeth J., Cheng S.H., Bankiewicz K.S., Shihabuddin L.S. Widespread AAV1- and AAV2-mediated transgene expression in the nonhuman primate brain: implications for Huntington's disease. Molecular Ther. Method. Clinic. Develop. 2016;3 doi: 10.1038/mtm.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haery L., Deverman B.E., Matho K.S., Cetin A., Woodard K., Cepko C., Guerin K.I., Rego M.A., Ersing I., Bachle S.M., Kamens J., Fan M. Adeno-associated virus technologies and methods for targeted Neuronal manipulation. Front. Neuroanat. 2019;13:93. doi: 10.3389/fnana.2019.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe toxicity in Nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 2018;29(3):285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O., Yao Z., Eggermont J., Höllt T., Levi B.P., Shehata S.I., Aevermann B., Beller A., Bertagnolli D., Brouner K., et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573(7772):61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley D., Fox A.S. The central extended amygdala guides survival-relevant tradeoffs: implications for understanding common psychiatric disorders. Neurosci. Biobehav. Rev. 2022;142 doi: 10.1016/j.neubiorev.2022.104879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux J., Wang Q., Katz N., Buza E.L., Bell P., Wilson J.M. The Neurotropic properties of AAV-PHP.B are limited to C57bl/6J mice. Mol. Ther. 2018;26(3) doi: 10.1016/j.ymthe.2018.01.018. Article 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux J., Yuan Y., Clark P.M., Wang Q., Martino R.A., Sims J.J., Bell P., Raymond A., Stanford W.L., Wilson J.M. The GPI-linked protein LY6A drives AAV-PHP.B transport across the blood-brain barrier. Mol. Ther. 2019;27(5) doi: 10.1016/j.ymthe.2019.02.013. Article 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux J., Buza E.L., Jeffrey B., Song C., Jahan T., Yuan Y., Zhu Y., Bell P., Li M., Chichester J.A., Calcedo R., Wilson J.M. MicroRNA-mediated inhibition of transgene expression reduces dorsal root ganglion toxicity by AAV vectors in primates. Sci. Transl. Med. 2020;12(569) doi: 10.1126/scitranslmed.aba9188. Article 569. [DOI] [PubMed] [Google Scholar]
- Horga G., Kaur T., Peterson B.S. Annual Research Review: current limitations and future directions in MRI studies of child- and adult-onset developmental psychopathologies. JCPP (J. Child Psychol. Psychiatry) 2014;55(6):659–680. doi: 10.1111/jcpp.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino C., Konno A., Hosoi N., Kaneko R., Mukai R., Nakai J., Hirai H. GABAergic neuron-specific whole-brain transduction by AAV-PHP.B incorporated with a new GAD65 promoter. Mol. Brain. 2021;14(1) doi: 10.1186/s13041-021-00746-1. Article 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S., Tzeng C.P., Nagy M.A., Stroud H., Koutsioumpa C., Wilcox O.F., Assad E.G., Green J., Harvey C.D., Griffith E.C., Greenberg M.E. A scalable platform for the development of cell-type-specific viral drivers. Elife. 2019;8 doi: 10.7554/eLife.48089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui Y., Zheng X., Zhang H., Li F., Yu G., Li J., Zhang J., Gong X., Guo G. Strategies for targeting Neural circuits: how to manipulate Neurons using virus vehicles. Front. Neural Circ. 2022;16 doi: 10.3389/fncir.2022.882366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington T.E., Srinivasan R. Adeno-associated virus expression of α-synuclein as a tool to model Parkinson's disease: current understanding and knowledge gaps. Aging and Disease. 2021;12(4):1120. doi: 10.14336/AD.2021.0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M.J., Coughlin G.M., Jackson C.R., Chen X., Chuapoco M.R., Vendemiatti J.L., Wang A.Z., Gradinaru V. Spatial transcriptomics for profiling the tropism of viral vectors in tissues. Nat. Biotechnol. 2023 doi: 10.1038/s41587-022-01648-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas J.H. Vol. 195. Elsevier; 2012. The evolution of neocortex in primates; pp. 91–102. (Progress in Brain Research). [DOI] [Google Scholar]
- Kalin N.H., Shelton S.E. Nonhuman primate models to study anxiety, emotion regulation, and psychopathology. Ann. N. Y. Acad. Sci. 2003;1008(1):189–200. doi: 10.1196/annals.1301.021. [DOI] [PubMed] [Google Scholar]
- Kamath T., Abdulraouf A., Burris S.J., Langlieb J., Gazestani V., Nadaf N.M., Balderrama K., Vanderburg C., Macosko E.Z. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease. Nat. Neurosci. 2022;25(5):588–595. doi: 10.1038/s41593-022-01061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplitt M.G., Feigin A., Tang C., Fitzsimons H.L., Mattis P., Lawlor P.A., Bland R.J., Young D., Strybing K., Eidelberg D., During M.J. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369(9579):2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- Katz P.S., Harris-Warrick R.M. The evolution of neuronal circuits underlying species-specific behavior. Curr. Opin. Neurobiol. 1999;9(5):628–633. doi: 10.1016/S0959-4388(99)00012-4. [DOI] [PubMed] [Google Scholar]
- Kells A.P., Forsayeth J., Bankiewicz K.S. Glial-derived neurotrophic factor gene transfer for Parkinson's disease: anterograde distribution of AAV2 vectors in the primate brain. Neurobiol. Dis. 2012;48(2):228–235. doi: 10.1016/j.nbd.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Harashima H. Non-invasive gene delivery across the blood-brain barrier: present and future perspectives. Neural Regenerat. Res. 2022;17(4):785. doi: 10.4103/1673-5374.320981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik D., Björklund A. Modeling CNS neurodegeneration by overexpression of disease-causing proteins using viral vectors. Trends Neurosci. 2003;26(7):386–392. doi: 10.1016/S0166-2236(03)00164-4. [DOI] [PubMed] [Google Scholar]
- Klapoetke N.C., Murata Y., Kim S.S., Pulver S.R., Birdsey-Benson A., Cho Y.K., Morimoto T.K., Chuong A.S., Carpenter E.J., Tian Z., Wang J., Xie Y., Yan Z., Zhang Y., Chow B.Y., Surek B., Melkonian M., Jayaraman V., Constantine-Paton M., et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11(3):338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P., Jahanshad N., Marcus D., Winkler A., Sprooten E., Nichols T.E., Wright S.N., Hong L.E., Patel B., Behrens T., Jbabdi S., Andersson J., Lenglet C., Yacoub E., Moeller S., Auerbach E., Ugurbil K., Sotiropoulos S.N., Brouwer R.M., et al. Heritability of fractional anisotropy in human white matter: a comparison of Human Connectome Project and ENIGMA-DTI data. Neuroimage. 2015;111:300–311. doi: 10.1016/j.neuroimage.2015.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körbelin J., Dogbevia G., Michelfelder S., Ridder D.A., Hunger A., Wenzel J., Seismann H., Lampe M., Bannach J., Pasparakis M., Kleinschmidt J.A., Schwaninger M., Trepel M. A brain microvasculature endothelial cell‐specific viral vector with the potential to treat neurovascular and neurological diseases. EMBO Mol. Med. 2016;8(6) doi: 10.15252/emmm.201506078. Article 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienen F.M., Goldman M., Zhang Q., del Rosario R C.H., Florio M., Machold R., Saunders A., Levandowski K., Zaniewski H., Schuman B., Wu C., Lutservitz A., Mullally C.D., Reed N., Bien E., Bortolin L., Fernandez-Otero M., Lin J.D., Wysoker A., et al. Innovations present in the primate interneuron repertoire. Nature. 2020;586(7828):262–269. doi: 10.1038/s41586-020-2781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensson K., Nennesmo I., Persson L., Lycke E. Neuron to neuron transmission of herpes simplex virus. J. Neurol. Sci. 1982;54(1):149–156. doi: 10.1016/0022–510X(82)90227-1. [DOI] [PubMed] [Google Scholar]
- Kumar M., Keller B., Makalou N., Sutton R.E. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001;12(15):1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- Kumar S.R., Miles T.F., Chen X., Brown D., Dobreva T., Huang Q., Ding X., Luo Y., Einarsson P.H., Greenbaum A., Jang M.J., Deverman B.E., Gradinaru V. Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat. Methods. 2020;17(5):541–550. doi: 10.1038/s41592-020-0799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin D.A., Shutova M.V., Johnston N.R., Smith O.P., Fedorin V.V., Kukushkin Y.S., van der Loo J.C.M., Johnstone E.C. The clinical landscape for AAV gene therapies. Nat. Rev. Drug Discov. 2021;20(3):173–174. doi: 10.1038/d41573-021-00017-7. [DOI] [PubMed] [Google Scholar]
- Lak A., Stauffer W.R., Schultz W. Dopamine prediction error responses integrate subjective value from different reward dimensions. Proc. Natl. Acad. Sci. USA. 2014;111(6):2343–2348. doi: 10.1073/pnas.1321596111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman D.S. Gene delivery and gene therapy with herpes simplex virus-based vectors. Gene. 2001;264(1):1–9. doi: 10.1016/S0378-1119(01)00322-5. [DOI] [PubMed] [Google Scholar]
- Lawler A.J., Ramamurthy E., Brown A.R., Shin N., Kim Y., Toong N., Kaplow I.M., Wirthlin M., Zhang X., Phan B.N., Fox G.A., Wade K., He J., Ozturk B.E., Byrne L.C., Stauffer W.R., Fish K.N., Pfenning A.R. Machine learning sequence prioritization for cell type-specific enhancer design. Elife. 2022;11 doi: 10.7554/eLife.69571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.T., Gee S.M., Vogt D., Patel T., Rubenstein J.L., Sohal V.S. Pyramidal Neurons in prefrontal cortex receive subtype-specific forms of excitation and inhibition. Neuron. 2014;81(1):61–68. doi: 10.1016/j.neuron.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.J., Guenther C.M., Suh J. Adeno-associated virus (AAV) vectors: rational design strategies for capsid engineering. Curr. Opinion Biomed. Eng. 2018;7:58–63. doi: 10.1016/j.cobme.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 2010;20(17):R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21(4):255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- Li X., Gutierrez D.V., Hanson M.G., Han J., Mark M.D., Chiel H., Hegemann P., Landmesser L.T., Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA. 2005;102(49):17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg J., Usoskin D., Bengtsson H., Gustafsson A., Kylberg A., Söderström S., Ebendal T. Transgenic expression of Cre recombinase from the tyrosine hydroxylase locus: transgenic expression of Cre recombinase. Genesis. 2004;40(2):67–73. doi: 10.1002/gene.20065. [DOI] [PubMed] [Google Scholar]
- Liu Q., Wu Y., Wang H., Jia F., Xu F. Viral tools for Neural circuit tracing. Neurosci. Bull. 2022;38(12):1508–1518. doi: 10.1007/s12264-022-00949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis Jeune V., Joergensen J.A., Hajjar R.J., Weber T. Pre-existing anti–adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods. 2013;24(2):59–67. doi: 10.1089/hgtb.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus C.J., Lee P.H., Atasoy D., Su H.H., Looger L.L., Sternson S.M. Chemical and genetic engineering of selective ion channel–ligand interactions. Science. 2011;333(6047):1292–1296. doi: 10.1126/science.1206606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiesen S.N., Lock J.L., Schoderboeck L., Abraham W.C., Hughes S.M. CNS transduction benefits of AAV-PHP.eB over AAV9 are dependent on administration route and mouse strain. Molecular Therapy. Methods & Clinical Development. 2020;19:447–458. doi: 10.1016/j.omtm.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Oue M., Hirai H. Generation of a neurodegenerative disease mouse model using lentiviral vectors carrying an enhanced synapsin I promoter. J. Neurosci. Methods. 2014;223:133–143. doi: 10.1016/j.jneumeth.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y., Konno A., Mochizuki R., Shinohara Y., Nitta K., Okada Y., Hirai H. Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci. Lett. 2018;665:182–188. doi: 10.1016/j.neulet.2017.11.049. [DOI] [PubMed] [Google Scholar]
- Mattar C.N., Waddington S.N., Biswas A., Johana N., Ng X.W., Fisk A.S., Fisk N.M., Tan L.G., Rahim A.A., Buckley S.M.K., Tan M.H., Lu J., Choolani M., Chan J.K.Y. Systemic delivery of scAAV9 in fetal macaques facilitates neuronal transduction of the central and peripheral nervous systems. Gene Ther. 2013;20(1):69–83. doi: 10.1038/gt.2011.216. [DOI] [PubMed] [Google Scholar]
- Mehta P., Kreeger L., Wylie D.C., Pattadkal J.J., Lusignan T., Davis M.J., Turi G.F., Li W.-K., Whitmire M.P., Chen Y., Kajs B.L., Seidemann E., Priebe N.J., Losonczy A., Zemelman B.V. Functional access to Neuron subclasses in rodent and primate forebrain. Cell Rep. 2019;26(10) doi: 10.1016/j.celrep.2019.02.011. Article 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard C., Hodes G.E., Russo S.J. Pathogenesis of depression: insights from human and rodent studies. Neuroscience. 2016;321:138–162. doi: 10.1016/j.neuroscience.2015.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich J.K., Graybuck L.T., Hess E.E., Mahoney J.T., Kojima Y., Ding Y., Somasundaram S., Miller J.A., Kalmbach B.E., Radaelli C., Gore B.B., Weed N., Omstead V., Bishaw Y., Shapovalova N.V., Martinez R.A., Fong O., Yao S., Mortrud M., et al. Functional enhancer elements drive subclass-selective expression from mouse to primate neocortex. Cell Rep. 2021;34(13) doi: 10.1016/j.celrep.2021.108754. Article 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.T., Freiwald W.A., Leopold D.A., Mitchell J.F., Silva A.C., Wang X. Marmosets: a neuroscientific model of human social behavior. Neuron. 2016;90(2):219–233. doi: 10.1016/j.neuron.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S. The current status of gene therapy for Parkinson's disease. Ann. Neurosci. 2010;17(2) doi: 10.5214/ans.0972-7531.1017209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R.R., Blankvoort S., Lagartos M.J., Kentros C. Enhancer-Driven gene expression (EDGE) enables the generation of viral vectors specific to Neuronal subtypes. iScience. 2020;23(3) doi: 10.1016/j.isci.2020.100888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson J.L. Short promoters in viral vectors drive selective expression in mammalian inhibitory neurons, but do not restrict activity to specific inhibitory cell-types. Front. Neural Circ. 2009;3 doi: 10.3389/neuro.04.019.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta K., Matsuzaki Y., Konno A., Hirai H. Minimal Purkinje cell-specific PCP2/L7 promoter virally available for rodents and Non-human primates. Molecular Therapy. Methods & Clinical Development. 2017;6:159–170. doi: 10.1016/j.omtm.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher M., Wang W., Child M.A., Ren X.-Q., Huang C., Ren A.Z., Tocci J., Chen Q., Bittner K., Tyson K., Pande N., Chung C.H.-Y., Paul S.M., Hou J. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Molecular Ther. Method. Clinic. Develop. 2021;20:366–378. doi: 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öngür D., Price J.L. The organization of networks within the orbital and medial prefrontal cortex of rats, monkeys and humans. Cerebr. Cortex. 2000;10(3):206–219. doi: 10.1093/cercor/10.3.206. [DOI] [PubMed] [Google Scholar]
- Park J.E., Zhang X.F., Choi S.-H., Okahara J., Sasaki E., Silva A.C. Generation of transgenic marmosets expressing genetically encoded calcium indicators. Sci. Rep. 2016;6(1) doi: 10.1038/srep34931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A.H., Baran A., Brehm Z.P., McCall M.N., Halushka M.K. Vol. 12. 2022. (A Curated Human Cellular microRNAome Based on 196 Primary Cell Types). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T., Cho J.R., Merten K., Howe M.W., Marley A., Xiong W.-H., Folk R.W., Broussard G.J., Liang R., Jang M.J., Zhong H., Dombeck D., von Zastrow M., Nimmerjahn A., Gradinaru V., Williams J.T., Tian L. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science. 2018;360(6396):eaat4422. doi: 10.1126/science.aat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez B.A., Shutterly A., Chan Y.K., Byrne B.J., Corti M. Management of neuroinflammatory responses to AAV-mediated gene therapies for Neurodegenerative diseases. Brain Sci. 2020;10(2):119. doi: 10.3390/brainsci10020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrides M., Pandya D.N. Dorsolateral prefrontal cortex: comparative cytoarchitectonic analysis in the human and the macaque brain and corticocortical connection patterns: dorsolateral prefrontal cortex in human and monkey. Eur. J. Neurosci. 1999;11(3):1011–1036. doi: 10.1046/j.1460-9568.1999.00518.x. [DOI] [PubMed] [Google Scholar]
- Petrides M., Pandya D.N. Comparative cytoarchitectonic analysis of the human and the macaque ventrolateral prefrontal cortex and corticocortical connection patterns in the monkey: ventrolateral prefrontal cortex in human and monkey. Eur. J. Neurosci. 2002;16(2):291–310. doi: 10.1046/j.1460-9568.2001.02090.x. [DOI] [PubMed] [Google Scholar]
- Phillips K.A., Bales K.L., Capitanio J.P., Conley A., Czoty P.W., ‘t Hart B.A., Hopkins W.D., Hu S.-L., Miller L.A., Nader M.A., Nathanielsz P.W., Rogers J., Shively C.A., Voytko M.L. Why primate models matter: why primate models matter. Am. J. Primatol. 2014;76(9):801–827. doi: 10.1002/ajp.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli M., Beyeler A. Valence coding in amygdala circuits. Curr. Opinion Behavioral Sci. 2019;26:97–106. doi: 10.1016/j.cobeha.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine D.S., Wise S.P., Murray E.A. Evolution, emotion, and episodic engagement. Am. J. Psychiatr. 2021;178(8):701–714. doi: 10.1176/appi.ajp.2020.20081187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouchelon G., Vergara J., McMahon J., Gorissen B.L., Lin J.D., Vormstein-Schneider D., Niehaus J.L., Burbridge T.J., Wester J.C., Sherer M., Fernandez-Otero M., Allaway K.C., Pelkey K., Chittajallu R., McBain C.J., Fan M., Nasse J.S., Wildenberg G.A., Fishell G., Dimidschstein J. A versatile viral toolkit for functional discovery in the nervous system. Cell Rep. Method. 2022;2(6) doi: 10.1016/j.crmeth.2022.100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S., Fang R., Huang H., Zhao Y., Raviram R., Gorkin D.U., Zhang Y., Sos B.C., Afzal V., Dickel D.E., Kuan S., Visel A., Pennacchio L.A., Zhang K., Ren B. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci. 2018;21(3):432–439. doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss T.M., Wise S.P. Evolution of prefrontal cortex. Neuropsychopharmacology. 2022;47(1):3–19. doi: 10.1038/s41386-021-01076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich E.L., Wallis J.D. Decoding subjective decisions from orbitofrontal cortex. Nat. Neurosci. 2016;19(7):973–980. doi: 10.1038/nn.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J.A., Koczot F. Adenovirus-associated virus multiplication. J. Virol. 1972;10 doi: 10.1128/jvi.10.1.1-8.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseboom P.H., Mueller S.A.L., Oler J.A., Fox A.S., Riedel M.K., Elam V.R., Olsen M.E., Gomez J.L., Boehm M.A., DiFilippo A.H., Christian B.T., Michaelides M., Kalin N.H. Evidence in primates supporting the use of chemogenetics for the treatment of human refractory neuropsychiatric disorders. Mol. Ther. 2021;29(12):3484–3497. doi: 10.1016/j.ymthe.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth B.L. DREADDs for neuroscientists. Neuron. 2016;89(4):683–694. doi: 10.1016/j.neuron.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin A.N., Malik R., Cho K.K.A., Lim K.J., Lindtner S., Robinson Schwartz S.E., Vogt D., Sohal V.S., Rubenstein J.L.R. Regulatory elements inserted into AAVs confer preferential activity in cortical interneurons. Eneuro. 2020;7(6) doi: 10.1523/ENEURO.0211-20.2020. ENEURO.0211-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudebeck P.H., Saunders R.C., Prescott A.T., Chau L.S., Murray E.A. Prefrontal mechanisms of behavioral flexibility, emotion regulation and value updating. Nat. Neurosci. 2013;16(8):1140–1145. doi: 10.1038/nn.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A. The marmoset as a model for the study of primate parental behavior. Neurosci. Res. 2015;93:99–109. doi: 10.1016/j.neures.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Samaranch L., Salegio E.A., San Sebastian W., Kells A.P., Foust K.D., Bringas J.R., Lamarre C., Forsayeth J., Kaspar B.K., Bankiewicz K.S. Adeno-associated virus serotype 9 transduction in the central Nervous system of Nonhuman primates. Hum. Gene Ther. 2012;23(4):382–389. doi: 10.1089/hum.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samelson-Jones B.J., Finn J.D., Favaro P., Wright J.F., Arruda V.R. Timing of intensive immunosuppression impacts risk of transgene antibodies after AAV gene therapy in Nonhuman primates. Molecular Ther. Method. Clinic. Develop. 2020;17:1129–1138. doi: 10.1016/j.omtm.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi S.P., Clarke J., Kinnecom C., Tamsett T.J., Li L., Stanek L.M., Passini M.A., Grabowski G.A., Schlossmacher M.G., Sidman R.L., Cheng S.H., Shihabuddin L.S. CNS expression of glucocerebrosidase corrects α-synuclein pathology and memory in a mouse model of Gaucher-related synucleinopathy. Proc. Natl. Acad. Sci. USA. 2011;108(29):12101–12106. doi: 10.1073/pnas.1108197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi S.P., Clarke J., Viel C., Chan M., Tamsett T.J., Treleaven C.M., Bu J., Sweet L., Passini M.A., Dodge J.C., Yu W.H., Sidman R.L., Cheng S.H., Shihabuddin L.S. Augmenting CNS glucocerebrosidase activity as a therapeutic strategy for parkinsonism and other Gaucher-related synucleinopathies. Proc. Natl. Acad. Sci. USA. 2013;110(9):3537–3542. doi: 10.1073/pnas.1220464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E., Suemizu H., Shimada A., Hanazawa K., Oiwa R., Kamioka M., Tomioka I., Sotomaru Y., Hirakawa R., Eto T., Shiozawa S., Maeda T., Ito M., Ito R., Kito C., Yagihashi C., Kawai K., Miyoshi H., Tanioka Y., et al. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459(7246):523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- Schmitz M.T., Sandoval K., Chen C.P., Mostajo-Radji M.A., Seeley W.W., Nowakowski T.J., Ye C.J., Paredes M.F., Pollen A.A. The development and evolution of inhibitory neurons in primate cerebrum. Nature. 2022;603(7903):871–877. doi: 10.1038/s41586-022-04510-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena-Esteves M., Saeki Y., Fraefel C., Breakefield X.O. HSV-1 amplicon vectors—simplicity and versatility. Mol. Ther. 2000;2(1):9–15. doi: 10.1006/mthe.2000.0096. [DOI] [PubMed] [Google Scholar]
- Shinohara Y., Konno A., Takahashi N., Matsuzaki Y., Kishi S., Hirai H. Viral vector-based dissection of marmoset GFAP promoter in mouse and marmoset brains. PLoS One. 2016;11(8) doi: 10.1371/journal.pone.0162023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaers J.B., Steele J., Case C.R., Cowper A., Amunts K., Zilles K. Primate prefrontal cortex evolution: human brains are the extreme of a lateralized ape trend. Brain Behav. Evol. 2011;77(2):67–78. doi: 10.1159/000323671. [DOI] [PubMed] [Google Scholar]
- Snyder A.C., Yu B.M., Smith M.A. A stable population code for attention in prefrontal cortex leads a dynamic attention code in visual cortex. J. Neurosci. 2021;41(44):9163–9176. doi: 10.1523/JNEUROSCI.0608-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R., Pekrun K., Khan T.A., Zhang F., Paşca S.P., Kay M.A. Selection of rAAV vectors that cross the human blood-brain barrier and target the central nervous system using a transwell model. Molecular Ther. Method. Clinic. Develop. 2022;27:73–88. doi: 10.1016/j.omtm.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016;21:75–80. doi: 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer W.R., Lak A., Yang A., Borel M., Paulsen O., Boyden E.S., Schultz W. Dopamine Neuron-specific optogenetic stimulation in rhesus macaques. Cell. 2016;166(6) doi: 10.1016/j.cell.2016.08.024. Article 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subiaul F., Cantlon J.F., Holloway R.L., Terrace H.S. Vol. 305. 2004. (Cognitive Imitation in Rhesus Macaques). [DOI] [PubMed] [Google Scholar]
- Sun J., Roy S. Gene-based therapies for neurodegenerative diseases. Nat. Neurosci. 2021;24(3):297–311. doi: 10.1038/s41593-020-00778-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M., Lagerborg K.A., Stanton A., King E.M., Ye S., Tellez L., Krunnfusz A., Tavakoli S., Widrick J.J., Messemer K.A., Troiano E.C., Moghadaszadeh B., Peacker B.L., Leacock K.A., Horwitz N., Beggs A.H., Wagers A.J., Sabeti P.C. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell. 2021;184(19):4919–4938.e22. doi: 10.1016/j.cell.2021.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B., Menon V., Nguyen T.N., Kim T.K., Jarsky T., Yao Z., Levi B., Gray L.T., Sorensen S.A., Dolbeare T., Bertagnolli D., Goldy J., Shapovalova N., Parry S., Lee C., Smith K., Bernard A., Madisen L., Sunkin S.M., et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016;19(2):335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B., Yao Z., Graybuck L.T., Smith K.A., Nguyen T.N., Bertagnolli D., Goldy J., Garren E., Economo M.N., Viswanathan S., Penn O., Bakken T., Menon V., Miller J., Fong O., Hirokawa K.E., Lathia K., Rimorin C., Tieu M., et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563(7729):72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka I., Nogami N., Nakatani T., Owari K., Fujita N., Motohashi H., Takayama O., Takae K., Nagai Y., Seki K. Generation of transgenic marmosets using a tetracyclin-inducible transgene expression system as a neurodegenerative disease model. Biol. Reprod. 2017;97(5):772–780. doi: 10.1093/biolre/iox129. [DOI] [PubMed] [Google Scholar]
- Tovote P., Esposito M.S., Botta P., Chaudun F., Fadok J.P., Markovic M., Wolff S.B.E., Ramakrishnan C., Fenno L., Deisseroth K., Herry C., Arber S., Lüthi A. Midbrain circuits for defensive behaviour. Nature. 2016;534(7606):206–212. doi: 10.1038/nature17996. [DOI] [PubMed] [Google Scholar]
- Tremblay S., Acker L., Afraz A., Albaugh D.L., Amita H., Andrei A.R., Angelucci A., Aschner A., Balan P.F., Basso M.A., Benvenuti G., Bohlen M.O., Caiola M.J., Calcedo R., Cavanaugh J., Chen Y., Chen S., Chernov M.M., Clark A.M., et al. An open resource for Non-human primate optogenetics. Neuron. 2020;108(6):1075–1090.e6. doi: 10.1016/j.neuron.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulusoy A., Decressac M., Kirik D., Björklund A. Vol. 184. Elsevier; 2010. Viral vector-mediated overexpression of α-synuclein as a progressive model of Parkinson's disease; pp. 89–111. (Progress in Brain Research). [DOI] [PubMed] [Google Scholar]
- Ungless M.A., Grace A.A. Are you or aren't you? Challenges associated with physiologically identifying dopamine neurons. Trends Neurosci. 2012;35(7):422–430. doi: 10.1016/j.tins.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. National Library of Medicine. (n.d https://www.clinicaltrials.gov [DOI] [PubMed]
- Visel A., Taher L., Girgis H., May D., Golonzhka O., Hoch R.V., McKinsey G.L., Pattabiraman K., Silberberg S.N., Blow M.J., Hansen D.V., Nord A.S., Akiyama J.A., Holt A., Hosseini R., Phouanenavong S., Plajzer-Frick I., Shoukry M., Afzal V., et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152(4):895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vormstein-Schneider D., Lin J.D., Pelkey K.A., Chittajallu R., Guo B., Arias-Garcia M.A., Allaway K., Sakopoulos S., Schneider G., Stevenson O., Vergara J., Sharma J., Zhang Q., Franken T.P., Smith J., Ibrahim L.A., Mastro K.J., Sabri E., Huang S., et al. Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat. Neurosci. 2020;23(12):1629–1636. doi: 10.1038/s41593-020-0692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Juggling safety and efficacy: finding ways to achieve both. Hum. Gene Ther. 2021;32(11–12):538–539. doi: 10.1089/hum.2021.29164.dwa. [DOI] [PubMed] [Google Scholar]
- Wang W., Kim C.K., Ting A.Y. Molecular tools for imaging and recording neuronal activity. Nat. Chem. Biol. 2019;15(2):101–110. doi: 10.1038/s41589-018-0207-0. [DOI] [PubMed] [Google Scholar]
- Wei J.-R., Hao Z.-Z., Xu C., Huang M., Tang L., Xu N., Liu R., Shen Y., Teichmann S.A., Miao Z., Liu S. Identification of visual cortex cell types and species differences using single-cell RNA sequencing. Nat. Commun. 2022;13(1):6902. doi: 10.1038/s41467-022-34590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker B.J. Open SUNY Textbooks; 2017. The History of Our Tribe, Hominini. [Google Scholar]
- Wittkopp P.J., Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012;13(1):59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18(1):80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Hu P., Li J., Chen J., Song W., Wang X.-J., Yang T., Dehaene S., Tang S., Min B., Wang L. Geometry of sequence working memory in macaque prefrontal cortex. Science. 2022;375(6581):632–639. doi: 10.1126/science.abm0204. [DOI] [PubMed] [Google Scholar]
- Xu H., Liu L., Tian Y., Wang J., Li J., Zheng J., Zhao H., He M., Xu T.-L., Duan S., Xu H. A disinhibitory microcircuit mediates conditioned social fear in the prefrontal cortex. Neuron. 2019;102(3):668–682.e5. doi: 10.1016/j.neuron.2019.02.026. [DOI] [PubMed] [Google Scholar]
- Yang W., Yuste R. In vivo imaging of neural activity. Nat. Methods. 2017;14(4):349–359. doi: 10.1038/nmeth.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Li S., Wang H., Guo Y., Gessler D.J., Cao C., Su Q., Kramer J., Zhong L., Ahmed S.S., Zhang H., He R., Desrosiers R.C., Brown R., Xu Z., Gao G. Global CNS transduction of adult mice by intravenously delivered rAAVrh.8 and rAAVrh.10 and Nonhuman primates by rAAVrh.10. Mol. Ther. 2014;22(7):1299–1309. doi: 10.1038/mt.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim Y.Y., Teague C.D., Nestler E.J. In vivo locus-specific editing of the neuroepigenome. Nat. Rev. Neurosci. 2020;21(9):471–484. doi: 10.1038/s41583-020-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.H., Park J.H., Kim Y.H., Min J., Hwang E., Lee C.J., Francis Suh J.-K., Hwang O., Jeon S.R. Optogenetic inactivation of the subthalamic Nucleus improves forelimb akinesia in a rat model of Parkinson disease. Neurosurgery. 2014;74(5):533–541. doi: 10.1227/NEU.0000000000000297. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Muñoz-Manchado A.B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C., Rolny C., Castelo-Branco G., Hjerling-Leffler J., Linnarsson S. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zerucha T., Stühmer T., Hatch G., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L.R., Ekker M. A highly conserved enhancer in the dlx5/dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 2000;20(2):709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang L.-P., Brauner M., Liewald J.F., Kay K., Watzke N., Wood P.G., Bamberg E., Nagel G., Gottschalk A., Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446(7136):633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhang H., Yang B., Mu X., Ahmed S.S., Su Q., He R., Wang H., Mueller C., Sena-Esteves M., Brown R., Xu Z., Gao G. Several rAAV vectors efficiently cross the blood–brain barrier and transduce Neurons and astrocytes in the Neonatal mouse central Nervous system. Mol. Ther. 2011;19(8):1440–1448. doi: 10.1038/mt.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C.-X., Wang S.-M., Bai Y.-H., Luo T.-T., Wang J.-Q., Dai C.-Q., Guo B.-L., Luo S.-C., Wang D.-H., Yang Y.-L., Wang Y.-Y. Lentiviral vectors and adeno-associated virus vectors: useful tools for gene transfer in pain research: lentiviral and adeno-associated virus vectors. Anat. Rec. 2018;301(5):825–836. doi: 10.1002/ar.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler T.E. Measuring peripheral oxytocin and vasopressin in nonhuman primates. Am. J. Primatol. 2018;80(10) doi: 10.1002/ajp.22871. [DOI] [PubMed] [Google Scholar]
- Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16(6):1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Zolotukhin S., Vandenberghe L.H. AAV capsid design: a Goldilocks challenge. Trends Mol. Med. 2022;28(3):183–193. doi: 10.1016/j.molmed.2022.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.