Abstract
Objectives
Erosive hand osteoarthritis (EHOA) is a severe subset of hand osteoarthritis (OA). It is unclear if EHOA is genetically different from other forms of OA. Sequence variants at ten loci have been associated with hand OA but none with EHOA.
Methods
We performed meta-analysis of EHOA in 1484 cases and 550 680 controls, from 5 populations. To identify causal genes, we performed eQTL and plasma pQTL analyses, and developed one zebrafish mutant. We analysed associations of variants with other traits and estimated shared genetics between EHOA and other traits.
Results
Four common sequence variants associated with EHOA, all with relatively high effect. Rs17013495 (SPP1/MEPE, OR=1.40, p=8.4×10−14) and rs11243284 (6p24.3, OR=1.35, p=4.2×10−11) have not been associated with OA, whereas rs11631127 (ALDH1A2, OR=1.46, p=7.1×10−18), and rs1800801 (MGP, OR=1.37, p=3.6×10−13) have previously been associated with hand OA. The association of rs1800801 (MGP) was consistent with a recessive mode of inheritance in contrast to its additive association with hand OA (OR homozygotes vs non-carriers=2.01, 95% CI 1.71 to 2.37). All four variants associated nominally with finger OA, although with substantially lower effect. We found shared genetic components between EHOA and other OA measures, grip strength, urate levels and gout, but not rheumatoid arthritis. We identified ALDH1A2, MGP and BMP6 as causal genes for EHOA, with loss-of-function Bmp6 zebrafish mutants displaying EHOA-like phenotypes.
Conclusions
We report on significant genetic associations with EHOA. The results support the view of EHOA as a form of severe hand OA and partly separate it from OA in larger joints.
Keywords: Osteoarthritis; Polymorphism, Genetic; Bone Density
WHAT IS ALREADY KNOWN ON THIS TOPIC
No genetic associations have been reported for erosive hand osteoarthritis (EHOA).
WHAT THIS STUDY ADDS
This study finds the first genetic association with EHOA at four loci that all confer relatively high risk of the disease, identifies candidate causal genes at three loci: ALDH1A2, MGP and BMP6, and strong candidates at one locus: SPP1, IBSP and MEPE.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study highlights EHOA as somewhat separate from osteoarthritis in the larger joints and points to potential drug targets for the disease.
Introduction
Erosive hand osteoarthritis (EHOA) is a severe form of hand osteoarthritis (OA), one of the most prevalent forms of OA.1–3 The clinical burden of EHOA is higher than for other types of hand OA (nodal hand OA or OA in the thumb base). It is characterised by abrupt onset with inflammation, radiographic features of central erosions and collapse of the subchondral bone, and rapid progression. Markers of inflammation and bone resorption are higher in EHOA patients than in other forms of hand OA. This can make it challenging to differentiate clinically from erosive rheumatoid arthritis (RA) and erosive gout in the small joints of the hand, two disease entities that have specific effective therapies on the market, while no disease-modifying drugs are yet available for EHOA. Between 5% and 20% of patients with symptomatic hand OA have EHOA which, as other OA types, predominantly affects females (reviewed in ref 3).3
Although EHOA is phenotypically different from nodal hand OA in the distal and proximal interphalangeal joints, it is not clear if EHOA represents a genetically distinct form of hand OA. Several studies have identified a genetic or familial component to EHOA4 5 and a few candidate genes and loci, such as HLA alleles and the IL1B gene, have been suggested.6–8
There is, however, no genome-wide association study (GWAS) of EHOA that has been reported, but ten loci have been described for hand/finger/thumb OA.9–12 The first and only meta-analysis of hand OA, which included 20 901 individuals with hand OA from 9 populations,12 found associations at the previously reported ALDH1A2,9 MGP 10 11 and WNT9A 11 loci, as well as at seven additional loci. None of the earlier studies separated EHOA from finger or hand OA, that is, EHOA patients were included in these analyses.
Here, based on five independent EHOA study populations, we identified four genetic loci that associate with EHOA. Two of these loci were previously associated with hand OA overall, at ALDH1A2 and MGP. We also discovered two new loci with candidate causal genes involved in bone biology, BMP6 and SPP1/MEPE/IBSP. Our data indicate that EHOA has substantial genetic overlap with finger OA, yet displays risk alleles that are associated with susceptibility of EHOA over that of finger or hand OA and of OA in other joints.
Methods
Details on the study populations and the methods used are given in online supplemental material to this publication.
ard-2022-223468supp002.pdf (512KB, pdf)
Study populations
Iceland: EHOA (918 cases) was diagnosed from conventional dorsopalmar radiographs taken of individuals with provisional diagnosis of hand OA and compared with 109 249 controls. The proximal and distal interphalangeal joints were scored according to Verbruggen-Veys (VV)13 and patients with at least one joint in the E phase (erosive) or R phase (remodelled) were classified as having EHOA. Individuals diagnosed with RA were excluded.
The Netherlands: The EHOA cases (N=145) were derived from the Hand OSTeoArthritis in Secondary care study,14 and the controls (N=5102) from the Nijmegen Biomedical Study.15 EHOA cases were classified according to VV,13 excluding RA.
UK: The UK Biobank resource (http://www.ukbiobank.ac.uk) includes data from 500 000 volunteers who were recruited between the age of 40 and 69 years in 2006–2010 across the United Kingdom. EHOA included those with the International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD10) code M15.4, excluding RA (63 EHOA cases/430 875 controls).
USA: The EHOA cases (N=145) included those with ICD-10 code M15.4, excluding RA, in the Utah Population Database16 and the Intermountain Healthcare HerediGene: Population Study (Utah, USA), compared with 5308 controls.
Spain: The EHOA cases (N=218) were derived from the PROspective COhort of A Coruña cohort,5 17 and the controls (N=164) were from other projects at A Coruña University Hospital who had not been diagnosed with hand OA on radiographs. EHOA cases were scored according to VV.13
All participants in this study were genetically determined to be of European descent.
Genotyping and association analysis
All the samples, except UK Biobank, were genotyped at deCODE genetics, using various Illumina chips, while UK Biobank genotyping used a custom-made Affimetrix chip. Imputation of all datasets was performed at deCODE genetics. Association analysis was done using logistic regression, adjusting for age, sex and principal components.
EHOA meta-analysis
We meta-analysed GWAS summary results from the additive model using a fixed-effects inverse variance method,18 including variants with info >0.8 and present in at least two datasets (N=46 million). For GWS thresholds we used the weighted Holm-Bonferroni method to allocate familywise error rate of 0.05 equally between five annotation-based classes of sequence variants.19 For the EHOA associated variants we also tested the recessive model, and the full genotype model.
Polygenic Risk Score and phenotype correlation analysis
We used Polygenic Risk Score (PRS) analysis based on a EHOA meta-analysis of Icelandic, Dutch, Spanish and US GWASs to investigate its correlation with about 5000 quantitative and case/control traits in the UK Biobank dataset. The PRSs was calculated using genotypes for about 600 000 autosomal markers included on the Illumina SNP chips to avoid uncertainty due to imputation quality.20
Genetic correlations
Using cross-trait linkage disequilibrium (LD) score regression method,21 we estimated the genetic correlation between EHOA and other OA subtypes in the Genetics of Osteoarthritis (GO) consortium dataset,12 and with other traits identified as correlated with EHOA in the PRS analysis in data from UK biobank, or associated with the EHOA variants, and RA (see online supplemental material for description of these phenotypes). In this analysis, we used results for about 1.2 million well-imputed variants, and for LD information, we used precomputed LD scores for European populations (downloaded from: https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2.
Phenoscan of public datasets
Associations of EHOA variants with other phenotypes was assessed using the Open Targets Genetics website (https://genetics.opentargets.org/), and a diverse set of phenotypes in UK Biobank that were generated at deCODE genetics. Associations with the lead EHOA variants, and variants in LD with the EHOA variants (r2>0.8), and p<1×10−6 were evaluated.
Functional annotation of sequence variants and enrichment of association signals
We determined if the lead sequence variant or correlated variants (r2>0.80) were located within candidate cis-regulatory elements (cCRE)22 or tissue-specific regulatory regions23 and looked for association signals in enhancer elements defined in EpiMap. We also determined their location within tissue-specific regulatory regions.23
Co-localisation of GWA signals with expression quantitative trait loci (eQTL) and protein quantitative trait loci (pQTL) signals
We analysed co-localisation of the EHOA associations with variation in gene transcription (eQTL) or variations in protein levels in plasma (plasma pQTL).24 For the eQTLs analysis, we used data from the publicly available Genotype-Tissue Expression (GTEx) project (https://www.gtexportal.org/), and deCODE genetics RNA sequence data from whole blood of 13 175 Icelanders and subcutaneous adipose tissue from 700 Icelanders.25 For plasma pQTL analysis, we used the dataset described in Ferkingstad et al,26 which tested association of 27.2 million variants with levels of 4719 proteins (adjusted and standardised levels) in plasma samples from 35 559 Icelanders.
Plasma protein levels
The dataset used for analysis of plasma protein levels is the same as for the plasma proteomics, restricted to those EHOA patients who had their sample taken within a year (±1 year) from the radiograph that was used to diagnose EHOA. Association between protein levels and EHOA was tested with logistic regression (R V.3.6.3), adjusting for age, sex and body mass index. Results are represented as OR of having EHOA per SD increase in standardised plasma protein levels.
Zebrafish experiments
The zebrafish (Danio rerio) Tu strain was used in all experiments. The generation of F0 and germline zebrafish lacking bmp6 gene function is described in detail in online supplemental material and shown schematically in online supplemental figure S1. Cartilage and bone staining was performed on 14 days post fertilisation (dpf) larvae.
Patient and public involvement statement
This research was done without direct patient involvement.
Results
GWAS and meta-analysis
To search for sequence variants that contribute to EHOA, we performed GWAS in samples from Iceland, The Netherlands, Spain, UK and USA (table 1), and subsequently meta-analysed the results from 1484 subjects with EHOA and 550 680 controls.
Table 1.
Characteristics of the study subjects
| N (% female) | Age, mean (±SD) | BMI, mean (±SD) | ||
| Iceland | EHOA | 918 (79) | 75.0 (11.2) | 27.3 (4.9) |
| Controls | 109 249 (46) | 66.5 (14.0) | 26.8 (5.3) | |
| UK Biobank | EHOA | 63 (79) | 61.3 (6.6) | 28.6 (6.4) |
| Controls | 430 875 (54) | 57.4 (8.0) | 27.4 (4.8) | |
| USA | EHOA | 145 (82) | 68.9 (12.1) | 27.5 (6.1) |
| Controls | 5308 (60) | 56.3 (18.2) | 29.6 (6.9) | |
| Spain | EHOA | 218 (84) | 61.1 (8.7) | 28.1 (5.3) |
| Controls | 164 (32) | 58.9 (12.6) | 27.5 (4.6) | |
| The Netherlands | EHOA | 139 (82) | 64.3 (8.4) | 27.5 (4.7) |
| Controls | 5102 (53) | 54.9 (18.2) | 25.2 (4.0) |
BMI, body mass index; EHOA, erosive hand osteoarthritis.
We found four independent associations which satisfied our GWS criteria (table 2, online supplemental table S1, figure 1 and online supplemental material): rs17013495 (4q22.1, between SPP1 and MEPE), rs11243284 (6p24.3), rs1800801 in 5’UTR of MGP (12p12.3) and rs11631127 (15q21.3, in ALDH1A2).
Table 2.
Genome wide significant associations with erosive hand osteoarthritis
| Variant | Chr:position | EA/NEA | Freq% | Closest gene | VA | P value | OR |
| rs17013495 | 4:87 885 460 | T/C | 59.6 | SPP1/MEPE | Intergenic | 8.40E-14 | 1.40 (1.28, 1.53) |
| rs11243284 | 6:8 945 086 | C/T | 28.9 | Intergenic | 4.20E-11 | 1.35 (1.23, 1.48) | |
| rs1800801 | 12:14 885 854 | T/C | 37.2 | MGP | 5'UTR | 3.60E-13 | 1.37 (1.26, 1.49) |
| rs11631127 | 15:57 977 811 | C/G | 57.6 | ALDH1A2 | Intron | 7.10E-18 | 1.46 (1.34, 1.59) |
Results are shown from the meta-analysis of the Icelandic, Dutch, Spanish, UK and US sets. Results for individual sample sets are shown in online supplemental table S1. Chr is chromosome, Pos is the position in build GRCh38, EA designate the effect allele (EA) and NEA the other allele (non effect allele). Freq. is the allelic frequency of the effect allele. Gene refers to the nearest gene and VA is variant annotation. 5'UTR is the 5 prime untranslated region. P values are two sided and derived from a likelihood ratio test.
Figure 1.
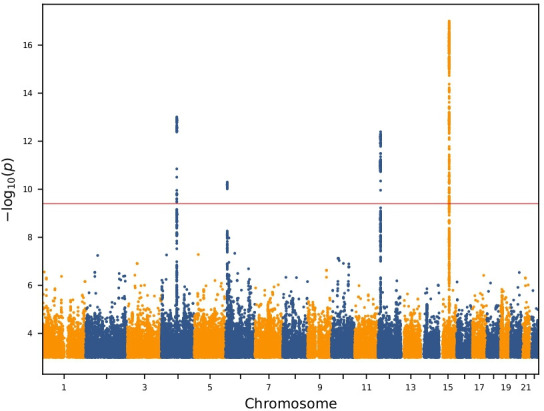
Manhattan plot of the genome-wide analysis of erosive hand osteoarthritis The p values (−log10) are plotted against their respective positions on each chromosome. Results are shown for all variants with significance level p<0.001 and imputation information greater than 0.8.
ard-2022-223468supp001.pdf (199.2KB, pdf)
The associations at MGP and ALDH1A2 have previously been reported for hand OA,9–11 whereas rs17013495 (SPP1/MEPE) and rs11243284 (6p24.3) have not, nor with any other forms of OA. Rs11243284 at 6p24.3 is not correlated with the recently identified association of rs12190551 with spine OA (r2=0.002).27
Rs1800801 in the 5’ UTR in MGP associated stronger with EHOA under a recessive model (OR=1.85 (95% CI 1.59 to 2.14), p=3.7 × 10−16), than under an additive/multiplicative model (OR=1.37 (1.26, 1.49), p=3.6 × 10−13) (online supplemental table S2). In the full genotype model, which assesses risk of heterozygous and homozygous genotypes compared with the homozygous wild-type, the OR for the heterozygotes (TC) was smaller than expected for the additive model, ORhet=1.15 (1.00, 1.32), p=0.047, while the OR for the homozygotes (TT) was larger, ORhom=2.01 (1.71, 2.37), p=1.1 × 10−16. The full model fits significantly better than the additive model for rs1800801 (p=0.0011) (online supplemental table S2), demonstrating the recessive nature of this association. As opposed to the association of rs1800801 with EHOA, the association of rs1800801 with hand, finger and thumb OA was consistent with the additive model rather than the recessive model (online supplemental table S3).
For the other three EHOA-associated variants, we did not observe deviation from the additive/multiplicative model for the genotype risk (online supplemental table S2).
Functional annotation of the EHOA-associated variants
We annotated the EHOA variants according to location in ENCODE’s encyclopaedia of cCRE,22 their tissue specificity,23 co-localisation with mRNA expression (eQTL) in various tissues and co-localisation with protein expression (pQTL) in plasma. We specifically note that bone, cartilage or other joint tissues are not available for eQTL/pQTL analysis in any public dataset.
The EHOA-associated variants (the lead variant or highly correlated variants, r2>0.8) at all four loci reside in enhancer-like sequences (online supplemental table S4), and the variants at MGP and ALDH1A2 also overlap with promoter-like sequences, suggesting a regulatory role of these variants in expression of nearby genes. The 12p12.3 (MGP), 15q21.3 (ALDH1A2) and 4q22.1 (SPP1/MEPE) signals are in cCREs found in many different tissues, whereas the 6p24.3 signal is restricted to few tissue types (online supplemental table S5), possibly suggesting tissue specific activity. Consistent with this observation we found co-localisation of the EHOA variants and/or mRNA expression or protein levels in plasma, at three of the loci: SPP1 at 4q22.1, MGP at 12p12.3 and ALDH1A2 at 15q21.3 (online supplemental table S6 and S7). MGP and ALDH1A2 are also predicted target genes in the EpiMap resource28 (online supplemental table S8). Furthermore, all of the four EHOA loci are within tissue-specific regulatory regions for vascular/endothelial cells which we estimate is 2.8-fold higher than expected by chance alone (expected overlap=35%; 95% CI 0% to 75%), but, as we tested for enrichment within 16 different tissue-specific groups,23 the enrichment was only nominally significant (p=0.011, online supplemental table S9).
The EHOA risk allele of rs11631127 co-localised with reduced expression of ALDH1A2 in cultured fibroblasts (online supplemental table S6), consistent with previous results in cartilage and other joint tissues,9 29 and rs1800801[T] in MGP co-localised with both reduced MGP eQTL (online supplemental table S6) in several tissues and with reduced matrix Gla protein (encoded by the MGP gene) pQTL in plasma (online supplemental table S7), also consistent with previous results.10 12 28 30 Since the MGP gene is expressed at a very low level in blood cells the protein in plasma primarily comes from other tissues. Furthermore, in our data, an increased plasma level of matrix Gla protein associated with lower odds of EHOA (OR=0.75 per SD, p=0.028, Nerosive=55, Ncontrols=27 083, online supplemental figure S2).
Rs17013495[T] at the 4q22.1 locus co-localised with reduced mRNA expression of the SPP1 gene in spleen (online supplemental table S6), and associated with decreased level of osteopontin (encoded by the SPP1 gene) in plasma (online supplemental table S7), although not the strongest cis-pQTL for this protein in plasma. Increased levels of bone sialoprotein 2, encoded by the IBSP gene at the 4q22.1 locus, associated with reduced odds of EOHA (OR=0.74 per SD, p=0.023, online supplemental figure S2), although pQTL or eQTL for this gene did not co-localise with the EHOA variants. However, we note that expression of the IBSP gene is mostly restricted to bone and cartilage, tissues without public eQTL/pQTL datasets.
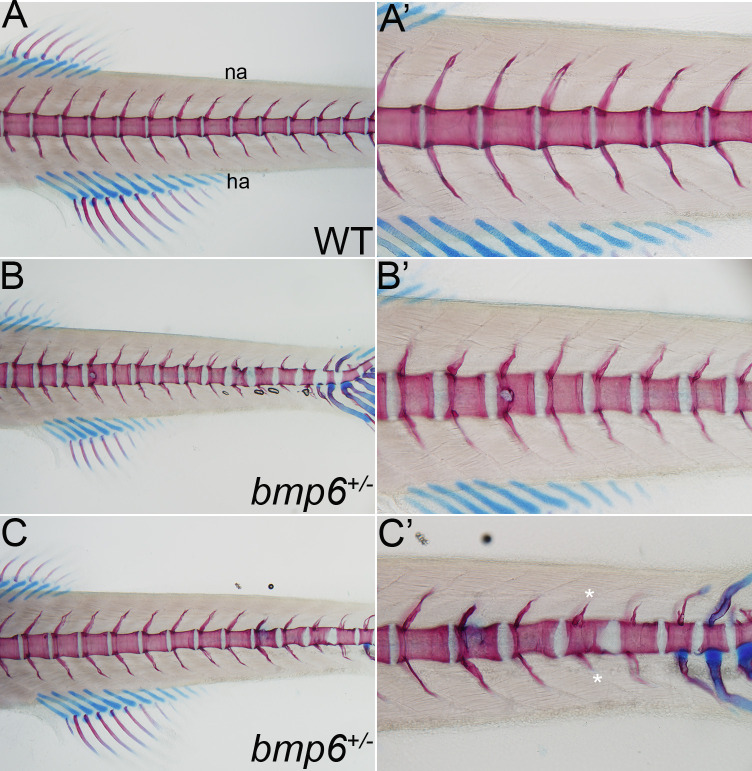
We did not detect eQTLs or pQTLs at the 6p24.3 locus. However, of the nine genes within 1.5 MB of rs11243284, BMP6 is the most likely candidate gene because of the known role of the BMP signalling pathway in skeletal formation and homeostasis.31–33 To uncover biological functions of BMP6 in vivo, we examined the consequences of complete loss of bmp6 function in the zebrafish. We used CRISPR-Cas9 methods to generate F0 and germline deletions of bmp6 (online supplemental figure S1). WT and bmp6+/ − have a normally segmented vertebral column indicating that Bmp6 does not affect the overall development or patterning of the larval skeleton (figure 2 and online supplemental figure S3). In contrast to WT or control larvae, bmp6+/− have multiple defects reminiscent of EHOA, including bone erosions, structural defects in the vertebral precursors and ectopic cartilage formation. These data support that BMP6 is a strong candidate gene in EHOA.
Figure 2.
Loss of bmp6 causes erosive-like phenotypes in the zebrafish vertebral precursors. (A–C’). Analysis of cartilage (blue) and bone (red) in the vertebral column of 14 days post fertilisation wild-type (WT) and bmp6+/- zebrafish larvae. (A, A’) WT larvae have a normally segmented and ossified centra (vertebral precursors) and neural (na) and hemal arches (ha), whereas (B, C’) bmp6+/ − have multiple defects, including bone erosions (arrow in B and B’), structural defects in the centra (arrowhead in B, C and C’), ectopic cartilage formation (arrow in C’), and disruptions in the neural and hemal arches (asterisks in C’). No defects are observed in the cartilaginous structures of the fins. All images are lateral views with anterior to the left.
All the above genes (ALDH1A2, MGP, BMP6, SPP1 and IBSP) are expressed in human cartilage,34 with relative expression from the 0.01st percentile (MGP) to the 12th percentile (BMP6).
Association of EHOA variants with other OA subtypes and relevant diseases or traits
To address association of the four EHOA variants with other OA subtypes and other diseases or traits, we used data from the GO consortium12 and public datasets (Open Targets Genetics and UK Biobank data). Furthermore, we generated EHOA PRS to run a non-hypothesis driven scan for genetic overlap with other diseases/traits in UK Biobank, and subsequently, assessed the genetic component shared by EHOA and other traits with LD score regression.
All four EHOA variants associated with finger OA in the GO consortium data (PBonferroni <0.0025) but with considerably lower OR estimate than for EHOA (table 3). All EHOA variants, except rs11243284 at 6p24.3, also associated nominally with thumb OA. Of special note is the opposite effect of rs1800801 (MGP) and rs11631127 (ALDH1A2) on knee OA compared with EHOA, that is, the EHOA risk alleles associated with reduced risk of knee OA, consistent with what was also observed in the GO consortium meta-analysis.12 None of the EHOA variants associated with spine OA.
Table 3.
Association of the four EHOA variants with other osteoarthritis in the GO consortium meta-analysis
| Finger OA (N=10 804 cases/255 814 controls) |
Thumb OA (N=10 536 cases/236 919 controls) |
Hip OA (N=36 520 cases/317 590 controls) |
Knee OA (N=63 498 cases/335 777 controls) |
Spine OA (N=28 731 cases/307 798 controls) |
|||||||
| Variant (allele) | Chr:position | OR | P value | OR | P value | OR | P value | OR | P value | OR | P value |
| rs17013495(T) | chr4:87 885 460 | 1.08 | 2.3E-05* | 1.05 | 7.9E-03* | 0.99 | 0.16 | 1.00 | 0.58 | 1.01 | 0.32 |
| rs11243284(C) | chr6:8 945 086 | 1.10 | 1.3E-06* | 1.00 | 0.85 | 1.00 | 0.84 | 1.00 | 0.59 | 0.99 | 0.25 |
| rs1800801(T) | chr12:14 885 854 | 1.16 | 8.6E-16* | 1.06 | 2.5E-04* | 0.97 | 5.5E-03 | 0.98 | 2.3E-03* | 1.01 | 0.27 |
| rs11631127(C) | chr15:57 977 811 | 1.09 | 3.7E-07* | 1.10 | 1.3E-08* | 1.02 | 0.079 | 0.97 | 1.3E-06* | 1.00 | 0.64 |
Results are shown for OA subsets phenotypes in the Genetics of Osteoarthritis Consortium meta-analysis.12 Chr is chromosome, Pos is the position in build GRCh38.
*Denotes significant associations after correction for multiple testing.
EHOA, erosive hand osteoarthritis; GO, genetics of osteoarthritis; OA, osteoarthritis.
Three of the EHOA signals, rs17013495 (SPP1), rs1800801 (MGP) and rs11631127 (ALDH1A2), showed some multitrait associations, although mostly with musculoskeletal measures; hand grip strength and bone density (online supplemental table S10). No disease or trait, except for OA, was shared by two or more of the EHOA loci. Of note is association of rs17013495[C] with increased levels of urate and risk of gout, another form of arthritis caused by uric acid crystal deposition, but severe gout can also result in bone erosions. Follow-up of these observations for all four EHOA variants in UK Biobank data and in our meta-analysis of bone density shows an association of all four EHOA risk alleles with reduced grip strength (online supplemental figure S4), but only rs17013495 (SPP1/MEPE) associated with urate (online supplemental table S11). All four EHOA variants also associated nominally with lumbar spine bone mineral density (LS-BMD), but the direction of effects was not consistent between the four variants. Only rs1800801 (MGP) associated with BMD estimated with heel ultrasound (eBMD).
Consistent with the above-described observations, the EHOA PRS scan was only significant (p<1.0 × 10−5, accounting for 5000 main phenotypes) for hand OA measures, other arthrosis diagnosis (ICD10:M19), polyarthrosis (ICD10:M15), pain due to OA and hand grip strength (online supplemental table S12).
We estimated the extent of shared genetics between EHOA and the other OA subtypes and the traits identified in the phenoscans through genetic correlation analysis. Although, not identified in the multitrait associations analysis nor in the PRS scan, we also included RA in this analysis because that it is another form of inflammatory arthritis that can result in bone erosions and is, as gout, a clinical differential diagnosis to EHOA.
We observed highest genetic correlation between EHOA and those types of OA of which EHOA is a subset, that is, finger OA and hand OA, followed by thumb OA, knee OA, hip OA and the weakest with spine OA (figure 3). Reduced grip strength, increased urate levels and gout were also nominally correlated genetically with EHOA, whereas measures of bone density and RA were not.
Figure 3.
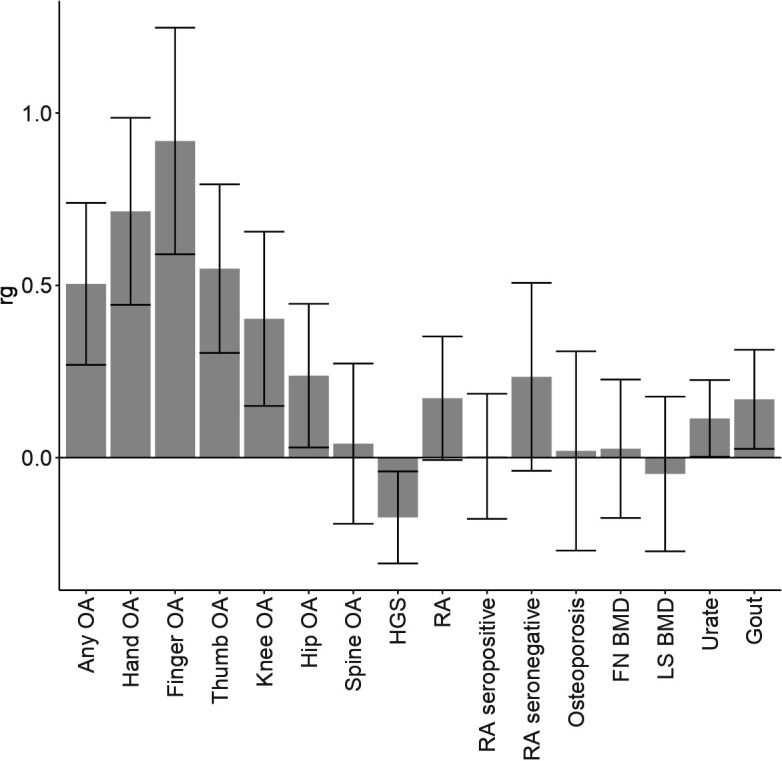
Genetic correlation between EHOA and other OA subtypes and diseases/traits The genetic correlation coefficient (rg) and SE, of genetic correlation between EHOA and other OA subtypes, any OA (which includes all types of OA), and several diseases/traits are shown. HGS is hand grip strength, FN_BMD is femoral neck bone mineral density, LS_BMD is lumbar spine bone mineral density, and RA is rheumatoid arthritis. BMD, bone mineral density; EHOA, erosive hand osteoarthritis; LS-BMD, lumbar spine BMD; OA, osteoarthritis.
The extent of genetic correlation between EHOA and other OA types is also reflected by the associations of GO consortium variants with EHOA12 (online supplemental table S13). Eight of the 10 GO independent associations with hand, finger or thumb OA, associated with EHOA under a false discovery rate of 5% in our data, whereas only 3 of the remaining 68 independent knee, hip, spine or any OA variants did so. The small sample size of our EHOA dataset may not be powered to detect associations with these variants, however, similar results were also observed for direct comparison of the ORs of EHOA and the other OA subsets, irrespective of the significance of the association (online supplemental figure S5). We note that as for the EHOA variants reported here, a majority of the finger/hand OA variants associated with EHOA with larger ORs than with finger/hand OA in the GO data, indicating that EHOA is a severe subset of finger OA.
Discussion
Here, we describe the first GWAS of EHOA. Despite a modest sample size of 1484 cases, we found 4 significant EHOA loci, all of which confer relatively high effect on EHOA risk.
Two of the associated loci, rs1800801 (MGP) and rs11631127 (ALDH1A2), have previously been associated with hand OA overall.9 10 12 Both of these loci also associated with knee OA with opposite effects to that of EHOA, that is, the EHOA risk alleles associate with protection of knee OA.12 The EHOA risk alleles at these loci co-localise with lower mRNA expression of ALDH1A2 and MGP in cartilage, other joint tissues as well as some other tissues,9 10 12 29 30 35 and the rs1800801 (MGP) risk allele also co-localises with lower levels of matrix Gla protein levels in plasma, indicating that ALDH1A2 and MGP genes are likely EHOA candidate causal genes at these loci.9 10 12 29 30 35 We also show that the matrix Gla protein in plasma is lower in EHOA patients than in controls, further supporting a causal role of the MGP gene in OA, with lower level of protein predisposing to the disease. rs1800801 (MGP) associated with EHOA under a recessive model, whereas the association with finger OA is consistent with an additive model. The matrix Gla protein is a vitamin K dependent inhibitor of ectopic tissue calcification, particularly of vascular and cartilage calcification.36 37 The function of the protein depends on the post-translational Ca++ binding γ-carboxyglutamic acid residues (Gla), mediated by vitamin K, but fully carboxylated form of matrix Gla protein has been shown to be lower in OA cartilage than in normal cartilage.38
We found two association signals for EHOA that have not been associated with OA before, rs17013495 (SPP1/MEPE) and rs11243284 (BMP6). Both variants associated nominally with finger OA in our data, although with lower effect. The SPP1/MEPE locus is a well-known locus for BMD39–41 and the EHOA risk variant also associated with increased LS-BMD in our data. We also observed association with increased levels of urate and risk of gout.
There are strong candidate genes at the SPP1/MEPE locus, that harbours a cluster of five genes that encode the SIBLNG (small integrin-binding ligand N-linked glycoprotein) family of extracellular matrix proteins, three of which are expressed in the relevant tissues of bone and/or cartilage: IBSP (bone sialoprotein 2), MEPE (matrix extracellular phosphoglycoprotein) and SPP1 (osteopontin). SPP1 is expressed in many tissues and cell types whereas expression of IBSP and MEPE is mostly restricted to bone, cartilage and teeth.42–44 We found co-localisation of rs17013495 EHOA risk variant and lower expression of SPP1 in spleen, and association with a secondary pQTL for plasma levels of osteopontin. We also observed lower levels of bone sialoprotein 2 in plasma of EHOA patients. The origin of this protein in plasma is most likely from bone as it constitutes approximately 12% of the non-collagenous proteins in human bone and is not expressed in other tissues than bone and/or cartilage. However, since no dataset is currently available to conduct well-powered eQTL or pQTL studies in joint tissues, the possible causal effect of these genes on EHOA cannot be differentiated at this stage. They all play key biological roles in the mineralisation of bone, form an integral part of the mineralised matrix and are involved in chondrocyte differentiation, bone formation and remodelling.45
The similarities in the bone phenotypes that we observed in the zebrafish bmp6 mutants we created with the clinical hallmarks of EHOA suggests that BMP6, that has a role in maintaining bone and joint homeostasis, is the candidate causative gene at the 6p24.3 locus. Although several studies have examined the function of BMP6 on bone formation, its precise role remains unclear possibly due to functional redundancy of other BMPs or genetic compensation.32 46–48 Recently, a GWAS found that an intronic variant in BMP6, rs12190551[C], uncorrelated with the EHOA signal, associated with spine OA. The spine OA risk allele correlated with reduced expression of BMP6 mRNA in the tibial nerve in the GTEx portal.27 Previous transcriptomic analysis of musculoskeletal tissue from bmp mutants has demonstrated that loss of bmp6 activated the NF-κB pathway, which inhibited development of osteoblasts and promoted osteoclast formation.46 Further, gain-of-function and loss-of-function studies in animal models are needed to delineate the precise role and mechanism of BMP6 function in OA.49
Our phenoscan of over 5000 different diseases and traits in UK Biobank using the EHOA PRS, as well as genetic correlation analysis using LD score regression, indicated that EHOA unsurprisingly shares genetics with different measures of OA, but also with decreased hand grip strength, increased urate concentrations and gout, but not RA. The genetic correlation with other OA subtypes shows, as expected, the most shared genetics between EHOA and finger and hand OA, of which EHOA is a subset. EHOA, gout and RA share the clinical features of joint inflammation, and erosions in the most severe cases of gout and RA. It should also be noted that it can be difficult to differentiate between EHOA and gout, both clinically and radiographically. In contrast to EHOA, there are several effective disease-modifying antirheumatic therapies available for RA that hinder progression to erosive disease, but those have not proven effective against EHOA, also indicating a different underlying pathogenesis.
Here, we describe the first robust loci for EHOA. All four loci conferred relatively high risk of the disease, with one locus, rs1800801 in MGP, associating with EHOA under recessive mode of inheritance with OR=2.0, compared with additive association with finger OA, thus differentiating EHOA from finger OA. All four risk variants associated with lowered hand grip strength. Two of the EHOA variants, rs17013495 (SPP1/MEPE) and rs11243284 (BMP6), only associated with EHOA and/or hand OA, and no other type of OA. Of special note is the opposite effect of rs1800801 in MGP and rs11631127 in ALDH1A2 on knee OA compared with EHOA, that is, the EHOA risk allele of these variants confer protection of knee OA. The likely EHOA candidate genes at these loci implicate roles of cartilage calcification (MGP), vitamin A (ALDH1A2) and bone/cartilage mineralisation/remodelling (BMP6, SPP1/IBSP/MEPE) pathways in EHOA. Moreover, our results support the notion that EHOA is a severe form of hand OA as evident by higher risk of the EHOA and reported hand OA variants in EHOA than of fingers/thumbs OA, as well as high genetic correlation. Our results also indicate some genetic, and or functional or biological, distinction between EHOA and OA in the larger joints, since the EHOA risk alleles either do not confer risk, or confer protection, of OA in these joints, and the lower genetic correlation.
Acknowledgments
We thank the study subjects for their valuable participation. This research has been conducted using the UK Biobank Recourse under application numbers 23359 and 56270.
Footnotes
Handling editor: Josef S Smolen
Twitter: @nacho_rego
Contributors: US, UT, GT, SS, HJ, SS, MJJ and KS designed the study and interpreted the results. DGF, LS, GT, SHL and US coordinated or performed statistical and bioinformatics analyses. OAS performed analyses of regulatory regions. MJJ, NK and KH coordinated, analysed or carried out zebrafish experiments. HJ, US, TR, SS, LAK, MK, TB, CH, FJB, NO, IR-P, MJJ, NK and KN carried out subject recruitment, ascertainment or managed phenotype data. US, UT, GT, SS and MJJ drafted the manuscript. MK, TB and FJB drafted the manuscript for intellectual content. All authors contributed to the final version of the manuscript. US and KS are responsible for the overall content as guarantors.
Funding: The study is sponsored by deCODE genetics/Amgen. The Hostas study is financially supported by the Dutch Arthritis Society, who did not have influence on the design, performance and analysis of the study.
Competing interests: The authors US, LS, GT, OAS, SS, SHL, DFG, UT and KS are affiliated with deCODE genetics/Amgen and declare competing interests as employees. The author MK reports consultancy/lecture fees outside the submitted work from Pfizer, Novartis, UCB, Galapagos, Flexion, Kiniksa, Jansen, Abbvie, CHDR, all paid to institution. Royalties from Wolters Kluwer and Springer Verlag, paid to institution.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
The summary results from the EHOA meta-analysis are available at https://www.decode.com/summarydata. Other data are available on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants. All Icelandic participants who donated samples gave informed consent and the National Bioethics Committee of Iceland approved the study (VSN_14–148, VSN_14–015 v8), which was conducted in agreement with conditions issued by the Data Protection Authority of Iceland. The Dutch subjects gave written informed consent and the study was approved by the Leiden University Medical Center medical ethical committee (CCMO reference NL26201.058.08) and the Institutional Review Board of the Radboud University Medical CenterCentre. All UK participants gave informed consent and UK Biobank’s scientific protocol and operational procedures were reviewed and approved by the North West Research Ethics Committee. US subjects were informed of the study protocol and procedures prior to providing consent and the study was approved by the Institutional Review Board at the University of Utah (IRB#: 79442) and Intermountain Healthcare (IRB#: 1051071). PROCOAC samples belong to the Sample Collection for Research on Rheumatic Diseases authorised by the Galician Research Ethics Committee (CAEIG) with registry code 2013/107 and inscribed in the National Registry of Biobanks - Collections Section (Code C.0000424). The patients have signed an informed consent agreement form prior to collection
References
- 1. Kloppenburg M, Kwok W-Y. Hand osteoarthritis -- a heterogeneous disorder. Nat Rev Rheumatol 2011;8:22–31. 10.1038/nrrheum.2011.170 [DOI] [PubMed] [Google Scholar]
- 2. Marshall M, Watt FE, Vincent TL, et al. Hand osteoarthritis: clinical phenotypes, molecular mechanisms and disease management. Nat Rev Rheumatol 2018;14:641–56. 10.1038/s41584-018-0095-4 [DOI] [PubMed] [Google Scholar]
- 3. Favero M, Belluzzi E, Ortolan A, et al. Erosive hand osteoarthritis: latest findings and outlook. Nat Rev Rheumatol 2022;18:171–83. 10.1038/s41584-021-00747-3 [DOI] [PubMed] [Google Scholar]
- 4. Bijsterbosch J, van Bemmel JM, Watt I, et al. Systemic and local factors are involved in the evolution of erosions in hand osteoarthritis. Ann Rheum Dis 2011;70:326–30. 10.1136/ard.2010.138230 [DOI] [PubMed] [Google Scholar]
- 5. Oreiro-Villar N, Raga AC, Rego-Pérez I. PROCOAC (prospective cohort of A coruñA) description: spanish prospective cohort to study osteoarthritis. Reumatol Clin (Engl Ed) 2020;18:100–4. 10.1016/j.reuma.2020.08.010 [DOI] [PubMed] [Google Scholar]
- 6. Pattrick M, Manhire A, Ward AM, et al. Hla-A, B antigens and alpha 1-antitrypsin phenotypes in nodal generalised osteoarthritis and erosive osteoarthritis. Ann Rheum Dis 1989;48:470–5. 10.1136/ard.48.6.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramonda R, Musacchio E, Campana C, et al. Immunogenetic aspects of erosive osteoarthritis of the hand in patients from northern Italy. Scand J Rheumatol 2011;40:139–44. 10.3109/03009742.2010.507216 [DOI] [PubMed] [Google Scholar]
- 8. Stern AG, de Carvalho MRC, Buck GA, et al. Association of erosive hand osteoarthritis with a single nucleotide polymorphism on the gene encoding interleukin-1 beta. Osteoarthritis Cartilage 2003;11:394–402. 10.1016/s1063-4584(03)00054-2 [DOI] [PubMed] [Google Scholar]
- 9. Styrkarsdottir U, Thorleifsson G, Helgadottir HT, et al. Severe osteoarthritis of the hand associates with common variants within the aldh1a2 gene and with rare variants at 1p31. Nat Genet 2014;46:498–502. 10.1038/ng.2957 [DOI] [PubMed] [Google Scholar]
- 10. den Hollander W, Boer CG, Hart DJ, et al. Genome-Wide association and functional studies identify a role for matrix Gla protein in osteoarthritis of the hand. Ann Rheum Dis 2017;76:2046–53. 10.1136/annrheumdis-2017-211214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boer CG, Yau MS, Rice SJ, et al. Genome-wide association of phenotypes based on clustering patterns of hand osteoarthritis identify wnt9a as novel osteoarthritis gene. Ann Rheum Dis 2021;80:367–75. 10.1136/annrheumdis-2020-217834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boer CG, Hatzikotoulas K, Southam L, et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell 2021;184:6003–5. 10.1016/j.cell.2021.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verbruggen G, Veys EM. Numerical scoring systems for the anatomic evolution of osteoarthritis of the finger joints. Arthritis Rheum 1996;39:308–20. 10.1002/art.1780390221 [DOI] [PubMed] [Google Scholar]
- 14. Damman W, Liu R, Kroon FPB, et al. Do comorbidities play a role in hand osteoarthritis disease burden? data from the hand osteoarthritis in secondary care cohort. J Rheumatol 2017;44:1659–66. 10.3899/jrheum.170208 [DOI] [PubMed] [Google Scholar]
- 15. Wetzels JFM, Kiemeney LALM, Swinkels DW, et al. Age- and gender-specific reference values of estimated GFR in Caucasians: the Nijmegen biomedical study. Kidney Int 2007;72:632–7. 10.1038/sj.ki.5002374 [DOI] [PubMed] [Google Scholar]
- 16. Kazmers NH, Meeks HD, Novak KA, et al. Familial clustering of erosive hand osteoarthritis in a large statewide cohort. Arthritis Rheumatol 2021;73:440–7. 10.1002/art.41520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oreiro-Villar N, Raga AC, Rego-Pérez I, et al. PROCOAC (prospective cohort of a coruñA) description: Spanish prospective cohort to study osteoarthritis. Reumatol Clin (Engl Ed) 2020:S1699-258X(20)30231-X. 10.1016/j.reuma.2020.08.010 [DOI] [PubMed] [Google Scholar]
- 18. MANTEL N, HAENSZEL W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- 19. Sveinbjornsson G, Albrechtsen A, Zink F, et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat Genet 2016;48:314–7. 10.1038/ng.3507 [DOI] [PubMed] [Google Scholar]
- 20. Kong A, Frigge ML, Thorleifsson G, et al. Selection against variants in the genome associated with educational attainment. Proc Natl Acad Sci U S A 2017;114:E727–32. 10.1073/pnas.1612113114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. Ld score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–5. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. ENCODE Project Consortium, Moore JE, Purcaro MJ, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020;583:699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meuleman W, Muratov A, Rynes E, et al. Index and biological spectrum of human DNase I hypersensitive sites. Nature 2020;584:244–51. 10.1038/s41586-020-2559-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014;10:e1004383. 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Styrkarsdottir U, Lund SH, Thorleifsson G, et al. Meta-Analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat Genet 2018;50:1681–7. 10.1038/s41588-018-0247-0 [DOI] [PubMed] [Google Scholar]
- 26. Ferkingstad E, Sulem P, Atlason BA, et al. Large-Scale integration of the plasma proteome with genetics and disease. Nat Genet 2021;53:1712–21. 10.1038/s41588-021-00978-w [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Grant RA, Shivakumar MK, et al. Genome-Wide association analysis across 16,956 patients identifies a novel genetic association between BMP6, NIPAL1, CNGA1 and spondylosis. Spine (Phila Pa 1976) 2021;46:E625–31. 10.1097/BRS.0000000000003880 [DOI] [PubMed] [Google Scholar]
- 28. Boix CA, James BT, Park YP, et al. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 2021;590:300–7. 10.1038/s41586-020-03145-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shepherd C, Zhu D, Skelton AJ, et al. Functional characterization of the osteoarthritis genetic risk residing at aldh1a2 identifies rs12915901 as a key target variant. Arthritis Rheumatol 2018;70:1577–87. 10.1002/art.40545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shepherd C, Reese AE, Reynard LN, et al. Expression analysis of the osteoarthritis genetic susceptibility mapping to the matrix Gla protein gene MGP. Arthritis Res Ther 2019;21:149. 10.1186/s13075-019-1934-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev 2003;24:218–35. 10.1210/er.2002-0023 [DOI] [PubMed] [Google Scholar]
- 32. Gitelman SE, Kobrin MS, Ye JQ, et al. Recombinant vgr-1/BMP-6-expressing tumors induce fibrosis and endochondral bone formation in vivo. J Cell Biol 1994;126:1595–609. 10.1083/jcb.126.6.1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu M, Chen G, Li YP. Tgf-Β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res 2016;4:16009. 10.1038/boneres.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ramos YFM, den Hollander W, Bovée JVMG, et al. Genes involved in the osteoarthritis process identified through genome wide expression analysis in articular cartilage; the raak study. PLoS ONE 2014;9:e103056. 10.1371/journal.pone.0103056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Houtman E, Coutinho de Almeida R, Tuerlings M, et al. Characterization of dynamic changes in matrix gla protein (MGP) gene expression as function of genetic risk alleles, osteoarthritis relevant stimuli, and the vitamin K inhibitor warfarin. Osteoarthritis Cartilage 2021;29:1193–202. 10.1016/j.joca.2021.05.001 [DOI] [PubMed] [Google Scholar]
- 36. Schurgers LJ, Uitto J, Reutelingsperger CP. Vitamin K-dependent carboxylation of matrix gla-protein: a crucial switch to control ectopic mineralization. Trends Mol Med 2013;19:217–26. 10.1016/j.molmed.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 37. Newman B, Gigout LI, Sudre L, et al. Coordinated expression of matrix Gla protein is required during endochondral ossification for chondrocyte survival. J Cell Biol 2001;154:659–66. 10.1083/jcb.200106040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wallin R, Schurgers LJ, Loeser RF. Biosynthesis of the vitamin K-dependent matrix Gla protein (MGP) in chondrocytes: a fetuin-MGP protein complex is assembled in vesicles shed from normal but not from osteoarthritic chondrocytes. Osteoarthritis Cartilage 2010;18:1096–103. 10.1016/j.joca.2010.05.013 [DOI] [PubMed] [Google Scholar]
- 39. Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-Wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 2012;44:491–501. 10.1038/ng.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Medina-Gomez C, Kemp JP, Trajanoska K, et al. Life-course genome-wide association study meta-analysis of total body BMD and assessment of age-specific effects. Am J Hum Genet 2018;102:88–102. 10.1016/j.ajhg.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morris JA, Kemp JP, Youlten SE, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet 2019;51:258–66. 10.1038/s41588-018-0302-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petersen DN, Tkalcevic GT, Mansolf AL, et al. Identification of osteoblast/osteocyte factor 45 (OF45), a bone-specific cDNA encoding an RGD-containing protein that is highly expressed in osteoblasts and osteocytes. J Biol Chem 2000;275:36172–80. 10.1074/jbc.M003622200 [DOI] [PubMed] [Google Scholar]
- 43. Fisher LW, Fedarko NS. Six genes expressed in bones and teeth encode the current members of the sibling family of proteins. Connect Tissue Res 2003;44 Suppl 1:33–40. [PubMed] [Google Scholar]
- 44. Gullard A, Gluhak-Heinrich J, Papagerakis S, et al. MEPE localization in the craniofacial complex and function in tooth dentin formation. J Histochem Cytochem 2016;64:224–36. 10.1369/0022155416635569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malaval L, Aubin JE, Vico L. Role of the small integrin-binding ligand N-linked glycoprotein (sibling), bone sialoprotein (BSP) in bone development and remodeling. Osteoporos Int 2009;20:1077–80. 10.1007/s00198-009-0869-2 [DOI] [PubMed] [Google Scholar]
- 46. Xu H, Tong G, Yan T, et al. Transcriptomic analysis provides insights to reveal the bmp6 function related to the development of intermuscular bones in zebrafish. Front Cell Dev Biol 2022;10:821471. 10.3389/fcell.2022.821471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beederman M, Lamplot JD, Nan G, et al. Bmp signaling in mesenchymal stem cell differentiation and bone formation. J Biomed Sci Eng 2013;6:32–52. 10.4236/jbise.2013.68A1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Solloway MJ, Dudley AT, Bikoff EK, et al. Mice lacking BMP6 function. Dev Genet 1998;22:321–39. [DOI] [PubMed] [Google Scholar]
- 49. Jurynec MJ, Gavile CM, Honeggar M, et al. The NOD/RIPK2 signaling pathway contributes to osteoarthritis susceptibility. Genetics [Preprint] 2022. 10.1101/2022.02.07.479420 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ard-2022-223468supp002.pdf (512KB, pdf)
ard-2022-223468supp001.pdf (199.2KB, pdf)
Data Availability Statement
The summary results from the EHOA meta-analysis are available at https://www.decode.com/summarydata. Other data are available on reasonable request.