Highlights
-
•
70 Human milk oligosaccharides (HMOs) were effectively identified by UPLC-QE-HF-MS.
-
•
14 Bovine milk oligosaccharides (BMOs) were effectively identified by UPLC-QE-HF-MS.
-
•
23 Goat milk oligosaccharides (GMOs) were effectively identified by UPLC-QE-HF-MS.
-
•
The nature and number of OS in human and animal milk were significantly different.
-
•
14 Neutral and 3 acidic oligosaccharides (OS) were reported at first time in rat milk.
-
•
The composition and abundance of RMOs were more similar to that of HMOs.
Keywords: Milk oligosaccharides, Liquid chromatography-Q exactive-HF hybrid quadrupole-Orbitrap-mass spectrometry, Identification, Composition
Abstract
The complex oligosaccharides (OS) in different milk are more difficult to detect and complicated to analyze as their enormous structural complexity. UPLC-QE-HF-MS was supposed to be a highly effective method for OS identification. In present study, 70 human milk oligosaccharides (HMOs), 14 bovine milk oligosaccharides (BMOs), 23 goat milk oligosaccharides (GMOs) and 24 rat milk oligosaccharides (RMOs) were detected by using UPLC-QE-HF-MS, respectively. There were highly differences in number and composition between the four milk OS. 14 neutral and 3 acidic OS were firstly found in rat milk. The composition and abundances of RMOs were might more similar to that of HMOs, comparing with BMOs and GMOs. The similarity between HMOs and RMOs might provide theoretical basis for better application of rats in biological/biomedical studies of HMOs as models. The BMOs and GMOs were expected to be suitable for applications in medical and functional foods as a promising bioactive molecular.
1. Introduction
Milk is a highly nutritional food containing various OS with complex structures. As a class of bioactive molecules, these OS are the third largest component after lactose and lipids in human milk (Lu, Zhang, Song, Zhang, Pang, Sari, et al., 2020). The milk OS play important roles in improving growth and development of newborns (Underwood, Gaerlan, De Leoz, Dimapasoc, Kalanetra, Lemay, et al., 2015). The OS are generally defined as carbohydrate polymers that contain 3 to 10 monosaccharide units covalently linked through glycosidic bonds (N Tao, DePeters, Freeman, German, Grimm, & Lebrilla, 2008), which are divided into (i) neutral OS, whose structures are mainly lactose linked with neutral monosaccharides such as glucose or galactose (Hex), N-acetylglucosamine or N-acetylgalactosamine (HexNAc) and fucose or deoxyhexose (Fuc) and (ii) acidic OS, containing acidic components such as N-acetylneuraminic (NeuAc) also known as sialic acid or N-glycolylneuraminic acid (NeuGc) (Martín-Ortiz, Salcedo, Barile, Bunyatratchata, Moreno, Martin-García, et al., 2016; Meyrand, Dallas, Caillat, Bouvier, Martin, & Barile, 2013).
There are large numbers of bioactive OS in human milk and animal milk, and the quite differences in contents and diversities of OS between human and animal milk are described according to an increasing number of reports (Li, Jiang, Zhou, Ding, Guo, Li, et al., 2021). The human milk contains more OS than non-human mammalian milk and there are more than 200 structures of HMOs have been identified so far (Albrecht, Lane, Marino, Al Busadah, Carrington, Hickey, et al., 2014; Ninonuevo, Park, Yin, Zhang, Ward, Clowers, et al., 2006). Currently, the number of GMOs have been found is up to 78 (Martín-Ortiz, et al., 2016), which is approximately 4 times higher than bovine milk, but the concentration is still much lower than that in human milk (Albrecht et al., 2014, Sousa et al., 2019). There are 50 compositions and 37 structures of BMOs have been determined until now (Aldredge, Geronimo, Hua, Nwosu, Lebrilla, & Barile, 2013). Rats have been used as a common tooling model in various biomedical studies (Dvorak, Halpern, Holubec, Dvorakova, Dominguez, Williams, et al., 2004) and the anatomy, physiology, genetics, basic biology and biochemistry of rats and mice are well-understood (Gosling, 2001), but there are very limited studies on RMOs comparing with HMOs, BMOs and GMOs. Only three acidic OS, including 3′-sialyllactose (3′-SL), 6′-sulphated lactose (6′-Su-L) and 6′-sulphate-3′-sialyllactose (6′-Su-3′-SL), have been identified in rat milk a few decades ago (Carubelli et al., 1961, Choi and Carubelli, 1968, Naccarato et al., 1975). In a recent study, 15 acidic RMOs that containing 9 monosialylated, 2 disialylated, 1 monosulphated, and 3 both monosulphated and monosialylated OS have been detected by a high-sensitivity online solid-phase extraction and hydrophilic interaction chromatography coupled with electrospray tandem mass spectrometry (HPLC-ESI-CID-MS/MS)(Li, et al., 2021). However, the neutral RMOs are still unclear so far. The well-understanding of completely basic information of bioactive RMOs are of great interest to biological/biomedical researches using rats as model.
To date, a large number of studies using different analytical methods to detect or quantify the OS in human and animal milk, such as high performance liquid chromatography (HPLC)(Tadasu Urashima, Asakuma, Leo, Fukuda, Messer, & Oftedal, 2012), microfluidic chips and mass spectrometry (Leo, Asakuma, Fukuda, Senda, & Urashima, 2010), HPLC-mass spectrometry (HPLC-MS)(Austin & Bénet, 2018), high performance anion exchange chromatography-pulsed amperometric detection (HPAEC-PAD)(Wang, Zhou, Gong, Chen, Feng, Liu, et al., 2020), nuclear magnetic resonance (NMR)(Urakami, Saeki, Watanabe, Kawamura, Nishizawa, Suzuki, et al., 2018), capillary electrophoresis(Monti, Cattaneo, Orlandi, & Curadi, 2015), ultra performance liquid chromatography coupled with Q-Exactive Focus mass spectrometry (UPLC-Q-Exactive Focus–MS/MS)(Lu, et al., 2020) and HPLC-ESI-CID-MS/MS (Li, et al., 2021). In recent years, the advanced liquid chromatography coupled with a triple quadrupole mass spectrometer (LC-QQQ-MS/MS) was used to detect and absolute quantify twelve OS in human milk (Zhang, et al., 2022). UPLC-QE-HF-MS was successfully adopted to identify the profiles of HMOs in breast milk of four mothers and fecal OS in the feces of their breast-fed infant in our previous study (Li, Zhou, & Xu, 2023). Although several methods have been reported to successfully identify and analyze OS in human and animal milk, the complex OS in different milk are more difficult to detect and more complicated to analyze due to their enormous structural complexity. The ultra-performance liquid chromatography-Q Exactive-HF hybrid quadrupole-Orbitrap-mass spectrometry (UPLC-QE-HF-MS) is an advance technique with higher resolutional, higher precise, and higher sensitive, compared with other existing methods in some previous reports stated above. The usage of UPLC‐QE‐HF‐MS for analyzing OS in animal milk has not been reported yet. The more accurate information of newly OS were speculated to be found using this technique. There aren't much OS data on animal milk from other species, especially for the rat milk, that show how many different oligosaccharide structures are typically present compared to human milk. More information is required, nevertheless, regarding the structural and analytical properties of oligosaccharides found in the milk of various animals. Meanwhile, the feasibility of the new method UPLC‐QE‐HF‐MS for HMOs identification has been verified in our previous research (Li, Zhou, & Xu, 2023). We supposed that UPLC‐QE‐HF‐MS could also be a highly effective method for animal OS identification, especially for the un-known neutral RMOs, since UPLC-QE-HF-MS has higher resolution than regular triple/four-stage quad or orbitrap system, and the results of identification will be more accurate. In general, the aim of this study was to investigate the composition of OS present in rats, goats, bovine and human milk and compare their differences using UPLC‐QE‐HF‐MS. In this work, we have firstly identified and analyzed the neutral and acidic OS in various milk by using LC-QE-HF-MS, and then comparative analyzed the differences in composition between human milk and animals milk, such as goat milk, bovine milk and rat milk. These complete knowledge of RMOs was regarded as basic information and should be helpful for further biological and biomedical researches using rats as models, especially for fundamental studies on OS that could not be performed in human beings. Furthermore, it was necessary for comprehensively comparing the distinctions of OS among human milk and animal milk so as to provide the theoretical basic for better understanding the potential values of animal milk as commercially nutritional substitutes for mother’s milk in food industry. Meanwhile, it was of great significance to clarify the composition of RMOs and their similarities or differences with HMOs as for further exploring the mechanism of healthy effects of OS.
2. Materials and methods
2.1. Reagents and materials
The rat milk sample was collected from mature breast rats in Health Science Center of Peking University. The bovine milk and goat milk were obtained from Lvneng farms (Shanxi, China). The puerperal women who underwent routine postpartum examination at postnatal 42 days were recruited in Maternal and Child Health Care Hospital of Changping District, Beijing, from January 1st, 2021 to June 30th, 2021. The inclusions were as follows: being with no diabetes, hypertension, abnormal thyroid function, abnormal liver function, abnormal kidney function and other diseases, no mastitis and other breast related diseases, no serious genetic defects and mental diseases; no history of long-term use of antibiotics; agreeing to participate in this study and signing the informed consent. 10 mL breast milk was collected with a sterile electric breast pump, and the nipple and areola were disinfected before collection. After collection, all milk samples were mixed together (n ≥ 5 in each group) and stored at −80℃ for further use. Methanol, methanoic acid, acetonitrile and ammonium formate were purchased from Beijing Chemical Reagent Co. (China). All reagents were of analytical grade or chromatographic grade.
2.2. Sample preparation
The methods of sample preparation were performed according to a previously published method (Lu, et al., 2020) and with some modifications. Samples were centrifuged at 10,000 g for 30 min at 10 °C to remove the upper fat. Two volumes of methanol were subsequently added and incubated at 4 °C for 2 h for removing the whey protein. The mixture solution was centrifuged at 4000 g for 30 min at 4 °C for complete phase separation. The supernatant contains OS fraction was carefully transferred to a new tube for later analysis.
2.3. OS detection by UPLC-QE-HF-MS
In current research, an ACQUITY UPLC BEH Amide Column (2.1 mm × 100 mm, 1.7 μm) was used to separate OS on Waters system. The basic parameters were listed as follows: The column temperature: 35 ℃; Flow rate: 0.3 mL/min; Mobile phase A: acetonitrile; Mobile phase B: 10 mM ammonium formate; Gradient elution condition: 0–20 min 95 %-78 % A; 20–35 min, 78 %-73 % A; 35–38 min, 73 %-62 % A; 38–45 min, 62 %-50 % A; The injection volume: 10 μL.
The OS was determined by UPLC-QE-HF-MS with an electrospray ionization (ESI) source. The mass spectrum conditions included the heated capillary of 320 ℃, the spray voltage was 3.8 kV in positive mode and 3.4 kV in negative mode, the curtain gas was at 35 psi, collision-activated dissociation was at medium. Each ion is scanned based on optimized declustering voltage and collision energy during detection. The analysis was operated with full scan (m/z 300–2000) in positive and negative mode for all OS. The matching and analysis of all possible OS was performed according to JCGG database (https://jcggdb.jp/idb/indexList.do?id=inchikey) and OS database (This database is a self-built database and the data was obtained from the literatures). The matching degree between the actual molecular weight and the theoretical molecular weight is controlled within 5 ppm. The mass range for MS was set to m/z 400–2000. The difference of retention time (RT) less than 0.5 min was used as a repeating substance.
3. Results and discussion
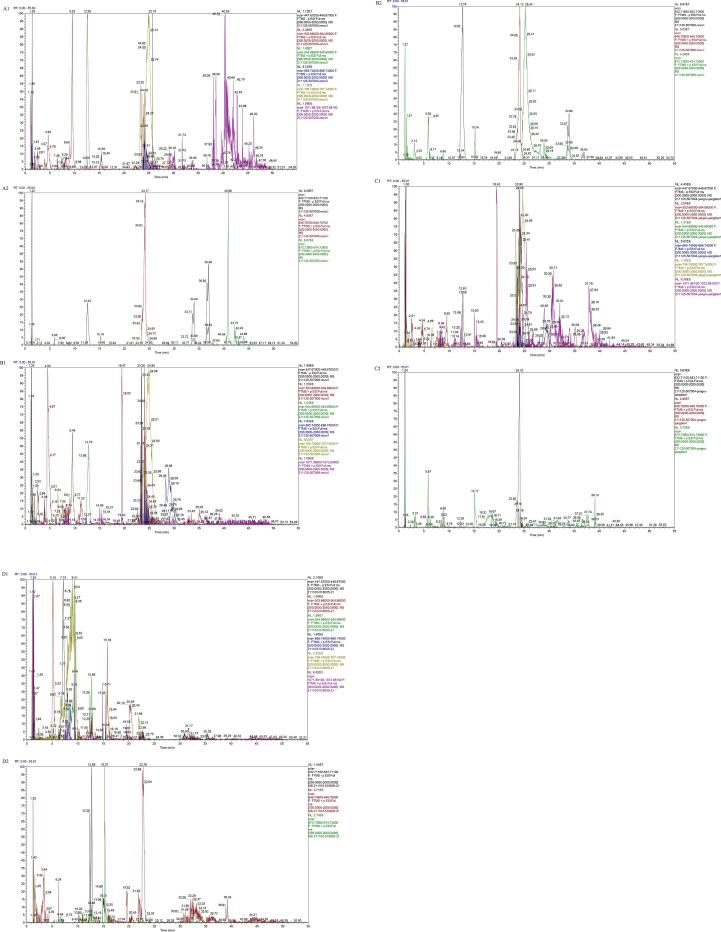
There were approximately 40 HMOs standards commercially available at the different suppliers available for determination of HMOs, but these standards were still insufficient for quantifying hundreds of founded HMOs. Therefore, the advanced UPLC-QE-HF-MS technique was applied for identifying the large number of OS in human, bovine, goat and rat milk. The total ion chromatogram (TIC) of different samples were shown in Fig. 1. The neutral OS and acidic OS at each peak of Fig. 1 were analyzed by two databases as seen in Table 1 and Table 2, each MS/MS spectra of LC fractions of OS in Table 1, Table 2 can be found in Supplementary Fig. S1 and S2, respectively.
Fig. 1.
Total ion chromatogram of OS from (A1&A2) Human milk, (B1&B2) Bovine milk, (C1&C2) Goat milk and (D1&D2) Rat milk.
Table 1.
Neutral OS identified and their peak area in various milk.
 |
 |
 |
 |
 |
 |
 |
 |
Table 2.
Acidic OS identified and their peak area in various milk.
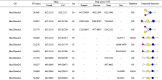 |
 |
 |
 |
 |
3.1. Identification and analysis of neutral OS in various milk
As seen in Table 1, Hex2HexNAc1Fuc1, Hex3HexNAc1dHex1, Hex4HexNAc2dHex1, Hex3HexNAc3dHex1, Hex5HexNAc3, Hex4HexNAc2dHex3, Hex5HexNAc2dHex2, Hex5HexNAc3dHex1, Hex5HexNAc3dHex2 and Hex6HexNAc4 were exclusively detected in human milk. Except bovine milk, the Hex2Fuc1 at m/z 488.1741 and its isomers had been detected in other three milks. All the milk samples contained Hex3, Hex4 and Hex2HexNAc1 and their isomers. Hex1HexNAc1Fuc1 at m/z 529.2007 and its isomers were detected in both human and goat milk. Hex2Fuc1, known as fucosyllactose, had the highest abundance in HMOs in present study, which was followed by Hex2NeuAc1 and Hex3HexNAc1. The finding was in consistent with previous reports that Hex2Fuc1 (m/z 488.1741) was one of the most abundant OS in human milk (Moreno & Sanz, 2014). It was worth noting that Hex3HexNAc1 (m/z 707.2484) and Hex4HexNAc2 (m/z 1072.3810) had been proved to have prebiotic activity, which can be preferentially consumed by some Bifidobacterium strains (LoCascio, Ninonuevo, Freeman, Sela, Grimm, Lebrilla, et al., 2007). As the third most abundant OS in HMOs, Hex3HexNAc1 at m/z 707.2484 and its isomers was found in human milk, but there was a slight amount of Hex3HexNAc1 present in rat milk. Hex4HexNAc2 at m/z 1072.3810 was also detected either in human milk or rat milk with a small abundance. However, Hex3HexNAc1 and Hex4HexNAc2 were not observed in bovine and goat milk. Hex1HexNAc1Fuc1 (m/z 529.2007), Hex4 (m/z 666.2219), Hex3HexNAc3dHex1 (m/z 1259.4650), Hex3HexNAc8 (m/z 2128.8042) and their isomers have not been detected in human milk in a previous study (Porfirio, Archer-Hartmann, Moreau, Ramakrishnan, Haque, Kirkpatrick, et al., 2020). Of all the 5 neutral OS identified in bovine milk of this study had the same compositions with HMOs. Hex4 was first found in human milk (Table 1). 6 fucosylated OS were identified in 23 GMOs and 3 fucosylated OS were detected in 24 RMOs. The most abundant OS in BMOs and GMOs was Hex4. All the neutral GMOs described in present work are reported in other studies (Lu et al., 2020, Martín-Ortiz et al., 2016) and also had the same compositions with HMOs. 11 of 14 neutral OS identified in rat milk were present in human milk. Hex3dHex (m/z 814.2953), Hex6 (m/z 1008.3379), Hex3HexNAc4dHex3 (m/z 1754.6602) were only observed in rat milk (Table 1). Hex3dHex2 and Hex3HexNAc4dHex3 were of particular interest that they were for the first time identified in milk. All the neutral OS were reported for the first time in rat milk.
3.2. Identification and analysis of acidic OS in various milk
Table 2 showed the full list of acidic OS at each peak which analyzed by two databases. The proportion of identified acidic OS in human milk was 22.39 %. Among the acidic HMOs detected, Hex1HexNAc1NeuAc1, Hex2NeuAc1dHex1, Hex5HexNAc3NeuAc1, Hex5HexNAc4NeuAc2 and Hex5HexNAc4NeuAc1dHex2 were newly identified in human milk comparing with other literature (Porfirio, et al., 2020). Hex2NeuAc1dHex1, Hex3HexNAc1NeuAc1dHex1 and some complex OS with large mass (greater than1000), such as Hex4HexNAc1NeuAc1, Hex3HexNAc1NeuAc2, Hex4HexNAc2NeuAc1, Hex3HexNAc1NeuAc2dHex1, Hex4HexNAc2NeuAc1dHex1, Hex4HexNAc2NeuAc1dHex1, Hex4HexNAc2NeuAc2, Hex4HexNAc2NeuAc2, Hex5HexNAc3NeuAc1, Hex5HexNAc3NeuAc1dHex1, Hex5HexNAc3NeuAc2, Hex5HexNAc3NeuAc1dHex2, Hex6HexNAc4NeuAc1, Hex5HexNAc3NeuAc2dHex1, Hex6HexNAc4NeuAc1dHex1, Hex5HexNAc4NeuAc2 and Hex5HexNAc4NeuAc1dHex2 were exclusively detected in human milk. It was particular Hex1HexNAc1NeuAc1 at m/z 674.2383, which is described in animal (e.g. bovine and goat) milk (Lu, et al., 2020; Sunds, Bunyatratchata, Robinson, Glantz, Paulsson, Leskauskaite, et al., 2021). Hex3HexNAc2 (m/z 692.2487) and Hex3NeuAc2 (m/z 1086.3600) were only present in bovine milk, as well as Hex2HexNAc1NeuAc1, Hex2HexNAc1NeuAc2 and their isomers were only present in rat milk. Hex1HexNAc1NeuGc1 (m/z 690.2331), Hex3NeuGc1 (m/z 811.2594), Hex2NeuAc1NeuGc1 (m/z 940.3020) and Hex2NeuGc2 (m/z 956.2969) and their isomers had only been identified in goat milk in present study. All the milk samples contained Hex2NeuAc1 and their isomers with m/z of 633.2116. The Hex2NeuAc1 was absolutely quantified as 3′-SL or its isomer 6′-SL in other reports (Lu et al., 2020, Macias Rostami et al., 2014), which was the most abundant acidic OS in human, goat, bovine and rat milk, and the results were in agreement with some previous studies (Barile, Marotta, Chu, Mehra, Grimm, Lebrilla, et al., 2010; Li et al., 2021, Lu et al., 2020; N Tao, DePeters, Freeman, German, Grimm, & Lebrilla, 2008). Hex1HexNAc1NeuAc1 at m/z 674.2383 and its isomers were identified in three milk samples except rat milk. Hex2NeuGc1 at m/z 649.2065 and its isomers were detected in both rat and goat milk, which was similar to Hex3NeuAc1 (m/z 795.2645) and Hex2NeuAc2 (m/z 924.3070), that were detected in both bovine and goat milk. Hex2NeuGc1, Hex3NeuGc1, Hex2NeuAc1NeuGc1 and Hex1HexNAc1NeuGc1 can be found in bovine milk previously (N Tao, DePeters, Freeman, German, Grimm, & Lebrilla, 2008), whereas these OS had not been identified in bovine milk in this work. Up until now, merely 15 acidic OS were identified in rat and mouse milk, which included three sulphated OS (Li, et al., 2021). However, there was no sulphated OS detected in current study. Among these 24 RMOs identified, 3 acidic OS, including Hex2NeuGc1 and its isomer and Hex2HexNAc1NeuAc2 were reported for the first time in rat milk. Noticeably, Hex2HexNAc1NeuAc1 (m/z 836.2910) was only detected in rat milk in present study, which have been identified in bovine and goat milk (Lu, et al., 2020; N Tao, DePeters, Freeman, German, Grimm, & Lebrilla, 2008). Hex3HexNAc1NeuAc1 (m/z 998.3438) was only detected in both human and rat samples in current research, which can be detected in bovine and goat milk previously (Barile et al., 2010, Lu et al., 2020). Hex2HexNAc1NeuAc2 at m/z 1127.3863 was particularly interest that it was first identified in rat milk, which is described in bovine milk before (Sunds, et al., 2021).
3.3. Comparative analysis of OS in various milk
In the present investigation, 70 HMOs, 14 BMOs, 23 GMOs and 24 RMOs were detected in human and animal milks using UPLC-QE-HF-MS technique, respectively. 45 neutral and 25 acidic OS were identified in human milk, 5 neutral and 9 acidic OS were found in bovine milk, 9 neutral and 14 acidic OS were detected in goat milk and RMOs include 14 neutral and 12 acidic OS. By analyzing the mass of OS in Table 1, Table 2, the neutral and acidic OS from bovine, goat and rat milk were mainly of short chain length comparing with HMOs, which was in agreement with a previous study (Albrecht, et al., 2014). It was obvious that the nature and number of OS were different between human and animal milk. The analysis of this study showed that many OS have relatively large amounts of their isomers. The abundance (peak area) and content of total, acidic and neutral OS in different milk were shown in Table 3. In current research, the numbers and abundance of HMOs were much higher than that of BMOs, GMOs and RMOs (Table 3). The concentrations and numbers in milk of most farm animals including cows, goats, sheep and pigs are much lower than that in human milk (Nannan Tao, Ochonicky, German, Donovan, & Lebrilla, 2010), which was in agreement with the results of present study. There were more than 200 different OS have been separated and described by HPLC-MS and at least 162 structures of HMOs have been determined (T. Urashima, Hirabayashi, Sato, & Kobata, 2018). The various analytical techniques, OS extraction methods, the genetic variation and decreased number and concentration of glycans during the course of lactation made the qualitative and quantitative composition of OS in animal milk variable and complex (Albrecht et al., 2014, Barile et al., 2010; N Tao, DePeters, German, Grimm, & Lebrilla, 2009). The number and composition of HMOs, BMOs and GMOs had been extensively studied before, and in current study, the number of OS identified in different milk samples were lower than previous studies. 71 HMOs, 14 BMOs and 23 GMOs have been detected in this work, which are much lower than 115, 40 (N Tao, DePeters, German, Grimm, & Lebrilla, 2009) and 64 (Lu, et al., 2020), respectively. These findings might be concerning with the applied databases, analytical techniques and the samples. The number of neutral OS was more than that of acidic OS in human and rat milk. There were more types of acidic OS than that of neutral OS in bovine and goat milk. As shown in Table 3, 77.61 % of HMOs was neutral OS, which was similar with the results reported by previous study (Albrecht, et al., 2014). Among all the milk samples investigated, human milk contained the highest percentage and the most abundant variety of neutral OS. The abundances of neutral OS were much higher than that of acidic OS in human, bovine and goat milk as depicted in Table 3. The content of acidic OS (59.47 %) was more than that of neutral OS (40.53 %) in rat milk (Table 3). To our knowledge, the acidic OS are the major components among the OS in animal milk (Li, et al., 2021). And the findings of this study were highly in contrast to previous studies that indicated the BMOs and GMOs contain higher percentage of acidic OS than neutral OS (Albrecht et al., 2014, Lu et al., 2020). It is particularly that a higher percentage of neutral OS observed to shift as time goes, especially for the enormous differences between colostrum and mature milk (Barile, et al., 2010). The controversial results of the abundance of BMOs and GMOs observed in present study might be result from the use of mature milk and the different variety of the animal. Furthermore, it was worth noting that the number and total abundance of HMOs and RMOs were greater than that of both BMOs and GMOs (Table 3). In present study, a total of 24 RMOs were detected, of which 14 neutral OS and 12 acidic OS. The rats share 90 % of the genome with humans (Dvorak, et al., 2004). In present work, 9 BMOs (included 5 neutral and 4 acidic OS), 11 GMOs (included 7 neutral and 4 acidic OS) and 13 RMOs (included 9 neutral and 4 acidic OS) had the same composition with HMOs. The composition and abundance of RMOs come more closest to that of HMOs comparing with BMOs and GMOs (Table 3), the results were in consistent with previous study reported that rat and mouse share more common OS with human when comparing with animals (such as cow, goat and sheep)(Li, et al., 2021). The similarity of composition and abundance between RMOs and HMOs might because of the dominance by gene in terms of evolution, which was deserved to be studied in the future.
Table 3.
The abundance (peak area) and relative content of total, acidic and neutral OS in different milk.
| Milk | Total OS | Neutral OS | Acidic OS | ||
|---|---|---|---|---|---|
| Abundance (×106) | Abundance (×106) | Content (%) | Abundance (×106) | Content (%) | |
| Human | 58630.9695 | 45500.6790 | 77.61 | 13130.2905 | 22.39 |
| Bovine | 15330.8885 | 10834.0522 | 70.67 | 4496.8363 | 29.33 |
| Goat | 12069.9302 | 8348.9513 | 69.17 | 3720.9789 | 30.83 |
| Rat | 46279.2930 | 18755.7231 | 40.53 | 27523.5699 | 59.47 |
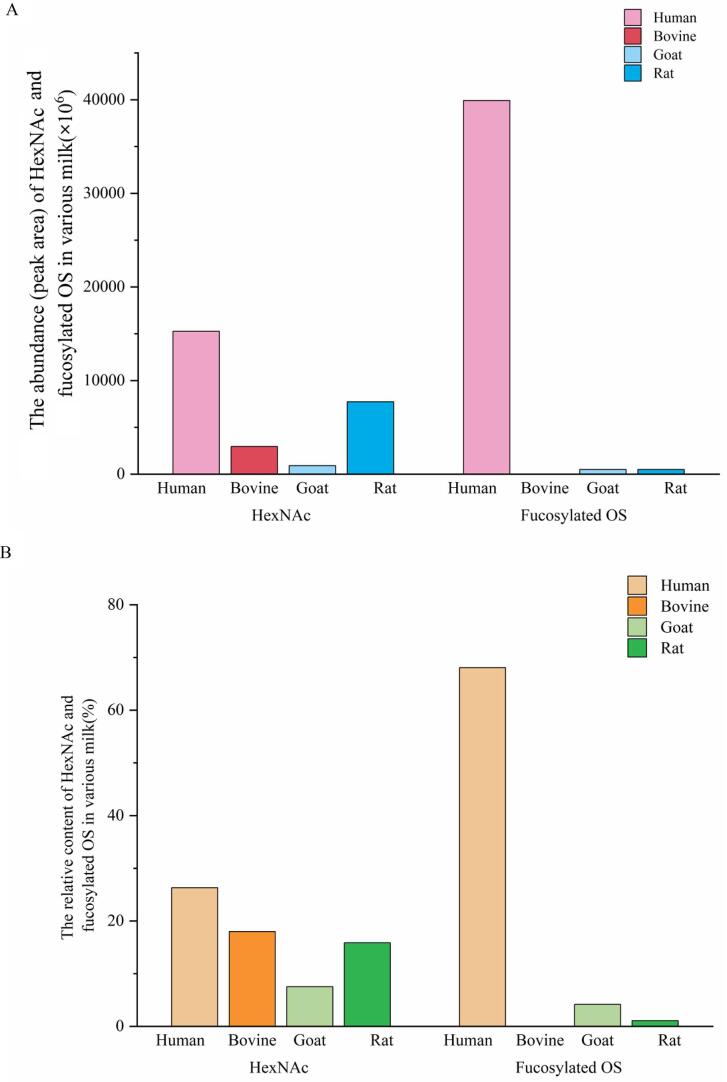
N-acetylhexosamine (HexNAc) is regarded as a component of so-called bifidus factor (Barile et al., 2010, GyÖRGY et al., 1974). Of the colostrum samples analyzed in the present work, 26.30 %, 17.99 %, 7.51 %, and 15.86 % of the HMOs, BMOs, GMOs and RMOs contained HexNAc, respectively (Fig. 2B). Obviously, the human milk included more HexNAc of OS than animal milk, and rat milk had the second most abundant of HexNAc after human milk (Fig. 2A). According to the composition of literature data (Porfirio, et al., 2020), the dHex in Table 1, Table 2 is speculated to represent Fuc in this work. In general, the main differences between HMOs and animal OS are that most HMOs are highly fucosylated OS (Mehra, Barile, Marotta, Lebrilla, Chu, & German, 2014). The neutral HMOs which containing N-acetylglucosamine and fucose monomers are regarded as substances for the development of the intestinal microbiota typical for breastfed infants (Simon, Goode, Mobasseri, & Zopf, 1997). Furthermore, the fucosylated HMOs are related to the lower risk of diarrhea and respiratory diseases in breast-fed infants (Stepans, Wilhelm, Hertzog, Rodehorst, Blaney, Clemens, et al., 2006). Different from neutral OS in animal milk, approximately 68.07 % of HMOs were fucosylated OS with large degrees of complexity (Fig. 2B). Numerous studies have revealed that no fucosylated species are described in BMOs (Mehra, Barile, Marotta, Lebrilla, Chu, & German, 2014; N Tao, DePeters, Freeman, German, Grimm, & Lebrilla, 2008; N Tao, DePeters, German, Grimm, & Lebrilla, 2009), and these findings were in consistent with the results of our study that there was no fucosylated OS detected in BMOs (Fig. 2). As shown in Fig. 2B, the contents of fucosylated OS in goat and rat milk were 4.17 % and 1.09 %, respectively, which were significantly lower than that in human milk. The abundance of fucosylated OS in goat milk was similar with that in rat milk (Fig. 2A).
Fig. 2.
The abundance (peak area) (A) and relative content (B) of HexNAc and fucosylated OS in various milk.
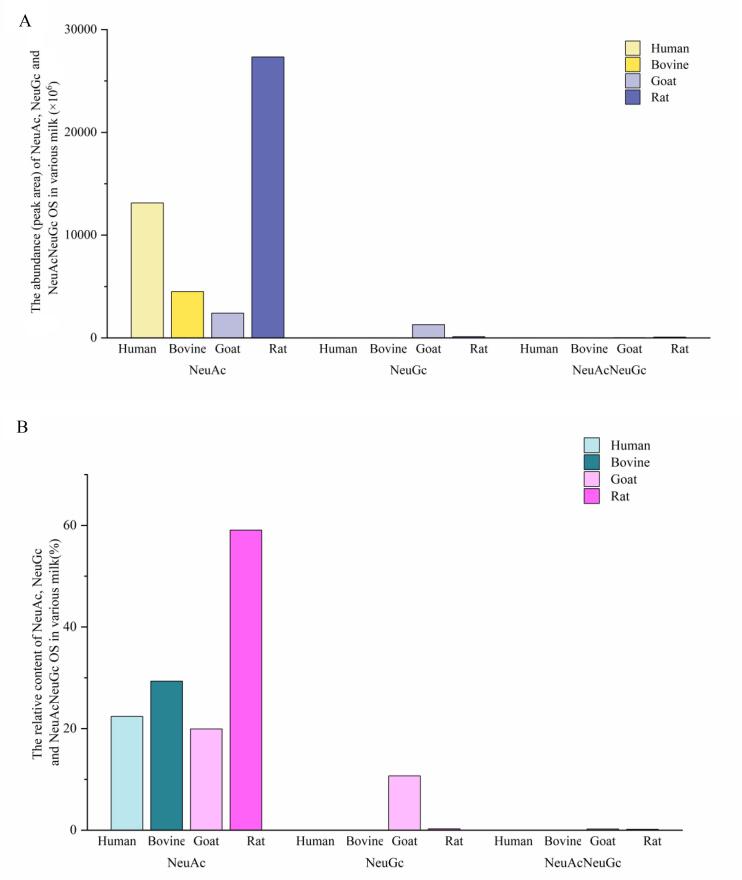
As seen in Fig. 3, all the acidic HMOs and BMOs only contained NeuAc, without NeuGc and NeuAcNeuGc. These findings were in consistent with some previous studies shown that no OS identified in both human and bovine milk contain NeuGc (Sunds et al., 2021, Wu et al., 2011). The NeuGc is not exist in human milk might due to the lack of ability to synthesize NeuGc arise from a mutation occurred in CMP-NeuAc hydroxylase gene which converts NeuAc to NeuGc (Lu, et al., 2020). It is mostly common that no OS contained monosaccharide NeuGc found in colostrum and early lactation milk (N Tao, DePeters, German, Grimm, & Lebrilla, 2009). Nevertheless, the absence of NeuGc are also been found in mid-lactation bovine milk (Robinson, Poulsen, Colet, Duchene, Larsen, & Barile, 2019) and mature (average lactation of 133 days) bovine milk (Sunds, et al., 2021). In present study, the bovine milk sample was mature milk, the result of the absence of NeuGc was might since the unique feature of the native breeds. The OS contained NeuGc are regarded as relationship with some negative health outcomes, such as possibly promoting inflammation and cancer progression (Samraj, Pearce, Läubli, Crittenden, Bergfeld, Banda, et al., 2015). Therefore, the absence of NeuGc in mature human and bovine milk was possible a beneficial trait. Out of 14 acidic GMOs identified in present study, 7 OS only contained NeuAc and 6 OS only contained NeuGc, while 1 OS contained both NeuAc and NeuGc (NeuAcNeuGc) with a slight abundance (Fig. 3A). In rat milk, 12 OS were acidic ones, in which 9 OS only contained NeuAc and 2 OS only contained NeuGc, while 1 OS contained both NeuAc and NeuGc (NeuAcNeuGc). The content of NeuGc-OS (0.25 %) and NeuAcNeuGc-OS (0.16 %) in rat milk were extremely small (Fig. 3B). The rat milk contained the most abundant NeuAc (Fig. 3A) with the content of 59.06 % (Fig. 3B), which was followed by human milk and bovine milk (Fig. 3A). Goat milk had the lowest abundance of NeuAc with the content of 19.92 % (Fig. 3B), comparing with other three milks. The sialylated OS (containing the monomer NeuAc) is benefit to preventing the adhesion of pathogenic bacteria to the intestinal epithelial surface (Simon, Goode, Mobasseri, & Zopf, 1997) and allergy (Eiwegger, Stahl, Haidl, Schmitt, Boehm, Dehlink, et al., 2010). On the other hand, sialylated OS is also shown to improve growth outcomes in undernourished infants and children (Charbonneau, O’Donnell, Blanton, Totten, Davis, Barratt, et al., 2016). As stated above, it was obvious to draw a conclusion that HMOs had the highest nutritional value among the OS in all the milk samples, and RMOs had higher nutritional value than BMOs and GMOs via analyzing the abundance of total OS, NeuAc OS, HexNAc OS and Fucosylated OS.
Fig. 3.
The abundance (peak area) (A) and relative content (B) of NeuAc, NeuGc and NeuAcNeuGc OS in various milk.
4. Conclusion
In conclusion, 70 HMOs, 14 BMOs, 23 GMOs and 24 RMOs were identified in human and animal milk via using UPLC-QE-HF-MS technique, respectively, which suggested that the method was effective and robust for detecting the OS in milks in present study. The number of identified OS in current research was relatively lower compared to previously reports might because of the applied databases, analytical techniques and breeds of samples. This work further verified the significant variations between the nature and number of OS in human and animal milk (e.g.bovine, goat and rat milk). Noticeably, many OS with distinct compositions, contents and abundances had been newly revealed in this work, in particular, the various contents of neutral and acidic OS in goat and bovine milk as well as 14 neutral OS and 3 acidic OS were found in rat milk for the first time. The compositions and abundances of RMOs were might more similar with that of HMOs comparing with BMOs and GMOs were of particular interests, which might due to the large similarity of genome with humans. The similarity between HMOs and RMOs might provide theoretical basis for better application of rats in biological/biomedical studies of HMOs as models. Based on the analysis of the abundances of total OS, NeuAc OS, HexNAc OS and Fucosylated OS, HMOs might have the highest nutritional value among the OS in different milk samples, which was followed by RMOs. Due to the existence of nutritional structure in BMOs and GMOs, the potential value of animal milk OS for promoting human health should be concerned. What’s more, rat milk was able to be used for different mechanism studies, bovine and goat milk could be considered as source of bioactive OS which were able to be further applied in medical and functional foods due to the existence of functional structures in BMOs and GMOs.
CRediT authorship contribution statement
Rui Li: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Formal analysis. Yalin Zhou: Software, Visualization, Resources, Writing – review & editing. Yajun Xu: Data curation, Funding acquisition, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the grants from National Natural Science Foundation of China (NSFC, No. 82173500) and the National Key Research and Development Program of China (No. 2022YFF1100104 and No. 2022YFD2101505).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2023.100705.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Albrecht S., Lane J.A., Marino K., Al Busadah K.A., Carrington S.D., Hickey R.M., Rudd P.M. A comparative study of free oligosaccharides in the milk of domestic animals. British Journal of Nutrition. 2014;111(7):1313–1328. doi: 10.1017/S0007114513003772. [DOI] [PubMed] [Google Scholar]
- Aldredge D.L., Geronimo M.R., Hua S., Nwosu C.C., Lebrilla C.B., Barile D. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology. 2013;23(6):664–676. doi: 10.1093/glycob/cwt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S., Bénet T. Quantitative determination of non-lactose milk oligosaccharides. Analytica Chimica Acta. 2018;1010:86–96. doi: 10.1016/j.aca.2017.12.036. [DOI] [PubMed] [Google Scholar]
- Barile D., Marotta M., Chu C., Mehra R., Grimm R., Lebrilla C.B., German J. Neutral and acidic oligosaccharides in Holstein-Friesian colostrum during the first 3 days of lactation measured by high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Journal of dairy science. 2010;93(9):3940–3949. doi: 10.3168/jds.2010-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carubelli R., Ryan L.C., Trucco R.E., Caputto R. Neuramin-lactose sulfate, a new compound isolated from the mammary gland of rats. Journal of Biological Chemistry. 1961;236(9):2381–2388. [PubMed] [Google Scholar]
- Charbonneau M.R., O’Donnell D., Blanton L.V., Totten S.M., Davis J.C., Barratt M.J.…Bain J.R. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. 2016;164(5):859–871. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.U., Carubelli R. Neuramine-lactose, neuramine-lactose sulfate, and lactose sulfate from rat mammary glands. Isolation, purification, and permethylation studies. Biochemistry. 1968;7(12):4423–4430. doi: 10.1021/bi00852a039. [DOI] [PubMed] [Google Scholar]
- Dvorak, B., Halpern, M. D., Holubec, H., Dvorakova, K., Dominguez, J. A., Williams, C. S., & Meza, Y. G. (2004). Rat milk decreases necrotizing enterocolitis in a rat model. In Protecting Infants through Human Milk, (pp. 471-473): Springer. [DOI] [PubMed]
- Eiwegger T., Stahl B., Haidl P., Schmitt J., Boehm G., Dehlink E.…Szépfalusi Z. Prebiotic oligosaccharides: In vitro evidence for gastrointestinal epithelial transfer and immunomodulatory properties. Pediatric Allergy and Immunology. 2010;21(8):1179–1188. doi: 10.1111/j.1399-3038.2010.01062.x. [DOI] [PubMed] [Google Scholar]
- Gosling S.D. From mice to men: What can we learn about personality from animal research? Psychological bulletin. 2001;127(1):45. doi: 10.1037/0033-2909.127.1.45. [DOI] [PubMed] [Google Scholar]
- GyÖRGY P., Jeanloz R.W., von Nicolai H., Zilliken F. Undialyzable growth factors for Lactobacillus bifidus var. pennsylvanicus: Protective effect of sialic acid bound to glycoproteins and oligosaccharides against bacterial degradation. European journal of biochemistry. 1974;43(1):29–33. doi: 10.1111/j.1432-1033.1974.tb03380.x. [DOI] [PubMed] [Google Scholar]
- Leo F., Asakuma S., Fukuda K., Senda A., Urashima T. Determination of sialyl and neutral oligosaccharide levels in transition and mature milks of Samoan women, using anthranilic derivatization followed by reverse phase high performance liquid chromatography. Bioscience, biotechnology, and biochemistry. 2010 doi: 10.1271/bbb.90614. [DOI] [PubMed] [Google Scholar]
- Li J., Jiang M., Zhou J., Ding J., Guo Z., Li M.…Liang X. Characterization of rat and mouse acidic milk oligosaccharides based on hydrophilic interaction chromatography coupled with electrospray tandem mass spectrometry. Carbohydrate polymers. 2021;259 doi: 10.1016/j.carbpol.2021.117734. [DOI] [PubMed] [Google Scholar]
- Li R., Zhou Y., Xu Y. Comparative Analysis of Oligosaccharides in Breast Milk and Feces of Breast-Fed Infants by Using LC-QE-HF-MS: A Communication. Nutrients. 2023;15(4):888. doi: 10.3390/nu15040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio R.G., Ninonuevo M.R., Freeman S.L., Sela D.A., Grimm R., Lebrilla C.B.…German J.B. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. Journal of agricultural and food chemistry. 2007;55(22):8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- Lu J., Zhang Y., Song B., Zhang S., Pang X., Sari R.N.…Lv J. Comparative analysis of oligosaccharides in Guanzhong and Saanen goat milk by using LC–MS/MS. Carbohydrate polymers. 2020;235 doi: 10.1016/j.carbpol.2020.115965. [DOI] [PubMed] [Google Scholar]
- Macias Rostami S., Bénet T., Spears J., Reynolds A., Satyaraj E., Sprenger N., Austin S. Milk oligosaccharides over time of lactation from different dog breeds. PLoS One1. 2014;9(6):e99824. doi: 10.1371/journal.pone.0099824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Ortiz A., Salcedo J., Barile D., Bunyatratchata A., Moreno F.J., Martin-García I.…Ruiz-Matute A.I. Characterization of goat colostrum oligosaccharides by nano-liquid chromatography on chip quadrupole time-of-flight mass spectrometry and hydrophilic interaction liquid chromatography-quadrupole mass spectrometry. Journal of Chromatography A. 2016;1428:143–153. doi: 10.1016/j.chroma.2015.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R., Barile D., Marotta M., Lebrilla C.B., Chu C., German J.B. Novel high-molecular weight fucosylated milk oligosaccharides identified in dairy streams. PLoS One1. 2014;9(5):e96040. doi: 10.1371/journal.pone.0096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyrand M., Dallas D., Caillat H., Bouvier F., Martin P., Barile D. Comparison of milk oligosaccharides between goats with and without the genetic ability to synthesize αs1-casein. Small Ruminant Research. 2013;113(2–3):411–420. doi: 10.1016/j.smallrumres.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti L., Cattaneo T.M.P., Orlandi M., Curadi M.C. Capillary electrophoresis of sialylated oligosaccharides in milk from different species. Journal of Chromatography A. 2015;1409:288–291. doi: 10.1016/j.chroma.2015.07.076. [DOI] [PubMed] [Google Scholar]
- Moreno F.J., Sanz M.L. John Wiley & Sons; 2014. Food oligosaccharides: Production, analysis and bioactivity. [Google Scholar]
- Naccarato W., Ray R., Wells W. Characterization and tissue distribution of 6-O-beta-D-galactopyranosyl myo-inositol in the rat. Journal of Biological Chemistry. 1975;250(5):1872–1876. [PubMed] [Google Scholar]
- Ninonuevo M.R., Park Y., Yin H., Zhang J., Ward R.E., Clowers B.H.…Grimm R. A strategy for annotating the human milk glycome. Journal of agricultural and food chemistry. 2006;54(20):7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- Porfirio S., Archer-Hartmann S., Moreau G.B., Ramakrishnan G., Haque R., Kirkpatrick B.D.…Azadi P. New strategies for profiling and characterization of human milk oligosaccharides. Glycobiology. 2020;30(10):774–786. doi: 10.1093/glycob/cwaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R.C., Poulsen N.A., Colet E., Duchene C., Larsen L.B., Barile D. Profiling of aminoxyTMT-labeled bovine milk oligosaccharides reveals substantial variation in oligosaccharide abundance between dairy cattle breeds. Scientific reports. 2019;9(1):1–10. doi: 10.1038/s41598-019-41956-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samraj A.N., Pearce O.M., Läubli H., Crittenden A.N., Bergfeld A.K., Banda K.…Diaz S.L. A red meat-derived glycan promotes inflammation and cancer progression. Proceedings of the National Academy of Sciences. 2015;112(2):542–547. doi: 10.1073/pnas.1417508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P., Goode P., Mobasseri A., Zopf D. Inhibition of Helicobacter pylori binding to gastrointestinal epithelial cells by sialic acid-containing oligosaccharides. Infection and immunity. 1997;65(2):750–757. doi: 10.1128/iai.65.2.750-757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa Y.R., Medeiros L.B., Pintado M.M.E., Queiroga R.C. Goat milk oligosaccharides: Composition, analytical methods and bioactive and nutritional properties. Trends in Food Science & Technology. 2019;92:152–161. [Google Scholar]
- Stepans M.B.F., Wilhelm S.L., Hertzog M., Rodehorst T.K.C., Blaney S., Clemens B.…Newburg D.S. Early consumption of human milk oligosaccharides is inversely related to subsequent risk of respiratory and enteric disease in infants. Breastfeeding Medicine. 2006;1(4):207–215. doi: 10.1089/bfm.2006.1.207. [DOI] [PubMed] [Google Scholar]
- Sunds A.V., Bunyatratchata A., Robinson R., Glantz M., Paulsson M., Leskauskaite D.…Vegarud G.E. Comparison of bovine milk oligosaccharides in native North European cattle breeds. International Dairy Journal. 2021;114 doi: 10.1016/j.idairyj.2020.104917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao N., DePeters E., Freeman S., German J., Grimm R., Lebrilla C.B. Bovine milk glycome. Journal of dairy science. 2008;91(10):3768–3778. doi: 10.3168/jds.2008-1305. [DOI] [PubMed] [Google Scholar]
- Tao N., DePeters E., German J., Grimm R., Lebrilla C.B. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. Journal of dairy science. 2009;92(7):2991–3001. doi: 10.3168/jds.2008-1642. [DOI] [PubMed] [Google Scholar]
- Tao N., Ochonicky K.L., German J.B., Donovan S.M., Lebrilla C.B. Structural determination and daily variations of porcine milk oligosaccharides. Journal of agricultural and food chemistry. 2010;58(8):4653–4659. doi: 10.1021/jf100398u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood M.A., Gaerlan S., De Leoz M.L.A., Dimapasoc L., Kalanetra K.M., Lemay D.G.…Lebrilla C.B. Human milk oligosaccharides in premature infants: Absorption, excretion, and influence on the intestinal microbiota. Pediatric research. 2015;78(6):670–677. doi: 10.1038/pr.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakami H., Saeki M., Watanabe Y., Kawamura R., Nishizawa S., Suzuki Y.…Ajisaka K. Isolation and assessment of acidic and neutral oligosaccharides from goat milk and bovine colostrum for use as ingredients of infant formulae. International Dairy Journal. 2018;83:1–9. [Google Scholar]
- Urashima T., Asakuma S., Leo F., Fukuda K., Messer M., Oftedal O.T. The predominance of type I oligosaccharides is a feature specific to human breast milk. Advances in Nutrition. 2012;3(3):473S–482S. doi: 10.3945/an.111.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urashima T., Hirabayashi J., Sato S., Kobata A. Human Milk Oligosaccharides as Essential Tools for Basic and Application Studies on Galectins. Trends in Glycoscience and Glycotechnology. 2018;30(172):SJ11-SJ24. [Google Scholar]
- Wang Y., Zhou X., Gong P., Chen Y., Feng Z., Liu P.…Song L. Comparative major oligosaccharides and lactose between Chinese human and animal milk. International Dairy Journal. 2020;108 [Google Scholar]
- Wu S., Grimm R., German J.B., Lebrilla C.B. Annotation and structural analysis of sialylated human milk oligosaccharides. Journal of proteome research. 2011;10(2):856–868. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Vervoort J., Pan J., Gao P., Zhu H., Wang X.…Lyu J. Comparison of twelve human milk oligosaccharides in mature milk from different areas in China in the Chinese Human Milk Project (CHMP) study. Food Chemistry. 2022;395 doi: 10.1016/j.foodchem.2022.133554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.