Abstract
Regenerative medicine is a highly advanced medical field that aims to restore tissues and organs lost due to diseases and injury using a person's own cells or those of others. Direct cellular reprogramming is a promising technology that can directly induce cell-fate conversion from terminally differentiated cells to other cell types and is expected to play a pivotal role in applications in regenerative medicine. The induction of direct cellular reprogramming requires one or more master transcription factors with the potential to reconstitute cell type-specific transcription factor networks. The set of master transcription factors may contain unique transcription factors called pioneer factors that can open compacted chromatin structures and drive the transcriptional activation of target genes. Therefore, pioneer factors may play a central role in direct cellular reprogramming. However, our understanding of the molecular mechanisms by which pioneer factors induce cell-fate conversion is still limited. This review briefly summarizes the outcomes of recent findings and discusses future perspectives, focusing on the role of pioneer factors in direct cellular reprogramming.
Keywords: Cell-fate conversion, Differentiation, Regenerative medicine, Transcription, Chromatin
1. Direct cellular reprogramming: a technology for inducing cell-fate conversion
In regenerative medicine, tissues and organs lost owing to disease and injury are expected to be restored using cells harvested from patients or others, utilizing the inherent plasticity and ability of cells to self-organize. Induced pluripotent stem cells (iPSCs) are powerful tools for regenerative medicine [1]. Nonetheless, the risk of tumor formation from residual undifferentiated cells and the time and cost inefficiencies associated with obtaining a sufficient number of differentiated cells for medical applications remain significant challenges.
Direct cellular reprogramming is an emerging technology that directly converts terminally differentiated cells into cells of other lineages without going through an intermediate pluripotent state [2]. This technology could potentially overcome the aforementioned issues associated with iPSCs and is gaining interest as a complementary approach to iPSCs in regenerative medicine. Moreover, recent advances in the study of cell-fate conversion have demonstrated that the technology of in vivo cellular reprogramming will be developed as a means to regenerate tissues and organs by directly inducing in situ reprogramming of cells in the vicinity of damaged and diseased areas rather than relying on transplantation [3].
2. Transcription factor sets used in direct cellular reprogramming
Research on direct cellular reprogramming began attracting much attention in 1987, with a landmark study on the cell-fate conversion of mouse fibroblasts to myoblasts induced by forced expression of the transcription factor MyoD [4]. Almost 20 years after its discovery, multiple combinations of transcription factors have been introduced into cells to screen for cell-type-specific sets of transcription factors, as in the case of iPSC induction. After much effort, defined transcription factors that enable the direct induction of various cell types, including cells characterized as neurons (induced neuronal cells: iNCs [5]), cardiomyocytes (induced cardiomyocytes: iCMs [6]), and hepatocytes (induced hepatocyte-like cells: iHepCs [7,8]), were identified. Subsequently, these cell type-specific transcription factors can be inferred using computational approaches [[9], [10], [11]], which are required for validation experiments using cells. Consequently, more than 40 different types of differentiated cells have been generated from other types of differentiated cells [2,12]. Recent studies have challenged the direct induction of tissue-specific stem cells and progenitor cells from fully differentiated cells [13,14]. These directly induced stem and progenitor cells may be preferred over terminally differentiated cells because of their potential for propagation and differentiation in culture and after transplantation into injured tissues and organs. In addition, cell reprogramming technology has enabled the direct induction of tumor-forming cells from normal somatic cells [15,16] and has evolved to induce stable inhibition of tumor cell proliferation and functional differentiation of tumor cells using a defined set of transcription factors [[17], [18], [19]]. Direct reprogramming technology, which uses specific combinations of transcription factors, is expanding into many fields beyond regenerative medicine.
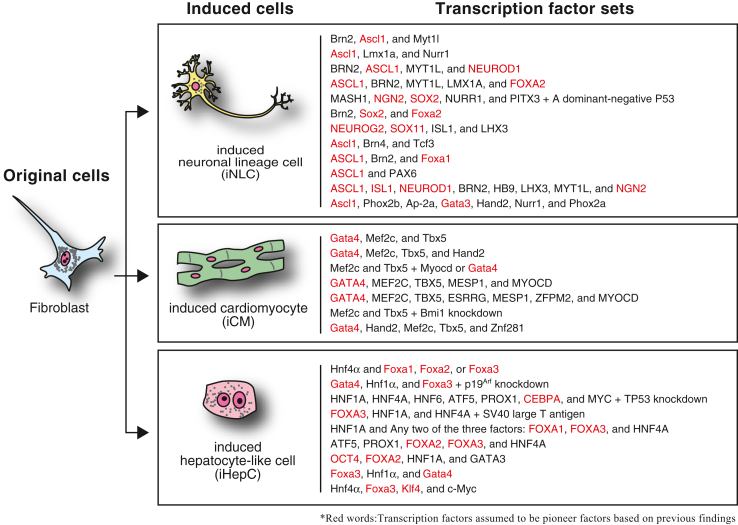
In direct cellular reprogramming methods using two or more defined transcription factors, some of these transcription factors can be replaced with other transcription factors to induce different but related types of cells (Fig. 1). For example, induction of dopaminergic neuron-like cells from mouse and human fibroblasts requires Ascl1, Lmx1a, and Nurr1, whereas replacement of Lmx1a and Nurr1 with other transcription factors, such as Isl1, Neurod1, Brn2, Hb9, Lhx3, Myt1l, and Ngn2, results in the induction of motor neuron-like cells [20,21]. Thus, it is suggested that the cell-type-specific sets of transcription factors contain at least two kinds: one is fundamentally required to initiate the induction of cell-fate conversion, and the others are transcription factors involved in acquiring the target cell properties. Our previous study demonstrated that prior chromatin binding of Foxa protein family members (Foxa1, Foxa2, and Foxa3) leads to subsequent Hnf4α binding to similar regions during the reprogramming of mouse fibroblasts to iHepCs [22]. The combination of transcription factors required to induce cell-fate conversion may also depend on the original cell types [2]. If the original cells endogenously express reprogramming factors, introducing these factors may not be necessary to induce cell-fate conversion. Indeed, direct cellular reprogramming between related cell types is often induced using a smaller number of transcription factors than between different cell types [2,12].
Fig. 1.
Representative sets of transcription factors used to induce direct cellular reprogramming. Three different types of differentiated cells directly induced from fibroblasts using defined transcription factors are shown. In this figure, neuronal subtypes induced by different sets of transcription factors are collectively called ‘induced neuronal lineage cell (iNLC).’ Pioneer factors in the list of transcription factor sets are highlighted in red.
3. Molecular mechanisms of transcriptional and chromatin regulation during direct cellular reprogramming
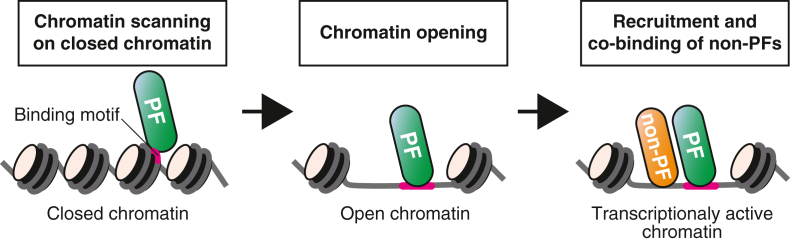
Ascl1 and Foxa protein family members act as pioneer factors that are transcription factors responsible for relaxing closed chromatin and recruiting co-binding factors (Fig. 2) [[23], [24], [25], [26], [27], [28]]. These pioneer factors may be critical for creating the capacity of cells to accept and initiate direct cellular reprogramming. Previous studies have shown that pioneer factors are present in almost all sets of transcription factors used to induce direct cellular reprogramming (Fig. 1) [29]. Our analyses and those of other groups revealed that pioneer factors play a central role in the induction of cell-fate conversion [22,26]. Only pioneer factors can make chromatin competent for recruiting other transcription factors associated with cell differentiation [25,30]. By contrast, recent studies have suggested that all transcription factors can act as pioneer factors depending on their expression levels and abundance ratios [31,32]. This idea is partially supported by other studies showing that the outcome of cellular reprogramming is influenced by the expression levels and abundance ratios of reprogramming-inducing transcription factors [16,33,34]. It has also been reported that transcription factors introduced and exogenously expressed in cells can either promote or inhibit chromatin binding to each other [35]. Moreover, the pioneering activity of pioneer factors may depend on the type of transcription factors introduced into the cells [36,37]. These findings indicate that understanding the molecular mechanisms underlying direct cellular reprogramming requires further investigation.
Fig. 2.
Molecular function of pioneer factors. A pioneer factor (PF) recognizes its binding motif within closed chromatin in chromatin scanning results and opens the chromatin structure in this locus. Subsequently, non-PFs recruited to a similar region bind to that region and induce transcriptional activation of a target gene with the PF.
Once pioneer factors bind to and open chromatin, chromatin accessibility is preserved by binding other transcription factors recruited to these regions [38]. Although this phenomenon has also been observed in direct cellular reprogramming [39], chromatin binding of pioneer factors persists even after the completion of cell reprogramming [22,26]. Thus, it has been suggested that pioneer factors continuously bind to chromatin to maintain chromatin accessibility and binding of other transcription factors. The ablation of all Foxa protein family members in the liver decreases chromatin accessibility and induces the dissociation of Hnf4α [40]. In addition to the regulation by direct chromatin binding, pioneer factors may also epigenetically contribute to the maintenance of cell identity after the induction of direct cellular reprogramming because pioneer factors could regulate DNA methylation and mitotic gene bookmarking [[41], [42], [43], [44]]. Future studies will be useful for a better understanding the relationship between the role of pioneer factors and the maintenance of induced cell identity.
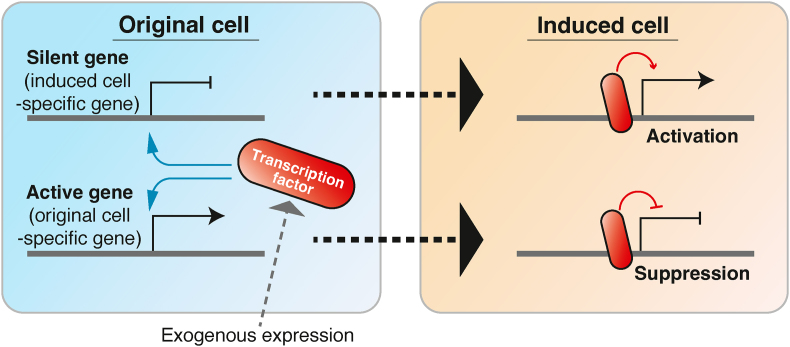
Direct cellular reprogramming involves both the acquisition of target cell traits and the elimination of the original cell traits. Recent studies have shown that transcription factors exogenously introduced into cells act as transcriptional activators or suppressors, depending on the context of the target genes (Fig. 3) [45,46]. Our previous study demonstrated that the Foxa protein family of pioneer factors might be involved in both the activation and suppression of target gene transcription [22]. Interaction with a REST complex and regulation of the repressive histone mark H3K27me3 with a Polycomb complex may be important for suppressing target gene expression in direct cellular reprogramming [[47], [48], [49], [50], [51], [52], [53], [54], [55], [56]]. It would be interesting to understand how pioneer factors select which genes to activate and suppress during direct cellular reprogramming.
Fig. 3.
Dual functions of the transcription factors exogenously expressing in cells. Transcription factors used for inducing direct cellular reprogramming act as suppressors and activators in expressing original and induced cell-specific genes, respectively.
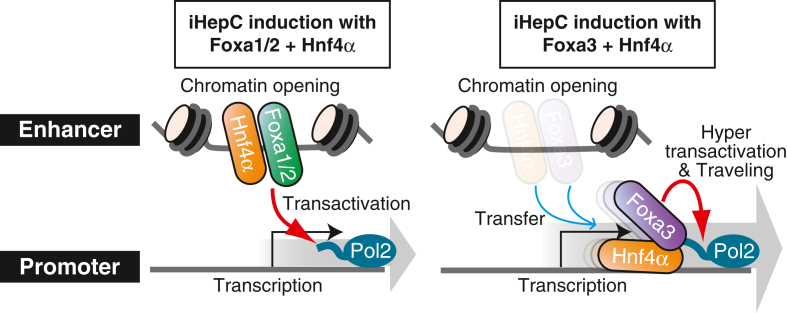
In our previous study, Foxa1/Foxa2 and Foxa3 exhibited diverse molecular dynamics and unforeseen behaviors (Fig. 4) [22]. Only Foxa3 has a specific property in which it binds to and co-moves with RNA polymerase II (Pol2) and Hnf4α on the target genes, in addition to characteristic pioneering activities such as chromatin opening, recruitment of a co-factor Hnf4α, enhancer activation, and stimulation and suppression of the transcription of target genes. Compared with the dynamic behavior of Foxa3, Foxa1 and Foxa2 exhibit static behavior in the enhancer regions far from the transcription start sites. Notably, all members of the Foxa protein family can induce similar transcriptomic states by controlling the expression of common gene sets. Similar to Foxa3, other pioneer factors have additional functions required for cellular programming and reprogramming.
Fig. 4.
Molecular dynamics and behaviors of Foxa protein family members. Foxa1 and Foxa2 bind to and open the chromatin of enhancer regions and recruit Hnf4α to similar areas to activate target gene transcription. Foxa3 also binds to and opens the chromatin of enhancer regions. However, Foxa3 promptly translocates from enhancer regions to the vicinity of the transcriptional start sites (TSS), binds to Pol2, and activates target gene transcription by moving on DNA together with Pol2 and Hnf4α.
4. Conclusion
Although the molecular mechanisms underlying direct cellular reprogramming are currently being elucidated, many aspects require clarification. Further investigations are required to unveil the crucial involvement of pioneer factors in direct cellular reprogramming. For this purpose, emerging analytical technologies may be useful, including single-cell transcriptome and epigenome analyses, three-dimensional genome analysis, proteomics, and single-molecule imaging. In addition, traditional research fields such as developmental biology, cell biology, and biochemistry should be incorporated to analyze the molecular mechanisms underlying direct cellular reprogramming more comprehensively for a deeper understanding.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported in part by the JSPS KAKENHI (Grant Numbers: JP16K08592 and JP23K11851 to K.H.; JP18H05102, JP19H01177, JP19H05267, JP20H05040, JP21K19916, JP22H05634, JP22H04698, and JP22H00592 to A.S.), the Program for Basic and Clinical Research on Hepatitis of the Japan Agency for Medical Research and Development (AMED) (JP23fk0210116 to A.S.), the Research Center Network for Realization of Regenerative Medicine of AMED (JP23bm1123005 to A.S.), the Medical Research Center Initiative for High Depth Omics (to K.H. and A.S.), the Takeda Science Foundation (to A.S.), the Uehara Memorial Foundation (to K.H. and A.S.), the Kato Memorial Trust for Nambyo Research (to A.S.), the Suzuken Memorial Foundation (to A.S.), and the Naito Foundation (to A.S.).
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
This article is written by the recipient of The JSRM Awards (Basic Researches) 2021.
References
- 1.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Horisawa K., Suzuki A. Direct cell-fate conversion of somatic cells: toward regenerative medicine and industries. Proceedings of the Japan Academy, Series B. 2020;96:131–158. doi: 10.2183/pjab.96.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srivastava D., DeWitt N. In vivo cellular reprogramming: the next generation. Cell. 2016;166:1386–1396. doi: 10.1016/j.cell.2016.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis R.L., Weintraub H., Lassar A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-X. [DOI] [PubMed] [Google Scholar]
- 5.Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekiya S., Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 8.Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 9.Cahan P., Li H., Morris S.A., Lummertz da Rocha E., Daley G.Q., Collins J.J. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rackham O.J.L., Firas J., Fang H., Oates M.E., Holmes M.L., Knaupp A.S., et al. A predictive computational framework for direct reprogramming between human cell types. Nat Genet. 2016;48:331–335. doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- 11.Eguchi R., Hamano M., Iwata M., Nakamura T., Oki S., Yamanishi Y. TRANSDIRE: data-driven direct reprogramming by a pioneer factor-guided trans-omics approach. Bioinformatics. 2022;38:2839–2846. doi: 10.1093/bioinformatics/btac209. [DOI] [PubMed] [Google Scholar]
- 12.Wang H., Yang Y., Liu J., Qian L. Direct cell reprogramming: approaches, mechanisms and progress. Nat Rev Mol Cell Biol. 2021;22:410–424. doi: 10.1038/s41580-021-00335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura S., Suzuki A. Generation of mouse and human organoid-forming intestinal progenitor cells by direct lineage reprogramming. Cell Stem Cell. 2017;21:456–471.e5. doi: 10.1016/j.stem.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Inada H., Udono M., Matsuda-Ito K., Horisawa K., Ohkawa Y., Miura S., et al. Direct reprogramming of human umbilical vein- and peripheral blood-derived endothelial cells into hepatic progenitor cells. Nat Commun. 2020;11:5292. doi: 10.1038/s41467-020-19041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L., Wang Y., Cen J., Ma X., Cui L., Qiu Z., et al. Modelling liver cancer initiation with organoids derived from directly reprogrammed human hepatocytes. Nat Cell Biol. 2019;21:1015–1026. doi: 10.1038/s41556-019-0359-5. [DOI] [PubMed] [Google Scholar]
- 16.Goya T., Horisawa K., Udono M., Ohkawa Y., Ogawa Y., Sekiya S., et al. Direct conversion of human endothelial cells into liver cancer-forming cells using nonintegrative episomal vectors. Hepatology Communications. 2022;6:1725–1740. doi: 10.1002/hep4.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Z., He Z., Cai Y., Zhang C., Fu G., Li H., et al. Conversion of hepatoma cells to hepatocyte-like cells by defined hepatocyte nuclear factors. Cell Res. 2019;29:124–135. doi: 10.1038/s41422-018-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takashima Y., Horisawa K., Udono M., Ohkawa Y., Suzuki A. Prolonged inhibition of hepatocellular carcinoma cell proliferation by combinatorial expression of defined transcription factors. Cancer Sci. 2018;109:3543–3553. doi: 10.1111/cas.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong L., Yan Q., Zhang Y., Fang X., Liu B., Guan X. Cancer cell reprogramming: a promising therapy converting malignancy to benignity. Cancer Commun. 2019;39:48. doi: 10.1186/s40880-019-0393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caiazzo M., Dell'Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q.J., Li J.J., Lin X., Lu Y.Q., Guo X.X., Dong E.L., et al. Modeling the phenotype of spinal muscular atrophy by the direct conversion of human fibroblasts to motor neurons. Oncotarget. 2017;8:10945–10953. doi: 10.18632/oncotarget.14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horisawa K., Udono M., Ueno K., Ohkawa Y., Nagasaki M., Sekiya S., et al. The dynamics of transcriptional activation by hepatic reprogramming factors. Mol Cell. 2020;79:660–676.e8. doi: 10.1016/j.molcel.2020.07.012. [DOI] [PubMed] [Google Scholar]
- 23.Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/S1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 24.Heinrich C., Blum R., Gascón S., Masserdotti G., Tripathi P., Sánchez R., et al. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaret K.S., Carroll J.S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wapinski O.L., Vierbuchen T., Qu K., Lee Q.Y., Chanda S., Fuentes D.R., et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwafuchi-Doi M., Donahue G., Kakumanu A., Watts J.A., Mahony S., Pugh B.F., et al. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez Garcia M., Moore C.D., Schulz K.N., Alberto O., Donague G., Harrison M.M., et al. Structural features of transcription factors associating with nucleosome binding. Mol Cell. 2019;75:921–932.e6. doi: 10.1016/j.molcel.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris S.A. Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks. Development (Camb) 2016;143:2696–2705. doi: 10.1242/dev.138263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwafuchi-Doi M., Zaret K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen J.L., Loell K.J., Cohen B.A. A test of the pioneer factor hypothesis using ectopic liver gene activation. Elife. 2022;11 doi: 10.7554/eLife.73358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen J.L., Cohen B.A. A quantitative metric of pioneer activity reveals that HNF4A has stronger in vivo pioneer activity than FOXA1. Genome Biol. 2022;23:221. doi: 10.1186/s13059-022-02792-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris S.A., Cahan P., Li H., Zhao A.M., San Roman A.K., Shivdasani R.A., et al. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Liu Z., Yin C., Asfour H., Chen O., Li Y., et al. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ Res. 2015;116:237–244. doi: 10.1161/CIRCRESAHA.116.305547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stone N.R., Gifford C.A., Thomas R., Pratt K.J.B., Samse-Knapp K., Mohamed T.M.A., et al. Context-specific transcription factor functions regulate epigenomic and transcriptional dynamics during cardiac reprogramming. Cell Stem Cell. 2019;25:87–102.e9. doi: 10.1016/j.stem.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H., Keepers B., Qian Y., Xie Y., Colon M., Liu J., et al. Cross-lineage potential of Ascl1 uncovered by comparing diverse reprogramming regulatomes. Cell Stem Cell. 2022;29:1491–1504.e9. doi: 10.1016/j.stem.2022.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geusz R.J., Wang A., Lam D.K., Vinckier N.K., Alysandratos K.-D., Roberts D.A., et al. Sequence logic at enhancers governs a dual mechanism of endodermal organ fate induction by FOXA pioneer factors. Nat Commun. 2021;12:6636. doi: 10.1038/s41467-021-26950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spitz F., Furlong E.E.M. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 39.Mayran A., Khetchoumian K., Hariri F., Pastinen T., Gauthier Y., Balsalobre A., et al. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat Genet. 2018;50:259–269. doi: 10.1038/s41588-017-0035-2. [DOI] [PubMed] [Google Scholar]
- 40.Reizel Y., Morgan A., Gao L., Lan Y., Manduchi E., Waite E.L., et al. Collapse of the hepatic gene regulatory network in the absence of FoxA factors. Genes Dev. 2020;34:1039–1050. doi: 10.1101/gad.337691.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balsalobre A., Drouin J. Pioneer factors as master regulators of the epigenome and cell fate. Nat Rev Mol Cell Biol. 2022:1–16. doi: 10.1038/s41580-022-00464-z. [DOI] [PubMed] [Google Scholar]
- 42.Caravaca J.M., Donahue G., Becker J.S., He X., Vinson C., Zaret K.S. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27:251–260. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y., Pelham-Webb B., Di Giammartino D.C., Li J., Kim D., Kita K., et al. Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep. 2017;19:1283–1293. doi: 10.1016/j.celrep.2017.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellec M., Dufourt J., Hunt G., Lenden-Hasse H., Trullo A., Zine El Aabidine A., et al. The control of transcriptional memory by stable mitotic bookmarking. Nat Commun. 2022;13:1176. doi: 10.1038/s41467-022-28855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhee C., Lee B.K., Beck S., Le Blanc L., Tucker H.O., Kim J. Mechanisms of transcription factor-mediated direct reprogramming of mouse embryonic stem cells to trophoblast stem-like cells. Nucleic Acids Res. 2017;45:10103–10114. doi: 10.1093/nar/gkx692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mall M., Kareta M.S., Chanda S., Ahlenius H., Perotti N., Zhou B., et al. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature. 2017;544:245–249. doi: 10.1038/nature21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masserdotti G., Gillotin S., Sutor B., Drechsel D., Irmler M., Jørgensen H.F., et al. Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell. 2015;17:74–88. doi: 10.1016/j.stem.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rastegar-Pouyani S., Khazaei N., Wee P., Mohammadnia A., Yaqubi M. Role of hepatic-Specific transcription factors and polycomb repressive complex 2 during induction of fibroblasts to hepatic fate. PLoS One. 2016;11 doi: 10.1371/journal.pone.0167081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y., Wang L., Vaseghi H.R., Liu Z., Lu R., Alimohamadi S., et al. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell. 2016;18:382–395. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drouin-Ouellet J., Lau S., Brattås P.L., Rylander Ottosson D., Pircs K., Grassi D.A., et al. REST suppression mediates neural conversion of adult human fibroblasts via microRNA-dependent and -independent pathways. EMBO Mol Med. 2017;9:1117–1131. doi: 10.15252/emmm.201607471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dal-Pra S., Hodgkinson C.P., Mirotsou M., Kirste I., Dzau V.J. Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by MIR combo. Circ Res. 2017;120:1403–1413. doi: 10.1161/CIRCRESAHA.116.308741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y., Yu C., Daley T.P., Wang F., Cao W.S., Bhate S., et al. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell. 2018;23:758–771.e8. doi: 10.1016/j.stem.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y., Alimohamadi S., Wang L., Liu Z., Wall J.B., Yin C., et al. A loss of function screen of epigenetic modifiers and splicing factors during early stage of cardiac reprogramming. Stem Cell Int. 2018;2018 doi: 10.1155/2018/3814747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuda T., Irie T., Katsurabayashi S., Hayashi Y., Nagai T., Hamazaki N., et al. Pioneer factor NeuroD1 rearranges transcriptional and epigenetic profiles to execute microglia-neuron conversion. Neuron. 2019;101:472–485.e7. doi: 10.1016/j.neuron.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 55.Elhanani O., Salame T.M., Sobel J., Leshkowitz D., Povodovski L., Vaknin I., et al. REST inhibits direct reprogramming of pancreatic exocrine to endocrine cells by preventing PDX1-mediated activation of endocrine genes. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107591. [DOI] [PubMed] [Google Scholar]
- 56.Riching A.S., Danis E., Zhao Y., Cao Y., Chi C., Bagchi R.A., et al. Suppression of canonical TGF-β signaling enables GATA4 to interact with H3K27me3 demethylase JMJD3 to promote cardiomyogenesis. J Mol Cell Cardiol. 2021;153:44–59. doi: 10.1016/j.yjmcc.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]