Abstract
The aim of this study was to predict tyrosine kinase inhibitors (TKI) plus anti-PD-1 antibodies (TKI-PD-1) efficacy as second-line treatment in advanced hepatocellular carcinoma (HCC) using radiomics analysis. From November 2018 to November 2019, a total of 55 patients were included. Radiomic features were obtained from the CT images before treatment and filtered using intraclass correlation coefficients (ICCs) and least absolute shrinkage and selection operator (LASSO) methods. Subsequently, ten prediction algorithms were developed and validated based on radiomic characteristics. The accuracy of the constructed model was measured through area under the receiver operating characteristic curve (AUC) analysis; survival analysis was performed via Kaplan–Meier and Cox regression analyses. Overall, 18 (32.7%) out of 55 patients had progressive disease. Through ICCs and LASSO, ten radiomic features were entered into the algorithm construction and validation. Ten machine learning algorithms showed different accuracies, with the support vector machine (SVM) model having the highest AUC value of 0.933 in the training cohort and 0.792 in the testing cohort. The radiomic features were associated with overall survival. In conclsion, the SVM algorithm is a useful method to predict TKI-PD-1 efficacy in patients with advanced HCC using images taken prior to treatment.
Subject terms: Cancer, Cancer imaging, Gastrointestinal cancer, Immunology, Immunotherapy, Tumour immunology, Cancer imaging
Introduction
Hepatocellular carcinoma (HCC) was the sixth most prevalent cancer and the fourth leading cause of cancer death worldwide in 20181. HCC imposes a heavy disease burden in developing countries, especially in Eastern Asia and sub-Saharan Africa, with a majority of patients suffering from advanced disease2.
Systematic treatments, including tyrosine kinase inhibitors (TKIs) and immune checkpoint inhibitors (ICIs), are recommended for patients with advanced HCC. Prior to 2020, sorafenib3,4 and lenvatinib5 were the primary first-line treatments for advanced HCC. However, in 2020, the publication of the IMbrave 150 trial demonstrated that the combination of atezolizumab and bevacizumab (T + A) resulted in superior outcomes to sorafenib6. Additionally, in 2022, the combination of tremelimumab and durvalumab (D + T) also proved more effective than sorafenib7. As such, T + A and D + T are now the preferred first-line treatments for advanced HCC, while sorafenib, lenvatinib, durvalumab, and pembrolizumab are other recommended regimens according to NCCN guidelines8. However, for patients who have failed first-line therapy, there is no established optimal course of treatment under NCCN guidelines8. The Barcelona Clinic Liver Cancer (BCLC) guidelines still regard treatment after first-line failure as an area of ongoing research9. Between July 2018 and July 2021, Lei et al. conducted a study involving patients with unresectable HCC (uHCC) who had failed sorafenib treatment10. They discovered that a combination of TKI and PD-1 inhibitors (TKI-PD-1) was more advantageous than TKI monotherapy for these patients, indicating that TKI-PD-1 therapy could be a promising treatment option for advanced HCC after first-line failure.
However, a subset of patients still progress during TKI-PD-1 therapy, and thus the identification of biomarkers to predict each patient’s response to combination therapy is essential. To date, serum and tissue sample analyses have not clearly defined the subpopulation most likely to progress2. In addition to collecting invasive serum and tissue samples, noninvasive imaging is routinely performed during routine medical examinations. Radiomics comprise valuable information sources for prognostication11. Machine learning can automatically construct models to interpret medical images based on their radiomic features dataset. A predictive approach using CT-based radiomics is a noninvasive, cost-effective way to identify patients at high risk of PD during TKI-PD-1 therapy. However, such machine learning methods need further study before applying radiomics in the clinic. Therefore, this study was carried out to construct and compare machine learning models based on radiomics for predicting the response of advanced HCC to combined TKI-PD-1 treatment as second-line treatment.
Results
Patient population
A total of 55 patients were enrolled in the study, including 37 (67.3%) patients with non-progression disease (non-PD) and 18 (32.7%) patients with PD. The PD patients experienced a shorter overall survival (OS) than the non-PD patients (9.35 months versus not reached, P < 0.001, Fig. 1a). The median patient age in the overall setting was 53.0 years (range, 43.0–55.0), and most patients were male (90.9%). The etiology of hepatitis B HCC accounted for 94.5%, and 67.3% of patients had an alpha fetoprotein level > 200 ng/mL. Their mean albumin-bilirubin (ALBI) score was − 2.60, which was relatively higher in PD patients than in non-PD patients (− 2.29 versus − 2.76, P < 0.001). Median platelet (PLT), aspartate transaminase (AST), alanine transaminase (ALT), and alkaline phosphatase (ALP) levels were 194.00*109/L, 60.30 U/L, 44.50 U/L, and 134.10 U/L, respectively. At baseline, 61.8% and 38.2% of patients had multiple tumors and a single nodule, respectively. The tumor diameter was > 5 cm in 81.8% of patients and < 5 cm in 18.2%. The proportions of patients with macroscopic vascular invasion and extrahepatic spread at baseline were 80% and 34.5%, respectively. Characteristic differences were not reported for these variables (P > 0.05) except for ALBI (Table 1).
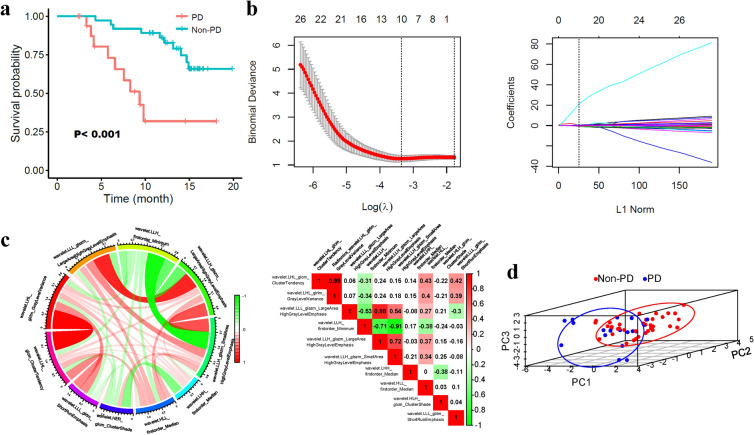
Figure 1.
Radiomics features of the selection process for predicting progressive disease (PD). (a) PD and non-PD stratified the overall survival (OS) of HCC patients treated with TKI-PD-1. (b) LASSO method to find the 10 most meaningful features of PD. (c) A chord diagram and heatmap illustrate the correlation matrix among all 10 features. In the left chord diagram, the wider the bands are, the closer the correlation. In the right heatmap (generated by R software version 3.6.0), correlated paired features are exhibited with color backgrounds. Red indicates a positive correlation; green indicates a negative correlation. (d) Principal component analysis of the features correlated with PD.
Table 1.
Patients’ demographic characteristics.
| Overall (n = 55) | PD (progressive disease, n = 18) | Non-PD (n = 37) | P-value | |
|---|---|---|---|---|
| Sex (%) | 0.160 | |||
| Female | 5 (9.1) | 0 (0.0) | 5 (13.5) | |
| Male | 50 (90.9) | 18 (100.0) | 32 (86.5) | |
| Age (median [interquartile range, IQR]) | 53.00 [43.00, 55.00] | 51.50 [48.50, 55.00] | 53.00 [43.00, 57.00] | 0.336 |
| Hepatitis B virus infection (%) | 0.247 | |||
| No | 3 (5.5) | 2 (11.1) | 1 (2.7) | |
| Yes | 52 (94.5) | 16 (88.9) | 36 (97.3) | |
| Platelets (median [IQR]) | 194.00 [162.50, 282.50] | 198.50 [160.50, 319.00] | 192.00 [164.00, 281.00] | 0.851 |
| Albumin-bilirubin (mean (standard deviation, SD)) | − 2.60 (0.44) | − 2.29 (0.38) | − 2.76 (0.38) | < 0.001 |
| Aspartate aminotransferase (median [IQR]) | 60.30 [36.90, 95.00] | 64.85 [40.17, 116.58] | 60.30 [36.90, 84.10] | 0.379 |
| Alanine transaminase (median [IQR]) | 44.50 [30.45, 66.30] | 48.75 [28.85, 71.20] | 40.90 [30.70, 66.00] | 0.713 |
| Alkaline phosphatase (median [IQR]) | 134.10 [102.90, 182.50] | 137.75 [101.62, 219.65] | 131.00 [106.60, 173.60] | 0.446 |
| Alpha fetoprotein level (%) | 0.761 | |||
| < 200 ng/ml | 18 (32.7) | 5 (27.8) | 13 (35.1) | |
| ≥ 200 ng/ml | 37 (67.3) | 13 (72.2) | 24 (64.9) | |
| Number (%) | > 0.99 | |||
| Single | 21 (38.2) | 7 (38.9) | 14 (37.8) | |
| Multiple | 34 (61.8) | 11 (61.1) | 23 (62.2) | |
| Diameter (%) | 0.713 | |||
| < 5 cm | 10 (18.2) | 4 (22.2) | 6 (16.2) | |
| ≥ 5 cm | 45 (81.8) | 14 (77.8) | 31 (83.8) | |
| Vascular invasion (%) | > 0.99 | |||
| No | 11 (20.0) | 3 (16.7) | 8 (21.6) | |
| Yes | 44 (80.0) | 15 (83.3) | 29 (78.4) | |
| Distant spread (%) | 0.764 | |||
| No | 36 (65.5) | 11 (61.1) | 25 (67.6) | |
| Yes | 19 (34.5) | 7 (38.9) | 12 (32.4) |
Radiomics feature selection and analysis
In total, 2,458 features were acquired for every patient, among which 568 features were excluded with intraclass correlation coefficients (ICCs) < 0.90, leaving 1,890 features for further analysis. Then, 10 discriminatory wavelet-related features between the non-PD and PD patients assessed by RECIST 1.1 were identified via least absolute shrinkage and selection operator (LASSO) feature selection (Fig. 1b, Supplementary Table S1).
After obtaining these 10 features, we further explored their relationships using Spearman correlation analysis. The Chord diagram shows links between the features (shown in Fig. 1c). Obviously, warm red wide bands indicated feature 1 and feature 2, feature 3 and feature 5, and feature 5 and feature 6 were significantly positively related to each other in the network. Meanwhile, features 4 and 6 were negatively related to the green wide band connection. Overall, the detailed relationships among features are shown in heatmap Fig. 1c, and feature 1 and feature 2 had the highest correlation coefficient (0.99) in Spearman analysis. We next performed a principal component analysis (PCA) process to compress the features and reduce the dimensionality. As visualized in Fig. 1d, the PD patients (left bottom cluster) could be separated from the non-PD patients (right top cluster).
Differential values were distributed between PD and non-PD patients for all radiomics features (Supplementary Figure S1). Higher expression was noted for features 1, 2, 4, 7, and 10 in non-PD patients, and the difference for feature 1 was significant (P < 0.05). In contrast, PD patients showed relatively high expression of features 3, 5, 6, 8, and 9, but the difference was not significant (P > 0.05). The area under the receiver operating characteristic curve (AUC) for the prediction of PD ranged from 0.572 to 0.664 for the ten features, as shown in Supplementary Figure S2. Among these features, the AUC of feature 4 was 0.661 (95% CI: 0.500–0.822) and the AUC of 6 was 0.664 (95% CI: 0.509- 0.818), both of which reached significance in ROC analysis (P < 0.05).
Radiomics-based machine learning algorithm predicts the response
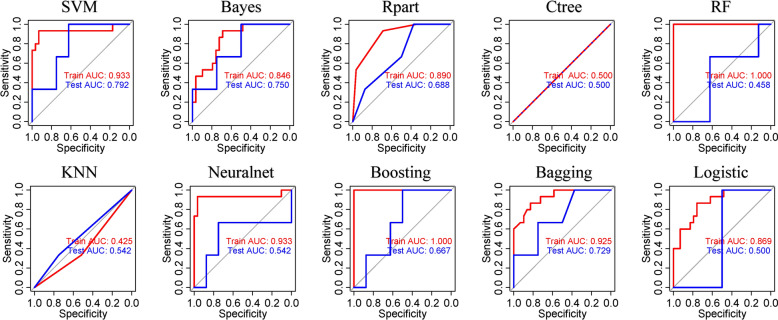
Ten algorithms distinguished PD from non-PD disease with varied efficacy. Overall, better prediction results were observed in the training cohort than in the test cohort (Fig. 2). All ten algorithms, including support vector machine (SVM), naïve Bayes (NB), recursive partitioning and regression trees (Rpart), conditional inference trees (Ctree), random forests (RF), k-nearest neighbors (KNN), neuralnet, boosting, bagging, and logistics, obtained mean F1 scores of 0.88, 0.80, 0.85, 0.80, 1.00, 0.89, 0.96, 1.00, 0.91, and 0.82 in the training cohort and 0.80, 0.69, 0.70, 0.80, 0.79, 0.76, 0.75, 0.77, 0.74, and 0.74 in the testing cohort, respectively. Overall, the classification model using the SVM algorithm achieved the best F1 score. Moreover, the SVM algorithm predicted non-PD and PD with a mean accuracy, sensitivity, specificity and precision of 81.8%, 100.0%, 43.7%, and 78.8%, respectively, in the training cohort and 69.1%, 95.0%, 20.0%, and 70.6%, respectively, in the testing cohort (Supplementary Table S2).
Figure 2.
The box plot shows the F1-score, accuracy, sensitivity, specificity, and precision of ten machine learning models in the training and testing cohorts.
The AUC values of each model were generally higher in the training cohort than in the test cohort (Fig. 3). Different accuracies of the models were exhibited, among which the SVM model had the highest AUC score of 0.933 in the training cohort and 0.792 in the testing cohort. The SVM, Bayes, and bagging algorithms all achieved AUCs > 0.7 in the test cohort.
Figure 3.
The overall accuracy of ten machine learning algorithms was assessed by area under the receiver operating characteristic curves in the training and testing cohorts.
Survival analysis
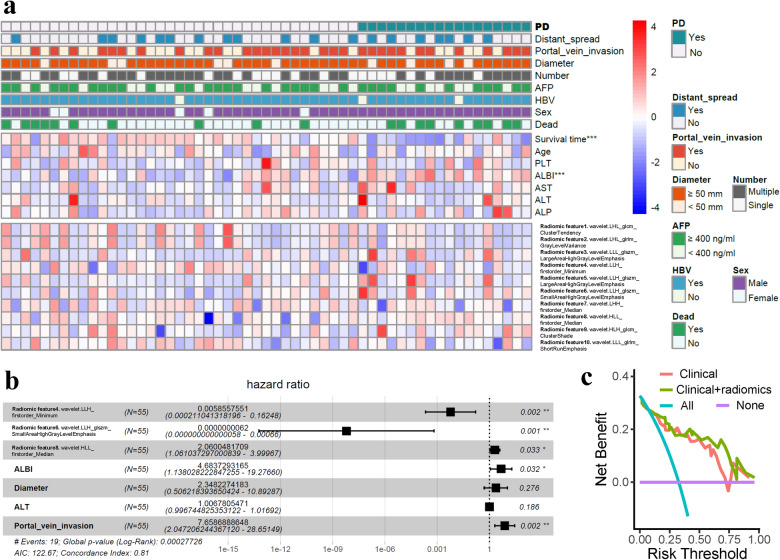
Overall, 19 patients died during the follow-up. OS was not reached in all patients, and the OS in subgroups based on the drugs used is presented in Supplementary Figure S3, which shows no significant difference among subgroups according to the drugs used (P = 0.990). The log-rank test was used to compare high versus low radiomic features with survival. Four out of ten features were found to be significantly associated with OS, including feature 4 (P = 0.039), feature 5 (P = 0.049), feature 8 (P = 0.018), and feature 9 (P = 0.041), as shown in Supplementary Figure S4. Cox regression analysis incorporating radiomic features and clinical variables was conducted to identify survival-associated factors (Fig. 4a). A total of 7 factors (features 4, 6, 8, ALBI, tumor diameter, ALT, and portal vein invasion) generated in the univariate Cox regression model were further selected to be included in the multivariate Cox regression model. Finally, feature 4 (P = 0.002) and feature 6 (P = 0.001) were favorable survival factors, while feature 8 (P = 0.033), ALBI (P = 0.032), and portal vein invasion (P = 0.002) were hazardous survival factors in the multivariate Cox regression model with a c-index of 0.81 (Fig. 4b). Figure 4c shows the more net benefit of using a model comprising clinical characteristics plus radiomics than using a clinical model.
Figure 4.
The landscape of the radiomic features and clinical characteristics. (a) The progressive disease (PD) patients had shorter survival times and higher albumin-bilirubin (ALBI) scores; their radiomic features exhibited differential expression levels between PD and non-PD patients. (b) Radiomic features 4, 6, and 8, ALBI, and portal vein invasion were survival-associated factors in univariable and multivariable Cox regression survival analysis. (c) Additional net benefit of radiomics in decision curve analysis. ***P < 0.001; **P < 0.01; *P < 0.05.
Discussion
The present study demonstrated that radiomic features extracted from initial CT images before TKI-PD-1 treatment could predict the HCC treatment response. The radiomics-based SVM machine learning algorithm using ten LASSO filtered features predicted the PD response with a mean accuracy, sensitivity, specificity, precision, and F1 score of 81.8%, 100.0%, 43.7%, 78.8%, and 88.0%, respectively, in the training cohort and 69.1%, 95.0%, 20.0%, 70.6%, and 80.0%, respectively, in the testing cohort.
Treatment options for advanced HCC have rapidly evolved over the past several years. The optimal course of treatment for advanced HCC patients who do not respond to first-line therapy is currently not well established12,13. Since TKIs target vascular endothelial growth factor receptors (VEGFRs) and modulate the immune microenvironment, the treatment efficacy of PD-1 inhibitors would be theoretically improved when combined with TKIs14. Indeed, emerging clinical data suggest that although a single agent has demonstrated disease control with manageable toxicity, improved synergistic outcomes have been shown. Lenvatinib combined with pembrolizumab resulted in a DCR of > 85% in a recent phase Ib study2. In a phase II trial RESCUE, the DCR was > 77% in the first-line and 75% in the second-line advanced HCC cohort treated with camrelizumab combined with apatinib15. Additionally, a recent study demonstrated that a combination of TKI-PD-1 was found to be more advantageous than TKI monotherapy for HCC patients who had previously failed sorafenib treatment10. This suggests that TKI-PD-1 therapy could be a promising treatment option for advanced HCC after first-line therapy failure.
The additional refinement of the subpopulation of patients most likely to benefit from this combination therapy would be of clinical significance. However, reliable prediction tools to support the precise therapy of TKI-PD-1 are not currently available and are urgently needed in the era of immunotherapy. To date, predictive markers of response or resistance using serum or tissue samples as biomarkers for advanced HCC have not been clearly defined, including PD-L1 expression and tumor mutation burden (TMB) levels16,17. Recently, Yang et al. found that copy number variations (CNVs) in plasma cell-free DNA (cfDNA) could predict the clinical result of combined PD-1 inhibitor and lenvatinib therapy and other immune checkpoint inhibitor-based therapies in hepatobiliary cancers18. However, further study is warranted to verify the clinical value of plasma cfDNA in HCC. Thus, further research to identify new possible methods to predict the treatment response to TKI-PD-1 is necessary.
Radiomics involves applications of computer vision and artificial intelligence to investigate the hidden characteristics of radiographic images in a quantitative manner19. CT images of tumors contain a large amount of useful information that generally cannot be recognized by physicians simply looking at them20. The expression of immunotherapy target PD-L1 at the protein level, as well as PD-1 and CTLA4 at the mRNA level, was found by Hectors et al. to be correlated with radiomic features of HCC21. Radiomics also showed a powerful ability to predict CD8+ T-cell infiltration of HCC in the study of Liao et al.22. Therefore, radiomics could be useful in identifying HCC patients who can benefit from immunotherapies. Acquired from routine clinical images, radiomics is a noninvasive, cost-effective method to predict the patient response, and its dynamic changes could be monitored during therapy23. Furthermore, the entire three-dimensional tumor landscape is captured by radiomics instead of a small portion of the tumor with spatial heterogeneity as occurs in the biopsy method24. Moreover, there is increasing research attention on the importance of the peritumor microenvironment25. Results have shown that the fusion rad-score, which consists of features from the peritumoral area, exhibited better performance than the tumor rad-score26,27. This indicates that the combination of peritumoral features provides more information on the tumor microenvironment, which can better reflect the biological behavior of the tumor. Therefore, in our study, we examined the entire tumor and its surrounding area, allowing us to collect more information on the tumor and its microenvironment27,28.
Machine learning has achieved tremendous success in recent decades. Machine learning is an effective method in the high-throughput era due to the vast amount of data that cannot be directly calculated by our human brains. In 2018, radiomics was demonstrated to be associated with clinical outcomes of cancer patients treated with anti-PD-1 and anti-PD-L1 monotherapy in a retrospective multicohort study29 by Sun et al. Recently, Colen et al. found that a radiomics-based signature could predict the response to pembrolizumab in 57 patients with rare types of late-stage cancer with 94.7% accuracy, 97.3% sensitivity, and 90% specificity30. Therefore, radiomics-based machine learning may be useful for the development of models to predict TKI-PD-1 efficacy. However, a radiomics signature extracted from baseline CTs of patients with advanced HCC treated with immune checkpoint blockade-based therapy to distinguish patients at risk of progression has not previously been reported. Thus, in this study, following the LASSO selection of 10 radiomic features, ten machine learning algorithms were applied for model construction, with SVM achieving the best performance in both the training and testing cohorts. The preliminary results showed that the SVM method was preferred regarding model construction using radiomics for advanced HCC patients treated with TKI-PD-1, while its efficacy needs to be verified in more studies.
The present study had some inherent limitations. First, whether MRI could provide more information and is more suitable for predicting TKI-PD-1 treatment efficacy needs to be determined. Second, the nature of the study was retrospective with a relatively small sample size of 55 patients because combined TKI-PD-1 therapy (different combinations) has only been applied in recent years. The overfitting problem of machine learning algorithms should be noted, and multicenter studies with large populations and subgroup analyses are warranted in the future. Third, certain tumors could not be delineated due to their infiltrative growth type, and in cases of qualified tumor imaging, only the largest tumor was segmented. Fourth, the performance of the proposed SVM model for prediction was moderate and it needs to be optimized before clinical application. Other machine learning or deep learning methods may be worthwhile in future research. Last, the integration of longitudinal multiomics (genomics, pathomics) data would be clinically significant.
In conclusion, our preliminary study found that radiomics could predict TKI-PD-1 outcomes in patients with advanced HCC. Due to its noninvasive and cost-effective nature, radiomics is a promising approach to pretreatment prediction and decision-making. However, additional prospective studies with a large population obtained from multiple centers are needed to translate the present study into clinical applications.
Methods
The Ethical Review Committee of Sun Yat-sen University Cancer Center approved the study, and informed consent was obtained from all patients. The study was conducted in accordance with the principles of the Declaration of Helsinki.
Study design and patient population
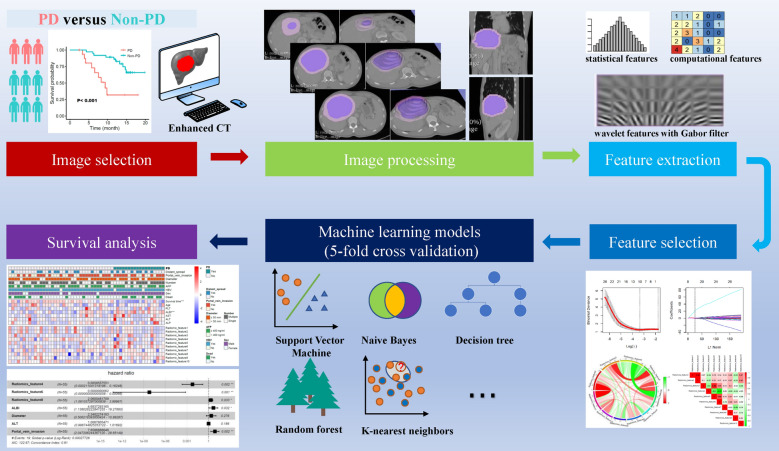
The entire study design is shown in Fig. 5. Patients with advanced HCC who received TKIs orally once daily combined with a PD-1 inhibitor given intravenously (TKI-PD-1) from November 2018 to November 2019 were enrolled in this study. Participants were followed from enrollment to the date of death, loss to follow-up, or April 15, 2021.
Figure 5.
General design of the present study.
The inclusion and exclusion criteria of patients and treatment protocols were in Supplementary method 1.1 and 1.2. The flowchart of our study is illustrated in Fig. 6.
Figure 6.
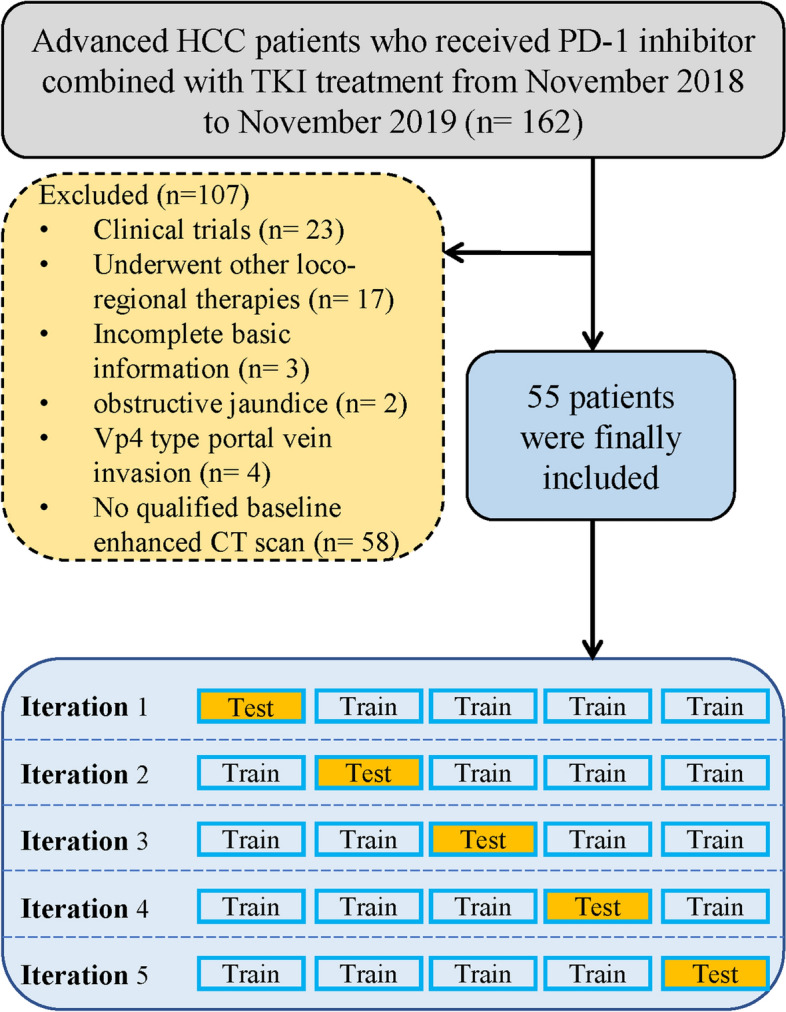
Flowchart of the enrolled patients.
Clinical information collection and response evaluation
Data from all enrolled patients were collected, including age, sex, virus infection, PLT, ALBI, ALT, AST, ALP, alpha-fetoprotein (AFP), and survival. The ALBI of each patient was calculated according to the following formula: ALBI = (log10 bilirubin × 0.66) − (albumin × 0.085)31. In addition, imaging data, including the number and size of tumors, vascular invasion, and extrahepatic metastasis, were collected based on the abdominal CT. All best imaging responses were assessed during treatment on available follow-up images. Progressive disease (PD) was classified into the PD group, while stable disease (SD), partial response (PR), or a complete response (CR) were classified into the non-PD group according to the Response Evaluation Criteria in Solid Tumors V.1.1 guidelines32.
Model construction
Imaging collection and radiomic feature extractions process can be found in Supplementary method 1.3 and 1.4. After the radiomic features were obtained and filtered, ten common machine learning methods were applied to construct possible models, including SVM, NB, Rpart, Ctree, RF, KNN, neuralnet, boosting, bagging, and logistics. Due to the relatively small population, K-fold cross-validation was applied. K-fold cross-validation comprises the following steps: split the data set randomly into 5 folds (11 patients in each fold); construct the model on 4 folds, test the model on the remaining onefold, calculate the error (E) on the observations in the 1 remaining fold; and calculate the average error (AE). The equation is as follows, where k represents the number of folds and Ei indicates the error on the ith iteration:
Statistical analysis
Continuous and categorical variables were compared through Student’s t/Mann–Whitney U test and χ2 or Fisher’s exact test. The Spearman method was applied to evaluate the correlations between features. PCA was used to visualize LASSO selected radiomic features in three-dimensional space. The AUC were generated through the R package “pROC”. Radiomics-based machine learning algorithms were developed and compared using the metrics of accuracy, sensitivity, specificity, precision, F1-score, and AUC33. Kaplan–Meier curves were plotted to assess OS, and survival differences between two groups were calculated via the log-rank test. A multivariate Cox regression model incorporating clinical and radiomics features was carried out to predict OS. The factors generated in the univariate Cox regression model with a P value less than 0.1 were entered into the multivariate Cox regression model. Decision curve analysis was performed to explore the adding value of radiomic features to clinical variables. For all tests, two-sided P < 0.05 was considered statistically significant. All statistical analyses were performed using R software (version 3.6.0).
Supplementary Information
Author contributions
Q.C. designed this study. Y.J., W.D. and S.P. performed the search and collected data. Y.J., W.D. and S.P. performed analysis. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the WUJIEPING Medical Foundation (320.6750.2021-02-126).
Data availability
Data available on request from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wei Dong and Ye Ji.
Contributor Information
Shan Pi, Email: pish@mail.sysu.edu.cn.
Qi-Feng Chen, Email: chenqf25@sysucc.org.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-37862-y.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Finn RS, et al. Phase Ib study of lenvatinib plus pembrolizumab in patients with unresectable hepatocellular carcinoma. J. Clin. Oncol. 2020;38:2960–2970. doi: 10.1200/JCO.20.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 4.Cheng AL, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 5.Kudo M, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 6.Finn RS, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 2020;382:1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 7.Kudo M. Durvalumab plus tremelimumab in unresectable hepatocellular carcinoma. Hepatobiliary Surg. Nutr. 2022;11:592–596. doi: 10.21037/hbsn-22-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines). Hepatocellular Carcinoma. Version 1.2023. https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1514. (2023).
- 9.Reig M, et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022;76:681–693. doi: 10.1016/j.jhep.2021.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei J, et al. TKI or TKI combined with PD-1 inhibitors as second-line treatment for HCC patients after sorafenib failure. Front. Pharmacol. 2022;13:1026337. doi: 10.3389/fphar.2022.1026337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambin P, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PubMed] [Google Scholar]
- 13.Faivre S, Rimassa L, Finn RS. Molecular therapies for HCC: Looking outside the box. J. Hepatol. 2020;72:342–352. doi: 10.1016/j.jhep.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Jain RK. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell. 2014;26:605–622. doi: 10.1016/j.ccell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, et al. Camrelizumab in combination with apatinib in patients with advanced hepatocellular carcinoma (RESCUE): A nonrandomized, open-label, phase II trial. Clin. Cancer Res. 2021;27:1003–1011. doi: 10.1158/1078-0432.CCR-20-2571. [DOI] [PubMed] [Google Scholar]
- 16.Shrestha R, et al. Monitoring immune checkpoint regulators as predictive biomarkers in hepatocellular carcinoma. Front. Oncol. 2018;8:269. doi: 10.3389/fonc.2018.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marabelle A, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–1365. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 18.Yang X, et al. Cell-free DNA copy number variations predict efficacy of immune checkpoint inhibitor-based therapy in hepatobiliary cancers. J. Immunother. Cancer. 2021 doi: 10.1136/jitc-2020-001942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bera K, Velcheti V, Madabhushi A. Novel quantitative imaging for predicting response to therapy: Techniques and clinical applications. Am. Soc. Clin. Oncol. Educat. Book Am. Soc. Clin. Oncol. Ann. Meet. 2018;38:1008–1018. doi: 10.1200/EDBK_199747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hectors SJ, et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur. Radiol. 2020;30:3759–3769. doi: 10.1007/s00330-020-06675-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao H, et al. Preoperative radiomic approach to evaluate tumor-infiltrating CD8(+) T cells in hepatocellular carcinoma patients using contrast-enhanced computed tomography. Ann. Surg. Oncol. 2019;26:4537–4547. doi: 10.1245/s10434-019-07815-9. [DOI] [PubMed] [Google Scholar]
- 23.Mu W, et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat. Commun. 2020;11:5228. doi: 10.1038/s41467-020-19116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinn PO, et al. A coclinical radiogenomic validation study: Conserved magnetic resonance radiomic appearance of periostin-expressing glioblastoma in patients and xenograft models. Clin. Cancer Res. 2018;24:6288–6299. doi: 10.1158/1078-0432.CCR-17-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia TY, et al. Predicting microvascular invasion in hepatocellular carcinoma using CT-based radiomics model. Radiology. 2023;307:222729. doi: 10.1148/radiol.222729. [DOI] [PubMed] [Google Scholar]
- 26.Zhang R, et al. A nomogram based on bi-regional radiomics features from multimodal magnetic resonance imaging for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Quant Imaging Med Surg. 2019;9:1503–1515. doi: 10.21037/qims.2019.09.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan G, et al. Development and validation of a contrast-enhanced CT-based radiomics nomogram for prediction of therapeutic efficacy of anti-PD-1 antibodies in advanced HCC patients. Front. Immunol. 2020;11:613946. doi: 10.3389/fimmu.2020.613946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braman N, et al. Association of Peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)-positive breast cancer. JAMA Netw. Open. 2019;2:e192561. doi: 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun R, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 30.Colen RR, et al. Radiomics analysis for predicting pembrolizumab response in patients with advanced rare cancers. J. Immunother. Cancer. 2021 doi: 10.1136/jitc-2020-001752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinato DJ, et al. The ALBI grade provides objective hepatic reserve estimation across each BCLC stage of hepatocellular carcinoma. J. Hepatol. 2017;66:338–346. doi: 10.1016/j.jhep.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz LH, et al. RECIST 1.1 -Update and clarification: From the RECIST committee. Eur. J. Cancer. 2016;62:132–137. doi: 10.1016/j.ejca.2016.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, et al. Deep learning-based six-type classifier for lung cancer and mimics from histopathological whole slide images: A retrospective study. BMC Med. 2021;19:80. doi: 10.1186/s12916-021-01953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request from the corresponding author.