Abstract
Phytosulfokine (PSK) is a danger-associated molecular pattern recognized by PHYTOSULFOKINE RECEPTOR 1 (PSKR1) and initiates intercellular signaling to coordinate different physiological processes, especially in the defense response to the necrotrophic fungus Botrytis cinerea. The activity of peptide receptors is largely influenced by different posttranslational modifications, which determine intercellular peptide signal outputs. To date, the posttranslational modification to PHYTOSULFOKINE RECEPTOR 1 (PSKR1) remains largely unknown. Here, we show that tomato (Solanum lycopersicum) PSKR1 is regulated by the ubiquitin/proteasome degradation pathway. Using multiple protein–protein interactions and ubiquitylation analyses, we identified that plant U-box E3 ligases PUB12 and PUB13 interacted with PSKR1, among which PUB13 caused PSKR1 ubiquitylation at Lys-748 and Lys-905 sites to control PSKR1 abundance. However, this posttranslational modification was attenuated upon addition of PSK. Moreover, the disease symptoms observed in PUB13 knock-down and overexpression lines demonstrated that PUB13 significantly suppressed the PSK-initiated defense response. This highlights an important regulatory function for the turnover of a peptide receptor by E3 ligase-mediated ubiquitylation in the plant defense response.
Phytosulfokine inhibits plant U-box E3 ligase-mediated ubiquitylation of PHYTOSULFOKINE RECEPTOR 1 (PSKR1) to trigger plant defense against Botrytis cinerea in tomato.
Introduction
Plants possess a sophisticated immune system to cope with microbial pathogen invasion, which is responsible for plant growth and productivity in crop fields. During plant-pathogen battles, pathogens often injure plant cells, causing the release of host-derived molecules, such as ATP, DNA, and peptides (Boutrot and Zipfel 2017). Some of these ubiquitous self-molecules are able to serve as danger-associated molecular patterns (DAMPs) triggering intercellular immune responses. The perception of DAMPs by cell surface pattern recognition receptors activates pattern-triggered immunity (PTI), restricting the duplication of potential pathogens (Zhou and Zhang 2020). Notably, plant endogenous peptides deriving from cleaved and degraded proteins can function as DAMPs with consequent activation of the intracellular secondary immune signals, such as Ca2+ influx, reactive oxygen speciesROS, mitogen-activated protein kinases/calcium-dependent protein kinases, and downstream hormone signals (Tanaka and Heil 2021). In particular, phytosulfokine (PSK) is one of the most extensively studied sulfated peptides with regards to defense functions.
PSK is a disulfated pentapeptide of Tyr(SO3H)-Ile-Tyr(SO3H)-Thr-Gln, which appears to be ubiquitous in land plants, such as Arabidopsis thaliana, asparagus (Asparagus officinalis), rice (Oryza sativa), and tomato (Solanum lycopersicum) (Sauter 2015). The mature PSK is processed from 80 to 120 amino acid prepropeptides by the subtilase-mediated proteolysis and the tyrosylprotein sulfotransferase-mediated sulfonation (Kaufmann and Sauter 2019; Reichardt et al. 2020). The major understanding of PSK signaling was dependent on the identification of its plasma membrane-localized receptor PHYTOSULFOKINE RECEPTOR (PSKR) (Ding et al. 2023; Matsubayashi et al. 2002; Wang et al. 2015b; Zhang et al. 2018). PSKR belongs to a typical leucine-rich repeat receptor-like kinase type receptor, which contained extracellular LRRs embracing an island domain for PSK binding, a transmembrane domain, and an intercellular kinase domain overlapping with canonical guanylate cyclase core for the generation of cyclic guanylic acid (Kwezi et al. 2011). The recognition and biological activities of PSK largely relied on PSKR1, and its putative paralog PSKR2 functioned as an alternative but less active PSK receptor (Matsubayashi et al. 2006; Zhang et al. 2018). PSKR1 acted differentially in plant defense based on different types of pathogen infection. Arabidopsis psrk1 mutants enhanced their resistance against (hemi)-biotrophs, including the bacterium Pseudomonas syringae and Ralstonia solanacearum, the oomycete Hyaloperonospora arabidopsidis, the fungus Fusarium oxysporum, and the root-knot nematode Meloidogyne incognita (Mosher et al. 2013; Shen and Diener 2013; Rodiuc et al. 2016). In contrast, blocking or suppressing the PSK perception in plants promoted the disease susceptibility caused by pathogens with a necrotrophic lifestyle. Arabidopsis psrk1 mutants were more susceptible to the fungi Alternaria brassicicola (Mosher et al. 2013). Similarly, RNAi-based silencing of PSKR1 in tomato plants leads to impaired disease resistance against necrotrophic fungus Botrytis cinerea (Zhang et al. 2018).
The recognition of PSK by PSKR1 is a master switch for downstream signaling. In planta, PSKR1 was associated with the promiscuous co-receptor BAK1 to form a heterodimer since the perception of PSK ligand (Wang et al. 2015b). Mutual phosphorylation events mediated by PSKR1 and co-receptor were regarded as the initiation of intercellular PSK signaling. Moreover, PSKR1 itself also displayed auto- and transphosphorylation activities (Hartmann et al. 2015; Kaufmann et al. 2017). In addition, PSKR1 associated with the plasma membrane-localized H+-ATPases AHA1 and AHA2 to form a functional complex with BAK1 and Ca2+ channel CNGC17, mediating PSK-induced cell expansion (Ladwig et al. 2015). Previous study in tomato also revealed that PSKR1 mediated PSK signal to trigger Ca2+ influx, which initiated the auxin biosynthesis (Zhang et al. 2018). However, how PSKR1 is dynamically regulated at the protein level remains unclear.
The selective degradation of a target protein by the ubiquitin-proteasome system has been implicated in a wide range of plant physiological processes, including immune responses (Trujillo 2018). Ubiquitination is a common posttranslational modification and occurred via the sequential activation of an enzymatic cascade, in which, the ubiquitin is transferred to a ubiquitin-conjugating enzyme by a ubiquitin-activating enzyme (E1), and then E2 is associated with a ubiquitin ligase (E3) to further deliver the ubiquitin to target proteins (Sadanandom et al. 2012). During the protein ubiquitination process, E3 ligases determine the substrate specificity of the 26S proteasome system for distinct target proteins (Smalle and Vierstra 2004). PLANT U-BOX (PUB) is one of the three major E3 ligase classes (homologous to E6AP C-terminus, RING-finger, and U-box) in plants. Total of 66 and 62 PUB gene family members have been identified in Arabidopsis and tomato genomes, respectively (Wiborg et al. 2008; Sharma and Taganna 2020). Several PUB proteins have been reported to be involved in plant innate immunity by manipulating the ubiquitination process of receptor-like kinases (RLKs). AtPUB13 not only targeted the chitin receptor LYK5 but also bound to a bacterium flagellin receptor AtFLS2 to regulate ligand-induced protein degradation, and eventually attenuated early PTI response in Arabidopsis (Lu et al. 2011; Liao et al. 2017). A similar ubiquitylation event of MtPUB1-mediated MtLYK3 degradation was also discovered in Medicago truncatula (Mbengue et al. 2010). SPL11, a putative ortholog of Arabidopsis PUB13 in rice, ubiquitinated an RLK-type protein SDS2 to reduce plant resistance to the blast fungus Magnaporthe oryzae (Fan et al. 2018). Rice PUB15 directly targeted the kinase domain of another RLK-type protein PID2 and activated the basal immune responses against M. oryzae (Wang et al. 2015a). Therefore, we speculate that protein turnover may also occur in PSKR1 through the PUBs-mediated ubiquitination, in regulating downstream defense responses.
B. cinerea causes gray mold and rot diseases in a broad host range of crops, resulting in devastating economic losses during cultivation and postharvest stages. As a B. cinerea host, tomato is not only an economically important crop worldwide but also serves as a classic model plant species to study plant defense against different pathogen infection. In this study, we generated stable transgenic tomato PSKR1 knock-out and overexpression lines to demonstrate its positive function in plant immunity against B. cinerea. We also showed that PSKR1 protein abundance was regulated by the ubiquitin/proteasome degradation pathway in absence of pathogen attacks, and PUB12 and PUB13 acted as a direct target of PSKR1 to promote its ubiquitylation. In response to B. cinerea infection, PSK reduced PUB12/13-mediated interaction and ubiquitylation, which activated intracellular PSK defense signaling. Therefore, we propose that PSK inhibits continuous protein degradation of PSKR1 by PUB12 and PUB13, initiating the subsequent defense gene expression in response to B. cinerea infection.
Results
PSKR1 confers plant immunity against B. cinerea in tomato
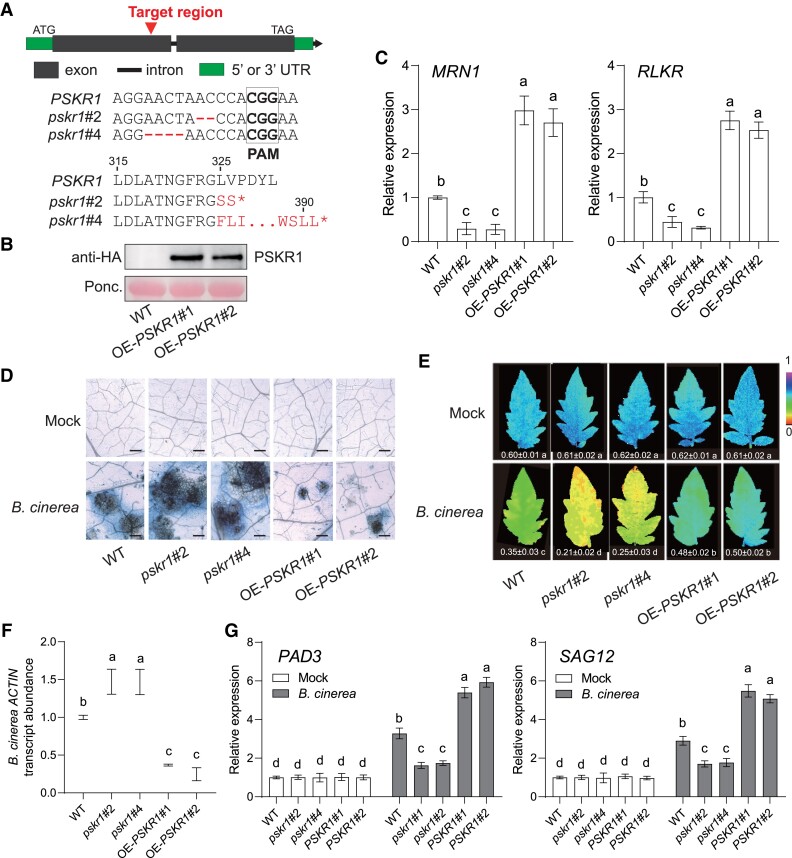
To confirm PSKR1 functions in plant immunity with genetic evidence, we generated PSKR1 knock-out and overexpression lines via CRISPR-Cas9 editing and transgenic overexpression approaches, respectively. Two homozygous CRISPR-Cas9 edited lines pskr1#2 and pskr1#4 (carrying 2 and 4 bp deletion in the first exon that led to an early stop codon of protein translation at 326 and 391 sites, respectively) (Fig. 1A), and two overexpression lines OE-PSKR1#1 and OE-PSKR1#2 (Fig. 1B) were isolated for further experiments. First of all, we checked the transcript abundance of PSK-responsive MRN1 and RLKR genes based on a previous study (Kaufmann et al. 2021), and found that these two genes were suppressed in pskr1 mutants but significantly induced in OE-PSKR1 lines (Fig. 1C). For the disease assay, the pskr1 mutants exhibited more severe disease symptoms than the wild-type (WT) plants at 3 d postinoculation (dpi) with B. cinerea, whereas both OE-PSKR1 lines promoted plant defense against the pathogens (Fig. 1D). Consistently, the pskr1 mutants showed a significant decrease in ΦPSII (photosystem II) values (Fig. 1E) and increases in leaf B. cinerea actin transcript abundance (Fig. 1F), which indicated the damage in plant photosystem and the fungal growth, respectively. By contrast, both OE-PSKR1 lines reduced fungal growth and foliar damage compared with the WT plants (Fig. 1, E and F). Moreover, B. cinerea-induced gene expression of PAD3 and SAG12 was inhibited in PSKR1 knock-out mutants but promoted in overexpression lines (Fig. 1G). These observations provide solid genetic evidence that plant peptide receptor PSKR1 enhanced the defense response of tomato plants against B. cinerea.
Figure 1.
Role of PSKR1 in defense of tomato plants against B. cinerea. A) Schematic illustration of the sgRNA target site (red arrows) in WT PSKR1 and two alleles (pskr1#2 and pskr1#4) from CRISPR/Cas 9 edited T2 mutant lines. The deleted nucleotide sequences of each line are labeled in red. pskr1#2 and pskr1#4 contained a premature stop codon at 326th and 391th amino acid of the PSKR1 protein, respectively. B) Identification of PSKR1 overexpression lines by immunoblot with an anti-HA antibody. Ponceau S (Ponc.) staining was used as a protein loading control. C) Effects of PSKR1 on the transcript abundance of PSK-responsive genes MRN1 and RLKR. The transcript abundance of each gene in WT plants was defined as 1. D to G) Disease symptoms in PSKR1 mutated and overexpressed lines inoculated with B. cinerea. Five-wk-old tomato plants were sprayed with B. cinerea spore suspension. D) Representative images of trypan blue staining for cell death in indicated tomato leaves at 3 d postinoculation with B. cinerea (dpi). Bar = 250 μm. E) Representative chlorophyll fluorescence imaging of ΦPSII at 3 dpi. The value below each individual image indicates the degree of ΦPSII. F) Relative B. cinerea ACTIN transcript abundance in infected tomato leaves at 1 dpi. The transcript abundance of B. cinerea ACTIN in WT plants was defined as 1. G) Effects of PSKR1 on the gene expression of B. cinerea-induced gene PAD3 and SAG12 in indicated tomato plants at 1 dpi. The transcript abundance of each gene under mock treatment in WT plants was defined as 1. Data are presented in (C, E to G) as the means of three biological replicates (±SD, n = 3), and different letters indicate significant differences (P < 0.05) according to Tukey's test.
PSKR1 proteins are degraded by 26S proteasomes
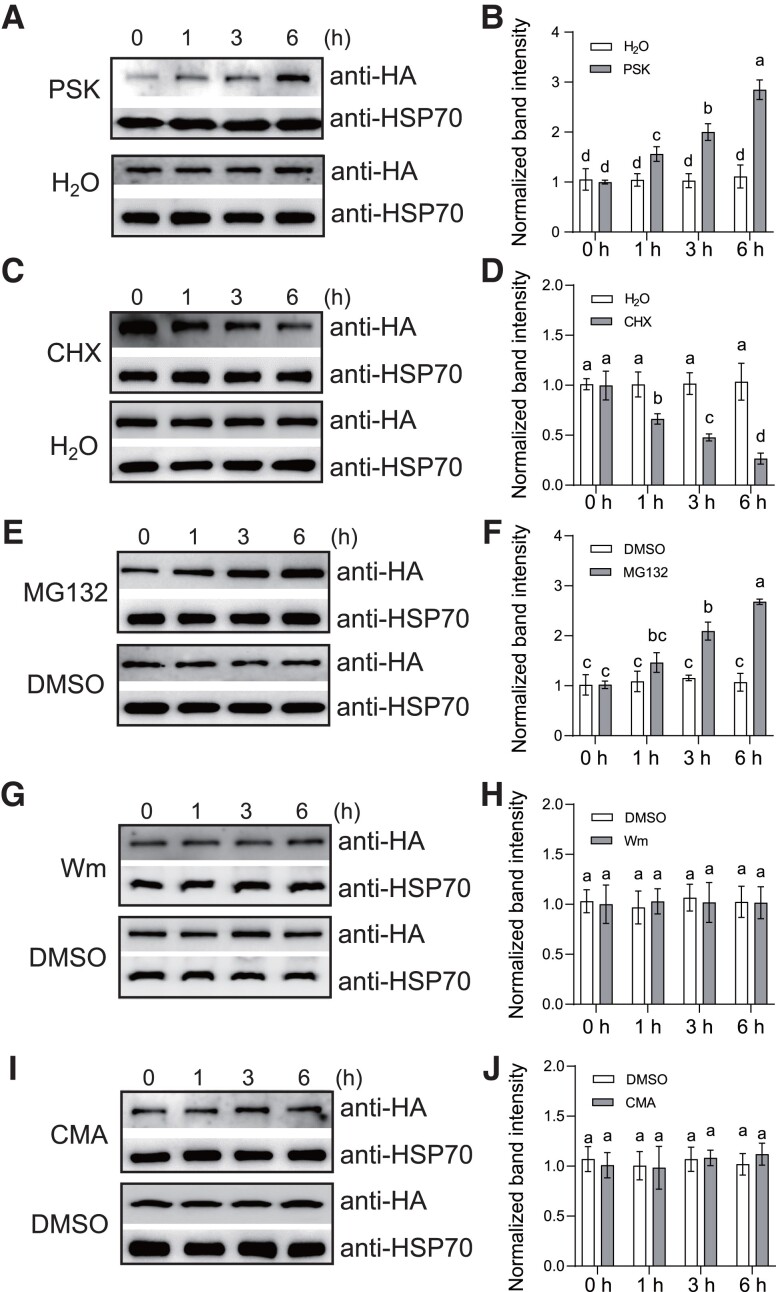
Previous studies revealed multiple RLKs were able to be ubiquitinated and subsequently degraded by 26S proteasomes (Lu et al. 2011; Liao et al. 2017; Zhou et al. 2018; Chen et al. 2021). To explore the regulation of PSKR1 protein abundance, the OE-PSKR1(#1) lines of tomato plants were used to quantify the PSKR1 protein level by immunoblots with anti-HA antibody. Interestingly, we found that exogenous PSK gradually increased the PSKR1 protein accumulation from 1 to 6 h after treatment (Fig. 2, A and B). It is noteworthy that protein abundance is tightly governed by its biosynthesis and degradation. To investigate whether PSKR1 protein is regulated by protein degradation, the OE-PSKR1 plants were treated with the synthesis inhibitor cycloheximide (CHX). As shown in Fig. 2, C and D, CHX treatment led to large reduction in PSKR1 protein abundance, implying that the protein degradation occurred in HA-tagged PSKR1. In plants, protein degradation usually relied on either the ubiquitin/proteasome degradation pathway or the endocytic lysosomal/vacuolar degradation pathway. When the OE-PSKR1 plants were treated with the 26S proteasome inhibitor MG132, the endogenous protein abundance of PSKR1 continuously increased within the next 6 h (Fig. 2, E and F). By contrast, neither the vacuolar degradation inhibitor Wortmannin (Wm) nor the vacuolar protease inhibitor Concanamycin A (CMA) treatment affected PSKR1 protein abundance (Fig. 2, G to J), which suggested that PSKR1 degradation was likely independent of the vacuolar degradation pathway. Taken together, our results indicated that the degradation of PSKR1 protein is mediated by the ubiquitin/proteasome pathway.
Figure 2.
PSKR1 protein abundance is regulated by the degradation pathway. A) PSK-induced PSKR1 protein abundance. Five-wk-old tomato OE-PSKR1 plants were treated with 1 μM PSK or dH2O for the indicated times. Then the total protein was extracted and subjected to immunoblotting with an anti-HA antibody. Anti-HSP70 was used as a sample loading control. B) Quantification of relative protein intensity in A). The protein abundance of PSKR1 in 0 h of each treatment was defined as 1. C) The protein synthesis inhibitor CHX inhibited PSKR1 protein accumulation. Five-wk-old tomato OE-PSKR1 plants were treated with 200 μM CHX or dH2O for the indicated times. Then the total protein was extracted and subjected to immunoblotting with an anti-HA antibody. Anti-HSP70 was used as a sample loading control. D) Quantification of relative protein intensity in (C). The protein abundance of PSKR1 in 0 h of each treatment was defined as 1. E) The 26S proteasome inhibitor MG132 promoted PSKR1 protein abundance. Five-wk-old tomato OE-PSKR1 plants were treated with 50 μM MG132 or DMSO for the indicated times. Then the total protein was extracted and subjected to immunoblotting with an anti-HA antibody. Anti-HSP70 was used as a sample loading control. F) Quantification of relative protein intensity in (E). The protein abundance of PSKR1 in 0 h of each treatment was defined as 1. G) The vacuolar degradation inhibitor wortmannin (Wm) did not influence PSKR1 protein abundance. Five-wk-old tomato OE-PSKR1 plants were treated with 33 μM Wm or DMSO for the indicated times. Then the total protein was extracted and subjected to immunoblotting with an anti-HA antibody. Anti-HSP70 was used as a sample loading control. H) Quantification of relative protein intensity in (G). The protein abundance of PSKR1 in 0 h of each treatment was defined as 1. I) The vacuolar protease inhibitor CMA did not affect PSKR1 protein abundance. Five-wk-old tomato OE-PSKR1 plants were treated with 0.1 μM CMA or DMSO for the indicated times. Then the total protein was extracted and subjected to immunoblotting with an anti-HA antibody. Anti-HSP70 was used as a sample loading control. J) Quantification of relative protein intensity in (I). The protein abundance of PSKR1 in 0 h of each treatment was defined as 1. Data are presented in (B, D, F, H, J) as the means of three biological replicates (± SD, n = 3), and different letters indicate significant differences (P < 0.05) according to Tukey's test.
E3 ligases PUBs interact with PSKR1
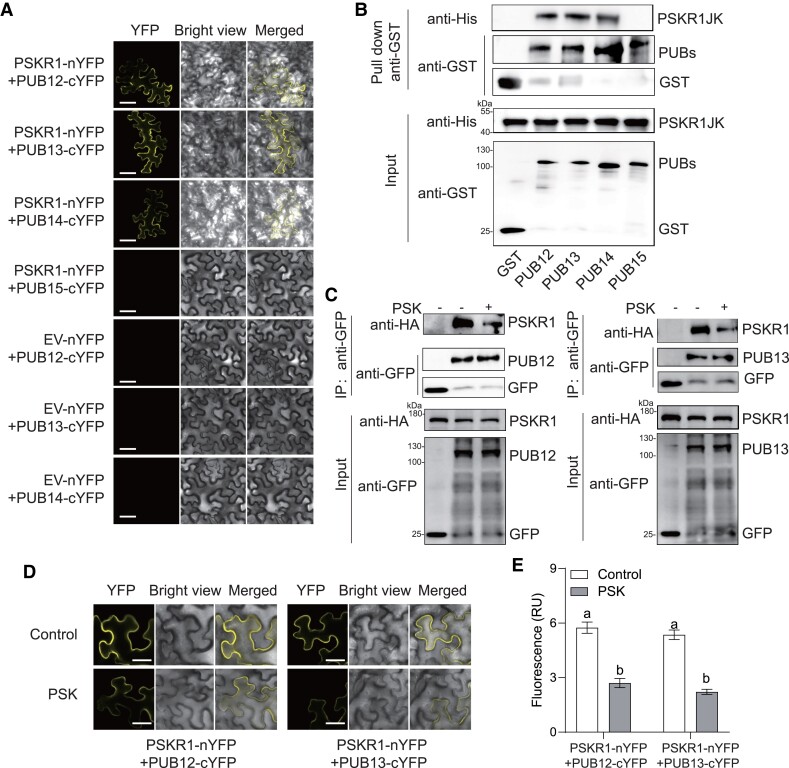
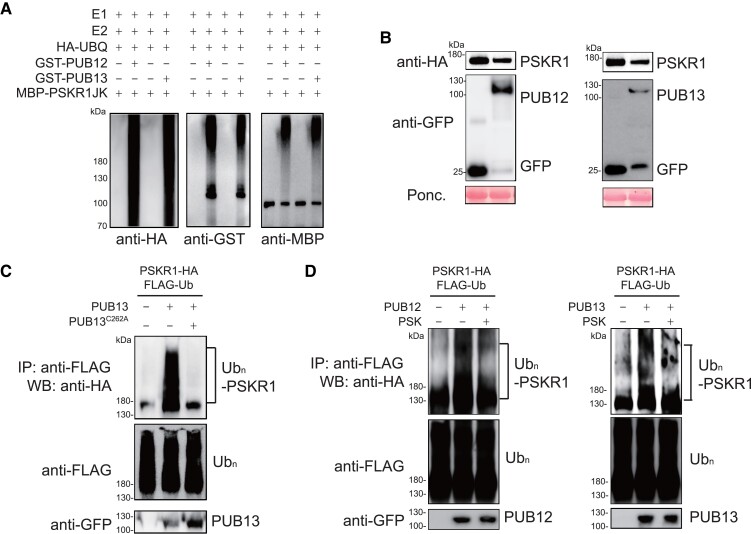
It has been identified that AtPUB12/13 mediated the protein ubiquitination of multiple RLKs in Arabidopsis, including AtFLS2, AtBRI1, AtLRR1, AtCERK1, AtLYK5, etc. (Lu et al. 2011; Liao et al. 2017; Yamaguchi et al. 2017; Zhang et al. 2018; Chen et al. 2021). Hence, we assumed a similar ubiquitination event also happened in post-transcriptional modification of tomato PSKR1 protein. Then, we screened the putative PUBs (PUB12-15), several putative orthologs of AtPUB12/13 in tomato (Supplemental Fig. S1), to investigate if any PUB isoform interacts with PSKR1 via the BiFC assays. Consequently, the YFP fluorescence signal was observed at the cell membrane of N. benthamiana leaves with the expression of PSKR1 with PUB12, PUB13, and PUB14, but not PUB15, suggesting PSKR1 could associate with PUB12/13/14 rather than PUB15 (Fig. 3A). To further confirm whether PSKR1 directly interacts with PUB12/13/14, we performed an in vitro GST-pull-down assay. Consistently, MBP-PSKR1JK could be specifically pulled down by the glutathione beads immobilizing with GST-PUB12, GST-PUB13, and GST-PUB14 rather than GST-PUB15 or GST only (Fig. 3B). To determine whether the binding affinities of specific PUB-PSKR1 interaction is affected by PSK, the Co-IP assays were performed in N. benthamiana leaves, showing that GFP-tagged PUB12 and PUB13 interact with PSKR1-HA in the resting state, whereas the interactions of PUB12-PSKR1 and PUB13-PSKR1 but not PUB14-PSKR1were largely inhibited in the presence of PSK (Fig. 3C; Supplemental Fig. S2). Consistently, the BiFC fluorescence intensity of PUB12-PSKR1 and PUB13-PSKR1 reduced largely with the supplement of PSK (Fig. 3, D and E). Taken together, our results suggest that PUB12/13 is constitutively associate with PSKR1 to maintain its protein abundance, while PSK reduced these interactions.
Figure 3.
PSK induced the dissociation of PSKR1 from PUB12/13. A) BiFC analyses of the binding between PSKR1 and PUBs. Agrobacterium carrying the indicated P2YN/C constructs were inoculated to 5-wk-old fully expanded N. benthamiana leaves. YFP fluorescence was visualized by confocal microscopy at 48 h after inoculation. Bar = 50 μm. B) PSKR1 interacted with PUB12/13/14 in an in vitro GST-pull-down assay. His-fusion PSKR1JK protein was incubated with glutathione beads coupled with GST-PUB12, GST-PUB13, GST-PUB14, GST-PUB15, and GST control for 3 h. Then, the beads were collected and washed for anti-His and anti-GST immunoblots. The input fusion proteins were indicated by immunoblots with anti-His antibody and anti-GST antibody, respectively. C) Co-IP analyses of the associations between HA-tagged PSKR1 and GFP-tagged PUB12/13 with or without application of 1 μM PSK. Five-wk-old fully expanded N. benthamiana leaves were contransfected with PSKR1-HA and PUB12-GFP, PUB13-GFP, or GFP control. After 48 h of inoculation, half of the leaves were infiltrated with 1 μM PSK, and the other parts were infiltrated with dH2O control for 1 h before the sample collection for Co-IP. The associations were detected by anti-HA and anti-GFP immunoblots. The input proteins before IP were detected by anti-HA immunoblot and anti-GFP antibody, respectively. D) Changes in the BiFC fluorescence signal of PSKR1-PUB12 and PSKR1-PUB13 interaction with or without application of 1 μM PSK for 2 h. Bar = 25 μm. E) The fluorescence signal intensity from (D) from three independent repeats was quantified and the data are shown as means of three biological replicates (± SD, n = 3), and different letters indicate significant differences (P < 0.05) according to Tukey's test.
PUB12/13 negatively regulates plant defense against B. cinerea
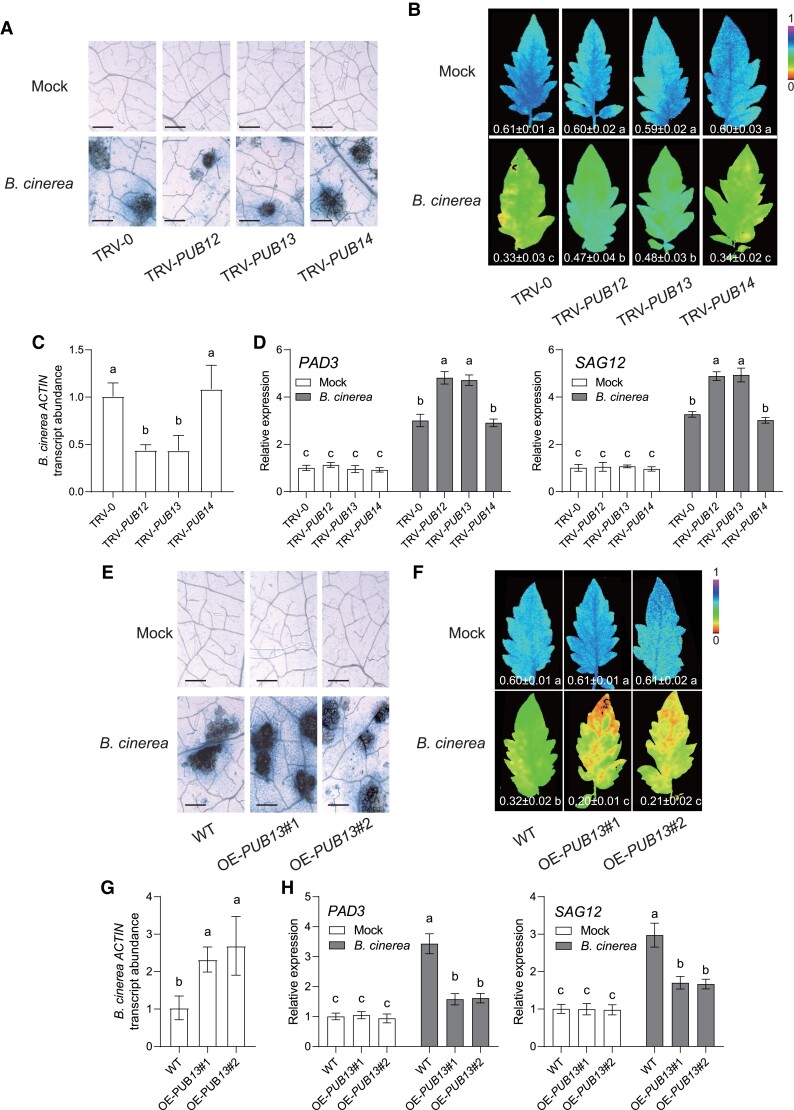
To determine whether PUBs participate in plant immune response, we quantified the transcript abundance of these genes in response to B. cinerea. The gene expressions of PUB12 and PUB13 were greatly reduced in B. cinerea-infected plants, whereas B. cinerea inoculation did not affect PUB14 transcript expression (Supplemental Fig. S3). Then, VIGS assays were conducted for silencing these genes in tomato plants, respectively. This approach reduced the transcript abundance of target genes by up to 70% compared with the empty tobacco rattle virus vector inoculated (TRV-0) control plants (Supplemental Fig. S4). Upon B. cinerea inoculation, the disease susceptibility of TRV-PUB12 and TRV-PUB13 plants was distinctly enhanced compared with TRV-0 plants, as shown by the less dead cell accumulation, the significant increase in ΦPSII values, and decreases in foliar B. cinerea ACTIN mRNA accumulation, whereas silencing target gene in the TRV-PUB14 plants did not affect plant defense against B. cinerea (Fig. 4, A to C). In addition, silencing PUB12 or PUB13 in tomato plants lead to a further increase in transcript abundance of B. cinerea-induced genes PAD3 and SAG12 (Fig. 4D). We next generated and isolated two OE-PUB13 stable lines of tomato plants. Consistent with the disease phenotype of transient expression, PUB13 overexpression lines reduced defense response to B. cinerea (Fig. 4, E to H).
Figure 4.
PUB12 and PUB13 negatively regulated disease resistance against B. cinerea in tomato. A to D) Disease symptoms in tomato PUB-silenced plants inoculated with B. cinerea. A) Representative images of trypan blue staining for cell death in indicated tomato VIGS leaves at 3 d postinoculation with B. cinerea (dpi). Bar = 250 μm. B) Representative chlorophyll fluorescence imaging of ΦPSII at 3 dpi. The value below each individual image indicates the degree of ΦPSII, and the false color code depicted below the image ranges from 0 (black) to 1.0 (purple). C) Relative B. cinerea ACTIN transcript abundance in infected tomato leaves at 1 dpi. The transcript abundance of B. cinerea ACTIN in TRV-0 plants was defined as 1. D) Effects of PUB silencing on the gene expression of B. cinerea-induced genes PAD3 and SAG12 in indicated tomato plants at 1 dpi. The transcript abundance of each gene under mock treatment in TRV-0 plants was defined as 1. E to H) Disease symptoms in tomato OE-PUB13 plants inoculated with B. cinerea. E) Representative images of trypan blue staining for cell death in OE-PUB13 lines and WT plants at 3 dpi. Bar = 250 μm. F) Representative chlorophyll fluorescence imaging of ΦPSII at 3 dpi. The value below each individual image indicates the degree of ΦPSII. G) Relative B. cinerea ACTIN transcript abundance in infected tomato leaves at 1 dpi. The transcript abundance of B. cinerea ACTIN in WT plants was defined as 1. H) Effects of PUB13 overexpression on the gene expression of B. cinerea-induced genes PAD3 and SAG12 in indicated tomato plants at 1 dpi. The transcript abundance of each gene under mock treatment in WT plants was defined as 1. Data are presented in (B to D, F to H) as the means of three biological replicates (±SD, n = 3), and different letters indicate significant differences (P < 0.05) according to Tukey's test.
PSK inhibits PUB12/13-mediated protein ubiquitination of PSKR1
Because PUB12 and PUB13 belong to PLANT U-BOX proteins, it was reasonable to presume that both PUB12 and PUB13 may have E3 ligase activity. According to the ladder-like smear shown above PUB12 and PUB13 proteins using anti-HA antibody, both GST-PUB12 and GST-PUB13 were able to be auto-ubiquitinated in the presence of recombinant UBA1 (E1), UBC10 (E2), ATP, and HA-tagged ubiquitin (HA-UBQ), suggesting PUB12 and PUB13 indeed function as E3 ubiquitin ligases, and can be auto-ubiquitinated (Supplemental Fig. S5). Since PUB12 and PUB13 interacted with PSKR1, we next investigated whether PSKR1 could be a substrate ubiquitinated by PUB12 and PUB13. The in vitro reactions showed the detected ubiquitinated forms of MBP-PSKR1JK only in the presence of GST-PUB12 or GST-PUB13, whereas GST alone failed to ubiquitinate PSKR1JK (Fig. 5A). In planta, we observed a reduced PSKR1 protein abundance when it was co-expressed with PUB12 or PUB13 in N. benthamiana leaves (Fig. 5B). Furthermore, the ubiquitination of PSKR1 by PUB13 was detected in planta, which showed a ladder-like smear band of PSKR1 after western-blot (WB) with anti-HA antibody followed with an anti-FLAG antibody of immunoprecipitation (Fig. 5C). When the conserved E2-binding cysteine residue was mutated to alanine (C262A) in PUB13, the ubiquitination of PKSR1 would be remarkably inhibited (Fig. 5C). Because PSK induced the dissociation of PUB12/13 from PSKR1, we further examined whether PSK treatment modulates PSKR1 ubiquitination in vivo. Apparently, PSK treatment reduced PSKR1 ubiquitination by PUB12 and PUB13 as shown by the decreased ladder-like smear formation (Fig. 5D). Taken together, our results showed that PUB12 and PUB13-mediated PSKR1 ubiquitination in vitro and in vivo, but these ubiquitination events were largely reduced in the presence of PSK.
Figure 5.
PSK inhibited PUB12/13-meidated PSKR1 ubiquitination. A) PUB12/13 ubiquitinates PSKR1 in vitro. Ubiquitination of MBP-PSKR1JK was analyzed in the presence of E1, E2, ubiquitin, and GST-PUB12 or GST-PUB13 by immunoblots with an anti-HA antibody and anti-GST antibody, respectively. B) PUB12/13 reduced PSKR1 protein abundance. Agrobacterium carrying the indicated binary vectors were inoculated to 5-wk-old fully expanded N. benthamiana leaves. After 2-d of inoculation, samples were collected for immunoblots with anti-HA antibody and anti-GFP antibody, respectively. Ponceau S (Ponc.) staining was used as a protein loading control. C) PUB13 ubiquitinates PSKR1 in vivo. Five-wk-old fully expanded N. benthamiana leaves were cotransfected with FLAG-tagged ubiquitin (FLAG-Ub), HA-tagged PSKR1, and together with GFP-tagged PUB13, PUB13C262A (blocking E2-binding mutant), or a control GFP vector. After 2 d of transient expression, the samples were treated with 2 μM MG132 for 3 h and collected. The ubiquitinated PSKR1 was detected with an anti-HA WB after anti-FLAG immunoprecipitation (top). The total ubiquitinated proteins were evaluated by an anti-HA WB (middle) and the input PUB13 proteins were detected by an anti-GFP WB (Bottom). D) PSK suppressed PUB12/13-mediated ubiquitination of PSKR1 in planta. Five-wk-old N. benthamiana leaves were cotransfected with FLAG-Ub, PSKR1-HA, and together with PUB12-GFP, PUB13-GFP, or GFP control. After 2 d of transient expression, the leaves were further infiltrated with 1 μM PSK for 3 h in the presence of 2 μM MG132 before sample collection. The ubiquitinated PSKR1 was detected by an anti-HA antibody after immunoprecipitation with an anti-FLAG antibody (top). The total ubiquitinated proteins (middle) and input GFP-tagged PUB proteins (bottom) were indicated with the immunoblot with an anti-FLAG antibody and an anti-GFP antibody, respectively.
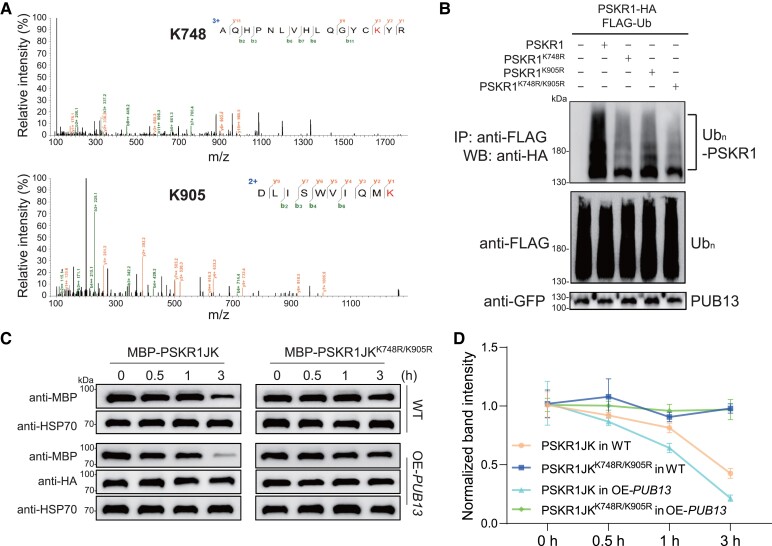
To identify the potential PSKR1 residue(s) ubiquitinated, we performed LC-MS/MS analysis after a ubiquitination reaction. According to the mass spectrum results, we identified two modified peptides (735-AQHPNLVHLQGYCKYR-750 and 896-DLISWVIQMK-905) ubiquitinated at Lys748 and Lys905 residues, respectively (Fig. 6A). Both Lys sites are located in the kinase domain of PSKR1 (Supplemental Fig. S6). To reveal the function of these Lys sites in PSKR1 ubiquitination by PUB13, we mutagenized Lys to Arg and found the ubiquitination of PSKR1 was diminished, especially in PSKR1K748R/K905R. These results suggest that these two Lys sites are critical for PUB13-mediated PSKR1 ubiquitination (Fig. 6B). To explore whether ubiquitination affects PSKR1 protein stability, we examined the degradation of recombinant MBP-PSKR1JK and MBP-PSKR1JKK748R/K905R incubated with total protein extracted from OE-PUB13 and WT plants in a cell-free system. Recombinant MBP-PSKR1JK incubated with OE-PUB13 extracted protein showed a faster degradation rate than that with WT extracts, whereas recombinant MBP- PSKR1JKK748R/K905R were quite stable either incubated with total protein extracted from OE-PUB13 lines or WT extracts (Fig. 6, C and D). These results indicate that ubiquitination at the K748 and K905 sites determines the protein stability of PSKR1.
Figure 6.
PSKR1K748/K905 was critical for PUB13-mediated ubiquitination. A) Lys-748 and Lys-905 residues of PSKR1 were ubiquitinated by PUB13. An LC-MS/MS analysis of the ubiquitination reaction carried out by GST-PUB13 and MBP-PSKR1JK showed the ubiquitination of Lys-748 and Lys-905 in PSKR1 by PUB13. B) PSKR1K748/K905R showed reduced ubiquitination by PUB13. Five-wk-old N. benthamiana leaves were cotransfected with FLAG-Ub, PUB13-GFP, and together with PSKR1-HA, PSKR1K748R-HA, PSKR1K905R-HA, PSKR1K748R/K905R-HA, or empty vector control. After 2 d of transient expression, the leaves were further infiltrated with 2 μM MG132 for 3 h before sample collection. The ubiquitinated PSKR1 was detected by an anti-HA antibody after immunoprecipitation with an anti-FLAG antibody (top). The total ubiquitinated proteins (middle) and input GFP-tagged PUB proteins (bottom) were indicated with the immunoblot with an anti-FLAG antibody and an anti-GFP antibody, respectively. C) Degradation of MBP-PSKR1JK and MBP-PSKR1JKK748R/K905R in cell-free assays from OE-PUB13 and WT protein extracts. Recombinant purified MBP-PSKR1JK and MBP-PSKR1JKK748R/K905R were incubated with OE-PUB13 and WT protein extracts for 0.5, 1, and 3 h. Recombinant protein abundance was evaluated by immunoblot with an anti-MBP antibody. The immunoblot with anti-HSP70 antibody was used as a loading control and the immunoblot with anti-HA antibody was used to indicate PUB13 overexpression. D) Quantification of relative protein intensity in (C). The protein abundance of PSKR1 in 0 h of each treatment was defined as 1. Data are presented in (D) as the means of three biological replicates (± SD, n = 3).
PUB13 inhibits PSK-induced defense against B. cinerea in tomato
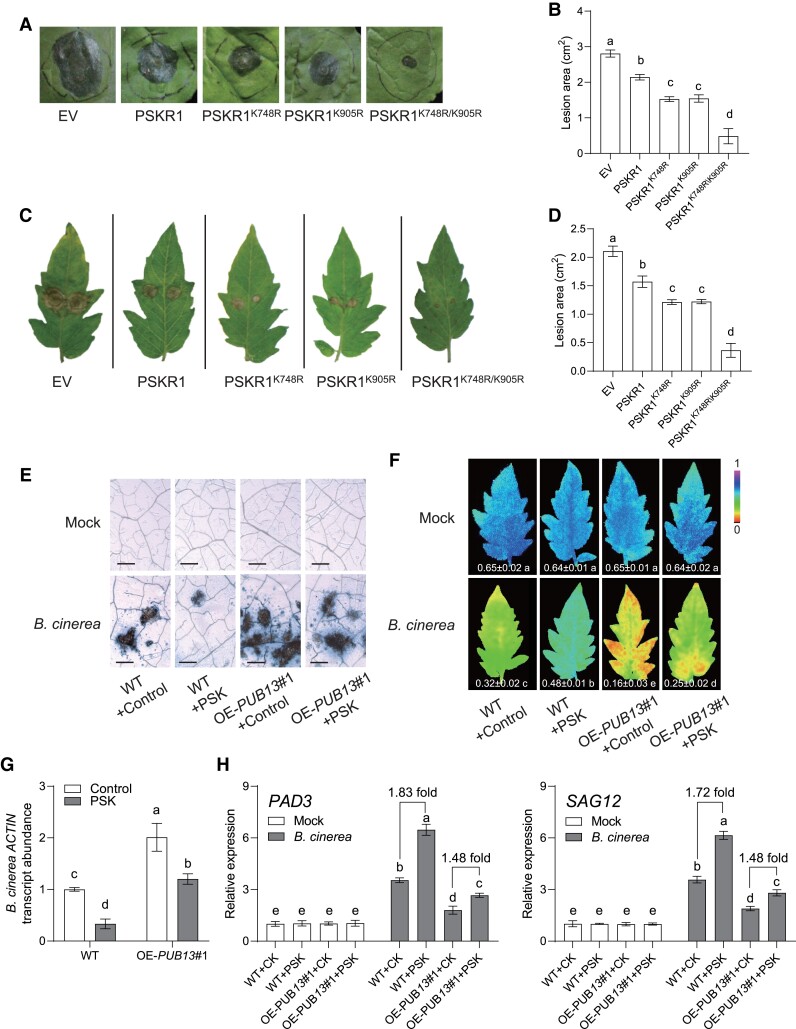
To gain further insight into PUB13-mediated PSKR1 ubiquitination on plant defense against B. cinerea, different types of PSKR1 were transiently expressed in N. benthamiana leaves. Compared with the WT PSKR1, the Lys-to-Arg mutated forms of PSKR1, in particular PSKR1K748R/K905R, presented higher resistance to B. cinerea (Fig. 7, A and B). Consistently, transiently expressing different types of PSKR1 in tomato OE-PUB13 lines also indicated that blocking PUB13-mediated ubiquitination was preferer to increase defense against B. cinerea attacks (Fig. 7, C and D).
Figure 7.
PUB13-mediated PSKR1 ubiquitination affected PSK- initiated tomato defense against B. cinerea. A, B) Effect of Lys-to-Arg mutated PSKR1 variants on plant defense to B. cinerea in N. benthamiana leaves. A) Representative images of disease symptoms on N. benthamiana leaves at 5 d postinoculation with B. cinerea (dpi). Images shown were digitally extracted and scaled for comparison. B) Quantification of B. cinerea-induced lesion area of N. benthamiana leaves at 5 dpi. Agrobacterium carrying the indicated binary vectors were transiently expressed in N. benthamiana leaves for 2 d, and then leaves were used for B. cinerea dipping inoculation. C, D) Effect of Lys-to-Arg mutated PSKR1 variants on plant defense to B. cinerea in tomato OE-PUB13 lines. C) Representative images of disease symptom on tomato leaves at 3 dpi. Images shown were digitally extracted and scaled for comparison. D) Quantification of B. cinerea-induced lesion area of indicated tomato leaves at 3 dpi. Agrobacterium carrying the indicated binary vectors were transiently expressed in tomato OE-PUB13 lines for 2 d, and then leaves were used for B. cinerea dipping inoculation. E to H) Effects of PSK on defense of OE-PUB13 and WT plants to B. cinerea infection. E) Representative leaf images for disease symptoms at 3 d postinoculation with B. cinerea (dpi). Bar = 250 μm. F) The representative chlorophyll fluorescence imaging of ΦPSII at 3 dpi. The value below each individual image indicates the degree of ΦPSII. G) Relative B. cinerea actin transcript abundance in infected tomato leaves at 1 dpi. Five-wk-old indicated tomato plants were treated with 10 μM PSK or dH2O control 12 h before B. cinerea inoculation. H) Effects of PSK application on the gene expression of B. cinerea-induced genes PAD3 and SAG12 in tomato OE-PUB13 and WT plants at 1 dpi. The transcript abundance of each gene under mock treatment in WT plants was defined as 1. Five-wk-old tomato OE-PUB13 and WT plants pretreated with 10 μM PSK or dH2O control for 1 d were sprayed with B. cinerea spore suspension. Data are presented in (B, D, F to H) as the means of three biological replicates (± SD, n = 6 in B, D; 3 in F to H), and different letters indicate significant differences (P < 0.05) according to Tukey's test.
To examine whether PUB13 affects the PSK-initiated defense response of tomato plants to B. cinerea, we examined the defense response in OE-PUB13 plants after PSK elicitation. PSK-initiated defense was suppressed significantly in OE-PUB13 lines compared with WT plants (Fig. 7, E to G). Consistently, PSK further increased 1.83-fold PAD3 gene expression and 1.72-fold SAG12 gene expression, while that PSK-mediated induction was dropped to 1.48- and 1.48-fold in OE-PUB13 lines in response to B. cinerea (Fig. 7H). Our data collectively suggested that PUB13 acts as a critical regulator of PSKR1 to modulate PSK-induced defense response of tomato plants.
Discussion
PSK was originally identified as a plant growth-promoting factor and has since been shown to be involved in multiple physiological processes, including plant growth, development, and defense response to environmental stress (Matsubayashi and Sakagami 1996; Sauter 2015). Previously, we identified the tomato PSK peptide receptor PSKR1 and further revealed PSKR1 played a master role in the signal transduction cascade by which PSK triggers defense against the necrotrophic pathogen B. cinerea (Zhang et al. 2018). Consistent with these previous findings, we generated the stable PSKR1 knock-out and overexpression tomato plants and demonstrated that PSKR1 played a positive role in the activation of defense response to B. cinerea infection (Fig. 1). As a typical plant peptide receptor, PSKR1 could be a good paradigm to understand peptide-mediating signaling in plants. The activity of PSKR1 is controlled by different posttranslational modifications. For instance, the phosphorylation of AtPSKR1 cytoplasmic kinase domain resulted in reduced transphosphorylation activity, impairing shoot growth promotion (Kaufmann et al. 2017). In this study, we found that there was another regulation mechanism of PSKR1 through E3 ligases-mediated degradation by the ubiquitin/proteasome pathway. PUB12/13 continuously bound to PSKR1 with moderate ubiquitylation of PSKR1 in the absence of pathogen infection, whereas PSK perception promoted PSKR1 and PUB12/13 dissociation, contributing to the activation of PSK-mediated intercellular defense signaling.
Protein accumulation and stability of plant peptide receptors determined peptide signal outputs. We mutated or overexpressed PSKR1 in tomato plants, leading to significant changes in plant defense against B. cinerea (Fig. 1). Consistent with our findings, overexpression of OsPSKR1 in rice enhanced resistance to bacterial leaf streak caused by Xanthomonas oryzae pv. oryzicola (Yang et al. 2019). AtPSKR1-overexpressed lines of Arabidopsis plants had an effect on resistance to A. brassicicola (Mosher et al. 2013). Multiple danger-associated peptides, such as PSK and Plant elicitor peptides (Peps) could induce their receptors at transcript and/or protein levels (Jing et al. 2019) (Fig. 2). On the other hand, the protein accumulation of plant membrane-localized receptors was also modulated by different posttranslational modification. Ubiquitylation-mediated protein degradation occurred in nearly all aspects of plant physiological processes, which was also commonly observed in plasma membrane-associated RLKs (Trenner et al. 2022). Inhibition of 26S proteasome activity by MG132 not only promoted the PSKR1 protein abundance we showed here (Fig. 2), but also increased the protein abundance of other RLK-type receptors elsewhere, including fungal chitin receptor AtLYK5, bacterial flagellin receptor AtFLS2, and RGF1 peptide receptor AtRGFR1 (Lu et al. 2011; Liao et al. 2017; An et al. 2018).
Although dozens of plant peptide receptors have been identified, the E3 ligase-mediated posttranslational modifications of plant peptide receptors have been poorly discovered. In this study, we found that PUB12/13 directly targeted and regulated PSKR1 protein degradation via ubiquitination modification (Figs. 3 and 5). PUB13 ubiquitinated PSKR1 intracellular kinase domain at Lys-748 and Lys-905 sites (Fig. 6). Recently, a finding indicated another plant peptide EPF/EPFL activated AtPUB30/31 ubiquitinating the EPF/EPFL receptor ERECTA for degradation to ensure optimal signaling outputs in turn (Chen et al. 2022). In line with prior studies with Arabidopsis, AtPUB12/13 ubiquitylated bacterial flagellin receptor AtFLS2 and brassinosteroid (BR) receptor AtBRI1 in the presence of its ligand, resulting in receptors endocytosis and degradation (Lu et al. 2011; Zhou et al. 2018). However, these protein turnover in Arabidopsis were not coincident with the PSK ligand activation of PSKR1. In Arabidopsis, both flg22 and BR ligands promoted the formation of the PUB-RLK complex to induce RLK receptor protein degradation (Lu et al. 2011; Zhou et al. 2018). In contrast, PSK induced the dissociation of PUB12/13 from PSKR1 and attenuated PUB12/13-mediated ubiquitylation on PSKR1, implying PUB12/13 continuously targeted PSKR1 to moderately modulate its protein abundance in the absence of PSK activation (Figs. 3 and 5). Chitooctaose induced the dissociation of AtPUB13 and AtLYK5, reducing ubiquitylation-mediated protein degradation of AtLYK5 (Liao et al. 2017). These ubiquitylation events had the effect of controlling a proper and effective receptor abundance in steady status, avoiding spurious and harmful consequences caused by excessive receptor signal outputs. The PUB-RLK interactions not only contributed to PUB-mediated ubiquitylation of RLKs but also might result in the phosphorylation of PUBs in turn (Mbengue et al. 2010; Zhou et al. 2018). Further study will be required to explore whether PSKR1 could phosphorylate PUB12/13 to regulate the in turn ubiquitylation and downstream defense responses.
As plant-specific E3 ligases, plant PUB proteins have been studied extensively in connection with plant defense against diverse pathogens with different traits (Trenner et al. 2022). For example, Atpub22/23/24 mutants showed broad resistance to diverse pathogen infection, including the bacteria P. syringae pv. tomato, the oomycete H. arabidopsidis, and the fungus F. oxysporum (Trujillo et al. 2008; Chen et al. 2014). In this study, we found the expression of PUB13 together with PUB12 was suppressed in response to B. cinerea inoculation, and silencing of PUB12 or PUB13 significantly enhanced leaf defense, leading to alleviated disease symptoms (Fig. 4). Similarly, Atpub25, Atpub26 single mutants and Atpub12/13, Atpub25/26 double mutants in Arabidopsis enhanced disease resistance against B. cinerea (Wang et al. 2018; Zhou et al. 2018). Besides, both transient overexpression of PUB13 in N. benthamiana and transgenic OE-PUB13 tomato lines decreased plant defense responses against B. cinerea, which provided the genetic evidence that PUB13 participated in plant defense against B. cinerea as a negative regulator (Fig. 4). To date, majority of PUBs identified in different plant species played a negative role in defense response to diverse pathogen infection (Trenner et al. 2022). Apart from the negative function of PUB13 in plant defense, this study indicates that PUB13-mediated ubiquitylation suppressed PSK-initiated defense response, and inhibiting the ubiquitylation of PSKR1 enhanced plant resistance to B. cinerea attack (Fig. 7).
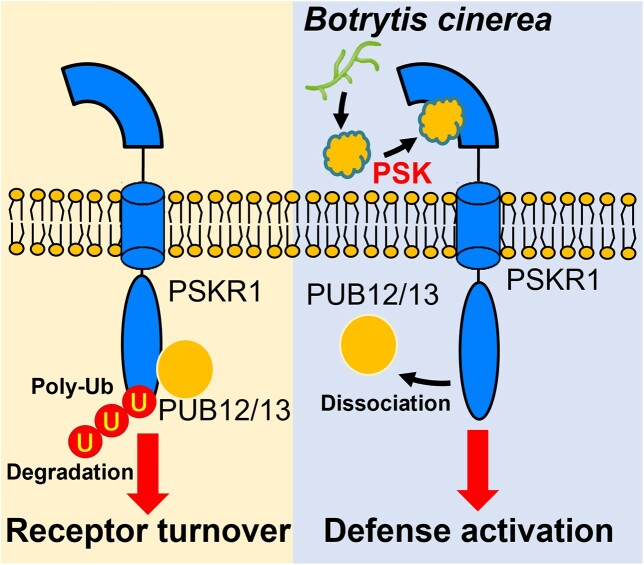
In conclusion, the data presented here showed that the protein abundance of PSK receptor PSKR1 was modulated by the E3 ligase-mediated ubiquitylation. PUB12 and PUB13 substantially interacted with PSKR1, leading to its moderate protein turnover in absence of pathogen attacks. In presence of B. cinerea infection, the E3 ligases-peptide receptor interactions were attenuated by the perception of PSK, which potentially contributed to enhance intercellular defense outputs (Fig. 8). Our finding provides a basis for understanding the sophisticated regulatory network of peptide signaling.
Figure 8.
A proposed working model for the role of PUB12/13 in PSK-induced defense against B. cinerea in tomato plants. In the absence of a pathogen attack, PUB12/13 moderately ubiquitinates PSKR1 for eventual degradation. Upon B. cinerea infection, PSK, acting as an inducible DAMP, promotes the dissociation of PUB12/13 from PKSR1, triggering downstream defense response.
Materials and methods
Plasmid construction and transformation
To generate the CRISPR/Cas9 vectors, the target mutagenesis sequences of PSKR1 were selected using the CRISPR-P program (available at http://crispr.hzau.edu.cn/CRISPR2/). The synthesized oligos were cloned into the AtUb-sgRNA-AtUBQ-Cas9 intermediate vector, and the sgRNA expression cassettes were assembled into the pCambia1301 binary vector. To generate the constructs for transgenic overexpression lines, PCR-amplified coding sequence (CDS) without stop codon of PSKR1 and PUB13 from tomato cDNA were inserted into the CaMV 35S-driven pFGC1008 vector with an HA tag at C-terminus. The confirmed vectors were transformed into tomato plants by Agrobacterium-mediated cotyledon tissue culture. The stable tomato knock-out and over-expression lines were identified as previously described (Hu et al. 2021b). To generate the virus-induced gene silencing (VIGS) vectors, a 200–500 bp fragment of CDS in target genes was amplified and cloned into the EcoR I-BamH I sites of pTRV2. Then, VIGS vectors were electroporated into A. tumefaciens strain GV3101.The Agrobacterium-mediated VIGS assays in tomato plants were performed as previously described (Zhang et al. 2018). The tomato VIGS plants with higher than 70% silencing efficiency were selected for experiments.
For partial binary vectors used in transient overexpression assays in Nicotiana benthamiana and tomato, CDS without stop codon of each gene were amplified from tomato cDNA and cloned into pDONR-ZEO intermediate vector via GATEWAY BP reaction. After Sanger sequencing confirmation, the fragment cassette from the pDONR-ZEO vector was assembled into CaMV 35S-driven destination vectors: pGWB505 with C-terminal GFP tag, pGWB514 with C-terminal HA tag. For BiFC assay, full-length CDS without stop codon of PSKR1 and PUB12/13/14 were constructed into CaMV 35S-driven vectors of p2YN and p2YC, respectively.
For Escherichia coli expression vectors, the protein-coding regions of E1 and E2, PUB12/13/14, and PSKR1JK (the juxtamembrane and kinase domains of PKSR1) were recombined into pET28a, pGEX-4T-1, and pMAL-c2x. Then, the vectors were transformed into E. coli BL21(DE3) strain for recombinant protein expression.
The primers used for vector plasmid construction were listed in Supplemental Table S1.
Plant growth and chemical treatments
The tomato (Solanum lycopersicum) cultivar condine red and N. benthamiana were used in this study. Germinated seeds were grown in 100 cm3 plastic pots (one plant per pot) containing peat and vermiculite (7:3, v/v), receiving Hoagland nutrient solution. The growth conditions were maintained at 23/21 °C (day/night) and a photoperiod of 12 h with 400 µmol m−2 s−1 photosynthetic photon flux density.
Five-wk-old tomato plants were sprayed with 10 μM PSK (Iris Biotech) or dH2O as control 12 h before pathogen inoculation, and N. benthamiana leaves were infiltrated with 1 μM PSK (Iris Biotech) or dH2O as control 1 h before sample collection. For the protein abundance assay, the uppermost one or two fully expanded leaves from 5-wk-old OE-PSKR1 tomato plants were inoculated with 1 μM PSK, 200 μM CHX (MedChemExpress), 50 μM MG132 (MedChemExpress), 33 μM wortmannin (Wm) (Sigma-Aldrich), 0.1 μM CMA (MedChemExpress), and the corresponding control dH2O or dimethyl sulfoxide (DMSO).
Pathogen inoculation and disease symptom assays
The B. cinerea B05.10 strain was used for the pathogen assay. B. cinerea culture and isolation were performed as previously described (Hu et al. 2018). For pathogen inoculation in tomato plants, approximate 2 × 105 mL−1 of B. cinerea spores were uniformly sprayed to 5-wk-old tomato plants. For pathogen inoculation in N. benthamiana leaves, 2.5 μL B. cinerea spores were dipped on the upper surface of the detached N. benthamiana leaves. The disease symptoms were evaluated via quantifying B. cinerea actin transcript abundance by RT-qPCR and analyzing the chlorophyll fluorescence with an Image-pulse amplitude modulation chlorophyll fluorometer using the saturation pulse method (Hu et al. 2018).
Agrobacterium-mediated transient expression in N. benthamiana and tomato
The transient overexpression assays in N. benthamiana and tomato were performed as previously described with minor modification (Reichardt et al. 2018). The binary vectors were introduced into A. tumefaciens strain GV3101 for transient expression in N. benthamiana, and into A. tumefaciens strain C58C1 for transient expression in tomato, respectively. After 2-d growth on the LB plates with appropriate antibiotics, the bacterial colonies were washed by the infiltration buffer (10 mM MES, 10 mM MgCl2, 0.15 mM acetosyringone, pH 5.6), and mixed to result in a final OD600 = 0.8 for syringe infiltration in N. benthamiana, OD600 = 0.3 for vacuum infiltration in tomato, respectively. A needleless syringe was used to infiltrate the bacterial suspension into the leaves of 6-wk-old N. benthamiana. A large desiccator was used for vacuum (75 mbar) infiltration of agrobacteria in 3-wk-old tomato plants. Three days after infiltration, the leaves of N. benthamiana and tomato were harvested for further analysis. Transient protein expression was detected by the immunoblots with indicated antibodies as previously described (Liao et al. 2017).
Protein–protein interaction assays
For BiFC assays, Agrobacterium suspension (OD600 = 1) carrying p2YN-PSKR1 and p2YC-PUBs was infiltrated into fully expanded N. benthamiana leaves using a needleless syringe. After 48 h of inoculation, the fluorescence images were captured using a confocal laser scanning microscope (Zeiss LSM 780).
For GST-pull-down assays, the His-, GST-fused recombinant proteins were expressed in E. coli BL21(DE3) strain, and purified with Ni-NTA Agarose (Qiagen), or Pierce Glutathione Agarose (Thermo), according to each manufacturer's instructions. GST-PUBs were used for pulling down interacted proteins as previously described (Hu et al. 2021a). The pulled-down proteins were detected by immunoblotting with anti-His antibody (Sigma).
For Co-IP assays, Agrobacterium suspension (OD600 = 1) carrying different binary vectors was syringe-infiltrated into 5-wk-old N. benthamiana leaves. The leaf samples were collected at 48-h postinoculation (hpi) for Co-IP. The Co-IP assays were performed as previously described (Hu et al. 2021a). Briefly, the GFP-tagged proteins were immunoprecipitated with 10 μL GFP-Trap Agarose (Chromotek) in 300 μL Co-IP buffer [50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100 (v/v), 2 mM NaF, 2 mM Na3VO4, 1 mM DTT, and protease inhibitor from Roche]. After 3-h incubation and 4 times washing, the immunoprecipitated proteins were analyzed by immunoblot with appropriate antibodies.
In vitro and in vivo ubiquitination assays
In addition to His-, GST-fused purified recombinant proteins, the MBP-tagged proteins were also purified from E. coli BL21(DE3) strain by Amylose Resin High Flow (New England Biolabs). The in vitro ubiquitination assay was performed as previously described with minor modifications (Liao et al. 2017). The 30 μL ubiquitination reaction mixtures contain 0.25 μg of His-E1, 0.5 μg of His-E2, 1.25 μg of HA-UBQ (Boston Biochem), 1 μg of GST-PUBs as E3, and 1 μg of MBP-tagged substrate MBP-PSKR1JK or MBP protein control in the ubiquitination buffer (0.1 M Tris–HCl, pH 7.5, 25 mM MgCl2, 2.5 mM DTT, and 10 mM ATP). After 3-h of incubation at 30 °C, the reactions were stopped by adding 4×SDS loading buffer and then boiling for 5 min. The ubiquitinated proteins were analyzed by WB with anti-HA (Thermo-Fisher), anti-GST (Cell Signaling Technology), or anti-MBP (Cell Signaling Technology) antibodies, respectively.
For in vivo ubiquitination assay, 5-wk-old N. benthamiana leaves were transfected with a mixture of agrobacterium suspension carrying FLAG-UBQ, PSKR1-HA, and GFP-tagged PUB12, PUB13, or GFP alone as a control, respectively. The ubiquitinated proteins were immunoprecipitated by the anti-FLAG M2 Affinity Gel (Sigma-Aldrich, 2220), and then detected by WB with an anti-HA antibody.
Mass spectrometry analysis of ubiquitination sites
The ubiquitination sites were identified as previously described (Ma et al. 2020). In vitro ubiquitination reactions with GST-PUB13 and MBP-PSKR1JK were performed as mentioned above with overnight incubation. Reactions were loaded on an SDS-PAGE gel and ran for a relatively short time until the ubiquitinated bands could be separated from the original MBP-PSKR1JK. Ubiquitinated bands were sliced and trypsin-digested for 20 h at 37 °C before LC-MS/MS analysis on an LTQ-Orbitrap hybrid mass spectrometer (Thermo-Fisher). The MS/MS spectra were analyzed and the images were exported with Mascot2.2 software.
Cell-free protein degradation assay
Cell-free protein degradation assays were performed as previously described with minor modifications (Chong et al. 2022). Total proteins from the leaves of 5-wk-old OE-PUB13 and WT plants were extracted by the extraction buffer (25 mM Tris–HCl, pH 5.6, 10 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP, and 1 mM phenylmethylsulfonyl fluoride). The indicated recombinant proteins were then added to total protein extracts. After incubation for indicated times, the proteins were separated by SDS-PAGE. The recombinant proteins were determined by immunoblot with anti-MBP antibody.
Transcript analysis
Total RNA was isolated with an RNA prepure plant kit (Tiangen, Beijing, China) and reverse-transcribed to cDNA using a ReverTra Ace qPCR RT kit (Vazyme, Nanjing, China). RT-qPCR assays were performed using the SYBR Green PCR master mix kit (Vazyme, Nanjing, China) on a LightCycler 480 II detection system (Roche, Germany). ACTIN and UBQ were used as internal references to calculate the relative expression of target genes. Gene-specific primers are listed in Supplemental Table S1.
Statistical analysis
At least three independent biological replicates were sampled for each determination. Unless otherwise stated, each biological replicate consisted of an independent sample that was pooled of two leaves, each taken from a different plant. The experiments were independently performed twice or three times. The obtained data were subjected to analysis of variance using SAS 8.0 software (SAS Institute), and means were compared using Tukey's test at the 5% level.
Accession numbers
Sequence data from this article can be found in the Sol Genomics Network (https://solgenomics.net/) database and GenBank database under the following accession numbers: PUB12 (Solyc11g066040), PUB13 (Solyc06g076040), PUB14 (Solyc11g008390), PUB15 (Solyc04g082440), PSKR1 (Solyc01g008140), MRN1 (Solyc12g006530), RLKR (Solyc02g079990), SAG12 (Solyc02g076910), PAD3 (Solyc09g092600), ACTIN (Solyc03g078400), UBQ (Solyc01g096290) and B. cinerea ACTIN (XM_001553318).
Supplemental data
The following materials are available in the online version of this article.
The following supporting data are available for this article.
Supplemental Figure S1 . Phylogenetic analysis of partial PUBs from tomato and Arabidopsis.
Supplemental Figure S2 . Effect of PSK application on PSKR1-PUB14 interaction.
Supplemental Figure S3 . Effects of B. cinerea inoculation on the transcript expression of PUBs in tomato plants.
Supplemental Figure S4 . Gene silence efficiency of VIGS tomato plants.
Supplemental Figure S5 . PUB12 and PUB13 have E3 ligase enzyme activity.
Supplemental Figure S6 . Schematic diagram showing Lys-748 and Lys-905 ubiquitination sites of PSKR1.
Supplemental Table S1 . Primers used in this study.
Supplementary Material
Contributor Information
Zhangjian Hu, Department of Horticulture, Zhejiang University, Hangzhou 310058, China; Hainan Institute, Zhejiang University, Yazhou Bay Science and Technology City, Sanya 572025, China.
Hanmo Fang, Department of Horticulture, Zhejiang University, Hangzhou 310058, China.
Changan Zhu, Department of Horticulture, Zhejiang University, Hangzhou 310058, China.
Shaohan Gu, Department of Horticulture, Zhejiang University, Hangzhou 310058, China.
Shuting Ding, Department of Horticulture, Zhejiang University, Hangzhou 310058, China.
Jingquan Yu, Department of Horticulture, Zhejiang University, Hangzhou 310058, China; Hainan Institute, Zhejiang University, Yazhou Bay Science and Technology City, Sanya 572025, China.
Kai Shi, Department of Horticulture, Zhejiang University, Hangzhou 310058, China; Hainan Institute, Zhejiang University, Yazhou Bay Science and Technology City, Sanya 572025, China.
Funding
This work was supported by the Key Research and Development Program of Zhejiang Province (2021C02040), the National Natural Science Foundation of China (32172650), and the Starry Night Science Fund of the Zhejiang University Shanghai Institute for Advanced Study (SN-ZJU-SIAS-0011).
Data availability
The authors confirm that all experimental data are available and accessible via the main text and/or the supplemental data.
References
- An ZC, Liu YL, Ou Y, Li J, Zhang BW, Sun DY, Sun Y, Tang WQ. Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc Natl Acad Sci U S A. 2018:115(5):1123–1128. 10.1073/pnas.1714177115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol. 2017:55(1):257–286. 10.1146/annurev-phyto-080614-120106 [DOI] [PubMed] [Google Scholar]
- Chen L, Cochran AM, Waite JM, Shirasu K, Bemis SM, Torii KU. Direct attenuation of Arabidopsis ERECTA signalling by a pair of U-box E3 ligases. Nat Plants. 2022:9(1):112–127. 10.1038/s41477-022-01303-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XX, Wang TT, Rehman AU, Wang Y, Qi JS, Li Z, Song CP, Wang BS, Yang SH, Gong ZZ. Arabidopsis U-box E3 ubiquitin ligase PUB11 negatively regulates drought tolerance by degrading the receptor-like protein kinases LRR1 and KIN7. J Integr Plant Biol. 2021:63(3):494–509. 10.1111/jipb.13058 [DOI] [PubMed] [Google Scholar]
- Chen YC, Wong CL, Muzzi F, Vlaardingerbroek I, Kidd BN, Schenk PM. Root defense analysis against Fusarium oxysporum reveals new regulators to confer resistance. Sci Rep. 2014:4(1):5584. 10.1038/srep05584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L, Xu R, Huang PC, Guo PC, Zhu MK, Du H, Sun XL, Ku LX, Zhu J-K, Zhu YF. The tomato OST1-VOZ1 module regulates drought-mediated flowering. Plant Cell. 2022:34(5):2001–2018. 10.1093/plcell/koac026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Lv J, Hu Z, Wang J, Wang P, Yu J, Foyer CH, Shi K. Phytosulfokine peptide optimizes plant growth and defense via glutamine synthetase GS2 phosphorylation in tomato. EMBO J. 2023:42(6):e111858. 10.15252/embj.2022111858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JB, Bai PF, Ning YS, Wang JY, Shi XT, Xiong YH, Zhang K, He F, Zhang CY, Wang RY, et al. The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe. 2018:23(4):498–510. 10.1016/j.chom.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann J, Linke D, Bonniger C, Tholey A, Sauter M. Conserved phosphorylation sites in the activation loop of the Arabidopsis phytosulfokine receptor PSKR1 differentially affect kinase and receptor activity. Biochem J. 2015:472(3):379–391. 10.1042/BJ20150147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Li J, Ding S, Cheng F, Li X, Jiang Y, Yu J, Foyer CH, Shi K. The protein kinase CPK28 phosphorylates ascorbate peroxidase and enhances thermotolerance in tomato. Plant Physiol. 2021a:186(2):1302–1317. 10.1093/plphys/kiab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Ma Q, Foyer CH, Lei C, Choi HW, Zheng C, Li J, Zuo J, Mao Z, Mei Y, et al. High CO2- and pathogen-driven expression of the carbonic anhydrase βCA3 confers basal immunity in tomato. New Phytol. 2021b:229(5):2827–2843. 10.1111/nph.17087 [DOI] [PubMed] [Google Scholar]
- Hu Z, Shao S, Zheng C, Sun Z, Shi J, Yu J, Qi Z, Shi K. Induction of systemic resistance in tomato against Botrytis cinerea by N-decanoyl-homoserine lactone via jasmonic acid signaling. Planta. 2018:247(5):1217–1227. 10.1007/s00425-018-2860-7 [DOI] [PubMed] [Google Scholar]
- Jing Y, Zheng X, Zhang D, Shen N, Wang Y, Yang L, Fu A, Shi J, Zhao F, Lan W, et al. Danger-associated peptides interact with PIN-dependent local auxin distribution to inhibit root growth in Arabidopsis. Plant Cell. 2019:31(8):1767–1787. 10.1105/tpc.18.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann C, Motzkus M, Sauter M. Phosphorylation of the phytosulfokine peptide receptor PSKR1 controls receptor activity. J Exp Bot. 2017:68(7):1411–1423. 10.1093/jxb/erx030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann C, Sauter M. Sulfated plant peptide hormones. J Exp Bot. 2019:70(16):4267–4277. 10.1093/jxb/erz292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann C, Stuhrwohldt N, Sauter M. Tyrosylprotein sulfotransferase-dependent and -independent regulation of root development and signaling by PSK LRR receptor kinases in Arabidopsis. J Exp Bot. 2021:72(15):5508–5521. 10.1093/jxb/erab233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwezi L, Ruzvidzo O, Wheeler JI, Govender K, Iacuone S, Thompson PE, Gehring C, Irving HR. The phytosulfokine (PSKα) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J Biol Chem. 2011:286(25):22580–22588. 10.1074/jbc.M110.168823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwig F, Dahlke RI, Stuhrwohldt N, Hartmann J, Harter K, Sauter M. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell. 2015:27(6):1718–1729. 10.1105/tpc.15.00306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Cao Y, Sun X, Espinoza C, Nguyen CT, Liang Y, Stacey G. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 2017:214(4):1646–1656. 10.1111/nph.14472 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011:332(6036):1439–1442. 10.1126/science.1204903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Claus LAN, Leslie ME, Tao K, Wu Z, Liu J, Yu X, Li B, Zhou J, Savatin DV, et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature. 2020:581(7807):199–203. 10.1038/s41586-020-2210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Kihara H, Niwa M, Sakagami Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006:142(1):45–53. 10.1104/pp.106.081109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002:296(5572):1470–1472. 10.1126/science.1069607 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci U S A. 1996:93(15):7623–7627. 10.1073/pnas.93.15.7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Herve C, et al. The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell. 2010:22(10):3474–3488. 10.1105/tpc.110.075861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S, Seybold H, Rodriguez P, Stahl M, Davies KA, Dayaratne S, Morillo SA, Wierzba M, Favery B, Keller H, et al. The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 2013:73(3):469–482. 10.1111/tpj.12050 [DOI] [PubMed] [Google Scholar]
- Reichardt S, Piepho HP, Stintzi A, Schaller A. Peptide signaling for drought-induced tomato flower drop. Science. 2020:367(6485):1482–1485. 10.1126/science.aaz5641 [DOI] [PubMed] [Google Scholar]
- Reichardt S, Repper D, Tuzhikov AI, Galiullina RA, Planas-Marquès M, Chichkova NV, Vartapetian AB, Stintzi A, Schaller A. The tomato subtilase family includes several cell death-related proteinases with caspase specificity. Sci Rep. 2018:8(1):10531. 10.1038/s41598-018-28769-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodiuc N, Barlet X, Hok S, Perfus-Barbeoch L, Allasia V, Engler G, Seassau A, Marteu N, de Almeida-Engler J, Panabieres F, et al. Evolutionarily distant pathogens require the Arabidopsis phytosulfokine signalling pathway to establish disease. Plant Cell Environ. 2016:39(7):1396–1407. 10.1111/pce.12627 [DOI] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S. The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 2012:196(1):13–28. 10.1111/j.1469-8137.2012.04266.x [DOI] [PubMed] [Google Scholar]
- Sauter M. Phytosulfokine peptide signalling. J Exp Bot. 2015:66(17):5161–5169. 10.1093/jxb/erv071 [DOI] [PubMed] [Google Scholar]
- Sharma B, Taganna J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci Rep. 2020:10(1):9581. 10.1038/s41598-020-66553-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Diener AC. Arabidopsis thaliana RESISTNACE TO FUSARIUM OXYSPORUM 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet. 2013:9(5):e1003525. 10.1371/journal.pgen.1003525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004:55(1):555–590. 10.1146/annurev.arplant.55.031903.141801 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Heil M. Damage-associated molecular patterns (DAMPs) in plant innate immunity: applying the danger model and evolutionary perspectives. Annu Rev Phytopathol. 2021:59(1):53–75. 10.1146/annurev-phyto-082718-100146 [DOI] [PubMed] [Google Scholar]
- Trenner J, Monaghan J, Saeed B, Quint M, Shabek N, Trujillo M. Evolution and functions of plant U-box proteins: from protein quality control to signaling. Annu Rev Plant Biol. 2022:73(1):93–121. 10.1146/annurev-arplant-102720-012310 [DOI] [PubMed] [Google Scholar]
- Trujillo M. News from the PUB: plant U-box type E3 ubiquitin ligases. J Exp Bot. 2018:69(3):371–384. 10.1093/jxb/erx411 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol. 2008:18(18):1396–1401. 10.1016/j.cub.2008.07.085 [DOI] [PubMed] [Google Scholar]
- Wang JL, Grubb LE, Wang JY, Liang XX, Li L, Gao CL, Ma MM, Feng F, Li M, Li L, et al. A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell. 2018:69(3):493–504. 10.1016/j.molcel.2017.12.026 [DOI] [PubMed] [Google Scholar]
- Wang JZ, Li HJ, Han ZF, Zhang HQ, Wang T, Lin GZ, Chang JB, Yang WC, Chai JJ. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature. 2015b:525(7568):265–268. 10.1038/nature14858 [DOI] [PubMed] [Google Scholar]
- Wang J, Qu B, Dou S, Li L, Yin DD, Pang ZQ, Zhou ZZ, Tian MM, Liu GZ, Xie Q, et al. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 2015a:15(1):49. 10.1186/s12870-015-0442-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiborg J, O'Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem J. 2008:413(3):447–457. 10.1042/BJ20071568 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Mezaki H, Fujiwara M, Hara Y, Kawasaki T. Arabidopsis ubiquitin ligase PUB12 interacts with and negatively regulates Chitin Elicitor Receptor Kinase 1 (CERK1). PLoS One. 2017:12(11):e0188886. 10.1371/journal.pone.0188886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zhang B, Qi G, Shang L, Liu H, Ding X, Chu Z. Identification of the phytosulfokine receptor 1 (OsPSKR1) confers resistance to bacterial leaf streak in rice. Planta. 2019:250(5):1603–1612. 10.1007/s00425-019-03238-8 [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu ZJ, Lei C, Zheng CF, Wang J, Shao SJ, Li X, Xia XJ, Cai XZ, Zhou J, et al. A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. Plant Cell. 2018:30(3):652–667. 10.1105/tpc.17.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JG, Liu DR, Wang P, Ma XY, Lin WW, Cheng SX, Mishev K, Lu DP, Kumar R, Vanhoutte I, et al. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc Natl Acad Sci USA. 2018:115(8):E1906–E1915. 10.1073/pnas.1712251115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Zhang Y. Plant immunity: danger perception and signaling. Cell. 2020:181(5):978–989. 10.1016/j.cell.2020.04.028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that all experimental data are available and accessible via the main text and/or the supplemental data.