Abstract
As the main fungal etiologic agent of apple (Malus domestica) replant disease (ARD), Fusarium solani seriously damages apple roots. Ethylene response factors (ERFs) play an important role in plant resistance to biotic stress. Here, we show that MdERF114 is expressed during F. solani infections and positively regulates the resistance of apple roots to F. solani. Yeast one-hybrid, dual-luciferase, electrophoretic mobility shift assays and determinations of lignin content indicated that MdERF114 directly binds the GCC-box of the MdPEROXIDASE63 (MdPRX63) promoter and activates its expression, resulting in lignin deposition in apple roots and increased resistance to F. solani. We identified a WRKY family transcription factor, MdWRKY75, that binds to the W-box of the MdERF114 promoter. Overexpression of MdWRKY75 enhanced resistance of apple roots to F. solani. MdMYB8 interacted with MdERF114 to enhance resistance to F. solani by promoting the binding of MdERF114 to the MdPRX63 promoter. In summary, our findings reveal that the MdWRKY75-MdERF114-MdMYB8-MdPRX63 module is required for apple resistance to F. solani and the application of this mechanism by Agrobacterium rhizogenes-mediated root transformation provides a promising strategy to prevent ARD.
Introduction
Apple (Malus domestica) replant disease (ARD) is a soil-borne disease that severely inhibits the growth of young apple trees and reduces yield during apple replanting (Grunewaldt-Stöcker et al. 2019). The etiology of ARD is complex yet the accumulation of harmful soil fungi is hypothesized as its main cause (Mazzola 1998; Manici et al. 2003; van Schoor et al. 2009). Indeed, Fusarium species are the causal agents of ARD in the Bohai Gulf, South Africa, and Italy (van Schoor et al. 2009; Kelderer et al. 2012; Wang et al. 2018). A current study showed that Fusarium solani destroyed the reactive oxygen species scavenging system, causing both oxidative damage and growth inhibition of apple rootstocks (Xiang et al. 2021b). Moreover, F. solani blocked water transport, resulting in water deficit stress and disruption of the apple photosystem (Yan et al. 2018). Despite them posing a threat to both the environment and human health, fumigation and fungicide use are the current main methods of controlling ARD. The most effective strategy for prevention and control of ARD is to breed ARD-resistant rootstocks using molecular biological techniques (Zhu et al. 2014). However, the mechanisms by which apples defend themselves against F. solani remain unclear. This thus necessitates investigations into the mechanism of interactions between apples and F. solani.
To mitigate the adverse effects of harmful fungi, plants evolved a series of effective mechanisms to defend against fungal infections. Relatedly, transcription factors (TFs) contribute to biotic stress defenses by activating the expression of target genes via binding to specific DNA sequences (Reboledo et al. 2022). The ethylene response factors (ERFs) TF is involved in plant defense responses to biotic stress (Feng et al. 2020). ERFs belong to APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) superfamily and thus contain a conserved AP2 domain (Nakano et al. 2006). ERFs specifically bind to GCC-boxes to activate downstream defense genes involved in the plant response to biological stress. For example, in Arabidopsis (Arabidopsis thaliana), ERF11 activated BTB AND TAZ DOMAIN PROTEIN 4 (BT4) transcription to regulate immunity to Pseudomonas syringae (Zheng et al. 2019). AtERF114 enhanced the resistance of Arabidopsis to PevD1 by increasing lignin content via binding to the PAL1 promoter (Li et al. 2022). ZmERF105 positively regulated maize (Zea mays L.) resistance to Exserohilum turcicum by regulating the expression of several pathogenesis-related (PR) genes (Zang et al. 2020). Relatedly, the apple rootstock G.935, which was resistant to ARD, had numerous highly expressed ERFs (Zhu et al. 2016). Apple roots responding to F. solani had many highly expressed ERFs (Xiang et al. 2021a). However, the function of ERFs in defending the apple against ARD is unclear, necessitating the determination of the specific transcriptional regulation mechanisms of ERFs in ARD.
Other families of TFs contribute to plant resistance to biotic stress, such as WRKY and MYB TFs (Jiang et al. 2017; Ma et al. 2022). The Gossypium hirsutum WRKY genes GhWRKY39-1 and GhWRKY40 mediated the resistance of Nicotiana benthamiana to Ralstonia solanacearum (Shi et al. 2014). In Arabidopsis, WRKY8 regulated their susceptibility to P. syringae and Botrytis cinerea, to confer resistance to TMC-cg by coordinating abscisic acid and ethylene signaling pathways (Chen et al. 2013). NtWRKY12 regulated the transcription of PR-1a, which is a salicylic acid (SA)-inducible defense gene (van Verk et al. 2008). MYB TFs also contributed to plant defenses against biotic stress. Heterologous expression of MdMYB30 in Arabidopsis enhanced resistance to various bacterial pathogens (Zhang et al. 2019). AtMYB96 enhanced Arabidopsis resistance to P. syringae by regulating SA-signaling pathway defense genes (Seo and Park 2010). Several WRKY and MYB TFs were involved in genetic responses to F. solani infections (Xiang et al. 2021a).
Lignin is an important phenolic polymer in plants with resistance to biotic and abiotic stresses (Mottiar et al. 2023). The biosynthesis of lignin occurs in plants via phenylpropanoid pathway. Three monolignol precursors (p-coumaryl, coniferyl, and sinapyl alcohols) are formed from phenylalanine through a series of catalytic reactions and then oxidized and polymerized by cross-linking reaction triggered to form lignin (Mottiar et al. 2023). Peroxidases (PRXs) play an important role in the oxidative polymerization of three monolignol precursors to form lignin (Barceló et al. 2004; Marjamaa et al. 2009). In Arabidopsis, the suppression of AtPRX72 lead to the decrease of lignin content (Fernandez-Perez et al. 2015). Overexpression of OsPRX38 promotes the deposition of lignin in Arabidopsis which confer the tolerance to arsenic (Kidwai et al. 2019). The seeds from prx2 prx25 double mutant plants shows changes in lignin content which leads to shortened seed longevity (Renard et al. 2020). However, there are few studies on the function and regulatory relationship of PRXs in interaction between roots and pathogenic fungi.
In this study, the ERF TF MdERF114 positively regulated apple resistance to F. solani. MdERF114 bound directly to the MdPRX63 promoter to increase the deposition of lignin, which resulted in resistance to F. solani. MdWRKY75 enhanced apple resistance to F. solani by binding to the MdERF114 promoter. Moreover, MdMYB8 participated in the apple defense against F. solani by interacting with MdERF114 to enhance its binding to the MdPRX63 promoter. Our study reveals a molecular mechanism by which MdWRKY75-MdERF114-MdMYB8-MdPRX63 mediates apple resistance to F. solani. These insights will aid the breeding of resistant rootstocks. It provides an important means to obtain ARD-resistance rootstock by Agrobacterium rhizogenes-mediated root transformation producing overexpression transgenic plant.
Results
The expression pattern and subcellular localization of MdERF114
The RNA-seq results (Xiang et al. 2021a) showed that F. solani up-regulated MdERF114 (MD06G1130400) (ERF TF). Further analysis showed that MdERF114 contains an 807 bp open reading frame and encodes a protein with 269 amino acids. RT-qPCR showed that F. solani induced MdERF114 expression, peaking at 72 h (Supplemental Fig. S1A). MeJA also induced MdERF114 expression (Supplemental Fig. S1B). However, SA treatment did not significantly change MdERF114 expression (Supplemental Fig. S1C). MdERF114 was highly expressed in roots and stems (Supplemental Fig. S1D). Analysis of the MdERF114 promoter sequence revealed that it contains several cis-acting elements related to stress and hormonal responses (Supplemental Fig. S1E). These results indicate that MdERF114 may play an essential role in root response to stress.
The ORF sequence of MdERF114 was cloned and inserted into pBIN-GFP vector, and transformed into Agrobacterium tumefaciens strain GV3101, then observed using a confocal laser scanning microscope (Leica SP5, Leica Microsystems, Buffalo Grove, IL). Results revealed that MdERF114 was localized in the nucleus (Supplemental Fig. S2).
MdERF114 positively regulates apple resistance to F. solani
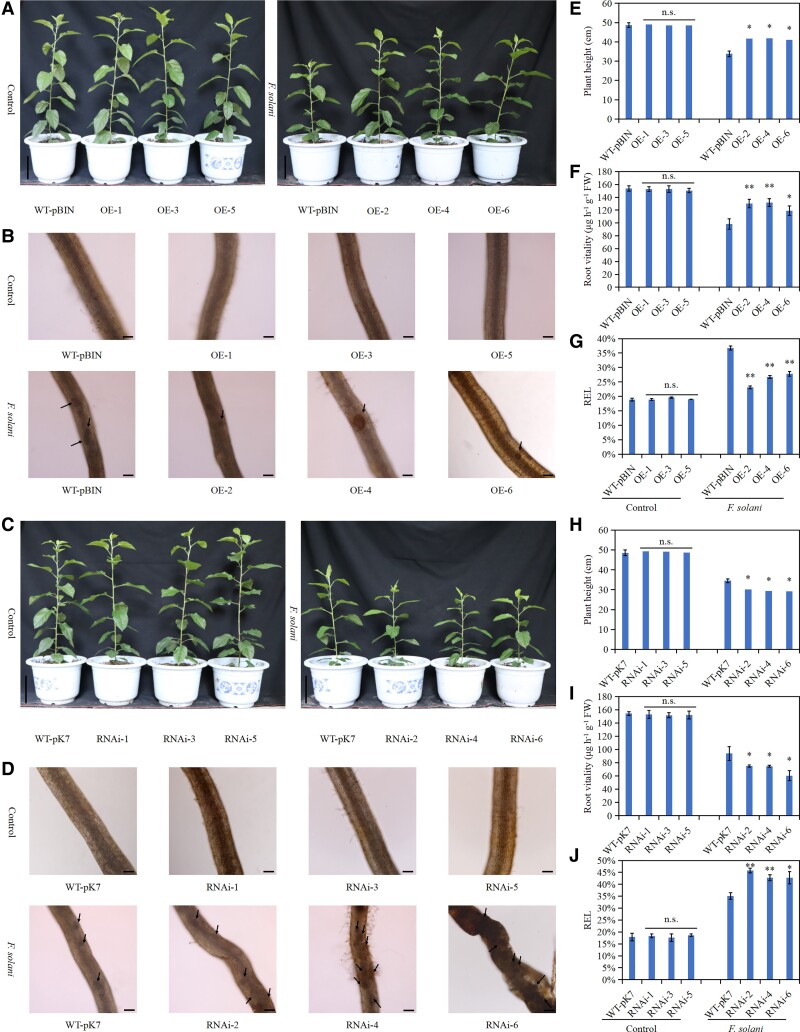
The MdERF114-overexpressing and MdERF114-RNAi roots were obtained via A. rhizogenes-mediated transformation (Supplemental Fig. S3), then inoculated with F. solani to explore the function of MdERF114 in apple response to F. solani. There was no obvious difference in growth between MdERF114-overexpressing lines and WT-pBIN line or MdERF114-RNAi lines and WT-pK7 line before F. solani inoculation. However, F. solani inoculation inhibited the growth of all the plants (Fig. 1A). Fusarium solani inoculation significantly inhibited the growth of plants with WT-pBIN roots than those with MdERF114-overexpressing roots. The plant heights of OE-2, OE-4, and OE-6 were 23.41%, 24.06%, and 21.43% higher than those of WT-pBIN, respectively (Fig. 1E). Moreover, F. solani inoculation significantly inhibited the growth of plants with MdERF114-RNAi roots than WT-pK7 plants. The plant heights of RNAi-2, RNAi-4, and RNAi-6 were 87.57%, 84.98%, and 84.56% of those of WT-pK7 plants, respectively (Fig. 1H). A microscope showed that no damage occurred in the roots before F. solani treatment. However, the MdERF114-overexpressing roots were slightly injured after F. solani treatment, with a few brown spots and minor wounds (Fig. 1B). Besides, dense brown lesions were observed on MdERF114-RNAi roots, some of which were necrotic (Fig. 1D). The root vitality and relative electrolytic leakage (REL) were also measured to evaluate the root damage. The MdERF114-overexpressing roots had higher root vitality and lowered REL after F. solani treatment than WT-pBIN (Fig. 1, F and G). However, MdERF114-RNAi roots had lower root vitality and higher REL than WT-pK7 (Fig. 1, I and J). These results indicate that MdERF114 positively regulates apple resistance to F. solani.
Figure 1.
MdERF114 positively regulates the resistance of apple to Fusarium solani. A) Phenotypic analysis of plants overexpressing MdERF114 in the roots under F. solani treatment. Scale bar = 10 cm. B) Observation of MdERF114-overexpressing roots under F. solani treatment. The black arrow indicates the wound. Scale bar = 100 μm. C) Phenotypic analysis of plants with MdERF114-RNAi roots under F. solani treatment. Scale bar = 10 cm. D) Observation of MdERF114-RNAi roots under F. solani treatment. The black arrow indicates the wound. Scale bar = 100 μm. E) Height of plants overexpressing MdERF114 in the roots under F. solani treatment. F) Root vitality of MdERF114-overexpressing roots under F. solani treatment. G) REL of MdERF114-overexpressing roots under F. solani treatment. H) Height of plants with MdERF114-RNAi roots under F. solani treatment. I) Vitality of MdERF114-RNAi roots under F. solani treatment. J) Root REL of MdERF114-RNAi roots under F. solani treatment. WT-pBIN, GL3 transformed with an pBIN-GFP empty vector; OE 1–6, different MdERF114-overexpressing root lines. WT-pK7, GL3 transformed with an pK7GWIWG2 empty vector; RNAi 1–6, different MdERF114-RNAi root lines. Data are shown as means ± standard deviation (Sd) with three biological replicates. Different letters indicate significant differences between treatments based on Student's t-test (*P ≤ 0.05; **P ≤ 0.01).
MdERF114 directly binds to the MdPRX63 promoter
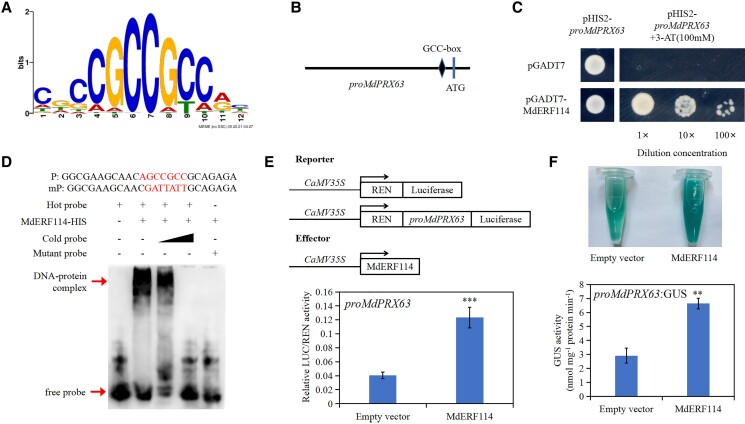
The downstream target genes of MdERF114 were predicted via DNA affinity purification sequencing (DAP-seq) to explore the regulatory network of MdERF114 under F. solani treatment. A total of 2,483 potential downstream target genes and the highest scoring cis-acting element, GCC-box (GCCGCC), were identified (Fig. 2A). A previous study found that AtERF114 is involved in lignin synthesis in Arabidopsis (Li et al. 2022). Herein, DAP-seq found several genes related to lignin synthesis (MD17G1040100, MD04G1101700, MD16G1085600, MD13G1086200, MD09G1226600, MD03G1059000, MD09G1039100, MD16G1062800). However, the identification of the promoter of these genes revealed that only the peroxidase MdPRX63 (MD17G1040100) had a GCC-box (Fig. 2B), indicating that MdPRX63 might function as a downstream target gene of MdERF114. Yeast one-hybrid assay (Y1H), electrophoretic mobility shift assay (EMSA), dual-luciferase assay, and β-glucuronidase (GUS) staining assay were then performed to further verify this hypothesis. Y1H assay showed that the Y187 yeast strain co-transformed with pGADT7-MdERF114 and pHIS2-proMdPRX63 could grow on the medium supplemented with 100 mM 3-amino-1, 2, 4-triazole (3-AT), while the yeast strain transformed with pHIS2-proMdPRX63 could not grow on this medium (Fig. 2C). EMSA results also showed that MdERF114 could bind to the promoter of MdPRX63 while the mutant probe could not be bind by MdERF114 (Fig. 2D). In addition, luciferase assay was conducted by constructing the ORF sequence of MdERF114 in pGreenII 62-SK vector and the MdPRX63 promoter in pGreenII 0800-LUC vector. The N. benthamiana co-injected with the MdERF114 and MdPRX63 promoters had higher firefly luciferase (LUC) activity than control (Fig. 2E). Similarly, GUS staining assay showed that apple callus co-expressing MdERF114 and MdPRX63 promoters had higher GUS activity than callus expressing MdPRX63 promoter (Fig. 2F). Taken together, these results suggest that MdERF114 directly binds to the promoter of MdPRX63.
Figure 2.
MdERF114 directly binds to the MdPRX63 promoter. A) Potential binding elements of MdERF114 identified by DNA affinity purification sequencing (DAP- seq). B) MdPRX63 promoter sequence analysis revealed the presence of GCC-box binding element. C) Yeast one-hybrid (Y1H) assay showed that MdERF114 binds to the MdPRX63 promoter. The yeast strain co-transformed with pGADT7 and pHIS2-proMdPRX63 was used as the control. The screening concentration of 3-AT was 100 mM. D) EMSA indicates that MdERF114 binds to the GCC-box in MdPRX63 promoter. The “+” represents the presence of relevant probes or proteins and “-” represents the absence of relevant probes or proteins. The 5'-AGCCGCC-3' is the motif bound by MdERF114 in “P” and the 5'-AGCCGCC-3' motif was replaced by 5'-GATTATT-3' in “mP”. E) Relative LUC/REN activity in Nicotiana benthamiana co-expressing MdERF114 and proMdPRX63. F) GUS staining assay showing that MdERF114 can activate proMdPRX63 expression. Data are shown as means ± standard deviation (Sd) with three biological replicates in E and F. Different letters indicate significant differences between treatments based on Student's t-test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Detection of lignin content in MdERF114 transgenic roots under F. solani treatment
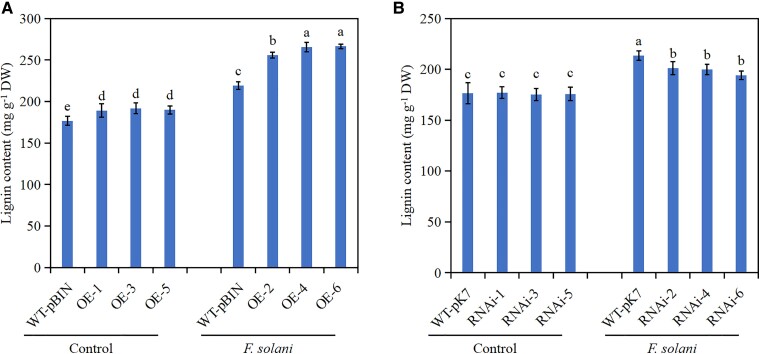
Previous studies have shown that PRX genes are associated with lignin synthesis. In this study, we determined the lignin content of MdERF114 transgenic roots under F. solani treatment, and the results showed that the lignin content of MdERF114-overexpressing roots (OE-2, OE-4, and OE-6) was significantly higher under F. solani treatment than that of WT-pBIN roots by 1.16, 1.21 and 1.22 times, respectively (Fig. 3A). However, the lignin content of MdERF114-RNAi roots under F. solani treatment was lower than that of WT-pK7 roots (Fig. 3B). These results indicate that MdERF114 affects root lignin deposition under F. solani treatment by regulating MdPRX63 transcription.
Figure 3.
Determination of lignin content in MdERF114 transgenic roots under Fusarium solani treatment. A) Determination of lignin content in MdERF114-overexpressing roots. B) Determination of lignin content in MdERF114-RNAi roots. WT-pBIN, GL3 transformed with an pBIN-GFP empty vector; OE 1–6, different MdERF114-overexpressing root lines. WT-pK7, GL3 transformed with an pK7GWIWG2 empty vector; RNAi 1–6, different MdERF114-RNAi root lines. Data are shown as means ± standard deviation (Sd) with three biological replicates. Different letters indicate significant differences between treatments based on Tukey's test (P < 0.05).
MdPRX63 overexpression enhances the resistance of apple roots to F. solani
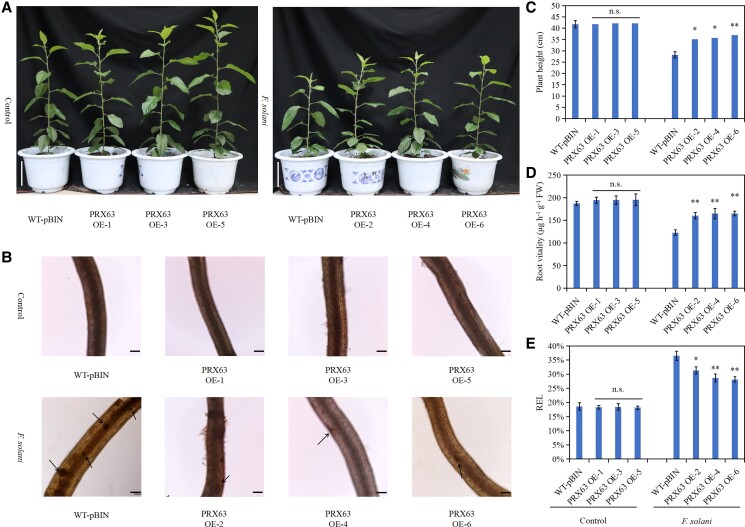
The MdPRX63-overexpressing roots (Supplemental Fig. S4) were inoculated with F. solani to explore the function of MdPRX63 in apple root response to F. solani. The results showed that plants with MdPRX63-overexpressing roots had better growth status and higher plant height under F. solani treatment than WT-pBIN plants (Fig. 4, A and C). Moreover, MdPRX63-overexpressing roots had higher root vitality and lower REL after F. solani treatment (Fig. 4, D and E). In addition, microscopic observation showed that the brown wound in MdPRX63-overexpressing roots was much less than that of WT-pBIN roots after F. solani inoculation (Fig. 4B). These results indicate that MdPRX63 overexpression enhances root resistance to F. solani.
Figure 4.
Overexpression of MdPRX63 enhances apple resistance to Fusarium solani. A) Phenotype of plants overexpressing MdPRX63 in the roots under F. solani treatment. Scale bar = 10 cm. B) Observation of MdPRX63-overexpressing roots under F. solani treatment. The black arrow indicates the wound. Scale bar = 100 μm. C) Height of plants overexpressing MdPRX63 in the roots under F. solani treatment. D) Vitality of MdPRX63-overexpressing roots under F. solani treatment. E) REL of MdPRX63-overexpressing roots under F. solani treatment. WT-pBIN, GL3 transformed with an empty vector containing the GFP tag; PRX63 OE 1–6, different MdPRX63-overexpressing root lines. Data are shown as the means ± standard deviation (Sd) with three biological replicates. Different letters indicate significant differences between treatments based on Student's t-test (*P ≤ 0.05; **P ≤ 0.01).
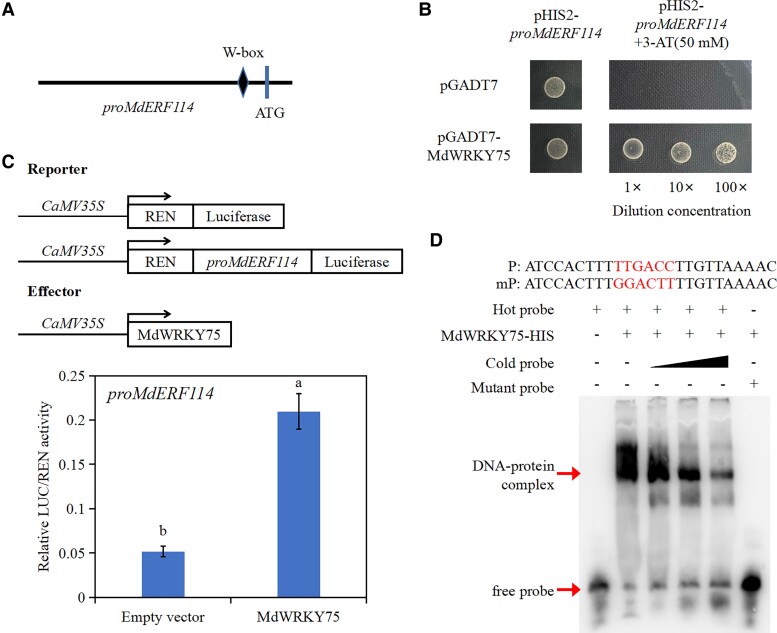
MdWRKY75 binds to the MdERF114 promoter
The MdERF114 promoter was constructed in pHIS2 vector as bait for screening upstream proteins of MdERF114 using Y1H system to further understand the signaling pathway of MdERF114 involved in apple root defense against F. solani. Results showed that a WRKY family TF, MdWRKY75 (MD09G1008800), could bind the MdERF114 promoter. From the analysis of the MdERF114 promoter, it was found that there is a W-box (TTGACC) element on the MdERF114 promoter (Fig. 5A). Y1H assay was performed to further verify this result. The yeast strain co-transformed with pGADT7-MdWRKY75 and pHIS2-proMdERF114 could grow on the medium supplemented with 50 mM 3-AT, while that co-transformed with pGADT7 and pHIS2-proMdERF114 could not grow on this medium (Fig. 5B). Meanwhile, the MdWRKY75-GST fusion protein was purified, then the probe containing W-box (TTGACC) was synthesized for EMSA. The results showed that MdWRKY75-GST fusion protein could bind to the MdERF114 promoter, while the mutant probe could not be bound by MdWRKY75-GST (Fig. 5D). Dual-luciferase assay was performed to further verify when MdWRKY75 can directly regulate MdERF114 in vivo. The results showed that the LUC activity of N. benthamiana leaves co-expressing MdWRKY75 and MdERF114 promoter was significantly increased compared with the negative control (Fig. 5C). These results suggest that MdWRKY75 binds to the MdERF114 promoter.
Figure 5.
MdWRKY75 directly binds to the MdERF114 promoter. A) MdERF114 promoter sequence analysis reveals the presence of W-box binding element. B) Yeast one-hybrid (Y1H) assay shows that MdWRKY75 binds to the MdERF114 promoter. The yeast strain co-transformed with pGADT7 and pHIS2-proMdERF114 was used as the control. The screening concentration of 3-AT was 50 mM. C) Relative LUC/REN activity of Nicotiana benthamiana co-expressing MdWRKY75 and proMdERF114. Data are shown as means ± standard deviation (Sd) with three biological replicates in C. Different letters indicate significant differences between treatments based on Tukey's test (P < 0.05). D) EMSA indicates that MdWRKY75 binds to the W-box in promoter of MdERF114. The “+” represents the presence of relevant probes or proteins and “-” represents the absence of relevant probes or proteins. The 5'-TTGACC-3' is the motif bound by MdWRKY75 in “P” and the 5'-TTGACC-3' motif was replaced by 5'-GGACTT-3' in “mP”.
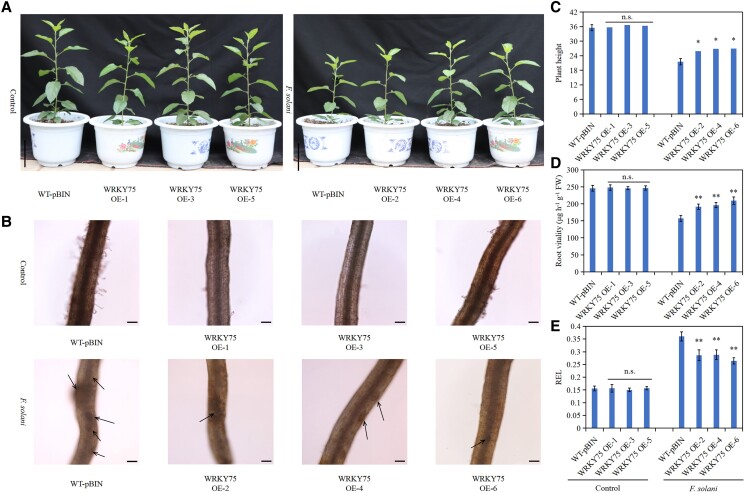
MdWRKY75 overexpression enhances the resistance of apple roots to F. solani
The expression of MdWRKY75 in roots under F. solani was assessed to investigate whether MdWRKY75 is involved in the resistance of apple roots to F. solani. RT-qPCR results showed that F. solani induced MdWRKY75 expression, peaking at 48 h (Supplemental Fig. S5A). Further results showed that the growth of plants with MdWRKY75-overexpressing roots was better than that of WT-pBIN. Although F. solani inhibited the growth of all plants, the height of plants with MdWRKY75-overexpressing roots was significantly higher than that of WT-pBIN plants (Fig. 6, A and C). Furthermore, WT-pBIN roots had dense wounds and even necrosis, while MdWRKY75-overexpressing roots only had fewer brown wounds (Fig. 6B). Meanwhile, MdWRKY75-overexpressing roots (WRKY75 OE-2, WRKY75 OE-4, and WRKY75 OE-6) had higher root vitality than WT-pBIN roots by 1.22, 1.25, and 1.33 times, respectively (Fig. 6D). However, MdWRKY75-overexpressing roots had a significantly lower REL than WT-pBIN roots (Fig. 6E). These results indicate that MdWRKY75 overexpression increases the resistance of apple roots to F. solani.
Figure 6.
Overexpression of MdWRKY75 enhances apple resistance to Fusarium solani. A) Phenotype of plants overexpressing MdWRKY75 in the roots under F. solani treatment. Scale bar = 10 cm. B) Observation of MdWRKY75-overexpressing roots under F. solani treatment. The black arrow indicates the wound. Scale bar = 100 μm. C) Height of plants overexpressing MdWRKY75 in the roots under F. solani treatment. D) Vitality of MdWRKY75-overexpressing roots under F. solani treatment. E) REL of MdWRKY75-overexpressing roots under F. solani treatment. WT-pBIN, GL3 transformed with an empty vector containing the GFP tag; WRKY75 OE 1–6, different MdWRKY75-overexpressing root lines. Data are shown as means ± standard deviation (Sd) with three biological replicates. Different letters indicate significant differences between treatments based on Student's t-test (*P ≤ 0.05; **P ≤ 0.01).
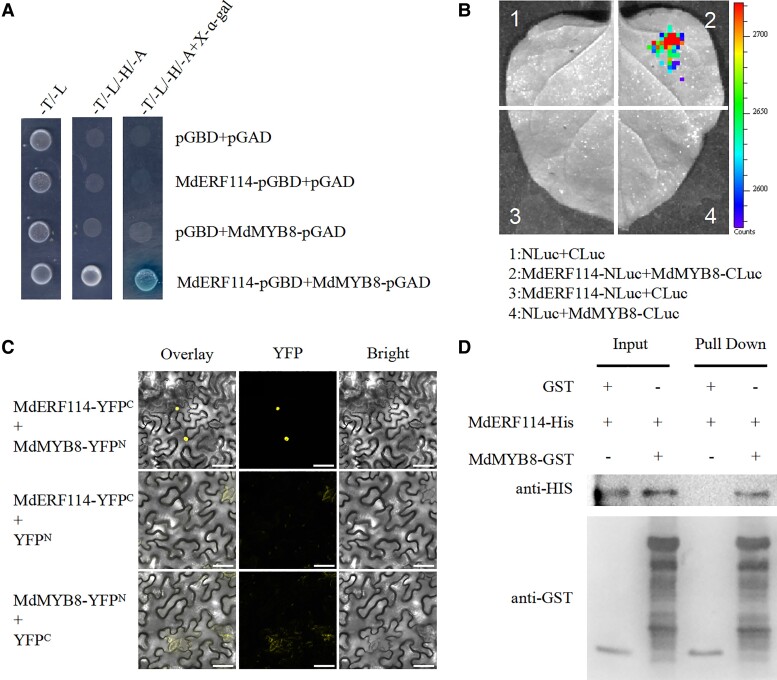
MdMYB8 interacts with MdERF114
The MdERF114 interacting proteins were screened using a yeast two-hybrid (Y2H) system to further analyze the regulatory network of MdERF114 involved in apple defense against F. solani. The results showed that an MYB family TF MdMYB8 (MD06G1217200) could interact with MdERF114. The Y2H assay was performed to elucidate whether MdMYB8 interacts with MdERF114. The results showed that the yeast strain co-expressing MdERF114-pGBD and MdMYB8-pGAD could grow normally in SD/-L/-H/-T/-A, while the control could not grow in SD/-L/-H/-T/-A (Fig. 7A). Meanwhile, luciferase complementarity assay was performed to verify the interaction between MdMYB8 and MdERF114. Fluorescence signal was detected in sites co-injected with MdERF114-Nluc and MdMYB8-Cluc (Fig. 7B). Bimolecular fluorescence complementation (BiFC) assay was further used to verify the interaction between MdMYB8 and MdERF114. The MdERF114-YFPC and MdMYB8-YFPN vectors were constructed and transferred into A. tumefaciens, then injected into N. benthamiana. Confocal microscope showed that MdMYB8 interacted with MdERF114 in the nucleus (Fig. 7C). In addition, pull-down assay was conducted using the fusion proteins of GST, MdERF114-HIS, and MdMYB8-GST to verify the interaction between MdMYB8 and MdERF114. The MdERF114-HIS could be captured by MdMYB8-GST, but could not be captured by GST (Fig. 7D). Taken together, these results indicate that MdMYB8 can interact with MdERF114, indicating that MdMYB8 may affect the binding of MdERF114 to the MdPRX63 promoter.
Figure 7.
MdMYB8 interacts with MdERF114. A) Yeast two-hybrid (Y2H) assay showing that MdMYB8 interacts with MdERF114 in yeast. B) Luciferase complementarity assay showing that fluorescence signal was only observed in sites co-injected with MdERF114-Nluc and MdMYB8-Cluc. C) BiFC assay showing that MdMYB8 interacts with MdERF114 in the nucleus. Scale bar = 50 μm. D) Pull-down assay showed that MdERF114-HIS could be captured by MdMYB8-GST and not Glutathione S-transferase (GST). The “+” represents the presence of relevant proteins and “-” represents the absence of relevant proteins.
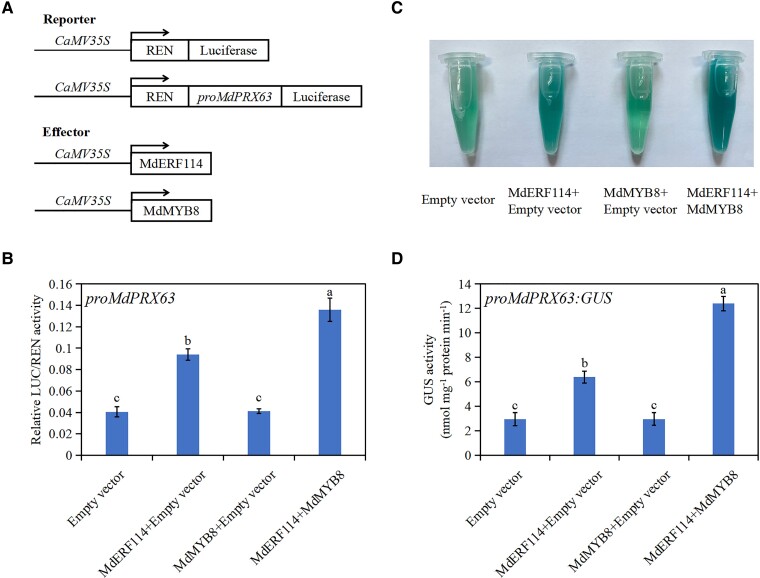
MdMYB8 promotes the binding of MdERF114 to the MdPRX63 promoter
Luciferase assay was performed to verify whether MdMYB8 can affect the binding of MdERF114 to the MdPRX63 promoter. The ORF of MdMYB8 and MdERF114 were inserted into pGreenII 62-SK vector, while the MdPRX63 promoter was inserted into pGreenII 0800-LUC vector. The luciferase activity of N. benthamiana leaves co-expressing MdMYB8, MdERF114, and MdPRX63 promoter was significantly higher than that of N. benthamiana leaves co-expressing MdERF114 and MdPRX63 promoter (Fig. 8, A and B). GUS staining assay was performed to further confirm this result. Three vectors (35S::MdMYB8, 35S::MdERF114, and proMdPRX63-GUS) were constructed, then transformed into apple callus. The callus co-expressing MdMYB8, MdERF114, and proMdPRX63 had higher GUS activity than the callus co-expressing MdERF114 and proMdPRX63 (Fig. 8, C and D). These results indicate that the interaction between MdMYB8 and MdERF114 promotes the binding of MdERF114 to the MdPRX63 promoter.
Figure 8.
MdMYB8 promotes MdERF114 binding to the MdPRX63 promotor. A, B) Relative LUC/REN activity of Nicotiana benthamiana co-expressing MdMYB8, MdERF114 and proMdPRX63. C, D) Relative GUS activity analysis. The 35s::MdMYB8, 35s::MdERF114 and proMdPRX63-GUS were co-transformed into apple calli and stained. The apple calli co-expressing 35s::MdERF114 and proMdPRX63-GUS, 35s::MdMYB8 and proMdPRX63-GUS, proMdPRX63-GUS were used as control. Data are shown as means ± standard deviation (Sd) with three biological replicates in B and D. Different letters indicate significant differences between treatments based on Tukey's test (P < 0.05).
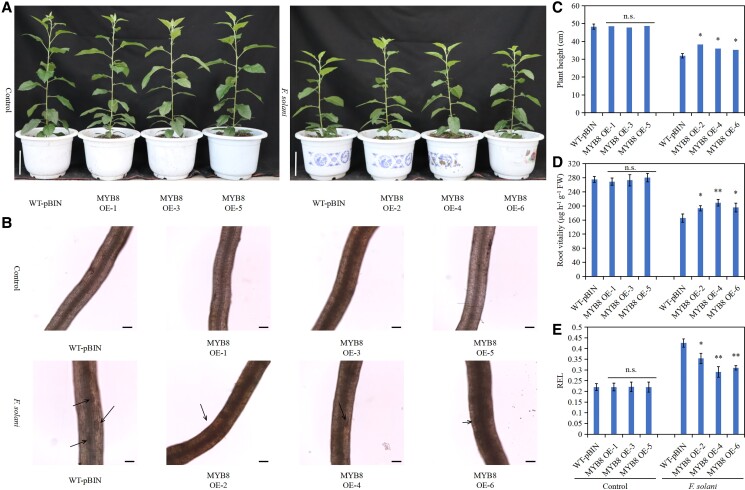
MdMYB8 overexpression enhances the resistance of apple roots to F. solani
The above results indicated that MdMYB8 may be involved in the mechanism of apple root resistance to F. solani. The expression pattern of MdMYB8 in apple roots was determined under F. solani treatment to assess this hypothesis. RT-qPCR results showed that F. solani induced MdMYB8 expression (Supplemental Fig. S6A). The MdMYB8-overexpressing roots were then inoculated with F. solani to explore the function of MdMYB8 (Supplemental Fig. S6, B and C). The growth status of plants with MdMYB8-overexpressing roots was better than that of WT-pBIN plants under F. solani treatment (Fig. 9A). Furthermore, plant height and root vitality of plants with MdMYB8-overexpressing roots were significantly higher than those of WT-pBIN, while REL was lower in plants with MdMYB8-overexpressing roots than in WT-pBIN plants (Fig. 9, C–E). In addition, F. solani caused less damage to MdMYB8-overexpressing roots than WT-pBIN roots (Fig. 9B). These results indicate that MdMYB8 overexpression increases the resistance of apple roots to F. solani.
Figure 9.
Overexpression of MdMYB8 enhances apple resistance to Fusarium solani. A) Phenotype of plants overexpressing MdMYB8 in the roots under F. solani treatment. Scale bar = 10 cm. B) Observation of MdMYB8-overexpressing roots under F. solani treatment. The black arrow indicates the wound. Scale bar = 100 μm. C) Height of plants overexpressing MdMYB8 in the roots under F. solani treatment. D) Vitality of MdMYB8-overexpressing roots under F. solani treatment. E) REL of MdMYB8-overexpressing roots under F. solani treatment. WT-pBIN, GL3 transformed with an empty vector containing the GFP tag; MYB8 OE1–6, different MdMYB8-overexpressing root lines. Data are as means ± standard deviation (Sd) with three biological replicates. Different letters indicate significant differences between treatments based on Student's t-test (*P ≤ 0.05; **P ≤ 0.01).
Discussion
MdERF114 enhances the resistance of apples to F. solani by regulating lignin biosynthesis
Fusarium solani, the main pathogenic fungus associated with ARD, causes substantial damage to apple roots (Yan et al. 2018; Xiang et al. 2021b). ERF TFs are plant-specific TFs involved in plant growth, development, and response to biotic and abiotic stresses (Licausi et al. 2013; Feng et al. 2020). A previous study reported numerous ERF TFs after conducting RNA-seq of apple rootstock M9T337 in response to F. solani (Xiang et al. 2021a). A combination of the above RNA-seq data and expression pattern analysis revealed that MdERF114 was upregulated by F. solani, suggesting that MdERF114 might play a role in the interaction of apple roots with F. solani. Herein, MdERF114 transgenic roots, obtained by A. rhizogenes-mediated transformation, were inoculated with F. solani to explore the function of MdERF114. Results demonstrated that overexpression of MdERF114 enhanced the resistance of apple roots to F. solani, whereas MdERF114-RNAi roots exhibited higher susceptibility. These results suggest that MdERF114 mediates the defense response of apple roots against to F. solani.
Lignin, the main component of plant cell wall, plays a critical role in determining the cell wall's mechanical strength and oxidation resistance (Xie et al. 2018; Cesarino 2019; Vanholme et al. 2019). When pathogenic microorganisms infect plants, lignin is rapidly deposited in the cell wall, thereby limiting the diffusion of mycelia and toxins into the plant cell and preventing pathogens from absorbing nutrients from the plant cell. This process provides a physical barrier for plants to resist the invasion of pathogenic microorganisms (Ma et al. 2018; Dong and Lin 2021). Here, a PRX gene, MdPRX63, was identified as the target gene of MdERF114, and MdERF114 was further verified to bind to the MdPRX63 promoter by Y1H, luciferase, and EMSA assay. A previous study showed that PRX genes are involved in the last step of the lignin biosynthesis pathway, which mediates the final synthesis of Guajacyl lignin, Syringyl lignin, and Hydroxy-phenyl lignin (Quan et al. 2019). The result of the lignin content determination in MdERF114 transgenic roots showed that the transcriptional regulation of MdERF114 on MdPRX63 affected the lignin biosynthesis under F. solani treatment. Similar to the function of ERF114 in Arabidopsis (Li et al. 2022), ERF114 affected lignin content by regulating the expression of PAL1. The deposition of lignin had a substantial effect on the disease resistance of apple roots (Zhu et al. 2021). In addition, we also found that overexpression of MdPRX63 enhanced the resistance of apple roots to F. solani. Thus, the resistance of apple roots to F. solani mediated by MdERF114 may be achieved by regulating the transcription of MdPRX63 to influence lignin biosynthesis.
MdWRKY75 increased apple resistance to F. solani by regulating MdERF114
WRKY TF, one of the largest family of TFs in land plants, modulate plant response to biotic and abiotic stress through the hormone signal pathway (Jiang et al. 2017). Accumulating evidence has revealed that WRKY TFs are involved in plant defense pathways to pathogens, growth and development, senescence, biosynthesis, and regulation of hormone signaling (Rushton et al. 2010; Bakshi and Oelmuller 2014). In addition, they play an important role in plant disease resistance. A previous study showed that nine WRKY TFs in Brochypodium distachyon were induced upon infection with Fusarium (Wen et al. 2014), and MdWRKY74 in apple is involved in the resistance to F. solani (Xiang et al. 2022). Interestingly, the Y1H system performed in this study found that MdWRKY75, a WRKY TF, binds the MdERF114 promoter, and the result was verified by Y1H, luciferase, and EMSA assays. A previous report has revealed that MdWRKY75e, a homolog of MdWRKY75, could confer elevated apple resistance to Alternaria alternata through regulating lignin biosynthesis (Hou et al. 2021). Interestingly, our data also showed that MdWRKY75 was induced by F. solani, and MdWRKY75 overexpression apple roots led to enhanced apple resistance to F. solani. We speculate that MdWRKY75 may confer enhanced resistance to F. solani by regulating the transcription of MdERF114, considering that MdWRKY75 can bind to the MdERF114 promoter. However, future work is needed to determine the underlying mechanism by which MdWRKY75 manipulates pathway for apple resistance to F. solani.
MdMYB8 enhances resistance to F. solani by interacting with MdERF114
It is well known that ERF TFs can interact with other proteins to play a role in the plant disease resistance process (Meng et al. 2013). For example, the interaction between VaERF16 and VaMYB306 improves the resistance of grapevine to Botrytis cinerea (Zhu et al. 2022). MdERF100 interacts with MdbHLH92 to enhance apple resistance to powdery mildew (Zhang et al. 2021). In this study, a Y2H was employed to screen the interaction between MdMYB8 and MdERF114, and the results were verified by Y2H, BiFC, luciferase complementation, and pull-down assays. Previous studies have shown that MYB8 is involved in the synthesis of secondary metabolites in Nicotiana attenuate and hop (Humulus lupulus L.) (Onkokesung et al. 2012; Schäfer et al. 2017; Kocábek et al. 2018). Our results also demonstrated that the interaction between MdMYB8 and MdERF114 enhanced the binding of MdERF114 to the MdPRX63 promoter. Moreover, under F. solani treatment, MdMYB8 was induced to up-regulate and enhance the resistance of apple to F. solani. Therefore, we speculate that MdMYB8 participates in the defense response of apple roots against F. solani by interacting with MdERF114 to promote the binding of MdERF114 to the MdPRX63 promoter.
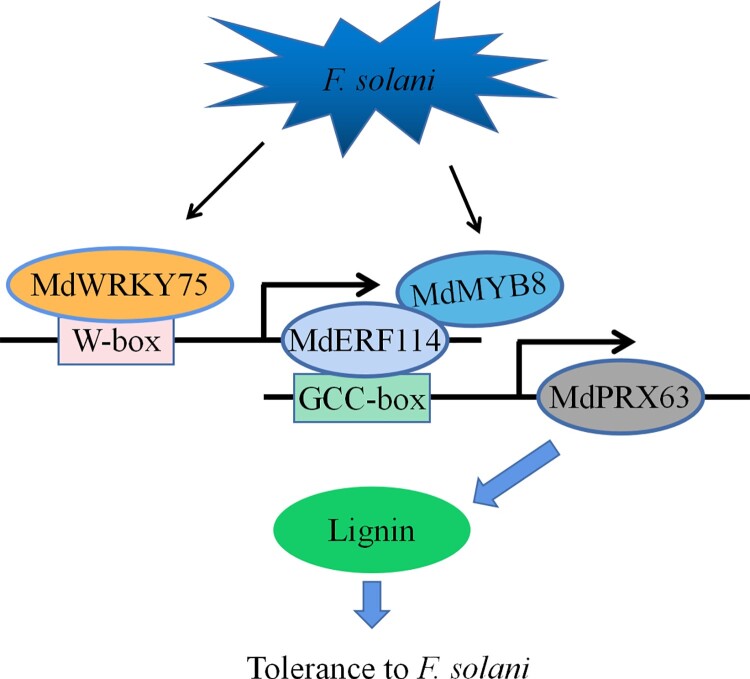
In conclusion, this study identified a defense mechanism against F. solani infection in apple roots mediated by MdERF114 (Fig. 10). Specifically, MdERF114 promotes lignin deposition in apple roots by binding to the promoter of MdPRX63 and regulating its transcription, thereby enhancing the resistance to F. solani. We also found that the expression of MdWRKY75 was upregulated by F. solani infection. Subsequently, MdWRKY75 regulates the expression of MdERF114 by binding to the W-box element of the MdERF114 promoter, ultimately conferring resistance to F. solani. In addition, results revealed that MdMYB8 is involved in the MdERF114-mediated defense network against F. solani by interacting with MdERF114 to promote binding of MdERF114 to the MdPRX63 promoter. The application of the A. rhizogenes-mediated root transformation makes it possible to obtain transgenic rootstocks with resistance genes quickly and efficiently. Transgenic roots with defense mechanisms were obtained by A. rhizogenes-mediated root transformation and applied to grafting, providing a promising strategy to prevent ARD. Overall, this study provides a reference on the MdERF114-mediated root defense response to pathogens and lays the theoretical foundation for breeding resistant rootstocks and prevention ARD.
Figure 10.
A working model illustrating the role of MdWRKY75-MdERF114-MdMYB8-MdPRX63 in apple response to Fusarium solani. Fusarium solani induces the expression of MdWRKY75, which directly binds to the W-box motif in the promoter of MdERF114. MdERF114 directly binds to the GCC-box motif of the MdPRX63 promoter and activates its expression, resulting in lignin deposition. The expression of MdMYB8 is also induced by F. solani. The interaction between MdMYB8 and MdERF114 enhances the binding of MdERF114 to the MdPRX63 promoter. The deposition of lignin increases the resistance to F. solani in apple roots.
Materials and methods
Plant materials and growth conditions
Tissue culture seedlings of apple cultivar GL-3 (Malus × domestica cv. Gala) were used for apple root transformation. The seedlings were cultured on MS basal medium containing 30 gL–1 sucrose, 0.3 mgL–1 6-BA, and 0.2 mgL–1 IAA under a 14 h photoperiod with 60 μmol m−2 s−1 light intensity for 1 month. The 35S::MdERF114-GFP, MdERF114-RNAi 35S::MdPRX63-GFP, 35S::MdWRKY75-GFP, and 35S::MdMYB8-GFP fusion vectors were constructed. Then, A. rhizogenes K599 (Weidi Biotechnology, Shanghai, China) was transformed with pBIN-GFP, pK7GWIWG2, and the fusion vectors to obtain transgenic roots as described by Meng et al. (2019). The primers are listed in Supplemental Table S1.
Positive transgenic root transformants were used to inoculate F. solani as per the method of Liu et al. (2022a). Briefly, potato glucose liquid medium was inoculated with F. solani strain (MG836251.1) and incubated at 24°C in the dark for 48 h. The spore suspension was obtained by filtering the medium through eight layers of sterile gauze. Its concentration was adjusted to 105 cells/mL–1 using sterile water. Treatments and control plants were watered with 200 mL of spore suspension and water only, respectively.
Physiological analysis
Root vitality was measured using a root vitality analysis kit from Suzhou Grace Biotechnology Co. Ltd (Suzhou, China). Three biological replicates were performed for each treatment.
According to the method of Liu et al. (2022a), fresh roots were used to determine the REL of roots. Three biological replicates were performed for each treatment.
The lignin content of roots was determined using a lignin content analysis kit (Suzhou Comin Biotechnology Co., Ltd., Suzhou, China). Three biological replicates were performed for each treatment.
Y1H assay
Inserts containing the ORFs of MdERF114 and MdWRKY75 were cloned into the pGADT7 vector. Likewise, inserts containing the promoters of MdPRX63 and MdERF114 were cloned into the pHIS2 vector. The 3-AT was used to suppress the expression of pHIS2-proMdPRX63 and pHIS2-proMdERF114. Then, according to the method of Liu et al. (2022b), the yeast strain Y187 was transformed with fusion vectors for Y1H assays. The primers are listed in Supplemental Table S1.
Dual-luciferase assays
Each of the ORF sequences for MdERF114, MdMYB8, and MdWRKY75 were individually cloned into pGreenII 62-SK vectors. Each of the promoters for MdERF114 and MdPRX63 was individually cloned into pGreenII 0800-LUC vectors. Agrobacterium tumefaciens strain GV3101 (Weidi Biotechnology, Shanghai, China) was transformed with fusion vectors. Then, LUC and renilla luciferase (REN) activities were assessed using a dual-luciferase reporter gene assay kit (Yeasen, Shanghai, China). Three biological replicates were performed for each treatment. The primers are listed in Supplemental Table S1.
EMSA assay
The ORFs of MdERF114 and MdWRKY75 were cloned into the pET-32a (+) vector and upon expression, MdERF114-HIS and MdWRKY75-HIS fusion proteins were obtained. Biotin-labeled probes for MdPRX63 and MdERF114 were synthesized by Sangon Biotech (Shanghai, China). The EMSA assay was done with a LightShift Chemiluminescent EMSA kit (Thermo Scientific, Waltham, MA, USA). The primers are listed in Supplemental Table S1.
GUS analysis
The promoter of MdPRX63 was cloned into the pC0390-GUS vector, and apple callus was co-transformed with 35S::MdERF114 and proMdPRX63-GUS (35S::MdERF114, 35S::MdMYB8, and proMdPRX63-GUS) using a vacuum method. Histochemical staining was performed to measure the GUS activity in the transformants (An et al. 2018). Three biological replicates were performed for each treatment. The primers are listed in Supplemental Table S1.
Y2H assay
The ORFs of MdERF114 and MdMYB8 were cloned into the pGBKT7 and pGADT7 vectors, respectively. Y2H Gold yeast (Weidi Biotechnology, Shanghai, China) was transformed with both plasmids. Positive transformants were selected for by cultivation on media lacking Trp and Leu, and also media lacking Ade, Trp, Leu, and His. The primers are listed in Supplemental Table S1.
Pull-down assay
The ORFs of MdERF114 and MdMYB8 were cloned into the pET-32a (+) and pGEX-4T-1 vectors, respectively. Pull-down assays were carried out using a Pierce GST Protein Interaction Pull-Down Kit (Thermo Scientific, Waltham, MA, USA). The primers are listed in Supplemental Table S1.
BiFC assay
The ORFs of MdERF114 and MdMYB8 were cloned into the pSPYCE-35S and pSPYNE-35S vectors, respectively. Agrobacterium tumefaciens strain GV3101 (Weidi Biotechnology, Shanghai, China) was transformed with the two vectors, which were also co-injected into N. benthamiana. Yellow fluorescent protein (YFP) fluorescence was observed under a confocal laser scanning microscope (Leica SP5, Leica Microsystems, Buffalo Grove, IL, USA). The excitation wavelength was 514 nm and the fluorescence was detected with a 522–560-nm band-pass filter. The primers are listed in Supplemental Table S1.
Luciferase complementation assay
MdERF114-NLuc and MdMYB8-CLuc vectors were constructed. Agrobacterium tumefaciens strain GV3101 (Weidi Biotechnology, Shanghai, China) was then transformed with these two vectors, which were also co-injected into N. benthamiana. The fluorescence intensity was measured with an in vivo imaging system (Xenogen, Boston, USA) (Jiang et al. 2021). The primers are listed in Supplemental Table S1.
Statistical analysis
Statistical analysis was conducted using SPSS Statistics version 18.0 (SPSS Inc., Chicago, IL, USA). One-way ANOVA and Student's t-test were used to determine the statistical significance of differences between various treatments.
Accession numbers
Sequence data from this article can be found in the NCBI data libraries under accession numbers MdERF114 (XM_017328133.2), MdPRX63 (XM_008343268.3), MdWRKY75 (XM_008380876.3), and MdMYB8 (DQ267899.1).
Supplementary Material
Acknowledgments
We are grateful to Prof. Zhiquan Mao (Shandong Agricultural University) for providing strains of F. solani and Prof. Zhihong Zhang (Shenyang Agricultural University) for providing tissue-cultured GL-3 apple plants. We also sincerely thank Dr. Jing Zhang and Yangyang Yuan (Horticulture Science Research Center, Northwest A&F University, Yangling, China) for providing professional technical assistance with LC–MS/MS analysis and laser confocal microscopy imaging.
Contributor Information
Yusong Liu, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A & F University, Yangling 712100, China.
Qianwei Liu, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A & F University, Yangling 712100, China.
Xuewen Li, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A & F University, Yangling 712100, China.
Zhijun Zhang, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A & F University, Yangling 712100, China.
Shukang Ai, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A & F University, Yangling 712100, China.
Cheng Liu, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A & F University, Yangling 712100, China.
Author contributions
C.Li., F.M., and Y.L. conceived and designed the study. Y.L., Q.L., X.L., Z.Z., S.A. and C.Liu. performed the analyses. Y.L. drafted the manuscript. F.M. and C.Li. supervised the process of this research. F.M. and C.Li. provided financial support for the study. All authors critically revised and provided final approval of this manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Fig. S1 . Analysis of MdERF114 expression patterns under different conditions and cis-acting elements of MdERF114 promoter.
Supplemental Fig. S2 . Subcellular localization of MdERF114.
Supplemental Fig. S3 . Determination of MdERF114 transgenic roots.
Supplemental Fig. S4 . Determination of MdPRX63 transgenic roots.
Supplemental Fig. S5 . Analysis of MdWRKY75 expression pattern under Fusarium solani treatment and determination of MdWRKY75 transgenic roots.
Supplemental Fig. S6 . Analysis of MdMYB8 expression pattern under Fusarium solani treatment and determination of MdMYB8 transgenic roots.
Supplemental Table S1 . The primers used in this study.
Funding
This work was supported by the National Natural Science Foundation of China (31972389) and the earmarked fund for the China Agriculture Research System of MOF and MARA (CARS-27).
References
- An JP, Li R, Qu FJ, You CX, Wang XF, Hao YJ. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018:96(3): 562–577 10.1111/tpj.14050 [DOI] [PubMed] [Google Scholar]
- Bakshi M, Oelmuller R. WRKY Transcription factors: jack of many trades in plants. Plant Signal Behav. 2014:9(2):e27700. 10.4161/psb.27700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló AR, Gómez Ros LV, Gabaldón C, López-Serrano M, Pomar F, Carrión JS, Pedreño MA. Basic peroxidases: the gateway for lignin evolution? Phytochem Rev. 2004:3(1–2):61–78 10.1023/B:PHYT.0000047803.49815.1a [DOI] [Google Scholar]
- Cesarino I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr Opin Biotechnol. 2019:56:209–214 10.1016/j.copbio.2018.12.012 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang L, Li D, Wang F, Yu D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2013:110(21): E1963–E1971 10.1073/pnas.1221347110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong NQ, Lin HX. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J Integr Plant Biol. 2021:63(1):180–209 10.1111/jipb.13054 [DOI] [PubMed] [Google Scholar]
- Feng K, Hou XL, Xing GM, Liu JX, Duan AQ, Xu ZS, Li MY, Zhuang J, Xiong AS. Advances in AP2/ERF super-family transcription factors in plant. Crit Rev Biotechnol. 2020:40(6):750–776 10.1080/07388551.2020.1768509 [DOI] [PubMed] [Google Scholar]
- Fernandez-Perez F, Pomar F, Pedreno MA, Novo-Uzal E. Suppression of Arabidopsis peroxidase 72 alters cell wall and phenylpropanoid metabolism. Plant Sci. 2015:239:192–199 10.1016/j.plantsci.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Grunewaldt-Stöcker G, Mahnkopp F, Popp C, Maiss E, Winkelmann T. Diagnosis of apple replant disease (ARD): microscopic evidence of early symptoms in fine roots of different apple rootstock genotypes. Sci Hortic. 2019:243:583–594 10.1016/j.scienta.2018.09.014 [DOI] [Google Scholar]
- Hou Y, Yu X, Chen W, Zhuang W, Wang S, Sun C, Cao L, Zhou T, Qu S. MdWRKY75e enhances resistance to Alternaria alternata in Malus domestica. Hortic Res. 2021:8(1):225. 10.1038/s41438-021-00701-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J. WRKY transcription factors in plant responses to stresses. J Integr Plant Biol. 2017:59(2):86–101 10.1111/jipb.12513 [DOI] [PubMed] [Google Scholar]
- Jiang H, Ma QJ, Zhong MS, Gao HN, Li YY, Hao YJ. The apple palmitoyltransferase MdPAT16 influences sugar content and salt tolerance via an MdCBL1-MdCIPK13-MdSUT2.2 pathway. Plant J. 2021:106(3):689–705 10.1111/tpj.15191 [DOI] [PubMed] [Google Scholar]
- Kelderer M, Manici LM, Caputo F, Thalheimer M. Planting in the ‘inter-row’ to overcome replant disease in apple orchards: a study on the effectiveness of the practice based on microbial indicators. Plant Soil. 2012:357(1–2):381–393 10.1007/s11104-012-1172-0 [DOI] [Google Scholar]
- Kidwai M, Dhar YV, Gautam N, Tiwari M, Ahmad IZ, Asif MH, Chakrabarty D. Oryza sativa class III peroxidase (OsPRX38) overexpression in Arabidopsis thaliana reduces arsenic accumulation due to apoplastic lignification. J Hazard Mater. 2019:362:383–393 10.1016/j.jhazmat.2018.09.029 [DOI] [PubMed] [Google Scholar]
- Kocábek T, Mishra AK, Matoušek J, Patzak J, Lomnická A, Khare M, Krofta K. The R2R3 transcription factor HlMYB8 and its role in flavonoid biosynthesis in hop (Humulus lupulus L.). Plant Sci. 2018:269:32–46 10.1016/j.plantsci.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ren J, Jia F, Zeng H, Li G, Yang X. Ethylene-responsive factor ERF114 mediates fungal pathogen effector PevD1-induced disease resistance in Arabidopsis thaliana. Mol Plant Pathol. 2022:23(6):819–831 10.1111/mpp.13208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 2013:199(3):639–649 10.1111/nph.12291 [DOI] [PubMed] [Google Scholar]
- Liu YJ, An JP, Gao N, Wang X, Chen XX, Wang XF, Zhang S, You CX. MdTCP46 interacts with MdABI5 to negatively regulate ABA signalling and drought response in apple. Plant Cell Environ. 2022b:45(11):3233–3248 10.1111/pce.14429 [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu Q, Li X, Tang Z, Zhang Z, Gao H, Ma F, Li C. Exogenous dopamine and MdTyDC overexpression enhance apple resistance to Fusarium solani. Phytopathology. 2022a:112(12):2503–2513. 10.1094/PHYTO-04-22-0142-R [DOI] [PubMed] [Google Scholar]
- Ma R, Liu B, Geng X, Ding X, Yan N, Sun X, Wang W, Sun X, Zheng C. Biological function and stress response mechanism of MYB transcription factor family genes. J Plant Growth Regul. 2022:42(1):83–95. 10.1007/s00344-021-10557-2 [DOI] [Google Scholar]
- Ma QH, Zhu HH, Qiao MY. Contribution of both lignin content and sinapyl monomer to disease resistance in tobacco. Plant Pathol. 2018:67(3):642–650 10.1111/ppa.12767 [DOI] [Google Scholar]
- Manici LM, Ciavatta C, Kelderer M, Erschbaumer G. Replant problems in South Tyrol: role of fungal pathogens and microbial population in conventional and organic apple orchards. Plant Soil. 2003:256(2):315–324 10.1023/A:1026103001592 [DOI] [Google Scholar]
- Marjamaa K, Kukkola EM, Fagerstedt KV. The role of xylem class III peroxidases in lignification. J Exp Bot. 2009:60(2):367–376 10.1093/jxb/ern278 [DOI] [PubMed] [Google Scholar]
- Mazzola M. Elucidation of the microbial complex having a causal role in the development of apple replant disease in Washington. Phytopathology. 1998:88(9):930–938 10.1094/PHYTO.1998.88.9.930 [DOI] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 2013:25(3): 1126–1142 10.1105/tpc.112.109074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D, Yang Q, Dong B, Song Z, Niu L, Wang L, Cao H, Li H, Fu Y. Development of an efficient root transgenic system for pigeon pea and its application to other important economically plants. Plant Biotechnol J. 2019:17(9): 1804–1813 10.1111/pbi.13101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y, Smith RA, Karlen SD, Ralph J, Mansfield SD. Evolution of p-coumaroylated lignin in eudicots provides new tools for cell wall engineering. New Phytol. 2023:237(1):251–264 10.1111/nph.18518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006:140(2):411–432 10.1104/pp.105.073783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onkokesung N, Gaquerel E, Kotkar H, Kaur H, Baldwin IT, Galis I. MYB8 controls inducible phenolamide levels by activating three novel hydroxycinnamoyl-coenzyme A: polyamine transferases in Nicotiana attenuata. Plant Physiol. 2012:158(1):389–407 10.1104/pp.111.187229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan M, Du Q, Xiao L, Lu W, Wang L, Xie J, Song Y, Xu B, Zhang D. Genetic architecture underlying the lignin biosynthesis pathway involves noncoding RNAs and transcription factors for growth and wood properties in Populus. Plant Biotechnol J. 2019:17(1):302–315 10.1111/pbi.12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboledo G, Agorio A, De Leon IP. Moss transcription factors regulating development and defense responses to stress. J Exp Bot. 2022:73(13):4546–4561 10.1093/jxb/erac055 [DOI] [PubMed] [Google Scholar]
- Renard J, Martinez-Almonacid I, Sonntag A, Molina I, Moya-Cuevas J, Bissoli G, Munoz-Bertomeu J, Faus I, Ninoles R, Shigeto J, et al. PRX2 and PRX25, peroxidases regulated by COG1, are involved in seed longevity in Arabidopsis. Plant Cell Environ. 2020:43(2):315–326 10.1111/pce.13656 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010:15(5):247–258 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Schäfer M, Brütting C, Xu S, Ling Z, Steppuhn A, Baldwin IT, Schuman MC. NaMYB8 regulates distinct, optimally distributed herbivore defense traits. J Integr Plant Biol. 2017:59(12):844–850 10.1111/jipb.12593 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park CM. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010:186(2):471–483 10.1111/j.1469-8137.2010.03183.x [DOI] [PubMed] [Google Scholar]
- Shi W, Hao L, Li J, Liu D, Guo X, Li H. The Gossypium hirsutum WRKY gene GhWRKY39-1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Rep. 2014:33(3):483–498 10.1007/s00299-013-1548-5 [DOI] [PubMed] [Google Scholar]
- Vanholme R, De Meester B, Ralph J, Boerjan W. Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol. 2019:56:230–239 10.1016/j.copbio.2019.02.018 [DOI] [PubMed] [Google Scholar]
- van Schoor L, Denman S, Cook NC. Characterisation of apple replant disease under South African conditions and potential biological management strategies. Sci Hortic. 2009:119(2):153–162 10.1016/j.scienta.2008.07.032 [DOI] [Google Scholar]
- van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJM. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008:146(4):1983–1995 10.1104/pp.107.112789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Yin C, Pan F, Wang X, Xiang L, Wang Y, Wang J, Tian C, Chen J, Mao Z. Analysis of the fungal community in apple replanted soil around Bohai Gulf. Hortic Plant J. 2018:4(5):175–181 10.1016/j.hpj.2018.05.003 [DOI] [Google Scholar]
- Wen F, Zhu H, Li P, Jiang M, Mao W, Ong C, Chu Z. Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res. 2014:21(3):327–339 10.1093/dnares/dst060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Wang M, Huang J, Jiang W, Yan Z, Chen X, Yin C, Mao Z. MdWRKY74 is involved in resistance response to apple replant disease. Plant Growth Regul. 2022:96(1):145–156 10.1007/s10725-021-00766-w [DOI] [Google Scholar]
- Xiang L, Wang M, Pan F, Wang G, Jiang W, Wang Y, Chen X, Yin C, Mao Z. Transcriptome analysis Malus domestica ‘M9T337’ root molecular responses to Fusarium solani infection. Physiol Mol Plant Pathol. 2021a:113101567. 10.1016/j.pmpp.2020.101567 [DOI] [Google Scholar]
- Xiang L, Zhao L, Wang M, Huang J, Chen X, Yin C, Mao Z. Physiological responses of apple rootstock M.9 to infection by Fusarium solani. HortScience. 2021b:56(9):1104–1111 10.21273/HORTSCI15945-21 [DOI] [Google Scholar]
- Xie M, Zhang J, Tschaplinski TJ, Tuskan GA, Chen JG, Muchero W. Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front Plant Sci. 2018:9:1427. 10.3389/fpls.2018.01427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Han G, Ren C, Zhao S, Wu X, Bian T. Fusarium solani infection depressed photosystem performance by inducing foliage wilting in apple seedlings. Front Plant Sci. 2018:9:479. 10.3389/fpls.2018.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Z, Lv Y, Liu S, Yang W, Ci J, Ren X, Wang Z, Wu H, Ma W, Jiang L, et al. A novel ERF transcription factor, ZmERF105, positively regulates maize resistance to Exserohilum turcicum. Front Plant Sci. 2020:11:850. 10.3389/fpls.2020.00850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang L, Ma H, Zhang Y, Zhang X, Ji M, van Nocker S, Ahmad B, Zhao Z, Wang X, et al. Overexpression of the apple (Malus × domestica) MdERF100 in Arabidopsis increases resistance to Powdery mildew. Int J Mol Sci. 2021:22(11):5713. 10.3390/ijms22115713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Zhang CL, Wang GL, Wang YX, Qi CH, Zhao Q, You CX, Li YY, Hao YJ. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 2019:19(1):362. 10.1186/s12870-019-1918-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Xing J, Zhang K, Pang X, Zhao Y, Wang G, Zang J, Huang R, Dong J. Ethylene response factor ERF11 activates BT4 transcription to regulate immunity to Pseudomonas syringae. Plant Physiol. 2019:180(2):1132–1151 10.1104/pp.18.01209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Fazio G, Mazzola M. Elucidating the molecular responses of apple rootstock resistant to ARD pathogens: challenges and opportunities for development of genomics-assisted breeding tools. Hortic Res. 2014:1(1):14043. 10.1038/hortres.2014.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li G, Singh J, Khan A, Fazio G, Saltzgiver M, Xia R. Laccase directed lignification is one of the major processes associated with the defense response against Pythium ultimum infection in apple roots. Front Plant Sci. 2021:12:629776. 10.3389/fpls.2021.629776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Shin S, Mazzola M. Genotype responses of two apple rootstocks to infection by Pythium ultimum causing apple replant disease. Can J Plant Pathol. 2016:38(4):483–491 10.1080/07060661.2016.1260640 [DOI] [Google Scholar]
- Zhu Y, Zhang X, Zhang Q, Chai S, Yin W, Gao M, Li Z, Wang X. The transcription factors VaERF16 and VaMYB306 interact to enhance resistance of grapevine to Botrytis cinerea infection. Mol Plant Pathol. 2022:23(10):1415–1432 10.1111/mpp.13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.