Abstract
S-RNase-mediated self-incompatibility (SI) prevents self-fertilization and promotes outbreeding to ensure genetic diversity in many flowering plants, including pear (Pyrus sp.). Brassinosteroids (BRs) have well-documented functions in cell elongation, but their molecular mechanisms in pollen tube growth, especially in the SI response, remain elusive. Here, exogenously applied brassinolide (BL), an active BR, countered incompatible pollen tube growth inhibition during the SI response in pear. Antisense repression of BRASSINAZOLE-RESISTANT1 (PbrBZR1), a critical component of BR signaling, blocked the positive effect of BL on pollen tube elongation. Further analyses revealed that PbrBZR1 binds to the promoter of EXPANSIN-LIKE A3 (PbrEXLA3) to activate its expression. PbrEXLA3 encodes an expansin that promotes pollen tube elongation in pear. The stability of dephosphorylated PbrBZR1 was substantially reduced in incompatible pollen tubes, where it is targeted by ARIADNE2.3 (PbrARI2.3), an E3 ubiquitin ligase that is strongly expressed in pollen. Our results show that during the SI response, PbrARI2.3 accumulates and negatively regulates pollen tube growth by accelerating the degradation of PbrBZR1 via the 26S proteasome pathway. Together, our results show that an ubiquitin-mediated modification participates in BR signaling in pollen and reveal the molecular mechanism by which BRs regulate S-RNase-based SI.
The brassinosteroid-activated transcription factor BRASSINAZOLE-RESISTANT1 regulates pollen tube growth during self-incompatibility signaling in pear.
Introduction
The majority of flowering plants have both male and female organs in the same flower (Charnov et al. 1976; Lloyd 1982; Christopher et al. 2019), which greatly increases the probability of self-fertilization. Because inbreeding commonly reduces the adaptability of their offspring, hermaphroditic plants have evolved many effective reproductive strategies to prevent it from occurring (Barrett 1998). One such strategy is self-incompatibility (SI), which relies on self-recognition between the pistil and pollen (de Nettancourt 2001; Qiao et al. 2004). The SI response allows the pistil of a flower to distinguish between self- (genetically related) and nonself- (genetically unrelated) pollen and is mainly controlled by a single highly polymorphic S locus (Kao and Tsukamoto 2004). Compared with nonself-pollen, self-pollen deposited on the surface of the stigma shows a reduced germination rate and inhibited pollen tube elongation in the style (Kao and Tsukamoto 2004). Thus, incompatible pollen tubes are unable to successfully transport sperm cells to the ovary.
S-RNase-based SI widely occurs in the Plantaginaceae, Rosaceae, and Solanaceae and is one of the most common forms of genetically controlled mate selection (Anderson et al. 1989; Xue et al. 1996; Wang et al. 2003). During fertilization, S-RNases, ribonuclease proteins encoded by the female determinant, are specifically expressed in the style (Kao and Tsukamoto 2004). They are indiscriminately transported to the growing compatible and incompatible pollen tubes and are recognized by the male determinant-encoded S-locus F-box protein (Anderson et al. 1986; Luu et al. 2000; Meng et al. 2014; Wei et al. 2014). Nonself S-RNases can be modified by ubiquitin and degraded via the 26S proteasome pathway (Lai et al. 2002; Sijacic et al. 2004), while the protected self S-RNases exhibit cytotoxic properties, leading to the arrest of pollen growth (Gray et al. 1991).
Phytohormones are important endogenous signaling substances that affect plant reproduction (Santner and Estelle 2009; Sankaranarayanan et al. 2013). For example, gene ontology terms associated with gibberellins (GAs), auxin, abscisic acid (ABA), and ethylene were found to be enriched in the pre- and post-pollinated papilla cells of Arabidopsis (Arabidopsis thaliana) (Matsuda et al. 2015). Similar results were also found in transcriptome analyses of compatible and self-incompatible pollination in pear (Pyrus bretschneideri) and Brassica napus (Shi et al. 2017; Zhang et al. 2017). Furthermore, the production of jasmonic acid (JA) was found to be induced by S-RNases in apple (Malus × domestica) pollen tube, and JA acts as a resistance substance in the immune response triggered by S-RNases (Gu et al. 2019). Pollination with incompatible pollen promotes the synthesis of ethylene and senescence in Dendrobium flowers (Ketsa et al. 2001). Recently, phosphoproteomic analyses of pollination in B. napus identified a multihormone crosstalk mechanism involving brassinosteroids (BRs), ABA, and ethylene during stigmatic papillae rejection of donor pollen (Duan et al. 2020).
The BRs are a class of naturally occurring steroid phytohormones that promote cell elongation (Divi and Krishna 2009; Yamamuro et al. 2000). They are perceived through the cell surface receptor-like kinase BRASSINOSTEROID-INSENSITIVE 1 (BRI1), and downstream signal transduction results in the dephosphorylation of 2 closely related transcription factors (TFs); BRASSINAZOLE-RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1) (Wang et al. 2002; Yin et al. 2002). Subsequently, the dephosphorylated BZR1/BES1 mediates various BR-related responses through binding to the promoters of downstream target genes (He et al. 2005; Yin et al. 2005). Several studies have shown that BZR1 can integrate signals from multiple pathways to coordinate cell growth (Bai et al. 2012; Li et al. 2018). In Arabidopsis, BZR1 functions as an upstream regulator of genes encoding hydroxyproline-rich glycoproteins related to pollen tube elongation (Li et al. 2020). Although the SI response in brassicas is not regulated by S-RNases, BRI1 KINASE INHIBITOR1 acts as a repressor of BZR1/BES1 in B. napus, and negatively regulates pollen germination and pollen tube growth (Duan et al. 2020). Therefore, a signaling cascade mediated by BZR1 likely functions in SI, but the detailed mechanism is still unclear.
Here, we show that the TF PbrBZR1 positively regulates pollen tube growth in pear by directly binding to the promoter of PbrEXLA3 and activating its expression. A pollen-specific E3 ubiquitin ligase, PbrARI2.3, captures dephosphorylated PbrBZR1 and mediates its degradation via the ubiquitin/26S proteasome pathway. PbrARI2.3 accumulates in the pollen tube of incompatible pollen, leading to a reduction in the levels of dephosphorylated PbrBZR1, thereby inhibiting pollen tube growth by downregulating the expression of PbrEXLA3. Therefore, our results show how an E3 ubiquitin ligase-mediated BR pathway regulates pollen tube growth, and demonstrate the mechanism by which the PbrBZR1–PbrARI2.3 module participates in the SI response in pear.
Results
Exogenous application of BL promotes incompatible pollen tube growth
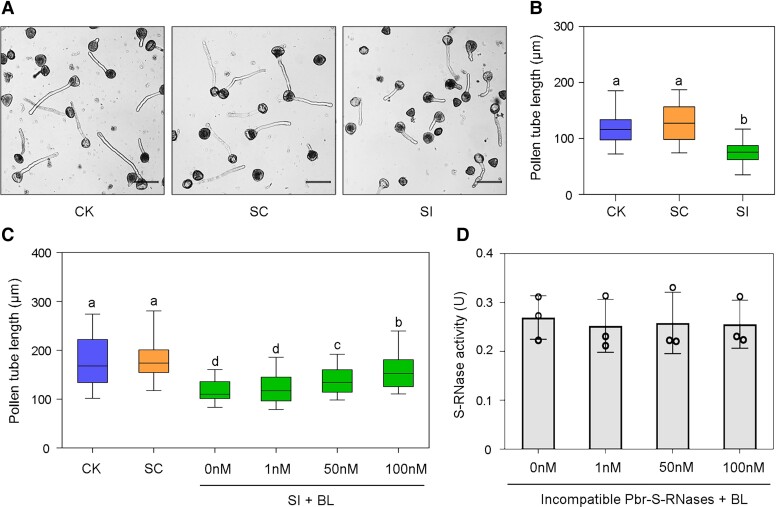
Because the SI reaction mainly occurs in pollen tubes, we simulated this process by treating pear pollen tubes with PbrS-RNases produced in vitro using a prokaryotic system (Chen et al. 2018). Two combinations of recombinant S-RNase proteins (PbrS3-RNase and PbrS5-RNase) and (PbrS7-RNase and PbrS34-RNase) were incubated with “Cuiguan” pollen with the S haplotype (S3S5), and constituted the incompatible rPbrS-RNases (SI) treatment and the compatible rPbrS-RNases (SC) treatment, respectively. As expected, the SI treatment significantly inhibited pollen tube growth compared with CK (control, untreated pollen) and the SC treatment (Fig. 1, A and B). Because BRs are important regulators of cell elongation, we explored the role of brassinolide (BL) in pollen tube growth during SI signaling. Liquid germination medium supplemented with BL at different concentrations was used to enhance BR signaling in pollen tubes (Wang et al. 2021). The BL alleviated the inhibitory effect of the SI treatment on pear pollen tube elongation in a concentration-dependent manner (Fig. 1C). To detect whether BR directly interacts with incompatible S-RNases, an in vitro S-RNase activity assay was performed using yeast (Saccharomyces cerevisiae) RNA as the substrate. Increasing the concentration of BL did not affect the activity of incompatible S-RNase (Fig. 1D). Thus, BR appears to be a positive regulator of incompatible pollen tube elongation.
Figure 1.
Effect of exogenous BL on incompatible pollen tube growth. A) “Cuiguan” pollen tube growth at 2 h after SC and SI treatments. Bars = 50 µm. The experiments were performed 3 times with similar results, and a representative picture is shown. B) Length of pollen tubes shown in (A). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 × interquartile range; points, outliers. C) Length of SC- and SI-treated pollen tubes on medium with BL at indicated concentrations. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 × interquartile range; points, outliers. For (B) and (C), at least 100 pollen tubes were measured per replicate in each treatment. All treatments were performed in 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). D) Activity of incompatible S-RNases (PbrS3-RNase + PbrS5-RNase) incubated with exogenous BL. Data are mean ± SD of 3 independent biological replicates. CK, liquid germination medium; SC, compatible PbrS-RNase treatment; SI, incompatible PbrS-RNase treatment;
PbrBZR1 is essential for BR-modulated pollen tube elongation
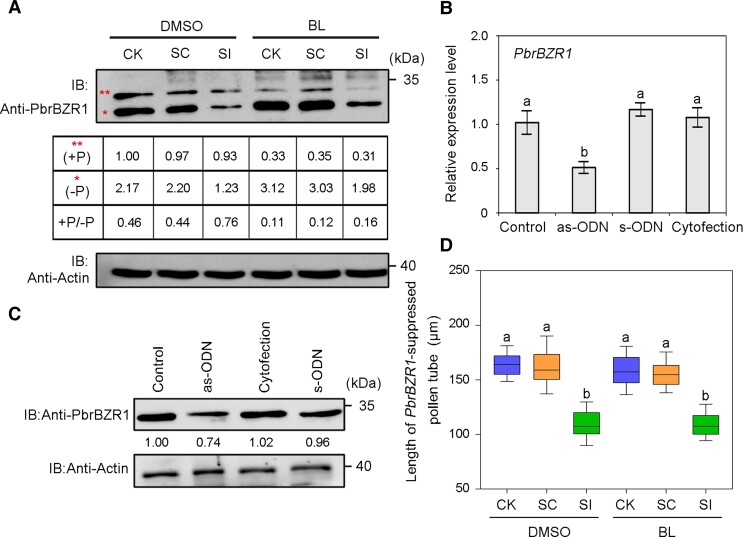
To explore how BR regulates SI, we first examined the transcript profile of PbrBZR1 in pollen tubes by RT-qPCR. The SI treatment did not affect the transcript level of PbrBZR1 (Supplemental Fig. S1). Enhancing BR signaling can induce the conversion of phosphorylated (deactivated) BZR1/BES1 to dephosphorylated (active) BZR1/BES1 to trigger various BR responses (He et al. 2019). Because phosphorylated proteins can reversibly combine with Phos-tags fixed in the gel during electrophoresis, their migration rate is slower than that of dephosphorylated proteins, so they can be distinguished by differences in protein mobility (Kinoshita et al. 2009). Here, the total proteins extracted from pear pollen tubes were separated on a Phos-tag gel and detected with anti-PbrBZR1 antibody. The amount of dephosphorylated PbrBZR1 protein was much lower in the SI treatment than in the CK and SC treatment, leading to a substantial increase in the ratio of phosphorylated to dephosphorylated PbrBZR1 in incompatible pollen tubes (Fig. 2A). We also examined the protein dephosphorylation status and abundance of PbrBZR1 in incompatible pollen tubes in response to BL. Although BL application greatly reduced the ratio of phosphorylated to dephosphorylated PbrBZR1, dephosphorylated PbrBZR1 accumulated to higher levels in the CK and SC treatment than in the SI treatment (Fig. 2A). Thus, SI reduces the stability of the active (dephosphorylated) form of PbrBZR1.
Figure 2.
PbrBZR1 functional assay in pear pollen tubes. A) Protein level of PbrBZR1 in SC- and SI-treated pollen tubes after addition of 100 nM BL or DMSO, as determined by western blot analysis. Total proteins extracted from pollen tubes were separated on SDS-PAGE gels supplemented with Phos-tag prior to immunoblotting with anti-PbrBZR1 antibody. Amount of phosphorylated PbrBZR1 protein in untreated pollen tubes (CK) grown on medium containing DMSO was set to 1.00. Two asterisks represent phosphorylated form of PbrBZR1 (+P), whereas one asterisk represents dephosphorylated form of PbrBZR1 (−P). The experiments were performed 3 times with similar results, and a representative picture is shown. B) Transcript levels of PbrBZR1 in untreated pollen (control) and pollen transfected with antisense oligodeoxynucleotide (as-ODN), sense oligodeoxynucleotide of PbrBZR1 (s-ODN), or transfection agent alone (cytofection), as determined by RT-qPCR assays. Data are mean ± SD of 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). C) Protein level of PbrBZR1 in pollen treated with as-ODN as determined by western blot analysis using anti-PbrBZR1 antibody. Amount of PbrBZR1 protein in control was set to 1.00. The experiments were performed 3 times with similar results, and a representative picture is shown. D) Length of PbrBZR1-suppressed pollen tubes grown on medium containing 100 nM BL or DMSO under SC and SI treatments. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 × interquartile range; points, outliers. At least 100 pollen tubes were measured per replicate in each treatment. All treatments were performed in 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). CK, liquid germination medium; SC, compatible PbrS-RNase treatment; SI, incompatible PbrS-RNase treatment; DMSO, add 100 nM DMSO to the pollen medium; BL, add 100 nM BL to the pollen medium; IB, immunoblot.
To investigate the function of PbrBZR1 in relation to BR-regulated pollen tube growth, antisense oligodeoxynucleotide (as-ODN) silencing technology was used to suppress the expression of PbrBZR1 in pollen tubes. Transfection with a PbrBZR1-antisense oligodeoxynucleotide (as-ODN-PbrBZR1) remarkably reduced PbrBZR1 transcript levels (Fig. 2B) and PbrBZR1 protein levels (Fig. 2C) in pollen tubes compared those in the control (nothing added), the cytofection treatment (transfection agent only), and the sense oligodeoxynucleotide transfection (s-ODN-PbrBZR1) treatment. This also led to significantly slower pollen tube growth on germination medium containing exogenous BL (Supplemental Fig. S2A), indicating that PbrBZR1 may positively modulate BR-controlled pollen tube growth. Specific knockdown of PbrBZR1 did not affect the growth of SI-treated pollen tubes (Supplemental Fig. S2B) but suppressed the induction of SI-treated pollen tube growth by exogenous BL (Fig. 2D), further confirming that PbrBZR1 is necessary for BR signaling to regulate incompatible pollen tube growth.
PbrBZR1 enhances the transcription of PbrEXLA3 by binding to its promoter
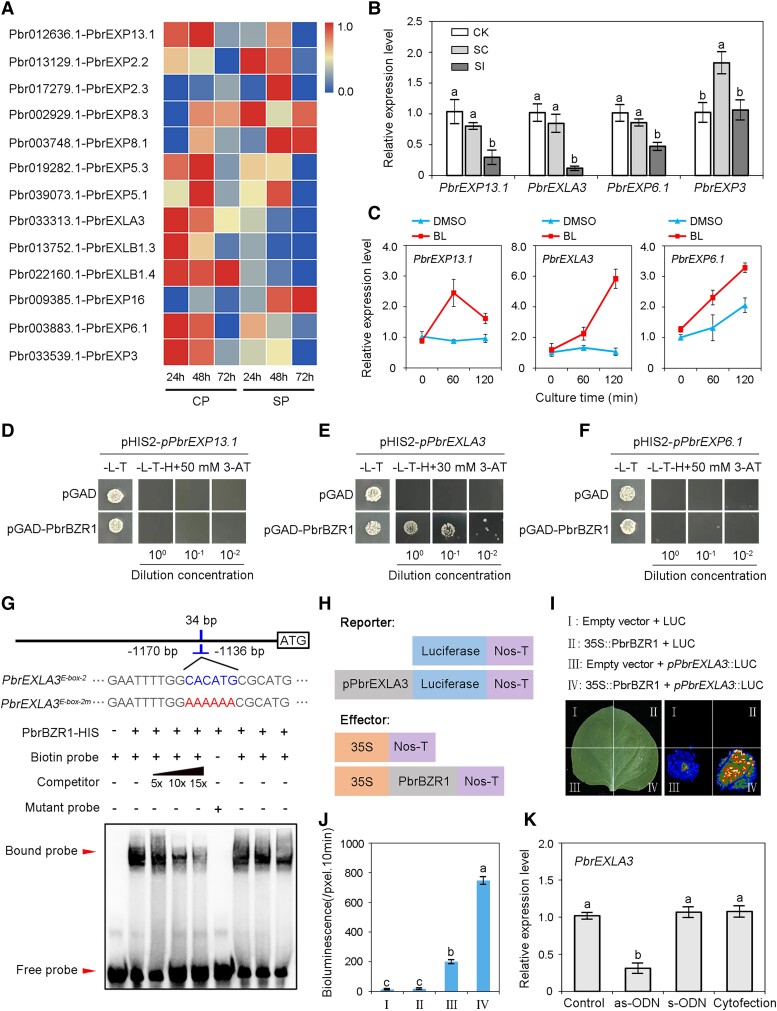
Expansins are a class of important cell growth modulators that are involved in plant growth and development (Cosgrove 2000). Given that SI causes growth arrest in incompatible pollen tubes, we speculated that expansin proteins may be involved in this process. By comparing the reverse transcription quantitative PCR (RT-qPCR) results of cross-pollination (CP; compatible reaction) and self-pollination (SP; incompatible reaction) in pear pistils (Supplemental Table S1), we identified 13 differentially expressed expansin genes in pear (Fig. 3A). Because we used in vitro pollen culture to simulate the SI reaction in vivo, the expansin genes that are highly expressed in pollen were preferentially selected. Of the 4 expansin genes expressed in pollen tubes (Supplemental Fig. S3), namely PbrEXP13.1, PbrEXLA3, PbrEXP6.1, and PbrEXP3, 3 were downregulated during the SI response (all except PbrEXP3) (Fig. 3B). Subsequently, we detected the transcript levels of these genes in BL-treated pollen tubes and found that all 3 were induced by the BR signal (Fig. 3C).
Figure 3.
PbrBZR1 binds to the promoter of PbrEXLA3. A) Heat map illustrating transcript levels of 13 differentially expressed pear expansin genes in CP- vs. SP-pollinated pistils. Heat map was scaled by row using the zero-to-one method. The color on the scale indicates the gene expression level from 0.0 to 1.0. B) Transcript levels of PbrEXP13.1, PbrEXLA3, PbrEXP6.1, and PbrEXP3 in SC- and SI-treated pollen tubes as determined by RT-qPCR assays. Data are mean ± SD of 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). C) Transcript levels of PbrEXP13.1, PbrEXLA3, and PbrEXP6.1 in BL-treated pollen tubes as determined by RT-qPCR assays. DMSO, add 100 nM DMSO to the pollen medium; BL, add 100 nM BL to the pollen medium. Data are mean ± SD of 3 independent biological replicates. D) to (F) Yeast one-hybrid (Y1H) assays verifying interaction between PbrBZR1 and promoters of PbrEXP13.1 (D), PbrEXLA3 (E), and PbrEXP6.1 (F). Screening marker was 3-amino-1,2,4-triazole (3-AT), which prevents leaky expression of HIS3 encoded in the pHIS2 vector. Transformed yeast cells were grown on SD/−Leu/−Trp (−L/−T) and SD/−Leu/−Trp/−His (−L/−T/−H) 30 or 50 mM 3-AT media in plates for 3 d. The experiments were performed 3 times with similar results, and a representative picture is shown. G) Position of E-box-2 sequence in the PbrEXLA3 promoter. EMSA results indicating binding of PbrBZR1-HIS fusion protein to PbrEXLA3 promoter. Labeled probes were incubated with PbrBZR1-HIS, after which free and bound DNA fragments were separated by acrylamide gel electrophoresis. The experiments were performed 3 times with similar results, and a representative picture is shown. E-box sequence (CACATG); Mutant E-box sequence (AAAAAA). H) Schematic representation of LUC reporter vector containing the promoter of PbrEXLA3 and the effector vector containing PbrBZR1. I) Transient expression assays showing that PbrBZR1 activates the expression of pPbrEXLA3::LUC. The experiments were performed 3 times with similar results, and a representative picture is shown. J) Quantitative analysis of luminescence intensity as indicated in (I). The Y-axis unit is defined as the bioluminescence per pixel over 10 min. K) Transcript level of PbrEXLA3 in PbrBZR1-suppressed pollen tubes as determined by RT-qPCR assay. For (J) and (K), data are mean ± SD of 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). CP, compatible reaction; CK, liquid germination medium; SC, compatible PbrS-RNase treatment; SI, incompatible PbrS-RNase treatment; SP, incompatible reaction.
Previous studies have shown that expansin genes encoding cell wall proteins are tightly regulated by BZR1 in Arabidopsis (Bai et al. 2012). We detected several putative E-box (a specific recognition site of BZR1) elements in the promoters of PbrEXP13.1, PbrEXLA3, and PbrEXP6.1 (Supplemental Fig. S4A), so yeast one-hybrid (Y1H) assays were carried out to detect whether PbrBZR1 can interact with their promoters. In these analyses, the yeast cells containing pGAD-PbrBZR1 and pHIS2-pPbrEXLA3 could grow on SD/−Leu/−Trp/−His/(−L/−T/−H) medium, but yeast cells harboring the empty vector or other constructs could not (Fig. 3, D, E, and F). We therefore deduced that PbrEXLA3 is a potential target of PbrBZR1. Subsequently, an electrophoretic mobility shift assay (EMSA) was conducted to investigate potential binding. As shown in Supplemental Fig. S4B, the putative E-box elements in the PbrEXLA3 promoter were distributed in the regions of −340 to −355 bp (E-box-1), −1,151 to −1157 bp (E-box-2), and −1,412 to −1418 bp (E-box-3) from the start codon. PbrBZR1 protein bound the pPbrEXLA3 sequence (E-box-2) (Supplemental Fig. S4C and Fig. 3G). With the addition of unlabeled probes (competitors), the ability of PbrBZR1 to interact with the promoter of PbrEXLA3 was gradually weakened (Fig. 3G). The band indicative of binding was no longer visible when the E-box-2 site was mutated (Fig. 3G). These findings show that PbrEXLA3 is a direct target of PbrBZR1.
Next, we studied the transcriptional regulation of PbrEXLA3 by PbrBZR1 using a luciferase (LUC) reporter assay. Specifically, the promoter region of PbrEXLA3 was fused with the coding sequence (CDS) of firefly LUC (pPbrELA3::LUC), and the PbrBZR1 CDS was fused with the 35S promoter to generate the effector construct (35S:: PbrBZR1) (Fig. 3H). Significant induction of LUC activity was observed in Nicotiana benthamiana leaves co-infiltrated with 35S::PbrBZR1 and pPbrEXLA3::LUC (Fig. 3, I and J), indicating that PbrBZR1 could activate the expression of PbrEXLA3. Similarly, knockdown of PbrBZR1 in pollen tubes led to decreased transcript levels of PbrEXLA3 but not PbrEXP6.1 or PbrEXP13.1 (Fig. 3K and Supplemental Fig. S5). Thus, PbrBZR1 functions as an activator upstream of PbrEXLA3.
PbrEXLA3 positively regulates pollen tube growth in pear
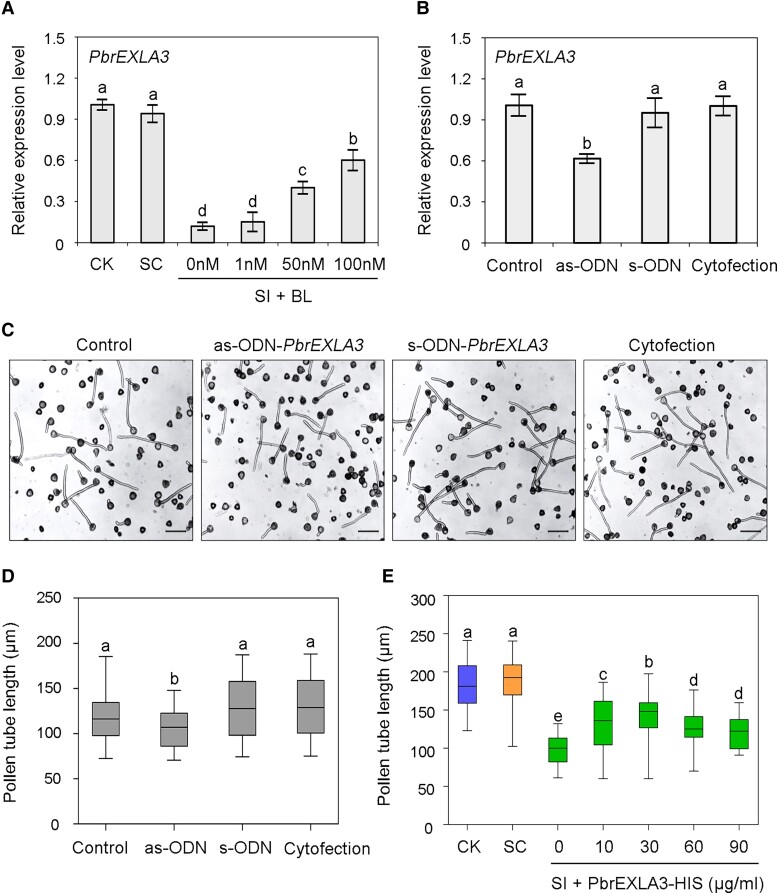
Next, RT-qPCR assays were performed to analyze the transcript profile of PbrEXLA3 in the crosstalk between SI and BR signaling. The addition of exogenous BL induced PbrEXLA3 transcription in SI-treated pollen tubes, suggesting that PbrEXLA3 is antagonistically regulated by SI and BR signals (Fig. 4A). To explore the function of PbrEXLA3 in pollen tubes, an as-ODN method was used to silence the expression of its encoding gene (Fig. 4B). Downregulation of PbrEXLA3 hindered the elongation of pollen tubes (Fig. 4, C and D). We obtained purified PbrEXLA3-HIS fusion protein using a prokaryotic expression system. Then, the PbrEXLA3-HIS fusion protein was added at different concentrations (10, 30, 60, 90 µg/ml) to germination medium to determine its effect on the elongation of compatible and incompatible pollen tubes. As shown in Supplemental Fig. S6, the addition of PbrEXLA3-HIS fusion protein accelerated pollen tube growth under SC treatment. Furthermore, the self S-RNase-mediated inhibition of pear pollen tube elongation was remarkably reduced in the presence of PbrEXLA3 (Fig. 4E). Thus, PbrEXLA3 positively regulates pollen tube growth in response to SI and BR signals.
Figure 4.
PbrEXLA3 functional assay in pear pollen tubes. A) Transcript levels of PbrEXLA3 in SC- and SI-treated pollen tubes growing on medium with BL at indicated concentrations, as determined by RT-qPCR assays. SI + BL, different concentrations of exogenous BL are added to incompatible PbrS-RNase treatment. B) Transcript levels of PbrEXLA3 in untreated pollen (control) and pollen transfected with antisense oligodeoxynucleotide (as-ODN), sense oligodeoxynucleotide of PbrEXLA3 (s-ODN), or transfection agent alone (cytofection), as determined by RT-qPCR assays. For (A) and (B), data are mean ± SD of 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). C) “Cuiguan” pollen treated with as-ODN for 2 h. Bars = 50 µm. The experiments were performed 3 times with similar results, and a representative picture is shown. D) Length of pollen tubes shown in (C). Center line, median; Box limits, upper and lower quartiles; whiskers, 1.5 × interquartile range; points, outliers. E) Length of SC- and SI-treated pollen tubes on medium with PbrEXLA3-HIS fusion protein at indicated concentrations. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 × interquartile range; points, outliers. SI + PbrEXLA3-HIS, different amounts of PbrEXLA3 fusion proteins are added to incompatible PbrS-RNase treatment. For (D) and (E), at least 100 pollen tubes were measured per replicate in each treatment. All treatments were performed in 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). CK, liquid germination medium; CK, liquid germination medium; SC, compatible PbrS-RNase treatment.
PbrARI2.3 interacts with PbrBZR1 to promote its degradation
The results described above show that SI affects the stability of dephosphorylated PbrBZR1 (Fig. 2A). A yeast 2-hybrid screening assay was carried out to study the post-transcriptional modulation of PbrBZR1 during the SI response. As a result, Pbr020065.1, which is highly expressed in pollen and has a full-length CDS of 1,587 bp, was obtained (Supplemental Fig. S7, A and B). Phylogenetic analysis indicated that its predicted protein shows high sequence similarity with the ubiquitin RING E3 ligase AtARI2 (Arabidopsis ARIADNE 2) (Supplemental Fig. S7C; Mladek et al. 2003), which contains a conserved C3HC4 RING-finger motif (124–168 aa), a central RING-finger domain (IBR, 215–257 aa), and a second C3HC4 RING-finger motif (284–312 aa) (Supplemental Fig. S7D). Thus, we designated Pbr020065.1 as PbrARI2.3 and named the 2 proteins in the same clade PbrARI2.1 (Pbr017853.1) and PbrARI2.2 (Pbr006197.1).
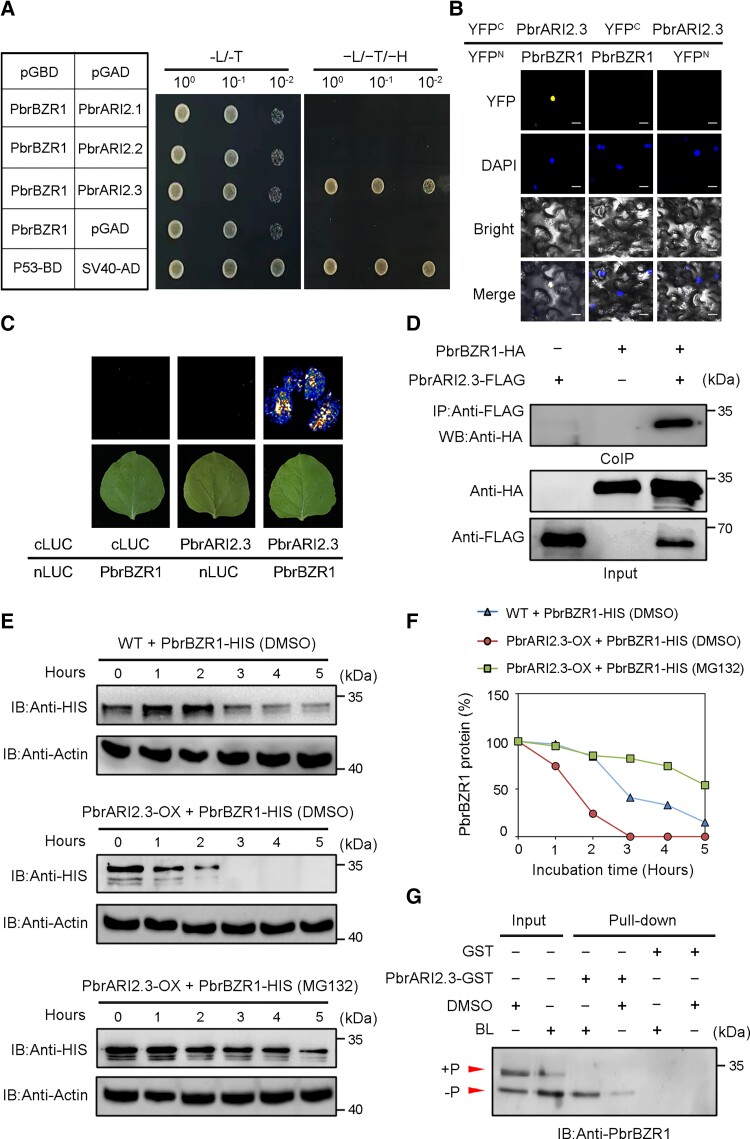
To confirm this interaction, a series of protein–protein interaction assays was conducted. First, the CDSs of 3 PbrARI genes were fused with pGAD vector to generate PbrARI2.1-AD, PbrARI2.2-AD, and PbrARI2.3-AD, and the CDS of PbrBZR1 was fused with the pGBD vector to generate PbrBZR1-BD. In yeast 2-hybrid (Y2H) assays, only the yeast cells coexpressing PbrBZR1-BD and PbrARI2.3-AD could grow on selective medium (Fig. 5A), suggesting that PbrBZR1 specifically interacts with PbrARI2.3. Next, bimolecular fluorescence complementation (BiFC) assays were carried out to determine the site of the PbrBZR1–PbrARI2.3 interaction in planta. In these analyses, the PbrBZR1 CDS was fused with pSPYNE-35S to generate PbrBZR1-YFPN, and the PbrARI2.3 CDS was fused with pSPYCE-35S to generate PbrARI2.3-YFPC. A YFP signal was found in the nuclei of Nicotiana benthamiana cells when both fusion proteins were present (Fig. 5B). Consistent with this result, in the luciferase complementation imaging (LCI) assays, we detected substantial LUC activity when tobacco leaves were coinfiltrated with PbrARI2.3-cLUC and PbrBZR1-nLUC, and no LUC activity in the controls (Fig. 5C). This interaction was confirmed by coimmunoprecipitation (Co-IP) assays (using the anti-HA antibody) of pear calli protoplasts cotransfected with PbrBZR1-HA and PbrARI2.3-Flag (Fig. 5D).
Figure 5.
PbrARI2.3 interacts with and degrades PbrBZR1 protein. A) Y2H assays showing interaction between PbrBZR1 and PbrARI2.3. SV40 and P53 were used as a positive control, and pGAD (empty vector) and pPbrBZR1-BD plasmids were used as a negative control. B) BiFC assay demonstrating interaction between PbrBZR1 and PbrARI2.3 in vivo. DAPI field indicates locations of nuclei. Scale bar = 20 µm. YFPN, N-terminal fragments of yellow fluorescent protein; YFPC, C-terminal fragments of yellow fluorescent protein. C) LCI assay showing interaction between PbrBZR1 and PbrARI2.3 in vivo. D) Co-IP assay showing interaction between PbrBZR1 and PbrARI2.3 in vivo. E) Protein degradation assays of PbrBZR1-HIS fusion proteins in the total protein extract of wild-type (WT) and PbrARI2.3-overexpressing (PbrARI2.3-OX) transgenic calli. For MG132 treatment, samples were incubated in degradation buffer containing 50 µM MG132 or DMSO for indicated times. PbrBZR1-HIS protein levels were detected by western blotting using anti-HIS antibody. F) Degradation curve of PbrBZR1-HIS proteins generated using ImageJ software. G) Semi in vivo pull-down assay showing interaction between dephosphorylated form of PbrBZR1 and PbrARI2.3. Total proteins extracted from 2-week-old WT pear calli treated 1 µM BL and DMSO were used in pull-down assays. cLUC, C-terminal fragments of firefly luciferase; IP, immunoprecipitation; IB, immunoblot; nLUC, N-terminal fragments of firefly luciferase.
Because PbrARI2.3 is a putative ubiquitin E3 ligase, we suspected that PbrARI2.3 may affect the stability of the PbrBZR1 protein. We conducted an in vitro protein degradation assay of the purified PbrBZR1-HIS fusion proteins using plant proteins extracted from PbrARI2.3-overexpressing (PbrARI2.3-OX) transgenic pear calli (Supplemental Fig. S8). The PbrBZR1-HIS protein degraded faster in the presence of proteins extracted from PbrARI2.3-OX calli than in the presence of proteins extracted from WT, and all the PbrBZR1-HIS protein had completely disappeared after 3 h of incubation (Fig. 5, E and F). However, when the proteasome inhibitor MG132 was added to the mixture, the PbrARI2.3-mediated degradation of PbrBZR1-HIS was substantially inhibited. These findings show that PbrARI2.3 promotes the degradation of the PbrBZR1 protein through the 26S proteasome pathway.
Next, semi-in vivo pull-down assays were conducted to test the affinity of PbrARI2.3 for the 2 forms (phosphorylated and dephosphorylated) of PbrBZR1. Total proteins from DMSO-treated or BL-treated WT calli were incubated with the PbrARI2.3-GST fusion protein, and the resulting protein complex was immunoassayed with the specific PbrBZR1 antibody. After the obtained protein complexes were separated by Phos-tag acrylamide gel electrophoresis, only the dephosphorylated form of PbrBZR1 was pulled-down by PbrARI2.3 (Fig. 5G), suggesting that PbrARI2.3 mainly captures the dephosphorylated form of PbrBZR1. Therefore, PbrARI2.3 interacts with dephosphorylated PbrBZR1 and promotes its degradation.
PbrARI2.3 negatively regulates pollen tube growth via ubiquitination of PbrBZR1
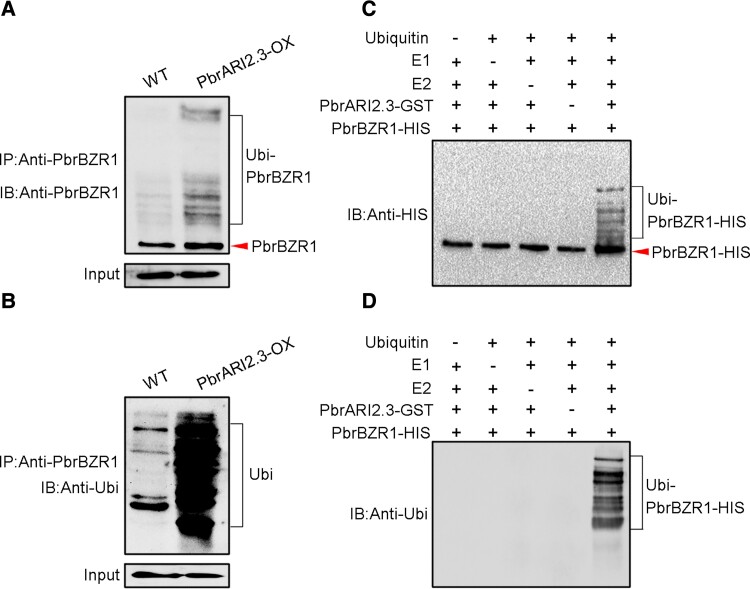
Next, the ubiquitin levels of PbrBZR1 in WT and PbrARI2.3-OX transgenic calli were detected in an in vivo ubiquitination experiment. When the anti-PbrBZR1 antibody was used to test the status of PbrBZR1 proteins, more of the high-molecular-weight ubiquitinated form of PbrBZR1 (Ubi-PbrBZR1) was detected in PbrARI2.3-OX transgenic calli than in WT controls (Fig. 6A). This result was supported by immunoblotting assays using an anti-ubiquitin antibody. Specifically, a stronger ubiquitin signal was detected in PbrARI2.3-OX than in WT calli (Fig. 6B). Thus, PbrBZR1 is ubiquitinated, and this requires PbrARI2.3. Subsequently, in vitro ubiquitination assays using purified PbrBZR1-HIS fusion protein and PbrARI2.3-GST fusion protein were conducted. In these analyses, PbrBZR1 was ubiquitinated when the reaction factors were all present (Fig. 6, C and D), confirming that PbrBZR1 is a direct substrate of PbrARI2.3 for ubiquitination.
Figure 6.
PbrBZR1 is a substrate of PbrARI2.3. A and B) Ubiquitination of PbrBZR1 by PbrARI2.3 in vivo. PbrBZR1 and ubiquitinated PbrBZR1 were detected by immunoprecipitation using anti-PbrBZR1 (A) and anti-ubiquitin (Ubi) antibodies (B) from the wild-type (WT) and PbrARI2.3-overexpressing (PbrARI2.3-OX) transgenic calli, respectively. C and D) Ubiquitination of PbrBZR1 by PbrARI2.3 in vitro. C) Anti-HIS and (D) anti-Ubi antibodies were used to detect PbrBZR1-HIS and ubiquitinated PbrBZR1-HIS, respectively. E1, ubiquitin activating enzyme; E2, ubiquitin conjugating enzyme; IP, immunoprecipitation, IB, immunoblot.
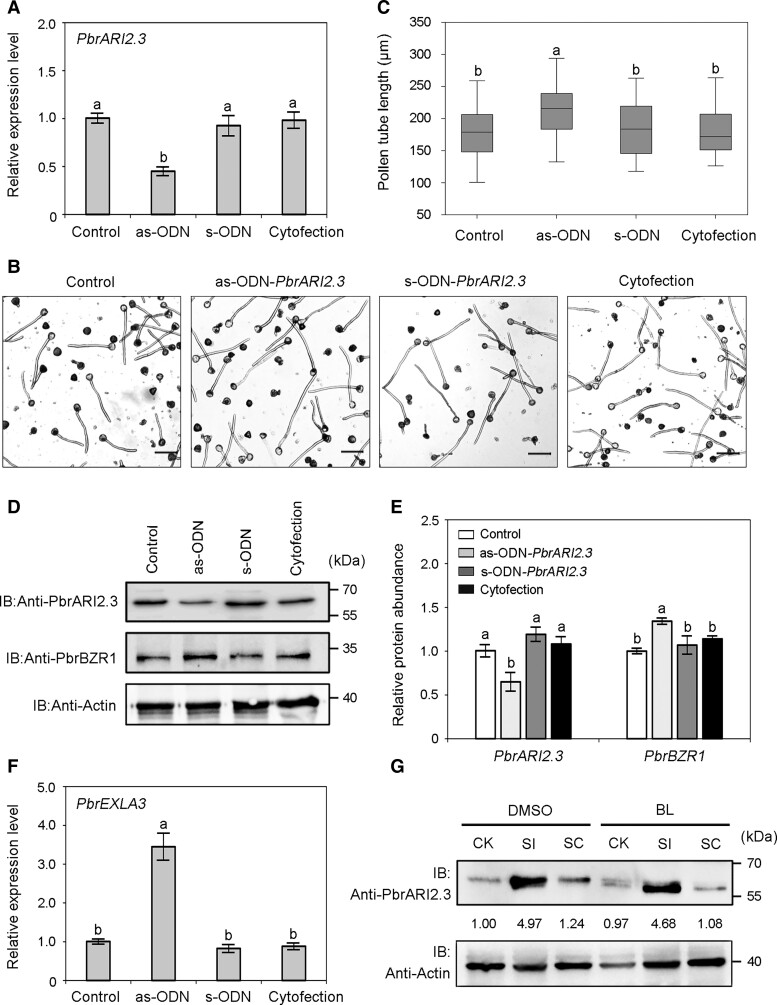
Next, we used as-ODN silencing technology to verify the role of PbrARI2.3. Transfection of pollen with an antisense PbrARI2.3 oligodeoxynucleotide (as-ODN-PbrARI2.3) resulted in significantly decreased PbrARI2.3 transcript levels (Fig. 7A) and accelerated pollen tube growth (Fig. 7, B and C). Considering the ubiquitin modification of PbrBZR1 by PbrARI2.3, we speculated that PbrARI2.3 may regulate the expression of PbrBZR1-downstream genes by affecting the stability of PbrBZR1. As hypothesized, compared with the control groups, pollen treated with as-ODN-PbrARI2.3 produced less PbrARI2.3 protein and consequently accumulated more PbrBZR1 protein (Fig. 7, D and E). The transcript level of PbrEXLA3 showed the same trend as the protein level of PbrBZR1 (Fig. 7F), suggesting that the ability of PbrARI2.3 to regulate PbrEXLA3 depends on PbrBZR1.
Figure 7.
PbrARI2.3 functional assay in pear pollen tubes. A) Transcript levels of PbrARI2.3 in untreated pollen (control) and pollen transfected with antisense oligodeoxynucleotide (as-ODN), sense oligodeoxynucleotide of PbrARI2.3 (s-ODN), or transfection agent alone (cytofection), as determined by RT-qPCR assays. Each sample contained 0.1 g of pollens, and the data are mean ± SD of 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). B) “Cuiguan” pollen treated with as-ODN for 2.5 h. Bars = 50 µm. The experiments were performed 3 times with similar results, and a representative picture is shown. C) Length of pollen tubes shown in (B). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 × interquartile range; points, outliers. At least 100 pollen tubes were measured per replicate in each treatment. All treatments were performed in 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). D) Protein levels of PbrARI2.3 and PbrBZR1 in pollen tubes shown in (B). Anti-PbrARI2.3 and anti-PbrBZR1 antibodies were used to detect PbrARI2.3 and PbrBZR1, respectively. The experiments were performed 3 times with similar results, and a representative picture is shown. E) Relative quantitative analysis of PbrARI2.3 and PbrBZR1 protein levels by western blotting. Data are mean ± SD of 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). F) Transcript level of PbrEXLA3 in pollen tubes shown in (B). Each sample contained 0.1 g of pollens, and the data are mean ± SD of 3 independent biological replicates. Different letters indicate significant differences as determined by Tukey's HSD test (P < 0.05). G) Protein levels of PbrARI2.3 in SC- and SI-treated pollen tubes after the addition of 100 nM BL or DMSO, as determined by western blot analysis. Total proteins extracted from pollen tubes were immunoprecipitated with anti-PbrARI2.3 antibody. DMSO, add 100 nM DMSO to the pollen medium; BL, add 100 nM BL to the pollen medium. The experiments were performed 3 times with similar results, and a representative picture is shown. CK, liquid germination medium; SC, compatible PbrS-RNase treatment; SI, incompatible PbrS-RNase treatment.
To determine whether PbrARI2.3 is involved in SI, its transcript and protein levels were detected. The transcript level of PbrARI2.3 was increased in SI-treated pollen tubes (Supplemental Fig. S9). The abundance of PbrARI2.3 protein in pollen tubes was quantified by immunoprecipitation with a specific anti-PbrARI2.3 antibody. As shown in Fig. 7G, there was substantially more PbrARI2.3 protein in SI-treated pollen tubes than in the CK and SC-treated pollen tubes. However, the addition of exogenous BL did not affect the stability of PbrARI2.3 protein. Thus, PbrARI2.3 responds to the SI signal both at the transcriptional and protein levels. Further functional verification analyses showed that knockdown of PbrARI2.3 in pollen through as-ODN treatment could inhibit the effect of S-RNase on the elongation of incompatible pollen tubes (Supplemental Fig. S10). Since pollen tube growth arrest and pollen cell death are both important hallmarks of S-RNase-based SI, we detected pear pollen viability after different as-ODN treatments (as-ODN-PbrARI2.3, as-ODN-PbrBZR1, and as-ODN-PbrEXLA3) by fluorescein diacetate (FDA) staining. The staining intensity of as-ODN-transfected pollen was similar to that in the control groups (Supplemental Fig. S11), indicating that the PbrARI2.3-PbrBZR1-PbrEXLA3 pathway might not directly participate in the regulation of pollen cell death.
Discussion
The SI reproductive mechanism in plants allows for increased opportunities for outcrossing events, ensuring continuous genetic variation within the population and reducing the risk of inbreeding depression (Nasrallah 2000). During this process, S-RNase has a toxic effect on incompatible pollen that lands on the pistil, making it unable to grow and reach the ovule (Hiratsuka and Tezuka 1980; Chen et al. 2018). Ongoing studies have identified many hormonal molecules, such as ABA (Wang et al. 2020), auxin (Chen and Zhao 2008; He et al. 2018), GAs (Singh et al. 2002; Cheng et al. 2004), and BRs (Ye et al. 2010; Vogler et al 2014) that appear to control the fertilization process by influencing pollen tube elongation. However, the exact roles of these plant hormones in mediating SI and the molecular mechanisms of the regulatory responses remain to be elucidated.
A recent study has established that female SI in heterostylous Primula is controlled by the BR-inactivating cytochrome P450 CYP734A50 (Huu et al. 2016, 2022). The deletion or inactivity of CYP734A50 or excessive accumulation of BRs within the flower promotes the extension of the style and changing of the female SI type (Huu et al. 2022). Here, we explored the effect of BL, an active BR, on pollen tube elongation and the molecular mechanisms underlying SI in pear. Application of exogenous BL promoted the growth of incompatible pollen tubes in a dose-dependent manner (Fig. 1C), suggesting that the BR signal functions as an activator in the SI reaction of pear. Whereas some ions and small molecules are known to affect protein activity and/or affinity (Wu et al. 2003; Cao et al. 2009), the BL treatments did not directly affect the activity of the S-RNases (Fig. 1D). Thus, we speculate that BRs may regulate the SI pathway by triggering its downstream signal components.
It is well known that BZR1 is involved in the regulation of pollen development (Huang et al. 2013; Zhu et al. 2015; Kim et al. 2021), but its specific role remains unclear. In apple pollen tubes, the transcription of MdMYC2 induced by S-RNases depends on the JA signaling pathway (Gu et al. 2019). In the current study, the SI treatment promoted the degradation of dephosphorylated PbrBZR1 in pollen tubes even after the addition of BL (Fig. 2A), suggesting that ubiquitin participates in the SI-mediated posttranscriptional modification of PbrBZR1 through a BR-independent signaling pathway. In Arabidopsis, the DELLA-BZR1-PHYTOCHROME-INTERACTING FACTOR4 (PIF4) transcription module mediates the coordinated modulation of growth by GA, BR, and light signals (Bai et al. 2012). In this ternary complex, BZR1 acts as an important regulator that integrates signals from multiple pathways to control cell elongation and seedling etiolation. In Brassica napus, a transcriptional cascade involving BZR1 is important for BR to regulate SI-mediated pollen tube elongation (Duan et al. 2020), although the SI mechanism in Brassica differs from the S-RNase-based SI mechanism in pear. Here, knockdown of PbrBZR1 reduced the length of pollen tubes, compared with those in the controls (Supplemental Fig. S2), similar to the positive role of OsBZR1 in pollen maturation in rice (Oryza sativa) (Zhu et al. 2015). Furthermore, downregulation of PbrBZR1 in pollen reduced the positive effect of BL on incompatible pollen tube elongation (Fig. 2D), suggesting that PbrBZR1 is essential for BR-regulated SI.
Expansins are extracellular proteins that loosen the cell wall in a pH-dependent manner during plant growth. They function in cell enlargement (Cosgrove 2000) and cell wall disassembly during fruit ripening (Rose and Bennett 1999), abscission, and other cell separation events (Cho and Cosgrove 2000). Emerging evidence has indicated that expansins are related to the hormonal regulation of plant tolerance to various stresses (Ding et al. 2008; Zhao et al. 2012). Here, we observed that PbrEXLA3 expression was modulated by both SI and BR signals, and BR appeared to function as an activator upstream of the SI signal (Fig. 4A). In Arabidopsis, 2 hypocotyl elongation-related expansin genes, EXP1 and EXP8, are the common downstream targets of PIF4 and BZR1 (Bai et al. 2012). Consistent with this, we found that PbrBZR1 transcriptionally activated PbrEXLA3 through directly binding to its promoter (Fig. 3). Two other BR-induced expansin genes, PbrEXP6.1 and PbrEXP13.1, which are not the direct targets of PbrBZR1, may be regulated by other BR signaling factors, such as homologs of BZR1 (Wang et al. 2013). Although expansins have unique physical effects on the cell wall, there is no direct data to prove that they participate in pollen tube growth during SI. Here, knockdown of PbrEXLA3 resulted in reduced elongation of pollen tubes (Fig. 4, C and D), and the addition of prokaryotic-expressed PbrEXLA3 greatly weakened the effect of incompatible S-RNases on pollen tubes (Fig. 4E). Therefore, PbrEXLA3 positively regulates the growth of pollen tubes and may act as a defensive agent after self S-RNase enters pollen tubes, just like MdD1 in apple (Gu et al. 2019).
Ubiquitination is an important post-translational modification by which cellular proteins are targeted for degradation. This process is implicated in the control of almost all biological events (Pickart 2001). Previous studies have reported that BZR1/BES1 proteins undergo ubiquitination in the crosstalk between multiple signaling pathways (Wang et al. 2013; Kim et al. 2014; Yang et al. 2017). Here, we found that PbrARI2.3 interacted with the dephosphorylated form of PbrBZR1 to mediate its degradation through the 26S proteasome pathway (Figs. 5 and 6). ARI proteins belong to a large family of E3 ubiquitin ligases with RING-between-RING fingers domains (Mladek et al. 2003). Most information about the function of ARI proteins has been obtained from studies on the immune response of animals (Wenzel et al. 2011; Dove et al. 2016; Xiong et al. 2022). The transcription of some members of the ARI gene family is affected by diverse external stresses (Mladek et al. 2003), so it is possible that ARI genes in plants are involved in responses to specific environmental conditions. Here, SI treatment induced pollen-specific expression of PbrARI2.3 at the transcript and protein levels (Supplemental Fig. S9 and Fig. 7G), indicating that PbrARI2.3 plays an important role in the SI response. Functional verification analyses showed that PbrARI2.3 inhibits pollen tube elongation by reducing the stability of PbrBZR1 (Fig. 7B−E). Correspondingly, the increased abundance of PbrBZR1 led to the up-regulation of PbrEXLA3 expression (Fig. 7G). This result further confirms that the PbrARI2.3-PbrBZR1 module functions upstream of PbrEXLA3.
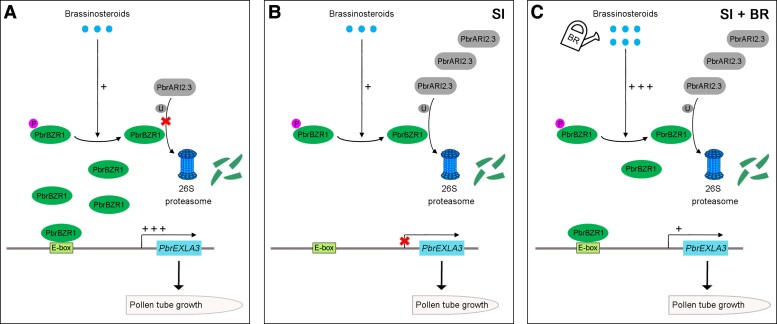
In conclusion, we propose a model to explain how BRs regulate the growth of incompatible pollen tubes in pear (Fig. 8). Under normal conditions, BR-induced dephosphorylated PbrBZR1 promotes pollen tube growth by transcriptionally activating PbrEXLA3. When self S-RNase enters the pollen tube, a large amount of PbrARI2.3 captures dephosphorylated PbrBZR1 and promotes its degradation, which eventually leads to the growth arrest of incompatible pollen tubes. When the incompatible pollen tubes are treated with exogenous BR, most PbrBZR1 is present in the dephosphorylated form, which greatly increases the probability of its binding to the PbrEXLA3 promoter, thus partially alleviating the inhibitory effect of incompatible S-RNases on pollen tube growth. In addition to direct degradation of RNA in self-pollen, S-RNases regulate the growth of pollen tubes by triggering other pathways including depolymerization of the actin cytoskeleton and scavenging of tip-localized reactive oxygen species (Wang et al. 2010; Chen et al. 2018). A previous study showed that BR can change the configuration of the actin cytoskeleton and the localization of the auxin transporter PIN-FORMED2 (PIN2) to control plant growth and gene transcription (Lanza et al. 2012). The results of the present study show that BR participates in the SI response through the PbrARI2.3–PbrBZR1–PbrEXLA3 pathway, but it is not clear whether BR regulates SI-mediated cytoskeletal depolymerization in a similar manner. Overall, these results provide information on the role of BRs in S-RNase-based SI and demonstrate how a ubiquitin modification mechanism participates in controlling plant cell growth.
Figure 8.
Model of participation of PbrBZR1-PbrARI2.3 complex in BR-mediated pollen tube growth during SI in pear. A) Under normal conditions, PbrARI2.3 barely accumulates, leading to the inability to degrade the BR-activated dephosphorylated (active) form of PbrBZR1. Dephosphorylated PbrBZR1 promotes pollen tube growth by activating PbrEXLA3 transcription. B) Under SI conditions, PbrARI2.3 accumulates and captures dephosphorylated PbrBZR1 to mediate its degradation through the 26S proteasome pathway. Thus, little PbrBZR1 is available to bind to the PbrEXLA3 promoter and regulate its transcription. As a result, pollen tube growth is inhibited. C) Under BR-sufficient and SI conditions, most PbrBZR1 protein exists in the dephosphorylated (active) form, so it cannot be completely degraded by PbrARI2.3. Thus, the dephosphorylated PbrBZR1 can act on PbrEXLA3 to alleviate the SI-induced inhibition of pollen tube growth. Circle “P” represents phosphorylation modification; Circle “U” represents ubiquitination modification; plus sign represents the ability of PbrBZR1 to activate PbrEXLA3 expression (the more plus signs, the stronger the ability); Cross symbol represents the inhibition of PbrEXLA3 transcription.
Materials and methods
Plant materials and culture conditions
Pollen was collected from “Cuiguan” pear (Pyrus pyrifolia) trees growing in the Fruit Experimental Yard of Nanjing Agricultural University, China. Pollen grains were isolated from mature flowers and cultured in vitro as previously described (Xia et al. 2023). In vivo pollination experiments were conducted with a cross-compatible pair, with “Cuiguan” as the male parent and “Dangshansuli” as the female parent. Treated pollen tubes were examined under a light microscope, and pollen tube length was measured using ImageJ software. Each treatment was repeated 3 times, and at least 100 pollen tubes were counted each time. The pear calli used for genetic transformation were cultivated on medium as previously described (Ming et al. 2022). The pear calli were grown in the dark at 25 °C and subcultured every 15 d.
PbrS-RNase and BR treatments
For the SC and SI treatments, prokaryon-expressed PbrS-RNase fusion proteins with a HIS-tag (SC, PbrS7-Rnase and PbrS34-Rnase; SI, PbrS3-Rnase and PbrS5-Rnase) were added to pollen cultures (“Cuiguan”; S3 or S5 cultures, respectively). For the BR treatments, BL at different concentrations (1, 50, and 100 nM) was added to pollen cultures containing incompatible PbrS-RNases. The final concentration of recombinant PbrS-RNases was 0.15 U.
S-RNase activity analysis
Torula yeast RNA was used as a substrate to measure S-RNase activity as previously described (Gu et al. 2019). The recombinant PbrS3-RNase and PbrS5-RNase (30 µg/mL) were incubated in reaction buffer containing BL at different concentrations (1, 50, and 100 nM) at 37 °C for 15 min. After adding the termination solution, the absorbance of the supernatant at 260 nm was measured using a UV-2450 spectrophotometer (Shimadzu, Kyoto, Japan).
FDA staining assays
The FDA staining solution was prepared by dissolving 5 mg FDA in 1 mL acetone. The stain solution was sealed and stored at 4°C. One pollen suspension cultured for 2.5 h was mixed with 10 µl FDA solution to give a final concentration of 1 µM and then the mixture was incubated in the dark for 5 min. A 40-µL aliquot of the mixture was removed from the bottom of the tube, dropped onto a slide, and observed and photographed under a LSM800 confocal microscope (Carl Zeiss, Jena, Germany). The changes and development status of pollen were observed, and the number of surviving pollen grains (indicated by green fluorescence) was counted (laser wavelength, 488 nm; detection wavelength, 481–549 nm; Master gain, 650; Digital gain, 1.0). The total number of pollen grains was counted under visible light. Five randomly selected fields of view were analyzed for each treatment, and the data were used to calculate the pollen staining rate.
Extraction of total RNA and RT-qPCR analysis
Total RNA was extracted and RT-qPCR assays were performed as described by Wang et al. (2021). Pyrus UBQ was used as the internal control gene. The primers used are shown in Supplemental Table S2.
Antisense oligodeoxynucleotide experiment
Both phosphorothioated as-ODN and phosphorothioated sense ODN (s-ODN) were designed using RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The candidate as-ODN sequences of the target regions were evaluated using Soligo software (http://sfold.wadsworth.org/soligo.pl). The as-ODN treatment was conducted as described by Chen et al. (2018). Before the as-ODN treatment, we first pre-cultured the pollen to ensure the consistency of its germination rate. The phosphorothioated ODN sequences are listed in Supplemental Table S2.
Phylogenetic analysis
Phylogenetic analysis was performed using MEGA version 7.0 as previously described (Sun et al. 2021). The sequence data of the ARIADNE (ARI) proteins in pear and Arabidopsis can be obtained from the Pear Genome Project database (http://dx.doi.org/10.5524/100083) and the Arabidopsis Information Resource (https://www.arabidopsis.org), respectively.
Generation of transgenic pear calli
To generate overexpression pear calli, the PbrBZR1 CDS was cloned and inserted into pRI101-AN-GFP. Transgenic pear calli were obtained by Agrobacterium tumefaciens (LBA4404)-mediated genetic transformation as previously described (Wang et al. 2021). The primers used are listed in Supplemental Table S2.
Protein extraction and immunoblotting assays
The total plant proteins were extracted from pear calli or pollen tubes using the Plant Protein Extraction Kit (CWBIO, Beijing, China). Briefly, 1 g samples were ground in liquid nitrogen and then incubated in 5 ml Plant Protein Extraction Agent at 0 °C for 0.5 h. The supernatant was collected by centrifugation and then detected by a western blot assay. For the Phos-tag gel electrophoresis assay, the proteins were subjected to 12% (w/v) SDS-PAGE supplemented with 50 μM Phos-tag and 10 mM Mn2+. The separated proteins were transferred to a polyvinylidene difluoride membrane, and PbrBZR1 protein was detected using an anti-PbrBZR1 antibody. Monoclonal antibodies (anti-actin, anti-HIS, anti-GST, anti-Ubi, anti-FLAG, anti-HA, and anti-GFP) were prepared by Abmart (Shanghai, China). Polyclonal antibodies (anti-PbrBZR1 and anti-PbrARI2.3) were prepared by the ABclonal Co. Ltd. (Wuhan, China). Protein abundance was analyzed using ImageJ software.
Y1H assays
The promoter fragments of PbrEXP13.1, PbrEXLA3, and PbrEXP6.1 were separately inserted into the pHIS2 vector, while the PbrBZR1 CDS was inserted into the pGADT7 vector. The self-activation of promoters was suppressed by 3-amino-1,2,4-triazole (3-AT). The Y187 yeast (Saccharomyces cerevisiae) cells carrying the recombinant pHIS2 and pGADT7 plasmids were grown on selective medium lacking Leu, Trp, and His (−L/−T/−H) to detect interactions. The empty pGADT7 vector was used as the control. The primers used are listed in Supplemental Table S2.
EMSAs
The EMSAs were conducted using a LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific, Waltham, MA, USA). The PbrBZR1 CDS was inserted into the pET-32a vector with a HIS tag. The PbrBZR1-HIS protein was mixed with the biotin-labeled E-box probe, and incubated at 24 °C for 20 min. The free and bound probes were separated by acrylamide gel electrophoresis. The probe sequences are listed in Supplemental Table S3.
Dual-LUC reporter assays
The promoter fragment of PbrEXPLA3 was inserted into the pGreenII 0800-LUC vector to generate the reporter construct (pPbrEXLA3::LUC), and the PbrBZR1 CDS was inserted into the pGreenII 62-SK vector to generate the effector construct (35S::PbrBZR1). A. tumefaciens (GV3101) cells harboring 35S::PbrBZR1 and pPbrEXLA3::LUC were coinfiltrated into Nicotiana benthamiana leaves and then the change in LUC activity was detected using a charge-coupled device camera (Andor Technology, Belfast, Ireland, UK). The primers used are listed in Supplemental Table S2.
Y2H assays
The Y2H experiments were conducted using a Y2H kit (Clontech, Mountain View, CA, USA) following the manufacturer's protocol. The PbrBZR1 CDS was inserted into the pGBDT7 vector, while the PbrARI2.1, PbrARI2.2, and PbrARI2.3 CDSs were each inserted into the pGADT7 vector. Different combinations were transferred into Y2H yeast cells. The yeast cells were then grown on selective medium lacking Trp, Leu, His (−T/−L/−H) to detect possible interactions. The primers used are listed in Supplemental Table S2.
BiFC assays
The PbrBZR1 and PbrARI2.3 CDSs were inserted into the pSPYNE and pSPYCE vectors to generate PbrBZR1-YFPN and PbrARI2.3-YFPC, respectively. Then, A. tumefaciens (GV3101) cells harboring PbrBZR1-YFPN and PbrARI2.3-YFPC were coinfiltrated into N. benthamiana leaves. The YFP signal in infiltrated N. benthamiana leaves was detected (laser wavelength, 514 nm; detection wavelength, 490–540 nm; Master gain, 700; Digital gain, 1.0) using a LSM800 confocal microscope (Carl Zeiss, Jena, Germany). Cells were also stained with DAPI (Thermo Fisher Scientific) as a nucleus-specific marker. The primers used are listed in Supplemental Table S2.
LCI assays
The PbrBZR1 and PbrARI2.3 CDSs were inserted into the pCAMBIA1300-CLuc and pCAMBIA1300-NLuc vectors to generate PbrBZR1-NLuc and PbrARI2.3-CLuc, respectively. Then, A. tumefaciens (GV3101) cells harboring PbrBZR1-NLuc and PbrARI2.3-CLuc were coinfiltrated into N. benthamiana leaves. The LUC activity in the N. benthamiana leaves was detected using a charge-coupled device camera (Andor Technology). The primers used are listed in Supplemental Table S2.
Co-IP assays
The PbrBZR1 and PbrARI2.3 CDSs were inserted into the pEarlyGate201 and pEarlyGate202 vectors to generate PbrBZR1-HA and PbrARI2.3-FLAG, respectively. The PbrBZR1-HA and PbrARI2.3-FLAG plasmids were co-transfected into pear calli protoplasts, which were then incubated for 5 h at 25 °C. The supernatant of protoplasts and anti-FLAG agarose beads were incubated in test tubes for 3 h. The beads were washed 5 times with immunoprecipitation buffer and then HA and FLAG were detected using anti-HA and anti-FLAG antibodies, respectively. The primers used are listed in Supplemental Table S2.
Semi-in vivo pull-down assays
The PbrARI2.3 CDS was inserted into the pGEX-4T-1 vector to generate PbrARI2.3-GST fusion protein. Total protein extracts from 10-day-old WT pear calli were incubated with glutathione-Sepharose beads containing PbrARI2.3-GST for 2 h at 4 °C. The PbrBZR1 protein pulled down by PbrARI2.3-GST was detected using an anti-PbrBZR1 antibody.
In vitro protein degradation assays
The PbrBZR1 CDS was inserted into the pET-32a vector to generate the PbrBZR1-HIS fusion protein. Total protein extracts from 10-day-old WT and PbrARI2.3-OX pear transgenic calli were incubated with PbrBZR1-HIS for indicated times at 22 °C. The residual protein levels were detected with the anti-HIS antibody. For the MG132 treatment, the pear calli were pretreated with 50 µM MG132 for 1 h.
In vitro and in vivo ubiquitination assays
For in vitro ubiquitination assays, calli extracts were incubated with a reaction buffer containing recombinant PbrBZR1-HIS fusion protein at 30 °C for 16 h and then the incubation solution was immunoprecipitated with anti-HIS and anti-Ubi antibodies. For in vivo ubiquitination assays, WT and transgenic calli pear transgenic calli were prepared. The PbrBZR1 proteins were immunoprecipitated with an anti-PbrBZR1 antibody using the Pierce classic IP kit (Thermo Fisher Scientific) following the manufacturer's protocol. The eluted proteins were detected with an anti-Ubi antibody.
Statistical analysis
ANOVA and Tukey's honestly significant difference (HSD) tests were used to determine statistical significance. In figures and tables, different letters indicate significant differences (P < 0.05).
Accession numbers
Sequences of genes mentioned in this article are listed in the Pear Genome Project database (http://dx.doi.org/10.5524/100083) under the following accession numbers: PbrBZR1 (Pbr020081.1), PbrARI2.1 (Pbr017853.1), PbrARI2.3 (Pbr006197.1), PbrARI2.3 (Pbr020065.1), PbrEXP13.1 (Pbr012636.1), PbrEXP2.2 (Pbr013129.1), PbrEXP2.3 (Pbr017279.1), PbrEXP8.3 (Pbr002929.1), PbrEXP8.1 (Pbr003748.1), PbrEXP5.3 (Pbr019282.1), PbrEXP5.1 (Pbr039073.1), PbrEXLA3 (Pbr033313.1), PbrEXLB1.3 (Pbr013752.1), PbrEXLB1.4 (Pbr022160.1), PbrEXP16 (Pbr009385.1), PbrEXP6.1 (Pbr003883.1), PbrEXP3 (Pbr033539.1).
Supplementary Material
Acknowledgments
The bioinformatic analysis was supported by the Bioinformatics Center of Nanjing Agricultural University.
Contributor Information
Yicheng Wang, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Panpan Liu, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Yiling Cai, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Yu Li, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Chao Tang, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Nan Zhu, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Peng Wang, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Shaoling Zhang, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Juyou Wu, Sanya Institute of Nanjing Agricultural University, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement, Jiangsu Academy of Agricultural Sciences, Nanjing 210014, China.
Author contributions
J.W. and Y.W. designed the experiments; Y.W. and P.W. performed the experiments. P.L., Y.C., and N.Z. helped complete the experiments. Y.L. and C.T. performed the bioinformatics analysis. Y.W., J.W., and S.Z. wrote the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. PbrBZR1 does not respond to SI at the transcription level.
Supplemental Figure S2. Functional verification of PbrBZR1 in BR- and SI-mediated pollen tube growth.
Supplemental Figure S3. Transcript levels of expansin genes in pistil and pollen.
Supplemental Figure S4. PbrBZR1 binds to the promoter of PbrEXLA3.
Supplemental Figure S5. Transcript levels of PbrEXP6.1 and PbrEXP13.1 in PbrBZR1-suppressed pollen tubes.
Supplemental Figure S6. Length of SC-treated pollen tubes grown on medium containing different concentrations of PbrEXLA3-HIS fusion protein.
Supplemental Figure S7. Identification of PbrARI2.3 in pear.
Supplemental Figure S8. Identification of PbrARI2.3-overexpressing transgenic pear calli.
Supplemental Figure S9. PbrARI2.3 responds to SI at the transcription level.
Supplemental Figure S10. Length of PbrARI2.3-suppressed pollen tubes grown under SC treatment.
Supplemental Figure S11. Detection of pollen viability after as-ODN treatments.
Supplemental Table S1. Differentially expressed expansin genes after pollination.
Supplemental Table S2. Primers used to analyze gene expression and construct vectors.
Supplemental Table S3. Primers used to synthesize EMSA probes.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (grant no. BK20221006), the Jiangsu Agriculture Science and Technology Innovation Fund (grant no. CX(22)3044), the China Postdoctoral Science Foundation (grant no. 2021M701743), the National Natural Science Foundation of China (grant no. 32172543), and the Earmarked Fund for China Agriculture Research System (grant no. CARS-28).
Data availability
All relevant data can be found within the manuscript and its supporting materials..
References
- Anderson MA, Cornish EC, Mau SL, Williams EG, Hoggart R, Atkinson A, Bonig I, Grego B, Simpson R, Roche PJ, et al. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature. 1986:321(6065):38–44. 10.1038/321038a0 [DOI] [Google Scholar]
- Anderson MA, Mcfadden GI, Bernatzky R, Atkinson A, Orpin T, Dedman H, Tregear G, Fernley R, Clarke AE. Sequence variability of three alleles of the self-incompatibility gene of Nicotiana alata. Plant Cell. 1989:1(5):483–491. 10.1105/tpc.1.5.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012:14(8):810–817. 10.1038/ncb2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SC. The evolution of mating strategies in flowering plants. Trends Plant Sci. 1998:3(9):335–341. 10.1016/S1360-1385(98)01299-0 [DOI] [Google Scholar]
- Cao H, Wu DH, Wang HX, Xu M. Effect of the glycosylation of flavonoids on interaction with protein. Spectrochim Acta Part A. 2009:73(5):972–975. 10.1016/j.saa.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Charnov EL, Bull JJ, Maynard Smith J. Why be an hermaphrodite? Nature. 1976:263(5573):125–126. 10.1038/263125a0 [DOI] [Google Scholar]
- Chen J, Wang P, de Graaf BHJ, Zhang H, Jiao H, Tang C, Zhang S, Wu J. Phosphatidic acid counteracts S-RNase signaling in pollen by stabilizing the actin cytoskeleton. Plant Cell. 2018:30(5):1023–1039. 10.1105/tpc.18.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhao J. Free IAA in stigmas and styles during pollen germination and pollen tube growth of Nicotiana tabacum. Physiol Plant. 2008:134(1):202–215. 10.1111/j.1399-3054.2008.01125.x [DOI] [PubMed] [Google Scholar]
- Cheng H, Qin LJ, Lee S, Fu XD, Richards DE, Cao DN, Luo D, Harberd NP, Peng JR. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development. 2004:131(5):1055–1064. 10.1242/dev.00992 [DOI] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Nat Acad Sci USA. 2000:97(17):9783–9788. 10.1073/pnas.160276997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher DA, Mitchell RJ, Trapnell DW, Smallwood PA, Semski WR, Karron JD. Hermaphroditism promotes mate diversity in flowering plants. Am J Bot. 2019:106(8):1131–1136. 10.1002/ajb2.1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. New genes and new biological roles for expansins. Curr Opin Plant Biol. 2000:3(1):73–78. 10.1016/S1369-5266(99)00039-4 [DOI] [PubMed] [Google Scholar]
- De Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin (Germany): Springer-Verlag; 2001. [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. Activation of the indole-3-acetic acid–amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008:20(1):228–240. 10.1105/tpc.107.055657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi UK, Krishna P. Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. Nat Biotechnol. 2009:26(3–4):131–136. 10.1016/j.nbt.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016:17(8):1221–1235. 10.15252/embr.201642641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan ZQ, Dou SW, Liu ZQ, Li B, Yi B, Shen JX, Tu JX, Fu TD, Dai C, Ma CZ. Comparative phosphoproteomic analysis of compatible and incompatible pollination in Brassica napus L. Acta Biochim Biophys Sin (Shanghai). 2020:52(4):446–456. 10.1093/abbs/gmaa011 [DOI] [PubMed] [Google Scholar]
- Gray JE, McClure BA, Bonig I, Anderson MA, Clarke AE. Action of the style product of the self-incompatibility gene of Nicotiana alata (S-RNase) on in vitro-grown pollen tubes. Plant Cell. 1991:3(3):271–283. 10.2307/3869367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu ZY, Li W, Doughty J, Meng D, Yang Q, Yuan H, Li Y, Chen QJ, Yu J, Liu CS, et al. A gamma-thionin protein from apple, MdD1, is required for defence against S-RNase-induced inhibition of pollen tube prior to self/non-self recognition. Plant Biotechnol J. 2019:17(11):2184–2198. 10.1111/pbi.13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005:307(5715):1634–1638. 10.1126/science.1107580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SL, Hsieh HL, Jauh GY. Small auxin up RNA62/75 are required for the translation of transcripts essential for pollen tube growth. Plant Physiol. 2018:178(2):626–640. 10.1104/pp.18.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Liu J, Dong H, Sun J. The blue-light receptor CRY1 interacts with BZR1 and BIN2 to modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol Plant. 2019:12(5):689–703. 10.1016/j.molp.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Hiratsuka S, Tezuka T. Changes in proteins in pistils after self- and cross-pollination in Japanese pear. J Jpn Soc Hortic Sci. 1980:49(1):57–64. 10.2503/jjshs.49.57 [DOI] [Google Scholar]
- Huang HY, Jiang WB, Hu YW, Wu P, Zhu JY, Liang WQ, Wang ZY, Lin WH. BR Signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol Plant. 2013:6(2):456–469. 10.1093/mp/sss070 [DOI] [PubMed] [Google Scholar]
- Huu CN, Kappel C, Keller B, Sicard A, Takebayashi Y, Breuninger H, Nowak MD, Bäurle I, Himmelbach A, Burkart M, et al. Presence versus absence of CYP734A50 underlies the style-length dimorphism in primroses. eLife. 2016:5:e17956. 10.7554/eLife.17956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huu CN, Plaschil S, Himmelbach A, Kappel C, Lenhard M. Female self-incompatibility type in heterostylous Primula is determined by the brassinosteroid-inactivating cytochrome P450 CYP734A50. Curr Biol. 2022:32(3):671. 10.1016/j.cub.2021.11.046 [DOI] [PubMed] [Google Scholar]
- Kao TH, Tsukamoto T. The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell. 2004:16(suppl_1):S72–S83. 10.1105/tpc.016154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketsa S, Bunya-atichart K, van Doorn WG. Ethylene production and post-pollination development in Dendrobium flowers treated with foreign pollen. Aust J Plant Physiol. 2001:28(5): 409–415. 10.1071/PP00048 [DOI] [Google Scholar]
- Kim B, Jeong YJ, Corvalan C, Fujioka S, Cho S, Park T, Choe S. Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 2014:77(5):737–747. 10.1111/tpj.12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim SH, Shin DM, Kim SH. ATBS1-INTERACTING FACTOR 2 negatively modulates pollen production and seed formation in Arabidopsis. Front Plant Sci. 2021:12:1548. 10.3389/fpls.2021.704958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using phos-tag SDS-PAGE. Nat Protoc. 2009:4(10):1513–1521. 10.1038/nprot.2009.154 [DOI] [PubMed] [Google Scholar]
- Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol. 2002:50(1):29–41. 10.1023/A:1016050018779 [DOI] [PubMed] [Google Scholar]
- Lanza M, Garcia-Ponce B, Castrillo G, Catarecha P, Sauer M, Rodriguez-Serrano M, Páez-García A, Sanchez-Bermejo E, Mohan TC, del Puerto YL, et al. Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev Cell. 2012:22(6):1275–1285. 10.1016/j.devcel.2012.04.008 [DOI] [PubMed] [Google Scholar]
- Li QF, Lu J, Yu JW, Zhang CQ, He JX, Liu QQ. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim Biophys Acta Gene Regul Mech. 2018:1861(6):561–571. 10.1016/j.bbagrm.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Li Y, Mullin M, Zhang Y, Drews F, Welch LR, Showalter AM. Identification of cis-regulatory sequences controlling pollen-specific expression of hydroxyproline-rich glycoprotein genes in Arabidopsis thaliana. Plants. 2020:9(12):1751. 10.3390/plants9121751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DG. Selection of combined versus separate sexes in seed plants. Am Nat. 1982:120(5):571–585. 10.1086/284014 [DOI] [Google Scholar]
- Luu DT, Qin X, Morse D, Cappadocia M. S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature. 2000:407(6804):649–651. 10.1038/35036623 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Matsushima M, Nabemoto M, Osaka M, Sakazono S, Masuko-Suzuki H, Takahashi H, Nakazono M, Iwano M, Takayama S, et al. Transcriptional characteristics and differences in Arabidopsis stigmatic papilla cells pre- and post-pollination. Plant Cell Physiology. 2015:56(4):663–673. 10.1093/pcp/pcu209 [DOI] [PubMed] [Google Scholar]
- Meng D, Gu Z, Li W, Wang A, Yuan H, Yang Q, Li T. Apple MdABCF assists in the transportation of S-RNase into pollen tubes. Plant J. 2014:78(6):990–1002. 10.1111/tpj.12524 [DOI] [PubMed] [Google Scholar]
- Ming M, Long HJ, Ye ZC, Pan CT, Chen JL, Tian R, Sun CR, Xue YS, Zhang YX, Li JM, et al. Highly efficient CRISPR systems for loss-of-function and gain-of-function research in pear calli. Hortic Res. 2022:9:uhac148. 10.1093/hr/uhac148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladek C, Guger K, Hauser MT. Identification and characterization of the ARIADNE gene family in Arabidopsis. A group of putative E3 ligases. Plant Physiol. 2003:131(1):27–40. 10.1104/pp.012781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah JB. Cell–cell signaling in the self-incompatibility response. Curr Opin Plant Biol. 2000:3(5):368–373. 10.1016/S1369-5266(00)00098-4 [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001:70(1):503–533. 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Qiao H, Wang HY, Zhao L, Zhou JL, Huang J, Zhang YS, Xue YB. The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell. 2004:16(3):582–595. 10.1105/tpc.017673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Bennett AB. Cooperative disassembly of the cellulose–xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci. 1999:4(5):176–183. 10.1016/S1360-1385(99)01405-3 [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan S, Jamshed M, Deb S, Chatfield-Reed K, Kwon EJG, Chua G, Samuel MA. Deciphering the stigmatic transcriptional landscape of compatible and self-incompatible pollinations in Brassica napus reveals a rapid stigma senescence response following compatible pollination. Mol Plant. 2013:6(6):1988–1991. 10.1093/mp/sst066 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009:459(7250):1071–1078. 10.1038/nature08122 [DOI] [PubMed] [Google Scholar]
- Shi D, Tang C, Wang R, Gu C, Wu X, Hu S, Jiao J, Zhang SL. Transcriptome and phytohormone analysis reveals a comprehensive phytohormone and pathogen defence response in pear self-/cross-pollination. Plant Cell Rep. 2017:36(11):1785–1799. 10.1007/s00299-017-2194-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao TH. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004:429(6989):302–305. 10.1038/nature02523 [DOI] [PubMed] [Google Scholar]
- Singh DP, Jermakow AM, Swain SM. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell. 2002:14(12):3133–3147. 10.1105/tpc.003046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Lu J, Bai M, Chen Y, Wang W, Fan C, Liu J, Ning G, Wang C. Phytochrome-interacting factors interact with transcription factor CONSTANS to suppress flowering in rose. Plant Physiol. 2021:186(2):1186–1201. 10.1093/plphys/kiab109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler F, Schmalzl C, Englhart M, Bircheneder M, Sprunck S. Brassinosteroids promote Arabidopsis pollen germination and growth. Plant Reprod. 2014:27(3):153–167. 10.1007/s00497-014-0247-x [DOI] [PubMed] [Google Scholar]
- Wang YC, Mao ZL, Jiang HY, Zhang ZY, Wang N, Chen XS. Brassinolide inhibits flavonoid biosynthesis and red-flesh coloration via the MdBEH2.2–MdMYB60 complex in apple. J Exp Bot. 2021:72(18):6382–6399. 10.1093/jxb/erab284 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He JX, Chen M, Vafeados D, Yang YL, Fujioka S, Yoshida S, Asami T, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002:2(4):505–513. 10.1016/S1534-5807(02)00153-3 [DOI] [PubMed] [Google Scholar]
- Wang HH, Qiu Y, Yu Q, Zhang Q, Li XY, Wang JM, Li XF, Zhang Y, Yang Y. Close arrangement of CARK3 and PMEIL affects ABA-mediated pollen sterility in Arabidopsis thaliana. Plant Cell Environ. 2020:43(11):2699–2711. 10.1111/pce.13871 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell. 2013:27(6):681–688. 10.1016/j.devcel.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang X, Skirpan AL, Kao TH. S-RNase-mediated self-incompatibility. J Exp Bot. 2003:54(380):115–122. 10.1093/jxb/erg008 [DOI] [PubMed] [Google Scholar]
- Wang C, Wu J, Xu G, Gao Y, Chen G, Wu J, Wu H, Zhang S. S-RNase disrupts tip-localized reactive oxygen species and induces nuclear DNA degradation in incompatible pollen tubes of Pyrus pyrifolia. J Cell Sci. 2010:123(24):4301–4309. 10.1242/jcs.075077 [DOI] [PubMed] [Google Scholar]
- Wei L, Fan J, Li J, Song Y, Li Q, Zhang YE, Xue Y. SCFSLF-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida. Front Genet. 2014:5:228. 10.3389/fgene.2014.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011:474(7349):105–108. 10.1038/nature09966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Suen SY, Chen SC, Tzeng JH. Analysis of protein adsorption on regenerated cellulose-based immobilized copper ion affinity membranes. J Chromatogr A. 2003:996(1–2): 53–70. 10.1016/S0021-9673(03)00531-4 [DOI] [PubMed] [Google Scholar]
- Xia ZH, Wen BX, Shao J, Zhang TC, Hu MM, Lin L, Zheng YP, Shi ZX, Dong XL, Song JJ, et al. The transcription factor PbrbZIP52 positively affects pear pollen tube longevity by promoting callose synthesis. Plant Physiol. 2023:191(3):1734–1750. 10.1093/plphys/kiad002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong TC, Wei MC, Li FX, Shi M, Gan H, Tang Z, Dong HP, Liuyu T, Gao P, Zhong B, et al. The E3 ubiquitin ligase ARIH1 promotes antiviral immunity and autoimmunity by inducing mono-ISGylation and oligomerization of cGAS. Nat Commun. 2022:13(1):5973. 10.1038/s41467-022-33671-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Carpenter R, Dickinson HG, Coen ES. Origin of allelic diversity in antirrhinum S locus RNases. Plant Cell. 1996:8(5): 805–814. 10.2307/3870283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive 1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000:12(9):1591–1605. 10.1105/tpc.12.9.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MR, Li CX, Cai ZY, Hu YM, Nolan T, Yu FF, Yin YH, Xie Q, Tang GL, Wang XL. SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev Cell. 2017:41(1):47–58.e4. 10.1016/j.devcel.2017.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci USA. 2010:107(13):6100–6105. 10.1073/pnas.0912333107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yokoda T, Asami T, Chory J. A new class of transcription factors mediate brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005:120(2):249–259. 10.1016/j.cell.2004.11.044 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002:109(2):181–191. 10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
- Zhang T, Gao C, Yue Y, Liu Z, Ma C, Zhou G, Yang Y, Duan Z, Li B, Wen J, et al. Time-course transcriptome analysis of compatible and incompatible pollen-stigma interactions in Brassica napus L. Front Plant Sci. 2017:8:682. 10.3389/fpls.2017.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MR, Han YY, Feng YN, Li F, Wang W. Expansins are involved in cell growth mediated by abscisic acid and indole-3-acetic acid under drought stress in wheat. Plant Cell Rep. 2012:31(4):671–685. 10.1007/s00299-011-1185-9 [DOI] [PubMed] [Google Scholar]
- Zhu XL, Liang WQ, Cui X, Chen MJ, Yin CS, Luo ZJ, Zhu JY, Lucas WJ, Wang ZY, Zhang DB. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of carbon starved anther, a MYB domain protein. Plant J. 2015:82(4):570–581. 10.1111/tpj.12820 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials..