Abstract
Enhancing fruit sugar contents, especially for high-flavonoid apples with a sour taste, is one of the main goals of horticultural crop breeders. This study analyzed sugar accumulation and the underlying mechanisms in the F2 progenies of a hybridization between the high-sugar apple (Malus × domestica) variety “Gala” and high-flavonoid apple germplasm “CSR6R6”. We revealed that MdSWEET9b (sugars will eventually be exported transporter) helps mediate sugar accumulation in fruits. Functional characterization of MdSWEET9b in yeast mutants lacking sugar transport as well as in overexpressing and CRISPR/Cas9 knockdown apple calli revealed MdSWEET9b could transport sucrose specifically, ultimately promoting normal yeast growth and accumulation of total sugar contents. Moreover, MdWRKY9 bound to the MdSWEET9b promoter and regulated its activity, which responded to abscisic acid (ABA) signaling. Furthermore, MdWRKY9 interacted with MdbZIP23 (basic leucine zipper) and MdbZIP46, key ABA signal transducers, at the protein and DNA levels to enhance its regulatory effect on MdSWEET9b expression, thereby influencing sugar accumulation. Based on the contents of ABA in lines with differing sugar contents and the effects of ABA treatments on fruits and calli, we revealed ABA as one of the main factors responsible for the diversity in apple fruit sugar content. The results of this study have clarified how MdSWEET9b influences fruit sugar accumulation, while also further elucidating the regulatory effects of the ABA-signaling network on fruit sugar accumulation. This work provides a basis for future explorations of the crosstalk between hormone and sugar metabolism pathways.
MdbZIP23/46–MdWRKY9–MdSWEET9b promotes fruit sugar accumulation under ABA signaling conditions.
Introduction
The soluble solid contents and composition are the key determinants of fruit flavor quality. Sugars, as the main soluble solid component, are important nutrients and a key factor influencing fruits flavor quality, but they are also crucial signals that regulate the expression of related genes as well as fruit (plant) growth and development, stress responses, and other developmental processes (Rolland et al. 2006; Ruan et al. 2010; Tsai and Gazzarrini 2014; Ljung et al. 2015; Han et al. 2022). Therefore, clarifying the mechanism mediating the accumulation of sugar is critical for improving fruit quality.
Sugars are important photosynthates produced in leaves and then transported through the phloem system to the storage organs, including fruits (Veerle et al. 2013; Ren et al. 2020). Substantial studies have explored fruit sugar accumulation and the “source–sink” sugar transport pathways, which involve key sugar transporters, including sucrose transporters (SUTS, sucrose transporter; SUCS, sucrose carriers), monosaccharide transporters (STPS, sugar transporter protein; HT, hexose transporter; TMTS, tonoplast monosaccharide transporter; PMT, Polyol monosaccharide transporters; ERD, early response to drought resembles proteins; INT, inositol transporter; pGlcT, plastidic glucose translocator), polysaccharide transporters (SOT, sorbitol transporter; MAT, monoamine transporter; PLT, phospholipid transporter), and SWEET, as well as key enzymes, such as sucrose synthase (SS, sucrose synthase; SPS, sucrose phosphate synthase), invertase (SAI, soluble acid invertase; SNI, soluble neutral invertase) and hexokinase (HKS, hexokinase). These proteins regulate the intensity and speed of the sugar transport from the source to the sink and affect the ability of fruits to store sugars (Patrick 1997; Sturm and Tang 1999; Wormit et al. 2006; Braun and Slewinski 2009; Cho et al. 2009; Chen et al. 2010; Turgeon 2010; Wingenter et al. 2010; Patrick 2013; Chen 2014; Yuan et al. 2014). Notably, SWEET is a recently discovered sugar transporter family. Since they were identified in Arabidopsis (Arabidopsis thaliana) by Chen et al. (2010), SWEET family members have also been detected and characterized in other plant species, including rice (Oryza sativa), soybean (Glycine max), grape (Vitis vinifera), sorghum (Sorghum bicolor), and pear (Pyrus bretschneideri) (Yuan and Wang 2013; Chong et al. 2014; Wei et al. 2014; Patil et al. 2015; Mizuno et al. 2016; Li et al. 2017), and exert essential functions related to plant growth and development, stress responses, and the regulation of sugar metabolism (Chardon et al. 2013; Klemens et al. 2013; Guo et al. 2014; Durand et al. 2016; Ma et al. 2017a; Gao et al. 2018; Wang et al. 2019; An et al. 2019). For example, Ko et al. (2021) confirmed that in tomato (Solanum lycopersicum), SlSWEET15 was localized in vascular tissues and seed coats, which were the main sites of sucrose unloading in fruits. Subsequent knockout experiments confirmed that SlSWEET15-mediated sucrose efflux was likely required for sucrose unloading from the seed coats to the developing embryos. Shammai et al. (2018) also identified a gene encoding SWEET protein in tomato, namely, SlFgr (fructose to glucose ratio). Overexpression of SlFgr reduced glucose levels and promoted fructose accumulation in fruit, ultimately altering the fructose/glucose ratio in ripe tomato fruits. Moreover, SlSWEET7a and SlSWEET14, which were located on the plasma membrane, promoted sugar accumulation and affected fruit development (Zhang et al. 2021). In the fleshy cucumber (Cucumis sativus) fruit, CsSWEET7a was localized in the plasma membrane in the companion cells of the phloem and involved in carbohydrate distribution in sink organs, thus affecting fruit development (Li et al. 2021). In apple, Zhen et al. (2018) generated and applied molecular markers to explore apple SWEET genes related to the sugar accumulation in fruits. They identified MdSWEET9b and MdSWEET15a as candidate genes encoding proteins that regulate sugar accumulation in apples. However, few studies have investigated how SWEET family members regulate sugar accumulation in apple fruits. Thus, MdSWEET family members will need to be more thoroughly characterized to explore their roles in sugar accumulation in apple fruits.
The function of sugar transporters is often affected by external factors, especially exogenous hormones (Miranda Rossetto et al. 2003; Ma et al. 2017b; Farcuh et al. 2018; Wei et al. 2018; Yuan et al. 2018). Yuan et al. (2018) have revealed that overexpression of the auxin response factor gene SlARF10 (auxin response factor10) in tomato, could increase the fruit starch, fructose, and sucrose contents, while downregulation of its expression substantially decreases sugar accumulation in fruits. In another study, an exogenous GA3 treatment delayed sucrose accumulation in banana fruits for at least 2 d, which was mainly due to the SPS abnormal expression (Miranda Rossetto et al. 2003). Wei et al. (2018) reported that FaMYB44.2 negatively regulates the accumulation of soluble sugars and malic acid in strawberry fruits via inhibiting FaSPS expression. Additionally, Farcuh et al. (2018) found that in respiratory climacteric fruits, treatment with ethylene, an important signaling factor, reduces the key sucrose catabolism and induces the expression of sucrose biosynthesis-related genes. However, as a crucial factor involved in nonclimacteric fruit ripening, abscisic acid (ABA) was also very important for fruit sugar accumulation (Adriana et al. 2011; Jia et al. 2016). Recent research indicated that ABA has a major role in the process mediating the accumulation of sugars in climacteric fruits (Afoufa-Bastien et al. 2010; Ma et al. 2017b; Murcia et al. 2017). For example, Ma et al. (2017b) observed that ABA can induce the expression of MdAREB2, which encodes a transcription factor that was phosphorylated by the protein kinase MdCIPK22, leading to the accumulation of soluble sugars in apples, and an increase in the stress resistance and fruit quality of transgenic plants. However, the mechanism underlying ABA regulating on fruit sugar accumulation is still unknown. Our results demonstrated that ABA contents increased rapidly during apple fruits' late developmental stages. In particular, the ABA accumulation rate and concentration were substantially higher in the high-sugar lines than in the low-sugar lines. Additionally, our study further elucidated the regulatory relationship between ABA signals and sugar accumulation in apple fruits, providing researchers with useful information for future investigations of the crosstalk between hormone and sugar metabolism pathways.
Globally, apple is an important succulent fruit crop because of its health benefits. Red-fleshed apples are a rich source of anthocyanins, flavonols, and other substances. Therefore, breeders using red-fleshed apples accessions as the parents to develop new apple varieties with enhanced characteristics (e.g. high-flavonoid contents), with implications for the efficient development of the apple industry. However, Hu et al. (2016) revealed that the MdMYB1 transcription factor promoted the accumulation of anthocyanins in apple fruits, while also promoted the transport of malic acid into vacuoles, resulting in red-fleshed apple fruits that were more acidic than white-fleshed apple fruits. Therefore, improving the sugar contents in red-fleshed apple fruits is critical for increasing the fresh flavor quality of high-flavonoid apples, as well as for clarifying the mechanism underlying sugar transport, which will promote the breeding of varieties producing high-flavonoid apple fruits. In our laboratory, we have previously obtained the F2 hybrid progenies of a cross between the high-sugar apple variety “Gala” and the high-flavonoid apple germplasm “CSR6R6” (‘Red Fuji’ × Xinjiang red-fleshed apple). The 2-yr analysis of the sugar component contents revealed abundant differences in the progenies (Supplemental Fig. S1). We screened three stable lines with extremely high-sugar levels (B20/C7/A6, which are subsequently referred to as H1/H2/H3, respectively) and three lines with extremely low-sugar levels (B13/D14/D8, which are subsequently referred to as L1/L2/L3, respectively) and characterized MdSWEET functions in these lines. Furthermore, we explored the molecular mechanism controlling the sugar accumulation in red-fleshed apple hybrid progenies, aiming to provide a basis for future attempts at improving the transport of sugars in apple and for breeding novel high-flavonoid (red-fleshed) apple varieties.
Results
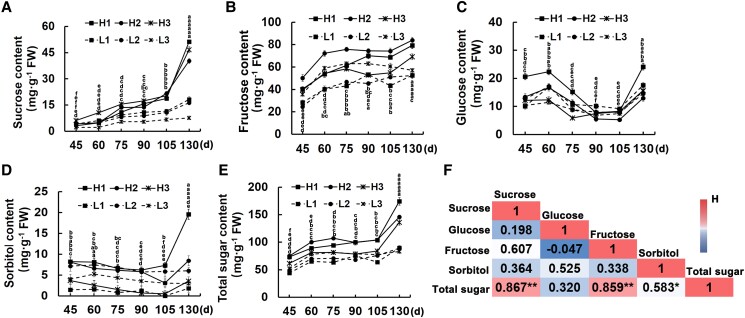
Content of sugar components in fruits of different apple lines
To explore the sugar components and total sugar accumulation during the fruit developmental stage of lines with different sugar contents, we sampled their fruits of their development stage in 2020 (Supplemental Fig. S2) and determined sugar components (sucrose, glucose, fructose, and sorbitol) using high-performance liquid chromatography (HPLC). The results revealed substantial differences in individual sugar components and the total sugar contents among the fruit developmental stages. In particular, sugar accumulates rapidly from the fifth stage (105 d after full bloom) to the fruit maturation stage (Fig. 1, A–E). Additionally, the total sugar contents were significantly higher in the high-sugar lines than low-sugar lines (Fig. 1E). We also analyzed the correlation between the contents of different sugar components and total sugar, and observed that the highest correlation was between sucrose and the total sugar contents (sucrose > fructose > sorbitol > glucose), with a correlation coefficient of as high as 0.867 (i.e. extremely significant positive correlation) (Fig. 1F). Moreover, we examined the correlation between the sugar components and the total sugar contents in hybrid progenies collected in 2018 and 2019. The 2-year data indicated that the correlation was the highest between sucrose and the total sugar contents, with correlation coefficients of 0.859 in 2018 and 0.813 in 2019, implying that sucrose accumulation has an important effect on the total sugar contents of red-fleshed apple hybrid progenies (Supplemental Table S1).
Figure 1.
Analysis of sugar components in fruit of different lines at developing stage. The contents of A) sucrose, B) fructose, C) glucose, D) sorbitol, and E) total sugar in different lines during development were determined and analyzed. FW, fresh weight. The numbers below the X-axis indicated the days after full bloom (d). Error bars represent the averages of three biological replicates ± SD. Different letters represent differences in the fruit development process. From top to bottom, they are H1, H2, H3, L1, L2, L3, respectively. Significance was defined at P < 0.05 (Student's t-test). F) Correlation analysis of sugar components and total sugar contents in fruit during development. Asterisks indicate statistical significance (**P < 0.01, *P < 0.05, Student's t-test).
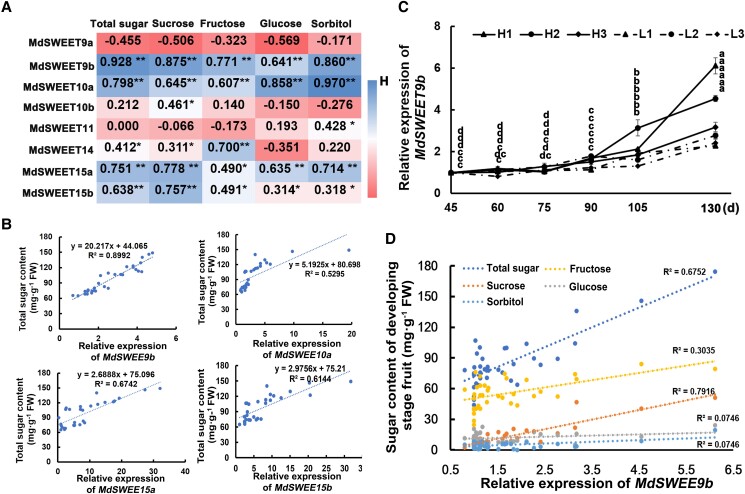
Expression analysis of MdSWEET III family members in different apple lines
It is well known that the SWEET III subfamily (AtSWEET9-15) of the model plant A. thaliana preferentially transports sucrose (Chen et al. 2010; Klemens et al. 2013). Therefore, we compared the amino acid sequences encoded by A. thaliana SWEET III genes in the NCBI apple database (https://blast.ncbi.nlm.nih.gov/Blast.cgi), which includes eight members of the MdSWEET III family (MdSWEET9a/9b/10a/10b/11/14/15a/15b) (Supplemental Fig. S3), and analyzed their expression in different lines using reverse transcription quantitative PCR (RT-qPCR). The results showed that the expression levels of MdSWEET9b/10a/15a/15b were significantly correlated with the total sugar contents. Among them, the expression of MdSWEET9b had the strongest correlation with total sugar content (the correlation coefficient was 0.928) (Fig. 2A; Supplemental Fig. S4). Subsequently, we explored the expression levels of MdSWEET9b/10a/15a/15b and their correlation with total sugar contents in 30 hybrid progeny lines with varying sugar contents. We found their expression levels differed among hybrid progenies (Supplemental Fig. S5). The correlation analysis also showed that MdSWEET9b expression had the highest correlation with the total sugar content (the correlation coefficient was 0.8992; Fig. 2B). Therefore, we further measured the expression level of MdSWEET9b in fruits of different lines during development, and found a similar sugar accumulation trend and a substantially correlation with total sugar accumulation (Fig. 2, C and D). These results suggest that MdSWEET9b may be the main factor regulating fruit sugar accumulation. Furthermore, its expression level was also linearly correlated with the contents of specific sugar components, especially sucrose, both in the hybrid progenies and during fruit development (Fig. 2D; Supplemental Fig. S6).
Figure 2.
Analysis of MdSWEET III family expression in different lines and correlation with sugar contents. A) Correlation analysis between MdSWEET III family gene expression level and the content of total sugar and sugar components in different lines. Asterisks indicate statistical significance (**P < 0.01, *P < 0.05, Student's t-test). B) Association analysis of MdSWEET9b/10a/15a/15b expression level and total sugar content in hybrid progenies. C, D) Expression level of MdSWEET9b(C) and its correlation with total sugar and sugar components content (D) in different sugar content lines during development. FW, fresh weight. Analyzed and drawn using Microsoft Excel. The larger the R2, the higher the correlation between the data. Error bars represent the averages of three biological replicates ± SD. Different letters represent differences in the fruit development process. Significance was defined at P < 0.05 (Student's t-test).
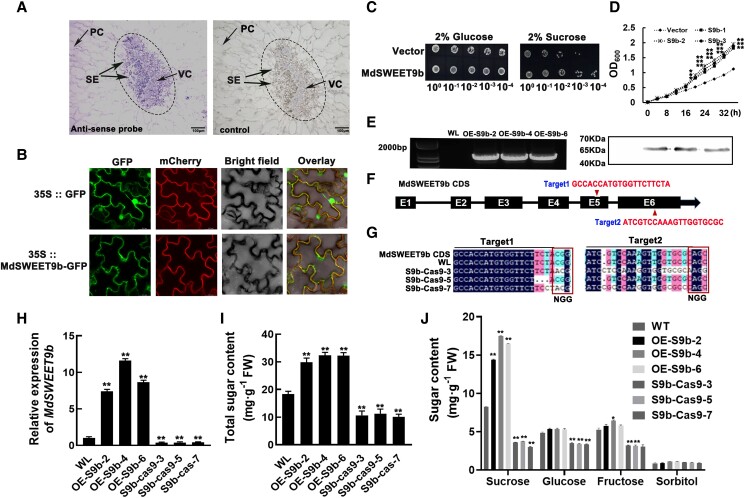
MdSWEET9b localization and its function analysis in sugar transport-deficient yeast and apple calli
We first examined the tissue-specific localization of MdSWEET9b transcripts in apple fruits by in situ hybridization with gene-specific probes. The MdSWEET9b-mRNA signals mainly concentrated in the vascular bundle sieve elements (SE) and its surrounding parenchyma cells (VC) (Fig. 3A; Supplemental Fig. S7A; where the site was stained blue-purple with dye), which was reported to be the main site of sucrose offloading in the fruit (Ko et al. 2021). In addition, there was also a weak signal in pulp cells (PC), and this localization is accurate because we did not detect its mRNA signal when the MdSWEET9b sense probe as a control (Fig. 3A; Supplemental Fig. S7B). Subsequently, we transiently expressed a construct encoding MdSWEET9b fused to the green fluorescent protein (GFP) in Nicotiana tabacum leaves, which revealed the fusion protein was localized in the cytoplasmic membrane (Fig. 3B). Together, the subcellular localization and in situ hybridization analyses suggest MdSWEET9b mainly functions on the cytoplasmic membrane of the vascular bundle sieve element and its surrounding parenchyma cells.
Figure 3.
Subcellular localization and functional analysis of MdSWEET9b in sugar transport deficient yeast mutants and apple calli. A) Cell-specific localization of MdSWEET9b transcripts analyzed by in situ hybridization. Cross-sections of apple fruits were hybridized with MdSWEET9b-specific antisense probes, and the sense probe was used as control. The vascular bundle and surrounding parenchyma cells were outlined by dotted lines. The location containing the target gene was stained blue-purple. SE, sieve elements; VC, vascular bundle parenchyma cells; PC, pulp cells. B) Image of MdSWEET9b-GFP subcellular localization in N. benthamiana leaves. An mCherry-labeled plasma membrane marker (AtPIP2A) was co-expressed to visualize the plasma membrane. C) Functional validation of MdSWEET9b in SUSY7/ura3 yeast mutants with sucrose transport deficiency. Empty vector was transformed as a growth control, the numbers under the panel indicate the dilution fold. D) Growth curves of SUSY7/ura3 yeast mutants containing MdSWEET9b-PYES or vector (as a negative control). The number below the X-axis indicates the measurement time (h). E)MdSWEET9b overexpression in “Orin” calli (OE-S9b-2/4/6) verified by PCR amplification and western blotting. F, G)MdSWEET9b knockdown target design (F) and transgenic sequencing results (G). Sequences were aligned using DNAMAN. Before NGG was the target sequence. The dark region was the target sequence, and the other colored region was the difference in sequence among the lines indicated. There was no difference between wild type (wild-type “Orin”) calli and target sequence, and multiple base mutations appeared in the knockdown calli (S9b-Cas9-3/5/7). H) Expression level of MdSWEET9b in overexpression and knockdown calli. I, J) Contents of total sugar (I) and individual sugar components (J) in transgenic calli. Error bars represent the averages of three biological replicates ± SD. Asterisks indicate statistical significance (**P < 0.01, *P < 0.05, Student's t-test).
To explore its function, we cloned the MdSWEET9b coding sequence (CDS) into the pYES-DEST2 vector and expressed it in hexose-transport-deficient yeast mutant EBY.VW4000 and sucrose-transport-deficient yeast mutant SUSY7/ura3. On the medium supplemented with 2% glucose as carbon source medium, yeast mutant SUSY7/ura3 cells transformed with the empty vector (pYES-DEST2) or MdSWEET9b-pYES grew normally (reflecting normal yeast activity). By contrast, on the medium supplemented with 2% sucrose, yeast mutant SUSY7/ura3 cells containing the empty vector grew slowly, whereas the strain containing MdSWEET9b-pYES restored the growth of SUSY7/ura3 yeast mutant (Fig. 3C). Additionally, the yeast growth curve indicated that the mutant strain containing MdSWEET9b-pYES grew rapidly on the sucrose medium (Fig. 3D). These results indicate that MdSWEET9b can transport sucrose into yeast mutants and restore normal yeast growth. However, the mutant EBY.VW4000 containing MdSWEET9b-pYES was unable to grow on medium containing hexose (fructose, glucose, and galactose), but could grow normally on medium containing 2% maltose (reflecting normal yeast activity) (Supplemental Fig. S8). This suggests that MdSWEET9b specifically transports sucrose to the cytosol of yeast cells, thereby restoring the growth of yeast mutants.
To further functionally characterize the contribution of MdSWEET9b to fruit sugar accumulation, we obtained three stably transformed MdSWEET9b-overexpression (OE-S9b-2/4/6) and MdSWEET9b-knockdown (S9b-Cas9-3/5/7) “Orin” calli (Supplemental Fig. S9A), which are useful for the rapid evaluation of the functions of metabolism-related genes in apple fruits (Zhu et al. 2021). In OE-S9b overexpression calli, target genes and protein bands were detected, which indicated that the MdSWEET9b overexpression was successful (Fig. 3E). In the knockdown calli, multiple deletions, insertions and substitutions occurred in the MdSWEET9b target sequences and its subsequent base sequences, indicating that MdSWEET9b knockdown was successful (Fig. 3, F and G). Furthermore, RT-qPCR analysis showed that the MdSWEET9b expression level increased significantly in the MdSWEET9b-overexpression calli, while it was at a very low level in the knockdown calli (Fig. 3H). Finally, analysis of the total sugar contents showed that compared to the wild-type “Orin” calli (WL), MdSWEET9b-overexpression calli had significantly increased total sugar contents, whereas the knockdown of MdSWEET9b had the opposite effect (Fig. 3I). In addition, analysis of each sugar component revealed that the sucrose was rapidly accumulated rapidly in the MdSWEET9b-overexpression calli, but decreased significantly in the MdSWEET9b-knockdown calli. Hence, MdSWEET9b can positively regulate sucrose transport into fruit cells, leading to an increase in the total sugar contents in fruits (Fig. 3J). These results were consistent with the findings of the yeast mutant analyses. Moreover, we found that the contents of glucose and fructose in MdSWEET9b-knockdown calli were significantly reduced, but not changed significantly in MdSWEET9b-overexpression calli, indicating that MdSWEET9b may indirectly regulate glucose and fructose accumulation, but its regulation of sucrose was direct.
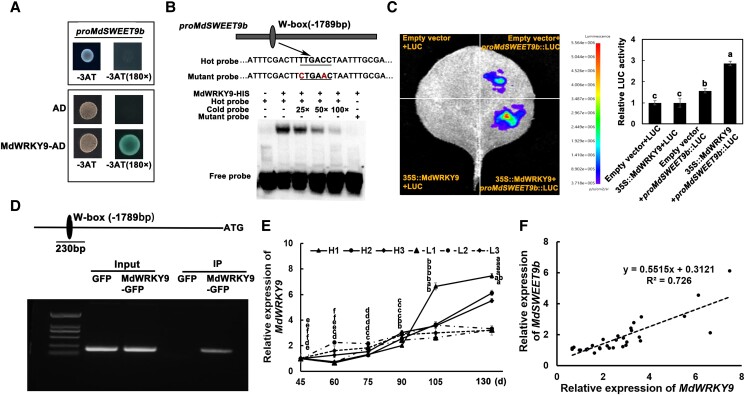
MdSWEET9b expression is regulated by MdWRKY9 in vitro and in vivo
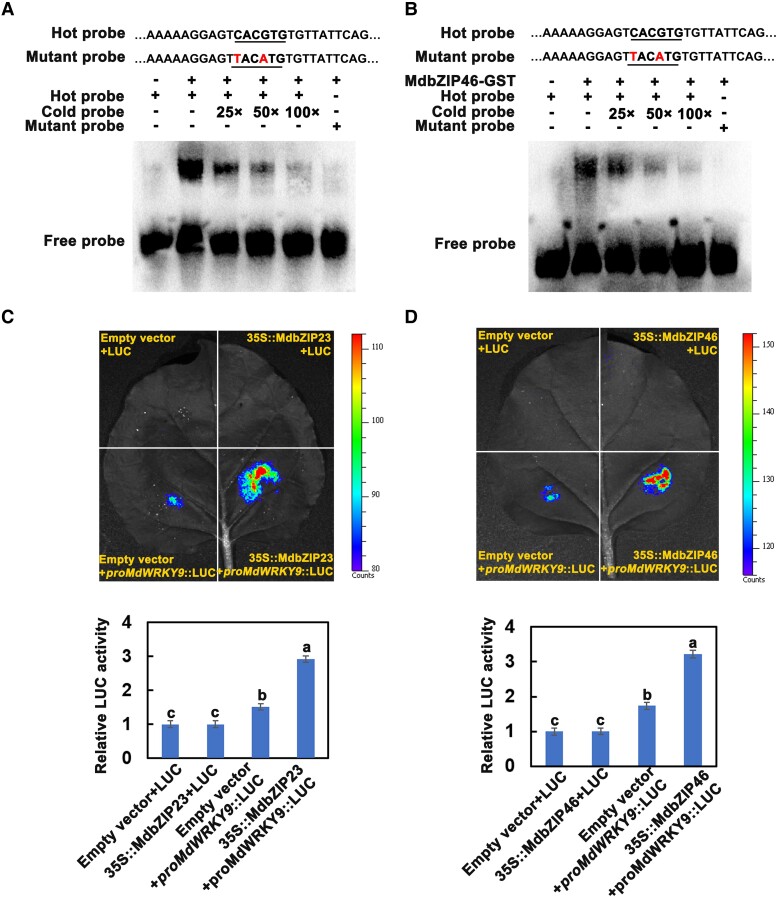
To further investigate whether MdSWEET9b expression is regulated by other signals, we used its promoter as the bait for yeast one-hybrid (Y1H) assay. Several proteins, including F-box protein, ATP synthase, NAD(P)H dehydrogenase, and MdWRKY9 were identified as putative trans-acting factors for the MdSWEET9b promoter (Supplemental Table S3). Based on the previous reports and gene annotations, we selected the MdWRKY9 transcription factor genes for further analysis. We inserted MdWRKY9 CDS into the pGADT7 vector and the 2000-bp MdSWEET9b promoter was incorporated into the pHIS2 vector. These recombinant plasmids were inserted into Y187 cells to specifically validate the Y1H assay. Yeast cells co-transformed with pGADT7 and proMdSWEET9b-PHIS2 could not grow on the −T−H−L medium containing 180 mM 3-AT. By contrast, yeast cells co-transformed with MdWRKY9-PGAD and proMdSWEET9b-PHIS2 grew normally on the same medium, with blue colonies after the addition of X-α-gal. The results reflected an interaction between MdWRKY9 and the MdSWEET9b promoter (Fig. 4A). We also analyzed the MdSWEET9b promoter and detected a W-box site at −1,789 bp. Subsequent electrophoretic mobility shift assays (EMSA) confirmed that MdWRKY9 could bind to the W-box motif in the MdSWEET9b promoter. Adding competitive cold substantially inhibited the radioactive signals reflecting the binding of MdWRKY9 to the MdSWEET9b promoter, and the binding was also blocked by the probe containing two mutated nucleotides (Fig. 4B).
Figure 4.
MdWRKY9 protein directly binds to the promoter region of MdSWEET9b.A) Y1H assays. The empty pGADT7 vector (AD) served as a negative control. The growth of the strain indicate the interaction between MdWRKY9 and the MdSWEET9b promoter. B) EMSA experiments showing the binding of MdWRKY9 to the MdSWEET9b promoter. Hot probes represented biotin-conjugated promoter fragments that contained specific W-box motifs of MdSWEET9b. The cold probe was an unlabeled competitive probe. The mutant probe was a marker fragment containing the mutant candidate W-box motifs of MdSWEET9b. Cold probes were added in increasing amounts (25×, 50×, and 100× fold probe concentration). The “+” and “−” indicate the presence and absence of the indicated probe or protein, respectively. C) LUC experiment showing the binding of MdWRKY9 to the MdSWEET9b promoter in vivo. D) Binding of MdWRKY9 to the MdSWEET9b promoter in vivo in ChIP-PCR assay. Those DNA fragments enriched in every ChIP served as the biological replicate in PCR. E, F) The expression level of MdWRKY9(E) and its association analysis with MdSWEET9b expression level (F) in different sugar content lines during development. Error bars represent the averages of three biological replicates ± SD. Different letters represent differences in the fruit development process. Significance was defined at P < 0.05 (Student's t-test). The statistical analysis described here was applicable to all data that requires statistical analysis.
To further confirm the relationship between MdWRKY9 and MdSWEET9b, we performed in vivo luciferase reporter (LUC) and ChIP-PCR assays. The luminescence intensity of Nicotiana benthamiana epidermal cells simultaneously expressing 35S::MdWRKY9 and proMdSWEET9b::LUC was stronger than that of cells expressing proMdSWEET9b::LUC alone or the negative control (empty vector), indicative of the interaction between MdWRKY9 and the MdSWEET9b promoter (Fig. 4C). The ChIP-PCR assays results also showed the interaction between them, the MdSWEET9b promoter fragments containing the W-box site were substantially enriched in the MdWRKY9-GFP “Orin” calli (relative to the control level; Fig. 4D). Subsequently, we conducted RT-qPCR analysis of MdWRKY9 expression during fruit development and found that the expression level was consistent with the trend of MdSWEET9b and substantially correlated with MdSWEET9b expression, further suggesting that MdWRKY9 could regulate the transcriptional activity of MdSWEET9b (Fig. 4, E and F).
MdWRKY9 positively regulates MdSWEET9b expression and promotes sugar accumulation in fruits
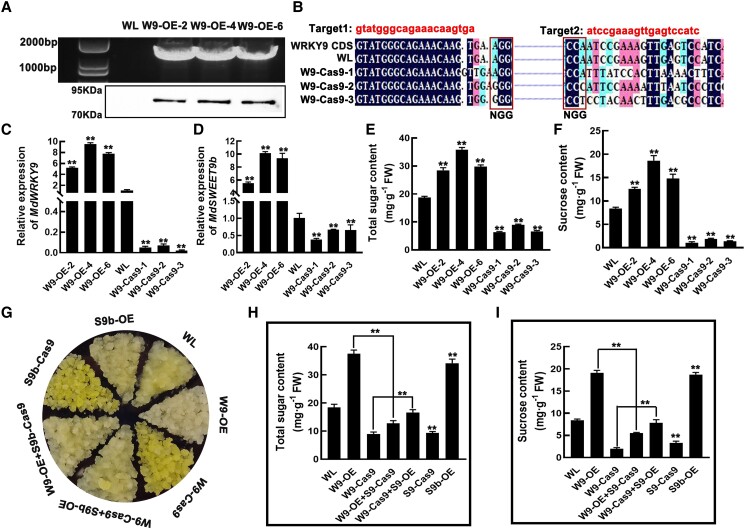
To explore the regulatory effect of MdWRKY9 on MdSWEET9b expression and sugar accumulation, we detected sugar contents and MdSWEET9b expression levels in MdWRKY9-overexpression (W9-OE-2/4/6, the target gene and protein bands were detected in these overexpressed calli.) and MdWRKY9-knockdown (W9-Cas9-1/2/3, their target sequence and its subsequent base sequence show multiple positions of deletion, insertion, and replacement; Fig. 5, A and B; Supplemental Fig. S9B). In the MdWRKY9-overexpression calli, MdSWEET9b expression level was significantly upregulated, and the total sugar contents increased significantly. In contrast, the opposite trend was observed in the MdSWEET9b-knockdown calli, in which the MdWRKY9 and MdSWEET9b expression levels decreased and sugar accumulation was significantly inhibited (Fig. 5, C–E). These results indicated that MdWRKY9 could positively regulate MdSWEET9b expression and promote sugar accumulation in fruits. Analyses of sugar components in transgenic calli indicated that the overexpression and knockdown of MdWRKY9, respectively, resulted in a significant increase and decrease in the content of each examined sugar component, especially sucrose. More specifically, compared with WL calli, sucrose content was approximately two to three times higher in the MdWRKY9-overexpression calli, but about four to eight times lower in the MdWRKY9-knockdown calli (Fig. 5F; Supplemental Fig. S10, A–C).
Figure 5.
MdWRKY9 positively regulates MdSWEET9b expression and promotes sugar accumulation in apple calli. A)MdWRKY9 overexpression (W9-OE-2/4/6) in “Orin” calli verified by PCR amplification and western blotting. B)MdWRKY9 CRISPR/Cas9 knockdown sites and first-generation sequencing of MdWRKY9-Cas9 knockdown (W9-Cas9-1/2/3) calli. Sequences were aligned using DNAMAN. Before NGG was the target sequence. The dark region was the target sequence, and the other colored region was the difference in sequence among the lines indicated. There was no difference between wild-type (wild-type “Orin”) calli and target sequence, and multiple base mutations appeared in the knockdown calli. C, D) Expression analysis of MdWRKY9(C) and MdSWEET9b(D) in MdWRKY9 transgenic calli. E, F) Total sugar (E) and sucrose contents (F) in MdWRKY9 transgenic calli. G) Calli phenotypes of wild type, MdWRKY9 and MdSWEET9b single and co-transferable calli (overexpression and knockdown). H, I) Total sugar (H) and sucrose (I) in MdWRKY9 and MdSWEET9b single and co-transferable calli. FW, fresh weight. Error bars represent the ±SD of three independent biological replicates. Asterisks indicate statistical significance (**P < 0.01, *P < 0.05, Student's t-test). The statistical analysis described here was applicable to all data that requires statistical analysis.
To further elucidate the regulatory effects of MdWRKY9–MdSWEET9b on sugar compositions and contents, we incorporated 35S::MdSWEET9b into MdWRKY9-knockdown calli and knockdown MdSWEET9b in MdWRKY9-overexpressing calli to obtain the MdWRKY9-Cas9 + MdSWEET9b-OE and MdWRKY9-OE + MdSWEET9b-Cas9 calli (Fig. 5G; Supplemental Fig. S11, A and B). Quantification of sugar contents showed that MdSWEET9b overexpression partially alleviated the effect of knocking down MdWRKY9 on the sugar accumulation in calli. Its addition increased the total sugar contents of MdWRKY9-Cas9 calli. However, MdSWEET9b knockdown in the MdWRKY9-overexpression calli significantly decreased in total sugar contents (Fig. 5H), indicating that regulating MdWRKY9 expression significantly affected sugar contents by modulating MdSWEET9b expression. Among them, the sugar component contents of the co-transposition calli revealed the largest changes were in the sucrose contents, while the content of other sugar components did not change, implying that the effects of MdWRKY9–MdSWEET9b on sugar accumulation were primarily associated with the sucrose contents (Fig. 5I; Supplemental Fig. S12, A–C).
ABA signaling positively regulates sugar accumulation in the hybrid progenies
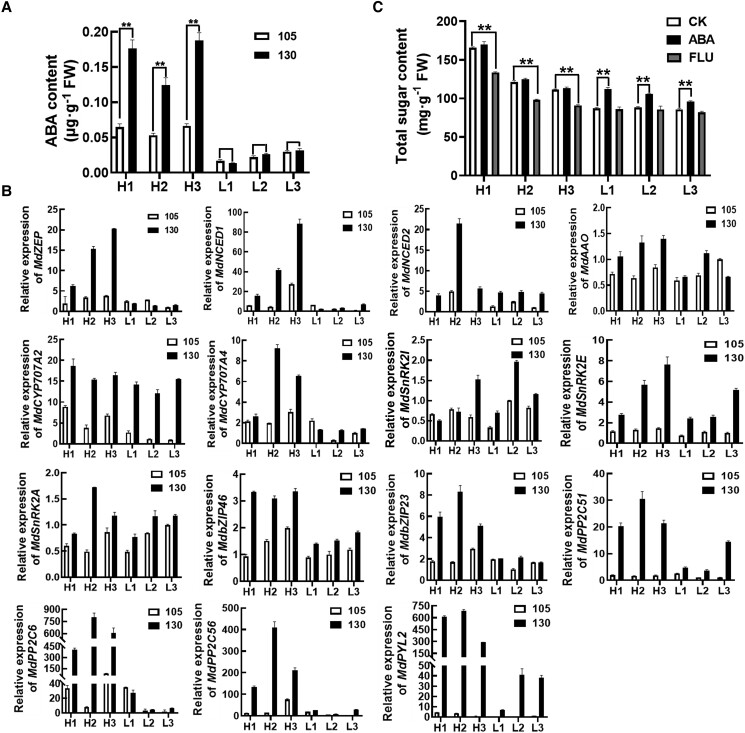
MdWRKY9 positively regulates MdSWEET9b expression, thereby promoting sugar accumulation in fruits. To better understand the differences mechanism in sugar accumulation among different apple lines, we first examined whether the MdWRKY9 CDS and promoter are different among different apple lines. Therefore, we cloned the MdWRKY9 CDS and promoter in various lines, but there were no differences in the examined sequences (Supplemental Fig. S13, A and B). Next, we considered whether MdWRKY9 and MdSWEET9b expression varies among different apple lines due to variations in specific signals. Since the plant hormone ABA plays an important role in the accumulation of sugar in fruits, we quantitatively analyzed the hormone contents in the hybrid progenies and observed that the ABA contents increased rapidly in the late fruit developmental stage (105 and 130 d after full bloom, the period sugar accumulation rapid), and showed substantially differences among different apple lines. Interestingly, the ABA contents increased more in the high-sugar lines than in the low-sugar lines (Fig. 6A), indicating that ABA at a high concentration may be responsible for high sugar accumulation in the high-sugar lines.
Figure 6.
Detection of ABA signal in fruit of hybrid progenies and its effect on fruits sugar contents. A) ABA contents in fruits of different lines during late fruit development. B) Expression levels of genes related to ABA synthesis (MdZEP/MdNCED1/MdNCED2/MdAAO), metabolism (MdCYP707A2/4), and signal transduction (MdSnRK2I/SnRK2E/SnRK2A/MdbZIP46/MdbZIP23/PYL2/PP2C51/PP-2C6/PP2C56) in fruits of different lines during late fruit development. 105 and 130 represent the days after full bloom, respectively. C) Effects of ABA and FLU on sugar contents in fruit of different lines. FW, fresh weight. Error bars represent the ±SD of three independent biological replicates. Asterisks indicate statistical significance by SPSS statistical 22 software (**P < 0.01, *P < 0.05, Student's t-test). The statistical analysis described here was applicable to all data that requires statistical analysis.
A recent study showed that ABA can self-catalyze its biosynthesis in the late fruit ripening stage (Li et al. 2022). In order to further explore the difference of ABA signal in different lines with different sugar content, we analyzed the expression of genes related to ABA synthesis (MdZEP/MdNCED1/MdNCED2/MdAAO), metabolism (MdCYP707A2/4), and signal transduction (MdSnRK2I/SnRK2E/SnRK2A/MdbZIP46(AREB1)/MdbZIP23 (ABF3)/PYL2/PP2C51/PP2C6/PP2C56) during the late fruit developmental stage. The expression levels of genes associated with ABA synthesis and signal transduction were higher in the high-sugar lines than in the low-sugar lines, which may help to explain the high ABA contents (signal) in the high-sugar lines (Fig. 6B). Although the expression of the ABA metabolism-related genes MdCYP707A2/4 was inconsistent with the changes in fruit sugar accumulation and ABA contents, the expression levels of the ABA synthesis-related genes and the ABA contents were clearly higher in the high-sugar lines than in the low-sugar lines, which is directly related to the stronger ABA signal in the high-sugar lines than in the low-sugar lines.
To verify that the ABA signal may be responsible for the differences in the fruit sugar contents among the examined lines, we treated the low-sugar lines with ABA during the late fruit developmental stage and then analyzed the fruit sugar contents. The results showed that the exogenous ABA treatment significantly promoted fruit sugar accumulation in the low-sugar lines, whereas the same treatment did not significantly increase sugar accumulation in fruits of the high-sugar lines (Fig. 6C). These results suggested that ABA can promote sugar accumulation in fruits, and indicated that ABA contents are high enough in the high-sugar lines to promote sugar accumulation whereas the ABA contents in low-sugar lines are insufficient for inducing the accumulation of excessive sugar in fruits. Moreover, FLU (Fluridone, ABA inhibitor) treatment significantly inhibited sugar accumulation in the high-sugar lines, but not in the low-sugar lines, indirectly reflecting the low ABA levels in the low-sugar lines (Fig. 6C). Furthermore, this low ABA contents in the low-sugar lines were not due to immature fruit ripening, as indicated by the detection of fully mature fruits on the basis of starch staining results (Supplemental Fig. S14). We showed that the ABA contents at mature fruits were indeed low in the low-sugar lines. Hence, differences in the fruit ABA contents were the important causes of the observed diversity in the fruit sugar contents.
MdWRKY9–MdSWEET9b positively regulates sugar accumulation in the hybrid progenies in response to ABA signals
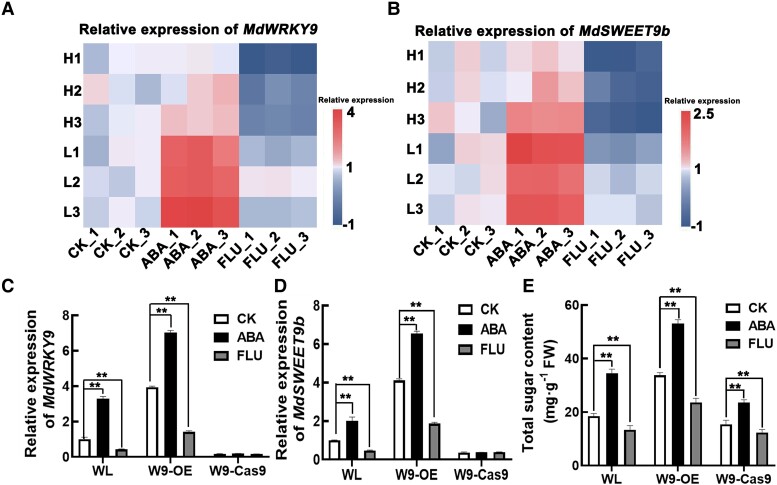
We determined the expression levels of MdWRKY9 and MdSWEET9b in fruits treated with ABA and FLU, and found that their expression levels were highly induced by ABA in the low-sugar lines and substantially inhibited by FLU in the high-sugar lines, consistent with changes in sugar accumulation in fruits under the same treatment (Fig. 7, A and B). These results suggest that ABA may affect fruit sugar accumulation through regulating the MdWRKY9–MdSWEEET9b pathway.
Figure 7.
ABA signal enhanced the regulation of MdWRKY9 on MdSWEET9b and increased the sugar content in fruit. A, B) Expression level analysis of MdWRKY9 and MdSWEET9b in different lines under different treatments (CK, water solution, ABA, abscisic acid solution, FLU, fluridone solution). The redder the color, the higher the gene expression level, the bluer the lower the gene expression level. C–E) Effects of ABA on MdWRKY9(C), MdSWEET9b(D) gene expression and sugar contents (E) in transgenic calli (overexpression calli: W9-OE and knockdown calli: W9-Cas9). WL (wild-type “Orin”) as a control. After 14 d of treatment, the related indexes were determined. Error bars represent the ±SD of three independent biological replicates. Asterisks indicate statistical significance (**P < 0.01, *P < 0.05, Student's t-test).
To more precisely investigate the effects of ABA on the regulation of MdSWEET9b expression by MdWRKY9 and on sugar accumulation in fruits, we treated wild type and transgenic calli with ABA and FLU. The ABA treatment increased the MdWRKY9 and MdSWEET9b expression levels as well as total sugar contents in wild-type calli, while FLU treatment led to the opposite effects. In addition, ABA treatment did not significantly alter the expression levels of MdWRKY9 and MdSWEET9b in MdWRKY9-knockdown calli compared with wild-type calli (Fig. 7, C and D). These results suggest that, on the one hand, MdWRKY9 can regulates sugar accumulation via ABA signaling, and on the other hand, MdSWEET9b should plays a role in the downstream of MdWRKY9. By comparing the sugar content in calli, we found although ABA treatment increased sugar accumulation in MdWRKY9-knockdown calli, it did not completely compensate the effect of MdWRKY9 knockdown on the inhibition of sugar accumulation (Fig. 7E), suggesting that MdWRKY9 was important in the process of ABA signaling-mediated sugar accumulation in fruits. Additionally, we found that FLU significantly inhibited sugar accumulation in calli, but this effect was partially compensated by MdWRKY9 overexpression. That is, following the FLU treatment, the sugar contents were significantly higher in MdWRKY9-overexpression than in wild-type calli (Fig. 7E). Accordingly, ABA signaling regulated MdWRKY9 expression thereby promoting MdSWEET9b expression and ultimately positively regulating sugar accumulation.
MdWRKY9 interacts with the ABA signal transduction factors MdbZIP23/46 to further regulate MdSWEET9b expression
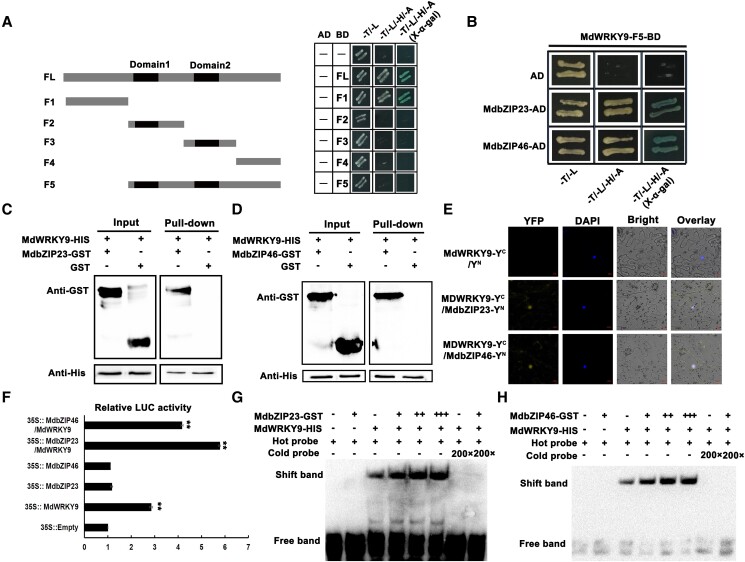
Our RT-qPCR data indicated that the expression levels of multiple genes in the ABA signal transduction pathway increased rapidly during the sugar accumulation period in fruits (Fig. 6B). We further assessed the potential interactions between the proteins encoded by these genes and MdWRKY9 were assessed in a yeast two-hybrid (Y2H) assay, which demonstrated that the ABA signal transduction factors MdbZIP23 and MdbZIP46 could interact with MdWRKY9 (Fig. 8, A and B), whereas interactions were not detected between the other proteins (MdSnRK2A/2E/2I and MdPP2C6/51/56) and MdWRKY9 (Supplemental Fig. S15). Subsequent pull-down assay also showed that MdbZIP23/46-GST was pulled down by MdWRKY9-HIS, but not by GST, reflecting an in vitro interaction between MdWRKY9 and MdbZIP23/46 (Fig. 8, C and D). Additionally, strong YFP (Yellow fluorescent protein) signals were detected in N. benthamiana epidermal cells co-expressing MdbZIP23/46-YFPN and MdWRKY9-YFPC (Fig. 8E), indicative of the in vivo interaction between MdWRKY9 and MdbZIP23/46. In addition, MdbZIP23 and MdbZIP46 also showed similar expression patterns to MdWRKY9/MdSWEET9b during fruit development, which were significantly expressed in the period of rapid sugar accumulation, indicating that they played an important role in the process of sugar accumulation (Supplemental Fig. S16). To determine whether these transcription factors affect the transcriptional regulation of MdSWEET9b by MdWRKY9, we conducted LUC reporter assays, which revealed that the MdSWEET9b promoter activity in response to the co-expression of MdbZIP23/46 and MdWRKY9 was significantly higher than that after the expression of MdWRKY9 alone, indicating that the MdbZIP23/46–MdWRKY9 interaction significantly promotes the ability of MdWRKY9 to activate the MdSWEET9b promoter (Fig. 8F). In the EMSA experiments, increases in the MdbZIP23/46 protein concentrations enhanced the strength of the binding of MdWRKY9 to the MdSWEET9b promoter (Fig. 8, G and H). Therefore, the ABA-signal transduction factors MdbZIP23/46 interacted with MdWRKY9 to enhance the regulatory effect of MdWRKY9 on the expression of the downstream gene MdSWEET9b.
Figure 8.
ABA signal transduction factors MdbZIP23 and MdbZIP46 interact with MdWRKY9 in vivo and vitro to further regulate the activity of MdSWEET9b. A, B) Y2H assays. (A) Screening for self-activating regions of MdWRKY9-BD vector that F1 segment was self-activated. Thus, we used MdWRKY9-F5-PGBKT7 as the bait for subsequent Y2H assays. (B) MdWRKY9 interacted with MdbZIP23 and MdbZIP46. The empty pGADT7 vector (AD) served as a negative control. Blue lines indicate interactions between MdWRKY9 and MdbZIP23/46. C, D) MdWRKY9 interacted with MdbZIP23 and MdbZIP46 in pull-down assays. The “+” and “−” indicate the presence and absence of the indicated protein, respectively. E) MdWRKY9 interacted with MdbZIP23 and MdbZIP46 in BiFC assays. F) LUC experiments showed that MdbZIP23 and MdbZIP46 interacted with MdWRKY9 to promote the regulation of MdWRKY9 on MdSWEET9b. Error bars represent the ±SD of three independent biological replicates. Asterisks indicate statistical significance (**P < 0.01, Student's t-test). G, H) EMSA experiments showed that MdbZIP23 and MdbZIP46 interacted with MdWRKY9 to promote the regulation of MdWRKY9 on MdSWEET9b. The “+” and “−” indicate the presence and absence of the indicated probe or protein, respectively.
The ABF (bZIP) family transcription factors directly influence plant growth and development downstream of ABA signaling. The bZIP family also often regulates downstream gene activity at the DNA level. Therefore, we analyzed the MdWRKY9 promoter and found that two bZIP-binding sites (G-box1 and G-box2). To verify the relationship between MdbZIP23/46 and MdWRKY9 at the protein-DNA level, we designed a specific hot probe for an EMSA experiment. The results indicated that MdbZIP23/46 could bind to G-box2, but not to G-box1, in the MdWRKY9 promoter (Supplemental Fig. S17). Subsequently, we designed mutant and cold probes for G-box2 and conducted another EMSA experiments, which confirmed that MdbZIP23/46 bind specifically to the G-box2 site in the MdWRKY9 promoter, as the concentration of cold probe increases, its binding strength weakens, and the binding of MdWRKY9 was blocked by the mutant probe (Fig. 9, A and B). Furthermore, the luminescence intensity was stronger in N. benthamiana epidermal cells co-expressing 35S::MdbZIP23/46 and proMdWRKY9::LUC than that of the cells expressing proMdWRKY9::LUC alone or the negative control (no load) cells, reflecting the interaction between MdbZIP23/46 and the MdWRKY9 promoter and confirming that 35S::MdbZIP23/46 enhanced proMdWRKY9::LUC activity (Fig. 9, C and D).
Figure 9.
ABA signal transduction factor MdbZIP23 and MdbZIP46 protein directly binds to the promoter region of MdWRKY9.A, B) EMSA experiments showed that the binding of MdbZIP23 (A) and MdbZIP46 (B) to the MdWRKY9 promoter. Hot probes represented biotin-conjugated promoter fragments that contained specific G-box motifs of MdWRKY9. The “+” and “−” indicate the presence and absence of the indicated probe or protein, respectively. C, D) LUC experiment showed that the binding of MdbZIP23 (C) and MdbZIP46 (D) to the MdWRKY9 promoter in vivo. Error bars represent the averages of three biological replicates ± SD. Different letters represent differences in the fruit development process. Significance was defined at P < 0.05 (Student's t-test).
Overall, our results indicated that the key transduction factors MdbZIP23/46 in the ABA-signaling pathway interact with MdWRKY9 at both the protein and DNA levels, thereby promoting the regulation of MdSWEET9b expression by MdWRKY9.
Discussion
For fruit crops, the variety and content of soluble sugars (such as fructose, sucrose, glucose, sorbitol, etc.) in the fruit largely determine the quality and commercial value of fresh fruit (Li et al. 2012). In the present study, we investigated soluble sugar accumulation in the red-fleshed apple hybrid progenies and found that sucrose accumulation was the major factor influencing their sugar contents. Wang et al. (2022b) also recently described the contribution of sucrose to the total sugar content in apple fruits, and found that sucrose was also a critical factor associated with fruit sugar contents in the “Honeycrisp” × “Qinguan” F1 generation hybrid progenies. Moreover, studies have also shown that the accumulation of sucrose, an important source of sweetness and flavor of fleshy fruits (Ayre 2011; Braun et al. 2014), is closely related to fruit sugar content and fruit ripening in strawberry (Fait et al. 2008; Merchante et al. 2013; Vallarino et al. 2015; Jia et al. 2016). Thus, it is important to explore the internal mechanism of sucrose accumulation in fruits for fruit tree breeding. Of course, in addition to sucrose, fructose, glucose, and sorbitol are also important sugar components in fruits. There is still a long way to go to clarify the underlying mechanism of the accumulation of various soluble sugars in fruits and the crosstalk between them.
Plants have evolved sophisticated mechanisms to regulate sugar source–sink transport, loading, and unloading (Veerle et al. 2013). This study identified a SUTS, designated MdSWEET9b, that can specifically transport sucrose and ultimately promoted fruit sugar accumulation. As one of the sugar transporters, the SWEET family is highly conserved in bacteria, fungi, plants, and animals (Jia et al. 2017). In plants, AtSWEET1 (AT1G21460) was characterized as a glucose bidirectional uniporter/facilitator (Chen et al. 2010). VvSWEET10 can transport hexose in grapes, and its overexpression substantially increased glucose, fructose, and the total sugar contents in grape calli and tomato (Zhang et al. 2019). The SlSWEET15 transporter of tomato helps regulate the release of sucrose from phloem cells and the subsequent uptake by storage cells (Ko et al. 2021). Additionally, SlSWEET7a and SlSWEET14 overexpression increases fruit sugar contents and modulates fruit and plant growth and development (Zhang et al. 2021). In addition, many studies had shown that SWEET family members were mainly localized in the cytoplasmic membrane (Shammai et al. 2018; Zhang et al. 2021). We found that MdSWEET9b was localized to the cytoplasmic membrane, indicating its role in sugar transmembrane transport. Moreover, our in situ hybridization results showed that MdSWEET9b was mainly located in the vascular bundle sieve element and its surrounding parenchyma cells. Ko et al. (2021) also confirmed that SlSWEET15 was localized in vascular tissues and seed coats, which were the main sites of sucrose unloading in fruits. In conclusion, we believe that MdSWEET9b plays a crucial role in the sugar unloading process, accelerating the transmembrane transport of sugar and ultimately promoting the accumulation of sugar in fruit. However, the functions of SWEET proteins are multifaceted. In addition to sugar transport function, they also play essential roles in nectar secretion (Lin et al. 2014), seed development (Ko et al. 2021), resistance to pathogens (Lecourieux et al. 2014), and abiotic stresses (Conde et al. 2018a, 2018b). Additional functions of SWEET9b in hybrid progenies need further investigation.
WRKY transcription factors are one of the largest families of transcription factors in plants (Eulgem et al. 2000; Zhang and Wang 2005; Singh et al. 2017; Cui et al. 2019). They activate sugar transporters by binding to the promoters of sugar-responsive genes (Chen et al. 2019). In this study, we observed that MdSWEET9b's effect on sucrose content was mainly regulated by MdWRKY9, which could bind specifically to the W-box motif of its promoter and positively regulate its activity, thereby promoting the accumulation of sucrose and total sugar content in fruits. Consistently, Li et al. (2021) found that WRKY31 activates SWEET15 transcription, which is the key to the high sugar content of “Nanguo” pear bud sport fruit. A recent study also confirmed that MdWRKY75 interacts with SWEET1 to mediate sucrose accumulation in apple fruits and the occurrence of fruit bitter blight (Sun et al. 2022). In addition, we also found that MdWRKY9 itself could increase glucose and fructose contents, while MdWRKY9 knockdown decreases glucose and fructose contents (Supplemental Fig. S12), which made the sugar content of MdWRKY9-Cas9 + MdSWEET9b-OE material significantly higher than that of MdWRKY9-Cas9 material, but still lower than that of wild-type calli (Fig. 5H). Although the effect of MdWRKY9 on other sugar components was not as obvious as that on sucrose (Fig. 5, H and I; Supplemental Fig. S14), this result provides a basis for further exploring the possibility of its regulatory mechanism of sugar content in fruits.
As an important hormone in plants, ABA plays an important role in fruit sugar accumulation. Many early reports suggested that ABA could promote the movement of photocontractual compounds to sink organs (Dewdney and McWha 1979; Ackerson 1985). Kobashi et al. (1999) showed that exogenous application of ABA increased the accumulation of sugar in peach (Amygdalus persica) fruits. ABA also promoted sorbitol absorption rate in apple (Berüter et al. 1983). In addition, it can also induce the expression of sugar transport-related genes, promote the accumulation of soluble sugar, and improve fruit quality (Ma et al. 2017b). Similarly, our results showed that ABA was an important factor promoting sugar accumulation in the red-fleshed apple hybrid progenies. Moreover, we showed that ABA treatment of transgenic calli, increased MdWRKY9 expression, promoted MdSWEET9b activity, and finally induced sugar accumulation in fruits. Furthermore, we revealed that the ABF family members MdbZIP23 and MdbZIP46 can interact with MdWRKY9 at the protein and DNA levels. These interactions enhance the regulatory effect of MdWRKY9 on MdSWEET9b expression. The ABF family is well known as a key factor in the ABA-signaling pathway. When ABA is present, the receptor RCAR/PYR/PYLs can sense ABA and form ternary complexes with PP2Cs and SnRK2s to inhibit the PP2C enzyme activity, and dissociate the PP2Cs–SnRK2s complex at the same time. The dissociated SnRK2s are autophosphorylated to activate ABF transcription factors through phosphorylation (Choi et al. 2000; Fujita et al. 2013; Joo et al. 2019; Antoni et al. 2021). Adriana et al. (2011) found that tomato SlAREB1 was involved in hexose accumulation in fruits, and SLAREB1-overexpression lines had high hexose content and increased expression of genes encoding vacuolar invertase (EC 3.2.1.26) and sucrose synthase (EC 2.4.1.13). This study confirmed that ABA signals could promote fruit sugar accumulation by regulating the MdWRKY9–MdSWEET9b activity, further elucidating the regulatory relationship between ABA signaling and fruit sugar metabolism.
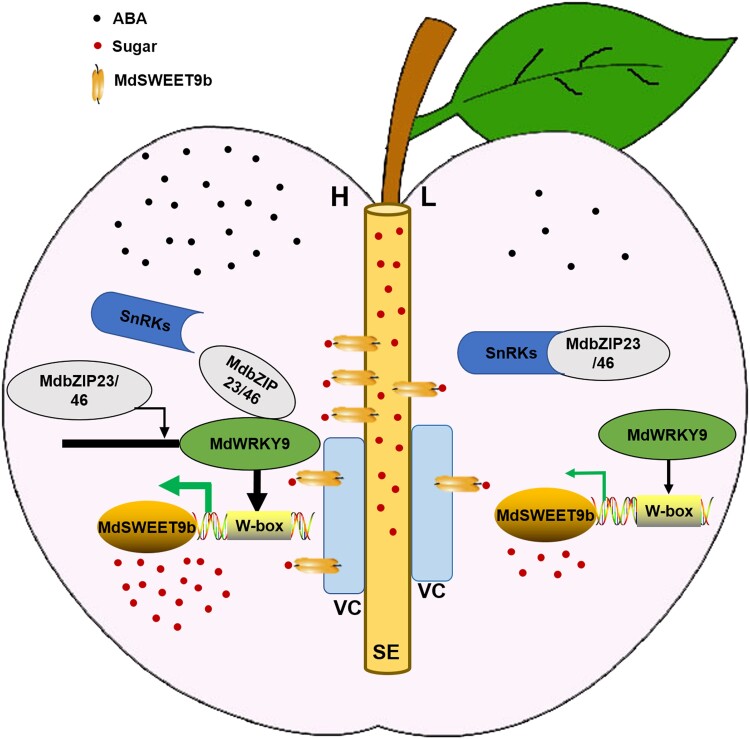
Taken together, our findings indicate that difference in sugar content among red-fleshed apple hybrid progenies was mainly due to the differences in their sucrose accumulation, and the differences in MdSWEET9b expression were the main contributors to the differences in the sucrose content and total sugar contents of fruits. MdSWEET9b was located in the vascular bundle SE and its surrounding parenchyma cells (VC) and functions to promote sugar unloading into fruits. Its activity was regulated by MdWRKY9, which can be induced by ABA signaling. ABA-signal transduction factors MdbZIP23 and MdbZIP46 could interact with MdWRKY9, thereby promoting the regulatory effect of MdWRKY9 on MdSWEET9b and ultimately enhancing the effects of this pathway on sugar accumulation in fruits. Furthermore, MdbZIP23 and MdbZIP46 could bind to the MdWRKY9 promoter to increase related gene expression and sugar accumulation in fruits (Fig. 10). In the red-fleshed apple hybrid progenies, the difference of ABA content resulted in the difference of sugar content in fruits. This study can provide guidance for future research on the mechanism of the relationship between hormone and sugar metabolism.
Figure 10.
Proposed model for MdWRKY9 to mediate its regulation of MdSWEET9b and promote apple fruits sugar accumulation under ABA-signaling conditions. MdSWEET9b was located in the SE and its VC to promote sugar unloading into the fruit. Its activity was regulated by MdWRKY9, which can be induced by ABA signaling. The key ABA signal transducers MdbZIP23 and MdbZIP46 were interacted with MdWRKY9 at the protein and DNA levels. The high concentration of ABA in high-sugar lines (H) promoted this pathway and made the fruit show high sugar content. MdbZIP23/46 represents that these two proteins interact with WRKY9, respectively.
Materials and methods
Plant materials and treatments
The apple (Malus × domestica) F2 hybrid progenies were grown at the Guanxian Fruit Tree Breeding Base, Liaocheng city, Shandong province (36°29′N, 115°27′E). All trees were managed according to the standard commercial practices recommended for the region. In 2018 and 2019, fruit samples were collected from all F2 hybrid progenies (mature stage) to measure the sugar component contents (145 lines in 2018 and 132 lines in 2019). Three stably inherited high-sugar lines (B20/C7/A6) and three low-sugar lines (B13/D14/D8) were screened. In 2020, we analyzed the fruit development stage of the different lines (from 45 d after full bloom until fruits ripened; six times in total). On each sampling date, we randomly collected five apple fruits from each line. The fruits were peeled and the pulp was frozen in liquid nitrogen and stored at −80 °C for the subsequent experiments.
For the hormone treatment of apple fruits, we injected fruits (into the heart cavity from the central mouth of the calyx end) with the following solutions using a medical syringe when the fruits were about to enter the ripening stage (105 d after full bloom): ABA (0.5 mmol·L−1), FLU (50 μmol·L−1), and water (control, CK). For each treatment, 10 fruits with a smooth surface and no pests were injected with 1.0 mL solution. After 20 d of treatment (fruit ripening), the fruits were peeled and the pulp was frozen in liquid nitrogen and stored at −80 °C for the subsequent experiments.
The “Orin” calli used for genetic transformation in this study were cultured on Murashige and Skoog (MS) medium containing 0.8 mg·L−1 6-benzylaminopurine (6-BA) and 1.5 mg·L−1 2,4-dichlorophenoxyacetic acid (2,4-D) in the dark at 24 °C. All materials were subcultured every 15 d. For the hormone treatment of calli, wile-type and transgenic calli were, respectively, grown on MS medium and the MS medium containing 50 μmol·L−1 ABA and 50 μmol·L−1 FLU. After 14 d, samples were collected and stored at −80 °C for the subsequent analyses of sugar compositions and gene expression levels. The experiment was repeated three times.
The N. benthamiana plants used in this study were grown in a plant growth chamber at 23 °C with a 16-h light/8-h dark cycle and 70 ± 5% relative humidity.
Total RNA extraction and RT-qPCR analysis
Total RNA was extracted from samples using the Total RNA Rapid Extraction Kit (Zomanbio, Beijing, China) and then reverse transcribed to generate first-strand cDNA using the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). We used SYBR Premix Ex Taq II (Takara, Dalian, China) for RT-qPCR analyses. Forward and reverse primers were designed for the MdActin internal reference gene. The RT-qPCR primers are listed in Supplemental Table S2. Gene expression levels were analyzed according to the 2−ΔΔCT method (Livak and Schmittgen 2001), with three replicates per sample. The experiment was repeated three times.
Subcellular localization of MdSWEET9b
MdSWEET9b was inserted into the pRI101-GFP vector (Wang et al. 2020). After verifying the accuracy of the cloned sequence, Agrobacterium tumefaciens strain LBA4404 cells were transformed with the recombinant plasmid using the freeze–thaw method. The MdSWEET9b-GFP cells were resuspended (OD600 = 0.6 to 0.8) in the infiltration buffer (10 mM MgCl2, 10 mM MES, and 150 μM acetosyringone) and injected into N. benthamiana leaves. The leaves were observed using a confocal laser microscope LSM880 (Zeiss, https://www.zeiss.com) at 2.5 d after the injection. The mCherry labeled plasma membrane (PM) marker (AtPIP2A) was co-expressed to visualize the plasma membrane. The excitation of GFP signal was performed using a 488-nm solid-state laser, fluorescence was detected at 498 to 540 nm, and the intensity and gain were 4.9% and 800, respectively. The mCherry signal excitation was performed using a 543 nm solid-state laser, and fluorescence was detected at 590 to 640 nm, and the intensity and gain were 9.1% and 800, respectively. Pinholes were adjusted to 1 Airy Unit for each wavelength. The primers are listed in Supplemental Table S2.
In situ hybridization
RNA in situ hybridization was performed using standard protocols (Kouchi and Hata 1993) with slight modifications. Briefly, specific SWEET9b Digoxigenin-labeled sense and antisense RNA probes were synthesized following the manufacturer's instructions (Roche Applied Science). The primers used are in Supplemental Table S2 (sense and antisense). The fruit samples were the fruit of hybrid progeny H2 line (105 d after full bloom). After the sample was fixed, dehydrated, waxed, and embedded, it was sliced with MICROM 315R microtome (Thermo Scientific) with a thickness of 10 μm. The probe concentration for hybridization was 800 ng μL−1. After the glycerol gelatin film was sealed, the images were collected and analyzed with Olympus BX63 fluorescence microscope (Japan). The experiment was completed in College of Horticulture, Northwest A&F University.
Heterologous expression of MdSWEET9b in yeast
We inserted the MdSWEET9b CDS into the pYES-DEST2 vector, which was then inserted into yeast strains SUSY7/ura3, which is deficient in sucrose transport, and EBY.VW4000, in which hexose transport is defective. Ten microliter of recombinant plasmid was added to 50 μL of yeast competent state, and 500 μL PEG/LIAC was added for mixing. After 30 min of water bath at 30 °C, 20 μL DMSO was added for mixing, followed by 42 °C water bath for 15 min and ice bath for 3 min. The supernatant was removed by centrifugation and 1 mL YPM liquid medium was added, and the strain was incubated by shaking at 28 °C for 90 min. Then, the strain was resuspended with 0.9% (w/v) NaCl, and coated on the corresponding selective medium with different sugar sources. The SUSY7/ura3 yeast strain containing MdSWEET9b was grown on uracil-deficient selective medium (SD/−Ura medium) containing glucose as the carbon source. The EBY.VW4000 yeast strain containing MdSWEET9b was grown on SD/−Ura medium containing maltose as the carbon source. After a 2.5-d growth period, individual colonies were verified by PCR, after which positive monoclonal strains were subdivided in the corresponding culture medium for growth and preservation. The obtained positive strains were cultured in the corresponding liquid medium. When the OD value reached 1.0, the cultures were diluted (10−1, 10−2, 10−3, and 10−4 times) and then used to inoculate the SD/−Ura medium supplemented with different sugar components. After a 3 to 5 d incubation, the growth of the strains was examined. To prepare growth curves, the positive and control strains were cultured in liquid medium and the OD values were recorded every 4 h. All yeast strains were grown in a constant-temperature incubator at 30 °C. The vector control and mutant yeast strains used in this study were provided by Li Mingjun (College of Horticulture, Northwest A&F University).
Construction of overexpression and CRISPR/Cas9 knockout vectors and the genetic transformation of calli
Similar to the subcellular localization experiment, the MdWRKY9 CDS was inserted into the pRI101-GFP vector. An online tool (http://crispr.hzau.edu.cn/CRISPR2/) was used for designing the MdSWEET9b and MdWRKY9 CRISPR/Cas9 knockout targets and the corresponding primers. We used a two-step method to obtain the final knockout vector. First, we used the primers in Supplemental Table S2 and the PCBC-DT1DT2 vector as a template to clone guide RNA (sgRNA). The obtained sgRNA was ligated to pHSE401 vector by T4 enzyme after Bsa1 single digestion (Xing et al. 2014; Ko et al. 2021). The target sequences were inserted into the pHSE401 vector. A. tumefaciens LBA4404 cells were transformed with the generated recombinant plasmids and then stored at −80 °C prior to the calli transformations, which were performed as described by Fang et al. (2019). The transformed calli were verified by PCR, western blot, and RT-qPCR analyses. The positive calli were cultured for the subsequent experiments.
Y1H screening of apple fruit cDNA libraries
Y1H screening was implemented as described by Tran et al. (2004). The AD-cDNA (AD represents PGADT7-Rec) library from Gala apple fruit at various stages of development was assembled by Oebiotech Biomedical Technology Co., LTD (Shanghai), using the Clontech Matchmaker onehybrid system. The reporter plasmid pABAi-ProMdSWEET9b was constructed using the 2000-bp fragment of the MdSWEET9b promoter, and the recombinant plasmid was then transformed into Y1H susceptible yeast. The AD-cDNA library was transfected into susceptible yeast containing pABAi-ProMdSWEET9b to identify the binding specific transcription factors. The primers used are listed in Supplemental Table S2.
Yeast one-hybrid and two-hybrid assays
The Y1H assays were performed as previously described (Wang et al. 2017). The MdWRKY9 CDS was inserted into the pGADT7 vector, whereas the 2000-bp MdSWEET9b promoter sequence was incorporated into the pHIS2 vector. The self-activation of the resulting pHIS2 plasmids was inhibited using 3-AT. Yeast strain Y187 cells (Clontech) carrying the recombinant pGADT7 and pHIS2 plasmids were used to inoculate SD medium lacking Trp, His, and Leu to examine potential interactions. The empty pGADT7 vector served as the control. Primer details are listed in Supplemental Table S2.
For the Y2H assay, the MdWRKY9 CDS was inserted into pGBKT7. The recombinant plasmid and the empty pGADT7 vector were used for the co-transformation of Y2H yeast strain cells. Because of the observed self-activation in the transformed cells, we divided the MdWRKY9 CDS into five fragments. The results showed that F1 segment is self-activated. Thus, we used MdWRKY9-F5-PGBKT7 as the bait to screen for key interacting proteins in the ABA-signaling pathway (MdSnRK2I/MdSnRK2A/MdSnRK2E/MdPP2C51/MdPP2C6/MdPP2C56/MdbZIP23/MdbZIP46). The sequences encoding the potential interacting partners were inserted into the pGADT7 vector. The Y2H yeast cells co-transformed with the generated recombinant plasmids and MdWRKY9-C-PGBKT7 were grown on selective medium lacking Trp, Leu, His, and Ade (−T/−L/−H/−A). To confirm the positive interactions, X-α-gal was used to measure the β-galactose activity (Zhang et al. 2020). Blue plaque indicates interaction between proteins. Primer information is listed in Supplemental Table S2.
Bimolecular fluorescence complementation assay
The BiFC assay was implemented as previously described (Jia et al. 2018). The CDS of MdWRKY9 was cloned into the pSPYCE vector, whereas the CDS of MdbZIP23 and MdbZIP46 were cloned into the pSPYNE vector. The transformation of N. benthamiana and the analysis of YFP fluorescence were performed as described by Wang et al. (2018). The cells with recombinant plasmids were cultured in a 28 °C constant temperature incubator to make their OD value reach 1.0. MdWRKY9-YC and MdbZIP23/46-YN bacterial solution was mixed in 1:1 equal volume, suspended with infection solution and injected into N. benthamiana leaves. The fluorescence was observed using a Zeiss LSM 880 confocal microscope with a 10× air objective or 20× air objective. For imaging DAPI- and YFP-signal together, excitation lines of a solid-state laser of 405 nm for DAPI and a laser of 514 nm for YFP were used alternately with the sequential scanning mode of the microscope. Fluorescence was detected using a 415 to 480 nm bandpass filter for DAPI and a 522 to 560 nm bandpass filter for YFP, the gains are less than 800. In this way, any crosstalk and bleed-through of fluorescence were eliminated. Pinholes were adjusted to 1 Airy Unit for each wavelength. The primers used for this assay are listed in Supplemental Table S2.
Protein extraction and western blotting
Protein extraction and western blotting analysis were conducted as previously described (Jia et al. 2018). Total protein was extracted from the samples using a Plant Protein extraction kit (KANGWSJI, Jiangsu, China). The anti-GFP antibody was obtained from Abmart Biomedicine Co., LTD (Shanghai, China).
Pull-down assay
MdWRKY9 was inserted into the pET-32a (+) vector for the expression of a His-tagged fusion protein. Both MdbZIP23 and MdbZIP46 were inserted into separate pGEX-4T-1 vectors for the production of GST-tagged fusion proteins. The recombinant plasmids were inserted into separate BL21 cells to obtain MdWRKY9-His, MdbZIP23-GST, and MdbZIP46-GST proteins as described by Wang et al. (2018). Proteins were purified using a commercial protein purification kit (Beyotime Biotechnology, Shanghai, China). The eluted protein samples were analyzed in a western blot using anti-GST and anti-His antibodies (Abmart, Shanghai, China). Primer details are listed in Supplemental Table S2.
Electrophoretic mobility shift assay
The MdWRKY9-His fusion protein was expressed and purified as described above. The MdSWEET9b, MdbZIP23, and MdbZIP46 promoter sequences were used to design specific hot, cold, and mutant probes. All probes for the promoter fragments were labeled and synthesized by Sangon Biotechnology Co., Ltd. (Supplemental Table S2). Double-stranded probes were synthesized using the DNA Oligos annealing buffer (Beyotime Biotechnology, Shanghai, China). The protein-probe reaction mixtures were incubated at 24 °C for 30 min in the dark and then separated on 6% (v/v) native polyacrylamide gels that had been pre-electrophoresed for 1 h in 0.5× TBE buffer. Then, the gels were electroblotted onto Hybond N+ (Amersham, Amenham, UK) nylon membranes in 0.5 × TBE for 3.5 h, and the labeled probes were detected following the manufacturer's instructions. Please refer to Fang et al. (2019) for specific details regarding the completion of the EMSA experiments.
LUC reporter assay
The MdWRKY9, MdbZIP23, and MdbZIP46 CDSs were inserted into separate pGreenII 62-SK vectors, whereas the MdSWEET9b and MdWRKY9 promoters were inserted into separate pGreenII 0800-LUC vectors (Wang et al. 2020). The recombinant plasmids were integrated into A. tumefaciens LBA4404 cells, with P19 used as a helper plasmid. The transformed A. tumefaciens cells were used for the transient transformation of N. benthamiana leaves. The LUC activity was measured using the NightOwl II LB983 in vivo imaging system (Berthold Technologies, Germany). The primers used are listed in Supplemental Table S2.
ChIP-PCR analysis
Calli stably transformed with the MdWRKY9-GFP construct were used for the ChIP-PCR analysis. Specifically, the Ez-chip 244 Chromatin Immunoprecipitation Kit (Upstate, Waltham, MA, USA) was used (Wang et al. 2022a). Calli containing empty GFP vector were used as the negative control. Refer to the instructions for specific operation. The samples were cross-linked in vacuum, frozen in liquid nitrogen, ground, and then SDS lysis buffer was added for ultrasonic crushing (crushing five times: beating for 5 s, stopping for 5 s). Immunoprecipitation was performed with the corresponding antibody. Primers were designed to target the W-box region (230 bp) in the MdSWEET9b promoter. Specific DNA fragments were amplified by PCR using the primers listed in Supplemental Table S2.
Analyses of sugar components and total sugar contents
The sugar components of fruits and calli were determined using an HPLC system (Agilent 393 Technologies 1260 Series). Extracts were prepared from 5.0 g evenly mixed samples and then analyzed as described by Li et al. (2020). Briefly, the sample was ground to a fine powder in liquid nitrogen, followed by the addition of 10 mL 80% (v/v) ethanol, and the samples were broken by ultrasound at 80 °C for 30 min. The supernatant was collected by centrifugation at 10,000 × g for 5 min. The procedure was repeated twice, mixing the supernatant and evaporating in boiling water. After drying in a 50 mL centrifuge tube, the samples were dissolved in 1 mL ultrapure water and filtered through a 0.45 μm membrane to determine the soluble sugar content in the filtrate. HPLC (Agilent 1260) was performed with the following components and parameters: 7.8 × 300 mm Carbomix CA-NP column (Sepax); ultrapure water was used as the mobile phase at a flow rate of 1 mL min−1. The column temperature was 80 °C; the temperature of the refractive index detector was 35 °C; the injection volume was 10 μL. The experiment was completed at the College of Horticulture, Northwest A&F University. Total sugar contents were determined using the Total Carbohydrate Assay kit (Solarbio Ltd, Beijing, China). The experiment was repeated three times.
Statistical analysis
All experiments were performed in triplicate. Error bars represent the standard deviation of three replicates. Values are presented as the mean ± standard deviation of three independent biological replicates. Linear regression analysis was performed using Microsoft Excel and significant differences (P < 0.05 and P < 0.01) were detected according to Tukey one-way ANOVA with SPSS Statistics 22 (IBM Corporation, Armonk, NY, USA).
Accession numbers
Sequence data from this article can be found in the National Center for Biotechnology Information databases under the following accession numbers: MdSWEET9a (XM_008372549), MdSWEET9b (XM_029090187), MdSWEET10a (XM_008358349), MdSWEET10b (XM_008394505), MdSWEET11 (XM_029093209), MdSWEET14 (XM_029104208), MdSWEET15a (XM_008391699), MdSWEET15b (XM_0083548300), MdAAO (XM_008363703), MdCYP707A2 (XM_008358695), MdCYP707A4 (XM_008374924), MdSRK2I (NM_001328806), MdSRK2E (NM_001328724); MdSRK2A (NM_001320014), MdbZIP46 (XM_029106874), MdbZIP23 (XM_008374912), MdPP2C51 (XM_029106210), MdPP2C6 (NC_041790), and MdPYL2 (XM_008353730).
Supplementary Material
Acknowledgments
We thank the laboratory of Li Mingjun of Northwest A&F University for providing plasmid and yeast strains. We thank the Experimental Center platform of College of Horticulture, Northwest A&F University for providing technical support.
Contributor Information
Shuhui Zhang, State Key Laboratory of Crop Biology, College of Horticulture Sciences and Engineering, Shandong Agricultural University, Tai’an 271018, Shandong, China.
Hui Wang, College of Horticulture, Northwest A&F University, Yangling 712100, Shanxi, China.
Tong Wang, State Key Laboratory of Crop Biology, College of Horticulture Sciences and Engineering, Shandong Agricultural University, Tai’an 271018, Shandong, China.
Jing Zhang, State Key Laboratory of Crop Biology, College of Horticulture Sciences and Engineering, Shandong Agricultural University, Tai’an 271018, Shandong, China.
Wenjun Liu, State Key Laboratory of Crop Biology, College of Horticulture Sciences and Engineering, Shandong Agricultural University, Tai’an 271018, Shandong, China.
Hongcheng Fang, State Forestry and Grassland Administration Key Laboratory of Silviculture in the Downstream Areas of the Yellow River, College of Forestry, Shandong Agricultural University, Tai’an 271018, Shandong, China.
Zongying Zhang, State Key Laboratory of Crop Biology, College of Horticulture Sciences and Engineering, Shandong Agricultural University, Tai’an 271018, Shandong, China.
Xuesen Chen, State Key Laboratory of Crop Biology, College of Horticulture Sciences and Engineering, Shandong Agricultural University, Tai’an 271018, Shandong, China.
Nan Wang, State Key Laboratory of Crop Biology, College of Horticulture Sciences and Engineering, Shandong Agricultural University, Tai’an 271018, Shandong, China.
Author contributions
X.C., N.W., and S.Z. conceived and designed the experiments. S.Z., H.W., and T.W. performed the experiments. S.Z., H.W., J.Z., W.L., H.F., and Z.Z. contributed reagents, materials, and data analysis. S.Z., H.W., F.P., and N.W. wrote the paper.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . The sugar components analysis in fruit ripening stage of F2 hybrid progenies in 2018 and 2019.
Supplemental Figure S2 . Phenotypes of fruit development of different lines.
Supplemental Figure S3 . Evolutionary tree analysis of SWEET III subfamily in Arabidopsis thaliana and apple.
Supplemental Figure S4 . RT-qPCR analysis of SWEET III subfamily expression in different sugar content lines.
Supplemental Figure S5 . Expression levels of MdSWEET9b/10a/15a/15b in 30 hybrid progenies.
Supplemental Figure S6 . Analysis of the relationship between MdSWEET9b expression level and the contents of sugar components (sucrose, fructose, glucose and sorbitol) in 30 hybrid progenies.
Supplemental Figure S7 . Cell-specific localization of MdSWEET9b transcripts analyzed by in situ hybridization.
Supplemental Figure S8 . Functional validation of MdSWEET9b in yeast mutants EBY.VW4000 lacking hexose transport.
Supplemental Figure S9 . Phenotypes of MdSWEET9b and MdWRKY9 transgenic calli.
Supplemental Figure S10 . Content analysis of sugar components in MdWRKY9 transgenic calli.
Supplemental Figure S11 . The expression levels of MdSWEET9b and MdWRKY9 in MdWRKY9 and MdSWEET9b single and co-transferable calli.
Supplemental Figure S12 . Content analysis of sugar components in MdWRKY9 and MdSWEET9b single and co-transferable calli.
Supplemental Figure S13 . The promoter and CDS sequences of MdWRKY9 in differential lines.
Supplemental Figure S14 . Images of fruit starch staining treated with ABA and ABA inhibitor FLU.
Supplemental Figure S15 . The interaction between ABA signal transduction-related proteins (MdSnRK2A/2E/2I, MdbZIP23/46, MdPP2C51/6/56) and MdWRKY9 were verified by Y2H assay.
Supplemental Figure S16 . Analysis of the expression level of MdbZIP23 and MdbZIP46 in different sugar content lines at development stage.
Supplemental Figure S17 . The EMSA experiment was used to detect the binding of MdbZIP23 and MdbZIP46 to G-box1 and G-box2 of MdWRKY9 promoter.
Supplemental Table S1 . Correlation analysis of total sugar and sugar components in F2 hybrid progenies in 2018 and 2019.
Supplemental Table S2 . Primers used in this study.
Supplemental Table S3 . List of genes identified from Y1H screen using MdSWEET9b promoter as bait.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32172533) and the Agricultural Variety Improvement Project of Shandong Province (Grant Nos. 2021LZGC024, 2022LZGC010).
Data availability
All relevant data can be found within the manuscript and its supporting materials.
References
- Ackerson RC. Invertase activity and abscisic acid in relation to carbohydrate status in developing soybean reproductive structures. Crop Sci. 1985:25(4):615–618. 10.2135/cropsci1985.0011183X002500040009x [DOI] [Google Scholar]
- Adriana B, María LC, Mercedes V, Salvador R, Aurelio GC, Jose AC. Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor. Physiol Plantarum. 2011:141(3):215–226. 10.1111/j.1399-3054.2010.01435.x [DOI] [PubMed] [Google Scholar]
- Afoufa-Bastien D, Medici A, Jeauffre J, Coutos-Thévenot P, Lemoine R, Atanassova R, Laloi M. The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 2010:10(1):245–271. 10.1186/1471-2229-10-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Zeng T, Ji C, De Graaf S, Zheng Z, Xiao TT, Pan Z. A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol. 2019:224(1):396–408. 10.1111/nph.15975 [DOI] [PubMed] [Google Scholar]
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Rodrigues A, Pizzio GA, Rodriguez PPL. Selective inhibition of clade a phosphatases type 2c by PYR/PYLl/RCAR abscisic acid receptors. Plant Physiol. 2012:158(2):970–980. 10.1104/pp.111.188623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayre BG. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol plant. 2011:4(3):377–394. 10.1093/mp/ssr014 [DOI] [PubMed] [Google Scholar]
- Berüter J. Effect of abscisic acid on sorbitol uptake in growing apple fruits. J Exp Bot. 1983:34(6):737–743. 10.1093/jxb/34.6.737 [DOI] [Google Scholar]
- Braun DM, Slewinski TL. Genetic control of carbon partitioning in grasses: roles of sucrose transporters and tie-dyed loci in phloem loading. Plant Physiol. 2009:149(1):71–81. 10.1104/pp.108.129049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Wang L, Ruan YL. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J Exp Bot. 2014:65(7):1713–1735. 10.1093/jxb/ert416 [DOI] [PubMed] [Google Scholar]
- Chardon F, Bedu M, Calenge F, Klemens PAW, Spinner L, Clement G, Léran S, Ferrand M, Lacombe B, Loudet O, et al. . Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr Biol. 2013:23(8):697–702. 10.1016/j.cub.2013.03.021 [DOI] [PubMed] [Google Scholar]
- Chen LQ. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014:201(4):1150–1155. 10.1111/nph.12445 [DOI] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. . Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010:468(7323):527–532. 10.1038/nature09606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QS, Xu XY, Xu D, Zhang HS, Zhang CK, Li G. WRKY18 and WRKY53 coordinate with HISTONE ACETYLTRANSFERASE1 to regulate rapid responses to sugar. Plant Physiol. 2019:180(4):2212–2226. 10.1104/pp.19.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JI, Ryoo N, Eom JS, Lee DW, Kim HB, Jeong SW, Lee YH, Kwon YK, Cho MH, Bhoo SH, et al. . Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 2009:149(2):745–759. 10.1104/pp.108.131227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HI, Hong JH, Ha JO, Kang JY, Kim SY. ABFs, a family of aba-responsive element binding factors. J Biol Chem. 2000:275(3):1723–1730. 10.1074/jbc.275.3.1723 [DOI] [PubMed] [Google Scholar]
- Chong JL, Piron MC, Meyer S, Merdinoglu D, Bertsch C, Mestre P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J Exp Bot. 2014:65(22):6589–6601. 10.1093/jxb/eru375 [DOI] [PubMed] [Google Scholar]
- Conde A, Neves A, Breia R, Pimentel D, Dinis LT, Bernardo S, Correia CM, Cunha A, Gerós H, Moutinho-Pereira J. Kaolin particle film application stimulates photoassimilate synthesis and modifies the primary metabolome of grape leaves. J Plant Physiol. 2018a:223(10):47–56. 10.1016/j.jplph.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Conde A, Soares F, Breia R, Gerós H. Postharvest dehydration induces variable changes in the primary metabolism of grape berries. Food Res Int. 2018b:105(03):261–270. 10.1016/j.foodres.2017.11.052 [DOI] [PubMed] [Google Scholar]
- Cui X, Yan Q, Gan S, Xue D, Wang H, Xing H, Zhao J, Guo N. GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae. BMC Plant Biol. 2019:19(1):1–15. 10.1186/s12870-019-2132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewdney SJ, McWha JA. Abscisic acid and the movement of photosynthetic assimilates towards developing wheat (Triticum aestivum L.) grains. Z Planzein Physiol. 1979:92(2):183–186. 10.1016/S0044-328X(79)80110-5 [DOI] [Google Scholar]
- Durand M, Porcheron B, Hennion N, Maurousset L, Lemoine R, Pourtau N. Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol. 2016:170(3):1460–1479. 10.1104/pp.15.01926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000:5(5):199–206. 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A. Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol. 2008:148(2):730–750. 10.1104/pp.108.120691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang HC, Dong YH, Yue XX, Hu JF, Jiang SH, Xu HF, Wang YC, Su MY, Zhang J, Zhang ZY, et al. . The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019:42(7):2090–2104. 10.1111/pce.13552 [DOI] [PubMed] [Google Scholar]
- Farcuh M, Rivero RM, Sadka A, Blumwald E. Ethylene regulation of sugar metabolism in climacteric and non-climacteric plums. Postharvest Biol Tec. 2018:139(03):20–30. 10.1016/j.postharvbio.2018.01.012 [DOI] [Google Scholar]
- Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plantarum. 2013:147(1):15–27. 10.1111/j.1399-3054.2012.01635.x [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang C, Han X, Wang ZY, Ma L, Yuan P, Wu JN, Zhu XF, Liu JM, Li DP, et al. . Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol Plant Pathol. 2018:19(9):2149–2161. 10.1111/mpp.12689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WJ, Nagy R, Chen HY, Pfrunder S, Yu YC, Santelia D, Frommer WB, Martinoia E. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol. 2014:164(2):777–789. 10.1104/pp.113.232751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Qiao Y, Yao L, Hao W, Liu Y, Shi W, Fan M, Bai MY. TOR And SnRK1 fine tune SPEECHLESS transcription and protein stability to optimize stomatal development in response to exogenously supplied sugar. New Phytol. 2022:234(1):107–121. 10.1111/nph.17984 [DOI] [PubMed] [Google Scholar]
- Hu DG, Yu JQ, Han PL, Xie XB, Sun CH, Zhang QY, Wang JH, Hao YJ. The regulatory module MdPUB29-MdbHLH3 connects ethylene biosynthesis with fruit quality in apple. New Phytol. 2019:221(4):1966–1982. 10.1111/nph.15511 [DOI] [PubMed] [Google Scholar]
- Jia B, Zhu XF, Pu ZJ, Duan YX, Hao LJ, Zhang J, Chen LQ, Jeon CO, Xuan YH. Integrative view of the diversity and evolution of SWEET and semi SWEET sugar transporters. Front Plant Sci. 2017:8(12):2178. 10.3389/fpls.2017.02178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia DJ, Shen F, Wang Y, Wu T, Xu XF, Zhang XZ, Han ZH. Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes MdSAUR37, MdPP2CH and MdALMTII. Plant J. 2018:95(3):427–443. 10.1111/tpj.13957 [DOI] [PubMed] [Google Scholar]
- Jia HF, Jiu ST, Zhang C, Wang C, Tariq P, Liu ZJ, Wang BJ, Cui LW, Fang JG. Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress-ripening transcription factor. Plant Biotechnol J. 2016:14(10):2045–2065. 10.1111/pbi.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H, Lim CW, Lee SC. Roles of pepper bZIP transcription factor CaATBZ1 and its interacting partner RING-type E3 ligase CaASRF1 in modulation of ABA signalling and drought tolerance. Plant J. 2019:100(2):399–410. 10.1111/tpj.14451 [DOI] [PubMed] [Google Scholar]
- Klemens PA, Patzke K, Deitmer JW, Spinner L, Le-Hir R, Bellini C, Bedu M, Chardon F, Krapp A, Neuhaus E. Overexpression of the vacuola rsugar carrier AtSWEET16 modifies germination, growth and stress tolerance in Arabidopsis. Plant Physiol. 2013:163(3):1338–1352. 10.1104/pp.113.224972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HY, Ho LH, Ekkehard NH, Guo WJ. Transporter SlSWEET15 unloads sucrose from phloem and seed coat for fruit and seed development in tomato. Plant Physiol. 2021:187(4):2230–2245. 10.1093/plphys/kiab290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashi K, Gemma H, Iwahori S. Sugar accumulation in peach fruit as affected by abscisic acid (ABA) treatment in relation to some sugar metabolizing enzymes. Engei Gakkai zasshi. 1999:68(3):465–470. 10.2503/jjshs.68.465 [DOI] [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet. 1993:238(1–2):106–119. 10.1007/BF00279537 [DOI] [PubMed] [Google Scholar]
- Lecourieux F, Kappel C, Lecourieux D, Serrano A, Torres E, Arce-Johnson P, Delrot S. An update on sugar transport and signaling in grapevine. J Exp Bot. 2014:65(3):821–832. 10.1093/jxb/ert394 [DOI] [PubMed] [Google Scholar]
- Li J, Qin M, Qiao X, Cheng Y, Li X, Zhang H, Wu J. A new insight into the evolution and functional functional divergence of SWEET transporters in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2017:58(4):839–850. 10.1093/pcp/pcx025 [DOI] [PubMed] [Google Scholar]
- Li MJ, Feng FJ, Cheng LL. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS ONE. 2012:7(3):e33055. 10.1371/journal.pone.0033055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TY, Dai ZR, Zeng BZ, Li J, Ouyang JY, Kang L, Wang W, Jia WS. Autocatalytic biosynthesis of abscisic acid and its synergistic action with auxin to regulate strawberry fruit ripening. Hortic Res. 2022:9(1):076. 10.1093/hr/uhab076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Guo W, Li JC, Yue PT, Bu HD, Jiang J, Liu WT, Xu YX, Yuan H, Li T, et al. . Histone acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in pear fruit. Plant Physiol. 2020:182(4):2035–2046. 10.1104/pp.20.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YX, Liu H, Yao XH, Wang J, Feng S, Sun LL, Ma S, Xu K, Chen LQ, Sui XL. Hexose transporter CsSWEET7a in cucumber mediates phloem unloading in companion cells for fruit development. Plant Physiol. 2021:186(1):640–654. 10.1093/plphys/kiab046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin IW, Sosso D, Chen LQ, Gase K, Kim SG, Kessler D, Klinkenberg PM, Gorder MK, Hou BH, Qu XQ, et al. . Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature. 2014:508(7497):546–549. 10.1038/nature13082 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and 2−ΔΔCT method. Methods. 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ljung K, Nemhauser JL, Perata P. New mechanistic links between sugar and hormone signalling networks. Curr Opin Plant Biol. 2015:25(6):130–137. 10.1016/j.pbi.2015.05.022 [DOI] [PubMed] [Google Scholar]
- Ma L, Zhang DC, Miao QS, Yang J, Xuan YH, Hu YB. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 2017a:58(5):863–873. 10.1093/pcp/pcx040 [DOI] [PubMed] [Google Scholar]
- Ma QJ, Sun MH, Lu J, Liu YJ, Hu DG, Hao YJ. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 2017b:174(4):2348–2362. 10.1104/pp.17.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Vallarino JG, Osorio S, Araguez I, Villarreal N, Ariza MT, Martinez GA, Medina-Escobar N, Civello MP, Fernie AR, et al. . Ethylene is involved in strawberry fruit ripening in an organ-specific manner. J Exp Bot. 2013:64(14):4421–4439. 10.1093/jxb/ert257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda Rossetto MR, Purgatoo E, Oliveria doNascimento JR, Lajolo FM, Cordenunsi BR. Effects of gibberellic acid on sucrose accumulation and sucrose biosynthesizing enzymes activity during banana ripening. Plant Growth Regul. 2003:41(3):207–214. 10.1023/B:GROW.0000007508.91064.8c [DOI] [Google Scholar]
- Mizuno H, Kasuga S, Kawahigashi H. The sorghum SWEET gene family: stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol Biofuels. 2016:9(1):127. 10.1186/s13068-016-0546-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia G, Pontin M, Piccoli P. Role of ABA and Gibberellin A3 on gene expression pattern of sugar transporters and invertases in Vitis vinifera cv. Malbec during berry ripening. Plant Growth Regul. 2018:84(2):275–283. 10.1007/s10725-017-0338-4 [DOI] [Google Scholar]
- Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao M, Sonah H, Song L, Lin L, Chaudhary J, et al. . Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics. 2015:16(1):520. 10.1186/s12864-015-1730-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JW. Phloem unloading: sieve element unloading and post-sieve element transport. Annu Rev Plant Physiol Plant Mol Biol. 1997:48(1):191–222. 10.1146/annurev.arplant.48.1.191 [DOI] [PubMed] [Google Scholar]
- Patrick JW. Does don Fisher's high-pressure manifold model account for phloem transport and resource partitioning? Front Plant Sci. 2013:4(6):184. 10.3389/fpls.2013.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Sun HH, Zong M, Guo SG, Ren ZJ, Zhao JY, Li MY, Zhang J, Tian SW, Wang JF, et al. . Localization shift of a sugar transporter contributes to phloem unloading in sweet watermelons. New Phytol. 2020:227(6):1858–1871. 10.1111/nph.16659 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006:57(1):675–709. 10.1146/annurev.arplant.57.032905.105441 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Jin Y, Yang YJ, Li GJ, Boyer JS. Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol Plant. 2010:3(6):942–955. 10.1093/mp/ssq044 [DOI] [PubMed] [Google Scholar]
- Shammai A. P, YeselsonY M, Faigenboim A, Moy-Komemi M, Cohen S, Cohen D, Besaulov E, Efrati A, Houminer N, Bar M, et al. . Natural genetic variation for expression of a SWEET transporter among wild species of Solanum lycopersicum (tomato) determines the hexose composition of ripening tomato fruit. Plant J. 2018:96(2):343–357. 10.1111/tpj.14035 [DOI] [PubMed] [Google Scholar]
- Singh AK, Kumar SR, Dwivedi V, Rai A, Pal S, Shasany AK, Nagegowda DA. A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 2017:215(3):1115–1131. 10.1111/nph.14663 [DOI] [PubMed] [Google Scholar]
- Sturm A, Tang GQ. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999:4(10):401–407. 10.1016/S1360-1385(99)01470-3 [DOI] [PubMed] [Google Scholar]
- Sun C, Zhang WW, Qu HY, Yan LF, Li LX, Zhao YQ, Yang HQ, Zhang H, Yao GF, Hu KD. Comparative physiological and transcriptomic analysis reveal MdWRKY75 associated with sucrose accumulation in postharvest ‘Honeycrisp’ apples with bitter pit. BMC Plant Biol. 2022:22(1):71. 10.1186/s12870-022-03453-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004:16(9):2481–2498. 10.1105/tpc.104.022699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AY-L, Gazzarrini S. Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front Plant Sci. 2014:5(4):119. 10.3389/fpls.2014.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R. The role of phloem loading reconsidered. Plant Physiol. 2010:152(4):1817–1823. 10.1104/pp.110.153023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallarino JG, Osorio S, Bombarely A, Casanal A, Cruz-Rus E, Sanchez-Sevilla JF, Amaya I, Giavalisco P, Fernie AR, Botella MA. Central role of FaGAMYB in the transition of the strawberry receptacle from development to ripening. New Phytol. 2015:208(2):482–496. 10.1111/nph.13463 [DOI] [PubMed] [Google Scholar]
- Veerle DS, Tom DS, Ingvar B, Kathy S. Phloem transport: a review of mechanisms and controls. J Exp Bot. 2013:64(16):4839–4850. 10.1093/jxb/ert302 [DOI] [PubMed] [Google Scholar]
- Wang N, Liu WJ, Yu L, Guo ZW, Chen ZJ, Jiang SH, Xu HF, Fang HC, Wang YC, Zhang ZY, et al. . HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiol. 2020:184(3):1273–1290. 10.1104/pp.20.01106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Qu CZ, Jiang SH, Chen ZJ, Xu HF, Fang HC, Su MY, Zhang J, Wang YC, Liu WJ, et al. . The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 2018:96(1):39–55. 10.1111/tpj.14013 [DOI] [PubMed] [Google Scholar]
- Wang N, Xu HF, Jiang SH, Zhang ZY, Lu NL, Qiu HR, Qu CZ, Wang YC, Wu SJ, Chen XS. MYB12 And MYB22 play essential roles in proanthocyanidin andflavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 2017:90(2):276–292. 10.1111/tpj.13487 [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang Z, Li LX, Wang HB, Zhou H, Chen XS, Feng SQ. Apple MdMYB306-like inhibits anthocyanin synthesis by directly interacting with MdMYB17 and MdbHLH33. Plant J. 2022a:110(4):1021–1034. 10.1111/tpj.15720 [DOI] [PubMed] [Google Scholar]
- Wang SD, Yokosho K, Guo RZ, Whelan J, Ruan YL, Ma JF, Shou HX. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol. 2019:180(4):2133–2141. 10.1104/pp.19.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Ma BQ, Yang NX, Jin L, Wang L, Ma SY, Ruan YL, Ma FW, Li MJ. Variation in the promoter of the sorbitol dehydrogenase gene MdSDH2 affects binding of the transcription factor MdABI3 and alters fructose content in apple fruit. Plant J. 2022b:109(5):1183–1198. 10.1111/tpj.15624 [DOI] [PubMed] [Google Scholar]
- Wei LZ, Mao WW, Jia MR, Xing SN, Ali U, Zhao YY, Chen YT, Cao ML, Dai ZR, Zhang KK, et al. . FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J Exp Bot. 2018:69(20):4805–4820. 10.1093/jxb/ery249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei XY, Liu FL, Chen C, Ma FW, Li MJ. The Malus domestica sugar transporter gene family: identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front Plant Sci. 2014:5(11):569. 10.3389/fpls.2014.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingenter K, Schulz A, Wormit A, Wic S, Trentmann O, Hoermiller II, Heyer AG, Marten I, Hedrich R, Neuhaus HE. Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiol. 2010:154(2):665–677. 10.1104/pp.110.162040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormit A, Trentmann O, Feifer I, Lohr C, Tjaden J, Meyer S, Schmidt U, Martinoia E, Neuhaus HE. Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell. 2006:18(12):3476–3490. 10.1105/tpc.106.047290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014:14(1):327. 10.1186/s12870-014-0327-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Wang SP. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant. 2013:6(3):665–674. 10.1093/mp/sst035 [DOI] [PubMed] [Google Scholar]
- Yuan M, Zhao JW, Huang RY, Li XH, Xiao JH, Wang SP. Rice MtN3/saliva/SWEET gene family: evolution, expression profiling, and sugar transport. J Integr Plant Biol. 2014:56(6):559–570. 10.1111/jipb.12173 [DOI] [PubMed] [Google Scholar]
- Yuan YJ, Mei LH, Wu MB, Wei W, Shan W, Gong Z, Zhang Q, Yang F, Yan F, Zhang Q, et al. . SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J Exp Bot. 2018:69(11):5507–5518. 10.1093/jxb/ery328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SH, Peng FT, Xiao YS, Wang WR, Wu XL. Peach PpSnRK1 participates in sucrose-mediated root growth through auxin signaling. Front in Plant Sci. 2020:11(4):409. 10.3389/fpls.2020.00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XS, Feng CY, Wang MN, Li TL, Liu X, Jiang J. Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits. Hortic Res. 2021:8(1):186. 10.1038/s41438-021-00624-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Wang LJ. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 2005:5(1):1. 10.1186/1471-2148-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zou LM, Ren C, Wang Y, Fan PG, Li SH, Liang ZC. Vvsweet10 mediates sugar accumulation in grapes. Genes (Basel). 2019:10(4):255. 10.3390/genes10040255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen QL, Fang T, Peng Q, Liao L, Zhao L, Owiti A, Han YP. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic Res. 2018:5(1):14. 10.1038/s41438-018-0024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LC, Li BY, Wu LM, Li HX, Wang ZY, Wei XY, Ma BQ, Zhang YF, Ma FW, Ruan YL, et al. . MdERDL6-mediated glucose efflux to the cytosol promotes sugar accumulation in the vacuole through up-regulating TSTs in apple and tomato. Proc Natl Acad Sci USA. 2021:118(1):e2022788118. 10.1073/pnas.2022788118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.