Abstract
Plant somatic embryogenesis (SE) is an in vitro biological process wherein bipolar structures are induced to form somatic cells and regenerate into whole plants. MicroRNA (miRNA) is an essential player in plant SE. However, the mechanism of microRNA408 (miR408) in SE remains elusive. Here, we used stable transgenic technology in longan (Dimocarpus longan) embryogenic calli to verify the mechanism by which miR408 promotes cell division and differentiation of longan early SE. dlo-miR408-3p regulated riboflavin biosynthesis by targeting nudix hydrolase 23 (DlNUDT23), a previously unidentified gene mediating N6-methyladenosine (m6A) modification and influencing RNA homeostasis and cell cycle gene expression during longan early SE. We showed that DlMIR408 overexpression (DlMIR408-OE) promoted 21-nt miRNA biosynthesis. In DlMIR408-OE cell lines, dlo-miR408-3p targeted and downregulated DlNUDT23, promoted riboflavin biosynthesis, decreased flavin mononucleotide (FMN) accumulation, promoted m6A level, and influenced miRNA homeostasis. DNA replication, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, the pentose phosphate pathway, and taurine and hypotaurine metabolism were also closely associated with riboflavin metabolism. In a riboflavin feeding assay, dlo-miR408-3p and pre-miR408 were upregulated and DlNUDT23 was downregulated, increasing the m6A level and cell division and differentiation in longan globular embryos. When riboflavin biosynthesis was inhibited, dlo-miR408-3p was downregulated and DlNUDT23 was upregulated, which decreased m6A modification and inhibited cell division but did not inhibit cell differentiation. FMN artificial demethylated m6A modification affected the homeostasis of precursor miRNA and miRNA. Our results revealed a mechanism underlying dlo-miR408-3p-activated riboflavin biosynthesis in which DlNUDT23 is targeted, m6A modification is dynamically mediated, and cell division is affected, promoting early SE in plants.
MicroRNA408 targets a previously unknown gene, regulates riboflavin biosynthesis, and mediates m6A modification and RNA homeostasis, promoting cell division and early somatic embryogenesis in longan.
Introduction
Plant regeneration can be achieved through somatic embryogenesis (SE) without fertilization. This highlights the outstanding totipotent potential of plant somatic cells (Koltunow and Grossniklaus 2003). In seed plants, the development of the early zygotic embryo (ZE) determines seed quality (Radoeva et al. 2019). In horticultural crops, embryo development directly determines fruit quality and yield. Because the various stages of plant SE are extremely similar to those of ZE at morphological, physiological, biochemical, transcriptional, and protein levels, SE is a perfect model system for investigating the molecular mechanisms underlying plant cell totipotency (Pramanik et al. 1992; Zimmerman 1993; Goldberg et al. 1994). Globally, longan (Dimocarpus longan Lour.) is among the most important tropical and subtropical woody fruit trees. Longan fruit is of high nutritional and medical value and is, therefore, regarded as a treasure among fruits. The longan SE system is an excellent model among woody plants (Lai et al. 2000). Lai et al. conducted considerable intensive research into the mechanism of cytology and molecular biology during longan SE (Lai et al. 1997; Lin et al. 2017; Chen et al. 2020a). Elucidating the molecular mechanism underpinning cell totipotency in woody plants is of great importance. However, the embryogenic genes that are activated during early SE and the molecular mechanism underpinning cellular reprogramming remain poorly understood (Kennedy 2005; Vogel 2005; Li et al. 2022).
Increasing evidence demonstrates that microRNA (miRNA) has a crucial role in plant SE and responds to variations in the extracellular environment (Katarzyna et al. 2017; Long et al. 2018; Wang et al. 2018). RNA polymerase II transcribes MIRNA loci to primary miRNAs (pri-miRNAs), which then forms precursor miRNAs (pre-miRNAs). These pre-miRNAs are processed into 21- to 24-nt mature miRNA duplexes through multiple and sequential cuts (Moro et al. 2018; Tomassi et al. 2020). Subsequently, target genes are downregulated through cleavage or translation at the transcription or posttranscription level. miR408 is a class of 21-nt miRNA and is highly conserved in terrestrial plants. pre-miR408 is less conserved in different species, and miR408 may target different genes to realize completely different biological functions (Liu et al. 2017; Gao et al. 2022). Because of these characteristics, miR408 plays diverse roles in plant development. However, the target genes of miR408 remain poorly understood. Studies have shown that miR408 targets Uclacyanins (OsUCLs) (Zhang et al. 2017), Plantacyanin (PCY) (Jiang et al. 2021a; Hao et al. 2022), and Laccase 13 (LAC13) (Song et al. 2018), affects copper homeostasis in vivo, and participates in photosynthesis regulation. It also targets Vacuolar invertase 1 (VIN1), thus regulating glucose and fructose synthesis (Ma 2012). Moreover, miR408-3p targets RNA polymerase II in rice (Oryza sativa) callus morphogenesis (Solanki et al. 2019). The miR408 promoter also has a functional and distinctive role in the embryo (Yang et al. 2021). dlo-miR408-3p negatively regulates DlLAC12 to participate in globular embryo (GE) development and abiotic stress in longan (Xu et al. 2022a). Transient dlo-miR408-3p overexpression (dlo-miR408-3p-agomir) promotes cell proliferation and GE morphogenesis, which may affect the cell cycle (Xu et al. 2022b). Thus, we aimed to determine the molecular mechanism of miR408 in regulating cell division and differentiation in longan. To address this question, we thoroughly investigated the biological function of miR408 using the SE model system and stable genetic transformation technology in longan.
mRNA and miRNA metabolism finely controls RNA quality and quantity mainly through multiple RNA degradation pathways. m6A methylation is the most abundant RNA reversible modification in eukaryotes, widely extensive in mRNA and noncoding RNA (ncRNA). m6A primarily occurs in the adenine of “RRACH” site. The expression of “writers, erasers, and readers” in the m6A pathway directly determines the level of m6A (Yue et al. 2019; Zheng et al. 2020). In mRNA adenosine methylase (MTA) mutants of Arabidopsis (Arabidopsis thaliana), MTA introduces m6A methylation modification on pri-miRNA, and the secondary structure of the stem-loop is substantially reduced, leading to substantial pri-miRNA accumulation, but miRNA levels are reduced (Bhat et al. 2020). MTA protein inactivation affects GE development and eventually leads to embryo death (Zhong et al. 2008). Two previous studies have shown that chemical modification or oxidative stress also regulates m6A modifications. In HeLa cells, flavin mononucleotide (FMN) acts as an artificial demethylase to specifically oxidize N6-methyl-substituted adenosine after blue light absorption and effectively downregulates m6A expression (Xie et al. 2017; Xie et al. 2019). These studies imply that m6A methylation of mRNA and pri-miRNA influences mRNA and miRNA biosynthesis. This process may also be affected by powerful small molecules, further regulating RNA homeostasis.
FMN and flavin adenine dinucleotide (FAD) are riboflavin (vitamin B2, VB2) co-factors and act as co-substrates for many epigenetic processes, thereby integrating nutrition, metabolism, and gene expression (Shi et al. 2004; Kaelin and McKnight 2013). Riboflavin metabolism is ancestrally conserved, and its synthesis in plants is de novo (Bacher et al. 1999). Purine metabolism and the pentose phosphate pathway (PPP) offer the initial precursors of GTP and ribulose 5-phosphate required for riboflavin biosynthesis (Hasnain et al. 2013). Tian et al. (2022) revealed the molecular mechanism underpinning riboflavin, which was involved in integrating cell energy and cell cycle, and regulating maize seed development and embryonic development. FMN and FAD have pivotal roles in ATP generation and metabolic homeostasis in the mitochondrial electron transfer chain (Sharma et al. 2019). FAD serves as a cofactor for lysine-specific demethylase (LSD) in the catalysis of oxidation of H3-K4 N-methyl (Forneris et al. 2006). In early embryos, H3K4me3 participates in the reprogramming of zygotic epigenomes (Xia et al. 2019). Moreover, H3K4me3 regulates DNA replication and damage, thus influencing the cell cycle (Serra-Cardona et al. 2022). These studies demonstrate that riboflavin biosynthesis is essential for the epigenetic regulation of cell division and differentiation in plant SE.
However, no evidence is available reporting that miRNA regulates riboflavin biosynthesis in plants. NUDIX belongs to the pyrophosphate hydrolase family. It mainly regulates FMN synthesis from FAD and influences riboflavin homeostasis (Maruta et al. 2012). In addition, NUDIX can uncap the small regulatory RNA linked to the NAD “cap”. NUDIX plays an essential role in RNA stability through the initiation of RNase E-mediated RNA degradation (Cahova et al. 2015; Sharma et al. 2020). In mammals, NAD-capped and FAD-capped RNAs can be uncapped by Nudix to regulate cell cycle and proliferation (Sharma et al. 2020). Through experiments of transient transformation in Nicotiana tabacum and longan SE and the treatment with riboflavin and an inhibitor (diphenyleneiodonium chloride, DPI) in vivo and in vitro, we show that dlo-miR408-3p negatively regulates a previously unidentified gene, DlNUDT23. Therefore, we show that the molecular mechanism of riboflavin metabolism-mediated m6A modification is targeted by dlo-miR408-3p-DlNUDT23, and consequently, this affects RNA homeostasis and cell proliferation during longan early SE.
In a previous study, the author found that dlo-miR408-3p is upregulated during longan early SE (Xu et al. 2020). These results suggested that dlo-miR408-3p overexpression may promote GE development. Moreover, studies have found that the expression patterns of miR408 located on the 3′-end arm and 5′-end arm of pre-miR408 were completely varied under different abiotic stresses (Xu et al. 2022a). We further investigated the mechanism involved in pre-miR408 processing to miR408. SE is applied for regenerating genetically modified plants during transformation. Notably, transgenic technology of the Sapindaceae plant has only been performed by Xu (2010), Zhang (2004), and Zeng (2002) for gene function validation. However, the regeneration pathway of longan SE is of endogenous origin and is sensitive to Agrobacterium tumefaciens and antibiotics, and is, therefore, difficult to regenerate. None of the aforementioned scholars have achieved stable transgenic regenerative cell lines, and hence, SE has not been successfully used to study gene functions in the past 12 yr. In our study, we focused on the function of DlMIR408 during longan early SE. Longan stable transgenic technology was used to construct DlMIR408 overexpression (DlMIR408-OE) cell lines by infecting longan embryogenic calli (EC). DlMIR408-regulated pathways were analyzed through whole-transcriptome high-throughput sequencing. We also performed cell morphology analysis and m6A modification level analysis. HPLC was used to detect the physiological content of riboflavin and taurine. Our work provides new insights into the molecular action of miR408 in longan early SE and closes a long-standing gap in understanding miRNA-mediated regulation of riboflavin synthesis and plant embryonic development.
Results
Expression patterns of the dlo-miR408 family in different longan tissues
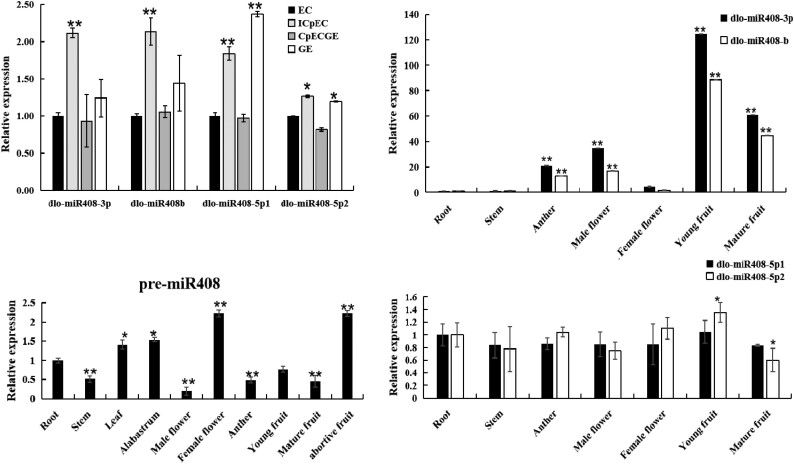
The longan genome contained 1 pre-miR408 and 4 mature dlo-miR408s. The relative expression analysis showed that dlo-miR408-3p and dlo-miR408b had the same expression pattern in different longan early somatic embryogenic stages and tissues, whereas dlo-miR408-5p1 and dlo-miR408-5p2 had the same expression pattern (Fig. 1). Compared with roots, pre-miR408 expression was relatively higher in the female flower and abortive fruit, followed by leaf and alabastrum, and the expression was lower in the stem, male flower, and anther. The expression of dlo-miR408-3p and dlo-miR408b was highly significantly upregulated in young and mature fruits. The expression of dlo-miR408-5p1 and dlo-miR408-5p2 was stable in different longan tissues (Fig. 1). These results indicated that the expression patterns of pre-miR408 and dlo-miR408 were different. dlo-miR408-3p and dlo-miR408-5p, which were located at different pre-miR408 arms, had different expression patterns. Therefore, the regulation mechanism during pre-miR408 processing remains unclear and needs to be further investigated.
Figure 1.
The relative expression patterns of pre-miR408 and dlo-miR408 members in different early SE and different tissues of longan. EC, embryogenic callus; ICpEC, incomplete compact pro-embryogenic cultures; CpECGE, compact pro-embryogenic cultures globular embryos; GE, globular embryo. The different early SE of longan are induced according to Lai (2003). Error bars indicate means ± SDs, n = 3. *P ≤ 0.05, **P ≤ 0.01.
DlMIR408-OE cell lines obtained in longan EC
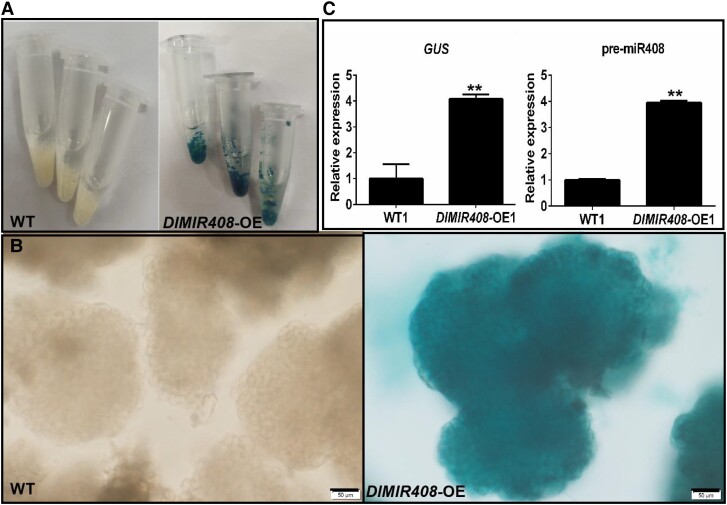
To further validate the functions of dlo-miR408 in longan. A. tumefaciens-mediated transformation was performed in longan. To generate the DlMIR408-OE vector, we amplified and cloned the full-length DlMIR408 cDNA (475 bp) into the pCAMBIA1301 (1301) backbone vector by using Pst1 and Sal1 (Supplemental Fig. S1, A to D). The receptor cell line was infected by Agrobacterium (Supplemental Fig. S1E). The reverse transcription quantitative PCR (RT-qPCR) result showed that pre-miR408 expression was significantly higher in the DlMIR408-OE-transformed transgenic callus (Supplemental Fig. S1, F to I). The presence of pri-miR408 and GUS DNA in the wild type (WT) and DlMIR408-OE cell lines was validated using PCR. pri-miR408 was present in all WT and DlMIR408-OE cell lines (Supplemental Fig. S1, J and K), whereas GUS was only amplified in DlMIR408-OE cell lines (OE1−3) (Supplemental Fig. S1L). Moreover, in subcultured generation six (S6), GUS histochemical staining was used to detect the positive homozygous transgenic GE (Fig. 2, A and B), GUS and pre-miR408 was overexpressed in the homozygous GE of DlMIR408-OE cell lines (Fig. 2, C). This result demonstrated that the DlMIR408 gene sequence was stably overexpressed in the longan genome.
Figure 2.
The expression pattern of GUS and the pre-miR408, and the cell morphology in WT and DlMIR408-OE cell line for the whole-transcriptome sequencing. A) The WT and DlMIR408-OE cell lines after GUS staining, and the cell lines with independent 3 biological replicates. B) The positive homozygous transgenic GE of DlMIR408-OE under Olympus microscope observation, scale bars = 50 μm. C) The expression patterns of GUS and pre-miR408 in WT and DlMIR408-OE1. Error bars indicate means ± SDs, n = 3. **Highly significant difference (Duncan’s post hoc test, P ≤ 0.01).
Whole-transcript sequencing of DlMIR408-OE transgenic cell lines
To further reveal the molecular mechanism of DlMIR408 during longan early SE, whole-transcriptional sequencing was performed in the WT and DlMIR408-OE cell lines. The RNA integrity number values for WT and DlMIR408-OE were larger than 8 and 6, respectively. In total, 27.86 Gb of clean reads were obtained from 2 samples. The lncRNA database of each sample generated at least 12 Gb of clean data. A total of 21.63 Mb was obtained for miRNA, with each sample miRNA generating no less than 10 M clean reads (Supplemental Table S1). A Q20 ≥ 97.9% and Q30 ≥ 94% for each sample indicated that the data quality was high, with the data having a very low probability of base mismatch and being very reliable for subsequent analysis. For the reference longan third-generation genome, the miRNA database had a high quality (98%). The mRNA and lncRNA database contained over 99% clean reads, of which 75% were mapped, and 99% unmatched to ribosomal sequences, which were used for subsequent ncRNA identification (Supplemental Table S2).
Overexpression of DlMIR408-regulated longan SE by affecting mRNAs, miRNAs, and lncRNAs
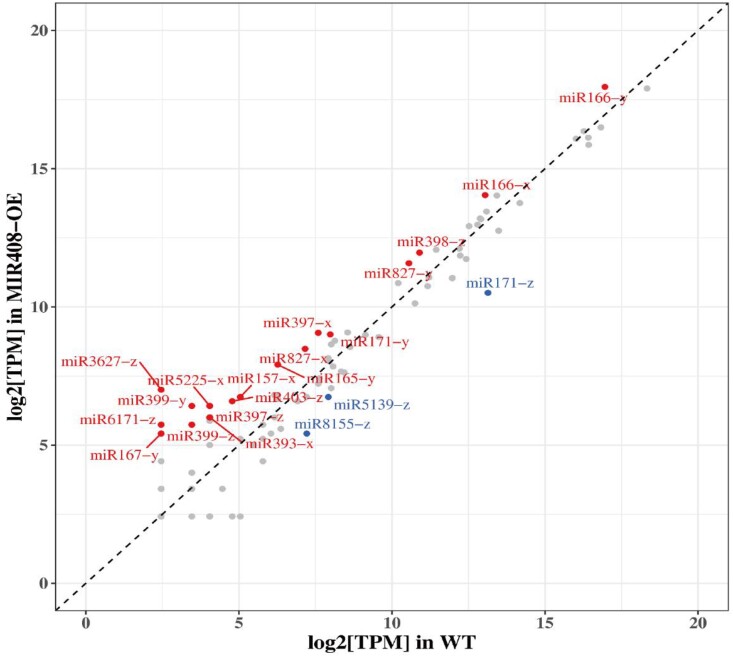
We next conducted the differential expression (DE) analysis of mRNAs, miRNAs, and lncRNAs after DlMIR408-OE. A total of 6,253 DE mRNAs were screened, of them, 2,642 were upregulated and 3,611 were downregulated. In total, 1,301 DE mRNAs were specifically upregulated in the DlMIR408-OE cell line, and 641 DE mRNAs were specifically downregulated in the DlMIR408-OE. A total of 65 known pre-miRNAs and 17 previously unknown pre-miRNAs were identified. In total, 114 known miRNAs belonged to 73 MIRNA gene families, with 107 in WT and 101 in DlMIR408-OE. A total of 21 DE miRNAs were screened, and 85.7% of the miRNAs (18/21) were upregulated. Of them, 61.1% were 21-nt miRNAs (11/18) (Supplemental Table S3; Fig. 3). Compared with WT, 17 previously unknown DE miRNAs were 24 nt in DlMIR408-OE (Supplemental Table S3). A total of 3,060 DE lncRNAs were screened, of them, 2,278 and 782 were upregulated and downregulated, respectively. A total of 8,230 previously unknown lncRNAs were obtained, including 6,761 cis-lncRNAs and 1,285 antisense-lncRNAs, targeting 9,740 and 1,157 mRNAs, to obtain 14,704 and 1,327 lncRNA-mRNA pairs, respectively. These results indicated that DlMIR408-OE promoted 21-nt miRNA biosynthesis in longan. These DE RNAs were used for constructing regulatory models of mRNAs with miRNAs and lncRNAs.
Figure 3.
Known DE miRNAs in WT and DlMIR408-OE cell lines. The red label represents upregulated DE miRNAs, the blue label represents downregulated DE miRNAs, x represents the 5p, y represents 3p, and z represents not only maps to 5p but also maps to 3p after comparison with miRNA in other species.
Expression pattern analysis of DE mRNAs in the DlMIR408-OE cell lines
We further analyzed the functional annotation of DE RNAs affected by DlMIR408-OE. The results showed that DE mRNAs contained 2 major categories of transcription factors (TFs) and functional genes. In total, 58 TF gene families were identified, and the most differentially expressed members included v-myb avian myeloblastosis viral oncogene homolog (MYB), basic helix-loop-helix (bHLH), ethylene responsive factor (ERF), and NAM-ATAF1/2-CUC2 (NAC) (Supplemental Fig. S2), followed by C2H2, M-type, WRKYGQK domain zinc finger motif (WAKY), FAR1, B3 domain transcription factor (B3), basic leucine zipper (bZIP), and other TF gene families, of them, MYB73 was predicted to be a target gene of the miR408 family. In DlMIR408-OE, the genes most highly upregulated were involved in the TCA cycle, RNA metabolism, riboflavin metabolism, GPI signaling, plant pathogen response pathway, plant hormone metabolism, calcium transport, and lipid metabolism (Supplemental Tables S4-1). The number of downregulated genes was higher than that of upregulated genes in DlMIR408-OE cell lines. The downregulated genes were mainly involved in RNA transport and metabolism, miRNA-mediated mRNA degradation, sugar metabolism, ion transport, amino acid metabolism, folate biosynthesis, cell cycle, and cell polarity (Supplemental Table S4-2). Importantly, dlo-miR408-3p was predicted to target a previously unidentified gene, DlNUDT23. These results suggested that DlMIR408 was involved in RNA metabolism and regulated RNA stability, miRNA processing pathways, riboflavin biosynthesis, ion channel transport, and cell cycle regulation (Supplemental Fig. S3).
Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses of DE mRNAs, miRNAs, and lncRNAs in the DlMIR408-OE cell lines
To investigate the potential role of the mRNA–miRNA–lncRNA model in longan early SE, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed. Compared with the WT, GO analysis revealed that mRNAs, miRNAs target genes, and lncRNAs target genes were significantly enriched in purine nucleotide binding, ion binding and transport, and cofactor binding (Supplemental Fig. S4, A, B, D, and F; Supplemental Table S5). KEGG analysis revealed that mRNAs, miRNAs target genes, and lncRNA target genes were significantly enriched in DNA replication, mismatch repair, cell cycle metabolism, amino acid metabolism, fatty acid metabolism, m7G cap structure formation, RNA capping, NAD binding, tRNA binding, and transcription cofactor activity in the DlMIR408-OE cell line (Supplemental Fig. S4, A, C, and F; Supplemental Table S5). Analysis of the top 20 KEGG pathways of mRNAs revealed significant enrichment in DNA replication, riboflavin metabolism, mismatch repair, GPI-anchor biosynthesis, taurine and hypotaurine metabolism, sulfur metabolism, fatty acid degradation, phenylalanine metabolism, and homologous recombination (Supplemental Fig. S4A; Supplemental Table S5). Moreover, analysis of the top 20 KEGG pathways of target genes of known DE miRNAs and lncRNAs revealed significant enrichment in amino acid metabolism, energy metabolism, and carbohydrate metabolism (Supplemental Fig. S4, C and F; Supplemental Table S5). The target genes of previously unknown DE miRNAs were involved in glyoxylate decarboxylation metabolism, glycine–serine–threonine, and carbon metabolism (Supplemental Fig. S4C).
Association among riboflavin metabolism, GPI-anchor biosynthesis, and PPP in the DlMIR408-OE cell lines
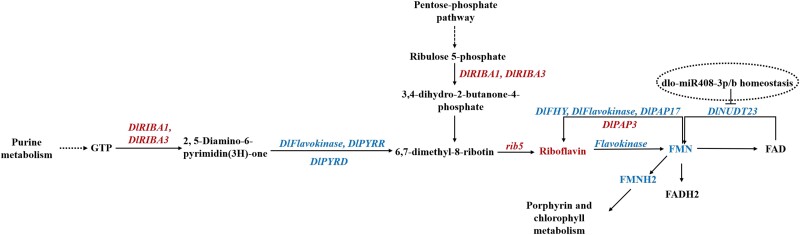
Riboflavin is a cofactor for enzymes such as succinate dehydrogenase in the TCA cycle and acyl-CoA dehydrogenase in fatty acid oxidation (Kaelin and McKnight 2013). To further comprehend the importance of riboflavin metabolism during early SE, the association among the significantly enriched metabolic pathways of riboflavin metabolism, GPI metabolism, and the PPP were analyzed in the DlMIR408-OE cell lines. The purine metabolism pathway and the PPP provide GTP and ribulose 5-phosphate, which act as synthesis precursors, for riboflavin metabolism. In DlMIR408-OE cell lines, DlRIBA1, DlRIBA3, and Dlrib5 were significantly upregulated, thereby promoting the synthesis of 3, 4-dihydro2-butane-4-phosphate and 2, 5-diamino-pyrimidine. Intriguingly, DlPYRR, DlPYRD, and DlFlavokinase were significantly downregulated, leading to 5-amino-6-uracil synthesis. The phosphate metabolism branch was activated, riboflavin accumulation increased, DlFlavokinase was downregulated, and FMN accumulation was decreased. Simultaneously, DlNUDT23 (Dlo029903) downregulation inhibited FAD transformation into FMN (Fig. 4). Furthermore, lncRNA transcriptome data revealed that lncRNA positively or negatively regulated riboflavin metabolism in DlMIR408-OE cell lines. For example, DlNUDT23 was positively regulated by lncRNAs (MSTRG.7391.1, HSP40), and both were significantly downregulated. Meanwhile, DlNUDT23 was also predicted to be negatively regulated by dlo-miR408-3p. DlRIBA3 (Dlo026341) was positively regulated by lncRNA (MSTRG.4152.1), and both were significantly upregulated. DlPYRD was negatively regulated by lncRNA (MSTRG.19600.1). MSTRG.19600.1 was significantly upregulated, whereas DlPYRD was significantly downregulated. DlPAP17 was negatively regulated by lncRNA (MSTRG.8602.1). MSTRG.8602.1 was significantly upregulated, whereas DlPAP17 was significantly downregulated (Supplemental Table S4-3).
Figure 4.
The association analysis of riboflavin metabolism, purine metabolism, and PPP in DlMIR408-OE cell line. The deep red color represents upregulation, and the blue color represents downregulation. The dotted circle indicates the regulatory model identified in DlMIR408-OE cell line and needs to be further verified.
In addition, in the PPP, the gene Dlo004745 was significantly upregulated, leading to the synthesis of D-6-phosphate glucose, which acted as a synthesis precursor of GPI-anchor biosynthesis, and promoted phosphatidylinositol synthesis. By contrast, PIG C/H/P and PIG X/N/B/O/S were significantly downregulated. Most genes in taurine and hypotaurine metabolism were significantly downregulated, thereby inhibiting the synthesis of 5-glutamine-taurine. Thus, our results demonstrated that the PPP was activated, riboflavin metabolism was significantly activated, phosphatidylinositol metabolism was affected, and taurine content was significantly decreased in DlMIR408-OE cell lines.
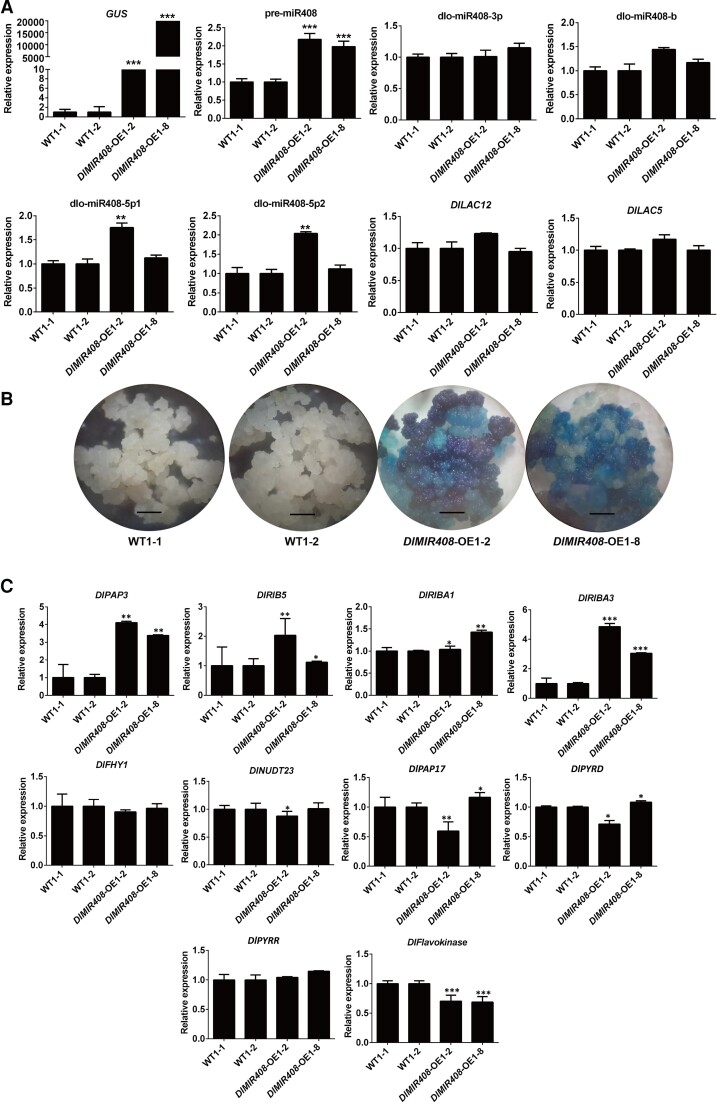
The transcriptional pattern validation of dlo-miR408/DlLAC12 and the mRNA–miRNA–lncRNA model in the DlMIR408-OE cell line
To validate the accuracy of whole-transcriptome data, RT-qPCR was performed to verify the transcriptional expression patterns of pre-miR408, dlo-miR408-3p, dlo-miR408b, dlo-miR408-5p1, dlo-miR408-5p2, and known target gene (DlLAC12) and its alternative splice transcript DlLAC5 in the S9 of DlMIR408-OE, which has been identified as a positive homozygous transgenic cell line through GUS histochemical staining. According to the results, the expression of GUS and pre-miR408 was highly significantly upregulated in DlMIR408-OE 1-2 and OE 1-8 cell lines (Fig. 5, A and B; Supplemental Table S6). Intriguingly, dlo-miR408-5p1 and dlo-miR408-5p2 were localized in the 5′-end arm and were significantly upregulated in the DlMIR408-OE 1-2 cell lines. However, dlo-miR408-3p localized in the 3′-end arm was stably expressed. No significant difference was observed in the expression of the target genes DlLAC12 and DlLAC5 in DlMIR408-OE 1-2 cell lines. These results indicated that the expression patterns of dlo-miR408 differed between the 3′-end and 5′-end arms in pre-miR408. This result was consistent with the expression patterns of dlo-miR408 in different tissues (Fig. 5A; Supplemental Table S6) and under different abiotic stresses (Xu et al. 2022a). Based on the results of GO analysis that revealed DE mRNA enriched in RNA processing and metabolism, we further questioned whether they were related to posttranscriptional modifications at the RNA level during pre-miR408 processing into mature miR408.
Figure 5.
The transcriptional level validation of dlo-miR408/DlLAC12 and mRNA–miRNA–lncRNA models in DlMIR408-OE stable transgenic cell lines. A) The expression patterns of GUS, pre-miR408, miR408-3p/b/5p1/5p2, and DlLAC12/DlLAC5.B) The GUS histochemical staining of WT (1-1 and 1-2) and DlMIR408-OE (1-2 and 1-8) transgenic cell lines. The scale bar represents 1 mm. C) The expression patterns of riboflavin metabolism-related genes in WT (1-1 and 1-2) and DlMIR408-OE (1-2 and 1-8). D) The expression patterns of the mRNA–miRNA–lncRNA models which participate in riboflavin and phosphate metabolism. Error bars indicate means ± SDs, n = 3 (Duncan’s post hoc test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
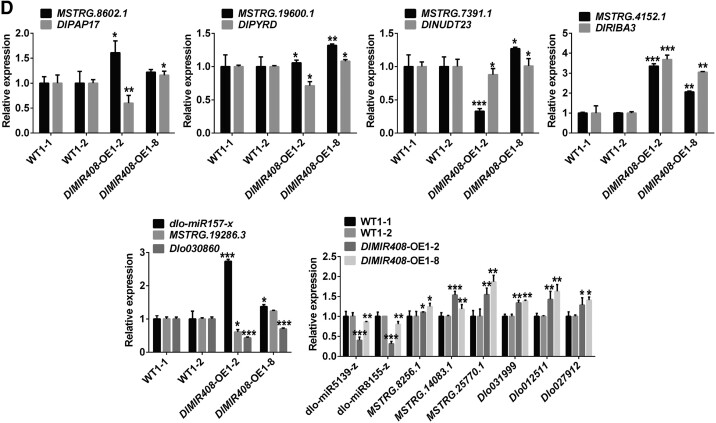
Furthermore, the expression patterns of riboflavin metabolism-related genes and mRNA–miRNA–lncRNA models were verified through RT-qPCR. The results revealed that DlPAP3, DlRIB5, DlRIBA1, and DlRIBA3 were significantly upregulated, whereas DlFHY1, DlNUDT23, DlPAP17, DlPYRD, DlPYRR, and DlFlavokinase were significantly downregulated in DlMIR408-OE cell lines (Fig. 5C; Supplemental Table S6). Remarkably, MSTRG 7391.1 (HSP40) positively regulated DlNUDT23, and both of them were downregulated in DlMIR408-OE. In addition, we identified that the Dlo030860-miR157-X-MSTRG.19286.3 model participated in inorganic phosphate transport, UVB photosensitivity regulation, and actin depolymerization. The Dlo027912 (SBT1.7)/Dlo031999/Dlo012511-miR5139-z/miR8155-z-MSTRG.8256.1/MSTRG.25770.1/MSTRG.14083.1 model influenced the activities of RNA–DNA hybrid ribonuclease and serine-type endopeptidase. The expression patterns of these models were also consistent with the transcriptome data (Fig. 5D; Supplemental Table S6). The aforementioned data indicated that lncRNA–miRNA–mRNA models participated in riboflavin metabolism, inorganic phosphate transport, UVB photosensitivity regulation, actin depolymerization, and serine-type endopeptidase activity after DlMIR408-OE.
dlo-miR408-3p degrades DlNUDT23, inhibits FMN biosynthesis, and regulates riboflavin biosynthesis
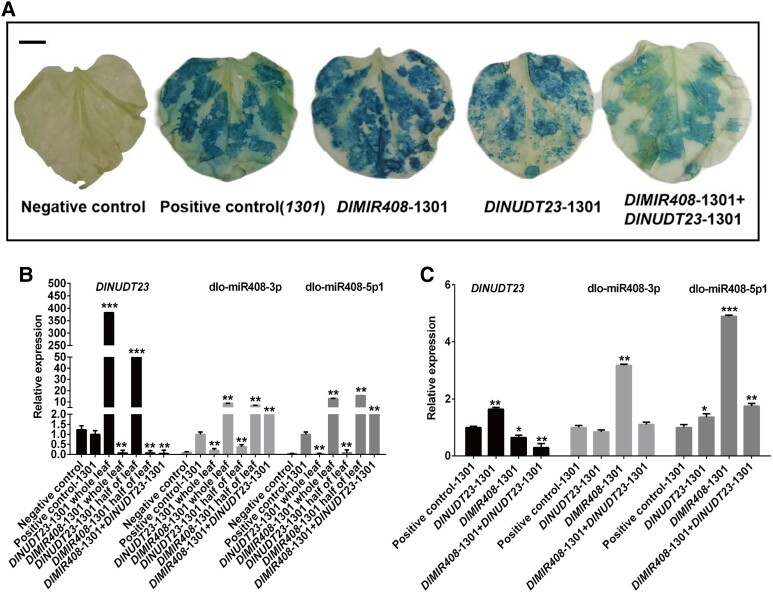
In the riboflavin biosynthesis pathway, DlNUDT23 was predicted to be a target of dlo-miR408-3p in the DlMIR408-OE cell lines (Fig. 4). To further investigate whether dlo-miR408-3p specifically and negatively regulated DlNUDT23 in vivo. DlMIR408-1301 and DlNUDT23-1301 overexpression vectors were constructed and transiently transformed in Nicotiana benthamiana and longan EC. We then performed GUS histochemical staining and quantitative expression analysis of dlo-miR408 and DlNUDT23 was performed through RT-qPCR. In the N. benthamiana transient transformation experiment, compared with the negative control, GUS staining of N. benthamiana leaves transformed with both DlMIR408-1301+ DlNUDT23-1301 was lighter than that of those transformed with the 1,301 empty vectors, DlMIR408-1301 and DlNUDT23-1301 alone (Fig. 6A). Compared with the negative control and positive control-1301, the relative expression of dlo-miR408-3p and dlo-miR408-5p1, from high to low, was “DlMIR408-1301 whole leaf” > “DlMIR408-1301 half of leaf” > “DlMIR408-1301 + DlNUDT23-1301” > “DlNUDT23-1301 half of leaf” > “DlNUDT23-1301 whole leaf” (Fig. 6B; Supplemental Table S7). By contrast, the DlNUDT23 expression changed from low to high for the aforementioned combinations (Supplemental Table S7). In summary, the results of the transient expression assay in N. benthamiana revealed that dlo-miR408-3p could degrade DlNUDT23. Next, the transient transformation of longan EC assay showed that DlNUDT23 was significantly downregulated in DlMIR408-1301 or DlMIR408-1301 + DlNUDT23-1301 cell lines and only significantly upregulated in the DlNUDT23-1301 cell lines. Conversely, dlo-miR408-3p was only significantly upregulated in the DlMIR408-1301 EC cell line and downregulated in the DlNUDT23-1301 EC cell line (Fig. 6C; Supplemental Table S7). The aforementioned experimental results supported that dlo-miR408-3p negatively regulated DlNUDT23. This is also consistent with the finding that dlo-miR408-3p was upregulated, and DlNUDT23 was downregulated during longan early SE (Supplemental Fig. S5).
Figure 6.
The regulatory relationship of dlo-miR408-3p and DlNUDT23 in N. tabacum and longan EC. A) The N. tabacum leaf after infecting the DlMIR408-1301 and DlNUDT23-1301 recombination vectors. The scale bar represents 1 cm. B) The expression patterns of DlNUDT23, dlo-miR408-3p, and dlo-miR408-5p1 infecting with DlMIR408-1301 and DlNUDT23-1301 recombination vector in different groups of tobacco leaf. C) The expression patterns of DlNUDT23, dlo-miR408-3p, and dlo-miR408-5p1 in longan EC. Error bars indicate means ± SDs, n = 3 (Duncan’s post hoc test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
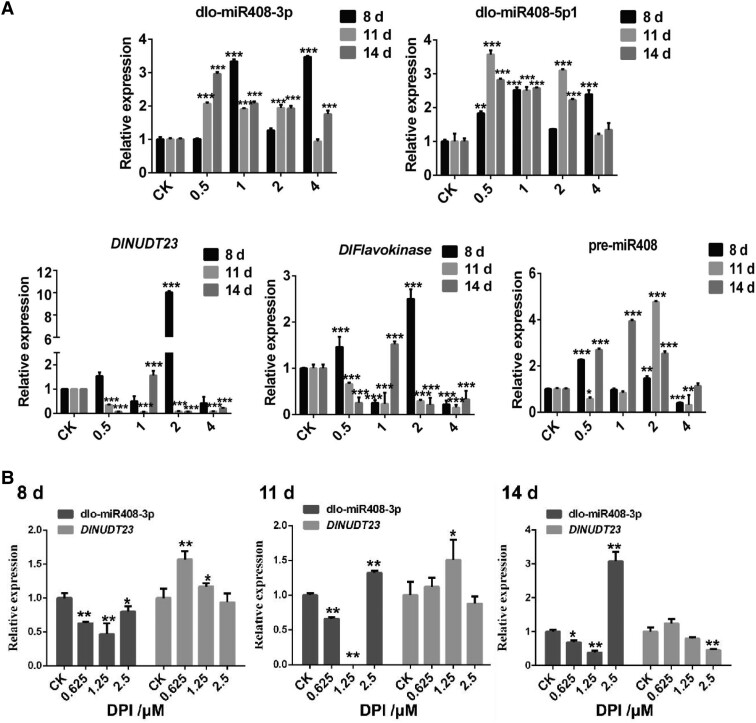
In addition, in vitro feeding assays with different concentrations of riboflavin and DPI were used to further investigate the expression patterns of miR408 and the target DlNUDT23. At 8 d (early GE stage), with 1.0 and 4.0 mM riboflavin supplementation, dlo-miR408-3p was significantly upregulated, and DlNUDT23 was significantly downregulated, and both of them exhibited a typical negative regulation model. This model also appeared during culture treatment for 11 and 14 d (middle and late stages of GE), with 0.5 to 4.0 mM riboflavin supplementation. The expression pattern of DlFlavokinase was the same as that of DlNUDT23. The expression pattern of dlo-miR408-5p1 was the same as that of dlo-miR408-3p during culture treatment for approximately 11 and 14 d with 0.5 to 4.0 mM riboflavin supplementation. However, there was no obvious rule about the expression pattern of pre-miR408 in different riboflavin treatments (Fig. 7A; Supplemental Table S7). By contrast, when supplemented with 0.625 and 1.25 μM DPI, dlo-miR408-3p was significantly downregulated, whereas DlNUDT23 was significantly upregulated during culture treatment for 8 to 14 d (Fig. 7B; Supplemental Table S7). The results supported our hypothesis that dlo-miR408-3p negatively regulated DlNUDT23 and responded to riboflavin metabolism.
Figure 7.
The expression patterns of dlo-miR408 and the target gene, DlNUDT23, in the treatment of differential concentration of riboflavin and DPI. A) The relative expression of dlo-miR408-3p, dlo-miR408-5p1, DlNUDT23, DlFlavokinase, and pre-miR408 in different concentrations of riboflavin treated on different days (8, 11, and 14 d). CK means control check, 0.5, 1, 2, and 4 mean different concentrations of riboflavin treatment, and the unit is mM. B) The relative expression of dlo-miR408-3p and DlNUDT23 in different concentrations of DPI treated on different days (8, 11, and 14 d), CK means control check, 0.625, 1.25, and 2.5 mean different concentrations of DPI treatment, and the unit is μM. Error bars indicate means ± SDs, n = 3 (Duncan’s post hoc test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
Together, these data further revealed that dlo-miR408-3p specifically targeted DlNUDT23 degradation, thereby affecting FMN accumulation and promoting riboflavin biosynthesis in longan.
Riboflavin promotes longan early SE
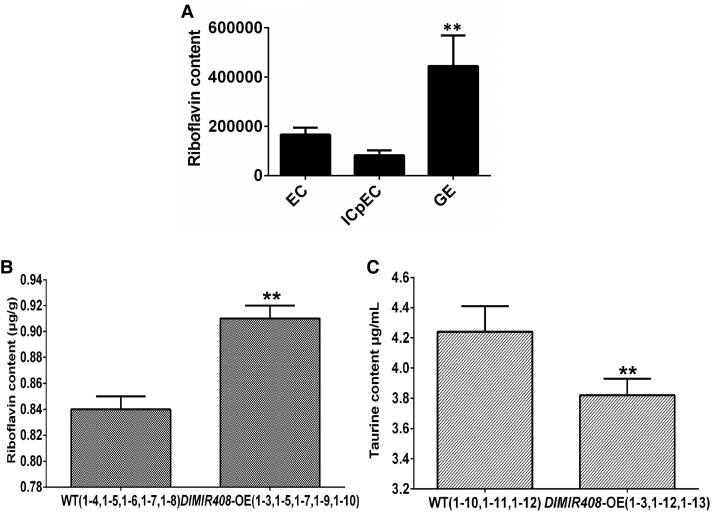
We further confirmed the hypothesis that riboflavin accumulation promotes longan early SE. First, the riboflavin content was significantly increased in metabolomics data of longan early SE. Moreover, compared with the WT, the riboflavin content was also significantly increased in DlMIR408-OE cell lines through HPLC determination (Fig. 8; Supplemental Table S8). The taurine content was significantly decreased, which was consistent with the transcriptome data (Fig. 8; Supplemental Table S8). The results of physiological content determination preliminarily supported our speculation, that is, DlMIR408-OE increased riboflavin accumulation and might promote early SE in longan.
Figure 8.
Riboflavin content and taurine content in longan early SE and DlMIR408-OE transgenic cell lines. A) The riboflavin content in metabolome during longan early SE, EC represents embryogenic callus, ICpEC represents incomplete compact pro-embryogenic cultures, GE represents globular embryo. The Y-axis represents the relative peak area. B) The riboflavin content in DlMIR408-OE (1-3, 1-5, 1-7, 1-9, and 1-10) and WT (1-4, 1-5, 1-6, 1-7, and 1-8), which were measured by HPLC. C) The taurine content in DlMIR408-OE (1-3, 1-12, and 1-13) and WT (1-10, 1-11, and 1-12), which were measured by HPLC. Error bars indicate means ± SDs, n = 3. *Significant difference (Duncan’s post hoc test, *P ≤ 0.05).
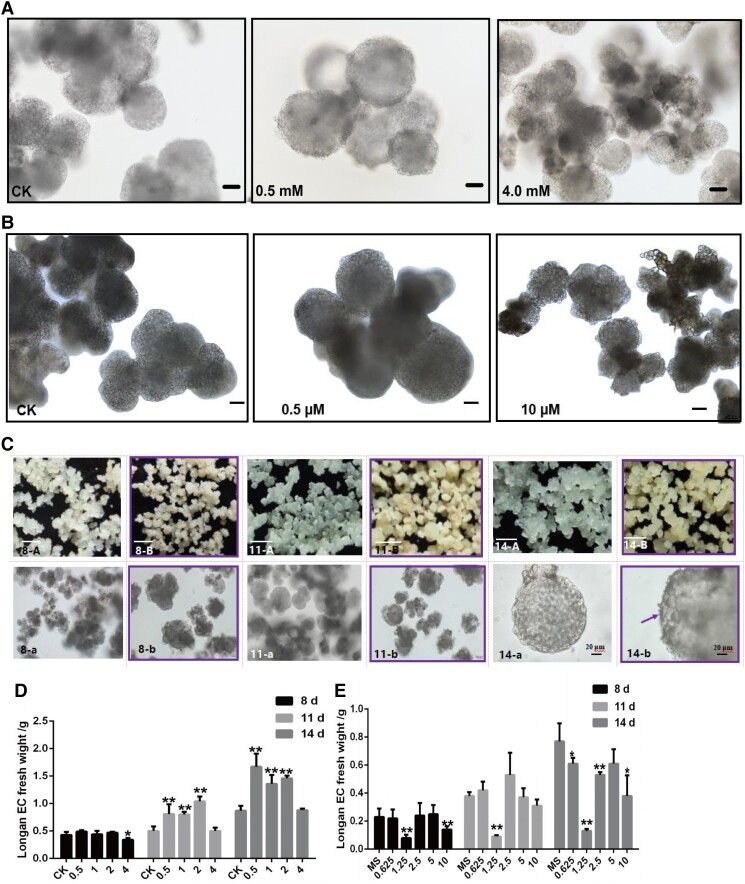
Next, we tested 3 in vitro feeding assays with different concentrations of riboflavin, FMN, and DPI in the dark. First, the results of cell morphology analysis revealed that during culture treatment for 14 d, in the presence of 0.5 mM riboflavin, the GE size was generally larger than that in the control check (CK) (0 mM). Nevertheless, with 4.0 mM riboflavin supplementation, the GE size was generally smaller than that in the CK (0 mM) (Fig. 9A). Cell morphology after treatment with other riboflavin concentrations is presented in Supplemental Fig. S6(a). The result of FMN treatment showed that 0.5 μM FMN resulted in larger GE than those in the CK, while 10 μM FMN inhibited GE differentiation (Fig. 9B). This was consistent with riboflavin-mediated changes in cell morphology during early SE and further supported the conclusion that riboflavin promoted longan early SE. Simultaneously, compared with CK, the fresh weight of longan GE was significantly increased with 0.5 to 2.0 mM riboflavin treatment (Fig. 9D; Supplemental Table S9). Therefore, these cell morphology data supported our idea that activated riboflavin promotes cell division and differentiation during longan early SE.
Figure 9.
Cell division and differentiation of longan EC after treating with different concentrations of riboflavin and DPI. A) The cell morphology of longan EC which is treated with CK (0), 0.5, and 4.0 mM riboflavin, culturing at 14 d, respectively, and the scale bars = 100 μm. B) The cell morphology of longan EC treated with CK (0), 0.5, and 10 μM FMN, culturing at 14 d, respectively, the scale bars = 100 μm. C) The morphology of callus and somatic cells which are treated with CK (0 μM) (A, a) and 1.25 μM (B, b) DPI, and culturing at 8 d (8-A, 8-a; 8-B, 8-b), 11 d (11-A, 11-a; 11-B, 11-b), and 14 d (14-A, 14-a; 14-B, 14-b), respectively. In 8-A, 8-B, 11-A, 11-B, 14-A and 14-B, the scale bars = 1 mm. In 8-a, 8-b, 11-a and 11-b, scale bars = 100 μm. In 14-a and 14-b, the scale bars = 20 μm. The purple frame indicates longan EC after 1.25 μM DPI treatment, the purple arrow indicates the surface of the protoderm in GE of longan. D) Longan EC fresh weight with different concentrations of riboflavin treatments, the unit is mM. E) The longan EC fresh weight of different concentrations of DPI treatments, the unit is μM. Error bars indicate means ± SDs, n = 3. Duncan’s post hoc test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
To determine cell division and differentiation during riboflavin deficiency, we supplemented with different DPI concentrations. With 1.25 μM DPI supplementation, GE was more yellow and compact, and exhibited obvious differentiation on different treatment days (Fig. 9C). Cell morphology after treatment with other DPI concentrations is presented in Supplemental Fig. S6(b). Interestingly, during the 14 d, rough material accumulated on the protoderm surface in GE (Fig. 9C). However, compared with CK, the fresh weight of longan GE was significantly decreased with 1.25 μM DPI treatment (Fig. 9E; Supplemental Table S9). This was contrary to the effect of riboflavin treatment on cell proliferation.
Taken together, the in vitro feeding assay with different concentrations of riboflavin, FMN, and DPI in the dark revealed that the low concentration of riboflavin (0.5 mM) promoted the expansion of GE and cell proliferation, whereas higher riboflavin concentration (4.0 mM) had no significant effect on cell proliferation and GE development. However, when the riboflavin biosynthesis pathway was inhibited after 1.25 μM DPI treatment, GE proliferation was inhibited, but differentiated normally, and some rough material appeared on the surface of protoepidermal cells in GE. This was contrary to the result that a low concentration of riboflavin promoted GE proliferation in longan. Based on these data, our study evidenced that riboflavin accumulation promoted cell division and differentiation during longan early SE through in vitro positive and negative validation tests.
DlMIR408-OE increases riboflavin content and promotes m6a modification
As aforementioned, in DlMIR408-OE cell lines, pre-miR408 and dlo-miR408-5p1/5p2 were significantly upregulated and dlo-miR408-3p was not significantly upregulated (Fig. 5A). In total, 61.1% of the 21-nt miRNA exhibited upregulated expression (Fig. 3). Moreover, dlo-miR408-3p negatively downregulated DlNUDT23 expression, inhibiting FMN biosynthesis. As illustrated, FMN can be used as an artificial demethylase to specifically oxidize N6-methyl-substituted adenosine and effectively downregulate m6A level with high sensitivity and selectivity (Xie et al. 2017; Xie et al. 2019). Simultaneously, the gene expression patterns of the m6A methylation pathway showed that the expression of “reader” genes (DlFIP37, DlECT2, and DlVIR) and “eraser” genes (DlALKBH2, DlALKBH6, and DlALKBH9B) was downregulated, whereas the expression of “writer” genes (DlMTA16, 21, 23) was upregulated (Supplemental Fig. S3). We further speculated that the pre-miR408 structure might be modified by m6A, and DlMIR408-OE decreased FMN content, then increased the m6A modification level, which in turn affected 21-nt miRNA homeostasis and the expression levels of miRNAs localized on different pre-miRNA arms. Further supporting this assumption, online prediction of the pre-miR408 structure showed that “AGACA” and “GAACA” m6A modification sites were present at the 5′-end arm of pre-miR408, in which “GAACA” happened to fall on dlo-miR408-5p, while no m6A modification site was observed on dlo-miR408-3p (Supplemental Fig. S7A). In addition, whole-transcriptome data analysis revealed that no m6A modification sites were present on 95% (20/21) of the 21-nt DE miRNA precursors, whereas the miRNA precursors that were not differentially expressed contained moderate or high-confidence m6A modification sites (Supplemental Fig. S7, C and D). Thus, we further proposed the hypothesis that miR408 overexpression negatively regulated DlNUDT23 to affect the level of FMN synthase-mediated m6A modification and thus affect the expression levels of miRNA and pre-miRNA processing.
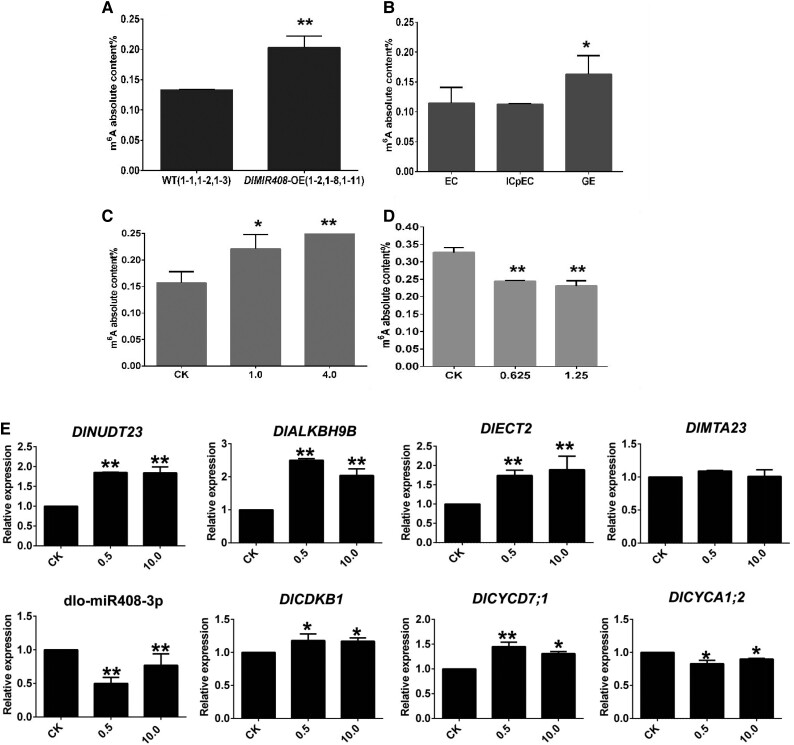
Subsequently, to further show the dlo-miR408-3p-DlNUDT23-FMN-m6a model regulated longan early SE, we performed m6A absolute quantification (Supplemental Table S10). The results showed that the m6A modification level increased in longan early SE and DlMIR408-OE cell lines (Fig. 10, A and B). Moreover, when treated for approximately 11 d, m6A absolute quantity increased with increasing riboflavin concentration (Fig. 10C). When riboflavin biosynthesis was inhibited, the m6A modification level decreased with increasing DPI concentration (Fig. 10D). To the contrary, with increasing concentration of FMN treated 14 d, dlo-miR408-3p was significantly downregulated and DlNUDT23 was upregulated. dlo-miR408-3p and DlNUDT23 also maintained a typical negative regulatory model. DlALKBH9B (Dlo017784) and DlECT2 (Dlo018976) were upregulated, while DlMTA23 had no significant difference among treatments (Fig. 10E). Cell cycle-related genes (DlCDKB1 and DlCYCD7;1) were upregulated, and DlCYCA1;2 was downregulated. These results demonstrated that when we promoted FMN biosynthesis, m6A modification level might be decreased. The cell cycle was also promoted by FMN (Supplemental Table S10). These results initially supported our hypothesis that DlMIR408-OE increased riboflavin, and promoted m6A accumulation and GE development in longan. dlo-miR408-3p downregulated DlNUDT23 expression, decreasing FMN accumulation, and increasing riboflavin content in the positive and negative in vitro feeding assays. Hence, our data supported the hypothesis that the dlo-miR408-3p-DlNUDT23-FMN-m6A model promoted somatic cell division and differentiation in longan.
Figure 10.
dlo-miR408-3p-DlNUDT23-FMN-m6A model regulated longan early SE. A) The m6A absolute content in DlMIR408-OE cell lines. B) The m6A absolute content during different stages of longan SE (EC, ICpEC, and GE). C) The m6A absolute content in different concentrations of riboflavin (CK(0), 1.0, and 4.0 mM) treated 11 d. D) The m6A absolute content in different concentrations of DPI (CK(0), 0.625, and 1.25 μM) treated 11 d. E) The expression patterns of dlo-miR408-3p, DlNUDT23, m6A pathway-related genes, and cell cycle-related genes during different concentrations of FMN (CK(0), 0.5, and 10.0 μM) treated 14 d, the unit is μM. Error bars indicate means ± SDs, n = 3. Duncan’s post hoc test, *P ≤ 0.05, **P ≤ 0.01.
Discussion
Plant regeneration highlights the outstanding totipotent potential of plant somatic cells without fertilization. The development of somatic embryos directly determined the fruit quality and quantity. miRNA responds to variations in the extracellular environment and plays a crucial role in plant SE. Although miR408 targets different genes to regulate different biological processes in plant development, the mechanism of miR408 remains elusive in plant SE. Moreover, this must be accompanied by nutrient regulation during SE. FMN and FAD are riboflavin co-factors and act as co-substrates for many epigenetic processes, thereby integrating nutrition, metabolism, and gene expression. Whether miRNA regulates riboflavin metabolism and early somatic embryo development remains largely unknown. Here, we provide genetic, physiological, and whole-transcriptome high-throughput sequencing evidence supporting the conclusion that dlo-miR408-3p targets a previously unidentified gene, DlNUDT23, activates riboflavin metabolism, mediates m6A modification by influencing miRNA and RNA homeostasis and cell cycle gene expression, and promotes cell division and differentiation, and finally promotes early SE of longan.
DlMIR408-OE activates riboflavin biosynthesis and promotes early SE in longan
Our data demonstrated that DlMIR408-OE activated riboflavin biosynthesis-related gene upregulation (DlPAP3, DlRIB5, DlRIBA1, and DlRIBA3), significantly downregulated the genes of DlPYRD, DlFHY, DlFlavokinase, and DlNUDT23, and reduced FMN synthesis. Riboflavin metabolism was enriched in KEGG (Fig. 4). This result indicated riboflavin metabolism played an essential role during longan early SE. This result is similar to previous studies in which riboflavin directly regulates the normal operation of the cell cycle, affects cell proliferation and differentiation by influencing changes in gene expression and protein abundance, and determines cell fate, lack of riboflavin leads to early embryo abortion (Hilary et al. 2012; Hesami et al. 2020). However, until now, only 1 study showed that the riboflavin transport gene (RFVT3) was targeted by miRNAs in posttranscriptional regulation in human intestinal epithelial cells (Lakhan et al. 2017). The specific molecular mechanism of miRNA-regulated riboflavin biosynthesis in plants remains unclear. Notably, our functional experiments evidenced that dlo-miR408-3p targeted DlNUDT23 and regulated riboflavin accumulation and promoted longan early SE. Unlike animals, riboflavin can be de novo produced through an ancestrally conserved pathway. The molecular regulation of riboflavin biosynthesis substantiates a key role in coordinating cellular energy and cell cycle for modulating endosperm development in seed plant (Tian et al. 2022). In our study, physiological content determination tests displayed that the riboflavin content was increased in GE and DlMIR408-OE. Simultaneously, riboflavin biosynthesis was also influenced by GPI-anchor biosynthesis and PPP pathway, and positively or negatively regulated by the mRNA–miRNA–lncRNA models (MSTRG 7391.1 (HSP40): DlNUDT23, MSTRG.4152.1: DlRIBA3, MSTRG.8602.1: DlPAP17, MSTRG.19600.1: DlPYRD) (Supplemental Table S4-3; Fig. 5D). Three in vitro feeding assays proved that dlo-miR408-3p targeted DlNUDT23 and regulated FMN synthesis and riboflavin accumulation (Supplemental Figure S7, A and B; Fig. 10E). Low concentration of riboflavin promoted cell proliferation and embryo enlargement of early GE in longan (Fig. 9, A and D). Low concentration of FMN upregulated cell cycle gene expression (DlCDKB1 and DlCYCD7;1), downregulated DlCYCA1;2, and promoted GE enlargement (Figs. 9B and 10E). Our previous study showed that DlCYCD7;1 upregulated during longan early SE (Zhao et al. 2022). In Arabidopsis, CYCD7;1 overexpression promoted overgrowth of the embryo and endosperm (Carl et al. 2012). However, in longan early SE, a high concentration of riboflavin inhibited cell proliferation and embryo differentiation. In contrast, when riboflavin biosynthesis inhibited (1.25 μM DPI treatment), cell proliferation was significantly inhibited but the differentiation of GE did not influence. This further implies that DlMIR408 activates riboflavin biosynthesis and promotes early SE by regulating cell cycle gene expression in longan. This mechanism was verified in plants using the longan SE model system and longan stable transgenic technology. Theoretical insights into the regulation of longan embryo development were gained in vivo by spraying riboflavin.
Riboflavin metabolism mediates m6a modification targeted through dlo-miR408-3p-mediated degradation of DlNUDT23
Studies have shown that miR408 targeted different genes in plants. Among them, the genes for plastocyanin, blue copper oxidase, and laccase were the most commonly reported. In rice, miR408 targeted VIN, UCL, and RNA pol II (Ma 2012; Zhang et al. 2017; Solanki et al. 2019). In wheat, miR408 targeted TOC1 (Zhao et al. 2016). While miR408 targeted 3-ketoacyl-CoA synthase 4 (lbKCS), plantacyanin (lbPCL), and galacturonosyltransferase 7-like (lbGAUT) in Pachyrhizus erosus (Kuo et al. 2019). Notably, dlo-miR408-3p negatively regulated DlNUDT23 and inhibited FMN synthesis in longan through predictive analysis of the whole transcriptome, in vitro feeding assays, and transient transformation assay. A further study revealed that riboflavin typically serves as a precursor of FMN and FAD for diverse metabolic processes, and generates cofactors (Kaelin and McKnight 2013). FMN acted as an artificial demethylase, a m6A-modified “eraser” to effectively downregulate m6A modification after absorbing blue light (Xie et al. 2017). The specific mechanism of this chemical modification is possibly that FMN acts similarly through the single-electron transport mechanism to naturally prevent RNA degradation. In the presence of light, FMN is excited and gets a proton and an electron from N6-methyl in m6A. FMN deprotonates amine cation and semi-quinone FMNH was obtained through the proton-coupled electron transfer pathway (Xie et al. 2019). In our study, dlo-miR408-3p negatively regulated DlNUDT23, reduced FMN synthesis, and enhanced the absolute content of m6A modifications, activating the cell cycle gene expression, and finally promoting early SE as determined through the in vitro positive and negative feeding assays and in vivo transient transformation assays (Figs. 10 and 11).
Figure 11.
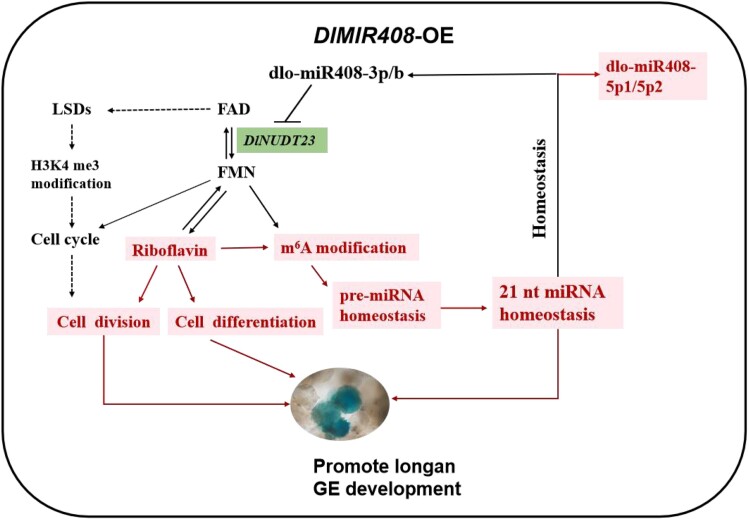
The schematic diagram of the molecular mechanism of dlo-miR408 that regulates riboflavin metabolism and mediates m6A modification. The solid line represents the empirical regulatory relationship. The dotted line represents the regulatory relationship, which was supported by previous evidence (Tian et al. 2022). The red color represents upregulation. The solid lines represent the regulatory relationships verified in this study. GE, globular embryo.
The m6A methylation level is directly determined by the expression of the m6A methylation pathway-related genes (Yue et al. 2019; Zheng et al. 2020). Riboflavin generates FAD, which also acts as a co-substrate for many epigenetic processes, thereby integrating nutrition, metabolism, and gene expression (Shi et al. 2004). In our study, the m6A methylation pathway-related genes differentially expressed in the DlMIR408-OE cell lines (Supplemental Fig. S3). During FMN treatment, DlNUDT23, DlALKBH9B, and DlECT2 were upregulated, dlo-miR408-3p was downregulated, cell cycle-related genes (DlCDKB1 and DlCYCD7;1) were upregulated, and DlCYCA1;2 was downregulated (Fig. 10). This demonstrated that FMN also influenced the expression of m6A pathway genes and decreased the m6A level in longan early SE, and regulated cell division and differentiation by affecting the cell cycle. In addition, FAD acts as a prosthetic group of LSDs and activates H3K4me3 methylation modification, and DNA methylation modifications to affect the expression of cell cycle-related genes and gene transcriptional repression (Venkatachalam et al. 2008; Tian et al. 2022). Studies have shown that double-strand breaks mostly occur at preferred sites, which were marked by H3K4me3 (Chen et al. 2020b). In our study, the whole-transcriptome analysis of DlMIR408-OE cell lines revealed that genes associated with the TCA cycle, RNA metabolism, flavin-binding enzymes, and cryptochrome blue receptor were significantly upregulated (Supplemental Table S4-1). The DEAD-box family members, CNGC family, and most miRNA biosynthesis-related genes were downregulated. Hence, we speculated that there might be another mechanism, and riboflavin metabolism might affect H3K4me3 methylation, thus regulating the cell cycle and influencing early embryonic development of longan (Fig. 11). We here revealed that the dlo-miR408-3p-DlNUDT23-FMN model may act as an artificial demethylation mechanism for regulating m6A methylation modification during longan SE. Thus, the scientific question of the mechanism of dlo-miR408-3p that promotes cell proliferation and differentiation during early SE has been addressed.
DlMIR408-OE influences homeostasis of RNA metabolism during longan early SE
Many factors affect the homeostasis of RNA metabolism, including miRNA processing, RNA degradation in transcription and posttranscriptional regulation, lncRNA regulation, m6A modification, m7G capping modification, and NAD+ capping modification. In our study, DlMIR408-OE substantially promoted the expression of 61.1% 21-nt miRNA (Fig. 3). Subsequently, DE mRNA annotation analysis was mainly related to RNA metabolism (Supplemental Table S4-1), such as mRNA splicing, processing and transport, miRNA synthesis, and DlCRY2 and m6A methylation (Supplemental Table S4-2; Supplemental Fig. S3). Moreover, m6A modification sites were predicted on the pre-miR408 structure and known targets (DlLAC12), and exhibited no sites on the DE miRNA precursor (Supplemental Fig. S7, A and B). Thus, we speculated that DlMIR408-OE promotes m6A methylation and affected RNA homeostasis during longan SE. A study noted that the CRY2 receptor affected the m6A encoder complex activity under blue light and regulated the circadian rhythm of plants (Wang et al. 2021). DlCRY2 was significantly upregulated in the DlMIR408-OE cell lines (Supplemental Table S4-1). These results suggested that FMN might also absorb blue light to regulate an increase in the m6A methylation modification level, or the DlCRY2 receptor might directly affect the m6A encoder activity to affect m6A modification during longan early SE.
In addition, the m7G cap protects RNA from degradation, maintains transcript stability, recruits related proteins, and ultimately affects the spatial and temporal distribution of genes and proteins (Malbec et al. 2019). In embryonic stem (ES) cells of mammals, m7G-modified miRNAs tend to form G-quadruplexes, which affected G-quadruplexes complex formation and promoted pre-miRNA processing. Mettl1/WDR4-mediated m7G tRNA methylation was conducive to normal mRNA translation and self-renewal and differentiation of the ES cells (Lin et al. 2018). In our study, the m7G cap structure, RNA cap, and magnesium ion transmembrane transport activity were significantly enriched (Supplemental Fig. S4A). This implies that DlMIR408-OE may regulate m7G modification and influence RNA or miRNA homeostasis. Moreover, NAD+ capping modification exists during eukaryotic RNA 5′ modification. This is similar to the m7G capping process and widely exists in animals and plants. After transcription initiation, NAD+ reduces NAD+ (NADH) and dpCoA caps are added to RNA. This capping mainly maintains RNA stability. Nudix or DXO hydrolyzes the diphosphate bond to remove the NAD cap and produce nicotinamide mononucleotide and 5′-phosphate ribonucleic acid. In mammals and bacteria, Nudt12 removes NAD capping in vitro so that, RNA decay accelerated, which is contrary to the effect of m7G caping modification. This mechanism is relatively conserved in animals, plants, and microorganisms (Grudzien-Nogalska et al. 2019). In Arabidopsis, NAD tagSeq revealed that NAD+ cap RNA was mainly produced by numerous protein-coding genes and particularly gathered at specific regulatory small RNAs (sRNAs) and some mRNA 5′ fragments (Cahova et al. 2015; Zhang et al. 2019; Hu et al. 2021). Consistently, in our study, previously unknown miRNA target genes were mainly enriched in the oxidation–reduction reaction involving NAD binding and the NAD or NADP receptor (Supplemental Fig. S4B). Meanwhile, dlo-miR408-3p was proved to target DlNUDT23. Thus, we preliminarily speculated that DlMIR408 may be involved in the 5′ NAD+ capping modification during RNA processing and may regulate the homeostasis of RNA metabolism in the longan early SE. Thus, we propose that DlMIR408-OE not only mediates m6A methylation modification, but also may affect m7G and NAD+ capping as well as miRNA and RNA processing to maintain the RNA homeostasis.
Taken together, our data indicate that overexpression DlMIR408 activated riboflavin biosynthesis, reduced FMN accumulation, mediated m6A modification, and promoted early SE in longan. Riboflavin accumulation promoted cell division and differentiation during longan early SE via the model of dlo-miR408-3p-DlNUDT23-FMN and regulating cell cycle gene expression. 21-nt miRNA and RNA homeostasis might be influenced through overexpressing DlMIR408 mediating m6A modification, m7G modification, and NAD+ capping. Schematic diagram of the molecular mechanism of dlo-miR408 that regulates riboflavin metabolism and mediates m6A modification in Fig. 11.
Conclusions
Our work revealed a previously unexplored molecular mechanism of dlo-miR408: it regulates riboflavin biosynthesis and dynamically mediates m6A modification, affecting RNA homeostasis in longan early SE. This molecular mechanism provides insights regarding riboflavin biosynthesis and m6A modification targeted by miRNA, which regulate cell division and cell differentiation in plants. In this study, the DlMIR408-OE cell lines were constructed through stable genetic transformation of longan EC, and this approach was combined with the whole-transcriptome high-throughput sequencing. We speculated that dlo-miR408 targeted DlNUDT23, activated riboflavin biosynthesis, and influenced miRNA and RNA homeostasis. This was confirmed by the in vitro positive or negative feeding assay and the in vivo transient transformation assay. Moreover, we showed that riboflavin homeostasis mediated the m6A modification level, and influenced cell division and cell differentiation during longan early SE through physiological content determination and cell morphology and growth observation after the feeding assay. In addition, we revealed that the model of DlNUDT23-miR408-3p-MSTRG 7391.1 participates in riboflavin biosynthesis. All the results identified here provide the basis for the development of testable hypotheses and experimental approaches. Our work provided insights into the molecular mechanism of miR408 in longan early SE and showed that miRNA regulates riboflavin synthesis and influences plant SE.
Materials and methods
Plant materials
EC of longan (D. longan Lour.) “Honghezi” were used as the genetically transformed receptor cell line. The EC were induced using a ZE (2 to 3 mm) (Lai et al. 1997; Fang 2012). Then, the EC were cultured with MS medium supplemented with 1.0 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D) and cultured for 10 to 15 d at 25 °C. The WT EC had no vector infection. The pCAMBIA1301:GUS (1301) empty vector was modified by Feng (2016). The EHA105 Agrobacterium competent was supplied by Weidi company (Shanghai, China). Kanamycin (Kan) was purchased from Luoen. Acetosyringone (AS), rifampicin, and cefotaxime sodium salt were purchased from Thermo and YeaSen (Fuzhou, China).
Construction and transformation of vectors
The DlMIR408 genomic DNA sequence was PCR amplified from the EC, and the sequence length was 475 bp. Using PstI and SalI, the PCR product of DlMIR408 was cloned into the multiple cloning sites of the 35S-pCAMBIA1301:GUS vector in T4 ligase assay. The vectors were then mobilized into Agrobacterium EHA105 through freeze-thawing with liquid nitrogen. A. tumefaciens was used to transform the longan EC (refer to Xu 2010). Normally, longan EC were subcultured for 20 d as 1 generation (Lai et al. 1997). The longan calli with Agrobacterium were co-cultured for approximately 6 to 7 d, and this was referred to as subcultured generation 0 (S0), and GUS staining was performed using a GUS staining kit (Huayueyang, China). The DlMIR408-OE cell line was selected on MS medium containing no antibiotic by using the physical separation inoculation method. Subcultured generation one (S1) and subcultured generation two (S2) were obtained. S0 was used for analyzing transient gene expression, subcultured generation three (S3) was used for GUS gene and pri-miR408 gDNA PCR amplification, S2 and S6 were used for phenotypic characterization, and S6 was used for the whole-transcriptome high-throughput sequencing.
Whole-transcriptome sequencing, assembly, and analysis
LncRNA and sRNA libraries were constructed from the DlMIR408-OE cell line and WT and were sequenced using Illumina Novaseq-6000 PE150 by Gene Denovo Biotechnology Co. (Guangzhou, China). Agilent 2100 and ABI StepOnePlus Real-Time PCR System (Life Technologies) were used to detect the quality and production, respectively. The 2 libraries were constructed as described by Lyu et al. (2020) and Jiang et al. (2022). Next, hisat2 was used to align the libraries with the longan third-generation genome (NCBI: Bio project PRJNA792504). Stringtie was used to reconstruct the transcripts and obtain known and previously unknown transcripts. Low-quality reads and sequences containing adapters and poly-N tails were removed to obtain clean data. The unmapped reads of previously unidentified predicted lncRNAs in the ribosome database and high-quality sRNA clean reads were used for the transcriptome analysis.
To evaluate gene expression levels, transcript abundances were calculated and normalized to Fragments Per Kilobase of exon model per Million mapped fragments (FPKM). The false discovery rate (FDR) < 0.05 and |log2FC| > 1 indicated significant DE of lncRNAs and mRNAs. The read count was standardized using edgeR (Robinson et al. 2010). The FPKMs of lncRNAs and mRNAs were analyzed after correcting deep sequencing and transcript length. The known miRNAs were identified through blasting in the miRBase database of plants. miRNAs and pre-miRNAs were predicted using miRPara of SVM (version 6.3). The miRNAs were predicted and identified with hairpin structures, and the classes and abundance of tags were counted. The miRNA family was further analyzed. The identified miRNAs were used to determine the tag per million (TPM), TPM = T*10^6/N (T represents miRNA tags and N is the total miRNA tags). The dispersion was set up as 0.01 by edgeR (Robinson et al. 2010), the selection criterion of DE miRNA was the expression level change of more than 2 times, and P < 0.05.
The DE mRNAs, miRNAs, and lncRNAs were then subjected to enrichment analysis of GO and KEGG pathways. The mRNA–miRNA–lncRNA model was constructed in DlMIR408-OE cell lines. The FDR < 0.05 and |log2FC| > 1 were used as the difference threshold for significant DE of mRNA and lncRNA. P < 0.05 and |log2FC| > 1 were used as the difference threshold for significant DE miRNA. The principle was followed (Salmena et al. 2011). The bubble diagrams were used to analyze the top 20 pathways of GO and KEGG enrichment for mRNAs, miRNA targets, and lncRNA targets. The subcultured generation nine (S9) DlMIR408-OE transgenic cell lines and RT-qPCR were used to validate the expression patterns of the mRNA–miRNA–lncRNA model.
Physiological determination
The DlMIR408-OE cell lines (1-3, 1-5, 1-7, 1-9, and 1-10) and WT cell lines (1-4, 1-5, 1-6, 1-7, and 1-8) were used as materials to extract riboflavin. The riboflavin content was measured according to Pan et al. (2001) using HPLC, and the parameters for this determination are shown in Supplemental Table S11. The taurine content was measured according to Zhang et al. (2009) in the DlMIR408-OE (1-3, 1-12, and 1- 13) and WT (1-10, 1-11, and 1-12) cell lines. The HPLC parameters used for measuring taurine content are listed in Supplemental Table S11. The global m6A methylation ratio was measured. First, SRAMP (http://www.cuilab.cn/sramp) was used to predict m6A modification of the pri-miR408 sequence and differentially expressed pre-miRNAs in the whole-transcriptome database of the DlMIR408-OE cell lines. Second, total RNA was extracted using the TransZol Up reagent (TransGen Biotech) from longan early SE (embryonic callus (EC), incomplete compact pro-embryogenic cultures (ICpEC), compact pro-embryogenic cultures globular embryos (CpECGE), GE) (Lai 2003) and from DlMIR408-OE cell lines (1-2 and 1-8) and WT lines (1-1 and 1-2) that had been subjected to riboflavin and DPI treatments. According to the manufacturer's instructions, the colorimetric method was used to detect the global m6A methylation level by using the EpiQuik m6A RNA Methylation Quantification Kit (Epigentek, Farmingdale, NY, USA).
A. tumefaciens-mediated transient transformation
Sequence information of DlNUDT23 was retrieved from the longan third-generation genome. NcoI and BglII were selected as the double restriction enzymes. DlNUDT23 gDNA was PCR amplified from EC, ligated downstream of the CMV 35S promoter in pCAMBIA1301, confirmed through sequencing, and cloned into the multiple cloning sites of the 35S-pCAMBIA1301:GUS vector in the T4 ligase assay. The vector was imported into Agrobacterium EHA105 through freeze-thawing with liquid nitrogen. The recombinant Agrobacterium of DlNUDT23-1301 and DlMIR408-1301 was used to transform N. benthamiana leaves using A. tumefaciens (refer to Wang 2013). For transformation in N. benthamiana, Agrobacterium was used as the negative control and 1301-GUS was used as the positive control. The DlMIR408-1301 plasmid was injected into the whole leaf and into one-half of a leaf, with the other half of the leaf left uninfected. The DlNUDT23-1301 plasmid was injected into the whole leaf and into one-half of a leaf, with the other half of the leaf left uninfected. DlMIR408-1301 + DlNUDT23-1301 was injected into the whole leaf simultaneously. For transformation in longan EC, Agrobacterium was used as a negative control and 1301-GUS as the positive control. Then, the calli were simultaneously infected with DlMIR408-1301, DlNUDT23-1301, and DlMIR408-1301 + DlNUDT23-1301.
In vitro feeding assays
Riboflavin and DPI with a purity of >98% were purchased from Baiquan Biotechnology Co. (Fuzhou, China). The riboflavin concentration was set at 0 (CK), 0.5, 1.0, 2.0, and 4.0 mM (refer to Guo et al. 2017; Jiang et al. 2021b). The FMN concentration was set at 0 (CK), 0.5, and 10 μM (refer to Xie et al. 2019). The DPI concentrations were set at 0 (CK), 0.625, 1.25, 2.5, 5, and 10 μM (refer to Chae and Lee 2001 ; Kanamori et al. 2012). In the assay, deionized water was used to dissolve riboflavin and DPI powder and to prepare 0.1 and 1 mM stock solutions, respectively. MS was used as the basic medium. Longan EC were cultured in the medium supplied with 1.0 mg L−1 2, 4-D for 20 d, and inoculated to the culture medium supplied with different concentrations of riboflavin treatment on 90 mm disposable petri dishes. Every plate was inoculated with 4 clumps of EC of uniform size exhibiting vigorous growth. After 8, 11, and 14 d, cell proliferation was measured on an electronic balance, and cell morphology of EC was observed under the microscope (Olympus, Japan). The samples were collected and quickly frozen in liquid nitrogen and stored at −80 °C in a refrigerator for RNA extraction and RT-qPCR analysis.
RNA isolation and RT-qPCR
Total RNA was extracted from the DlMIR408-OE and WT cell lines by using the TransZol Up reagent (TransGen Biotech) according to the manufacturer's protocol. Reverse transcription was performed using the HifairTM II 1st Strand cDNA Synthesis SuperMix (Yeasen, Shanghai, China) for gene RT-qPCR, and TransScriptGreen miRNA Two-Step qRT-PCR SuperMix (TransGen Biotech, Fuzhou, China) and universal miRNA RT-qPCR primer were used for miRNA RT-qPCR. Specific primers were designed for DlMIR408, DlLAC12, and DlLAC5. RT-qPCR was performed using the Hieff UNICON qPCR SYBR Green Master Mix (Yeasen, Shanghai, China) in a 96-well plate on a LightCycler 480 instrument (Roche Applied Science, Switzerland). Each reaction was performed in triplicate. For each gene, the RT-qPCR system consisted of 10 μL of SYBR, 2 μL of cDNA template, 0.8 μL primer, and 6.4 μL of ddH2O. Each miRNA RT-qPCR system consisted of 10 μL of 2×TransStarTop/Tip Green qPCR mix, 2 μL of cDNA template, 0.4 μL primer, and 7.2 μL of ddH2O. The RT-qPCR procedure was as follows: initial denaturation for 30 s at 95 °C, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s, and extension at 72 °C for 15 s. DlElf-4a, DlFSD, and DlEF-1a were used as the single internal controls for gene relative expression (Lin and Lai 2010). 5.8S and U6 were used as the single internal controls for miRNA relative expression. The 2−△△CT method was used to calculate the relative expression (Lin and Lai 2013a; Lin and Lai 2013b). All primers used in this research are listed in Supplemental Table S12.
Statistical analysis
Statistical analyses were conducted using IBM SPSS 19.0 statistical software. One-way ANOVA was performed followed by Duncan's test. Error bars show means ± SD of 3 replicates (n = 3). Asterisks indicate a significant difference (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
Accession numbers
The whole-transcriptome sequencing raw and processed sequencing data of WT and DlMIR408-OE cell line generated in this study have been submitted to the NCBI SRA (https://dataview.ncbi.nlm.nih.gov/object/PRJNA894314?reviewer=jnl7npj065er4hd8sqn2m5uk7m), under accession number: PRJNA894314. The sRNA raw data and processed sequencing data of WT and DlMIR408-OE have been submitted to NCBI SRA (https://dataview.ncbi.nlm.nih.gov/object/PRJNA895752?reviewer=pa8n87rfgvn0jlgoc8mscg97s0), under accession number: PRJNA895752.
Supplementary Material
Acknowledgments
We would like to thank all the research workers at the Institute of Horticultural Biotechnology for their contribution to our study. We also thank MJEditor (www.mjeditor.com) and Elsevier's Language Editing services for their linguistic assistance during the preparation of this manuscript.
Contributor Information
Xiaoping Xu, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China; Institute of Biotechnology, Fujian Academy of Agricultural Sciences, Fuzhou, Fujian 350003, China.
Chunyu Zhang, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Xiaoqiong Xu, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Roudi Cai, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Qingxu Guan, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Xiaohui Chen, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Yukun Chen, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Zihao Zhang, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Xu XuHan, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China; Institut de la Recherche Interdisciplinaire de Toulouse, IRIT-ARI, 31300 Toulouse, France.
Yuling Lin, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Zhongxiong Lai, Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China.
Author contributions
Z.X.L., Y.L.L., and X.P.X. designed and coordinated the work. X.P.X. constructed transgenic cell lines, analyzed whole-transcriptome data, performed functional verification experiment, and wrote and revised the manuscript. C.Y.Z. and X.Q.X. performed FMN feeding assay and RT-qPCR. C.Y.Z., X.Q.X., R.D.C., Q.X.G., X.H.C., Y.K.C., and Z.H.Z. prepared the materials. Y.L.L., X.X.H., and Z.X.L. advised the experiment design, data analyzed, and revised the manuscript. All authors read and approved the final manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. 1301: DlMIR408: GUS recombinant vector constructed and overexpression DlMIR408 in longan embryogenic callus.
Supplemental Figure S2. The types and numbers of DE transcriptional factors (TF) in the longan DlMIR408-OE cell lines compared with WT cell lines.
Supplemental Figure S3. The differential expression of mRNA in DlMIR408-OE cell lines.
Supplemental Figure S4. The GO and KEGG functional annotation of the mRNA, known and novel microRNA (miRNA) target genes, antisense-lncRNA target gene, cis-lncRNA target gene, and cis-lncRNA and antisense-lncRNA in WT and DlMIR408-OE of longan.
Supplemental Figure S5. The relative expression and FPKM of dlo-miR408 and DlNUDT23 during longan early SE.
Supplemental Figure S6. Cell morphology of longan embryogenic callus (EC) treated with different concentrations of riboflavin and riboflavin inhibitor (DPI).
Supplemental Figure S7. The prediction of m6A modification sites on the stem-loop structure of pre-miR408, DlLAC12 mRNA, and the precursor miRNA (pre-miRNA) structure of differential expression and no differential expression microRNA (miRNA).
Supplemental Table S1. The whole-transcriptional sequencing data analysis in DlMIR408-OE cell lines.
Supplemental Table S2. miRNA transcriptome data analysis in DlMIR408-OE cell lines of longan.
Supplemental Table S3. The differential expression analysis of 21 known miRNAs and 17 previously unknown miRNAs. x represents the 5p, y represents 3p, and z represents not only maps to 5p but also maps to 3p after comparison with miRNA in other species.
Supplemental Table S4. The specific upregulated and downregulated DE mRNA (|log2(fc)|≥10) in DlMIR408-OE cell lines, and the gene expression of mRNA–miRNA–lncRNA model in DlMIR408-OE cell line by the whole-transcriptome analysis.
Supplemental Table S5. The top 20 enrichment of GO and KEGG analysis in mRNAs, miRNAs, and lncRNAs transcriptome.
Supplemental Table S6. The expression pattern of miR408 family members and riboflavin pathway-related genes by RT-qPCR in DlMIR408-OE cell lines.
Supplemental Table S7. The relative expression of miR408 family and DlNUDT23 and DlFlavokinase in Nicotiana benthamiana and longan EC transient system, and different concentrations of riboflavin and DPI treatment.
Supplemental Table S8. Content of riboflavin and taurine in longan early SE and transgenic DlMIR408-OE cell lines.
Supplemental Table S9. The cell fresh weight during different concentrations of riboflavin and DPI treatment on different days.
Supplemental Table S10. m6A content detection in different early SE, DlMIR408-OE, and different concentrations of riboflavin and DPI treatment and the relative expression of dlo-miR408-3p, DlNUDT23, RNA methylation, and cell cycle-related genes during different concentrations of FMN treatment.
Supplemental Table S11. The liquid condition of High-Performance Liquid Chromatography (HPLC).
Supplemental Table S12. All the primers used in this study.
Funding
This work was funded by the National Natural Science Foundation of China (31572088), funding for Plateau Discipline Construction in Fujian Province (102/71201801101), special fund for scientific and technological innovation of Fujian Agricultural and Forestry University Applied Basic Research (KF2015108, CXZX2017189, CXZX2018076).
Data availability
All data are provided along with the manuscript and its supplemental material.
References
- Bacher A, Eberhardt S, Fischer M, Kis K, Richter G. Biosynthesis of vitamin b2 (riboflavin). Annu Rev Nutr. 1999:20(1):153. 10.1146/annurev.nutr.20.1.153 [DOI] [PubMed] [Google Scholar]
- Bhat SS, Bielewicza D, Gulanicz T, Bodi Z, Yu X, Anderson SJ, Szewc L, Bajczyk M, Dolata J, Grzelak N, et al. mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. PNAS. 2020:117(35):21785–21795. 10.1073/pnas.2003733117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahova H, Winz ML, Höfer K, Nübel G, Jäschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015:519(7543):374–377. 10.1038/nature14020 [DOI] [PubMed] [Google Scholar]
- Carl C, Walter D, James AHM. D-type cyclins control cell division and developmental rate during Arabidopsis seed development. J Exp Bot. 2012:63(10):3571–3586. 10.1093/jxb/ers015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Lee WS. Ethylene-and enzyme-mediated superoxide production and cell death in carrot cells grown under carbon starvation. Plant Cell Rep. 2001:20(3):256–261. 10.1007/s002990000307 [DOI] [Google Scholar]
- Chen YK, Lin XY, Lai ZX. Advances in somatic embryogenesis of longan. J Trop Crops. 2020a:41(10):1990–2002. 10.3969/j.issn.1000-2561 [DOI] [Google Scholar]
- Chen Y, Lyu R, Rong B, Zheng Y, Lin Z, Dai R, Zhang X, Xie N, Wang S, Tang F, et al. Refined spatial temporal epigenomic profiling reveals intrinsic connection between PRDM9-mediated H3K4me3 and the fate of double-stranded breaks. Cell Res. 2020b:30(3):256–268. 10.1038/s41422-020-0281-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZZ. Studies on proteomics during the middle stage of somatic embryogenesis and the expression and regulation of Ran family genes during somatic embryogenesis in Dimocarpus longan Lour. [PhD thesis]. Fuzhou: Fujian Agriculture and Forest University;2012.
- Feng X. Genome-wide identification and function analysis of the superoxide dismutase gene family in Musa spp. [PhD thesis]. Fuzhou: Fujian Agriculture and Forest University;2016.
- Forneris F, Binda C, Dall’Aglio A, Fraaije MW, Battaglioli E, Mattevi A. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J Biol Chem. 2006:281(46):35289–35295. 10.1074/jbc.M607411200 [DOI] [PubMed] [Google Scholar]
- Gao Y, Feng BH, Gao CX, Zhang HQ, Wen FT, Tao LX, Fu GF, Xiong J. The evolution and functional roles of miR408 and its targets in plants. Int J Mol Sci. 2022:23(1):530. 10.3390/ijms23010530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg BR, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994:266(5185):605–614. 10.1126/science.266.5185.605 [DOI] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, Wu YX, Jiao XF, Cui HJ, Mateyak MK, Hart RP, Tong L, Kiledjian M. Structural and mechanistic basis of the mammalian Nudt12 RNA deNADding. Nat Chem Biol. 2019:15(6):575–582. 10.1038/s41589-019-0293-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LL, Xi FF YUYH, Wang ZG, Zhang GH, Guo DL. Studies of the riboflavin treatment for promoting the early ripening of “Kyoho” grape berry. Acta Hortic. 2017:44(10):1861–1870. 10.16420/j.issn.0513-353x [DOI] [Google Scholar]
- Hao C, Yang Y, Du J, Deng XW, Li L. The PCY-SAG14 phytocyanin module regulated by PIFs and miR408 promotes dark-induced leaf senescence in Arabidopsis. PNAS. 2022:119(3):e2116623119. 10.1073/pnas.2116623119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain G, Frelin O, Roje S, Ellens KW, Ali K, Guan JC, Garrett T J, Crécy-Lagard V, Gregory JF 3rd, McCarty DR, et al. Identification and characterization of the missing pyrimidine reductase in the plant riboflavin biosynthesis pathway. Plant Physiol. 2013:161(1):48–56. 10.1104/pp.112.208488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesami M, Naderi R, Tohidfar M. Introducing a hybrid artificial intelligence method for high-throughput modeling and optimizing plant tissue culture processes: the establishment of a new embryogenesis medium for chrysanthemum, as a case study. Appl Microbiol Biot. 2020:104(23):10249–10263. 10.1007/s00253-020-10978-1 [DOI] [PubMed] [Google Scholar]
- Hilary JP, Corfe B M, Nakano E. Riboflavin in development and cell fate. Subcell Biochem. 2012:56(12):229–245. 10.1007/978-94-007-2199-9_12 [DOI] [PubMed] [Google Scholar]
- Hu H, Flynn N, Zhang HL, You CJ, Hang RL, Wang XF, Zhong H, Chan ZL, Xia YJ, Chen XM. SPAAC-NAD-seq, a sensitive and accurate method to profile NAD+-capped transcripts. PNAS. 2021:118(13):e2025595118. 10.1073/pnas.2025595118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang AL, Guo ZL, Pan JW, Yang YZ, Zhuang Y, Zuo DQ, Hao C, Gao ZX, Xin PY, Chu JF, et al. The PIF1-MIR408-plantacyanin repression cascade regulates light dependent seed germination. Plant Cell. 2021a:33(5):1506–1529. 10.1093/plcell/koab060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CN, Li CY, Li YH, Kou CC, Ge YH. Effects of riboflavin treatment on blue mould and reactive oxygen species and phenylpropanoid metabolisms in postharvest apples. Storage Process. 2021b:21(3):1–7. 10.3969/j.issn.1009-6221.2021.03.001 [DOI] [Google Scholar]
- Jiang YN, Shen XY, Dong CR, Zhi FN, Gao Y, Shi CP, Chao YQ, Xu JC, Shang DS, Xu J, et al. The whole transcriptome analysis and the circRNA-lncRNA network construction in arsenic trioxide-treated mice myocardium. Biomed Pharmacother. 2022:151:113183. 10.1016/j.biopha.2022.113183 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013:153(1):56–69. 10.1016/j.cell.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori M, Mizuta H, Yasui H. Effects of ambient calcium concentration on morphological form of callus-like cells in Saccharina japonica (Phaeophyceae) sporophyte. J Appl Phycol. 2012:24(4):701–706. 10.1007/s10811-011-9688-x [DOI] [Google Scholar]
- Katarzyna S, Dawid B, Jakub D, Wójcik AM, Nowak K, Szczygieł-Sommer A, Szweykowska-Kulinska Z, Jarmolowski A. MicroRNAs are intensively regulated during induction of somatic embryogenesis in Arabidopsis. Front. Plant Sci. 2017:8(18):1–16. 10.3389/fpls.2017.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D. What don’t we know? Science. 2005:309(5731):75. 10.1126/science.309.5731.75 [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Grossniklaus U. A developmental perspective. Annu Rev Plant Biol. 2003:54(1):547–574. 10.1146/annurev.arplant.54.110901.160842 [DOI] [PubMed] [Google Scholar]
- Kuo YW, Shane LJ, Li YC, Jhu MY, King YC, Jeng ST. Micror408 regulates defense response upon wounding in sweet potato. J Exp Bot. 2019:70(2):469–483. 10.1093/jxb/ery381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZX. Research on Biotechnology of longan. 1st ed. Fuzhou: Fujian Science & Technology Publishing House; 2003. [Google Scholar]
- Lai ZX, Chen CL, Zeng LH, Chen ZG. Somatic embryogenesis in longan [Dimocarpus longan Lour.](Chapter 13). In: Jain SM, Gupta PK, Newton RJ., editors Somatic embryogenesis in woody plants (Forestry Sciences, Volume 67). Netherlands: Springer; 2000. p. 415–431. [Google Scholar]
- Lai ZX, Pan LZ, Chen ZG. Establishment and maintenance of longan embryogenic cell lines. J Fujian Agric Univ. 1997:2(26):33–40. http://www.cnki.com.cn/Article/CJFDTotal-FJND702.006.htm [Google Scholar]
- Lakhan R, Subramanian VS, Said HM. Role of MicroRNA-423-5p in posttranscriptional regulation of the intestinal riboflavin transporter-3. Am J Physiol Gastrointest Liver Physiol. 2017:313(6):589–598. 10.1152/ajpgi.00238.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MF, Justyna WM, Iris H, Anneke H, Chen BJ, Ricardo R, Gerco CA, Kim B. Auxin biosynthesis maintains embryo identity and growth during BABY BOOM-induced somatic embryogenesis. Plant Physiol. 2022:188(2):1095–1110. 10.1093/plphys/kiab558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Lai ZX. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010:178(4):359–365. 10.1016/j.plantsci.2010.02.005 [DOI] [Google Scholar]
- Lin YL, Lai ZX. Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.). PLoS One. 2013a:8(4):e60337. 10.1371/journal.pone.0060337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Lai ZX. Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant Physiol Biochem. 2013b:66(20):e25. 10.1016/j.plaphy.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Lin SB, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell. 2018:71(2):244–255. 10.1016/j.molcel.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Min JM, Lai RL, Wu Z, Chen Y, Yu L, Cheng C, Jin Y, Tian Q, Liu Q, et al. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. GigaScience. 2017:6(5):1–14. 10.1093/gigascience/gix023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WH, Ni SS, Lin ZC, Lin YL, Lai ZX. Evolution and molecular characteristics of the miR408 family in plants. Chin J Appl Environ Biol. 2017:23(6):1042–1051. 10.3724/SP.J.1145.2017.06021 [DOI] [Google Scholar]
- Long JM, Liu CY, Feng MQ, Liu Y, Wu XM, Guo WW. miR156-SPLs module regulates somatic embryogenesis induction in Citrus callus. J Exp Bot. 2018:69(12):2979–2993. 10.1093/jxb/ery132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu K, Li Y, Xu Y, Yue H, Wen Y, Liu T, Chen S, Liu Q, Yang W, Zhu X, et al. Using RNA sequencing to identify a putative lncRNA-associated ceRNA network in laryngeal squamous cell carcinoma. RNA Biol. 2020:17(7):977–989. 10.1080/15476286.2020.1741282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SY. Expression pattern and function of miR408 in seed development of Rice (Orvza sativa) [MS thesis]. [Hangzhou]: Zhe jiang University;2012.
- Malbec L, Zhang T, Chen YS, Zhang Y, Sun BF, Shi BY, Zhao YL, Yang Y, Yang YG. Dynamic methylome of internal mRNA N7-methylguanosine and its regulatory role in translation. Cell Res. 2019:29(11):927–941. 10.1038/s41422-019-0230-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Yoshimoto T, Ito D, Ogawa T, Tamoi M, Yoshimura K, Shigeoka S. An Arabidopsis FAD pyrophosphohydrolase, AtNUDX23, is involved in flavin homeostasis. Plant Cell Physiol. 2012:53(6):1106–1116. 10.1093/pcp/pcs054 [DOI] [PubMed] [Google Scholar]
- Moro B, Chorostecki U, Arikit S, Suarez IP, Höbartner C, Rasia RM, Meyers BC, Palatnik JF. Efficiency and precision of microRNA biogenesis modes in plants. Nucleic Acids Res. 2018:46(20):10709–10723. 10.1093/nar/gky853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JG, Wang KF, Zhang YL, Liu YH. Method for determination of water soluble vitamin in pollen by high performance liquid chromatography. J Tongji Univ. 2001:29(5):581–583. https://wap.cnki.net/qikan-TJDZ200105018.html [Google Scholar]
- Pramanik KS, Krochko JE, Bewley JD. Distribution of cytosolic mRNAs between polysomal and ribonucleoprotein complex fractions in Alfalfa embryos: stage-specific translational repression of storage protein synthesis during early somatic embryo development. Plant Physiol. 1992:99(4):1590–1596. 10.1104/pp.99.4.1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoeva T, Vaddepalli P, Zhang Z, Weijers D. Evolution, initiation, and diversity in early plant embryogenesis. Dev Cell. 2019:50(5):533–543. 10.1016/j.devcel.2019.07.011 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. Edger: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010:26(1):139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011:146(3):353–358. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Cardona A, Duan S, Yu C, Zhang Z. H3k4me3 recognition by the COMPASS complex facilitates the restoration of this histone mark following DNA replication. Sci Adv. 2022:8(18):eabm6246. 10.1126/sciadv.abm6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Grudzien-Nogalska E, Hamilton K, Jiao XF, Yang J, Tong L, Kiledjian M. Mammalian Nudix proteins cleave nucleotide metabolite caps on RNAs. Nucleic Acids Res. 2020:48(12):6788–6798. 10.1093/nar/gkaa402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Maklashina E, Cecchini G, Iverson TM. Maturation of the respiratory complex II flavoprotein. Curr Opin Struct Biol. 2019:59(1):38–46. 10.1016/j.sbi.2019.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004:119(7):941–953. 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Solanki M, Sinha A, Shukla LI. The miR408 expression in scutellum derived somatic embryos of Oryza sativa L. ssp. indica varieties: media and regenerating embryos. Plant Cell Tissue Org. 2019:138(1):53–66. 10.1007/s11240-019-01602-w [DOI] [Google Scholar]
- Song ZQ, Zhang LF, Wang YL, Li HX, Li S, Zhao HJ, Zhang HY. Constitutive expression of miR408 improves biomass and seed yield in Arabidopsis. Front Plant Sci. 2018:8:2114. 10.3389/fpls.2017.02114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian QZ, Wang G, Ma XX, Shen QW, Ding ML, Yang XY, Luo XL, Li RR, Wang ZH, Wang XY, et al. Riboflavin integrates cellular energetics and cell cycle to regulate maize seed development. Plant Biotechnol J. 2022:20(8):1487–1501. 10.1111/pbi.13826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassi AH, Re DA, Romani F, Cambiagno DA, Gonzalo L, Moreno JE, Arce AL, Manavella PA. The intrinsically disordered protein CARP9 bridges HYL1 to AGO1 in the nucleus to promote microRNA activity. Plant Physiol. 2020:184(1):316–329. 10.1104/pp.20.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam P, De Toledo S M, Pandey BN, Tephly LA, Carter A B, Little J B, Spitz DR, Azzam EI. Regulation of normal cell cycle progression by flavin-containing oxidases. Oncogene. 2008:27(1):20. 10.1038/sj.onc.1210634 [DOI] [PubMed] [Google Scholar]
- Vogel G. How does a single somatic cell become a whole plant? Science. 2005:309(5731):86. 10.1126/science.309.5731.86 [DOI] [PubMed] [Google Scholar]
- Wang YT. Studies on cloning of the primary transcripts and function analyses of some microRNAs from embryonic callus in Dimocarpus longan Lour.[MS thesis]. [Fuzhou]: Fujian Agriculture and Forestry University;2013.
- Wang X, Jiang BC, Gu LF, Chen YD, Mora M, Zhu M, Noory E, Wang Q, Lin CT. A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat Plants. 2021:7(10):1397–1408. 10.1038/s41477-021-01002-z [DOI] [PubMed] [Google Scholar]
- Wang LC, Liu N, Wang TY, Li JY, Wen TW, Yang XY, Lindsey K, Zhang XL. The GhmiR157a/GhSPL10 regulatory module controls initial cellular dedifferentiation and callus proliferation in cotton by modulating ethylene-mediated flavonoid biosynthesis. J Exp Bot. 2018:69(5):1081–1093. 10.1093/jxb/erx475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Xu J, Yu G, Yao GD, Xu K, Ma XS, Zhang N, Liu BF, Li T, Lin ZL, et al. Resetting histone modifications during human parental-to-zygotic transition. Science. 2019:365(6451):353–360. 10.1126/science.aaw5118 [DOI] [PubMed] [Google Scholar]
- Xie LJ, Wang RL, Wang D, Liu L, Cheng L. Visible-light-mediated oxidative demethylation of N6-methyl adenines. Chem Commun. 2017:53(77):10734–10737. 10.1039/C7CC05544G [DOI] [PubMed] [Google Scholar]
- Xie LJ, Yang XT, Wang RL, Cheng HP, Li ZY, Liu L, Mao LQ, Wang M, Cheng L. Identification of flavin mononucleotide as cell-active artificial N6-methyladenosine RNA demethylase. Angew Chem Int Ed Engl. 2019:58(15):5028–5032. 10.1002/anie.201900901 [DOI] [PubMed] [Google Scholar]
- Xu QF. The optimization of somatic embryogenesis system and its applications to germplasm conservation and transformation in Dimocarpus longan Lour. [MS thesis]. [Fuzhou]: Fujian Agriculture and Forestry University;2010.
- Xu XP, Cao QY, Cai RD, Guan QX, Zhang ZH, Chen YK, Xu XH, Lin YL, Lai ZX. Gene cloning and expression analysis of dlo-miR408 and its target DlLAC12 in Dimocarpus longan Lour. Acta Hortic. 2022a:49(9):1–17. 10.16420/j.issn.0513-353x.2021-0573 [DOI] [Google Scholar]
- Xu XP, Cao QY, Guan QX, Mohammad AM, Cai RD, Chen XH, Zhang ZH, Chen YK, Xu XH, Lin YL, et al. Genome-wide identification of miRNAs and targets associated with cell wall biosynthesis: differential roles of dlo-miR397a and dlo-miR408-3p during early somatic embryogenesis in longan. Plant Sci. 2022b:323:111372. 10.1016/j.plantsci.2022.111372 [DOI] [PubMed] [Google Scholar]
- Xu XP, Chen XH, Chen Y, Zhang QL, Su LY, Chen X, Chen YK, Zhang ZH, Lin YL, Lai ZX. Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour. Sci Rep. 2020:10(1):4626. 10.1038/s41598-020-60946-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Zhao XL, Dai ZY, Ma FL, Miao XX, Shi ZY. Osmir396/growth regulating factor modulate rice grain size through direct regulation of embryo-specific miR408. Plant Physiol. 2021:186(1):519–533. 10.1093/plphys/kiab084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Nie XJ, Yan ZG, Weining S. N6-methyladenosine regulatory machinery in plants: composition, function and evolution. Plant Biotechnol. J. 2019:17(7):1194–1208. 10.1111/pbi.13149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LH. Agrobacterium tumefaciens-mediated genetic transformation of Litchi (Litchi chinensis Sonn.) with LEAFY gene [PhD thesis]. Fuzhou: Fujian Agriculture and Forestry University;2002.
- Zhang MX. Establishment of banana and longan transgenic receptor system and the preliminary study on the transformation of the PEAS gene [MS thesis]. Fuzhou: Fujian Agriculture and Forest University;2004.
- Zhang L, Geng Y, Tu WL, Zhang JJ. Determination of taurine in Neverita didyma (Roding) and Asterias rollestoni Bell by HPLC. Marine Sciences. 2009:33(4):6–8. https://www.cnki.com.cn/Article/CJFDTOTAL-HYKX200904002.htm [Google Scholar]
- Zhang JP, Yu Y, Feng YZ, Zhou YF, Zhang F, Yang YW, Lei MQ, Zhang YC, Chen YQ. Mir408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiol. 2017:175(3):1175–1185. 10.1104/pp.17.01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Zhong H, Zhang SD, Shao XJ, Ni M, Cai ZW. NAD Tagseq reveals that NAD+-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis. PNAS. 2019:116(24):12072–12077. 10.1073/pnas.1903683116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Hong P, Wu JY, Chen XB, Ye XG, Pan YY, Wang J, Zhang XS. The tae-miR408-mediated control of TaTOC1 genes transcription is required for the regulation of heading time in Wheat. Plant Physiol. 2016:170(3):1578–1594. 10.1104/pp.15.01216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao PC, Zhang CY, Song YY, Xu XQ, Wang JY, Wang JH, Zheng TY, Lin YL, Lai ZX. Genome-wide identification, expression and functional analysis of the core cell cycle gene family during the early somatic embryogenesis of Dimocarpus longan Lour. Gene. 2022:821:146286. 10.1016/j.gene.2022.146286 [DOI] [PubMed] [Google Scholar]
- Zheng HX, Sun X, Zhang XS, Sui N. M6a editing: new tool to improve crop quality? Trends Plant Sci. 2020:25(9):859–867. 10.1016/j.tplants.2020.04.005 [DOI] [PubMed] [Google Scholar]
- Zhong SL, Li HY, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA Is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008:20(5):1278–1288. 10.1105/tpc.108.058883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JL. Somatic embryogenesis: a model for early development in higher plants. Plant Cell. 1993:5(10):1411–1423. 10.2307/3869792 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided along with the manuscript and its supplemental material.