Abstract
Sugars are fundamental to plant developmental processes. For fruits, the accumulation and proportion of sugars play crucial roles in the development of quality and attractiveness. In citrus (Citrus reticulata Blanco.), we found that the difference in sweetness between mature fruits of “Gongchuan” and its bud sport “Youliang” is related to hexose contents. Expression of a SuS (sucrose synthase) gene CitSUS5 and a SWEET (sugars will eventually be exported transporter) gene CitSWEET6, characterized by transcriptome analysis at different developmental stages of these 2 varieties, revealed higher expression levels in “Youliang” fruit. The roles of CitSUS5 and CitSWEET6 were investigated by enzyme activity and transient assays. CitSUS5 promoted the cleavage of sucrose to hexoses, and CitSWEET6 was identified as a fructose transporter. Further investigation identified the transcription factor CitZAT5 (ZINC FINGER OF ARABIDOPSIS THALIANA) that contributes to sucrose metabolism and fructose transportation by positively regulating CitSUS5 and CitSWEET6. The role of CitZAT5 in fruit sugar accumulation and hexose proportion was investigated by homologous transient CitZAT5 overexpression, -VIGS, and -RNAi. CitZAT5 modulates the hexose proportion in citrus by mediating CitSUS5 and CitSWEET6 expression, and the molecular mechanism explained the differences in sugar composition of “Youliang” and “Gongchuan” fruit.
The transcription factor CitZAT5 participates in sugar metabolism and transport by regulating CitSUS5 and CitSWEET6, promoting sugar accumulation and increasing hexose proportion in citrus fruit.
Introduction
Sugars are synthesized through photosynthesis and represent mobile and accessible materials available for metabolism, providing energy and a source of carbon skeletons for plant growth and development. Numerous studies have confirmed that sugars also play important roles in signaling, osmotic homeostasis, and response to abiotic stress (Bhargava et al. 2013; Smeekens and Hellmann 2014; Ma et al. 2018). For fruits, soluble sugars serve as direct contributors to sweetness and a supply of basic carbon skeleton for the synthesis of vitamins, aromatic substances, and other metabolites, which are critical for fruit development and quality development (Zhang et al. 2018a, Liu et al. 2020; Wu et al. 2021). The soluble sugar contents of fruits depend mainly on the production of photosynthate in green leaves, with sugars subsequently being transported through the phloem to fruit cells where they participate in metabolism and storage. Sugars can also be synthesized in unripe green fruits before ripening and from the degradation of stored starch during ripening. The accumulation of sugars is, therefore, determined by the joint function of sugar transporters and enzymes involved in metabolism during fruit developing and is of crucial importance to the growth and maintenance of fruits and their final quality when ripe (Katz et al. 2007; Bush 2020). Exploring the mechanism of sugar accumulation is, therefore, critical for understanding the composition and concentrations of sugars, which determines their sweetness and is an important component of fruit quality.
Substantial progress has been made in understanding sugar metabolism and transport patterns and how the process of sugar metabolism powers and supports the growth of sinks, providing an energy source and precursors for synthesis of essential polymers and maintaining carbon flow throughout the whole plant level (Ruan 2012). For most fruits, a sucrose cycle contributes to core sugar metabolism (Chen et al. 2021). Three key enzymes, sucrose-phosphate synthase (SPS, EC 2.4.1.14), sucrose synthase (SuS, EC 2.4.1.13), and invertase (INV, beta-fructofuranosidase, EC 3.2.1.26), have been widely studied (Stein and Granot 2019; Ru et al. 2021). SuS and INV are mainly responsible for sucrose decomposition. Sucrose is hydrolyzed by apoplastic invertase into glucose and fructose upon phloem unloading in the extracellular space (Chen et al. 2021). Apple (Malus domestica Borkh.) hexose transporter (HT) controlling cell wall invertase (CWINV) activity was confirmed to influence carbohydrate partitioning by regulating apoplastic hexose levels (Wang et al. 2020b). Sucrose transported into the cytosol is degraded by SuS and cytosolic INV, or by acidic INV in the vacuole (Liu et al. 2018). For example, a substantially lower fructose level had been predicted due to the dramatically reduced sorbitol in transgenic apple fruit with antisense suppression of aldose-6-phosphate reductase gene A6PR. However, increased transcript levels of MdNINV1, MdNINV3, and MdSUSY1-3 and higher activities of NINV and SUSY help compensate for the reduced level of fructose, which is probably realized through the higher availability of sucrose in the cytosol (Li et al. 2018). SPS catalyzes the conversion of fructose-6-phosphate and uridine diphosphate glucose (UDPG) into sucrose-6-phosphate, a substrate of sucrose synthesis (Liu et al. 2018). Higher SPS expression level and activity were verified to be important contributors to increased superior sucrose content in melon (Cucumis melo var. makuwa Makino) (Gao et al. 2021) and pineapple (Ananas comosus cv. Comte de paris) (Zhang et al. 2012). The processes of sugar transportation into nonphotosynthetically active sinks such as seeds and fruits are of crucial importance to growth, adaptation to the environment, and resistance to adversity (Xu et al. 2018; Bush 2020; Mathan et al. 2020; Li et al. 2021a) and require a battery of transporters. Several sugar transporters have been characterized and considered to participate in apoplastic transportation in plants, such as sucrose transporters (SUTs), monosaccharide transporters (MSTs), and sugars will eventually be exported transporters (SWEETs) (Chen et al. 2010; Doidy et al. 2012; Jian et al. 2016; Julius et al. 2017). For example, CsSWEET7a is responsible for sugar unloading into fruit by removing hexoses from companion cells to the apoplastic space in cucumber (Cucumis sativus L.) (Li et al. 2021c). In tomato (Solanum lycopersicum) fruit, SlSWEET15 unloads sucrose from phloem cells to apoplasm and subsequently transport into parenchyma cells for storage (Ko et al. 2021). In watermelon (Citrullus lanatus) fruit, ClSWEET3 helps transport hexose from the intercellular space into fruit storage cells (Ren et al. 2021). Sugar accumulation and transportation are transcriptionally regulated by transcription factors (TFs), which have been characterized in model plants and fruit crops (Wei et al. 2018; Peng et al. 2019), and upstream transcriptional regulation of sugar-related genes has been increasingly investigated in recent years. In rice (Oryza sativa), OsDOF11 modulates sugar transport by regulating both the SUT and SWEET genes (Wu et al. 2018). In apple fruit, MdAREB2 is involved in ABA-induced sugar accumulation by activating the sugar transport genes MdSUT2 and MdTMT1 (Ma et al. 2017). In grape (Vitis vinifera), VvWRKY22 has been shown to modulate sugar accumulation by interacting with VvSnRK1.1/VvSnRK1.2 (Huang et al. 2021).
Citrus (Citrus reticulata Blanco.) is one of the most important cultivated fruit crops in the world, with a large planting area and high yield. Moreover, the abundant nutrient materials and excellent flavor quality make it highly favored by consumers (Wu et al. 2021). The proportion and content of sugars confer sweetness to fruits and are, to some extent, important indicators of citrus quality (Kelebeka and Selli 2011). In recent years, studies on sugar in citrus fruit have mainly focused on the analysis of the mode of accumulation, the expression pattern of genes in the sucrose metabolic pathway, and the effect of exogenous treatment on sugar levels (Katz et al. 2007; Deng et al. 2019; Wu et al. 2021). However, research on the regulation of sugar accumulation and the function of transcription factors in sugar accumulation has rarely been reported in citrus.
In this study, the differences in hexose content during fruit development in 2 citrus varieties “Gongchuan” (GC) and “Youliang” (YL) (a bud sport variety of “GC’) were analyzed. Transcriptome analysis, heterologous expression in yeast cells, and transient assays in citrus fruit identified the sucrose synthase isozyme gene CitSUS5 and SWEET family member CitSWEET6 as candidate genes contributing to the high hexose content in “YL.” A zinc finger protein CitZAT5, which positively regulates both CitSUS5 and CitSWEET6, was characterized and yeast two-hybrid assay and BiFC indicated that CitZAT5 could interact with CitNAC47 to enhance the activation effect on CitSUS5. Transient overexpression, RNAi, and VIGS provided additional evidence for the vital role of CitZAT5 in sugar accumulation and modulating the proportion of hexose.
Results
The accumulation pattern of soluble sugar during “YL” and “GC” fruit development
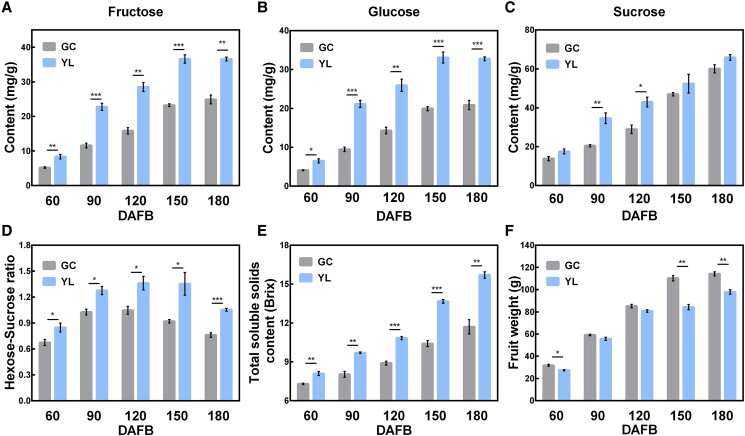
The soluble sugar contents were analyzed during growth and development of “YL” and “GC” citrus fruit (Fig. 1). The results showed that the hexose contents of “YL” were invariably higher than those of “GC” throughout fruit development (Fig. 1, A and B). Further analysis revealed that these differences in hexose accumulation were already evident at the early stages of fruit development. The fructose contents in “YL” fruit were 3.2, 11.2, and 12.7 mg g−1 higher, while the glucose contents were 2.4, 11.7, and 11.7 mg g−1 higher, than in “GC” fruit at 60, 90, and 120 DAFB (days after full bloom) (Fig. 1, A and B). “YL” fruit also accumulated more sucrose than “GC” fruit during the fruit expansion period (between 90 and 120 DAFB) but showed no significant difference at the fruit ripening stage (150 and 180 DAFB) (Fig. 1C). Analysis of the sugar composition between the 2 varieties (Fig. 1D) showed that the hexose–sucrose ratio of “YL” was invariably higher than that of “GC” throughout fruit development, and at the mature stage (180 DAFB), the ratio of hexose to sucrose was 1.05 in “YL” and 0.76 in “GC.” The total soluble solids (TSS) content of “YL” was invariably higher than those of “GC” throughout fruit development, which was consistent with the hexose accumulation results. The TSS contents in “YL” fruit were 0.8, 1.7, and 1.9 (%) higher than those in “GC” fruit at 60, 90, and 120 DAFB (Fig. 1E). The fruit weights of “GC” were higher than those of “YL” at 60, 150, and 180 DAFB (Fig. 1F). Collectively, the difference in soluble sugar contents between “YL” and “GC” fruits was mainly reflected in the difference in hexose.
Figure 1.
Sugar profiles in “GC” and “YL” fruits during development. Fructose A), glucose B), and sucrose C) contents of “GC” and “YL” fruits. D) Hexose/sucrose ratio of “GC” and “YL” fruits at different stages of fruit development. Changes in total soluble solids (TSS) E) and fruit fresh weight F). DAFB, days after full bloom. Error bars represent the SE (standard error, n = 3). Asterisks denote significant differences using Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
Identification of potential genes involved in sugar accumulation
At the early stages of fruit development, “YL” and “GC” fruits already exhibited differences in hexose content (Fig. 1). Transcriptome data from 60, 90, and 120 DAFB were analyzed to identify genes likely to be involved in differential accumulation patterns of soluble sugars between these 2 varieties. Putative sugar-related candidate genes were identified from transcriptome data, and close attention was paid to several gene families, including those encoding enzymes in sucrose metabolic pathways and sugar transporters. The differential expression patterns in “GC” and “YL” of all 45 genes from SPS, SUS, INV, and SWEET families are shown as a heatmap of fold changes (FC, FPKM ratio of “YL”/“GC”) (Supplemental Fig. S1). According to this analysis, 6 candidate genes exhibited higher FPKM ratio of “YL”/“GC,” with a fold change at any 2 stages of over 2-fold, including 3 genes encoding SuS, SPS, and INV enzymes involved in sucrose metabolism (CitSUS5 and CitSPS4, CitCWIN) and 3 SWEET family members (CitSWEET6, CitSWEET8, and CitSWEET11d), which may be involved in sugar transportation (Supplemental Fig. S2). The INV family is also known to participate in sucrose conversion to glucose and fructose, and CitCWIN also showed a substantial fold change, but it showed excessively low transcript abundance so that we preferred other dominant factors. The function of CitSWEET8 was detected via heterologous expression in a hexose transport-deficient yeast strain (EBY.VW4000) and in a sucrose transport-deficient yeast strain (SUSY7/ura3), and neither hexose nor sucrose transport capacity was identified (Supplemental Fig. S3). SPS is involved in sucrose synthesis, and CitSWEET11d has also been shown to specifically promote sucrose accumulation (Hu et al. 2021). Based on the differences in hexose accumulation between the 2 varieties, we selected CitSUS5 and CitSWEET6 as candidate genes for further study.
CitSUS5 is involved in hexose accumulation
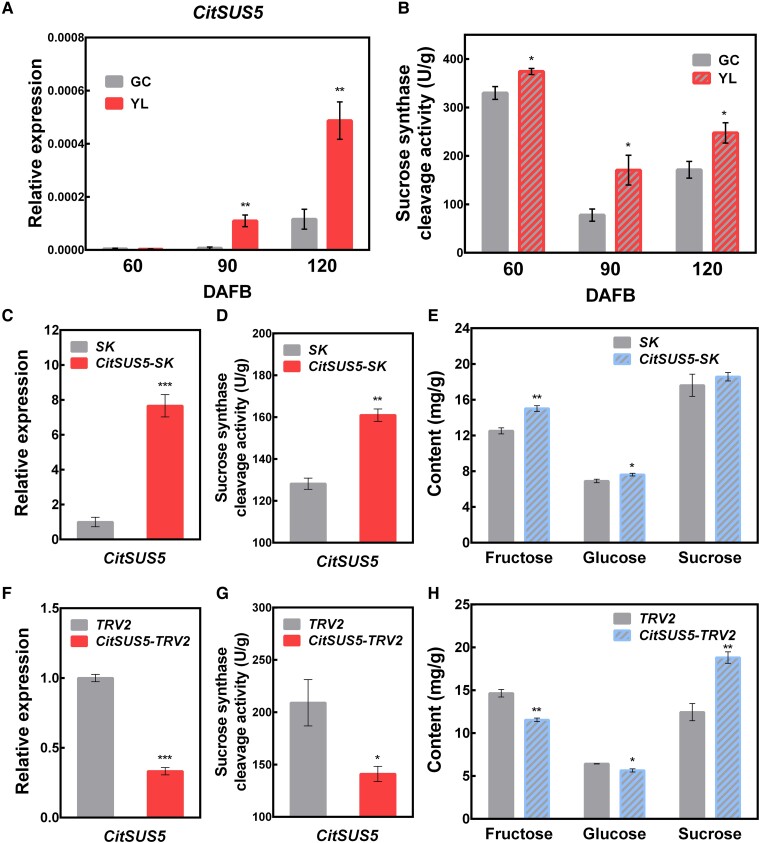
Sucrose synthase enzyme (SuS) can promote the accumulation of hexoses by cleaving sucrose to glucose and fructose. The transcript levels of CitSUS5 were higher in “YL” fruit than in “GC” fruit at the early stages of fruit development (Fig. 2A). In agreement with the expression pattern of CitSUS5, the enzyme activity of sucrose synthase was significantly higher in “YL” fruit than in “GC” fruit (Fig. 2, A and B), which confirmed the association of CitSUS5 with the accumulation of hexoses by cleaving sucrose. To further explore the potential role of CitSUS5 in hexose accumulation in citrus fruit, CitSUS5 was both transiently overexpressed driven by the 35S promoter and silenced by VIGS in citrus fruit. The results showed that CitSUS5 overexpressing (OE) fruit had significantly higher fructose and glucose concentrations than the control fruit with the level of CitSUS5 transcripts and the sucrose synthase cleavage activity significantly increased (Fig. 2, C–E). Compared with the control plants (transformed with empty vector TRV2), the expression level of CitSUS5 was significantly decreased to 33.2% and the sucrose synthase cleavage activity was decreased to 67.5%, resulting in a significant decrease in hexose content and a significant increase in sucrose content (Fig. 2, F–H). Collectively, our results suggest that CitSUS5 participates in promoting the cleavage of sucrose to hexoses.
Figure 2.
Functional characterization of CitSUS5. A) Expression pattern of CitSUS5 in “GC” and “YL” fruit. B) Analysis of total sucrose synthase cleavage activity in “GC” and “YL” fruit. DAFB, days after full bloom. Transcript levels of CitSUS5 verified by RT-qPCR C), total sucrose synthase cleavage activity D), and sugar content E) were analyzed in citrus transiently overexpressed CitSUS5; SK refers to empty vector. Transcript levels of CitSUS5 verified by RT-qPCR F), total sucrose synthase cleavage activity G), and sugar content H) were analyzed by transient VIGS of CitSUS5; TRV2 refers to the empty vector. Error bars represent SE based on 3 biological replicates. Asterisks denote significant differences using Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
To investigate the cellular location, a subcellular localization assay of CitSUS5 was performed by expressing in Nicotiana benthamiana leaves, and GFP as a control. Strong green fluorescence of CitSUS5 was observed in the cytosol, plasma membrane, and cell nucleus of N. benthamiana leaves (Supplemental Fig. S4).
CitSWEET6 acts as a fructose transporter
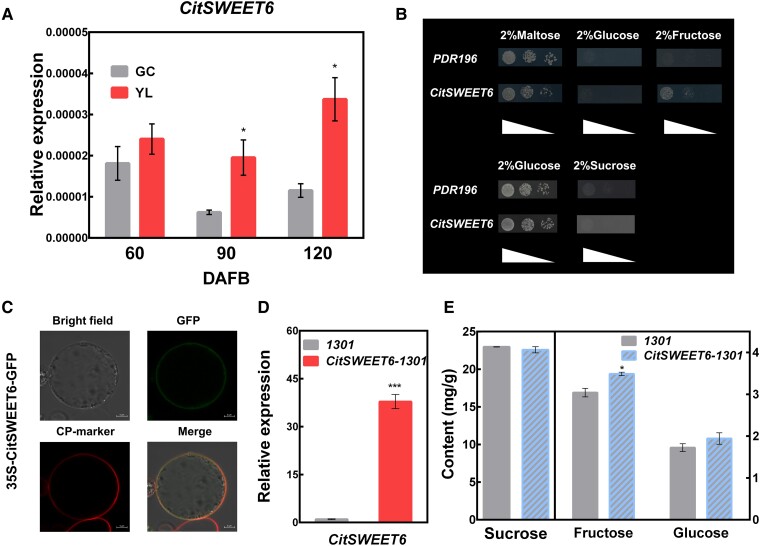
CitSWEET6 belongs to the SWEET family, members of which have recognized roles in sugar transport. The analysis of its expression pattern indicated that CitSWEET6 was significantly more highly expressed in “YL” fruit than in “GC” fruit during the early stages of fruit development (Fig. 3A). The function of CitSWEET6 was studied via heterologous expression in EBY.VW4000 and SUSY7/ura3 (Fig. 3B). CitSWEET6-EBY.VW4000 transformants grew well on media supplemented with fructose but not glucose as the sole carbon source, indicating CitSWEET6 has an effective fructose transport capacity, while CitSWEET6-SUSY7/ura3 transformants failed to survive on media with sucrose as the solo carbon source, indicating that CitSWEET6 may not transport sucrose efficiently. These results suggest that CitSWEET6 function as a fructose transporter.
Figure 3.
Functional characterization of CitSWEET6. A) Expression pattern of CitSWEET6 transcripts in “GC” and “YL” fruits. DAFB, days after full bloom. B) Analysis of the transport activity of CitSWEET6 in yeast cells. Yeast mutant strain EBY.VW4000 expressing CitSWEET6 grown in SD (-Ura) media supplemented with 2% maltose, 2% fructose, or 2% glucose (upper half). The yeast mutant strain SUSY7/ura3 expressing CitSWEET6 grew in SD (-Ura) media supplemented with 2% glucose or 2% sucrose (bottom half). An empty vector (pDR196) was used as a negative control. C) Subcellular localization of CitSWEET6-GFP fusion protein in rice protoplasts together with a plasma membrane (PM) marker. Bars = 5 µm. Transcript levels of CitSWEET6 verified by RT-qPCR D) and sugar content E) were analyzed in citrus transiently overexpressed CitSWEET6; 1,301 refers to an empty vector. Error bars represent SE based on 3 biological replicates. Asterisks denote significant differences using Student's t-test (*P < 0.05; ***P < 0.001).
To determine the subcellular localization of CitSWEET6, the CitSWEET6-GFP fusion protein driven by the CaMV 35S promoter was transiently expressed in rice protoplasts (Fig. 3C), with the GFP vector as a control (Supplemental Fig. S5A). CitSWEET6-GFP fluorescent signals were ubiquitously expressed in the plasma membrane, as the signal aligned with that of FM4-64, a plasma membrane (PM) marker (Fig. 3C). A strong green fluorescence of CitSWEET6 was also observed in the plasma membrane in N. benthamiana leaves (Supplemental Fig. S5B).
A transient overexpression assay was also performed in citrus fruit to further illustrate the role of CitSWEET6 in fructose accumulation. The results showed that CitSWEET6 OE fruit had a significantly higher fructose concentration than the control (Fig. 3, D and E). An additional 30 SWEET proteins of various species were collected from previous studies and were identified to participate in soluble sugar transport. A phylogenetic tree of these SWEET genes was constructed by aligning the full-length amino acid sequences of CitSWEET6 with these proven SWEETs. CitSWEET6 clustered with several SWEET members that have been verified to be fructose transporters (Supplemental Fig. S6).
In vivo regulatory effects of transcription factors on the CitSUS5 and CitSWEET6 promoters
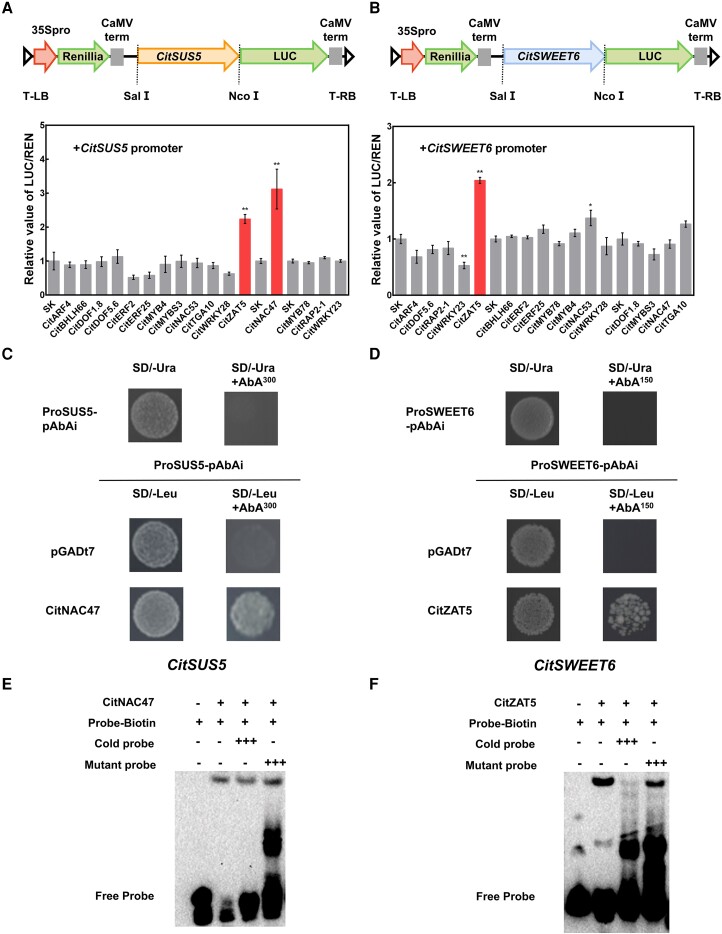
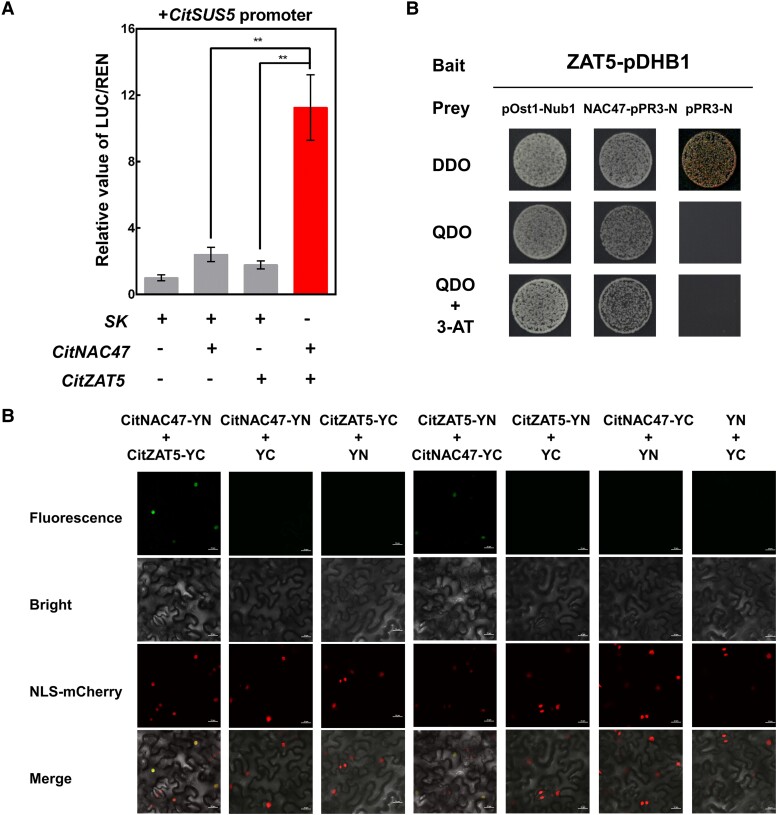
To understand the regulatory mechanism governing expression of CitSUS5 and CitSWEET6, transcription factors (TFs) with increased transcript levels were selected based on the association between their expression profiles with sugar contents (Supplemental Fig. S7). Sixteen TFs were identified and tested for transcriptional activation of CitSUS5 and CitSWEET6 promoters by dual-luciferase assays. The results showed that the CitSUS5 promoter was activated by both CitNAC47 and CitZAT5 by 3.1- and 2.2-fold, respectively, while the transcriptional activity of the CitSWEET6 promoter was activated 2.04-fold by CitZAT5 (Fig. 4, A and B). Transient expression in N. benthamiana leaves of both 35S-CitZAT5-GFP and 35S-CitNAC47-GFP showed green fluorescence in the nucleus, overlapping with red fluorescence of the mCherry nuclear marker, indicating that CitZAT5 and CitNAC47 are both nuclear-localized proteins (Supplemental Fig. S8). The transcript levels of CitNAC47 and CitZAT5 increased with the accumulation of sugar during the early fruit developmental stages, and both were significantly higher in “YL” fruit than in “GC” fruit (Supplemental Fig. S8).
Figure 4.
Regulatory effects of transcription factors on the promoter activities of CitSUS5 and CitSWEET6. Schematic map of the CitSUS5-LUC construct A) and CitSWEET6-LUC construct B). The ratio of firefly luciferase (LUC) and Renilla luciferase (REN) of the empty vector (SK) plus promoter was set at 1. Error bars represent SE based on 4 biological replicates. Asterisks denote significant differences using Student's t-test (*P < 0.05; **P < 0.01). In C and D), yeast one-hybrid analyses. Autoactivation of proSUS5-pAbAi and proSWEET6-pAbAi was tested on SD/-Ura in the presence of 300 and 150 ng mL−1 aureobasidin A (AbA), respectively. Physical interaction was determined on SD medium lacking Leu in the presence of the corresponding amount of AbA. The empty pGADT7 vector was used as negative controls. E and F) In vitro binding of CitNAC47 to the promoter of CitSUS5 and binding of CitZAT5 to CitSWEET6 performed by electrophoretic mobility shift assay (EMSA). Wild-type probes containing the recognition motives (Supplemental Table S3) were biotin labeled, while competition for binding was performed with unlabeled probes and mutant probes. The symbol + or − represents presence or absence, respectively; +++ indicates increasing amounts.
Dual-luciferase assays indicated that CitZAT5 was able to transactivate the promoters of both target genes, implying its positive role in both sucrose cleavage and fructose transportation. Yeast one-hybrid assays indicated that the CitZAT5 protein directly bound to the CitSWEET6 promoter, and CitNAC47 protein directly bound to the CitSUS5 promoter (Fig. 4, C and D). This was confirmed by EMSA analysis which showed that both CitZAT5 and CitNAC47 TF proteins could directly bind in vitro to the specific probes from the CitSWEET6 and CitSUS5 promoters, respectively (Fig. 4, E and F).
Synergistic effect of CitZAT5 and CitNAC47 on the CitSWEET6 promoter
Since both CitNAC47 and CitZAT5 positively activated the CitSUS5 promoter, a possible synergistic effect between these 2 transcription factors was tested by dual-luciferase assays (Fig. 5A). The combination of CitNAC47 and CitZAT5 resulted in an 11.3-fold synergistic activation effect of the CitSUS5 promoter compared to the much lower individual CitNAC47 or CitZAT5 activations (2.4- and 1.8-fold, respectively). Protein–protein interactions between CitNAC47 and CitZAT5 were verified by a yeast two-hybrid assay (dual-hunter system) and bimolecular fluorescence complementation assay (BiFC) (Fig. 5, B and C). These results suggest that CitNAC47 and CitZAT5 act synergistically to activate the CitSUS5 promoter activity.
Figure 5.
Synergistic interactions of CitNAC47 and CitZAT5 on activation of the CitSUS5 promoter. A) Effect of the CitNAC47 and CitZAT5 individually and in combination on the activation of the CitSUS5 promoter. The LUC/REN ratio of the empty vector plus promoter was used as a calibrator (set as 1). Error bars represent the SE based on 4 biological replicates (**P < 0.01). B) Interaction between CitNAC47 and CitZAT5 in yeast two-hybrid assays. The transformants were cultivated on different media: (i) DDO (SD medium-Trp-Leu); (ii) QDO (SD medium-Trp-Leu-His-Ade); and (iii) QDO+3-AT (QDO medium supplemented with 1 mm 3-amino-1,2,4-triazole). Protein–protein interactions were determined by the growth of yeast cells on QDO and QDO+3AT. pOst1-NubI, positive control; pPR3-N, negative control. C) The in vivo interaction between CitNAC47 and CitZAT5 was determined using BiFC in N. benthamiana. N- and C-terminal fragments of YFP were fused to the C- and N-termini of CitNAC47 and CitZAT5, respectively. Combinations of YFPN and YFPC with the corresponding CitNAC47 and CitZAT5 constructs were used as negative controls. The fluorescence of YFP represents protein–protein interactions. Bars = 25 mm.
Functional analysis of CitZAT5
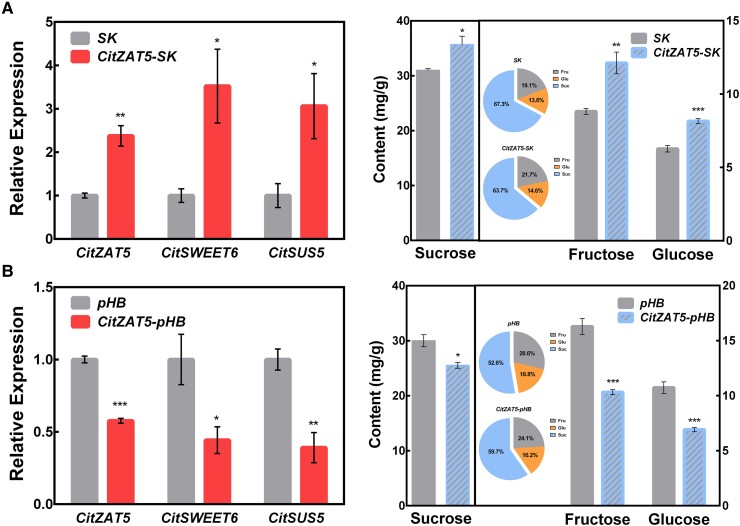
CitZAT5 was indicated to indirectly transactivate the promoter of CitSUS5 by interacting with CitNAC47 and directly bind and transactivate the promoter of CitSWEET6, implying its potential important roles in both hexose accumulation and fructose transportation. We performed a series of experiments to explore the effect of CitZAT5 on sugar accumulation. Transient overexpression of CitZAT5 in citrus fruit promoted the accumulation of fructose, glucose, and sucrose with increasing amounts of rates at 37%, 30%, and 15%, respectively, compared to the control fruit. In addition, the expression levels of the downstream targets CitSUS5 and CitSWEET6 were significantly increased, which is consistent with a mechanism involving their upregulation (Fig. 6A).
Figure 6.
Functional characterization of CitZAT5. A) Transient overexpression of CitZAT5 in citrus fruit. RT-qPCR verification of transcript levels of CitZAT5, CitSWEET6, and CitSUS5 (left); sugar contents were measured (right); SK refers to empty vector, n = 4. Sugar proportions in control and CitZAT5 OE fruits are shown in the 2 pie charts. B) Transcript levels of CitZAT5 and the 2 target genes were analyzed (left). Transient RNAi of CitZAT5 reduced sugar contents in citrus fruit (right); pHB refers to empty vector, n = 4. Sugar proportions in control and CitZAT5-RNAi fruit are compared in pie charts. Error bars represent the Se. Asterisks denote significant differences using Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
To better understand the role of CitZAT5 in sugar accumulation in citrus fruit, an RNA interference (RNAi) assay was performed by infiltrating the vectors into citrus fruit. The results showed that the transcript level of CitZAT5 in CitZAT5-RNAi fruit was reduced to 58% of that in control fruit, and the expression levels of CitSUS5 and CitSWEET6 were significantly reduced (Fig. 6B). The sugar content of CitZAT5-RNAi fruit was also significantly reduced, with the concentrations of fructose, glucose, and sucrose decreasing by 37%, 36%, and 15%, respectively, compared to the control fruit (Fig. 6B).
Additional VIGS of CitZAT5 was performed in citrus seedlings. Compared with the control plants (transformed with empty vector TRV2), the expression levels of CitZAT5, CitSUS5, and CitSWEET6 were all significantly decreased, resulting in a significant decrease in sugar content (Supplemental Fig. S9). The concentrations of fructose, glucose, and sucrose were decreased to 15.8%, 38.2%, and 48.3%, respectively, compared to the control plants (Supplemental Fig. S9). Collectively, our results suggest that CitZAT5 positively regulates sugar accumulation in citrus fruit through modulating the transcription of CitSUS5 and CitSWEET6.
Association between CitZAT5 transcripts and sugar content across citrus cultivars
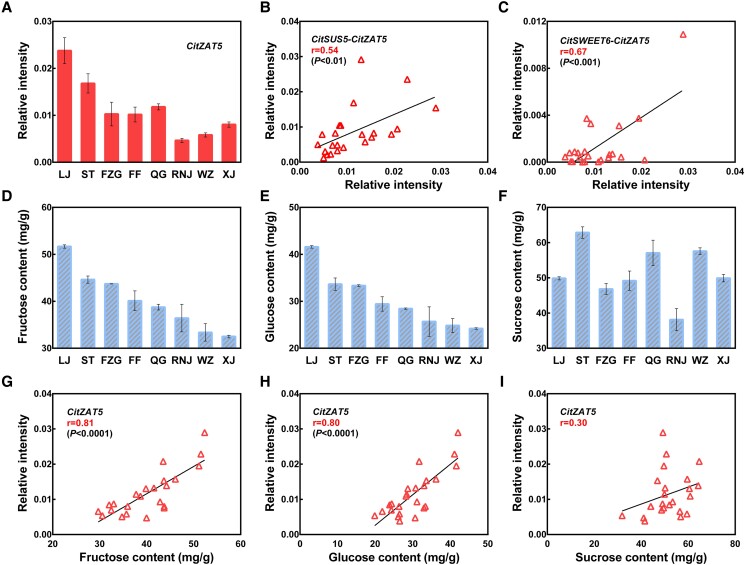
To further validate the regulatory effects of CitZAT5 on sugar accumulation, gene expression patterns (Fig. 7A) and sugar contents (Fig. 7, D–F) were analyzed in 8 varieties of Satsuma mandarin (Citrus unshiu Marcov.). The correlation between the transcript level of CitZAT5 and the sugar content was obtained using linear regression analysis (Fig. 7, G–I). Strongly positive correlations were observed between the expression levels of CitZAT5 and fructose (r = 0.81, P < 0.0001; Fig. 7G) and glucose (r = 0.80, P < 0.0001; Fig. 7H) in different citrus varieties, while there was a lower level of correlation with sucrose content across cultivars (Fig. 7I). CitZAT5 expression was significantly positively correlated with CitSUS5 (r = 0.54, P < 0.01; Fig. 7B) and CitSWEET6 (r = 0.67, P < 0.001; Fig. 7C). Taken together, these results indicate that the expression of CitZAT5 is positively correlated with hexose accumulation, which is consistent with the role of CitZAT5 examined.
Figure 7.
Relationship between CitZAT5 transcripts and sugar contents in 8 citrus varieties. A) Expression level of CitZAT5 in 8 varieties. Linear regression analysis between expression of CitSUS5B), CitSWEET6C), and CitZAT5. Fructose D), glucose E), and sucrose F) contents in 8 citrus varieties. G, H, and I) Linear regression analysis between CitZAT5 expression and fructose, glucose, and sucrose content. Error bars represent the Se.
Discussion
Hexoses make a greater contribution to sweetness in “YL” than in “GC”
Sucrose is regarded as a predominant soluble sugar and acts as the main transported form of carbohydrate (Li et al. 2020; Braun 2022), and most studies of sugar accumulation in plants have focused on sucrose metabolism and regulation (Baker et al. 2016; Ma et al. 2018; Braun 2022). The importance of hexoses in plants has also been gradually recognized (Xu et al. 2012; Sosso et al. 2015; Li et al. 2021c), and the quality and flavor of fleshy fruits depend on the proportions of several soluble sugars (Shammai et al. 2018; Liu et al. 2020). Hexose contents are tightly linked with the growth of cucumber (Li et al. 2021c), and elevated hexose and low sucrose contents in tomato accompany a series of advantageous traits, such as larger size, thinner pericarp, dwarfness, and early flowering (Wang et al. 2020b). In addition, the importance of hexose in the ripening of grape is widely accepted, and several hexose-related genes have been identified as being involved in regulating hexose accumulation and berry ripening (Chong et al. 2014; Zhang et al. 2019).
In citrus fruit, the main soluble sugars are sucrose, fructose, and glucose. In the majority of cultivars, sucrose contributes the highest proportion and has therefore attracted most attention for research (Hu et al. 2021; Wu et al. 2021). However, the relative sweetness of fructose to sucrose is approximately twice, so fruits containing more fructose taste sweeter (Shammai et al. 2018). In this study, higher sugar contents were observed in “YL,” a bud sport variety of “GC,” and the sugar profile indicated a higher level of hexose accumulation at the ripening stage of “YL” than “GC,” while there was no difference in sucrose (Fig. 1), suggesting that hexose is the key factor influencing the sweetness of “YL” and “GC.” Similar results were found in strawberry (Fragaria × ananassa Duchesne) fruits, with some excellent strawberry strains showing a particular predominance of hexoses (Liu et al. 2020). It is worth mentioning that the transcript levels of CitSPS4 and CitSWEET11d were notably higher in “YL” fruit than in “GC” fruit at 90 and 120 DAFB, which might explain why “YL” fruit did not exhibit a lower sucrose content as a result of the more active sucrose decomposition (Supplemental Fig. S10). Taken together, our results indicate that increasing the proportion of hexoses by modulating transcript levels of appropriate genes such as CitSUS5 and CitSWEET6 may be a good strategy for improving citrus fruit sweetness and quality.
CitSUS5 is associated with hexose accumulation in citrus fruit
SuS catalyzes reversible sucrose decomposition to produce fructose and UDPG and is generally considered a vital contributor to sucrose catabolism and hexose accumulation in sink tissue (Coleman et al. 2009; Xu et al. 2012), and its activity tightly parallels sink tissue development in plants (Coleman et al. 2009; Xu et al. 2012; Liu et al. 2018). For example, in tobacco (Nicotiana tabacum cv. Xanthi), SuSy-transformed lines showed increased glucose and fructose contents (Coleman et al. 2006). In the present study, significantly more CitSUS5 transcripts were detected in “YL,” which contains a higher content of hexoses. Consistently, higher SuS cleavage enzyme activity was detected in “YL” (Fig. 2) and it is believed that this favors conversion of sucrose to hexoses. In addition, transient overexpression of CitSUS5 significantly increased the hexose content while transient VIGS of CitSUS5 significantly decreased the hexose content and increased the sucrose content in citrus fruit (Fig. 2), which provided direct evidence for its role in hexose accumulation in vivo. Notably, the sucrose content of CitSUS5-OE lines did not decrease as predicted (Fig. 2E). Considering the homeostasis between sucrose synthesis and decomposition in fruit, exogenous sucrose might be absorbed from sucrose supplemented in medium or endogenous sucrose might be synthesized to compensate sucrose, which is consumed by CitSUS5. A positive correlation between the expression level of CitSUS5 and hexose content among 8 citrus cultivars was noticed, which also supports the suggestion that CitSUS5 contributes to hexose accumulation (Supplemental Fig. S11A). Studies in blueberry (Vaccinium spp.) and pitaya (Hylocereus) showed that VcSS1 induced by ethylene positively stimulates the increase in fructose and glucose (Wang et al. 2019c), and the expression of HpSuSy1 was tightly associated with the elevated accumulation of glucose and fructose during fruit maturation (Wei et al. 2019). Overall, these results support the potential role of CitSUS5 in hexose accumulation in citrus fruit.
CitSWEET6 contributes to fructose transport in citrus fruit
The definite function of SWEET family members in plant sugar transport was first identified by Chen, and AtSWEET11 and AtSWEET12 were functionally characterized as being involved in unloading sucrose in the phloem of Arabidopsis leaves (Chen et al. 2012). Subsequently, additional SWEET family members were characterized and shown to be involved in sucrose, glucose, and fructose transport in different species, including cucumber (C. sativus L.), tomato (S. lycopersicum), soybean (Glycine max), and grapevine (V. vinifera) (Chong et al. 2014; Wang et al. 2019b; Zhang et al. 2021b; Hu et al. 2022). ZmSWEET4c in maize (Zea mays L. ssp. mays) could transport both glucose and fructose and has been involved in regulating hexose distribution during the seed domestication process (Sosso et al. 2015). SlSWEET7a and SlSWEET14 have both been shown to transport hexose and sucrose to influence the sugar content in tomato (Zhang et al. 2021b). Here, we identified a SWEET transporter, CitSWEET6, located at the plasma membrane that functions as a fructose transporter (Fig. 3, Supplemental Fig. S5), which is consistent with its homology to rice OsSWEET4 (O. sativa) (Sosso et al. 2015). In addition, transient overexpression of CitSWEET6 significantly increased the fructose content in citrus fruit, which provided pivotal evidence for the involvement of CitSWEET6 in fructose transportation (Fig. 3). A positive correlation between the expression level of CitSWEET6 and hexose content among 8 citrus cultivars also corroborates the importance of CitSWEET6 in hexose accumulation (Supplemental Fig. S11B). Our previous study identified CitSWEET11d, which promotes sucrose accumulation in citrus fruit (Hu et al. 2021), and the characterization of CitSWEET6 broadens the range of sugar transporters in citrus fruit.
CitZAT5 participates in hexose accumulation in citrus fruit by upregulating CitSUS5 and CitSWEET6
The role of transcription factors in regulating structural genes involved in sugar accumulation has been widely recognized in various species (Chen et al. 2017; Ma et al. 2017; Huang et al. 2021), and regulators upstream of SuS and SWEET members have been identified (Deng et al. 2020; Mathan et al. 2020). HpWRKY3 was verified to modulate HpSuSy (Wei et al. 2019), and OsERF2 affected sucrose and UDPG contents by transcriptionally regulating OsSUS3 and OsSUS6 (Xiao et al. 2016). Additionally, regulators of SWEETS, such as GhMYB212, could function in facilitating sucrose transport during cotton fiber elongation by controlling the expression of GhSWEET12 (Sun et al. 2019). In pear (Pyrus ussuriensis) fruits, PuWRKY31 targets PuSWEET15, which is associated with differences in sucrose content between 2 pear varieties (Li et al. 2020).
Here, we identified a C2H2-type transcription factor, CitZAT5, which transactivates the promoter activity of CitSUS5 and CitSWEET6, suggesting that unlike the TFs described above, CitZAT5 could participate in both sugar metabolism and transportation. Phenotypic and molecular analysis of CitZAT5-RNAi, CitZAT5-VIGS, and CitZAT5 overexpression lines showed that CitZAT5 facilitates not only hexose accumulation but also sucrose accumulation, indicating that CitZAT5 might also be involved in other pathways in sucrose accumulation, and the specific regulatory mechanism needs to be further studied (Fig. 6, Supplemental Fig. S9). Besides, sugar proportion analysis in CitZAT5-RNAi, CitZAT5-VIGS, and CitZAT5 overexpression lines proved that CitZAT5 plays an important role in hexose accumulation more than sucrose accumulation (Fig. 6, Supplemental Fig. S9). In addition, there was higher positive correlations between the expression level of CitZAT5 and hexose content than sucrose content in 8 citrus cultivars, which supported our inference that CitZAT5 might function in improving the proportion of hexose rather than sucrose (Fig. 7). Notably, C2H2-type zinc finger proteins were known to function as key transcriptional regulators involved in plant growth and development (Xiao et al. 2009; Feurtado et al. 2011; Černý et al. 2018) and respond to a wide range of adverse stresses, such as low temperature (Vogel et al. 2005), salt (Rehman et al. 2021), drought (Huang et al. 2008), and oxidative stress (Kiełbowicz-Matuk, 2012; Wang et al. 2019a). However, there have been few reports on the role of ZAT in fruit quality, although a recent study in kiwifruit (Actinidia spp.) revealed that AdZAT5 is a key factor in pectin degradation and fruit softening (Zhang et al. 2021a). Our study suggested that CitZAT5 participates in determining hexose accumulation and proportion in citrus fruit by simultaneously participating in fructose transport and sucrose metabolism, which enriches the known biological functions of the C2H2-type zinc finger family and can provide ideas for subsequent research. In addition, the discovery of the function of CitZAT5 may improve our understanding of the transcriptional network of SWEET in fruits.
Usually, genes are transcriptionally regulated by several TFs collaboratively (Zhang et al. 2018b; Tang et al. 2021). We characterized another TF, CitNAC47, which could also transactivate the promoter of CitSUS5 (Fig. 4A). The combination of CitNAC47 and CitZAT5 showed a synergistic effect on the transcriptional activation of the CitSUS5 promoter and this was due to the protein–protein interaction, confirmed via BiFC and yeast two-hybrid assays (Fig. 5), suggesting that CitZAT5 might contribute to sucrose metabolism by interacting with CitNAC47 to synergistically regulate the expression of CitSUS5. Although previous studies in strawberry fruit have shown that the coordination between FaMYB10 and FabHLH3 may affect sucrose accumulation by reversing the repressive effect of FaMYB44.2 on FaSPS3 (Wei et al. 2018), such an interaction between ZAT-NAC has not been reported for the regulation of sugar accumulation, which provides clues about the regulation of sugar accumulation.
Conclusion
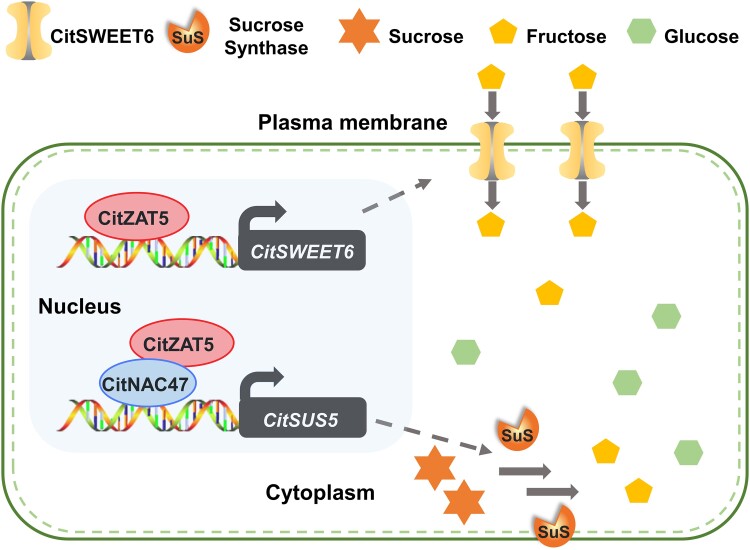
In this study, CitSUS5 and CitSWEET6 were shown to be important for the accumulation of hexose in citrus fruit. In addition, a zinc finger transcription factor, CitZAT5, was characterized, which functions as a key regulator in both sucrose metabolism and fructose transport. On the one hand, CitZAT5 activates the transcription of CitSUS5 in conjunction with CitNAC47, which promotes the degradation of sucrose into hexoses; on the other hand, it also regulates the expression of CitSWEET6, which participates in fructose transport (Fig. 8). CitZAT5 transcript levels exhibited a significant correlation with hexose content in different citrus cultivars, which supports its crucial role in hexose accumulation across citrus cultivars. These findings enhance our understanding of the regulatory mechanism of sugar accumulation and provide insights for the improvement of fruit flavor quality and modulation of the hexose proportion.
Figure 8.
A working model showing the molecular mechanism regulating sugar accumulation in citrus fruit. CitZAT5 can interact with CitNAC47, and the complex binds to the promoter of CitSUS5 to promote its expression. Increased sucrose synthase degrades more sucrose into hexoses. In addition, CitZAT5 directly binds to the promoter of CitSWEET6 to induce its expression, resulting in a high level of fructose.
Materials and methods
Plant materials
Satsuma mandarin “Gongchuan” (C. unshiu Marcov. Variety Miyagawa wase) and “Youliang” (C. unshiu Marcov. Variety Yura) were both obtained from Linhai, Zhejiang province, China. Fruits of uniform size and appearance were harvested at 5 time points, 60, 90, 120, 150, and 180 DAFB (days after full bloom), with 3 biological replicates. Fruit flesh was frozen in liquid nitrogen and stored at −0°C for further analysis.
Eight varieties of satsuma mandarin were obtained from Taizhou, Zhejiang province, China. “LJ,” “ST,” “FZG,” “FF,” “QG,” “RNJ,” “WZ,” and “XJ” represent the names of these 8 varieties: “LiJian,” “ShanTian,” “FeiZhiGuang,” “FengFu,” “QiuGuang,” “RiNanJi,” “WeiZhang,” and “XingJin,” respectively. The fruits of these 8 varieties were harvested at the mature stage and stored at −80°C.
Sugar measurement
The sugars of peeled citrus fruit were extracted and measured according to Li et al. (2017b) with some modification. Samples (0.1 g) were ground in liquid nitrogen, extracted with 1.4 mL methanol at 70°C for 15 min, and then centrifuged at 10,000 × g. The upper phase was removed and mixed with 1.5 mL Milli-Q water and 0.75 mL trichloromethane. The mixture was centrifuged at 2,200 × g for 10 min. The upper phase was removed and stored at −80°C until analysis. Aliquots of 100 µL upper phase were dried in a vacuum. The residue was dissolved in 60 µL 20 mg ml−1 pyridine methoxyamine hydrochloride and incubated at 37°C for 1.5 h. The samples were then treated with 40 µL Bis (trimethylsilyl) trifluoroacetamide (1% v/v trimethylchlorosilane) at 37°C for 0.5 h. Ribitol (20 µL, 0.2 mg mL−1) was added to each sample as an internal standard. A 1 µL aliquot of each sample was absorbed with a split ratio of 10 : 1 and injected into a GC-MS fitted with a fused silica capillary column (30 m × 0.25 mm internal diameter, 0.25 µm DB-5 MS stationary phase). The injector temperature was 250°C and the helium carrier gas had a flow rate of 10.0 mL min−1. The column temperature was held at 100°C for 1 min, increased to 185°C at a rate of 2.5°C min−1, then increased to 190°C at a rate of 0.35°C min−1, subsequently increased to 250°C at a rate of 8°C min−1, and held for 5 min, and then finally increased to 280°C at a rate of 5°C min−1 and held for 3 min. The MS operating parameters were as follows: ion source temperature was 230°C and interface temperature was 280°C. By comparing with the retention time (RT) of available authentic standards, compounds could be distinguished and notarized: percentages were preliminarily obtained by normalizing the data using the internal standard method. The absolute amounts of the sugars were determined by comparison with calibration standard curves.
Total soluble solids (TSS) measurement
TSS were measured using a digital hand-held refractometer (PR101-α, Atago, Japan). Three drops of juice were measured, and an average calculated for 1 replicate and 3 biological replicates was used to obtain a final TSS value.
Dual-luciferase assays
Full-length coding sequences (CDS) of transcription factors were amplified and inserted into the pGreen II 0029 62-SK vector (Hellens et al. 2005). The promoters of CitSWEET6 and CitSUS5 were constructed in the pGreen II 0800-LUC vector (Hellens et al. 2005). Primers for vector construction are listed in Supplemental Table S1. All constructs were electroporated into Agrobacterium tumefaciens GV3101, and the cultures were adjusted to an OD600 of 0.75 with infiltration buffer (10 mM MES, 10 mM MgCl2, 150 mM acetosyringone, pH 5.6). For the investigation of the activity of each transcription factor with 1 of the gene promoters, a mixture of A. tumefaciens containing TF (1 mL) and promoter (100 μL) was infiltrated into N. benthamiana leaves by a needleless syringe. N. benthamiana plants were grown in a greenhouse with a light/dark cycle of 16 : 8 h at 24°C. Three days after infiltration, discs from the N. benthamiana leaves were collected, and the enzyme activities of firefly luciferase (LUC) and Renilla luciferase (REN) were measured using the Dual-Luciferase Reporter Assay Kit (Promega, America) on a GloMax 96 Microplate Luminometer (Promega, America). The LUC/REN value of the empty vector SK on the promoter was set as 1, as a calibrator. At least 4 biological replicates were performed for each individual experiments.
RNA extraction and RT-qPCR
Total RNA was extracted according to Wang et al. 2020a with some modification. For cDNA synthesis, 1 µg of total RNA from each sample was treated with a gDNA wiper (Vazyme, Nanjing, China) to remove genomic DNA and HiScript II qRT SuperMix (Vazyme) was used to synthesize first-strand cDNA. All gene sequences were obtained using the Citrus Genome Database (http://www.citrusgenomedb.org), and primer sequences were designed by the NCBI website (https://www.ncbi.nlm.nih.gov/). RT-qPCR was performed using LightCycler 480 (Roche) and ChamQ Universal SYBR qPCR Master Mix (Vazyme). The PCR mixture (20 µL) included 10 µL ChamQ Universal SYBR qPCR Master Mix, 2 µL diluted cDNA, 0.4 µL of each gene-specific primer (10 µ M), and 7.2 µL DEPC-treated water. The reaction was performed using the following program: RT-qPCR initiated for 5 min at 95°C, followed by 50 cycles of 95°C for 10 s, 60°C for 10 s, and 75°C for 15 s, followed by an automatic melting curve analysis. Citrus actin (XM_006464503) was used as a control to quantify cDNA abundance (Li et al. 2017b). The expression levels were analyzed by the 2−ΔCT method. Primers for reverse transcription quantitative PCR are listed in Supplemental Table S2.
Heterologous expression of CitSWEET6 and CitSWEET8 in yeast
For complementation assays in yeast (Saccharomyces cerevisiae) cells, the full-length coding sequences (CDS) of CitSWEET6 and CitSWEET8 were inserted into the PDR196 vector. Primers for vector construction are listed in Supplemental Table S1. The constructs were transformed into the hexose transport-deficient yeast strain EBY.VW4000 (Schneidereit et al. 2003) and the sucrose uptake-deficient yeast strain SUSY7/ura3 (Riesmeier et al. 1992). The empty pDR196 vector (Rentsch et al. 1995; Meyer et al. 2006) was used as a negative control. The transformants were cultured on SD (synthetic-deficient)/-uracil media supplemented with a specific sugar as the sole carbon source and were diluted to OD600 values of 0.06, 0.006, and 0.0006. For yeast hexose mutant cells, serial dilutions of yeast cell suspensions were dropped onto solid SD/-uracil media consisting of either 2% w/v maltose (control) or 2% glucose/fructose. For yeast sucrose uptake-deficient cells, serial dilutions of yeast cell suspensions were added dropwise onto solid SD/-uracil media containing 2% glucose (control) or 2% sucrose. Yeast cells were grown on medium for 3 to 4 d at 30°C.
Subcellular localization analysis
The CDS of CitSUS5, CitSWEET6, CitZAT5, and CitNAC47 without the stop codon was fused to the pCAMBIA1300-sGFP vector (Chalfie et al. 1994) at the C-terminus and expressed transiently in transgenic N. benthamiana plants which expressed H2B-RFP as a nuclear marker (Li et al. 2017a) by A. tumefaciens infiltration (GV3101) using the same method as mentioned for the dual-luciferase assay. Two days after injection, the fluorescence was imaged with a confocal laser scanning microscope (Nikon A1-SHS, Tokyo, Japan). The excitation of the GFP signal was performed using a 488-nm solid-state laser, fluorescence was detected at 498 to 520 nm, and the intensity and gain were 2% and 630, respectively. The RFP signal excitation was performed using a 587 nm solid-state laser, and fluorescence was detected at 580 to 610 nm, and the intensity and gain were 4% and 630, respectively. The primers for GFP construction are listed in Supplemental Table S1.
The 35S::CitSWEET6-GFP fusion construct was transformed into rice protoplasts. Rice protoplast preparation and transformation were performed according to a previous study (Chen et al. 2011). A 63× oil immersion objective was used for confocal imaging. N-(3-Triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (FM4-64; cat. No. T3166, Thermo Fisher Scientific) was a cell membrane dye used as a plasma membrane (PM) marker (Griffing 2008). FM4-64 was finally added on the slides to stain the prepared protoplasts before being observed. Fluorescence in protoplasts was observed using a Zeiss LSM710 confocal laser scanning microscope. The excitation of the GFP signal was performed using a 488-nm solid-state laser, fluorescence was detected at 498 to 520 nm, and the intensity and gain were 60% and 700, respectively. The PM marker signal excitation was performed using a 514 nm solid-state laser, and fluorescence was detected at 560 to 635 nm, and the intensity and gain were 60% and 650, respectively.
Bimolecular fluorescence complementation assay
Full-length CitZAT5 and CitNAC47 were cloned into either C-terminal or N-terminal fragments of yellow fluorescent protein (YFP) vectors (Lv et al. 2014). The constructs were transformed into A. tumefaciens (strain GV3101) and then transiently expressed in H2B-RFP transgenic N. benthamiana leaves (Li et al. 2017a). Two days after injection, the YFP fluorescence of N. benthamiana leaves was detected by a confocal laser scanning microscope (Nikon A1-SHS). The excitation of GFP signal was performed using a 488-nm solid-state laser, fluorescence was detected at 498 to 520 nm, and the intensity and gain were 2% and 640, respectively. The YFP signal excitation was performed using a 488 nm solid-state laser, and fluorescence was detected at 515 to 545 nm, and the intensity and gain were 30% and 640, respectively. The primers used for vector construction are listed in Supplemental Table S1.
Enzyme activity assays
Sucrose synthase (SuS) activity was assayed in the degradative direction by a sucrose synthase (decomposition direction SS-I) activity assay kit (Solarbio, Beijing, China). Samples (0.1 g) were ground in liquid nitrogen and prepared for extraction according to the manufacturer's instructions. SuS is a key enzyme in plant sugar metabolism, which catalyzes the reversible synthesis of sucrose from fructose and UDPG and the cleavage of sucrose, in the presence of UDP, to fructose and UDPG. Fructose can react with 3,5-dinitrosalicylic acid to produce a brown-red substance with a characteristic absorption peak at 540 nm. The activity of SS-I can be calculated by measuring the change of absorbance value at 540 nm. After extraction, the activity determination was performed by a Synergy H1 microplate reader (BioTek, America).
Yeast one-hybrid assay
Yeast one-hybrid assays were performed to test the interaction between transcription factors and the promoters of 2 structural genes according to the Matchmaker Gold Yeast One-hybrid Library Screening System (Clotech, Japan). The promoter sequence of CitSUS5 was amplified and inserted into the pAbAi vector (Clotech, Japan). When testing the AbA resistance of the pCitSWEET6-AbAi, even a concentration of 800 ng mL−1 Aureobasidin A failed to suppress basal expression of AbAr in either bait strain. Three tandem copies of potential target element of CitSWEET6 were synthesized and inserted into the pAbAi vector. The recombinant vectors were then linearized and transformed into Y1HGold yeast strain. The full-length CitNAC47 and CitZAT5 were cloned into the pGADT7 vector. The Y1HGold strain carrying the CitSUS5 promoter was transfected with the CitNAC47-pGADT7 plasmid and the empty vector pGADT7, with the latter as negative control. The Y1HGold strain carrying potential target element from the CitSWEET6 promoter (listed in Supplemental Table S1) was transfected with the CitZAT5-pGADT7 plasmid and also with the empty vector pGADT7 as negative control. The probes are listed in Supplemental Table S1.
Recombinant protein purification and EMSA
Electrophoretic mobility shift assay (EMSA) was used to analyze the interaction of TFs and the promoters. The full-length CitZAT5 and CitNAC47 were inserted into pET-32a (Clontech, Japan) to generate the recombinant N-terminal TFs His fusion protein. The constructs were purified and transformed into Escherichia coli strain Rosetta 2 (DE3) pLysS (Novagen, Germany). The transformed cells were incubated in LB medium until OD600 reached 0.6 and then incubated at 16°C for 20 h induced by 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The cells were collected by centrifugation and resuspended in buffer (20 mM Tris-HCl, pH = 8.0, 0.5 M NaCl), after which they were subjected to sonication on ice with 3-s/2-s on/off cycle for 15 min and centrifuged at 10,000 rpm for 20 min at 4°C. Subsequently, Ni-NTA resin (Transgen, China) was added to the supernatant to combine His-tagged protein at 4°C for 1 h, and the target protein was eluted with elution buffers containing gradient imidazole (100, 125, and 150 mM). EMSA was performed by using a LightShift Chemiluminescent EMSA kit (Thermo, America) according to the manufacturer's instructions. Single-strand oligonucleotides were synthesized and 30-biotin-end-labelled by HuaGene. The EMSA probes are listed in Supplemental Table S3.
Yeast two-hybrid assay
Protein–protein interaction was investigated in yeast using the DUAL hunter system (Dualsystems Biotech, Switzerland). The full-length coding sequence of CitZAT5 was cloned into the pDHB1 vector as bait, and the full length of CitNAC47 was cloned into the pPR3N vector as prey. The primers used for vector construction are described in Supplemental Table S1. All constructs were transformed into the yeast strain NMY51 according to the manufacturer's instructions. Liquid cultures of double transformants were plated at OD600 = 0.02 dilutions on elective media: (i) SD medium lacking Trp and Leu (DDO); (ii) SD medium lacking Trp, Leu, His, and Ade (QDO); and (iii) SD medium lacking Trp, Leu, His, and Ade and supplemented with 1 mM 3-amino-1,2,4-triazole (QDO+3AT). Autoactivation was judged according to the cotransformation of empty pPR3-N vectors and target genes with pDHB1. Protein–protein interaction assays were performed by cotransformation of CitNAC47 in pPR3N and CitZAT5 in pDHB1. The presence of colonies in QDO and QDO+3AT indicated a protein–protein interaction. Yeast cells were grown on medium for 3 to 4 d at 30°C.
Transient overexpression, RNAi, and VIGS assays
The full-length CDS of CitZAT5 and CitSUS5 was inserted into the pGreenII 0029 62-SK vector, acting as the overexpression construct. Forward and reverse PCR-amplified cDNA fragments of CitZAT5 were inserted into the 2×CaMV35S-driven vector pHB to produce the CitZAT5-RNAi construct. The constructs were electroporated into A. tumefaciens strain GV3101. The cultures were adjusted to an OD600 of 0.6 to 0.8 with infiltration buffer (10 mM MES, 10 mM MgCl2, 150 mM acetosyringone, pH 5.6). For transient overexpression and RNAi, citrus pulp was soaked in the A. tumefaciens suspension carrying empty vector or target gene and then cultivated on MS medium for 7 d.
The full-length of CitSWEET6 was cloned downstream of the CaMV 35S promoter of a modified pCAMBIA1301 vector (Khanna and Raina 1999). The recombinant vector was then introduced into A. tumefaciens strain EHA105 (Lv et al. 2014). The method of infection and cultivation was the same as described above.
Virus-induced gene silencing (VIGS) was performed based on the TRV-RNA1 (pTRV1) and TRV-RNA2 (pTRV2) vectors (Liu et al. 2002). A 395-bp CitSUS5 fragment was inserted into pTRV2. The pTRV1, pTRV2, and pTRV2-CitSUS5 constructs were injected into strain EHA105. The method of infection and cultivation was the same as described above. For VIGS, the pTRV1 and pTRV2-CitSUS5 constructs in infiltration buffer were mixed in a 1:1 ratio, with pTRV1 and pTRV2 acted as the control. Satsuma mandarin (Citrus unshiu Marcov.) at 120 to 130 DAFB was used for transient CitSUS5-OE and CitSUS5-VIGS, and Huyou (Citrus changshanensis) fruit at 210 to 240 DAFB was used for transient CitZAT5-OE, CitZAT5-RNAi, and CitSWEET6-OE.
VIGS of CitZAT5 was performed according to Li et al. (2021b) with some modification. A 370-bp CitZAT5 fragment was inserted into pTRV2. The pTRV1, pTRV2, and pTRV2-CitZAT5 constructs were injected into strain EHA105. The bacterial cells were cultivated in LB broth and adjusted to an OD600 of 0.8 with infiltration buffer as mentioned above. For VIGS, the pTRV1 and pTRV2-CitZAT5 constructs in infiltration buffer were mixed in a 1:1 ratio, with pTRV1 and pTRV2 also mixed to act as the control. The infiltration mixtures were infiltrated into germinated seedlings of “Ponkan” (C. reticulata Blanco cv. Ponkan) by the vacuum method. After infiltration, the seedlings were grown in darkness for 2 d and then moved to a growth chamber for 40 d. The sugar contents and gene expression were determined as described above. The probes are listed in Supplemental Table S1.
Statistical analysis
Data analysis was performed with Microsoft Excel, GraphPad Prism 6, and IBM SPSS Statistics 26. Figures were drawn with GraphPad Prism 6, Cytoscape 3.9.0, Microsoft PowerPoint, and Photoshop. The heatmap was drawn with TBtools. The phylogenetic trees were constructed using MEGA 7.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XM_006439225 (CitSWEET6), XM_006430338 (CitSUS5), XM_006424377 (CitZAT5), XM_006443288 (CitNAC47), and XM_006464503 (Citrus Actin).
Supplementary Material
Acknowledgments
We would like to thank Professor Xueren Yin for comments and suggestions for the manuscript. We thank Prof. Zhenghe Li (Zhejiang University, PR China) for the seeds of H2B-RFP transgenic line, and Prof. Xiaolei Sui and Prof. Zhenxian Zhang (China Agricultural University, PR China) for the yeast strains EBY.VW4000 and SUSY7/ura3.
Contributor Information
Heting Fang, College of Agriculture & Biotechnology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China.
Yanna Shi, College of Agriculture & Biotechnology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; The State Agriculture Ministry Laboratory of Horticultural Plant Growth, Development and Quality Improvement, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China.
Shengchao Liu, College of Agriculture & Biotechnology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China.
Rong Jin, Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; Department of Horticulture and Agricultural Experiment Station, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China.
Jun Sun, Zhejiang Agricultural Technology Extension Center, Hangzhou 310029, China.
Donald Grierson, Division of Plant and Crop Sciences, School of Biosciences, University of Nottingham, Nottingham, UK.
Shaojia Li, College of Agriculture & Biotechnology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; The State Agriculture Ministry Laboratory of Horticultural Plant Growth, Development and Quality Improvement, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China.
Kunsong Chen, College of Agriculture & Biotechnology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China; The State Agriculture Ministry Laboratory of Horticultural Plant Growth, Development and Quality Improvement, Zhejiang University, Zijingang Campus, Hangzhou 310058, PR China.
Author contributions
S.J.L. and K.C. conceived the research plans; S.J.L. and K.C. supervised the experiments; H.F. and S.C.L. performed most of the experiments; R.J. and J.S. provided technical assistance to H.F.; S.J.L. and K.C. designed the experiments and analyzed the data; S.C.L., H.F., and Y.S. wrote the article with contributions of all the authors, and D.G. revised the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Heatmap of fold changes (FC, FPKM ratio of “YL”/“GC”) in transcripts of all SPS, SUS, INV, and SWEET family members in citrus fruit.
Supplemental Figure S2 . Expression pattern of 6 candidate genes in transcriptome of “GC” and “YL” fruits.
Supplemental Figure S3 . Analysis of the transport activity of CitSWEET8 in EBY.VW4000 and SUSY7/ura3 yeast mutant strain.
Supplemental Figure S4 . Subcellular localization assay was performed with GFP vector and CitSUS5 in Nicotiana benthamiana leaves transformed by agroinfiltration (bars = 20 μm).
Supplemental Figure S5 . Subcellular localization assays.
Supplemental Figure S6 . Phylogenetic analysis of CitSWEET6 and 30 reported SWEET proteins.
Supplemental Figure S7 . Expression pattern of 16 transcription factors in transcriptome of “GC” and “YL” fruits.
Supplemental Figure S8 . Expression pattern and subcellular localization assays of TFs.
Supplemental Figure S9 . VIGS of CitZAT5 reduced sugar contents in citrus plants.
Supplemental Figure S10 . Expression pattern of CitSPS4 and CitSWEET11d in “GC” and “YL” fruits.
Supplemental Figure S11 . Correlation analysis between hexose content and CitSUS5, CitSWEET6 transcripts.
Supplemental Table S1 . Primers used for vector construction.
Supplemental Table S2 . Primers used for reverse transcription quantitative PCR.
Supplemental Table S3 . Primers used for EMSA.
Funding
This work was supported by the National Key Research and Development Program of China (2022YFD2100102), the National Natural Science Foundation of China (31801591), the 111 Project (B17039), and Zhejiang Provincial Cooperative Extension Project of Agricultural Key Technology (2022XTTGGP01).
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
- Baker RF, Leach KA, Boyer NR, Swyers MJ, Benitez-Alfonso Y, Skopelitis T, Luo A, Sylvester A, Jackson D, Braun DM. Sucrose transporter ZmSut1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiol. 2016:172(3):1876–1898. 10.1104/pp.16.00884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava S, Sawant K, Tuberosa R. Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant Breed. 2013:132(1):21–32. 10.1111/pbr.12004 [DOI] [Google Scholar]
- Braun DM. Phloem loading and unloading of sucrose: what a long, strange trip from source to sink. Annu Rev Plant Biol. 2022:73(1):15.1–15.32. 10.1146/annurev-arplant-070721-083240 [DOI] [PubMed] [Google Scholar]
- Bush DR. Identifying the pathways that control resource allocation in higher plants. Proc Natl Acad Sci USA. 2020:117(16):8669–8671. 10.1073/pnas.2002581117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Černý M, Habánová H, Berka M, Luklová M, Brzobohatý B. Hydrogen peroxide: its role in plant biology and crosstalk with signalling networks. Int J Mol Sci. 2018:19(9):2812. 10.3390/ijms19092812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu G, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994:263(5148):5148. 10.1126/science.8303295 [DOI] [PubMed] [Google Scholar]
- Chen J, Liu Y, Ni J, Wang Y, Bai Y, Shi J, Gan J, Wu Z, Wu P. OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 2011:157(1):269–278. 10.1104/pp.111.181669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010:468(7323):527–532. 10.1038/nature09606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012:335(6065):207–211. 10.1126/science.1213351 [DOI] [PubMed] [Google Scholar]
- Chen T, Zhang Z, Li B, Qin G, Tian S. Molecular basis for optimizing sugar metabolism and transport during fruit development. aBIOTECH. 2021:2(3):330–340. 10.1007/s42994-021-00061-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Chao YC, Tseng TW, Huang CK, Lo PC, Lu CA. Two MYB-related transcription factors play opposite roles in sugar signaling in Arabidopsis. Plant Mol Biol. 2017:93(3):299–311. 10.1007/s11103-016-0562-8 [DOI] [PubMed] [Google Scholar]
- Chong J, Piron MC, Meyer S, Merdinoglu D, Bertsch C, Mestre P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J Exp Bot. 2014:65(22):6589–6601. 10.1093/jxb/eru375 [DOI] [PubMed] [Google Scholar]
- Coleman HD, Ellis DD, Gilbert M, Mansfield SD. Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotechnol J. 2006:4(1):87–101. 10.1111/j.1467-7652.2005.00160.x [DOI] [PubMed] [Google Scholar]
- Coleman HD, Yan J, Mansfield SD. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci USA. 2009:106(31):13118–13123. 10.1073/pnas.0900188106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Mai Y, Niu J. Fruit characteristics, soluble sugar compositions and transcriptome analysis during the development of Citrus maxima “seedless”, and identification of SUS and INV genes involved in sucrose degradation. Gene. 2019:689:131–140. 10.1016/j.gene.2018.12.016 [DOI] [PubMed] [Google Scholar]
- Deng Y, Wang J, Zhang Z, Wu Y. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol J. 2020:18(9):1897–1907. 10.1111/pbi.13349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doidy J, Grace E, Kühn C, Simon-Plas F, Casieri L, Wipf D. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 2012:17(7):413–422. 10.1016/j.tplants.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Feurtado JA, Huang D, Wicki-Stordeur L, Hemstock LE, Potentier MS, Tsang EW, Cutler AJ. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell. 2011:23(5):1772–1794. 10.1105/tpc.111.085134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Duan X, Jiang H, Yang F, Qi H. CmMYB113 regulates ethylene-dependent sucrose accumulation in postharvest climacteric melon fruit. Postharvest Biol Technol. 2021:181:111682. 10.1016/j.postharvbio.2021.111682 [DOI] [Google Scholar]
- Griffing LR. FRET Analysis of transmembrane flipping of FM4-64 in plant cells: is FM4-64 a robust marker for endocytosis?. J Microsc. 2008:231(2):291–298. 10.1111/j.1365-2818.2008.02042.x [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005:1(1):1–14. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Zhang F, Song S, Yu X, Ren Y, Zhao X, Liu H, Liu G, Wang Y, He H. CsSWEET2, a hexose transporter from cucumber (Cucumis sativus L.), affects sugar metabolism and improves cold tolerance in Arabidopsis. Int J Mol Sci. 2022:23(7):3886. 10.3390/ijms23073886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Li S, Lin X, Fang H, Shi Y, Grierson D, Chen K. Transcription factor CitERF16 is involved in citrus fruit sucrose accumulation by activating CitSWEET11d. Front Plant Sci. 2021:12:809619. 10.3389/fpls.2021.809619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wu W, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot. 2008:59(11):2991–3007. 10.1093/jxb/ern155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Yu D, Wang X. VvWRKY22 transcription factor interacts with VvSnRK1.1/VvSnRK1.2 and regulates sugar accumulation in grape. Biochem Biophys Res Commun. 2021:554:193–198. 10.1016/j.bbrc.2021.03.092 [DOI] [PubMed] [Google Scholar]
- Jian H, Lu K, Bo Y, Wang T, Zhang L, Zhang A, Wang J, Liu L, Qu C, Li J. Genome-Wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in oilseed rape (Brassica napus L.). Front Plant Sci. 2016:7:1464. 10.3389/fpls.2016.01464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius BT, Leach KA, Tran TM, Mertz RA, Braun DM. Sugar transporters in plants: new insights and discoveries. Plant Cell Physiol. 2017:58(9):1442–1460. 10.1093/pcp/pcx090 [DOI] [PubMed] [Google Scholar]
- Katz E, Fon M, Lee YJ, Phinney BS, Sadka A, Blumwald E. The citrus fruit proteome: insights into citrus fruit metabolism. Planta. 2007:226(4):989–1005. 10.1007/s00425-007-0545-8 [DOI] [PubMed] [Google Scholar]
- Kelebeka H, Selli S. Determination of volatile, phenolic, organic acid and sugar components in a Turkish cv. Dortyol (Citrus sinensis L. Osbeck) orange juice. J Sci Food Agric. 2011:91(10):1855–1862. 10.1002/jsfa.4396 [DOI] [PubMed] [Google Scholar]
- Khanna HK, Raina SK. Agrobacterium-mediated transformation of indica rice cultivars using binary and superbinary vectors. Aust J Plant Physiol. 1999:26(4):311–324. 10.1071/PP98160 [DOI] [Google Scholar]
- Kiełbowicz-Matuk A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 2012:185–186:78–85. 10.1016/j.plantsci.2011.11.015 [DOI] [PubMed] [Google Scholar]
- Ko HY, Ho LH, Neuhaus HE, Guo WJ. Transporter SlSWEET15 unloads sucrose from phloem and seed coat for fruit and seed development in tomato. Plant Physiol. 2021:187(4):2230–2245. 10.1093/plphys/kiab290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhao N, Li Z, Xu X, Wang Y, Yang X, Liu S, Wang A, Zhou X. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 2017a:13(2):1006213. 10.1371/journal.ppat.1006213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li P, Ma F, Dandekar AM, Cheng L. Sugar metabolism and accumulation in the fruit of transgenic apple trees with decreased sorbitol synthesis. Hortic Res. 2018:5(1):60. 10.1038/s41438-018-0064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Wang L, Liu H, Yuan M. Impaired SWEET-mediated sugar transportation impacts starch metabolism in developing rice seeds. Crop J. 2021a:10(1):98–108. 10.1016/j.cj.2021.04.012 [DOI] [Google Scholar]
- Li SJ, Liu SC, Lin XH, Grierson D, Yin XR, Chen KS. Citrus heat shock transcription factor CitHsfA7-mediated citric acid degradation in response to heat stress. Plant Cell Environ. 2021b:45(1):95–104. 10.1111/pce.14207 [DOI] [PubMed] [Google Scholar]
- Li SJ, Yin XR, Wang WL, Liu XF, Chen KS. Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J Exp Bot. 2017b:68(13):3419–3426. 10.1093/jxb/erx187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Guo W, Li J, Yue P, Bu H, Jiang J, Liu W, Xu Y, Yuan H, Li T, et al. Histone acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in pear fruit. Plant Physiol. 2020:182(4):2035–2046. 10.1104/pp.20.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu H, Yao X, Wang J, Feng S, Sun L, Ma S, Xu K, Chen LQ, Sui X. Hexose transporter CsSWEET7a in cucumber mediates phloem unloading in companion cells for fruit development. Plant Physiol. 2021c:186(1):640–654. 10.1093/plphys/kiab046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HT, Ji Y, Liu Y, Tian SH, Gao QH, Zou XH, Yang J, Dong C, Tan JH, Ni DA. The sugar transporter system of strawberry: genome-wide identification and expression correlation with fruit soluble sugar-related traits in a Fragaria × ananassa germplasm collection. Hortic Res. 2020:7(1):1303–1321. 10.1038/s41438-020-00359-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002:30(4):415–429. 10.1046/j.1365-313X.2002.01297.x [DOI] [PubMed] [Google Scholar]
- Liu YJ, Wang GL, Ma J, Xu ZS, Wang F, Xiong AS. Transcript profiling of sucrose synthase genes involved in sucrose metabolism among four carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biol. 2018:18(1):8. 10.1186/s12870-017-1221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, et al. SPX4 Negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell. 2014:26(4):1586–1597. 10.1105/tpc.114.123208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QJ, Sun MH, Lu J, Kang H, You CX, Hao YJ. An apple sucrose transporter MdSUT2.2 is a phosphorylation target for protein kinase MdCIPK22 in response to drought. Plant Biotechnol J. 2018:17(3):625–637. 10.1111/pbi.13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QJ, Sun MH, Lu J, Liu YJ, Hu DG, Hao YJ. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 2017:174(4):2348–2362. 10.1104/pp.17.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan J, Singh A, Ranjan A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol Plant. 2020:171(4):620–637. 10.1111/ppl.13210 [DOI] [PubMed] [Google Scholar]
- Meyer A, Eskandari S, Grallath S, Rentsch D. AtGAT1, a high affinity transporter for γ-aminobutyric acid in Arabidopsis thaliana. J Biol Chem. 2006:281(11):7197–7204. 10.1074/jbc.M510766200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Wang Q, Wang Y, Cheng B, Zhao Y, Zhu S. A maize NAC transcription factor, ZmNAC34, negatively regulates starch synthesis in rice. Plant Cell Rep. 2019:38(12):1473–1484. 10.1007/s00299-019-02458-2 [DOI] [PubMed] [Google Scholar]
- Rehman A, Wang N, Peng Z, He S, Zhao Z, Gao Q, Wang Z, Li H, Du X. Identification of C2H2 subfamily ZAT genes in Gossypium species reveals GhZAT34 and GhZAT79 enhanced salt tolerance in Arabidopsis and cotton. Int J Biol Macromol. 2021:184:967–980. 10.1016/j.ijbiomac.2021.06.166 [DOI] [PubMed] [Google Scholar]
- Ren Y, Li M, Guo S, Sun H, Zhao J, Zhang J, Liu G, He H, Tian S, Yu Y, et al. Evolutionary gain of oligosaccharide hydrolysis and sugar transport enhanced carbohydrate partitioning in sweet watermelon fruits. Plant Cell. 2021:33(5):1554–1573. 10.1093/plcell/koab055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB. NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 1995:370(3):264–268. 10.1016/0014-5793(95)00853-2 [DOI] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 1992:11(13):4705–4713. 10.1002/j.1460-2075.1992.tb05575.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru L, Chen B, Li Y, Wills RBH, Lv Z, Lu G, Yang H. Role of sucrose phosphate synthase and vacuolar invertase in postharvest sweetening of immature sweetpotato tuberous roots (Ipomoea batatas (L.) Lam cv ‘Xinxiang’). Sci Hortic. 2021:282:110007. 10.1016/j.scienta.2021.110007 [DOI] [Google Scholar]
- Ruan YL. Signaling role of sucrose metabolism in development. Mol Plant. 2012:5(4):763–765. 10.1093/mp/sss046 [DOI] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Büttner M. Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiol. 2003:133(1):182–190. 10.1104/pp.103.026674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammai A, Petreikov M, Yeselson Y, Faigenboim A, Moy-Komemi M, Cohen S, Cohen D, Besaulov E, Efrati A, Houminer N, et al. Natural genetic variation for expression of a SWEET transporter among wild species of Solanum lycopersicum (tomato) determines the hexose composition of ripening tomato fruit. Plant J. 2018:96(2):343–357. 10.1111/tpj.14035 [DOI] [PubMed] [Google Scholar]
- Smeekens S, Hellmann HA. Sugar sensing and signaling in plants. Front Plant Sci. 2014:5:113. 10.3389/fpls.2014.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, Mccarty DR, Chourey PS, et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet. 2015:47(12):1489–1493. 10.1038/ng.3422 [DOI] [PubMed] [Google Scholar]
- Stein O, Granot D. An overview of sucrose synthases in plants. Front Plant Sci. 2019:10:95. 10.3389/fpls.2019.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Gao Z, Wang J, Huang Y, Chen Y, Li J, Lv M, Wang J, Luo M, Zuo K. Cotton fiber elongation requires the transcription factor GhMYB212 to regulate sucrose transportation into expanding fibers. New Phytol. 2019:222(2):864–881. 10.1111/nph.15620 [DOI] [PubMed] [Google Scholar]
- Tang H, Bi H, Liu B, Lou S, Song Y, Tong S, Chen N, Jiang Y, Liu J, Liu H. WRKY33 Interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 2021:229(1):106–125. 10.1111/nph.17020 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005:41(2):195–211. 10.1111/j.1365-313X.2004.02288.x [DOI] [PubMed] [Google Scholar]
- Wang K, Ding Y, Cai C, Chen Z, Zhu C. The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol Plant. 2019a:165(4):690–700. 10.1111/ppl.12728 [DOI] [PubMed] [Google Scholar]
- Wang S, Yokosho K, Guo R, Whelan J, Ruan YL, Ma JF, Shou H. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol. 2019b:180(4):2133–2141. 10.1104/pp.19.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhou Q, Zhou X, Zhang F, Ji S. Ethylene plays an important role in the softening and sucrose metabolism of blueberries postharvest. Food Chem. 2019c:310:125965. 10.1016/j.foodchem.2019.125965 [DOI] [PubMed] [Google Scholar]
- Wang WQ, Wang J, Wu YY, Li DW, Allan AC, Yi XR. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 2020a:225(4):1618–1634. 10.1111/nph.16233 [DOI] [PubMed] [Google Scholar]
- Wang Z, Wei X, Yang J, Li H, Ma B, Zhang K, Zhang Y, Cheng L, Ma F, Li M. Heterologous expression of the apple hexose transporter MdHT2.2 altered sugar concentration with increasing cell wall invertase activity in tomato fruit. Plant Biotechnol J. 2020b:18(2):540–552. 10.1111/pbi.13222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Mao W, Jia M, Xing S, Ali U, Zhao Y, Chen Y, Cao M, Dai Z, Zhang K, et al. FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J Exp Bot. 2018:69(20):4805–4820. 10.1093/jxb/ery249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Cheng MN, Ba LJ, Zeng RX, Luo DL, Qin YH, Liu ZL, Kuang JF, Lu WJ, Chen JY. Pitaya HpWRKY3 is associated with fruit sugar accumulation by transcriptionally modulating sucrose metabolic genes HpINV2 and HpSuSy1. Int J Mol Sci. 2019:20(8):1890. 10.3390/ijms20081890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Li M, Zhang C, Tan Q, Yang X, Sun X, Pan Z, Deng X, Hu C. Effects of phosphorus on fruit soluble sugar and citric acid accumulations in citrus. Plant Physiol Biochem. 2021:160:73–81. 10.1016/j.plaphy.2021.01.015 [DOI] [PubMed] [Google Scholar]
- Wu Y, Lee SK, Yoo Y, Wei J, Kwon SY, Lee SW, Jeon JS, An G. Rice transcription factor OsDOF11 modulates sugar transport by promoting expression of Sucrose Transporter and SWEET genes. Mol Plant. 2018:11(6):833–845. 10.1016/j.molp.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Xiao G, Qin H, Zhou J, Quan R, Lu X, Huang R, Zhang H. OsERF2 controls rice root growth and hormone responses through tuning expression of key genes involved in hormone signaling and sucrose metabolism. Plant Mol Biol. 2016:90(3):293–302. 10.1007/s11103-015-0416-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Tang J, Li Y, Wang W, Li X, Jin L, Xie R, Luo H, Zhao X, Meng Z, et al. STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J. 2009:59(5):789–801. 10.1111/j.1365-313X.2009.03913.x [DOI] [PubMed] [Google Scholar]
- Xu Q, Chen S, Ren Y. Regulation of sucrose transporters and phloem loading in response to environmental cues. Plant Physiol. 2018:176(1):930–945. 10.1104/pp.17.01088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SM, Brill E, Llewellyn DJ, Furbank RT, Ruan YL. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol Plant. 2012:5(2):430–441. 10.1093/mp/ssr090 [DOI] [PubMed] [Google Scholar]
- Zhang C, Bian Y, Hou S, Li X. Sugar transport played a more important role than sugar biosynthesis in fruit sugar accumulation during Chinese jujube domestication. Planta. 2018a:248(5):1187–1199. 10.1007/s00425-018-2971-1 [DOI] [PubMed] [Google Scholar]
- Zhang QY, Ge J, Liu XC, Wang WQ, Liu XF, Yin XR. Consensus co-expression network analysis identifies AdZAT5 regulating pectin degradation in ripening kiwifruit. J Adv Res. 2021a:11:19. 10.1016/j.jare.2020.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng C, Wang M, Li T, Liu X, Jiang J. Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits. Hortic Res. 2021b:8(1):186. 10.1038/s41438-021-00624-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Wang W, Du LQ, Xie JH, Yao YL, Sun GM. Expression patterns, activities and carbohydrate-metabolizing regulation of sucrose phosphate synthase, sucrose synthase and neutral invertase in pineapple fruit during development and ripening. Int J Mol Sci. 2012:13(8):9460–9477. 10.3390/ijms13089460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yin X, Xiao Y, Zhang Z, Li S, Liu X, Zhang B, Yang X, Grierson D, Jiang G, et al. An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiol. 2018b:178(1):189–201. 10.1104/pp.18.00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zou L, Ren C, Ren F, Wang Y, Fan P, Li S, Liang Z. VvSWEET10 mediates sugar accumulation in grapes. Genes (Basel). 2019:10(4):255. 10.3390/genes10040255 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).