Abstract
Grapevine (Vitis vinifera L.) is one of the most widely cultivated fruit crops because the winemaking industry has huge economic relevance worldwide. Uncovering the molecular mechanisms controlling the developmental progression of plant organs will prove essential for maintaining high-quality grapes, expressly in the context of climate change, which impairs the ripening process. Through a deep inspection of transcriptomic data, we identified VviNAC60, a member of the NAC transcription factor family, as a putative regulator of grapevine organ maturation. We explored VviNAC60 binding landscapes through DNA affinity purification followed by sequencing and compared bound genes with transcriptomics datasets from grapevine plants stably and transiently overexpressing VviNAC60 to define a set of high-confidence targets. Among these, we identified key molecular markers associated with organ senescence and fruit ripening. Physiological, metabolic, and promoter activation analyses showed that VviNAC60 induces chlorophyll degradation and anthocyanin accumulation through the upregulation of STAY-GREEN PROTEIN 1 (VviSGR1) and VviMYBA1, respectively, with the latter being upregulated through a VviNAC60–VviNAC03 regulatory complex. Despite sharing a closer phylogenetic relationship with senescence-related homologs to the NAC transcription factor AtNAP, VviNAC60 complemented the nonripening(nor) mutant phenotype in tomato (Solanum lycopersicum), suggesting a dual role as an orchestrator of both ripening- and senescence-related processes. Our data support VviNAC60 as a regulator of processes initiated in the grapevine vegetative- to mature-phase organ transition and therefore as a potential target for enhancing the environmental resilience of grapevine by fine-tuning the duration of the vegetative phase.
The transcriptional factor VviNAC60 regulates organ maturation initiation in grapevine, promoting anthocyanin synthesis, chlorophyll degradation, cell death, and other senescence-related processes.
Introduction
Plant lifespans are marked by ordered entry and exit to different developmental phases. Complex signaling networks, involving time-dependent interactions among hormones, transcription factors, microRNAs and environmental stimuli, govern phase transitions, ensuring dynamic and fine-tuning adjustments of the plant developmental program also in the context of biotic and abiotic external factors (Ma et al. 2020; Manuela and Xu 2020). At the end of vegetative phase plant organs enter maturation phases such as senescence, ripening, and secondary growth. These irreversible developmental processes transform organs from nutrient assimilation to quality formation or nutrient remobilization. Senescence is a finely regulated deterioration process, involving ordered physiological, biochemical, and metabolic changes, crucial for recycling of materials and energy (Lim et al. 2007), whereas ripening is an active presenescence biological process during which fruit undergo dramatic changes to became attractive for animals that will promote the dispersion of seeds.
Senescence and ripening are both accomplished by chlorophyll degradation, secondary metabolite accumulation and cell wall breakdown and dismantling and involve at least some common signaling and regulatory factors including ethylene and transcription factors (Gapper et al. 2013).
At the molecular level, it has been shown that members of the NAC (NAM, ATAF, and CUC) transcription factor family play key roles in the regulation of leaf senescence and fruit ripening by interacting with other transcription factors, hormones, and environmental signals (Forlani et al. 2021; Kou et al. 2021). Tomato (Solanum lycopersicum) NON-RIPENING (NOR) was the first NAC TF described as a master regulator of fruit ripening (White 2002; Kumar et al. 2018; Gao et al. 2020), and more recently shown to play a key role in leaf senescence (Ma et al. 2019).
NAC genes act as both transcriptional activators and repressors and this duality may be achieved by recruiting or interacting with different transcriptional partners (Kim et al. 2016). In peach (Prunus persica), 2 NAC TFs, BL and PpNAC1, have been shown to work in a heterodimeric complex for the activation of the R2R3 MYB gene PpMYB10.1, a regulator of anthocyanin levels in fruit flesh during maturation (Zhou et al. 2015). NAC TFs are also involved in ripening of nonclimacteric fruits, as the NAC gene FaRIF which controls critical ripening-related processes in strawberry (Fragaria × ananassa Duch.; Martin-Pizarro et al. 2021), and FcrNAC22 which mediates red light-induced fruit coloration by enhancing the expression of carotenoid biosynthetic genes in citrus (Fortunella crassifolia Swingle; Gong et al. 2021).
Grapevine (Vitis vinifera L.) is one of the oldest and most widely cultivated perennial nonclimacteric fruit crops in the world. Grape berry development consists of 2 phases of growth: the first, namely “berry formation” or “vegetative/herbaceous phase”, involves pericarp growth due to rapid cell division and elongation; while, the second, “ripening”, involves physical and metabolic changes, including accumulation of sugars, loss of organic acids, softening, skin pigmentation, and synthesis of volatile aromas (Coombe 1992). The quality of grapes is extremely sensitive to environmental factors and global warming is negatively affecting the maturation process (Webb et al. 2007; Kuhn et al. 2014). Clearer understanding of the molecular mechanisms controlling plant organ development and developmental phase transitions is essential in developing strategies for optimizing the grape ripening process and related fruit quality traits in the context of climate change and additional abiotic stresses.
A clear transcriptional distinction between vegetative/green and mature/woody tissues has been shown (Fasoli et al. 2012) and co-expression network analyses have led to the identification of a set of candidate genes, called switch genes, as putative key regulators of the organ phase transition to mature growth in grapevine (Palumbo et al. 2015; Massonnet et al. 2017). Two NAC family members, VviNAC33 and VviNAC60, have been described as switches. We recently demonstrated that VviNAC33 induces leaf de-greening through activation of the STAY-GREEN PROTEIN 1 (VviSGR1) and growth cessation via inhibition of AUXIN EFFLUX FACILITATOR VviPIN1 and VviRopGEF1 expression and proposed this NAC as a key controller of senescence-associated processes both in leaf and berry (D’Inca et al. 2021).
Here, we present a functional characterization of VviNAC60, through combined DNA-binding, transcriptome activity, and functional analyses. We identified VviNAC60 high-confidence promoter targets, revealing its regulative role in secondary metabolism, cell death, and organ de-greening. Stable transgenic lines overexpressing VviNAC60 and its transgenic chimeric repressors showed clear alteration of senescence processes. In addition, the heterologous expression of the VviNAC60 in the tomato nor mutant rescued the nor mutation at the same level as the tomato NAC-NOR. Together, our results define VviNAC60 as a player of the regulatory network controlling processes related to organ senescence and ripening and a target for modification/breeding toward climate resilience and fruit quality in grapevine.
Results
VviNAC60 is activated during vegetative-to-mature phase transition in grapevine organs and is highly co-expressed with senescence and ripening-associated genes
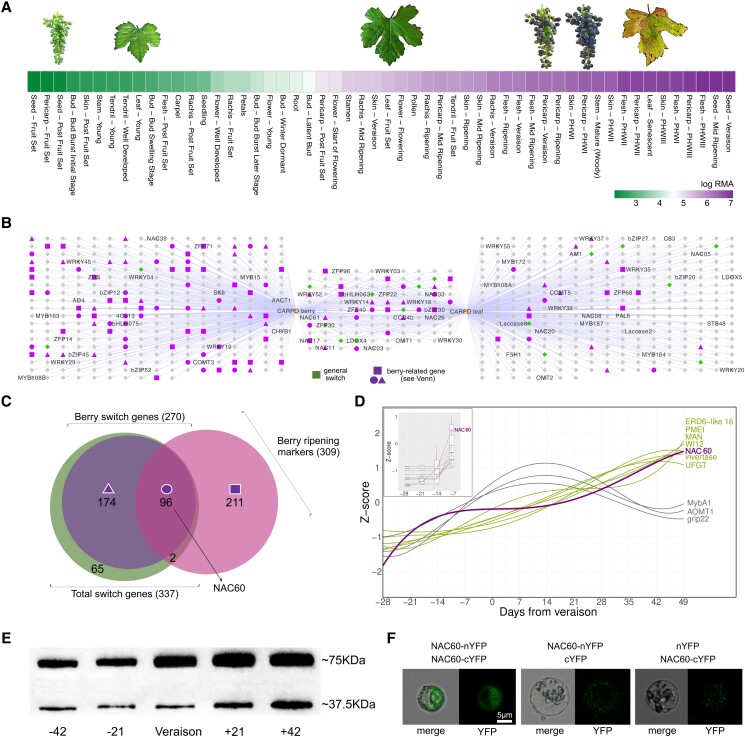
VviNAC60 was previously found among switch genes, putative master regulators of the vegetative-to-mature phase transition in grapevine organs (Palumbo et al. 2015). This gene shows an increase in expression from vegetative/growth to the senescence/ripening phase in all organs, especially in all berry tissues and leaves examined (Fig. 1A; Supplemental Figs. S1, S2A).
Figure 1.
VviNAC60 expression throughout organs development and self-interaction in grapevine. A) Gene expression behavior of VviNAC60 in grapevine organs throughout development. The data were sourced from the atlas transcriptomic dataset of cv. Corvina (Fasoli et al. 2012), RMA, robust multiarray average; PHW, postharvest withering. B) Aggregate co-expression networks of the TOP420 most highly co-expressed genes with VviNAC60 taken from berry and leaf condition-dependent analyses (Orduña et al. 2022). Gene names appearing in the network were cross-referenced from the Grape Gene Reference Catalogue (Navarro-Paya et al. 2022). Node colors and shapes represent the genes classified in C) resulting from the overlap of general and berry switch genes identified in (Palumbo et al. 2015; Massonnet et al. 2017) and berry transition marker genes (Fasoli et al. 2018). D) Expression profiles of VviNAC60 and literature ripening-associated genes (biomarkers) across a high-resolution series of berry developmental stages studied through RNA-seq (Fasoli et al. 2018). E) Western blot analysis of different stages of berry development. Total protein extracts were blotted using anti-VviNAC60 polyclonal antibody. Stages correspond to 42 and 21 d before veraison, veraison, 21- and 42-days post veraison, selected for association with VviNAC60 and biomarker expression profiles. The VviNAC60 protein has a molecular weight of ∼37.5 kDa, which is represented by the lower bands; the upper bands (∼75 kDa) reflect the VviNAC60 homodimer prevalence in all the samples. F) BiFC analysis in grapevine protoplasts showing VviNAC60/VviNAC60 protein interaction. Corresponding controls are also shown in full overexposition mode. Image on left panel shows a representative case of YFP signal being detected in the cell nucleus, by using confocal laser scanning.
While inspecting VviNAC60 gene co-expression networks (GCNs) constructed from publicly available berry or leaf RNA-seq datasets (more than 1,400 SRA runs; Orduña et al. 2022), several switch genes (Palumbo et al. 2015; Massonnet et al. 2017) and berry ripening transition markers (Fasoli et al. 2018) were found (Fig. 1B, C), including VviWRKY19 and VvibHLH75 identified as markers of the first transition during berry ripening in Fasoli et al. (2018; Supplemental Dataset S1). A high proportion of transcription factors from different families are highly co-expressed with VviNAC60 with several berry related “markers’ found in the berry-GCN. In the leaf-GCN a huge amount of genes involved in senescence process have been identified, such as the stay-green protein 1 (VviSGR1) involved in chlorophyll degradation (Park et al. 2007; Shimoda et al. 2016), 2 autophagy proteins, several ubiquitin-related proteins, 3 aspartic proteases and a nitrilase, involved in cellular protein degradation processes (Quirino et al. 2000; Gepstein and Glick 2013), 2 laccases, a peroxidase, an alpha dioxygenase and a glutathione reductase, involved in the regulation of oxidative processes and a number of genes involved in pathogen defense mechanisms.
Interestingly, many NAC family members were found in both GCNs; particularly interesting were VviNAC11, a berry specific switch (Massonnet et al. 2017), VviNAC33, recently characterized as a master regulator of senescence (D’Inca et al. 2021), VviNAC26, likely involved in regulating fruit and seed development (Zhang et al. 2021), and VviNAC61, described as a postharvest withering-related gene (Zenoni et al. 2016).
Analysis of VviNAC60 expression during berry development revealed that its induction starts before the expression of several well-known ripening-associated genes (Fig. 1D), including a berry sugar invertase, the berry anthocyanin regulator VviMYBA1 and its targets VviUFGT and VviAOMT1, and the cell wall metabolism-related VviGRIP22.
VviNAC60 protein accumulation throughput berry development was assessed using a specific polyclonal antibody showing its presence in all 5 stages surveyed, increasing from preripening to fully ripe stages. Though the VviNAC60 monomer (∼37.5 kDa) was detected, the VviNAC60 homodimer (∼75 kDa) was the prevalent form (Fig. 1E). Moreover, a deeper inspection at the earlier stages of berry development indicates the VviNAC60 protein begins to accumulate around 42 d before the onset of ripening (i.e. veraison;Supplemental Fig. S2B), in full association with the gene expression profile previously observed in the grapevine Atlas (Fig. 1A).
The capacity of VviNAC60 to interact with itself has been demonstrated by performing bimolecular fluorescence complementation (BiFC) analysis which also highlighted VviNAC60 homodimer localization into the nucleus (Fig. 1F). In fact, VviNAC60 protein contains a monopartite N-terminal nuclear localization signal (PRDRKYP; Supplemental Fig. S3). These data indicate that VviNAC60 regulatory activity is mainly achieved through the conformation of a VviNAC60–VviNAC60 complex potentially interacting with open chromatin.
We amplified the full-length coding region of VviNAC60 from cultivar Syrah berry cDNA. The deduced amino acid sequence (335 aa) showed 99% identity to the predicted putative ortholog in PN40024 based on the 12X.2 reference genome assembly (VCost.v3 annotation; Canaguier et al. 2017), with only 2 amino acid substitutions in the highly divergent C terminus at positions 223 and 256 (Supplemental Fig. S3).
We constructed a full NAC family phylogenetic tree using 74 grapevine and 93 tomato sequences, adding different NAC proteins that have been functionally characterized in Arabidopsis and other plant species (Supplemental Dataset S2). We show that VviNAC60 belongs to the NAP-clade, including OsNAP (Liang et al. 2014), SlNAC1, AtNAP (Kou et al. 2012), CmNAC60 (Cao et al. 2019), and GhNAP (Fan et al. 2015), all involved in the regulation of senescence. The VviNAC60 most similar protein is AtANAC047, induced upon senescence in leaf petioles (Rauf et al. 2013). Moreover, in the same clade, we found SlNAP2 that regulates the expression of NOR (Ma et al. 2018) and VviNAC26. Interestingly, the NAP-clade is close to the NOR-clade, including SlNOR-like 1 (Gao et al. 2018), PpNAC1 that regulates phenylalanine biosynthesis in pine (Pascual et al. 2018), the recently characterized FvRIF, involved in fruit ripening control in strawberry (Martin-Pizarro et al. 2021), and 2 grapevine NACs, VviNAC03 and VviNAC18 (Supplemental Fig. S4).
VviNAC60-overexpressing plants accelerate the senescence program with increased chlorophyll degradation and cell death
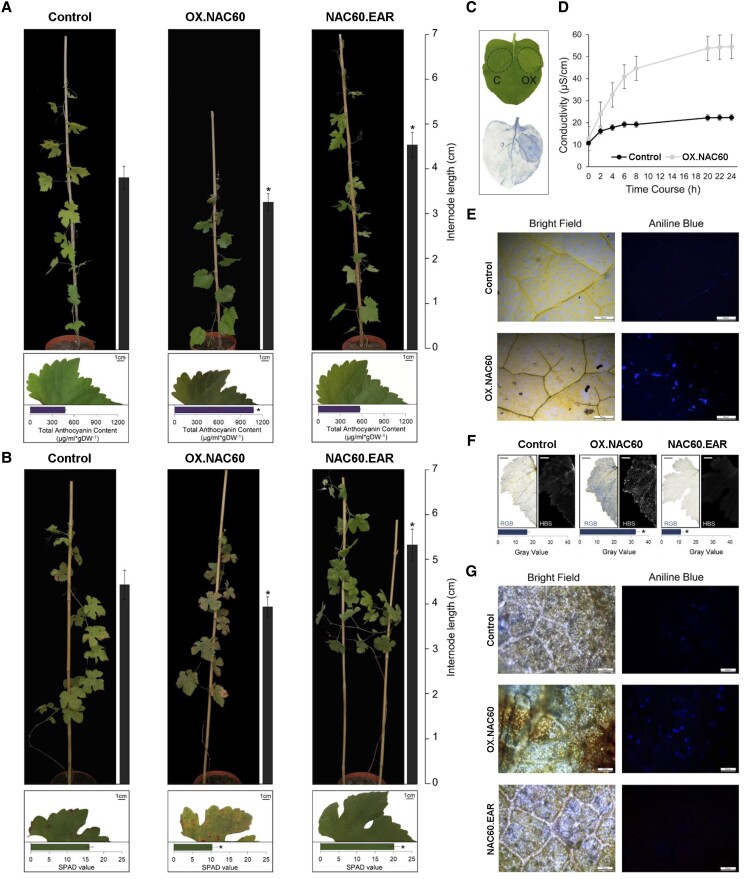
VviNAC60 was overexpressed in grapevine cv. Syrah in 3 independent transgenic lines (OX#1, OX#2, and OX#3; Supplemental Fig. S5A, B).
The overexpression of VviNAC60 led to slightly stunted growth compared to the control, due to a significant reduction in internode length (Fig. 2A), reduced leaf area (c. 40%; Supplemental Fig. S5C) and accumulation of anthocyanin in young leaves (Fig. 2A, Supplemental Fig. S5D). We selected the line #1 (hereafter OX.NAC60) displaying the highest expression for further analysis. We also generated a dominant suppressor which overcomes the activity of endogenous VviNAC60, by fusing the EAR-repression motif (SRDX) to the carboxyl terminus of the protein and by placing the chimeric repressor under the control of the endogenous VviNAC60 promoter (Hiratsu et al. 2002).
Figure 2.
Phenotypic changes in transgenic grapevine plants with altered expression of VviNAC60. A) Whole 2 mo plant and leaf phenotype caused by the ectopic expression of VviNAC60 in selected OX and EAR lines compared to vector control. Internode lengths on plants and anthocyanins contents on leaves are indicated by the bars next to each picture. The data are expressed as mean ± SD (n = 4). Asterisks indicate significant differences (*, P < 0.01; t-test) in the OX.NAC60 lines compared to the control. SD, standard deviation. B) Whole 10 mo plant and leaf phenotype caused by the ectopic expression of VviNAC60 in selected OX and EAR lines compared to vector control. Internode lengths on plants and chlorophyll contents on leaves are indicated by the bars next to each picture. Asterisks indicate significant differences (*, P < 0.01; t-test) in the OX.NAC60 and NAC60.EAR lines compared to the control. C) Phenotype showed by N. benthamiana leaf after 72 h from the agroinfiltration with OX. NAC60 (OX) and the control C; upper site) and cell death visualized by trypan blue staining (lower site). D) Ion leakage assay on N. benthamiana leaves after 24 h from the agroinfiltration with OX.NAC60 and the control. The data are expressed as mean ± SD (n = 5). E) Callose deposition in N. benthamiana leaves after 72 h from the agroinfiltration with OX.NAC60 and the control visualized by aniline blue staining. The same leaves were photographed at the same time under 2 light fields. F) Cell death on OX and EARNAC60 grapevine leaves compared to the control visualized by trypan blue staining. The same leaves were photographed at the same time under 2 light fields. The data are expressed as mean ± SD (n = 3). Asterisks indicate significant differences (*, P < 0.01; t-test) in the OX.NAC60 and NAC60.EAR lines compared to the control. SD, standard deviation. Bars = 1 cm. G) Callose deposition was visualized by aniline blue staining in OX and EAR grapevine leaves and the control in a bright field (on the left) and under an epifluorescence microscope (on the right). Magnification 10×. The same leaves were photographed at the same time under 2 light fields.
Three independent transgenic lines of cv. Syrah transformed with the Pro VviNAC60: VviNAC60-SRDX construct were obtained (EAR#1, EAR#2, and EAR#3). The transgene expression was confirmed by RT-PCR in fully expanded leaves, when the expression of endogenous VviNAC60 begins to increase (Supplemental Fig. S6). The plants in which VviNAC60-SRDX is under the control of the endogenous promoter displayed normal growth with increases in internode length (Fig. 2A) and leaf area (∼35%; Supplemental Fig. S6C), but with similar leaf anthocyanin content as compared to the control (Fig. 2A). We selected the line #3 (hereafter NAC60.EAR) for further analysis.
At present, neither the VviNAC60 transgenic plants nor the controls flower under our growing conditions, hindering our ability to determine the effects of OX.NAC60 or NAC60.EAR in transgenic berries.
Ten months old OX.NAC60 and NAC60.EAR transgenic plants showed opposite phenotypes in terms of symptoms of senescence compared to the control: the overexpression of VviNAC60 led to premature leaf senescence, while the dominant repressor showed delayed senescence (Fig. 2B). Chlorophyll content was lower in OX.NAC60 senescing leaves, while higher in NAC60.EAR, compared to the control (Fig. 2B). We further investigated the influence of VviNAC60 on cell death in Nicotiana benthamiana leaves agroinfiltrated with VviNAC60 under the control of a 35S promoter. At 72 h post-infection, in contrast to the control, leaves expressing VviNAC60 displayed browning necrotic regions (Fig. 2C, Supplemental Fig. S7A). In line with macroscopic observations, trypan blue staining (Fig. 2C, Supplemental Fig. S7B) and ion leakage measurement (Fig. 2D) clearly confirmed a substantial increase of cell death when VviNAC60 is ectopically expressed. Moreover, besides cell death induction, aniline blue staining revealed callose deposition in VviNAC60-agroinfected leaves (Fig. 2E, Supplemental Fig. S7C). Similarly, in grapevine, trypan blue and aniline blue staining highlighted cell death and callose deposition in areas showing clearer senescence symptoms in OX. NAC60 and milder responses in NAC60.EAR leaves as compared to wild type (Fig. 2F, G), consistent with the high transgene expression and the chimeric repressor activity, respectively.
Identification of VviNAC60 high confident targets
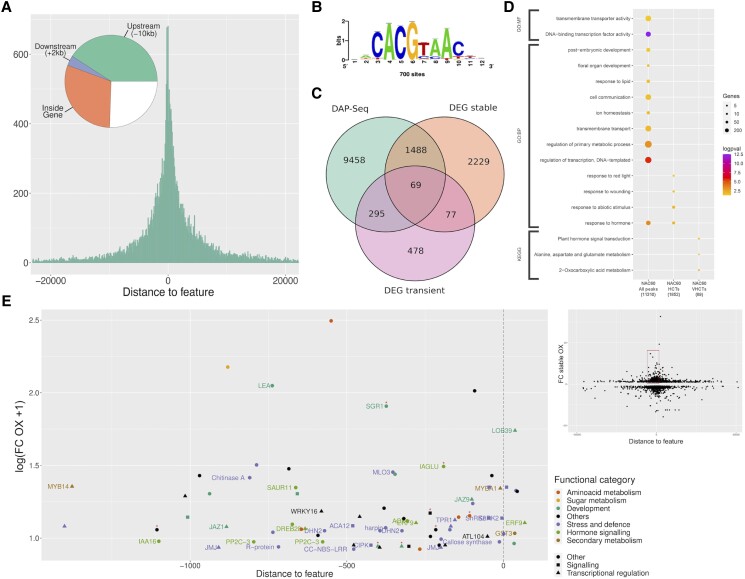
We carried out DNA affinity purification followed by sequencing (DAP-seq) to inspect the binding landscape of VvNAC60. Using 4 biological replicates and genomic DNA from leaves and berries we identified 27,715 binding events (i.e. enriched peaks compared to input), assigned to 11,310 genes, for which their position related to the transcription start site (TSS) of each gene showed a preferential distribution in proximal promoter regions (27.65% of peaks being assigned between −3 kb and the TSS; Fig. 3A; Supplemental Dataset S3). Retrieving the TOP600 highest-scored peaks allowed us to identify one major binding motif (CACGTAAC) in their center (Fig. 3B). This strongly significant consensus motif was compared to Arabidopsis NAC phylogenetic binding footprints (O’Malley et al. 2016) showing its closest resemblance to AtANAC047's motif (kaCACGtAACyt compared to NAC_tnt.ANAC047_col_m1; Pearson correlation of 0.945).
Figure 3.
Identification of VviNAC60 targets by DAP-seq and transcriptomic analysis. A) Distribution of VviNAC60 DNA binding events (27,715 peaks, assigned in at least 1 of the 3 analyses: 500 ng leaf, 1,000 ng leaf, and 500 ng berry) with respect to their position from the transcription start sites (TSS) of their assigned genes (n: 11,310). B) De novo forward binding motif obtained from the inspection of the top 600-scoring peaks of VviNAC60 500 ng leaf library using Regulatory Sequence Analysis Tools (RSAT; k-mer sig = 2.61; e-value = 0.0025; number of peaks with at least one predicted site: 733 = 24.43%). C) Prediction of VviNAC60 targets based on the overlap of DAP-seq assigned genes (sum of the 3 analyses) and DEGs detected in stable and transient VviNAC60-overexpressing plants. D) Functional enrichment analysis of VviNAC60 bound genes (assigned from all peaks) and selected targets (HCTs, high-confidence targets; VHCTs: very high-confidence targets). VHCTs were selected by focusing on peaks in the −1.5 kb and + 100 bp region and on genes with FC ≥ 1.3 in either the stable or transient overexpressing lines. As additional criteria for VHCTs, we only considered peaks present at the same position from the TSS in at least 2 of the 3 analyses. Gene ontology terms and KEGG pathways shown were filtered based on significance (Benjamini–Hochberg adjusted P-value < 0.05) and biological redundancy (complete list of terms can be found in Supplemental Table S4). The size of each dot represents the number of genes in the input query that are annotated to the corresponding term, and the color represents the significance. Total number of genes in each list is shown in parenthesis. GO:BP, Gene Ontology: Biological Process; GO:MF, Gene Ontology: Molecular Function; KEGG, Kyoto Encyclopedia of Genes and Genomes. E, Relationship between peak distances assigned to each gene and differential expression in stable and/or transient VviNAC60 overexpression (OX) lines in selected VHCTs. Fold Change values correspond to the expression in OX lines versus the control lines, with a positive threshold of FC ≥ 1.3. Node color depicts biological processes for each gene, whereas the shape is depicted by its molecular function. Red asterisks depict those genes upregulated in the transient overexpressing lines.
To identify VvNAC60 high confident targets (HCTs), we combined our DAP-seq data with differentially expressed genes (DEGs) induced by VvNAC60 stable and transient overexpression. On the one hand, the transcriptomic analysis performed on young leaves of the OX. VvNAC60 transgenic line revealed 3,863 DEGs (2,136 up- and 1,727 downregulated) compared with the control (P < 0.05; t-test; Supplemental Dataset S4), while the transient VvNAC60 overexpression in cv. Sultana (Supplemental Fig. S8) identified 919 DEGs (415 up- and 504 downregulated; P < 0.05; t-test; Supplemental Dataset S5).
The overlap of DAP-seq data with at least one of the DEGs lists shows 1,852 HCT genes (Fig. 3C; Supplemental Dataset S6), those which are mainly enriched in “abiotic stimuli”, “response to hormone”, “wounding”, and “red-light” gene ontology categories (Fig. 3D). By using a more stringent criteria (e.g. excluding binding events located before −1.5 kb and beyond 100 bp after the TSS in at least 2 replicates and selecting DEGs with fold change (FC) > 1.3), we narrowed down our gene set to 89 very-high-confidence targets (VHCTs; Supplemental Table S1), enriched in “plant hormone signal transduction”, “amino acid metabolism” and “2-oxocarboxylic acid metabolism” descriptors (Fig. 3D; Supplemental Table S2; some of them being validated by RT-qPCR, Supplemental Fig. S9). The expression of VHCT genes were profiled by exploring the cv. Corvina atlas dataset revealing that most of them follows an activation pattern throughout organ development (Supplemental Fig. S10). In particular, several genes were induced in ripening berry at higher expression levels compared to the other organs.
Among the VHCTs list we found many genes involved in plant development, such as LATERAL ORGAN BOUNDARIES 39 (VviLOB39); in chlorophyll and photosystem degradation, such as STAY GREEN 1 (VviSGR1; Park et al. 2007; Shimoda et al. 2016); in hormone signaling, such as VviIAA16, VviIAGLU, and VviSAUR11; in secondary metabolism, such as the stilbene-related VviMYB14 (Höll et al. 2013; Orduña et al. 2022) and the anthocyanin-related VviMYBA1 (Walker et al. 2007), and in response to biotic and abiotic stresses, such as resistance protein-encoding genes, VviHIN1 (harpin inducing protein 1-like 9), VviACA12, VviJAZ1, VviJAZ9, and VviATL104 as well as VviCHITA and a callose synthase-encoding gene corroborating the phenotype observed in VviNAC60-overexpressing plants (Fig. 3E).
VviNAC60 directly controls the expression of genes related to senescence and ripening
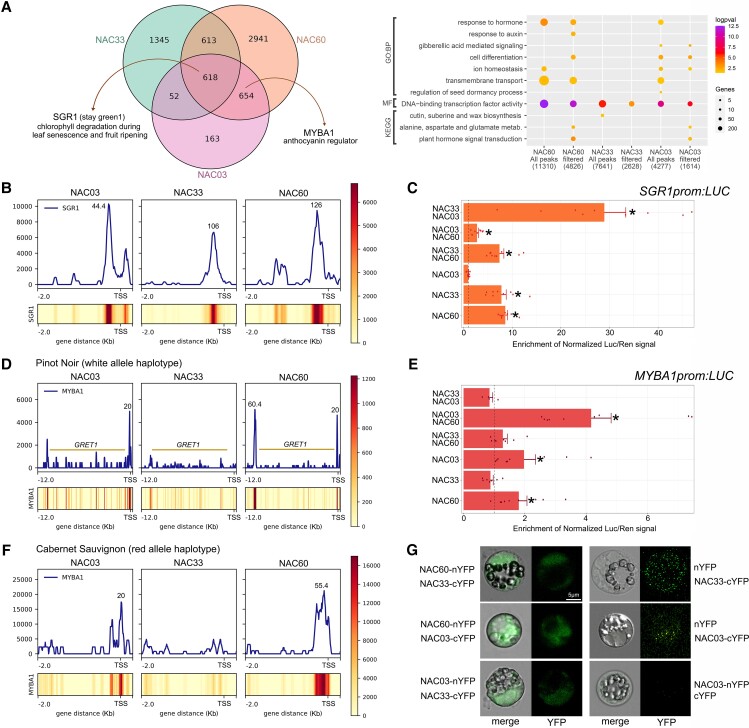
To unravel the regulatory mechanisms of VviNAC60 we compared its targets with DAP-seq data generated for 2 other grapevine NAC family members, namely VviNAC33 (D'Inca et al. 2021) and VviNAC03. VviNAC03 was selected for its highly similar expression profile with VviNAC60 during flowering and berry ripening (Supplemental Fig. S1) and because it is the most similar grapevine NAC to the tomato SlNOR (Supplemental Fig. S4). DAP-seq performed on VviNAC03 led to 4,277 peaks (Supplemental Dataset S7) with the consensus sequence CACGCAAC as the most frequent binding motif (Supplemental Fig. S11A), a footprint matching with AtNAM and AtNAP binding motifs. For VviNAC33, with a consensus motif ACA(A/C)GCAAC matching that of AtANAC017, AtNAM and AtCUC2, a total of 7,641 peaks were obtained (Supplemental Fig. S11B; Supplemental Dataset S8). We focused on the filtered DAP-seq bound genes (i.e. with peak distances between −5 kb and +100 bp), corresponding to 4,826, 1,614, and 2,628 putative targets identified for VviNAC60, VviNAC03, and VviNAC33, respectively. All these groups of genes were mainly enriched in “DNA-binding transcription factor activity”, suggesting a high regulatory hierarchy for these VviNAC TFs. VviNAC60 and VviNAC03 putative target genes were enriched in “plant hormone signal transduction”, “amino acid metabolism”, and “cell differentiation”, while VviNAC60 specific putative target genes were enriched in “transmembrane transport” and “response to auxin” (Fig. 4A). VviNAC60 showed the highest number of specific targets (2,941) and a similar number of shared genes with both VviNAC03 and VviNAC33. In contrast, fewer genes were shared between VviNAC33 and VviNAC03 (Fig. 4A).
Figure 4.
Similarities among grape VviNAC60 and other VviNAC-TFs cistromes. A) Overlap of NAC03, NAC33, and NAC60 filtered DAP-seq bound genes (positional filtering, with peaks ranging between −5,000 bp from Transcriptional Start Site up to 100 bp from the end of the gene; left panel); enrichment analysis of significant (P < 0.05; t-test) gene ontology terms and KEGG pathways for VviNAC-bound genes (right panel). B) VviNAC DNA binding landscapes in the proximal promoter region of VviSGR1 gene. DAP-seq binding signals, shown as density plots and heatmaps and delimited between −2 kb and + 2 kb from the TSS. Binding events identified by GEM are pointed out with their corresponding signal score. C) VviSGR1 promoter activation tested by dual-luciferase reporter assay in infiltrated N. benthamiana leaves. The single and combined activity of VviNAC03, VviNAC33, and VviNAC60 were tested. Firefly LUCIFERASE (LUC) activity values are reported relative to the RENILLA (REN) value and normalized against the control (empty-effector vector). Each value represents the mean of 3 biological replicates, each with 3 technical replicates (± SD). Asterisks indicate significant differences in promoter activation compared with the control (*, P < 0.05; t-test). D) VviNAC DNA binding events in promoter regions of VviMYBA1 gene. Upper panel: sequencing reads were mapped in the cv. PN40024 white allele haplotype that presents the insertion of the GRET1 retrotransposon in VviMYBA1 proximal upstream region (histograms and heatmaps plotted between −12 kb and the TSS of the VviMYBA1 gene). Bottom panel: mapping conducted in the red allele haplotype of cv. Cabernet Sauvignon (no GRET1 insertion). (B–D: scale represents RPKM-normalized data). E) VviMYBA1 promoter activation tested by dual-luciferase reporter assay in infiltrated N. benthamiana leaves. The single and combined activity of all VviNACs were tested. Firefly LUCIFERASE (LUC) activity values are reported relative to the RENILLA (REN) value and normalized against the control (empty-effector vector). Each value represents the mean of 3 biological replicates, each with 3 technical replicates (± SD). Asterisks indicate significant differences in promoter activation compared with the control (*, P < 0.05; t-test). F) BiFC analysis showing VviNAC60/VviNAC33, VviNAC60/VviNAC03, and VviNAC03/VviNAC33 protein interactions. Images are confocal laser scanning micrographs of PEG-transformed grapevine protoplasts. Corresponding controls (right panel) are shown in full overexposition mode.
The comparison among the 3 VviNACs revealed 618 common target genes (Supplemental Dataset S9), supporting the hypothesis that these 3 VviNACs could participate in the regulation of common pathways. Interestingly, among them, we found VviSGR1. The binding landscape of VviNAC03, VviNAC33, and VviNAC60 on VviSGR1 promoter confirmed the binding signal of the 3 VviNACs (Fig. 4B). The dual-luciferase assay showed that VviNAC60 and VviNAC33 significantly activated the SGR1 promoter at the same level, while no VviSGR1 promoter activity was induced by VviNAC03 (Fig. 4C). The analysis of the 3 combinations, VviNAC60/VviNAC33, VviNAC60/VviNAC03, VviNAC33/VviNAC03, revealed that only the latter showed a strong positive synergic effect on VviSGR1 promoter activity (Fig. 4C).
Among bound genes in common between VviNAC60 and VviNAC03 we found the anthocyanin pathway regulator VviMYBA1, one of the 2 main genes of the berry color locus. As the reference PN40024 genome assembly only considers the white allele haplotype (with the presence of the VviMYBA1-inactivating GRET1 retrotransposon), we also mapped the reads from all 3 VviNAC libraries in the red allele haplotype of cv. Cabernet Sauvignon to evaluate the effect of GRET1 position in the binding of these TFs in VviMYBA1 promoter. The analysis highlighted a very proximal binding signal for VviNAC60 and VviNAC03 in both genotypes, while a further upstream binding of VviNAC60 before the GRET1 insertion was also found in the white allele haplotype. The presence of close binding events in both VviNAC60 and VviNAC03 suggests a positive interaction between these 2 TFs (Fig. 4E). No binding signal was identified for VviNAC33 (Fig. 4D). Interestingly, the binding motif recognized by VviNAC60 on the VviMYBA1 promoter diverged from the major binding motif identified in the TOP600 peaks. This consensus motif (AATGGGAC; Supplemental Fig. S12) was also present in the peaks identified in the promoter regions of the other berry color locus gene VviMYBA2 (Walker et al. 2007) and the 3 VviMYBA genes from the vegetative color locus (VviMYBA5, VviMYBA6, and VviMYBA7; Matus et al. 2017). We show that VviNAC60 and VviNAC03, but not VviNAC33, significantly activate VviMYBA1 promoter at the same level, and such activation further increased in the presence of both TFs.
VviMYB14, one of the main regulators of stilbene synthesis, was found among VviNAC60-bound genes at −1,377 bp from TSS, respectively. The dual-luciferase assay confirmed that VviNAC60 significantly induced VviMYB14 expression (Supplemental Fig. S13).
The BiFC highlighted the formation of heterodimer combinations among the 3 VviNACs (Fig. 4F), suggesting that their action is exerted by an interaction with different VviNAC partners. The multispiked binding profile of VviNAC60 in cv. Cabernet Sauvignon VviMYBA1 promoter suggests that the 2 peaks separated by GRET1 are at a very close distance in the red-allele and could form part of a large binding of VviNAC03–VviNAC60 and VviNAC60–VviNAC60 hetero and homodimers, respectively.
VviNAC60 is able to complement the tomato nor mutation
To investigate the ability of VviNAC60 to regulate fruit ripening in the absence of fruit from transgenic grapevines, the gene was overexpressed in the nor tomato mutant. We overexpressed also VviNAC33 and VviNAC03 (Supplemental Table S3). The heterologous expression level of each transgene was measured in T3 fruits at breaker (Br) + 7 by RT-qPCR (Supplemental Fig. S14). We noted an association between the level of transgene expression and the level of phenotypic rescue. VviNAC60#1, VviNAC03#12, and VviNAC33#3 lines (hereafter 35S:VviNAC60, 35S:VviNAC03, and 35S:VviNAC33, respectively) were selected for further analyses based on their higher transgene expression compared with the other lines (Supplemental Fig. S14). 35S:VviNAC60 plants showed a stunted growth in comparison to nor plants, confirmed by reduced internode length, while no evident alterations were observed for 35S:VviNAC03 and 35S:VviNAC33 plants (Fig. 5A).
Figure 5.
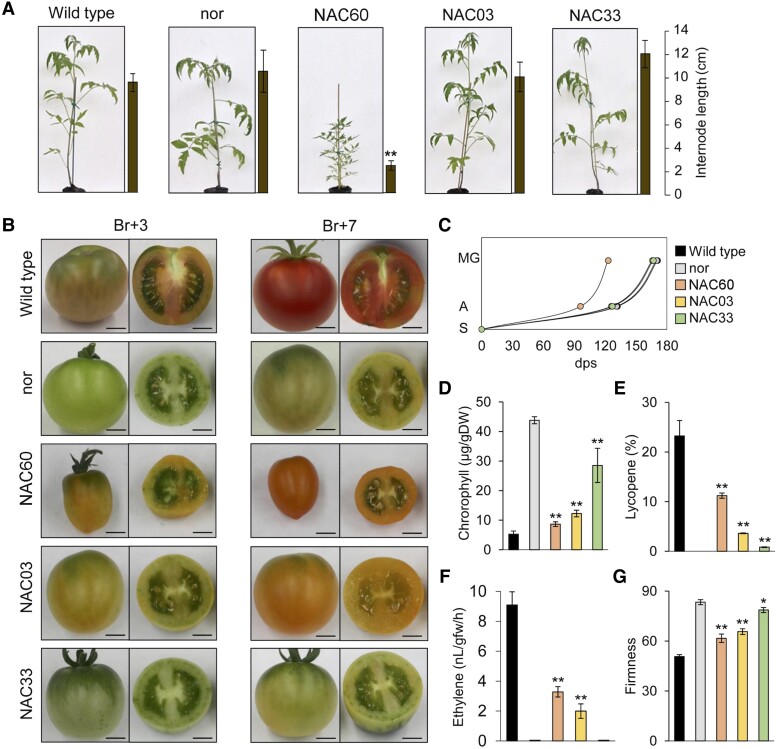
Phenotypic evaluation of VviNAC60, VviNAC03, and VviNAC33 heterologous expression in nor tomato mutant background. A) Whole tomato plant phenotype corresponding to wild type (Solanum lycopersicum cv. Ailsa Craig), nor and T3 fruit transformed with 35S: VviNAC60, 35S:VviNAC03, and 35S:VviNAC33 in nor tomato mutant background. Internode lengths on plants are indicated by the bars next to each picture. The data are expressed as mean ± SD (n = 4). Asterisks indicate significant differences (**, P < 0.01; t-test) compared to the nor. B) Phenotype of tomato fruits corresponding to wild type, nor and T3 fruit transformed with 35S: VviNAC60, 35S:VviNAC03, and 35S:VviNAC33 in nor tomato mutant background. They were collected at the breaker (Br) + 3 and +7. Bar, 2 cm. C) Growth curve of VviNAC60, VviNAC03, and VviNAC33 transgenic plants in nor background, wild type and nor (n = 9) from seedling to mature green stage. S, seedling; A, anthesis; MG, mature green; dps, day post seedling. D) Chlorophyll content in VviNAC60, VviNAC03, and VviNAC33 transgenic fruits in nor background, wild type and nor at Br + 3. The data are expressed as mean ± SD (n = 4). Asterisks indicate significant differences (**, P < 0.01; t-test) compared to nor. E) Lycopene content in VviNAC60, VviNAC03, and VviNAC33 transgenic fruits in nor background, wild type and nor at Br + 3. The data are expressed as mean ± SD (n = 4). Asterisks indicate significant differences (**, P < 0.01; t-test) compared to the nor. F) Ethylene production in VviNAC60, VviNAC03, and VviNAC33 transgenic fruits in nor background, wild type and nor at Br +5. Each value represents the mean ± standard deviation (SD) of 3 biological replicates. Asterisks indicate significant differences (**, P < 0.01; t-test) compared to nor. G) Fruits firmness of VviNAC60, VviNAC03, and VviNAC33 transgenic fruits in nor background, wild type and nor at Br +3. The data are expressed as mean ± SD (n = 4). Asterisks indicate significant differences (**, P < 0.01; *, P < 0.05; t-test) compared to nor.
35S:VviNAC60 and 35S:VviNAC03 pericarps showed different degrees of yellowness at Br + 3 with 35S: VviNAC60 fruits redder than 35S:VviNAC03 at Br + 7. 35S:VviNAC33 transgenic fruits were indistinguishable from nor in both analyzed stages (Fig. 5B). 35S: VviNAC60 fruits were smaller than the other same-age fruits (Fig. 5B) and displayed earlier ripening: they flowered 1 mo earlier than the other same-age plants and showed the interval between flowering and fruit mature green stage 2-wk shorter than the others lines (Fig. 5C).
Pigment content analysis revealed a significant decrease in chlorophyll in all transgenic fruits (Fig. 5D) as well as a significant increase in lycopene (Fig. 5E), the most abundant carotenoid in ripe tomato, in both 35S:VviNAC60 and 35S:VviNAC03 in comparison to nor. In particular, 35S:VviNAC33 showed the highest chlorophyll content and 35S: VviNAC60 the highest lycopene content, supporting the phenotypic observations.
Regarding ethylene production, we analyzed Br + 5 stage since 35S:VviNAC60 fruit showed the highest ethylene production at this stage (Supplemental Fig. S15). Nor and 35S:VviNAC33 fruits failed to undergo an increase in ethylene production, while ethylene was significantly higher in 35S:VviNAC60 and 35S:VviNAC03 fruits compared to nor at the same age (Fig. 5F). Fruit softening in 35S:VviNAC60 and 35S:VviNAC03 fruits were significantly increased in comparison to same-age nor fruits, while no difference was measured in 35S:VviNAC33 fruits (Fig. 5G).
RT-qPCR analyses performed on the 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE (SlACS4), involved in ethylene biosynthesis, the POLYGALACTURONASE 2A (SlPG2a), involved in softening pathway, the PHYTOENE SYNTHASE 1 (SlPSY1), involved in carotenoid pathway, and the STAY-GREEN PROTEIN 1 (SlSGR1), involved in chlorophyll and photosystem degradation, showed that all these genes were significantly upregulated in 35S:VviNAC60 and 35S:VviNAC03 transgenic fruits compared to nor, while in 35S:VviNAC33 fruits only SlSGR1 was significantly upregulated (Supplemental Fig. S16).
Discussion
Several grapevine transcription factors have been isolated and studied in the past decade as regulators of berry metabolism and environmental and pathogen responses. Many of these characterizations have been enhanced by the use of heterologous systems, while only recently by means of systems biology approaches (Falchi et al. 2019; Matus et al. 2019). However, central mediators of developmental transitions involving all these biological processes have not been identified to date.
By using a wide repertoire of molecular and computational methods, we show here that the grape transcription factor VviNAC60 controls many different processes activated during senescence and ripening indicating its role as master regulator of the vegetative-to-mature phase organ transition in grapevine.
VviNAC60 belongs to a subclade of NAC genes previously known as related to senescence in Arabidopsis and other plant species. VviNAC60 expression shows a clear increase from green-to-mature stages in several grapevine organs including leaves and fruits. Our aggregate GCN analyses in both leaves and berries revealed that VviNAC60 was tightly connected with previously identified developmental transition genes (i.e. switch and ripening markers genes). In addition, VviNAC60 was highly co-expressed with VviNAC33, a recently characterized master regulator of leaf senescence (D’Inca et al. 2021) and VviNAC03, the still uncharacterized most similar gene to the tomato NON-RIPENING (SlNOR) regulator (Vrebalov et al. 2002). In this work, we found that in fact the 3 VviNACs share several putative targets and are able to form heterodimers with each other, suggesting that VviNAC60 and these other 2 VviNACs co-operate in the same regulatory networks. Moreover, several other lines of evidence support that each regulator may be specialized in one particular process, with VviNAC33 being more specialized in leaf senescence whereas presumably VviNAC03 has a major impact in controlling in fruit ripening and VviNAC60 influencing both developmental processes.
VviNAC60 controls several molecular events related to senescence-related processes
VviNAC60 overexpression triggered senescence symptoms, such as chlorophyll degradation, the cessation of organ growth and cell death induction, whereas the chimeric repressors delayed senescence. We demonstrated that VviNAC60 directly induces the expression of VviSGR1, a key regulator of leaf yellowing that interacts with chlorophyll catabolic enzymes (Zhou et al. 2011) at the same level of VviNAC33. While VviNAC60 and VviNAC33 did form a heterodimer, no additive effects were observed when analyzed in combination by luciferase assay. In contrast, VviSGR1 expression was highly induced by the VviNAC33/VviNAC03 combination.
Moreover, we observed a reduction in leaf size when VviNAC60 was overexpressed, both in grapevine and the tomato nor mutant, and an increase in leaf size in response to chimeric repressor expression in grapevine. The same phenotypic effect was observed in transgenic grapevines with altered expression of VviNAC33, which affected the expression of genes involved in auxin metabolism (D’Inca et al. 2021). We found that putative targets of VviNAC60 were mainly enriched in “response to auxin” (Fig. 4A) and that many auxin-related genes were downregulated in both transient and stable VviNAC60 transgenic grapevines (Supplemental Datasets S4 and S5), strongly supporting that VviNAC60 is involved in the control of the cessation of organ growth and initiation of senescence by affecting auxin metabolism, likely through its interaction with VviNAC33. Consistently, among VviNAC60 positively regulated targets, we found genes encoding components of the ABA transduction pathway, such as RGLG1, PP2CA, and SnRK2, suggesting a role for VviNAC60 in ABA-mediated gene regulation of senescence (Chen et al. 2020). A clear induction of cell death was observed in VviNAC60 overexpressing lines as part of the general senescence symptoms. Several lines of evidence suggest that a cross-talk might exist between signaling pathways of leaf senescence and pathogen-induced defense responses (Yoshida et al. 2002; Espinoza et al. 2007; Huysmans et al. 2017). Though still unclear, these 2 types of programmed cell death may share common regulators (Yoshida et al. 2002). Interestingly, among the VviNAC60 VHCTs identified in overlapping of DAP-seq and transcriptomics data, we found several genes involved in stress and pathogen response, including a gene encoding the ARABIDOPSIS TÓXICOS EN LEVADURA (ATL) E3 UBIQUITIN LIGASE VviATL104 (Ariani et al. 2016). This gene belongs to a module of grapevine ATL co-expressed genes related to biotic stress (i.e. CC6). This CC6 module also includes VviATL156, whose function in grapevine resistance to Plasmopara viticola was recently demonstrated (Vandelle et al. 2021). Interestingly, VviNAC60 is upregulated in VviATL156-overexpressing Vitis vinifera plants. The further comparison of available datasets highlighted 13 VviNAC60 targets upregulated in VviATL156-OE plants and 8 targets belonging to the CC6 biotic stress-related module (Supplemental Fig. S17). In addition, 2 VviNAC60 targets were common to all 3 datasets, namely VviACA12- and HARPIN INDUCING PROTEIN 1-LIKE 9-encoding genes, 2 key players in plant defense responses, in particular related to hypersensitive cell death (Zhang et al. 2011; Yu et al. 2018). Additionally, ABA might promote plant defense by increasing callose deposition (Chitarra et al. 2018) and, accordingly, a gene encoding a callose synthase was also found among the high confident targets of the transcription factor and callose deposition was demonstrated in VviNAC60-OX plants. Based on these observations we speculate that VviNAC60 may also play a role in grapevine defense implementation. Thus, in line with the potential relationship between senescence and disease development, suggested by the induction of a common set of genes by these 2 processes (Quirino et al. 2000; Gepstein and Glick 2013), VviNAC60 may represent a key node in the regulation of the broad-sense programmed cell death.
A relationship between senescence and disease de-velopment is suggested by the induction of a common set of genes by these 2 processes. Some SAGs code for proteins related to defence-associated processes [e.g. pathogenesis-related (PR) proteins] that are induced during the hypersensitive response (HR) against avirulent pathogens (Quirino et al. 2000; Gepstein and Glick 2013).
VviNAC60 is a putative regulator of fruit ripening
By different experimental and computational approaches we have shown that VviNAC60 is also induced during fruit ripening, regulates ripening-related genes and is co-expressed with ripening marker genes, including genes related to ABA, a berry ripening trigger (Pilati et al 2017). Moreover, the stable VviNAC60 overexpression in grapevine induced a significant anthocyanin accumulation in young leaves by activating the expression of VviMYBA1, a positive regulator of anthocyanin synthesis acting at the onset of berry ripening (Walker et al. 2007). Interestingly, VviNAC60 and VviNAC03 are co-expressed during fruit ripening and act synergically in the activation of VviMYBA1 promoter by forming heterodimers. The ability of VviNACs to activate VviMYB genes has been demonstrated in peach, where the NAC TF blood (BL) activates the transcription of PpMYB10.1 by acting as a heterodimer with PpNAC1 (Zhou et al. 2015). Similarly, in apple, MdNAC52 regulates anthocyanin biosynthesis by activating MdMYB9 and MdMYB11 (Sun et al. 2019). Here, we ascertained VviNAC-to-VviMYB regulation in grapevine, demonstrating that VviNAC03 and VviNAC60 bind to the same VviMYBA1 promoter region, close to the TSS, and that they form a heterodimeric complex. Considering this evidence and the co-expression of VviNAC60 along with VviMYBA1 during berry development, we speculate that VviNAC60 might control VviMYBA1 activation together with VviNAC03, as a one of the first molecular events hallmarking the onset of berry ripening.
Such VviNAC-to-VviMYB regulation scenario is further supported by the fact that VviNAC60 also activates VviMYB14, a known regulator of stilbene metabolism, whose expression is part of the berry ripening/post-ripening program (Höll et al. 2013; Zenoni et al. 2016) and of berry response to pathogens (Lovato et al. 2019). This suggests that VviNAC60, which is induced at the onset of ripening, might also contribute to specific processes featured during ripening progression by co-operating with other factor(s), becoming available only in after the onset of ripening, to activate this VviMYB.
The functional investigation of VviNAC60 in grape berry ripening, as occurring for the study of genes involved in grapevine fruit development, was hindered by the difficulty to obtain berries from transgenic plants that cannot be grown in the field, due to GMO legal restrictions. Indeed, up-to-date, neither VviNAC60 transgenic plants nor control plants were able to flower under greenhouse conditions, precluding the opportunity to figure out the effects of OX.VviNAC60 or VviNAC60.EAR in transgenic grape berries. In this context the use of a heterologous tomato system represents the best alternative available for testing VviNAC60 function. Despite tomato undergoes a different ripening progression and associated changes as compared to grape, it offers several positive aspects: (i) it is a berry, (ii) VviNAC03 and VviNAC60 are highly similar to the tomato ripening-regulator NOR, (iii) both the grape and tomato genes are strongly induced during ripening and (iv) tomato harbors a mutation in the abovementioned homologous gene that can be exploited through complementation.
It was recently shown that the nor mutation is not a knock-out but it has a dominant negative repressive effect due to expression of a truncated SlNOR protein. However, the dominant effects of the nor mutant protein can be overcome by overexpression of a normal functioning allele (Gao et al. 2020). With this system, we showed that VviNAC60 heterologous expression indeed rescues the nor maturation process in tomato, just as SlNOR when overexpressed in the nor mutant background (Gao et al. 2020). Accordingly, 35S:VviNAC60 fruits showed a smaller dimension than control same-age fruits, as reported for 35S:NAC-NOR. Also, VviNAC60 significantly activated key genes involved in tomato fruit ripening, such as SlACS4, SlPG2a, SlPSY1, and SlSGR1, though their expression level did not reach the same level as in the wild type. We hypothesize that the transgene-derived VviNAC60 can compete with the truncated NOR protein in binding to the promoters of the target genes determining only a partial activation.
In addition, the complementation analysis showed that VviNAC03 is also involved in ripening progression control, activating the previously mentioned tomato ripening genes, albeit at a lower level compared with VviNAC60.
Conversely, the heterologous expression of VviNAC33 in nor mutant was not able to rescue ripening phenotype, though significant chlorophyll degradation was observed (associated with an increase of SGR1 expression), confirming its role in organ de-greening (D’Incà et al. 2021). Despite this observation, it is worth mentioning that VviNAC33 expression in nor mutant fruits was lower in comparison with VviNAC60 and VviNAC03. It is known that VviNAC33 is a target of the miRNA164, which plays conserved roles in regulating organ ageing, such as leaf senescence and fruit ripening, in a variety of plants (Kim et al. 2009; Wang et al. 2020). In grapevine, it was shown that the expression of miRNA164 is high in small fruits, while it decreases during ripening (Sun et al. 2019). Though the way in which miRNA164 family members exert their function in tomato remains to be elucidated, a possible effect of tomato miRNA164 on VviNAC33 transgene expression cannot be ruled out. The complementation of the mutant phenotype was achieved with VviNAC60 and its interacting partner (VviNAC03) but not the third and similar NAC we tested (VviNAC33), indicating specificity in the ripening effects.
VviNAC60 is part of a regulatory network controlling senescence and ripening-related processes
Overall, we showed that VviNAC60 is a regulator of organ phase transition through the control of genes involved in diverse developmental processes. Besides an ascertained role in senescence, several lines of evidence support the possible role of VviNAC60 in ripening (Fig. 6), such as (i) it's mRNA is strongly induced at the onset of berry ripening and correlated with expression of known ripening “marker” genes, (ii) a large part of its VHCTs are highly expressed in ripening berry, (iii) the ability to directly activate VviSGR1, VviMYBA1, and VviMYB14 ripening genes, and (iv) the capacity of VviNAC60 to complement the tomato nor mutation similarly to the endogenous NAC SlNOR gene.
Figure 6.
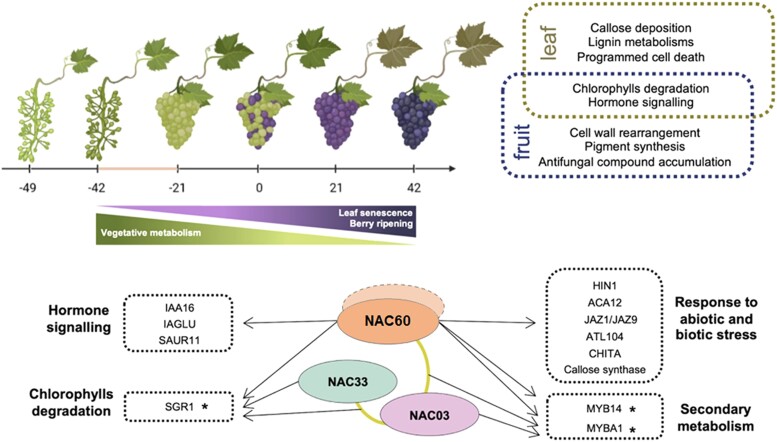
Leaf and fruit developmental progression with the respective and common VviNAC60-regulated gene categories and a proposed regulatory mechanism model for the action of the 3 studied VviNAC TFs on selected targets. Asterisks (*) indicate genes validated by dual-luciferase assays, the gold line indicates the interaction validated by BiFC analyses and the dotted NAC60 represents the possible homodimerization. The grape images were created with BioRender.com.
The presented results allowed us to propose a model of regulatory action in which VviNAC60 and the other 2 VviNACs activate specific targets involved in many key ripening- and senescence-related processes in grapevine in the same regulatory network (Fig. 6). In details, VviMYB1 and VviMYBA14 involved in berry secondary metabolism, VviSGR1 in chlorophyll degradation, VviIAA16, VviIAGLU and VviSUAR11 in hormone signaling and VviHIN1, VviACA12, VviJAZ/VviJAZ9, VviATL104, VviCHITA and CALLOSE SYNTHASE involved in response to abiotic and biotic stress. Considering that VviNAC03 and VviNAC33 are more specialized in ripening and senescence, respectively, the specific interaction of VviNAC60 with one or the other TF, as demonstrated in this work, would allow it to play a role in both biological processes.
The functional characterization reported here defines VviNAC60 as a key component of the molecular mechanisms governing organ maturation in grapevine. Nevertheless, not being available at this time fruit from plants with altered expression of VviNAC60, further investigations will be necessary to define the precise role of VviNAC60 in grape ripening.
Moreover, additional studies describing time-evolving regulatory networks will be necessary to better clarify the regulatory interactions of VviNAC60 and other grapevine NACs, considering also possible environment-specific behaviors, for instance depending on temperature conditions. In this regard, a preliminary inspection of publicly available transcriptomic data, reporting gene expression changes in berries under different temperature regimes, revealed a nonconsistent expression of VviNAC60 in response to increasing temperatures. However, this point deserves further attention in the context of global climate change: uncovering and modifying the molecular components governing maturation will be essential to maintain high-quality grapes and wine. This may include altering the duration of the vegetative growth phase to modify the timing of ripening onset with the aim to help to reverse the current trend toward earlier ripening. This would allow, in the field, to avoid the harvesting of grapes under unfavorable temperature conditions, a currently emerging problem leading to poor-quality grape products.
Materials and methods
Plant material
Nicotiana benthamiana plants, Vitis vinifera cv. Sultana plantlets, cv. Syrah embryogenic calli for the genetic transformation, 35S:VviNAC60 and VviNAC60-EAR transgenic plants were grown as previously described (Amato et al. 2019).
For the DAP-seq analysis, cv. Syrah fruiting cuttings were propagated as previously described (Mullins and Rajasekaran 1981).
Embryogenic calli of V. vinifera cv. Sultana for protoplast isolation and transfection were initiated in the 2016 season from leaf disks as previously described (Bertini et al. 2019). Embryogenic calli were maintained and subcultured (Iocco et al. 2001).
The abovementioned different cultivars of grapevine have been chosen because the original protocols have been specifically developed, e.g. cv Syrah has been proven easy to transform and for fruiting cuttings cultivation system and cv Sultana has been proven suitable for transient expression studies in grapevine.
Wild-type tomato (Solanum lycopersicum) cv. Ailsa Craig (AC), nor mutant and 35S: VviNAC60, 35S:VviNAC03 and 35S:VviNAC33 transgenic plants were grown under greenhouse condition, with day temperature ranging between 25 °C and 30 °C, night temperature ranging between 20 °C and 24 °C, and natural light integrated with artificial light to obtain a day length of 16 h.
Gene co-expression network construction
Berry and leaf GCNs were produced as in Orduña et al. (2022) with some modifications explained in Supplemental Methods S1. A total of 35 and 42 Transcriptomic RNA-seq Sequence Read Archive (SRA) studies were explored, encompassing 807 and 670 runs from fruit/fw and leaf samples, respectively. The top 420 most highly co-expressed genes for VviNAC60 were used to generate its gene-centered co-expression networks. The expression profiles of grapevine NAC genes were analyzed across 807 transcriptomic datasets (i.e. SRA runs, used for generating the GCNs) that were manually classified in 3 tissue categories (veraison/postveraison berry, preveraison berry and inflorescence/flower). These data have been integrated in the EXHARA tool available in the Vitis Visualization Platform (VitViz; available at http://www.vitviz.tomsbiolab.com/).
Phylogenetic analysis
Details for phylogenetic analysis are included in Supplemental Methods S2.
Isolation and cloning
The VviNAC60 coding sequence (with 3′ UTR) was amplified from cv. Syrah mid-ripening and ripening berry skin, the VviNAC03 coding sequence (with 3′ UTR) was amplified from cv. Corvina mid-ripening (Venturini et al. 2013) and the VviNAC33 coding sequence (with 3′UTR) was amplified as described previously (D’Inca et al. 2021). All the amplifications were performed using KAPA HiFi DNA polymerase (KAPA Biosystems, Wilmington, MA, USA) and the primers listed in Supplemental Table S4. The PCR products were directionally cloned into the Gateway entry vector pENTR/D-TOPO (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA).
Details for agroinfiltration in N. benthamiana are included in Supplemental Methods S3.
Transcription factors in vitro translation
Genomic DNA extraction and the Illumina genomic DNA libraries preparation were performed as previously described (D’Inca et al. 2021).
The VviNAC60 and VviNAC03 sequences were recombined from the pENTR/D-TOPO entry vector to the Gateway-compatible pIX-HALO destination vector (Bartlett et al. 2017) using LR Clonase II (Life Technologies). The HALO-NAC60, HALO-VviNAC03, and GST-HALO (used as negative control) fusion proteins were in vitro translated using 1,000 ng of pIX-HALO-TF plasmid in 3 different TNTR SP6 coupled reticulocyte lysate system (Promega) reactions.
DAP-seq
DNA affinity purification sequencing was performed as previously described (D’Inca et al. 2021) and a total of 3.5 million reads were obtained for each sample. Four DAP-seq library samples per NAC TF were generated and analyzed to study VVINAC60 and VviNAC33 cistromes: 3 libraries were obtained from in vitro translated TFs challenged with genomic DNA from young leaves (one using 1,000 ng and 2 replicates using 500 ng) and one library generated from the interaction of each TF with 500 ng of genomic DNA from green berries. One sample was used for studying VviNAC03 cistrome, obtained from the interaction of this TF with 500 ng of genomic DNA from young leaves.
DAP-seq reads were mapped to the “PN40024” 12X.v2 and “Cabernet Sauvignon” clone 08 v1.1 genome (the best references known to date) and TF-associated peaks were identified as in (Orduña et al. 2022) with specific parameters found in Supplemental Methods S4. Metagene plots were produced using Deeptools suite v.3.3.2, computing and normalizing the coverage for each BAM file with a BinSize = 10 and RPKM normalization. RPKM value for each bin is the average between the 10 positions that define each bin. BigWig files for the individual replicates for each TF were merged using bigWigMerge v.2 and bedGraphToBigWig v.4. De novo motif discovery was performed by retrieving 200 bp sequences, centered at GEM-identified binding events, for the 600 most enriched peaks for each TF and running the RSAT software (http://rsat.eead.csic.es/plants/peak-motifs_form.cgi), with default parameters.
Total protein extraction and quantification
Total protein extracts were extracted from V. vinifera cv. Syrah berries (Wu et al. 2014). The 6 berry samples were harvested at 49-, 42- and 21-days before veraison, at veraison, 21- and 42-days post veraison. The total protein extracts were quantified using the Bradford Reagent (SIGMA).
Western blot analysis
Details for western blot analysis are included in Supplemental Methods S5.
Transient expression
For N. benthamiana, pK7WG2.0 vectors containing 35S: VviNAC60 or a noncoding sequence (negative control) were transferred to Agrobacterium tumefaciens strain C58C1 by electroporation. Three fully expanded leaves were syringe infiltrated and the phenotypic analysis was carried out 3 d after agroinfiltration.
The same vectors were used in 5-wk-old in vitro plantlets of cv. Sultana (7 plants for VviNAC60 overexpression and 5 for the control) as previously described (Amato et al. 2017). The material for RNA extraction and transcriptomic analysis was collected 7 d after the agroinfiltration.
Transgenic plants
In grapevine, the pK7WG2.0 vectors containing 35S: VviNAC60, VviNAC60-EAR or the eGFP sequence (negative control) were transferred to A. tumefaciens strain EHA105 by electroporation. The genetic transformations were performed as previously described (Amato et al. 2019).
In tomato, the pK7WG2.0 vectors containing 35S: VviNAC60, 35S:VviNAC03, and 35S:VviNAC33 were transferred to A. tumefaciens strain LBA4404 by electroporation. Transformation of nor mutant tomato cv. AC cotyledon explants was performed as previously described (Frary and Van Eck 2005). Details are included in Supplemental Methods S6 and in Supplemental Figs. S18 and S19.
Internode length, leaf area, and SPAD measurement
Details for internode length, leaf area, and SPAD measurement are included in Supplemental Methods S7.
Pigment analysis
Details for pigment analysis are included in Supplemental Methods S8.
Ethylene and firmness measurement
Details for ethylene and firmness measurement are included in Supplemental Methods S9.
Reverse-transcription quantitative PCR (RT-qPCR)
Total RNA was isolated from grapevine apical leaves (for transient expression), from fully expanded leaves (for OX.VviNAC60 transgenic plants overexpressing VviNAC60 and for VviNAC60.EAR transgenic plants expressing the chimeric repressor), from berries at 5 different ripening stages, fruit set, post fruit set, veraison, mid-ripening, and ripening (for VviNAC60 expression level in berries) and from tomato leaves and fruits pericarp using the Spectrum Plant Total RNA kit. Gene expression analysis by RT-qPCR was performed as previously described (Dal Santo et al. 2016) using the primers listed in Supplemental Table S4. Each value corresponds to the mean ± SD of 3 technical replicates relative to the VviUBIQUITIN1 (VIT_16s0098g01190) and the SlACTIN (Solyc03g078400) internal controls in grapevine and tomato, respectively.
Transcriptomic analysis
The microarray analysis was performed with the RNA isolated for RT-qPCR. For transient expression, the 4 most highly overexpressing plants were selected and used as biological replicates, while for OX. VviNAC60 transgenic lines, the OX1, 2 and 3 lines were used as biological replicates. The cDNA synthesis, labeling, hybridization, washing, scanning, and feature extraction and statistical analysis were performed as previously described (Amato et al. 2017). DEGs were identified by Student's t-test (α = 0.05), assuming equal variance among samples, and standard Bonferroni correction.
Dual luciferase reporter assay
The VviMYBA1 (1237 bp) and VviMYB14 (1569 bp) putative regulative regions were amplified by Nested PCR from cv. Syrah genomic DNA by using KAPA HiFi DNA polymerase and primers listed in Supplemental Table S1. Purified PCR product was cloned into the pPGWL7.0 reporter vector (https://gatewayvectors.vib.be/collection/ppgwl7) to control the firefly luciferase gene (LUC).
The VviMYBA1pro:LUC, VviMYB14pro:LUC, and VviSGR1pro:LUC (previously engineered by D’Inca et al. 2021) pK7WG2 reporter vectors, the 35S: VviNAC60, 35S:VviNAC33, 35S:ViNAC03 effector vectors, and the Renilla reniformis reference vector (previously engineered by Amato et al. 2019) were transferred to A. tumefaciens strain C58C1 by electroporation. Dual Luciferase Reporter Assay was carried out in N. benthamiana leaves (Cavallini et al. 2015). Firefly and R. reniformis luminescence were detected using a Tecan Infinite M200 PLEX instrument.
Grapevine protoplast transfection and BiFC analysis
VviNAC60, VviNAC33, VviNAC03 coding sequences were cloned into the pGWcY Gateway vector. VviNAC60 and VviNAC03 coding sequences were also cloned into the pnYGW gateway vector. As a positive control, the nuclear homodimerization of AtbZIP63 was tested (Walter et al. 2004), by using the pUC-SPYNEGW and pUC-SPYCEGW vectors. V. vinifera cv. Sultana protoplasts were isolated from embryogenic calli and transfected (Bertini et al. 2019), cultured into multiwell plates in the dark at 25 °C and analyzed 1 d after transfection. The YFP signal was detected using a Leica TCS SP5 AOBS confocal microscope (Argon laser, 514 nm excitation source, 550–570 nm collection bandwidth, auto gain).
Electrolyte leakage assay
Electrolyte leakage assay was performed to quantify cell death. Details for the analysis are included in Supplemental Methods S10.
Trypan blue and aniline blue staining
Details for trypan blue and aniline blue staining are included in Supplemental Methods S11.
Accession numbers
Microarray data for the transient expression experiments are available at GEO under the accession GSE185447. Microarray data for the transgenic plants are available at GEO under the accession GSE185448. DAP-seq raw data have been submitted to GEO, including metadata of samples and conducted analysis (bioinformatic parameters) according to the FAIR principles. DAP-seq results on NAC60, NAC03, and NAC33 can be visualized in the DAPBrowse tool available at the Vitis Visualization Platform (http://www.vitviz.tomsbiolab.com/). Identity codes for the main genes/proteins mentioned in the manuscript are reported in Supplemental Table S4 and Supplemental Fig. S4. The roles for NAC60, NAC33, and NAC03 have been deposited in the Gene Reference Catalogue found at the Grape Genomics Encyclopedia portal (http://grapedia.org/).
Supplementary Material
Contributor Information
Erica D’Incà, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
Chiara Foresti, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
Luis Orduña, Institute for Integrative Systems Biology (I2SysBio), Universitat de València-CSIC, Paterna 46908, Valencia, Spain.
Alessandra Amato, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
Elodie Vandelle, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
Antonio Santiago, Institute for Integrative Systems Biology (I2SysBio), Universitat de València-CSIC, Paterna 46908, Valencia, Spain.
Alessandro Botton, Department of Agronomy, Food, Natural Resources, Animals and Environment, University of Padova, Padova 35122, Italy.
Stefano Cazzaniga, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
Edoardo Bertini, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
Mario Pezzotti, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
James J Giovannoni, USDA-ARS Robert W. Holley Center and Boyce Thompson Institute for Plant Research, Tower Road, Cornell Campus, Ithaca, NY 14853, USA.
Julia T Vrebalov, USDA-ARS Robert W. Holley Center and Boyce Thompson Institute for Plant Research, Tower Road, Cornell Campus, Ithaca, NY 14853, USA.
José Tomás Matus, Institute for Integrative Systems Biology (I2SysBio), Universitat de València-CSIC, Paterna 46908, Valencia, Spain.
Giovanni Battista Tornielli, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
Sara Zenoni, Department of Biotechnology, University of Verona, 37134 Verona, Italy.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Fig. S1 . Expression of NAC genes throughout organ development in grapevine.
Supplemental Fig. S2 . Levels of VviNAC60 gene and protein expression in berries.
Supplemental Fig. S3 . Alignment of predicted VviNAC60 amino acid sequences from Pinot Noir and Syrah cultivars.
Supplemental Fig. S4 . Phylogenetic relationships of NAC proteins in different plant species.
Supplemental Fig. S5 . Phenotypic changes in transgenic grapevine plants overexpressing VviNAC60.
Supplemental Fig. S6 . Phenotypic changes in transgenic grapevine plants expressing the repressor VviNAC60-EAR.
Supplemental Fig. S7 . Effects of transient heterologous expression of VviNAC60 in N. benthamiana leaves.
Supplemental Fig. S8 . VviNAC60 expression level determined by RT-qPCR in transgenic grapevine cv. Sultana leaves.
Supplemental Fig. S9 . Expression level of VviMYBA1, VviMYB14, VviWRKY16, and VviSGR1 determined by RT-qPCR in transgenic grapevine cv. Syrah leaves overexpressing VviNAC60.
Supplemental Fig. S10 . Expression profiles of VviNAC60 VHCT genes by exploring the cv. Corvina atlas dataset.
Supplemental Fig. S11 . Distribution of VviNAC03 and VviNAC33 DNA binding events.
Supplemental Fig. S12 . Identification of a VviNAC60 binding motif in the proximal promoter regions of VviMYBA genes from chromosomes 2 and 14.
Supplemental Fig. S13 . VviNAC60 DNA binding landscapes in the proximal promoter region of the VviMYB14 gene, and VviMYB14 promoter activation assessed by dual-luciferase reporter assay in infiltrated N. benthamiana leaves.
Supplemental Fig. S14 . Expression level of VviNAC60, VviNAC03, and VviNAC33 transgenes determined by RT-qPCR in T3 fruits in nor mutant background at Br + 7.
Supplemental Fig. S15 . Ethylene production during ripening of tomato in wild type, nor, and T3 fruit transformed with 35S:VviNAC60 in nor tomato mutant background.
Supplemental Fig. S16 . Expression levels of tomato ripening-related genes (SlACS4; SlPG2a; SlPSY1; SlSGR1) determined by RT-qPCR in wild type, nor, and T3 fruit transformed with 35S:VviNAC60, 35S:VviNAC03 and 35S:VviNAC33 in nor tomato mutant background at Br + 3.
Supplemental Fig. S17 . VviNAC60 VHCTs found in the module of VviATL co-expressed genes specifically related to biotic stress (CC6) and/or upregulated in grapevine plants overexpressing VviATL156 (L1mvsWTm).
Supplemental Fig. S18 . Phenotype of T0 tomato fruits (Solanum lycopersicum cv. Ailsa Craig) of the 2 selected lines carrying 35S: VviNAC60, 35S:VviNAC03, or 35S:VviNAC33 in the nor tomato mutant background.
Supplemental Fig. S19 . Expression level of each transgene determined by RT-qPCR in T1 leaves in nor tomato mutant background.
Supplemental Table S1 . The 89 genes identified as very-high-confidence targets (VHCTs) by combining DAP-seq with transcriptomics data.
Supplemental Table S2 . Complete list of Gene Ontology terms for Fig. 3D.
Supplemental Table S3 . Number of T1 tomato plants obtained from T0 generation.
Supplemental Table S4 . List of primers used.
Supplemental Dataset S1 . List of genes showing highest co-expression with VviNAC60.
Supplemental Dataset S2 . Protein sequences of NAC transcription factors from grapevine, tomato, and Arabidopsis as well as all those characterized in any other species.
Supplemental Dataset S3 . DAP-seq_All peaks VviNAC60.
Supplemental Dataset S4 . Differentially expressed genes in transgenic plants stably overexpressing VviNAC60.
Supplemental Dataset S5 . Differentially expressed genes in transgenic plants transiently overexpressing VviNAC60.
Supplemental Dataset S6 . High-confidence targets of VviNAC60 identified by combining DAP-seq data with transcriptomic analysis.
Supplemental Dataset S7 . DAP-seq_All peaks VviNAC03.
Supplemental Dataset S8 . DAP-seq_All peaks VviNAC33.
Supplemental Dataset S9 . Commonly bound genes identified in VviNAC03, VviNAC33, and VviNAC60 filtered datasets.
Supplemental Methods S1 . Gene co-expression network construction.
Supplemental Methods S2 . Phylogenetic analysis.
Supplemental Methods S3 . Isolation and cloning.
Supplemental Methods S4 . DAP-seq.
Supplemental Methods S5 . Western blot analysis.
Supplemental Methods S6 . Transgenic plants.
Supplemental Methods S7 . Internode length, leaf area and SPAD measurement.
Supplemental Methods S8 . Pigment analysis.
Supplemental Methods S9 . Ethylene and firmness measurement.
Supplemental Methods S10 . Electrolyte leakage assay.
Supplemental Methods S11 . Trypan blue and aniline blue staining.
Author contributions
E.D., S.Z., and G.B.T. designed the research; E.D., C.F., L.O., A.A., E.V., S.C., E.B., and J.V. performed research and analyzed data; A.B. contributed with analytic tools; J.T.M. designed the bioinformatic procedures; C.F., L.O., and A.S. performed the DAP-seq analysis and its integration to transcriptomic data, network construction and phylogenetics, contributed with the computational tools and analyzed data; E.D., S.Z., J.T.M., J.G., M.P., and G.B.T. wrote the paper. All authors reviewed the final version.
Funding
This work was supported by Grant Ricerca di Base “Definition of master regulator genes of fruit ripening in grapevine”, University of Verona, awarded to S.Z., by PRIN 2017 “Regulation of gene expression in grapevine: analysis of genetic and epigenetic determinants” to M.P., by National Science Foundation grant IOS-1855585 and the United States Department of Agriculture – Agricultural Research Service to J.J.G. and by grants PGC2018-099449-A-I00, PID2021-128865NB-I00, RYC-2017-23645, and PRE2019-088044 to J.T.M. and L.O. from the Ministerio de Ciencia, Innovación y Universidades (MCIU, Spain), Agencia Estatal de Investigación (AEI, Spain), and Fondo Europeo de Desarrollo Regional (FEDER, European Union), respectively. This article is based upon work from COST Action CA 17111 INTEGRAPE, supported by COST (European Cooperation in Science and Technology).
References
- Amato A, Cavallini E, Walker AR, Pezzotti M, Bliek M, Quattrocchio F, Koes R, Ruperti B, Bertini E, Zenoni S, et al. The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 2019:99(6): 1220–1241. 10.1111/tpj.14419 [DOI] [PubMed] [Google Scholar]
- Amato A, Cavallini E, Zenoni S, Finezzo L, Begheldo M, Ruperti B, Tornielli GB. A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front Plant Sci. 2017:7: 1979. 10.3389/fpls.2016.01979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariani P, Regaiolo A, Lovato A, Giorgetti A, Porceddu A, Camiolo S, Wong D, Castellarin S, Vandelle E, Polverari A. Genome-wide characterisation and expression profile of the grapevine ATL ubiquitin ligase family reveal biotic and abiotic stress-responsive and development-related members. Sci Rep. 2016:6(1): 38260. 10.1038/srep38260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett A, O’Malley RC, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc. 2017:12(8): 1659–1672. 10.1038/nprot.2017.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini E, Tornielli GB, Pezzotti M, Zenoni S. Regeneration of plants from embryogenic callus-derived protoplasts of garganega and sangiovese grapevine (Vitis vinifera L.) cultivars. Plant Cell Tissue and Organ Culture. 2019:138(2): 239–246. 10.1007/s11240-019-01619-1 [DOI] [Google Scholar]
- Canaguier A, Grimplet J, Di Gaspero G, Scalabrin S, Duchene E, Choisne N, Mohellibi N, Guichard C, Rombauts S, Le Clainche I, et al. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genom Data. 2017:14: 56–62. 10.1016/j.gdata.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Zhang Z, Wang C, Li X, Guo C, Yang L, Guo Y. Identification of a novel melon transcription factor CmNAC60 as a potential regulator of leaf senescence. Genes (Basel). 2019:10(8): 584. 10.3390/genes10080584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 2015:167(4): 1448–1470. 10.1104/pp.114.256172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y. Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol. 2020:62(1): 25–54. 10.1111/jipb.12899 [DOI] [PubMed] [Google Scholar]
- Chitarra W, Cuozzo D, Ferrandino A, Secchi F, Palmano S, Perrone I, Boccacci P, Pagliarani C, Gribaudo I, Mannini F, et al. Dissecting interplays between Vitis vinifera L. and grapevine virus B (GVB) under field conditions. Mol Plant Pathol. 2018:19(12): 2651–2666. 10.1111/mpp.12735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe BG. Research on development and ripening of the grape berry. Am J Enol Vitic. 1992:43(1): 101–110. 10.5344/ajev.1992.43.1.101 [DOI] [Google Scholar]
- Dal Santo S, Fasoli M, Negri S, D’Inca E, Vicenzi N, Guzzo F, Tornielli GB, Pezzotti M, Zenoni S. Plasticity of the berry ripening program in a white grape variety. Front Plant Sci. 2016:7: 970. 10.3389/fpls.2016.00970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Inca E, Cazzaniga S, Foresti C, Vitulo N, Bertini E, Galli M, Gallavotti A, Pezzotti M, Tornielli GB, Zenoni S. VviNAC33 promotes organ de-greening and represses vegetative growth during the vegetative-to-mature phase transition in grapevine. New Phytol. 2021:231(2): 726–746. 10.1111/nph.17263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C, Medina C., Somerville S, Arce-Johnson P.. Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J Exp Bot. 2007:58(12): 3197–3212. 10.1093/jxb/erm165 [DOI] [PubMed] [Google Scholar]
- Falchi R, Wong DCJ, Yan Y, Savoi S, Gambetta GA, Castellarin SD. The genomics of grape berry ripening. In: Cantu D, Walker A, editors. The Grape Genome. 1st ed. Switzerland AG, Cham: Springer International Publishing; 2019. p. 246–274 [Google Scholar]
- Fan K, Bibi N, Gan S, Li F, Yuan S, Ni M, Wang M, Shen H, Wang X. A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J Exp Bot. 2015:66(15): 4669–4682. 10.1093/jxb/erv240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoli M, Dal Santo S, Zenoni S, Tornielli GB, Farina L, Zamboni A, Porceddu A, Venturini L, Bicego M, Murino V, et al. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell. 2012:24(9): 3489–3505. 10.1105/tpc.112.100230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoli M, Richter CL, Zenoni S, Bertini E, Vitulo N, Dal Santo S, Dokoozlian N, Pezzotti M, Tornielli GB. Timing and order of the molecular events marking the onset of berry ripening in grapevine. Plant Physiol. 2018:178(3): 1187–1206. 10.1104/pp.18.00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlani S, Mizzotti C, Masiero S. The NAC side of the fruit: tuning of fruit development and maturation. BMC Plant Biol. 2021:21(1): 238. 10.1186/s12870-021-03029-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Van Eck J. Organogenesis from transformed tomato explants. Methods Mol Biol. 2005:286: 141–150. 10.1385/1-59259-827-7:141 [DOI] [PubMed] [Google Scholar]
- Gao Y, Wei W, Fan Z, Zhao X, Zhang Y, Jing Y, Zhu B, Zhu H, Shan W, Chen J, et al. Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. J Exp Bot. 2020:71(12): 3560–3574. 10.1093/jxb/eraa131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wei W, Zhao X, Tan X, Fan Z, Zhang Y, Jing Y, Meng L, Zhu B, Zhu H, et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic Res. 2018:5: 75. 10.1038/s41438-018-0111-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper NE, McQuinn RP, Giovannoni JJ. Molecular and genetic regulation of fruit ripening. Plant Mol Biol. 2013:82(6): 575–591. 10.1007/s11103-013-0050-3 [DOI] [PubMed] [Google Scholar]
- Gepstein S, Glick BR. Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol Biol. 2013:82(6): 623–633. 10.1007/s11103-013-0038-z [DOI] [PubMed] [Google Scholar]
- Gong J, Zeng Y, Meng Q, Guan Y, Li C, Yang H, Zhang Y, Ampomah-Dwamena C, Liu P, Chen C, et al. Red light-induced kumquat fruit coloration is attributable to increased carotenoid metabolism regulated by FcrNAC22. J Exp Bot. 2021:72(18): 6274–6290. 10.1093/jxb/erab283 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Letters. 2002:514: 351–354. 10.1016/S0014-5793(02)02435-3 [DOI] [PubMed] [Google Scholar]
- Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, Rausch T, Lucchin M, Boss PK, Dry IB, Bogs J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell. 2013:25(10): 4135–4149. 10.1105/tpc.113.117127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans Marlies, Lema A Saul, Coll Nuria S, Nowack Moritz K. Dying two deaths — programmed cell death regulation in development and disease. Curr Opin Plant Biol. 2017:35: 37–44. 10.1016/j.pbi.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Iocco P, Franks T, Thomas MR. Genetic transformation of major wine grape cultivars of Vitis vinifera L. Transgenic Res. 2001:10(2): 105–112. 10.1023/a:1008989610340 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Nam HG, Lim PO. Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol. 2016:33: 48–56. 10.1016/j.pbi.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, et al. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009:323(5917): 1053–1057. 10.1126/science.1166386 [DOI] [PubMed] [Google Scholar]
- Kou X, Watkins CB, Gan SS. Arabidopsis AtNAP regulates fruit senescence. J Exp Bot. 2012:63(17): 6139–6147. 10.1093/jxb/ers266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou X, Zhou J, Wu CE, Yang S, Liu Y, Chai L, Xue Z. The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review. Plant Mol Biol. 2021:106(3): 223–238. 10.1007/s11103-021-01128-w [DOI] [PubMed] [Google Scholar]
- Kuhn N, Abello C, Godoy F, Delrot S, Arce-Johnson P. Differential behavior within a grapevine cluster: decreased ethylene-related gene expression dependent on auxin transport is correlated with low abscission of first developed berries. PLoS One. 2014:9(11): e111258. 10.1371/journal.pone.0111258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Tamboli V, Sharma R, Sreelakshmi Y. NAC-NOR mutations in tomato penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chem. 2018:259: 234–244. 10.1016/j.foodchem.2018.03.135 [DOI] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci U S A. 2014:111(27): 10013–10018. 10.1073/pnas.1321568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007:58(1): 115–136. 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- Lovato A, Zenoni S, Tornielli GB, Colombo T, Vandelle E, Polverari A. Specific molecular interactions between Vitis vinifera and Botrytis cinerea are required for noble rot development in grape berries. Postharvest Biol Technol. 2019:156: 110924. 10.1016/j.dib.2019.104150 [DOI] [Google Scholar]
- Ma X, Balazadeh S, Mueller-Roeber B. Tomato fruit ripening factor NOR controls leaf senescence. J Exp Bot. 2019:70(10): 2727–2740. 10.1038/nbt.2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Y, Tureckova V, Xue GP, Fernie AR, Mueller-Roeber B, Balazadeh S. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol. 2018:177(3): 1286–1302. 10.1104/pp.18.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Zhao P, Liu S, Yang Q, Guo H. The control of developmental phase transitions by microRNAs and their targets in seed plants. Int J Mol Sci. 2020:21(6): 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuela D, Xu M. Juvenile leaves or adult leaves: determinants for vegetative phase change in flowering plants. Int J Mol Sci. 2020:21(24): 9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Pizarro C, Vallarino JG, Osorio S, Meco V, Urrutia M, Pillet J, Casanal A, Merchante C, Amaya I, Willmitzer L, et al. The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell. 2021:33(5): 1574–1593. 10.1093/plcell/koab070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet M, Fasoli M, Tornielli GB, Altieri M, Sandri M, Zuccolotto P, Paci P, Gardiman M, Zenoni S, Pezzotti M. Ripening transcriptomic program in red and white grapevine varieties correlates with berry skin anthocyanin accumulation. Plant Physiol. 2017:174(4): 2376–2396. 10.1104/pp.17.00311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Cavallini E, Loyola R, Höll J, Finezzo L, Dal Santo S, Vialet S, Commisso M, Roman F, Schubert A, et al. A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J. 2017:91(2): 220–236. 10.1111/tpj.13558 [DOI] [PubMed] [Google Scholar]
- Matus JT, Ruggieri V, Romero F, Moretto M, Wong D. Status and prospects of systems biology in grapevine research. In: Cantu D, Walker A, editors. The Grape Genome. Vol. 1. Switzerland AG, Cham: Springer International Publishing; 2019. p. 137–166 [Google Scholar]
- Mullins MG, Rajasekaran K. Fruiting cuttings: revised method for producing test plants of grapevine cultivars. Am J Enol Vitic. 1981:32(1): 35. 10.5344/ajev.1981.32.1.35 [DOI] [Google Scholar]
- Navarro-Paya D, Santiago A, Orduna L, Zhang C, Amato A, D’Inca E, Fattorini C, Pezzotti M, Tornielli GB, Zenoni S, et al. The grape gene reference catalogue as a standard resource for gene selection and genetic improvement. Front Plant Sci. 2022:12: 803977. 10.3389/fpls.2021.803977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016:165(5): 1280–1292. 10.1016/j.cell.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orduña L, Li M, Navarro-Paya D, Zhang C, Santiago A, Romero P, Ramsak Z, Magon G, Höll J, Merz P, et al. Orchestration of the stilbene synthase gene family and their regulators by subgroup 2 MYB genes. bioRxiv. 2020 [Google Scholar]
- Orduña L, Li M, Navarro-Paya D, Zhang C, Santiago A, Romero P, Ramsak Z, Magon G, Höll J, Merz P, et al. Direct regulation of shikimate, early phenylpropanoid, and stilbenoid pathways by subgroup 2 R2R3-MYBs in grapevine. Plant J. 2022:110(2): 529–547. 10.1111/tpj.15686 [DOI] [PubMed] [Google Scholar]
- Palumbo MC, Zenoni S, Fasoli M, Massonnet M, Farina L, Castiglione F, Pezzotti M, Paci P. Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell. 2015:26(12): 4617–4635. 10.1105/tpc.114.133710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell. 2007:19(5): 1649–1664. 10.1105/tpc.106.044891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual MB, Llebres MT, Craven-Bartle B, Canas RA, Canovas FM, Avila C. PpNAC1, a main regulator of phenylalanine biosynthesis and utilization in maritime pine. Plant Biotechnol J. 2018:16(5): 1094–1104. 10.1111/pbi.12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati S, Bagagli G, Sonego P, Moretto M, Brazzale D, Castorina G, Simoni L, Tonelli C, Guella G, Engelen K, et al. Abscisic acid is a major regulator of grape berry ripening onset: new insights into ABA signaling network. Front Plant Sci. 2017:8: 1093. 10.3389/fpls.2017.01093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM. Molecular aspects of leaf senescence. Trends Plant Sci. 2000:5(7): 278–282. 10.1016/s1360-1385(00)01655-1 [DOI] [PubMed] [Google Scholar]
- Rauf M, Arif M, Fisahn J, Xue GP, Balazadeh S, Mueller-Roeber B. NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. Plant Cell. 2013:25(12): 4941–4955. 10.1105/tpc.113.117861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y, Ito H, Tanaka A. Arabidopsis STAY-GREEN, Mendel's Green cotyledon gene, encodes magnesium-dechelatase. Plant Cell. 2016:28(9): 2147–2160. 10.1105/tpc.16.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Jiang S, Zhang T, Xu H, Fang H, Zhang J, Su M, Wang Y, Zhang Z, Wang N, et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019:289: 110286. 10.1016/j.plantsci.2019.110286 [DOI] [PubMed] [Google Scholar]
- Vandelle E, Colombo T, Regaiolo A, Maurizio V, Libardi T, Puttilli MR, Danzi D, Polverari A. Transcriptional profiling of three Pseudomonas syringae pv. Actinidiae biovars reveals different responses to apoplast-like conditions related to strain virulence on the host. Mol Plant Microbe Interact. 2021:34(4): 376–396. 10.1094/MPMI-09-20-0248-R [DOI] [PubMed] [Google Scholar]
- Venturini L, Ferrarini A, Zenoni S, Tornielli GB, Fasoli M, Dal Santo S, Minio A, Buson G, Tononi P, Zago ED, et al. De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics. 2013:14(1): 41. 10.1186/1471-2164-14-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002:296(5566): 343–346. 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DA, Thomas MR, Robinson SP. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007:49(5): 772–785. 10.1111/j.1365-313X.2006.02997.x [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant Journal. 2004:40(3): 428–438. 10.1111/j.1365-313X.2004.02219.x [DOI] [PubMed] [Google Scholar]
- Wang WQ, Wang J, Wu YY, Li DW, Allan AC, Yin XR. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 2020:225(4): 1618–1634. 10.1111/nph.16233 [DOI] [PubMed] [Google Scholar]
- Webb LB, Whetton PH, Barlow EWR. Modelled impact of future climate change on the phenology of winegrapes in Australia. Austral J Grape Wine Res. 2007:13(3): 165–175. 10.1111/j.1755-0238.2007.tb00247.x [DOI] [Google Scholar]
- White PJ. Recent advances in fruit development and ripening: an overview. J Exp Bot. 2002:53(377): 1995–2000. 10.1093/jxb/erf105 [DOI] [PubMed] [Google Scholar]
- Wu X, Gong F, Wang W. Protein extraction from plant tissues for 2DE and its application in proteomic analysis. Proteomics. 2014:14(6): 645–658. 10.1002/pmic.201300239 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A. Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Pant J. 2002:29(4): 427–437. 10.1046/j.0960-7412.2001.01228.x [DOI] [PubMed] [Google Scholar]
- Yu H, Yan J, Du X, Hua J. Overlapping and differential roles of plasma membrane calcium ATPases in Arabidopsis growth and environmental responses. J Exp Bot. 2018:69(10): 2693–2703. 10.1093/jxb/ery073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Fasoli M, Guzzo F, Dal Santo S, Amato A, Anesi A, Commisso M, Herderich M, Ceoldo S, Avesani L, et al. Disclosing the molecular basis of the postharvest life of berry in different grapevine genotypes. Plant Physiol. 2016:172(3): 1821–1843. 10.1038/nbt.2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Dong R, Wang Y, Li X, Ji M, Wang X. NAC domain gene VvNAC26 interacts with VvMADS9 and influences seed and fruit development. Plant Physiol Biochem. 2021:164: 63–72. 10.1016/j.plaphy.2021.04.031 [DOI] [PubMed] [Google Scholar]
- Zhang L, Xiao S, Li W, Feng W, Li J, Wu Z, Gao X, Liu F, Shao M. Overexpression of a harpin-encoding gene hrf1 in rice enhances drought tolerance. J Exp Bot. 2011:62(12): 4229–4238. 10.1093/jxb/err131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Pislariu C, Nakashima J, Fu C, Jiang Q, Quan L, Blancaflor EB, Tang Y, Bouton JH, et al. From model to crop: functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011:157(3): 1483–1496. 10.1104/pp.111.185140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin-Wang K, Wang H, Gu C, Dare AP, Espley RV, He H, Allan AC, Han Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015:82(1): 105–121. 10.1111/tpj.12792 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.