Key Points
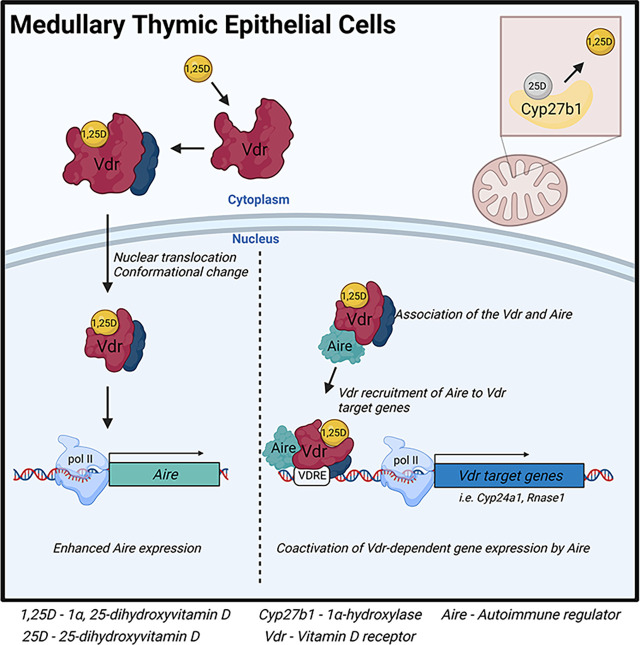
The Vdr is expressed in Aire+ medullary thymic epithelial cells.
Thymic vitamin D signaling stimulates Aire expression and Aire-dependent gene transcription.
Aire interacts with the Vdr and is a coactivator of Vdr-dependent transcription.
Visual Abstract
Abstract
Vitamin D deficiency is associated with the development of autoimmunity, which arises from defects in T cell tolerance to self-antigens. Interactions of developing T cells with medullary thymic epithelial cells, which express tissue-restricted Ags, are essential for the establishment of central tolerance. However, vitamin D signaling in the thymus is poorly characterized. We find that stromal and hematopoietic cells in the mouse thymus express the vitamin D receptor (Vdr) and Cyp27b1, the enzyme that produces hormonal 1,25-dihydroxyvitamin D (1,25D). Treatment of cultured thymic slices with 1,25D enhances expression of the critical medullary thymic epithelial cell transcription factor autoimmune regulator (Aire), its colocalization with the Vdr, and enhances tissue-restricted Ag gene expression. Moreover, the Vdr interacts with Aire in a 1,25D-dependent manner and recruits Aire to DNA at vitamin D response elements, where it acts as a Vdr coactivator. These data link vitamin D signaling directly to critical transcriptional events necessary for central tolerance.
Introduction
Vitamin D signaling is widespread, including in cells of the innate and adaptive immune systems (1). Although vitamin D can be obtained from the diet, supplements, or cutaneous exposure to adequate UVB irradiation (1), vitamin D–poor diets, sun avoidance, and conservative dress lead to widespread vitamin D deficiency (2). Profound deficiency in children causes rickets, a diagnosis associated with increased risk of diseases unrelated to poor calcium status (3), including autoimmune disorders (1, 4). Vitamin D is activated by primarily hepatic 25-hydroxylation followed by peripheral 1α-hydroxylation catalyzed by Cyp27b1 to generate 1,25-dihydroxyvitamin D (1,25D). 1,25D binds to the vitamin D receptor (Vdr), a nuclear receptor and ligand-regulated transcription factor (1), which binds to cognate vitamin D response elements (VDREs). In the periphery, 1,25D attenuates T cell activation and proliferation and suppresses T cell–driven inflammation while enhancing suppressive regulatory T cells (5). However, the links between vitamin D deficiency in infancy and childhood and the subsequent risk of autoimmunity suggest that vitamin D signaling also contributes to thymic central tolerance.
Central tolerance is established by elimination of developing thymocytes with overly self-reactive Ag receptors via negative selection or diversion toward immunoregulatory lineages (6). Transcription of tissue-restricted Ag (TRA) genes in medullary thymic epithelial cells (mTECs) is critical for central tolerance. The transcription factor autoimmune regulator (Aire) is necessary for expression of most TRAs (7), and its loss in humans causes autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED), characterized by multiorgan autoimmunity (7). Very little is known about the role of 1,25D in thymic development, cellularity, and function. One study provided evidence that invariant NKT (iNKT) cells fail to mature in the thymus of Vdr-null mice (8). However, the molecular mechanisms of thymic Vdr action have not been addressed. We show that Vdr signaling is active in thymic hematopoietic and stromal cells, including mTECs, and that 1,25D induces Aire and TRA expression. Moreover, Aire is recruited to Vdr target genes and acts as a coactivator. This implicates 1,25D signaling in transcriptional events necessary for central tolerance.
Materials and Methods
Mice
Six- to 10-wk-old male or female C57BL/6J mice (The Jackson Laboratory) were maintained in a specific pathogen-free environment at the Maisonneuve-Rosemont Hospital Research Center. All animal protocols were approved by the local Animal Care Committee in accordance with the Canadian Council on Animal Care guidelines.
Thymic slice preparation and culture
Thymic slices were prepared as described (9). Harvested thymi were placed into a 4% low-melt agarose solution at <40°C. After solidification, 500-µm-thick sections were prepared. Slices (3–10/condition) were cultured in six-well plates and treated with RPMI 1640 containing 100 nM 1,25D or 1 µl of vehicle (100% EtOH), for up to 24 h at 37°C (5% CO2).
Preparation of single-cell suspensions
Tissue was cut into 1-mm pieces and digested in RPMI 1640 (Wisent) containing 10% FBS (MilliporeSigma) (R10) and collagenase D (250 µg/ml; Sigma-Aldrich), papain (250 µg/ml; Worthington Biochemical), and DNase I (200 µg/ml; Sigma-Aldrich) for 30 min at 37°C with shaking. Cells were further released by pipetting up and down. Digestion buffer was freshly prepared for each experiment.
Flow cytometry
Cells (2–5 × 106) were incubated with fixable viability dye diluted in PBS at 4°C for 20 min. Cells were washed and resuspended with PBS containing 1% FBS, 2 mM EDTA (FACS buffer), fluorophore-conjugated Abs, and Fc Block (Supplemental Table I) and incubated for 30 min at 4°C. For intracellular proteins, cells were fixed and permeabilized using a Foxp3 Fix/Perm kit (Thermo Fisher Scientific) for 30 min at room temperature (RT). Cells were then resuspended in 1× Perm/Wash buffer containing fluorophore-conjugated Abs and incubated for 30 min at 4°C. Cells and UltraComp eBeads (Thermo Fisher Scientific) were used for compensation controls. Samples were acquired on an LSRFortessa (BD Biosciences) and analyzed by FlowJo (BD Biosciences). Gating was as follows (see Supplemental Fig. 3): mTEChi (EpCAM+, UEA-1+/−, Ly51−, MHC class II [MHC-II]hi, CD80+), mTEClo (EpCAM+, UEA-1+/−, Ly51−, MHC-II+/−, CD80−), cortical TECs (cTECs; EpCAM+, Ly51+), B cells (CD45+, CD19+, MHC-II+, CD11c−), dendritic cells (CD45+, CD11c+, MHC-II+, CD19−), and thymocytes (CD45+, TCRβ+/−, MHC-II−, CD19−, CD11c−).
Cell sorting
TECs were enriched by positively selecting for EpCAM+ cells by MACS (Miltenyi Biotec). Flow cytometric staining was performed as described above. Total TECs (EpCAM+, CD45−) or mTECs (EpCAM+, CD45−, UEA-1+, Ly51−) were sorted with a BD FACSAria Fusion into cold FACS buffer.
Immunofluorescence microscopy
Samples were fixed with 4% paraformaldehyde and incubated with sucrose (BioShop) solutions (10–30%) for ∼8 h each and embedded using base molds (Fisher Scientific) containing OCT compound (Fisher Scientific), frozen on dry ice, and stored at −80°C. Five- to 20-µm sections were obtained with a Leica CM3050 S cryostat and SuperFrost Plus slides (Fisher Scientific). A hydrophobic barrier was applied (MilliporeSigma), and slides were blocked with PBS containing 2% BSA (w/v), 0.3% Triton X-100, 1% FBS, and 10% goat serum (blocking buffer) for 1 h at RT. Slides were incubated in a humidified chamber with primary Abs diluted in blocking buffer (Supplemental Table I) overnight at 4°C, secondary Abs conjugated to fluorophores diluted in blocking buffer for 1 h at RT, fluorophore-conjugated primary Abs for 1 h at RT, and DAPI diluted in PBS for 5 min at RT. Slides were washed three times with 100 µM Tris buffer between each step, mounted with Fluoromount-G (Thermo Fisher Scientific) and coverslips (no. 1.5; Fisher Scientific), and imaged with a Zeiss LSM 710 confocal microscope with ×20–100 objectives. Images were analyzed with Fiji (ImageJ). For colocalization analysis, images were acquired with the same settings and JaCoP was used on split channels without prior contrast changes and automatic thresholds to quantify overlap.
Gene expression
Cells (5 × 106) were lysed and RNA was extracted using a FavorPrep tissue total RNA mini kit (Favorgen) and quantified with a NanoDrop 2000c (Thermo Fisher Scientific). Five hundred nanograms of RNA was converted to cDNA using Advantech 5× master mix. Oligonucleotide sequences (available upon request) were designed with Primer3, and primer specificity was verified by basic local alignment search tool. Ten nanograms of cDNA was used per quantitative PCR (qPCR) reaction and BlasTaq 2× qPCR MasterMix (Applied Biological Materials). qPCR reactions and melting curves were done with a Light Cycler 96 (Roche). Gene expression was normalized to 18S. Fold changes for thymic slices were calculated by comparing gene expression between treatment conditions from individual mice.
Plasmids
VDR expression vector pcDNA 3.1-VDR was generated by inserting the human VDR cDNA as a BamH1-Xho1 fragment. The AIRE cDNA expression vector (pGMV3-Aire-myc) was from Sino Biological (HG17322-UT). The 1,25D-responsive luciferase reporter plasmid pVDRE2-pGL4.24 was constructed by insertion of tandem 31-bp oligonucleotides containing consensus VDREs upstream of the minimal promoter of pGL4.24 (Promega). Cells were transfected using CalFectin transfection reagent (SL100478; SignaGen Laboratories).
Immunoprecipitation and Western analysis
Cells were lysed with 20 mmol/l Tris (pH 7.5), 100 mmol/l NaCl, 0.5% Nonidet P-40, 0.5 mmol/l EDTA, and 0.5 mmol/l PMSF. For co-IP assays, 4 ng of AIRE Ab (Supplemental Table I) was prebound for 2 h to Dynabeads protein A, washed with lysis buffer, added to the lysate, and immunoprecipitated overnight. Beads were washed 5× with washing buffer (20 mmol/L Tris, pH 7.5, 200 mmol/L NaCl, 1% Nonidet P-40, 0.5 mmol/L EDTA, 0.5 mmol/L PMSF) and processed for Western blotting.
Chromatin immunoprecipitation
Sorted mTECs were prepared as described above. Chromatin immunoprecipitation (ChIP) assays were performed as described (10). After fixation, cells were recovered in 1% IGEPAL, 20 mM Tris (pH 8), 0.5 mM EDTA, and 100 mM NaCl + RNase, pelleted at 6000 rpm, resuspended in 1% SDS, NaCl 50 mM, and 20 mM Tris (pH 8) + RNase, sonicated to obtain fragments <500 bp, vortexed, and centrifuged at 10,000 rpm for 10 min at 4°C. One percent of each sample was stored at −20°C for input controls. The remainder was divided into IgG control and immunoprecipitation tests. A protease inhibitor mixture and 1 µl of Dynabeads protein G (Thermo Fisher Scientific, 10003D) per 2 µg of Ab was added, and samples were incubated overnight at 4°C. The supernatant was removed while samples were on a magnet, and proteinase K with 1% IGEPAL buffer was added to each sample, including inputs, and shaken at 1000 rpm for 2 h at 45°C. Crosslinking was reversed at 65°C for 4 h. The supernatant was removed while samples were on a magnet, and the FavorPrep GEL/PCR purification mini kit (Favorgen) was used to collect DNA.
Luciferase reporter assay
HEK293 cells were cultured in DMEM with heat-inactivated FBS and Normocin and transfected when 60% confluent (luciferase expression vector only [with and without 1,25D 100 nM], luciferase + VDR expression vector + increasing amounts of AIRE [with and without 100 nM 1,25D; 0–100 ng of AIRE]). Plates were incubated for 4–5 h prior to incubation overnight with 1,25D (100 nM). Two hundred microliters of Glo Lysis Buffer (Promega, Madison, WI) was added and plates were left for 5 min on the shaker. One hundred microliters of each lysate and luciferase substrate was transferred to a round-bottom tube and luminescence was immediately read with a luminometer.
Human single-cell RNA sequencing data analysis
Raw data (GSM4466784/5) were analyzed with Seurat and filtered by unique feature counts and mitochondrial genes. Samples were integrated, scaled, and normalized. Dimensionality reduction was conducted using the first 20 principal components. Uniform manifold approximation and projections were constructed and clusters were identified using the shared nearest neighbor algorithm (resolution = 0.5).
Statistical analyses
Statistics were calculated using GraphPad Prism version 8 with parametric unpaired t tests, as well as nonparametric tests or parametric paired t tests for thymic slice data. Statistical significance is indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results and Discussion
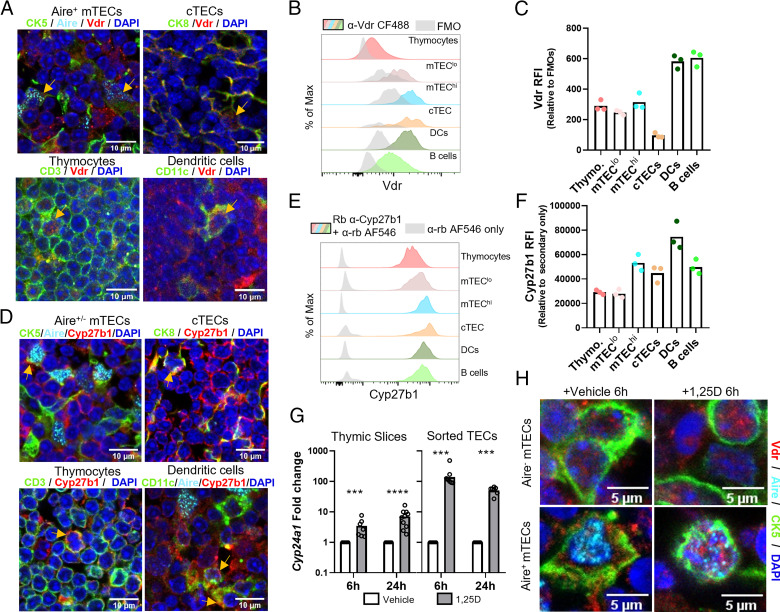
Although the Vdr is expressed in thymocytes and thymic iNKT cells (8, 11), receptor function in other important cell types, notably TECs, has not been examined. The Vdr and Cyp27b1 mRNAs are expressed in mature mouse mTECs at higher levels than in thymocytes and iNKT cells (Supplemental Fig. 1A, 1B), and Vdr mRNA in mouse mTECs is abundant relative to other nuclear receptor transcripts (Supplemental Fig. 2A) (12). Human VDR expression was also detected in TEC samples from two independent single-cell RNA sequencing studies, along with expression of CYP24A1, indicative of active vitamin D signaling (Supplemental Fig. 2B, 2C) (13, 14). We detected Vdr and Cyp27b1 mRNA in whole mouse thymi and in sorted total TECs (Supplemental Fig. 1C). Immunofluorescence (IF) of thymic sections revealed abundant Vdr protein in the thymic cortex and medulla (Supplemental Fig. 1D). Vdr expression was also confirmed in multiple cell types, including Aire+ TECs, using cell-specific markers (Fig. 1A; see also Supplemental Fig. 1E for images of Vdr channels only).
FIGURE 1.
Active 1,25D signaling in the thymus. (A) IF microscopy of thymic sections showing Vdr staining in Aire+ mTECs, cTECs, thymocytes, and dendritic cells. (B and C) Flow cytometric analysis of Vdr expression in multiple thymic cell types (fluorescence minus one [FMO] was gated on respective populations). RFI, relative fluorescence intensity. (D) Cytoplasmic Cyp27b1 staining in Aire+/− mTECs, cTECs, thymocytes, and dendritic cells. (E and F) Flow cytometric analysis of Cyp27b1 expression in multiple thymic cell types. (G) Induction of Cyp24a1 mRNA (log scale) by 1,25D in mouse thymic slices and in sorted TECs (CD45−EpCAM+). ***p < 0.001, ****p < 0.0001. (H) Vdr staining in Aire− and Aire+ mTECs in vehicle or 1,25D-treated thymic slices. Data are individual mice (n ≥ 3, two or more independent experiments).
Vdr expression was also validated in hematopoietic and stromal cells by flow cytometry (Fig. 1B, 1C, gating strategy in Supplemental Fig. 3). Its expression in both cortical TECs (cTECs) and mTECs suggests a role for 1,25D signaling in multiple thymic processes. Moreover, the relatively high Vdr expression in medullary APCs suggests that its signaling may contribute to central tolerance. We also detected Cyp27b1 protein in multiple thymic cell types by IF (Fig. 1D) and flow cytometry (Fig. 1E, 1F), providing evidence for the thymic production of 1,25D.
We analyzed the 1,25D-dependent induction of the Vdr target gene Cyp24a1 in sorted TECs and in cultured thymic slices, which maintain tissue integrity and important cell–cell interactions (9). 1,25D induced Cyp24a1 5- to 8-fold in thymic slices (Fig. 1G). Given that TECs represent only ∼1% of thymic cellularity, we treated sorted TECs and found that Cyp24a1 was induced 50- to 100-fold by 1,25D (Fig. 1G). This observation is also consistent with the levels of mTEC Cyp24a1 mRNA observed by expression profiling (Supplemental Fig. 1F). Finally, the Vdr was mostly cytoplasmic in thymic slices cultured in vehicle, whereas it was more nuclear in the presence of 1,25D (Fig. 1H). This suggests that the largely nuclear Vdr staining seen in multiple cell types in freshly isolated thymus (Supplemental Fig. 1D) is consistent with active 1,25D signaling.
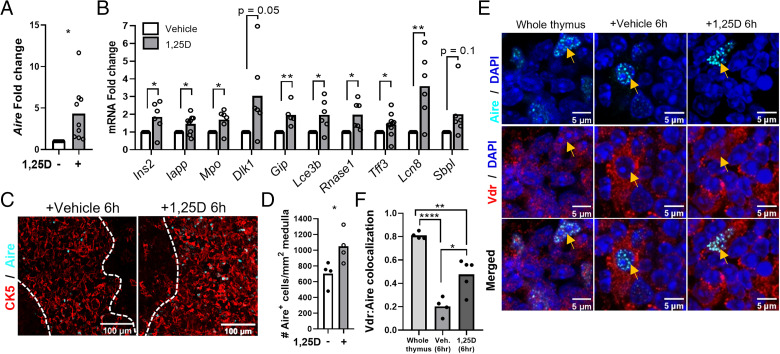
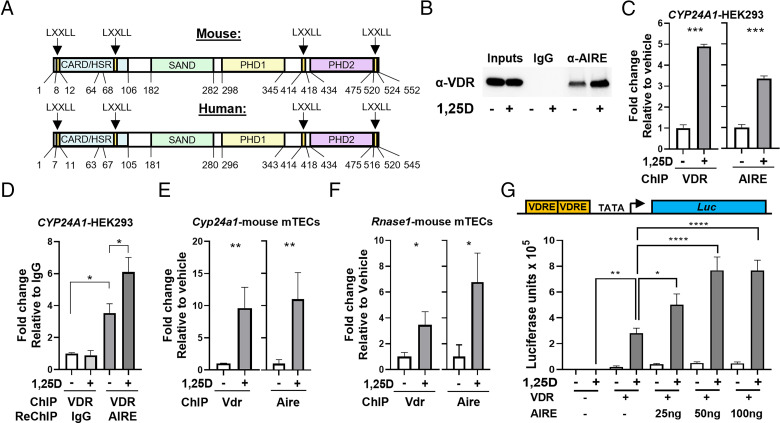
Due to the enrichment of 1,25D signaling in TECs and the coexpression of the Vdr and Aire, we investigated the potential functional relationship of the two proteins. Expression of Aire and Aire-dependent TRA mRNAs was induced in thymic slices treated with 1,25D (Fig. 2A, 2B). Moreover, 1,25D treatment of thymic slices increased the number of Aire+ cells (Fig. 2C, 2D). 1,25D also enhanced the colocalization of the Vdr with Aire in mTECs (Fig. 2E, 2F). Although the Vdr regulates transcription by pleiotropic mechanisms (15), its partial overlap with Aire suggests that the two proteins may interact functionally. This notion is supported by the presence of four LXXLL motifs in mouse and human AIRE (Fig. 3A) (16), which are found in nuclear receptor coactivators (17). We observed a 1,25D-dependent co-IP of the human VDR with human AIRE expressed in HEK293 cells (Fig. 3B). In addition, AIRE expressed in HEK293 cells was recruited in a 1,25D-dependent manner to a VDRE in the human CYP24A1 gene in ChIP assays (Fig. 3C). Re-ChIP experiments at the same VDRE confirmed the formation of a VDR–AIRE complex (Fig. 3D).
FIGURE 2.
1,25D signaling regulates Aire and Aire-dependent gene expression. (A) Induction of Aire mRNA by 1,25D in mouse thymic slices (24 h). (B) Aire-dependent gene expression in 6-h vehicle versus 1,25D-treated thymic slices. (C and D) Representative images (C) of Aire+ mTECs in vehicle versus 1,25D-treated thymic slices and quantification (D). (E) Representative images of Vdr localization with Aire in whole thymus and vehicle- or 1,25D-treated thymic slices. (F) Quantification of Vdr colocalization with Aire (Manders’ M1 coefficient). Data are individual mice (n ≥ 2, two or more experiments). *p < 0.05, **p < 0.01, ****p < 0.0001.
FIGURE 3.
The Vdr and Aire interact in a 1,25D-dependent manner. (A) Domain structures of mouse and human Aire with LXXLL motifs indicated. (B) 1,25D enhanced co-IP of the human VDR with AIRE in HEK293-transfected cells. (C) 1,25D-dependent ChIP of the human VDR (left) or AIRE (right) on CYP24A1 VDREs in transfected HEK293 cells. (D) Re-ChIP of VDR ChIP on CYP24A1 VDREs with IgG or anti-AIRE showing that the VDR and AIRE are present on CYP24A1 VDREs together. (E) ChIP of the mouse Vdr (left) or mouse Aire (right) on Cyp24a1 VDREs from sorted primary mouse mTECs. (F) ChIP of the mouse Vdr (left) or mouse Aire (right) on Rnase1 VDREs from sorted primary mouse mTECs. (G) Coactivation of 1,25D-dependent luminescence by AIRE in transfected HEK293 cells. Schematic showing minimal promoter construct (top) and summary data from two experiments (bottom). Data are mean± SD (n ≥ 3, two experiments). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Next, we examined the 1,25D-dependent recruitment of Aire to VDREs in mouse mTECs. We observed 1,25D-dependent binding of endogenous Vdr and Aire to a Cyp24a1 VDRE in sorted mTECs (Fig. 3E). A screen of mouse Vdr ChIP-seq datasets identified a peak of 1,25D-dependent Vdr binding adjacent to the Rnase1 gene in an intestinal epithelial dataset (Supplemental Fig. 1G) (18). As shown above, Rnase1 expression was induced in thymic slices by 1,25D (Fig. 2B). We detected 1,25D-dependent recruitment of the Vdr and Aire to the same site in the Rnase1 gene in sorted mTECs (Fig. 3F). The potential function of AIRE as a VDR cofactor was further addressed using a minimal promoter composed of tandem VDREs inserted adjacent to a binding site for RNA polymerase II (TATA box). This arrangement was used to minimize the potential intervention of other classes of transcription factors on more complex promoters. Coexpression of increasing amounts of AIRE enhanced 1,25D-dependent induction of a luciferase reporter gene (Fig. 3G) in HEK293 cells. Collectively, these data reveal the 1,25D-dependent recruitment of AIRE to VDREs, and that AIRE functions as a VDR coactivator.
In conclusion, we find that 1,25D signaling, which has been implicated in autoimmunity (19), is active in thymic hematopoietic and stromal compartments, and it induces the expression of Aire mRNA and protein in mTECs as well as the expression of a number of TRA genes. Moreover, AIRE is an interaction partner of the VDR and acts as a VDR coactivator. As Aire does not appear to be a site-specific binding protein (7), our data suggest a novel mechanism by which Aire can be recruited to chromatin. Induction of TRA gene expression by 1,25D could occur by direct binding of the Vdr to some genes or indirectly via induced Aire expression. This provides evidence that 1,25D signaling increases the amount or diversity of rare self-peptides presented to thymocytes during induction of central tolerance.
Mouse and human Aire contain four LXXLL motifs found in nuclear receptor coregulators (17). The presence of four such motifs suggests they may act at least partially redundantly in VDR–AIRE interactions. It also raises the possibility that Aire can be recruited to chromatin by other nuclear receptors expressed in mTECs. Available sequence data reveal expression of several nuclear receptors in sorted MHC-IIhi mTECs (Supplemental Fig. 2). Notably, these include the gene encoding the androgen receptor (Ar). Androgens enhance Aire transcription in mTECs by inducing Ar binding to Aire promoter regions (20, 21), raising the possibility that Aire may also be a cofactor of the Ar. Collectively, these findings suggest that Aire may be recruited to chromatin by multiple agonist-bound nuclear receptors.
Supplementary Material
Acknowledgments
We acknowledge the Advanced Bioimaging Facility at McGill University for help with imaging experiments.
Footnotes
This work was supported by the Canadian Institutes of Health Research Grant CIHR PJT-180271 (to J.H.W. and H.J.M.).
The online version of this article contains supplemental material.
- Aire
- autoimmune regulator
- Ar
- androgen receptor
- ChIP
- chromatin immunoprecipitation
- co-IP
- co-immunoprecipitation
- cTEC
- cortical TEC
- 1,25D
- 1,25-dihydroxyvitamin D
- IF
- immunofluorescence
- iNKT
- invariant NKT
- MHC-II
- MHC class II
- mTEC
- medullary thymic epithelial cell
- qPCR
- quantitative PCR
- RT
- room temperature
- TRA
- tissue-restricted Ag
- Vdr
- vitamin D receptor
- VDRE
- vitamin D response element
Disclosures
The authors have no financial conflicts of interest.
References
- 1. Bouillon, R., Marcocci C., Carmeliet G., Bikle D., White J. H., Dawson-Hughes B., Lips P., Munns C. F., Lazaretti-Castro M., Giustina A., Bilezikian J.. 2019. Skeletal and extraskeletal actions of vitamin D: current evidence and outstanding questions. Endocr. Rev. 40: 1109–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arabi, A., El Rassi R., El-Hajj Fuleihan G.. 2010. Hypovitaminosis D in developing countries-prevalence, risk factors and outcomes. Nat. Rev. Endocrinol. 6: 550–561. [DOI] [PubMed] [Google Scholar]
- 3. Pettifor, J. M., Thandrayen K., Thacher T. D.. 2018. Vitamin D deficiency and nutritional rickets in children. In Vitamin D, 4th Ed. Feldman D., ed. Academic Press, Cambridge, MA, p. 179–201. [Google Scholar]
- 4. Cantorna, M. T., Mahon B. D.. 2004. Mounting evidence for vitamin D as an environmental factor affecting autoimmune disease prevalence. Exp. Biol. Med. (Maywood) 229: 1136–1142. [DOI] [PubMed] [Google Scholar]
- 5. Bhalla, A. K., Amento E. P., Serog B., Glimcher L. H.. 1984. 1,25-Dihydroxyvitamin D3 inhibits antigen-induced T cell activation. J. Immunol. 133: 1748–1754. [PubMed] [Google Scholar]
- 6. Klein, L., Robey E. A., Hsieh C.-S.. 2019. Central CD4+ T cell tolerance: deletion versus regulatory T cell differentiation. Nat. Rev. Immunol. 19: 7–18. [DOI] [PubMed] [Google Scholar]
- 7. Mathis, D., Benoist C.. 2009. Aire. Annu. Rev. Immunol. 27: 287–312. [DOI] [PubMed] [Google Scholar]
- 8. Yu, S., Cantorna M. T.. 2008. The vitamin D receptor is required for iNKT cell development. Proc. Natl. Acad. Sci. USA 105: 5207–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sood, A., Dong M., Melichar H. J.. 2016. Preparation and applications of organotypic thymic slice cultures. J. Vis. Exp. (114): 54355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Memari, B., Bouttier M., Dimitrov V., Ouellette M., Behr M. A., Fritz J. H., White J. H.. 2015. Engagement of the aryl hydrocarbon receptor in mycobacterium tuberculosis-infected macrophages has pleiotropic effects on innate immune signaling. J. Immunol. 195: 4479–4491. [DOI] [PubMed] [Google Scholar]
- 11. Arora, J., Wang J., Weaver V., Zhang Y., Cantorna M. T.. 2022. Novel insight into the role of the vitamin D receptor in the development and function of the immune system. J. Steroid Biochem. Mol. Biol. 219: 106084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heng, T. S., Painter M. W.; Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9: 1091–1094. [DOI] [PubMed] [Google Scholar]
- 13. Jones, R. C., Karkanias J., Krasnow M. A., Pisco A. O., Quake S. R., Salzman J., Yosef N., Bulthaup B., Brown P., Harper W., et al. Tabula Sapiens Consortium . 2022. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376: eabl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bautista, J. L., Cramer N. T., Miller C. N., Chavez J., Berrios D. I., Byrnes L. E., Germino J., Ntranos V., Sneddon J. B., Burt T. D., et al. 2021. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun. 12: 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. White, J. H., Salehi-Tabar R., Dimitrov V., Bouttier M.. 2018. Diverse mechanisms of transcriptional regulation by the vitamin D receptor. In Vitamin D, 4th Ed. Feldman D., ed. Academic Press, Cambridge, MA, p. 175–187. [Google Scholar]
- 16. Nagamine, K., Peterson P., Scott H. S., Kudoh J., Minoshima S., Heino M., Krohn K. J., Lalioti M. D., Mullis P. E., Antonarakis S. E., et al. 1997. Positional cloning of the APECED gene. Nat. Genet. 17: 393–398. [DOI] [PubMed] [Google Scholar]
- 17. Savkur, R. S., Burris T. P.. 2004. The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 63: 207–212. [DOI] [PubMed] [Google Scholar]
- 18. Lee, S. M., Riley E. M., Meyer M. B., Benkusky N. A., Plum L. A., DeLuca H. F., Pike J. W.. 2015. 1,25-Dihydroxyvitamin D3 controls a cohort of vitamin D receptor target genes in the proximal intestine that is enriched for calcium-regulating components. J. Biol. Chem. 290: 18199–18215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dankers, W., Colin E. M., van Hamburg J. P., Lubberts E.. 2017. Vitamin D in autoimmunity: molecular mechanisms and therapeutic potential. Front. Immunol. 7: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilhelmson, A. S., Lantero Rodriguez M., Johansson I., Svedlund Eriksson E., Stubelius A., Lindgren S., Fagman J. B., Fink P. J., Carlsten H., Ekwall O., Tivesten Å.. 2020. Androgen receptors in epithelial cells regulate thymopoiesis and recent thymic emigrants in male mice. Front. Immunol. 11: 1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu, M. L., Bakhru P., Conley B., Nelson J. S., Free M., Martin A., Starmer J., Wilson E. M., Su M. A.. 2016. Sex bias in CNS autoimmune disease mediated by androgen control of autoimmune regulator. Nat. Commun. 7: 11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.