Abstract
Sleep and circadian rhythms are observed broadly throughout animal phyla and influence neural plasticity and cognitive function. However, the few phylogenetically conserved cellular and molecular pathways that are implicated in these processes are largely focused on neuronal cells. Research on these topics has traditionally segregated sleep homeostatic behavior from circadian rest-activity rhythms. Here we posit an alternative perspective, whereby mechanisms underlying the integration of sleep and circadian rhythms that affect behavioral state, plasticity, and cognition reside within glial cells. The brain-type fatty acid binding protein, FABP7, is part of a larger family of lipid chaperone proteins that regulate the subcellular trafficking of fatty acids for a wide range of cellular functions, including gene expression, growth, survival, inflammation, and metabolism. FABP7 is enriched in glial cells of the central nervous system and has been shown to be a clock-controlled gene implicated in sleep/wake regulation and cognitive processing. FABP7 is known to affect gene transcription, cellular outgrowth, and its subcellular localization in the fine perisynaptic astrocytic processes (PAPs) varies based on time-of-day. Future studies determining the effects of FABP7 on behavioral state- and circadian-dependent plasticity and cognitive processes, in addition to functional consequences on cellular and molecular mechanisms related to neural-glial interactions, lipid storage, and blood brain barrier integrity will be important for our knowledge of basic sleep function. Given the comorbidity of sleep disturbance with neurological disorders, these studies will also be important for our understanding of the etiology and pathophysiology of how these diseases affect or are affected by sleep.
Keywords: BBB, synaptic plasticity, homeostasis, glycolysis, transcytosis, endocytosis, astroctye, β-oxidation
Fatty-acid binding proteins
Fatty-acid binding proteins are a family of small ∼15 kDa lipid-binding proteins that belong to the calycin superfamily, which include avidins and lipocalins. FABPs share β-barrel structural motifs that bind small hydrophobic molecules (Schaap et al., 2002), despite low primary sequence similarity (Agellon, 2023). FABPs bind the hydrophobic region of fatty acids and their metabolites, particularly long-chain polyunsaturated fatty acids (PUFAs) in higher order species, and transport them to various subcellular locations, which affect a wide range of cellular processes, including signal transduction, oxidation, membrane synthesis, transcription, fat storage, autocrine/paracrine function, inflammation, and metabolism (Furuhashi and Hotamisligil, 2008; Storch and Corsico, 2008). FABPs also bind xenobiotics, including cannabinoids, benzodiazepines, antinociceptives, non-steroidal anti-inflammatory drugs, and peroxisome proliferators, and are involved in xenobiotic absorption, distribution, and metabolism in various organs (Yabut and Isoherranen, 2023). FABPs are present across phylogeny, from invertebrates such as Caenorhabditis elegans and the fruit fly, Drosophila melanogaster, to rodents and other mammals, including humans (Zheng et al., 2013; Zhang et al., 2020). Phylogenetic studies suggest FABPs likely evolved from a common ancestor via tandem gene duplication, with the first gene duplication dated ∼930 million years ago (Schaap et al., 2002; Zheng et al., 2013; Zhang et al., 2020). Recruitment of FABPs during the evolution of animals from fungi and plants is thought to facilitate increased subcellular trafficking of ligands and mitochondrial oxidation of long-chain fatty acids (Schaap et al., 2002). FABPs were initially discovered in the cytosol of intestinal mucosa, liver, and myocardial tissues (Ockner et al., 1972). FABPs can be differentially expressed in various tissues and cell types (Furuhashi and Hotamisligil, 2008). For example, in mammals, heart-type FABP (H-FABP/FABP3), epidermal-type FABP (E-FABP/FABP5), and brain-type FABP (B-FABP/FABP7) are all present within the adult central nervous system (CNS) (Veerkamp and Zimmerman, 2001), with FABP3 primarily expressed in neurons, FABP5 in neurons and glia, and FABP7 in astrocytes and precursor cells (Owada et al., 1996b; Furuhashi and Hotamisligil, 2008; Storch and Corsico, 2008).
Fatty acid binding protein 7 in brain development and proliferation
Fatty acid binding protein 7, also known as mammary derived growth inhibitor-related gene (MRG) and brain lipid binding protein (BLBP), is an ontogenically expressed FABP with elevated expression early in development that decreases over the lifespan in mammals (Bennett et al., 1994; Gerstner et al., 2008; Clarke et al., 2018). FABP7 was first identified in radial glial cells of embryonic brain and neural progenitors of mature brain (Bennett et al., 1994; Feng et al., 1994; Kurtz et al., 1994). FABP7-expressing progenitors in early development are thought to contribute to most adult neural cell populations throughout the mammalian CNS (Anthony et al., 2004). FABP7 was identified as the first predominantly specific Notch target gene in the CNS (Anthony et al., 2005), its developmental expression is dependent on Pax6 (Arai et al., 2005) and POU/Pbx (Josephson et al., 1998) transcription factors. Following development, FABP7 expression appears to be pluripotent as it is found in multiple cells in nervous tissue, including astrocytes, radial glia, oligodendrocyte progenitor cells (OPCs), Bergman glia, Müller glia, and satellite glia of the spinal cord (Kurtz et al., 1994; Owada et al., 1996b; Yanase et al., 2002). In postnatal hippocampal neurogenesis, FABP7 is expressed in neural stem cells (NSCs) of the dentate gyrus, and proliferation of these NSCs is decreased with subsequent reduction in their survival in FABP7 knockout (KO) mice (Matsumata et al., 2012). FABP7 expression was also detected in NG2 (+) OPCs, and cultured OPCs showed a significant decrease in proliferation/differentiation in the population of FABP7- KO OPCs compared with wild-type (WT) OPCs (Sharifi et al., 2013). Following forebrain ischemia, FABP7 expression in neural stem/progenitor cells increased 7–10 days post-ischemia, consistent with peak hippocampal neurogenesis (Kato et al., 2020). FABP7 expression associated with hippocampal neurogenesis following ischemic insult was also observed in non-human primates (Ma et al., 2010; Boneva et al., 2011). In FABP7-KO mice, neurogenesis was significantly decreased compared to WT mice under both normal and ischemic conditions, suggesting that FABP7 regulates the proliferation of neuronal stem/progenitor cells. Together these findings provide compelling evidence that FABP7 is a key regulator in the growth and organization of multiple CNS cell types.
Integrated model for FABP7 in sleep, circadian rhythms, plasticity, and metabolic function
Sleep is a characteristic behavior that is exhibited broadly throughout the animal kingdom, including invertebrate and vertebrate species (Allada and Siegel, 2008; Lesku et al., 2008; Cirelli, 2009). Despite this, we know relatively little of what cellular and molecular mechanisms are fundamental to sleep drive. A long-standing hypothesis in the sleep field maintains two-processes that contribute to sleep behavior: a circadian (time-of-day) component, which is driven by phylogenetically conserved core-clock system, and a “sleep homeostasis” component, which is driven by prior time spent awake (Borbely and Achermann, 1999; Borbely et al., 2016). In one perspective, sleep homeostasis is generated via reciprocal switching between wake- and sleep-promoting neurons to inhibit each other (Strecker et al., 2000; Saper et al., 2001, 2005; Szymusiak et al., 2007; Eban-Rothschild et al., 2018), but this model is challenging for species that lack similar anatomical circuits or neurochemistries (Artiushin and Sehgal, 2017; Donlea et al., 2017; Ly et al., 2018). In another view, sleep drive is thought to occur in an independent fashion within neurons throughout brain, based on their prior history of excitation and use (Tononi and Cirelli, 2006; Krueger et al., 2008; Krueger and Tononi, 2011; Havekes and Aton, 2020; Krueger, 2020). Alterations in neuronal activity in both perspectives represent the fundamental driving force behind sleep homeostasis. Recently glial cells have received more attention for their relevance in sleep and circadian rhythm behaviors (Halassa et al., 2009; Halassa and Haydon, 2010; Barca-Mayo and López, 2021; Damulewicz et al., 2022a; Ingiosi and Frank, 2022; Hastings et al., 2023) and their contributions are evidenced across phylogenetically disparate species (Poskanzer and Yuste, 2016; Stahl et al., 2018; Artiushin and Sehgal, 2020; Ingiosi et al., 2020; Jackson et al., 2020; Blum et al., 2021; Vaidyanathan et al., 2021; Reitman et al., 2023). Exactly how circadian and homeostatic processes interact to organize sleep behavior remains unclear and are likely not operating independently (Deboer et al., 2003, 2007; Easton et al., 2004; Laposky et al., 2005; Wright et al., 2012; Deboer, 2018). Further, the likely involvement of other processes such as energy metabolism (Benington and Heller, 1995; Scharf et al., 2008; Franken and Dijk, 2009; Dash et al., 2013; Bellesi et al., 2018; Malik et al., 2020), lipid signaling and storage (Thimgan et al., 2010, 2015; Yurgel et al., 2018; Ioannou et al., 2019a; Li Y. et al., 2023), astrocyte-neurometabolic coupling through glymphatics (Jessen et al., 2015; Haydon, 2017; Lundgaard et al., 2017), autophagy (Xie et al., 2020; Bedont et al., 2021; Damulewicz et al., 2022b; Guo et al., 2022), and the astrocyte-neuron lactate shuttle (ANLS) (Scharf et al., 2008; Petit et al., 2013) reflect a complex and multivariable network ripe for exploration.
Fatty acid binding protein 7 mRNA expression is enriched in dendritic layers of hippocampus (Zhong et al., 2006) and induced following kainate injection known to increase neural activity (Owada et al., 1996a). In addition cyclic AMP response element binding protein, CREB, a transcription factor widely associated with synaptic plasticity, memory, sleep, and circadian rhythms (Nguyen and Woo, 2003; Gerstner and Yin, 2010; Havekes et al., 2015, 2016; Kreutzmann et al., 2015; Xia and Storm, 2017; Lisman et al., 2018), elicits a persistent form of hippocampal long-term potentiation with only a weak stimulus when made constitutively active (Barco et al., 2002). Constitutive CREB-induced hippocampal FABP7 mRNA expression mirrors the temporal profile of CREB-induced BDNF mRNA expression in hippocampus (Barco et al., 2005), suggesting common pathways may exist in neural plasticity-related processes coupled to astrocyte function (Stellwagen and Malenka, 2006; Ota et al., 2013; Perez-Catalan et al., 2021; Lawal et al., 2022). FABP7 is enriched in astrocytes and is involved in lipid signaling cascades that regulate changes in cell growth, morphology, and motility (Feng et al., 1994; Arai et al., 2005; Mita et al., 2007, 2010), and regulates dendritic morphology and neuronal excitatory synapse formation, and synaptic transmission (Ebrahimi et al., 2016). Neuronal activity is known to initiate lipid peroxidation, lipoprotein export, and peroxidized lipid storage of lipid droplets (LDs) in astrocytes (Ioannou et al., 2019b). LDs are lipid storage organelles consisting of a layer of polarized lipids with a neutral lipid core mostly composed of triglycerides and esterified cholesterol (Welte, 2015; Olzmann and Carvalho, 2019). Following stress, astrocytes accumulate LDs, which protects cells from lipotoxicity, reactive oxygen species (ROS)-mediated lipid peroxidation, and can be used as fuel in mitochondrial β-oxidation (Smoliè et al., 2021). The ANLS has been suggested to play a role in promoting ROS waste removal tied to LD formation in glia via apolipoproteins (Liu et al., 2017). FABP7 protects astrocytes from ROS toxicity through increased LD formation (Islam et al., 2019). Following hypoxia, FABP7 induction by HIF-1α also led to LD accumulation via fatty-acid uptake to protect against ROS and support cellular survival (Bensaad et al., 2014). Interestingly, knock-down of FABP7 increased ROS and upregulated uncoupling protein 1 (UCP1), which depolarized mitochondrial membranes, increased proton leakage, and glycolysis (Kawashima et al., 2020). Therefore, mechanisms underlying use-dependent neural-glial interactions together with lipid storage and metabolic function may provide a key mediator for coupling sleep homeostasis with circadian rhythms (Figure 1).
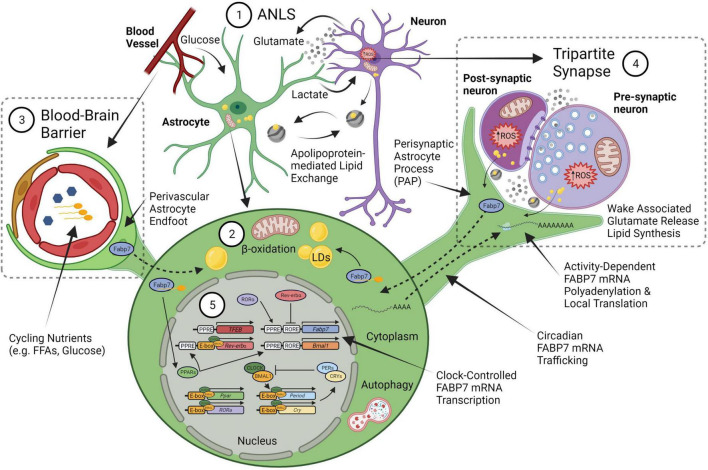
FIGURE 1.
An integrated neural-glial metabolic clock-sleep model. (1) The astrocyte-neuron lactate shuttle (ANLS) hypothesis proposes that activity-dependent glutamate release at synapses triggers astrocyte glucose uptake from blood, which is then converted to lactate and sent to neurons to support the increased metabolic demand. Sleep pressure (homeostasis) would be linked to the increased energy produced by this lactate in neuronal mitochondria from wake-associated glutamate release, which generates reactive oxygen species (ROS) production and subsequent lipid formation that are transferred back to glia via apolipoproteins (i.e., ApoE). (2) These lipids will bind fatty acid transport proteins, including FABP7, to form lipid droplets (LDs) in astrocytes. The lipid stores can be used as fuel in astrocyte mitochondria via β-oxidation to produce ketone bodies. (3) Astrocyte endfeet surround the brain vasculature as one of the cellular components of the blood brain barrier (BBB). Circulating nutrients and metabolic constituents such as free fatty acids (FFA) and glucose are taken up by astrocytes and used as energy for the brain. (4) This wake-associated glutamate release would also be tied to local translation of FABP7 mRNA in the fine perisynaptic astrocytic process (PAP) of the tripartite synapse, which consists of an astrocyte ensheathment along with pre- and post-synaptic neuronal compartments, to couple on-site FABP7 protein demand with newly synthesized lipids derived from local excitatory synaptic activity in neurons. (5) The timing of FABP7 mRNA expression is regulated by the circadian clock via Rev-erbα, a transcriptional repressor that binds to RORE cis-elements in the promoters of FABP7 gene and the core-clock transcriptional activator BMAL1. Following transcription, FABP7 mRNA is trafficked to PAPs where it is locally translated upon behavioral state-dependent changes in neural activity. Therefore, clock-controlled expression of FABP7 relays the circadian timing of sleep with changes in sleep pressure through mechanisms underlying local translation at PAPs. FABP7 may in turn feedback on transcription of the core-clock via nuclear localization and activation of peroxisome proliferator-activated receptor (PPARs), for example through FFAs, such as omega-3 polyunsaturated fatty-acids known to oscillate in the peripheral vasculature, or those from PAPs. PPAR—mediated transcription of Transcription Factor EB (TFEB) may in turn initiate autophagy (Soto-Avellaneda and Morrison, 2020), or alternatively, autophagy could be stimulated via other BMAL1-dependent mechanisms in astrocytes (McKee et al., 2023). Circadian FABP7 may also regulate BBB permeability over the course of the day to influence the transmission of peripheral signals and metabolic constituents to (or from) the brain. Created with BioRender.com.
Here we propose FABP7 as a glial-derived molecule which integrates sleep and circadian rhythms, activity-dependent neural plasticity with lipid signaling and metabolism. FABP7 mRNA expression cycles in synchrony throughout the brain over a 24-h rhythm, including in sleep, wake, and circadian controlling centers (Panda et al., 2002; Ueda et al., 2002; Gerstner et al., 2006, 2008, 2011a). In mammals, the circadian core clock transcriptional translational feedback loop consists of the transcription factors, circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like protein (BMAL1), which heterodimerize in the nucleus to promote the expression of numerous cis-acting E-box promoter element containing genes, including period (PER) and cryptochrome (CRY) genes (Bass and Takahashi, 2010; Mohawk et al., 2012). FABP7 mRNA circadian oscillation is disrupted in arrhythmic BMAL1 KO mice compared to WT mice, and its baseline level of expression was elevated with no effect on either FABP3 or FABP5 transcripts (Lananna et al., 2018; Gerstner and Paschos, 2020). However, E-box elements were not detected bioinformatically in the murine FABP7 promoter, while multiple Rev-erbα (NR1D1) binding sites (called Rev-erbα response element, RORE), a nuclear receptor/transcriptional repressor and component of the metabolic arm of the clock (Bugge et al., 2012; Cho et al., 2012; Zhang et al., 2015), were identified (Vanderheyden et al., 2021). The promoter of the FABP7 gene is a direct target of Rev-erbα (Schnell et al., 2014; Vanderheyden et al., 2021), and regulates FABP7 transcription across multiple brain areas, with baseline FABP7 mRNA expression elevated ∼6–10 fold compared in Rev-erbα mutants over WT, similar to what was observed in the BMAL1 KO (Gerstner and Paschos, 2020). This suggests that the alterations in FABP7 mRNA may be indirectly regulated by BMAL1 via changes in the expression of Rev-erbα (Figure 1).
Murine FABP7 brain transcript levels are maximal after the normal waking phase and begin to decline at circadian times of day that correspond with the normal discharge of sleep pressure (Gerstner et al., 2006, 2008; Schnell et al., 2014; Gerstner and Paschos, 2020; Vanderheyden et al., 2021). Previous work has demonstrated that sleep disruption reduced FABP7 mRNA levels in brain tissue of multiple species, including birds and mammals (Cirelli et al., 2006; Jones et al., 2008; Guindalini et al., 2009; Hor et al., 2019). We have shown that FABP7 in turn regulates sleep in flies, mice, and humans (Gerstner et al., 2011b,2017; Vanderheyden et al., 2022). Transgenic flies that overexpress the mouse FABP7 or Drosophila FABP7 homologue, dFABP, increase sleep compared to non-transgenic control flies (Gerstner et al., 2011b). Drosophila glia dFABP is also associated with LD formation (Kis et al., 2015), and dFABP overexpression enhanced memory in flies (Gerstner et al., 2011a,b). FABP7 KO mice show fragmented sleep compared to WT mice, similar to what is observed in human carriers of the FABP7 T61M mutation compared to non-carriers (Gerstner et al., 2017). Interestingly, flies that overexpress the human FABP7 T61M mutation compared to non-mutant human FABP7 specifically in astrocytes also show fragmented sleep (Gerstner et al., 2017). A more recent study showed that flies that overexpress dFABP in glia have normal circadian rhythmicity, while RNAi against dFABP incurred more arrhythmic flies, compared to controls (Jang et al., 2022). Together these studies suggest that glial FABP7 is a well-conserved integrated modulator of sleep and circadian behavior.
The cellular and molecular mechanisms that integrate the circadian timing of sleep/wake cycles with sleep homeostasis may be linked to the patency of neuronal-glial interactions, which may occur via Perisynaptic Astrocytic Processes (PAPs) (Halassa et al., 2009; Frank, 2013). Astrocytes can extend these fine, peripheral, filamentous structures around the pre- and post-synaptic areas, collectively called the tripartite synapse (Araque et al., 1999; Perea et al., 2009; Santello et al., 2012; Figure 1). PAPs have been shown to influence synaptic activity by several mechanisms, including neurotransmitter uptake, metabolism, and the release of gliotransmitters (Reichenbach et al., 2010; Frank, 2013). Astrocytes can extend or retract (Bernardinelli et al., 2014a) to morphologically adjust neuron-astrocyte interactions at synapses with concomitant localization of the actin-binding protein, ezrin (Derouiche and Frotscher, 2001; Derouiche et al., 2002), which is activated by phosphorylation (Tsukita and Yonemura, 1997). We previously characterized sleep loss-induced reductions in neural-glial interactions in flies (Vanderheyden et al., 2019), however, to our knowledge PAPs have not been characterized in the adult Drosophila melanogaster brain. PAPs have been shown to expand with increases in synaptic activity and with heightened glutamatergic tone (Theodosis et al., 2008; Perez-Alvarez et al., 2014), and is correlated with wake behavior (Naylor et al., 2012). Ultrastructural studies report astrocytic interfaces increase near synapses and are associated with wakefulness in rodents (Bellesi et al., 2015). PAPs are known to change dynamically with circadian rhythm (Lavialle et al., 2011) and with activity-dependent synaptic plasticity (Genoud et al., 2006; Bernardinelli et al., 2014b). We also documented the circadian rhythm of mammalian FABP7 mRNA trafficking to PAPs, which coincides with cycling FABP7 PAP protein levels, and is maximal during the wake phase of the day and reduced in the sleep phase (Gerstner et al., 2012). It follows that this process in astrocytes is modulated by cytoplasmic polyadenylation element binding proteins (CPEBs) (Gerstner et al., 2012), which are known to regulate subcellular trafficking, localization, and translation of neuronal synaptic plasticity-related transcripts such as αCaMKII (Wu et al., 1998; Huang et al., 2002, 2003). Since FABP7 is regulated by the circadian clock, affects sleep behavior, and its PAP-enrichment oscillates over the light-dark cycle, it is a strong candidate molecule for the integration of the circadian timing of sleep with sleep-need via changes in neuronal-glial interactions. We propose that changes in neuronal-glial interactions and PAPs integrate circadian processes with sleep/wake behavior via FABP7 (Figure 1).
Fatty acid binding protein 7 in injury and disease
Fatty acid binding protein 7 has been shown to be involved in reactive gliosis of the CNS. Cortical FABP7-positive (+) astrocytes increased in response to a stab injury in WT mice, and the number of reactive astrocytes was decreased in FABP7-KO mice (Sharifi et al., 2011). In normal, uninjured cortex, FABP7 was localized to glial fibrillary acidic protein (GFAP) + astrocytes (21% of FABP7 + cells) and Neural/glial antigen-2 (NG2) + oligodendrocyte progenitor cells (62%). However, in injured cortex there was a significant increase in FABP7 + /GFAP + cells but no change was detected in FABP7 + /NG2 + cells (Sharifi et al., 2011). In the stab-injured cortex of FABP7-KO mice there was also a decrease in the number of the proliferation marker bromodeoxyuridine/5-bromo-2′-deoxyuridine (BrdU) + astrocytes compared with WT mice, further implicating FABP7 in repair (Sharifi et al., 2011). Using a scratch-injury model in primary cultured astrocytes, increased FABP7 was observed at the peri-injury borders compared to intact astrocytes. Moreover, FABP7-KO astrocytes showed a slower proliferation compared with WT astrocytes by BrdU + immunochemistry (Hara et al., 2020). FABP7-assisted CNS repair extends beyond the parenchyma. In a mouse spinal cord compression model, FABP7 was primarily upregulated in proliferative astrocytes compared to non-injured control mice (Senbokuya et al., 2019). In this model, FABP7-KO mice had significantly lower surviving ventral neurons 28 days post-injury compared to WT mice, suggesting that astrocytic FABP7 has a neuroprotective role (Senbokuya et al., 2019). This is recapitulated with several reports of elevated FABP7 expression following traumatic brain injury (TBI) (Halford et al., 2017; Rui et al., 2019; Mao et al., 2020). FABP7 expression was also associated with reactive astroglial hypertrophy in spinal cord autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis (MS) (Bannerman et al., 2007) and in astrocytes of lesions in early stage MS patients (Kipp et al., 2011). In demyelinating regions of EAE mice increased astrocytic FABP7 expression relative to non-EAE mice was observed and compared to WT mice, FABP7-KO mice manifest with early onset of EAE symptoms (Kamizato et al., 2019). The clinical score, however, was significantly reduced in the late phase of EAE, indicating a differential role for FABP7 in early versus late stages of MS. Together, these data demonstrate astrocytic FABP7 expression is integrally connected with reactive gliosis and brain injury.
Many diseases, nervous system dysfunction, and neurological disorders are associated with alterations in FABP7 expression. FABP7 has been implicated multiple cancers, Down syndrome, schizophrenia, and various neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS), Parkinson’s disease (PD), and Alzheimer’s disease (AD) (Cheon et al., 2003; Sánchez-Font et al., 2003; Watanabe et al., 2007; Teunissen et al., 2011; Guttula et al., 2012; Matsumata et al., 2016; Kagawa et al., 2019; Killoy et al., 2020; Young, 2020; Asaro et al., 2021; Koga et al., 2021; Cheng et al., 2022; Needham et al., 2022; Tandon et al., 2023). In an unbiased proteomics screen, hippocampal FABP7 was elevated in a mouse model of Alexander disease (AxD), and in AxD patient brain tissue (Heaven et al., 2022). In the adult rat brain, FABP7 in gomori-positive astrocytes is enriched at cytoplasmic granules that originate from damaged mitochondria (Young et al., 1996). Moreover, aging-related mitochondrial pathology occurs in FABP7 + astrocytes, which can hinder cell function, is speculated to be linked to AD etiology (Young, 2020). FABP7 levels are elevated in serum of 29, 35, and 24% of the patients with AD, PD, and other cognitive disorders, respectively, and in 2% of the healthy donors (Teunissen et al., 2011). FABP7 levels in serum of psoriatic patients is elevated compared to controls without dermatoses and is currently being considered as a putative index of neurodegenerative processes linked to psoriasis (Nowowiejska et al., 2022a,b). A quantitative trait loci analysis for low pre-pulse inhibition (PPI), a phenotypic marker of schizophrenia, revealed a strong association with the FABP7 locus in mice, and FABP7 is significantly upregulated in human postmortem brains of schizophrenics compared to controls (Watanabe et al., 2007). FABP7-KO mice exhibit altered anxiety-like behavior (Owada et al., 2006; Vanderheyden et al., 2022) and deficits in PPI (Watanabe et al., 2007). Plasma FABP7 concentrations also correlated with Positive and Negative Syndrome Scale clinical scores, particularly in severities of depression/anxiety, cognition, and positive symptoms of schizophrenia patients (Koga et al., 2021). Together, these studies implicate FABP7 in a broad array of illnesses and disorders, underscoring the importance of its role in pathophysiological conditions associated with disease. Given that sleep and circadian disturbances are comorbid with many diseases and disorders (Wulff et al., 2010; Musiek et al., 2013; Depner et al., 2014; Musiek and Holtzman, 2016; Abbott et al., 2020; Colwell, 2021; Grandner, 2022; Harvey, 2022; Nassan and Videnovic, 2022), and coupled with a growing literature touting chronotherapy to optimize treatment for diseases (Fu and Kettner, 2013; Cederroth et al., 2019; Sulli et al., 2019; Lee et al., 2021; Van Drunen and Eckel-Mahan, 2022), the relevance of FABP7 expression in the context of sleep/wake and circadian regulation is poised for future translational studies and has clinical utility in the development of therapeutic strategies to treat a wide range of CNS-related disorders.
Conclusion and future directions
Incorporation of other inputs, such as metabolism and psycho-social behavior, into the two-process model of sleep/wake regulation has clear conceptual improvements for sleep and circadian research (Borbely et al., 2016). While most studies remain focused on neuronal function, alternative work has examined the role of glia, other cells, tissues, peripheral systems, and microbiota in sleep behavior (Anafi et al., 2013; Frank, 2013; Arnardottir et al., 2014; Ehlen et al., 2017; Matenchuk et al., 2020; Patke et al., 2020; Cable et al., 2021; Withrow et al., 2021). The proposition that FABP7 represents a molecular “node” that integrates circadian and sleep behavior with changes in neuronal-glial interactions is inherently colligated in metabolic processes.
The pleiotropic nature of FABP7 cellular signaling offers a plethora of empirical opportunities for determining functional roles of sleep and circadian rhythm biology. FABP7 has especially high binding affinity to long-chain polyunsaturated fatty-acids, especially the omega-3 docosahexaenoic acid (DHA) (Balendiran et al., 2000). DHA in blood oscillates over the day despite changes in homeostatic sleep pressure in humans (Dallmann et al., 2012). DHA is the most abundant omega-3 in the brain, makes up 10-20% of total lipids, and is implicated in many diseases, including cancer, neurodegenerative diseases, and various neurological and psychiatric disorders (Weiser et al., 2016; Sun et al., 2018; Montecillo-Aguado et al., 2020; von Schacky, 2021). Upon binding DHA a conformational shift signals nuclear localization in FABP7 to mediate peroxisome proliferator-activated receptor-gamma (PPARγ)-dependent transcription (Tripathi et al., 2017). Circadian oscillations in levels of circulating glucose and lipids are diametrically opposed, and are in opposite phases, between nocturnal and diurnal species (Kumar Jha et al., 2015; Figure 2). Oscillations in these metabolic nutrients are likely closely tied to transport systems at the blood brain barrier (BBB) linked to differential bioenergetics (e.g., lipid oxidation and glycolysis), recycling and waste clearance mechanisms (e.g., autophagy and glymphatics) that incorporate behavioral state with circadian rhythms. Taken together, FABP7 may integrate peripheral lipid circadian oscillations in the brain vasculature at the BBB with molecular transcriptional processes within the clock feedback loop balanced against energetic demands from wake-dependent synaptic activity and energy supply (Figures 1, 2).
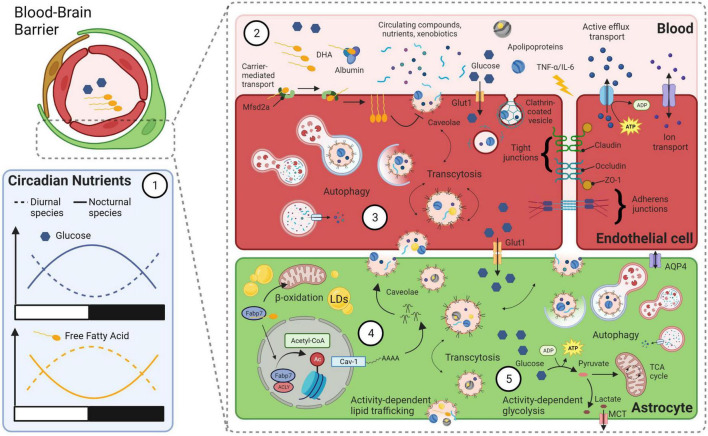
FIGURE 2.
A model linking bioenergetics with integrated clock-state-dependent fluctuations in metabolism and BBB permeability. (1) Diurnal and nocturnal species have diametrically opposed rhythms in circulating levels of glucose and free fatty acids (FFAs). Within species, levels of glucose and FFAs oscillate over the day in opposite phases. (2) Various transport mechanisms across the BBB can influence permeability and are closely tied to circadian rhythms and behavioral state. For example, circadian rhythms of circulating omega-3 fatty-acid, DHA, may bind to endothelial transport protein major facilitator superfamily domain containing 2a (Mfsd2a), which flips DHA from the outer-membrane to inner-membrane surface. This is known to block caveolae formation and transcytosis at the BBB (Andreone et al., 2017). Other lipid structures, such as apolipoproteins, may bind to receptors for endocytosis. Circadian changes in circulating glucose are trafficked across the BBB by transporters (e.g., Glut1). Circulating cytokines such as TNF-α and IL-6, whose levels increase with sleep loss, can disrupt tight junction proteins and increase BBB permeability. Clock-state dependent changes in efflux and ion transport can also occur at the BBB. (3) Autophagy of endocytosed materials and cellular damage following BBB disruption due to extended wakefulness may protect endothelial cells and increase their survival. (4) Astrocyte caveolae formation is driven by FABP7-mediated acetylation of histones via an ATP-citrate lyase (ACLY) interaction that increases nuclear Acetyl-CoA levels and activates the promoter of the caveolin-1 gene. Caveolae transcytosis in astrocytes may therefore be integrated between circadian oscillation in circulating DHA levels in the vasculature and the clock-controlled expression of FABP7. Astrocyte uptake of cycling lipids and DHA may in turn influence the formation of lipid droplets (LDs) and subsequent β-oxidation in mitochondria. (5) A balance between activity-dependent energy demands and circulating nutrient availability integrates the circadian clock with behavioral state in astrocytes. For example, local use-dependent neural activity will increase the ANLS, to convert glucose into lactate that is delivered to neurons from astrocytes via MCTs. This wake-dependent neural activity is coupled to increased lipid synthesis in neurons, that is taken up by astrocytes and stored in LDs for later use as fuel (see Figure 1). Metabolic byproducts of this bioenergetics pathway are cleared via autophagy, exocytosis and/or by the glymphatic system, via water channels such as Aquaporin 4 (AQP4). Created with BioRender.com.
The BBB is a dynamic structure composed of many cell types, including endothelial cells, astrocytes, pericytes, neurons, and microglia that form the neurovascular unit, which has been implicated in many neurological disorders, as well as sleep and circadian processes (He et al., 2014; Pan and Kastin, 2017; Artiushin et al., 2018; Hurtado-Alvarado et al., 2018; Cuddapah et al., 2019; Zhang et al., 2021; Li F. et al., 2023; Schurhoff and Toborek, 2023). FABP7 has recently been shown to regulate opioid-mediated disruption of BBB integrity, which permits infiltration of fragile-like regulatory T cells into the nucleus accumbens, a process that leads to synaptic instability and withdrawal symptoms (Zhu et al., 2023). Acute cocaine administration produces a transient increase in BBB permeability (Barr et al., 2020), and FABP7 has been implicated in cocaine-seeking behavior under stressful conditions, where WT mice showed stress-induced conditioned place preference for cocaine, FABP 5/7 double KO mice did not (Hamilton et al., 2018). A reduction in FABP7 protein and transcript levels in the nucleus accumbens was also observed in a juvenile mouse model for stress-induced cocaine seeking behavior (Lo Iacono et al., 2016). Compared to acute social stress, chronic social stress had lower levels of FABP7 mRNA in hippocampus (Stankiewicz et al., 2015). Chronic mild stress reduces brain glucose metabolism in many brain regions, including hippocampus, of WT mice but not in FABP7 KO mice (Hamilton et al., 2022). Increases in glucose transporter-1 were observed in the BBB in frontal cortex and hippocampus of rats exposed to restraint stress (Sántha et al., 2015), and chronically stressed mice show increased BBB permeability (Lee et al., 2018). Following single-prolonged stress, a rodent model for post-traumatic stress disorder, we observed disrupted unconditioned anxiety in FABP7 KO mice compared to WT mice, which was also associated with abnormal stress-dependent sleep suppression. Following TBI, FABP7 also protects BBB integrity through a caveolin-1 signaling pathway (Rui et al., 2019) and nuclear FABP7 is known to interact with ATP-citrate lyase (ACLY) to drive acetyl-coA-mediated histone regulation of caveolin-1 gene expression (Kagawa et al., 2020; Figure 2). Given BBB circadian disruptions occur following stress responses and in several neurological disorders, including brain metastasis, epilepsy, AD, and PD (Schurhoff and Toborek, 2023), future studies determining the relationship between FABP7 signaling, brain injury, BBB permeability, stress, and sleep/circadian rhythms will be important for the treatment of neurological disorders and diseases. The integrated glial metabolic clock-sleep model provides a conceptual framework to both appreciate and investigate these collective biologies and systems. Recently it was shown that β-oxidation in glial mitochondria provide ketone bodies to fuel neurons in the absence of glycolysis in Drosophila, supporting our model (McMullen et al., 2023). Further studies determining the phylogenetically conserved mechanisms within the model will be important for our understanding of the fundamental properties of sleep.
Data availability statement
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
JG wrote the manuscript with input from co-authors. All authors approved the final version of the manuscript.
Funding Statement
This work was supported by NIH grant R35GM133440.
Conflict of interest
JG was founder of Blood Brain Biotechnology, LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abbott S. M., Malkani R. G., Zee P. C. (2020). Circadian disruption and human health: A bidirectional relationship. Eur. J. Neurosci. 51 567–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agellon L. B. (2023). Importance of fatty acid binding proteins in cellular function and organismal metabolism. J. Cell Mol. Med. 1–11. 10.1111/jcmm.17703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R., Siegel J. M. (2008). Unearthing the phylogenetic roots of sleep. Curr. Biol. 18 R670–R679. 10.1016/j.cub.2008.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anafi R. C., Pellegrino R., Shockley K. R., Romer M., Tufik S., Pack A. I. (2013). Sleep is not just for the brain: Transcriptional responses to sleep in peripheral tissues. BMC Genomics 14:362. 10.1186/1471-2164-14-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreone B. J., Chow B. W., Tata A., Lacoste B., Ben-Zvi A., Bullock K., et al. (2017). Blood-brain barrier permeability is regulated by lipid transport-dependent suppression of caveolae-mediated transcytosis. Neuron 94 581–594.e5. 10.1016/j.neuron.2017.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony T. E., Klein C., Fishell G., Heintz N. (2004). Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron 41 881–890. 10.1016/s0896-6273(04)00140-0 [DOI] [PubMed] [Google Scholar]
- Anthony T. E., Mason H. A., Gridley T., Fishell G., Heintz N. (2005). Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 19 1028–1033. 10.1101/gad.1302105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y., Funatsu N., Numayama-Tsuruta K., Nomura T., Nakamura S., Osumi N. (2005). Role of Fabp7, a downstream gene of Pax6, in the maintenance of neuroepithelial cells during early embryonic development of the rat cortex. J. Neurosci. 25 9752–9761. 10.1523/jneurosci.2512-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araque A., Parpura V., Sanzgiri R. P., Haydon P. G. (1999). Tripartite synapses: Glia, the unacknowledged partner. Trends Neurosci. 22 208–215. 10.1016/s0166-2236(98)01349-6 [DOI] [PubMed] [Google Scholar]
- Arnardottir E. S., Nikonova E. V., Shockley K. R., Podtelezhnikov A. A., Anafi R. C., Tanis K. Q., et al. (2014). Blood-gene expression reveals reduced circadian rhythmicity in individuals resistant to sleep deprivation. Sleep 37 1589–1600. 10.5665/sleep.4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G., Sehgal A. (2017). The Drosophila circuitry of sleep-wake regulation. Curr. Opin. Neurobiol. 44 243–250. 10.1016/j.conb.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G., Sehgal A. (2020). The glial perspective on sleep and circadian rhythms. Annu. Rev. Neurosci. 43 119–140. 10.1146/annurev-neuro-091819-094557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G., Zhang S. L., Tricoire H., Sehgal A. (2018). Endocytosis at the Drosophila blood-brain barrier as a function for sleep. Elife 7:e43326. 10.7554/eLife.43326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaro A., Sinha R., Bakun M., Kalnytska O., Carlo-Spiewok A. S., Rubel T., et al. (2021). ApoE4 disrupts interaction of sortilin with fatty acid-binding protein 7 essential to promote lipid signaling. J. Cell Sci. 134:jcs258894. 10.1242/jcs.258894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balendiran G. K., Schnutgen F., Scapin G., Borchers T., Xhong N., Lim K., et al. (2000). Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J. Biol. Chem. 275 27045–27054. 10.1074/jbc.M003001200 [DOI] [PubMed] [Google Scholar]
- Bannerman P., Hahn A., Soulika A., Gallo V., Pleasure D. (2007). Astrogliosis in EAE spinal cord: Derivation from radial glia, and relationships to oligodendroglia. Glia 55 57–64. 10.1002/glia.20437 [DOI] [PubMed] [Google Scholar]
- Barca-Mayo O., López M. (2021). Astrocyte clocks and glucose homeostasis. Front. Endocrinol. (Lausanne) 12:662017. 10.3389/fendo.2021.662017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A., Alarcon J. M., Kandel E. R. (2002). Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell 108 689–703. 10.1016/s0092-8674(02)00657-8 [DOI] [PubMed] [Google Scholar]
- Barco A., Patterson S. L., Alarcon J. M., Gromova P., Mata-Roig M., Morozov A., et al. (2005). Gene expression profiling of facilitated L-LTP in VP16-CREB mice reveals that BDNF is critical for the maintenance of LTP and its synaptic capture. Neuron 48 123–137. 10.1016/j.neuron.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Barr J. L., Brailoiu G. C., Abood M. E., Rawls S. M., Unterwald E. M., Brailoiu E. (2020). Acute cocaine administration alters permeability of blood-brain barrier in freely-moving rats- Evidence using miniaturized fluorescence microscopy. Drug Alcohol. Depend. 206:107637. 10.1016/j.drugalcdep.2019.107637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J., Takahashi J. S. (2010). Circadian integration of metabolism and energetics. Science 330 1349–1354. 10.1126/science.1195027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont J. L., Toda H., Shi M., Park C. H., Quake C., Stein C., et al. (2021). Short and long sleeping mutants reveal links between sleep and macroautophagy. Elife 10:e64140. 10.7554/eLife.64140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellesi M., de Vivo L., Koebe S., Tononi G., Cirelli C. (2018). Sleep and wake affect glycogen content and turnover at perisynaptic astrocytic processes. Front. Cell Neurosci. 12:308. 10.3389/fncel.2018.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellesi M., de Vivo L., Tononi G., Cirelli C. (2015). Effects of sleep and wake on astrocytes: Clues from molecular and ultrastructural studies. BMC Biol. 13:66. 10.1186/s12915-015-0176-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benington J. H., Heller H. C. (1995). Restoration of brain energy metabolism as the function of sleep. Prog. Neurobiol. 45 347–360. [DOI] [PubMed] [Google Scholar]
- Bennett E., Stenvers K. L., Lund P. K., Popko B. (1994). Cloning and characterization of a cDNA encoding a novel fatty acid binding protein from rat brain. J. Neurochem. 63 1616–1624. 10.1046/j.1471-4159.1994.63051616.x [DOI] [PubMed] [Google Scholar]
- Bensaad K., Favaro E., Lewis C. A., Peck B., Lord S., Collins J. M., et al. (2014). Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 9 349–365. 10.1016/j.celrep.2014.08.056 [DOI] [PubMed] [Google Scholar]
- Bernardinelli Y., Muller D., Nikonenko I. (2014a). Astrocyte-synapse structural plasticity. Neural Plast. 2014:232105. 10.1155/2014/232105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardinelli Y., Randall J., Janett E., Nikonenko I., Konig S., Jones E. V., et al. (2014b). Activity-dependent structural plasticity of perisynaptic astrocytic domains promotes excitatory synapse stability. Curr. Biol. 24 1679–1688. 10.1016/j.cub.2014.06.025 [DOI] [PubMed] [Google Scholar]
- Blum I. D., Keleş M. F., Baz E. S., Han E., Park K., Luu S., et al. (2021). Astroglial calcium signaling encodes sleep need in drosophila. Curr. Biol. 31 150–162.e7. 10.1016/j.cub.2020.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneva N. B., Kaplamadzhiev D. B., Sahara S., Kikuchi H., Pyko I. V., Kikuchi M., et al. (2011). Expression of fatty acid-binding proteins in adult hippocampal neurogenic niche of postischemic monkeys. Hippocampus 21 162–171. 10.1002/hipo.20732 [DOI] [PubMed] [Google Scholar]
- Borbely A. A., Achermann P. (1999). Sleep homeostasis and models of sleep regulation. J. Biol. Rhythms 14 557–568. [DOI] [PubMed] [Google Scholar]
- Borbely A. A., Daan S., Wirz-Justice A., Deboer T. (2016). The two-process model of sleep regulation: A reappraisal. J. Sleep Res. 25 131–143. [DOI] [PubMed] [Google Scholar]
- Bugge A., Feng D., Everett L. J., Briggs E. R., Mullican S. E., Wang F., et al. (2012). Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26 657–667. 10.1101/gad.186858.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cable J., Schernhammer E., Hanlon E. C., Vetter C., Cedernaes J., Makarem N., et al. (2021). Sleep and circadian rhythms: Pillars of health-a Keystone Symposia report. Ann. N. Y. Acad. Sci. 1506 18–34. 10.1111/nyas.14661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederroth C. R., Albrecht U., Bass J., Brown S. A., Dyhrfjeld-Johnsen J., Gachon F., et al. (2019). Medicine in the fourth dimension. Cell Metab. 30 238–250. 10.1016/j.cmet.2019.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Wang Y. F., Shinoda Y., Kawahata I., Yamamoto T., Jia W. B., et al. (2022). Fatty acid-binding protein 7 triggers α-synuclein oligomerization in glial cells and oligodendrocytes associated with oxidative stress. Acta Pharmacol. Sin. 43 552–562. 10.1038/s41401-021-00675-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon M. S., Kim S. H., Fountoulakis M., Lubec G. (2003). Heart type fatty acid binding protein (H-FABP) is decreased in brains of patients with Down syndrome and Alzheimer’s disease. J. Neural Transm. Suppl. 67, 225–234. 10.1007/978-3-7091-6721-2_20 [DOI] [PubMed] [Google Scholar]
- Cho H., Zhao X., Hatori M., Yu R. T., Barish G. D., Lam M. T., et al. (2012). Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485 123–127. 10.1038/nature11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C. (2009). The genetic and molecular regulation of sleep: From fruit flies to humans. Nat. Rev. Neurosci. 10 549–560. 10.1038/nrn2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Faraguna U., Tononi G. (2006). Changes in brain gene expression after long-term sleep deprivation. J. Neurochem. 98 1632–1645. 10.1111/j.1471-4159.2006.04058.x [DOI] [PubMed] [Google Scholar]
- Clarke L. E., Liddelow S. A., Chakraborty C., Münch A. E., Heiman M., Barres B. A. (2018). Normal aging induces A1-like astrocyte reactivity. Proc. Natl. Acad. Sci. U.S.A. 115 E1896–E1905. 10.1073/pnas.1800165115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell C. S. (2021). Defining circadian disruption in neurodegenerative disorders. J. Clin. Invest. 131:e148288. 10.1172/jci148288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah V. A., Zhang S. L., Sehgal A. (2019). Regulation of the blood-brain barrier by circadian rhythms and sleep. Trends Neurosci. 42 500–510. 10.1016/j.tins.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann R., Viola A. U., Tarokh L., Cajochen C., Brown S. A. (2012). The human circadian metabolome. Proc. Natl. Acad. Sci. U.S.A. 109 2625–2629. 10.1073/pnas.1114410109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M., Doktór B., Baster Z., Pyza E. (2022a). The role of glia clocks in the regulation of sleep in Drosophila melanogaster. J Neurosci. 42 6848–6860. 10.1523/jneurosci.2340-21.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M., Szypulski K., Pyza E. (2022b). Glia-neurons cross-talk regulated through autophagy. Front. Physiol. 13:886273. 10.3389/fphys.2022.886273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash M. B., Bellesi M., Tononi G., Cirelli C. (2013). Sleep/wake dependent changes in cortical glucose concentrations. J. Neurochem. 124 79–89. 10.1111/jnc.12063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboer T. (2018). Sleep homeostasis and the circadian clock: Do the circadian pacemaker and the sleep homeostat influence each other’s functioning? Neurobiol. Sleep Circadian Rhythms 5 68–77. 10.1016/j.nbscr.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboer T., Détári L., Meijer J. H. (2007). Long term effects of sleep deprivation on the mammalian circadian pacemaker. Sleep 30 257–262. 10.1093/sleep/30.3.257 [DOI] [PubMed] [Google Scholar]
- Deboer T., Vansteensel M. J., Détári L., Meijer J. H. (2003). Sleep states alter activity of suprachiasmatic nucleus neurons. Nat. Neurosci. 6 1086–1090. 10.1038/nn1122 [DOI] [PubMed] [Google Scholar]
- Depner C. M., Stothard E. R., Wright K. P., Jr. (2014). Metabolic consequences of sleep and circadian disorders. Curr. Diab. Rep. 14:507. 10.1007/s11892-014-0507-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouiche A., Frotscher M. (2001). Peripheral astrocyte processes: Monitoring by selective immunostaining for the actin-binding ERM proteins. Glia 36 330–341. 10.1002/glia.1120 [DOI] [PubMed] [Google Scholar]
- Derouiche A., Anlauf E., Aumann G., Muhlstadt B., Lavialle M. (2002). Anatomical aspects of glia-synapse interaction: The perisynaptic glial sheath consists of a specialized astrocyte compartment. J. Physiol. Paris 96 177–182. 10.1016/s0928-4257(02)00004-9 [DOI] [PubMed] [Google Scholar]
- Donlea J. M., Alam M. N., Szymusiak R. (2017). Neuronal substrates of sleep homeostasis; lessons from flies, rats and mice. Curr. Opin. Neurobiol. 44 228–235. 10.1016/j.conb.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Easton A., Meerlo P., Bergmann B., Turek F. W. (2004). The suprachiasmatic nucleus regulates sleep timing and amount in mice. Sleep 27 1307–1318. 10.1093/sleep/27.7.1307 [DOI] [PubMed] [Google Scholar]
- Eban-Rothschild A., Appelbaum L., de Lecea L. (2018). Neuronal mechanisms for sleep/wake regulation and modulatory drive. Neuropsychopharmacology 43 937–952. 10.1038/npp.2017.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi M., Yamamoto Y., Sharifi K., Kida H., Kagawa Y., Yasumoto Y., et al. (2016). Astrocyte-expressed FABP7 regulates dendritic morphology and excitatory synaptic function of cortical neurons. Glia 64 48–62. 10.1002/glia.22902 [DOI] [PubMed] [Google Scholar]
- Ehlen J. C., Brager A. J., Baggs J., Pinckney L., Gray C. L., DeBruyne J. P., et al. (2017). Bmal1 function in skeletal muscle regulates sleep. Elife 6:e26557. 10.7554/eLife.26557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Hatten M. E., Heintz N. (1994). Brain lipid-binding protein (BLBP): A novel signaling system in the developing mammalian CNS. Neuron 12 895–908. 10.1016/0896-6273(94)90341-7 [DOI] [PubMed] [Google Scholar]
- Frank M. G. (2013). Astroglial regulation of sleep homeostasis. Curr. Opin. Neurobiol. 23 812–818. 10.1016/j.conb.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Franken P., Dijk D. J. (2009). Circadian clock genes and sleep homeostasis. Eur. J. Neurosci. 29 1820–1829. 10.1111/j.1460-9568.2009.06723.x [DOI] [PubMed] [Google Scholar]
- Fu L., Kettner N. M. (2013). The circadian clock in cancer development and therapy. Prog. Mol. Biol. Transl. Sci. 119 221–282. 10.1016/b978-0-12-396971-2.00009-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M., Hotamisligil G. S. (2008). Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 7 489–503. 10.1038/nrd2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud C., Quairiaux C., Steiner P., Hirling H., Welker E., Knott G. W. (2006). Plasticity of astrocytic coverage and glutamate transporter expression in adult mouse cortex. PLoS Biol. 4:e343. 10.1371/journal.pbio.0040343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Paschos G. K. (2020). Circadian expression of Fabp7 mRNA is disrupted in Bmal1 KO mice. Mol. Brain 13:26. 10.1186/s13041-020-00568-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Yin J. C. (2010). Circadian rhythms and memory formation. Nat. Rev. Neurosci. 11 577–588. 10.1038/nrn2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Bremer Q. Z., Vander Heyden W. M., Lavaute T. M., Yin J. C., Landry C. F. (2008). Brain fatty acid binding protein (Fabp7) is diurnally regulated in astrocytes and hippocampal granule cell precursors in adult rodent brain. PLoS One 3:e1631. 10.1371/journal.pone.0001631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Perron I. J., Riedy S. M., Yoshikawa T., Kadotani H., Owada Y., et al. (2017). Normal sleep requires the astrocyte brain-type fatty acid binding protein FABP7. Sci. Adv. 3:e1602663. 10.1126/sciadv.1602663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Vander Heyden W. M., Lavaute T. M., Landry C. F. (2006). Profiles of novel diurnally regulated genes in mouse hypothalamus: Expression analysis of the cysteine and histidine-rich domain-containing, zinc-binding protein 1, the fatty acid-binding protein 7 and the GTPase, ras-like family member 11b. Neuroscience 139 1435–1448. 10.1016/j.neuroscience.2006.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Vanderheyden W. M., LaVaute T., Westmark C. J., Rouhana L., Pack A. I., et al. (2012). Time of day regulates subcellular trafficking, tripartite synaptic localization, and polyadenylation of the astrocytic Fabp7 mRNA. J. Neurosci. 32 1383–1394. 10.1523/jneurosci.3228-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Vanderheyden W. M., Shaw P. J., Landry C. F., Yin J. C. (2011a). Cytoplasmic to nuclear localization of fatty-acid binding protein correlates with specific forms of long-term memory in Drosophila. Commun. Integr. Biol. 4 623–626. 10.4161/cib.4.5.16927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner J. R., Vanderheyden W. M., Shaw P. J., Landry C. F., Yin J. C. (2011b). Fatty-acid binding proteins modulate sleep and enhance long-term memory consolidation in Drosophila. PLoS One 6:e15890. 10.1371/journal.pone.0015890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandner M. A. (2022). Sleep, health, and society. Sleep Med. Clin. 17 117–139. 10.1016/j.jsmc.2022.03.001 [DOI] [PubMed] [Google Scholar]
- Guindalini C., Andersen M. L., Alvarenga T., Lee K., Tufik S. (2009). To what extent is sleep rebound effective in reversing the effects of paradoxical sleep deprivation on gene expression in the brain? Behav. Brain Res. 201 53–58. 10.1016/j.bbr.2009.01.027 [DOI] [PubMed] [Google Scholar]
- Guo L., Reed K. M., Carter A., Cheng Y., Roodsari S. K., Martinez Pineda D., et al. (2022). Sleep-disturbance-induced microglial activation involves CRH-mediated Galectin 3 and autophagy dysregulation. Cells 12:160. 10.3390/cells12010160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttula S. V., Allam A., Gumpeny R. S. (2012). Analyzing microarray data of Alzheimer’s using cluster analysis to identify the biomarker genes. Int. J. Alzheimers Dis. 2012:649456. 10.1155/2012/649456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa M. M., Haydon P. G. (2010). Integrated brain circuits: Astrocytic networks modulate neuronal activity and behavior. Annu. Rev. Physiol. 72 335–355. 10.1146/annurev-physiol-021909-135843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa M. M., Florian C., Fellin T., Munoz J. R., Lee S. Y., Abel T., et al. (2009). Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron 61 213–219. 10.1016/j.neuron.2008.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford J., Shen S., Itamura K., Levine J., Chong A. C., Czerwieniec G., et al. (2017). New astroglial injury-defined biomarkers for neurotrauma assessment. J. Cereb. Blood Flow Metab. 37 3278–3299. 10.1177/0271678x17724681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J., Marion M., Figueiredo A., Clavin B. H., Deutsch D., Kaczocha M., et al. (2018). Fatty acid binding protein deletion prevents stress-induced preference for cocaine and dampens stress-induced corticosterone levels. Synapse 72:e22031. 10.1002/syn.22031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J., Roeder N., Richardson B., Hammond N., Sajjad M., Yao R., et al. (2022). Unpredictable chronic mild stress differentially impacts resting brain glucose metabolism in fatty acid-binding protein 7 deficient mice. Psychiatry Res. Neuroimaging 323:111486. 10.1016/j.pscychresns.2022.111486 [DOI] [PubMed] [Google Scholar]
- Hara T., Abdulaziz Umaru B., Sharifi K., Yoshikawa T., Owada Y., Kagawa Y. (2020). Fatty acid binding protein 7 is involved in the proliferation of reactive astrocytes, but not in cell migration and polarity. Acta Histochem. Cytochem. 53 73–81. 10.1267/ahc.20001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A. G. (2022). Treating sleep and circadian problems to promote mental health: Perspectives on comorbidity, implementation science and behavior change. Sleep 45:zsac026. 10.1093/sleep/zsac026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M. H., Brancaccio M., Gonzalez-Aponte M. F., Herzog E. D. (2023). Circadian rhythms and astrocytes: The good, the bad, and the ugly. Annu. Rev. Neurosci. 46:1. 10.1146/annurev-neuro-100322-112249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havekes R., Aton S. J. (2020). Impacts of sleep loss versus waking experience on brain plasticity: Parallel or orthogonal? Trends Neurosci. 43 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havekes R., Meerlo P., Abel T. (2015). Animal studies on the role of sleep in memory: From behavioral performance to molecular mechanisms. Curr. Top. Behav. Neurosci. 25 183–206. 10.1007/7854_2015_369 [DOI] [PubMed] [Google Scholar]
- Havekes R., Park A. J., Tudor J. C., Luczak V. G., Hansen R. T., Ferri S. L., et al. (2016). Sleep deprivation causes memory deficits by negatively impacting neuronal connectivity in hippocampal area CA1. Elife 5:e13424. 10.7554/eLife.13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon P. G. (2017). Astrocytes and the modulation of sleep. Curr. Opin. Neurobiol. 44 28–33. 10.1016/j.conb.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Hsuchou H., He Y., Kastin A. J., Wang Y., Pan W. (2014). Sleep restriction impairs blood-brain barrier function. J. Neurosci. 34 14697–14706. 10.1523/jneurosci.2111-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaven M. R., Herren A. W., Flint D. L., Pacheco N. L., Li J., Tang A., et al. (2022). Metabolic enzyme alterations and astrocyte dysfunction in a murine model of Alexander disease with severe reactive gliosis. Mol. Cell Proteomics 21:100180. 10.1016/j.mcpro.2021.100180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor C. N., Yeung J., Jan M., Emmenegger Y., Hubbard J., Xenarios I., et al. (2019). Sleep-wake-driven and circadian contributions to daily rhythms in gene expression and chromatin accessibility in the murine cortex. Proc. Natl. Acad. Sci. U.S.A. 116 25773–25783. 10.1073/pnas.1910590116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. S., Carson J. H., Barbarese E., Richter J. D. (2003). Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 17 638–653. 10.1101/gad.1053003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. S., Jung M. Y., Sarkissian M., Richter J. D. (2002). N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 21 2139–2148. 10.1093/emboj/21.9.2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Alvarado G., Becerril-Villanueva E., Contis-Montes de Oca A., Domínguez-Salazar E., Salinas-Jazmín N., Pérez-Tapia S. M., et al. (2018). The yin/yang of inflammatory status: Blood-brain barrier regulation during sleep. Brain Behav. Immun. 69 154–166. 10.1016/j.bbi.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Ingiosi A. M., Frank M. G. (2022). Goodnight, astrocyte: Waking up to astroglial mechanisms in sleep. FEBS J. 290 2553–2564. 10.1111/febs.16424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingiosi A. M., Hayworth C. R., Harvey D. O., Singletary K. G., Rempe M. J., Wisor J. P., et al. (2020). A role for astroglial calcium in mammalian sleep and sleep regulation. Curr. Biol. 30 4373–4383.e7. 10.1016/j.cub.2020.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou M. S., Jackson J., Sheu S. H., Chang C. L., Weigel A. V., Liu H., et al. (2019a). Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell 177 1522–1535.e14. 10.1016/j.cell.2019.04.001 [DOI] [PubMed] [Google Scholar]
- Ioannou M. S., Liu Z., Lippincott-Schwartz J. (2019b). A Neuron-glia co-culture system for studying intercellular lipid transport. Curr. Protoc. Cell Biol. 84:e95. 10.1002/cpcb.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A., Kagawa Y., Miyazaki H., Shil S. K., Umaru B. A., Yasumoto Y., et al. (2019). FABP7 protects astrocytes against ROS toxicity via lipid droplet formation. Mol. Neurobiol. 56 5763–5779. 10.1007/s12035-019-1489-2 [DOI] [PubMed] [Google Scholar]
- Jackson F. R., You S., Crowe L. B. (2020). Regulation of rhythmic behaviors by astrocytes. Wiley Interdiscip. Rev Dev. Biol. 9 e372. 10.1002/wdev.372 [DOI] [PubMed] [Google Scholar]
- Jang S., Choi B., Lim C., Lee B., Cho K. S. (2022). Roles of Drosophila fatty acid-binding protein in development and behavior. Biochem. Biophys. Res. Commun. 599 87–92. 10.1016/j.bbrc.2022.02.040 [DOI] [PubMed] [Google Scholar]
- Jessen N. A., Munk A. S., Lundgaard I., Nedergaard M. (2015). The glymphatic System: A Beginner’s guide. Neurochem Res 40 2583–2599. 10.1007/s11064-015-1581-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Pfister-Genskow M., Benca R. M., Cirelli C. (2008). Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J. Neurochem. 105 46–62. 10.1111/j.1471-4159.2007.05089.x [DOI] [PubMed] [Google Scholar]
- Josephson R., Müller T., Pickel J., Okabe S., Reynolds K., Turner P. A., et al. (1998). POU transcription factors control expression of CNS stem cell-specific genes. Development 125 3087–3100. 10.1242/dev.125.16.3087 [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Umaru B. A., Ariful I., Shil S. K., Miyazaki H., Yamamoto Y., et al. (2019). Role of FABP7 in tumor cell signaling. Adv. Biol. Regul. 71 206–218. 10.1016/j.jbior.2018.09.006 [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Umaru B. A., Shima H., Ito R., Zama R., Islam A., et al. (2020). FABP7 regulates Acetyl-CoA metabolism through the interaction with ACLY in the nucleus of astrocytes. Mol. Neurobiol. 57 4891–4910. 10.1007/s12035-020-02057-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizato K., Sato S., Shil S. K., Umaru B. A., Kagawa Y., Yamamoto Y., et al. (2019). The role of fatty acid binding protein 7 in spinal cord astrocytes in a mouse model of experimental autoimmune encephalomyelitis. Neuroscience 409 120–129. 10.1016/j.neuroscience.2019.03.050 [DOI] [PubMed] [Google Scholar]
- Kato T., Yoshioka H., Owada Y., Kinouchi H. (2020). Roles of fatty acid binding protein 7 in ischemic neuronal injury and ischemia-induced neurogenesis after transient forebrain ischemia. Brain Res. 1736:146795. 10.1016/j.brainres.2020.146795 [DOI] [PubMed] [Google Scholar]
- Kawashima M., Bensaad K., Zois C. E., Barberis A., Bridges E., Wigfield S., et al. (2020). Disruption of hypoxia-inducible fatty acid binding protein 7 induces beige fat-like differentiation and thermogenesis in breast cancer cells. Cancer Metab. 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killoy K. M., Harlan B. A., Pehar M., Vargas M. R. (2020). FABP7 upregulation induces a neurotoxic phenotype in astrocytes. Glia 68 2693–2704. 10.1002/glia.23879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipp M., Gingele S., Pott F., Clarner T., van der Valk P., Denecke B., et al. (2011). BLBP-expression in astrocytes during experimental demyelination and in human multiple sclerosis lesions. Brain Behav. Immun. 25 1554–1568. 10.1016/j.bbi.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Kis V., Barti B., Lippai M., Sass M. (2015). Specialized cortex glial cells accumulate lipid droplets in Drosophila melanogaster. PLoS One 10:e0131250. 10.1371/journal.pone.0131250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M., Nakagawa S., Sato A., Oka M., Makikhara K., Sakai Y., et al. (2021). Plasma fatty acid-binding protein 7 concentration correlates with depression/anxiety, cognition, and positive symptom in patients with schizophrenia. J. Psychiatr. Res. 144 304–311. 10.1016/j.jpsychires.2021.10.028 [DOI] [PubMed] [Google Scholar]
- Kreutzmann J. C., Havekes R., Abel T., Meerlo P. (2015). Sleep deprivation and hippocampal vulnerability: Changes in neuronal plasticity, neurogenesis and cognitive function. Neuroscience 309 173–190. 10.1016/j.neuroscience.2015.04.053 [DOI] [PubMed] [Google Scholar]
- Krueger J. M. (2020). Sleep and circadian rhythms: Evolutionary entanglement and local regulation. Neurobiol. Sleep Circadian Rhythms 9:100052. 10.1016/j.nbscr.2020.100052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger J. M., Tononi G. (2011). Local use-dependent sleep; synthesis of the new paradigm. Curr. Top. Med. Chem. 11 2490–2492. 10.2174/156802611797470330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger J. M., Rector D. M., Roy S., Van Dongen H. P., Belenky G., Panksepp J. (2008). Sleep as a fundamental property of neuronal assemblies. Nat. Rev. Neurosci. 9 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Jha P., Challet E., Kalsbeek A. (2015). Circadian rhythms in glucose and lipid metabolism in nocturnal and diurnal mammals. Mol. Cell Endocrinol. 418(Pt 1) 74–88. 10.1016/j.mce.2015.01.024 [DOI] [PubMed] [Google Scholar]
- Kurtz A., Zimmer A., Schnütgen F., Brüning G., Spener F., Müller T. (1994). The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development 120 2637–2649. 10.1242/dev.120.9.2637 [DOI] [PubMed] [Google Scholar]
- Lananna B. V., Nadarajah C. J., Izumo M., Cedeño M. R., Xiong D. D., Dimitry J., et al. (2018). Cell-autonomous regulation of astrocyte activation by the circadian clock protein BMAL1. Cell Rep. 25 1–9.e5. 10.1016/j.celrep.2018.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposky A., Easton A., Dugovic C., Walisser J., Bradfield C., Turek F. (2005). Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep 28 395–409. 10.1093/sleep/28.4.395 [DOI] [PubMed] [Google Scholar]
- Lavialle M., Aumann G., Anlauf E., Prols F., Arpin M., Derouiche A. (2011). Structural plasticity of perisynaptic astrocyte processes involves ezrin and metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 108 12915–12919. 10.1073/pnas.1100957108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal O., Ulloa Severino F. P., Eroglu C. (2022). The role of astrocyte structural plasticity in regulating neural circuit function and behavior. Glia 70 1467–1483. 10.1002/glia.24191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kang B. M., Kim J. H., Min J., Kim H. S., Ryu H., et al. (2018). Real-time in vivo two-photon imaging study reveals decreased cerebro-vascular volume and increased blood-brain barrier permeability in chronically stressed mice. Sci Rep 8:13064. 10.1038/s41598-018-30875-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Field J. M., Sehgal A. (2021). Circadian rhythms, disease and chronotherapy. J. Biol. Rhythms 36 503–531. 10.1177/07487304211044301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesku J. A., Roth T. C., Rattenborg N. C., Amlaner C. J., Lima S. L. (2008). Phylogenetics and the correlates of mammalian sleep: A reappraisal. Sleep Med. Rev. 12 229–244. 10.1016/j.smrv.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Li F., Artiushin G., Sehgal A. (2023). Modulation of sleep by trafficking of lipids through the Drosophila blood-brain barrier. Elife 12:e86336. 10.7554/eLife.86336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Haynes P., Zhang S. L., Yue Z., Sehgal A. (2023). Ecdysone acts through cortex glia to regulate sleep in Drosophila. Elife 12:e81723. 10.7554/eLife.81723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J., Cooper K., Sehgal M., Silva A. J. (2018). Memory formation depends on both synapse-specific modifications of synaptic strength and cell-specific increases in excitability. Nat. Neurosci. 21 309–314. 10.1038/s41593-018-0076-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., MacKenzie K. R., Putluri N., Maletiæ-Savatiæ M., Bellen H. J. (2017). The glia-neuron lactate shuttle and elevated ROS promote lipid synthesis in neurons and lipid droplet accumulation in glia via APOE/D. Cell Metab. 26 719–737.e 10.1016/j.cmet.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Iacono L., Valzania A., Visco-Comandini F., Viscomi M. T., Felsani A., Puglisi-Allegra S., et al. (2016). Regulation of nucleus accumbens transcript levels in mice by early-life social stress and cocaine. Neuropharmacology 103 183–194. 10.1016/j.neuropharm.2015.12.011 [DOI] [PubMed] [Google Scholar]
- Lundgaard I., Lu M. L., Yang E., Peng W., Mestre H., Hitomi E., et al. (2017). Glymphatic clearance controls state-dependent changes in brain lactate concentration. J. Cereb. Blood Flow Metab. 37 2112–2124. 10.1177/0271678x16661202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly S., Pack A. I., Naidoo N. (2018). The neurobiological basis of sleep: Insights from Drosophila. Neurosci Biobehav Rev 87 67–86. 10.1016/j.neubiorev.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Zhang M., Mori Y., Yao C., Larsen C. P., Yamashima T., et al. (2010). Cellular localization of epidermal-type and brain-type fatty acid-binding proteins in adult hippocampus and their response to cerebral ischemia. Hippocampus 20 811–819. 10.1002/hipo.20682 [DOI] [PubMed] [Google Scholar]
- Malik D. M., Paschos G. K., Sehgal A., Weljie A. M. (2020). Circadian and sleep metabolomics across species. J. Mol. Biol. 432 3578–3610. 10.1016/j.jmb.2020.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W., Yi X., Qin J., Tian M., Jin G. (2020). CXCL12 promotes proliferation of radial glia like cells after traumatic brain injury in rats. Cytokine 125:154771. 10.1016/j.cyto.2019.154771 [DOI] [PubMed] [Google Scholar]
- Matenchuk B. A., Mandhane P. J., Kozyrskyj A. L. (2020). Sleep, circadian rhythm, and gut microbiota. Sleep Med. Rev. 53 101340. 10.1016/j.smrv.2020.101340 [DOI] [PubMed] [Google Scholar]
- Matsumata M., Inada H., Osumi N. (2016). Fatty acid binding proteins and the nervous system: Their impact on mental conditions. Neurosci. Res. 102 47–55. 10.1016/j.neures.2014.08.012 [DOI] [PubMed] [Google Scholar]
- Matsumata M., Sakayori N., Maekawa M., Owada Y., Yoshikawa T., Osumi N. (2012). The effects of Fabp7 and Fabp5 on postnatal hippocampal neurogenesis in the mouse. Stem Cells 30 1532–1543. 10.1002/stem.1124 [DOI] [PubMed] [Google Scholar]
- McKee C. A., Polino A. J., King M. W., Musiek E. S. (2023). Circadian clock protein BMAL1 broadly influences autophagy and endolysosomal function in astrocytes. Proc. Natl. Acad. Sci. U.S.A. 120:e2220551120. 10.1073/pnas.2220551120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen E., Hertenstein H., Strassburger K., Deharde L., Brankatschk M., Schirmeier S. (2023). Glycolytically impaired Drosophila glial cells fuel neural metabolism via β-oxidation. Nat. Commun. 14:2996. 10.1038/s41467-023-38813-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita R., Beaulieu M. J., Field C., Godbout R. (2010). Brain fatty acid-binding protein and omega-3/omega-6 fatty acids: Mechanistic insight into malignant glioma cell migration. J. Biol. Chem. 285 37005–37015. 10.1074/jbc.M110.170076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita R., Coles J. E., Glubrecht D. D., Sung R., Sun X., Godbout R. (2007). B-FABP-expressing radial glial cells: The malignant glioma cell of origin? Neoplasia 9 734–744. 10.1593/neo.07439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk J. A., Green C. B., Takahashi J. S. (2012). Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35 445–462. 10.1146/annurev-neuro-060909-153128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecillo-Aguado M., Tirado-Rodriguez B., Tong Z., Vega O. M., Morales-Martínez M., Abkenari S., et al. (2020). Importance of the role of ω-3 and ω-6 polyunsaturated fatty acids in the progression of brain cancer. Brain Sci. 10:381. 10.3390/brainsci10060381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek E. S., Holtzman D. M. (2016). Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science 354 1004–1008. 10.1126/science.aah4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek E. S., Lim M. M., Yang G., Bauer A. Q., Qi L., Lee Y., et al. (2013). Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J. Clin. Invest. 123 5389–5400. 10.1172/jci70317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassan M., Videnovic A. (2022). Circadian rhythms in neurodegenerative disorders. Nat. Rev. Neurol. 18 7–24. 10.1038/s41582-021-00577-7 [DOI] [PubMed] [Google Scholar]
- Naylor E., Aillon D. V., Barrett B. S., Wilson G. S., Johnson D. A., Harmon H. P., et al. (2012). Lactate as a biomarker for sleep. Sleep 35 1209–1222. 10.5665/sleep.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham H., Torpey G., Flores C. C., Davis C. J., Vanderheyden W. M., Gerstner J. R. (2022). A dichotomous role for FABP7 in sleep and Alzheimer’s disease pathogenesis: A hypothesis. Front. Neurosci. 16:798994. 10.3389/fnins.2022.798994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P. V., Woo N. H. (2003). Regulation of hippocampal synaptic plasticity by cyclic AMP-dependent protein kinases. Prog. Neurobiol. 71 401–437. 10.1016/j.pneurobio.2003.12.003 [DOI] [PubMed] [Google Scholar]
- Nowowiejska J., Baran A., Flisiak I. (2022a). Fatty acid-binding proteins in psoriasis-A review. Metabolites 12:833. 10.3390/metabo12090833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowowiejska J., Baran A., Hermanowicz J. M., Sieklucka B., Krahel J. A., Kiluk P., et al. (2022b). Fatty acid-binding protein 7 (FABP-7), Glutamic acid and neurofilament light chain (NFL) as potential markers of neurodegenerative disorders in psoriatic patients-a pilot study. J. Clin. Med. 11:2430. 10.3390/jcm11092430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockner R. K., Manning J. A., Poppenhausen R. B., Ho W. K. (1972). A binding protein for fatty acids in cytosol of intestinal mucosa, liver, myocardium, and other tissues. Science 177 56–58. 10.1126/science.177.4043.56 [DOI] [PubMed] [Google Scholar]
- Olzmann J. A., Carvalho P. (2019). Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20 137–155. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota Y., Zanetti A. T., Hallock R. M. (2013). The role of astrocytes in the regulation of synaptic plasticity and memory formation. Neural Plast. 2013:185463. 10.1155/2013/185463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owada Y., Abdelwahab S. A., Kitanaka N., Sakagami H., Takano H., Sugitani Y., et al. (2006). Altered emotional behavioral responses in mice lacking brain-type fatty acid-binding protein gene. Eur. J. Neurosci. 24 175–187. 10.1111/j.1460-9568.2006.04855.x [DOI] [PubMed] [Google Scholar]
- Owada Y., Yoshimoto T., Kondo H. (1996b). Spatio-temporally differential expression of genes for three members of fatty acid binding proteins in developing and mature rat brains. J. Chem. Neuroanat. 12 113–122. 10.1016/s0891-0618(96)00192-5 [DOI] [PubMed] [Google Scholar]
- Owada Y., Yoshimoto T., Kondo H. (1996a). Increased expression of the mRNA for brain- and skin-type but not heart-type fatty acid binding proteins following kainic acid systemic administration in the hippocampal glia of adult rats. Brain Res. Mol. Brain Res. 42 156–160. 10.1016/s0169-328x(96)00182-9 [DOI] [PubMed] [Google Scholar]
- Pan W., Kastin A. J. (2017). The blood-brain barrier: Regulatory roles in wakefulness and sleep. Neuroscientist 23 124–136. 10.1177/1073858416639005 [DOI] [PubMed] [Google Scholar]
- Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., et al. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109 307–320. [DOI] [PubMed] [Google Scholar]
- Patke A., Young M. W., Axelrod S. (2020). Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21 67–84. 10.1038/s41580-019-0179-2 [DOI] [PubMed] [Google Scholar]
- Perea G., Navarrete M., Araque A. (2009). Tripartite synapses: Astrocytes process and control synaptic information. Trends Neurosci. 32 421–431. 10.1016/j.tins.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Perez-Alvarez A., Navarrete M., Covelo A., Martin E. D., Araque A. (2014). Structural and functional plasticity of astrocyte processes and dendritic spine interactions. J. Neurosci. 34 12738–12744. 10.1523/jneurosci.2401-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Catalan N. A., Doe C. Q., Ackerman S. D. (2021). The role of astrocyte-mediated plasticity in neural circuit development and function. Neural Dev. 16:1. 10.1186/s13064-020-00151-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J. M., Gyger J., Burlet-Godinot S., Fiumelli H., Martin J. L., Magistretti P. J. (2013). Genes involved in the astrocyte-neuron lactate shuttle (ANLS) are specifically regulated in cortical astrocytes following sleep deprivation in mice. Sleep 36 1445–1458. 10.5665/sleep.3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poskanzer K. E., Yuste R. (2016). Astrocytes regulate cortical state switching in vivo. Proc. Natl. Acad. Sci. U.S.A. 113 E2675–E2684. 10.1073/pnas.1520759113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach A., Derouiche A., Kirchhoff F. (2010). Morphology and dynamics of perisynaptic glia. Brain Res. Rev. 63 11–25. [DOI] [PubMed] [Google Scholar]
- Reitman M. E., Tse V., Mi X., Willoughby D. D., Peinado A., Aivazidis A., et al. (2023). Norepinephrine links astrocytic activity to regulation of cortical state. Nat. Neurosci. 26 579–593. 10.1038/s41593-023-01284-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Q., Ni H., Lin X., Zhu X., Li D., Liu H., et al. (2019). Astrocyte-derived fatty acid-binding protein 7 protects blood-brain barrier integrity through a caveolin-1/MMP signaling pathway following traumatic brain injury. Exp. Neurol. 322:113044. 10.1016/j.expneurol.2019.113044 [DOI] [PubMed] [Google Scholar]
- Sánchez-Font M. F., Bosch-Comas A., Gonzàlez-Duarte R., Marfany G. (2003). Overexpression of FABP7 in Down syndrome fetal brains is associated with PKNOX1 gene-dosage imbalance. Nucleic Acids Res. 31 2769–2777. 10.1093/nar/gkg396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santello M., Cali C., Bezzi P. (2012). Gliotransmission and the tripartite synapse. Adv. Exp. Med. Biol. 970 307–331. 10.1007/978-3-7091-0932-8_14 [DOI] [PubMed] [Google Scholar]
- Sántha P., Veszelka S., Hoyk Z., Mészáros M., Walter F. R., Tóth A. E., et al. (2015). Restraint stress-induced morphological changes at the blood-brain barrier in adult rats. Front. Mol. Neurosci. 8:88. 10.3389/fnmol.2015.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper C. B., Chou T. C., Scammell T. E. (2001). The sleep switch: Hypothalamic control of sleep and wakefulness. Trends Neurosci. 24 726–731. [DOI] [PubMed] [Google Scholar]
- Saper C. B., Scammell T. E., Lu J. (2005). Hypothalamic regulation of sleep and circadian rhythms. Nature 437 1257–1263. [DOI] [PubMed] [Google Scholar]
- Schaap F. G., van der Vusse G. J., Glatz J. F. (2002). Evolution of the family of intracellular lipid binding proteins in vertebrates. Mol. Cell Biochem. 239 69–77. [PubMed] [Google Scholar]
- Scharf M. T., Naidoo N., Zimmerman J. E., Pack A. I. (2008). The energy hypothesis of sleep revisited. Prog. Neurobiol. 86 264–280. 10.1016/j.pneurobio.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell A., Chappuis S., Schmutz I., Brai E., Ripperger J. A., Schaad O., et al. (2014). The nuclear receptor REV-ERBalpha regulates Fabp7 and modulates adult hippocampal neurogenesis. PLoS One 9:e99883. 10.1371/journal.pone.0099883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurhoff N., Toborek M. (2023). Circadian rhythms in the blood-brain barrier: Impact on neurological disorders and stress responses. Mol. Brain 16:5. 10.1186/s13041-023-00997-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senbokuya N., Yoshioka H., Yagi T., Owada Y., Kinouchi H. (2019). Effects of FABP7 on functional recovery after spinal cord injury in adult mice. J. Neurosurg. Spine 31, 291–297. 10.3171/2019.2.spine18844 [DOI] [PubMed] [Google Scholar]
- Sharifi K., Ebrahimi M., Kagawa Y., Islam A., Tuerxun T., Yasumoto Y., et al. (2013). Differential expression and regulatory roles of FABP5 and FABP7 in oligodendrocyte lineage cells. Cell Tissue Res. 354 683–695. 10.1007/s00441-013-1730-7 [DOI] [PubMed] [Google Scholar]
- Sharifi K., Morihiro Y., Maekawa M., Yasumoto Y., Hoshi H., Adachi Y., et al. (2011). FABP7 expression in normal and stab-injured brain cortex and its role in astrocyte proliferation. Histochem. Cell Biol. 136 501–513. 10.1007/s00418-011-0865-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoliè T., Zorec R., Vardjan N. (2021). Pathophysiology of lipid droplets in neuroglia. Antioxidants (Basel) 11:22. 10.3390/antiox11010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Avellaneda A., Morrison B. E. (2020). Signaling and other functions of lipids in autophagy: A review. Lipids Health Dis. 19:214. 10.1186/s12944-020-01389-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl B. A., Peco E., Davla S., Murakami K., Caicedo Moreno N. A., van Meyel D. J., et al. (2018). The taurine transporter Eaat2 functions in ensheathing glia to modulate sleep and metabolic rate. Curr. Biol. 28 3700–3708.e4. 10.1016/j.cub.2018.10.039 [DOI] [PubMed] [Google Scholar]
- Stankiewicz A. M., Goscik J., Majewska A., Swiergiel A. H., Juszczak G. R. (2015). The effect of acute and chronic social stress on the hippocampal transcriptome in mice. PLoS One 10:e0142195. 10.1371/journal.pone.0142195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen D., Malenka R. C. (2006). Synaptic scaling mediated by glial TNF-alpha. Nature 440 1054–1059. 10.1038/nature04671 [DOI] [PubMed] [Google Scholar]
- Storch J., Corsico B. (2008). The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu. Rev. Nutr. 28 73–95. 10.1146/annurev.nutr.27.061406.093710 [DOI] [PubMed] [Google Scholar]
- Strecker R. E., Morairty S., Thakkar M. M., Porkka-Heiskanen T., Basheer R., Dauphin L. J., et al. (2000). Adenosinergic modulation of basal forebrain and preoptic/anterior hypothalamic neuronal activity in the control of behavioral state. Behav. Brain Res. 115 183–204. 10.1016/s0166-4328(00)00258-8 [DOI] [PubMed] [Google Scholar]
- Sulli G., Lam M. T. Y., Panda S. (2019). Interplay between circadian clock and cancer: New frontiers for cancer treatment. Trends Cancer 5 475–494. 10.1016/j.trecan.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. Y., Simonyi A., Fritsche K. L., Chuang D. Y., Hannink M., Gu Z., et al. (2018). Docosahexaenoic acid (DHA): An essential nutrient and a nutraceutical for brain health and diseases. Prostaglandins Leukot Essent. Fatty Acids 136 3–13. 10.1016/j.plefa.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymusiak R., Gvilia I., McGinty D. (2007). Hypothalamic control of sleep. Sleep Med. Adv. Sleep Med. 8 291–301. [DOI] [PubMed] [Google Scholar]
- Tandon R., Levey A. I., Lah J. J., Seyfried N. T., Mitchell C. S. (2023). Machine learning selection of most predictive brain proteins suggests role of sugar metabolism in Alzheimer’s disease. J. Alzheimers Dis. 92 411–424. 10.3233/jad-220683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen C. E., Veerhuis R., De Vente J., Verhey F. R., Vreeling F., van Boxtel M. P., et al. (2011). Brain-specific fatty acid-binding protein is elevated in serum of patients with dementia-related diseases. Eur. J. Neurol. 18 865–871. 10.1111/j.1468-1331.2010.03273.x [DOI] [PubMed] [Google Scholar]
- Theodosis D. T., Poulain D. A., Oliet S. H. (2008). Activity-dependent structural and functional plasticity of astrocyte-neuron interactions. Physiol. Rev. 88 983–1008. 10.1152/physrev.00036.2007 [DOI] [PubMed] [Google Scholar]
- Thimgan M. S., Seugnet L., Turk J., Shaw P. J. (2015). Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila. Sleep 38 801–814. 10.5665/sleep.4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimgan M. S., Suzuki Y., Seugnet L., Gottschalk L., Shaw P. J. (2010). The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 8:e1000466. 10.1371/journal.pbio.1000466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G., Cirelli C. (2006). Sleep function and synaptic homeostasis. Sleep Med. Rev. 10 49–62. [DOI] [PubMed] [Google Scholar]
- Tripathi S., Kushwaha R., Mishra J., Gupta M. K., Kumar H., Sanyal S., et al. (2017). Docosahexaenoic acid up-regulates both PI3K/AKT-dependent FABP7-PPARgamma interaction and MKP3 that enhance GFAP in developing rat brain astrocytes. J. Neurochem. 140 96–113. 10.1111/jnc.13879 [DOI] [PubMed] [Google Scholar]
- Tsukita S., Yonemura S. (1997). ERM proteins: Head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem. Sci. 22 53–58. 10.1016/s0968-0004(96)10071-2 [DOI] [PubMed] [Google Scholar]
- Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., et al. (2002). A transcription factor response element for gene expression during circadian night. Nature 418 534–539. 10.1038/nature00906 [DOI] [PubMed] [Google Scholar]
- Vaidyanathan T. V., Collard M., Yokoyama S., Reitman M. E., Poskanzer K. E. (2021). Cortical astrocytes independently regulate sleep depth and duration via separate GPCR pathways. Elife 10:e63329. 10.7554/eLife.63329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Drunen R., Eckel-Mahan K. (2022). Circadian rhythms as modulators of brain health during development and throughout aging. Front. Neural Circuits 16:1059229. 10.3389/fncir.2022.1059229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheyden W. M., Fang B., Flores C. C., Jager J., Gerstner J. R. (2021). The transcriptional repressor Rev-erbα regulates circadian expression of the astrocyte Fabp7 mRNA. Curr. Res. Neurobiol. 2:100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheyden W. M., Lefton M., Flores C. C., Owada Y., Gerstner J. R. (2022). Fabp7 Is required for normal sleep suppression and anxiety-associated phenotype following single-prolonged stress in mice. Neuroglia 3 73–83. 10.3390/neuroglia3020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheyden W. M., Van Dongen H. P., Frank M. G., Gerstner J. R. (2019). Sleep pressure regulates mushroom body neural-glial interactions in Drosophila. Matters Sel. 1–7. 10.19185/matters.201903000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerkamp J. H., Zimmerman A. W. (2001). Fatty acid-binding proteins of nervous tissue. J. Mol. Neurosci. 16 133–142; discussion151–137. 10.1385/jmn:16:2-3:133 [DOI] [PubMed] [Google Scholar]
- von Schacky C. (2021). Importance of EPA and DHA blood levels in brain structure and function. Nutrients 13:1074. 10.3390/nu13041074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., Toyota T., Owada Y., Hayashi T., Iwayama Y., Matsumata M., et al. (2007). Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol. 5:e297. 10.1371/journal.pbio.0050297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser M. J., Butt C. M., Mohajeri M. H. (2016). Docosahexaenoic acid and cognition throughout the Lifespan. Nutrients 8:99. 10.3390/nu8020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M. A. (2015). Expanding roles for lipid droplets. Curr. Biol. 25 R470–R481. 10.1016/j.cub.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withrow D., Bowers S. J., Depner C. M., González A., Reynolds A. C., Wright K. P., Jr. (2021). Sleep and circadian disruption and the gut microbiome-possible links to dysregulated metabolism. Curr. Opin. Endocr. Metab. Res. 17 26–37. 10.1016/j.coemr.2020.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. P., Lowry C. A., Lebourgeois M. K. (2012). Circadian and wakefulness-sleep modulation of cognition in humans. Front. Mol. Neurosci. 5:50. 10.3389/fnmol.2012.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Wells D., Tay J., Mendis D., Abbott M. A., Barnitt A., et al. (1998). CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron 21 1129–1139. 10.1016/s0896-6273(00)80630-3 [DOI] [PubMed] [Google Scholar]
- Wulff K., Gatti S., Wettstein J. G., Foster R. G. (2010). Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat. Rev. Neurosci. 11 589–599. 10.1038/nrn2868 [DOI] [PubMed] [Google Scholar]
- Xia Z., Storm D. (2017). Role of circadian rhythm and REM sleep for memory consolidation. Neurosci. Res. 118 13–20. 10.1016/j.neures.2017.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Ba L., Wang M., Deng S. Y., Chen S. M., Huang L. F., et al. (2020). Chronic sleep fragmentation shares similar pathogenesis with neurodegenerative diseases: Endosome-autophagosome-lysosome pathway dysfunction and microglia-mediated neuroinflammation. CNS Neurosci. Ther. 26 215–227. 10.1111/cns.13218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabut K. C. B., Isoherranen N. (2023). Impact of intracellular lipid binding proteins on endogenous and xenobiotics ligand metabolism and disposition. Drug Metab. Dispos. 51, 700–717. 10.1124/dmd.122.001010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanase H., Shimizu H., Yamada K., Iwanaga T. (2002). Cellular localization of the diazepam binding inhibitor in glial cells with special reference to its coexistence with brain-type fatty acid binding protein. Arch. Histol. Cytol. 65 27–36. 10.1679/aohc.65.27 [DOI] [PubMed] [Google Scholar]
- Young J. K. (2020). Neurogenesis makes a crucial contribution to the neuropathology of Alzheimer’s disease. J. Alzheimers Dis. Rep. 4 365–371. 10.3233/adr-200218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. K., Baker J. H., Muller T. (1996). Immunoreactivity for brain-fatty acid binding protein in gomori-positive astrocytes. Glia 16 218–226. [DOI] [PubMed] [Google Scholar]
- Yurgel M. E., Shah K. D., Brown E. B., Burns C., Bennick R. A., DiAngelo J. R., et al. (2018). Ade2 functions in the Drosophila fat body to promote sleep. G3 (Bethesda) 8 3385–3395. 10.1534/g3.118.200554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. L., Lahens N. F., Yue Z., Arnold D. M., Pakstis P. P., Schwarz J. E., et al. (2021). A circadian clock regulates efflux by the blood-brain barrier in mice and human cells. Nat. Commun. 12:617. 10.1038/s41467-020-20795-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Fang B., Emmett M. J., Damle M., Sun Z., Feng D., et al. (2015). Gene regulation. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348 1488–1492. 10.1126/science.aab3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang J., Ren Y., Lu R., Yang L., Nie G. (2020). Tracing the evolution of fatty acid-binding proteins (FABPs) in organisms with a heterogeneous fat distribution. FEBS Open Biol. 10 861–872. 10.1002/2211-5463.12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Blair D., Bradley J. E. (2013). Phyletic distribution of fatty acid-binding protein genes. PLoS One 8:e77636. 10.1371/journal.pone.0077636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Zhang T., Bloch L. M. (2006). Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci 7:17. 10.1186/1471-2202-7-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yan P., Wang R., Lai J., Tang H., Xiao X., et al. (2023). Opioid-induced fragile-like regulatory T cells contribute to withdrawal. Cell 186 591–606.e3. 10.1016/j.cell.2022.12.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.