Abstract
Anoplophora glabripennis (Coleoptera: Cerambycidae: Lamiinae) is an invasive wood borer pest that has caused considerable damage to forests. Gut bacteria are of great importance in the biology and ecology of herbivores, especially in growth and adaptation; however, change in the gut bacterial community of this pest feeding on different hosts is largely unknown. In this study, we investigated the gut bacterial communities of A. glabripennis larvae fed on different preferred hosts, Salix matsudana and Ulmus pumila, using 16S rDNA high-throughput sequencing technology. A total of 15 phyla, 25 classes, 65 orders, 114 families, 188 genera, and 170 species were annotated in the gut of A. glabripennis larvae fed on S. matsudana or U. pumila using a 97% similarity cutoff level. The dominant phyla were Firmicutes and Proteobacteria and the core dominant genera were Enterococcus, Gibbsiella, Citrobacter, Enterobacter, and Klebsiella. There was significantly higher alpha diversity in the U. pumila group than in the S. matsudana group, and principal co-ordinate analysis showed significant differences in gut bacterial communities between the two groups. The genera with significant abundance differences between the two groups were Gibbsiella, Enterobacter, Leuconostoc, Rhodobacter, TM7a, norank, Rhodobacter, and Aurantisolimonas, indicating that the abundance of larval gut bacteria was affected by feeding on different hosts. Further network diagrams showed that the complexity of the network structure and the modularity were higher in the U. pumila group than in the S. matsudana group, suggesting more diverse gut bacteria in the U. pumila group. The dominant role of most gut microbiota was related to fermentation and chemoheterotrophy, and specific OTUs positively correlated with different functions were reported. Our study provides an essential resource for the gut bacteria functional study of A. glabripennis associated with host diet.
Keywords: Anoplophora glabripennis, intestinal microbial communities, different hosts, 16S rDNA, adaptation
1. Introduction
Gut-associated bacteria are vital mediators of plant and insect interactions (Sugio et al., 2015; Berasategui et al., 2017; Bozorov et al., 2019; Chen et al., 2020; Xiong, 2022). Over a long period of coevolution, insect gut bacteria have acquired the ability to quickly adapt to changes in insect diet (Pérez-Cobas et al., 2015; Su et al., 2017), allowing the insect hosts to adapt to different kinds of food resources (Lv et al., 2021; Yuan et al., 2021; Chen et al., 2022). For instance, the gut microbes derived from Diaphorina citri (Meng et al., 2022), Leptinotarsa decemlineata (Chung et al., 2017), Phthorimaea operculella (Zheng et al., 2020), Henosepilachna vigintioctopunctata (Lü et al., 2019), and Curculio chinensis (Zhang et al., 2020) assist insect hosts in adapting to different host plants by providing nutrients and degrading toxic secondary compounds (Chu et al., 2013; Zhang et al., 2020).
Wood-boring beetles live in a nutrient-poor environment and must cope with large amounts of toxic secondary compounds and plant cell walls mostly comprising macromolecules that are difficult to utilize, such as lignin, cellulose, hemicellulose, and pectin (Mattson, 1980; Geib et al., 2008; Shin et al., 2021). The symbiotic microbial community is involved in insect adaptation to natural stress (Mattson, 1980; Anand et al., 2010; Scully et al., 2013; Ayayee et al., 2014). For example, the gut bacteria of two Apriona species contribute to the degradation of plant toxic secondary compounds (Zhang S. K. et al., 2022). Anoplophora glabripennis larvae harbor a diversity of bacteria in their gut that have putative roles in nutrient provisioning, lignocellulose metabolism, and allelochemical metabolism (Geib et al., 2009; Mason et al., 2019; Wang et al., 2023).
Urban street tree biodiversity plays a crucial role in the function and stability of urban ecosystems (Jim and Zhang, 2013; Zhang and Jim, 2014). Salix matsudana and Ulmus pumila are common urban street trees of high ecological value in the city of Changchun, China (Zhang et al., 2015; Yang et al., 2022a). The Asian long-horned beetle (A. glabripennis) is a serious pest of S. matsudana and U. pumila (Haack et al., 2006; Hu et al., 2009; Faccoli and Favaro, 2016; Liu et al., 2016); it is native to China and Korea and is largely polyphagous on woody broadleaf trees (Wang, 2004; Haack et al., 2006; Van der Gaag and Loomans, 2014; Meng et al., 2015). The female adult mainly feeds on bark and branches to provide essential nutrition for laying eggs. Newly hatched larvae first feed on the phloem, and the second instar larvae bore into the xylem and heartwood to feed (Luo et al., 2003). Larval feeding destroys vascular tissue that surrounds the tree, eventually leading to its death, thereby seriously threatening the consolidation of afforestation efforts and affecting the sustainable development of forestry (Nowak et al., 2001; Haack et al., 2010; Zhou et al., 2021). Host plants carry a distinct nutritional formula that promotes diverse gut environments in insects that will further shape the structural and functional variations in the gut bacteria communities (Zhang J. et al., 2022). However, there are no studies regarding gut bacterial communities of A. glabripennis when reared on S. matsudana and U. pumila host plants. In this study, we used bacterial 16S-rRNA sequencing to characterize the structure of the bacterial community in the gut of A. glabripennis after feeding on two different hosts and to analyze the potential function of this flora. This study aims to assess the effects of host plants on A. glabripennis and to provide a basis for the development of efficient and green control measures for the insect.
2. Materials and methods
2.1. Sample collection
The A. glabripennis larvae feeding in naturally infested S. matsudana and U. pumila were collected in the field in the city of Baoding, Hebei province, China. In August, trees of the two species were selected, with similar diameters at breast height and degrees of damage. The trunks of the two host trees were split and A. glabripennis larvae were removed and, respectively, put into autoclave centrifuge tubes and sealed with absorbent cotton balls. Six replicates were taken for each sample from six different trunks. The larvae were identified as fourth instar according to the head capsule width (Li et al., 2011).
To eliminate the effects of microbial residues on insect epidermis on the diversity of intestinal bacteria, the larvae were soaked in 75% ethanol for 3 min and then washed with sterile water three times before dissecting the gut. During the procedure of dissecting the gut, we first used sterile tweezers to place the larvae on sterile petri dishes (diameter 90 mm) and dissected the whole body under a microscope (LEICA M250C). We then removed the fat inside until the gut was exposed. We finally used sterile tweezers to remove the whole gut of the larvae. The dissected gut was transferred to a 1.5-mL centrifuge tube, frozen with liquid nitrogen, and stored in an ultra-low temperature refrigerator (Li et al., 2021).
2.2. High-throughput sequencing and analysis
Six larvae from each host were dissected for gut microbial community investigation. The DNA of gut bacteria was extracted using MagPure Stool DNA KF kit B (Magen, China) according to the manufacturer’s protocol. The V3–V4 variable regions of 16S rDNA were amplified with the primers 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR cycling conditions were as follows: 94°C for 3 min, 30 cycles of 94°C for 30 s, 56°C for 45 s, 72°C for 45 s, and final extension at 72°C for 10 min. The PCR products were purified with AmpureXP beads and eluted in elution buffer. Libraries were qualified using an Agilent 2100 Bioanalyzer (Agilent, USA). The validated libraries were used for sequencing on an Illumina MiSeq platform (BGI, Shenzhen, China) following the standard Illumina pipelines (Li L. Y. et al., 2022).
2.3. Quality control and operational taxonomic unit (OTU) identification
Raw reads were filtered according to the following criteria: (i) overlap longer than 10 bp and a mismatch rate less than 0.02 were removed and (ii) nucleotide sequences with average quality scores lower than 20 were removed using the sliding window trimming approach. Paired-end reads were merged by the Fast Length Adjustment of Short reads program (FLASH, v1.2.11) (Magoc and Salzberg, 2011). Chimeric sequences and chloroplast and mitochondria sequences were identified and removed using UCHIME (Edgar et al., 2011). All effective assembled reads were clustered into OTUs with a cutoff value of 97% using UPARSE software (v7.0.1090) (Edgar, 2013). The OTU representative sequences were constructed and further annotated based on RDP Classifier (version 11.5) against the 16S rRNA database Silva SSU138.1 using a confidence threshold of 0.8 (Quast et al., 2013).
2.4. Diversity analysis
Alpha and beta diversity were estimated at the OTU level using MOTHUR (Schloss et al., 2009) and UniFrac (Lozupone et al., 2011), respectively. We used R-forge to compare the results of principal coordinates analysis (PCoA). Permutational MANOVA was conducted to further confirm the observed differences (Clarke, 1993), with p < 0.05 considered significant. Statistical analysis of metagenomic profiles (STAMP) was used to assess the differences between OTU groups (Parks et al., 2014; Lü et al., 2019). To understand the relationships among the genera, Spearman’s correlation coefficients were used for network analyses. The network topological properties were calculated using Gephi (Jiang et al., 2017). Functional Annotation of Prokaryotic Taxa (FAPROTAX) was used to predict the function of the microbial community in different samples (Louca et al., 2016). A correlation heatmap was used to analyze OTUs and functional abundance correlation.
3. Results
3.1. 16S rRNA sequencing data quality analysis
The gut bacterial 16S rRNA sequencing of A. glabripennis larvae collected on S. matsudana and U. pumila resulted in 770,189 clean reads. Each sample contained approximately an average of 64,000 reads, with a range of 59,304–66,418. The range of average sequence length was 425.89–429.86 bp (Table 1). The Good’s coverage of each sample was greater than 0.99, indicating that sequencing analysis of 16S rDNA was representative to a certain extent (Table 1). Rarefaction curves suggested that the data covered most of the microbial information in the sample (Figure 1).
TABLE 1.
Diversity and abundance of gut bacteria communities of A. glabripennis larvae reared on S. matsudana and U. pumila.
| Sample | Sequences | Chao | Richness | Shannon | Simpson | ACE | Evenness | Coverage |
| U. pumila_1 | 66280 | 163.83 | 153 | 1.96 | 0.35 | 166.47 | 0.27 | 0.99 |
| U. pumila_2 | 64607 | 76.5 | 51 | 2.01 | 0.31 | 72.39 | 0.35 | 0.99 |
| U. pumila_3 | 63868 | 90.5 | 58 | 2.05 | 0.31 | 103.14 | 0.35 | 0.99 |
| U. pumila_4 | 60720 | 65.13 | 52 | 2.47 | 0.27 | 65.86 | 0.43 | 0.99 |
| U. pumila_5 | 63957 | 90.69 | 80 | 2.16 | 0.29 | 96.42 | 0.34 | 0.99 |
| U. pumila_6 | 66418 | 210.25 | 199 | 3.18 | 0.31 | 202.55 | 0.42 | 0.99 |
| S. matsudana_1 | 65864 | 165.04 | 143 | 1.42 | 0.45 | 170.83 | 0.20 | 0.99 |
| S. matsudana_2 | 65428 | 156.18 | 136 | 1.62 | 0.51 | 163.57 | 0.23 | 0.99 |
| S. matsudana_3 | 59304 | 111 | 104 | 0.86 | 0.82 | 106.95 | 0.13 | 0.99 |
| S. matsudana_4 | 61509 | 194.2 | 169 | 3.13 | 0.18 | 197.73 | 0.42 | 0.99 |
| S. matsudana_5 | 66032 | 120 | 113 | 0.91 | 0.69 | 120.19 | 0.13 | 0.99 |
| S. matsudana_6 | 66202 | 117.75 | 109 | 0.47 | 0.89 | 115.40 | 0.07 | 0.99 |
FIGURE 1.
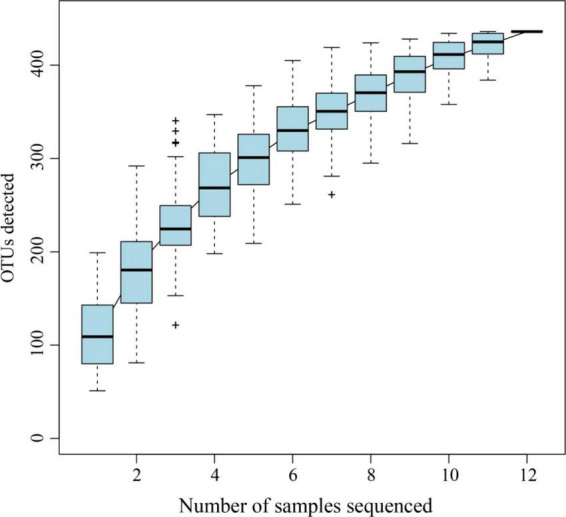
Rarefaction curve of A. glabripennis larvae reared on S. matsudana and U. pumila.
3.2. Composition of A. glabripennis gut bacterial community
A total of 436 OTUs were obtained by clustering with 97% similarity, including 15 phyla, 25 classes, 65 orders, 114 families, 188 genera, and 170 species. There were 152 OTUs unique to the gut of the larvae feeding on S. matsudana (S. matsudana group), 127 OTUs were unique to the gut of larvae feeding on U. pumila (U. pumila group), and 157 OTUs were shared between those feeding on either host (Figure 2A). At the phylum level, Firmicutes and Proteobacteria were dominant bacteria for both plant hosts, accounting for 93.7% and 98.17% of the total sequences (Figure 2B). At the genus level, the top five OTUs of the S. matsudana group were annotated as Enterococcus (55.34%), Gibbsiella (26.83%), Dysgonomonas (5.7%), Citrobacter (5.28%), and Enterobacter (3.82%), while the top four OTUs of the U. pumila group were Gibbsiella (48.54%), Enterococcus (29.92%), Enterobacter (7.84%), and Citrobacter (6.67%) (Figure 2C). Five core gut bacteria genera were present in larvae from both hosts: Enterococcus (52.83%), Gibbsiella (34.01%), Citrobacter (7.84%), Enterobacter (4.73%), and Klebsiella (0.59%) (Figure 2C). Collectively, the results showed that Proteobacteria and Firmicutes were the most abundant phyla for the two hosts, while the dominant genera changed according to the host.
FIGURE 2.
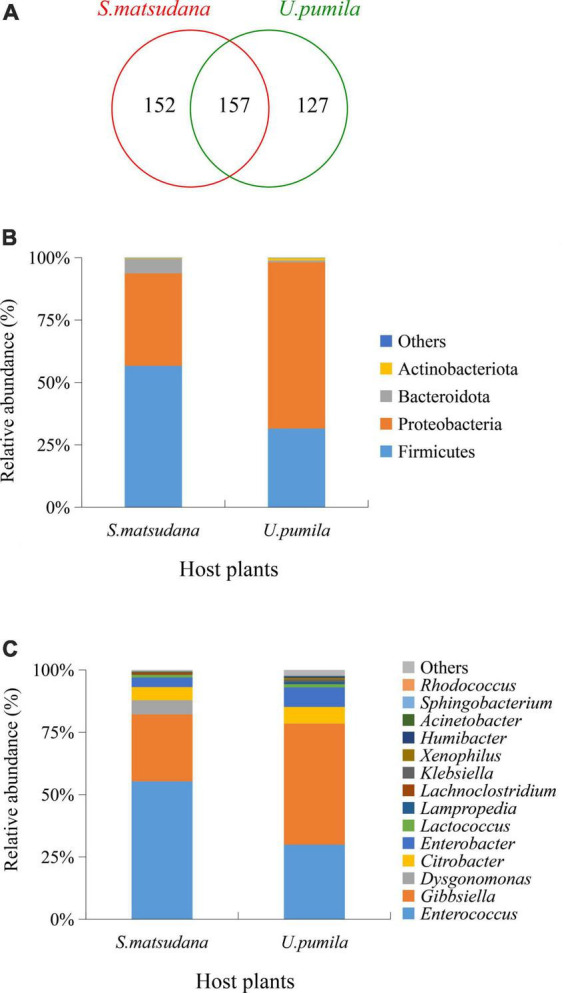
Gut bacteria composition identified in A. glabripennis larvae reared on S. matsudana and U. pumila. Venn diagram represents the number of shared and specific OTUs (A). Relative abundances of bacterial communities at phylum (B) and genus (C) levels for A. glabripennis are illustrated. Columns represent different samples, different colors represent different annotated information, and “others” represents all species except those annotated above.
3.3. Diversity comparison of gut bacteria
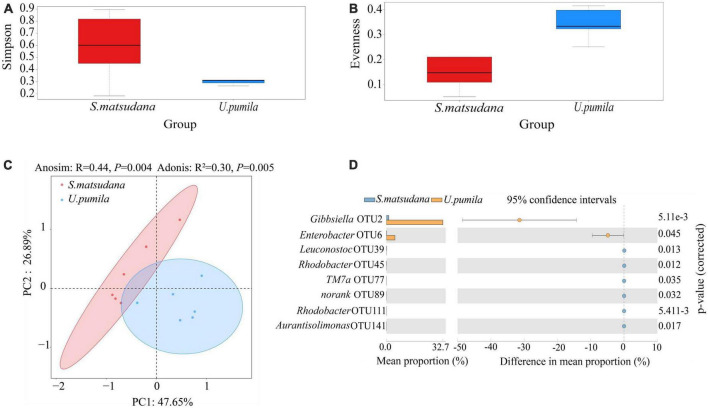
We then compared the diversity of the gut bacterial community derived from the two hosts. The microbial Simpson and Evenness indices of gut bacterial microbiota were significantly higher for the U. pumila group than for the S. matsudana group (Table 1 and Figures 3A, B). The microbial communities obtained from the different hosts were significantly clustered into two different groups in the PCoA plot (Figure 3C), with all three variables together explaining 74.54% of the total variance (Anosim, R = 0.44, P = 0.004; Adonis, R2 = 0.30, P = 0.005).
FIGURE 3.
Diversity of the gut bacteria communities of A. glabripennis larvae reared on S. matsudana and U. pumila via Simpson (A), Evenness (B), PCoA (C), and analysis of differences between groups of OTUs (D).
In order to further understand the microbial communities of A. glabripennis larvae from the two host plants, the OTUs/species with significant differences were compared (Figure 3D). Eight OTUs were identified with significant differences between the samples from the two groups. The relative abundances of Gibbsiella (OTU2) and Enterobacter (OTU6 were greater for the U. pumila group than for the S. matsudana group. The relative abundances of Leuconostoc (OTU39), Rhodobacter (OTU45), TM7a (OTU77), norank (OTU89), Rhodobacter (OTU111), and Aurantisolimonas (OTU141) were higher for the S. matsudana group than for the U. pumila group. Thus, the host plants influenced the intestinal microbial community of A. glabripennis larvae.
3.4. Construction of ecological network of intestinal flora
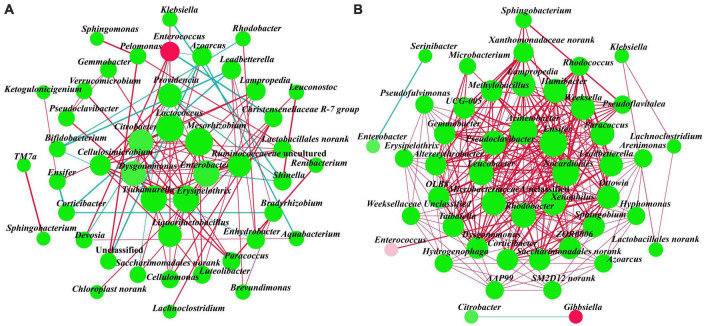
Two association networks were constructed to determine the patterns of gut bacterial communities of A. glabripennis fed with S. matsudana and U. pumila (Figures 4A, B). The network diagram for the S. matsudana group included 45 nodes and 100 edges (85 positive and 15 negative correlations) and the U. pumila group included 47 nodes and 262 edges (260 positive and two negative correlations). This indicated that the complexity of the network structure and the modularity were higher in the U. pumila group than in the S. matsudana group. We found that in the S. matsudana group, the degrees of Lactococcus and Mesorhizobium were 11; Enterobacter, Erysipelothrix, and Tsukamurella were 10; and Dysgonomonas and Citrobacter were 9, and there were positive correlations among them. Degrees equal to or larger than 18 in U. pumila were as follows: Acinetobacter and Ensifer with 23, Nocardioides with 22 and Xenophilus, Rhodobacter, Microbacteriaceae_Un classified, Leucobacter, and Pseudoclavibacter. Xenophilus, Rhodobacter, Microbacteriaceae_Unclassified, Leucobacter, and Pseudoclavibacter with 18. There were positive correlations among these genera. Overall, the results indicated that there were more cooperation and exchange events among most bacterial genera during the adaptation of A. glabripennis larvae to different hosts.
FIGURE 4.
Interaction networks of gut bacteria genera from the S. matsudana (A) and U. pumila (B) groups based on correlation analysis. For each panel, the node represents unique genera, the size of each node represents degree, the red line represents positive correlation, and the blue line represents negative correlation. When a correlation coefficient exceeds 0.6 and p < 0.05, the relationships are kept.
3.5. Functional prediction of gut bacteria
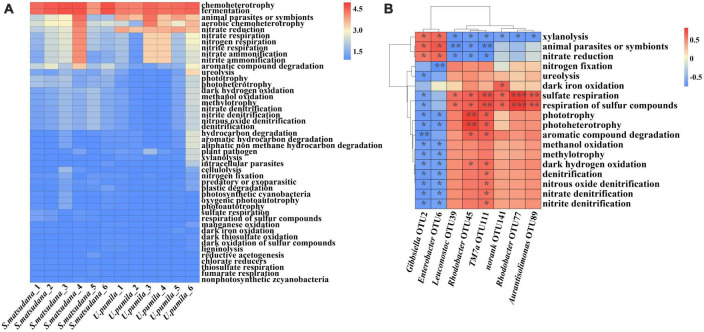
Comparative functional analysis showed similar function patterns among bacterial communities of the two host plants. A total of 46 putative functions were identified from the two host plants. Most OTUs were annotated with fermentation and chemoheterotrophy functions. The other eight main functions were nitrate ammonification, nitrite ammonification, nitrite respiration, nitrate respiration, nitrogen respiration, nitrate reduction, aromatic compound degradation, and aerobic chemoheterotrophy (Figure 5A). To further investigate the relationship between OTUs and functions, the functions with significance that were correlated with specific OTUs were analyzed (Figure 5B). Gibbsiella (OTU2) and Enterobacter (OTU6) were positively correlated with nitrate reduction animal parasites and symbiots and xylanolysis, and the relative abundance of these two OTUs was greater for the U. pumila group than for the S. matsudana group. Leuconostoc (OTU39), Rhodobacter (OTU45), TM7a (OTU77), norank (OTU89), Rhodobacter (OTU111), and Aurantisolimonas (OTU141) were positively correlated with sulfate respiration and respiration of sulfur compounds. Additionally, Rhodobacter (OTU45) was positively correlated with aromatic compound degradation, dark hydrogen oxidation, phototrophy and photoheterotrophy. Aurantisolimonas (OTU141) was positively correlated with dark iron oxidation. Rhodobacter (OTU111) was positively correlated with nitrate denitrification, nitrite denitrification, nitrous oxide denitrification, denitrification, dark hydrogen oxidation, aromatic compound degradation, phototrophy and photoheterotrophy. The relative abundances of Leuconostoc (OTU39), Rhodobacter (OTU45), TM7a (OTU77), norank (OTU89), Rhodobacter (OTU111), and Aurantisolimonas (OTU141) were higher for the S. matsudana group than for the U. pumila group. Overall, the dominant roles of most gut bacteria were related to fermentation and chemoheterotrophy, and OTUs that were positively correlated with different functions were identified.
FIGURE 5.
Functional predictions (A) and the relationship between OTUs and functional abundance (B) of the gut bacteria communities of A. glabripennis larvae reared on S. matsudana and U. pumila. The color scale represents the normalized values of relative abundances by log10 (A), blue indicates negative correlation, red indicates positive correlation, and a darker color indicates a stronger correlation (*p < 0.05, **p < 0.01, ***p < 0.001) (B).
4. Discussion
Gut bacteria in insects play important and diverse roles in host digestion (Jing et al., 2020), detoxification (Blanton and Peterson, 2020; Siddiqui et al., 2022), development (Duplais et al., 2021), pathogen resistance (Khan et al., 2021), immune response (Engel and Moran, 2013), and the production of essential vitamins and amino acids (Bisch et al., 2018). In contrast, the bacterial diversity, structure, or composition of the gut in many insects (Montagna et al., 2015; Li et al., 2021) could be influenced by host diets. In this study, the composition and diversity of the gut bacteria of A. glabripennis from two hosts were studied. The bacterial Simpson and Evenness indices (alpha diversity) were higher for U. pumila larvae than S. matsudana larvae, and beta diversity significantly differed between the two hosts. The abundance of Enterococcus was higher in the S. matsudana group than in the U. pumila group, while Gibbsiella was more abundant in the U. pumila group than in the S. matsudana group. Consistent with previous findings in Spodoptera frugiperda (Jones et al., 2019), Grapholita molesta (Liu et al., 2020), and Cnaphalocrocis medinalis (Yang et al., 2022b) that fed on different food sources, we found high variability in gut bacterial composition and abundance. Moreover, the observed variations in the gut of A. glabripennis larvae could be attributed to the diverse secondary metabolites of the different plants or the different compositions of endophytic bacteria in plants (Afzala et al., 2019; Guerrieri et al., 2019).
Our findings revealed that the bacterial communities of A. glabripennis larvae fed on S. matsudana and U. pumila were similar at the phylum level, consisting of Proteobacteria and Firmicutes, which is consistent with the results of Schloss et al. (2006), who identified the same two abundant bacteria phyla in A. glabripennis. Similar to Wang et al. (2022), who identified that genera Enterococcus and Gibbsiella were dominant in A. glabripennis larvae fed on Populus gansuensis, we found that the same genera were abundant in gut bacteria of A. glabripennis. Enterococcus was recognized as promoting the growth and development of host insects by synthesizing vitamins and amino acids (Sabo et al., 2020; Li C. M. et al., 2022), improving gut immunity (Ennahar et al., 1998; Mohamed and Huang, 2007; Ruiz-Rodriguez et al., 2012; Shao et al., 2017) and degrading carbohydrates and secondary compounds (Dantur et al., 2015; Vilanova et al., 2016; Li C. M. et al., 2022). Thus, we speculated that Enterococcus contributes to the adaptation of wood borer beetles to different hosts. Gibbsiella can degrade lignocellulosic compounds and fix nitrogen (Rizzi et al., 2013). Interestingly, Gibbsiella is present in many wood-boring beetles, such as Apriona germari and Apriona swainsoni (Zhang J. et al., 2022; Zhang S. K. et al., 2022), Agrilus biguttatus (Brown et al., 2015), and Anoplophora chinensis (Rizzi et al., 2013), suggesting that Gibbsiella may play an important role in the adaptation of wood-boring insects to different food sources.
The association network revealed that different hosts influenced the bacterial networks. The higher numbers of network topology properties, such as the number of nodes, positive correlations, negative correlations, and degree observed in the U. pumila group indicated a complex network for this group. This further suggests that compared with S. matsudana, the U. pumila group may be superior at enhancing gut bacterial network complexity and systemic resistance to the external environment. We also found that there were cooperation and exchange events among most bacterial groups. For example, Dysgonomonas and Citrobacter showed positive correlations in the S. matsudana group and have been reported to possess the ability to degrade cellulose (Li X. Y. et al., 2022), indicating that these two genera might work together to improve degradation efficiency. In the U. pumila group, Acinetobacter and Rhodobacter showed positive correlations, and the former is capable of degrading cellulose and hydrocarbons (Pourramezan et al., 2012; Rehman et al., 2018), while the latter can fix nitrogen (Liu et al., 2018; Wang et al., 2019; Xu et al., 2022). In the treatment of rural wastewater in Dianchi Lake, Rhodobacter contributed to biofilm formation and the degradation of pollutants in the early stage of adding microbial carriers, and Acinetobacter contributed to nitrogen removal in the stable stage of the microbial community (Chen et al., 2019). Thus, it seems that there is a cooperative relationship between these two genera. However, there is also competition among gut bacteria (Umu et al., 2017; Cantu-Jungles and Hamaker, 2020). We found a negative correlation between Enterococcus and Christensenellaceae R-7 group in the S. matsudana group, which is consistent with the results of Zhang et al. (2019), who found that the abundance of Christensenellaceae R-7 group in the gut decreased after Enterococcus faecalis was added into the diet of Hy-Line Brown laying hens.
There were 46 functions of gut bacteria predicted and shared between the two hosts. Consistent with the findings for Nilaparvata lugens (Wang et al., 2021), Apriona germari (Zhang J. et al., 2022), and A. glabripennis (Wang et al., 2023), fermentation and chemoheterotrophy functions were annotated as the main functions. These two functions were mainly contributed by the abundance of Proteobacteria and Firmicutes (Ugwu et al., 2022). Similarly, Enterobacter, Klebsiella, and Citrobacter species (belonging to Proteobacteria) are capable of fermentation (Ogilvie et al., 1997; Dong et al., 2020). In addition, Gibbsiella, also a member of Proteobacteria, can ferment glucose (Campos et al., 2014), while glucose can induce lipid accumulation (Li et al., 2015). This indicates that Gibbsiella might participate in fermenting glucose into lipids that further promote the growth and development of A. glabripennis larvae. Citrobacter and Klebsiella can degrade aromatic compounds (Ammar et al., 2005; Kraigher and Mandic-Mulec, 2020; Zhou et al., 2022). Some functions of gut bacteria are related to the nitrogen cycle, which is essential for insect growth and development. Enterobacter, Klebsiella, and Citrobacter are capable of nitrate reduction and/or nitrate respiration (Ogilvie et al., 1997; Dong et al., 2020; Jiang et al., 2022). Citrobacter is commonly associated with denitrification processes (Meng et al., 2019). Furthermore, Enterobacter, Klebsiella, and Citrobacter have been identified as being associated with detoxification (Zhang et al., 2013; Cheng et al., 2017; Francoeur et al., 2020; Li Z. et al., 2022). The relationship of OTUs and functional reanalysis showed that Enterobacter (OTU6) was related to nitrate reduction and xylanolysis, which is consistent with the results of Zhao et al. (2020) and Xia (2014). Leuconostoc (OTU39), Rhodobacter (OTU45), TM7a (OTU77), norank (OTU89), Rhodobacter (OTU111), and Aurantisolimonas (OTU141) were predicted to be positively correlated with nitrogen, sulfur, and hydrogen cycles, indicating that these OTUs were closely related to A. glabripennis growth and development. However, one problem with this study is that all outcomes were from bioinformatic predictions, and the functions of the detected OTUs need to be verified in the future.
5. Conclusion
This study demonstrates that (1) the gut bacterial communities of larvae that fed on S. matsudana differed from those that fed on U. pumila, (2) there were many events of cooperation and communication among gut bacteria, and (3) OTUs that positively correlated with different functions in the adaptation to different hosts were identified. Our study significantly contributes to the understanding of the relationship between gut microbes of A. glabripennis and insect–host plant interactions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA949278 https://www.ncbi.nlm.nih.gov/, SUB12998666.
Author contributions
HL designed the project. XW, HW, and JZ analyzed the data and wrote the manuscript. XW, XS, and ZC performed the experiments. SG, XS, and FZ embellished the picture. All authors contributed to the article and approved the submitted version.
Acknowledgments
Statistical analysis and constructing figures were performed using the Biozeron Cloud Platform (http://www.cloud.biomicroclass.com/CloudPlatform) and Wekemo Bioincloud (http://bioincloud.tech).
Funding Statement
This work was funded by the National Natural Science Foundation of China (Grant No. 32171799) and the Key R&D Plan of Hebei Province (Grant No. 20326514D).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Afzala I., Shinwari Z. K., Sikandar S., Shahzad S. (2019). Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221 36–49. 10.1016/j.micres.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Ammar E., Nasri M., Medhioub K. (2005). Isolation of Enterobacteria able to degrade simple aromatic compounds from the wastewater from olive oil extraction. World J. Microbiol. Biotechnol. 21 253–259. 10.1007/s11274-004-3625-y [DOI] [Google Scholar]
- Anand A. A. P., Vennison S. J., Sankar S. G., Prabhu D. I. G., Vasan P. T., Raghuraman T., et al. (2010). Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect Sci. 10:107. 10.1673/031.010.10701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayayee P., Rosa C., Saunders M., Felton G. W., Ferry G., Hoover K. (2014). Gut microbes contribution to nitrogen provisioning in a wood-feeding Cerambycid. Environ. Entomol. 43 903–912. 10.1603/EN14045 [DOI] [PubMed] [Google Scholar]
- Berasategui A., Salem H., Paetz C., Santoro M., Gershenzon J., Kaltenpoth M., et al. (2017). Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol. Ecol. 26 4099–4110. 10.1111/mec.14186 [DOI] [PubMed] [Google Scholar]
- Bisch G., Neuvonen M. M., Pierce N. E., Russell J. A., Koga R., Sanders J. G., et al. (2018). Genome evolution of Bartonellaceae symbionts of ants at the opposite ends of the trophic scale. Genome Biol Evol. 10 1687–1704. 10.1093/gbe/evy126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton A. G., Peterson B. F. (2020). Symbiont-mediated insecticide detoxification as an emerging problem in insect pests. Front. Microbiol. 11:547108. 10.3389/fmicb.2020.547108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozorov T. A., Rasulov B. A., Zhang D. Y. (2019). Characterization of the gut microbiota of invasive Agrilus mali Matsumara (Coleoptera:Buprestidae) using high-throughput sequencing: Uncovering plant cell-wall degrading bacteria. Sci. Rep. 9:4923. 10.1038/s41598-019-41368-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. D., Inward J., Jeger M., Denman S. (2015). A review of Agrilus biguttatus in UK forests and its relationship with acute oak decline. Forestry 88 53–63. 10.1093/forestry/cpu039 [DOI] [Google Scholar]
- Campos C. R., Mesquita V. A., Silva C. F., Schwan R. F. (2014). Efficiency of physicochemical and biological treatments of vinasse and their influence on indigenous microbiota for disposal into the environment. Waste Manag. 34 2036–2046. 10.1016/j.wasman.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Cantu-Jungles T. M., Hamaker B. R. (2020). New view on dietary fiber selection for predictable shifts in gut microbiot. mBio 11 e2179–19. 10.1128/mBio.02179-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. S., Xie S., Zhang X. C., Zhang N., Feng H. H., Sun C., et al. (2020). Gut microbiota metabolic potential correlates with body size between mulberry-feeding lepidopteran pest species. Pest Manag. Sci. 76 1313–1323. 10.1002/ps.5642 [DOI] [PubMed] [Google Scholar]
- Chen G. K., Huang J., Fang Y., Zhao Y. G., Tain X. P., Jin Y. L., et al. (2019). Microbial community succession and pollutants removal of a novel carriers enhanced duckweed treatment system for rural wastewater in Dianchi lake basin. Bioresour. Technol. 276 8–17. 10.1016/j.biortech.2018.12.102 [DOI] [PubMed] [Google Scholar]
- Chen Y. P., Li Y. H., Sun Z. X., Du E. W., Lu Z. H., Li H., et al. (2022). Effects of host plants on bacterial community structure in larvae midgut of Spodoptera frugiperda. Insects 13:373. 10.3390/insects13040373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D., Guo Z., Riegler M., Xi Z. Y., Liang G. W., Xu Y. J. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5:13. 10.1186/s40168-017-0236-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C. C., Spencer J. L., Curzi M. J., Zavala J. A., Seufferheld M. J. (2013). Gut bacteria facilitate adaptation to crop rotation in the western corn rootworm. Proc. Natl. Acad. Sci. U. S. A. 110 11917–11922. 10.1073/pnas.1301886110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. H., Scully E. D., Peiffer M., Geib S. M., Rosa C., Hoover K., et al. (2017). Host plant species determines symbiotic bacterial community mediating suppression of plant defenses. Sci. Rep. 7:39690. 10.1038/srep39690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R. (1993). Non-parametric multivariate analysis of changes in community structure. Aust. Ecol. 18 117–143. 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- Dantur K. I., Enrique R., Welin B., Castagnaro A. P. (2015). Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express 5:15. 10.1186/s13568-015-0101-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. B., Yao T., Zhang Z. Y., Chen W. H., Liang J. D., Han Y. F., et al. (2020). Structure and function of bacterial microbiota in Eucommia ulmoides bark. Curr. Microbiol. 77 3623–3632. 10.1007/s00284-020-02157-2 [DOI] [PubMed] [Google Scholar]
- Duplais C., Sarou-Kanian V., Massiot D., Hassan A., Perrone B., Estevez Y., et al. (2021). Gut bacteria are essential for normal cuticle development in herbivorous turtle ants. Nat. Commun. 12:676. 10.1038/s41467-021-21065-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial ampliconreads. Nat. Methods 10 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P., Moran N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37 699–735. 10.1111/1574-6976.12025 [DOI] [PubMed] [Google Scholar]
- Ennahar S., Aoude-Werner D., Assobhei O., Hasselmann C. (1998). Antilisterial activity of enterocin 81, a bacteriocin produced by Enterococcus faecium WHE 81 isolated from cheese. J. Appl. Microbiol. 85 521–526. 10.1046/j.1365-2672.1998.853528.x [DOI] [PubMed] [Google Scholar]
- Faccoli M., Favaro R. (2016). Host preference and host colonization of the Asian long-horned beetle, Anoplophora glabripennis (Coleoptera Cerambycidae), in Southern Europe. Bull. Entomol. Res. 106 359–367. 10.1017/S0007485315001157 [DOI] [PubMed] [Google Scholar]
- Francoeur C. B., Khadempour L., Moreira-Soto R. D., Gotting K., Book A. J., Pinto-Tomás A. A., et al. (2020). Bacteria contribute to plant secondary compound degradation in a generalist herbivore system. mBio 11 e2146–20. 10.1128/mBio.02146-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geib S. M., Filley T. R., Hatcher P. G., Hoover K., Carlson J. E., Jimenez-Gasco M. D. M., et al. (2008). Lignin degradation in wood-feeding insects. Proc. Natl. Acad. Sci. U. S. A. 105 12932–12937. 10.1073/pnas.0805257105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geib S. M., Jimenez-Gasco M. M., Carlson J. E., Tien M., Hoover K. (2009). Effect of host tree species on cellulase activity and bacterial community composition in the gut of larval Asian longhorned beetle. Environ. Entomol. 38 686–699. 10.1603/022.038.0320 [DOI] [PubMed] [Google Scholar]
- Guerrieri A., Dong L., Bouwmeester H. J. (2019). Role and exploitation of underground chemical signaling in plants. Pest Manag. Sci. 75 2455–2463. 10.1002/ps.5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack R. A., Bauer L. S., Gao R. T., McCarthy J. J., Miller D. L., Petrice T. R., et al. (2006). Anoplophora glabripennis with in tree distribution, seasonal development, and host suitability in China and Chicago. Great Lakes Entomol. 39 169–183. [Google Scholar]
- Haack R. A., Hérard F., Sun J. H., Turgeon J. J. (2010). Managing invasive populations of Asian longhorned beetle and citrus longhorned beetle: A worldwide perspective. Annu. Rev. Entomol. 55 521–546. 10.1146/annurev-ento-112408-085427 [DOI] [PubMed] [Google Scholar]
- Hu J. F., Angeli S., Schuetz S., Luo Y. Q., Hajek A. E. (2009). Ecology and management of exotic and endemic Asian longhorned beetle Anoplophora glabripennis. Agric. For. Entomol. 11 359–375. 10.1111/j.1461-9563.2009.00443.x [DOI] [Google Scholar]
- Jiang Y., Li S., Li R., Zhang J., Liu Y., Lü L., et al. (2017). Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 109 145–155. 10.1016/j.soilbio.2017.02.010 [DOI] [Google Scholar]
- Jiang Z., Shen X., Shi B., Cui M., Wang Y., Li P. (2022). Arsenic mobilization and transformation by ammonium-generating bacteria isolated from high arsenic groundwater in hetao plain, China. Int. J. Environ. Res. Public Health 19:9606. 10.3390/ijerph19159606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jim C. Y., Zhang H. (2013). Species diversity and spatial differentiation of old-valuable trees in urban Hong Kong. Urban For. Urban Green. 12 171–182. 10.1016/j.ufug.2013.02.001 [DOI] [Google Scholar]
- Jing T. Z., Qi F. H., Wang Z. Y. (2020). Most dominant roles of insect gut bacteria: Digestion, detoxification, or essential nutrient provision? Microbiome 8:38. 10.1186/s40168-020-00823-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. G., Mason C. J., Felton G. W., Hoover K. (2019). Host plant and population source drive diversity of microbial gut communities in two polyphagous insects. Sci. Rep. 9:2792. 10.1038/s41598-019-39163-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I., Bai Y. R., Zha L. J., Ullah N., Ullah H., Shah S. R. H., et al. (2021). Mechanism of the gut microbiota colonization resistance and enteric pathogen infection. Front. Cell. Infect. Microbiol. 11:716299. 10.3389/fcimb.2021.716299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraigher B., Mandic-Mulec I. (2020). Influence of diclofenac on activated sludge bacterial communities in fed-batch reactors. Food Technol. Biotechnol. 58 402–410. 10.17113/ftb.58.04.20.6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. M., Han G. J., Sun J., Huang L. X., Lu Y. R., Xia Y., et al. (2022). The gut microbiota composition of Cnaphalocrocis medinalis and their predicted contribution to larval nutrition. Front. Microbiol. 13:909863. 10.3389/fmicb.2022.909863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Y., Han J., Wu L., Fang C., Li W. G., Gu J. M., et al. (2022). Alterations of gut microbiota diversity, composition and metabonomics in testosterone-induced benign prostatic hyperplasia rats. Mil. Med. Res. 9:12. 10.1186/s40779-022-00373-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., Mei C., Luo X. Y., Wulamu D., Zhan S., Huang Y. P., et al. (2022). Dynamics of the intestinal bacterial community in black soldier fly larval guts and its influence on insect growth and development. Insect Sci. 10.1111/1744-7917.13095 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Li Z., Gao J., Tian Y., Wang B., Xu J., Fu X., et al. (2022). ElNFS1, a nitroreductase gene from Enterobacter ludwigii, confers enhanced detoxification and phytoremediation of 4-nitrobenzaldehyde in rice. Environ. Pollut. 314:120292. 10.1016/j.envpol.2022.120292 [DOI] [PubMed] [Google Scholar]
- Li H. W., Zhao C. W., Yang Y., Zhou Z. X., Qi J. W., Li C. R. (2021). The influence of gut microbiota on the fecundity of Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae). J. Insect Sci. 21:15. 10.1093/jisesa/ieab061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li M. S., Zheng L. Y., Liu Y. S., Zhang Y. L., Yu Z. N., et al. (2015). Simultaneous utilization of glucose and xylose for lipid accumulation in black soldier fly. Biotechnol. Biofuels. 8:117. 10.1186/s13068-015-0306-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. J., Luo Y. Q., Yan X. F., Tian G. F., Nian Y. J., Sun H. (2011). Cellulase in larvae of Asian longhorn beetle Anoplophora glabripennis. Sci. Silv. Sin. 47 204–207. [Google Scholar]
- Liu H. P., Bauer L. S., Zhao T. H., Gao R. T., Poland T. M. (2016). Seasonal abundance and development of the Asian longhorned beetle and natural enemy prevalence in different forest types in China. Biol. Control 103 154–164. 10.1016/j.biocontrol.2016.08.010 [DOI] [Google Scholar]
- Liu J. X., Li C., Jing J. H., Zhao P. Y., Luo Z. M., Cao M. W., et al. (2018). Ecological patterns and adaptability of bacterial communities in alkaline copper mine drainage. Water Res. 133 99–109. 10.1016/j.watres.2018.01.014 [DOI] [PubMed] [Google Scholar]
- Liu Y. J., Shen Z. J., Yu J. M., Li Z., Liu X. X., Xu H. L. (2020). Comparison of gut bacterial communities and their associations with host diets in four fruit borers. Pest Manag. Sci. 76 1353–1362. 10.1002/ps.5646 [DOI] [PubMed] [Google Scholar]
- Louca S., Parfrey L. W., Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353 1272–1277. 10.1126/science.aaf4507 [DOI] [PubMed] [Google Scholar]
- Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R. (2011). UniFrac: An effective distance metric for microbial community comparison. ISME J. 5 169–172. 10.1038/ismej.2010.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü J., Guo W., Chen S., Guo M., Qiu B., Yang C., et al. (2019). Host plants influence the composition of the gut bacteria in Henosepilachna vigintioctopunctata. PLoS One 14:e0224213. 10.1371/journal.pone.0224213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. Q., Wen J. B., Xu Z. C. (2003). “Poplar longhorned beetle,” in Major Forest Disease and Insect Pests in China, eds Zhang X., Luo Y. (China: Forest Press; ). [Google Scholar]
- Lv D. B., Liu X. Y., Dong Y. L., Yan Z. Z., Zhang X., Wang P., et al. (2021). Comparison of gut bacterial communities of fall armyworm (Spodoptera frugiperda) reared on different host plants. Int. J. Mol. Sci. 22:11266. 10.3390/ijms222011266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason C. J., Campbell A. M., Scully E. D., Hoover K. (2019). Bacterial and fungal midgut community dynamics and transfer between mother and brood in the Asian longhorned beetle (Anoplophora glabripennis), an invasive xylophage. Microb. Ecol. 77 230–242. 10.1007/s00248-018-1205-1 [DOI] [PubMed] [Google Scholar]
- Mattson W. J. (1980). Herbivory in relation to plant nitrogen content. Annu. Rev. Ecol. Evol. Sci. 11 119–161. 10.1146/ANNUREV.ES.11.110180.001003 [DOI] [Google Scholar]
- Meng F. Q., Bar-Shmuel N., Shavit R., Behar A., Segoli M. (2019). Gut bacteria of weevils developing on plant roots under extreme desert conditions. BMC Microbiol. 19:311. 10.1186/s12866-019-1690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L. X., Xia C. X., Jin Z. X., Zhang H. Y. (2022). Investigation of gut bacterial communities of Asian Citrus Psyllid (Diaphorin acitri) reared on different host plants. Insects 13:694. 10.3390/insects13080694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng P. S., Hoover K., Keena M. A. (2015). Asian longhorned beetle (Coleoptera: Cerambycidae), an introduced pest of maple and other hardwood trees in North America and Europe. J. Integr. Pest Manag. 6 84–88. 10.1093/jipm/pmv003 [DOI] [Google Scholar]
- Mohamed J. A., Huang D. B. (2007). Biofilm formation by Enterococci. J. Med. Microbiol. 56 1581–1588. 10.1099/jmm.0.47331-0 [DOI] [PubMed] [Google Scholar]
- Montagna M., Chouaia B., Mazza G., Prosdocimi E. M., Crotti E., Mereghetti V., et al. (2015). Effects of the diet on the microbiota of the red palm weevil (Coleoptera: Dryophthoridae). PLoS One 10:e0117439. 10.1371/journal.pone.0117439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D. J., Pasek J. E., Sequeira R. A., Crane D. E., Mastro V. C. (2001). Potential effect of Anoplophora glabripennis (Coleoptera: Cerambycidae) on urban trees in the United States. J. Econ. Entomol. 94 116–122. 10.1603/0022-0493-94.1.116 [DOI] [PubMed] [Google Scholar]
- Ogilvie B. G., Rutter M., Nedwell D. B. (1997). Selection by temperature of nitrate-reducing bacteria from estuarine sediments: Species composition and competition for nitrate. FEMS Microbiol. Ecol. 23 11–22. 10.1016/S0168-6496(97)00004-4 [DOI] [Google Scholar]
- Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30 3123–3124. 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cobas A. E., Maiques E., Angelova A., Carrasco P., Moya A., Latorre A. (2015). Diet shapes the gut microbiota of the omnivorous cockroach Blattella germanica. FEMS Microbiol. Rev. 91:fiv022. 10.1093/femsec/fiv022 [DOI] [PubMed] [Google Scholar]
- Pourramezan Z., Ghezelbash G. R., Romani B., Ziaei S., Hedayatkhah A. (2012). Screening and identification of newly isolated cellulose-degrading bacteria from the gut of xylophagous termite Microcerotermes diversus (Silvestri). Microbiology 81 736–742. 10.1134/S0026261712060124 [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman K., Imran A., Amin I., Afzal M. (2018). Inoculation with bacteria in floating treatment wetlands positively modulates the phytoremediation of oil field waste water. J. Hazard. Mater. 349 242–251. 10.1016/j.jhazmat.2018.02.013 [DOI] [PubMed] [Google Scholar]
- Rizzi A., Crotti E., Borruso L., Jucker C., Lupi D., Colombo M., et al. (2013). Characterization of the bacterial community associated with larvae and adults of Anoplophora chinensis collected in Italy by culture and culture-independent methods. Biomed. Res. Int. 2013:420287. 10.1155/2013/420287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Rodriguez M., Valdivia E., Martin-Vivaldi M., Martin-Platero A. M., Martinez-Bueno M., Mendez M., et al. (2012). Antimicrobial activity and genetic profile of Enteroccoci isolated from hoopoes uropygial gland. PLoS One 7:e41843. 10.1371/journal.pone.0041843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo S. D., Mendes M. A., Araujo E. D., Muradian L. B. D., Makiyama E. N., LeBlanc J. G., et al. (2020). Bioprospecting of probiotics with antimicrobial activities against Salmonella Heidelberg and that produce B-complex vitamins as potential supplements in poultry nutrition. Sci. Rep. 10:7235. 10.1038/s41598-020-64038-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D., Delalibera I., Handelsman J., Raffa K. F. (2006). Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae). Environ. Entomol. 35 625–629. 10.1603/0046-225X-35.3.625 33044624 [DOI] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E. D., Geib S. M., Hoover K., Tien M., Tringe S. G., Barry K. W., et al. (2013). Metagenomic profiling reveals lignocellulose degrading system in a microbial community associated with a wood-feeding beetle. PLoS One 8:e73827. 10.1371/journal.pone.0073827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Chen B., Sun C., Ishida K., Hertweck C., Boland W. (2017). Symbiont-derived antimicrobials contribute to the control of the Lepidopteran gut microbiota. Cell Chem. Biol. 24 66–75. 10.1016/j.chembiol.2016.11.015 [DOI] [PubMed] [Google Scholar]
- Shin N. R., Shin S., Okamura Y., Kirsch R., Lombard V., Svacha P., et al. (2021). Larvae of longhorned beetles (Coleoptera; Cerambycidae) have evolved a diverse and phylogenetically conserved array of plant cell wall degrading enzymes. Syst. Entomol. 46 784–797. 10.1111/syen.12488 [DOI] [Google Scholar]
- Siddiqui J. A., Khan M. M., Bamisile B. S., Hafeez M., Qasim M., Rasheed M. T., et al. (2022). Role of insect gut microbiota in pesticide degradation: A review. Front. Microbiol. 13:870462. 10.3389/fmicb.2022.870462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L. J., Yang L. L., Huang S., Li Y., Su X. Q., Wang F. Q., et al. (2017). Variation in the gut microbiota of termites (Tsaitermes ampliceps) against different diets. Appl. Biochem. Biotechnol. 181 32–47. 10.1007/s12010-016-2197-2 [DOI] [PubMed] [Google Scholar]
- Sugio A., Dubreuil G., Giron D., Simon J. C. (2015). Plant-insect interactions under bacterial influence: Ecological implications and underlying mechanisms. J. Exp. Bot. 66 467–478. 10.1093/jxb/eru435 [DOI] [PubMed] [Google Scholar]
- Ugwu J. A., Ren W. Z., Asiegbu F. O. (2022). Monocot diet sources drive diversity of gut bacterial communities in Spodoptera frugiperda (Lepidoptera: Noctuidae) larvae. J. Appl. Entomol. 146 942–956. 10.1111/jen.13022 [DOI] [Google Scholar]
- Umu Ö. C. O., Rudi K., Diep D. B. (2017). Modulation of the gut microbiota by prebiotic fibres and bacteriocins. Microb. Ecol. Health Dis. 28:1348886. 10.1080/16512235.2017.1348886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Gaag D. J., Loomans A. J. M. (2014). Host plants of Anoplophora glabripennis, a review. Bull. OEPP 44 518–528. 10.1111/epp.12151 [DOI] [Google Scholar]
- Vilanova C., Baixeras J., Latorre A., Porcar M. (2016). The generalist inside the specialist: Gut bacterial communities of two insect species feeding on toxic plants are dominated by Enterococcus sp. Front. Microbiol. 7:1005. 10.3389/fmicb.2016.01005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. X., Li C. C., Wang X., Wang G. J., Shang S. Q., Dou Z. P., et al. (2022). Gut lignocellulose activity and microbiota in Asian longhorned beetle and their predicted contribution to larval nutrition. Front. Microbiol. 13:899865. 10.3389/fmicb.2022.899865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. F., Wang H. L., Su X. Y., Zhang J., Bai J. W., Zeng J. Y., et al. (2023). Dynamic changes of gut bacterial communities present in larvae of Anoplophora glabripennis collected at different developmental stages. Arch. Insect Biochem. Physiol. 112:e21978. 10.1002/arch.21978 [DOI] [PubMed] [Google Scholar]
- Wang Y. Q., Fan L. J., Huang J. T., Liang J. J., Wang X. N., Ren Y. C., et al. (2019). Evaluation of chemical composition, antioxidant activity, and gut microbiota associated with pumpkin juice fermented by Rhodobacter sphaeroides. Food Chem. 401:134122. 10.1016/j.foodchem.2022.134122 [DOI] [PubMed] [Google Scholar]
- Wang Z. G. (2004). Study on the occurrence dynamics of Anoplophora glabripennis (Coleoptera: Cerambycidae) and its control measures. Ph.D. thesis. China: Northeast Forestry University. [Google Scholar]
- Wang Z. L., Pan H. B., Wu W., Li M. Y., Yu X. P. (2021). The gut bacterial fora associated with brown planthopper is afected by host rice varieties. Arch. Microbiol. 203 325–333. 10.1007/s00203-020-02013-8 [DOI] [PubMed] [Google Scholar]
- Xia X. F. (2014). Organizational diversity and functional characterization of microbiota in the midgut of diamondback moth, Plutella xylostella (L.). Ph.D. thesis. China: Fujian Agriculture and Forestry University. [Google Scholar]
- Xiong W. H. (2022). Intestinal microbiota in various animals. Integr. Zool. 17 331–332. 10.1111/1749-4877.12633 [DOI] [PubMed] [Google Scholar]
- Xu Y. K., Li L. S., Lou S., Tian J. S., Sun S. H., Li X. D., et al. (2022). Effects of nano-aerators on microbial communities and functions in the water, sediment, and shrimp intestine in Litopenaeus vannamei aquaculture ponds. Microorganisms 10:1302. 10.3390/microorganisms10071302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sheng X. W., Zhai C., Wang Z. H., Wu J. J., Zhang D. (2022a). Species composition and diversity of middle-aged trees among different urban green space types and tree age classes in Changchun, Northeast China. Forests 13:1997. 10.3390/f13121997 [DOI] [Google Scholar]
- Yang Y., Liu X. G., Xu H. X., Liu Y. H., Lu Z. X. (2022b). Effects of host plant and insect generation on shaping of the gut microbiota in the rice leaffolder, Cnaphalocrocis medinalis. Front. Microbiol. 13:824224. 10.3389/fmicb.2022.824224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X. Q., Zhang X., Liu X. Y., Dong Y. L., Yan Z. Z., Lv D. B., et al. (2021). Comparison of gut bacterial communities of Grapholita molesta (Lepidoptera: Tortricidae) reared on different host plants. Int. J. Mol. Sci. 22:6843. 10.3390/ijms22136843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zheng H. F., He X. Y., Ren Z. B., Zhai C., Yu X. Y., et al. (2015). Effects of forest type and urbanization on species composition and diversity of urban forest in Changchun. Northeast China. Urban Ecosyst. 19 455–473. 10.1007/s11252-015-0473-5 [DOI] [Google Scholar]
- Zhang D. X., Ong Y. L., Li Z., Wu J. C. (2013). Biological detoxification of furfural and 5-hydroxyl methyl furfural in hydrolysate of oil palm empty fruit bunch by Enterobacter sp. FDS8. Biochem. Eng. J. 77 77–82. 10.1016/j.bej.2013.01.003 [DOI] [Google Scholar]
- Zhang H., Jim C. Y. (2014). Contributions of landscape trees in public housing estates to urban biodiversity in Hong Kong. Urban For. Urban Green 13 272–284. 10.1016/j.ufug.2013.12.009 [DOI] [Google Scholar]
- Zhang J., Wang H., Su X., Wang X., Yang M., Bai J., et al. (2022). Similar gut bacteria composition in Apriona germarion two preferred host plants. Arch. Insect Biochem. Physiol. 10:e21899. 10.1002/arch.21899 [DOI] [PubMed] [Google Scholar]
- Zhang S. K., Wang Y., Li Z. K., Xue H. J., Zhou X. D., Huang J. H. (2022). Two Apriona species sharing a host niche have different gut microbiome diversity. Mol. Ecol. 83 1059–1072. 10.1007/s00248-021-01799-4 [DOI] [PubMed] [Google Scholar]
- Zhang S. K., Shu J. P., Xue H. J., Zhang W., Zhang Y. B., Liu Y. N., et al. (2020). The gut microbiota in Camellia weevils are influenced by plant secondary metabolites and contribute to saponin degradation. mSystems 5 e692–19. 10.1128/mSystems.00692-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. W., Ma W. F., Zhang Z. D., Liu F. Y., Wang J., Yin Y. L., et al. (2019). Effects of Enterococcus faecalis on egg production, egg quality and caecal microbiota of hens during the late laying period. Arch. Anim. Nutr. 73 208–221. 10.1080/1745039x.2019.1591128 [DOI] [PubMed] [Google Scholar]
- Zhao B., Dan Q., Guo L. J., An Q., Guo J. S. (2020). Characterization of an aerobic denitrifier Enterobacter cloacae strain HNR and its nitrate reductase gene. Arch. Microbiol. 202 1775–1784. 10.1007/s00203-020-01887-y [DOI] [PubMed] [Google Scholar]
- Zheng Y. Q., Xiao G. L., Zhou W. W., Gao Y. L., Li Z. Y., Du G. Z., et al. (2020). Midgut microbiota diversity of potato tuber moth associated with potato tissue consumed. BMC Microbiol. 20:58. 10.1186/s12866-020-01740-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Zhang X. D., Yu L. F., Ren L. L., Luo Y. Q. (2021). Combining WV-2 images and tree physiological factors to detect damage stages of Populus gansuensis by Asian longhorned beetle (Anopolophora glabripennis) at the tree level. For. Ecosyst. 8:35. 10.21203/rs.3.rs-226465/v1 [DOI] [Google Scholar]
- Zhou X. R., Lei D., Tang J., Wu M., Ye H., Zhang Q. (2022). Whole genome sequencing and analysis of fenvalerate degrading bacteria Citrobacter freundii CD-9. AMB Express 12:51. 10.1186/s13568-022-01392-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA949278 https://www.ncbi.nlm.nih.gov/, SUB12998666.