Abstract
KAI1/CD82, a membrane tetraspanin protein, can prevent various cancers and retinal disorders through its anti-angiogenic and anti-metastatic capacity. However, little is known about its anti-inflammatory effect and molecular mechanism. Therefore, the present study aimed to inLPSvestigate effect of a recombinant protein of the large extracellular domain of human KAI1 (Gly 111-Leu 228, rhKAI1) on lipopolysaccharides (LPS)-stimulated RAW264.7 macrophage-like cells and mouse bone marrow-derived macrophages (BMDM) and to identify its underlying mechanism. Our data showed that rhKAI1 suppressed expression levels of classically macrophages (M1) phenotype-related surface markers F4/80+CD86+ in LPS-stimulated BMDM and RAW264.7 cells. In addition, LPS markedly increased mRNA expression and release levels of pro-inflammatory cytokines and mediators such as interleukin (IL)-1β, IL-6, tumor necrosis factor-α, cyclooxygenase-2, nitric oxide and prostaglandin E2, whereas these increases were substantially down-regulated by rhKAI1. Furthermore, LPS strongly increased expression of NF-κB p65 in the nuclei and phosphorylation of ERK, JNK, and p38 MAPK. However, nuclear translocation of NF-κB p65 and phosphorylation of JNK were greatly reversed in the presence of rhKAI1. Especially, rhKAI1 markedly suppressed expression of toll-like receptor (TLR4) and prevented binding of LPS with TLR4 through molecular docking predict analysis. Importantly, Glu 214 of rhKAI1 residue strongly interacted with Lys 360 of TLR4 residue, with a binding distance of 2.9 Å. Taken together, these findings suggest that rhKAI1 has an anti-inflammatory effect on LPS-polarized macrophages by interacting with TLR4 and down-regulating the JNK/NF-κB signaling pathway.
Keywords: Inflammation, JNK/NF-κB signaling pathway, KAI1, M1 macrophage polarization, TLR4
INTRODUCTION
Macrophages can act as effector defensive cells against pathogens and play a key regulator role in innate and adaptive immune responses (1). Under the influence of various environmental signals, macrophages present two functionally distinct phenotypes, classically activated macrophages (M1) and alternatively activated macrophages (M2) (2). M1-like macrophages are polarized by lipopolysaccharide (LPS) either alone or together with T helper type 1 (Th1) cytokines such as interferon gamma, granulocyte-macrophage colony-stimulating factor (GM-CSF) to promote T helper type 1 (Th1) response (3, 4). M1 macrophages are mainly involved in pro-inflammatory responses characterized by production of pro-inflammatory and Th1 cytokines including tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, and chemokines (3). In contrast, M2-like polarized macrophages contribute to tissue remodeling, repair, angiogenesis, and homeostasis through Th2 immune response (5). M2 macrophages are involved in anti-inflammatory responses by producing Th2 cytokines (such as IL-4 and IL-10) and transforming growth factor beta (TGF-β) (6). In this regard, numerous studies have reported that imbalance of macrophage polarization contributes to inflammation and dysregulation of the immune system, affecting various diseases such as obesity, cancer, and rheumatoid arthritis (4, 7, 8). Therefore, modulating the activation state of macrophages might be one of the effective therapeutic strategies.
KAI1, also known as CD82, is a protein belonging to the tetraspanin protein family. It has four transmembrane domains, two extracellular loops, a very small intracellular loop, and short N‐ and C‐terminal cytoplasmic tails (9, 10). KAI1 can interact with integrins, epidermal growth factor receptor, protein kinase C, Src kinase, and small GTPase. It is involved in cancer progression through regulation of cellular events including cell adhesion, migration, and survival (9, 10). Several tetraspains including KAI1 can interact with pattern recognition receptors such as C-type lectin receptors, TLRs, and Fc receptors (11, 12). In particular, both macrophages and dendritic cells can specifically recruit KAI1 to phagosomes upon phagocytosis of various fungi (13). In 2019, Tam et al. suggested that a lack of KAI1 reduced macrophage fungal killing and its single-nucleotide polymorphisms modulated cytokine production (11). Recently, it has been reported that KAI1 can restrain phagocyte migration in inflammation murine models and that it is required for normal morphology and activation of M2 macrophages (14). Furthermore, several studies have demonstrated that the large extracellular loop (LEL) of KAI1 is necessary for biological activities of tetraspanins with several pharmaceutical benefits (15-17). In 2021, Lee et al. demonstrated that KAI1 could directly bind to vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF) and block their signaling mechanism, ultimately having an anti-angiogenic effect (15). They also reported that recombinant human LEL protein of KAI1 (rhKAI1) could suppress tumor metastasis in B16 melanoma cells-incorporated mice and prevent neovascularization in oxygen-induced retinopathy mice. More recently, we have uncovered that rhKAI1 could partially interact with TGF-β binding sites of TGF receptor I/II, integrin β1, and integrin αv (16). Additionally, we have found that rhKAI1 could suppress TGF-β-induced epithelial-mesenchymal transition (EMT) of retinal pigment epithelial cells by blocking Smad signaling pathway (16). Although several studies have suggested that KAI1 could prevent various cancers and retinal disorders through its anti-angiogenic and anti-metastatic capacity, few studies have reported effects of KAI1 on macrophage polarization and its potential anti-inflammatory efficacy. Therefore, the present study aimed to determine effects of rhKAI1 on M1 polarized inflammatory response and identify the underlying molecular mechanism using LPS-stimulated mouse RAW 264.7 macrophage-like cells and primary bone marrow-derived macrophages (BMDM).
RESULTS AND DISCUSSION
rhKAI1 suppresses expression of M1 macrophage phenotype-related surface markers in LPS-stimulated BMDM and RAW264.7 cells
Cytotoxic effects of rhKAI1 on primary BMDM and RAW 264.7 cells were examined with a WST-1 assay. At concentrations up to 800 ng/ml, rhKAI1 had no cytotoxic effect on BMDM or RAW 264.7 cells with or without LPS stimulation (Data not shown). Furthermore, a cell morphology comparison showed that both unstimulated-BMDM and -RAW 264.7 cells had a round shape, whereas LPS-stimulated cells had an irregular shape with polygonal spindle-shaped pseudopodia (Fig. 1A). However, such morphological change induced by LPS was diminished by rhKAI1 treatment in RAW264.7 cells, while no apparent change was observed in BMDM cells. To investigate effects of rhKAI1 on expression levels of M1 macrophage phenotype-related surface markers in LPS-stimulated condition, populations of F4/80+CD86+ cells and F4/80+CD80+ cells were examined by flow cytometry. Results of flow cytometry indicated that 100 ng/ml LPS significantly increased F4/80+CD86+ cells to 4.60-fold of those in the control BMDM. However, such increase in the population of LPS-induced F4/80+CD86+ cells was markedly decreased by rhKAI1 pre-treatment in a concentration-dependent manner (Fig. 1B-D). In addition, the population of F4/80+CD86+ and F4/80+CD80+ cells was substantially enhanced by LPS treatment to 90.47-fold of that in control RAW 264.7 cells, while such increase was significantly suppressed by pretreatment with rhKAI1 (Fig. 1E, F). Quantitative mean fluorescence intensity results were similar to results of population analysis using M1 surface markers (Fig. 1G). Results of morphological observation and flow cytometry for F4/80+CD86+ cells and F4/80+CD80+ cells in BMDM and RAW264.7 cells revealed that RAW 264.7 cells were more sensitive to rhKAI1 than BMDM. Therefore, RAW264.7 cells were selected and used in subsequent experiments to investigate the protective effect of rhKAI1 on LPS-induced M1 polarization. To re-verify the inhibitory effect of rhKAI1 on the increase of F4/80+CD86+ cells induced by LPS treatment, we carried out an immunofluorescence analysis for CD86 in RAW 264.7 cells. As shown in Fig. 1H, the expression of CD86, a surface marker of M1 macrophage polarization, was markedly increased by LPS stimulation. However, such increase of CD86 induced by LPS was noticeably suppressed in the presence of rhKAI1, consistent with results obtained by flow cytometry.
Fig. 1.
rhKAI1 reduces the expression of LPS-stimulated M1 macrophage phenotype-related surface markers in primary mouse bone marrow-derived macrophages (BMDM) and RAW 264.7 mouse macrophage-like cells. Cells were stimulated with various concentrations of rhKAI1 for 24 h or pre-treated with or without rhKAI1 for 2 h, following by 100 ng/ml LPS treatment for 24 h. (A) Representative microscopy images of the cell. Cell morphology was observed using an inverted-phage contrast microscope, and acquired at 100× magnification. (B) Representative flow cytometry plots of F4/80+CD86+ in BMDM cells. (C) Quantification for M1 phenotype are displayed as the percentages of F4/80+CD86+ cells from CD11b-PE-gated cells in BMDM. (D) Quantitative mean fluorescence intensity (MFI) of F4/80+CD86+ cells in BMDM. (E) Representative flow cytometry plots of F4/80+CD86+ and F4/80+CD80+ in RAW 264.7 cells. (F) Quantification for M1 phenotype are displayed as the percentages of F4/80+CD86+ cells and F4/80+CD80+ cells from total live-gated cells in RAW 264.7 cells. (G) Quantitative MFI of F4/80+CD86+ cells and F4/80+CD80+ cells in RAW264.7 cells. Data are expressed as the mean ± SD (n = 5). ***P < 0.001 compared to control. #P < 0.05, ##P < 0.01 and ###P < 0.001 compared to LPS-stimulated cells. (H) Representative images were acquired using a confocal laser scanning microscopy. Confocal images show the expression of CD86 (green) and DAPI (blue). Scale bar = 50 μm.
rhKAI1 inhibits LPS-induced inflammatory response in RAW 264.7 cells
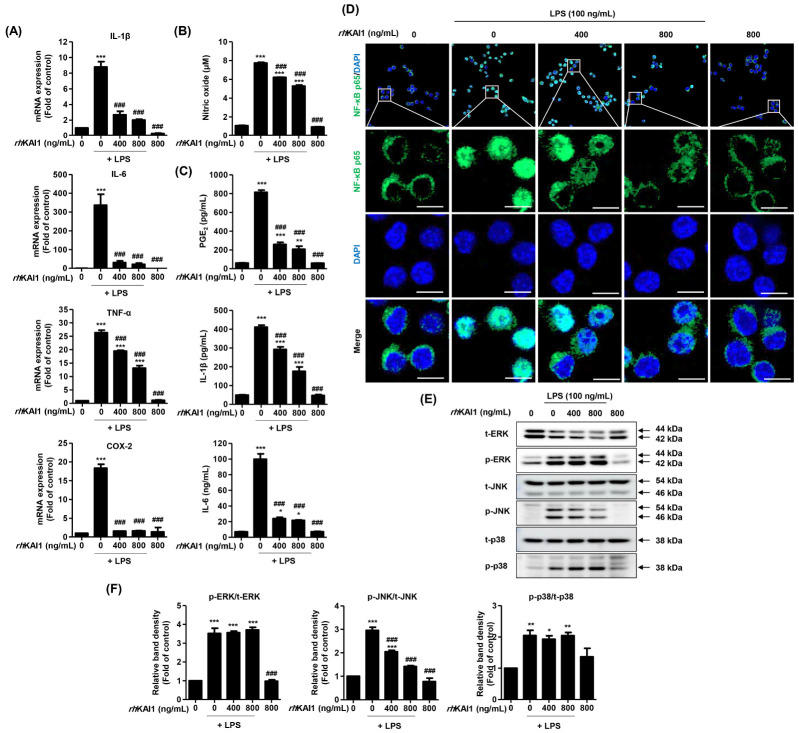
LPS is considered an initiator of M1 macrophages polarization. It can induce the production of key inflammatory mediators such as inflammatory cytokines and chemokines (3, 4). Extensive studies have shown that LPS can enhance pro-inflammatory cytokines (such as IL-1β, IL-6, IL-12 and TNF-α) and representative inflammatory mediators (e.g., NO and PGE2) in macrophages, ultimately promoting an inflammatory response (18). Inducible NO synthase (iNOS) and cyclooxygenase-2 (COX-2) are enzymes responsible for the production of NO and PGE2-that are also positively correlated with pro-inflammatory cytokines (19). Therefore, levels of pro-inflammatory cytokines and inflammatory mediators have been applied as indicators of anti-inflammatory efficacy (18, 19). In the present study, we assessed effects of rhKAI1 on expression levels of inflammatory cytokines in LPS-stimulated RAW 264.7 cells. Real-time qPCR results (Fig. 2A) revealed that LPS remarkably increased mRNA expression levels of pro-inflammatory cytokines and mediators including IL-1β, IL-6, TNF-α, and COX-2. However, such increases were substantially down-regulated by rhKAI1 pre-treatment. Especially, expression levels of IL-1β, IL-6, and COX-2 following rhKAI1 treatment were similar to those in untreated control cells. Next, to determine whether rhKAI1 could also regulate the release of inflammatory cytokines and mediators, secretion levels of NO, IL-1β, IL-6, and PGE2 in cell supernatants were analyzed. As shown in Fig. 2B, secretion levels of NO were markedly increased in LPS-stimulated cells (10.90 ± 1.13 μM, P < 0.001) compared to those in untreated cells (1.69 ± 0.47 μM). However, the secretion of NO was significantly reduced in 800 ng/ml rhKAI1 pre-treated cells (8.39 ± 0.76 μM, P < 0.05) compared with that in LPS-stimulated cells. Release levels of other inflammatory mediators such as PGE2, IL-1β, and IL-6 were also greatly increased by LPS, whereas such increases were suppressed by rhKAI1 pre-treatment before LPS treatment (Fig. 2C). These results suggest that rhKAI1 can suppress LPS-induced inflammatory response by modulating the production of pro-inflammatory cytokines and mediators. Findings of the present study are consistent with multiple previous studies suggesting that some members of the tetraspanin family, CD9, CD53, CD63, and CD81 can suppress inflammatory responses (20).
Fig. 2.
rhKAI1 suppress LPS-induced inflammatory response in RAW 264.7 macrophages via blocking of JNK/NF-κB signaling pathway. (A) Relative levels of mRNA expression for IL-1β, IL-6, TNF-α, and COX-2 were expressed as fold of control. (B) In the supernatants, NO concentration was measured by the Griess reaction. (C) The secretion levels of PGE2, IL-1β and IL-6 levels were determined using commercial ELISA kits. Data are expressed as the mean ± SD (n = 4). ***P < 0.001 compared to control. ###P < 0.001 compared to LPS-stimulated cells. (D) The cells subjected to immunofluorescence staining with NF-κB p65 antibody and representative images were acquired using a confocal laser scanning microscopy. Green fluorescence indicates the localization of NF-κB p65, and blue fluorescence by DAPI allows the nuclei. Scale bar = 50 μm. (E) The protein expression of MAPK signaling molecules including ERK, p-ERK, JNK, p-JNK, p38 MAPK and p-p38 MAPK. (F) Bar diagram showing the relative protein density after normalization with actin based on Western blot analysis. *P < 0.05, **P < 0.01, and ***P < 0.001 compared to control. ###P < 0.001 compared to LPS-stimulated cells.
rhKAI1 attenuates JNK/NF-κB p65 signaling pathway in LPS-stimulated RAW 264.7 cells
Inhibiting nuclear factor kappa-light-chain-enhancer of activated B (NF-κB) and mitogen-activated protein kinases (MAPK) pathways has been suggested as the main mechanism underlying the attenuation of LPS-induced inflammatory cytokine production (19, 21). Among various intracellular signaling pathways, NF-κB has been identified as the key transcription factor in regulating the expression of inflammatory factors under LPS stimulation (19). Inactive NF-κB heterodimer p65/50 can interact with IκB-α in the cytosol to form a complex. Upon stimulation with LPS, IκB-α is phosphorylated and degraded subsequently. Released NF-κB dimers then translocate to the nucleus to initiate inflammatory gene transcription (21). Therefore, we further assessed whether the anti-inflammatory effect of rhKAI1 involved the NF-κB signaling pathway through immunofluorescence analysis. As shown in Fig. 2D, NF-κB p65 expression was significantly increased in the nuclei of LPS-stimulated RAW 264.7 cells than in control cells. However, such increase of NF-κB p65 expression induced by LPS was greatly reversed in the presence of rhKAI1. Next, to investigate the molecular mechanism by which rhKAI1 regulated LPS-induced inflammation, MAPKs signaling pathways including extracellular-signal-regulated kinase (ERK), c-Jun N-terminal kinases (JNK), and p38 MAPK were analyzed. MAPKs can phosphorylate different intracellular proteins and transcription factors, subsequently regulating gene expression. ERK proteins are activated by endogenous or exogenous mitogens, cytokines, and growth factors to regulate cell proliferation (22). JNK and p38 MAPK are predominantly activated by inflammatory cytokines and bacterial LPS (23). In the present study, we found that phosphorylation levels of ERK, JNK, and p38 MAPK were substantially increased by LPS treatment (Fig. 2E, F). However, phosphorylation level of JNK was significantly reduced by rhKAI1 pre-treatment in a dose-dependent manner, while phosphorylation levels of ERK and p38 were not affected by rhKAI1. This result indicates that rhKAI1 can attenuate LPS-induced inflammatory response by blocking the JNK/NF-κB p65 signaling pathway.
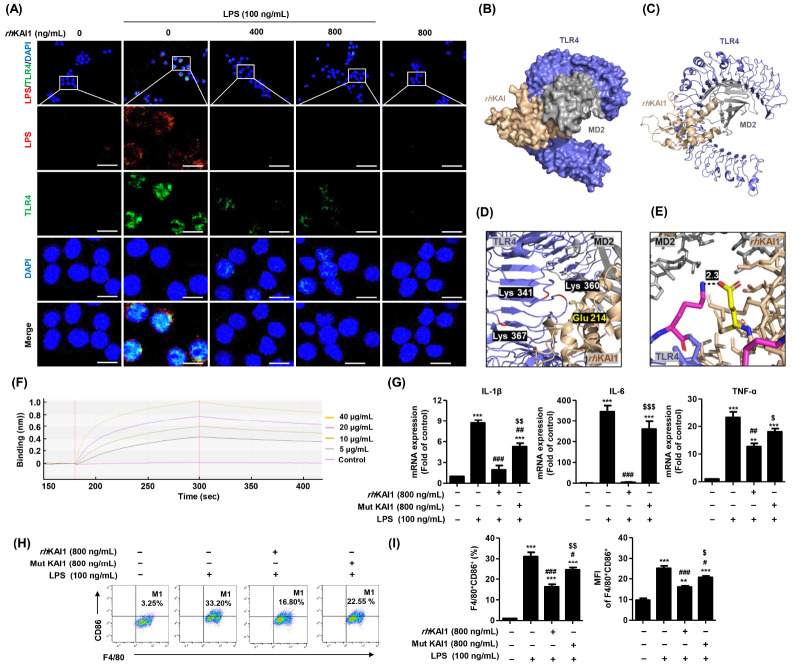
rhKAI1 prevents binding of LPS to TLR4s on the cell surface
TLRs expressed on the plasma and endosomal membranes of immune cells including macrophages are of interest to immunologists because of their front-line role in the initiation of innate immunity against invading pathogens (24, 25). Among various TLRs, TLR4 plays a crucial role in inflammatory and immune responses. It can efficiently sense Gram-negative bacterial infections through recognition of LPS, a bacterial membrane component (26). LPS binding to TLR4 leads to the activation and translocation of NF-κB into the nucleus, which then triggers the production of pro-inflammatory cytokines and type-I interferons (26, 27). To determine whether rhKAI1 could interact with TLR4 or prevent binding of LPS with TLR4, an immunofluorescence analysis was carried out to calculate molecular docking of each element. As shown in Fig. 3A, increment of TLR4 expression by LPS was markedly suppressed in the presence of rhKAI1. Subsequently, to predict the interaction between TLR4 and rhKAI1, 3-D structures of TLR4 and rhKAI1 were obtained from the PDB (PDB ID code of TLR4: 3VQ2) and Alphafold (AF ID code of hKAI1: P277701). Binding affinity was then analyzed and visualized with PyMOL molecular graphics system. As a result, we found that LPS could bind with four TLR4 residues: Glu 214, Lys341, Lys360, Lys367, and Arg434 (28). Among them, Glu 214 of rhKAI1 residue could strongly interact with Lys 360 of TLR4 residue. The distance between them was 2.9 Å (Fig. 3B-E). To validate this finding, we assessed the effect of rhKAI1 mutant (Glu 214) protein on LPS-stimulated macrophage polarization in RAW 264.7 cells. As shown in Fig. 1G, pretreatment with rhKAI1 mutant (Glu 214) protein markedly reversed down-regulation of mRNA expression of pro-inflammatory cytokines by wild type rhKAI1. The population and MFI of F4/80+CD86+ M1 surface marker were also reversed in rhKAI1 mutant (Glu 214) protein-treated cells (Fig. 3H, I). These results demonstrate that mutation of Glu 214 in rhKAI1 can decrease the anti-inflammatory effect of rhKAI1. Furthermore, we carried out a surface plasmon resonance binding analysis using BLItz, an optical technique for measuring interactions between proteins, peptides, nucleic acids, small molecules, and/or lipids in real time (29). Results of BLItz analysis showed that rhKAI1 strongly bound to recombinant human TLR4 in a highly specific fashion (Fig. 3F). These results indicate that rhKAI1 can directly interact with TLR4, which is involved in the suppression of LPS/TLR4-mediated M1 macrophage polarization. In a previous study, Khan et al. have demonstrated that KAI1 could interact with TLR9, an endosomal innate immune receptor, and modulate TLR9-dependent NF-κB nuclear translocation as a response to cytosine-phosphate-guanine stimulation in immune cells (30). Khan et al.’s results and our findings suggest that KAI1 could directly interact with TLR9 and TLR4 to block TLR-dependent downstream signaling pathway. We further determine whether down-regulation of TLR4 was mediated by the anti-inflammatory effect of rhKAI1 in LPS-stimulated cells. We found that the protective ability of rhKAI1 on increased cytokine secretion and TLR4 expression was enhanced in the presence of TAK-242, an inhibitor of TLR4 (Supplementary Fig. 1). These results demonstrate that KAI1 is associated with the TLR4 signaling pathway in the inhibition of inflammatory responses.
Fig. 3.
rhKAI1 interrupt interaction of LPS/TLR4 complex in LPS-stimulated RAW 264.7 macrophages. (A) The cells were subjected to immunofluorescence staining with LPS and TLR4 specific antibodies. Representative images were acquired using a confocal laser scanning microscopy. Confocal images show the expression of LPS (red), TLR4 (green), and DAPI (blue). Scale bar = 50 μm. The docking conformations of rhKAI1/TLR4 complex are illustrated by the surface (B) and cartoon (C). (D) The interacting residues of rhKAI1/TLR4 complex are shown in cartoon representation. (E) Atoms N and O were marked with blue and pink, respectively. The covalent bound of the rhKAI1 (yellow, Glu 214)-TLR4 (pink, Lys 360) complex and is illustrated by a black dotted line, and its distance is presented. (F) Binding affinity determination for rhKAI1 and rhTLR4 using BLItz label-free biolayer interferometry system. Binding (nm) refers to changes in optical interference. (G-I) Cells were pre-treated with rhKAI1 or mutant (Glu 214) protein of rhKAI1 for 2 h, following by 100 ng/ml LPS treatment for 24 h. (G) Relative levels of mRNA expression for IL-1β, IL-6, and TNF-α was expressed as fold of control. (H) Representative flow cytometry plots of F4/80+CD86+ in RAW 264.7 cells. (I) Quantification and MFI for F4/80+CD86+ cells and F4/80+CD80+ cells from total live-gated cells in RAW 264.7 cells. Data are expressed as the mean ± SD (n = 3). **P < 0.01 and ***P < 0.001 compared to control. #P < 0.05, ##P < 0.01 and ###P < 0.001 compared to LPS-stimulated cells. $P < 0.05, $$P < 0.01 and $$$P < 0.001 compared with LPS+rhKAI1 treatment cells.
In conclusion, the current study showed that rhKAI1 could attenuate LPS-induced inflammatory responses in RAW 264.7 macrophages-like cells and primary mouse BMDMs. The anti-inflammatory effect of rhKAI1 was supported by reduced M1 macrophage phenotype-related surface markers and inhibited production of pro-inflammatory cytokines and mediators. However, many questions remain unsolved regarding macrophage polarization. More efforts are needed to establish specific markers and steps of the differentiation process leading to different subpopulation of macrophages (31). We noted that rhKAI1 also suppressed nuclear translocation of NF-κB p65 and phosphorylation of JNK in LPS-stimulated RAW 264.7 cells. In addition, rhKAI1 inhibited the expression of TLR4 on the cellular surface and bound strongly to TLR4. Taken together, these findings suggest that rhKAI1 has an anti-inflammatory effect in LPS-polarized macrophages through its interaction with TLR4 and down-regulation of the JNK/NF-κB signaling pathway (Fig. 4). Although further studies are required to determine the effect of rhKAI1 in vivo, our findings indicate a pharmacological potential of rhKAI1 in the prevention of various inflammatory diseases.
Fig. 4.
Schematic summarizing the effect of rhKAI1/CD82 against LPS-stimulated inflammatory response in macrophages. rhKAI1/CD82 attenuates M1 macrophage polarization on LPS-stimulated RAW264.7 cells via blocking TLR4/JNK/NF-κB signal pathway.
MATERIALS AND METHODS
Details on the used methods are provided in the expanded Materials and Methods section in the online data supplement.
Funding Statement
ACKNOWLEDGEMENTS This research was supported by the Korean Fund for Regenerative Medicine (KFRM) grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Health & Welfare; grant No. KFRM 21A0502L1-12) and by Research institute for Convergence of biomedical science and technology of Pusan National University Yangsan Hospital. Fig. 4 was created in part with BioRender.com.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.van Rooijen N, Wijburg OL, van den Dobbelsteen GP, Sanders A. Macrophages in host defense mechanisms. Curr Top Microbiol Immunol. 1996;210:159–165. doi: 10.1007/978-3-642-85226-8_16. [DOI] [PubMed] [Google Scholar]
- 2.Yunna C, Mengru H, Lei W, Weidong C. Macrophage M1/M2 polarization. Eur J Pharmacol. 2020;877:173090. doi: 10.1016/j.ejphar.2020.173090. [DOI] [PubMed] [Google Scholar]
- 3.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 5.Biswas SK, Allavena P, Mantovani A. Tumor-associated macrophages: functional diversity, clinical significance, and open questions. Semin Immunopathol. 2013;35:585–600. doi: 10.1007/s00281-013-0367-7. [DOI] [PubMed] [Google Scholar]
- 6.Duluc D, Corvaisier M, Blanchard S, et al. Interferon-gamma reverses the immunosuppressive and protumoral properties and prevents the generation of human tumor-associated macrophages. Int J Cancer. 2009;125:367–373. doi: 10.1002/ijc.24401. [DOI] [PubMed] [Google Scholar]
- 7.Kwon DH, Lee H, Park C, et al. Glutathione induced immune-stimulatory activity by promoting M1-like macrophages polarization via potential ROS scavenging capacity. Antioxidants. 2019;8:413. doi: 10.3390/antiox8090413.7a629d2fc4a64c07accbc6226f1ca13a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu XQ, Dai Y, Yang Y, et al. Emerging role of microRNAs in regulating macrophage activation and polarization in immune response and inflammation. Immunology. 2016;148:237–248. doi: 10.1111/imm.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 10.Lee MS, Lee J, Kim YM, Lee H. The metastasis suppressor CD82/KAI1 represses the TGF-β 1 and Wnt signalings inducing epithelial-to-mesenchymal transition linked to invasiveness of prostate cancer cells. Prostate. 2019;79:1400–1411. doi: 10.1002/pros.23837. [DOI] [PubMed] [Google Scholar]
- 11.Tam JM, Reedy JL, Lukason DP, et al. Tetraspanin CD82 organizes dectin-1 into signaling domains to mediate cellular responses to Candida albicans. J Immunol. 2019;202:3256–3266. doi: 10.4049/jimmunol.1801384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Spriel AB, Figdor CG. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect. 2010;12:106–112. doi: 10.1016/j.micinf.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Hammond C, Denzin LK, Pan M, et al. The tetraspan protein CD82 is a resident of MHC class II compartments where it associates with HLA-DR, -DM, and -DO molecules. J Immunol. 1998;161:3282–3291. doi: 10.4049/jimmunol.161.7.3282. [DOI] [PubMed] [Google Scholar]
- 14.McGowan ENS, Wong O, Jones E, et al. Tetraspanin CD82 restrains phagocyte migration but supports macrophage activation. iScience. 2022;25:104520. doi: 10.1016/j.isci.2022.104520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JW, Hur J, Kwon YW, et al. KAI1(CD82) is a key molecule to control angiogenesis and switch angiogenic milieu to quiescent state. J Hematol Oncol. 2021;14:148. doi: 10.1186/s13045-021-01147-6.791b651888294fddb7d39ceb1e7fbdc8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H, Han JH, Kang YJ, et al. CD82 attenuates TGF-β1-mediated epithelial-mesenchymal transition by blocking smad-dependent signaling in ARPE-19 cells. Front Pharmacol. 2022;13:991056. doi: 10.3389/fphar.2022.991056.f964cb4eb52748f1922e05382e2e820c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hur J, Choi JI, Lee H, et al. CD82/KAI1 maintains the dormancy of long-term hematopoietic stem cells through Interaction with DARC-expressing macrophages. Cell Stem Cell. 2016;18:508–521. doi: 10.1016/j.stem.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Aleem D, Tohid H. Pro-inflammatory cytokines, biomarkers, genetics and the immune system: a mechanistic approach of depression and psoriasis. Rev Colomb Psiquiatr (Engl Ed) 2018;47:177–186. doi: 10.1016/j.rcpeng.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Park C, Cha HJ, Lee H, et al. The regulation of the TLR4/NF-κB and Nrf2/HO-1 signaling pathways is involved in the inhibition of lipopolysaccharide-induced inflammation and oxidative reactions by morroniside in RAW 264.7 macrophages. Arch Biochem Biophys. 2021;706:108926. doi: 10.1016/j.abb.2021.108926. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Bae S, Jang J, et al. CD53, a suppressor of inflammatory cytokine production, is associated with population asthma risk via the functional promoter polymorphism -1560 C>T. Biochim Biophys Acta. 2013;1830:3011–3018. doi: 10.1016/j.bbagen.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 21.Gasparini C, Feldmann M. NF-κB as a target for modulating inflammatory responses. Curr Pharm Des. 2012;18:5735–5745. doi: 10.2174/138161212803530763. [DOI] [PubMed] [Google Scholar]
- 22.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: is subcellular localization the answer? Cell Cycle. 2009;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono K, Han J. The p38 signal transduction pathway: activation and function. Cell Signal. 2000;12:1–13. doi: 10.1016/S0898-6568(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 24.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Swanson L, Katkar GD, Tam J, et al. TLR4 signaling and macrophage inflammatory responses are dampened by GIV/Girdin. Proc Natl Acad Sci USA. 2020;117:26895–26906. doi: 10.1073/pnas.2011667117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Ohto U, Fukase K, Miyake K, Shimizu T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc Natl Acad Sci U S A. 2012;109:7421–7426. doi: 10.1073/pnas.1201193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sultana A, Lee JE. Measuring protein-protein and protein-nucleic acid interactions by biolayer interferometry. Curr Protoc Protein Sci. 2015;79:19.25.1–19.25.26. doi: 10.1002/0471140864.ps1925s79. [DOI] [PubMed] [Google Scholar]
- 30.Khan NS, Lukason DP, Feliu M, et al. CD82 controls CpG-dependent TLR9 signaling. FASEB J. 2019;33:12500–12514. doi: 10.1096/fj.201901547R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chávez-Galán L, Olleros ML, Vesin D, Garcia I. Much more than M1 and M2 macrophages, there are also CD169(+) and TCR(+) macrophages. Front Immunol. 2015;6:263. doi: 10.3389/fimmu.2015.00263.edee05485c8f4e889c43bd809ef6644a [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.