Abstract
BACKGROUND:
Hereditary leiomyomatosis and renal cancer (HLRCC) is a cancer syndrome associated with a germline mutation in fumarate hydratase (FH). The syndrome is associated with cutaneous and uterine leiomyomas, and some patients develop a lethal form of kidney cancer. This study provides estimates for the FH carrier frequency and kidney cancer penetrance.
METHODS:
Data sets containing sequencing data for the FH gene were used: the 1000 Genomes Project (1000GP) and the Exome Aggregation Consortium (ExAC). Alterations in the FH gene were characterized on the basis of different variant risk tiers: 1) ClinVar annotated variants, 2) loss-of-function alterations, and 3) highly impactful missense alterations. The cumulative incidence of FH alterations overall and by different world populations was evaluated in 1000GP and ExAC. A lifetime penetrance of HLRCC kidney cancer risk was generated with 3 estimates of the annual incidence.
RESULTS:
The overall allele frequencies of tier 1 to 3 FH alterations in the ExAC and 1000GP data sets were 2.54 × 10–3 (1 in 393) and 1.20 × 10–3 (1 in 835), respectively. There were differences in the allele frequencies of FH alterations between world populations. Based on various estimates of the percentage of kidney cancers with FH alterations, the lifetime kidney cancer penetrance for carrier estimate 3 in ExAC was 1.7% to 5.8%.
CONCLUSIONS:
FH alterations are common and are carried by approximately 1 in 1000 individuals according to the more conservative estimates. The lifetime kidney cancer penetrance appears lower than previously estimated. Although databases are not population cohorts, they provide a useful quantitative estimate of rare variants with low penetrance.
Keywords: fumarate hydratase (FH), hereditary, hereditary leiomyomatosis and renal cancer (HLRCC), renal cell carcinom
INTRODUCTION
More than 50 cancer syndromes have been linked to specific germline alterations. In renal cell carcinoma (RCC), more than a dozen syndromes have been recognized since the first description of von Hippel–Lindau disease more than a century ago.1,2 Hereditary cutaneous leiomyoma (Reed’s syndrome) was first reported in the dermatology community in the 1970s.3 Since that time, the condition has been associated with RCC,4 uterine fibroids,5 and adrenal nodular hyperplasia,6 and it has been renamed hereditary leiomyomatosis and renal cancer (HLRCC). The gene associated with HLRCC was first linked to chromosome 1q42.3-q43 and was later identified as the Krebs cycle enzyme fumarate hydratase (FH).7 The condition is inherited in an autosomal dominant fashion, and individuals with 1 altered allele are considered affected. A fumarase deficiency can result with 2 altered alleles and is lethal in childhood. Because only 200 families have been described in the literature,8 there is still limited information on the penetrance of manifestations such as RCC.
Recognition of both a syndrome’s population incidence and the disease penetrance should influence clinical suspicion and ultimately referrals to genetic counseling/testing. For hereditary papillary renal cancer, for which there are fewer than 30 known families,9 screening broadly would have limited value. For some cancer syndromes such as von Hippel–Lindau disease, clinical recognition is less of a challenge because of the high disease penetrance (nearly 90%) of multiple manifestations not often seen sporadically and the high population frequency (approximately 1 in 27,000 individuals).10,11 As for less penetrant, nonsyndromic conditions with manifestations that occur sporadically, we may be grossly underestimating how common they are. For HLRCC, the population frequency is unknown, but it is believed to be a very rare condition.12 Given how common the benign and often asymptomatic manifestations of HLRCC are in the general population (uterine fibroids, cutaneous leiomyomas, and adrenal nodules), we may be significantly underestimating the population frequency. Even for individuals affected by RCC, an opportunity for diagnosis may be missed because of the challenging pathologic recognition of HLRCC. However, with aggressive screening of affected individuals, renal tumors may be found with localized and curable disease.13
We sought to estimate FH alterations in publicly available databases containing whole exome and genome data. We hypothesized that HLRCC is a fairly common disease with low RCC penetrance.
MATERIALS AND METHODS
Utilization of Data Sets
We used 2 publicly available data sets containing high-quality, high-coverage germline sequencing data for the FH gene: the 1000 Genomes Project (1000GP) and the Exome Aggregation Consortium (ExAC). 1000GP is a population-scale database of human variation containing the whole genome sequences of 2504 individuals.14 For 1000GP, because of the freely available nature of the data, no phenotype information was collected for any of the samples. All donors were older than 18 years and declared themselves to be healthy at the time of collection. There are 26 populations included in the phase 3 data set, and they form 5 continental superpopulations: African (AFR; 661 individuals), Ad Mixed American (AMR; 347 individuals), East Asian (EAS; 504 individuals), European (503 individuals), and South Asian (SAS; 489 individuals). ExAC (v.0.3.1) contains exome sequencing results from 60,706 individuals.15 It combines exome sequencing data from 14 consortium projects evaluating patients affected by diseases such as inflammatory bowel disease, schizophrenia and bipolar disorder, and diabetes (for a detailed cohort/study breakdown, please see http://exac.broadinstitute.org). The patients included in the ExAC project may be further subdivided according to sex and according to 6 world populations: African/African American (AFR [AFR populations in 1000GP and ExAC slightly differ in definition]; 5203 individuals), Latino (AMR; 5789 individuals), EAS (4327 individuals), Finnish (FIN; 3307 individuals), non-Finnish European (NFE; 33,370 individuals), SAS (8256 individuals), and Other (OTH; 454 individuals). These populations are separated by principal component analysis. The OTH population includes individuals who cannot cluster unambiguously. We excluded 7601 patients with cancer sequenced in conjunction with The Cancer Genome Atlas (TCGA). ExAC contains 1000GP in its data set (approximately 4% of the overall cohort), and because of privacy policies, it is not feasible to identify and exclude specific patients. Together, both databases provide unique estimates of the prevalence of germline variants from a large cohort of healthy individuals (1000GP) and a broader cohort including individuals with a diversified genetic background (ExAC). Data from kidney cancer cases at Yale New Haven Hospital was reviewed using an institutional review board approved protocol (#0805003787).
We used common bioinformatic programs, Polymorphism Phenotyping (PolyPhen; version 2.2.2) and Sorting Intolerant From Tolerant (SIFT; version 5.2.2), to predict variant pathogenicity based on sequence homology across species and similarity of alternate amino acids. PolyPhen and SIFT are 2 well-established algorithms for predicting the effects of variants in coding regions.16,17 Although both algorithms rely on sequence/homology features, PolyPhen also uses additional structure-based predictors such as amino acid residue contacts. Both tools have been also used by the ExAC consortium and integrated into its data release. For validation purposes, we also ran 2 ensemble methods, Variant Effect Scoring Tool 4 (VEST4) and Rare Exome Variant Ensemble Learner (REVEL), and we reported the results for our variants.18,19
Identifying Deleterious FH Variants
To isolate variants of likely deleterious consequences, we built inclusion criteria associated with 3 variant risk tiers of putative impact (Fig. 1 and Supporting Table 1). These 3 tiers correspond to separate confidence levels (CIs) according to the likelihood of deleterious consequences. Variant tier 1 (VT1) includes “pathogenic” and “likely pathogenic” variants in ClinVar (v2018.4.1), a publicly available database annotating the relationship of genomic variation to human health (http://www.ncbi.nlm.nih.gov/clinvar/). VT1 alterations have been annotated by experts and submitted to ClinVar; thus, they are most likely to have functional impact. Variant tier 2 (VT2) includes all alterations that are also likely pathogenic because they result in an imputative gene loss of function (LOF), including premature stop codons, stop loss, and start loss. Variant tier 3 (VT3) expands our estimate by including all missense alterations likely to have functional consequence. VT3 missense variants were stratified according to expected impact, and only those with a PolyPhen score > 0.9 (>90% probability of being damaging) and a SIFT classification as “deleterious” were retained. Two ensemble-based methods were used to confirm pathogenic variant classification, and those variants considered to be benign were to be excluded from further analyses.18,19
FIGURE 1.
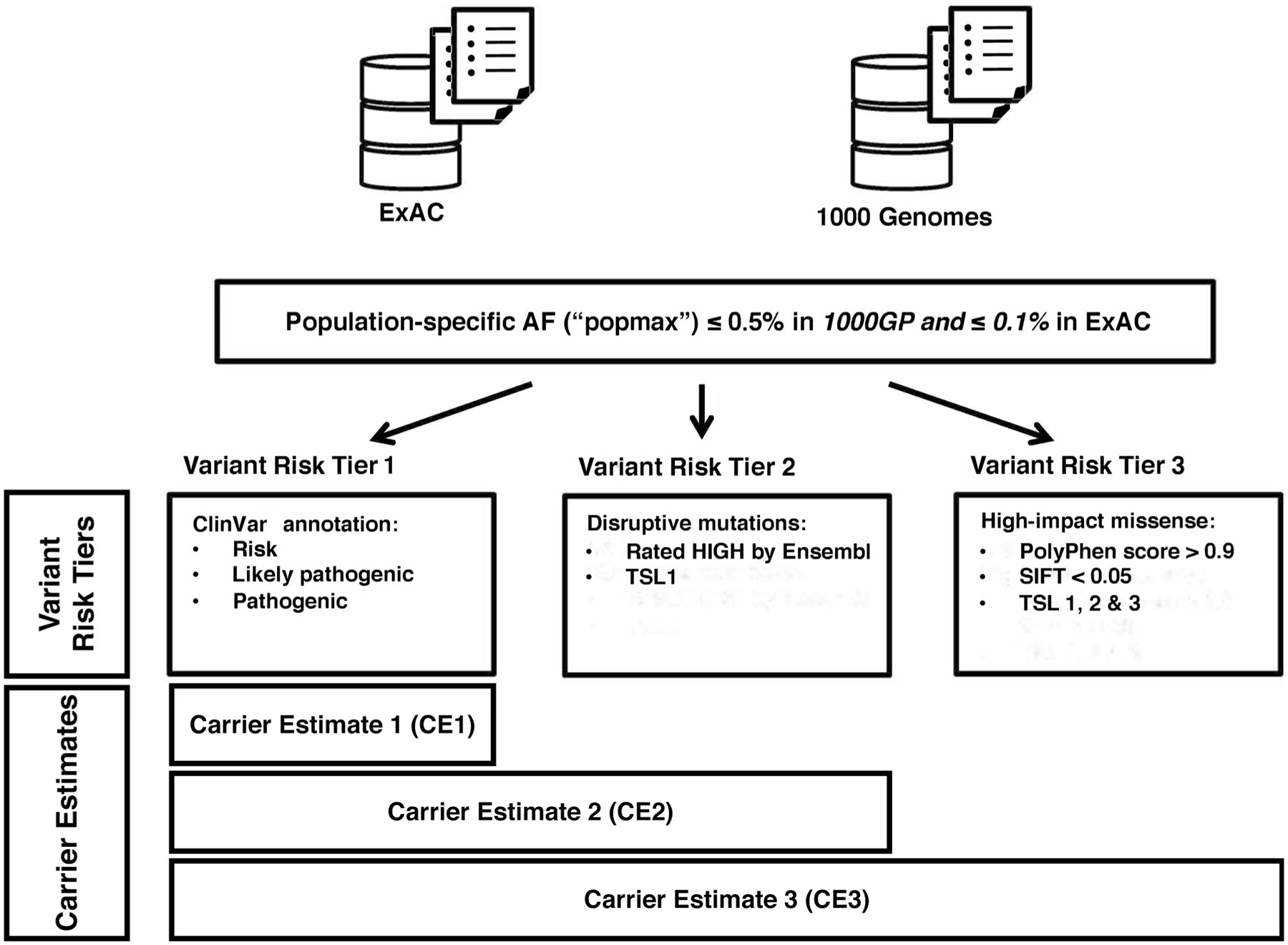
Fumarate hydratase variant classification scheme. 1000GP indicates 1000 Genomes Project; AF, allele frequency; ExAC, Exome Aggregation Consortium; PolyPhen, Polymorphism Phenotyping; SIFT, Sorting Intolerant From Tolerant; TSL, transcript support level.
Filtering based on the allele frequency was also applied according to the principle that common variants are unlikely to be deleterious. Any common individual variant with a minor allele frequency higher than 0.005 (in 1000GP)20 or 0.001 (in ExAC)15 in any of the subpopulations and with at least 3 observed allele counts was excluded. This population-level filter mitigated confounding differences in the population genetic background. This included a common FH variant, 1431_1433dupAAA, which had an allele frequency of 0.00136 in the NFE population (n = 74). Ensembl transcript support levels were used to ensure that variants affected actually expressed transcripts. Transcripts with higher support levels had more evidence for being truly expressed (eg, they were supported by nonsuspect messenger RNA). Only variants affecting transcript support level 1 (the most confident one) were included in VT2. In VT3, we expanded this to include all alterations affecting transcript support levels 1 to 3.
Estimation of the Underlying Allele Frequency
We statistically modeled each variant as a Bernoulli process, and our observed allele frequencies followed a binomial distribution with parameter , which is the underlying allele frequency in the population. The estimator is given by
To estimate the total allele frequency , we simply added our estimated value. Under the assumption of independency, the variance estimator was the sum of . We formed the by taking a standard deviance of ±1.96 around the estimation:
This interval should be always nonnegative.
Testing Allele Frequency Differences Across Populations
Our variance estimation was lower than expected because it assigned zero variance to unobserved mutations; this could be due to a lack of power caused by the small sample size. Therefore, we designed a more conservative combined Fisher exact test to evaluate the allele frequency differences between populations.
For each allele that occurred at least once in any population, we performed two 1-sided Fisher exact tests between 2 populations on the raw counts of wild-type and alternative alleles. Then, we combined all the P values from the same side with the Fisher method. We ran tests on every variation separately because they had different numbers of reliably observed alleles due to sequencing coverage and errors.
Estimates of the Percentage of RCC Attributable to HLRCC
There are no comprehensive estimates of kidney cancers attributable to HLRCC. It is assumed that HLRCC does not account for a large number of kidney cancers. Available cohorts of patients who have been counseled, have been sequenced, or have undergone close clinical evaluations could provide an estimate. Because institutions and practice patterns vary, there may be a wide range of estimates. To provide a range, 3 estimates were obtained from separate sources. Estimate 1 was generated from the Yale Urology and Genitourinary Cancer Genetics and Prevention Program (2013–2017), which screens all patients with kidney cancer for hereditary risk on the basis of common clinical factors and suspicious pathology.21–23 Estimate 2 was obtained from the combination of data from published papillary RCC cohorts that had genomic profiling (TCGA and a Foundation Medicine cohort; medium estimate).24,25 Estimate 3 was taken from a published Memorial Sloan Kettering Cancer Center cohort of 254 advanced (stage III/IV) RCC cases that underwent clinical sequencing.26 The stage distribution of HLRCC and the distribution of stage III/IV RCC were used to adjust the overall RCC attribution to HLRCC. The stage distribution of HLRCC kidney cancer was based on Grubb et al,26 and the distribution of stage III/IV cases (up to 28%) was based on data from the National Cancer Database.27,28 Brief clinical characteristics of the cohorts are presented in Supporting Table 2.
Estimating Allele/Carrier Frequency and Lifetime RCC Risk
Individuals with fumarase deficiency (a loss of both alleles) rarely survive past early childhood. Therefore the likelihood of individuals being homozygous for two deleterious variants is low and unlikely to significantly contribute to the population frequency. When allele frequency p is small, the higher order p2 can be neglected. On the basis of the total minor allele frequency of this type of dominant homozygous-lethal variant (ie, p), we estimated the carrier frequency to be 2p.
We calculated the annual incidence of HLRCC kidney cancer with the following information: 1) the US population (325 million),29 2) the incidence of RCC (15.1 per 100,000 individuals),30 3) the 3 different estimates of HLRCC identification in RCC cohorts, and 4) the carrier frequency estimates (cumulative) of the 3 variant risk tiers of FH alterations. For carrier estimates (CEs), different scenarios were created, including the addition of estimates from the more stringent variant risk tier (Fig. 1). For example, CE3 included the estimates from VT1, VT2, and VT3.
The penetrance or lifetime risk of RCC in HLRCC was determined from the number of years at risk starting with the age of screening (10 years). Because the life expectancy in the United States is nearly 80 years (79 years from the Centers for Disease Control and Prevention/National Center for Health Statistics), the number of at-risk years is 70. The lifetime risk estimates were calculated with the following formula:
With various estimates of 1) the carrier frequency and 2) the HLRCC percentage of total RCC, ranges were used to provide the penetrance or lifetime kidney cancer risk.
Using the lifetime risk estimates as well as the CE3 frequency and the HLRCC/RCC estimates (lowest to highest), we estimated the number of individuals with HLRCC who needed to be screened to identify a kidney cancer (100/lifetime risk of RCC). To develop a simple model of cost in the United States, we estimated how costly surveillance would be if all potential carriers (CE3) were identified. Currently, screening begins around the age of 10 years with annual magnetic resonance imaging (MRI). On the basis of the current cost for abdominal MRI (approximately $2600 according to CMS.gov), we estimated the annual cost if all carriers of a likely damaging allele were identified in childhood (age < 10 years) by using population size estimates (325 million).29 An estimated 70 years of screening from the age of 10 years to the median life expectancy (approximately 80 years) was used for lifetime health care costs per individual.
RESULTS
Multiple FH alterations were identified within the ExAC and 1000GP databases (Fig. 2A and Table 1). The 2 databases had different frequencies of tier 1 to 3 estimates (Fig. 2B). The ExAC registry contained 11 distinct known pathogenic/likely pathogenic FH variants found in ClinVar. The overall carrier frequency in ExAC of VT1 alterations was 7.44 × 10–4 (1 in 1344; Table 1). We did not identify any VT1 alterations in 1000GP. VT2 alterations were found in both ExAC and 1000GP data sets. CE2 (including the rates for VT1 and VT2) was 1.11 × 10–3 (1 in 901) and 0.0008 (1 in 1252) for the ExAC and 1000GP data sets, respectively (Table 1). There were multiple impactful VT3 missense alterations. The overall CE3 values in the ExAC and 1000GP data sets were 0.00254 (1 in 393) and 0.00120 (1 in 835), respectively. The frequency differences between ExAC and 1000GP were not statistically significant (all P values >.8 [combined 1-sided Fisher exact test]). These variants were widely distributed across the gene (Supporting Fig. 1). All identified variants were confirmed with VEST4 and REVEL. VEST4 was highly consistent with our PolyPhen 0.9 cutoff (all P values <.1 except for 1 variant with P = .119). REVEL further confirmed our variant classification because all scores were >0.794, a score higher than the threshold of >0.75, which produced >96% specificity.19 A list of classified pathogenic variants can be found in Supporting Table 3.
FIGURE 2.
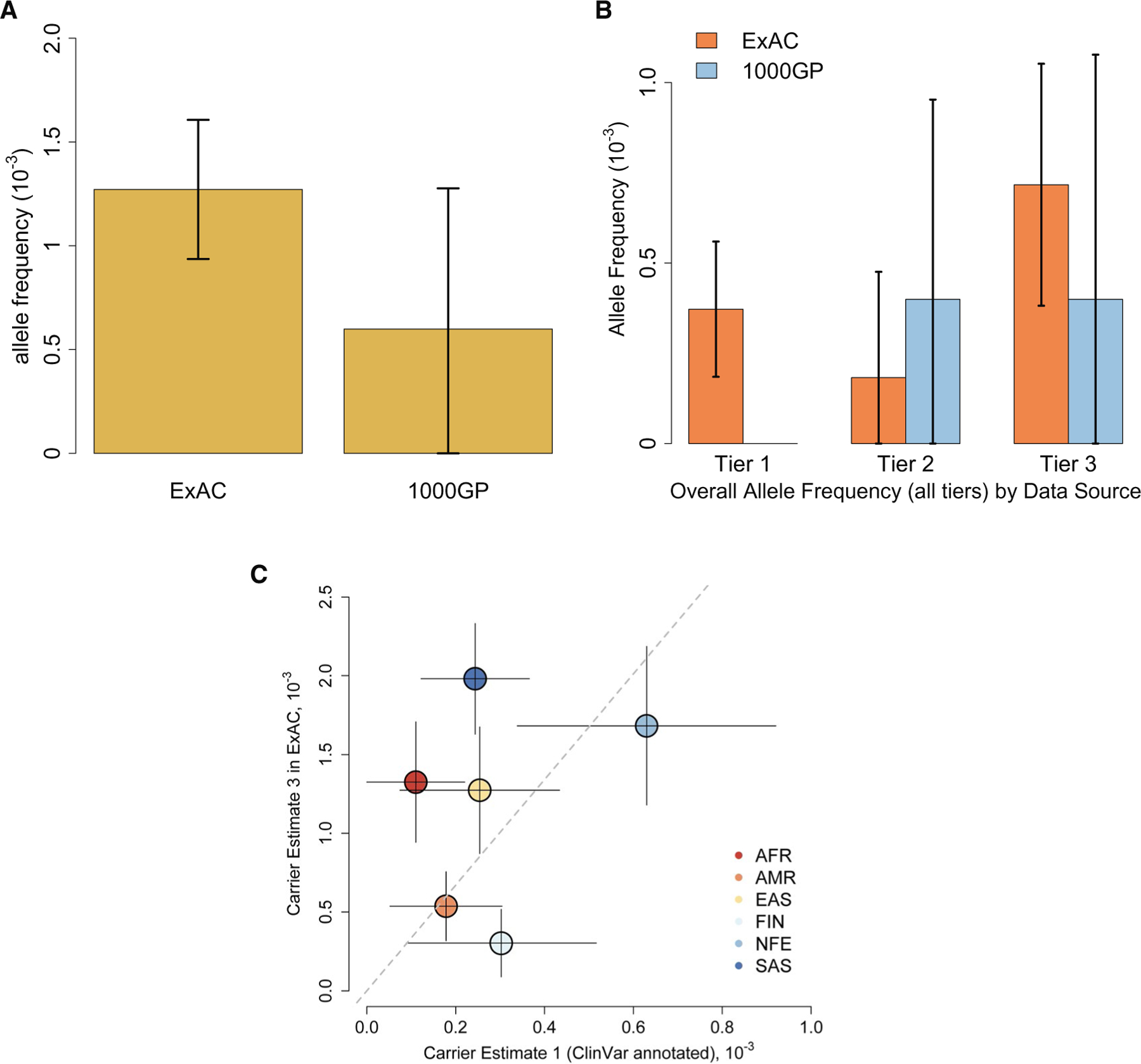
Fumarate hydratase variant allele frequencies by (A) databases and (B) databases and tiers. Error bars indicate 95% confidence intervals. (C) Frequencies of all impactful mutations (scenario 3) and ClinVar annotated pathogenic mutations in subpopulations. Solid lines indicate 95% confidence intervals; the dashed line is the zero-intercept linear fitting (coefficient = 3.35). 1000GP indicates 1000 Genomes Project; AFR, African/African American (AFR populations in 1000GP and ExAC slightly differ in definition); AMR, Ad Mixed American; EAS, East Asian; ExAC, Exome Aggregation Consortium; FIN, Finnish; NFE, non-Finnish European; SAS, South Asian.
TABLE 1.
FH Carrier Frequencies in the Exome Aggregation Consortium and 1000 Genomes Project Data Sets
| Exome Aggregation Consortium |
1000 Genomes Project |
|||||
|---|---|---|---|---|---|---|
| Scenario 1 (Tier 1) | Scenario 2 (Tiers 1 + 2) | Scenario 3 (Tiers 1–3) | Scenario 1 (Tier 1) | Scenario 2 (Tiers 1 + 2) | Scenario 3 (Tiers 1–3) | |
| FH allele frequency estimates | 0.00074 | 0.00111 | 0.00254 | — | 0.00080 | 0.00120 |
Abbreviation: FH, fumarate hydratase.
Because of the limited sample size of 1000GP, FH alterations were compared only by ExAC world populations. The OTH category was the smallest (<500 individuals), and because it represented a heterogeneous population in the registry, it was excluded when we were comparing populations. There were differences in the FH alterations between world populations (Fig. 3). The SAS population had the highest allele frequency of FH variants, but it was not statistically significant (0.00198; P = .357 in comparison with the other populations unified [combined 1-sided Fisher exact test]); the FIN population had the lowest (0.00030), and this may have been due to the small population sample size. There were also notable differences in the distributions of VT1, VT2, and VT3 alterations between groups. The SAS population had the highest frequency of VT3 alterations (P = .0067 in comparison with the other populations unified [combined 1-sided Fisher exact test]). The NFE population had higher but not statistically significant frequencies of VT1 and VT2 alterations (P = .73 and P = .45, respectively [combined 1-sided Fisher exact test]), and this was perhaps related to ClinVar reporting practices. Indeed, when plotting the carrier frequencies (CE3) against ClinVar annotated pathogenic mutations in subpopulations (Fig. 2C), we found that the SAS, AFR, and, to a lesser extent, EAS populations shifted to the upper left part of the plot; this indicated possible underannotation in those population.
FIGURE 3.

Allele frequencies of Exome Aggregation Consortium world populations. Error bars indicate 95% confidence intervals. AFR indicates African; AMR, Ad Mixed American; EAS, East Asian; FIN, Finnish; NFE, non-Finnish European; SAS, South Asian.
The annual number of cases of RCC attributed to HLRCC (HLRCC/RCC) varied significantly, although all the numbers reinforced that this is a less common histology. Estimate 1 was based on the number of cases seen at our institution. Despite a Clinical Laboratory Improvement Amendments–approved FH immunostain to test tumors with suspicious morphology and a dedicated cancer genetics program, only 3 of 741 new kidney cancer diagnoses (0.4%) were due to HLRCC. Estimate number 2 was derived from the identification of 23 FH-altered tumors (presumed germline) from a pooled cohort of 330 papillary RCCs from Foundation Medicine and TCGA. Because papillary kidney cancers constitute approximately 15% of kidney cancers,31 the estimate was 1.05%. Estimate 3 was based on 254 patients with stage III/IV RCC undergoing genomic testing; 7 had an FH alteration. Using the stage distribution of HLRCC26 and the known distribution of stages of kidney cancer (as high as 28% for stage III/IV),27,28 we obtained our highest estimate of 1.41%.
Based on the population and the incidence of kidney cancer, the annual estimates of HLRCC kidney cancer in the United States were 202, 523, and 703 from HLRCC/RCC estimates 1, 2, and 3, respectively. The annual RCC risk for FH carriers, based on HLRCC/RCC estimates 1, 2, and 3 for CE3, ranged from 0.024% to 0.085% for the ExAC registry and from 0.052% to 0.181% for 1000GP. The penetrance or lifetime risk based on a 70-year at-risk life expectancy varied significantly according to the CE used, but all remained significantly lower than previous estimates (Fig. 4). Supporting Table 4 provides a detailed list of the penetrance or lifetime risk for each CE based on our estimations. A very conservative estimate of the carrier frequency (based on CE2) would provide a lifetime penetrance ranging from 3.9% to 12.8% for ExAC and from 5.3% to 17.3% for 1000GP according to the estimated HLRCC frequencies. A broader carrier frequency definition (using CE3) expanded upon the population by including likely pathogenic variants (missense + ClinVar + LOF) confirmed with 4 predictive models. CE3 provided a lifetime penetrance ranging from 1.7% to 5.8% for ExAC and from 3.6% to 11.9% for 1000GP.
FIGURE 4.
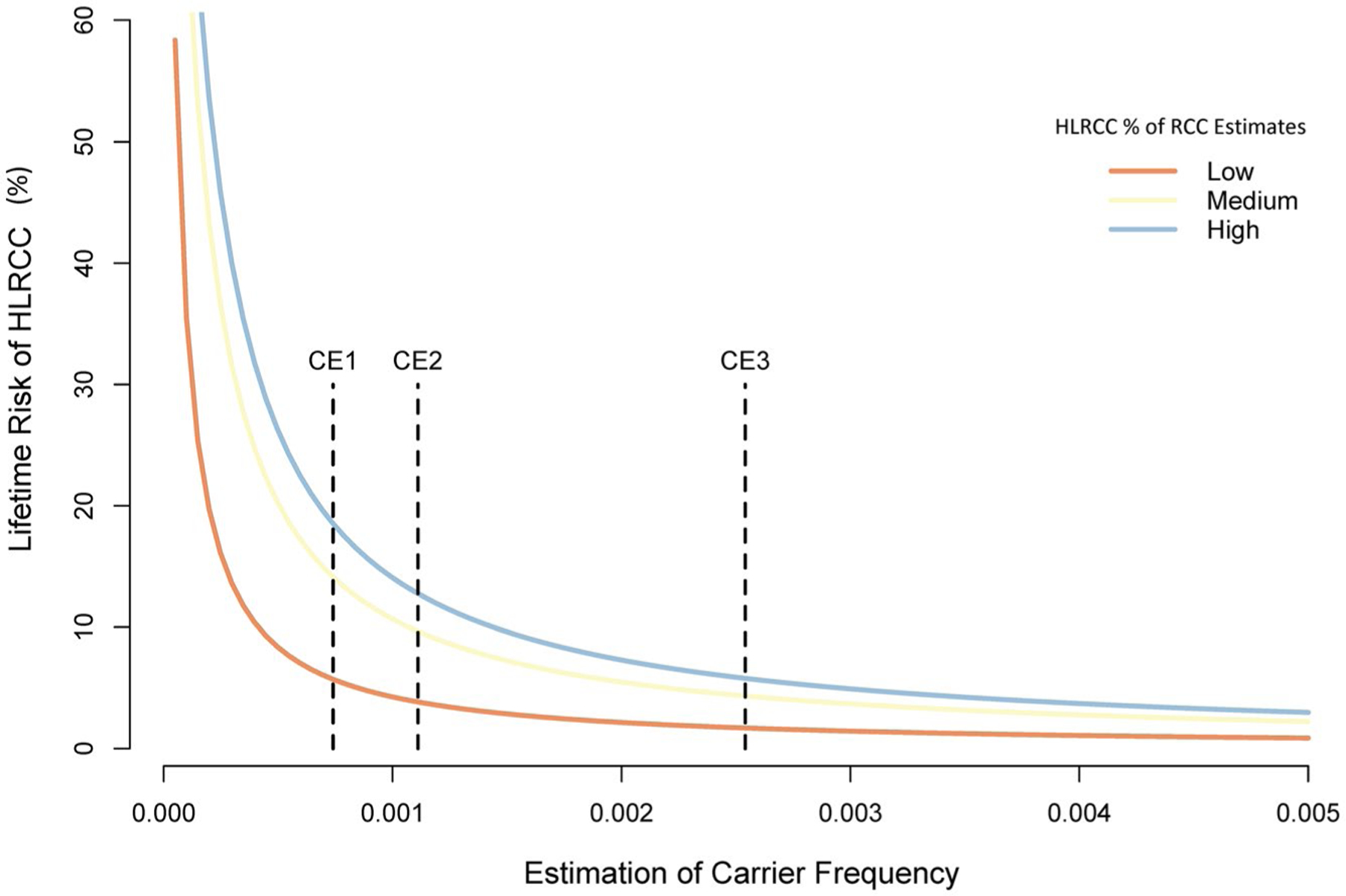
Estimated lifetime risk of HLRCC kidney cancer based on the carrier frequency estimates and the HLRCC kidney cancer annual incidence (low to high). Three dotted lines indicate CEs (1–3) based on the Exome Aggregation Consortium. CE indicates carrier estimate; HLRCC, hereditary leiomyomatosis and renal cancer; RCC, renal cell carcinoma.
The number needed to screen all individuals (CE3) to detect 1 individual with RCC would be 17 to 59 (approximately $3 million to $10 million in health care costs to identify a case). Based on all carriers (CE3) in the United States, the annual cost of screening in the United States would be approximately $3 billion.
DISCUSSION
We examined 2 large cohorts of whole exome and genome databases to estimate the population frequency of HLRCC, which is defined as carrying 1 deleterious variant in FH. Variants affecting FH were prioritized into 3 variant tiers. These 3 scenarios provide useful information: VT1 (ClinVar)/CE1 is the most conservative estimate because these variants are reported to cause the HLRCC phenotype. From ExAC, we found a carrier frequency of 1 in 1344 individuals, which was higher than expected for what has been considered a rare disease.12 In 1000GP, we did not identify a known pathogenic variant. Because HLRCC is a newer hereditary cancer syndrome, it is possible that ClinVar does not yet have a comprehensive list. Currently, there are only several hundred reported families with HLRCC, mostly from North America and Europe; therefore, we expanded our evaluation of FH variants. VT2 (LOF variants) includes likely pathogenic alterations that affect FH function and are probably yet to be recognized as disease-causing. In ExAC and 1000GP, CE2, including VT1 and VT2, was 1 in 901 individuals and 1 in 1252 individuals, respectively. Although CE2 is a useful estimate of the carrier frequency, it still may represent a conservative estimate. The inclusion of highly impactful missense variants (VT3) expands upon the likely pathogenic variants. Computational algorithms predict whether missense variants alter protein function by determining whether these are highly conserved residues within a critical regulatory region or there are mechanistic studies evaluating the influence of positional variants. The CE3 estimates are 1 in 393 individuals and 1 in 835 individuals from ExAC and 1000GP, respectively. Not all FH missense variants will cause a phenotype in affected individuals; however, we used very stringent cutoffs (maximum subpopulation frequency, PolyPhen score > 0.9 and SIFT score < 0.05). Strict cutoffs such as a REVEL score of 0.75 (>0.794 for all our variants) would have a sensitivity of 52% for the identification of disease-causing variants.19 Therefore, our conservative estimates may underestimate the allele prevalence in the population by misclassifying some pathogenic variants as benign.
Because HLRCC is the most aggressive kidney cancer syndrome, recognition of an affected individual’s risk is critical for counseling and screening. Current estimates of kidney cancer risk among individuals with HLRCC range from 15% to 32%12,26 and have been developed by risk assessment from single-institution case series. However, these numbers are strongly influenced by specialty, referral pattern, and the availability of genetic counseling. This is best evidenced by the highest estimate (32%) coming from a urologic oncology program focusing on hereditary kidney cancer.10 Affected individuals with cutaneous and uterine leiomyomas but without kidney cancer may not seek out medical care for these benign/sporadically occurring conditions. Although the initial reports of cutaneous leiomyomas mentioned pain in 89% of lesions,32 we have noted in our experience with cascade testing that many unaffected carriers have asymptomatic lesions found only with a total body skin examination. Similarly, for HLRCC-associated uterine fibroids, although these occur approximately 10 years earlier than sporadic forms, most women with early-onset fibroids will not be found with HLRCC.33–35 Although there are some characteristic pathologic features, these are subtle and are not always present.35 The variable penetrance, the difficulty in distinguishing these hereditary variants, and our data suggest a much larger reservoir of at-risk individuals.
The carrier frequencies from ExAC and 1000GP provide a useful reference overall, and our most comprehensive CE3 of the continental superpopulations grossly agrees, but there are some differences across various racial/ethnic groups. In particular, we observed that the SAS population had higher estimates (especially for tier 3 mutations), whereas those for the AMR and FIN populations were lower. The significant variability may be related to inherent structural factors such as founder effects, testing and reporting bias (as demonstrated in Fig. 2C), and regional genetic heterogeneity. In particular, we noticed that ClinVar overannotated the NFE and FIN populations and underannotated the AFR and SAS populations; this highlights the bias in our current clinical practice and reporting. It is also likely that the true reference sequences vary in less well-studied populations and that this influences the incidence of missense variants. A better understanding of structural bias and patterns helps with more accurate risk assessments of individuals from various genetic backgrounds and raises awareness to include more underannotated races in future studies. Moreover, some of the mutation frequency variability also may account for the frequency of HLRCC kidney cancer in different regions, and this is perhaps responsible for the high frequency recently observed in a large referral center in Shanghai, China (Dr. Jin Zhang, Renji Hospital, communication, written communication 10/2018). In the United States, the number of kidney cancer cases attributable to HLRCC is currently unknown. To provide an estimate, we used 3 resources to provide 3 different estimates (HLRCC/RCC). Although the estimates vary, they all confirm that HLRCC makes up a small percentage of kidney cancers, and they provide a useful range of 0.4% to 1.8%. With an improved understanding of the FH carrier frequency and annual estimates of HLRCC kidney cancer, we provide the first estimate of the lifetime RCC penetrance (see Supporting Table 4 and Fig. 4). Based on ExAC, this risk among carriers is significantly lower than previously believed because of the expected higher disease prevalence. With the broadest definition of who would be a carrier (CE3), the lifetime risks estimated from 1000GP and ExAC range from 3.6% to 11.9% and from 1.7% to 5.8%, respectively. The lower risk may be reassuring to individuals coming in for routine screening.
The much lower lifetime risk estimates also call into question whether current screening practices are appropriate and cost-effective. The estimated number needed to be screened in a lifetime ranges from 1 in 17 individuals to 1 in 59 individuals according to the aforementioned ExAC estimates using CE3. The current recommendations call for annual MRI starting around the age of 10 years. The costs to identify 1 case are significant ($3 million to $10 million), and although theoretical, the cost for HLRCC population screening ($3 billion) would further contribute to the ongoing health care crisis. Because HLRCC kidney cancer is uniformly lethal with dissemination, we strongly advocate screening; however, we must also consider whether there could be alternative, cost-effective screening strategies. The current guidelines for annual imaging frequency or modality are based not on evidence but on anecdotal experiences. Further investigation into the sensitivity of less expensive modalities such as renal ultrasound (used in other hereditary kidney cancer syndromes) could be considered. Extending imaging frequency beyond the annual recommendations also deserves further investigation, especially for those without any family history of RCC. However, until further data emerge from well-designed studies, we advocate for adherence to expert recommendations. A better understanding of the genotype-phenotype will also be critical for further refining which individuals are at greatest risk for kidney cancer. First understanding the functional impact of common missense alterations would be useful for determining which are truly deleterious and would allow us to individualize kidney cancer screening.
Our methods for calculating the allele frequency are not without limitations. The ExAC registry, though useful for evaluating genetic variants, is not an unbiased population database. We excluded the ExAC TCGA cases, which may actually lower the estimate of FH variants. The studies entered into ExAC try to exclude individuals with syndromic manifestations and pediatric genetic conditions, but how effective this is remains unknown. Whole exome sequencing is generally not reliable in reporting large structural variants, so we could not consider larger deletions affecting FH36; this may potentially make our estimates of the allele frequency slightly lower and our estimates of the penetrance higher. However, large deletions in FH are believed to be rare because this gene is under strong selection, and a recent report has found that it makes only up a minority of HLRCC cases.37 Although this database may not be useful for understanding highly penetrant conditions, which could be screened, ExAC remains a useful resource for assessing less penetrant conditions. Meanwhile, for rare diseases, the sample size of either database might be too low to capture many rare and impactful mutations, and thus the allele frequency might be underestimated. For example, we were unable to detect any VT1 in 1000GP, and the CEs from 1000GP suffer from very wide CIs. On the other hand, differences in sample sizes across populations might bias our estimate. In ExAC, the NFE population was several times bigger than any other superpopulation; thus, we had greater power to detect rare events (eg, we detected VT2 only in the NFE population). With such a rare genetic condition, detecting power is often a significant concern. We expect that more and diversified sequencing results in the future will mitigate this issue.
Our estimation of the HLRCC contribution to RCC may not be representative of the actual value seen clinically. The cohorts that contributed to estimates 2 and 3 (TCGA25 and Foundation Medicine38) were mainly from specialized academic centers. Similarly, estimate 3 was from a highly specialized center with a large hereditary cancer program and systemic therapy trials for less common subtypes, and perhaps a referral bias led to an enrichment of advanced disease and an overestimation of the frequency.39 Although it is chronologically and geographically unlikely that the same patients could have been included in the different cohorts, this could have influenced the data. Other factors could have led to an underestimation of the frequency because often HLRCC tumors can resemble other histologic morphologies, such as collecting duct morphology.21 Our estimate 1 (Yale) routinely evaluated these types of tumors for possible HLRCC and still had the lowest incidence. However, this was a consecutive series from a single hospital system including tertiary and several community affiliates, so perhaps this could be less biased by referral patterns. Finally, our estimation of RCC risk is simplistic and does not account for known environmental factors contributing to RCC, and it assumes a constant annual incidence of RCC from the time of screening to the current life expectancy. Also, within each variant tier, the allele frequencies are summarized together. This assumes that all risk is similar; however, there currently is no recognized genotype/phenotype association in HLRCC, but that may change with further research. Nonetheless, our estimates are meaningful until future studies further characterize additional molecular epidemiology risk factors.
In conclusion, we provide detailed estimates of carrier frequencies of FH variants by using those annotated in ClinVar, LOF, and high-impact missense mutations (using 4 predictive models). We demonstrate that the carrier frequency is very high but varies across populations. The higher reservoir of carriers suggests that the lifetime risk of RCC is lower than previously believed. Further research is needed to define risk, establish how genotype influences phenotype, and determine the optimal surveillance strategies for at-risk individuals.
Supplementary Material
FUNDING SUPPORT
This work was supported by the National Institutes of Health (1K08CA207845–01, R01 HG008126, and UM1 HG006504).
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The authors made no disclosures.
REFERENCES
- 1.Adeniran AJ, Shuch B, Humphrey PA. Hereditary renal cell carcinoma syndromes: clinical, pathologic, and genetic features. Am J Surg Pathol 2015;39:e1–e18. [DOI] [PubMed] [Google Scholar]
- 2.von Hippel E Ueber eine sehr seltene Erkrankung der Netzhaut. Graefes Arh Ophthalmol 1904;59:86–106. [Google Scholar]
- 3.Reed WB, Walker R, Horowitz R. Cutaneous leiomyomata with uterine leiomyomata. Acta Derm Venereol 1973;53:409–416. [PubMed] [Google Scholar]
- 4.Kiuru M, Launonen V, Hietala M, et al. Familial cutaneous leiomyomatosis is a two-hit condition associated with renal cell cancer of characteristic histopathology. Am J Pathol 2001;159:825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Launonen V, Vierimaa O, Kiuru M, et al. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci U S A 2001;98:3387–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuch B, Ricketts C, Vocke C, et al. Adrenal nodular hyperplasia in hereditary leiomyomatosis and renal cell cancer. J Urol 2013;189:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 2002;30:406–410. [DOI] [PubMed] [Google Scholar]
- 8.Patel VM, Handler MZ, Schwartz RA, Lambert WC. Hereditary leiomyomatosis and renal cell cancer syndrome: an update and review. J Am Acad Dermatol 2017;77:149–158. [DOI] [PubMed] [Google Scholar]
- 9.Shuch B, Singer EA, Bratslavsky G. The surgical approach to multifocal renal cancers: hereditary syndromes, ipsilateral multifocality, and bilateral tumors. Urol Clin North Am 2012;39:133–148. [DOI] [PubMed] [Google Scholar]
- 10.Lonser RR, Glenn GM, Walther M, et al. von Hippel-Lindau disease. Lancet 2003;361:2059–2067. [DOI] [PubMed] [Google Scholar]
- 11.Binderup ML, Galanakis M, Budtz-Jorgensen E, Kosteljanetz M, Luise BisgaardM. Prevalence, birth incidence, and penetrance of von Hippel–Lindau disease (vHL) in Denmark. Eur J Hum Genet 2017;25:301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menko FH, Maher ER, Schmidt LS, et al. Hereditary leiomyomatosis and renal cell cancer (HLRCC): renal cancer risk, surveillance and treatment. Fam Cancer 2014;13:637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan MMY, Barnicoat A, Mumtaz F, et al. Cascade fumarate hydratase mutation screening allows early detection of kidney tumour: a case report. BMC Med Genet 2017;18:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012;40:W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics 2013;14(suppl 3):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ioannidis NM, Rothstein JH, Pejaver V, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet 2016;99:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Southam L, Gilly A, Suveges D, et al. Whole genome sequencing and imputation in isolated populations identify genetic associations with medically-relevant complex traits. Nat Commun 2017;8:15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino MJ, Torres-Cabala C, Pinto P, Linehan WM. The morphologic spectrum of kidney tumors in hereditary leiomyomatosis and renal cell carcinoma (HLRCC) syndrome. Am J Surg Pathol 2007;31:1578–1585. [DOI] [PubMed] [Google Scholar]
- 22.Kopp RP, Stratton KL, Glogowski E, et al. Utility of prospective pathologic evaluation to inform clinical genetic testing for hereditary leiomyomatosis and renal cell carcinoma. Cancer 2017;123:2452–2458. [DOI] [PubMed] [Google Scholar]
- 23.Shuch B, Zhang J. Genetic predisposition to renal cell carcinoma: implications for counseling, testing, screening, and management. J Clin Oncol 2018;36:3560–3566. [DOI] [PubMed] [Google Scholar]
- 24.Pal S, Geynisman DM, Ali SM, et al. Comprehensive genomic profiling (CGP) of advanced papillary renal cell carcinoma (PRCC) to reveal distinctions from TCGA dataset [abstract 4517]. J Clin Oncol 2017;35(15 suppl):4517. [Google Scholar]
- 25.Linehan WM, Spellman PT, Ricketts CJ, et al. ; Cancer Genome Atlas Research Network. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 2016;374:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grubb RL III, Franks ME, Toro J, et al. Hereditary leiomyomatosis and renal cell cancer: a syndrome associated with an aggressive form of inherited renal cancer. J Urol 2007;177:2074–2079; discussion 2079–2080. [DOI] [PubMed] [Google Scholar]
- 27.Kane CJ, Mallin K, Ritchey J, Cooperberg MR, Carroll PR. Renal cell cancer stage migration: analysis of the National Cancer Data Base. Cancer 2008;113:78–83. [DOI] [PubMed] [Google Scholar]
- 28.Patel HD, Gupta M, Joice GA, et al. Clinical stage migration and survival for renal cell carcinoma in the United States. Eur Urol Oncol 2019;2:343–348. [DOI] [PubMed] [Google Scholar]
- 29.US Census Bureau. https://www.census.gov/
- 30.National Cancer Institute. Cancer Stat Facts: Kidney and Renal Pelvis Cancer https://seer.cancer.gov/statfacts/html/kidrp.html
- 31.Shuch B, Amin A, Armstrong AJ, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol 2015;67:85–97. [DOI] [PubMed] [Google Scholar]
- 32.Alam NA, Barclay E, Rowan AJ, et al. Clinical features of multiple cutaneous and uterine leiomyomatosis: an underdiagnosed tumor syndrome. Arch Dermatol 2005;141:199–206. [DOI] [PubMed] [Google Scholar]
- 33.Stewart L, Glenn GM, Stratton P, et al. Association of germline mutations in the fumarate hydratase gene and uterine fibroids in women with hereditary leiomyomatosis and renal cell cancer. Arch Dermatol 2008;144:1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinek P, Grossmann P, Hes O, et al. Genetic testing of leiomyoma tissue in women younger than 30 years old might provide an effective screening approach for the hereditary leiomyomatosis and renal cell cancer syndrome (HLRCC). Virchows Arch 2015;467:185–191. [DOI] [PubMed] [Google Scholar]
- 35.Harrison WJ, Andrici J, Maclean F, et al. Fumarate hydratase–deficient uterine leiomyomas occur in both the syndromic and sporadic settings. Am J Surg Pathol 2016;40:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Gerstein MB. Next-generation sequencing to diagnose suspected genetic disorders. N Engl J Med 2019;380:200. [DOI] [PubMed] [Google Scholar]
- 37.Vocke CD, Ricketts CJ, Merino MJ, et al. Comprehensive genomic and phenotypic characterization of germline FH deletion in hereditary leiomyomatosis and renal cell carcinoma. Genes Chromosomes Cancer 2017;56:484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal SK, Ali SM, Yakirevich E, et al. Characterization of clinical cases of advanced papillary renal cell carcinoma via comprehensive genomic profiling. Eur Urol 2018;73:71–78. [DOI] [PubMed] [Google Scholar]
- 39.Carlo MI, Mukherjee S, Mandelker D, et al. Prevalence of germline mutations in cancer susceptibility genes in patients with advanced renal cell carcinoma. JAMA Oncol 2018;4:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


