Abstract
CAG repeat expansions in exon 1 of the AR gene on the X chromosome cause spinal and bulbar muscular atrophy, a male-specific progressive neuromuscular disorder associated with a variety of extra-neurological symptoms. The disease has a reported male prevalence of approximately 1:30 000 or less, but the AR repeat expansion frequency is unknown. We established a pipeline, which combines the use of the ExpansionHunter tool and visual validation, to detect AR CAG expansion on whole-genome sequencing data, benchmarked it to fragment PCR sizing, and applied it to 74 277 unrelated individuals from four large cohorts. Our pipeline showed sensitivity of 100% [95% confidence interval (CI) 90.8–100%], specificity of 99% (95% CI 94.2–99.7%), and a positive predictive value of 97.4% (95% CI 84.4–99.6%). We found the mutation frequency to be 1:3182 (95% CI 1:2309–1:4386, n = 117 734) X chromosomes—10 times more frequent than the reported disease prevalence. Modelling using the novel mutation frequency led to estimate disease prevalence of 1:6887 males, more than four times more frequent than the reported disease prevalence. This discrepancy is possibly due to underdiagnosis of this neuromuscular condition, reduced penetrance, and/or pleomorphic clinical manifestations.
Keywords: androgen receptor, whole-genome sequencing, bioinformatics, population genetics, spinal and bulbar muscular atrophy
CAG repeat expansions in the AR gene cause spinal and bulbar muscular atrophy. Zanovello et al. establish a pipeline to detect AR CAG expansion from whole-genome sequencing data, and find the mutation frequency to be much higher than the reported disease prevalence, possibly due to underdiagnosis or pleomorphic manifestations.
Introduction
Spinal and bulbar muscular atrophy (SBMA), also known as Kennedy’s disease, occurs when the CAG repeat coding for a polyglutamine tract in exon 1 of the androgen receptor (AR) gene expands beyond 37 repeats.1 SBMA fully manifests only in males, with a mean age at onset of 43 years, which is partially influenced by CAG repeat size2 and is characterized by progressive muscular weakness induced by the degeneration of the lower motor neurons and primary muscular damage.1 Importantly, SBMA is also associated with a variety of non-neurological conditions, including insulin resistance, fatty liver disease, and metabolic syndrome.3
The information on the frequency of repeat expansion disorders has relied on epidemiology studies or PCR screening of selected populations. Epidemiological studies report a 1:30 303 or less prevalence amongst male populations,4–6 but SBMA is often reported to be underdiagnosed. However, an epidemiological study in the Vasa region of Finland reported 13 cases in a population of 85 000 males (1:6538), although this was attributed to a founder effect7; two studies based on PCR sizing in selected populations reported an unexpected high frequency of this genetic defect, namely a PCR screening of a European population, which found the mutation frequency to be 1:6888 X chromosomes8; and a meta-analysis of 86 datasets based on PCR sizing reported a population frequency of 1:3703.9
Although next-generation sequencing and public genomic data repository technologies have allowed the frequency of single nucleotide variants to be estimated precisely across very large populations,10 the inability to reliably size short tandem repeats (STRs) from whole-genome sequencing (WGS) has not permitted the same information to be gathered for STR expansions, which are a major cause of neurogenetic disorders including SBMA. Recently developed bioinformatics tools, such as ExpansionHunter, allow the sizing of STRs from WGS data.11
Given the unexpected findings from population studies and considering the limitation of PCR sizing and the use of selected populations, we sought to investigate the frequency of the genetic variant underlying SBMA in the general population by exploiting WGS and using clinically curated public genomic data repositories. We validated this approach, applied it to the 100,000 Genomes Project (100k GP) cohort12 and replicated it on three other large WGS datasets (Table 1 and Supplementary Table 1).
Table 1.
AR repeat expansion frequency
| Cohort | Gender | Phenotype category | Total participants | Total X chromosomes | Total expansions ≥38 | X chromosome frequency ≥38 (95% CI) |
|---|---|---|---|---|---|---|
| 100k GP | Male | Non–neuro | 13 072 | 13 072 | 2 | 1/6536 (1793–23 833) |
| Female | All | 20 400 | 40 800 | 11 | 1/3709 (2071–6642) | |
| gnomAD | Male | All | 14 947 | 14 947 | 5 | 1/2989 (1277–6998) |
| Female | All | 14 116 | 28 232 | 11 | 1/2567 (1433–4596) | |
| NIH | Male | Ctrl | 1529 | 1529 | 1 | 1/1529 (271–8661) |
| Female | All | 5176 | 10 352 | 2 | 1/5176 (1420–18 874) | |
| MinE | Male | Ctrl | 1272 | 1272 | 2 | 1/636 (175–2319) |
| Female | All | 3765 | 7530 | 3 | 1/2510 (854–7380) | |
| Summary | Male | – | 30 820 | 30 820 | 10 | 1/3082 (1674–5674) |
| Female | – | 43 457 | 86 914 | 27 | 1/3219 (2213–4683) | |
| All | – | 74 277 | 117 734 | 37 | 1/3182 (2309–4386) |
The summary result is highlighted in bold.
Materials and methods
Whole genome sequencing and AR genotyping
Whole-genome sequencing and cohort characterization
Supplementary Table 1 provides a summary of age and ethnicity of the cohorts assessed in this study. WGS data including chemistry, read length, coverage, alignment, genome build, and ExpansionHunter version from each cohort are summarized in Supplementary Table 2.
AR genotyping
ExpansionHunter (Illumina Inc., CA, USA) software was used to estimate repeat lengths of the AR CAG disease-causing expansions in samples that had undergone WGS. This algorithm has been validated using experimentally-confirmed samples carrying pathogenic expansions.13,14 Pathogenic alleles in the AR gene were defined as those containing 38 or more CAG repeats.1
Visual inspection
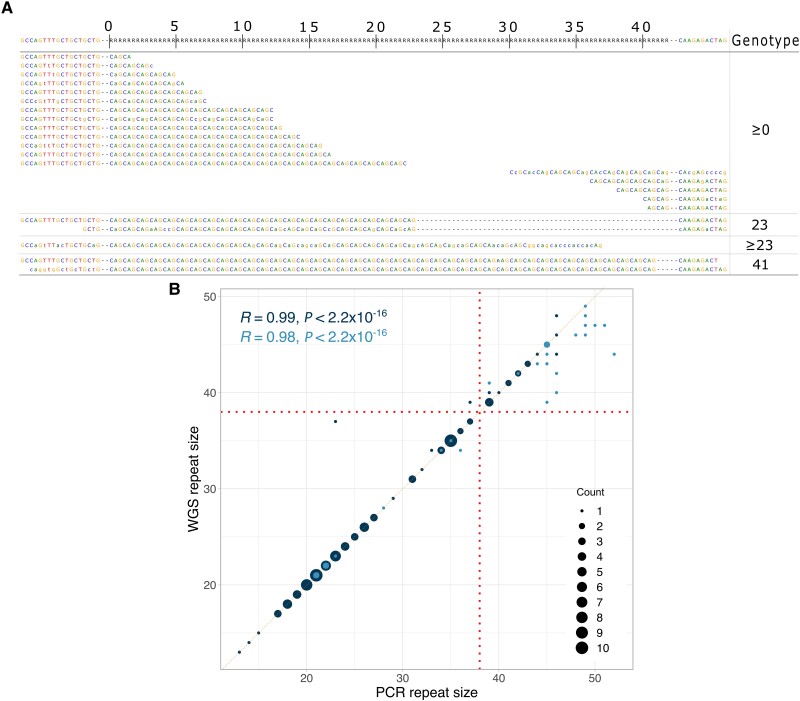
As previously validated,13,15 Expansion Hunter calls for AR CAG repeats underwent a blind quality check process by visual inspection. The ExpansionHunter calls can be visualized by generating ‘pileup’ graphs, which enable the reviewer to easily evaluate the number of reads and the sequences supporting each call, and therefore assess the length of the repeat expansion, as shown in Fig. 1A. A total of 486 pileups were checked, of which there were 282 from 100k GP cohort (≥34 repeats), 67 from NIH (≥34 repeats), 14 from Project MinE (≥37 repeats), and 123 from GnomAD (≥37 repeats). See Supplementary Table 1 for ExpansionHunter calls before and after the visual quality check in each cohort.
Figure 1.
Validation of the WGS pipeline. (A) Visualization of repeat expansion reads from ExpansionHunter shows reads revealing 23 and 41 CAG repeat alleles. (B) Comparison of repeat size estimation between WGS pipeline and PCR. n = 133 alleles. Dark points indicate length confirmed by reads spanning the whole repeat and both the flanking sides; light points indicate length confirmed by reads spanning part of the repeat and one flanking side.
AR detection by WGS benchmarking
To assess the performance of WGS to detect the CAG repeat in the AR gene, we benchmarked our WGS calls against PCR fragment analysis, obtained as follows.
WGS was obtained from 20 individuals with previously identified pathogenic expansion in AR by standard diagnostic PCR testing (i.e. positive control, Supplementary Fig. 1 and Supplementary Table 3, validation ID: NYGC 1–20; 22 alleles from 18 males and two females).
Furthermore, we obtained PCR fragment analysis results for 56 patients recruited to the 100k GP that had been tested previously for the AR expansion (i.e. negative controls, Supplementary Fig. 1 and Supplementary Table 3, validation id: GE_1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 30, 31, 32, 33, 35, 36, 37, 38, 39, 43, 45, 46, 47, 48, 51, 52, 54, 55, 59, 61, 63, 64, 66, 67, 69, 72, 73, 74, 75; 79 alleles from 33 males and 23 females).
We also assessed by PCR 21 DNA samples from patients recruited to the 100k GP, where WGS/ExpansionHunter predicted the presence of an expansion (Supplementary Fig. 1 and Supplementary Table 3, validation id: GE_28, 29, 34, 40, 41, 42, 44, 49, 50, 53, 56, 57, 58, 60, 62, 65, 68, 70, 71, 76, 77; 32 alleles from 10 males and 11 females).
PCR
The CAG trinucleotide repeat length in AR was quantified using a PCR method, where AR alleles were amplified by PCR using GoTaq DNA polymerase (Promega), with the forward primer (6FAM-GCCTGTTGAACTCTTCTGAGC) containing a fluorescein amidite (FAM)-label, used to enable fluorescence detection during the fragment analysis, and the reverse primer GCTGTGAAGGTTGCTGTTCCTC.16 PCR products were electrophoresed on an ABI 3730xl DNA analyser with a LIZ-500 size standard (Applied Biosystems). Fragment analysis was performed with GeneMapper software (version 5.0, Applied Biosystems), deriving numbers of repeats from a standard curve generated using samples of known repeat size ascertained by Sanger sequencing.
Statistical analysis
The statistical formulas used to assess the repeat expansion performance dataset have been taken from https://www.medcalc.org/calc/diagnostic_test.php. Considering TN = true negative; FP = false positive; TP = true positive; FN = false negative; PPV = positive predictive value:
| (1) |
| (2) |
| (3) |
The R correlation coefficient was calculated using Pearson’s equation:
| (4) |
where r = correlation coefficient; xi = values of the x-variable in a sample; x = mean of the values of the x-variable; yi = values of the y-variable in a sample; y = mean of the values of the y-variable.
95% CIs for the X chromosome frequencies were computed using the Wilson score method:
| (5) |
where p = confidence interval for the proportion; = estimated proportion; = statistical test; n = cohort numerosity.
Disease prevalence estimation
We tabulated the cumulative distribution of disease onset reported for 983 patients,9 binning them in 5-year age groups (Fig. 2C, top). We also plotted the distribution of the general English male population (n = 27 827 831),17 using the same 5-year age group bins (Fig. 2C, middle). We then multiplied the cumulative distribution of the disease onset by the corresponding general male count for each age group, to obtain the distribution of the disease by age group, which we then use to estimate the disease prevalence.
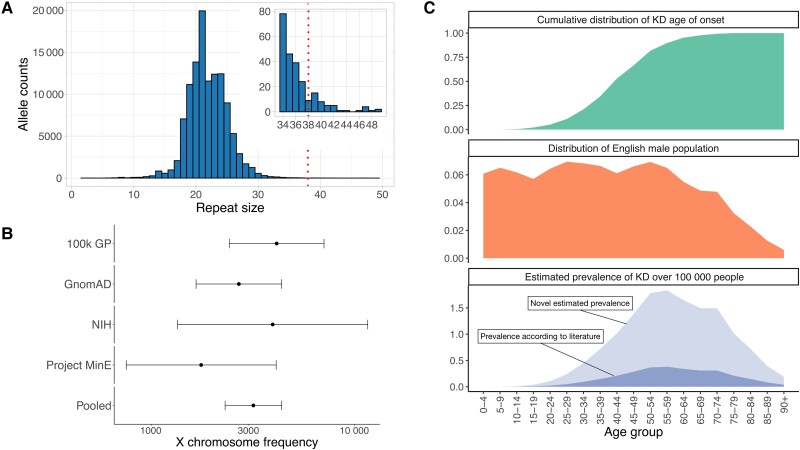
Figure 2.
WGS pipeline detects increased AR CAG expansion in four large cohorts. (A) Allele size distribution across 75 035 100k GP genomes; inset highlights the distribution of alleles containing ≥34 repeats. (B) Frequency estimation of AR CAG expansion. WGS pipeline detects 1:3182 AR expansion ≥38 repeats in the pooled 100k GP, gnomAD, NIH, and MinE cohorts. Error bars show 95% CI. (C) Top) Cumulative distribution of SBMA age of onset for n = 983 KD cases from the most recent KD meta-analysis; (middle) distribution of the English male population (n = 27 827 831); and (bottom) resulting estimated prevalence of SBMA by age group, considering the literature reported male prevalence of 1:30 303 or less (dark area), and the novel estimated prevalence according to our WGS result (1:6887 males, light area).
Haplotyping
Starting from the genomic variant call format (gVCF) files from the 100k GP individuals with more than 37 CAG repeats and a European genetic background (n = 24, of which 13 males and 11 females), we created merged VCFs for males and females, respectively. We then used gvcfgenotyper to select variants with a sex-adjusted minimum allele frequency (MAF) of 5% within the region comprising 579 kb before and 145 kb after the AR CAG repeat (ChrX:66 965 021–67 875 619, GRCh38).18 We repeated the process on n = 14 346 controls, of which there were 6631 males and 7715 females. Using plink, we created the case input files for Haploview, which were used to shortlist the variants using the tagger function. We then employed the resulting 31 variants to shortlist from a merged VCF file with data from both cases and controls (n = 14 370), creating the input files for the formal analysis, performed with Haploview. Within our cohort, we applied the following exclusion criteria: (i) Hardy-Weinberg equilibrium P-value for controls <0.001; (ii) genotyping rate >99%; and (iii) MAF >0.01.
Data availability
Primary data from the 100k GP, which are held in a secure Research Environment, are available to registered users. Please see https://www.genomicsengland.co.uk/about-gecip/for-gecip-members/data-and-data-access for further information.
Results
A sensitive and specific pipeline to detect AR CAG expansions
Our WGS analysis pipeline to analyse the AR expansion combines ExpansionHunter with visual validation of positive results, in accordance with recent guidelines from the American College of Medical Genetics (Fig. 1A).13,15
We benchmarked our pipeline against the gold standard diagnostic method, PCR fragment analysis. We used 133 alleles from 97 samples where the WGS pipeline identified PCR-confirmed expanded (n = 38) and normal (n = 94) alleles, resulting in a sensitivity of 100% (95% CI 90.8–100%), specificity 99% (95% CI 94.2–99.7%), and positive predictive value of 97.4% (95% CI 84.4–99.6%) (Table 2, Supplementary Fig. 1 and Supplementary Table 3).
Table 2.
Sensitivity, specificity, and positive predictive value for AR pathogenic expansion detection
| Parameter | Value (95% CI) |
|---|---|
| Sensitivity | 100% (90.8–100%) |
| Specificity | 99.0% (94.2–99.7%) |
| Positive predictive value | 97.4% (84.4–99.6%) |
Size estimation correlation yielded R = 0.99 (P < 2.2 × 10−16), with high accuracy in alleles with less than 38 repeats, whilst larger repeats were determined to be in the pathogenic range, but less accurately sized as previously shown (Fig. 1B and Supplementary Fig. 2).14
Unexpected frequency of pathogenic AR CAG expansions in the UK population
The 100k GP sequenced the whole genomes of people with a wide range of rare diseases and cancers in the National Health Service in England. Individuals were recruited with their family members where available.12 The AR allele size distribution in 75 035 individuals from this cohort showed a typical bell shape with a peak at 21 repeats (Fig. 2A and Supplementary Fig. 3).
Analysis of 40 412 unrelated individuals within this cohort identified 25 people carrying pathogenic repeats (≥38 repeats), including 11 females and 14 males. Clinical data available for each individual recruited to the 100k GP, including ICD-10 codes and Human Phenotype Ontology (HPO) terms, were reviewed. Of the 14 males, seven proved to have a clinically confirmed diagnosis of SBMA, whilst all remaining individuals were under 21 years of age, except for one recruited for retinal disorders (Supplementary Table 4). None of the female carriers, who can generally develop mild symptoms, had HPO terms associated with neuromuscular conditions.
To estimate the frequency of AR pathogenic expansions, we analysed the repeat size in all unrelated female and male individuals. To avoid overestimating the frequency due to individuals being recruited because of SBMA-related symptoms, we excluded all males recruited under ‘neurological disorders’. We found the X chromosome frequency of the pathogenic expansion to be 1:6536 (95% CI 1:1793–1:23 833, n = 13 072) and 1:3709 (95% CI 1:2071–1:6642, n = 40 800) in males and females respectively (Table 1 and Fig. 2B).
Multiple large cohorts confirm AR CAG expansion frequency
Given the surprisingly high frequency of the AR repeat expansion, we sought to carry out our analysis on replication datasets, using North American (NIH and gnomAD) and European (Project MinE) cohorts, where control and neurodegenerative diseases were sequenced with WGS10,19 (Supplementary Table 2). The AR expansion frequency was 1:2989 and 1:2567 X chromosomes in all males (n = 14 947) and all females (n = 28 232), respectively, in the gnomAD cohort, 1:1529 and 1:5176 X chromosomes in control males (n = 1529) and all females (n = 10 352), respectively, in the NIH cohort, and 1:636 and 1:2510 X chromosomes in control males (n = 1272) and all females (n = 7530), respectively, in the MinE cohort (Fig. 2B and Supplementary Fig. 4). Estimates of AR expansion frequency from these cohorts fall within the 95% CI of the frequency estimated in our 100k GP discovery cohort.
A pooled analysis resulted in an overall frequency of 1:3182 X chromosomes (95% CI 1:2309–1:4386, n = 117 734) (Table 1 and Supplementary Table 1). Notably, the results with a threshold of 37 repeats, which is known to cause SBMA, were even higher at 1:1899 X chromosomes (95% CI 1:1482–1:2434) (Supplementary Fig. 5 and Supplementary Table 1).
A discrepancy between expected disease prevalence and current diagnoses
The expected prevalence of the disease is lower than the mutation frequency, as SBMA is an adult-onset disease. We, therefore, used SBMA age of onset distribution9 and the general English male population age distribution17 with our genetic frequency data to estimate disease prevalence (Fig. 2C). Surprisingly, our results estimated SBMA prevalence at 1:6887 males, more than 4-fold more frequent than previous patient-based epidemiological studies.4–6 To rule out a founder effect, as seen in the Finnish study,7 we performed a haplotype analysis on European samples from the 100k GP, which resulted in non-significant associations (Supplementary Fig. 6).
Discussion
Overall, our work identifies an unexpected frequency of the AR pathogenic expansion in a UK cohort and confirms this finding using three other large European and North American datasets. Previous findings of an epidemiological study in the Vasa region and a meta-analysis are in line with our findings. Importantly, our use of WGS data allowed us to curate our dataset for relatedness and perform a haplotype analysis that rules out founder effects.
The discrepancy between patient numbers and the frequency of the genetic defect may be due to (i) underdiagnosis of this neuromuscular condition; (ii) variable disease expressivity/reduced penetrance; (iii) pleomorphic clinical manifestations; or (iv) a combination of these factors.
Underdiagnosis of the disease has frequently been suggested, and, whilst the classic disease manifestation with bulbar and limb weakness, highly elevated creatine kinase levels, and gynaecomastia is very typical, the disease can manifest with only certain symptoms and often with a negative family history due to its X-linked mode of transmission, favouring misdiagnosis.1,7
Differently from other STR expansion disorders showing incomplete penetrance for all the repeat lengths,20 SBMA is reported to be incompletely penetrant between 35 and 37 repeats, but fully penetrant from 38.1 Moreover, although strong variability in manifestations and severity of SBMA can occur within siblings, reports of incomplete penetrance within families of SBMA patients are lacking. A recent meta-analysis raised the hypothesis that the AR CAG repeat is partially penetrant up to 45 repeats,9 although the fact that in the 100k GP all the males older than 45 years, with more than 37 repeats, had an SBMA phenotype argues against reduced penetrance as being the main driver of the discrepancy between patient numbers and mutation frequency. Larger numbers and more targeted studies will be needed to fully clarify this.
Lastly, SBMA has been associated with a number of common non-neurological disorders such as insulin resistance, non-alcoholic fatty liver disease, and metabolic syndrome,3 and in light of the frequency of the genetic defect, it should likely be considered in people with these conditions.
In conclusion, we identified an unexpectedly high frequency of the SBMA genetic defect in European and North American populations, suggesting SBMA is underdiagnosed and highlighting how testing may be relevant not only to neuromuscular diseases.
Supplementary Material
Acknowledgements
This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly-owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. M.Z. would like to thank the EU Erasmus+ Programme.
Appendix 1
The Genomics England Research Consortium
John C. Ambrose, Prabhu Arumugam, Roel Bevers, Marta Bleda, Freya Boardman-Pretty, Christopher R. Boustred, Helen Brittain, Mark J. Caulfield, Georgia C. Chan, Greg Elgar, Tom Fowler, Adam Giess, Angela Hamblin, Shirley Henderson, Tim J. P. Hubbard, Rob Jackson, Louise J. Jones, Dalia Kasperaviciute, Melis Kayikci, Athanasios Kousathanas, Lea Lahnstein, Sarah E. A. Leigh, Ivonne U. S. Leong, Javier F. Lopez, Fiona Maleady-Crowe, Meriel McEntagart, Federico Minneci, Loukas Moutsianas, Michael Mueller, Nirupa Murugaesu, Anna C. Need, Peter O’Donovan, Chris A. Odhams, Christine Patch, Mariana Buongermino Pereira, Daniel Perez-Gil, John Pullinger, Tahrima Rahim, Augusto Rendon, Tim Rogers, Kevin Savage, Kushmita Sawant, Richard H. Scott, Afshan Siddiq, Alexander Sieghart, Samuel C. Smith, Alona Sosinsky, Alexander Stuckey, Mélanie Tanguy, Ana Lisa Taylor Tavares, Ellen R. A. Thomas, Simon R. Thompson, Arianna Tucci, Matthew J. Welland, Eleanor Williams, Katarzyna Witkowska, Suzanne M. Wood.
Project MinE ALS Sequencing Consortium
Wouter Van Rheenen, Sara L. Pulit, Annelot M. Dekker, Ahmad Al Khleifat, William J. Brands, Alfredo Iacoangeli, Kevin P. Kenna, Ersen Kavak, Maarten Kooyman, Russell L. McLaughlin, Bas Middelkoop, Matthieu Moisse, Raymond D. Schellevis, Aleksey Shatunov, William Sproviero, Gijs H. P. Tazelaar, Rick A. A. Van der Spek, Perry T. C. Van Doormaal, Kristel R. Van Eijk, Joke Van Vugt, A. Nazli Basak, Ian P. Blair, Jonathan D. Glass, Orla Hardiman, Winston Hide, John E. Landers, Jesus S. Mora, Karen E. Morrison, Stephen Newhouse, Wim Robberecht, Christopher E. Shaw, Pamela J. Shaw, Philip Van Damme, Michael A. Van Es, Naomi R. Wray, Ammar Al-Chalabi, Leonard H. Van den Berg, Jan H. Veldink.
Contributor Information
Matteo Zanovello, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK.
Kristina Ibáñez, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London EC1M 6BQ, UK.
Anna-Leigh Brown, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK.
Prasanth Sivakumar, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK.
Alessandro Bombaci, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK; ‘Rita Levi Montalcini’ Department of Neuroscience, University of Turin, Turin 10126, Italy.
Liana Santos, Neurogenetics Unit, National Hospital for Neurology and Neurosurgery, London WC1N 3BG, UK.
Joke J F A van Vugt, Department of Neurology, UMC Utrecht Brain Center, University Medical Center Utrecht, Utrecht University, Utrecht 3508, The Netherlands.
Giuseppe Narzisi, Center for Genomics of Neurodegenerative Disease, New York Genome Center, New York, NY 10013, USA.
Ramita Karra, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA; Department of Neurology, Brain Sciences Institute, Baltimore, MD 21287, USA.
Sonja W Scholz, Department of Neurology, Brain Sciences Institute, Baltimore, MD 21287, USA; Neurogenetics Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Jinhui Ding, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA.
J Raphael Gibbs, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA.
Adriano Chiò, ‘Rita Levi Montalcini’ Department of Neuroscience, University of Turin, Turin 10126, Italy.
Clifton Dalgard, Department of Anatomy, Physiology and Genetics, School of Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA.
Ben Weisburd, Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MT 02142, USA.
Michael G Hanna, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK.
Linda Greensmith, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK.
Hemali Phatnani, Center for Genomics of Neurodegenerative Disease, New York Genome Center, New York, NY 10013, USA.
Jan H Veldink, Department of Neurology, UMC Utrecht Brain Center, University Medical Center Utrecht, Utrecht University, Utrecht 3508, The Netherlands.
Bryan J Traynor, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA; Department of Neurology, Brain Sciences Institute, Baltimore, MD 21287, USA.
James Polke, Neurogenetics Unit, National Hospital for Neurology and Neurosurgery, London WC1N 3BG, UK.
Henry Houlden, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK; Neurogenetics Unit, National Hospital for Neurology and Neurosurgery, London WC1N 3BG, UK.
Pietro Fratta, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK.
Arianna Tucci, Department of Neuromuscular Diseases, Queen Square Institute of Neurology, UCL, London WC1N 3BG, UK; William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London EC1M 6BQ, UK.
The American Genome Center (TAGC) consortium, Genomics England Research Consortium, Project MinE ALS Sequencing Consortium, The NYGC ALS Consortium:
John C Ambrose, Prabhu Arumugam, Roel Bevers, Marta Bleda, Freya Boardman-Pretty, Christopher R Boustred, Helen Brittain, Mark J Caulfield, Georgia C Chan, Greg Elgar, Tom Fowler, Adam Giess, Angela Hamblin, Shirley Henderson, Tim J P Hubbard, Rob Jackson, Louise J Jones, Dalia Kasperaviciute, Melis Kayikci, Athanasios Kousathanas, Lea Lahnstein, Sarah E A Leigh, Ivonne U S Leong, Javier F Lopez, Fiona Maleady-Crowe, Meriel McEntagart, Federico Minneci, Loukas Moutsianas, Michael Mueller, Nirupa Murugaesu, Anna C Need, Peter O’Donovan, Chris A Odhams, Christine Patch, Mariana Buongermino Pereira, Daniel Perez-Gil, John Pullinger, Tahrima Rahim, Augusto Rendon, Tim Rogers, Kevin Savage, Kushmita Sawant, Richard H Scott, Afshan Siddiq, Alexander Sieghart, Samuel C Smith, Alona Sosinsky, Alexander Stuckey, Mélanie Tanguy, Ana Lisa Taylor Tavares, Ellen R A Thomas, Simon R Thompson, Arianna Tucci, Matthew J Welland, Eleanor Williams, Katarzyna Witkowska, Suzanne M Wood, Wouter Van Rheenen, Sara L Pulit, Annelot M Dekker, Ahmad Al Khleifat, William J Brands, Alfredo Iacoangeli, Kevin P Kenna, Ersen Kavak, Maarten Kooyman, Russell L McLaughlin, Bas Middelkoop, Matthieu Moisse, Raymond D Schellevis, Aleksey Shatunov, William Sproviero, Gijs H P Tazelaar, Rick A A Van der Spek, Perry T C Van Doormaal, Kristel R Van Eijk, Joke Van Vugt, A Nazli Basak, Ian P Blair, Jonathan D Glass, Orla Hardiman, Winston Hide, John E Landers, Jesus S Mora, Karen E Morrison, Stephen Newhouse, Wim Robberecht, Christopher E Shaw, Pamela J Shaw, Philip Van Damme, Michael A Van Es, Naomi R Wray, Ammar Al-Chalabi, Leonard H Van den Berg, and Jan H Veldink
Funding
A.T. is supported by a UK Medical Research Council Clinician Scientist Fellowship (MR/S006753/1). P.F. is supported by a UK Medical Research Council Senior Clinical Fellowship and Lady Edith Wolfson Fellowship (MR/M008606/1 and MR/S006508/1), the UCLH NIHR Biomedical Research Centre, the Neurological Research Trust and KDUK. This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No. 772376—EScORIAL). This study was supported by the ALS Foundation Netherlands. This work was supported, in part, by the Intramural Research Program of the National Institute on Aging (Z01-AG000949-02) and the National Institute of Neurological Disorders and Stroke.
Competing interests
Genomics England Ltd. is a wholly-owned Department of Health and Social Care company created in 2013 to introduce WGS into healthcare in conjunction with NHS England. All Genomics England affiliated authors are, or were, salaried by or seconded to Genomics England. J.H.V. received sponsored research agreements from Biogen. The other authors declare no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. La Spada A. Spinal and bulbar muscular atrophy. In: Adam MPArdinger HH and Pagon RA, et al., eds. GeneReviews®. University of Washington; 1999. https://www.ncbi.nlm.nih.gov/books/NBK1333/ [PubMed] [Google Scholar]
- 2. Fratta P, Nirmalananthan N, Masset L, et al. . Correlation of clinical and molecular features in spinal bulbar muscular atrophy. Neurology. 2014;82:2077–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manzano R, Sorarú G, Grunseich C, et al. . Beyond motor neurons: Expanding the clinical spectrum in SBMA. J Neurol Neurosurg Psychiatry. 2018;89:808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertolin C, Querin G, Martinelli I, Pennuto M, Pegoraro E, Sorarù G. Insights into the genetic epidemiology of spinal and bulbar muscular atrophy: Prevalence estimation and multiple founder haplotypes in the veneto Italian region. Eur J Neurol. 2019;26:519–524. [DOI] [PubMed] [Google Scholar]
- 5. Guidetti D, Sabadini R, Ferlini A, Torrente I. Epidemiological survey of X-linked bulbar and spinal muscular atrophy, or kennedy disease, in the province of reggio Emilia, Italy. Eur J Epidemiol. 2001;17:587–591. [DOI] [PubMed] [Google Scholar]
- 6. Zelinkova H, Kolejakova KL, Spalek P, Chandoga J, Konkolova J, Bohmer D. Molecular diagnosis of spinal and bulbar muscular atrophy in Slovakia. BLL. 2016;116:137–141. [DOI] [PubMed] [Google Scholar]
- 7. Udd B, Juvonen V, Hakamies L, et al. . High prevalence of SBMA in western Finland - is the syndrome underdiagnosed? Acta Neurol Scand. 1998;98:128–133. [DOI] [PubMed] [Google Scholar]
- 8. Gardiner SL, Boogaard MW, Trompet S, et al. . Prevalence of carriers of intermediate and pathological polyglutamine disease–associated alleles among large population-based cohorts. JAMA Neurol. 2019;76:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laskaratos A, Breza M, Karadima G, Koutsis G. Wide range of reduced penetrance alleles in spinal and bulbar muscular atrophy: A model-based approach. J Med Genet. 2021;58:385–391. [DOI] [PubMed] [Google Scholar]
- 10. Karczewski KJ, Francioli LC, Tiao G, et al. . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dolzhenko E, van Vugt JJFA, Shaw RJ, et al. . Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27:1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. The 100,000 Genomes Project Pilot Investigators . 100,000 Genomes pilot on rare-disease diagnosis in health care—preliminary report. N Engl J Med. 2021;385:1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ibanez K, Polke J, Hagelstrom T, et al. . Whole genome sequencing for diagnosis of neurological repeat expansion disorders. Lancet Neurol. 2022;21:234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dolzhenko E, Deshpande V, Schlesinger F, et al. . Expansionhunter: A sequence-graph-based tool to analyze variation in short tandem repeat regions. Birol I, ed. Bioinformatics. 2019;35:4754–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roy S, Coldren C, Karunamurthy A, et al. . Standards and guidelines for validating next-generation sequencing bioinformatics pipelines. J Mol Diagn. 2018;20:4–27. [DOI] [PubMed] [Google Scholar]
- 16. Fratta P, Collins T, Pemble S, et al. . Sequencing analysis of the spinal bulbar muscular atrophy CAG expansion reveals absence of repeat interruptions. Neurobiol Aging. 2014;35:443.e1–443.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Office for National Statistics. Population estimates for the UK, England and Wales, Scotland and Northern Ireland: mid-2019. Published 24 June 2020. https://www.ons.gov.uk/peoplepopulationandcommunity/populationandmigration/populationestimates/bulletins/annualmidyearpopulationestimates/mid2019estimates
- 18. Santos D, Pimenta J, Wong VC, Amorim A, Martins S. Diversity in the androgen receptor CAG repeat has been shaped by a multistep mutational mechanism. Am J Med Genet. 2014;165:581–586. [DOI] [PubMed] [Google Scholar]
- 19. Project MinE ALS Sequencing Consortium . Project MinE: Study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur J Hum Genet. 2018;26:1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murphy NA, Arthur KC, Tienari PJ, Houlden H, Chiò A, Traynor BJ. Age-related penetrance of the C9orf72 repeat expansion. Sci Rep. 2017;7:2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary data from the 100k GP, which are held in a secure Research Environment, are available to registered users. Please see https://www.genomicsengland.co.uk/about-gecip/for-gecip-members/data-and-data-access for further information.