Abstract
Neurofilament light chain, a putative measure of neuronal damage, is measurable in blood and CSF and is predictive of cognitive function in individuals with Alzheimer’s disease. There has been limited prior work linking neurofilament light and functional connectivity, and no prior work has investigated neurofilament light associations with functional connectivity in autosomal dominant Alzheimer’s disease. Here, we assessed relationships between blood neurofilament light, cognition, and functional connectivity in a cross-sectional sample of 106 autosomal dominant Alzheimer’s disease mutation carriers and 76 non-carriers. We employed an innovative network-level enrichment analysis approach to assess connectome-wide associations with neurofilament light.
Neurofilament light was positively correlated with deterioration of functional connectivity within the default mode network and negatively correlated with connectivity between default mode network and executive control networks, including the cingulo-opercular, salience, and dorsal attention networks. Further, reduced connectivity within the default mode network and between the default mode network and executive control networks was associated with reduced cognitive function. Hierarchical regression analysis revealed that neurofilament levels and functional connectivity within the default mode network and between the default mode network and the dorsal attention network explained significant variance in cognitive composite scores when controlling for age, sex, and education. A mediation analysis demonstrated that functional connectivity within the default mode network and between the default mode network and dorsal attention network partially mediated the relationship between blood neurofilament light levels and cognitive function.
Our novel results indicate that blood estimates of neurofilament levels correspond to direct measurements of brain dysfunction, shedding new light on the underlying biological processes of Alzheimer’s disease. Further, we demonstrate how variation within key brain systems can partially mediate the negative effects of heightened total serum neurofilament levels, suggesting potential regions for targeted interventions. Finally, our results lend further evidence that low-cost and minimally invasive blood measurements of neurofilament may be a useful marker of brain functional connectivity and cognitive decline in Alzheimer’s disease.
Keywords: NfL, neurodegeneration, functional connectivity, default mode network, resting state, enrichment
Wheelock et al. provide new evidence that low cost and minimally invasive measurements of serum neurofilament light are a useful marker of functional connectivity and cognitive decline in Alzheimer’s disease.
Introduction
In Alzheimer’s disease (AD), a cascading progression of events, including the spatially distributed accrual of amyloid-β (Aβ) plaques and neurofibrillary tau tangles, cortical thinning, hypometabolism, and disruptions in brain connectivity lead to characteristic and profound deficits in cognitive functioning.1–4 Recent studies have shown that blood-based assays of neurofilaments—structural proteins of neuron cytoskeletons that provide support and stability to axons—and neurofilament light chain (NfL) in particular, may provide an easy to index biomarker of neurodegeneration and disease progression in AD (Fig. 1).5 Given that concentrations of neurofilament proteins both in CSF and in blood increase in the setting of axonal damage,5–7 elevated NfL concentrations in the setting of AD may provide an indicator of disruption in neural communication within the brain. Further, AD-associated cognitive decline and the onset of dementia have been linked to the progressive decoupling of brain functional networks.8 Herein, we aim to investigate the direct relationship between blood-based NfL biomarkers of AD and brain functional network integrity.
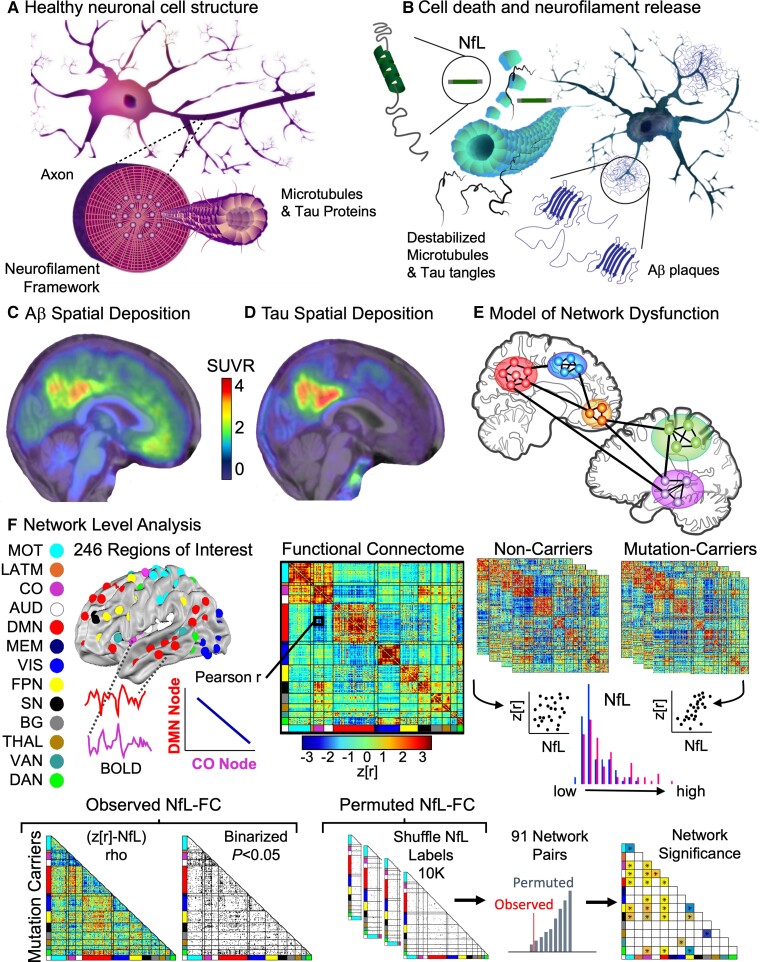
Figure 1.
Disruptions at the molecular level drive network level disruptions in the connectome. (A) Neurofilaments, surrounding an inner layer of microtubules and tau proteins, provide a structural framework to axons. (B) Axonal injury and neuronal death lead to the release of neurofilaments into the extracellular fluid. (C) Amyloid PET and (D) tau PET spatial localization converted to a regional SUVR for an individual with ADAD. (E) Regional neurodegeneration results in reduced communication among brain regions and resulting network dysfunction. (F) Network Level Analysis can be used to assess connectome-wide disruptions in FC and associations with biomarkers and clinical outcomes. BOLD time series were extracted from 246 spherical ROI. The non-parametric correlation between NfL and whole brain connectome was examined separately for each group. Significance was established by randomly permuting the NfL values 10 000 times and measuring the permuted connectome-NfL relationship. AUD = auditory; BG = basal ganglia; CO = cingulo-opercular; DAN, dorsal attention network; DMN, default mode network; FPN = frontoparietal network; LATM = lateral motor; MEM = memory; MOT = motor; SN = salience network; THAL = thalamus; VAN = ventral attention network; VIS = visual.
A powerful assay of brain health is functional connectivity (FC), which can be assessed using data from non-invasive resting state functional MRI (fMRI),9 a technique that calculates the temporal correlation of spontaneous fluctuations of the blood oxygen-level dependent (BOLD) signal between brain regions.10 Studies using these methods have largely focused on the default mode network (DMN),11 demonstrating that regions of early Aβ deposition overlap with reduced FC within brain networks.12–16 However, a growing literature has also demonstrated decline in FC within executive control networks17–21 as well as between the DMN and executive control networks such as the frontoparietal15 and dorsal attention networks17,18,22 with increasing disease severity. While prior research suggests reduced FC is predictive of disease onset,23 the temporal order of loss of connectivity within and between these networks is only partially understood.18,19,23–25 Though many studies have examined the relationship between FC and Aβ as indexed with CSF Aβ42 or deposits indexed with PET, only recently has research investigated possible disruptions in FC associated with CSF NfL in cognitively normal adults26 and individuals with mild cognitive impairment and dementia.27 Further, only one study to date has assessed blood plasma NfL associations with FC and found associations with FC between regions of the DMN and regions within frontal and parietal cortex in individuals with mild cognitive impairment and sporadic AD.28 Importantly, no extant research has assessed connectome-wide associations with blood-serum markers of NfL in either cognitively normal individuals or those with autosomal dominant AD.
Herein, we sought to examine the relationships between the progressive loss of brain connectivity characteristic of AD with cognition and a blood-based biomarker of neuronal integrity. To investigate the connections between blood-based NfL biomarker status, brain functional network health, and cognitive function, we use data from the Dominantly Inherited Alzheimer Network (DIAN), a unique international collaboration that has collected data from families with pathogenic mutations in the presenilin 1 (PSEN1), presenilin 2 (PSEN2), or amyloid precursor protein (APP) genes.29 Crucially, the relatively consistent age at symptom onset within families and mutation types allows participants to be staged relative to their estimated years before symptom onset (EYO). This feature of autosomal-dominant AD (ADAD) provides a unique opportunity to study how the temporal emergence of biomarkers covaries with the spatial progression of the disease within the brain. Indeed, prior research in ADAD has demonstrated that elevated blood NfL is associated with reduced white matter integrity30 and cortical thickness.31 Given that blood serum NfL is a direct measure of putative neuronal injury and given its prior associations with white and grey matter, we hypothesized that blood serum NfL levels would be associated with reduced neural circuit integrity, as indexed by a reduction in within- and between-network FC. To test this hypothesis, we assessed brain network FC as a function of disease stage in ADAD mutation carriers (MC) as compared to mutation non-carriers (NC). We then assessed connectome-wide brain network associations with serum NfL levels using an innovative network-level enrichment analysis. Specifically, we hypothesized that increasing NfL levels would be associated with increasing disruption of DMN within- and between-network connectivity, and that this loss of network integrity would be associated with poorer cognition among participants with ADAD. However, by employing a connectome-wide association approach we were able to probe all possible systems which may be impacted by neuronal cell death while controlling the family-wise error (FWE) rate with network-level analysis. Finally, we assessed the unique statistical contribution of NfL, FC, and PET Aβ deposits in explaining cognitive function in ADAD.
Materials and methods
Participants
Dominantly Inherited Alzheimer Network participants whose clinical, cognitive, neuroimaging, genetic, and blood draw assessment data passed quality control as a part of the 11th DIAN data release were considered for participation in this study. The institutional review board at Washington University in St. Louis provided supervisory review and human studies approval for secondary data analysis. Inclusion in this study was further restricted to participants with at least one serum NfL measurement and one resting state fMRI scan within one year of this serum NfL measurement. The average number of days between serum NfL and cognitive measurements and fMRI scan for each participant was [mean ± standard error (SE)] 4.49 ± 1.01 days. The final cohort consisted of 106 MC [81 (76.4%) PSEN1; 10 (9.4%) PSEN2; 15 (14.2%) APP] and 76 family member NC assessed cross-sectionally.
Cognitive assessment
Several clinical and cognitive assessments were collected in order to measure cognitive and functional status in DIAN participants. The Clinical Dementia Rating® (CDR®) Instrument32 is a five-point scale that characterizes six domains (memory, orientation, judgement and problem solving, community affairs, home and hobbies, and personal care). CDR = 0 indicates ‘cognitively normal’, and CDR > 0 indicates symptomatic AD (encompassing mild cognitive impairment due to AD and AD dementia). The Mini-Mental State Examination33 is an 11-item measure that tests five areas of cognitive function (orientation, registration, attention and calculation, recall, and language). A score of 23 or lower indicates cognitive impairment. Further, the Wechsler Adult Intelligence Scale (WAIS-R)34 was administered to measure intelligence and cognitive ability.
Cognitive Composite Score
We calculated a Cognitive Composite Score (CCS) to measure cognitive function for each individual using previously described methods.35–37 The cognitive composite score is a holistic summary of cognitive functions including episodic memory, executive functioning, processing speed, and mental status. The composite score consists of results from four tests: Mini-Mental State Exam,33 logical memory delay recall,38 the Digit Symbol Substitution test,34 and delayed recall of a 16-item word list (designed for the DIAN study by David A. Balota). A cognitive composite score is calculated by averaging each test’s normalized scores by an equal weight. The normalized test scores are obtained from standardizing each raw test score to the mean and standard deviation (SD) of previously reported values.37 Because of the ceiling effects of the Mini-Mental State Exam, the SD is small for the healthy population compared to other tests and hence an adjusted SD from a smoothing spline model was used for normalization.
Estimated years before symptom onset
To establish the EYO for all MC and NC participants, at each clinical evaluation, the participant’s current age was compared to the family mutation-specific expected age of onset of cognitive symptoms or, if onset for that specific mutation was unknown, to their parental age of first progressive cognitive decline.
Magnetic resonance imaging
All study sites used 3 T Siemens TIM Trio or Verio scanners which were required to pass regular quality control assessment. Resting state fMRI data were acquired using echo planar imaging (EPI). During resting state fMRI scans, participants were instructed to maintain visual fixation on a crosshair. T1-weighted magnetization-prepared rapid acquisition with gradient echo (MP-RAGE) images were acquired for all participants. Scan parameters can be found in Supplementary Table 8. T1 data were processed using FreeSurfer 5.34, and the Desikan atlas was used to produce regional estimates of grey matter for use in PET processing.
Positron emission tomography
Amyloid-β PET imaging with Pittsburgh Compound B was performed using a bolus injection of [11C] Pittsburgh Compound B. PET data were acquired using either a 70-min scan beginning at the start of the injection or a 30-min scan starting 40 min after the injection Data were converted to regional standardized uptake value ratios (SUVRs) relative to the cerebellar grey matter using regions of interest (ROI) generated in FreeSurfer39 and using a regional spread function with partial volume correction.40–42 A global measure of mean cortical uptake of Aβ burden was derived from cortical regions that have been shown to have elevated signal in AD.39
Neurofilament light
Detailed methods for collection and analysis of NfL in the DIAN cohort have previously been described.31 Briefly, fluids were collected in the morning under fasting conditions, and after collection, the tubes were left at room temperature for 30 min to allow clotting. The tubes were then centrifuged for 15 min, and the serum was placed into a single transfer tube, frozen, and shipped overnight on dry ice to the DIAN core laboratory at Washington University. Serum blood samples were then shipped to the University of Tübingen where they were assayed in duplicate for NfL using a Single Molecule Array (SIMOA HD-1) assay, the capture monoclonal antibody (mAB) 47:3, and the biotinylated detector antibody mAB 2:1 (Quanterix, Uman Diagnostics).
Functional connectivity methods
Preprocessing
Pre-processing generally followed previously described methods43,44 implemented in the 4dfp suite of tools (http://4dfp.readthedocs.io). Odd versus even slice intensity differences attributable to interleaved acquisition were corrected.45 Head motion was corrected within and across runs. Intensity inhomogeneity was corrected using the FAST module in FMRIB Software Library (FSL; 5.0.9)46 followed by intensity normalization to obtain a whole brain mode value of 1000. EPI distortion due to magnetization inhomogeneity was corrected using the mean field map method described by Gholipour et al.47 Atlas transformation was computed by registering the functional data to an atlas-representative template via the MP-RAGE (EPI_mean → MP-RAGE → template). The template was generated from a separate cohort of 12 cognitively normal healthy individuals. Volumetric time series were resampled in (3 mm)3 atlas space in a single step combining head motion correction, distortion correction, and atlas transformation. Frames corrupted by excessive head motion were identified on the basis of both DVARS (frame-to-frame signal change over the entire brain) and frame displacement (FD) measures.48 The DVARS censoring criterion was individually set to accommodate baseline shifts and the FD censoring criterion was 0.4 mm. Frames were censored if either criterion was exceeded. The time series were band-pass filtered to retain frequencies between 0.005 and 0.1 Hz. Censored frames were approximated by linear interpolation for purposes of band-pass filtering but excluded from all subsequent steps.
Denoising
Denoising was accomplished using a CompCor-like strategy.49 As previously described,50 nuisance regressors were derived from three compartments (white matter, ventricles, and extra-axial space) and were then dimensionality-reduced to create a metric for singular value decomposition (SVD). White matter and ventricular masks were segmented in each participant using FreeSurfer (5.3)51 and spatially resampled in register with FC data. All segmentations were visually inspected and manual intervention was applied when necessary by experienced trained technicians prior to inclusion in the processing stream. The final set of nuisance regressors also included the six parameters derived from rigid body head-motion correction, the global signal averaged over the (FreeSurfer-segmented) brain, and the global signal temporal derivative. As a final preprocessing step, the volumetric time series were non-linearly warped to Montreal Neurological Institute (MNI) 152 space (3 mm3 voxels) using FNIRT.52,53
Functional connectome
Regions of interest were selected from a subset of canonical 300 ROI54 to provide dense coverage of the entire cortical surface and subcortical structures (Fig. 1F). ROI in areas of fMRI signal drop out55 were excluded and the cerebellum was additionally excluded owing to inadequate coverage of this structure in some DIAN participants. Thus, FC was estimated using zero-lag Pearson correlations calculated between 246 ROI throughout the cortex and subcortical areas and organized into 12 canonical FC networks.54
Statistical analysis
Sample characteristics
Associations between sample characteristics were quantified using SPSS Statistics 27. We estimated normality for demographic and biological variables using the Kolmogorov–Smirnov test. Tests of normality indicated that serum NfL values were not normally distributed; thus, log values were calculated and used for further analyses.31 Given that log NfL values were still non-normal in the NC group, Spearman rank correlations were used for the Network Level Analysis with FC. Education, amyloid, and age were not normally distributed (Kolmogorov–Smirnov P < 0.05), while EYO and CCS were normal. Thus, for consistency, differences between MC and NC were characterized using Mann–Whitney U-tests, and associations among demographic and biological variables were quantified using Spearman rank correlation. Amyloid data were missing for 5 NC and 12 MC, while CCS were missing for 7 MC. Analyses with missing data were subject to pairwise exclusion.
Functional connectome associations with estimated years before symptom onset
MATLAB 2015a was used to quantify functional connectome differences as a function of mutation status and EYO. In order to increase reliability of FC estimates, connectome data were grouped according to mutation status and three EYO bins (EYO < −10; EYO ≥ −10 and ≤ 0; EYO > 0). In order to characterize the changing network topology over EYO, spring embedded plots were generated based on previously published methods.56 Differences in average within and between-network FC across mutation group and EYO was quantified using a one-way analysis of variance (ANOVA) with nested random effects for families. Bonferroni correction was used to control for multiple comparisons and significant effects were plotted based on mutation status and EYO using RStudio. Post hoc tests were conducted using Tukey’s Honest Significant Difference test was used to control for multiple comparisons.
Network Level Analysis
We examined associations between FC and NfL using a previously described Network Level Analysis approach in MATLAB (Fig. 1F).57–59 Given the strong association between NfL and age in both the MC and NC groups, age was regressed out of NfL and FC data prior to further analysis. Next, we calculated the partial Spearman rank correlation for each ROI pair against the NfL measurement across all participants within each group. Correlations with NfL were nominally thresholded (P < 0.05) and binarized for each group within and between network pairs. For each group, networks were tested for enrichment of NfL association using both a 1-degree-of-freedom χ2 test and hypergeometric tests. The χ2 test compares the observed number of strong associations within a functional network pair to that which would be expected if the overall number of strong associations was uniformly distributed across all possible network pairs. The statistic is large when the number of strong associations within a network pair is much less than (depletion) or much greater than (enrichment) expected. The hypergeometric statistic, as in Fisher’s exact test, assesses the likelihood of observing a given number of strong associations in a network pair, given (i) the total number of strong associations observed overall; and (ii) the total number of possible strong associations for that network pair (i.e. the total number of ROI pairs within a given network pair). Both the χ2 test and the hypergeometric tests had to pass the significance threshold for a given network pair’s enrichment to be considered significant. For both tests, the number of ROI-pairs within a network block passing a primary significance threshold were compared to the overall number of ROI-pairs passing the primary threshold across the entire connectome. Networks enriched for NfL in the MC group were compared to network enrichment in the NC group using the McNemar χ2 test. Within- and between-group network differences were determined by comparing measured NfL-FC network enrichment to a null distribution of network enrichment generated by randomization of NfL values (10 000 permutations). Network level false positive rate was controlled at P < 0.05 corrected for multiple comparisons.
Hierarchical regression analysis
We used hierarchical linear regression to test the unique variance explained in the CCS by network average FC, NfL, and Aβ PET in the MC group (SPSS Statistics 27). Demographic variables including age, sex, and education were included as covariates in all models. Variables were mean-centered and missing data were excluded listwise. Standardized β coefficients are reported.
Exploratory mediation analysis
To further understand the unique contribution of FC to variance in cognitive function, we ran a mediation analysis using the PROCESS macro for SPSS.60 Demographic variables including age, sex, and education were included as covariates in all models. Variables were mean-centred and missing data were excluded listwise. Standardized β coefficients are reported.
Code and data availability
Network Level Analysis methods can be found at https://github.com/WheelockLab/NetworkLevelAnalysisBeta. Data that support the findings of this study are available from DIAN at https://dian.wustl.edu/our-research/observational-study/dian-observational-study-investigator-resources/.
Results
Sample characteristics
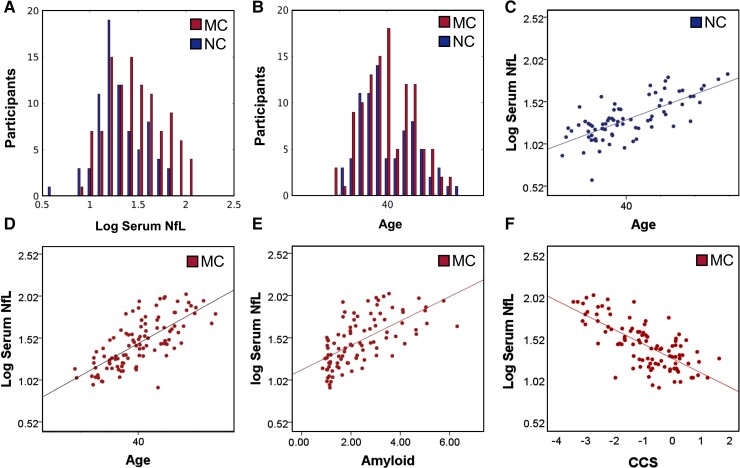
As expected, MC had greater serum NfL measurements than NC (P < 0.001) (Table 1). MC and NC did not differ by age, education, or sex. However, serum NfL was positively correlated with age in both groups (P < 0.001) (Fig. 2 and Table 2). Similarly, MC demonstrated higher CDR and lower (worse) CCS than NC (P < 0.001; Table 1). The EYO positively correlated with age and NfL in both MC and NC groups (P < 0.001; Table 2) while Aβ SUVR was correlated with CCS and NfL in MC (Fig. 2 and Table 2) but not NC.
Table 1.
Sample characteristics
| Measure | Mutation carriers (n = 106) |
Non-carriers (n = 76) |
Test statistic | df | P-value |
|---|---|---|---|---|---|
| χ2 | |||||
| Sex, female % | 49.1% | 61.8% | 2.92 | 1 | 0.088 |
| CDR 0 n(%) | 58 (54.7%) | 76 (100%) | 46.74 | 2 | <0.001 |
| CDR 0.5 | 30 (28.3%) | 0 (0%) | |||
| CDR ≥1 | 18 (17.0%) | 0 (0%) | |||
| – | Median | Median | Mann–Whitney U-test | z | – |
| Age, years | 40.19 | 37.47 | 3906.5 | −0.35 | 0.729 |
| Education, years | 14 | 15 | 3345.5 | −1.97 | 0.049 |
| EYO | −6.07 | −9.52 | 3564.0 | −1.32 | 0.186 |
| NfL, pg/ml | 28.35 | 19.25 | 2873.5 | −3.29 | <0.001 |
| PET Aβa | 2.01 | 1.05 | 614.0 | −8.96 | <0.001 |
| CCSb | −0.84 | 0.09 | 1706.5 | −6.19 | <0.001 |
Bold values indicate significance after Bonferroni correction for eight tests. df = degrees of freedom; PET Aβ = PET with Pittsburgh Compound B amyloid regional SUVRs.
Missing data for five NC (n = 71) and 12 MC participants (n = 94).
Missing data for seven MC participants (n = 99).
Figure 2.
Demographic characteristics and blood estimates of neurofilament light. (A) Mutation carriers had greater levels of serum NfL than NC (P < 0.05). (B) MC and NC did not differ in age. However, age positively correlated with serum NfL in both NC (C) and MC (D). (E–F) NfL was positively correlated with Aβ Pittsburgh Compound B SUVRs and CCS in MC but not NC.
Table 2.
Spearman correlations among sample characteristics
| Non-carriers n = 76 | Education | CCS | EYO | Age | log NfL |
|---|---|---|---|---|---|
| Education | – | – | – | – | – |
| CCS | 0.40 (<0.001) | – | – | – | – |
| EYO | −0.25 (0.029) | −0.25 (0.028) | – | – | – |
| Age | −0.21 (0.073) | −0.25 (0.029) | 0.85 (<0.001) | – | – |
| log NfL | 0.02 (0.855) | 0.01 (0.910) | 0.59 (<0.001) | 0.69 (<0.001) | – |
| PET Aβa | 0.24 (0.041) | 0.31 (0.008) | −0.03 (0.819) | 0.06 (0.627) | 0.17 (0.156) |
| Mutation carriers n = 106 | Education | CCS | EYO | Age | log NfL |
|---|---|---|---|---|---|
| Education | – | – | – | – | – |
| CCSb | 0.45 (<0.001) | – | – | – | – |
| EYO | −0.26 (0.007) | −0.75 (<0.001) | – | – | – |
| Age | −0.15 (0.136) | −0.66 (<0.001) | 0.82 (<0.001) | – | – |
| log NfL | −0.19 (0.049) | −0.69 (<0.001) | 0.78 (<0.001) | 0.74 (<0.001) | – |
| PET Aβc | −0.295 (0.004) | −0.59 (<0.001) | 0.66 (<0.001) | 0.53 (<0.001) | 0.67 (<0.001) |
Bold values indicate significance after Bonferroni correction for 30 tests. PET Aβ = PET with Pittsburgh Compound B amyloid regional SUVRs.
Missing five participants.
Missing seven participants.
Missing 12 participants.
Functional connectome associations with estimated years before symptom onset
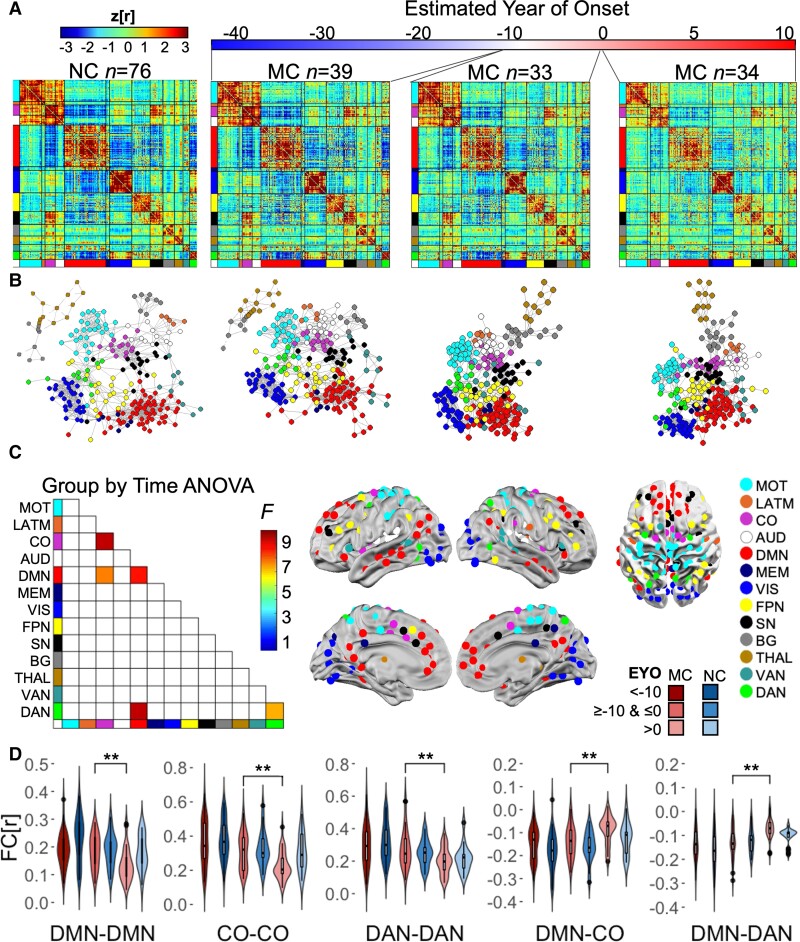
To assess connectome-wide changes associated with increasing disease burden, group average connectomes were calculated for NC and MC grouped into three different EYO bins (EYO < −10; EYO ≥ −10 and ≤ 0; EYO > 0) (Fig. 3A). Given that EYO and age are correlated in MC and NC, comparing FC networks between MC and NC within the same EYO time bin controls for age-related changes in FC. Spring embedded plots56 qualitatively illustrate FC differences between MC and NC and, more specifically, FC differences in MC across EYO, with a later stage EYO being associated with less distinct spatial topology (Fig. 3B). Spring embedded plots did not change across EYO in the NC (Supplementary Fig. 1). Further, one-way ANOVA with a nested random factor for families revealed five network pairs that differed as a function of group and EYO, including: DMN within network FC; cingulo-opercular (CO) within network FC; DMN-CO between-network FC; DMN and dorsal attention network (DAN) between-network FC; and DAN within network FC (Supplementary Table 1 and Fig. 3C). Post hoc tests with Tukey’s Honest Significant Difference revealed that FC was reduced post-EYO 0 in the MC group compared to NC (Fig. 3D) though these differences did not pass multiple comparisons (Supplementary Fig. 2 and Supplementary Table 2). Further, FC was reduced across all five network pairs when comparing pre- to post-EYO 0 within the MC group (P < 0.05 FWE corrected; Fig. 3D and Supplementary Table 3).
Figure 3.
Connectome changes as a function of EYO and carrier group. (A) Group average connectomes for NC compared to MC categorized into three EYO bins. MC demonstrated decreased FC compared to NC when approaching and upon surpassing their EYO. (B) Spring embedded plots for NC and MC with an EYO < −10 demonstrate a similar FC pattern, while plots for MC with an EYO > −10 exhibit reduced/less extensive network topology. (C) One-way ANOVA revealed five significant network pairs (FWE P < 0.001). (D) Post hoc tests revealed reduced FC in the MC group from pre- to post-EYO. MC had reduced FC compared to NC post-EYO (see Supplementary Tables 1–3). AUD = auditory; BG = basal ganglia; CO = cingulo-opercular; DAN, dorsal attention network; DMN, default mode network; FPN = frontoparietal network; LATM = lateral motor; MEM = memory; MOT = motor; SN = salience network; THAL = thalamus; VAN = ventral attention network; VIS = visual. *P < 0.05 FWE.
Connectome-wide associations with NfL
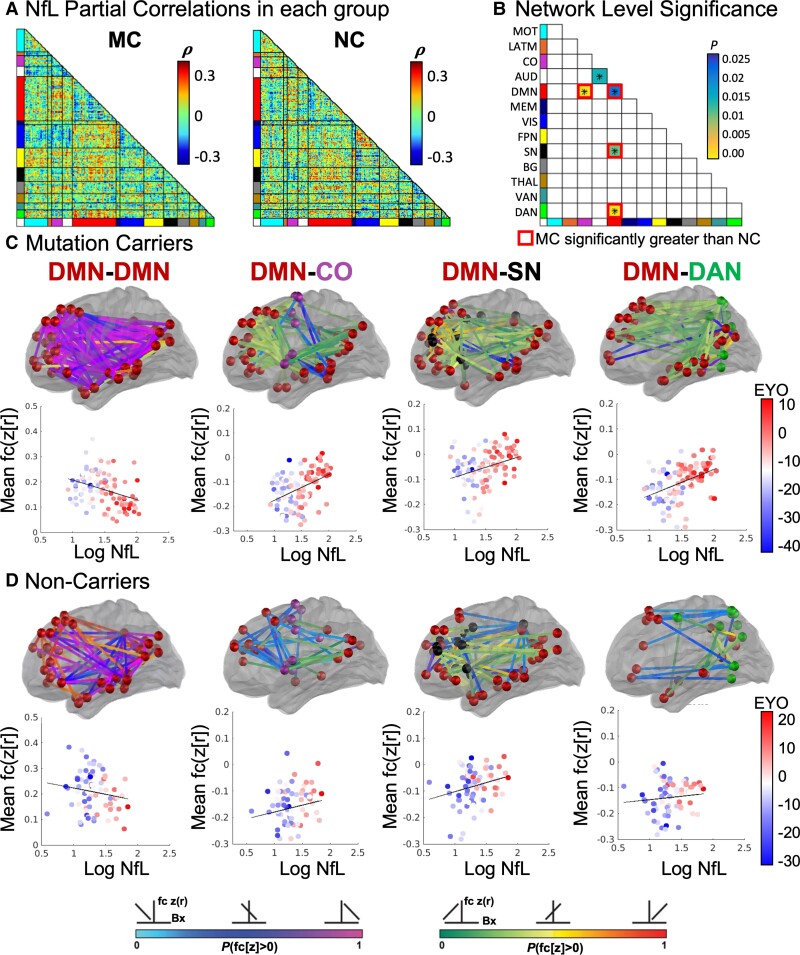
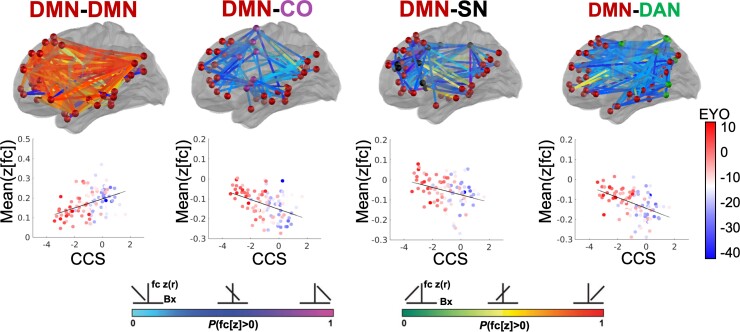
We next examined associations between FC and NfL using Network Level Analysis. We first calculated the partial Spearman rank correlation (regressing out age from FC and NfL) between FC of each ROI pair and the NfL measurement across all participants within each group (Fig. 4A). Connectivity among five networks were significantly correlated with NfL in MC including DMN and auditory network within-network connectivity, and between network connectivity of DMN with the CO network, salience network (SN), and DAN. Four of these network pairs were significantly associated with NfL above baseline levels of NfL-FC correlations in NC, namely the DMN within-network connectivity, and DMN connectivity with CO, SN, and DAN (Fig. 4B and Supplementary Table 4). Further examination of connectome correlations with NfL in the MC group revealed a negative correlation between NfL and DMN within-network connectivity. Specifically, individuals with the greatest levels of NfL had the lowest levels of DMN connectivity (Fig. 4C, Supplementary Fig. 3 and Supplementary Table 5), while a positive correlation was observed between NfL and DMN connectivity with CO, SN, and DAN networks (Fig. 4C and Supplementary Table 5). Individuals with the highest levels of NfL demonstrated reduced connectivity between DMN and executive control networks (CO, SN, and DAN). Given the correlation between blood NfL and Aβ standardized uptake value ratios in the brain, we further regressed out the effects of age and Aβ from correlations between NfL and FC. NfL was significantly associated with DMN-DAN connectivity above and beyond the effects of age and Aβ in the brain and relative to associations with NfL in NC (Supplementary Fig. 4 and Supplementary Table 6).
Figure 4.
Functional networks associated with NfL. (A) Partial correlations between NfL and FC (regressing out effects of age) in MC and NC groups. (B) MC demonstrated stronger associations with NfL than NC in four network pairs (P < 0.025). (C) For MC and (D) NC, pink and green lines indicate negative and positive correlations between NfL and FC, respectively, wherein individuals with the highest NfL have the lowest DMN FC along with reduced anti-correlation between DMN and SN, DAN, and CO networks. Scatter plots demonstrate correlation between NfL with average FC from within DMN and between DMN and SN, DAN, and CO networks.
Functional connectivity associations with cognitive function
Cognitive Composite Score associations with FC were examined in the network pairs identified from the NfL analysis in the MC group. CCS in the NC group had extremely restricted range and ceiling effects, so associations with FC were not examined in the NC group. The same networks that showed significant associations between FC and NfL in MC also showed significant FC associations with CCS. Specifically, DMN within-network FC was positively correlated with CCS (r = 0.42, P < 0.001) such that individuals with reduced DMN within-network FC had the poorest scores on the CCS. Similarly, FC between the DMN and executive control networks was negatively correlated with CCS [CO (r = −0.72, P < 0001), SN (r = −0.54, P < 0.001), and DAN (r = −0.71, P < 0.001)] such that individuals with between network FC closest to zero had the poorest CCS (Fig. 5 and Supplementary Table 7).
Figure 5.
Network FC associated with NfL is correlated with cognitive function in MC. Orange-red lines indicate a positive correlation between DMN within network FC and CCS, wherein individuals with the lowest CCS have the lowest DMN FC. Blue lines indicate a negative correlation between DMN FC with control networks and CCS, wherein individuals with the worse cognitive function have reduced anti-correlation between DMN and SN, DAN, and CO networks.
Biomarker prediction of cognitive function
Four hierarchical linear regressions were run to test for the predictive value of network average FC from the four networks identified with NfL association (DMN-DMN, DMN-SN, DMN-CO, DMN-DAN) in the MC group (Table 3) on CCS outcomes. To make sure that our assumptions for linear regressions are valid, we checked the normality of regression model residuals (Kolmogorov–Smirnov P > 0.05), and also visually inspected for violation of randomness and homoscedasticity with residual plots. In addition, we checked that the collinearity of variables with the variable inflation factors (VIF) and found that the VIF in all models ranged from 1.0–2.6, well below the problematic threshold for collinearity (VIF > 5).61 As an initial comparison, the first model included age, education, sex, and FC in the model. The second model included the Model 1 predictors + serum NfL. The third model included Model 2 predictors + PET amyloid. The explained variance was significantly improved in Model 2 (F change P < 0.001) but was not significantly improved in Model 3 with the addition of amyloid (F change P > 0.05 and AIC and adjusted R2 are similar for models with and without PET amyloid). In Model 2, education (β range 0.27–0.29) and serum NfL (β range 0.43–0.47) were consistently predictive of CCS. However, FC was also predictive of CCS across models, DMN–DMN (β = 0.24, P < 0.001), DMN-DAN (β = −0.26, P < 0.001), DMN-SN (β = −0.16, P = 0.023), and DMN-CO (β = −0.17, P = 0.015).
Table 3.
Hierarchical regression analysis predicting CCS
| Variable | Model 1 | Model 2 | Model 3 | |||
|---|---|---|---|---|---|---|
| β | P | β | P | β | P | |
| Age | −0.48 | <0.001*** | −0.20 | 0.021 | −0.20 | 0.019 |
| Education | 0.35 | <0.001*** | 0.28 | <0.001*** | 0.25 | <0.001*** |
| Sex (M) | −0.10 | 0.169 | −0.12 | 0.065 | −0.12 | 0.052 |
| DMN-DMN | 0.31 | <0.001*** | 0.24 | <0.001*** | 0.22 | 0.001** |
| log NfL | – | – | −0.46 | <0.001*** | −0.38 | <0.001*** |
| Amyloid | – | – | – | – | −0.15 | 0.069 |
| R2 adjusted | 0.57 | – | 0.67 | – | 0.68 | – |
| F | 29.93 | <0.001 | 36.58 | <0.001 | 31.95 | <0.001 |
| AIC | −40.80 | – | −63.24 | – | −64.86 | – |
| F change | – | – | 26.28 | <0.001 | 3.39 | 0.069 |
| Variable | Model 1 | Model 2 | Model 3 | |||
|---|---|---|---|---|---|---|
| β | P | β | P | β | P | |
| Age | −0.49 | <0.001*** | −0.21 | 0.022 | −0.21 | 0.019 |
| Education | 0.38 | <0.001*** | 0.29 | <0.001*** | 0.26 | <0.001*** |
| Sex (M) | −0.12 | 0.109 | −0.13 | 0.050 | −0.14 | 0.036 |
| DMN-SN | −0.26 | 0.001*** | −0.16 | 0.023 | −0.16 | 0.020 |
| log NfL | – | – | −0.47 | <0.001*** | −0.37 | <0.001*** |
| Amyloid | – | – | – | – | −0.18 | 0.027 |
| R2 adjusted | 0.54 | – | 0.65 | – | 0.66 | – |
| F | 26.23 | <0.001 | 32.25 | <0.001 | 29.08 | <0.001 |
| AIC | −34.17 | – | −55.79 | – | −59.16 | – |
| F change | – | – | 25.27 | <0.001 | 5.09 | 0.027 |
| Variable | Model 1 | Model 2 | Model 3 | |||
|---|---|---|---|---|---|---|
| β | P | β | P | β | P | |
| Age | −0.47 | <0.001*** | −0.19 | 0.035 | −0.19 | 0.031 |
| Education | 0.36 | <0.001*** | 0.28 | <0.001*** | 0.25 | <0.001*** |
| Sex (M) | −0.09 | 0.220 | −0.11 | 0.087 | −0.12 | 0.069 |
| DMN-CO | −0.26 | <0.001*** | −0.17 | 0.015 | −0.16 | 0.026 |
| log NfL | – | – | −0.47 | <0.001*** | −0.39 | <0.001*** |
| Amyloid | – | – | – | – | −0.16 | 0.053 |
| R2 adjusted | 0.54 | – | 0.65 | – | 0.66 | – |
| F | 26.38 | <0.001 | 32.71 | <0.001 | 28.87 | <0.001 |
| AIC | −34.44 | – | −56.61 | – | −58.72 | – |
| F change | – | – | 25.94 | <0.001 | 3.87 | 0.053 |
| Variable | Model 1 | Model 2 | Model 3 | |||
|---|---|---|---|---|---|---|
| β | P | β | P | β | P | |
| Age | −0.45 | <0.001*** | −0.20 | 0.021 | −0.20 | 0.019 |
| Education | 0.34 | <0.001*** | 0.27 | <0.001*** | 0.25 | <0.001*** |
| Sex (M) | −0.16 | 0.023 | −0.16 | 0.011 | −0.16 | 0.009 |
| DMN-DAN | −0.36 | <0.001*** | −0.26 | <0.001*** | −0.25 | <0.001*** |
| log NfL | – | – | −0.43 | <0.001*** | −0.35 | <0.001*** |
| Amyloid | – | – | – | – | −0.15 | 0.063 |
| R2 adjusted | 0.59 | – | 0.68 | – | 0.69 | – |
| F | 32.52 | <0.001 | 37.77 | <0.001 | 33.06 | <0.001 |
| AIC | −45.15 | – | −65.18 | – | −66.96 | – |
| F change | – | – | 23.35 | <0.001 | 3.55 | 0.063 |
After pairwise exclusion, Models 1 and 2 have n = 99 MCs, Model 3 has n = 87 MCs. Standardized β coefficients are reported, **P < 0.005, ***P < 0.001. AIC = Akaike information criterion.
Exploratory role of functional connectivity in mediating cognitive function
Given the joint contribution of NfL and FC in several network pairs in explaining variance in CCSs, we ran an exploratory mediation analysis to assess whether FC mediated the relationship between serum NfL levels and cognitive function. Because DMN within network FC was positively correlated with CCSs while DMN between network FC was negatively correlated, we chose to include within and between network FC in two separate models so that negative and positive effects would not cancel each other out in a multiple mediation model. We observed that DMN within network FC partially mediated the role of NfL levels on CCSs (Supplementary Fig. 5A). The DMN between network FC model also partially mediated the role of NfL levels on CCSs and this was driven by variance in DMN-DAN FC (Supplementary Fig. 5A).
Discussion
Assessing the functional connectivity within and between the brain’s resting state networks provides a method for localizing the functional ramifications of neurodegenerative diseases. The goal of this study was to investigate whether blood serum concentrations of NfL, a non-specific biomarker of axonal injury and neuronal death, corresponded with deterioration of FC networks, specifically in individuals with ADAD. Prior work suggests that NfL is a biomarker associated with neurodegeneration in ADAD,30,31,62–64 and that FC disruption is marginally predictive of CSF NfL measurements in cognitively normal adults.26 To investigate this association between NfL and FC in ADAD further, this study assessed patterns of FC degeneration and NfL blood levels in individuals carrying mutations for ADAD. Here, NfL measurements were negatively correlated with connectivity within the DMN and positively correlated with connectivity between DMN and executive control networks, including the CO, SN, and DAN. Further, DMN within and between network FC explained unique variance in CCS above and beyond the variance explained by NfL and demographic variables implicating the importance of FC networks in cognitive decline in ADAD.
Studies in sporadic AD have ubiquitously observed reduced FC within the DMN.9,17–21,24,65 Researchers have speculated that degeneration follows a spatio-temporal progression beginning in the posterior cingulate cortex, and then spreading forward to the anterior cingulate and prefrontal cortex.66 This pathological spreading through the DMN is thought to be a hallmark of the disease and mirrors metabolic disruption, amyloid deposition, and cortical atrophy.12–16,40 While a preponderance of prior AD research has focused on the DMN,11 a growing number of studies have also examined connectivity within DAN, SN, and FPN,18,23,26 and how the FC between networks impacts disease progression.19,21,22 The patterns that we observe here in ADAD with the DIAN cohort are consistent with this involvement of the DMN and pathology of the connections between the DMN and top-down cognitive control regions within the executive control networks reported in sporadic AD.
Within ADAD, previous work has reported a biphasic association between EYO and global FC using nonlinear modeling in the DIAN cohort. Specifically, the authors report that a decrease in whole brain FC as a function of EYO is observed both at EYO < −16.7 and then again EYO > 0.5 years, implicating a two-stage process in the decline of FC as a function of EYO.23 The present study builds on this prior work by additionally modeling EYO associations with between-network FC. In the present study, FC within the DMN, CO, and DAN and between the DMN-CO and DMN-DAN were apparent as a function of EYO, specifically when comparing pre-EYO 0 MC to post-EYO 0 MC. Consistent with prior work,18 these data suggest that degeneration of FC networks occurs relatively late in the disease course relative to changes in other biomarkers such as amyloid PET and CSF Aβ42, which are observable up to 20 years prior to EYO 0.40 It is notable that this degeneration in FC seems to demonstrate temporal correlation with atrophy and tau deposition, which occur near the onset of cognitive impairment,1,40 and spatial colocalization with peak levels of Aβ deposition most prominently within posterior portions of the DMN.12,15,65,67–69
In addition to disease-related variance in FC, we further investigated the association between FC and serum NfL. In AD, NfL levels are elevated with progressive cognitive impairment,31,70,71 altered brain metabolism,31 and atrophy.62–64,71 Additionally, NfL has been found to be positively correlated with age and with PET measurements of Aβ patterns in healthy adults72 and symptomatic individuals with AD.31,73 In the present study, on average, NfL levels were greater in ADAD MC than NC. However, in both groups, NfL was positively correlated with age, which is consistent with prior work demonstrating a positive correlation between age and increasing serum and CSF NfL levels in healthy adults.5,74 Given the relationship between age and NfL, we included age as a covariate in our models and used the mutation NC as an EYO matched comparison group and as a de facto control condition to compare the relative effects of NfL-FC associations in the MC group.
When regressing out variance due to age, we observed a negative correlation between NfL and DMN connectivity, such that higher NfL levels were associated with reduced within-network DMN FC for the ADAD MC group. Further, MC individuals with the highest levels of NfL demonstrated reduced magnitude of FC between DMN and executive control networks, including the CO, SN, and DAN. Even after controlling for Aβ, DMN-DAN between-network connectivity was significantly associated with NfL. Prior work has reported conflicting findings regarding the relationship between NfL and FC. Specifically, prior work in individuals ranging from cognitively normal to mild cognitive parament and dementia reported no association between CSF NfL and regions of the DMN27 while other work demonstrated correlations between blood plasma NfL and FC between regions of the DMN and frontal and parietal cortex.28 The present study lends further evidence that blood plasma NfL levels are associated with DMN and DMN-executive network FC in individuals with ADAD who range from cognitively normal to cognitively impaired. Similar to the present findings, prior research in the DIAN cohort has demonstrated that metrics of white matter integrity are reduced within posterior corpus callosum and superior longitudinal fasciculus, tracts connecting posterior DMN and executive control regions, in individuals with the highest levels of serum NfL.30 Taken together with these white matter integrity findings, the present results indicate that blood measurements of NfL track with increasing disruption of DMN within-network and DMN FC with executive control networks as individuals near and exceed their EYO year 0.
Executive control networks are necessary for top-down cognitive control and executive functions including inhibition, working memory, and cognitive flexibility.75,76 In the present study, CCS demonstrated a negative correlation with NfL in the MC and not the NC group, such that, for ADAD MC, higher levels of NfL were correlated with lower CCS, supporting prior work reporting these associations in sporadic AD and ADAD.31,62,64,77 Further, networks which were associated with NfL were also associated with CCS, such that CCS was positively correlated with DMN within-network connectivity and negatively correlated with DMN between-network connectivity. This is consistent with previous work in individuals with sporadic AD demonstrating a positive correlation between MMSE scores and DMN within-network FC,24,78 a negative correlation between MMSE scores and DMN and middle and inferior frontal gyrus between-network connectivity,79 and reduced FC anti-correlation between DMN-DAN with worsening CDR.17,18,22 Interestingly, previous studies have observed individuals with higher FC in executive control networks have a delay in cognitive decline.19 Prior research has often characterized the importance of DMN suppression for effective completion of executive and cognitive tasks and the loss of anti-correlation between the DMN and executive networks like the CO, SN, and DAN can lead to cognitive deficits observed in disease.80 Given that the DMN and executive control networks tend to be slightly anti-correlated on average, reduced FC magnitude between networks may be responsible for the decline in cognitive function exhibited in individuals with AD.
Further, we examined the relative contributions of demographics, serum NfL, functional connectivity, and PET amyloid in predicting concurrent cognitive function. The regression model adding PET amyloid did not significantly improve the variance explained in cognitive performance. In all four regression models, NfL and FC were predictive of CCSs with DMN within network and DMN-DAN having the largest standardized beta values of the FC network blocks. These results implicate the role of FC in these regions in cognitive decline associated with ADAD. The present findings suggest that, in addition to the significant roles of Aβ and tau in disease pathology, FC within the DMN and between the DMN and DAN also explains unique variance in worsening cognitive abilities in ADAD.
Finally, we examined the mediating role of within and between network FC on cognitive function. We observed that DMN within network FC and DMN between network FC partially mediated the association between NfL levels and cognitive function. Specifically, higher DMN within network FC and lower levels of DMN-DAN between network FC were associated with increased CCSs while higher blood levels of NfL were associated with reduced cognitive function. NfL is a cytoskeletal support protein and increased total blood levels are employed to be an indicator of cytoskeletal disruption and increased cell death. The mediation analysis suggests that cell death which occurs in specific FC network blocks (within the DMN and between DMN and DAN) likely plays an important role in disease progression and prognosis. Prior research suggests that there are subtypes of AD with distinct functional and structural patterns of degeneration81,82 and individual variability of DMN and DMN-DAN FC may play an important role in cognitive decline. The present study is consistent with prior work which observed that functional connectivity between DMN and regions of frontal cortex mediated the relationship between plasma NfL and measures of cognition in cognitively normal and symptomatic individuals with AD.28 Further, cognitive exercises or interventions (e.g. transcranial magnetic stimulation) which target these networks or regions within these networks such as the precuneus83 may have the potential to delay the onset or progression of dementia. Critically, while blood NfL correlates with cognitive function, the present study sheds light on the underlying systems biology elucidating the mechanism by which reduced neuronal integrity within functional brain systems is causing reduced cognitive function.
The present study employed global signal regression (GSR). The use of GSR has been the topic of much research and controversy. Global signal contains variance from cardiac, respiration, scanner, and head motion-related artifacts and GSR has been demonstrated to effectively mitigate head motion and improve quality control metrics.84–86 Here we have chosen to employ GSR to effectively control for head motion. However, GSR can impact the distribution of regional signal correlations and the changes in negative correlations between networks and associations with NfL and cognition in the present analysis should be considered in the context of GSR. Further, several researchers have raised concerns that GSR may remove potential signal of interest.86 Specifically, global signal changes with arousal, vigilance, and sleep, with higher arousal levels associated with lower global signal while states such as sleep are associated with increased global signal within sensorimotor regions including visual, auditory, and motor cortex.84,86 Further, increasing sleep depth is associated with increasing global signal due to decreased oxygen extraction, and removal of global signal during sleep reduces the BOLD estimates in regions such as the anterior and posterior cingulate (i.e. the DMN).87 While this poses a particular problem for fMRI studies collected during natural sleep (e.g. Developing Human Connectome Project, Baby Connectome Project, etc.), this may also pose challenges in Alzheimer’s disease research, where individuals with increasing levels of amyloid pathology have reduced sleep quality at night and increased daytime napping88 potentially leading participants with increasing severity of pathology to be more likely to fall asleep during MRI scans. The contribution of global signal to the pathophysiology of dementia has received limited research and is an important consideration for Alzheimer’s disease research.89 However, prior analysis of DIAN data with and without GSR has previously found minimal to no impact on reported findings.90
There are several limitations of this study to be considered. First, due to low signal to noise and magnetic susceptibility artifact within regions of entorhinal cortex, deep temporal lobe structures such as the hippocampus and amygdala are challenging to reliably sample with resting state fMRI. Given that these regions have been implicated in the spread of AD-related pathology, particularly tau pathology, future FC work targeting hippocampus and amygdala may shed further light on functional brain associations with NfL and cognitive outcomes. Second, although the focus on select regions and networks was guided by priori publications, we acknowledge that pre-registration is increasingly recognized in the neuroscience field as highly beneficial in raising confidence in results. Third, the cross-sectional analyses in the present study may not fully capture the spatiotemporal progression of FC network degeneration in ADAD. Our future work will assess within-subjects functional connectivity to characterize the spatiotemporal timing of FC network degeneration. Finally, it is possible that CSF measures of NfL may provide stronger associations, however CSF measurements were limited in number in this study thus precluding comparison. However, CSF measurements are more invasive and less accessible in low resource settings than blood-based biomarkers. The present findings indicate that blood assessments of NfL are associated with functional connectivity degeneration suggesting this low-cost and accessible fluid biomarker is a promising option for tracking neurodegeneration.
In conclusion, the present study demonstrates that FC within the DMN as well as between the DMN and executive control networks decline in individuals with ADAD mutations as they approach their estimated age of onset (EYO = 0). Further, DMN-DAN between-network FC disruption is associated with NfL blood estimates above and beyond the effects of normal aging and Aβ accumulation. Additionally, DMN within-network FC was predictive of CCS even when accounting for Aβ. In demonstrating that NfL, a marker of neural atrophy, correlates with reduced FC in ADAD, the present study provides further evidence for the accumulation of NfL as a possible neurodegenerative biomarker of AD and implicates this biomarker in the mechanisms of the degeneration of connectivity within the DMN and between the DMN and DAN.
Supplementary Material
Acknowledgements
Data collection and sharing for this project were supported by The Dominantly Inherited Alzheimer Network (DIAN, UF1AG032438) funded by the National Institute on Aging (NIA), the German Center for Neurodegenerative Diseases (DZNE), Raul Carrea Institute for Neurological Research (FLENI), generous funding from the Paula and Roger Riney Fund and the Brennan Family Fund, and partial support provided by the Research and Development Grants for Dementia from Japan Agency for Medical Research and Development, AMED, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI). Fig. 1 parts A and B were illustrated by Dr Andrea Scharf and Catie Newsom-Stewart in association with InPrint at Washington University in St. Louis. We would additionally like to thank Dr Evan Gordon for providing assistance and code for generating the spring embedded plots in Fig. 3 and Dr Andy Aschenbrenner for assistance with calculating the cognitive composite score. This manuscript has been reviewed by DIAN Study investigators for scientific content and consistency of data interpretation with previous DIAN Study publications. We acknowledge the altruism of the participants and their families and contributions of the DIAN research and support staff at each of the participating sites for their contributions to this study.
Appendix 1
Dominantly Inherited Alzheimer Network
Sarah Adams, Ricardo Allegri, Aki Araki, Nicolas Barthelemy, Randall Bateman, Jacob Bechara, Tammie Benzinger, Sarah Berman, Courtney Bodge, Susan Brandon, William (Bill) Brooks, Jared Brosch, Jill Buck, Virginia Buckles, Kathleen Carter, Dave Cash, Lisa Cash, Charlie Chen, Jasmeer Chhatwal, Patricio Chrem, Jasmin Chua, Helena Chui, Carlos Cruchaga, Gregory S. Day, Chrismary De La Cruz, Darcy Denner, Anna Diffenbacher, Aylin Dincer, Tamara Donahue, Jane Douglas, Duc Duong, Noelia Egido, Bianca Esposito, Anne Fagan, Marty Farlow, Becca Feldman, Colleen Fitzpatrick, Shaney Flores, Nick Fox, Erin Franklin, Nelly Friedrichsen, Hisako Fujii, Samantha Gardener, Bernardino Ghetti, Alison Goate, Sarah Goldberg, Jill Goldman, Alyssa Gonzalez, Brian Gordon, Susanne Gräber-Sultan, Neill Graff-Radford, Morgan Graham, Julia Gray, Emily Gremminger, Miguel Grilo, Alex Groves, Christian Haass, Lisa Häsler, Jason Hassenstab, Cortaiga Hellm, Elizabeth Herries, Laura Hoechst-Swisher, Anna Hofmann, David Holtzman, Russ Hornbeck, Yakushev Igor, Ryoko Ihara, Takeshi Ikeuchi, Snezana Ikonomovic, Kenji Ishii, Clifford Jack, Gina Jerome, Erik Johnson, Mathias Jucker, Celeste Karch, Stephan Käser, Kensaku Kasuga, Sarah Keefe, William (Bill) Klunk, Robert Koeppe, Deb Koudelis, Elke Kuder-Buletta, Christoph Laske, Jae-Hong Lee, Allan Levey, Johannes Levin, Yan Li, Oscar Lopez, Jacob Marsh, Rita Martinez, Ralph Martins, Neal Scott Mason, Colin Masters, Kwasi Mawuenyega, Austin McCullough, Eric McDade, Arlene Mejia, Estrella Morenas-Rodriguez, Hiroshi Mori, John Morris, James Mountz, Cath Mummery, Neelesh Nadkami, Akemi Nagamatsu, Katie Neimeyer, Yoshiki Niimi, James Noble, Joanne Norton, Brigitte Nuscher, Antoinette O’Connor, Ulricke Obermüller, Riddhi Patira, Richard Perrin, Lingyan Ping, Oliver Preische, Alan Renton, John Ringman, Stephen Salloway, Raquel Sanchez-Valle, Peter Schofield, Michio Senda, Nick Seyfried, Kristine Shady, Hiroyuki Shimada, Wendy Sigurdson, Jennifer Smith, Lori Smith, Beth Snitz, Hamid Sohrabi, Sochenda Stephens, Kevin Taddei, Sarah Thompson, Jonathan Vöglein, Peter Wang, Qing Wang, Elise Weamer, Chengjie Xiong, Jinbin Xu, Xiong Xu.
Contributor Information
Muriah D Wheelock, Department of Radiology, Washington University in St. Louis, St. Louis, MO, USA.
Jeremy F Strain, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Patricia Mansfield, School of Medicine, Saint Louis University, St. Louis, MO, USA.
Jiaxin Cindy Tu, Department of Radiology, Washington University in St. Louis, St. Louis, MO, USA.
Aaron Tanenbaum, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Oliver Preische, German Center for Neurodegenerative Diseases (DZNE), Tübingen, Germany.
Jasmeer P Chhatwal, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA.
David M Cash, Dementia Research Centre, UCL Queen Square Institute of Neurology, London, UK; UK Dementia Research Institute, UCL, London, UK.
Carlos Cruchaga, Department of Psychiatry, Washington University in St. Louis, St. Louis, MO, USA.
Anne M Fagan, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Nick C Fox, Dementia Research Centre, UCL Queen Square Institute of Neurology, London, UK; UK Dementia Research Institute, UCL, London, UK.
Neill R Graff-Radford, Department of Neurology, Mayo Clinic, Jacksonville, FL, USA.
Jason Hassenstab, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Clifford R Jack, Jr, Department of Radiology, Mayo Clinic, Rochester, MN, USA.
Celeste M Karch, Department of Psychiatry, Washington University in St. Louis, St. Louis, MO, USA.
Johannes Levin, Department of Neurology, Ludwig-Maximilians-Universität München, Munich, Germany; German Center for Neurodegenerative Diseases (DZNE), Munich, Germany; Munich Cluster for Systems Neurology (SyNergy), Munich, Germany.
Eric M McDade, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Richard J Perrin, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA; Department of Pathology & Immunology, Washington University in St. Louis, St. Louis, MO, USA.
Peter R Schofield, Neuroscience Research Australia, Sydney, NSW, Australia; School of Medical Sciences, University of New South Wales, Sydney, NSW, Australia.
Chengjie Xiong, Division of Biostatistics, Washington University in St. Louis, St. Louis, MO, USA.
John C Morris, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Randal J Bateman, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Mathias Jucker, German Center for Neurodegenerative Diseases (DZNE), Tübingen, Germany; Department of Cellular Neurology, Hertie Institute for Clinical Brain Research, University of Tübingen, Tübingen, Germany.
Tammie L S Benzinger, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Beau M Ances, Department of Neurology, Washington University in St. Louis, St. Louis, MO, USA.
Adam T Eggebrecht, Department of Radiology, Washington University in St. Louis, St. Louis, MO, USA.
Brian A Gordon, Department of Radiology, Washington University in St. Louis, St. Louis, MO, USA.
the Dominantly Inherited Alzheimer Network:
Sarah Adams, Ricardo Allegri, Aki Araki, Nicolas Barthelemy, Randall Bateman, Jacob Bechara, Tammie Benzinger, Sarah Berman, Courtney Bodge, Susan Brandon, William (Bill) Brooks, Jared Brosch, Jill Buck, Virginia Buckles, Kathleen Carter, Dave Cash, Lisa Cash, Charlie Chen, Jasmeer Chhatwal, Patricio Chrem, Jasmin Chua, Helena Chui, Carlos Cruchaga, Gregory S Day, Chrismary De La Cruz, Darcy Denner, Anna Diffenbacher, Aylin Dincer, Tamara Donahue, Jane Douglas, Duc Duong, Noelia Egido, Bianca Esposito, Anne Fagan, Marty Farlow, Becca Feldman, Colleen Fitzpatrick, Shaney Flores, Nick Fox, Erin Franklin, Nelly Friedrichsen, Hisako Fujii, Samantha Gardener, Bernardino Ghetti, Alison Goate, Sarah Goldberg, Jill Goldman, Alyssa Gonzalez, Brian Gordon, Susanne Gräber-Sultan, Neill Graff-Radford, Morgan Graham, Julia Gray, Emily Gremminger, Miguel Grilo, Alex Groves, Christian Haass, Lisa Häsler, Jason Hassenstab, Cortaiga Hellm, Elizabeth Herries, Laura Hoechst-Swisher, Anna Hofmann, David Holtzman, Russ Hornbeck, Yakushev Igor, Ryoko Ihara, Takeshi Ikeuchi, Snezana Ikonomovic, Kenji Ishii, Clifford Jack, Gina Jerome, Erik Johnson, Mathias Jucker, Celeste Karch, Stephan Käser, Kensaku Kasuga, Sarah Keefe, William (Bill) Klunk, Robert Koeppe, Deb Koudelis, Elke Kuder-Buletta, Christoph Laske, Jae-Hong Lee, Allan Levey, Johannes Levin, Yan Li, Oscar Lopez, Jacob Marsh, Rita Martinez, Ralph Martins, Neal Scott Mason, Colin Masters, Kwasi Mawuenyega, Austin McCullough, Eric McDade, Arlene Mejia, Estrella Morenas-Rodriguez, Hiroshi Mori, John Morris, James Mountz, Cath Mummery, Neelesh Nadkami, Akemi Nagamatsu, Katie Neimeyer, Yoshiki Niimi, James Noble, Joanne Norton, Brigitte Nuscher, Antoinette O’Connor, Ulricke Obermüller, Riddhi Patira, Richard Perrin, Lingyan Ping, Oliver Preische, Alan Renton, John Ringman, Stephen Salloway, Raquel Sanchez-Valle, Peter Schofield, Michio Senda, Nick Seyfried, Kristine Shady, Hiroyuki Shimada, Wendy Sigurdson, Jennifer Smith, Lori Smith, Beth Snitz, Hamid Sohrabi, Sochenda Stephens, Kevin Taddei, Sarah Thompson, Jonathan Vöglein, Peter Wang, Qing Wang, Elise Weamer, Chengjie Xiong, Jinbin Xu, and Xiong Xu
Funding
Additional support for this study was provided by NIH grants: R00 EB029343, T32 MH100019, K01 MH103594, K01 AG053474, P30 NS098577, P30 AG066444 Research Education Component, National Science Foundation (DGE-1745038), the National Institute for Health Research (NIHR) Queen Square Dementia Biomedical Research Centre, and the Medical Research Council Dementias Platform UK (MR/L023784/1 and MR/009076/1).
Competing interests
R.J.B. is Director of DIAN–TU and Principal Investigator of DIAN–TU-001. He receives research support from the NIA of the NIH, DIAN–TU trial pharmaceutical partners (Eli Lilly and Company, F. Hoffman-La Roche Ltd. and Avid Radiopharmaceuticals), Alzheimer’s Association, GHR Foundation, Anonymous Organization, DIAN–TU Pharma Consortium (active: Biogen, Eisai, Eli Lilly and Company, Janssen, F. Hoffmann-La Roche Ltd/Genentech; previous: AbbVie, Amgen, AstraZeneca, Forum, Mithridion, Novartis, Pfizer, Sanofi, United Neuroscience). He has been an invited speaker and consultant for AC Immune, F. Hoffman-La Roche Ltd. and Janssen and a consultant for Amgen and Eisai. A.M.F. has received research funding from the National Institute on Aging of the National Institutes of Health, Biogen, Centene, Fujirebio and Roche Diagnostics. She is currently a member of the scientific advisory boards for Roche Diagnostics and Genentech and consults for DiademRes, DiamiR and Siemens Healthcare Diagnostics. There are no conflicts. N.R.G.R. takes part in multicenter treatment studies sponsored by Biogen, Lilly, and AbbVie. J.L. reports personal fees from MODAG GmbH, personal fees from Bayer Vital, personal fees from Axon Neuroscience, non-financial support from AbbVie, personal fees from Thieme medical publishers, personal fees from W. Kohlhammer GmbH medical publishers, personal fees from Roche, personal fees from Biogen, outside the submitted work. P.R.S. reports grants from NIH (administered through Wash U), grants from Anonymous Foundation (administered through Wash U), and grants from Roth Charitable Foundation, during the conduct of the study. C.J. serves on an independent data monitoring board for Roche, has served as a speaker for Eisai, and consulted for Biogen, but he receives no personal compensation from any commercial entity. He receives research support from NIH, the GHR Foundation and the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Clinic. D.M.C. is supported by the UK Dementia Research Institute which receives its funding from DRI Ltd., funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK, as well as Alzheimer’s Research UK (ARUK-PG2017-1946) and the UCL/UCLH NIHR Biomedical Research Centre. T.L.S.B. has investigator-initiated research funding from the NIH, the Alzheimer’s Association, the Barnes-Jewish Hospital Foundation and Avid Radiopharmaceuticals (a wholly owned subsidiary of Eli Lilly). T.L.S.B. participates as a site investigator in clinical trials sponsored by Avid Radiopharmaceuticals, Eli Lilly, Biogen, Eisai, Jaansen, and Roche. She serves as an unpaid consultant to Eisai and Siemens. She is on the Speaker’s Bureau for Biogen.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Gordon BA, Blazey TM, Christensen J, et al. . Tau PET in autosomal dominant Alzheimer’s disease: Relationship with cognition, dementia and other biomarkers. Brain. 2019;142:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benzinger TL, Blazey T, JackCR, Jr., et al. . Regional variability of imaging biomarkers in autosomal dominant Alzheimer’s disease. Proc Natl Acad Sci U S A. 2013;110:E4502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buckner RL, Sepulcre J, Talukdar T, et al. . Cortical hubs revealed by intrinsic functional connectivity: Mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci. 2009;29:1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. La Joie R, Perrotin A, Barre L, et al. . Region-specific hierarchy between atrophy, hypometabolism, and β-amyloid (Aβ) load in Alzheimer’s disease dementia. J Neurosci. 2012;32:16265–16273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bridel C, van Wieringen WN, Zetterberg H, et al. . Diagnostic value of cerebrospinal fluid neurofilament light protein in neurology: A systematic review and meta-analysis. JAMA Neurol. 2019;76:1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petzold A. Neurofilament phosphoforms: Surrogate markers for axonal injury, degeneration and loss. J Neurol Sci. 2005;233(1–2):183–198. [DOI] [PubMed] [Google Scholar]
- 7. Gordon BA. Neurofilaments in disease: What do we know? Curr Opin Neurobiol. 2020;61:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brier MR, Thomas JB, Ances BM. Network dysfunction in Alzheimer’s disease: refining the disconnection hypothesis. Brain Connect. 2014;4:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang D, Raichle ME. Disease and the brain’s dark energy. Nat Rev Neurol. 2010;6:15–28. [DOI] [PubMed] [Google Scholar]
- 10. Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. [DOI] [PubMed] [Google Scholar]
- 11. Badhwar A, Tam A, Dansereau C, Orban P, Hoffstaedter F, Bellec P. Resting-state network dysfunction in Alzheimer’s disease: a systematic review and meta-analysis. Alzheimers Dement (Amst). 2017;8:73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drzezga A, Becker JA, Van Dijk KR, et al. . Neuronal dysfunction and disconnection of cortical hubs in non-demented subjects with elevated amyloid burden. Brain. 2011; 134:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mormino EC, Smiljic A, Hayenga AO, et al. . Relationships between beta-amyloid and functional connectivity in different components of the default mode network in aging. Cereb Cortex. 2011;21:2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sepulcre J, Sabuncu MR, Becker A, Sperling R, Johnson KA. In vivo characterization of the early states of the amyloid-beta network. Brain. 2013; 136:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Palmqvist S, Scholl M, Strandberg O, et al. . Earliest accumulation of beta-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nat Commun. 2017;8:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheline YI, Raichle ME, Snyder AZ, et al. . Amyloid plaques disrupt resting state default mode network connectivity in cognitively normal elderly. Biol Psychiatry. 2010;67:584–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brier MR, Thomas JB, Snyder AZ, et al. . Loss of intranetwork and internetwork resting state functional connections with Alzheimer’s disease progression. J Neurosci. 2012;32:8890–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomas JB, Brier MR, Bateman RJ, et al. . Functional connectivity in autosomal dominant and late-onset Alzheimer disease. JAMA Neurol. 2014;71:1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franzmeier N, Düzel E, Jessen F, et al. . Left frontal hub connectivity delays cognitive impairment in autosomal-dominant and sporadic Alzheimer’s disease. Brain. 2018;141:1186–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang K, Liang M, Wang L, et al. . Altered functional connectivity in early Alzheimer’s disease: A resting-state fMRI study. Hum Brain Mapp. 2007;28:967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dai Z, Lin Q, Li T, et al. . Disrupted structural and functional brain networks in Alzheimer’s disease. Neurobiol Aging. 2019;75:71–82. [DOI] [PubMed] [Google Scholar]
- 22. Meeker KL, Ances BM, Gordon BA, et al. . Cerebrospinal fluid Aβ42 moderates the relationship between brain functional network dynamics and cognitive intraindividual variability. Neurobiol Aging. 2021;98:116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith RX, Strain JF, Tanenbaum A, et al. . Resting-state functional connectivity disruption as a pathological biomarker in autosomal dominant Alzheimer disease. Brain Connect. 2021;11:239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao T, Quan M, Jia J. Functional connectivity of default mode network subsystems in the presymptomatic stage of autosomal dominant Alzheimer’s disease. J Alzheimers Dis. 2020;73:1435–1444. [DOI] [PubMed] [Google Scholar]
- 25. Chhatwal JP, Schultz AP, Johnson K, et al. . Impaired default network functional connectivity in autosomal dominant Alzheimer disease. Neurology. 2013;81:736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Millar PR, Ances BM, Gordon BA, et al. . Evaluating resting-state BOLD variability in relation to biomarkers of preclinical Alzheimer’s disease. Neurobiol Aging. 2020;96:233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pereira JB, Janelidze S, Ossenkoppele R, et al. . Untangling the association of amyloid-beta and tau with synaptic and axonal loss in Alzheimer’s disease. Brain. 2021;144:310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yao W, Zhang X, Zhao H, Xu Y, Bai F. Alzheimer’s disease neuroimaging I. Inflammation disrupts cognitive integrity via plasma neurofilament light chain coupling brain networks in Alzheimer’s disease. J Alzheimers Dis. 2022;89:505–518. [DOI] [PubMed] [Google Scholar]
- 29. Bateman RJ, Xiong C, Benzinger TL, et al. . Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schultz SA, Strain JF, Adedokun A, et al. . Serum neurofilament light chain levels are associated with white matter integrity in autosomal dominant Alzheimer’s disease. Neurobiol Dis. 2020;142:104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Preische O, Schultz SA, Apel A, et al. . Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nat Med. 2019;25:277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morris JC. The clinical dementia rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. [DOI] [PubMed] [Google Scholar]
- 33. Folstein MF, Folstein SE, McHugh PR. Mini-mental state. a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. [DOI] [PubMed] [Google Scholar]
- 34. Wechsler D. WAIS-R Manual: Wechsler adult intelligence scale-revised. Psychological Corporation; 1981. [Google Scholar]
- 35. Aschenbrenner AJ, James BD, McDade E, et al. . Awareness of genetic risk in the dominantly inherited Alzheimer network (DIAN). Alzheimers Dement. 2020;16:219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bateman RJ, Benzinger TL, Berry S, et al. . The DIAN-TU next generation Alzheimer’s prevention trial: adaptive design and disease progression model. Alzheimers Dement. 2017;13:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang G, Berry S, Xiong C, et al. . A novel cognitive disease progression model for clinical trials in autosomal-dominant Alzheimer’s disease. Stat Med. 2018;37:3047–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wechsler D. WMS-R: Wechsler memory scale-revised: manual. Psychological Corporation; 1987. [Google Scholar]
- 39. Su’ Y, D'Angelo GM, Vlassenko AG, et al. . Quantitative analysis of PiB-PET with FreeSurfer ROis. PLoS One. 2013;8:e73377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gordon BA, Blazey TM, Su Y, et al. . Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer's disease: a longitudinal study. The Lancet Neurology. 2018;17:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rousset OG, Ma Y, Evans AC. Correction for partial volume effects in PET: principle and validation. J Nucl Med. 1998;39:904–911. [PubMed] [Google Scholar]
- 42. Su Y, Blazey TM, Snyder AZ, et al. . Partial volume correction in quantitative amyloid imaging. Neuroimage. 2015;107:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laumann TO, Snyder AZ, Mitra A, et al. . On the stability of BOLD fMRI correlations. Cereb Cortex. 2017;27:4719–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gratton C, Koller JM, Shannon W, et al. . Emergent functional network effects in Parkinson disease. Cereb Cortex. 2019;29:2509–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hacker CD, Laumann TO, Szrama NP, et al. . Resting state network estimation in individual subjects. Neuroimage. 2013;82:616–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. [DOI] [PubMed] [Google Scholar]
- 47. Gholipour A, Kehtarnavaz N, Gopinath K, Briggs R, Panahi I. Average field map image template for echo-planar image analysis. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:94–97. [DOI] [PubMed] [Google Scholar]
- 48. Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Raut RV, Mitra A, Snyder AZ, Raichle ME. On time delay estimation and sampling error in resting-state fMRI. Neuroimage. 2019;194:211–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fischl B. Freesurfer. Neuroimage. 2012;62:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andersson JLR, Jenkinson M, Smith S. FMRIB technical report TR07JA2. https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FNIRT
- 53. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. [DOI] [PubMed] [Google Scholar]
- 54. Seitzman BA, Gratton C, Marek S, et al. . A set of functionally-defined brain regions with improved representation of the subcortex and cerebellum. Neuroimage. 2020;206:116290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ojemann JG, Akbudak E, Snyder AZ, McKinstry RC, Raichle ME, Conturo TE. Anatomic localization and quantitative analysis of gradient refocused echo-planar fMRI susceptibility artifacts. Neuroimage. 1997;6:156–167. [DOI] [PubMed] [Google Scholar]
- 56. Gordon EM, Laumann TO, Gilmore AW, et al. . Precision functional mapping of individual human brains. Neuron. 2017;95:791–807 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eggebrecht AT, Elison JT, Feczko E, et al. . Joint attention and brain functional connectivity in infants and toddlers. Cereb Cortex. 2017;27:1709–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wheelock MD, Lean RE, Bora S, et al. . Functional connectivity network disruption underlies domain-specific impairments in attention for children born very preterm. Cereb Cortex. 2021;31:1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wheelock MD, Austin NC, Bora S, et al. . Altered functional network connectivity relates to motor development in children born very preterm. NeuroImage. 2018;183:574–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hayes AF. Introduction to mediation, moderation, and conditional process analysis. Methodology in the social sciences series. 3rd ed. Guilford Press; 2022. [Google Scholar]
- 61. Doud JI. Multicollinearity and regression analysis. J Phys: Conf Ser. 2017;949:012009. [Google Scholar]
- 62. Mattsson N, Cullen NC, Andreasson U, Zetterberg H, Blennow K. Association between longitudinal plasma neurofilament light and neurodegeneration in patients with Alzheimer disease. JAMA Neurol. 2019;76:791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kang MS, Aliaga AA, Shin M, et al. . Amyloid-beta modulates the association between neurofilament light chain and brain atrophy in Alzheimer's disease. Mol Psychiatry. 2021;26:5989–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weston PSJ, Poole T, Ryan NS, et al. . Serum neurofilament light in familial Alzheimer disease: A marker of early neurodegeneration. Neurology. 2017;89:2167–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou Y, Dougherty JH Jr, Hubner KF, Bai B, Cannon RL, Hutson RK. Abnormal connectivity in the posterior cingulate and hippocampus in early Alzheimer's disease and mild cognitive impairment. Alzheimers Dement. 2008;4:265–270. [DOI] [PubMed] [Google Scholar]
- 66. Jones DT, Knopman DS, Gunter JL, et al. . Cascading network failure across the Alzheimer’s disease spectrum. Brain. 2016;139(Pt 2):547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brier MR, McCarthy JE, Benzinger TLS, et al. . Local and distributed PiB accumulation associated with development of preclinical Alzheimer's disease. Neurobiol Aging. 2016;38:104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goyal MS, Gordon BA, Couture LE, et al. . Spatiotemporal relationship between subthreshold amyloid accumulation and aerobic glycolysis in the human brain. Neurobiol Aging. 2020;96:165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Driscoll I, Troncoso JC, Rudow G, et al. . Correspondence between in vivo (11)C-PiB-PET amyloid imaging and postmortem, region-matched assessment of plaques. Acta Neuropathol. 2012;124:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Quiroz YT, Zetterberg H, Reiman EM, et al. . Plasma neurofilament light chain in the presenilin 1 E280A autosomal dominant Alzheimer's disease kindred: a cross-sectional and longitudinal cohort study. Lancet Neurol. 2020;19:513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dhiman K, Gupta VB, Villemagne VL, et al. . Cerebrospinal fluid neurofilament light concentration predicts brain atrophy and cognition in Alzheimer's disease. Alzheimers Dement (Amst). 2020;12:e12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Racine AM, Koscik RL, Nicholas CR, et al. . Cerebrospinal fluid ratios with Aβ42 predict preclinical brain beta-amyloid accumulation. Alzheimers Dement (Amst). 2016;2:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Meeker KL, Butt OH, Gordon BA, et al. . Cerebrospinal fluid neurofilament light chain is a marker of aging and white matter damage. Neurobiol Dis. 2022;166:105662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Khalil M, Pirpamer L, Hofer E, et al. . Serum neurofilament light levels in normal aging and their association with morphologic brain changes. Nat Commun. 2020;11:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Niendam TA, Laird AR, Ray KL, Dean YM, Glahn DC, Carter CS. Meta-analytic evidence for a superordinate cognitive control network subserving diverse executive functions. Cogn Affect Behav Neurosci. 2012;12:241–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Diamond A. Executive functions. Annu Rev Psychol. 2013;64:135–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lewczuk P, Ermann N, Andreasson U, et al. . Plasma neurofilament light as a potential biomarker of neurodegeneration in Alzheimer's disease. Alzheimers Res Ther. 2018;10:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liu J, Zhang X, Yu C, et al. . Impaired parahippocampus connectivity in mild cognitive impairment and Alzheimer's disease. J Alzheimers Dis. 2016;49:1051–1064. [DOI] [PubMed] [Google Scholar]
- 79. Cha J, Jo HJ, Kim HJ, et al. . Functional alteration patterns of default mode networks: comparisons of normal aging, amnestic mild cognitive impairment and Alzheimer's disease. Eur J Neurosci. 2013;37:1916–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Anticevic A, Cole MW, Murray JD, Corlett PR, Wang XJ, Krystal JH. The role of default network deactivation in cognition and disease. Trends Cogn Sci. 2012;16:584–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen P, Yao H, Tijms BM, et al. . Four distinct subtypes of Alzheimer's disease based on resting-state connectivity biomarkers. Biol Psychiatry. Published online 26 June 2022. 10.1016/j.biopsych.2022.06.019 [DOI] [PubMed] [Google Scholar]
- 82. Vogel JW, Hansson O. Subtypes of Alzheimer's disease: Questions, controversy, and meaning. Trends Neurosci. 2022;45:342–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Koch G, Casula EP, Bonni S, et al. . Precuneus magnetic stimulation for Alzheimer’s disease: a randomized, sham-controlled trial. Brain. 2022;145:3776–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Power JD, Laumann TO, Plitt M, Martin A, Petersen SE. On global fMRI signals and simulations. Trends Cogn Sci. 2017;21:911–913. [DOI] [PubMed] [Google Scholar]
- 85. Ciric R, Wolf DH, Power JD, et al. . Benchmarking of participant-level confound regression strategies for the control of motion artifact in studies of functional connectivity. Neuroimage. 2017;154:174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu TT, Nalci A, Falahpour M. The global signal in fMRI: nuisance or information? Neuroimage. 2017;150:213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. McAvoy MP, Tagliazucchi E, Laufs H, Raichle ME. Human non-REM sleep and the mean global BOLD signal. J Cereb Blood Flow Metab. 2019;39:2210–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ju YE, McLeland JS, Toedebusch CD, et al. . Sleep quality and preclinical Alzheimer disease. JAMA Neurol. 2013;70:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Han F, Chen J, Belkin-Rosen A, et al. . Reduced coupling between cerebrospinal fluid flow and global brain activity is linked to Alzheimer disease-related pathology. PLoS Biol. 2021;19:e3001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Strain JF, Brier MR, Tanenbaum A, et al. . Covariance-based vs. Correlation-based functional connectivity dissociates healthy aging from Alzheimer disease. Neuroimage. 2022;261:119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Network Level Analysis methods can be found at https://github.com/WheelockLab/NetworkLevelAnalysisBeta. Data that support the findings of this study are available from DIAN at https://dian.wustl.edu/our-research/observational-study/dian-observational-study-investigator-resources/.