Abstract
Cysteine-altering missense variants (NOTCH3cys) in one of the 34 epidermal growth-factor-like repeat (EGFr) domains of the NOTCH3 protein are the cause of NOTCH3-associated small vessel disease (NOTCH3-SVD). NOTCH3-SVD is highly variable, ranging from cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) at the severe end of the spectrum to non-penetrance. The strongest known NOTCH3-SVD modifier is NOTCH3cys variant position: NOTCH3cys variants located in EGFr domains 1–6 are associated with a more severe phenotype than NOTCH3cys variants located in EGFr domains 7–34. The objective of this study was to further improve NOTCH3-SVD genotype-based risk prediction by using relative differences in NOTCH3cys variant frequencies between large CADASIL and population cohorts as a starting point.
Scientific CADASIL literature, cohorts and population databases were queried for NOTCH3cys variants. For each EGFr domain, the relative difference in NOTCH3cys variant frequency (NVFOR) was calculated using genotypes of 2574 CADASIL patients and 1647 individuals from population databases. Based on NVFOR cut-off values, EGFr domains were classified as either low (LR-EGFr), medium (MR-EGFr) or high risk (HR-EGFr). The clinical relevance of this new three-tiered EGFr risk classification was cross-sectionally validated by comparing SVD imaging markers and clinical outcomes between EGFr risk categories using a genotype-phenotype data set of 434 CADASIL patients and 1003 NOTCH3cys positive community-dwelling individuals.
CADASIL patients and community-dwelling individuals harboured 379 unique NOTCH3cys variants. Nine EGFr domains were classified as an HR-EGFr, which included EGFr domains 1–6, but additionally also EGFr domains 8, 11 and 26. Ten EGFr domains were classified as MR-EGFr and 11 as LR-EGFr. In the population genotype–phenotype data set, HR-EGFr individuals had the highest risk of stroke [odds ratio (OR) = 10.81, 95% confidence interval (CI): 5.46–21.37], followed by MR-EGFr individuals (OR = 1.81, 95% CI: 0.84–3.88) and LR-EGFr individuals (OR = 1 [reference]). MR-EGFr individuals had a significantly higher normalized white matter hyperintensity volume (nWMHv; P = 0.005) and peak width of skeletonized mean diffusivity (PSMD; P = 0.035) than LR-EGFr individuals. In the CADASIL genotype–phenotype data set, HR-EGFr domains 8, 11 and 26 patients had a significantly higher risk of stroke (P = 0.002), disability (P = 0.041), nWMHv (P = 1.8 × 10−8), PSMD (P = 2.6 × 10−8) and lacune volume (P = 0.006) than MR-EGFr patients. SVD imaging marker load and clinical outcomes were similar between HR-EGFr 1–6 patients and HR-EGFr 8, 11 and 26 patients. NVFOR was significantly associated with vascular NOTCH3 aggregation load (P = 0.006), but not with NOTCH3 signalling activity (P = 0.88).
In conclusion, we identified three clinically distinct NOTCH3-SVD EGFr risk categories based on NFVOR cut-off values, and identified three additional HR-EGFr domains located outside of EGFr domains 1–6. This EGFr risk classification will provide an important key to individualized NOTCH3-SVD disease prediction.
Keywords: NOTCH3, SVD, CADASIL, risk classification, EGFr domain
Hack et al. report a novel three-tiered risk classification for NOTCH3 variants, which are known to be associated with a highly variable phenotype, ranging from CADASIL at the most severe end of the spectrum to non-penetrance. This risk classification provides an important key to individualized CADASIL disease prediction.
Introduction
NOTCH3-associated small vessel disease (NOTCH3-SVD) is defined as cerebral small vessel disease caused by distinct cysteine altering NOTCH3 missense variants in one of the the 34 epidermal growth-factor-like repeat (EGFr) domains of the NOTCH3 protein (NOTCH3cys).1 NOTCH3-SVD has a highly variable disease severity, of which the most severe end of the spectrum is known as cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL). However, NOTCH3cys variants have a high frequency (1:300) in the population worldwide where they are associated with a much milder later-onset NOTCH3-SVD, and even non-penetrance up to age 70 years.2–5
The strongest known NOTCH3-SVD modifier is NOTCH3cys variant position: NOTCH3cys variants located in one of the first six EGFr domains (EGFr 1–6) are associated with a more severe phenotype than NOTCH3cys variants located in one of EGFr domains 7–34 (EGFr 7–34).1,2,5–7 This is consistent with the fact that EGFr 7–34 variants are predominant in population databases,1,2 whereas EGFr 1–6 variants strongly predominate in CADASIL cohorts.5 However, several EGFr 7–34 NOTCH3cys variants are also frequent in CADASIL cohorts, implying that the genotype–phenotype correlation can be further delineated. However, studies so far have lacked the power to study associations between individual EGFr domains and disease severity.5–10
In this study, we set out to improve NOTCH3-SVD genotype-based risk stratification by comparing NOTCH3cys EGFr variant positions of 4221 NOTCH3cys positive individuals from CADASIL and population cohorts. Using this approach, we classified EGFr domains in three NOTCH3cys risk categories, namely high, medium and low risk. To validate the use of our new three-tiered EGFr risk classification, we studied the association between NOTCH3 EGFr risk categories and NOTCH3-small vessel disease severity in the largest data set so far of 1437 NOTCH3cys positive CADASIL patients and community-dwelling individuals. To assess underlying pathomechanisms, we studied the differences in vascular NOTCH3 aggregation load, NOTCH3 signalling activity and in silico 3D dimensional protein structures associated with these low, medium and high NOTCH3cys EGFr risk categories.
Materials and methods
NOTCH3 cys variant annotation
NOTCH3 cys variants were defined as heterozygous cysteine altering NOTCH3 missense variants in one of the 34 EGFr domains of the NOTCH3 protein. NOTCH3cys variants were annotated according to the Human Genome Variation Society (HGVS) nomenclature with reference transcript NM_000435.3. Uniprot was used to determine in which EGFr domain NOTCH3cys variants were located (https://www.uniprot.org/uniprot/Q9UM47). The p.Arg544Cys variant, which is predicted to be between EGFr domains 13 and 14, and the p.Arg886Cys variant, which is predicted to be between EGFr domains 22 and 23, were classified as being located in EGFr domains 13.5 and 22.5, respectively.
NOTCH3 cys variant frequencies in population databases
The frequency of NOTCH3cys variants in the population was assessed using the following whole-exome sequencing population databases: UK Biobank, gnomAD, DiscovEHR and BRAVO. All of these databases include community-dwelling individuals, which mainly comprise healthy volunteers, recruited as part of large population genetic studies or for various non-SVD and non-stroke disease studies. As we used a genotype-first approach we did not exclude individuals from these population databases based on their phenotype.
The UK Biobank is a prospective biobank study of ∼500 000 healthy volunteers aged 40–69 years recruited between 2006 and 2010 in the United Kingdom. A subset of 100 000 individuals also underwent a brain MRI, and of approximately 45 000 individuals brain MRIs were available at the time of analysis. Details on the UK Biobank study have been described previously.11 The NOTCH3cys variants were extracted from 454 805 whole-exome data using reference transcript ENST00000263388.7/NM_000435.3.
GnomAD is a database with 125 748 whole-exome and 15 708 whole-genome sequences from unrelated individuals sequenced as a part of population genetic and various non-SVD and non-stroke disease studies.12 Details about gnomAD can be found on https://gnomad.broadinstitute.org/. The non-TOPMed gnomAD v.2.1.1 database was used to exclude individuals who are also present in the BRAVO database.
The DiscovEHR collaboration between Regeneron Genetics Center and Geisinger Health Systems cohort is a project in which whole-exome sequencing data of >90.000 MyCode participants is combined with longitudinal electronic health records.2 MyCode participants were recruited in primary clinics and specialty clinics throughout the Geisinger Health System without regard to underlying diseases. Details about the DiscovEHR cohort, including a list of all identified NOTCH3cys variants, have been reported previously.2,13
BRAVO is a variant browser of a data set containing 132 345 whole-exome and genome-sequenced individuals from Trans-Omics for Precision Medicine (TOPMed) studies, which aim to improve the understanding of heart, lung, blood and sleep disorders. TOPMed data freeze 8 was used. Details about BRAVO can be found on https://bravo.sph.umich.edu/freeze8/hg38/.
NOTCH3 cys variant frequencies in CADASIL cohorts
To determine NOTCH3cys variant frequencies in CADASIL patients, we used genetic data from the scientific literature, as well as unpublished NOTCH3cys variant data available from databases from three European CADASIL referral centres. The expertise centre databases provided NOTCH3cys variant data for a total of 1184 CADASIL patients (950 Dutch patients from the Leiden University Medical Center, 139 German patients from the Ludwig-Maximilians-University and 95 Italian patients from the Fondazione IRCCS Instituto Neurologico Carlo Besta).
The scientific literature was queried using PubMed search term: ‘CADASIL’ OR ‘NOTCH3’. The last query was performed in November 2021. Inclusion criteria were: (i) the report included minimally 20 CADASIL patients; (ii) the frequency of the NOTCH3cys variants was reported and (iii) Sanger or next-generation sequencing was performed for all NOTCH3 exons (2–24) encoding the 34 EGFr domains. If an article reported the number of families rather than individuals with a specific NOTCH3cys variant, family count was used as a proxy for the NOTCH3cys variant frequency.14–16 The 1390 CADASIL patients in the 12 studies fulfilling the inclusion criteria were from the following (sovereign) countries: Germany,17 China,15,18,19 Italy,14 Japan,9 United Kingdom,16,20 Taiwan,21 Korea10,22 and France.23
Risk classification of EGFr domains based on NOTCH3cys variant frequency odds ratio
The relative difference in frequency of NOTCH3cys variants in CADASIL cohorts compared to the population databases was expressed by calculating the NOTCH3cys variant frequency odds ratio (NVFOR).
The NVFOR was calculated by dividing ‘the odds that a NOTCH3cys variant is located in a certain EGFr domain in CADASIL patients’ by ‘the odds that a NOTCH3cys variant is located in the same EGFr domain in NOTCH3cys-positive individuals from population databases’ (see Supplementary Fig. 1 for examples).
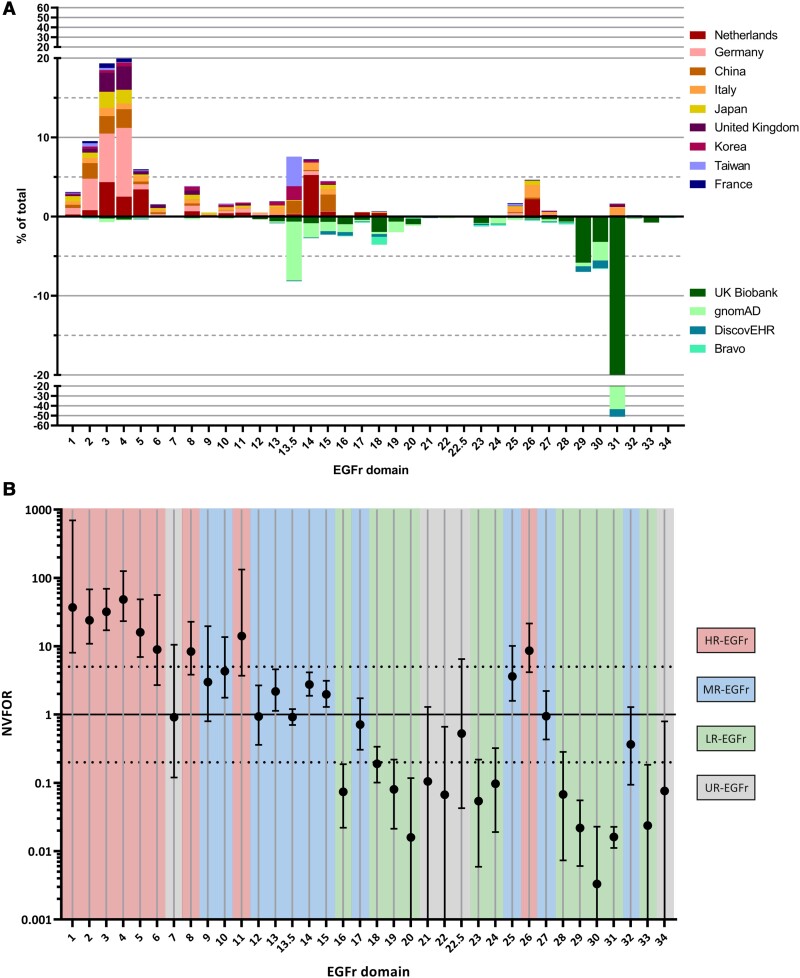
An NVFOR >1 for a specific EGFr domain indicates a relative higher frequency of NOTCH3cys variants in this domain in CADASIL databases than in population databases and conversely an NVFOR <1 indicates a relative higher frequency in population databases. Our goal was to classify each EGFr domain in one of the three EGFr risk categories, i.e. high-risk EGFr domains (HR-EGFr; relative high frequency in CADASIL cohorts), medium-risk EGFr domains (MR-EGFr; similar relative frequency in CADASIL cohorts and population databases) and low-risk EGFr domains (LR-EGFr; relative high frequency in population databases). For this purpose, the following cut-off values were selected prior to genotype–phenotype analysis: HR-EGFr = NVFOR > 5, MR-EGRr = NVFOR between 0.20 and 5 and LR-EGFr = NVFOR < 0.20. These NVFOR cut-off values were selected as they best reflect the differences in NVFOR between the 34 EGFr domains (Fig. 1). Also, the NVFOR cut-off values had to adhere to the following two rules: (i) the NVFOR cut-off values of HR-EGFr and LR-EGFr had to deviate with the same magnitude from NVFOR = 1 (in our case a factor 5 difference, i.e. NVFOR 5.0 and 0.2, respectively); and (ii) the NVFOR 95% confidence interval of each LR-EGFr and each HR-EGFr domain was not allowed to include NVFOR = 1. If an EGFr domain harboured less <10 NOTCH3cys variant counts, the power was considered too low and the domain was classified as having an unknown risk (UR-EGFr). Other cut-off values were deliberately not tested to prevent the need for multiple testing correction.
Figure 1.
The distribution of NOTCH3cys variants in CADASIL cohorts and in population databases and the NVFOR per EGFr domain. (A) Distribution of NOTCH3cys variants in CADASIL cohorts (top) and population databases (bottom) along the 34 EGFr domains. (B) Interval plot showing the NVFOR of each EGFr domain with error bars indicating 95% confidence intervals. Each EGFr domain is classified in an EGFr risk category, based on their respective NVFOR: HR-EGFr domains have NVFOR >5, MR-EGFr domains have an NVFOR between 0.20 and 5 and LR-EGFr domains have an NVFOR <0.20. If an EGFr domain included less than 10 NOTCH3cys variant counts, then it was classified as an UR-EGFr domain. The dotted lines indicate NVFOR 0.2 and 5.0, which are the cut-off points used for the EGFr risk categories.
Clinical and neuroimaging NOTCH3-SVD severity of EGFr risk categories in the CADASIL genotype–phenotype data set
The genotype–phenotype data set for analysing disease severity in CADASIL included 434 patients from the Dutch (n = 200), German (n = 139) and Italian (n = 95) CADASIL data sets. The clinical and neuroimaging data included age at first stroke (n = 430), disability (n = 431), normalized white matter hyperintensity (nWMHv; n = 334), normalized lacune volume (nLV; n = 313), brain parenchymal fraction (BPF; n = 334) and peak width of skeletonized mean diffusivity (PSMD; n = 292).
Stroke was defined as rapidly evolving focal symptoms lasting >24 h with no apparent cause other than of vascular origin. Disability was assessed using the modified Rankin Scale. Hypertension was defined as a previous diagnosis (>140 mmHg systolic or >90 mmHg diastolic) or use of an antihypertensive agent. Hypercholesterolemia was defined as a previous diagnosis. Diabetes was defined as a previous diagnosis of diabetes type 1 or 2 or use of a hypoglycaemic agent. Smoking was defined as current smoker.
Small vessel disease imaging markers were quantitatively assessed according to consensus criteria24 as described previously.6,25–27 BPF was defined as the ratio of brain parenchymal volume to the intracranial volume expressed as a percentage. WMH and lacune volume were also normalized to the intracranial volume: normalized volume = (unnormalized volume/intracranial volume) × 100. PSMD, an established and robust imaging marker for SVD,25 which is the difference between the 5th and 95th percentiles of the voxel-based mean diffusivity values within the white matter tract skeleton, was calculated by using a fully automated shell script (http://www.psmd-marker.com/) on our preprocessed diffusion weighted imaging sequences.
The 200 CADASIL patients and NOTCH3cys-positive family members from the Dutch CADASIL registry were participants in the prospective Disease Variability in NOTCH3-associated Small Vessel Disease (DiViNAS) study at the Leiden University Medical Center performed between November 2017 and December 2020. Participants were uniformly and comprehensively characterized, including a standardized brain MRI protocol, as described previously.6 Brain MRIs were made on a 3 T MR system (Philips Achieva TX, Philips Medical Systems); acquisition parameters are presented in Supplementary Table 1.
The 139 German CADASIL patients and NOTCH3cys-positive family members were participants in three different prospective cohort studies of the Ludwig-Maximilians-University, which were performed between 2007 and 2020.25–27 Brain MRIs were performed on three different MR systems: a 1.5 T Signa (GE Healthcare; n = 66), a 3 T Magnetom Verio (Siemens Healthineers; n = 52) and a 3 T Magnetom Skyra (Siemens Healthineers; n = 21). The acquisition parameters are presented in Supplementary Table 1.
The 95 Italian CADASIL patients and NOTCH3cys-positive family members were seen in the Fondazione IRCCS Istituto Neurologico Carlo Besta of Milan between March 2013 and March 2021. As patients with missing data were excluded, we included in the analyses 88 patients for the outcome age at first stroke and 86 for the outcome disability. Neuroimaging data were not available for the Italian CADASIL patients.
Analysis of clinical and neuroimaging NOTCH3-SVD severity of EGFr risk categories in the population genotype–phenotype data set
The genotype–phenotype data set for analysing disease severity in the population included 1003 NOTCH3cys-positive individuals who participated in the UK Biobank study. The clinical and neuroimaging data included age at first stroke (n = 1003), nWMHv (n = 94), nLV (n = 102), BPF (n = 97) and PSMD (n = 92).
History of stroke and cardiovascular risk factors were extracted from self-report, hospital and death records using ICD-9, ICD-10 and non-cancer illness codes (Supplementary Table 2). Hypertension was defined as a previous diagnosis or the use of antihypertensive medication. For analyses with SVD imaging markers, a mean systolic blood pressure >140 or mean diastolic pressure >90 on the day of the brain MRI was also defined as hypertension. Hypercholesterolemia was defined as a previous diagnosis. Diabetes was defined as a previous diagnosis of diabetes type I or II or the use of a hypoglycaemic agent. Smoking was defined as current smoker.
Brain MRI was available for 102 individuals. Fluid-attenuated inversion recovery (FLAIR) sequences of eight patients and diffusion-weighted imaging (DWI) sequences of 10 patients were excluded by the UK Biobank algorithm from quantitative analysis due to insufficient quality. The acquisition parameters of the brain MRI are presented in Supplementary Table 1. Brain volume was estimated using SIENAX. WMH were quantified using the Brain Intensity Classification Algorithm.28 PSMD was calculated after preprocessing the DWI sequences as described previously.6 Number and volume of lacunes was evaluated on T1-weighted images. FMRIB Software Library (FSL) v6.0.4 (Analysis Group, FMRIB, Oxford, UK) was used for volumetric analyses of lacunes.
NOTCH3-SVD severity of individual EGFr domains
In both the population and CADASIL genotype–phenotype data sets, we also compared NOTCH3-SVD severity between the individual EGFr domains within each of the three EGFr risk categories. For the analysis of SVD imaging markers, only EGFr domains with ≥10 NOTCH3cys-positive individuals were included, and for the analysis of clinical outcomes only EGFr domains with ≥20 NOTCH3cys-positive individuals were included.
Association between EGFr risk categories and vascular NOTCH3 aggregation
To assess association between NOTCH3 aggregation load and EGFr risk categories, the levels of vascular NOTCH3 aggregation, expressed as the immunohistochemical NOTCH3 score,29 was determined in skin vessels of the following groups: LR-EGFr, MR-EGFr, HR-EGFr 1–6 and HR-EGFr 26 patients. For each group, six patients were randomly selected from the Dutch CADASIL cohort with the following selection criteria: (i) age at research visit between 45 and 60 years; and (ii) patients could not have the same NOTCH3cys variant, except if no other patients were available (see Supplementary Table 3 for the NOTCH3cys variants of selected patients).
Skin biopsies from the lateral upper arm were taken using a 4-mm skin punch biopsy. Skin tissue samples were processed and stained with a monoclonal antibody targeting NOTCH3ECD (clone 1E4, Millipore), full-focus microscopy images were taken and NOTCH3ECD-positive area was quantified in all visible blood vessel walls (median 30, range 21–37) using ImageJ, as previously described.29
The NOTCH3ECD-positive area was expressed as percentage of vessel wall area. The NOTCH3 score was defined as the average NOTCH3ECD-positive area of the 10 vessels with the highest NOTCH3ECD-positive area per individual.29
Association between EGFr risk categories and NOTCH3 signalling
To determine whether variants in HR-, MR- and LR-EGFr domains differentially affect NOTCH3 signalling, NOTCH3 signalling activity was assessed in NIH 3T3 cells using a CBF1-responsive luciferase assay with and without Jagged1 stimulation, as described previously.30 The following NOTCH3 cDNA constructs were tested: wild-type, p.Arg141Cys (EGFr 3), p.Cys162Trp (EGFr 4), p.Cys183Arg (EGFr 4), p.Cys495Tyr (EGFr 12), p.Cys568Tyr (EGFr 14), p.Arg578Cys (EGFr 14), p.Cys1015Arg (EGFr 26), p.Arg1231Cys (EGFr 31), deletion of ligand binding domain (i.e. EGFr 10–11) and p.Cys455Arg (EGFr 11). NOTCH3 signalling was expressed as fold increase of luciferase activity upon Jagged1 stimulation (= ‘luciferase activity with Jagged1 stimulation’/‘luciferase activity without Jagged1 stimulation’).
Prediction of the 3D NOTCH3 protein structure
AlphaFold (https://alphafold.ebi.ac.uk/), a state-of-the-art artificial intelligence program, was used to predict the 3D structure of the extracellular domain of the NOTCH3 protein.31 The model with the highest confidence score (pLDDT = 68.2) was manually assessed using the ChimeraX software.32
Statistics
Normally distributed continuous variables were summarized as mean ± standard deviation (SD). Non-normally distributed continuous variables were summarized as median with interquartile range (IQR). The following variables were transformed to obtain a normal distribution, linear relation between dependent and independent variables and normally distributed residuals: nWMHv (square root), PSMD (natural log) and NVFOR (square root transformed). nLV was divided into five quantiles in the CADASIL genotype–phenotype data set to facilitate analysis.
NVFOR and 95% confidence intervals were calculated using binary logistic regression with Firth’s correction to account for rare event bias. To account for the effect of founder variants in the largest cohort/database on the calculation of NVFOR, the largest CADASIL cohort and the largest population database were weighted to the second largest CADASIL cohort and population database, respectively: ‘weight of largest data set’ = ‘number of NOTCH3cys variants in 2nd largest data set’/‘number of NOTCH3cys variants in largest data set’.
The CADASIL and population genotype–phenotype data sets were analysed separately. Multiple linear, logistic and Cox regression models were used to compare SVD imaging markers and clinical outcomes between EGFr risk categories. The following independent variables were always included in the models: age, sex, hypertension, diabetes mellitus type 1 or 2, hypercholesterolemia and packyears. However, as packyears were not available for Italian CADASIL patients, current smoking status was used in the analyses of clinical outcomes for the whole CADASIL genotype–phenotype data set. To account for clustering per study site, differences in MR protocols and quantification of SVD imaging markers between studies, ‘study’ was included as a fixed effect in all models.33
To assess the effect of NOTCH3cys variants leading to the gain of a cysteine residue (Cys-gain) or the loss of a cysteine residue (Cys-loss) on the SVD outcomes, a binary independent variable [Cys-gain (yes/no)] was added to the models.
Multiple linear regression models were used to compare NOTCH3 aggregation between groups. To compare NOTCH3 signalling activity between groups, linear mixed models were used where each independent experiment was added as a random effect.
To correct for multiple testing, Tukey’s correction was used for multiple linear regression models and linear mixed models and Bonferroni’s correction for ordinal logistic and Cox regression models.
All statistical analyses were performed in R (version 4.1.0). Two-tailed P < 0.05 was considered statistically significant.
Standard protocol approvals and patient consents
The prospective clinical study of Dutch CADASIL patients was approved by the medical ethics committee of Leiden Den Haag Delft (P18.164 and P17.170). The German CADASIL cohort was approved by the ethics committee of the Medical Faculty, LMU Munich (299/03 and 158–13). All participants gave written informed consent, which was obtained according to the Declaration of Helsinki.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
NOTCH3 cys variant frequency odds ratio
A total of 4221 individuals with 379 unique NOTCH3cys variant were included for the calculation of NVFOR, of which 2574 were diagnosed CADASIL patients (61.0%) and 1647 (39.0%) community-dwelling individuals (Supplementary Tables 4 and 7).
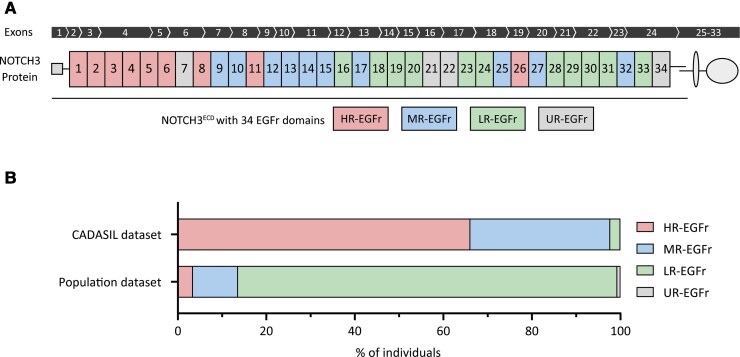
Based on the relative difference in NOTCH3cys variant frequency between CADASIL cohorts and population databases, we calculated the NVFOR for each EGFr domain (NVFOR range: 0.003–48.4). Based on our predefined cut-off values, we identified 9 EGFr domains with a high NVFOR (HR-EGFr), 10 EGFr domains with an intermediate NVFOR (MR-EGFr) and 11 EGFr domains with a low NVFOR (LR-EGFr) (Figs 1 and 2A). Four EGFr domains did not meet the minimum of 10 NOTCH3cys variant counts for NFVOR calculation and were therefore classified as having an undetermined NVFOR (UR-EGFr). The HR-EGFr domains were EGFr 1–6, 8, 11 and 26 (NVFOR range: 8.4–48.4); the MR-EGFr domains were EGFr 9, 10, 12–15, 17, 25, 27, 32 (NVFOR range: 0.36–4.3); and the LR-EGFr domains were EGFr 16, 18–20, 23, 24, 28–31, 33 (NVFOR range: 0.003–0.19). The UR-EGFr domains were 7, 21, 22, 22.5, 34.
Figure 2.
Distribution of high-risk, medium-risk and low-risk EGFr domains along the NOTCH3 protein and in CADASIL and population genotype–phenotype data sets. (A) Schematic representation of the transmembrane NOTCH3 protein. The NOTCH3 gene contains 33 exons, of which exons 2–24 encode the 34 EGFr domains located in the extracellular domain. Each EGFr domain is classified in an EGFr risk category based on their respective NVFOR. (B) Proportional bar charts showing the proportion of individuals with a NOTCH3cys variant for each EGFr risk category in the CADASIL and population genotype–phenotype data sets. Most CADASIL patients harbour an HR-EGFr variant (66.1%), whereas the majority of individuals in the population genotype–phenotype data set harbour an LR-EGFr variant (85.7%).
The relative difference in NOTCH3cys variant frequency between CADASIL cohorts and population databases was largely similar in individuals from European and Asian descent, with the exception of several founder NOTCH3cys variants (Supplementary Fig. 2).
Association between EGFr risk categories and NOTCH3-SVD severity in the genotype–phenotype data sets
The clinical and neuroimaging characteristics and the NOTCH3cys variant distribution of the CADASIL and population genotype–phenotype data sets can be found in Table 1, Supplementary Fig. 3 and Supplementary Table 8.
Table 1.
Characteristics of individuals with a NOTCH3cys variant in the CADASIL and population data sets
| CADASIL | Population | |
|---|---|---|
| N | 434 | 1003 |
| MRI available, n (%) | 334 (77.0) | 102 (10.2) |
| Demographics | ||
| Age at visit, mean (SD) | 51.7 (12.2) | 62.8 (10.0) |
| Men, n (%) | 195 (44.9) | 491 (49.0) |
| Cardiovascular risk factors | ||
| Hypertension, n (%) | 97 (22.7) | 351 (35.0) |
| Hypercholesterolemia, n (%) | 166 (38.9) | 228 (22.7) |
| Diabetes I or II, n (%) | 22 (5.1) | 113 (11.3) |
| Current smoker, n (%) | 83 (19.4) | 98 (9.8) |
| Pack years, median (IQR) | 1.8 (15) | - |
| Clinical symptoms | ||
| Stroke, n (%) | 159 (37.0) | 71 (7.1) |
| Age at first stroke, mean (SD) | 46.6 (10.4) | 61.2 (9.6) |
| mRS | ||
| 0–1 | 306 (71.0) | - |
| 2 | 87 (20.2) | - |
| 3–4 | 38 (8.8) | - |
| Neuroimaging SVD markers | ||
| nWMHv, median (IQR) | 3.63 (4.7) | 0.41 (0.7) |
| PSMD, median (IQR) [×10−4] | 4.19 (2.9) | 2.27 (0.7) |
| Presence of lacunes, n (%) | 186 (42.9) | 15 (14.7) |
| nLV, median (IQR) [×10−3] | 2.36 (23.3) | 0 (0) |
| BPF, mean (SD) | 74.6 (7.2) | 75.6 (2.8) |
mRS = modified Rankin scale.
Association between EGFr risk category and NOTCH3-SVD severity in the CADASIL genotype–phenotype data set
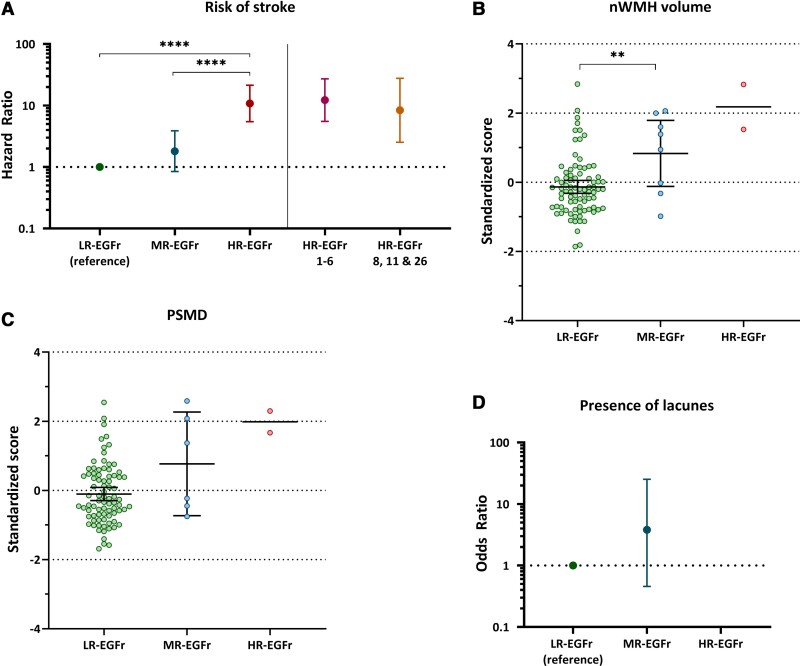
In the CADASIL genotype–phenotype data set, 287 patients harboured an HR-EGFr variant (66.1%), 137 an MR-EGFr variant (31.6%) and 10 an LR-EGFr variant (2.3%) (Fig. 2B). As there were only 10 CADASIL patients with an LR-EGFr variant, this group was not included in the analyses. Brain MRI was available for 77.0% of patients.
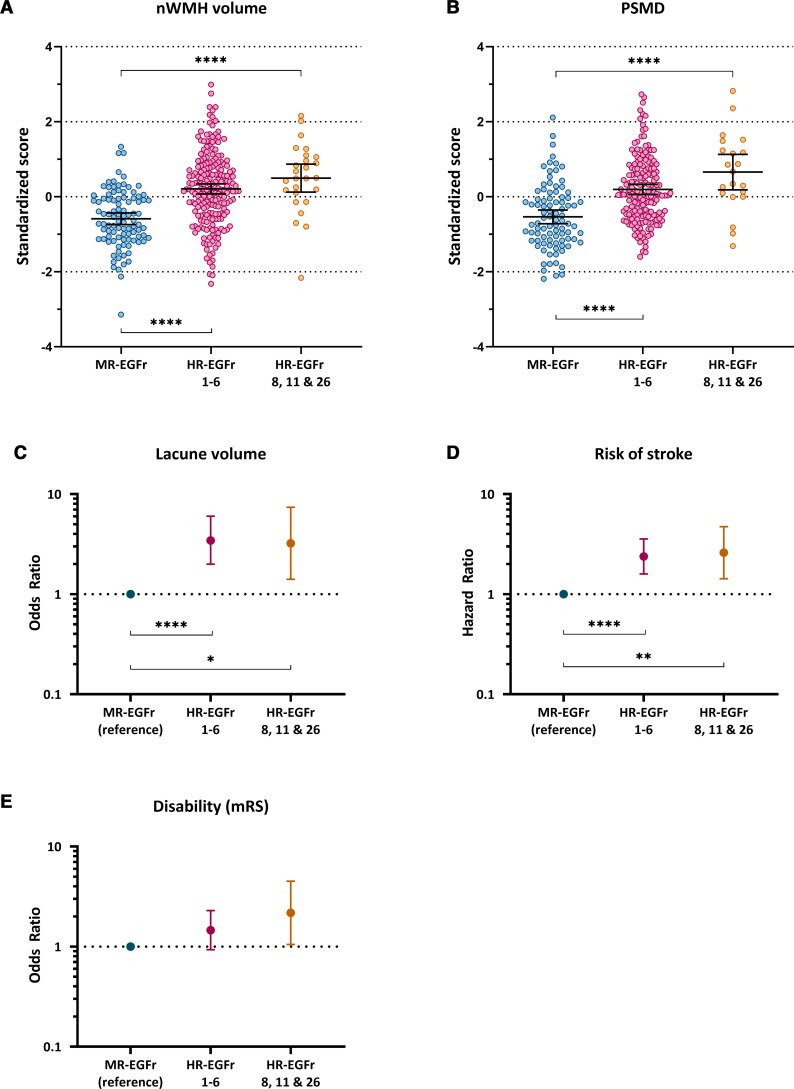
Patients with a NOTCH3cys variant in the newly identified HR-EGFR domains 8, 11 and 26 had a significantly higher load of neuroimaging SVD markers and were clinically significantly more severely affected than patients with MR-EGFr variants: nWMHv (P = 1.8 × 10−8), PSMD (P = 2.6 × 10−8), nLV (P = 0.006), risk of stroke (P = 0.002) and disability (P = 0.041) (Fig. 3). In line with this, patients with an HR-EGFr 8, 11 and 26 variant had a similar burden of SVD imaging markers and clinical outcomes as patients with HR-EGFr 1–6 variants: nWMHv (P = 0.27), PSMD (P = 0.087), nLV (P = 0.87), BPF (P = 0.65), disability (P = 0.26) and risk of stroke (P = 0.75). The BPF did not significantly differ between MR-EGFr and HR-EGFr EGFr domains 8, 11 and 26 (P = 0.19). Using NVFOR in multivariable models explained more of the clinical and neuroimaging variance than using the dichotomous classification ‘EGFr 1–6 versus EGFr 7–34’ (Supplementary Fig. 4).
Figure 3.
CADASIL patients with a NOTCH3cys variant located in the newly identified HR-EGFr domains 8, 11 and 26 are clinically and radiologically as severe as patients with HR-EGFr 1–6 variants. Two scatterplots and three interval plots showing nWMHv (A), PSMD (B), nLV (C), risk of stroke (D) and disability (E) stratified per EGFr risk category in the CADASIL genotype–phenotype data set after correction for cardiovascular risk factors and sex. CADASIL patients with NOTCH3cys variants located in the newly identified HR-EGFr domains 8, 11 and 26 had a significantly higher nWMHv (P = 1.8 × 10−8), PSMD (P = 2.6 × 10−8), nLV (P = 0.006), risk of stroke (P = 0.002) and disability (P = 0.041) than CADASIL patients with MR-EGFr variants. There were no significant differences between patients with HR-EGFr 1–6 variants and those with variants in HR-EGFR domains 8, 11 and 26 in nWMHv (P = 0.27), PSMD (P = 0.087), nLV (P = 0.87), disability (P = 0.26) and risk of stroke (P = 0.75). Error bars indicate 95% confidence interval of the group means. Statistical significance after correction for multiple testing: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
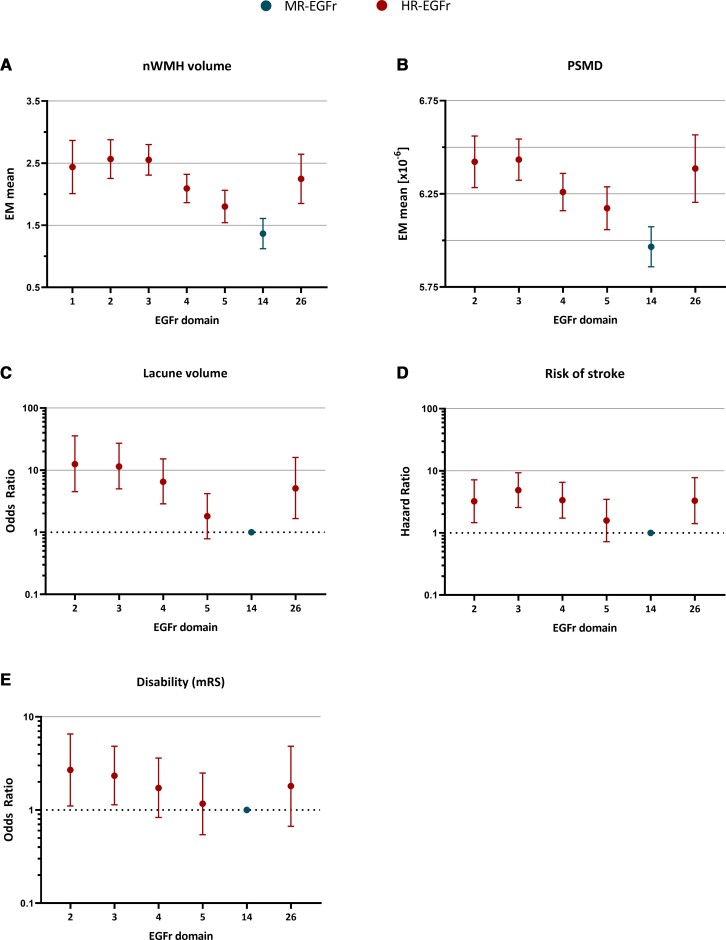
Of the most frequently mutated HR-EGFr domains in the CADASIL genotype–phenotype data set (i.e. EGFR 2–5 and 26), EGFr domains 2 and 3 were associated with the highest burden of nWMHv, PSMD, nLV, risk of stroke and disability (Fig. 4). There was an evident decrease in SVD imaging marker load and risk of stroke from EGFr domain 3 down through EGFr domain 5. EGFr domain 26 had as high a burden of SVD imaging markers and risk of stroke as HR-EGFr domains 2–5.
Figure 4.
NOTCH3 cys variants in EGFr domains 1–5, 14 and 26 are most frequent in the CADASIL genotype–phenotype data set: associations with disease severity. Five interval plots showing the estimated marginal means, odds ratios or hazard ratios of SVD imaging markers, risk of stroke and disability for the EGFr domains most frequently harbouring NOTCH3cys variants in the CADASIL genotype–phenotype data set, after correction for cardiovascular risk factors and sex. Only EGFr domains which had high NOTCH3cys variant counts (≥10 individuals for SVD imaging markers and ≥20 individuals for clinical outcomes) were included in the analyses. Error bars indicate 95% confidence intervals of the group means. (A–D) Of all frequently mutated HR-EGFr domains, EGFr domains 2 and 3 were associated with the highest burden of nWMHv, PSMD, nLV and risk of stroke. There was an evident decrease in the SVD imaging marker loads and risk of stroke in EGFr domain 3 through EGFr domain 5. EGFr domain 26 was associated with as high a burden of SVD imaging markers and risk of stroke as the other HR-EGFr domains. EGFr domain 14, which was the only MR-EGFr domain with a high frequency of NOTCH3cys variants in CADASIL patients, was associated with the lowest burden of SVD imaging markers and risk of stroke. (E) There were no significant differences in disability. Further details including statistically significant differences in the burden of SVD imaging markers and risk of stroke between EGFr domains are shown in Supplementary Tables 5 and 6.
Association between EGFr risk category and NOTCH3-SVD severity in the population genotype–phenotype data set
In the population genotype–phenotype data set, 34 individuals harboured an HR-EGFr variant (3.4%), 102 individuals an MR-EGFr variant (10.2%), 860 individuals an LR-EGFr variant (85.7%) and 7 an UR-EGFr variant (0.7%). Individuals with UR-EGFr variants were excluded from the analysis. Brain MRI was available for 10.2% of the individuals. The risk of stroke was the highest in individuals with an HR-EGFr variant (OR = 10.81, 95% CI: 5.46–21.37), followed by MR-EGFr individuals (OR = 1.81, 95% CI: 0.84–3.88), and was the lowest in LR-EGFr individuals [OR = 1 (reference)] (Fig. 5A). MR-EGFr individuals had a significantly higher nWMHv (P = 0.005) and PSMD (P = 0.035) than LR-EGFr individuals (Fig. 5B and C). There was no significant difference in the presence of lacunes (P = 0.17) and BPF (P = 0.27) between MR-EGFr and LR-EGFr individuals (Fig. 5D). The two HR-EGFr individuals with a brain MRI had a much higher mean nWMHv and PSMD than the MR-EGFr and LR-EGFr individuals.
Figure 5.
Association between small vessel disease imaging marker load and risk of stroke between EGFr risk categories in the population genotype–phenotype data set. Two scatterplots and two interval plots showing the risk of stroke, nWMHv, PSMD and nLV stratified per EGFr risk category in the population genotype–phenotype data set after correction for cardiovascular risk factors and sex. Error bars indicate 95% confidence intervals of the group means. (A) HR-EGFr individuals had a significantly higher risk of stroke than MR-EGFr (P = 1.5 × 10−4) and LR-EGFr individuals (P = 8.1 × 10−12). There was a trend to a higher risk of stroke for MR-EGFr individuals compared to LR-EGFr individuals (P = 0.13). Both HR-EGFr 1–6 and HR-EGFr 8, 11, 26 individuals had a much higher risk of stroke than MR-EGFr (P = 2.2 × 10−4 and P = 0.025, respectively) and LR-EGFr individuals (P = 7.4 × 10−10 and P = 5.0 × 10−4, respectively). (B and C) MR-EGFr individuals had a significantly higher nWMHv (P = 0.005) and PSMD (P = 0.035) than LR-EGFr individuals. Only two individuals in the population with a brain MRI harboured an HR-EGFr variant and both had a much higher mean nWMHv and PSMD than MR-EGFr and LR-EGFr individuals. (D) Only 15 of the 102 (14.7%) individuals with a brain MRI had lacunes on brain MRI. There was no significant difference in the presence of lacunes between MR-EGFr and LR-EGFr individuals (P = 0.16). As the two HR-EGFr individuals had no lacunes on brain MRI, the odds ratio for the presence of lacunes could not be calculated. Statistical significance after correction for multiple testing: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Of the most frequently mutated LR-EGFr domains in the population genotype–phenotype data set (EGFR 29, 30 and 31), EGFr domain 30 was associated with the lowest burden of nWMHv, PSMD and risk of stroke (Supplementary Fig. 5).
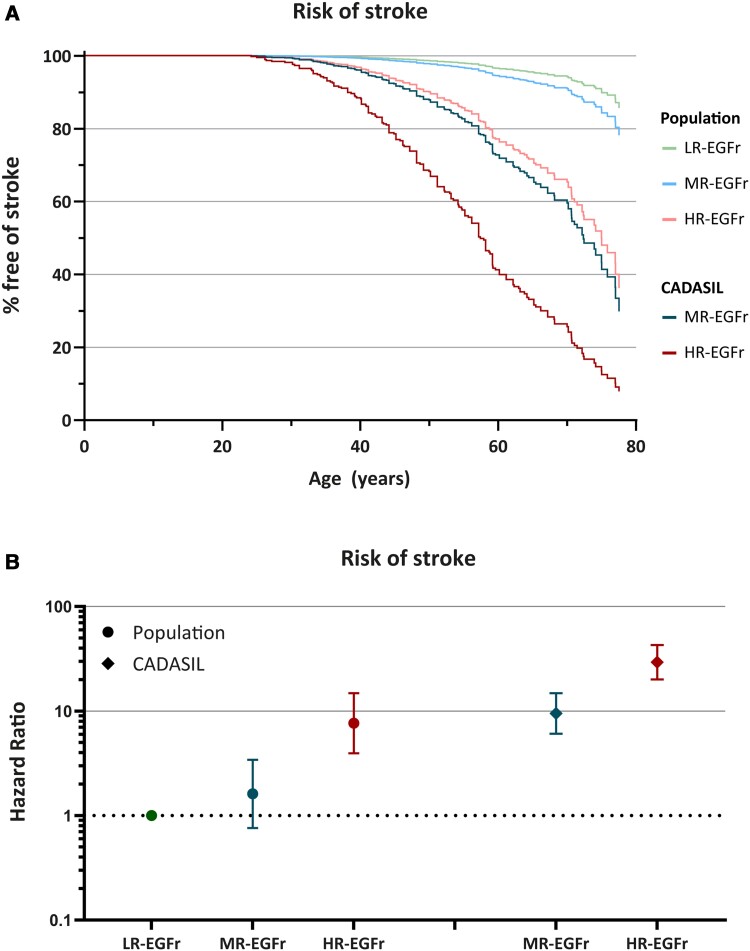
Differences in risk of stroke between EGFr risk categories in the CADASIL and the population genotype–phenotype data sets
Finally, we compared the risk of stroke between the EGFr risk categories in the CADASIL and population genotype–phenotype data sets. MR-EGFr and HR-EGFr patients in the CADASIL data set had a significantly earlier onset of stroke than individuals from the same EGFr risk category in the population data set [MR-EGFr: median 72 versus >78 years (P = 8.1 × 10−6); HR-EGFr: median 57 versus 74 years (P = 3.5 × 10−5); Fig. 6]. The risk of stroke for HR-EGFr individuals in the population data set and MR-EGFr patients from the CADASIL data set was similar (P = 0.54). SVD imaging markers could not be compared between the EGFr risk categories in the CADASIL and population data set due to differences in MR imaging protocols and quantification methods.
Figure 6.
The risk of stroke between the NOTCH3cys EGFr risk categories in the CADASIL and population genotype–phenotype data set. A survival curve (A) and interval plot (B), both corrected for cardiovascular risk factors and sex, showing that MR-EGFr and HR-EGFr patients in the CADASIL genotype–phenotype data set had a significant earlier onset of stroke than individuals from the same EGFr risk category in the population genotype–phenotype data set [MR-EGFr: median 72 versus >78 years (P = 8.1 × 10−6); HR-EGFr: median 57 versus 74 years (P = 3.5 × 10−5)]. The risk of stroke of HR-EGFr individuals in the population data set and MR-EGFr patients from the CADASIL data set was similar (P = 0.54).
The effect of the gain or the loss of a cysteine residue on NOTCH3-SVD severity
There were no evident differences between the distributions of Cys-gain and Cys-loss NOTCH3cys variants in CADASIL cohorts and population databases (data not shown). The percentage of Cys-gain variants was similar between CADASIL cohorts and the population databases (77.4% versus 76.1%; P = 0.35). In multivariable models, there were no significant differences between Cys-gain and Cys-loss variants in SVD imaging markers, risk of stroke and disability in the CADASIL (all P > 0.27) and population (all P > 0.14) genotype–phenotype data sets.
Mechanisms underlying EGFr risk categories
NOTCH3 aggregation
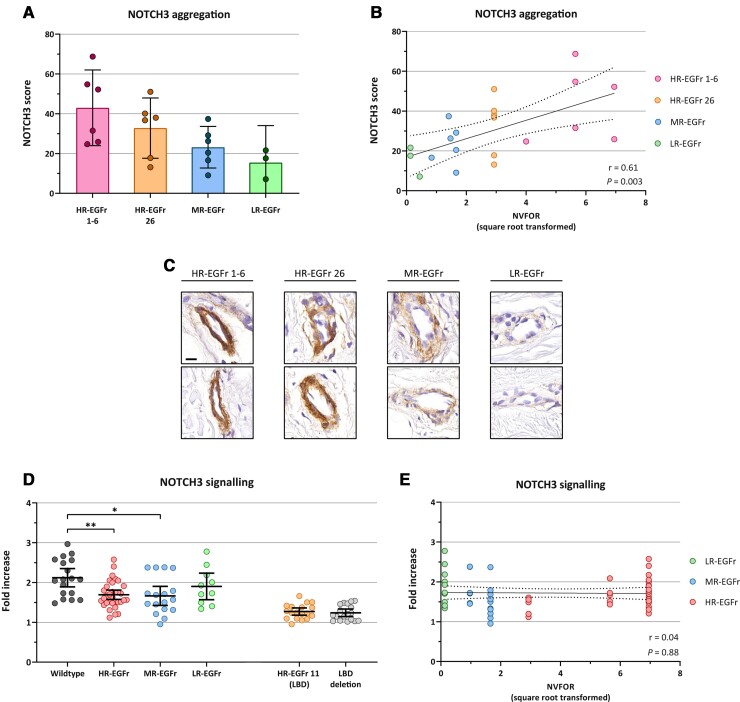
For immunohistochemistry, skin tissue samples were selected from six HR-EGFr 1–6, six HR-EGFr 26 and six MR-EGFr patients. From the LR-EGFr group, three skin tissue samples were available. HR-EGFr 1–6 CADASIL patients had a significantly higher NOTCH3 score than MR-EGFr patients: mean NOTCH3 score 43.0 (95% CI: 30.3–55.6) versus 23.2 (95% CI: 10.5–35.8) (P = 0.033; Fig. 7A). HR-EGFr 26 patients had a mean NOTCH3 score of 32.8 (95% CI: 20.1–45.5), which did not significantly differ from that of HR-EGFr 1–6 (P = 0.24) and MR-EGFr (P = 0.27) patients. The three LR-EGFr patients had a mean NOTCH3 score of 15.4 (95% CI: 0–32.6). NVFOR was significantly associated with NOTCH3 score (P = 0.003), also after correction for age (P = 0.006; Fig. 7B and C).
Figure 7.
Differences in NOTCH3 aggregation and signalling between EGFr risk categories. (A–C) NOTCH3 aggregation load, expressed as immunohistochemical NOTCH3 score,29 was determined in skin tissue samples of LR-EGFr (n = 3), MR-EGFr (n = 6) and HR-EGFr (n = 12) patients, of which six had an HR-EGFr 1–6 NOTCH3cys variant and six had a NOTCH3cys variant located in the newly identified HR-EGFr domain 26. (A) As there were only tissue samples available of three LR-EGFr patients, these patients were excluded from the group comparisons. HR-EGFr 1–6 CADASIL patients had a significantly higher NOTCH3 score than MR-EGFr patients: mean NOTCH3 score 43.0 (95% CI: 30.3–55.6) versus 23.2 (95% CI: 10.5–35.8) (P = 0.033). HR-EGFr 26 patients had a mean NOTCH3 score of 32.8 (95% CI: 20.1–45.5), which did not significantly differ from that of HR-EGFr 1–6 (P = 0.24) and MR-EGFr (P = 0.27) patients. The three LR-EGFr patients had a mean NOTCH3 score of 15.4 (95% CI: 0–32.6). (B) NVFOR was significantly associated with a NOTCH3 score (P = 0.003), also after correction for age (P = 0.006). (C) Representative images of NOTCH3-immunohistochemistry of skin biopsies of HR-EGFr 1–6, HR-EGFr 26, MR-EGFr and LR-EGFr patients, showing increasing granular NOTCH3 staining from low- to high-risk EGFr domains. Bar represents 20 µm. (D) NOTCH3 signalling activity was assessed in NIH 3T3 cells using a CBF1-responsive luciferase assay with and without Jagged1 stimulation. NOTCH3 signalling was expressed as fold increase of luciferase activity upon Jagged1 stimulation (= ‘luciferase activity with Jagged1 stimulation’/‘luciferase activity without Jagged1 stimulation’). Each dot represents an individual experiment. Signalling activity did not differ significantly between NOTCH3cys variants located in the other HR-EGFr domains and the MR-EGFr and LR-EGFr domains (all P > 0.58). However, NOTCH3cys variants in HR-EGFr and MR-EGFr showed a significantly lower NOTCH3 signalling activity compared to wild-type (P = 0.008 and P = 0.02). As expected, NOTCH3cys variants located in the HR-EGFr domain 11 had strongly impaired ligand-induced signalling activity, comparable to a deletion of the ligand binding domain (P = 0.53). There was no significant difference in NOTCH3 signalling activity between HR-EGFr domain 26 and other HR-EGFr domains, MR-EGFr domains and LR-EGFr domains (all P > 0.47). (E) NVFOR was not associated with NOTCH3 signalling activity (P = 0.88). Error bars indicate 95% confidence intervals of the group means. Statistical significance after correction for multiple testing: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
On visual inspection of the predicted Alphafold 3D protein structure, HR- and MR-EGFr domains seemed to be located predominantly at the surface of the protein and the newly identified HR-EGFr domain 26 was also located in a loop, in close proximity to the HR-EGFr domains 1–6 (Supplementary Fig. 6). The LR-EGFr domains were located either more internally or in a separate loop.
NOTCH3 signalling
Finally, we assessed whether variants in HR-, MR- and LR-EGFr domains differentially affect NOTCH3 signalling. The HR-EGFr domain 11 was analysed separately, as this domain is known to be involved in ligand binding. As expected, NOTCH3cys variants located in HR-EGFr domain 11 had strongly impaired ligand-induced signalling activity, comparable to a deletion of the ligand binding domain (P = 0.53). Signalling activity did not differ significantly between NOTCH3cys variants located in the other HR-EGFr domains, the MR-EGFr and LR-EGFr domains (all P > 0.58; Fig. 7D). However, NOTCH3cys variants in HR-EGFr and MR-EGFr showed a significantly lower NOTCH3 signalling activity compared to wild-type (P = 0.008 and P = 0.02, respectively). Signalling activity of LR-EGFr domains did not differ significantly from wild-type (P = 0.40). There was no significant difference in NOTCH3 signalling activity between HR-EGFr domain 26 and other HR-EGFr domains, MR-EGFr domains and LR-EGFr domains (all P > 0.47). NVFOR was not associated with NOTCH3 signalling activity (P = 0.88; Fig. 7E).
Discussion
By comparing the relative NOTCH3cys variant frequencies between 4221 NOTCH3cys-positive individuals from CADASIL and population cohorts, we classified NOTCH3cys variants into three EGFr risk categories and identified three novel HR-EGFr domains next to the known HR-EGFr domains 1–6. The clinical relevance of this new three-tiered EGFr risk classification was validated in a large genotype–phenotype data set of 1437 NOTCH3cys-positive CADASIL patients and community-dwelling individuals. Finally, we showed that EGFR risk categories were significantly associated with vascular NOTCH3 aggregation load, whereas NOTCH3 signalling activity did not differ significantly between the EGFr risk categories. This is in line with the prevailing theory that NOTCH3-SVD, with CADASIL at the severe end of the clinical spectrum, is primarily a vascular protein aggregation disorder, with HR-EGFr variants leading to high levels of NOTCH3 aggregation and clinically more severe small vessel disease.
Our results convincingly demonstrate that there are marked differences in NOTCH3-SVD severity between the EGFr risk categories, as there were significant differences in the most important clinical and neuroimaging outcomes in both the CADASIL and the population genotype–phenotype data sets.25,34 Although BPF has been reported to be an important SVD imaging marker in CADASIL,34 we did not observe significant differences in BPF between the EGFr risk categories. This is potentially due to the fact that severe WMH burden in CADASIL is known to be associated with brain swelling, which masks brain atrophy.6,35
The HR-, MR- and LR-EGFr risk categories can be used in the clinical setting as a starting point to predict disease severity in individuals in whom a NOTCH3cys variant is ascertained. However, as NOTCH3cys variants from the same EGFr risk category are associated with a more severe NOTCH3-SVD in individuals from CADASIL pedigrees than in community-dwelling individuals without a positive family history for CADASIL, it is essential to know whether a NOTCH3cys variant is ascertained because of a clinical suspicion of CADASIL or due to a positive family history, or if a NOTCH3cys variant is ascertained as an incidental finding in a patient who has had whole-exome sequencing for an unrelated disorder. In an individual from a CADASIL pedigree, a NOTCH3cys variant in an HR-EGFr domain will almost always lead to a classical severe CADASIL phenotype, whereas a variant in an MR-EGFr domain is associated with a much broader NOTCH3-SVD spectrum, including very mild subclinical phenotypes.36 Conversely, NOTCH3cys variants located in LR-EGFr and MR-EGFr domains have a relatively high population frequency (1:300 worldwide),1,4 and we are increasingly encountering these variants as incidental findings in patients with unrelated disorders now that whole-exome sequencing is becoming mainstream medical (genetic) practice. In these cases, a diagnosis of CADASIL should not be made unless there is a clear medical history of small vessel disease, as we found very low frequencies of stroke and lacunes in the MR- and LR-EGFr groups in the population genotype–phenotype data set and we have previously also reported non-penetrance in the population.1,2 The difference in NOTCH3-SVD severity between individuals from CADASIL pedigrees and community-dwelling individuals is striking and discovery of still largely unknown genetic and environmental factors underlying this difference may lead to new opportunities for secondary stroke prevention or therapeutic targets.
EGFr risk category was strongly positively associated with vascular NOTCH3 aggregation load. Although protein modelling using Alphafold showed some differences in the 3D localization of HR-, MR- and LR-EGFr domains, this could not convincingly clarify the difference in aggregation propensity. However, as patients with HR-EGFr 1–6 variants have been shown to not only have high NOTCH3 aggregation scores but also more granular osmiophilic material deposits (GOM),29 we hypothesize that HR-EGFr NOTCH3 proteins may be more prone to form complexes with matrisome proteins such as HTRA-1, TIMP3, vitronectin and LTBP-1.37–39 Alternatively, there may be a role for the two recently identified non-enzymatic cleavage sites at Asp–Pro bonds, which are located between HR-EGFr 1–2 and HR-EGFr 2–3,40,41 especially as we identified two additional Asp–Pro bonds close to HR-EGFr domain 26.
Although NOTCH3 signalling activity in the HR-EGFr and MR-EGFr categories was lower than for wild-type NOTCH3, we did not observe a significant association between EGFr risk categories and signalling activity. This suggests that, overall, impaired NOTCH3 signalling is not a determining factor in NOTCH3-SVD severity. NOTCH3cys variants located specifically in HR-EGFr domain 11, which is part of the NOTCH3 ligand binding domain, did show a strongly reduced NOTCH3 signalling activity. Interestingly, the vascular NOTCH3 aggregation load of the two CADASIL patients with an HR-EGFr 11 variant was relatively low for an HR-EGFr domain, but larger sample sizes are needed to draw any conclusions. Previous studies comparing the signalling properties of wildtype and mutant NOTCH3 have led to conflicting results: some studies observed (a trend to) lower signalling properties of mutant NOTCH3,42,43 whereas others found no differences between wild-type and mutant,44,45 or even higher signalling properties of mutant NOTCH3.46 These discrepancies between studies could be explained by several factors, including technical or batch differences and the used ligands. Whether and to what extent impaired signalling contributes to disease severity remains to be established.
Even when we accounted for all known NOTCH3-SVD modifying factors, i.e. NOTCH3cys variant position, sex and cardiovascular risk factors, there was still clinically relevant unexplained variability in CADASIL and NOTCH3-SVD severity. The gain or the loss of a cysteine residue by a NOTCH3cys variant did not explain part of the variability, as the percentage of Cys-gain and Cys-loss variants was similar in population and CADASIL cohorts and, moreover, there was no association between Cys-gain or Cys-loss NOTCH3cys variants and disease severity in both the CADASIL and population genotype–phenotype data sets. In our opinion this provides sufficient evidence to conclude that the loss or gain of a cysteine residue has no substantial effect on disease severity.
To avoid selection bias in this study, we used a genotype-first approach, meaning we did not exclude individuals based on their phenotype in either the CADASIL cohorts or the population databases. It is, therefore, possible that several (undiagnosed) CADASIL patients were included in the population databases,1 and vice versa, several individuals from a CADASIL pedigree with an extremely mild SVD phenotype were recruited in the CADASIL cohorts.36 However, these are rare exceptions to the rule and have only a negligible impact on the NVFOR estimates, as overall, the NOTCH3-SVD phenotype of patients from CADASIL cohorts is manifold more severe than the NOTCH3-SVD phenotype of community-dwelling individuals from population cohorts.
Within the HR-EGFr domains, HR-EGFr domains 2 and 3 were associated with a significantly more severe NOTCH3-SVD phenotype than the other HR-EGFr domains. Further fine-tuning of the EGFr risk classification will require even larger data sets, including data of individuals from various ethnic backgrounds and geographical regions. This will also diminish the effect of founder variants in the largest data sets on NVFOR estimates. For example, the frequency of NOTCH3cys variants in EGFr domains 5 and 14 in CADASIL patients from the Netherlands is higher than in the rest in the world due to two founder variants (p.Arg207Cys in EGFr 5; p.Arg578Cys in EGFr 14). Although the effect of founder variants on the estimates of NVFOR was taken into account by giving a lower weight to the largest data sets, an overestimation of the NVFOR in EGFr domains 5 and 14 cannot be excluded.
In conclusion, we have introduced a novel and clinically relevant three-tiered risk classification for NOTCH3cys variants, according to their location in the 34 NOTCH3 EGFr domains. We show substantial differences in NOTCH3-SVD severity between NOTCH3cys variants located in EGFr domains designated as low, medium or high risk. Using the NOTCH3cys EGFr risk classification will improve individualized NOTCH3-SVD disease prediction in both CADASIL patients and incidentally identified NOTCH3cys positive individuals, improve patient inclusion in clinical studies, facilitate identification of so far unknown disease modifiers and will guide the development of disease modifying therapies.
Supplementary Material
Acknowledgements
The authors would like to thank Hans Dauwerse,† Maurice Overzier and Max Trauernicht for performing the NOTCH3 signalling experiments and all the CADASIL patients who participated in our studies. This research was conducted using the UK Biobank Resource under Application Number 74162.
† Deceased 31 July 2020
Contributor Information
Remco J Hack, Department of Clinical Genetics, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands.
Gido Gravesteijn, Department of Clinical Genetics, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands.
Minne N Cerfontaine, Department of Clinical Genetics, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands.
Mark A Santcroos, Department of Human Genetics, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands.
Laura Gatti, Laboratory of Neurobiology, Fondazione IRCSS Istituto Neurologico Carlo Besta, 20133 Milan, Italy.
Anna Kopczak, Institute for Stroke and Dementia Research, LMU University Hospital Munich, 81377 Munich, Germany.
Anna Bersano, Cerebrovascular Unit, Fondazione IRCCS Istituto Neurologico Carlo Besta, 20133 Milan, Italy.
Marco Duering, Institute for Stroke and Dementia Research, LMU University Hospital Munich, 81377 Munich, Germany; Medical Image Analysis Center (MIAC) and Department of Biomedical Engineering, University of Basel, 4051 Basel, Switzerland.
Julie W Rutten, Department of Clinical Genetics, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands.
Saskia A J Lesnik Oberstein, Department of Clinical Genetics, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands.
Funding
This study and DiViNAS cohort was funded by the Netherlands Organisation for Health Research and Development (ZonMW 91717325) and The Netherlands Brain Foundation (HA2016-02-03).
ZOOM@SVDs is part of the SVDs@target project and was funded by the European Union’s Horizon 2020 research and innovative program under grant agreement No. 666,881.
Competing interests
Remco J. Hack is funded by the Netherlands Organisation for Health Research and Development (ZonMW 91717325). Minne N. Cerfontaine is funded by the Netherlands Organisation for Health Research and Development (ZonMW 91717325). Julie W. Rutten received financial research support from the Netherlands Organisation for Health Research and Development (ZonMW 91717325) and the Netherlands Brain Foundation (HA2016–02-03). Saskia A.J. Lesnik Oberstein has financial research support from the Netherlands Organisation for Health Research and Development (ZonMW 91717325) and the Netherlands Brain Foundation (HA2016-02-03). The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Rutten JW, Hack RJ, Duering M, et al. . Broad phenotype of cysteine altering NOTCH3 variants in UK biobank: CADASIL to non-penetrance. Neurology. 2020;95(13):e1835–e1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hack RJ, Rutten JW, Person TN, et al. . Cysteine-altering NOTCH3 variants are a risk factor for stroke in the elderly population. Stroke. 2020;51:3562–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG. CADASIL. Lancet Neurol. 2009;8:643–653. [DOI] [PubMed] [Google Scholar]
- 4. Rutten JW, Dauwerse HG, Gravesteijn G, et al. . Archetypal NOTCH3 mutations frequent in public exome: Implications for CADASIL. Ann Clin Transl Neurol. 2016;3:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rutten JW, Van Eijsden BJ, Duering M, et al. . The effect of NOTCH3 pathogenic variant position on CADASIL disease severity: NOTCH3 EGFr 1–6 pathogenic variant are associated with a more severe phenotype and lower survival compared with EGFr 7–34 pathogenic variant. Genet Med. 2019;21:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hack RJ, Cerfontaine MN, Gravesteijn G, et al. . Effect of NOTCH3 EGFr group, sex, and cardiovascular risk factors on CADASIL clinical and neuroimaging outcomes. Stroke. 2022;53:3133–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho BPH, Nannoni S, Harshfield EL, et al. . NOTCH3 variants are more common than expected in the general population and associated with stroke and vascular dementia: An analysis of 200 000 participants. J Neurol Neurosurg Psychiatry. 2021;92(7):694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adib-Samii P, Brice G, Martin RJ, Markus HS. Clinical spectrum of CADASIL and the effect of cardiovascular risk factors on phenotype: Study in 200 consecutively recruited individuals. Stroke. 2010;41:630–634. [DOI] [PubMed] [Google Scholar]
- 9. Mukai M, Mizuta I, Watanabe-Hosomi A, et al. . Genotype–phenotype correlations and effect of mutation location in Japanese CADASIL patients. J Hum Genet. 2020;65:637–646. [DOI] [PubMed] [Google Scholar]
- 10. Min JY, Park SJ, Kang EJ, Hwang SY, Han SH. Mutation spectrum and genotype–phenotype correlations in 157 Korean CADASIL patients: A multicenter study. Neurogenetics. 2022;23(1):45–58. [DOI] [PubMed] [Google Scholar]
- 11. Bycroft C, Freeman C, Petkova D, et al. . The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karczewski KJ, Francioli LC, Tiao G, et al. . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carey DJ, Fetterolf SN, Davis FD, et al. . The Geisinger MyCode community health initiative: An electronic health record-linked biobank for precision medicine research. Genet Med. 2016;18:906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bianchi S, Zicari E, Carluccio A, et al. . CADASIL in central Italy: A retrospective clinical and genetic study in 229 patients. J Neurol. 2015;262:134–141. [DOI] [PubMed] [Google Scholar]
- 15. Chen S, Ni W, Yin XZ, et al. . Clinical features and mutation spectrum in Chinese patients with CADASIL: A multicenter retrospective study. CNS Neurosci Ther. 2017;23:707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moreton FC, Razvi SS, Davidson R, Muir KW. Changing clinical patterns and increasing prevalence in CADASIL. Acta Neurol Scand. 2014;130:197–203. [DOI] [PubMed] [Google Scholar]
- 17. Opherk C, Peters N, Herzog J, Luedtke R, Dichgans M. Long-term prognosis and causes of death in CADASIL: A retrospective study in 411 patients. Brain. 2004;127(Pt 11):2533–2539. [DOI] [PubMed] [Google Scholar]
- 18. Hu Y, Sun Q, Zhou Y, et al. . NOTCH3 variants and genotype–phenotype features in Chinese CADASIL patients. Front Genet. 2021;12:705284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu X, Zuo Y, Sun W, et al. . The genetic spectrum and the evaluation of CADASIL screening scale in Chinese patients with NOTCH3 mutations. J Neurol Sci. 2015;354(1–2):63–69. [DOI] [PubMed] [Google Scholar]
- 20. Singhal S, Bevan S, Barrick T, Rich P, Markus HS. The influence of genetic and cardiovascular risk factors on the CADASIL phenotype. Brain. 2004;127(Pt 9):2031–2038. [DOI] [PubMed] [Google Scholar]
- 21. Liao YC, Hsiao CT, Fuh JL, et al. . Characterization of CADASIL among the Han Chinese in Taiwan: Distinct genotypic and phenotypic profiles. PLoS One. 2015;10:e0136501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim YE, Yoon CW, Seo SW, et al. . Spectrum of NOTCH3 mutations in Korean patients with clinically suspicious cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Neurobiol Aging. 2014;35:726.e1–6. [DOI] [PubMed] [Google Scholar]
- 23. Joutel A, Vahedi K, Corpechot C, et al. . Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 1997;350:1511–1515. [DOI] [PubMed] [Google Scholar]
- 24. Wardlaw JM, Smith EE, Biessels GJ, et al. . Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12:822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baykara E, Gesierich B, Adam R, et al. . A novel imaging marker for small vessel disease based on skeletonization of white matter tracts and diffusion histograms. Ann Neurol. 2016;80:581–592. [DOI] [PubMed] [Google Scholar]
- 26. Duering M, Csanadi E, Gesierich B, et al. . Incident lacunes preferentially localize to the edge of white matter hyperintensities: Insights into the pathophysiology of cerebral small vessel disease. Brain. 2013;136(Pt 9):2717–2726. [DOI] [PubMed] [Google Scholar]
- 27. van den Brink H, Kopczak A, Arts T, et al. . Zooming in on cerebral small vessel function in small vessel diseases with 7 T MRI: Rationale and design of the “ZOOM@SVDs” study. Cereb Circ Cogn Behav. 2021;2:100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Griffanti L, Zamboni G, Khan A, et al. . BIANCA (Brain intensity AbNormality Classification Algorithm): A new tool for automated segmentation of white matter hyperintensities. Neuroimage. 2016;141:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gravesteijn G, Hack RJ, Mulder AA, et al. . NOTCH3 Variant position is associated with NOTCH3 aggregation load in CADASIL vasculature. Neuropathol Appl Neurobiol. 2022;48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gravesteijn G, Dauwerse JG, Overzier M, et al. . Naturally occurring NOTCH3 exon skipping attenuates NOTCH3 protein aggregation and disease severity in CADASIL patients. Hum Mol Genet. 2020;29:1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jumper J, Evans R, Pritzel A, et al. . Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pettersen EF, Goddard TD, Huang CC, et al. . UCSF Chimerax: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McNeish D, Stapleton LM. Modeling clustered data with very few clusters. Multivariate Behav Res. 2016;51:495–518. [DOI] [PubMed] [Google Scholar]
- 34. Chabriat H, Herve D, Duering M, et al. . Predictors of clinical worsening in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy: Prospective cohort study. Stroke. 2016;47:4–11. [DOI] [PubMed] [Google Scholar]
- 35. Yao M, Jouvent E, During M, et al. . Extensive white matter hyperintensities may increase brain volume in cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Stroke. 2012;43:3252–3257. [DOI] [PubMed] [Google Scholar]
- 36. Hack RJ, Gravesteijn G, Cerfontaine MN, et al. . Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy family members with a pathogenic NOTCH3 variant can have a normal brain magnetic resonance imaging and skin biopsy beyond age 50 years. Stroke. 2022;53:1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Monet-Lepretre M, Haddad I, Baron-Menguy C, et al. . Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: A new pathomechanism in CADASIL. Brain. 2013;136(Pt 6):1830–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kast J, Hanecker P, Beaufort N, et al. . Sequestration of latent TGF-beta binding protein 1 into CADASIL-related Notch3-ECD deposits. Acta Neuropathol Commun. 2014;2:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zellner A, Scharrer E, Arzberger T, et al. . CADASIL brain vessels show a HTRA1 loss-of-function profile. Acta Neuropathol. 2018;136:111–125. [DOI] [PubMed] [Google Scholar]
- 40. Young KZ, Lee SJ, Zhang X, et al. . NOTCH3 is non-enzymatically fragmented in inherited cerebral small-vessel disease. J Biol Chem. 2020;295:1960–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang X, Lee SJ, Wang MM. Hydrolysis of a second Asp–Pro site at the N-terminus of NOTCH3 in inherited vascular dementia. Sci Rep. 2021;11:17246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arboleda-Velasquez JF, Manent J, Lee JH, et al. . Hypomorphic notch 3 alleles link notch signaling to ischemic cerebral small-vessel disease. Proc Natl Acad Sci U S A. 2011;108:E128–E135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peters N, Opherk C, Zacherle S, Capell A, Gempel P, Dichgans M. CADASIL-associated Notch3 mutations have differential effects both on ligand binding and ligand-induced Notch3 receptor signaling through RBP-Jk. Exp Cell Res. 2004;299:454–464. [DOI] [PubMed] [Google Scholar]
- 44. Cognat E, Baron-Menguy C, Domenga-Denier V, et al. . Archetypal Arg169Cys mutation in NOTCH3 does not drive the pathogenesis in cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy via a loss-of-function mechanism. Stroke. 2014;45:842–849. [DOI] [PubMed] [Google Scholar]
- 45. Joutel A, Monet M, Domenga V, Riant F, Tournier-Lasserve E. Pathogenic mutations associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy differently affect Jagged1 binding and Notch3 activity via the RBP/JK signaling pathway. Am J Hum Genet. 2004;74:338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baron-Menguy C, Domenga-Denier V, Ghezali L, Faraci FM, Joutel A. Increased Notch3 activity mediates pathological changes in structure of cerebral arteries. Hypertension. 2017;69:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.