Abstract
The absence of detergent and curvature makes nanodiscs excellent membrane mimetics. The lack of structural and mechanistic model of polymer-encapsulated lipid nanodiscs limits their use in the study of the structure, dynamics, and functions of membrane proteins. In this study, we parameterized and optimized the coarse-graining (CG) bead mapping for two differently charged and functionalized copolymers, containing styrene–maleic acid (SMAEA) and polymethacrylate (PMAQA), for the Martini force-field framework and showed nanodisc formation (<8 nm diameter) on a time scale of tens of microseconds using molecular dynamics (MD) simulations. Structural models of ~2.0 or 4.8 kDa PMAQA and ~2.2 kDa SMAEA polymer-based lipid nanodiscs highlight the importance of the polymer chemical structure, size, and polymer–lipid ratio in the optimization of the nanodisc structure. The ideal spatial arrangement of polymers in nanodiscs, nanodisc size, and thermal stability obtained from our MD simulation correlates well with the experimental observations. The polymer–nanodiscs were tested for the reconstitution of single-pass or multipass transmembrane proteins. We expect this study to be useful in the development of novel polymer-based lipid nanodiscs and for the structural studies of membrane proteins.
Graphical Abstract
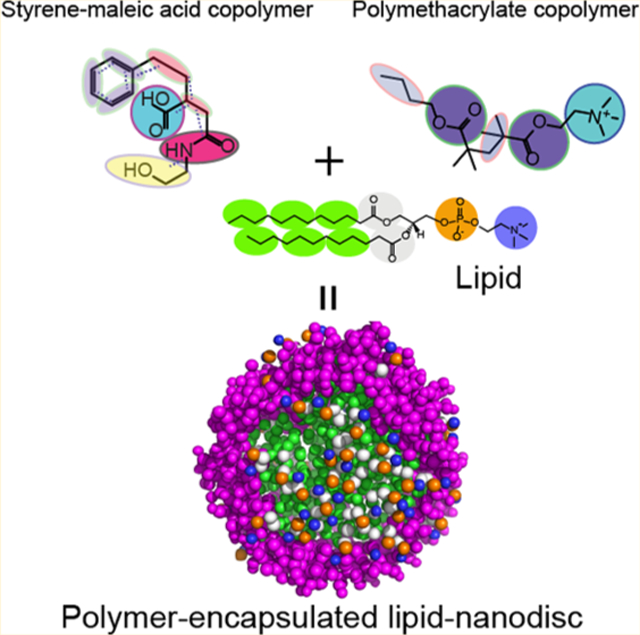
INTRODUCTION
Despite the recent advances in structural biology, membrane proteins continue to pose challenges to most biophysical techniques and biochemical approaches.1,2 The aggregation kinetics of membrane proteins outside lipid membrane3 impulse researchers to develop methods to study their native structures.4 As a result, reconstitution of membrane proteins using membrane mimetics such as bicelles, micelles, liposomes, or nanodiscs4 has significantly eased the interrogation of membrane-associated proteins. Particularly, there is considerable interest in the development and application of nanodiscs.5,6 Use of nanodiscs as a potential chemical tool for protein misfolded diseases like Alzheimer’s disease is bound to create avenues for exciting biomedical applications.7,8 These nanodiscs provide a detergent-free native-like lipid bilayer environment for functional reconstitution of a membrane protein or protein–protein complex. In addition, the size-tunable and lipid selectivity properties of nanodiscs have enabled researchers to design membrane protein selective nanodiscs for structural and functional studies.9
Recent studies have demonstrated the advantages of polymer-based nanodiscs that include extraction of membrane proteins directly from cells, tolerance against pH and divalent metal ions, size tunability by simply changing lipid–polymer ratio, formation of macro–nanodiscs and their magnetic-alignment, and feasibility of applying biophysical techniques including solution and solid-state NMR experiments.10–13 The challenges and difficulties in membrane protein solubilization and purification have thus recently been overcome in native functional states by polymers. As an example, a maleic acid-conjugated styrene or diisobutylene polymer (SMA or DIBMA) recently showed stable and functional extraction of a human protein called rhomboid proteases.14 We have also demonstrated the advantages of the use of polymer-based nanodiscs over peptide- or protein-based nanodiscs for the structural and functional investigation of the amyloidogenic peptide interaction with the lipid membrane at the molecular level using circular dichroism and NMR.15,16 In spite of the success, there are several disadvantages that need to be overcome for a wide spread application of polymer-based nanodiscs. For example, the amphiphilic polymer has been shown to interact with the membrane protein of interest, which can interfere with folding and induce nonnative conformational changes.17 On the other hand, nanodisc-forming polymers have recently been shown to have anti-amyloidogenic properties indicating their potential use in biomedical sciences.18 Synthetic modifications have been demonstrated to modulate the effect of the polymer belt on the targeted protein structural and functional characterization. However, there is a lack of understanding of spontaneous nanodisc formation by copolymers at an atomistic scale. Such information would enable the design of better suited polymer and polymer nanodiscs for structural and functional investigation of a given membrane protein.
While real-time monitoring of the self-assembly process to form nanodiscs at atomic resolution is challenging, here we report a coarse-grained (CG) molecular dynamics (MD) simulation approach and demonstrate its use to understand the formation of nanodiscs for two functionally different copolymers experimentally demonstrated to form nanodiscs. A similar approach in deciphering the molecular mechanism of nanodisc formation has recently been reported for a SMA copolymer nanodisc of size ≈ 7.4 kDa.19 All-atom and CG-MD simulations have also been employed to investigate the self-assembling of protein or peptide-based nanodiscs.20–26 Here specifically, we showed the effect of the size of the polymer and its chemical properties for lipid selectivity and polymer to lipid ratio on the formation of nanodiscs using multi-microseconds MD simulation. Specifically, parametrization and optimization of CG models of two functionally different copolymers, containing styrene–maleic acid (SMAEA) and polymethacrylate (PMAQA) in the Martini framework27 and their ability to self-assemble with lipids to form polymer nanodiscs are reported. Reconstitutions of three different membrane proteins including bacterial sensory rhodopsin (srII), amyloid precursor protein’s (APP) transmembrane domain, and integrin-β328–30 in these polymer nanodiscs are also demonstrated at an atomic scale using microsecond timescale CG MD simulations.
METHODS
Chemicals.
The PMAQA of ~4.7 kDa and SMAEA of ~2.2 kDa were synthesized and purified as reported elsewhere.15,31,32 A 1,2-Dimyristoyl-sn-glycero-3-phosphatidylcho-line (DMPC) lipid was purchased from Avanti Polar Lipids, Inc (Alabaster, AL). All other reagents were purchased from Sigma-Aldrich.
Dynamic Light Scattering.
Large unilamellar vesicles of DMPC were prepared as described elsewhere.16,33 Briefly, DMPC lipids were dissolved in a 1:1 chloroform and methanol and evaporated under the continuous steam of nitrogen gas and the lipid film was incubated overnight under vacuum. The DMPC lipid film was next hydrated in 10 mM sodium phosphate buffer (pH 7.4) followed by 5 min vortex mixing. The lipid mixture was suspended with a DMPC to PMAQA (w/w) ratio of 1:1.6 or 1:3.2 followed by 5 min vortex mixing and incubated for 15 min at 37 °C under shaking. The mixed solution was subjected to several freeze–thaw cycles to homogenize the samples and was incubated overnight at 37 °C with gentle shaking to generate nanodiscs. Next, dynamic light scattering (DLS) measurement (Wyatt Technology Corporation) was performed to check the size distribution of PMAQA–DMPC nanodiscs using a 1 μL quartz cuvette at 25 °C.
PMAQA and SMAEA CG Bead Mapping.
The 2D structure of PMAQA (~2.0 or 4.8 kDa) and SMAEA (~2.2 kDa) was generated using ChemBio Office Ultra 12.2 and exported to Chem3D 16.0. The chemical structures were energy-minimized using MMFF94 force field34 for 10 000 number of iterations with a minimum root-mean-square gradient of 0.1 in Chem3D. The energy-minimized structures were further subjected to MD simulation using Chem3D in MMFF94 for 100 000 steps at 300 K for structural refinement. The all-atom topology of the optimized model structures was obtained by inputting the structures to the Automated Topology Builder35 and CCPN NMR AcpyPE.36 The all-atom 3D structure and topology files were considered for the parameterization of CG bead mapping in the Martini force field framework as described elsewhere.19,27
The SMAEA polymer comprised of nine repeating units of styrene, ethanolamine, and carboxylic acid terminated with cumene. Each styrene group is represented with a three-bead mapping (referred to tyrosine),19 ethanolamine with two bead mapping, and the carboxyl group with one bead mapping for the Martini framework as shown in Figure 1. The bead selections were referred from the previous studies19 and the standard Martini models (Martini 2.2P37) that have been tested for other biomacromolecules including proteins. The ~2 kDa PMAQA polymer comprising six repeating units of quaternary ammonium (–N+R3) was assigned with one-bead referring to the Martini phosphatidylcholine lipid topology. One-bead mapping for the hydrophobic butyl (C4) chain and ester groups was considered for the PMAQA CG model building. A similar CG bead mapping was considered for ~4.8 kDa PMAQA to test the role of polymer length in the formation of nanodiscs. An optimal hydrophobicity (f) to hydrophilicity (1 – f) fraction (f = 0.5) was considered for PMAQA in reference to experimental results that showed nanodisc formation with the “f ” value ranging from ~0.3 to 0.6.32 The bonds, angles, and dihedral parameters were parameterized in reference to all-atom MD trajectories of the respective polymer obtained from 100 ns MD simulation in aqueous solution at 303.15 K. The carboxyl group in SMAEA and the quaternary ammonium group in PMAQA were assigned with one negative and positive charge, respectively.
Figure 1.
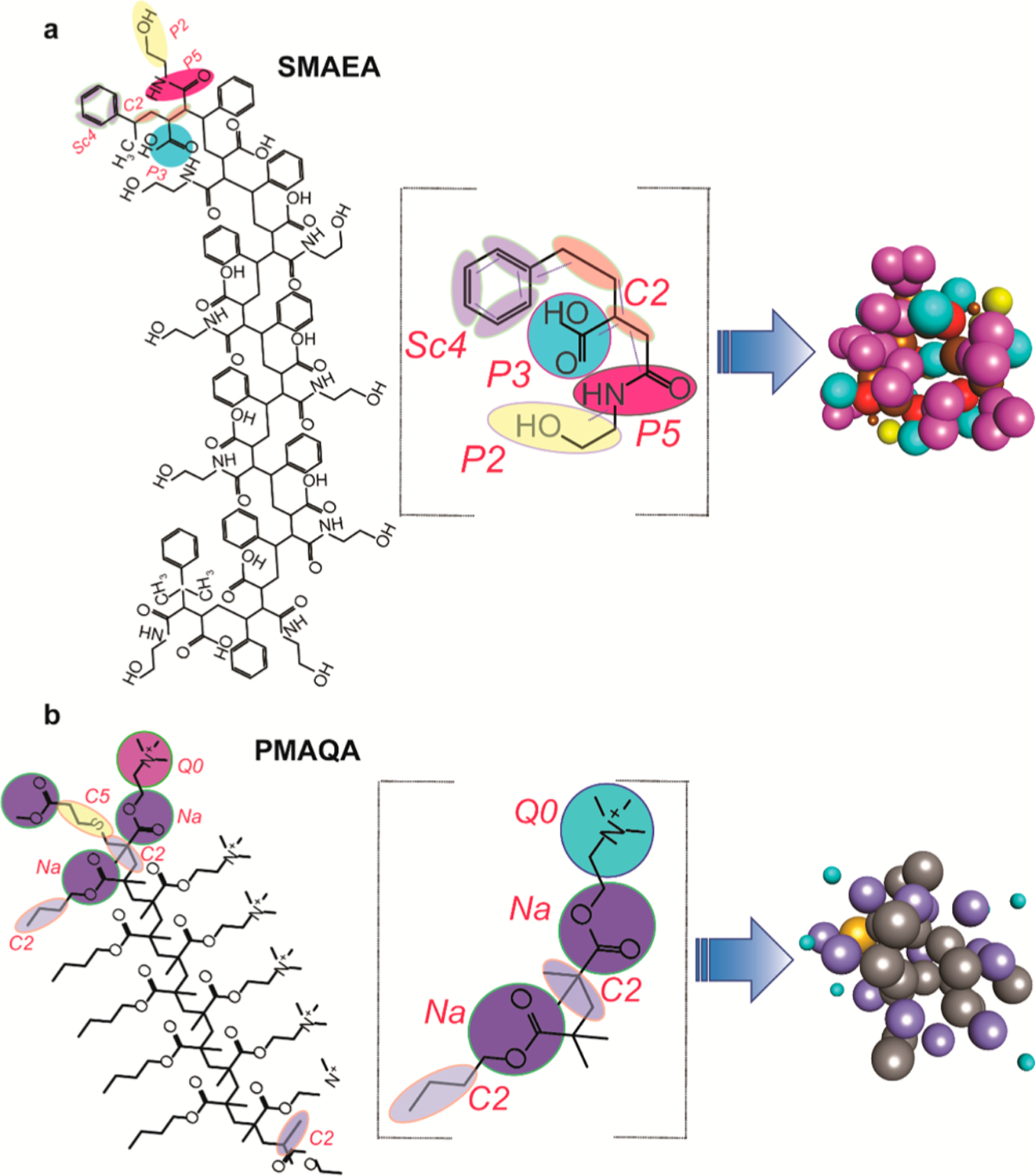
CG mapping for the parameterization of SMAEA (a) and PMAQA (b) nanodisc-forming copolymers. Chemical structures of SMAEA (top left) and PMAQA (bottom left) were built using ChemDraw. The selected CG groups are highlighted in colors (middle column), and the CG bead mapping of each repeated unit in SMAEA or PMAQA and their corresponding CG models are shown in the right-most panel.
MD Simulation.
The CG model for both PMAQA and SMAEA was generated from their all-atom MD simulation following the parametrizing of new molecule based on atomistic simulations documented in Martini (http://www.cgmartini.nl/index.php). Briefly, the polymers were simulated in aqueous solution with counter ions (Na+ or Cl–) for a production run of 100 ns using the all-atom topology and trajectory files were used to generate CG MD simulation inputs in GROMACS version 5.0.7.38 An in-house script was used to generate the indexing file of CG beads, angles, and bonds by inputting the all-atom topology information and our CG bead mapping approach is shown in Figure 1.
To monitor the binding of the polymer to lipid-bilayer, a CG MD system was built by placing SMAEA at a minimum distance ~1 nm from the 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DLPC) lipid bilayer in a cubic box system. The martini DLPC lipid was chosen for our study as it provides a general phosphatidylcholine lipid, corresponding to atomistic C12:0 dilauroyl (DLPC)–C14:0 dimyristoyl (DMPC) tails. In our previous experimental study,31,32 we demonstrated the spontaneous lipid nanodisc formation by both polymers selectively for phosphatidylcholine lipid with C14 tails. The DLPC lipid bilayer was built using the Martini insane python program (http://cgmartini.nl/index.php) and simulated for 2 μs prior to addition of SMAEA. The MD system was solvated using one-bead water and neutralized by adding counter Na+ ions under a periodic boundary condition. The SMAEA–DLPC lipid bilayer (SMAEA to DLPC = 1:4) MD system was next subjected to energy-minimization using the steepest descent method followed by constant volume and pressure equilibration as described elsewhere. The SMAEA–DLPC bilayer system was finally simulated for a production run of 4 μs at 303.15 K.
The CG models of the spontaneous assembling of the lipid and polymer were created by randomly placing the lipids and polymer at different ratios using gmx_insert. The molar concentration of PMAQA or SMAEA was directly referred from our experimental findings to build the MD systems. A polymer to lipid [DLPC or 1,2-dilauroyl-sn-glycero-3-phospho-l-serine (DLPS)] ratio of 1:4 was used for spontaneous MD simulation for ~2.0 kDa PMAQA and ~2.2 kDa SMAEA. A 1:4 or 1:8 polymer to DLPC ratio was used for ~4.8 kDa PMAQA MD simulation. All MD systems were solvated using a one-bead water model and neutralized using counter ions followed by energy minimization, equilibration, and production run for a time-scale of 10 μs at 303.15 K. For comparative analysis of lipid properties, a CG model structure of the membrane scaffold protein (MSP)–DLPC nanodisc was built using CHARMM-GUI39 and simulated for 1 μs at 303.15 K.
The protein reconstitution MD systems were generated by placing the protein ~1 nm away from the SMAEA–DLPC nanodiscs obtained as the end product of spontaneous assembly MD simulation (at 10 μs). Three different membrane proteins such as bacterial rhodopsin (srII; PDB ID: 1XIO),40 amyloid - precursor protein (PDB ID : 2LLM; GSQKLVFFAEDVGSNKGAIIGLMVGGVVIATVIVITLVMLKKK),41 and integrin-β3 (PDB ID: 2L91; PESPKGPDILVVLLSVMGAILLIGLAPLLIWALLITIHDRKEF)30 with single or multiple transmembrane domains were considered for reconstitution MD simulation. The CG beads of the targeted protein were generated using the martinize python script. The protein–nanodisc MD systems were solvated using the one-bead water model supplemented with 150 mM NaCl and the systems were neutralized with appropriate counter ions. Energy minimization, equilibration, and production MD run of 10 μs at 303.15 K were performed to monitor the reconstitution of all three membrane proteins into SMAEA–DLPC nanodiscs. The all-atom structures of reconstituted proteins in nanodiscs were obtained from their corresponding final CG MD snapshots using CHARMM-GUI.39
Bilayer Thickness Calculation.
The lipid-bilayer thickness from polymer-encased lipid nanodiscs was calculated using the GridMAT-MD program.42 The end structure (at 10 μs) of each MD system was retrieved and subjected to GridMAT-MD to calculate the bilayer thickness in a 20 × 20 matrix and plotted using OriginPro (academic license). For comparative bilayer thickness analysis, the bilayer thickness of the MSP–DLPC nanodisc built using CHARMM-GUI39 was calculated using GridMAT-MD. The protein and polymer–DLPC nanodisc lipid bilayer thickness were compared with each other and with the experimental values.43 The lateral diffusion rates of DLPC lipids were calculated from both polymer– and MSP–nanodiscs from the end 0.5 μs MD simulation.
MD trajectories were interpreted using VMD44 and the images were built using PyMOL (academic license, https://pymol.org/2/) and Discovery studio visualizer 3.5 (Accelrys).45 All CG MD simulations were carried out using Martini v 2.2P37 force field in GROMACS MD engine running parallel in SGI UV 3000. The list of MD simulation parameters is given in Table S1.
Simulated Annealing CG-MD.
The thermal stability of PMAQA– or SMAEA–DLPC nanodiscs was tested by performing simulated annealing CG-MD simulation by linearly increasing temperature over time as described elsewhere.46 A total of 14 annealing points was considered with coupling temperatures ranging from 298 up to 353 K on a time scale of 10 μs. The initial group was coupled to 298 K and the MD system was linearly heated up (increased in every 2 up to 6 μs). A constant temperature of 353 K was applied to both polymer–nanodisc systems from 6 to 10 μs to monitor nanodisc destabilization. The MD snapshots were retrieved in every 2 μs and superimposed using DSV. A reference atomic distance of 7.5 Å was defined in DSV and the nanodisc image was exported to ImageJ (NIH). Feret’s diameter, area, and perimeter of the discoidal-shaped nanodisc simulated at different temperature points were next analyzed using ImageJ and plotted in Origin.
RESULTS AND DISCUSSION
Construction of a Polymer CG Model.
Parameterization and construction of CG models to perform MD simulation using SMAEA (~2.2 kDa) and PMAQA (~2.0 or ~4.8 kDa) polymers are shown in Figure 1a,b. CG bead mapping was accomplished based on experimentally identified functional groups (styrene, −N+R3, COO–) that are crucial for the nanodisc formation by the polymers.31,32 The pseudo-bonds and bond lengths were determined from all-atom topology of the respective polymer (Figure 1). Two differently sized PMAQA (~2.0 and ~4.8 kDa) were tested with a similar CG bead mapping to understand the role of polymer length in the formation of nanodiscs. In addition, we employed two different lipids to evaluate the suitability of lipids for the nanodisc formation: zwitterionic lipid DLPC and anionic lipid DLPS.
Solubilization of Lipids by Polymers.
The solubilization of lipid-bilayers by a polymer has been shown to generate discoidal nanodiscs by light scattering, TIRF fluorescence and NMR experiments.31,32 Under the experimental conditions, such morphological transition happens spontaneously and sometimes require mechanical procedures such as repeated freeze and thaw cycles depending on the lipid composition.31,32 Here, we investigate such a transition using a DLPC lipid bilayer model in Martini force field.37 As illustrated in Figure S1, SMAEA was found to substantially destabilize the DLPC lipid bilayer (SMAEA–DLPC = 1:4) over a time period of 4 μS MD simulation. The self-assembly of SMAEA on the DLPC lipid bilayer facilitates membrane insertion and pore formation by SMAEA within ~2 μs. Segregation of upper and lower leaflet lipids with centrally bridged SMAEA was identified at the end of 4 μS MD simulation (Figure S1). Such membrane destabilization and transient pore formation by the SMA copolymer (~7.4 kDa) was recently reported.19 Even though SMAEA disrupts the DLPC lipid bilayer, we did not observe nanodisc formation within the microsecond timescale of MD simulation mentioned above. Therefore, these findings suggest that a longer timescale of MD simulation is essential to achieve enough disruption of the prestructured lipid bilayer by the polymer to form a discoidal nanodisc as reported from experimental studies.
Self-Assembly of Polymers and Lipids.
In order to overcome the limitation mentioned above, we next performed MD simulation to monitor the spontaneous assembly of the randomly distributed polymer (SMAEA or PMAQA) and free DLPC lipids (Figure S2) in solution at the 1:4 polymer–DLPC ratio. Remarkably, the randomly distributed polymers and DLPC lipids in aqueous solution exhibited the formation of discoidal shape nanodiscs within 10 μs simulation (Figure 2a,b). The microsecond scale MD simulation showed the formation of small-sized nanodiscs within several hundreds of nanoseconds followed by fusion to form a stable discoidal-shaped nanodisc in both polymers (Figure 2a,b). PMAQA and SMAEA copolymers with nearly equivalent size (~2.0 kDa), but differing in hydrophobic and hydrophilic functionalizations, generated nanodiscs of size ≈7.0 and 7.5 nm, respectively (Figure 2c,d). The polymers were found to be organized around the acyl chains of DLPC lipids exposing the lipid polar head groups to the solvent as reported from experimental observations.31,32 The distribution of PMAQA, in comparison to SMAEA, surrounding the lipid tails was found to be distorted with few molecules localized on the membrane surface (Figure 2c). Atomistic inspection showed that the −N+R3 groups of the PMAQA polymer were located close to (~0.5–0.7 nm) the anionic phosphate groups of DLPC lipids (Figure 2c). A very strong hydrophobic lipid–polymer interaction is required to overcome the abovementioned electrostatic polymer–lipid interaction energy barrier (between −N+R3 and PO4– groups) to uniformly assemble all the polymer molecules around the acyl chains of the lipid bilayer; this can be accomplished by enhancing the hydrophobicity of PMAQA. In contrast, SMAEA distribution was found to be uniform like a belt around DLPC acyl-chains (Figure 2d, see Video SV1).
Figure 2.
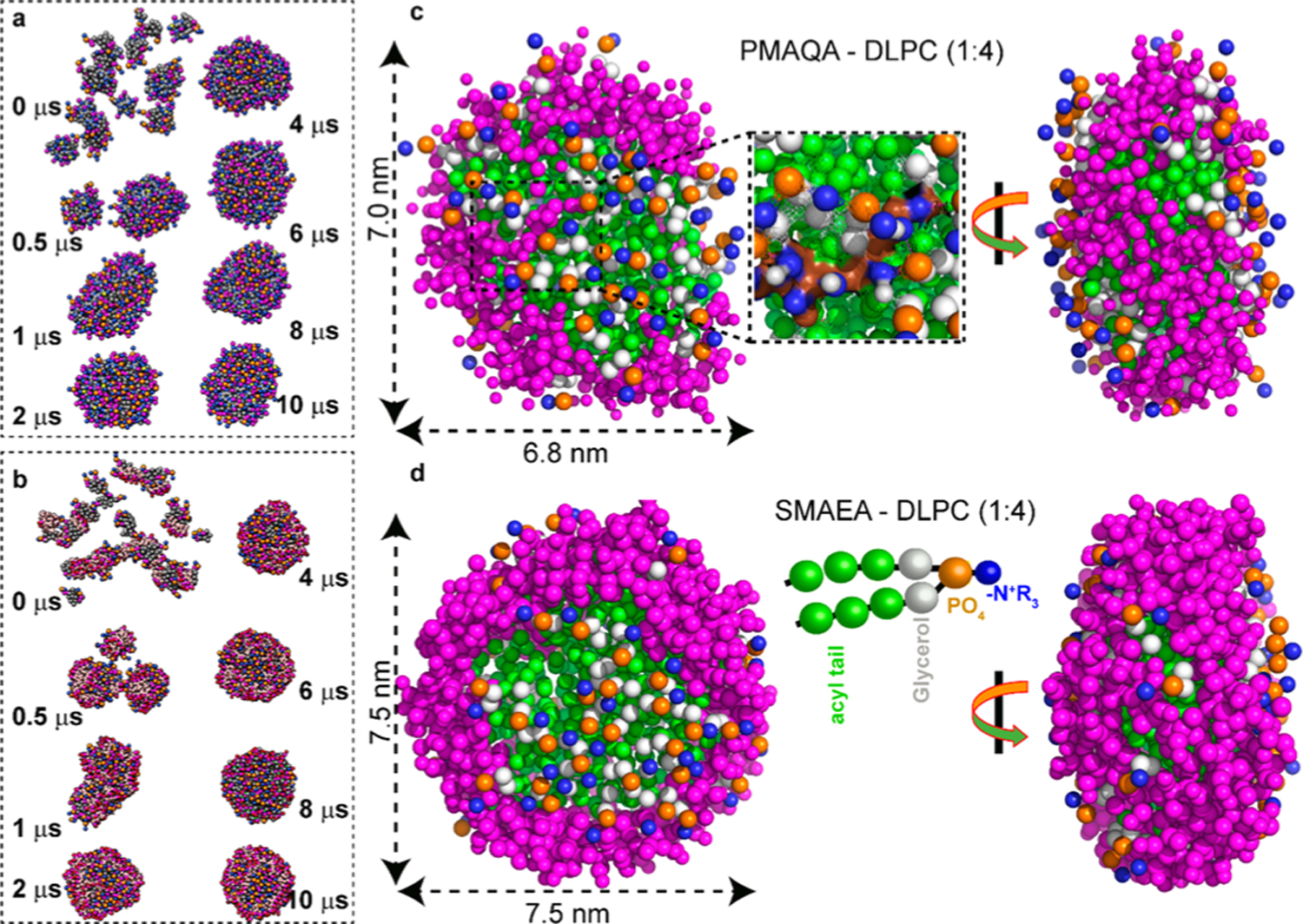
CG MD simulation showing the spontaneous formation of DLPC nanodiscs on a time scale of 10 μs. Time-lapsed MD snapshots showing the self-assembling of (a) PMAQA–DLPC and (b) SMAEA–DLPC nanodiscs at the 1:4 polymer–lipid ratio. Enlarged MD snapshots show the formation of discoidal DLPC nanodiscs of size ≈ 7 nm encased by PMAQA (c) or SMAEA (d); top–down (left) and side (right) views. The double-headed arrow indicates the average diameter of the nanodisc. The polymers are shown in violet and the CG atoms of DLPC in other colors (c,d). Water molecules and ions are not shown for transparency.
Evaluation of Lipid Bilayer Properties in Nanodiscs.
Lipid bilayer thickness of a membrane mimetic plays a very important role in the membrane interaction, folding, and topology of a membrane protein. Therefore, it is important to optimize the hydrophobic thickness of polymer-based nanodiscs for a successful reconstitution of a membrane protein. While this is a daunting task to achieve experimentally, MD simulations can be used to test different parameters and provide guidelines for the design of a near-ideal polymer and optimized lipid composition to be used in a polymer nanodisc. Therefore, after successfully monitoring the self-assembly process underlying the formation of lipid nanodiscs for two different polymers as described above (Figure 2), MD simulation results were used to analyze the influence of the orientations of polymer molecules in a nanodisc on the encased lipid bilayer’s thickness. As illustrated in Figure 3a, the SMAEA nanodisc exhibited a bilayer thickness of ≈3.5 nm at the center; whereas the boundary lipids surrounded by polymers depicted a low bilayer thickness. Such change in bilayer thickness for nanodiscs has been observed in previous MD simulations for MSP-encased nanodiscs.23 Further estimation of bilayer thickness using a 20 × 20 matrix by GridMAT-MD presented an average bilayer thickness of 2.57 ± 0.13 nm, which is in agreement with the experimentally determined hydrophobic bilayer thickness of 2.4 nm for DLPC (Figure 3c).42,43 In addition, for a comparative analysis, we calculated the DLPC lipid bilayer thickness from a preassembled MSP–DLPC nanodisc generated using CHARMM_GUI,39 and it was found to be 3.29 ± 0.26 nm (Figure S3). On the other hand, the lipid bilayer surface distribution and random orientation of PMAQA molecules (Figure 3b) exhibited a slightly lower average DLPC bilayer thickness of 2.08 ± 0.24 nm (Figure 3d). In a PMAQA–DLPC nanodisc, the thickness of several bilayer regions (blue regions in Figure 3d) was found to be <1.4 nm, indicating a partial surface adsorption of PMAQA molecules (Figure 3b). A notable difference is the inhomogeneity of polymer–nanodisc bilayer thickness when compared to the MSP-encased nanodiscs. This is due to the nonuniform stacking of polymer side chains across the lipid acyl chain unlike the uniform and azimuthal alignment of the chains of two MSP proteins.21,23,47 However, similar to MSP nanodiscs, polymer nanodiscs showed an edge effect on bilayer thickness along with a thicker center bilayer region.47 However, an optimized membrane thickness for polymer nanodiscs may require longer time-scale of simulation to unify the edge effect of the polymer belt. The lateral diffusion rates of DLPC lipids calculated from the microsecond MD simulations were further compared between protein- and polymer-encased nanodiscs. The MSP–DLPC nanodisc presented a lateral diffusion rate of (4.37 ± 0.78) × 10–7 cm2 s–1. The SMAEA– and PMAQA–DLPC nanodiscs showed a very little deviation in the lateral diffusion rates from that of MSP–DLPC with respective values of (4.33 ± 0.61) × 10–7 and (4.43 ± 0.15) × 10–7 cm2 s–1 at 303 K. The calculated diffusion rates for both polymer and MSP nanodiscs are of the same order of magnitude (10–7 cm2 s–1) as observed experimentally48 and previous MD simulations.49
Figure 3.
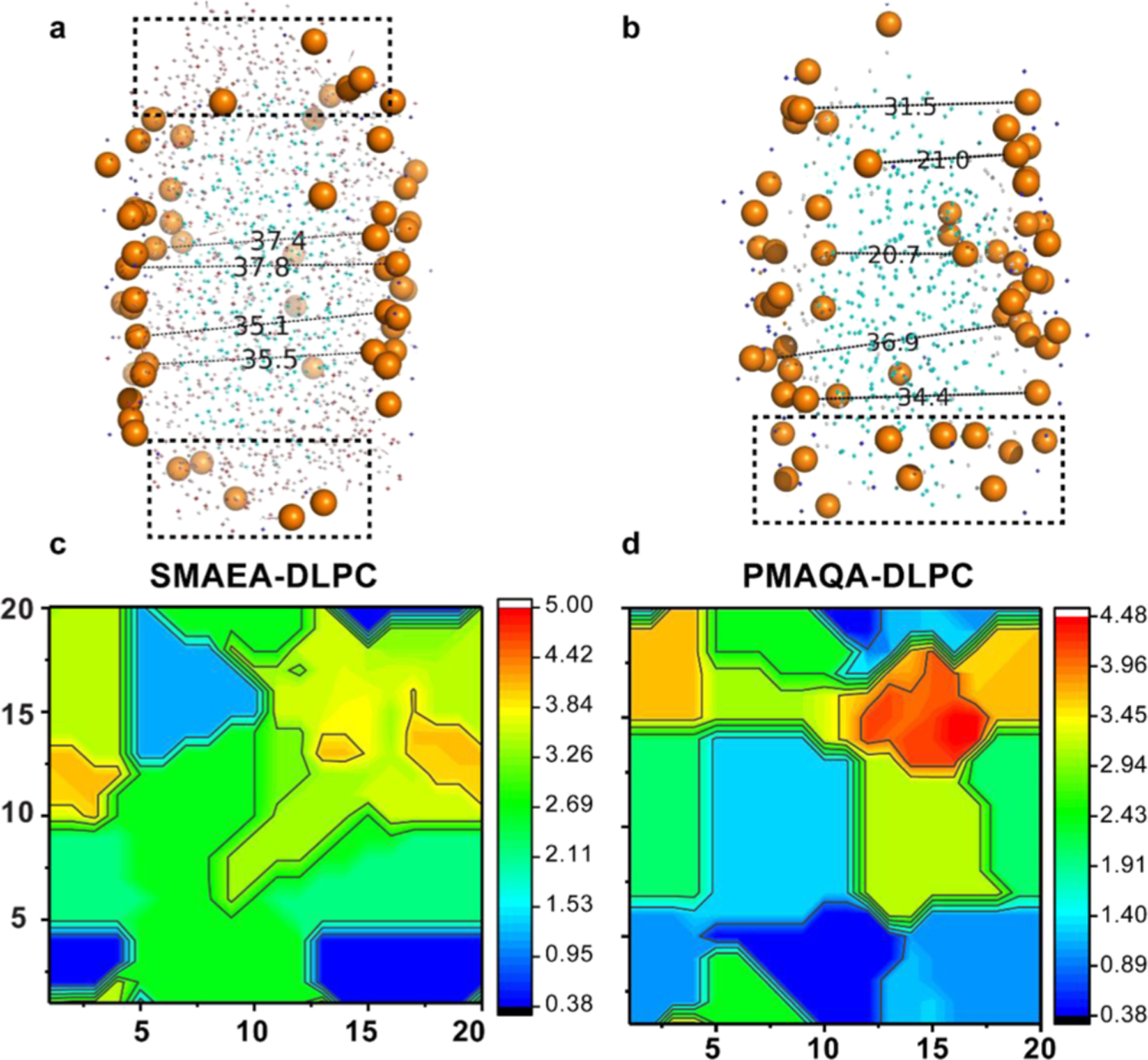
Lipid bilayer thickness in a polymer-encased DLPC nanodisc. PyMOL illustration of distance (in Å) between randomly selected upper and lower leaflet phosphate head groups in SMAEA (a) and PMAQA (b) nanodiscs retrieved at 10 μs MD simulation. The boxes in dashed lines represent the peripheral lipid heads closely located to the proximity of polymers. The representative bilayer thickness is calculated using GriDMAT-MD from SMAEA–DLPC (c) and PMAQA–DLPC (d) nanodiscs and plotted in a 20 × 20 matrix. The scale bar shown is in nanometer.
Role of Polymer Concentration and Lipid-Specificity in Nanodisc Formation.
Because it is experimentally challenging to screen and optimize polymers and their lipid specificity to form stable nanodiscs, we examined the role of polymer’s molecular size using MD simulation. Experimental results showed that PMAQA with an optimal size of ~4.8 kDa forms nanodiscs.32 Here, we tested the nanodisc-forming capability of ~4.8 kDa PMAQA for a variable polymer–lipid ratio as shown in Figure 4. At 1:8 PMAQA–DLPC ratio, we observed spontaneous nanodisc formation with a uniform distribution of polymer molecules surrounding the hydrophobic acyl chains of DLPC lipids (Figure 4a). Atomic inspection revealed that the hydrophobic butyl groups (C2) of the polymer are oriented toward the hydrophobic lipid core region, whereas the cationic −N+R3 groups (Q0) of the polymer are exposed to the solvent (Figure 4a). The ~4.8 kDa PMAQA forms a DLPC nanodisc that is nearly equal to the size of the SMAEA nanodisc (~7.5 nm diameter). The ~4.8 kDa PMAQA polymer nanodisc exhibited a lipid bilayer with a thickness of >3.5 nm for the central lipids (Figure 4c).
Figure 4.
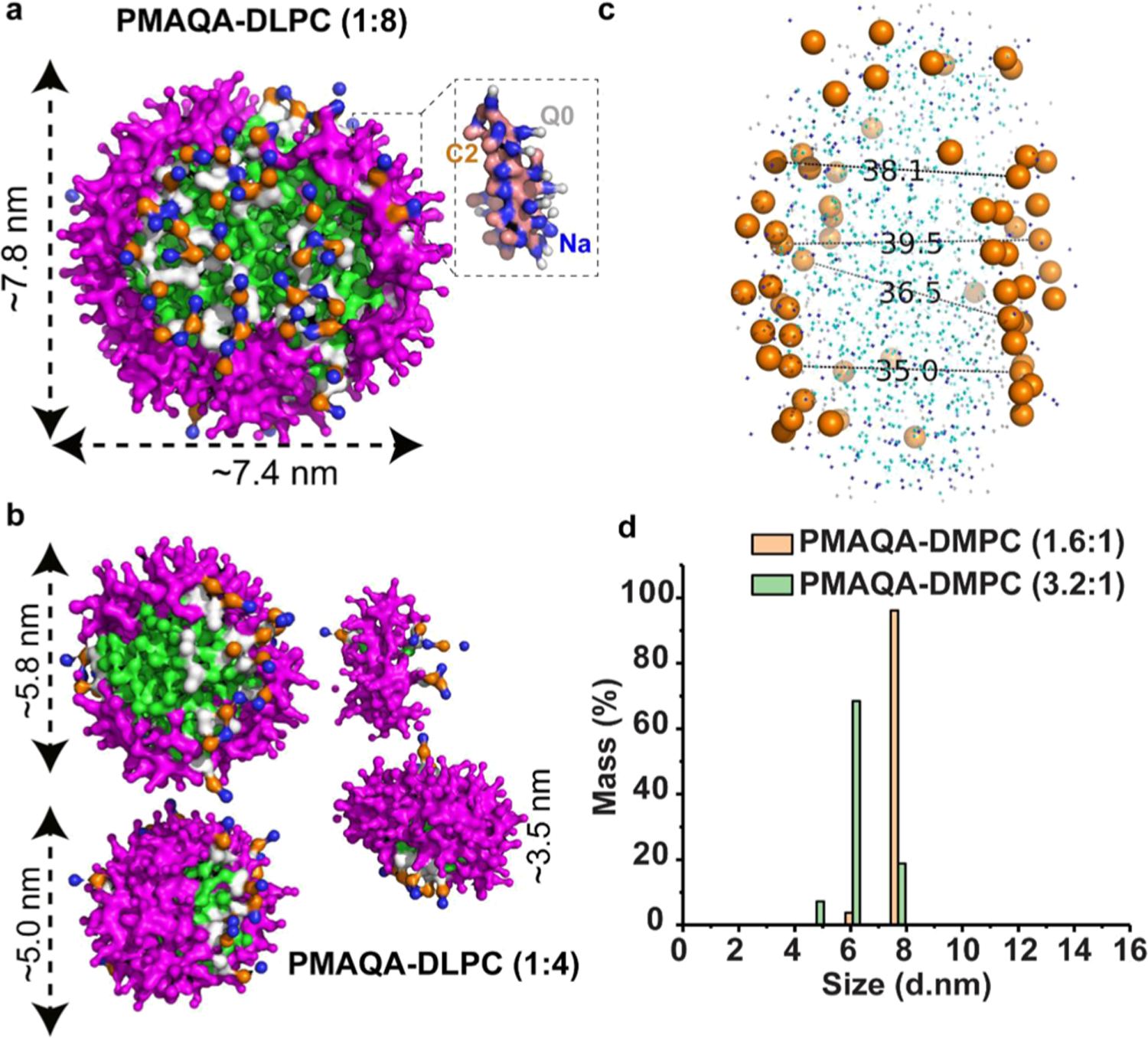
MD snapshots showing the formation of the DLPC nanodisc by ~4.8 kDa PMAQA at a variable polymer to lipid ratio. Spontaneous formation of nanodiscs by ~4.8 kDa PMAQA at 1:8 (a) and 1:4 (b) polymer to DLPC ratio at 10 μs MD simulation. The polymers are shown in violet and DLPC atoms in different colors as indicated in Figure 2. Water molecules and ions are not shown for transparency. (c) Distance between the lipid head groups (in Å) of PMAQA nanodiscs shown in (a) is measured using PyMOL. (d) DLS showing the size distribution of PMAQA–DLPC nanodiscs at the indicated polymer to lipid (w/w) ratio.
Next, we examined the effect of PMAQA polymer concentration on the size and morphology of nanodiscs formed. An increase in the concentration of PMAQA from 1:8 to a 1:4 (w/w) ratio of PMAQA–DLPC resulted in formation of smaller size nanodisc as expected from previously reported experimental results; in addition, nanodiscs with average diameters of ~5.8, 5.0, and 3.5 nm were also observed (Figure 4b). Unlike the PMAQA nanodiscs obtained from ~4.8 kDa at a 1:8 PMAQA–DLPC ratio (Figure 4a), the nanodiscs formed at higher concentrations of PMAQA were found to contain PMAQA in random orientations and tightly packed (Figure 4b). DLS measurements of nanodiscs designed at a PMAQA–DMPC ratio corresponding to that used in MD simulations supported the computationally observed nanodisc size mentioned above (Figure 4d). As shown in Figure 4d, the ~4.8 kDa PMAQA at the 1.6:1 polymer–DMPC (w/w) ratio exhibited ~6.0 and ~7.7 nm size nanodiscs with a mass percentage of ~3.7 and 96.1%, respectively. On the other hand, for the 3.2:1 PMAQA–DMPC (w/w) ratio, ~4.7, 6.0, and ~7.7 nm size nanodiscs with a mass percentage of ~7.2, 68.4, and 18.8%, respectively, were obtained (Figure 4d).
MD simulation results (Figure 4) show that the lateral packing of lipids in PMAQA nanodiscs depends on the size and concentration of the polymer as expected from experimental results.32 The ability of MD simulations to reveal the location of different molecules constituting the nanodisc can be utilized in the optimization of experimental conditions to achieve “fluid lamellar phase”-like lipid packing, which is crucial for functional reconstitution of a membrane protein or a protein–protein complex.17 Because experimental studies have reported the difficulties in reconstituting various lipid compositions in polymer nanodiscs,31,32 it is important to examine the suitability of the lipid composition for a given polymer to form stable nanodiscs using MD simulations. For this purpose, we simulated the self-assembly of PMAQA or SMAEA polymers in the presence of anionic DLPS lipids (with DLPC acyl chains). As shown in Figure S4, SMAEA forms a nanodisc within a time-scale of 10 μs, whereas PMAQA was found to be distributed on the membrane surface facilitated by its electrostatic interaction with negatively charged DLPS head groups.
Thermal Stability of Polymer–Nanodiscs.
Thermal stability of nanodiscs is important for various applications including the biophysical and structural studies of membrane proteins.50,51 Experimental studies have reported that peptide-or MSP-based nanodiscs can be stable up to ~60 °C.52 Thermal unfolding has been seen with further increase in temperature that destabilizes nanodisc’s size. In contrast to this, polymer-based nanodiscs are very stable and do not unfold even at 80 °C.31 To better understand the thermal stability of polymer-based nanodiscs, we investigated the thermal stability of PMAQA- and SMAEA–DLPC-based nanodiscs with respect to temperature increasing from 25 to ~80 °C as shown in Figure 5a. The simulated annealing experiment showed a minimal effect on the discoidal shape of polymer–nanodiscs with respect to temperature (Figure 5c,d). Quantitative analysis of the polymer–nanodisc size showed average Feret’s diameter of 8.17 ± 0.26 nm for PMAQA (8.55 nm at t = 0 μs) and 8.37 ± 0.32 nm for SMAEA–DLPC (7.87 nm at t = 0 μs) nanodiscs (Figure 5b). Increase in temperature from 25 to 37 °C showed substantial rearrangement of the polymers around the lipid molecules as revealed from the change in area and perimeters of the nanodiscs (Figure 5b). Further increase in temperature from 37 to 50 and 50 to 80 °C showed no significant change in Feret’s diameter; whereas periodic fluctuations in nanodisc perimeter and area were noticed indicating the effect of temperature on both polymer and lipid spatial rearrangement. However, unlike the peptide-or protein-based nanodiscs, both PMAQA and SMAEA nanodiscs exhibited stability even at 80 °C as shown in Figure 5c,d. An average area of 20.70 ± 1.56 and 24.23 ± 0.77 nm2 was calculated from MD snapshots taken at 2 μs interval of time for PMAQA (21.74 nm2 at t = 0 μs) and SMAEA–DLPC (24.02 nm2 at t = 0 μs) nanodiscs, respectively (Figure 5b). Overall, the simulated annealing MD simulation revealed the thermal stability of polymer nanodiscs up to 80 °C.
Figure 5.
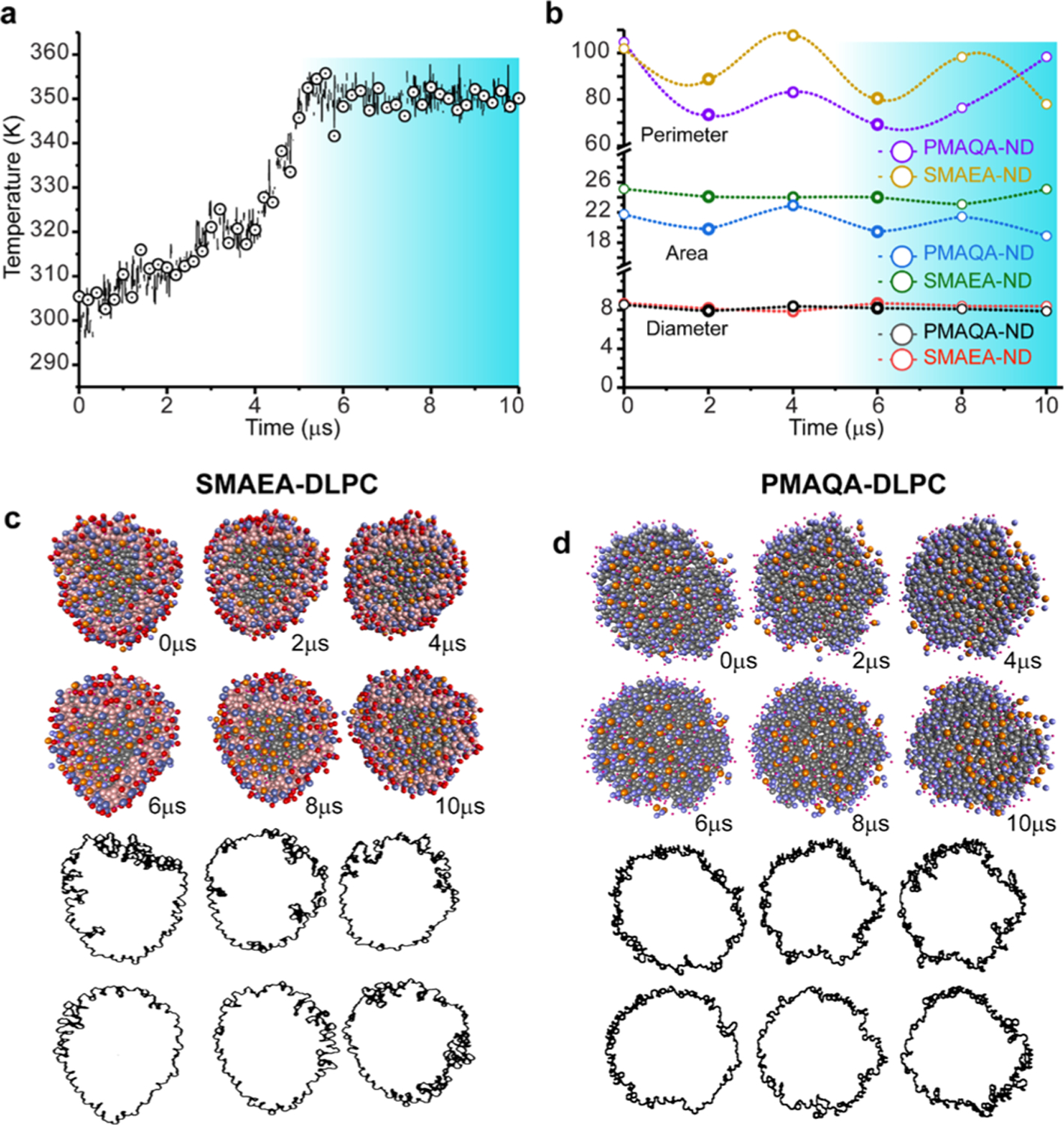
Thermal stability of SMAEA or PMAQA DLPC nanodiscs studied using simulated annealing MD simulation. (a) Polymer nanodiscs were subjected to a gradual increase in temperature from 298 to 353 K with respect to MD simulation time. (b) Feret’s diameter, perimeter, and area of nanodiscs calculated from MD snapshots retrieved from 1 μs time interval using ImageJ. The highlighted regions in (a,b) show the variation in the nanodisc size and shape with respect to temperature. (c,d) Illustration of polymer–nanodiscs analyzed using ImageJ (red) and the distribution of the polymer–belt as a function of indicated simulation times (in μs).
Reconstitution of Transmembrane Proteins in Polymer–Nanodiscs.
Next, we studied the spontaneous reconstitution of a seven transmembrane domain bacterial sensory rhodopsin (srII) (PDB ID: 1XIO)40 and a single transmembrane containing membrane proteins like integrin-β3 (PDB ID: 2L91)30 and APP (PDB ID: 2LLM)41 in polymer nanodiscs. Membrane proteins were initially placed in different orientations such as ≈1 nm away from the SMEA–DLPC nanodisc or partially inserted as shown in Figure 6 (top row). The srII protein was found to interact with the edge (~1 μs) of the nanodisc (SMAEA belt) and the complex remained stable for several microseconds followed by a change in the shape of the nanodisc from a discoidal to ellipsoidal shape (Figure S5). The lipid bilayer insertion of srII at 4 μs displaced several of SMAEA polymer molecules, and from 6 to 8 μs the seven transmembrane domains of the protein gradually oriented perpendicular to the plane of the lipid bilayer and remain inserted until the end of the simulation (Figure S5). CG to all-atom conversion showed that the srII transmembrane domains were well oriented within the lipid bilayer;28 however, the ~7.5 nm size nanodisc was found to be not efficient in maintaining the discoidal-shaped structural integrity of the SMAEA–DLPC nanodisc (Figures 6a and S5).
Figure 6.
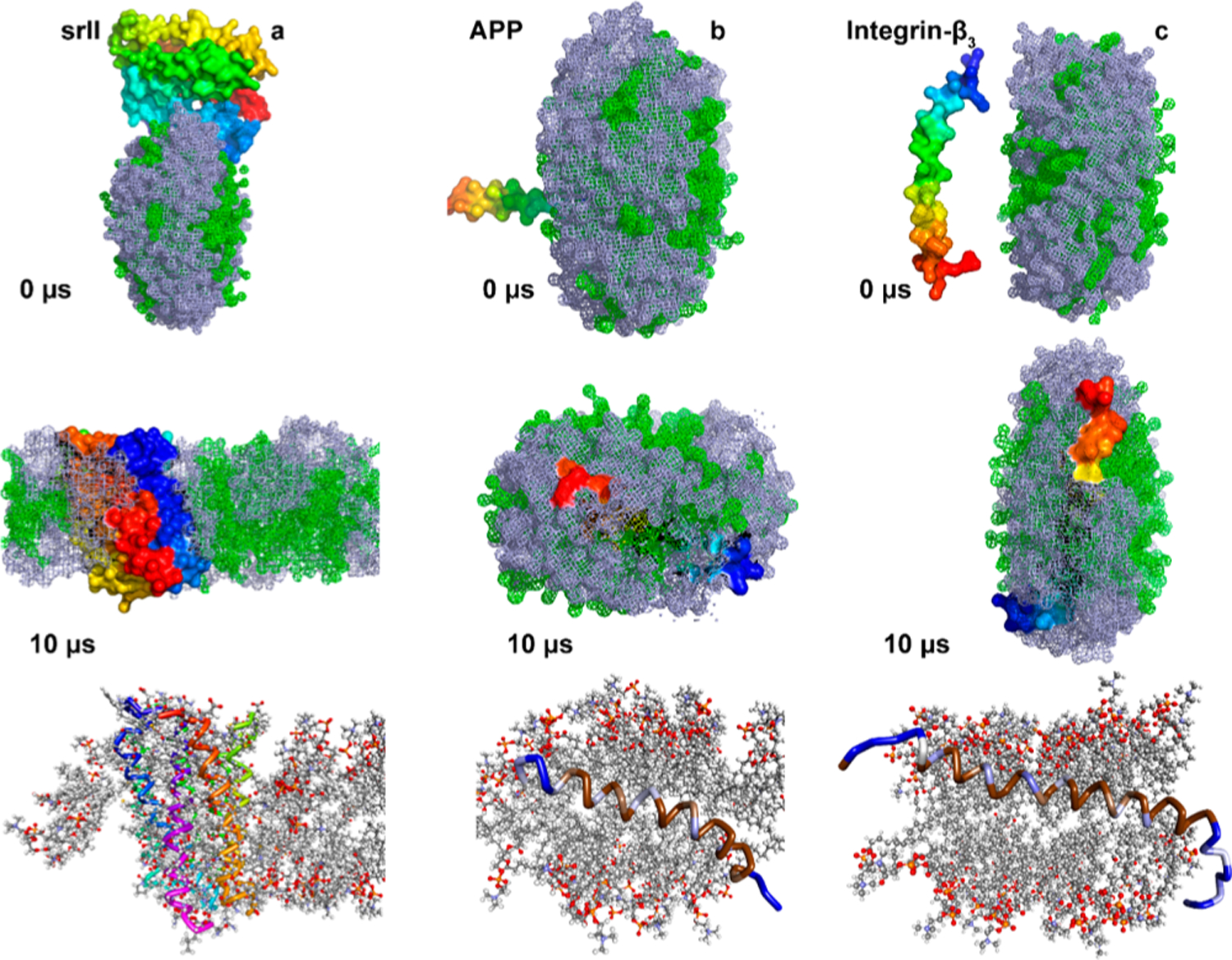
MD snapshots showing the reconstitution of membrane proteins in a SMAEA–DLPC nanodisc. Monitoring the reconstitution of seven transmembrane domain sensory rhodopsin-II (a), single-pass transmembrane amyloid-precursor protein (b) and integrin-β3 (c) into a SMAEA nanodisc (shown in Figure 2b) using 10 μs CG MD. Polymers (gray) and DLPC (green) are shown in mesh and proteins as the surface in PyMOL with N- and C-termini in blue and red, respectively. The corresponding all-atom models generated from the Martini CG model are shown in the bottom-most row. Membrane proteins are shown in ribbon and lipids in ball-stick. The zoomed structure of all-atom model structures is given in Figures S6 and S7.
We then studied the reconstitution of two different single-pass transmembrane proteins, namely APP and integrin-β3 (Figure 6b,c). Remarkably, unlike srII, APP and integrin-β3 were found to have little effect on the shape and size of SMAEA nanodiscs. Both APP and integrin-β3 were found to interact with nanodiscs within several hundreds of nanoseconds. The APP fragment that was marginally inserted into nanodisc’s lipid bilayer surface was found to incorporate its helix into the membrane and oriented at an angle ≈20° with respect to the bilayer normal as observed previously53 (Figures 6b and S6). The N- (GSNK) and C- (KKK) terminal lysine residues were exposed to the solvent, whereas the centrally located helix residues were packed inside the nanodisc facing one side to the lipids and other to the polymer belt (Figure 6b). Similarly, integrin-β3 was found to localize in the lipid bilayer with ≈30° tilt of the transmembrane helix with respect to the plane of the lipid bilayer, which is in agreement with previous reports that suggested an increase in the tilt angle in the absence of its other subunit’s TM domain (Figures 6c and S7).54,55
CONCLUSIONS
In conclusion, we have successfully demonstrated the formation of lipid-nanodiscs by two different amphiphilic copolymers consisting of styrene or PMAQA moieties using atomistic CG MD simulation. We expect that the parametrization of the polymers and methodology presented in this study will be useful in designing and screening new polymers that can efficiently form nanodiscs. The optimization of polymer length, lipid specificity, and stability would not only provide a better understanding of the polymer–belt interference with the target membrane protein and its function but also be useful in the development of nanomedicine or peptide membrane interaction studies as demonstrated in this work.15 We foresee that the computational approach employed here can be generalized by varying the polymer hydrophobic and hydrophilic moieties to design membrane protein selective nanodisc systems for a successful reconstitution. The CG parameters optimized for the Martini force-field framework in this study for polymers will create avenues for further development of CG systems for other polymers to be used in biological and chemical studies.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by funds from NIH (AG048934 to A.R.). We thank Dr. Charles L. Brooks III for fruitful discussion on the use of MD simulations to study nanodiscs. This work was (in part) performed under the International Collaborative Research Program of Institute for Protein Research, Osaka University, ICR-18-02. We thank Professor Toshimichi Fujiwara in the Institute for Protein Research, Osaka University, for providing parallel computing facility on SGI UV 3000.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.9b03681.
Parameters of CG MD simulation systems; MD snapshots showing interaction of SMAEA with the DLPC lipid bilayer at different time scales; random distribution of copolymers and DLPC lipids in solvent for spontaneous nanodisc generation; coarse-grained model of membrane scaffold protein (MSP)-encased DLPC nanodiscs; MD snapshots showing interaction or reconstitution of seven transmembrane domain bacterial sensory rhodopsin (srII) in SMAEA-DLPC nanodiscs at different time scales; representative coarse-grained (CG) to all-atom converted model structure of single transmembrane domain amyloid-precursor protein (APP) bound to SMAEA-DLPC nanodiscs and retrieved at 10 μs from MD simulation; and representative coarse-grained (CG) to all-atom converted model structure of integrin-β3 single transmembrane domain protein constituted in SMAEA-DLPC nanodiscs and retrieved at 10 μs from MD simulation (PDF)
Distribution of SMAEA around DLPC acyl-chains (MP4)
The authors declare no competing financial interest.
REFERENCES
- (1).Zorman S; Botte M; Jiang Q; Collinson I; Schaffitzel C Advances and Challenges of Membrane-Protein Complex Production. Curr. Opin. Struct. Biol 2015, 32, 123–130. [DOI] [PubMed] [Google Scholar]
- (2).Carpenter EP; Beis K; Cameron AD; Iwata S Overcoming the Challenges of Membrane Protein Crystallography. Curr. Opin. Struct. Biol 2008, 18, 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kucik DF; Elson EL; Sheetz MP Weak Dependence of Mobility of Membrane Protein Aggregates on Aggregate Size Supports a Viscous Model of Retardation of Diffusion. Biophys. J 1999, 76, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Frey L; Lakomek N-A; Riek R; Bibow S Micelles, Bicelles, and Nanodiscs: Comparing the Impact of Membrane Mimetics on Membrane Protein Backbone Dynamics. Angew. Chem., Int. Ed 2017, 56, 380–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Denisov IG; Sligar SG Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev 2017, 117, 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Barnaba C; Sahoo BR; Ravula T; Medina-Meza IG; Im S-C; Anantharamaiah GM; Waskell L; Ramamoorthy A Cytochrome-P450-Induced Ordering of Microsomal Membranes Modulates Affinity for Drugs. Angew. Chem., Int. Ed 2018, 57, 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Song Q; Huang M; Yao L; Wang X; Gu X; Chen J; Chen J; Huang J; Hu Q; Kang T; et al. Lipoprotein-Based Nanoparticles Rescue the Memory Loss of Mice with Alzheimer’s Disease by Accelerating the Clearance of Amyloid-Beta. ACS Nano 2014, 8, 2345–2359. [DOI] [PubMed] [Google Scholar]
- (8).Song H; Ma X; Xu J; Song Q; Hu M; Gu X; Zhang Q; Hou L; Chen L; Huang Y; et al. The Shape Effect of Reconstituted High-Density Lipoprotein Nanocarriers on Brain Delivery and Aβ Clearance. Nano Res. 2018, 11, 5615–5628. [Google Scholar]
- (9).Hagn F; Etzkorn M; Raschle T; Wagner G Optimized Phospholipid Bilayer Nanodiscs Facilitate High-Resolution Structure Determination of Membrane Proteins. J. Am. Chem. Soc 2013, 135, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Fiori MC; Jiang Y; Zheng W; Anzaldua M; Borgnia MJ; Altenberg GA; Liang H Polymer Nanodiscs: Discoidal Amphiphilic Block Copolymer Membranes as a New Platform for Membrane Proteins. Sci. Rep 2017, 7, 15227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Dörr JM; Scheidelaar S; Koorengevel MC; Dominguez JJ; Schäfer M; van Walree CA; Killian JA The Styrene–maleic Acid Copolymer: A Versatile Tool in Membrane Research. Eur. Biophys. J 2016, 45, 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Fiori MC; Jiang Y; Altenberg GA; Liang H Polymer-Encased Nanodiscs with Improved Buffer Compatibility. Sci. Rep 2017, 7, 7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hall SCL; Tognoloni C; Charlton J; Bragginton ÉC; Rothnie AJ; Sridhar P; Wheatley M; Knowles TJ; Arnold T; Edler KJ; et al. An Acid-Compatible Co-Polymer for the Solubilization of Membranes and Proteins into Lipid Bilayer-Containing Nanoparticles. Nanoscale 2018, 10, 10609–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Barniol-Xicota M; Verhelst SHL Stable and Functional Rhomboid Proteases in Lipid Nanodiscs by Using Diisobutylene/Maleic Acid Copolymers. J. Am. Chem. Soc 2018, 140, 14557–14561. [DOI] [PubMed] [Google Scholar]
- (15).Sahoo BR; Genjo T; Bekier M II; Cox SJ; Stoddard AK; Ivanova M; Yasuhara K; Fierke CA; Wang Y; Ramamoorthy A Alzheimer’s Amyloid-Beta Intermediates Generated by Polymer-Nanodiscs. Chem. Commun 2018, 54, 12883–12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sahoo BR; Genjo T; Cox SJ; Stoddard AK; Anantharamaiah GM; Fierke C; Ramamoorthy A Nanodisc-Forming Scaffold Protein Promoted Retardation of Amyloid-Beta Aggregation. J. Mol. Biol 2018, 430, 4230–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ravula T; Hardin NZ; Bai J; Im S-C; Waskell L; Ramamoorthy A Effect of Polymer Charge on Functional Reconstitution of Membrane Proteins in Polymer Nanodiscs. Chem. Commun 2018, 54, 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sahoo BR; Genjo T; Nakayama TW; Stoddard AK; Ando T; Yasuhara K; Fierke CA; Ramamoorthy A Cationic Polymethacrylate- Copolymer Acts as an Agonist for β - Amyloid and Antagonist for Amylin Fibrillation. Chem. Sci 2019, 10, 3976–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Xue M; Cheng L; Faustino I; Guo W; Marrink SJ Molecular Mechanism of Lipid Nanodisc Formation by Styrene Maleic Acid Copolymers. Biophys. J 2018, 115, 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Shih AY; Freddolino PL; Arkhipov A; Schulten K Assembly of Lipoprotein Particles Revealed by Coarse-Grained Molecular Dynamics Simulations. J. Struct. Biol 2007, 157, 579–592. [DOI] [PubMed] [Google Scholar]
- (21).Debnath A; Schäfer LV Structure and Dynamics of Phospholipid Nanodiscs from All-Atom and Coarse-Grained Simulations. J. Phys. Chem. B 2015, 119, 6991–7002. [DOI] [PubMed] [Google Scholar]
- (22).Vuorela T; Catte A; Niemelä PS; Hall A; Hyvönen MT; Marrink S-J; Karttunen M; Vattulainen I Role of Lipids in Spheroidal High Density Lipoproteins. PLoS Comput. Biol 2010, 6, No. e1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Siuda I; Tieleman DP Molecular Models of Nanodiscs. J. Chem. Theory Comput 2015, 11, 4923–4932. [DOI] [PubMed] [Google Scholar]
- (24).Ollila OHS; Lamberg A; Lehtivaara M; Koivuniemi A; Vattulainen I Interfacial Tension and Surface Pressure of High Density Lipoprotein, Low Density Lipoprotein, and Related Lipid Droplets. Biophys. J 2012, 103, 1236–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Popovic K; Holyoake J; Pomes R; Prive GG Structure of Saposin A Lipoprotein Discs. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 2908–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pourmousa M; Pastor RW Molecular Dynamics Simulations of Lipid Nanodiscs. Biochim. Biophys. Acta, Biomembr 2018, 1860, 2094–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Marrink SJ; Risselada HJ; Yefimov S; Tieleman DP; De Vries AH The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [DOI] [PubMed] [Google Scholar]
- (28).Sahoo BR; Fujiwara T Conformational States of HAMP Domains Interacting with Sensory Rhodopsin Membrane Systems: An Integrated All-Atom and Coarse-Grained Molecular Dynamics Simulation Approach. Mol. BioSyst 2017, 13, 193–207. [DOI] [PubMed] [Google Scholar]
- (29).Pantelopulos GA; Straub JE; Thirumalai D; Sugita Y Structure of APP-C991–99and Implications for Role of Extra-Membrane Domains in Function and Oligomerization. Biochim. Biophys. Acta, Biomembr 2018, 1860, 1698–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kim C; Schmidt T; Cho E-G; Ye F; Ulmer TS; Ginsberg MH Basic Amino-Acid Side Chains Regulate Transmembrane Integrin Signalling. Nature 2012, 481, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ravula T; Ramadugu SK; Di Mauro G; Ramamoorthy A Bioinspired, Size-Tunable Self-Assembly of Polymer–Lipid Bilayer Nanodiscs. Angew. Chem., Int. Ed 2017, 56, 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- (32).Yasuhara K; Arakida J; Ravula T; Ramadugu SK; Sahoo B; Kikuchi J.-i.; Ramamoorthy, A. Spontaneous Lipid Nanodisc Fomation by Amphiphilic Polymethacrylate Copolymers. J. Am. Chem. Soc 2017, 139, 18657–18663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Terakawa MS; Yagi H; Adachi M; Lee Y-H; Goto Y Small Liposomes Accelerate the Fibrillation of Amyloid β(1–40). J. Biol. Chem 2015, 290, 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Halgren TA Merck Molecular Force Field. I. Basis, Form, Scope, Parameterization, and Performance of MMFF94. J. Comput. Chem 1996, 17, 490–519. [Google Scholar]
- (35).Malde AK; Zuo L; Breeze M; Stroet M; Poger D; Nair PC; Oostenbrink C; Mark AE An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory Comput 2011, 7, 4026–4037. [DOI] [PubMed] [Google Scholar]
- (36).Da Silva AWS; Vranken WF ACPYPE - AnteChamber PYthon Parser InterfacE. BMC Res. Notes 2012, 5, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).de Jong DH; Singh G; Bennett WFD; Arnarez C; Wassenaar TA; Schäfer LV; Periole X; Tieleman DP; Marrink SJ Improved Parameters for the Martini Coarse-Grained Protein Force Field. J. Chem. Theory Comput 2013, 9, 687–697. [DOI] [PubMed] [Google Scholar]
- (38).Van Der Spoel D; Lindahl E; Hess B; Groenhof G; Mark AE; Berendsen HJC GROMACS: Fast, Flexible, and Free. J. Comput. Chem 2005, 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- (39).Jo S; Kim T; Iyer VG; Im W CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem 2008, 29, 1859–1865. [DOI] [PubMed] [Google Scholar]
- (40).Vogeley L; Sineshchekov OA; Trivedi VD; Sasaki J; Spudich JL; Luecke H Anabaena Sensory Rhodopsin: A Photochromic Color Sensor at 2.0 Å. Science 2004, 306, 1390–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Nadezhdin KD; Bocharova OV; Bocharov EV; Arseniev AS Structural and Dynamic Study of the Transmembrane Domain of the Amyloid Precursor Protein. Acta Naturae 2011, 3, 69–76. [PMC free article] [PubMed] [Google Scholar]
- (42).Allen WJ; Lemkul J. a.; Bevan DR GridMAT-MD: A grid-based membrane analysis tool for use with molecular dynamics. J. Comput. Chem 2009, 30, 1952–1958. [DOI] [PubMed] [Google Scholar]
- (43).Skar-Gislinge N; Simonsen JB; Mortensen K; Feidenhans’l R; Sligar SG; Lindberg Møller B; Bjørnholm T; Arleth L Elliptical Structure of Phospholipid Bilayer Nanodiscs Encapsulated by Scaffold Proteins: Casting the Roles of the Lipids and the Protein. J. Am. Chem. Soc 2010, 132, 13713–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Humphrey W; Dalke A; Schulten K VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- (45).Discovery Studio Modeling Environment, Release 3.5; Accelrys Software Inc.: San Diego, 2012. [Google Scholar]
- (46).Sahoo BR; Maharana J; Patra MC; Bhoi GK; Lenka SK; Dubey PK; Goyal S; Dehury B; Pradhan SK Structural and Dynamic Investigation of Bovine Folate Receptor Alpha (FOLR1), and Role of Ultra-High Temperature Processing on Conformational and Thermodynamic Characteristics of FOLR1-Folate Complex. Colloids Surf., B 2014, 121, 307–318. [DOI] [PubMed] [Google Scholar]
- (47).Schuler MA; Denisov IG; Sligar SG Nanodiscs as a New Tool to Examine Lipid-Protein Interactions. Methods Mol. Biol 2013, 974, 415–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Sheats JR; McConnell HM A photochemical technique for measuring lateral diffusion of spin-labeled phospholipids in membranes. Proc. Natl. Acad. Sci. U.S.A 1978, 75, 4661–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Marrink SJ; de Vries AH; Mark AE Coarse Grained Model for Semiquantitative Lipid Simulations. J. Phys. Chem. B 2004, 108, 750–760. [Google Scholar]
- (50).Nasr ML; Baptista D; Strauss M; Sun ZJ; Grigoriu S; Huser S; Plückthun A; Hagn F; Walz T; Hogle JM; et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 2017, 14, 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Wadsäter M; Maric S; Simonsen JB; Mortensen K; Cardenas M The Effect of Using Binary Mixtures of Zwitterionic and Charged Lipids on Nanodisc Formation and Stability. Soft Matter 2013, 9, 2329–2337. [Google Scholar]
- (52).Kariyazono H; Nadai R; Miyajima R; Takechi-Haraya Y; Baba T; Shigenaga A; Okuhira K; Otaka A; Saito H Formation of Stable Nanodiscs by Bihelical Apolipoprotein A-I Mimetic Peptide. J. Pept. Sci 2016, 22, 116–122. [DOI] [PubMed] [Google Scholar]
- (53).Lemmin T; Dimitrov M; Fraering PC; Dal Peraro M Perturbations of the Straight Transmembrane α-Helical Structure of the Amyloid Precursor Protein Affect Its Processing by γ-Secretase. J. Biol. Chem 2014, 289, 6763–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kalli AC; Campbell ID; Sansom MSP Multiscale Simulations Suggest a Mechanism for Integrin Inside-out Activation. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 11890–11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kalli AC; Campbell ID; Sansom MSP Conformational Changes in Talin on Binding to Anionic Phospholipid Membranes Facilitate Signaling by Integrin Transmembrane Helices. PLoS Comput. Biol 2013, 9, No. e1003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


