Abstract
Hummingbirds are very well adapted to sustain efficient and rapid metabolic shifts. They oxidize ingested nectar to directly fuel flight when foraging but have to switch to oxidizing stored lipids derived from ingested sugars during the night or long-distance migratory flights. Understanding how this organism moderates energy turnover is hampered by a lack of information regarding how relevant enzymes differ in sequence, expression, and regulation. To explore these questions, we generated a chromosome-scale genome assembly of the ruby-throated hummingbird (A. colubris) using a combination of long- and short-read sequencing, scaffolding it using existing assemblies. We then used hybrid long- and short-read RNA sequencing of liver and muscle tissue in fasted and fed metabolic states for a comprehensive transcriptome assembly and annotation. Our genomic and transcriptomic data found positive selection of key metabolic genes in nectivorous avian species and deletion of critical genes (SLC2A4, GCK) involved in glucostasis in other vertebrates. We found expression of a fructose-specific version of SLC2A5 putatively in place of insulin-sensitive SLC2A5, with predicted protein models suggesting affinity for both fructose and glucose. Alternative isoforms may even act to sequester fructose to preclude limitations from transport in metabolism. Finally, we identified differentially expressed genes from fasted and fed hummingbirds, suggesting key pathways for the rapid metabolic switch hummingbirds undergo.
Hummingbirds are distinguished by features of natural and evolutionary history, morphology, and physiology from mammalian model systems such as mice, rats, and humans. They are among the smallest vertebrate endotherms (2.5–3.5 g). They use hovering flight, displaying the highest wingbeat frequencies of any bird (and highest limb oscillation frequencies of any vertebrate, ∼50–60 Hz), and in doing so sustain the highest metabolic rates among all vertebrates (Suarez 1992). In addition, the ruby-throated hummingbird (Archilochus colubris) engages in an annual migratory journey from breeding grounds throughout eastern North America to wintering grounds as far south as Central America. If measured in terms of body lengths traveled, small North American hummingbirds engage in some of the longest distance aerial migrations of any species (Gass 1979). In doing so, they show a remarkable ability to sustain high rates of metabolism using endogenous lipids, an ability not shared by mice, rats, or humans (McCue and Pollock 2013).
To fuel these activities, hummingbirds oxidize fatty acids and carbohydrates in their flight muscles at rates faster than any other vertebrates thus far studied (Welch and Chen 2014). The dietary source of both fuels, carbohydrate and fat, is the same: simple sugars (glucose, fructose, sucrose) in floral nectar that provide >90% of the total calories they ingest (Baker et al. 1998). Once ingested, hummingbirds must either oxidize or convert them into energy-dense (and thus easier to carry) lipid depots. Remarkably, hummingbirds can switch between relying exclusively on oxidation of endogenous lipids to exclusive reliance on newly ingested sugars to fuel hovering flight over a period as short as 20–30 min (Welch et al. 2006; Welch and Suarez 2007; Chen and Welch 2014).
To keep up with the high energetic demands of hovering flight, in their flight muscles hummingbirds transport, take up, and oxidize circulating sugars at rates as much as 55× greater than the maximum rates observed in any nonflying mammals (Welch and Chen 2014). Once in circulation, the flux of sugar to, and oxidation in, exercising muscle is thought to be limited principally at each of three key steps: (1) delivery from capillaries to the extracellular space, (2) transport across the fiber membrane, and (3) phosphorylation in the muscle fiber (Rose and Richter 2005; Bertoldo et al. 2006; Wasserman et al. 2011; Welch and Chen 2014). Mechanistic understanding of steps 2 and 3 is poorly understood as SLC2A4 (previously known as GLUT4), the key glucose transporter in mammals, is absent in birds, and although hummingbird hexokinase activity is higher than other vertebrates, this alone cannot explain the rate of hummingbird glycolytic flux (Suarez et al. 2009).
The ability of hummingbirds to fuel hovering flight completely with fructose raises interesting fundamental questions about the enzymatic basis for rapid sugar flux. The same three key steps that regulate glucose uptake and oxidation by muscles presumably apply to fructose as well (Rose and Richter 2005; Wasserman et al. 2011; Welch and Chen 2014). In hummingbirds, there is ample evidence that at steps 1 and 2 the capacity for fructose uptake into flight muscle fibers is dramatically higher than in other vertebrates. However, the enzymatic basis for high rates of fructose phosphorylation (step 3) remains unknown.
Although common among migratory birds (Jenni and Jenni-Eiermann 1998; Guglielmo 2010), the ability to fuel flight exclusively or predominantly with endogenous lipid stores is itself something that distinguishes hummingbirds from model mammalian species. Many avian species build fat stores to power long-distance migratory flight using the fatty acids that are present in their diet (Guglielmo 2010). Some of these switch to or exploit seasonally available diets that are rich in specific lipid classes (Pierce et al. 2005; Guglielmo 2010). However, hummingbirds achieve high rates of de novo lipogenesis on a simple sugar diet and high rates of lipid accumulation to see them through both overnight fasts and migratory flight.
For these reasons, genomic studies of the ruby-throated hummingbird are warranted and necessary for further understanding of these fine-tuned metabolic systems. Here we produced a chromosome-level hybrid genome assembly of the ruby-throated hummingbird. We annotated the genome using a combination of Illumina and Oxford Nanopore cDNA sequencing from muscle and liver tissues in both the fasted and fed metabolic states to identify full coding sequences and multiple encoded isoforms. Finally, we performed differential expression analysis and differential alternative splicing analysis on fasted and fed birds in both muscle and liver tissues to fully characterize the mechanisms underlying high catalytic rates (high catalytic efficiency and/or high levels of enzyme expression) and control over metabolic flux. These results are crucial for understanding the hummingbirds’ exquisite control over rates of substrate metabolism and biosynthesis, which could give insight into metabolic control of orthologous pathways in humans.
Results
Chromosome-level genome assembly
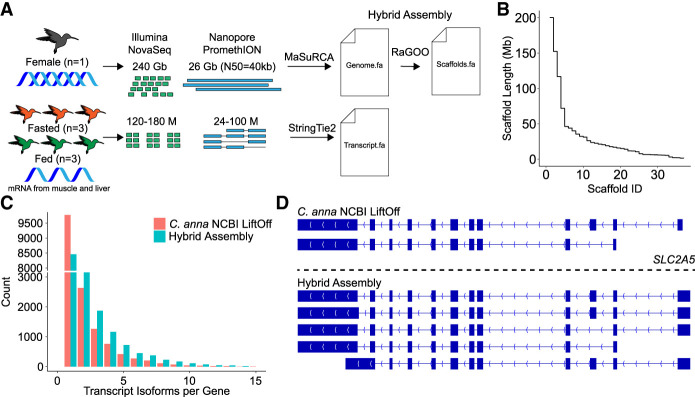
Using hummingbird brain tissue, we generated a total of 26 Gb of Nanopore sequencing data on the PromethION with a read-length N50 of 40 kb and 240 Gb of short-read Illumina NovaSeq data (Methods) (Fig. 1A). We performed hybrid assembly with MaSuRCA (Zimin et al. 2013, 2017), which assembles short Illumina reads into “super-reads,” which are then aligned to Nanopore reads to create “mega-reads.” These mega-reads are then assembled into contigs for assembly. Our resulting assembly had 1837 contigs with a contig N50 of 13.54 Mb. We found the assembly contained 1.13% heterozygous sequence (Supplemental Fig. S1). Using the assembly of a different hummingbird species, Anna's hummingbird (Calypte anna) (Rhie et al. 2021) as generated by the Vertebrate Genome Project (VGP) (Rhie et al. 2021), we performed reference-based scaffolding with RaGOO (Supplemental Fig. S2; Alonge et al. 2019). We used RaGOO without patching and MaSuRCA without gap filling, so no C. anna sequence was retained in our final assembly; it was only used for scaffolding. Our final assembly of the ruby-throated hummingbird had 33 chromosomes that contained 97.8% of the total sequence (Fig. 1B; Table 1). The total genome length was 1.1 Gb with a scaffold N50 of 46 Mb, a scaffold L50 of five chromosomes, with the largest scaffold (Chromosome 1) 200 Mb long (Table 1). However, it is important to recognize that reference-based scaffolding may lead to structural errors by masking variations that exist between the two species (Alonge et al. 2019).
Figure 1.
Experimental design and genome/transcriptome assembly overview. (A) Overview of experimental design and methods for producing a genome assembly and transcriptome of A. colubris. (B) Step plot of scaffold length for the 33 A. colubris chromosomes. (C) Number of isoforms per gene in the C. anna NCBI Liftoff annotation versus the StringTie2 hybrid transcriptome assembly. (D) The SLC2A5 gene locus in the C. anna NCBI Liftoff annotation and the StringTie2 hybrid transcriptome assembly.
Table 1.
Genome assembly and annotation statistics

We assessed the assembly for completeness using BUSCO for avian genomes (Marchi et al. 2017) and found it to be 96.6% complete (Supplemental Table S1). To further assess the accuracy and completeness of our assembly, we used Merqury (Rhie et al. 2020), which compares k-mers in an assembly to those found in the unassembled Illumina reads (Supplemental Fig. S3). A fraction of one-copy k-mers are found only in the reads (peak around 100×), representing heterozygous sequences missing from the assembly. This is common as heterozygous sequences are collapsed so the genome can be represented as a linear haploid assembly. From the k-mers, Merqury can also estimate an assembly consensus quality value (QV), which represents a log-scaled probability of error for the consensus base calls. Higher QVs indicate a more accurate consensus, where Q30 corresponds to 99.9% accuracy, Q40 to 99.99%. The QV for our assembly was 41.8, indicating an error rate of 6.6 × 10−5. Considering our QC metrics, we believe this to be a highly accurate and high-quality assembly as well as the only available assembly of the ruby-throated hummingbird, adding to our understanding of this important model organism.
Genome annotation
Avian genomes are the smallest of the amniotes, with smaller genomic elements (e.g., introns, exons, intergenic DNA) and fewer transposable elements compared with mammals (Zhang et al. 2014). With our new genome in hand, we examined the repetitive elements in the A. colubris genome assembly. We used RepeatModeler2 to generate repeat libraries (Flynn et al. 2020) and used them in combination with the curated avian library to perform homology-based repeat masking with RepeatMasker. Among vertebrates, birds show relatively low copy numbers and an overall reduced diversity of repetitive elements (International Chicken Genome Sequencing Consortium 2004; Dalloul et al. 2010; Warren et al. 2010; Sotero-Caio et al. 2017), with the exception of the woodpecker (Picoides pubescens), whose genome is 22.2% TEs, mostly contributed by the LINE/CR1 (Zhang et al. 2014). In A. colubris, we detected 163 Mb of repetitive sequence, representing 14.83% of the genome, including 116 Mb of TEs that make up 10.50% of the genome, consistent with the repeat content in other avian lineages (Table 1; Supplemental Table S2; Zhang et al. 2014). Among classified repeats, LINE/CR1 elements were the most abundant superfamily in the A. colubris genome, making up 6.95% of the sequence. Next were LTRs (2.57%) and repeats discovered by our libraries but not classified by RepeatClassifier (unknown; 2.50%).
Transcriptome assembly
To capture the complexity of differential splicing and the precision of splice junctions, including transcription start and end sites, we used a combination of short-read Illumina NovaSeq and long-read Oxford Nanopore cDNA sequencing. We collected mRNA from the muscle and liver of A. colubris hummingbirds that were fed sucrose ad libitum (fed; n = 3) or fasted for a time period of 1 h (fasted; n = 3) (Fig. 1A). Preliminary gene annotation was accomplished via a liftOver of the C. anna annotations from the NCBI annotation (GCF_003957555.1) with Liftoff, a tool that maps annotations between closely related species (Shumate and Salzberg 2021). The C. anna annotation Liftoff to A. colubris consisted of 15,879 genes and 31,163 transcripts for an average of two transcripts per locus. We then used the hybrid reference-based assembly pipeline from StringTie2 (Shumate et al. 2022) to expand our existing C. anna Liftoff annotation to a total of 17,878 genes and 43,348 transcripts. Our transcriptome assembly identified 8705 multitranscript loci with an average of 2.4 isoforms per gene, a large increase over the C. anna NCBI Liftoff annotation with 5909 multitranscript loci at 2.0 isoforms per gene (Fig. 1C). Additionally, our assembly identified 1999 novel loci that were not annotated in C. anna, 1051 of these receiving a functional annotation via BLASTing against the Swiss-Prot database. Eight hundred sixty-four of these 1999 were not only not annotated, but not present in the C. anna genome based on BLAST; 651 of those 864 did receive a functional annotation from Swiss-Prot. Included in these novel genes are genes critical to metabolism, including ALDOA, PFKM, G6PD, PGLS, PC, PCK2, PFKFB1, PYGM, and PLPPR1. Many of these genes (e.g., ALDOA and PYGM) have been historically “missing” from avian assemblies owing to being “hidden” in GC-rich areas that fail to assemble with short-read sequencing (Huttener et al. 2021; Rhie et al. 2021; Kim et al. 2022). The combination of hybrid genome and transcriptome assembly and deep sequencing of multiple tissues in the fasted and fed condition enabled assembly and annotation of these genes. Furthermore, the hybrid transcriptome assembly increases the number of isoform variants per gene as exemplified by the solute carrier family 2 member 5 (SLC2A5), previously known as GLUT5, where the C. anna NCBI annotation contained two isoforms and our new hybrid annotation contained five splice isoforms (Fig. 1D). Our expanded annotation allows us to understand gene expression changes at the transcriptome level during transitions between fuel use regimes, providing insights into potential mechanisms that make these organisms such flexible metabolic performers.
Positively selected genes in nectivory
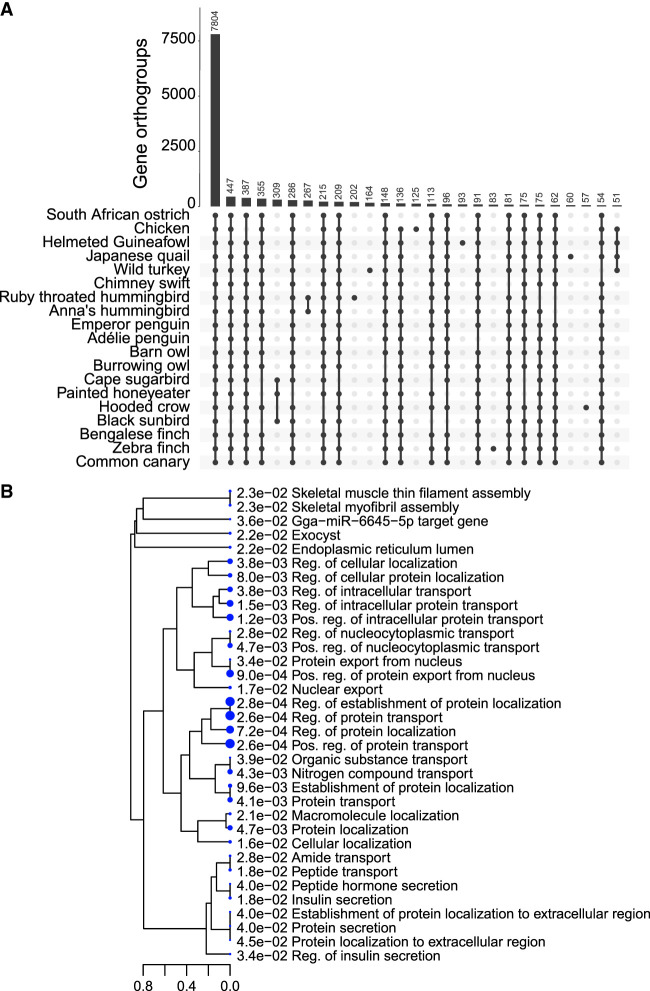
Nectar-feeding animals have among the highest recorded metabolic rates; incidentally, flight requires the highest metabolic rates of any form of locomotion known (Suarez 1992; Suarez et al. 2011). Using our new ruby-throated hummingbird genome assembly, we performed a phylogenetically informed branch-site test of positive selection (Yang 2007) of nectivorous avian species in a background of nonnectivorous species. We used 19 species with high-quality reference genomes available: chimney swift, Anna's hummingbird, helmeted guineafowl, chicken/red junglefowl, wild turkey, Japanese quail, zebra finch, Bengalese finch, common canary, painted honeyeater, black sunbird, cape sugarbird, emperor penguin, Adelie penguin, burrowing owl, barn owl, African ostrich, hooded crow, and ruby-throated hummingbird. Five of the included taxa are nectivorous and represent four separate lineages (Supplemental Fig. S4). We used OrthoFinder (v2.3.12) to identify orthologous gene clusters between all 19 species (Emms and Kelly 2019). OrthoFinder groups genes into orthogroups, sets of genes descended from a single gene in the species, the last common ancestor based on their sequence similarity. OrthoFinder assigned 97.9% of genes to orthogroups, generating 18,581 orthogroups that contained a total of 354,573 genes across all species; 7799 (42%) of these orthogroups were shared between all 19 species, and 1848 were shared and present as a single copy (Fig. 2A; Supplemental Fig. S5).
Figure 2.
Positive selection in nectivory. (A) Upset plot illustrating the number of shared orthogroups between the 19 species. Only bars with more than 50 orthogroups are shown. (B) ShinyGO Gene Ontology plot showing a hierarchical clustering tree to summarize the correlation among significant pathways. Pathways with many shared genes are clustered together. Dot size is representative of P-value significance.
We analyzed all these shared single-copy orthologous proteins for evidence of positive selection with PAML (Yang 2007). Of the 1848 shared single-copy orthologs, we found 148 (8.0%) genes with evidence of positive selection in the nectivorous branches (Supplemental Table S3). A Gene Ontology (GO) (FDR < .05) analysis revealed enrichment of positively selected genes in macromolecule (protein, amide, nitrogen, organic substance) transport, macromolecule localization, skeletal muscle assembly, and insulin secretion (Fig. 2B; Supplemental Table S4). Because metabolic flux occurs rapidly in nectar-feeding birds, it is likely that the flux is initially supported by rapid shuttling and sequestration of proteins and other metabolites as opposed to reliance on transcriptional changes. Many positively selected genes are enriched in transport processes, contributing to the rapid changes in the proteome and metabolome that occur in metabolic flux. We identified positive selection in genes involved in lipogenesis: beta-oxidation (HACD3) and ketone utilization (BDH2) (Guo et al. 2006). With an extremely low fat diet, nectivorous birds rely on rapid lipogenesis to generate endogenous fat storage and lipolysis for fueling flight in the fasted state. Additionally, muscles in these species are uniquely adapted to maintain hovering flight; therefore, positive selection of genes such as PROX1 and LMOD1 likely contribute to the ability of pectoralis muscles to support hovering flight (Nanda and Miano 2012; Kivelä et al. 2016). Finally, we observed an enrichment of positively selected genes in pathways related to insulin secretion (RBP4, GCG, RFX6, INHBB, SLC30A8, NOS2). Of these, RBP4 contributes to obesity and diabetes in humans, and its expression is elevated with SLC2A4 knockout (Steinhoff et al. 2021), highlighting this protein for further study. We also found GAPDH, encoding an enzyme crucial for oxidative sugar metabolism, is positively selected. GAPDH encodes the enzyme separating lower and upper glycolysis and is the rate limiting step in the pathway (Shestov et al. 2014). These data indicate that select genes (and the pathways they regulate) are positively selected in nectivorous birds to enable their highly energetically demanding lifestyle.
Hummingbird sugar transport and metabolism
A large body of literature points to the absence of SLC2A4 in birds (Seki et al. 2003; Sweazea and Braun 2006; Braun and Sweazea 2008; Welch et al. 2013; Huttener et al. 2021). We, too, were unable to detect the SLC2A4 gene in the A. colubris genome. However, it is possible that the SLC2A4 gene exists in a region of high GC content and was not fully assembled (Huttener et al. 2021). So to search for SLC2A4 transcripts, we generated a transcriptome assembly with Trinity using only the raw Illumina reads from the liver and muscle tissue of the fed hummingbirds according to the methods of Yin et al. (2019). Using BLAST, we searched for the putative chicken SLC2A4 gene in our hummingbird transcriptome but did not identify SLC2A4 transcripts in the ruby-throated hummingbird. We conclude that the SLC2A4 gene is likely not expressed in A. colubris.
Another key regulator of blood glucose homeostasis is glucokinase (GCK), which has a high Km and acts as a glucose sensor for both pancreatic β cells and the liver in mammals (Peter et al. 2011; Matschinsky and Wilson 2019). Our transcriptome assembly did not identify GCK in the assembled hummingbird transcriptome, and we did not identify any GCK sequence in any of the ruby-throated hummingbird RNA-seq reads. When we compared the ruby-throated hummingbird genome to the chicken reference genome, we determined that the region of the chicken genome containing the GCK gene is not syntenic to any of the hummingbird sequence (Supplemental Fig. S8).
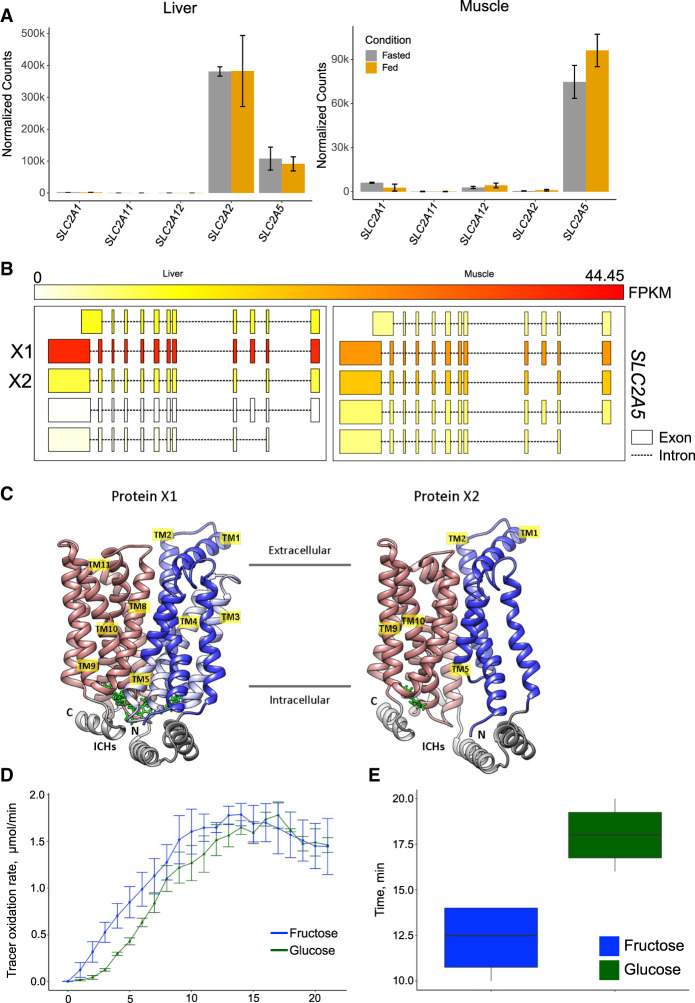
The absence of GCK and GLUT4 proteins leaves questions about how glucose enters hummingbird muscle cells. So, we investigated other highly expressed sugar transporters and their potential role in glucose uptake. In both the fasted and fed states, the hummingbird liver tissue has high expression of the SLC2A2 and SLC2A5 sugar transporters. SLC2A2 protein (previously known as GLUT2), which has a high Km, plays a stronger role in enteric (Karasov 2017) and hepatic (Mueckler and Thorens 2013) sugar transport, resulting in the expected higher expression we observe in liver over muscle samples. Chicken SLC2A1 and SLC2A3 share sequence homologies of ∼80% and ∼70%, respectively, with human GLUTs, but other isoforms such as SLC2A2 and SLC2A5 only share ∼65% and ∼64% sequence homology (Ali et al. 2020). A comparison of SLC2A2 sequences to 19 bird species reveals the loss of an N-linked glycosylation site in four of the 19 species (Workman et al. 2018). In the case of the African ostrich and the barn owl, this site is lost owing to truncation at the 5′ end of the protein. However, in the hummingbirds (Anna's hummingbird and the ruby-throated hummingbird), Asn-64 is replaced by Ser-64, eliminating the conserved N-linked glycosylation site present in the 16 avian species as well as humans and mice. In mice, the loss of this glycosylation site is coincident with increased SLC2A2 protein endocytosis and the onset of type 2 diabetes (Ohtsubo et al. 2005).
In the hummingbird muscle, in both the fasted and fed states, the most highly expressed sugar transporter is SLC2A5, which facilitates fructose uptake but is not highly expressed in mammals (Barone et al. 2009). Notably, in mammalian SLC2A5, a single-point mutation is enough to switch the substrate binding preference from fructose to glucose (Nomura et al. 2015). The abundance of SLC2A5 transcripts in hummingbird tissues, especially muscle tissue, is particularly interesting because it suggests this transporter is principally responsible for glucose/fructose transport into hummingbird tissues. There is considerable sequence divergence between hummingbird SLC2A5 and mammalian SLC2A5 (65.5% identity to human, 63.7% identity to mouse) and even from hummingbird to chicken (80.5% identity). We hypothesize that this form of hummingbird SLC2A5 has transport capacity for glucose, but at a lower affinity than its capacity for fructose. In bacterial GLUT transporters (e.g., XylE), a Trp residue at the floor of the sugar binding pocket displays two hydrogen bonds with the bound glucose (Sun et al. 2012). In the same position on rat SLC2A5, this residue moves to Alanine (Ala395) (Nomura et al. 2015), and in the hummingbird, this residue is serine (Ser403) (Supplemental Fig. S6). The Trp amino acid is well conserved among all human glucose transporters (SLC2A1–4); however, it moves to Ser in human SLC2A7 (also known as GLUT7), which is also a dual (glucose and fructose) transporter (Supplemental Fig. S6). With this information, we speculate that hummingbird SLC2A5 could be a dual glucose/fructose transporter.
Using long-read cDNA data, we quantified the relative abundance of SLC2A5 transcript isoforms in muscle and liver and identified differential alternative splicing occurring between muscle and liver tissues (Fig. 3B). The dominantly transcribed isoform translates to a protein highly similar in structure to mammalian SLC2A5 (Nomura et al. 2015). However, the muscle SLC2A5 has higher expression of the isoform that skips exon 3, resulting in missing transmembrane domains TM3, TM4, and the intracellular tip of TM5 (Fig. 3C). In this isoform, the salt bridges between the N- and C-terminal TM bundles are absent, and therefore, the outward facing state is likely not favored.
Figure 3.
Hummingbird sugar transporters. (A) Normalized gene count plots for ruby-throated hummingbird muscle and liver tissue. (B) Isoform expression of SLC2A5 in the liver (left) and muscle (right). FPKM of each isoform color-coded according to the top scale bar. (C) Ribbon representations of the two protein models for SLC2A5 isoforms predicted by AlphaFold2 based on the mammalian SLC2A5 ortholog. Left is the X1 isoform; right is the X2 isoform that is missing exon 3. In both atomic models, N- and C-terminal TM bundles are colored blue and red, respectively. Regions of the X1 isoform that are missing in the X2 isoform (TM3/TM4) are depicted in light blue. Arginine–glutamate salt bridges at the intracellular tips of TMs are green. (ICHs) Intracellular helices. (D) Tracer oxidation rate over 20 min when birds were fed 13C on either the glucose or the fructose portion of the sucrose disaccharide. (E) Peak oxidation time of glucose and fructose in minutes (P = 0.02, paired t-test).
Although the high expression of fructose transporter gene SLC2A5 strongly suggests that fructose uptake capacity may be sufficient to meet fructolytic and oxidative demand during hovering flight, the enzymatic basis for high rates of fructose phosphorylation is still unclear. The initial step of fructose metabolism in the liver in mammals is phosphorylation by the enzyme ketohexokinase (KHK) (Hayward and Bonthron 1998). However, in both humans and in the ruby-throated hummingbirds, the muscle mainly expresses hexokinase 2 (HK2), which is a glucose-specific kinase in humans (Fig. 3A). The hummingbird KHK and HK2 genes have 65% and 87% identity to their human orthologs, respectively; their substrate affinities could be different from their human orthologs.
Previous studies assessed A. colubris muscle total hexokinase activity and determined the Vmax to be 50% lower for fructose than glucose phosphorylation, which would not keep up with the calculated required rates of fructose oxidation by flight muscle during hovering flight (Myrka and Welch 2018). In previous work, we used a chronic stable isotope tracer methodology to examine the speed of glucose and fructose usage for de novo lipogenesis in the ruby-throated hummingbird (Dick et al. 2020). The vast majority of calories/carbon that hummingbirds (including in captivity) ingest is in the form of fructose and glucose, meaning that essentially all of the lipid stores hummingbirds accumulate are ultimately derived from the sugar in their diet. By using carbon isotope-enriched sugars, the relative contribution of carbon from different sugar sources to the endogenous lipid pool revealed a partial preference for glucose rather than fructose as a substrate for lipogenesis.
Using a similar approach, we traced the immediate oxidation of a sucrose-only meal in which the 13C on either the glucose or the fructose portion of the disaccharide was enriched. Immediate tracer oxidation rate was measured at 1-min intervals via the 13C signature of the expired CO, in combination with respirometry. We found that the respiratory exchange ratio (RER = V˙CO2/V˙O2) and tracer oxidation increased quickly with feeding (Fig. 3D; Supplemental Fig. S7A). RER rose to a ratio exceeding one, indicating the shift from fasted lipid fueled metabolism (RER ∼ 0.7) to sugar fueled metabolism (RER ∼ 1) and direct sugar oxidation co-occurring with lipogenesis from sugar (RER > 1) (Simonson and DeFronzo 1990). Peak tracer oxidation did not differ between the enriched sucrose solutions (P = 0.66) (Supplemental Fig. S7B). However, the time to peak oxidation differed, with the fructose-enriched sucrose solution reaching peak tracer oxidation faster than the glucose-enriched sucrose solution (P = 0.02) (Fig. 3E), indicating more rapid oxidation of the fructose portion of sucrose. Overall, these data support a hypothesis in which fructose is rapidly transported out of the blood and metabolized. In contrast, glucose remains in the bloodstream for longer and is used as a fuel source after blood fructose levels decline.
Identification of differentially expressed genes that respond to fasting
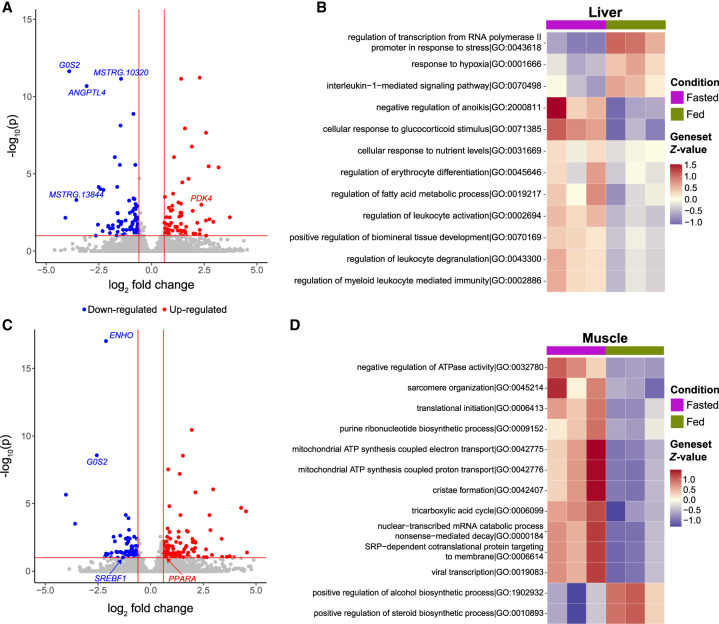
To identify differentially expressed genes (DEGs) that rapidly respond to fasting, we profiled the transcriptomes of total mRNA from the muscle and livers of A. colubris hummingbirds that were fed sucrose ad libitum (fed) or fasted for a time period of 1 h (fasted) (Fig. 1A). We analyzed three biological replicates for each metabolic condition (fasted vs. fed) with StringTie2 hybrid long- and short-read quantification and DESeq2 with our newly constructed reference and annotation (Love et al. 2014; Shumate et al. 2022). We identified 140 DEGs with adjusted P-values below 0.1 in the liver (Fig. 4A; Supplemental Table S5) and 191 DEGs in the muscle (Fig. 4C; Supplemental Table S6). To categorize these genes according to their GO, we used the Genetonic pipeline and generated functional gene-set enrichments for the A. colubris liver and muscle (Fig. 4B,D; Marini et al. 2021). This analysis yielded 200 statistically significant pathways (FDR < .05) in the liver and 106 in the muscle (Supplemental Tables S7, S8). The response to fasting in the muscle and liver influenced dramatically different metabolic and regulatory pathways in each tissue.
Figure 4.
Differential expression. (A) Volcano plot displaying differentially expressed genes (DEGs) from A. colubris liver tissue comparing fasted to fed. (B) Summary heatmap of top 12 enriched GO terms in the liver. (C) Volcano plot displaying DEGs from A. colubris muscle tissue comparing fasted to fed. (D) Summary heatmap of top 13 enriched GO terms in the muscle.
In the liver, the 1-h fast influenced many metabolic and homeostatic pathways (Fig. 4B), including coenzyme biosynthetic processes, cellular responses to nutrient levels, response to hypoxia, response to carbohydrate, fatty acid metabolic processes, homeostatic processes, and response to glucocorticoid stimulus (Fig. 4B). Genes particularly affected in these processes are key regulators of metabolic flux, including PDK4, G0S2, and ANGPTL4, which likely contribute to the rapid transition to lipid metabolism in the hummingbird liver during an acute food withdrawal. Induction of these genes occurs independently of PPAR signaling in the hummingbird liver as all the PPAR genes do not have any changes in expression between the fasted and fed state (Supplemental Table S9). Therefore, in hummingbirds, the PPAR pathway does not appear to control the expression of the metabolic switch genes in the liver, at least in an acute (1-h) fast. Many newly assembled genes were also differentially expressed, including MSTRG.10320 and MSTRG.13844, which we functionally annotated with Swiss-Prot as HRG1 and AT1B, respectively.
The most statistically significant pathway up-regulated in the fasted muscle was mitochondrial ATP synthesis coupled proton transport (GO:0042776, P = 1.90 × 10−6) (Fig. 4D). Other key genes regulating metabolic flux were affected such as ENHO, PPARA, G0S2, and SREBF1 (Fig. 4C). ENHO is associated with energy storage and metabolism as a precursor to the protein adropin and was down-regulated in fasted birds (Kumar et al. 2008; Aydin et al. 2013). Interestingly, in humans, adropin is generally associated with liver and brain expression as opposed to the skeletal muscle expression we observed in A. colubris. G0S2 is the only gene identified as differentially expressed in both the liver and muscle tissues. Although G0S2 is known to have a significant role in liver lipid transport, a definitive role for G0S2 within skeletal muscle has yet to be elucidated, although it appears that G0S2 is also present in mitochondria, with the speculation of several possible functions (Turnbull et al. 2016). These results point to the role of G0S2 in hummingbird rapid metabolic flux.
Discussion
The results of our study are critical in understanding the hummingbirds’ exquisite control over rates of substrate metabolism and biosynthesis. Our positive selection analysis points to a subset of 39 genes critical to the development of nectar-based lifestyle. Glycolysis, the tricarboxylic acid cycle, lipogenesis, and lipolysis pathways have to function rapidly to allow for the high energetic demands of flight and reliance on nectar as the only fuel source. This was evident in the positive selection of genes involved in these pathways (e.g., GAPDH, PDHA1, ACADL, HACD3, and BDH2) in nectivorous bird lineages.
We were specifically interested in glucose and fructose uptake into hummingbird tissue. In addition to the previously established lack of avian SLC2A4, we also identified the loss of GCK. Likely, the low levels of insulin secretion and high sustained blood glucose in hummingbirds are due in part to the lack of expression of GCK, a key regulator of insulin secretion and blood glucose homeostasis. With our new assembly, annotation, and expression data, we speculate that hummingbird SLC2A5 has transport affinity for both glucose and fructose with a higher affinity for fructose. Unlike most other animals, 50% of the hummingbirds’ diet consists of fructose (Baker et al. 1998), which studies show is much more cytotoxic than glucose (Horst et al. 2017). Consequently, hummingbirds rapidly sequester fructose into the muscle tissue, as evidenced by rapid declines in blood fructose levels upon fasting (Muhammad 2021). Further, we concluded from a tracer oxidation study that the birds are preferentially clearing fructose from circulation first and oxidizing it to CO2. Our data support the hypothesis that glucose and fructose are transported into the muscle cells via the SLC2A5 transporter, with fructose being favored first when concentrations of both are high.
Our improved assembly of the ruby-throated hummingbird genome and transcriptome allowed for isoform-level analysis of gene expression. This analysis revealed expression of a SLC2A5 protein variant in the hummingbird muscle with an internally facing active site, but a loss of exon three, possibly resulting in loss of transport activity. We speculate that a potential biological function of this SLC2A5 protein isoform is in sequestering fructose inside muscle cells rather than acting as a transporter, as fructose phosphorylation capacity is low and likely cannot keep up with the rapid import. Future biochemical and functional studies will show whether hummingbird SLC2A5 can transport fructose and glucose and how its variants differ from SLC2A5 found in other species.
Our characterization of the hummingbird liver and muscle expression profiles gave insights into the drivers of rapid metabolic flux in hummingbirds. For example, PDK4 was up-regulated in the fed-to-fasted transition, a protein kinase located in the matrix of the mitochondria that inhibits the pyruvate dehydrogenase complex (PDH) by phosphorylating one of its subunits. Because PDH is considered the gatekeeper of the TCA cycle, its inhibition in the fasted state would shut down complete glucose oxidation and promote gluconeogenesis and fat oxidation. It is possible that rapid switching from carbohydrate to fat oxidative catabolism in the fasted state is due in part to up-regulation of PDK4, suggesting it as a target for future study. Another example is the down-regulation of G0S2 in both liver and muscle; G0S2, the G0/G1 switch gene 2, is an inhibitor of adipose triglyceride lipase (ATGL), a rate-limiting enzyme that catalyzes the first step in triglyceride hydrolysis in adipocytes. Previous studies of G0S2 on chicken, turkey, and quail have revealed avian G0S2 has 50% to 52% homology to mammalian G0S2 and suggest its importance in regulation of ATGL-mediated lipolysis (Oh et al. 2011). Our results extend this role of G0S2 to rapid transition of fed-to-fasted metabolism across multiple tissues. Lastly, we observed changes in expression of genes controlling vessel dilation and constriction, likely important to osmoregulation (e.g., HRG1 and AT1B). Hummingbird kidneys are not designed to concentrate urine: When they are feeding, they must eliminate large quantities of water; but when not feeding, they are susceptible to dehydration (Bakken et al. 2004). So these changes in vessel dilation are likely necessary for preparing the splanchnic tissues for the osmotic shift that occurs during fasting. We noted only a slight increase in expression of SLC2A5 in the fed condition after the 1-h acute fast. However, this change was not statistically significant, and it is likely that a 1-h fast was not long enough to observe statistically significant changes in SLC2A5 expression. It is also possible that, as is the case for SLC2A1 and SLC2A4 in humans and other mammals, changes in expression are not the only, or even primary, way that variation in the activity of these transporters is achieved.
Our results leveraged cutting-edge long- and short-read sequencing technologies to generate a high-quality genome assembly and annotation of the ruby-throated hummingbird. Our analysis suggests multiple genes and gene pathways ripe for biochemical analysis, enabled through the references we have generated to allow rapid cloning and expression. Biochemical studies of enzymatic properties, for example, Kcat or Vmax, and structural biology via X-ray crystallography or cryoEM can be used to compare hummingbird genes to orthologs from other avian or mammalian species, giving us a greater understanding of these important metabolic pathways.
Methods
Animal use and ethics statement
This study was conducted under the authority, and adheres to the requirements, of the University of Toronto Laboratory Animal Care Committee (under protocol 20011649) and the guidelines set by the Canadian Council on Animal Care. Twelve adult male ruby-throated hummingbirds (A. colubris) were captured in the early summer at the University of Toronto Scarborough (UTSC) using modified box traps. The hummingbirds were individually housed in Eurocages at the UTSC vivarium on a 12 h:12 h light–dark cycle. The hummingbirds in these cages were provided with perches and were on an ad libitum diet of 18% weight to volume of NEKTON-Nektar-Plus (Keltern) for 2–3 mo until tissue sampling occurred.
One day before experiment day (23 h), 12 male birds were placed on a 33% sucrose solution ad libitum diet in place of the NEKTON-Nektar-Plus diet. Birds were then divided into a fed group (n = 6) and a fasted group (n = 6). One hour before sampling, birds from both conditions were placed in small glass jars that had perches. This restricted the birds’ ability to fly and was performed in hopes of reducing energy expenditure variation between individual birds. Birds in the fed group were then provided with ad libitum 1 M sucrose solution for 1 h up to sampling, which began at 10:00 h. The fasted group (n = 6) was deprived of food 1 h before sampling. The 1-h fast was chosen because previous work by Chen and Welch (2014) has shown via respirometry that this time is sufficient for the fasted hummingbird to shift from using circulating sugars to using fats for fueling metabolism.
Tissue samples were collected via terminal sampling of the hummingbirds. They were anesthetized via isoflurane inhalation and sacrificed using decapitation. Flight muscles (the pectoralis and supracoracoideus muscle) and liver were collected. Tissues were flash-frozen in liquid nitrogen and subsequently stored at −80°C. In addition, one female hummingbird was also captured and brain-harvested and stored as above. This sample was used for DNA isolation for genome assembly purposes and was not subject to any experimental conditions.
DNA sequencing
Genomic DNA was extracted from the hummingbird from two 25-mg pieces of brain tissue and two 25-mg pieces of pectoralis muscle tissue with the Nanobind CBB tissue kit Alpha Handbook v0.16d (4/2019) from Circulomics following the protocol for using the dounce homogenizer. DNA quality was assessed with the NanoDrop spectrophotometer. We generated a sheared Nanopore library and an ultra-long Nanopore library to enrich for both size and depth. For the sheared library, DNA was sheared to 10 kb with Covaris g-TUBE at 6 kRPM. For the ultra-long library, DNA was size-selected with the short read eliminator XS kit from Circulomics. Oxford Nanopore sequencing libraries were prepared using the ligation sequencing 1D kit (Oxford Nanopore SQK-LSK109) according to the manufacturer's instructions and sequenced for 72 h on 2 PromethION R9.4.1 flow cells. Nanopore reads were base-called with Guppy Software (version 3.0.6). Sequencing runs were pooled for genome assembly purposes. For shotgun Illumina sequencing, a paired-end (PE) library was prepared with the Nextera DNA flex library prep kit from Illumina and sequenced on the Illumina NovaSeq 6000 (Illumina).
Genome assembly
The genome was assembled using both Illumina and Nanopore sequencing data sets with MaSuRCA (Zimin et al. 2013) with FLYE_ASSEMBLY = 1 and all other parameters set as default. The genome was scaffolded with RaGOO using the C. anna assembly (GCA_003957555.2) as a reference (Alonge et al. 2019). Assembly similarity was first checked by aligning the two assemblies with NUCmer from the MUMmer package (Marçais et al. 2018), and assemblies were considered highly similar. Assembly completeness was checked with BUSCO using the Aves lineage (Manni et al. 2021). Assembly heterozygosity was quantified with the K-mer analysis toolkit (KAT) (Mapleson et al. 2017) GenomeScope (Vurture et al. 2017), and the assembly was determined to have 1.13% heterozygosity (Supplemental Fig. S1). Repeats were annotated by running RepeatModeler (v2.0.1) to generate a database of custom repeat annotations (Flynn et al. 2020). The assembly was first masked with RepeatMasker (v4.0.9) (Smit et al. 2013–2015) using the Aves database and then further masked using the custom generated RepeatModeler database.
RNA extraction
RNA was extracted from ∼40 to 50 mg of pectoralis tissue and 20 mg of liver using the Qiagen RNeasy fibrous tissue mini kit (Qiagen). RNA quality was assessed using NanoDrop and the presence of sharp 18S and 28S rRNA on an agarose gel. RNA quality was also assessed with the Agilent 2200 TapeStation system RNA high-sensitivity kit (Agilent) before and after poly(A) isolation with the NEBNext Poly(A) mRNA magnetic isolation module.
RNA sequencing
Poly(A) mRNA from all samples was supplemented with spike-in RNA variants (SIRV) set 3 from Lexogen. Libraries for Illumina sequencing were generated with NEBNext ultra RNA library prep kit for Illumina and sequenced on the Illumina NovaSeq 6000 (Illumina). Libraries for long-read sequencing were generated with the cDNA PCR sequencing kit (SQK-PCS109) from Oxford Nanopore Technologies according to the manufacturer's instructions. Libraries were each sequenced on a PromethION flow cell for 72 h.
Genome annotation and transcriptome assembly
Illumina RNA-seq reads were trimmed with Trimmomatic (v0.39) with the following parameters: SLIDINGWINDOW:4:20 LEADING:10 TRAILING:10 MINLEN:50 (Bolger et al. 2014). Trimmed reads were then aligned to the ruby-throated hummingbird reference genome with HISAT2 (v2.2.0) (Kim et al. 2019) with the parameters ‐‐score-min L,0,-0.5 -k 10 to account for the high heterozygosity in the wild hummingbirds and filtered for primary alignments with SAMtools (v1.9) (Li et al. 2009). Nanopore cDNA sequencing reads were aligned with deSALT (v1.5.4) and filtered for primary alignments with a mapping quality score greater than 50. Initial genome annotation was performed by lifting over the predicted annotations from the C. anna genome annotation (GCA_003957555.2) onto our ruby-throated hummingbird assembly with Liftoff (Shumate and Salzberg 2021). These lifted over annotations were used as a reference model for hybrid transcriptome assembly with StringTie2 (Shumate et al. 2022). We ran StringTie2 separately for each paired Illumina and Nanopore sample (n = 12) with the following command: StringTie ‐‐mix {short.bam} {long.bam} -G {LiftOff_annotation.gtf} ‐‐conservative -L -o {out.gtf} -p 10 -B -A {out.abun} -v.
To filter out low evidence assembled transcripts, we aligned the 12 StringTie2 GTFs with GffCompare (v0.11.2) (Pertea and Pertea 2020). The 12 GTF files were merged with StringTie merge and filtered to retain transcripts that had evidence from at least two of the 12 GTFs. Gene and transcript abundance measurements were computed against the final merged and filtered GTF file with the same command as above and the addition of the -e flag: stringtie ‐‐mix {short.bam} {long.bam} -G {filtered_merged.gtf} ‐‐conservative -L -o {out.gtf} -p 10 -B -A -e {out.abun} -v.
To correct for transcripts assigned to the incorrect gene locus during StringTie's merge function, we ran the R package IsoformSwitchAnalyzeR (Vitting-Seerup and Sandelin 2019) with fixStringTieAnnotationProblem = TRUE. We generated the transcript count matrix files using the prepDE.py3 script from StringTie2. The gene count matrix files were generated using the abundance measurements from the GTF and the gene-to-transcript associations from the IsoformSwitchAnalyzeR output. The resulting ruby-throated hummingbird transcriptome assembly is available on Zenodo (DOI:10.5281/zenodo.6363333). Protein predictions from the transcriptome were performed with TransDecoder (v5.5.0; https://github.com/TransDecoder/TransDecoder). Genes that were not annotated in the Anna's hummingbird reference were first confirmed to have functional open reading frames by identifying a corresponding protein prediction from the TransDecoder output. They were then functionally annotated by BLAST (v2.2.31+) (Camacho et al. 2009) to the Swiss-Prot database (Boeckmann 2003) and run through the InterProScan5 (v5.44-79.0) pipeline (Jones et al. 2014).
Differential expression
Differential gene expression was performed with DESeq2 (Love et al. 2014) filtering for genes with at least 10× coverage in at least four of the six samples per tissue. The three fasted samples and three fed samples were compared separately for the liver and muscle tissue, and significantly DEGs were determined with default setting from DESeq2 using adjusted P-values beneath 0.1; however, all DEGs and nonthresholded adjusted P-values can be found in Supplemental Tables S5 and S6. Isoform-level expression was quantified with Ballgown (Frazee et al. 2015), and isoform-level FPKM values were compared across the muscle and liver tissues. Significantly up-regulated pathways were determined with the GeneTonic R package (Marini et al. 2021) for both liver and muscle using a more conservative 0.05 cutoff.
Gene loss analysis
We did not identify the GCK gene in our transcriptome annotation or in the functional annotation of the predicted proteins. As further validation, we used BLAST (v2.2.31+) using the chimney swift (Chaetura pelagica) and chicken (Gallus gallus) GCK gene sequence and protein sequence to both the ruby-throated hummingbird predicted protein set and genome. The BLAST search did not uncover any hits we could determine to be open reading frames. We then aligned the chicken reference genome (GCA_000002315.5) to the ruby-throated hummingbird genome with minimap2 (Li 2016) with the following parameters: minimap2 -x asm20 -c ‐‐eqx. We noted that the region with the GCK gene in the chicken genome is nonsyntenic to any of the ruby-throated hummingbird DNA sequence. The paf output file was processed with rustybam (https://github.com/mrvollger/rustybam) and plotted with SafFire (https://mrvollger.github.io/SafFire/). To further ensure that there was no expression of the GCK gene in the ruby-throated hummingbird, we mapped all the RNA-seq reads to the chimney swift GCK gene sequence with Bowtie 2 (Langmead and Salzberg 2012), allowing for multiple mismatches by using the very-sensitive-local flag. None of the RNA-seq reads mapped to the chimney swift GCK sequence. Lastly, we also validated that the GCK gene was not present in the annotation of Anna's hummingbird either.
To further confirm missing genes (GCK, SLC2A4) in our assembly, we turned to the raw Illumina RNA-seq data and followed the methods outlined by Yin et al. (2019). In brief, we assembled the RNA-seq reads de novo with Trinity, without reliance on a genomic reference (Grabherr et al. 2011). Any transcript being expressed, regardless of whether or not the genomic sequence is in our assembly, should be captured in the Trinity transcriptome. We used TransDecoder (https://github.com/TransDecoder/TransDecoder/releases/tag/TransDecoder-v5.5.0) to identify open reading frames and convert the transcript sequences into protein sequences. We then used BLASTP to identify genes of interest. We BLASTed the Trinity protein assembly against the swift GCK protein sequence and the chicken putative SLC2A4 protein sequence from Huttener et al. (2021). We did not discover significant hits for either gene.
Positive selection analysis
For molecular evolution analyses, we used a consensus tree topology based on molecular phylogenies previously generated (Hackett et al. 2008; Prum et al. 2015; Oliveros et al. 2019). Species were chosen to give outgroups in multiple clades, as well as provide species as a sister lineage for each nectivorous lineage, where such a species existed in the publicly available genome databases. Additionally, species were added that evolved between nectivorous lineages to highlight the convergent nature of the phenotype. Adding lineages between nectivorous lineages allowed us to ensure that the branches we tested for positive selection, to the best of our ability, matched the branches in which the transition to nectivory happened in more species-rich phylogenies. Proteomes for all species selected were downloaded from NCBI and clustered with cd-hit (v4.8.1) with a sequence identity threshold of 98% to remove redundancy in the data sets (Li and Godzik 2006). Orthologous gene groups were generated by running the clustered proteomes through the OrthoFinder (v2.3.12) pipeline (Emms and Kelly 2019). One-to-one orthology groups (OGs) were determined by selecting all single-copy genes that were contained in all species.
For each one-to-one OG, the branch-site test of positive selection was performed using codeml in PAML v4.10 (Yang 2007; https://github.com/abacus-gene/paml) to detect genes under positive selection in nectivorous bird lineages. Using the phylogenetic tree reported in phylogenies previously generated (Hackett et al. 2008; Prum et al. 2015; Oliveros et al. 2019), the topology was unrooted using the ete3 toolkit (Huerta-Cepas et al. 2016), and foreground branches were assigned to the following nectivorous lineages: Grantiella picta, Promerops cafer, Leptocoma aspasia, and the clade of C. anna + A. colubris. A likelihood ratio test (LRT) was performed for each OG, with the branch-site model A (specified in Yang 2007) as the alternative model and model A with a fixed ω = 1 as the null model. LRT statistics were converted to P-values using pchisq in R v.3.5.0 (R Core Team 2018). To provide a conservative estimate of genes under positive selection among nectivorous lineages, each OG with a statistically significant LRT (P ≤ 0.05) was also required to possess at least one site under positive selection with a posterior probability ≥ 0.95 (according to the empirical Bayes analysis of positive selection included in the codeml branch-site test of positive selection). Genes under positive selection are reported in Supplemental Table S3. Intermediate files from this analysis are available on Zenodo (DOI:10.5281/zenodo.6363333).
Protein structure models
Structures for full-length SLC2A5 (isoform X1) and for the alternative spliced variant (isoform X2) were modeled with AlphaFold v2.01 (https://github.com/deepmind/alphafold) using default settings without templates to avoid model bias (Jumper et al. 2021). A reduced version of the BFD database (https://bfd.mmseqs.com/), optimized for speed and lower hardware requirements, was used during multisequence alignment (MSA). The overall confidence measure (predicted local-distance difference test [pLDDT]) for the generated models was >75, which generally indicates good backbone prediction. Atomic models with the highest scores at the overall confidence measure were selected (86.2 for isoform X1 and 84.5 for isoform X2). pLDDT is a per-residue confidence metric and, as such, can be used to monitor how the model confidence varies along the chain. Very low confidence regions (<50) included flexible N- and C-terminal ends that were removed from the models.
Tracer oxidation study
Four ruby-throated hummingbirds were fasted for 1 h and then placed in a 500-mL respirometry container for baseline fasting breath delta 13C breath stable isotope signature and RER recording (for respirometry and breath stable isotope set up, see Dick et al. 2020). After 5 min, the birds were then fed a 150 μL of a 20% sucrose solution with sucrose enriched with 13C on all six carbons of the glucose (sucrose [glucose-13C6, 98%], Cambridge Isotope Laboratories) or fructose (d-sucrose [fructose-13C6, 98%], Cambridge Isotope Laboratories) portion of the sucrose molecule. The birds were fed through a 1-mL syringe in the lid of the respirometry jar, which allowed for continuous breath measurements, and previous training allowed for quick consumption of the sucrose solutions. The time of feeding was recorded and used as t = 0. The respiratory measurements continued over the next 20 min to measure the rise and start of the fall of RER, representing the switch from a fasted, fat-fueled metabolism to a fed, sugar-fueled metabolism. The birds were then returned to their cages and repeated the process again 1 wk later with the other sucrose solution, with two birds starting with fructose-enriched, and two birds starting with the glucose-enriched. RER was analyzed following (Dick et al. 2020) and tracer oxidation rate analyzed following (McCue et al. 2010) and were averaged for each minute over the course of 20 min. The time to peak tracer oxidation rate was analyzed using a paired t-test.
Data access
All RNA and DNA sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA811496. The A. colubris assembly generated in this study has been submitted to the NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome/) under accession number GCA_023079485.1.
Supplementary Material
Acknowledgments
This study was supported by a grant from the Human Frontier Science Program (RGP0062 to M.V., G.W.W., K.C.W., and W.T.).
Author contributions: M.V., G.W.W., K.C.W., and W.T. conceived the study. K.C.W. and W.T. coordinated the collaboration. A.G., M.D., J.M.J., S.M., R.A., and R.E.W. acquired data. A.G., Q.H., M.T., X.A., M.V., K.C.W., and W.T. analyzed and interpreted the data. A.G., Q.H., M.V., G.W.W., K.C.W., and W.T. wrote the paper and interpreted the results.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276779.122.
Competing interest statement
W.T. has two patents (8,748,091 and 8,394,584) licensed to Oxford Nanopore Technologies.
References
- Ali RS, Dick MF, Muhammad S, Sarver D, Hou L, Wong GW, Welch KC Jr. 2020. Glucose transporter expression and regulation following a fast in the ruby-throated hummingbird, Archilochus colubris. J Exp Biol 223: jeb229989. 10.1242/jeb.229989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol 20: 224. 10.1186/s13059-019-1829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin S, Kuloglu T, Aydin S, Eren MN, Yilmaz M, Kalayci M, Sahin I, Kocaman N, Citil C, Kendir Y. 2013. Expression of adropin in rat brain, cerebellum, kidneys, heart, liver, and pancreas in streptozotocin-induced diabetes. Mol Cell Biochem 380: 73–81. 10.1007/s11010-013-1660-4 [DOI] [PubMed] [Google Scholar]
- Baker HG, Baker I, Hodges SA. 1998. Sugar composition of nectars and fruits consumed by birds and bats in the tropics and Subtropics1. Biotropica 30: 559–586. 10.1111/j.1744-7429.1998.tb00097.x [DOI] [Google Scholar]
- Bakken BH, McWhorter TJ, Tsahar E, Del Rio CM. 2004. Hummingbirds arrest their kidneys at night: diel variation in glomerular filtration rate in Selasphorus platycercus. J Exp Biol 207: 4383–4391. 10.1242/jeb.01238 [DOI] [PubMed] [Google Scholar]
- Barone S, Fussell SL, Singh AK, Lucas F, Xu J, Kim C, Wu X, Yu Y, Amlal H, Seidler U, et al. 2009. Slc2a5 (Glut5) is essential for the absorption of fructose in the intestine and generation of fructose-induced hypertension. J Biol Chem 284: 5056–5066. 10.1074/jbc.M808128200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoldo A, Pencek RR, Azuma K, Price JC, Kelley C, Cobelli C, Kelley DE. 2006. Interactions between delivery, transport, and phosphorylation of glucose in governing uptake into human skeletal muscle. Diabetes 55: 3028–3037. 10.2337/db06-0762 [DOI] [PubMed] [Google Scholar]
- Boeckmann B. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370. 10.1093/nar/gkg095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun EJ, Sweazea KL. 2008. Glucose regulation in birds. Comp Biochem Physiol B Biochem Mol Biol 151: 1–9. 10.1016/j.cbpb.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CCW, Welch KC Jr. 2014. Hummingbirds can fuel expensive hovering flight completely with either exogenous glucose or fructose. Funct Ecol 28: 589–600. 10.1111/1365-2435.12202 [DOI] [Google Scholar]
- Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Blomberg LA, Bouffard P, Burt DW, Crasta O, Crooijmans RPMA, et al. 2010. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol 8: e1000475. 10.1371/journal.pbio.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick MF, Alcantara-Tangonan A, Oghli YS, Welch KC. 2020. Metabolic partitioning of sucrose and seasonal changes in fat turnover rate in ruby-throated hummingbirds (Archilochus colubris). J Exp Biol 223: jeb212696. 10.1242/jeb.212696 [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20: 238. 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci 117: 9451–9457. 10.1073/pnas.1921046117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. 2015. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol 33: 243–246. 10.1038/nbt.3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass CL. 1979. Territory regulation, tenure, and migration in rufous hummingbirds. Can J Zool 57: 914–923. 10.1139/z79-112 [DOI] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmo CG. 2010. Move that fatty acid: fuel selection and transport in migratory birds and bats. Integr Comp Biol 50: 336–345. 10.1093/icb/icq097 [DOI] [PubMed] [Google Scholar]
- Guo K, Lukacik P, Papagrigoriou E, Meier M, Lee WH, Adamski J, Oppermann U. 2006. Characterization of human DHRS6, an orphan short chain dehydrogenase/reductase enzyme: a novel, cytosolic type 2 R-β-hydroxybutyrate dehydrogenase. J Biol Chem 281: 10291–10297. 10.1074/jbc.M511346200 [DOI] [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han K-L, Harshman J, et al. 2008. A phylogenomic study of birds reveals their evolutionary history. Science 320: 1763–1768. 10.1126/science.1157704 [DOI] [PubMed] [Google Scholar]
- Hayward BE, Bonthron DT. 1998. Structure and alternative splicing of the ketohexokinase gene. Eur J Biochem 257: 85–91. 10.1046/j.1432-1327.1998.2570085.x [DOI] [PubMed] [Google Scholar]
- Horst KT, Ter Horst K, Serlie M. 2017. Fructose consumption, lipogenesis, and non-alcoholic fatty liver disease. Nutrients 9: 981. 10.3390/nu9090981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33: 1635–1638. 10.1093/molbev/msw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttener R, Thorrez L, Veld TI, Granvik M, Van Lommel L, Waelkens E, Derua R, Lemaire K, Goyvaerts L, De Coster S, et al. 2021. Sequencing refractory regions in bird genomes are hotspots for accelerated protein evolution. BMC Ecol Evol 21: 176. 10.1186/s12862-021-01905-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Jenni L, Jenni-Eiermann S. 1998. Fuel supply and metabolic constraints in migrating birds. J Avian Biol 29: 521. 10.2307/3677171 [DOI] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov WH. 2017. Integrative physiology of transcellular and paracellular intestinal absorption. J Exp Biol 220: 2495–2501. 10.1242/jeb.144048 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee C, Ko BJ, Yoo DA, Won S, Phillippy AM, Fedrigo O, Zhang G, Howe K, Wood J, et al. 2022. False gene and chromosome losses in genome assemblies caused by GC content variation and repeats. Genome Biol 23: 204. 10.1186/s13059-022-02765-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivelä R, Salmela I, Nguyen YH, Petrova TV, Koistinen HA, Wiener Z, Alitalo K. 2016. The transcription factor Prox1 is essential for satellite cell differentiation and muscle fibre-type regulation. Nat Commun 7: 13124. 10.1038/ncomms13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KG, Trevaskis JL, Lam DD, Sutton GM, Koza RA, Chouljenko VN, Kousoulas KG, Rogers PM, Kesterson RA, Thearle M, et al. 2008. Identification of adropin as a secreted factor linking dietary macronutrient intake with energy homeostasis and lipid metabolism. Cell Metab 8: 468–481. 10.1016/j.cmet.2008.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32: 2103–2110. 10.1093/bioinformatics/btw152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38: 4647–4654. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapleson D, Garcia Accinelli G, Kettleborough G, Wright J, Clavijo BJ. 2017. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 33: 574–576. 10.1093/bioinformatics/btw663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14: e1005944. 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi F, Cirillo P, Mateo EC. 2017. Applications of RNA-seq and omics strategies: from microorganisms to human health. IntechOpen, London, UK. [Google Scholar]
- Marini F, Ludt A, Linke J, Strauch K. 2021. GeneTonic: an R/Bioconductor package for streamlining the interpretation of RNA-seq data. BMC Bioinformatics 22: 610. 10.1186/s12859-021-04461-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschinsky FM, Wilson DF. 2019. The central role of glucokinase in glucose homeostasis: a perspective 50 years after demonstrating the presence of the enzyme in islets of Langerhans. Front Physiol 10: 148. 10.3389/fphys.2019.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue MD, Pollock ED. 2013. Measurements of substrate oxidation using 13CO2-breath testing reveals shifts in fuel mix during starvation. J Comp Physiol B 183: 1039–1052. 10.1007/s00360-013-0774-z [DOI] [PubMed] [Google Scholar]
- McCue MD, Sivan O, McWilliams SR, Pinshow B. 2010. Tracking the oxidative kinetics of carbohydrates, amino acids and fatty acids in the house sparrow using exhaled 13CO2. J Exp Biol 213: 782–789. 10.1242/jeb.039842 [DOI] [PubMed] [Google Scholar]
- Mueckler M, Thorens B. 2013. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med 34: 121–138. 10.1016/j.mam.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad S. 2021. Fructose metabolism in the ruby-throated hummingbird (Archilochus colubris). University of Toronto, Toronto, Canada. [Google Scholar]
- Myrka AM, Welch KC Jr. 2018. Evidence of high transport and phosphorylation capacity for both glucose and fructose in the ruby-throated hummingbird (Archilochus colubris). Comp Biochem Physiol B Biochem Mol Biol 224: 253–261. 10.1016/j.cbpb.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Nanda V, Miano JM. 2012. Leiomodin 1, a new serum response factor-dependent target gene expressed preferentially in differentiated smooth muscle cells. J Biol Chem 287: 2459–2467. 10.1074/jbc.M111.302224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N, Verdon G, Kang HJ, Shimamura T, Nomura Y, Sonoda Y, Hussien SA, Qureshi AA, Coincon M, Sato Y, et al. 2015. Structure and mechanism of the mammalian fructose transporter GLUT5. Nature 526: 397–401. 10.1038/nature14909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S-A, Suh Y, Pang M-G, Lee K. 2011. Cloning of avian G(0)/G(1) switch gene 2 genes and developmental and nutritional regulation of G(0)/G(1) switch gene 2 in chicken adipose tissue. J Anim Sci 89: 367–375. 10.2527/jas.2010-3339 [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, Takamatsu S, Minowa MT, Yoshida A, Takeuchi M, Marth JD. 2005. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell 123: 1307–1321. 10.1016/j.cell.2005.09.041 [DOI] [PubMed] [Google Scholar]
- Oliveros CH, Field DJ, Ksepka DT, Barker FK, Aleixo A, Andersen MJ, Alström P, Benz BW, Braun EL, Braun MJ, et al. 2019. Earth history and the passerine superradiation. Proc Natl Acad Sci 116: 7916–7925. 10.1073/pnas.1813206116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G, Pertea M. 2020. GFF utilities: GffRead and GffCompare. F1000Res 9: 304. 10.12688/f1000research.23297.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter A, Stefan N, Cegan A, Walenta M, Wagner S, Königsrainer A, Königsrainer I, Machicao F, Schick F, Häring H-U, et al. 2011. Hepatic glucokinase expression is associated with lipogenesis and fatty liver in humans. J Clin Endocrinol Metab 96: E1126–E1130. 10.1210/jc.2010-2017 [DOI] [PubMed] [Google Scholar]
- Pierce BJ, McWilliams SR, O'Connor TP, Place AR, Guglielmo CG. 2005. Effect of dietary fatty acid composition on depot fat and exercise performance in a migrating songbird, the red-eyed vireo. J Exp Biol 208: 1277–1285. 10.1242/jeb.01493 [DOI] [PubMed] [Google Scholar]
- Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526: 569–573. 10.1038/nature15697 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol 21: 245. 10.1186/s13059-020-02134-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592: 737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AJ, Richter EA. 2005. Skeletal muscle glucose uptake during exercise: how is it regulated? Physiology 20: 260–270. 10.1152/physiol.00012.2005 [DOI] [PubMed] [Google Scholar]
- Seki Y, Sato K, Kono T, Abe H, Akiba Y. 2003. Broiler chickens (Ross strain) lack insulin-responsive glucose transporter GLUT4 and have GLUT8 cDNA. Gen Comp Endocrinol 133: 80–87. 10.1016/S0016-6480(03)00145-X [DOI] [PubMed] [Google Scholar]
- Shestov AA, Liu X, Ser Z, Cluntun AA, Hung YP, Huang L, Kim D, Le A, Yellen G, Albeck JG, et al. 2014. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. eLife 3: e03342. 10.7554/eLife.03342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumate A, Salzberg SL. 2021. Liftoff: accurate mapping of gene annotations. Bioinformatics 37: 1639–1643. 10.1093/bioinformatics/btaa1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumate A, Wong B, Pertea G, Pertea M. 2022. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comput Biol 18: e1009730. 10.1371/journal.pcbi.1009730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson DC, DeFronzo RA. 1990. Indirect calorimetry: methodological and interpretative problems. Am J Physiol 258: E399–E412. 10.1152/ajpendo.1990.258.3.E399 [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. Repeatmasker open-4.0. http://www.repeatmasker.org [accessed October 15, 2019].
- Sotero-Caio CG, Platt RN II, Suh A, Ray DA. 2017. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol Evol 9: 161–177. 10.1093/gbe/evw264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff JS, Lass A, Schupp M. 2021. Biological functions of RBP4 and its relevance for human diseases. Front Physiol 12: 659977. 10.3389/fphys.2021.659977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez RK. 1992. Hummingbird flight: sustaining the highest mass-specific metabolic rates among vertebrates. Experientia 48: 565–570. 10.1007/BF01920240 [DOI] [PubMed] [Google Scholar]
- Suarez RK, Welch KC Jr, Hanna SK, Herrera M LG. 2009. Flight muscle enzymes and metabolic flux rates during hovering flight of the nectar bat, Glossophaga soricina: further evidence of convergence with hummingbirds. Comp Biochem Physiol A Mol Integr Physiol 153: 136–140. 10.1016/j.cbpa.2009.01.015 [DOI] [PubMed] [Google Scholar]
- Suarez RK, Herrera M LG, Welch KC Jr. 2011. The sugar oxidation cascade: aerial refueling in hummingbirds and nectar bats. J Exp Biol 214: 172–178. 10.1242/jeb.047936 [DOI] [PubMed] [Google Scholar]
- Sun L, Zeng X, Yan C, Sun X, Gong X, Rao Y, Yan N. 2012. Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 490: 361–366. 10.1038/nature11524 [DOI] [PubMed] [Google Scholar]
- Sweazea KL, Braun EJ. 2006. Glucose transporter expression in English sparrows (Passer domesticus). Comp Biochem Physiol B Biochem Mol Biol 144: 263–270. 10.1016/j.cbpb.2005.12.027 [DOI] [PubMed] [Google Scholar]
- Turnbull PC, Longo AB, Ramos SV, Roy BD, Ward WE, Peters SJ. 2016. Increases in skeletal muscle ATGL and its inhibitor G0S2 following 8 weeks of endurance training in metabolically different rat skeletal muscles. Am J Physiol Regul Integr Comp Physiol 310: R125–R133. 10.1152/ajpregu.00062.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitting-Seerup K, Sandelin A. 2019. IsoformSwitchAnalyzeR: analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics 35: 4469–4471. 10.1093/bioinformatics/btz247 [DOI] [PubMed] [Google Scholar]
- Vurture GW, Sedlazeck FJ, Nattestad M, Underwood CJ, Fang H, Gurtowski J, Schatz MC. 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33: 2202–2204. 10.1093/bioinformatics/btx153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Künstner A, Searle S, White S, Vilella AJ, Fairley S, et al. 2010. The genome of a songbird. Nature 464: 757–762. 10.1038/nature08819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman DH, Kang L, Ayala JE, Fueger PT, Lee-Young RS. 2011. The physiological regulation of glucose flux into muscle in vivo. J Exp Biol 214: 254–262. 10.1242/jeb.048041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch KC Jr, Chen CCW. 2014. Sugar flux through the flight muscles of hovering vertebrate nectarivores: a review. J Comp Physiol B 184: 945–959. 10.1007/s00360-014-0843-y [DOI] [PubMed] [Google Scholar]
- Welch KC Jr, Suarez RK. 2007. Oxidation rate and turnover of ingested sugar in hovering Anna's (Calypte anna) and rufous (Selasphorus rufus) hummingbirds. J Exp Biol 210: 2154–2162. 10.1242/jeb.005363 [DOI] [PubMed] [Google Scholar]
- Welch KC Jr, Bakken BH, Martinez del Rio C, Suarez RK. 2006. Hummingbirds fuel hovering flight with newly ingested sugar. Physiol Biochem Zool 79: 1082–1087. 10.1086/507665 [DOI] [PubMed] [Google Scholar]
- Welch KC Jr, Allalou A, Sehgal P, Cheng J, Ashok A. 2013. Glucose transporter expression in an avian nectarivore: the ruby-throated hummingbird (Archilochus colubris). PLoS One 8: e77003. 10.1371/journal.pone.0077003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman RE, Myrka AM, Wong GW, Tseng E, Welch KC Jr, Timp W. 2018. Single-molecule, full-length transcript sequencing provides insight into the extreme metabolism of the ruby-throated hummingbird Archilochus colubris. GigaScience 7: 1–12. 10.1093/gigascience/giy009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yin Z-T, Zhu F, Lin F-B, Jia T, Wang Z, Sun D-T, Li G-S, Zhang C-L, Smith J, Yang N, et al. 2019. Revisiting avian “missing” genes from de novo assembled transcripts. BMC Genomics 20: 4. 10.1186/s12864-018-5407-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346: 1311–1320. 10.1126/science.1251385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. 2013. The MaSuRCA genome assembler. Bioinformatics 29: 2669–2677. 10.1093/bioinformatics/btt476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Puiu D, Luo M-C, Zhu T, Koren S, Marçais G, Yorke JA, Dvořák J, Salzberg SL. 2017. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res 27: 787–792. 10.1101/gr.213405.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.