Abstract
Thousands of unannotated small and alternative open reading frames (smORFs and alt-ORFs, respectively) have recently been revealed in mammalian genomes. While hundreds of mammalian smORF- and alt-ORF-encoded proteins (SEPs and alt-proteins, respectively) affect cell proliferation, the overwhelming majority of smORFs and alt-ORFs remain uncharacterized at the molecular level. Complicating the task of identifying the biological roles of smORFs and alt-ORFs, the SEPs and alt-proteins that they encode exhibit limited sequence homology to protein domains of known function. Experimental techniques for the functionalization of these gene classes are therefore required. Approaches combining chemical labeling and quantitative proteomics have greatly advanced our ability to identify and characterize functional SEPs and alt-proteins in high throughput. In this review, we briefly describe the principles of proteomic discovery of SEPs and alt-proteins, then summarize how these technologies interface with chemical labeling for identification of SEPs and alt-proteins with specific properties, as well as in defining the interactome of SEPs and alt-proteins.
Keywords: alt-protein, chemical biology, microprotein, proteogenomics, proteomics, smORF
Introduction
Small open reading frames (smORFs) shorter than 100 codons were previously excluded by genome annotation consortia in order to minimize false positives due to random ORF background in eukaryotic genomes [1]. Similarly, mammalian ORFs initiating at non-AUG start codons [2] or overlapping [3, 4] known protein coding sequences (the latter termed alternative ORFs, or alt-ORFs, which encode alternative proteins or alt-proteins), were not annotated [5]. However, thousands of smORFs and alt-ORFs are translated in mammalian cells [6, 7]. These previously unannotated genes are found in noncoding RNAs, in 5′ and 3′ untranslated regions of mRNAs, and in frame-shifted ORFs overlapping annotated protein-coding sequences [8]. Hundreds of mammalian smORF-encoded polypeptides (SEPs, also termed microproteins, small proteins, and micropeptides) regulate cell proliferation [9, 10], and the biological roles of several dozen human SEPs and alt-proteins have been defined at the molecular level [11]. Dysregulation of or mutations in multiple human smORFs have been shown to promote cancer [12]. However, due to their short lengths and lack of primary sequence homology to proteins of known function, the majority of smORFs and alt-ORFs remain uncharacterized [13].
Three high-throughput technologies have been developed for the identification of smORFs and alt-ORFs [5]: comparative genomics [15], ribosome profiling (RIBO-seq) [7, 9, 16–19], and proteomics coupled with transcriptomic/translatomic databases [13, 20, 21]. RIBO-seq [22, 23] and proteomics [24–26] can both be leveraged in quantitative mode to reveal changes in SEP and alt-protein translation under different conditions, revealing a degree of functional information. However, only proteomics can directly detect SEPs and alt-proteins at the protein level. Importantly, the combination of protein labeling technologies with proteomics can identify SEPs and alt-proteins with specific physical and chemical properties, a level of information inaccessible to genetic methods.
In this review, we summarize the principles of proteomic discovery of SEPs and alt-proteins, then discuss how these platforms can be utilized downstream of chemical labeling and enrichment to profile unannotated SEPs and alt-proteins that exhibit chemical reactivity, regulated synthesis, and subcellular localizations, as well as to identify interaction partners of SEPs and alt-proteins (Figure 1).
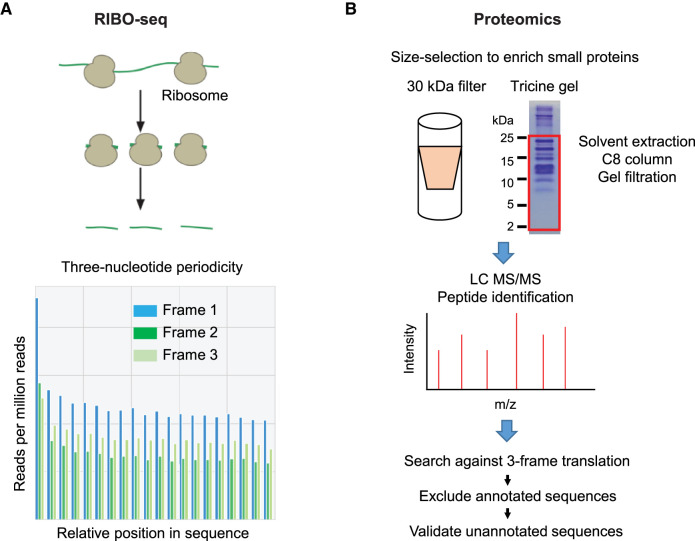
Figure 1. Overview of experimental methods for analysis of SEPs and alt-proteins.
(A) Schematic workflow of RIBO-seq for detection of smORFs and alt-ORFs. Ribosome protected RNA fragments are purified and sequenced. Those smORFs and alt-ORFs exhibiting three-nucleotide periodicity [7, 14] (representative ribosome profiling data shown) are most likely to be translated. (B) Schematic workflow of proteomics for detection of SEPs and alt-proteins. The small proteins are enriched by size selection (representative Coomassie-stained SDS–PAGE gel image shown), followed by proteomics, and the data are searched against a custom database containing canonical proteins as well as all candidate smORFs and alt-ORFs. The annotated tryptic peptide sequences are discarded, and the unannotated tryptic peptide-spectral matches can be validated and mapped to putative SEPs and alt-proteins.
Proteomic identification of SEPs and alt-proteins
In this section, we describe the basics of proteomic approaches for smORF and alt-ORF discovery. SEP and alt-protein proteomics has recently been reviewed [27, 28], and detailed protocols are available [29], so we provide only a brief overview here.
Due to their short lengths, SEPs and alt-proteins typically only generate one, or few, detectable tryptic peptides for liquid chromatography–tandem mass spectrometry (LC–MS/MS) detection, whereas larger proteins generate many peptides [13, 30]. To increase detection sensitivity for SEPs and alt-proteins, small proteins must be enriched after proteome extraction, using methods such as organic solvent extraction [31], peptide gel extraction [30], molecular weight cutoff filtration [13], or solid phase extraction [32]. Then the samples are enzymatically digested [33] and analyzed by LC–MS/MS; top-down LC–MS/MS identification of small proteins has also been reported [27]. Finally, MS/MS data must be searched against a database containing both annotated and unannotated proteins, such as in silico transcriptome translations [34, 35], RIBO-seq-derived translatomes [36, 37], or computationally predicted alt-ORF databases like OpenProt [38]. Peptide-spectral matches to annotated proteins and contaminants must next be discarded. Several methods for discarding hits matches to canonical proteins are available; our laboratory reported a script that removes peptides that are exact matches to annotated human protein sequences, which is followed by BLAST of each candidate peptide in order to ensure that it is at least two amino acids different from canonical proteins, and finally by manual validation of MS/MS spectral quality [13, 29]. An alternative approach is to filter peptide-spectral matches using PepQuery [39], which can exclude false positives due to isobaric amino acids, post-translational and chemical modifications to canonical peptides. It is critical to note that searches of expanded databases, coupled with the uncertainty of individual peptide-spectral matches, can lead to a high rate of false-positive identifications in SEP and alt-protein proteomic searches [40], which must be excluded by the experimenter, as previously described [13, 29]. After the exclusion of canonical proteins, the remaining peptide-spectral matches are candidate hits that can be mapped to SEPs and alt-proteins. Experimental (molecular) validation is ultimately required to confirm a novel SEP or alt-protein.
Principles of chemical proteomics in profiling of SEPs and alt-proteins
Approaches for chemical labeling of cellular proteins with ‘handles’ for proteomic analysis based on specific functions or properties can broadly be placed into three categories: (1) methods to reveal reactivity of amino acid residues using chemically tuned probe molecules (e.g. activity-based protein profiling/ABPP [41]); (2) for metabolic labeling (for example, bio-orthogonal non-canonical amino acid tagging or BONCAT [42]) that can install bio-orthogonal handles into cellular proteins for purification and analysis; and (3) proximity [43] approaches that label cellular proteins in accordance with their subcellular localizations and/or interactions. In the following sections, we will discuss how these tools have been adapted for analysis of SEPs and alt-proteins.
Reactive cysteine profiling for identification of nucleophilic SEPs
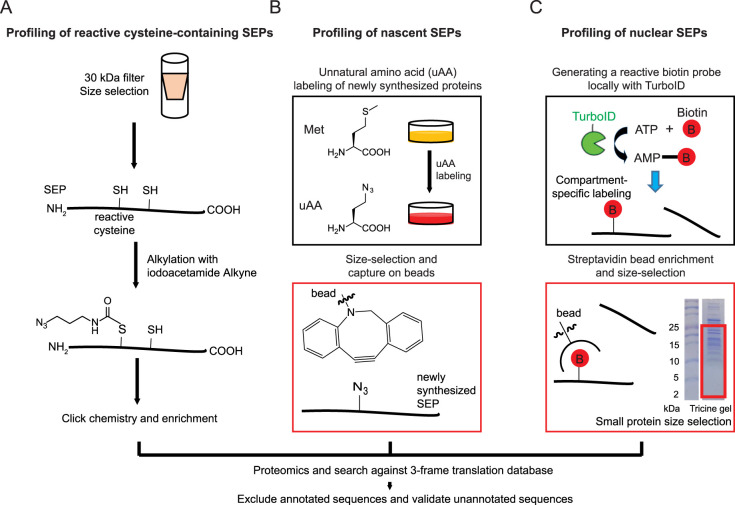
Proteomic identification of nucleophilic cysteine residues — a feature of the active sites of hydrolases and other enzyme classes, as well as many other proteins — using electrophilic probes was first developed for profiling of canonical proteins over a decade ago [44, 45]. Since then, this technique has been utilized to identify cysteine oxidation [46, 47] and post-translational modification [48] events, as well as to develop covalent ligands and inhibitors to cysteine-containing proteins [49, 50]. While SEP monomers are generally too small (<100 amino acids) to fold into enzyme-like structures with complex active sites, it is nonetheless possible that SEPs could harbor nucleophilic cysteine residues. To test this hypothesis, Saghatelian et al. [51] developed a modified method for reactive cysteine profiling in the small proteome. They began with isolating the peptidome by cell lysis and size selection using a 30 kDa molecular weight cutoff filter, then incubated the peptidome with an iodoacetamide (IA)-alkyne probe to label reactive cysteine-containing small proteins, which were then captured using Click chemistry with a biotin derivative and streptavidin pulldown (Figure 2A). The reactive small cysteinome thus captured was subjected to proteomics with custom database searching to identify 16 unannotated, nucleophilic SEPs in K562 cells. While the cellular functions of the SEPs identified in this work were not elucidated, this study provided the first evidence that chemical labeling can reveal reactive smORFs and alt-ORFs. It is possible that additional reactive SEPs remain to be discovered using new workflows, for example using varied size selection methods and electrophilic probes [52].
Figure 2. Chemical proteomic workflows to profile SEPs and alt-proteins.
(A) Schematic workflow of chemical proteomic profiling of reactive cysteine-containing SEPs and alt-proteins [51]. Small proteins were selected with a 30 kDa filter, followed by alkylation with iodoacetamide alkyne, which can specifically label reactive cysteine residues, to generate a handle for further Click chemistry-based enrichment. Coupled with custom smORF/alt-ORF database searching, nucleophilic cysteine-containing SEPs and alt-proteins were be identified. (B) Schematic workflow of bioorthogonal non-canonical amino acid tagging (BONCAT)-based proteomic profiling of SEPs and alt-proteins [53]. An unnatural amino acid (uAA) bearing a bioorthogonal azide moiety was metabolically incorporated into newly synthesized proteins, then the labeled small proteins were size-selected in solution with a C8 column. On-bead Click chemistry captures newly synthesized small proteins and enables removal of unlabeled proteins that did not undergo active synthesis during the labeling period, followed with trypsin digestion and proteomics to identify unannotated SEPs and alt-proteins [53]. (C) Schematic workflow of proximity labeling-based proteomic profiling of SEPs and alt-proteins [54]. TurboID, an engineered biotin ligase, biotinylates lysine residues on proximal proteins within the same subcellular region. To map SEPs and alt-proteins to subcellular compartments, TurboID was expressed in the compartment of interest as a genetic fusion to a localization sequence or protein. All proteins biotinylated by TurboID were enriched by streptavidin pulldown, followed with size-selection for small proteins and in-gel digestion (representative Coomassie-stained SDS–PAGE gel image shown). Finally, unannotated SEPs and alt-proteins were identified by mass spectrometry.
Metabolic labeling of newly synthesized SEPs and alt-proteins
Unnatural amino acids (uAA) that are nearly isosteric to the proteinogenic amino acids can be incorporated into proteins via the cellular protein synthesis machinery [55]. Briefly, the uAA is supplied to living cells, and cellular proteins synthesized during the labeling period incorporate the uAA at all codons corresponding to its natural analog. When the unnatural amino acid bears a bio-orthogonal functional group, the labeled proteome can be captured via Click chemistry with a biotin analog for proteomic identification. One such method, bio-orthogonal noncanonical amino acid tagging, or BONCAT, was developed in 2006 to enable identification of newly synthesized proteins in cells and neurons [42]. An approach for direct detection of newly synthesized SEPs and alt-proteins using a modified BONCAT workflow was recently reported by our group [53] (Figure 2B). Briefly, after uAA labeling, small proteins are enriched using a C8 column [32]. Labeled small proteins are then captured with cyclooctyne-derivatized beads [56], circumventing loss of small proteins and peptides during biotin/streptavidin-based capture [42]. On-bead digest is performed prior to LC–MS/MS analysis and custom database searching. This method revealed 22 SEPs, alternative proteins and N-terminal extensions of annotated proteins [53], stress-regulated translation of nine unannotated small proteins, and cell-cycle regulated synthesis of the alt-protein MINAS-60. We note that the sensitivity of the method is likely currently limited by covalent retention of the uAA residue-containing peptide on the beads. Development of capture-and-release strategies may solve this problem in the future.
Proximity biotinylation reveals subcellular localizations of SEPs and alt-proteins
Proximity labeling is a term describing a suite of technologies that utilize engineered enzymes (e.g. APEX/APEX2 [57–59] or BioID [60]/TurboID [61]) or photocatalysts (e.g. µMap [62]) to locally generate a reactive probe with a short half-life. These high-energy intermediates, which include phenoxy radicals (APEX/APEX2), esters (BioID/TurboID), and carbenes (µMap), can react with residues on nearby proteins and other biomolecules (aromatic sidechains, amines, and C-H bonds, respectively), or, over longer distances, with solvent, which quenches them. The probes bear a biotin or bioorthogonal moiety that permits isolation and proteomic identification of labeled proteins after cell lysis, thus retaining spatial information of where the identified proteins were localized in the intact cell. Depending on the half-life of the reactive probe generated, each proximity labeling technology is associated with a characteristic labeling radius, on the order of tens to hundreds of nanometers [63], a range spanning the sizes of protein complexes to organelles. These technologies have been applied to mapping subcellular and extracellular proteomes in cells and in vivo by targeting the enzyme or catalyst to specific regions of the cell [57, 64–66], as well as to identify protein–protein [43, 67] and protein–RNA [68, 69] interactions.
Because bioinformatic analyses of SEPs and alt-proteins are challenging due to their short lengths, it is difficult to predict their subcellular localizations. At the same time, SEPs and alt-proteins can exhibit specific subcellular localizations via binding to interaction partners despite lacking canonical localization signal sequences. For example, NBDY localizes to P-bodies as a result of its interaction with the mRNA decapping complex [70, 71] and alt-RPL36 partially localizes to endoplasmic reticulum (ER)-plasma membrane junctions due to its interaction with TMEM24 [72]. MRI/CYREN localizes to the nucleus as a result of its interaction with Ku [73–75]. (Since the size limit for passive diffusion through the nuclear pore is ∼30 kDa [76], most SEPs should be able to freely transit the nucleus and cytoplasm, and can be retained at sites of interactions.). Along the same lines, multiple secreted SEPs have been reported, but some do not bear classical signal sequences for secretion [77]. Counterexamples exist, like MINAS-60, which bears a poly-arginine motif near its C-terminus that may behave like a nucleolar localization signal [53, 78]. SEPs and alt-proteins localized to the ER [79], mitochondria [80–84], Golgi [9], and plasma membrane [85, 86] have also been reported in diverse species including yeast, insects, mouse and human, and some, but not all, of these SEPs and alt-proteins bear signal or transmembrane sequences that could allow prediction of the organelle in which they function. Furthermore, while some SEPs are conserved from flies to human (e.g. sarcolamban), and others are conserved in mammals (e.g. NBDY, PIGBOS [79], and many others), conservation of their subcellular localization has rarely been confirmed experimentally. Taken together, the rules that govern SEP and alt-protein subcellular localization may be complex, and experimental techniques may be required to improve the mapping of SEPs and alt-proteins.
Our group adapted TurboID proximity labeling for subcellular mapping of SEPs and alt-proteins, in a pipeline we termed MicroID [54]. To modify previously reported TurboID proteomic workflows for high-sensitivity detection of SEPs and alt-proteins, after proximity biotinylation and streptavidin enrichment, eluted proteins were separated on a Tricine gel, enabling isolation of the low molecular weight (2–25 kDa) proteome. In-gel digestion was followed by mass spectrometry and proteomic identification of unannotated tryptic peptides using a three-frame translated RNA-seq database. A total of 154 unannotated SEPs and alt-proteins were thus identified in the nucleus, nucleolus, nuclear envelope, and chromatin of HEK 293T cells via quantitative comparison to an untargeted (whole-cell) TurboID control. MicroID can also be applied in vivo: nuclear-targeted TurboID identified 96 SEPs and alt-proteins in multiple mouse tissues. In the future, application of MicroID in additional subcellular and extracellular regions, adaptation of other proximity labeling technologies with different residue specificity and labeling radii, and exploration of additional in vivo models may identify the localizations and physiological relevance of many more SEPs and alt-proteins.
Chemical approaches to identify interaction partners of SEPs and alt-proteins
Many SEPs and alt-proteins bind to and regulate the functions of canonical proteins [87, 88]. Identifying interaction partners of SEPs and alt-proteins has therefore proven important in understanding their cellular roles. However, for low-affinity or transient interactions, as well as for low-abundance or membrane-localized interaction partners, detection of SEP/alt-protein binding events can be challenging. Several chemical approaches to improve the identification of SEP and alt-protein interactions have been reported.
In a seminal study, APEX2 was utilized for the discovery of SEP interaction partners. APEX2 is an engineered ascorbate peroxidase that can activate biotin-phenol in the presence of hydrogen peroxide to generate a phenoxy radical, which cross-links to aromatic residues within a ∼20 nm labeling radius due to a 1 ms half-life of the radical [57]. Therefore, fusion of APEX2 to a SEP or alt-protein is expected to preferentially label their close interaction partners with biotin, enabling their purification and identification. Chu and colleagues [89] demonstrated this principle (Figure 3A) by fusing APEX2 to the SEP CYREN/MRI-2, which was previously reported to interact with Ku70/80 [74]. APEX2-mediated biotinylation demonstrated superior quantitative enrichment of known CYREN interaction partners over nonspecific proteins as compared with traditional co-immunopurification. Subsequently, APEX2 tagging was employed to show that the previously uncharacterized C11ORF98 SEP interacts with nucleolar proteins, suggesting that it may function in this organelle. While this study successfully demonstrated that proximity labeling can identify SEP–protein interactions with superior signal to noise relative to non-covalent pull-downs, it is important to note that proximity labeling enzymes are several times larger than most SEPs, and fusions may interfere with SEP localization, interactions and functions.
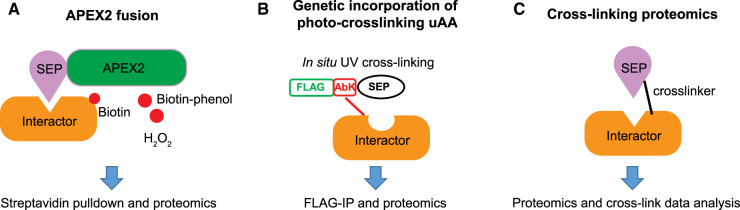
Figure 3. General workflow of chemical labeling methods to identify interaction partners of SEPs and alt-proteins.
(A) Schematic of APEX2 fusion-based proteomics to identify interaction partners of SEPs and alt-proteins. APEX2 is fused to the SEP of interest, and is expressed in cells. In the presence of biotin-phenol and H2O2, APEX2 generates biotin phenoxy radicals, which can label proximal proteins at aromatic amino acid sidechains, enabling their purification with streptavidin and identification with proteomics. (B) Schematic of photo-crosslinking uAA-based proteomics to identify interaction partners of SEPs and alt-proteins. A uAA such as AbK, a lysine analog bearing a diazirine photo-cross-linker, is incorporated into the SEP of interest using amber codon suppression technology in cells; the SEP is also fused to an epitope tag for purification of cross-linked complexes. After photo-irradiation, the diazirine is converted to a reactive carbene, which can insert into C-H bonds on proximal proteins to form a covalent bond. Immunopurification of the cross-linked complexes followed by trypsin digest and mass spectrometry enables identification of interaction partners. (C) Schematic of chemical cross-linking for identification of SEP interaction partners. Bivalent compounds containing two reactive chemical groups bridged by a linker are added to cells or lysates, enabling formation of covalent bonds to nucleophilic amino acid side chains on two interacting proteins, trapping them in a complex. Subsequent trypsin digest, followed by proteomics with smORF/alt-ORF database searching, enables global discovery of SEP and alt-protein complexes in an unbiased manner.
To circumvent artefacts due to tag size, Koh et al. [90] devised an approach to incorporate a photo-cross-linking amino acid into a specific position of a SEP (Figure 3B). This strategy, unnatural or noncanonical amino acid mutagenesis [91], utilizes an evolved mutant of Methanosarcina barkeri pyrrolysyl aminoacyl transfer RNA (tRNA) synthetase, along with an engineered variant of the M. barkeri tRNA that can be charged with an unnatural amino acid, and which bears an amber suppressor anticodon. This system is orthogonal in bacterial and eukaryotic cells, enabling specific incorporation of the unnatural amino acid uniquely at an amber codon strategically placed within the coding sequence of the target protein — in this case, a SEP. The amber suppressor system was used to genetically incorporate a diazirine amino acid, AbK, into multiple SEPs of unknown function. Photoactivation of AbK generates a reactive carbene intermediate, which can insert into C-H and unsaturated C-C bonds. A SEP bearing an AbK residue can be crosslinked to bound proteins; pulldown of the SEP via an affinity tag then permits digest and LC–MS/MS identification of cross-linked interaction partners. Using this approach, Koh and colleagues identified the interactomes of seven SEPs, including one that interacts with histone H2B and localizes to chromatin. This approach represents the minimal tag size that can be genetically incorporated for interaction partner identification, but presents the possible limitation that it requires overexpression of SEP coding sequences programmed with an amber codon. Despite the challenges, two subsequent studies applying unnatural amino acid mutagenesis to fluorescence imaging of SEPs and alt-proteins [72, 92] suggests that amber codon suppression may continue to find broader utility for minimally perturbative SEP and alt-protein labeling.
A third approach that enables covalent capture and identification of interaction partners without the need for overexpression or genetic fusion is cross-linking mass spectrometry (XL-MS) [93] (Figure 3C). In this approach, cell lysates are treated with a membrane-permeable crosslinker, such as a bis-succinimidyl ester, which reacts covalently with two groups (e.g. amines) on interacting proteins, and can contain a linker that fragments (such as sulfoxide) in the mass spectrometer. The cross-linked peptides can then be identified from their unique fragmentation spectra via shotgun proteomics and mapped to protein complexes via analysis with specialized software. This method is global and unbiased, and requires no enrichment of the proteins under study, offering a particularly high-throughput avenue to identify SEP and alt-protein interactions. Two studies have demonstrated that the XL-MS pipeline can be interfaced with proteogenomic database searching. In the first study [94], HeLa cell XL-MS data were reanalyzed against a database containing >200 000 predicted human alternative proteins in addition to the annotated human proteome. Nearly 300 candidate interactions between alt-proteins and canonical proteins were identified, including a candidate interaction between Alt-ATAD2 and ribosomal protein L10. While molecular validation was not provided, the study is notable for its quantitative comparison of database searches: searching the expanded database vs. the human proteome database decreased sensitivity for detection of annotated proteins, suggesting that near-matches to theoretical sequences can decrease peptide-spectral match scores, even for known proteins. In follow-up work [95], the same group subjected glioma cells to a timecourse of forskolin treatment, which promotes signaling associated with epithelial-to-mesenchymal transition. The cell extracts were subjected to XL-MS and complexes between alt-proteins and canonical proteins were dynamically mapped through the treatment. Again, >200 cross-links assigned to alt-protein-canonical protein interactions were identified, and the alt-proteins observed were dynamic at varied timepoints between 0 and 48 h of treatment. Candidate alt-protein-containing cross-links were identified with components of the translation machinery, the cytoskeleton and cell motility machinery. Of particular interest were three alt-proteins putatively interacting with tropomyosin 4: Alt-TRNAU1AP, Alt-EPHA5, and Alt-MAP2. Direct molecular evidence for the cellular expression of the alt-proteins implicated in these analyses will be of importance in the future; in addition, the simultaneity or exclusivity and cellular/phenotypic consequences of these candidate interactions will be interesting to probe using biochemical methods. These studies demonstrate that alt-proteins form complexes inside cells, an idea supported by similar observations in a recent reanalysis of a global interactomics dataset [96]. Overall, proximity labeling, photo-cross-linking, and chemical cross-linking offer complementary advantages to advance our understanding of the interactions of SEPs and alt-proteins.
Discussion
The study of SEPs and alt-proteins has begun to offer new insights into human biology and disease, and it is imperative to determine the molecular and organismal roles of thousands of these novel gene products that are currently uncharacterized.
Proteomics, particularly when coupled with chemical labeling, can provide information about the presence and abundance of SEPs and alt-proteins in the cell as well as their physical, chemical and functional properties. Such approaches have already been developed to report SEP and alt-protein reactivity, synthesis, localization, and interactions. In the future, there are clear avenues to improve on existing workflows for SEP and alt-protein functional proteomics, including application of new small protein extraction and enrichment methods up- or downstream of chemical labeling protocols. In addition, adaptation of other chemical labeling methods for nascent proteins, post-translational modifications and subcellular localizations, as well as analysis of secreted SEPs and alt-proteins, holds the potential to provide new insights into the functions of these species. Finally, while dysregulation and mutation of SEPs and alt-proteins contribute to human diseases, chemical labeling and proteomic analysis of SEPs and alt-proteins has not yet been applied in the disease context; doing so should provide key insights into the molecular mechanism of SEPs and alt-proteins in disease.
However, several key challenges remain. First, the proteoform [97] diversity of SEPs and alt-proteins is likely to be complex, as it is for canonical proteins. While multiple isoforms of some SEPs, like CYREN/MRI [74], have been documented, it is likely that alternative splicing of many other SEP and alt-protein transcripts generates isoform diversity that is currently almost entirely unexplored. Furthermore, while phosphorylation of multiple SEPs and alt-proteins has been reported [71, 72], few other SEP/alt-protein post-translational modifications have been examined, and the stoichiometry and site occupancy of post-translational modifications on intact SEP/alt-protein proteoforms is essentially unknown. While these questions are challenging to address even for canonical proteins, an understanding of the functional repertoire of SEPs and alt-proteins will require an accounting of all of their intact proteoforms in the cell. Intact proteoform and modification state diversity are impossible to fully map using bottom-up proteomics. Top-down proteomics is uniquely suited to addressing this question, and current efforts to advance this technology for SEP analysis offer an optimistic outlook for SEP proteoform identification in the future.
A second major issue for the field is the double-edged sword of sensitivity vs. false positive identifications. Proteomics experiments typically detect fewer alt-proteins and SEPs than ribosome profiling, likely a result of the stochastic nature of data-dependent acquisition, reliance on one or few tryptic peptides for detection of SEPs, and the challenge of detecting some classes of proteins [24] using bottom-up proteomics. However, a more fundamental challenge arises from peptide-spectral matching via database search. Expanded translatome or transcriptome databases are required for proteomic identification of SEPs and alt-proteins. These databases are several times larger than the canonical proteome, and contain many entries that do not correspond to bona fide cellular SEPs and alt-proteins; adding variable post-translational or chemical modifications to the search increases the size of the theoretical database yet again. As a result, false positive matches of experimentally acquired spectra to ORFs that are not really expressed occur [40, 98], a problem further exacerbated when modifications are included. At the same time, Fournier and colleagues showed that true positive identifications of canonical proteins are also decreased when searching expanded databases with stringent false discovery rates, perhaps due to near-matches to theoretical sequences that are not the reverse of real protein sequences but are present in the decoy database [95]. To solve these problems in the future, it will be essential to develop cell type specific translatome resources curated for expressed SEPs and alt-proteins in addition to the annotated proteome — or, better yet, to update the human proteome annotation with SEPs and alt-proteins with evidence for expression.
Perspectives
Small and alternative open reading frames (smORFs and alt-ORFs) were previously excluded from the human genome annotation, but are now known to encode thousands of small proteins with potential biological functionality and disease relevance. Identification and functional study of these unannotated small proteins represents a major opportunity to gain insights into biology.
Chemical labeling coupled with proteomic identification has begun to provide insight into the synthesis, localization, reactivity and interactions of smORF and alt-ORF-encoded proteins; some of these properties are challenging or impossible to investigate using genomic or computational technologies.
Proteomic detection of smORF and alt-ORF-encoded proteins still faces challenges including false positives, false negatives, and intact proteoform mapping, which can be solved with improved databases and mass spectrometric technologies in the future.
Acknowledgements
This work was supported by a Mark Foundation for Cancer Research Emerging Leader Award, a Paul G. Allen Frontiers Group Distinguished Investigator Award, and a Sloan Research Fellowship (FG-2022-18417) (to S.A.S.). X.C. was supported by Shanghai Pujiang Program (22PJ1402600), and in part by a Rudolph J. Anderson postdoctoral fellowship from Yale University. K.H.L. was supported in part by a NIH Pathway to Independence Award (4 R00DK129712-03).
Abbreviations
- BONCAT
bioorthogonal non-canonical amino acid tagging
- ER
endoplasmic reticulum
- MS
mass spectrometry
- smORFs
small and alternative open reading frames
- tRNA
transfer RNA
- uAA
unnatural amino acids
- SEP
smORF-encoded polypeptide
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Open Access
Open access for this article was enabled by the participation of Yale University in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with Individual.
References
- 1.Basrai, M.A., Hieter, P. and Boeke, J.D. (1997) Small open reading frames: beautiful needles in the haystack. Genome Res. 7, 768–771 10.1101/gr.7.8.768 [DOI] [PubMed] [Google Scholar]
- 2.Cao, X. and Slavoff, S.A. (2020) Non-AUG start codons: expanding and regulating the small and alternative ORFeome. Exp. Cell Res. 391, 111973 10.1016/j.yexcr.2020.111973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunet, M.A., Levesque, S.A., Hunting, D.J., Cohen, A.A. and Roucou, X. (2018) Recognition of the polycistronic nature of human genes is critical to understanding the genotype-phenotype relationship. Genome Res. 28, 609–624 10.1101/gr.230938.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright, B.W., Molloy, M.P. and Jaschke, P.R. (2022) Overlapping genes in natural and engineered genomes. Nat. Rev. Genet. 23, 154–168 10.1038/s41576-021-00417-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr, M.W., Mao, Y., Storz, G. and Qian, S.B. (2020) Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res. 48, 1029–1042 10.1093/nar/gkz734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mudge, J.M., Ruiz-Orera, J., Prensner, J.R., Brunet, M.A., Calvet, F., Jungreis, I.et al. (2022) Standardized annotation of translated open reading frames. Nat. Biotechnol. 40, 994–999 10.1038/s41587-022-01369-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez, T.F., Chu, Q., Donaldson, C., Tan, D., Shokhirev, M.N. and Saghatelian, A. (2020) Accurate annotation of human protein-coding small open reading frames. Nat. Chem. Biol. 16, 458–468 10.1038/s41589-019-0425-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couso, J.P. and Patraquim, P. (2017) Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 18, 575–589 10.1038/nrm.2017.58 [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., Brunner, A.D., Cogan, J.Z., Nunez, J.K., Fields, A.P., Adamson, B.et al. (2020) Pervasive functional translation of noncanonical human open reading frames. Science (1979) 367, 1140–1146 10.1126/science.aay0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prensner, J.R., Enache, O.M., Luria, V., Krug, K., Clauser, K.R., Dempster, J.M.et al. (2021) Noncanonical open reading frames encode functional proteins essential for cancer cell survival. Nat. Biotechnol. 39, 697–704 10.1038/s41587-020-00806-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright, B.W., Yi, Z., Weissman, J.S. and Chen, J. (2022) The dark proteome: translation from noncanonical open reading frames. Trends Cell Biol. 32, 243–258 10.1016/j.tcb.2021.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merino-Valverde, I., Greco, E. and Abad, M. (2020) The microproteome of cancer: from invisibility to relevance. Exp. Cell Res. 392, 111997 10.1016/j.yexcr.2020.111997 [DOI] [PubMed] [Google Scholar]
- 13.Slavoff, S.A., Mitchell, A.J., Schwaid, A.G., Cabili, M.N., Ma, J., Levin, J.Z.et al. (2013) Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64 10.1038/nchembio.1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazzini, A.A., Johnstone, T.G., Christiano, R., Mackowiak, S.D., Obermayer, B., Fleming, E.S.et al. (2014) Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 33, 981–993 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemm, M.R., Paul, B.J., Schneider, T.D., Storz, G. and Rudd, K.E. (2008) Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 70, 1487–1501 10.1111/j.1365-2958.2008.06495.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meydan, S., Marks, J., Klepacki, D., Sharma, V., Baranov, P.V., Firth, A.E.et al. (2019) Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell 74, 481–493.e6 10.1016/j.molcel.2019.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakahigashi, K., Takai, Y., Kimura, M., Abe, N., Nakayashiki, T., Shiwa, Y.et al. (2016) Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res. 23, 193–201 10.1093/dnares/dsw008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeremy, W., Fuad M, R.B.A. and Gisela, S. (2019) Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10, e02819-18 10.1128/mBio.02819-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia, N.T., Brar, G.A., Stern-Ginossar, N., Harris, M.S., Talhouarne, G.J., Jackson, S.E.et al. (2014) Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8, 1365–1379 10.1016/j.celrep.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffe, J.D., Berg, H.C. and Church, G.M. (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59–77 10.1002/pmic.200300511 [DOI] [PubMed] [Google Scholar]
- 21.Vanderperre, B., Lucier, J.F., Bissonnette, C., Motard, J., Tremblay, G., Vanderperre, S.et al. (2013) Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS ONE 8, e70698 10.1371/journal.pone.0070698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, R., Kroehling, L., Khitun, A., Bailis, W., Jarret, A., York, A.G.et al. (2018) The translation of non-canonical open reading frames controls mucosal immunity. Nature 564, 434–438 10.1038/s41586-018-0794-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sendoel, A., Dunn, J.G., Rodriguez, E.H., Naik, S., Gomez, N.C., Hurwitz, B.et al. (2017) Translation from unconventional 5′ start sites drives tumour initiation. Nature 541, 494–499 10.1038/nature21036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan, P., D'Lima, N.G. and Slavoff, S.A. (2018) Comparative membrane proteomics reveals a nonannotated E. coli heat shock protein. Biochemistry 57, 56–60 10.1021/acs.biochem.7b00864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Lima, N.G., Khitun, A., Rosenbloom, A.D., Yuan, P., Gassaway, B.M., Barber, K.W.et al. (2017) Comparative proteomics enables identification of nonannotated cold shock proteins in E. coli. J. Proteome Res. 16, 3722–3731 10.1021/acs.jproteome.7b00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao, X., Khitun, A., Na, Z., Dumitrescu, D.G., Kubica, M., Olatunji, E.et al. (2020) Comparative proteomic profiling of unannotated microproteins and alternative proteins in human cell lines. J. Proteome Res. 19, 3418–3426 10.1021/acs.jproteome.0c00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cassidy, L., Kaulich, P.T., Maaß, S., Bartel, J., Becher, D. and Tholey, A. (2021) Bottom-up and top-down proteomic approaches for the identification, characterization, and quantification of the low molecular weight proteome with focus on short open reading frame-encoded peptides. Proteomics 21, 2100008 10.1002/pmic.202100008 [DOI] [PubMed] [Google Scholar]
- 28.Ahrens, C.H., Wade, J.T., Champion, M.M. and Langer, J.D. (2022) A practical guide to small protein discovery and characterization using mass spectrometry. J. Bacteriol. 204, e0035321 10.1128/jb.00353-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khitun, A. and Slavoff, S.A. (2019) Proteomic detection and validation of translated small open reading frames. Curr. Protoc. Chem. Biol. 11, e77 10.1002/cpch.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, J., Ward, C.C., Jungreis, I., Slavoff, S.A., Schwaid, A.G., Neveu, J.et al. (2014) Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J. Proteome Res. 13, 1757–1765 10.1021/pr401280w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cassidy, L., Kaulich, P.T. and Tholey, A. (2019) Depletion of high-molecular-mass proteins for the identification of small proteins and short open reading frame encoded peptides in cellular proteomes. J. Proteome Res. 18, 1725–1734 10.1021/acs.jproteome.8b00948 [DOI] [PubMed] [Google Scholar]
- 32.Ma, J., Diedrich, J.K., Jungreis, I., Donaldson, C., Vaughan, J., Kellis, M.et al. (2016) Improved identification and analysis of small open reading frame encoded polypeptides. Anal. Chem. 88, 3967–3975 10.1021/acs.analchem.6b00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaulich, P.T., Cassidy, L., Bartel, J., Schmitz, R.A. and Tholey, A. (2021) Multi-protease approach for the improved identification and molecular characterization of small proteins and short open reading frame-encoded peptides. J. Proteome Res. 20, 2895–2903 10.1021/acs.jproteome.1c00115 [DOI] [PubMed] [Google Scholar]
- 34.Oyama, M., Itagaki, C., Hata, H., Suzuki, Y., Izumi, T., Natsume, T.et al. (2004) Analysis of small human proteins reveals the translation of upstream open reading frames of mRNAs. Genome Res. 14, 2048–2052 10.1101/gr.2384604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyama, M., Kozuka-Hata, H., Suzuki, Y., Semba, K., Yamamoto, T. and Sugano, S. (2007) Diversity of translation start sites may define increased complexity of the human short ORFeome. Mol. Cell. Proteomics 6, 1000–1006 10.1074/mcp.M600297-MCP200 [DOI] [PubMed] [Google Scholar]
- 36.Van Damme, P., Gawron, D., Van Criekinge, W. and Menschaert, G. (2014) N-terminal proteomics and ribosome profiling provide a comprehensive view of the alternative translation initiation landscape in mice and men. Mol Cell Proteomics 13, 1245–1261 10.1074/mcp.M113.036442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouspenskaia, T., Law, T., Clauser, K.R., Klaeger, S., Sarkizova, S., Aguet, F.et al. (2022) Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat Biotechnol. 40, 209–217 10.1038/s41587-021-01021-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunet, M.A., Lucier, J.F., Levesque, M., Leblanc, S., Jacques, J.F., Al-Saedi, H.R.H.et al. (2021) Openprot 2021: deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Res. 49, D380–D388 10.1093/nar/gkaa1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen, B., Wang, X. and Zhang, B. (2019) Pepquery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res. 29, 485–493 10.1101/gr.235028.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, K., Fu, Y., Zeng, W.-F., He, K., Chi, H., Liu, C.et al. (2015) A note on the false discovery rate of novel peptides in proteogenomics. Bioinformatics 31, 3249–3253 10.1093/bioinformatics/btv340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cravatt, B.F., Wright, A.T. and Kozarich, J.W. (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 10.1146/annurev.biochem.75.101304.124125 [DOI] [PubMed] [Google Scholar]
- 42.Dieterich, D.C., Link, A.J., Graumann, J., Tirrell, D.A. and Schuman, E.M. (2006) Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl Acad. Sci. U.S.A. 103, 9482–9487 10.1073/pnas.0601637103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin, W., Cho, K.F., Cavanagh, P.E. and Ting, A.Y. (2021) Deciphering molecular interactions by proximity labeling. Nat. Methods 18, 133–143 10.1038/s41592-020-01010-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weerapana, E., Wang, C., Simon, G.M., Richter, F., Khare, S., Dillon, M.B.D.et al. (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 10.1038/nature09472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weerapana, E., Simon, G.M. and Cravatt, B.F. (2008) Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 4, 405–407 10.1038/nchembio.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kovalyova, Y., Bak, D.W., Gordon, E.M., Fung, C., Shuman, J.H.B., Cover, T.L.et al. (2022) An infection-induced oxidation site regulates legumain processing and tumor growth. Nat. Chem. Biol. 18, 698–705 10.1038/s41589-022-00992-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng, X., Weerapana, E., Ulanovskaya, O., Sun, F., Liang, H., Ji, Q.et al. (2013) Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe 13, 358–370 10.1016/j.chom.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, C., Weerapana, E., Blewett, M.M. and Cravatt, B.F. (2014) A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85 10.1038/nmeth.2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crowley, V.M., Thielert, M. and Cravatt, B.F. (2021) Functionalized scout fragments for site-specific covalent ligand discovery and optimization. ACS Cent. Sci. 7, 613–623 10.1021/acscentsci.0c01336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Backus, K.M., Correia, B.E., Lum, K.M., Forli, S., Horning, B.D., González-Páez, G.E.et al. (2016) Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 10.1038/nature18002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwaid, A.G., Shannon, D.A., Ma, J., Slavoff, S.A., Levin, J.Z., Weerapana, E.et al. (2013) Chemoproteomic discovery of cysteine-containing human short open reading frames. J. Am. Chem. Soc. 135, 16750–3 10.1021/ja406606j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abbasov, M.E., Kavanagh, M.E., Ichu, T.-A., Lazear, M.R., Tao, Y., Crowley, V.M.et al. (2021) A proteome-wide atlas of lysine-reactive chemistry. Nat. Chem. 13, 1081–1092 10.1038/s41557-021-00765-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao, X., Khitun, A., Harold, C.M., Bryant, C.J., Zheng, S.J., Baserga, S.J.et al. (2022) Nascent alt-protein chemoproteomics reveals a pre-60S assembly checkpoint inhibitor. Nat. Chem. Biol. 18, 643–651 10.1038/s41589-022-01003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Na, Z., Dai, X., Zheng, S.-J., Bryant, C.J., Loh, K.H., Su, H.et al. (2022) Mapping subcellular localizations of unannotated microproteins and alternative proteins with MicroID. Mol. Cell 82, 2900–2911.e7 10.1016/j.molcel.2022.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saleh, A.M., Wilding, K.M., Calve, S., Bundy, B.C. and Kinzer-Ursem, T.L. (2019) Non-canonical amino acid labeling in proteomics and biotechnology. J. Biol. Eng. 13, 43 10.1186/s13036-019-0166-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jewett, J.C., Sletten, E.M. and Bertozzi, C.R. (2010) Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 132, 3688–3690 10.1021/ja100014q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hung, V., Udeshi, N.D., Lam, S.S., Loh, K.H., Cox, K.J., Pedram, K.et al. (2016) Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 10.1038/nprot.2016.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hung, V., Zou, P., Rhee, H.W., Udeshi, N.D., Cracan, V., Svinkina, T.et al. (2014) Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 55, 332–341 10.1016/j.molcel.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhee, H.W., Zou, P., Udeshi, N.D., Martell, J.D., Mootha, V.K., Carr, S.A.et al. (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science (1979) 339, 1328–1331 10.1126/science.1230593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roux, K.J., Kim, D.I., Raida, M. and Burke, B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Branon, T.C., Bosch, J.A., Sanchez, A.D., Udeshi, N.D., Svinkina, T., Carr, S.A.et al. (2018) Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 10.1038/nbt.4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geri, J.B., Oakley, J.V., Reyes-Robles, T., Wang, T., McCarver, S.J., White, C.H.et al. (2020) Microenvironment mapping via Dexter energy transfer on immune cells. Science (1979) 367, 1091–1097 10.1126/science.aay4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oakley, J.V., Buksh, B.F., Fernández, D.F., Oblinsky, D.G., Seath, C.P., Geri, J.B.et al. (2022) Radius measurement via super-resolution microscopy enables the development of a variable radii proximity labeling platform. Proc. Natl Acad. Sci. U.S.A. 119, e2203027119 10.1073/pnas.2203027119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loh, K.H., Stawski, P.S., Draycott, A.S., Udeshi, N.D., Lehrman, E.K., Wilton, D.K.et al. (2016) Proteomic analysis of unbounded cellular compartments: synaptic clefts. Cell 166, 1295–1307.e21 10.1016/j.cell.2016.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Go, C.D., Knight, J.D.R., Rajasekharan, A., Rathod, B., Hesketh, G.G., Abe, K.T.et al. (2021) A proximity-dependent biotinylation map of a human cell. Nature 595, 120–124 10.1038/s41586-021-03592-2 [DOI] [PubMed] [Google Scholar]
- 66.Youn, J.-Y., Dunham, W.H., Hong, S.J., Knight, J.D.R., Bashkurov, M., Chen, G.I.et al. (2018) High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e11 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
- 67.Liu, X., Salokas, K., Weldatsadik, R.G., Gawriyski, L. and Varjosalo, M. (2020) Combined proximity labeling and affinity purification−mass spectrometry workflow for mapping and visualizing protein interaction networks. Nat. Protoc. 15, 3182–3211 10.1038/s41596-020-0365-x [DOI] [PubMed] [Google Scholar]
- 68.Fazal, F.M., Han, S., Parker, K.R., Kaewsapsak, P., Xu, J., Boettiger, A.N.et al. (2019) Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178, 473–490.e26 10.1016/j.cell.2019.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaewsapsak, P., Shechner, D.M., Mallard, W., Rinn, J.L. and Ting, A.Y. (2017) Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. eLife 6, e29224 10.7554/eLife.29224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Na, Z., Luo, Y., Schofield, J.A., Smelyansky, S., Khitun, A., Muthukumar, S.et al. (2020) The NBDY microprotein regulates cellular RNA decapping. Biochemistry 59, 4131–4142 10.1021/acs.biochem.0c00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Na, Z., Luo, Y., Cui, D.S., Khitun, A., Smelyansky, S., Loria, J.P.et al. (2021) Phosphorylation of a human microprotein promotes dissociation of biomolecular condensates. J. Amer. Chem. Soc. 143, 12675–12687 10.1021/jacs.1c05386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao, X., Khitun, A., Luo, Y., Na, Z., Phoodokmai, T., Sappakhaw, K.et al. (2021) Alt-RPL36 downregulates the PI3K-AKT-mTOR signaling pathway by interacting with TMEM24. Nat. Commun. 12, 508 10.1038/s41467-020-20841-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hung, P.J., Johnson, B., Chen, B.-R., Byrum, A.K., Bredemeyer, A.L., Yewdell, W.T.et al. (2018) MRI is a DNA damage response adaptor during classical non-homologous end joining. Mol. Cell 71, 332–342.e8 10.1016/j.molcel.2018.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slavoff, S.A., Heo, J., Budnik, B.A., Hanakahi, L.A. and Saghatelian, A. (2014) A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 289, 10950–10957 10.1074/jbc.C113.533968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arnoult, N., Correia, A., Ma, J., Merlo, A., Garcia-Gomez, S., Maric, M.et al. (2017) Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 10.1038/nature24023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Timney, B.L., Raveh, B., Mironska, R., Trivedi, J.M., Kim, S.J., Russel, D.et al. (2016) Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 215, 57–76 10.1083/jcb.201601004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez, T.F., Lyons-Abbott, S., Bookout, A.L., De Souza E, V., Donaldson, C., Vaughan, J.M.et al. (2023) Profiling mouse brown and white adipocytes to identify metabolically relevant small ORFs and functional microproteins. Cell Metab. 35, 166–183.e11 10.1016/j.cmet.2022.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scott, M.S., Boisvert, F.-M., McDowall, M.D., Lamond, A.I. and Barton, G.J. (2010) Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. 38, 7388–7399 10.1093/nar/gkq653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chu, Q., Martinez, T.F., Novak, S.W., Donaldson, C.J., Tan, D., Vaughan, J.M.et al. (2019) Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 10, 4883 10.1038/s41467-019-12816-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rathore, A., Chu, Q., Tan, D., Martinez, T.F., Donaldson, C.J., Diedrich, J.K.et al. (2018) MIEF1 microprotein regulates mitochondrial translation. Biochemistry 57, 5564–5575 10.1021/acs.biochem.8b00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang, N., Li, F., Zhang, M., Zhou, H., Chen, Z., Ma, X.et al. (2021) An upstream open reading frame in phosphatase and tensin homolog encodes a circuit breaker of lactate metabolism. Cell Metab. 33, 128–144.e9 10.1016/j.cmet.2020.12.008 [DOI] [PubMed] [Google Scholar]
- 82.Zhang, S., Reljić, B., Liang, C., Kerouanton, B., Francisco, J.C., Peh, J.H.et al. (2020) Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat. Commun. 11, 1312 10.1038/s41467-020-14999-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Makarewich, C.A., Baskin, K.K., Munir, A.Z., Bezprozvannaya, S., Sharma, G., Khemtong, C.et al. (2018) MOXI is a mitochondrial micropeptide that enhances fatty acid β-oxidation. Cell Rep. 23, 3701–3709 10.1016/j.celrep.2018.05.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee, C.Q.E., Kerouanton, B., Chothani, S., Zhang, S., Chen, Y., Mantri, C.K.et al. (2021) Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. Nat. Commun. 12, 2130 10.1038/s41467-021-22397-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bi, P., Ramirez-Martinez, A., Li, H., Cannavino, J., McAnally, J.R., Shelton, J.M.et al. (2017) Control of muscle formation by the fusogenic micropeptide myomixer. Science (1979) 356, 323–327 10.1126/science.aam9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vakirlis, N., Acar, O., Hsu, B., Castilho Coelho, N., Van Oss, S.B., Wacholder, A.et al. (2020) De novo emergence of adaptive membrane proteins from thymine-rich genomic sequences. Nat. Commun. 11, 781 10.1038/s41467-020-14500-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saghatelian, A. and Couso, J.P. (2015) Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 11, 909–916 10.1038/nchembio.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sandmann, C.-L., Schulz, J.F., Ruiz-Orera, J., Kirchner, M., Ziehm, M., Adami, E.et al. (2023) Evolutionary origins and interactomes of human, young microproteins and small peptides translated from short open reading frames. Mol. Cell 83, 994–1011.e18 10.1016/j.molcel.2023.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chu, Q., Rathore, A., Diedrich, J.K., Donaldson, C.J., Yates, III, J.R. and Saghatelian, A. (2017) Identification of microprotein-protein interactions via APEX tagging. Biochemistry 56, 3299–3306 10.1021/acs.biochem.7b00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koh, M., Ahmad, I., Ko, Y., Zhang, Y., Martinez, T.F., Diedrich, J.K.et al. (2021) A short ORF-encoded transcriptional regulator. Proc. Natl Acad. Sci. U.S.A. 118, e2021943118 10.1073/pnas.2021943118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shandell, M.A., Tan, Z. and Cornish, V.W. (2021) Genetic code expansion: a brief history and perspective. Biochemistry 60, 3455–3469 10.1021/acs.biochem.1c00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lafranchi, L., Schlesinger, D., Kimler, K.J. and Elsässer, S.J. (2020) Universal single-residue terminal labels for fluorescent live cell imaging of microproteins. J. Am. Chem. Soc. 142, 20080–7 10.1021/jacs.0c09574 [DOI] [PubMed] [Google Scholar]
- 93.Yu, C. and Huang, L. (2018) Cross-linking mass spectrometry: an emerging technology for interactomics and structural biology. Anal. Chem. 90, 144–165 10.1021/acs.analchem.7b04431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cardon, T., Salzet, M., Franck, J. and Fournier, I. (2019) Nuclei of HeLa cells interactomes unravel a network of ghost proteins involved in proteins translation. Biochim. Biophys. Acta Gen. Subj. 1863, 1458–1470 10.1016/j.bbagen.2019.05.009 [DOI] [PubMed] [Google Scholar]
- 95.Cardon, T., Franck, J., Coyaud, E., Laurent, E.M.N., Damato, M., Maffia, M.et al. (2020) Alternative proteins are functional regulators in cell reprogramming by PKA activation. Nucleic Acids Res. 48, 7864–7882 10.1093/nar/gkaa277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leblanc, S., Brunet, M.A., Jacques, J.-F., Lekehal, A.M., Duclos, A., Tremblay, A.et al. (2022) Newfound coding potential of transcripts unveils missing members of human protein communities. Genomics Proteomics Bioinformatics 29, S1672-0229(22)00124-3 10.1016/j.gpb.2022.09.008 [DOI] [PubMed] [Google Scholar]
- 97.Aebersold, R., Agar, J.N., Amster, I.J., Baker, M.S., Bertozzi, C.R., Boja, E.S.et al. (2018) How many human proteoforms are there? Nat. Chem. Biol. 14, 206–214 10.1038/nchembio.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cargile, B.J., Bundy, J.L. and Stephenson, Jr, J.L. (2004) Potential for false positive identifications from large databases through tandem mass spectrometry. J. Proteome Res. 3, 1082–1085 10.1021/pr049946o [DOI] [PubMed] [Google Scholar]