Abstract
SARS-CoV-2 causes profound changes in the sense of smell, including total smell loss. Although these alterations are often transient, many patients with COVID-19 exhibit olfactory dysfunction that lasts months to years. Although animal and human autopsy studies have suggested mechanisms driving acute anosmia, it remains unclear how SARS-CoV-2 causes persistent smell loss in a subset of patients. To address this question, we analyzed olfactory epithelial samples collected from 24 biopsies, including 9 patients suffering from objectively quantified long-term smell loss following COVID-19. This biopsy-based approach revealed a diffuse infiltrate of T cells expressing interferon-gamma, and a shift in myeloid cell population composition including enrichment of CD207+ dendritic cells and depletion of anti-inflammatory M2 macrophages. Despite the absence of detectable SARS-CoV-2 RNA or protein, gene expression in the barrier supporting cells of the olfactory epithelium, termed sustentacular cells, appeared to reflect a response to ongoing inflammatory signaling, which was accompanied by a reduction in the number of olfactory sensory neurons relative to epithelial sustentacular cells. These data indicate that T-cell mediated inflammation persists in the olfactory epithelium long after SARS-CoV-2 has been eliminated from the tissue, suggesting a mechanism underlying long-term post-COVID smell loss.
One Sentence Summary:
Single cell analysis of olfactory tissue biopsies from patients with hyposmia suggest T cell-mediated mechanisms for post-acute COVID-19 sequelae.
Accessible Summary:
Hyposmia due to COVID-19 can last months to years. The pathobiology associated with lasting hyposmia is not clear, and effective treatment strategies are unavailable. Here, olfactory mucosa biopsies from subjects with persistent hyposmia following COVID-19 were obtained, and were analyzed using single cell RNA-sequencing and immunohistochemistry. Versus controls, post-COVID-19 hyposmic samples contained fewer olfactory sensory neurons and accompanying unresolved local immune cell alterations, including the presence of T cells expressing interferon-gamma, enrichment of CD207+ dendritic cells, and depletion of M2 macrophages. Our findings suggest mechanisms in the peripheral olfactory system underlying long-term post-COVID smell loss.
INTRODUCTION
Anosmia, the loss of the sense of smell, occurs in a majority of individuals with COVID-19, but may persist following recovery (1–5). It is thought that SARS-CoV-2 causes anosmia by affecting the olfactory epithelium, the peripheral organ for olfaction lining the olfactory cleft of the nasal cavity. The olfactory epithelium houses the primary olfactory sensory neurons responsible for detecting odors, a barrier supporting cell layer composed of sustentacular cells, and a population of basal stem or progenitor cells that continuously renew the olfactory epithelium (6–10). Commonly, patches of respiratory epithelium are interspersed within the olfactory cleft region, comprised of secretory cells, ciliated cells and basal cells. Olfactory sensory neurons detect volatile odors via olfactory receptors localized to the neuronal cilia in the nasal airspace (11). Transient gene expression changes in olfactory sensory neurons, alterations in the character of the mucus layer surrounding their cilia, and inflammation are thought to cause acute anosmia in animal models of SARS-CoV-2 infection (12). Importantly, work in both animal models and in human autopsy tissues demonstrates that sustentacular cells rather than neurons are infected by the virus (13, 14). Consistent with sustentacular cells representing a primary site of infection, polymorphisms in the UGT2A1/UGT2A2 locus, whose gene product is expressed in sustentacular cells, are associated with elevated risk of COVID-19-related acute loss of smell or taste (15). It is thought that, in most patients, after viral clearance the normal epithelial reparative processes reconstitute the sustentacular cell population (and any incidentally damaged neurons), restoring function (8).
However, it remains unclear what prevents recovery in the subset of individuals with COVID-19 with lasting olfactory function loss. There are several non-mutually-exclusive possibilities, including severe initial epithelial damage that diminishes or eliminates the basal stem cell pools that normally reconstitute the neuroepithelium. Other possibilities include infiltration of the olfactory epithelium by immune cell populations such that neuroinflammation or autoimmune phenomena perturb normal olfactory function and homeostasis through alterations in gene expression or other means, or central mechanisms that cause derangements in the olfactory bulbs of the brain or olfactory cortex. Examination of human autopsy tissue derived from patients who died from acute sequelae of COVID-19 reveal persistent infection of sustentacular cells, a lack of infection of olfactory sensory neurons, intact epithelial anatomy, and diverse molecular changes in olfactory sensory neurons that could lead to changes in smell detection, although smell was not assessed in any of these patients (12, 16). Whereas these findings suggest mechanisms relevant to acute COVID-19-related loss of smell, to date there has been no direct examination of olfactory tissue (including single cell RNA-sequencing) from humans suffering from long term olfactory dysfunction, which is one of the hallmark symptoms of post-acute sequelae of COVID-19 (PASC). Here, we obtained olfactory epithelium biopsies from 9 individuals with lasting PASC-related olfactory loss, defined by objective olfactory testing, and used immunohistochemistry and single cell RNA-sequencing (scRNA-seq) to identify cellular and transcriptional alterations associated with PASC-related olfactory dysfunction. Controls included newly obtained normosmic biopsies, mucus samples, and our published control scRNA-seq data sets for a total of 44 patient samples (table S1 and S2).
RESULTS
Histological assessment of PASC hyposmic olfactory mucosa suggests a role for T cell infiltrates
We performed an initial immunohistochemical assessment of olfactory epithelium biopsies from non-COVID normosmic controls, post-COVID normosmic controls, or post-COVID hyposmic individuals, obtained from patients undergoing trans-nasal endoscopic surgery as described previously (9). Biopsies lacking olfactory epithelium often occur if a disease process has destroyed the olfactory epithelium entirely, or if, due to sampling error, the tissue was harvested from an area normally lined by respiratory surface epithelium. In our samples, staining for the neuronal marker TUJ1 confirmed the presence of olfactory neurons, verifying the capture of olfactory epithelium rather than non-sensory respiratory epithelium (Fig. 1A).
Fig. 1. T cell infiltrates in olfactory epithelium of nasal biopsies from PASC hyposmic patients.
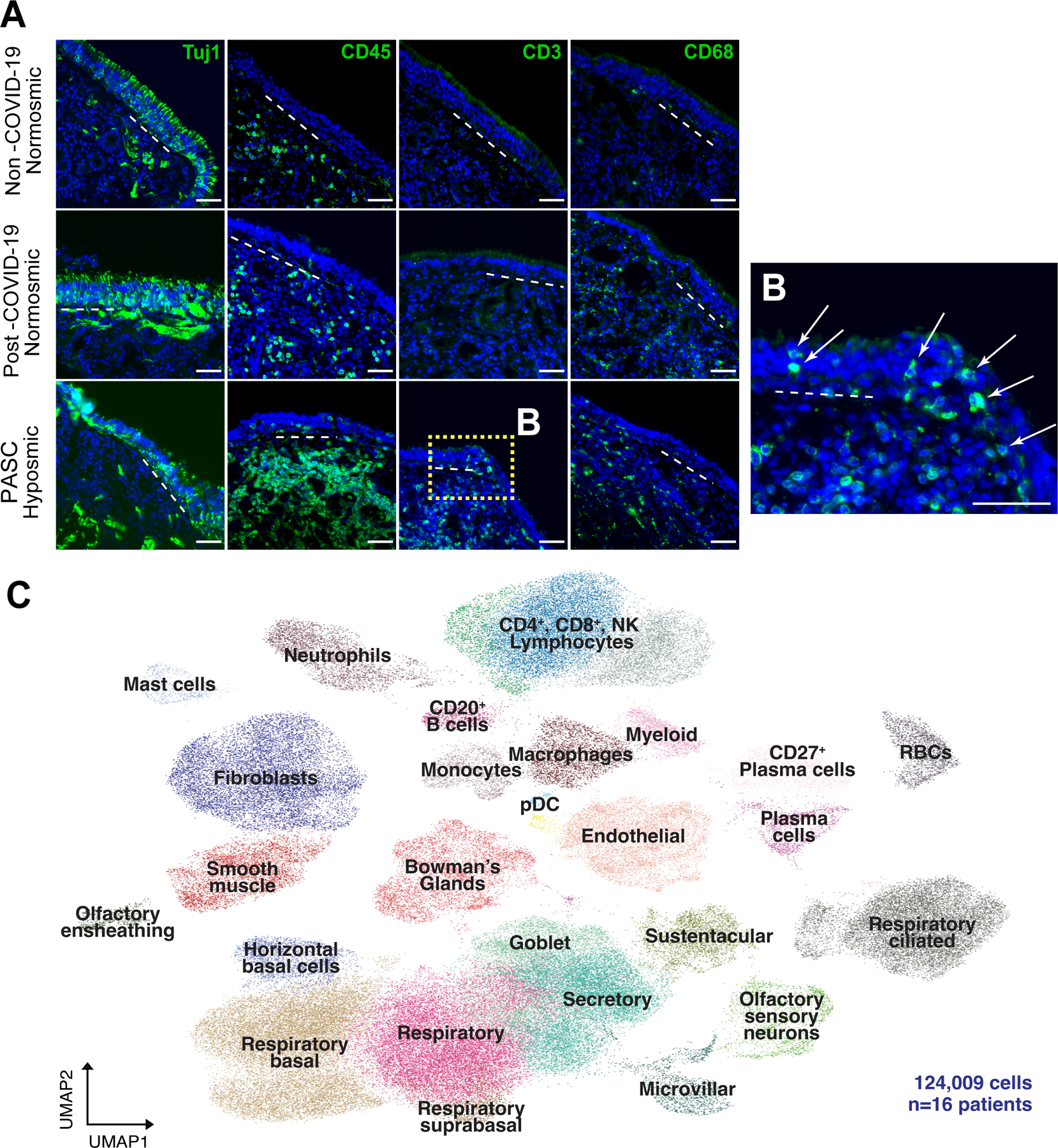
(A) Shown are representative immunohistochemistry images of nasal biopsy tissue from normosmic non-COVID-19, normosmic post-COVID-19 or PASC hyposmic individuals. Tissue sections were immunostained for the TUJ1 neuronal marker, CD45 pan-immune cell marker, CD3 T cell marker and CD68 myeloid cell marker. PASC hyposmic tissue showed dense CD45+ immune cell infiltration including prominent CD3+ lymphocytic infiltration, which was absent in the normosmic groups; scattered CD68+ cells were present in all conditions. (B) Enlarged area (dashed yellow box) shows CD3+ lymphocytes, with prominent epithelial infiltration (arrows); dashed white lines mark the basal lamina. Scale bar, 50 μm. (C) Because of these observations, additional biopsies were processed for scRNA-seq to permit quantitative analyses; uniform manifold approximation projection (UMAP) visualization of combined PASC hyposmic and control normosmic scRNA-seq datasets integrating n=16 human nasal biopsies permitted robust cell cluster analysis and annotation. Red blood cells (RBCs), plasmacytoid dendritic cell (pDC).
Despite our failure to detect SARS-CoV-2 by staining using a validated antibody to the nucleocapsid protein, we observed widespread infiltration of CD45+ immune cells in post-COVID hyposmic mucosa but not in either control group (table S1, Fig. 1A and fig. S1). Myeloid cells (as assessed by CD68 positivity) appeared similar in PASC hyposmic and normosmic control tissues. Interestingly, T cells, identified by CD3 expression, appeared more widespread in PASC hyposmic samples, and many of these were localized within the upper layers of the epithelium itself, rather than confined to the deeper stroma as was observed in control tissue (Fig. 1B). We failed to observe cells expressing these same markers in the post-COVID-19 normosmic samples or non-COVID-19 normosmic samples.
Olfactory biopsy analysis using scRNA-seq
Given the presence of T cell infiltration into the PASC epithelium, we obtained additional nasal biopsy samples from subjects reporting olfactory dysfunction persisting for at least 4 months since the onset of COVID-19 for analysis by single cell sequencing (scSeq) (table S1). We pre-operatively confirmed hyposmia in these patients using a well-validated smell identification test (5, 17, 18); many of our patients also reported subjectively some component of parosmia, or distorted odor perceptions. Endoscopic olfactory mucosa biopsies were obtained either in the otolaryngology clinic using a surgical or cytology brush technique, or in the operating room in subjects undergoing unrelated trans-sphenoidal procedures to access the pituitary for benign disease (fig. S2). Sinusitis or other known sinonasal disease was excluded by endoscopic exam and imaging, ruling out bacterial infection, edema, polyposis. None of the patients were acutely ill or subject to prior medical interventions e.g., prolonged intubation.
Biopsies were processed immediately for scRNA-seq analysis, as we have described previously (9, 19). Samples for scRNA-seq included biopsies from 6 PASC hyposmic individuals (age range 22–58 years; 5 female, 1 male, 4–16 months post-COVID-19 onset). We did not recover transcripts from SARS-CoV-2 in our biopsies, consistent with the absence of ongoing infection or persistent viral RNAemia (20, 21). For comparison, we analyzed 3 normosmic control samples (age range 51–71 years; 2 female, 1 male); to bolster these control data, we combined them with our additional previously published datasets from normosmic and presbyosmic patients to generate an integrated single-cell sequencing dataset from a total of 16 individuals, permitting robust cluster annotation from >124,000 cells (Fig. 1C). Uniform manifold approximation projection (UMAP) plots confirmed that the expected distribution of olfactory, respiratory and immune cells were captured for analysis.
Specific T cell subpopulations are enriched in PASC hyposmic olfactory samples
Given our observation of a T cell infiltrate in the olfactory epithelium samples, we asked whether there were any quantitative changes in specific immune cell populations residing in the PASC hyposmic olfactory epithelium. Based upon the expression of canonical markers (22), we were able to identify a wide variety of T cell subtypes present in both the control and PASC hyposmic olfactory epithelium (Fig. 2A). In particular there was enrichment of resident CD8+ T cells (CD8 Tres) belonging to cluster 5 (herein referred to as CD8 Tres 5) (Fig. 2B), which we identified based upon immune marker gene expression as γδ T cells (23) (Fig. 2C, D). γδ T cells, found in surface epithelia, exhibit diverse properties with functional specializations dependent upon their cytokine production, location, and activation state (24). Whereas specific roles for γδ T cells in COVID-19 remain unclear, potential functions in the PASC hyposmic olfactory epithelium may involve protection of epithelial barrier function, tissue remodeling, or an ongoing role in modulating immune responses in the context of severe prior damage and the need for epithelial repair (23–25).
Fig. 2. A CD8+ T cell subset is enriched in PASC hyposmic nasal biopsies.
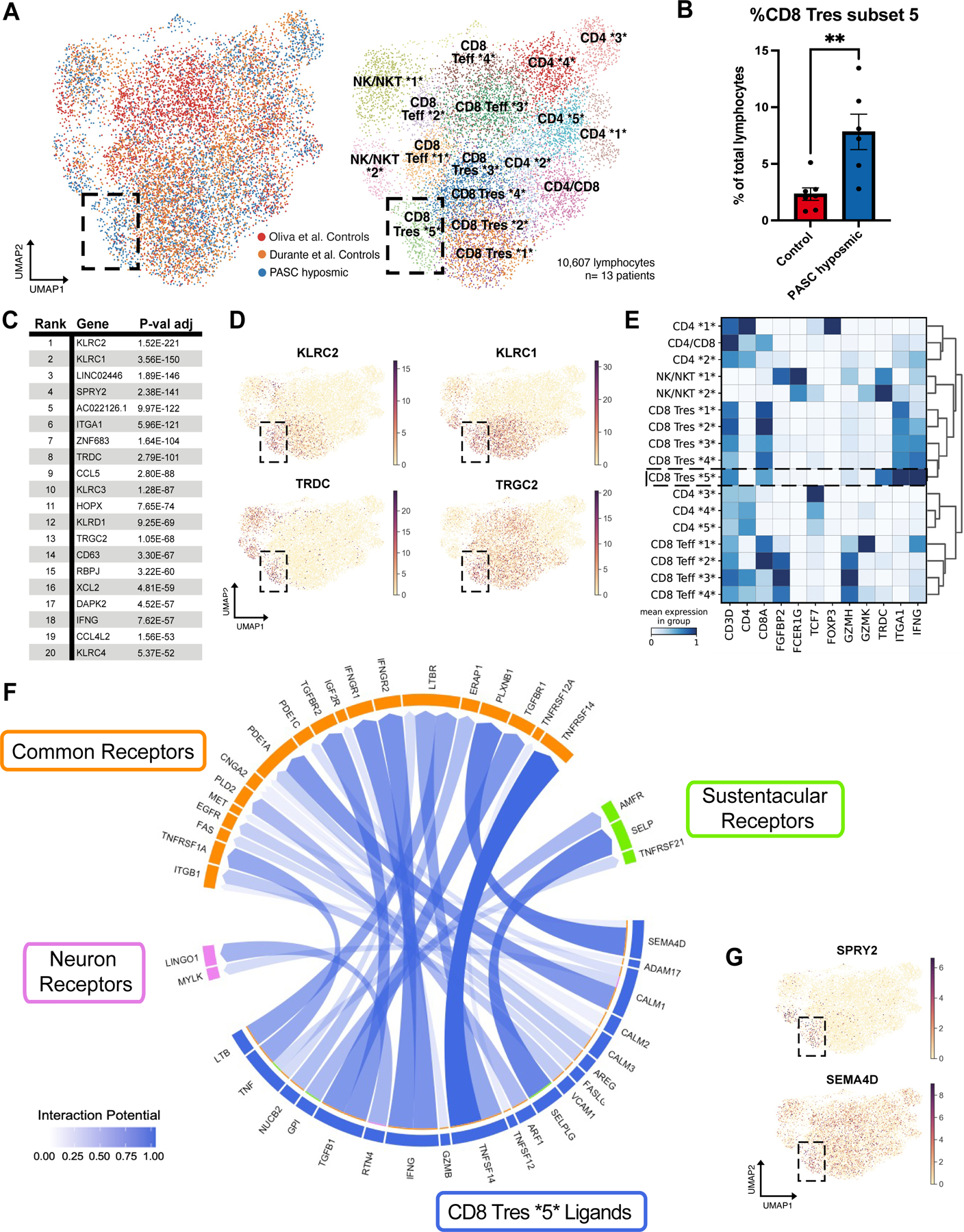
(A) UMAP visualization of all lymphocytes from CD8+, CD4+, and NK/NKT cell clusters from olfactory biopsy scRNA-seq data, comparing PASC hyposmic samples to 2 available normosmic control samples; “Oliva et al Controls” (19) were normosmic by Smell Identification Test scores, while the “Durante et al Controls” (9) were normosmic by subjective report. Black box denotes clusters enriched in PASC hyposmic samples. Teff, effector T cells; Tres, resident T cells; subsets within categories are designated by numbers (i.e. CD4 *1* thru CD4 *5*). (B) PASC hyposmic biopsies show enrichment for cell cluster CD8 Tres subset 5, compared to both control datasets (two-tailed t-test, P=0.0015). (C) Top ranked transcripts enriched in CD8 Tres subset 5 cluster (by adjusted P-value, Wilcoxon Rank-sum test, with Bonferroni correction). (D) Selected gene expression plots of significantly enriched genes in the PASC hyposmic-specific cluster CD8 Tres subset 5 (black box), including γδ T cell markers and associated genes (TRDC, TRGC2, KLRC1, KLRC2). (E) Selected gene expression and dendrogram clustering among lymphocyte subsets confirming annotations based on published marker genes. The IFNG gene is enriched in Tres subsets, especially the CD8 Tres subset 5. (F) Circos plot showing NicheNet analysis of CD8 Tres-derived ligands and their receptors in either sustentacular cells or olfactory sensory neurons, depicting interaction potential. Common receptors in orange are present in both sustentacular cells and neurons. (G) Additional plots confirm PASC hyposmic γδ T cells express T cell ligands identified by differential gene expression (SPRY2) or NicheNet analysis (SEMA4D).
Of interest, the γδ T cell cluster identified here expresses the inflammatory cytokine interferon-γ (Fig. 2E). Interferon-γ (type II interferon) mediates immunomodulatory responses and adaptive immunity. We did not identify any cell type that expressed type I interferons (interferon-α and interferon-β), which are associated with acute viral infection (fig. S3). Importantly, gene expression analysis of sustentacular cells and olfactory sensory neurons revealed expression of receptors for interferon-γ, in addition to expression of receptors for a number of other signaling ligands expressed by CD8 Tres 5 cells (Fig. 2F, G). Immunohistochemistry using antibody to γδ T cell receptor confirmed prominent γδ T cell infiltrates in PASC hyposmic olfactory epithelium (fig. S1). These observations demonstrate that the PASC olfactory epithelium harbors a unique population of γδ T cells that express interferon-γ, and that both sustentacular cells and olfactory sensory neurons express the relevant cognate receptors to respond to this (and other) inflammation-related ligands.
In a separate cohort of PASC hyposmic samples (n=13 patients, Table S2) and control samples (n=7 individuals), olfactory mucus was assayed to measure cytokines and chemokines (fig. S4). Consistent with a lack of severe cytotoxic inflammation, there were no markedly elevated changes in IL-1β or TNFα. Of interest, IFN-λ1, a pro-inflammatory cytokine, was upregulated in PASC hyposmic mucus (P<0.05, unpaired t test with Welch’s correction). Also, IFN-α2, a type I interferon involved in antiviral immunity trended towards upregulation in PASC hyposmic samples (P=0.0506, unpaired t test with Welch’s correction). IP-10 (CXCL10), which is directly stimulated by IFN-γ and was observed to be upregulated at 31 days post-infection in a hamster model of olfactory SARS-CoV-2 infection (21), was also upregulated in these samples (P <0.05, unpaired t test with Welch’s correction).
A shift in myeloid cell populations accompanies T cell alterations in PASC hyposmic olfactory epithelium
Given the enrichment of γδ T cells in PASC olfactory epithelium, we focused further attention on the myeloid lineage, which can coordinate alterations in lymphocyte populations. Anti-CD68 staining demonstrated the presence of myeloid cells in both control and PASC olfactory epithelium (Fig. 1A). Analysis of the myeloid clusters by scRNA-seq identified multiple subpopulations, which corresponded to different macrophage, monocyte and antigen-presenting dendritic cell (DC) subtypes (Fig. 3A–C). Dendritic cells can be segregated based on expression of CD207 (LANGERIN), with the CD207+ population exhibiting enriched expression of CCR6 and TLR10 (Fig. 3A, C); quantification revealed a shift towards CD207+ DCs in the PASC hyposmic olfactory epithelium, with a corresponding decrease in the CD207− population (Fig. 3D). Immunostaining confirmed the presence of CD207+ DCs in PASC hyposmic olfactory biopsies (Fig. 3E). Although CD207 is well known for its functional role within skin Langerhans cells, CD207+ DCs are broadly distributed, and are thought to survey tissues and coordinate immune responses including T cell activation (26, 27). Intriguingly, TLR10, a protein enriched in CD207+ DCs, plays a role in myeloid cell detection of influenza virus and subsequent organization of the immune response (28). Thus, the enrichment of CD207+ DC cells may help to orchestrate the immune infiltration seen in PASC hyposmic olfactory epithelium samples.
Fig. 3. PASC hyposmic olfactory samples show a myeloid cell shift with enriched dendritic cell subsets and decreased M2 macrophages.
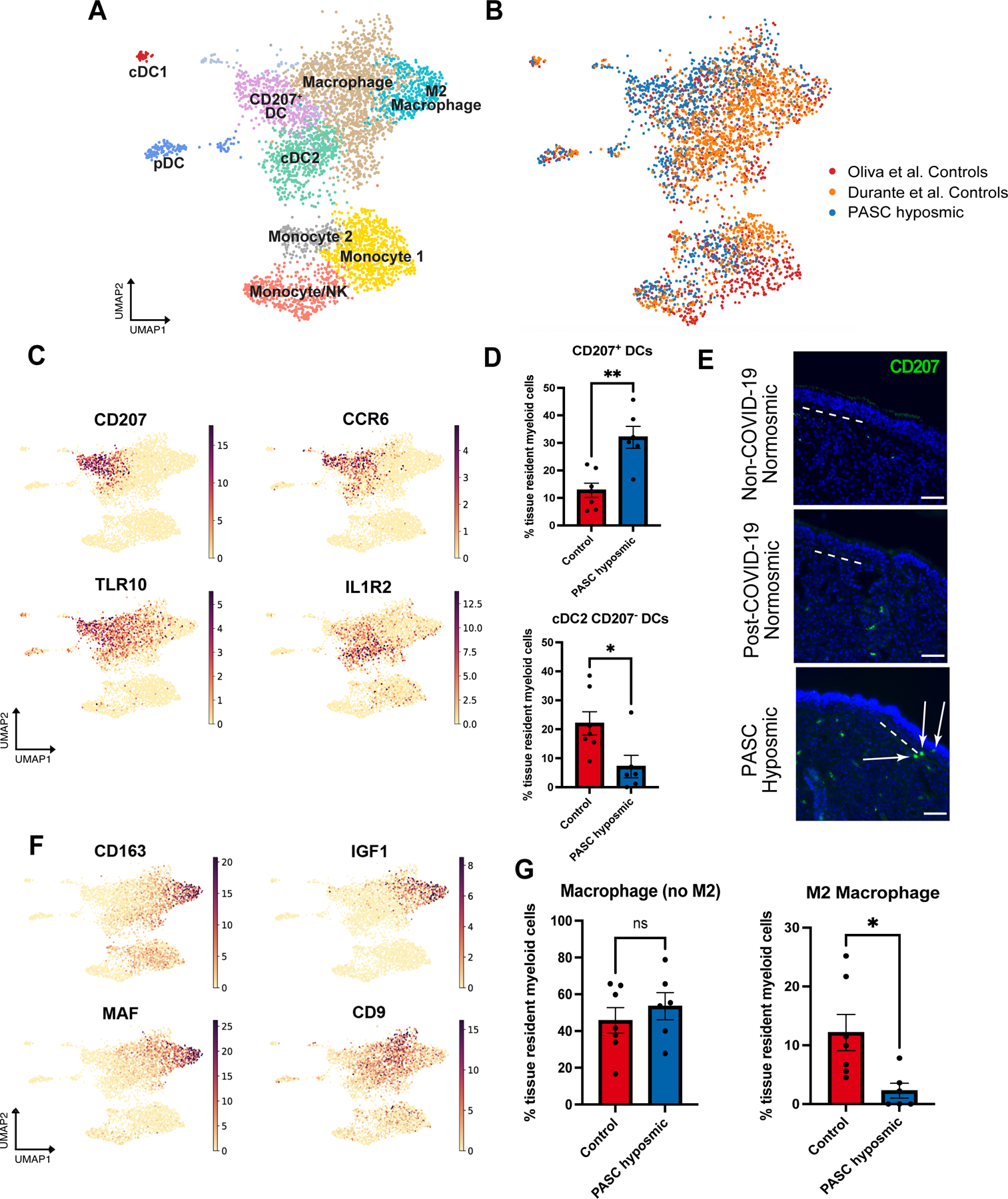
(A) UMAP visualization of olfactory biopsy scRNA-seq data for selected myeloid cell clusters, comparing PASC hyposmic to both normosmic control data sets. DC, dendritic cells; pDC, plasmacytoid dendritic cells (CLEC4C+); cDC1, conventional DC type 1 cells (CLEC9A+); cDC2, conventional DC type 2 cells (CLEC10A+). (B) UMAP plots in panel A, colored by sample contribution. (C) Selected gene expression plots confirm cluster annotation for CD207+ dendritic cells and additional marker genes obtained by differential gene expression analysis for the CD207+ dendritic cell cluster (CCR6 and TLR10) and the cDC2 cluster (IL1R2). (D) CD207+ dendritic cells (DCs) are enriched in PASC hyposmic biopsies (blue) compared to normosmic control (non-COVID-19) biopsies (red) (two-tailed t-test, P=0.0016; CD207− DCs are reduced, P=0.0236). (E) Anti-CD207 antibody staining confirmed CD207+ dendritic cells in PASC hyposmic nasal biopsies (white arrows); dashed white lines mark the basal lamina. Scale bar, 50 μm. (F) Gene expression plots indicate marker genes for M2 macrophage (CD163, MAF, IGF1) and total macrophage (CD9) clusters. (G) PASC hyposmic biopsies showed depletion of M2 macrophages relative to all resident myeloid cells (two-tailed t-test, P=0.0175).
Analysis of macrophage clusters revealed a distinct population of anti-inflammatory M2 macrophages marked by CD163 (Fig. 3A, F). Whereas there was no difference in the relative numbers of CD163− macrophages, we observed significantly fewer M2 macrophages in PASC hyposmic olfactory epithelium (Fig. 3G, P=0.0175, two-tailed T-test). A reduction in M2 macrophages is of interest, as this population can promote tissue repair via several mechanisms, especially given that the M2 macrophage population observed here produces IGF1, a known growth factor for olfactory sensory neurons (29, 30). This relative reduction in the M2 macrophages in the PASC hyposmic olfactory epithelium might also serve as a readout for the presence of ongoing pro-inflammatory signaling, biasing macrophages away from the M2 identity.
Sustentacular cells exhibit an immune response phenotype in PASC hyposmic olfactory samples
Sustentacular cells express coronavirus entry genes, and SARS-CoV-2 has been found to infect this cell population during acute COVID-19 (12–14). Sustentacular cells have multiple functions as the apical barrier cell lining the olfactory epithelium, including detoxification of harmful chemicals via robust expression of biotransformation enzymes, modulation of the ion content of the mucus layer in which OSN cilia are embedded, and feedback regulation of olfactory epithelium neuronal stem cells (31, 32). Based upon expression of canonical sustentacular markers such as ERMN (9), CYP2A13, and GPX6, we identified 779 high quality sustentacular cells from PASC hyposmic or control samples (Fig. 4A). We observed strong expression in sustentacular cells of UGT2A1, a gene shown by genome-wide association study (GWAS), to convey elevated risk of olfactory loss in COVID-19 (15).
Fig. 4. Sustentacular cell gene expression changes persist in PASC hyposmic olfactory epithelium.
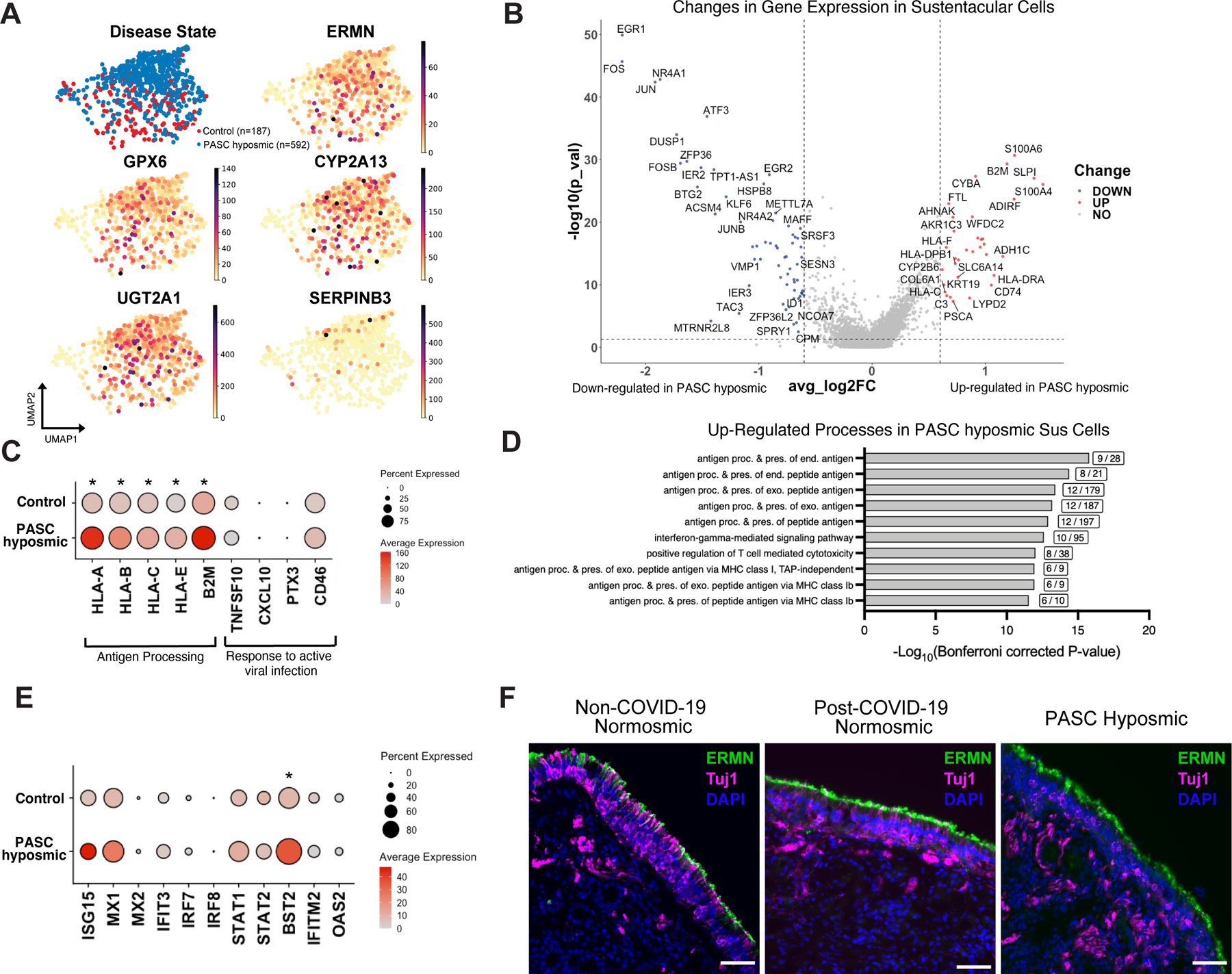
(A) UMAP visualization of sustentacular cell subset from olfactory biopsy scRNA-seq data sets including cells from PASC hyposmic (blue) and non-COVID-19 control normosomic (red) samples (n=9 biopsies; 6 PASC hyposmic, 3 non-COVID-19 normosmic). Gene expression plots show expression of selected canonical sustentacular cell markers (ERMN, GPX6, CYP2A13) and minimal expression of the respiratory marker SERPINB3. (B) Volcano plot showing differential gene expression in sustentacular cells in PASC hyposmic compared to control normosmic olfactory epithelium biopsies; red or blue indicates significant change >0.6 log2 fold change, P<0.05). (C) Visualization of expression of selected antigen presentation genes and genes normally involved in responses to active viral infection in sustentacular cells in PASC hyposmic and control normosmic (non-COVID-19) samples (all antigen presentation genes were significantly upregulated in PASC hyposmic samples, with adjusted P value *<0.05 by Wilcoxon Rank-Sum). (D) Gene set enrichment analysis of the transcripts upregulated in PASC hyposmic sustentacular cells in (C) identifies a variety of biological processes, including “antigen presentation” and “interferon gamma signaling” (boxed figures indicate number of altered genes per process term; only significant processes are included, based on −log10(Bonferroni corrected P value)); Sus, sustentacular cell. (E) Differential gene expression of pathogen response genes previously identified in sustentacular cells in hamsters 31 days after SARS-CoV-2 infection or in uninfected hamsters (21) (BST2, an interferon-induced host gene encoding a transmembrane protein with antiviral and inflammatory signaling activities, adjusted P value <0.05; STAT1, a transcription factor downstream of inflammatory signaling, adjusted P value <0.1, Wilcoxon Rank-Sum; see also fig. S5; *=P<0.05). (F) Immunohistochemistry depicting representative labeling comparing the sustentacular cell marker ERMN (green) and the neuronal marker TUJ1 (magenta) in normosmic (non-COVID-19) control biopsies and PASC hyposmic nasal biopsies. Nuclei are stained with DAPI (blue). Scale bar, 50 μm.
Differential gene expression analysis identified marked transcriptional alterations between PASC hyposmic and control sustentacular cells (Fig. 4B, fig. S5). Consistent with sustentacular cells mounting a response to inflammation, antigen presentation genes were enriched in sustentacular cells derived from PASC hyposmic samples (Fig. 4C and fig. S5B–D), with minimal changes in typical markers of active viral infection such as CXCL10, PTX3, or CD46. Consistent with the absence of acute viral responses, we did not detect SARS-CoV-2 transcripts in scRNA-seq biopsy samples from PASC hyposmic patients aligned to the viral reference genome. Gene set enrichment analysis of the transcripts significantly upregulated in PASC hyposmic samples (log2 fold change >0.6, P<0.05) identified several biological processes including interferon signaling and antigen presentation (Fig. 4D). Together, these findings suggest that sustentacular cells do not remain infected with SARS-CoV-2 during PASC, but rather appear to be responding to local pro-inflammatory cytokines in their micro-environment.
Previous bulk RNA-seq analysis of hamster olfactory epithelium after SARS-CoV-2 infection identified a set of pathogen response genes that were up- or down-regulated one month after infection in sustentacular cells and other cell types (21) (Fig. 4E). We therefore asked whether these same genes were altered in our human PASC olfactory epithelium samples, obtained four months or more after initial infection. Of the pathogenic response genes identified in hamsters, only BST2 was significantly enriched (adjusted P value <0.05, Wilcoxon rank-sum). BST2 (also known as Tetherin) is an interferon-induced host gene encoding a transmembrane protein with antiviral and inflammatory signaling activities (33, 34). We performed identical analysis on all other olfactory epithelium cell populations, which revealed significant changes only in BST2 expression specifically in horizontal basal cells (fig. S6, P<0.05, Wilcoxon rank-sum). Consistent with the absence of viral RNA in post-COVID-19 hyposmic olfactory epithelium samples, we observed no significant increase in any olfactory epithelium cell populations of ISG15, whose expression correlates with the presence of subgenomic nucleocapsid SARS-CoV-2 RNA (21).
We validated these findings through immunohistochemical staining of non-COVID-19 normosmic, post-COVID-19 normosmic, and PASC hyposmic biopsies (Fig. 4F). Anti-ERMN, which selectively labels the apical region of sustentacular cells but not respiratory epithelial cells, exhibited strong uniform signal in both controls or PASC hyposmic samples (Fig. 4F). In contrast, TUJ1 antibody, which stains immature OSN somata and neurites, exhibited abundant labeling in the control samples but less consistent labeling in PASC hyposmic samples (Fig. 4F; fig. S7).
The olfactory neuron population is reduced in PASC hyposmic samples
Despite the lymphocytic infiltrates in PASC hyposmic samples, pseudotime analysis confirmed the expected olfactory epithelium lineage relationships and marker gene expression within the olfactory epithelium, an adult neurogenic niche (Fig. 5A, B). Further analysis of the neuronal lineage subsets identified globose basal cells/neuronal precursors, immature olfactory sensory neurons and mature olfactory sensory neurons, suggesting that olfactory epithelium neurogenesis capacity was not exhausted in the PASC hyposmic samples (Fig. 5C, D). However, analysis of PASC hyposmic horizontal basal cells, which can express the SARS-CoV-2 receptor ACE2, revealed downregulation of subsets of transcripts involved in epithelial renewal (fig. S8), in line with observed horizontal basal cell changes in mouse models of localized inflammatory exposure (35). Analysis of olfactory sensory neuron-specific genes suggested modest alterations in PASC hyposmic samples. For instance, expression of signaling factors downstream of their olfactory receptors was broadly similar between PASC hyposmic and control groups (Fig. 5E, adjusted P value NS, Wilcoxon rank-sum). To quantify the number of olfactory sensory neurons based upon the scRNA-seq data, we normalized olfactory sensory neuron counts to sustentacular cell counts (Fig. 5F; n=5 PASC hyposmic, 5 control, P=0.034, 2-tailed t test). We used this approach as olfactory epithelium biopsies can be variable and because there are often patches of respiratory-like metaplasia in biopsies. When normalized in this manner, the number of olfactory sensory neurons was reduced relative to sustentacular cells in the PASC hyposmic samples compared to control samples. However, despite the reduction in olfactory sensory neuron number, we observed no differences in the frequency of cells expressing olfactory receptors, the mRNA expression of olfactory receptor genes, or the distribution of olfactory receptors expressed across olfactory sensory neurons (fig. S9). In contrast to widespread severe olfactory sensory neuron gene expression changes reported from human autopsy samples following fatal acute COVID-19 (12), the PASC hyposmic olfactory sensory neuron changes identified here were limited (Fig. 5E; fig. S9).
Fig. 5. Analysis of olfactory sensory neurons in PASC hyposmic samples.
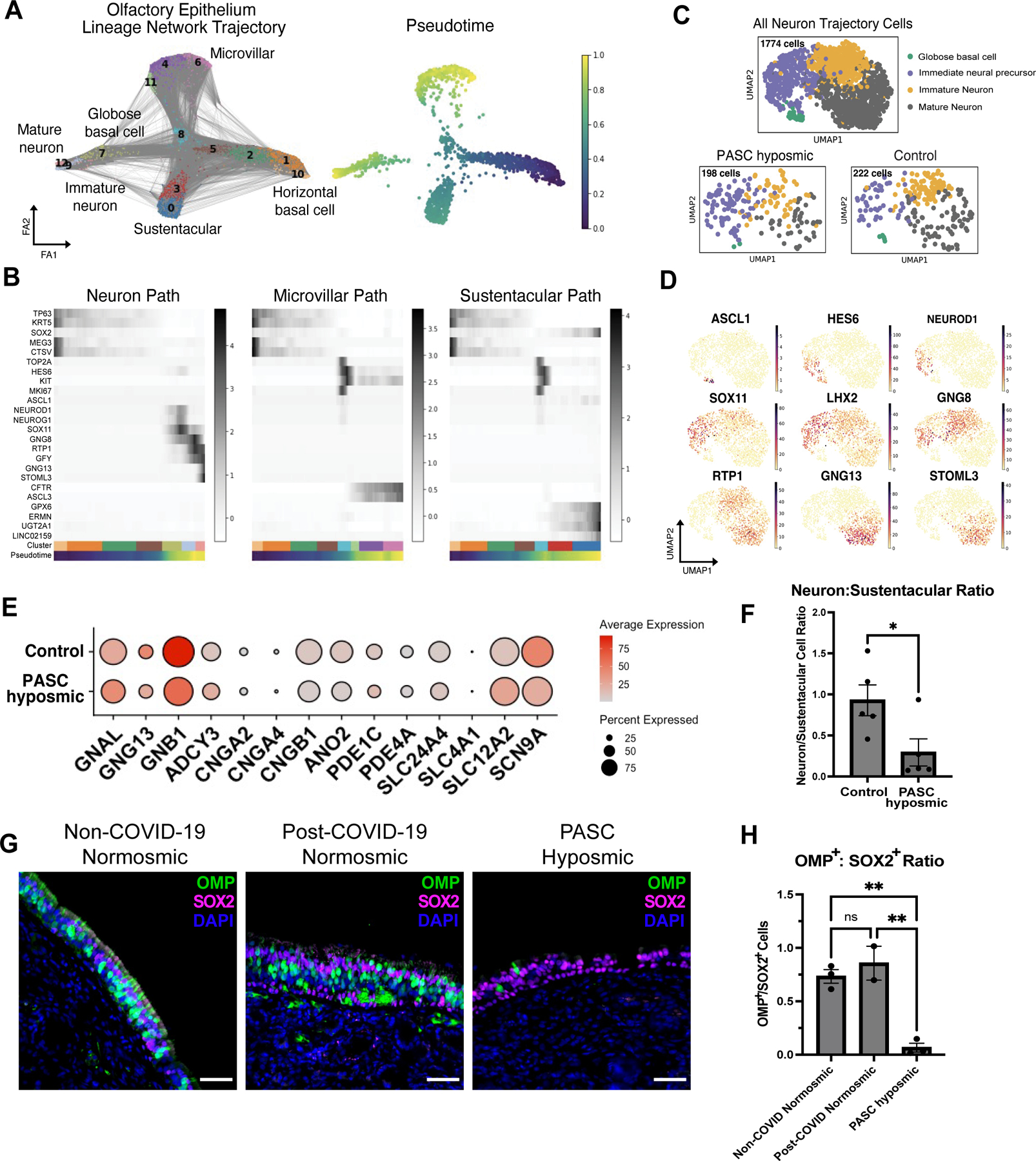
(A) Trajectory lineage analysis of cells in olfactory epithelium in PASC hyposmic biopsies (n=4) from COVID-19 patients compared to control normosmic (non-COVID-19) biopsies (n=3). Cells include horizontal basal cells, globose basal cells, olfactory sensory neurons, sustentacular cells, microvillar cells. (B) Heatmaps showing pseudotime progression for olfactory epithelial cell lineages, labeled based on UMAP relations in panel A. Representative transcript markers for each cellular differentiation state are shown on the y-axis. (C) UMAP showing annotations within the olfactory sensory neuron cluster from Fig. 1D. “All Neuron Trajectory Cells” includes neuron lineage cells in biopsies from 16 patients; “PASC hyposmic” includes 5 PASC hyposmic biopsies; “Control” includes 3 control normosmic biopsies confirmed by Smell Identification Test. (D) Gene expression plots showing established markers of the olfactory sensory neuron differentiation pathway. (E) Selected gene expression in PASC hyposmic versus control normosmic biopsy olfactory sensory neurons (mature olfactory sensory neurons and immature olfactory sensory neurons). (F) Ratio of olfactory sensory neurons (mature olfactory sensory neurons and immature olfactory sensory neurons) to sustentacular cells in PASC hyposmic (n=5) versus control normosmic (n=5) biopsies (error bars indicate SEM; two-tailed t test, P=0.034). (G, H) Immunohistochemistry confirmed a reduction in mature olfactory sensory neurons, labeled with an anti-OMP antibody in PASC hyposmic biopsies. Sustentacular cells were labeled by anti-SOX2 antibody; nuclei are stained with DAPI (blue) (P<0.01, one way ANOVA with Tukey test, Bonferroni correction).
To further validate the reduction in olfactory sensory neuron numbers identified by scRNA-seq, we performed immunohistochemical staining on additional samples, including non-COVID-19 normosmic (n=3), post-COVID-19 normosmic (n=2) and PASC hyposmic (n=3) biopsies. We visualized mature olfactory sensory neurons with anti-olfactory marker protein (OMP) antibody (36–38) and sustentacular cells with apical layer anti-SOX2 antibody (9, 38) (Fig. 5G; fig. S7). In both non-COVID-19 and post-COVID-19 normosmic groups, we identified prominent OMP+ neurons and well-organized SOX2+ apical sustentacular cell nuclei. In contrast, PASC hyposmic biopsies contained sparse OMP+ neurons, and the ratio of mature olfactory sensory neurons to sustentacular cells was significantly reduced (Fig 5G, H, P<0.01, one way ANOVA with Tukey test, Bonferroni correction). There was no difference in the OMP+ population in non-COVID-19 normosmic versus post-COVID-19 normosmic samples. Importantly, we verified that our analysis was performed on regions containing olfactory epithelium rather than respiratory metaplasia, based upon the presence of cells positive for the sustentacular-specific marker ERMN, and for the immature neuronal marker TUJ1 (fig. S7). These findings suggest a model in which immune infiltration, together with changes in stem cell and sustentacular cell function, converge to alter the number of mature olfactory sensory neurons, leading to persistent loss of smell.
DISCUSSION
SARS-CoV-2 infection can cause persistent dysfunction across many physiological systems, although the mechanisms that distinguish PASC from more acute pathophysiology remain to be determined (39, 40). Current evidence for COVID-19 damage within the human olfactory epithelium comes largely from autopsy studies involving patients who died from severe acute COVID-19; these studies lacked objective measurements of smell, and samples were obtained after major medical intervention (12, 16). Here, we provide an analysis of olfactory tissue biopsies from COVID-19 patients with PASC hyposmia, each of whom exhibited olfactory dysfunction following COVID-19 as documented by objective testing. Our results comparing scRNA-seq data between endoscopically-guided olfactory epithelium biopsies from PASC hyposmic patients and control normosmic individuals suggest a model in which altered interactions between immune cells and olfactory epithelium drive functional changes in sustentacular cells and olfactory sensory neurons.
Our findings are consistent with indolent localized immune cell responses driving phenotypic changes in sustentacular cells and olfactory sensory neurons. The changes observed in olfactory sensory neurons, including a relative reduction in cell number, especially of mature OMP+ neurons, could explain sensory dysfunction including hyposmia or parosmia. The absence of marked olfactory sensory neuron transcriptomic changes that we observed in the context of PASC-related hyposmia suggest some differences with findings in autopsy samples from acute COVID-19 cases (12). Acutely, local non-autonomous signals are thought to drive neuronal gene expression changes in the setting of severe inflammation. In PASC hyposmic olfactory epithelium, severe inflammation appears absent, and we instead identify interferon response signatures in the sustentacular cells, along with the presence of local lymphocyte populations expressing interferon-γ and γδ T cell markers, unique to the PASC hyposmic olfactory epithelium samples.
It is interesting to compare the phenotypes observed in humans with PASC-related smell loss and those observed previously in hamsters acutely infected with SARS-CoV-2 (12, 14). In the hamster model, a wide array of immune cells (including macrophages, neutrophils and monocytes) infiltrate the epithelium in the first several days after infection, before resolving nearly completely within two weeks. Our observation of a persistent infiltration of T cells in the human olfactory epithelium months after SARS-CoV-2 infection suggests that COVID-19 patients with PASC hyposmia may have a selective immunological response to prior infection that differs from the immunological responses generated acutely.
Our data are consistent with a provisional model in which a dysregulated axis among immune cells, horizontal basal/sustentacular cells, and olfactory sensory neurons arises in the PASC hyposmic olfactory epithelium, with a resultant sensory dysfunction. How and why this occurs in a subset of patients remains to be determined, but analysis of macrophages from patients with COVID-19 has shown that acute SARS-CoV-2 infection drives a pro-inflammatory reprogramming that is thought to induce long-term alterations in the function of other immune cells (41). In addition, in mouse models (35) and in presbyosmic humans (19), horizontal basal cells exhibit immune-responsive phenotypes, including signaling interactions that can recruit additional immune cells. Furthermore, unresolved immune responses, including activation of specific CD8+ T cell clonotypes during convalescence following SARS-CoV-2 infection, have been reported (42). It is tempting to speculate that these or similar processes may initiate the local olfactory epithelium immune cell alterations identified in our PASC hyposmic samples.
The data presented here also are relevant to several alternative mechanistic hypotheses about how SARS-CoV-2 infection might cause long term olfactory loss. One possibility suggested by work in animal models is that severe initial widespread cell damage might overwhelm the capacity of basal stem cells to reconstitute the olfactory epithelium, but our samples suggest that at least many areas of the human olfactory cleft harbor intact olfactory epithelium comprised of olfactory sensory neurons, sustentacular cells and basal cells (14). Persistent viral infection could also drive ongoing damage (43), but we find no evidence for active SARS-CoV-2 infection in our samples. Another possibility is that anosmia/parosmia is the consequence of severe ongoing mucosal inflammation, but our patient samples did not exhibit clinical inflammatory findings of local edema, polyposis or infection, and the molecular signatures identified in the olfactory epithelium were not consistent with broad inflammatory responses. It is important to note that central mechanisms may contribute to PASC-related smell loss, warranting further study. However, there is little evidence for SARS-CoV-2 infection of neurons in humans (16), and at least some of the observed imaging changes in the olfactory bulb or cortex (44) could reflect reduced peripheral input due to olfactory epithelium damage (the clear site of viral infection), or diffusion of inflammatory intermediates across the cribriform plate.
There are several limitations associated with our study. Given challenges related to the pandemic, it has been difficult to obtain samples from large numbers of patients with COVID-19, and thus our conclusions are driven by findings we observed in common across our limited set of patient samples. Furthermore, in our study, we merged patient samples obtained by two different methods, surgical excision and brush biopsy (fig. S10). We did not identify batch effects or gene expression changes related to biopsy technique (fig. S11). Finally, although we were careful to obtain biopsies from within the olfactory cleft region, the possibility of sample-to-sample variation in the specific contents of each olfactory epithelium biopsy was unavoidable.
The pandemic has highlighted the unmet need for new effective treatments for olfactory loss. The mechanistic insights provided here suggest potential new therapeutic strategies. For instance, selectively blocking local pro-inflammatory immune cells or directly inhibiting specific signaling nodes may interfere with a loop disrupting olfactory epithelium homeostasis or repair. The location of the olfactory epithelium, lining the olfactory cleft in the nose, is amenable to localized topical drug delivery, which may provide a means to avoid systemic or off-target effects of new therapeutic agents. Further studies testing therapeutics in animal models and humans, and longer follow up of patients with PASC olfactory dysfunction, will provide ongoing insights regarding the etiology and management of olfactory sensory dysfunction.
MATERIALS AND METHODS
Study Design
We performed a prospective analysis of olfactory epithelium tissue obtained from 9 patients suffering from post-COVID-19 hyposmia/anosmia. Olfactory function was measured using the Smell Identification Test (Sensonics, Inc). Biopsy samples were collected for either histology or scRNA-Seq analysis. Patient demographics, including length of hyposmia/anosmia and time since acute COVID-19 infection, were collected (table S1). Control samples included olfactory tissue biopsies obtained from normosmic individuals with no history of COVID-19 (3 patients) and from normosmic patients who had been previously diagnosed with COVID-19 (2 patients).
All human studies were performed under protocols approved by Institutional Review Boards of Duke University or University of California San Diego.
Biopsy Collection and Processing
All biopsy samples reported here were collected under Duke University School of Medicine IRB protocols 00088414 and 00105837. Patients were administered the Smell Identification Test prior to tissue collection to assess olfactory function. For surgical biopsies, olfactory mucosa was collected either in the operating room in patients undergoing transsphenoidal surgery for resection of a benign pituitary tumor or in the clinic. Briefly, using endoscopic visualization, olfactory cleft mucosa was sharply incised and elevated from underlying bone, and then excised with a through-cutting ethmoid forceps. For nasal cytology brush biopsies, tissue was collected in the clinic by gently positioning a cytology brush (Cat#4290, Hobbs Medical Inc, Stafford Springs, CT) in the olfactory cleft under endoscopic visualization (fig. S10). The brush was rotated briefly to collect surface mucosal cells. In all cases, samples were placed into collection solution [Hanks’ Balanced Salt Solution (HBSS) or Hibernate E medium, with 10% fetal bovine serum (FBS, all from Thermo Fisher, Waltham, MA)] on ice and processed immediately for analysis.
Surgical biopsy tissues were divided into smaller pieces sharply. All biopsies were digested for 15 minutes at 37°C with an enzyme cocktail comprised of Dispase/Collagenase A/EDTA mix, 2mg/mL Papain, and DNAse I (all from StemCell Tech, Vancouver, BC, Canada) with frequent gentle trituration. After 15 minutes, Accutase (StemCell Tech) was added, and samples were incubated for an additional 5 minutes at 37°C. At the end of 5 minutes, FBS was added. If samples still contained large pieces of tissue, they were filtered through a 250μm filter. All samples were then filtered through a 70μm filter and centrifuged 5 minutes at 400 × g. If abundant red blood cells were observed in the pellet, tissues were re-suspended in ACK Lysing Buffer (Thermo) and incubated at room temperature for 3–5 minutes with gently rocking. Samples were washed, spun and resuspended in HBSS or Hibernate-E containing non-acetylated bovine serum albumin 1 mg/ml (Thermo), anti-clumping reagent 0.5 μl/ml (Gibco), and N-acetyl cysteine 5 μg/ml (Sigma-Aldrich, St. Louis, MO) to a final concentration of 1 million cells/mL. Brush biopsies were processed similarly, but required slightly less time in dissociation enzymes and did not require an erythrocyte lysis step.
Single-Cell Sequencing
Samples were processed for single cell analysis as described previously (19). Briefly, cells were quantified with a viability stain on an automated counter (Cellaca MX, Nexcelom) and loaded onto a Chromium controller (10X Genomics, Pleasanton, CA) for cell capture and bar coding targeting 10,000 cells, per the 3’ v3.1 gene expression protocol per manufacturer’s instructions. Reverse transcription, amplification, library preparation, and sequencing (NovaSeq, Illumina) were performed per protocol.
Single-Cell RNA-Seq Analysis
Illumina base call files were converted to FASTQ files and processed through CellRanger Counts 6.1.2 (10X Genomics), aligned to either a human reference genome (GRCh38) or a combined reference genome containing human and SARS-CoV-2 genomes (45).
Starting from the raw cell by gene count matrices, data integration and preprocessing were performed using Scanpy (v1.8.2) and scvi-tools (v0.15.2). For accurate cell type identification, the data generated in this study were combined with our published human olfactory datasets (GSE139522, GSE184117) (9, 19). Highly-variable genes (HVGs) were identified using the scvi-tools “poisson_gene_selection” function (with patient id as the batch key), and the raw counts for these gene subsets were used as the input to the variational autoencoder (scVI) model. An scVI model (using the top 3000 HVGs) was trained for 500 unsupervised epochs with the default learning rate (with early stopping when the ELBO validation metric did not improve for at least 20 epochs) with the default parameters (10 latent dimensions, 128 nodes per hidden layer), a negative binomial observation model (gene_likelihood=“nb”), the percentage of mitochondrial genes as a continuous covariate, and categorical covariate keys for the patient condition and patient id categorical variables (which thus performed dataset integration and batch correction for the purposes of cell type identification). A k-nearest neighbor graph was constructed from the resulting 10-dimensional latent embedding (using k=15 neighbors). The knn graph which was used for cell type clustering via the Leiden algorithm (resolution=1.2) and as the input to the UMAP algorithm (with min_dist=0.5) for visualization. Clusters of dying cells containing high percentages of mitochondrial genes and low total counts as well as a cluster of cell doublets were removed, and the above procedure starting from the HVG identification was repeated (but with Leiden clustering resolution=1.6). The resulting cell type clusters were merged and manually annotated based on known cell type markers.
After identifying and annotating the broad clusters, cell types of interest were further subclustered in an iterative manner, using the same scVI embedding approach, starting from the reidentification of HVGs for each subset. After training a scVI model using only cell types in the olfactory epithelium (including olfactory horizontal basal cells, sustentacular cells, Bowman’s gland cells, microvillar cells, and olfactory sensory neurons), an additional cluster of olfactory sensory neuron-sustentacular cell doublets was identified and removed. Next, an scVI model was trained on the olfactory sensory neurons and microvillar cells (except using 2000 HVGs and 100 hidden nodes in the scVI model), and the resulting clusters from this model identified cell types of the olfactory sensory neuron lineage; a final scVI model (using 2000 HVGs and 100 hidden nodes) was used to embed and cluster these cells and the olfactory sensory neuron lineage cell types were manually annotated using known markers for globose basal cells (TOP2A, ASCL1), immediate neuron precursors (NEUROD1, SOX11), immature olfactory sensory neurons (GAP43, DCX, GNG8), and mature olfactory sensory neurons (GNG13, STOML3). The same approach was also used to further subcluster the broad lymphocyte cluster that contained the CD4+ T cells, CD8+ T cells and NK cells starting from the top 2000 HVGs from these cells and the resulting scVI embedding was then clustered (resolution=1.1) to identify the lymphocyte subtypes.
Trajectory and pseudotime analyses were performed using olfactory sensory neuron lineage cell types identified in the second iteration of the scVI model trained on these cells. Bowman’s gland cells were excluded from analysis. A new neighborhood graph was computed using n_neighbors=100 and n_pcs=20, and cells were reclustered using the default leiden algorithm with resolution=1.5. Cluster connectivity was then calculated using partition-based graph abstraction (PAGA) with default settings. PAGA plots were constructed using threshold=0.2. For plotting of pseudotime heatmaps, leiden clusters were ordered based on PAGA connectivity predictions.
Transcriptome distances were calculated from the pairwise correlation distance matrix of the embedding in the 10-dimensional scVI latent space embedding for cells from the olfactory sensory neuron lineage. Transcriptome distances were summarized for each olfactory sensory neuron cluster - condition (control vs PASC hyposmic) pair by taking the median pairwise transcriptome distance between cells of each pair.
For additional plots, such as differential expression analysis, filtered outputs were analyzed in R (v4.1.1) using the Seurat toolkit (v4.1.0) (46). Processed anndata objects from Scanpy were converted to R objects preserving all meta data (including scVI clusters) using the LoadH5Seurat function from SeuratDisk. Data were normalized using relative counts normalization prior to differential expression analysis. Differentially expressed genes were found using the FindMarkers function with default settings (Wilcoxon Rank-Sum) and plotted using ggplot2 (identifying significant DE genes with >log2 fold-change, adjusted P<0.05). Cluster markers of lymphocyte subsets were identified using FindAllMarkers with default settings. DotPlots were produced using relative normalized counts.
NicheNet analysis was conducted in R with the nichenetr package (V 1.1.0) using the default ligand-target prior model, ligand receptor network, and weight integrated networks (47). Specifically, cell populations of interest (i.e. lymphocyte clusters, olfactory sensory neurons, and sustentacular cells) with normalized gene expression were subset out from processed R objects (from anndata) and used as input for the appropriate receiver and sender populations. Circos plots were generated using Circlize (V 0.4.14).
We analyzed data sets from only-surgical or only-brush biopsy samples for individual comparisons (fig. S11), verifying no significant changes in the normalized olfactory epithelium cell population gene expression profiles.
Immunohistochemistry
Samples for histology were collected in Hanks’ Balanced Salt Solution (HBSS, Gibco) + 10% FBS. Tissues were fixed with 4% paraformaldehyde (Sigma, St. Louis) in phosphate buffered saline (PBS) for 4 hours at room temperature. Samples were washed with PBS and then incubated on a rocker at 4°C for 5–7 days in 30% sucrose, 250mM EDTA, and PBS. Samples were then flash frozen in OCT compound (VWR, Radnor, PA), sectioned at 10μm on a cryostat (CryoStar NX50, Thermo Fisher) and collected on Superfrost plus slides (Thermo Fisher).
Tissue sections were rehydrated in PBS and blocked in 5% normal goat serum in PBS with 0.1% Triton X-100. Antigen retrieval was performed on sections being stained for OMP by steaming tissue for 45 minutes in citrate-based antigen unmasking solution (Vector Laboratories, Newark, CA). Anti-Tubulin β3 (BioLegend, clone TUJ1, Cat# 801201; AB_2313773; 1:500), Anti-CD45 (BioLegend, clone HI30, Cat# 304001 AB_314389; 1:50), Anti-CD3 (BioLegend, clone HIT3a, Cat# 300301; AB_314037; 1:50), Anti-CD68 (BioLegend, clone BL13756, Cat# 375602; AB_2876705; 1:50), Anti-CD207(Langerin) (BioLegend, clone 4C7, Cat# 144201; AB_2562087; 1:50), Anti-TCR γ/δ (BioLegend, clone B1, Cat# 331202; AB_1089222; 1:50), anti-ERMN (Thermo, Cat# PA5–58327; AB_2641113; 1:100), anti-SARS-CoV-2 nucleocapsid (Novus NB100–56576; AB_838838; 1:250), anti-SOX2 (Invitrogen eBioscience Cat# 14–9811-82; AB_11219471; 1:50) or anti-OMP (Santa Cruz Cat# sc-365818; AB_108421641:500) primary antibodies diluted in blocking buffer were incubated on tissue sections for 1 hour at room temperature or overnight at 4 degrees. Following PBS washes, tissues were incubated with fluorescent conjugated secondary antibodies for 45 minutes (Jackson ImmunoResearch, West Grove, PA). Vectashield with DAPI (Vector Laboratories, Burlingame, CA) was applied to each section prior to coverslip. All images were acquired on a Leica DMi8 microscope system (Leica Microsystems). Images were analyzed using ImageJ software (V 2.3.0), and scale bars were applied using metadata from the original Leica acquisition software files.
For quantification of immunohistochemical labeling, Images were acquired with 40x objective and opened in ImageJ. Staining of adjacent sections with TUJ1/ERMN confirmed the presence of olfactory epithelium, rather than respiratory epithelium. Counts were conducted across a minimum of 500μm length of olfactory epithelium (average length counted per patient = 931μm). olfactory sensory neurons were counted based on the presence of an OMP+ cell soma and dendrite with associated DAPI+ nucleus. Sustentacular cells were counted based on SOX2+ nuclei. Only apical SOX2+ nuclei were counted as sustentacular cells, excluding SOX2+ horizontal basal cells, distinctly situated against the basement membrane with small flat morphology. For each patient, the total ratio of OMP+ cells to total SOX2+ apical cells was calculated. Adjacent sections were not included in counts for a given cell type-specific marker, avoiding a need for using Abercrombie correction.
Olfactory mucus assays
Mucus was obtained from the olfactory cleft using absorbent filter paper under endoscopic guidance per an approved IRB protocol at UC San Diego (#210078). Cohorts included PASC hyposmics (n=13 patients) or control normosmics (n=7), based on psychophysical testing using the Smell Identification Test. A fluorescent bead-based multiplex assay (LegendPlex, Biolegend) was used to quantify 13 cytokines/chemokines via flow cytometry.
Statistics
All sequencing data set analyses were performed in Python or R using the toolkits and packages described above. Plots were produced using Scanpy, Matplotlib, ggplot2 in associated R toolkits (48), or Graphpad Prism 9. Cell phenotype comparisons between PASC hyposmic and control samples were performed using unpaired 2-tailed t-test, with significance defined as P<0.05. For all parametric tests, Shapiro-Wilk tests were used to assess for normal distributions; appropriate significance tests were then used (either t-test or Mann-Whitney test). Differentially expressed genes were analyzed using Wilcoxon Rank-Sum. Error bars represent standard error of the mean. Differentially expressed gene sets were analyzed for gene ontology, cellular pathway or tissue output terms using ToppGene Suite (49). Immunohistochemistry quantification was compared using one way ANOVA with Tukey test, Bonferroni correction.
Supplementary Material
Acknowledgements:
We thank the patients who generously agreed to provide biopsy samples for this research. We appreciate the expert technical assistance of the Duke Molecular Genomics Core, and bioinformatics assistance from Vaibhav Jain. We also thank Clinical Research Coordinators, Amy Walker and Victoria Eifert, for expert assistance. Graphical schematics were created with Biorender.com.
Funding:
This study was supported by NIH DC018371, DC016859 (B.J.G.), NIH AG074324 (E.A.M.), NIH DC019956 (C.H.Y.) and funding from the Duke Department of Head and Neck Surgery & Communication Sciences.
Footnotes
List of Supplementary Materials
Fig S1 to S11
Table S1 and S2
Data File S1
Competing interests: B.J.G. has received consultant fees from Frequency Therapeutics and discloses unpaid consulting to Rhino Therapeutics. D.J. has received research support from Medtronic and Association for Migraine Disorders. H.M. has received royalties from Chemcom, has received research grants from Givaudan and has received consultant fees from Kao. Authors declare no other competing interests.
Data and Materials Availability
scRNA-seq data sets are deposited in GEO with accession number GSE201620. Code availability: Scripts involved in analysis are available in Zenodo (DOI 10.5281/zenodo.7102415) (50).
References and Notes
- 1.Carfi A, Bernabei R, Landi F, Persistent Symptoms in Patients After Acute COVID-19. JAMA 324, 603–605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schambeck SE, Crowell CS, Wagner KI, D’Ippolito E, Burrell T, Mijocevic H, Protzer U, Busch DH, Gerhard M, Poppert H, Beyer H, Phantosmia, Parosmia, and Dysgeusia Are Prolonged and Late-Onset Symptoms of COVID-19. J Clin Med 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karamali K, Elliott M, Hopkins C, COVID-19 related olfactory dysfunction. Curr Opin Otolaryngol Head Neck Surg 30, 19–25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boscolo-Rizzo P, Hummel T, Hopkins C, Dibattista M, Menini A, Spinato G, Fabbris C, Emanuelli E, D’Alessandro A, Marzolino R, Zanelli E, Cancellieri E, Cargnelutti K, Fadda S, Borsetto D, Vaira LA, Gardenal N, Polesel J, Tirelli G, High prevalence of long-term olfactory, gustatory, and chemesthesis dysfunction in post-COVID-19 patients: a matched case-control study with one-year follow-up using a comprehensive psychophysical evaluation. Rhinology 59, 517–527 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Boscolo-Rizzo P, Menegaldo A, Fabbris C, Spinato G, Borsetto D, Vaira LA, Calvanese L, Pettorelli A, Sonego M, Frezza D, Bertolin A, Cestaro W, Rigoli R, D’Alessandro A, Tirelli G, Da Mosto MC, Menini A, Polesel J, Hopkins C, Six-Month Psychophysical Evaluation of Olfactory Dysfunction in Patients with COVID-19. Chem Senses 46, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graziadei GA, Graziadei PP, Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J Neurocytol 8, 197–213 (1979). [DOI] [PubMed] [Google Scholar]
- 7.Leung CT, Coulombe PA, Reed RR, Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci 10, 720–726 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, Hewitt Coleman J, Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J Comp Neurol 525, 1034–1054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durante MA, Kurtenbach S, Sargi ZB, Harbour JW, Choi R, Kurtenbach S, Goss GM, Matsunami H, Goldstein BJ, Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat Neurosci 23, 323–326 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi R, Goldstein BJ, Olfactory epithelium: Cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope Investig Otolaryngol 3, 35–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck L, Axel R, A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Zazhytska M, Kodra A, Hoagland DA, Frere J, Fullard JF, Shayya H, McArthur NG, Moeller R, Uhl S, Omer AD, Gottesman ME, Firestein S, Gong Q, Canoll PD, Goldman JE, Roussos P, tenOever BR, Overdevest JB, Lomvardas S, Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell 185, 1052–1064 e1012 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brann DH, Tsukahara T, Weinreb C, Lipovsek M, Van den Berge K, Gong B, Chance R, Macaulay IC, Chou HJ, Fletcher RB, Das D, Street K, de Bezieux HR, Choi YG, Risso D, Dudoit S, Purdom E, Mill J, Hachem RA, Matsunami H, Logan DW, Goldstein BJ, Grubb MS, Ngai J, Datta SR, Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci Adv 6, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryche B, St Albin A, Murri S, Lacote S, Pulido C, Ar Gouilh M, Lesellier S, Servat A, Wasniewski M, Picard-Meyer E, Monchatre-Leroy E, Volmer R, Rampin O, Le Goffic R, Marianneau P, Meunier N, Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav Immun 89, 579–586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shelton JF, Shastri AJ, Fletez-Brant K, andMe C-T, Aslibekyan S, Auton A, The UGT2A1/UGT2A2 locus is associated with COVID-19-related loss of smell or taste. Nat Genet 54, 121–124 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Khan M, Yoo SJ, Clijsters M, Backaert W, Vanstapel A, Speleman K, Lietaer C, Choi S, Hether TD, Marcelis L, Nam A, Pan L, Reeves JW, Van Bulck P, Zhou H, Bourgeois M, Debaveye Y, De Munter P, Gunst J, Jorissen M, Lagrou K, Lorent N, Neyrinck A, Peetermans M, Thal DR, Vandenbriele C, Wauters J, Mombaerts P, Van Gerven L, Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell 184, 5932–5949 e5915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doty RL, Shaman P, Dann M, Development of the University of Pennsylvania Smell Identification Test: a standardized microencapsulated test of olfactory function. Physiol Behav 32, 489–502 (1984). [DOI] [PubMed] [Google Scholar]
- 18.Doty RL, Shaman P, Applebaum SL, Giberson R, Siksorski L, Rosenberg L, Smell identification ability: changes with age. Science 226, 1441–1443 (1984). [DOI] [PubMed] [Google Scholar]
- 19.Oliva AD, Gupta R, Issa K, Abi Hachem R, Jang DW, Wellford SA, Moseman EA, Matsunami H, Goldstein BJ, Aging-related olfactory loss is associated with olfactory stem cell transcriptional alterations in humans. J Clin Invest 132, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutmann C, Takov K, Burnap SA, Singh B, Ali H, Theofilatos K, Reed E, Hasman M, Nabeebaccus A, Fish M, McPhail MJ, O’Gallagher K, Schmidt LE, Cassel C, Rienks M, Yin X, Auzinger G, Napoli S, Mujib SF, Trovato F, Sanderson B, Merrick B, Niazi U, Saqi M, Dimitrakopoulou K, Fernandez-Leiro R, Braun S, Kronstein-Wiedemann R, Doores KJ, Edgeworth JD, Shah AM, Bornstein SR, Tonn T, Hayday AC, Giacca M, Shankar-Hari M, Mayr M, SARS-CoV-2 RNAemia and proteomic trajectories inform prognostication in COVID-19 patients admitted to intensive care. Nat Commun 12, 3406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frere JJ, Serafini RA, Pryce KD, Zazhytska M, Oishi K, Golynker I, Panis M, Zimering J, Horiuchi S, Hoagland DA, Møller R, Ruiz A, Kodra A, Overdevest JB, Canoll PD, Borczuk AC, Chandar V, Bram Y, Schwartz R, Lomvardas S, Zachariou V, tenOever BR, SARS-CoV-2 infection in hamsters and humans results in lasting and unique systemic perturbations post recovery. Science Translational Medicine 0, eabq3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, Cheng YL, Bush EC, Dogra P, Thapa P, Farber DL, Sims PA, Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun 10, 4706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pizzolato G, Kaminski H, Tosolini M, Franchini DM, Pont F, Martins F, Valle C, Labourdette D, Cadot S, Quillet-Mary A, Poupot M, Laurent C, Ysebaert L, Meraviglia S, Dieli F, Merville P, Milpied P, Dechanet-Merville J, Fournie JJ, Single-cell RNA sequencing unveils the shared and the distinct cytotoxic hallmarks of human TCRVdelta1 and TCRVdelta2 gammadelta T lymphocytes. Proc Natl Acad Sci U S A 116, 11906–11915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonneville M, O’Brien RL, Born WK, Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol 10, 467–478 (2010). [DOI] [PubMed] [Google Scholar]
- 25.von Massow G, Oh S, Lam A, Gustafsson K, Gamma Delta T Cells and Their Involvement in COVID-19 Virus Infections. Front Immunol 12, 741218 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen CE, Merad M, McClain KL, Langerhans-Cell Histiocytosis. N Engl J Med 379, 856–868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merad M, Ginhoux F, Collin M, Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat Rev Immunol 8, 935–947 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Lee SM, Kok KH, Jaume M, Cheung TK, Yip TF, Lai JC, Guan Y, Webster RG, Jin DY, Peiris JS, Toll-like receptor 10 is involved in induction of innate immune responses to influenza virus infection. Proc Natl Acad Sci U S A 111, 3793–3798 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alshoubaki YK, Nayer B, Das S, Martino MM, Modulation of the Activity of Stem and Progenitor Cells by Immune Cells. Stem Cells Transl Med 11, 248–258 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueha R, Kondo K, Ueha S, Yamasoba T, Dose-Dependent Effects of Insulin-Like Growth Factor 1 in the Aged Olfactory Epithelium. Front Aging Neurosci 10, 385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Getchell ML, Ding X, Getchell TV, Immunolocalization of two cytochrome P450 isozymes in rat nasal chemosensory tissue. Neuroreport 3, 749–752 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Herrick DB, Lin B, Peterson J, Schnittke N, Schwob JE, Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc Natl Acad Sci U S A 114, E5589–E5598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahauad-Fernandez WD, Okeoma CM, The role of BST-2/Tetherin in host protection and disease manifestation. Immun Inflamm Dis 4, 4–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberatore RA, Bieniasz PD, Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc Natl Acad Sci U S A 108, 18097–18101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M, Reed RR, Lane AP, Chronic Inflammation Directs an Olfactory Stem Cell Functional Switch from Neuroregeneration to Immune Defense. Cell Stem Cell 25, 501–513 e505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller A, Margolis FL, Immunological studies of the rat olfactory marker protein. J Neurochem 24, 1101–1106 (1975). [DOI] [PubMed] [Google Scholar]
- 37.Olender T, Keydar I, Pinto JM, Tatarskyy P, Alkelai A, Chien MS, Fishilevich S, Restrepo D, Matsunami H, Gilad Y, Lancet D, The human olfactory transcriptome. BMC Genomics 17, 619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitzek M, Patel PK, Solomon PD, Lin B, Hummel T, Schwob JE, Holbrook EH, Integrated age-related immunohistological changes occur in human olfactory epithelium and olfactory bulb. J Comp Neurol, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nalbandian A, Sehgal K, Gupta A, Madhavan MV, McGroder C, Stevens JS, Cook JR, Nordvig AS, Shalev D, Sehrawat TS, Ahluwalia N, Bikdeli B, Dietz D, Der-Nigoghossian C, Liyanage-Don N, Rosner GF, Bernstein EJ, Mohan S, Beckley AA, Seres DS, Choueiri TK, Uriel N, Ausiello JC, Accili D, Freedberg DE, Baldwin M, Schwartz A, Brodie D, Garcia CK, Elkind MSV, Connors JM, Bilezikian JP, Landry DW, Wan EY, Post-acute COVID-19 syndrome. Nat Med 27, 601–615 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havervall S, Rosell A, Phillipson M, Mangsbo SM, Nilsson P, Hober S, Thalin C, Symptoms and Functional Impairment Assessed 8 Months After Mild COVID-19 Among Health Care Workers. JAMA 325, 2015–2016 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bohnacker S HF, Henkel F, Quaranta A, Kolmert J, Priller A, Ud-Dean M, Giglberger J, Kugler LM, Pechtold L, Yazici S, Lechner A, Erber J, Protzer U, Lingor P, Knolle P, Chaker AM, Schmidt-Weber CB, Wheelock CE and Esser-von Bieren J, Mild COVID-19 imprints a long-term inflammatory eicosanoidand chemokine memory in monocyte-derived macrophages. Mucosal Immunology, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Y, Yuan D, Chen DG, Ng RH, Wang K, Choi J, Li S, Hong S, Zhang R, Xie J, Kornilov SA, Scherler K, Pavlovitch-Bedzyk AJ, Dong S, Lausted C, Lee I, Fallen S, Dai CL, Baloni P, Smith B, Duvvuri VR, Anderson KG, Li J, Yang F, Duncombe CJ, McCulloch DJ, Rostomily C, Troisch P, Zhou J, Mackay S, DeGottardi Q, May DH, Taniguchi R, Gittelman RM, Klinger M, Snyder TM, Roper R, Wojciechowska G, Murray K, Edmark R, Evans S, Jones L, Zhou Y, Rowen L, Liu R, Chour W, Algren HA, Berrington WR, Wallick JA, Cochran RA, Micikas ME, Unit IS-SC-B, Wrin T, Petropoulos CJ, Cole HR, Fischer TD, Wei W, Hoon DSB, Price ND, Subramanian N, Hill JA, Hadlock J, Magis AT, Ribas A, Lanier LL, Boyd SD, Bluestone JA, Chu H, Hood L, Gottardo R, Greenberg PD, Davis MM, Goldman JD, Heath JR, Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 185, 881–895 e820 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Melo GD, Lazarini F, Levallois S, Hautefort C, Michel V, Larrous F, Verillaud B, Aparicio C, Wagner S, Gheusi G, Kergoat L, Kornobis E, Donati F, Cokelaer T, Hervochon R, Madec Y, Roze E, Salmon D, Bourhy H, Lecuit M, Lledo PM, COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci Transl Med 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douaud G, Lee S, Alfaro-Almagro F, Arthofer C, Wang C, McCarthy P, Lange F, Andersson JLR, Griffanti L, Duff E, Jbabdi S, Taschler B, Keating P, Winkler AM, Collins R, Matthews PM, Allen N, Miller KL, Nichols TE, Smith SM, SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R, Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc Natl Acad Sci U S A 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browaeys R, Saelens W, Saeys Y, NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods 17, 159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Wickham H, ggplot2: Elegant Graphics for Data Analysis., (Springer-Verlag, New York, 2016). [Google Scholar]
- 49.Chen J, Bardes EE, Aronow BJ, Jegga AG, ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finlay JB, Brann DH, Datta SR, Goldstein BJ, Code from: “Persistent post-COVID-19 smell loss is associated with immune infiltration and altered olfactory epithelial gene expression” Zenodo; (2022); https://zenodo.org/record/7102415#.YyuFTiHMLao. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data sets are deposited in GEO with accession number GSE201620. Code availability: Scripts involved in analysis are available in Zenodo (DOI 10.5281/zenodo.7102415) (50).


