Abstract
Understanding factors that influence the spatial and temporal distributions of blood parasites is important to help predict how host species and their parasites may respond to global change. Factors that may influence parasite distributions are land cover and host dispersal patterns, which may result in exposure of a host to novel parasites, or escape from parasites of their origin. We screened golden-winged warblers from across the United States and Canada for blood parasites, and investigated whether land-use patterns or host dispersal affected the prevalence and composition of haemosporidian assemblages. Parasite prevalence varied strongly with study area, and areas with high agricultural cover had a significantly higher prevalence of Leucocytozoon and Parahaemoproteus parasites. Lineages of Parahaemoproteus and Leucocytozoon were genetically differentiated among study areas, and prevalence and composition of parasite assemblages indicated an increase in parasite prevalence and accumulation of unique parasite lineages from the southeast to the northwest. This matches the historical range expansion and natal dispersal patterns of golden-winged warblers, and suggests that golden-winged warblers may have been sensitive to novel parasites as they dispersed. The high prevalence and diversity of parasite lineages in the north-west extent of their breeding range (Manitoba) indicates that this population may face unique pressures.
Key words: Anthropogenic disturbance, biogeography, haemosporidia, host–parasite interactions, Vermivora
Introduction
The mechanisms that explain the composition of haemosporidian communities (here, composition is defined as the combination of parasite prevalence and diversity) within a host species are a complex and poorly understood component of haemosporidian biology. Two biogeographic factors may influence the composition of parasites: landscape-level human disturbance (Land Cover hypothesis) and host dispersal (Dispersal hypothesis). Parasite distributions are likely to vary with anthropogenic changes to the environment (e.g. Loiseau et al., 2012a). One of the most important determinants of blood parasite communities of birds is variation in the presence of vectors due to habitat or climate (see Sol et al., 2000; Kimura et al., 2006; Pagenkopp et al., 2008; Chasar et al., 2009). This may explain regional differences in parasite prevalence, as vector prevalence is strongly tied to environmental variables (Bishop et al., 1996; Samuel et al., 2011; LaPointe et al., 2012; Bernotiene and Bartkeviciene, 2013). While climate strongly influences parasite distribution, land cover variables can also be important (Pérez-Rodríguez et al., 2013). For example, agricultural irrigation can create damp earth and standing water, which provide habitat for Culicoides midges and mosquitoes, respectively. Additionally, though not all species of black fly can breed in irrigation ditches, they have been found in irrigation systems across the world (see Takoaka et al., 2000; Montagna et al., 2012; Córdoba Lloria et al., 2017), and irrigation areas in Alberta and Saskatchewan had high abundances of Leucocytozoon transmitting black flies (Fredeen and Shemanchuk, 1960). Some tropical studies have found anthropogenic disturbance to be associated with decreased parasite prevalence (Sehgal, 2010; Loiseau et al., 2012b; but see Okanga et al., 2013). However, further study is needed to better understand the effects of habitat type on blood parasites in temperate zones, as the different disturbance types in temperate areas will likely lead to a different mechanism for change to parasite composition.
It is likely that the biogeography of avian parasites is linked to the distribution and dispersal of their host species (the Dispersal hypothesis). There are two hypotheses about how birds may respond to parasite communities following host dispersal: the Enemy Release hypothesis, first proposed to explain patterns in invasive plants (Keane and Crawley, 2002) but recently applied to animals (Lewicki et al., 2015), and the Novel Enemies hypothesis (described in Møller and Szép, 2011, but named here). When a bird disperses into a new habitat, it may benefit from being released from its traditional suite of parasites (the Enemy Release hypothesis) (Lewicki et al., 2015), particularly if those parasites are known to have a negative impact on host fitness. Conversely, the Novel Enemies hypothesis is based on parasites being caught in an arms race with their hosts, where host and parasite co-evolve together in a specific geographic area (Møller and Szép, 2011). Thus, host populations become adapted to local parasites, while recent immigrants to an area are more susceptible to the local parasites given a lack of previous exposure (Møller and Szép, 2011; Sarquis-Adamson and MacDougall-Shackleton, 2016). Additionally, these hypotheses are not mutually exclusive, and birds may benefit from being released from old parasite sources while simultaneously being exposed to a suite of novel parasites, with unknown impacts.
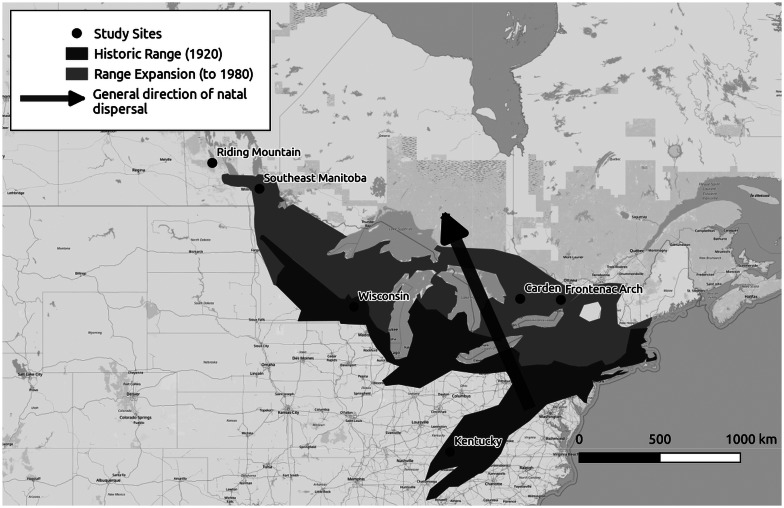
Golden-winged warblers (Vermivora chrysoptera) are an excellent candidate species to study host–parasite biogeography. Parasite prevalence varies strongly across this species' range (Vallender et al., 2012), and therefore provides an opportunity to investigate how changes in landscape structure across eastern North American relate to parasite prevalence. Observation and survey records of golden-winged warblers over the past 150 years indicate that the species has expanded its range in a north-northwesterly (NNW) direction, potentially driven by patterns of early successional habitat created by abandonment of cleared agricultural land, and climate change (Gill, 1980; Buehler et al., 2007; Confer et al., 2011; Fig. 1). While adult golden-winged warblers tend to have strong site philopatry, natal dispersal is high, and second-year individuals also tend to disperse to the north-northwest (López-Calderón et al., 2019, Fig. 1). Golden-winged warblers also display strong migratory connectivity, with northwestern birds overwintering in Central America, and southern birds overwintering in northern South America (Kramer et al., 2017, 2018). This connectivity suggests that parasites among different breeding populations should not become admixed on the wintering grounds; further, most haemosporidian parasite lineages are likely transmitted on the breeding grounds instead of the wintering grounds (Ricklefs et al., 2016). The combination of these factors allows us to test among competing dispersal-related parasite composition hypotheses. Golden-winged warblers are also threatened in Canada under the federal Species At Risk Act, and classifying the parasite composition across their range will help evaluate the risk that blood parasites could potentially pose to this species (Species At Risk Act, 2002, 11 2007).
Fig. 1.
Map depicting a historical range of golden-winged warblers and range expansion to 1980, accompanied by most likely natal dispersal patterns. The Riding Mountain, Manitoba population was colonized post-1980. Range data was adapted from Rosenberg et al. (2016), natal dispersal data is from López-Calderón et al. (2019). Map data is from openstreetmap.org.
The three common and widespread genera of haemosporidian parasites examined in this study are Plasmodium, Leucocytozoon and Parahaemoproteus, which are transmitted by mosquitoes, black flies and biting midges, respectively (Greiner et al., 1975; Valkiūnas, 2005; LaPointe et al., 2012). The potential impacts of these parasites on their avian hosts are diverse, and include lower mating success due to reduced plumage pigmentation, smaller body size or ability to produce mating calls (Figuerola et al., 1999; Freeman-Gallant et al., 2001); reduced clutch sizes, hatching rates and fledging rates (Marzal et al., 2005; Asghar et al., 2011) and reduced survival (Lachish et al., 2011). These fitness effects result in natural selection and cycles of host–parasite evolution, and alteration of these host–parasite interactions through anthropogenic change can pose new challenges to avian populations.
We described the prevalence and parasite lineages across the northwestern portion of the golden-winged warbler's range, with the intention of investigating which populations may be at the greatest risk from parasitism, and of estimating what factors shape trends in parasite prevalence. We compared support for two hypotheses that might explain geographic segregation in parasite composition: (1) the Land Cover hypothesis and (2) the Dispersal hypothesis. The latter may result from either (i) the presence of novel enemies or, conversely, (ii) enemy release. Under the Land Cover hypothesis, we predicted that regions that are highly anthropogenically disturbed would have higher parasite prevalence, because the dominant disturbance in the region is agriculture, and agricultural irrigation can create ideal vector habitat (see Patz et al., 2008). Under the Dispersal hypothesis, we predicted that prevalence of parasites would either be lowest in areas more recently colonized by golden-winged warblers (Enemy Release hypothesis) or be highest in the areas most recently colonized by golden-winged warblers (Novel Enemies hypothesis).
Methods
Field methods
We mist-netted and blood-sampled 257 golden-winged warblers (232 males, 24 females and one individual of unrecorded sex) from six study areas across their breeding range between 2005 and 2015 (Fig. 1). All samples were collected during the summer breeding season. Two of these study areas were in Manitoba: one in the west near Riding Mountain National Park (50.887°N, 99.686°W; 2009 n = 19, 2015 n = 31), and one in the southeast near the town of Richer (49.764°N, 96.528°W; 2009 n = 6, 2015 n = 44). Two study areas were in southern Ontario, a western site near the town of Carden (44.683°N, 79.057°W; 2015 n = 25), and an eastern site near Frontenac Arch Provincial Park (44.634°N, 76.332°W; 2015 n = 46). Our two final study areas were in the United States, in Kentucky, near the town of Pineville (36.810°N, 83.759°W; 2005 n = 6, 2006 n = 8, 2007 n = 20, 2008 n = 4) and Wisconsin, near the Sandhill State Wildlife area (44.295°N, 90.191°W; 2010 n = 48). We searched for golden-winged warblers in early successional aspen parkland or mixed forest ecosystems, and targeted the searches based on aerial surveys for available habitat and/or previous survey sightings (Moulton and Artuso, 2017; Rondel and Bird Studies Canada, unpublished results). This work was performed under permit by the University of Manitoba Animal Care Committee Protocol F2015-004 (AC11030).
Once a male golden-winged warbler was located, we identified the bird's territory and attracted the bird to 6 or 12-m mist nets using a conspecific territorial song playback. Upon capture, a ~20 µL blood sample was obtained from the brachial vein of each golden-winged warbler and stored in 85% ethanol or a tissue lysis buffer (White and Densmore, 1992), and the bird was released at the location of capture. To determine whether parasites might be transmitted on the breeding grounds, we also sampled resident black-capped chickadees (Poecile atricapillus) co-occurring in golden-winged warbler habitat (Meixell et al., 2016). We used the same method to capture black-capped chickadees. However, as we only caught 14 chickadees for this purpose, we interpret these results with caution, and also used a global database of avian haemosporidian parasites (MalAvi) to locate lineages from data that matched the lineages found in this study, and to determine whether these matching lineages were obtained in resident and migrant avian species (Bensch et al., 2009). A matching lineage in a resident species in North America provides evidence that a lineage can be transmitted on the breeding grounds, as resident species by definition do not leave the breeding grounds and therefore must have acquired parasites there. Further, we also screened 100 golden-winged warbler nestlings sampled in Manitoba in previous years, none of which revealed any infections (Enslow and Moulton, unpublished results). As the nestlings were only 5 days old, it is unlikely that they would have both acquired an infection and had it progress to a stage that is detectable within their blood.
Laboratory methods
To determine blood parasite composition, we extracted DNA from all 257 golden-winged warbler and 14 black-capped chickadee blood samples using a homemade kit (Ivanova et al., 2006). To screen for the presence of blood parasites, we used two primer sets that amplified overlapping regions of cytochrome b. Polymerase chain reaction (PCR) protocol A followed the methods of Vallender et al. (2012), which first looked at blood parasites in golden-winged warblers. In this protocol, primers L15183 (Szymanski and Lovette, 2005) and H15725 (Ricklefs and Fallon, 2002) amplified a 550 bp region in cytochrome b, typically covering the two parasite genera Plasmodium and Parahaemoproteus. PCR protocol B followed the nested PCR protocol outlined in Hellgren et al. (2004), which uses three PCR steps to detect parasite presence to a dilution of 1:10 000 parasite cell to blood cells. Under protocol B, the first two primers (HaemNF and HaemNR2) initially amplified a 682-bp region of cytochrome b. We then performed a second and third PCR, each using the product from the first PCR. In the second PCR, nested primers (HaemF and HaemR2) isolated a smaller 480-bp region that is present and distinguishable in Parahaemoproteus and Plasmodium species. In the third PCR, nested primers HaemFL and HaemR3L isolated Leucocytozoon species. We tested for the presence of successfully amplified parasite DNA by running each PCR product through an ethidium bromide-stained agarose TAE gel. We viewed these gels through UV light using a Kodak–Fisher Scientific Gel Logic 100 Imaging System and scored birds as infected or not based upon the presence or absence of PCR product, respectively. All PCR assays were run with positive and negative controls; false negatives resulted from failed PCRs and were re-run, while false positives indicated contamination or amplification of a non-target gene and resulted in refreshing reagents and re-running the PCR. A companion to this study determined that protocol B had higher success at detecting infections (Enslow, 2017); therefore, samples initially screened with protocol A were screened again using protocol B.
To identify the parasite to lineage, we sequenced positive blood parasite infections by purifying the PCR product and using a Big Dye sequencing method. Some samples were purified twice: in the first purification, we added 0.02 µL Exo, 0.2 µL of SAP (Applied Biosystems Canada, Burlington, Ontario) and 3.78 µL of DNA grade water (BioShop Canada, Burlington, Ontario) to the PCR product of each positively infected sample, and held this mixture at 37 °C for 15 min then at 80 °C for 15 min in a thermocycler (Eppendorf Mastercycler ep gradient S; Eppendorf Canada, Mississauga, Ontario). When we did not use the first purification step, we diluted the PCR product depending on the brightness of the band, with a minimum of 0× dilution (weak PCR bands) and a maximum of 5× dilution (strong PCR bands). Following the first purification, or dilution, we used each successfully amplified PCR product in two separate sequencing reactions. The first reaction combined 1 µL of PCR product with 1.5 µL ABI buffer, 0.5 µL of forward primer and 0.4 µL of Big Dye (Applied Biosystems Canada, Burlington, Ontario). The second reaction was identical but used the reverse primer to produce two sets of sequence data per sample; the second sequence was used to verify the data and minimize the number of ambiguous bases in the final sequence. We then purified and precipitated DNA from the sequencing reaction by adding 1 µL sodium hydroxide and 1 µL EDTA to each sample, washing and drying the samples with 70 and 90% ethanol, drying the precipitate in a Vacufuge™ (Eppendorf Canada, Mississauga, Ontario), and rehydrating the DNA with 15 µL of HI-DI™ Formamide (Applied Biosystems Canada, Burlington, Ontario). We sequenced the DNA-HIDI mixture on an ABI 3130 XL Automated Sequencer (Applied Biosystems Canada, Burlington, Ontario). We obtained the sequence data and cleaned and aligned sequences using Geneious version 10 (Kearse et al., 2012). We discovered the most similar reported parasite lineages using MalAvi BLAST (Bensch et al., 2009) or NCBI BLAST (Altschul et al., 1990), and used this information to assign each infection to the appropriate genus (Parahaemoproteus, Plasmodium or Leucocytozoon).
Statistical and spatial analysis
We tested for significant differences in parasite prevalence among study areas using generalized linear models with binomial distributions. The proportion of ‘successes’ (infections) exceeded 10% across the dataset. We ran models with each genus of parasite as the response variable and study site as the predictor variable, comparing differences among all regions, in ‘R’ 3.1.4 (R Core Team, 2015), using the glm (generalized linear model) function, and the brglm (bias-reduced generalized linear model) function in the package brglm (Kosmidis, 2013); brglm was used to test for significant differences when perfect separation occurred in the data (i.e. when any study area had zero infections of one parasite genus). We evaluated whether potentially biologically important variables (sex and capture year) should be included in the model using AIC and parsimony (Burnham and Anderson, 2002). Neither additional variable reduced the AIC by 2 points or more (see Table S1), so these models were not selected and for brevity, the results are not detailed here.
To test for genetic variation in haemosporidian parasite lineages among geographic areas, we used analysis of molecular variance (AMOVA), executed in Arlequin 3.5 (Excoffier and Lischer, 2010). We used Kimura-2 to calculate sequence distances (Kimura, 1980), and Monte Carlo simulation with 16 000 permutations to test for significance (as suggested by Guo and Thompson, 1992). Since Kentucky only contained one Plasmodium infection and zero Leucocytozoon infections it was only included in the Parahaemoproteus analysis. Ontario samples were also pooled because Carden and Frontenac had low sample sizes of Leucocytozoon (n = 1, Carden) and Plasmodium (n = 2, Frontenac), respectively, and successful sequences revealed that these regions contained identical parasite lineages. The genetic differentiation we present here is likely a minimum estimate because AMOVA tends to underestimate differentiation, especially with the relatively small sample sizes of infected birds found here (Fitzpatrick, 2009; Bird et al., 2011). We also used Arlequin to test for significance of paired FST values by permuting lineages among populations (Excoffier and Lischer, 2010). We compared the P value of pairwise FST values among the populations to determine which populations differed. Given that small differences in haemosporidian cytochrome b sequences have been associated with rapid speciation (Bensch et al., 2004; Ricklefs et al., 2014), we considered a lineage distinct if it was one base pair different from others, and counted any nucleotide difference, silent or non-silent (e.g. Lewicki et al., 2015).
Land cover was described at the site-level (‘landscape’) scale. We created a 50-km buffer around the centre-point of each of the six study areas, as that was the geographic extent that included all sample points for each study site, and determined the proportion of agricultural habitat within the resulting circular landscape, using ArcMap 10.2 (ESRI, 2011). The regional scale was chosen to encompass each population that to the best of our knowledge would have similar historical origins. Ontario and Manitoba study sites were analysed separately, a distinction was expected due to their different historical origins. Because we had six landscapes, we used a single variable to describe the amount of human disturbance within each landscape to reduce the risk of overparameterization. We chose proportion of agriculture as this index as it was the most extensive human disturbance across our study regions. To ensure this was a reasonable index of disturbance, we ran preliminary analyses to compare this model with those using alternate disturbance variables, including total human disturbance (agricultural plus other anthropogenic lands, such as mines and urban). Results were similar regardless of which fixed variable we used, but were strongest for the agriculture variable, so for conciseness we only show that model. We also included the study area as a random effect. For each Manitoba study area, our samples from 2015 were used to calculate this centre-point. Land cover sources differed depending on the broad study area; for Kentucky and Wisconsin we used the USGS Geological survey data (US Geological Survey, 2011); for Manitoba we used the Manitoba Land Initiative land use data (Manitoba Conservation, 2005); and for Ontario we combined two datasets from the Southern Ontario Land Resource Information System, using the older dataset in the geographic area that was not covered by the newer dataset (Ontario Ministry of Natural Resources 2008; Ontario Ministry of Natural Resources and Forestry, 2011). One sample from Wisconsin was excluded because an accurate GPS point was not recorded for this sample. We used binomial generalized linear mixed models in ‘R’ 3.1.4, package lme4, to test for an association between percent agriculture and occurrence of the three parasite genera. We included study area as a random effect, with percent agriculture as the fixed effect.
Results
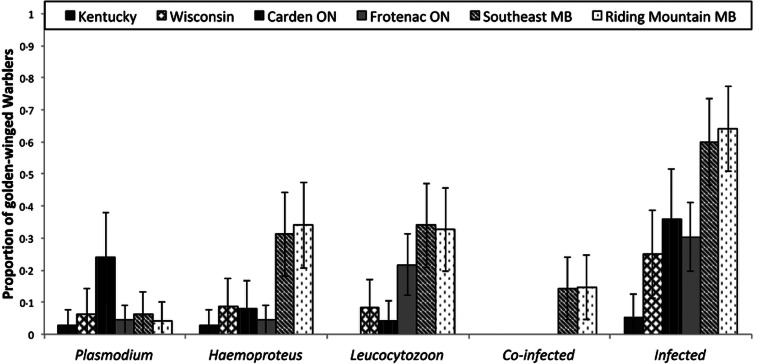
We detected 99 infected individuals out of 257 sampled golden-winged warblers. Five positive infections from Manitoba produced weak PCR bands that either corresponded to Parahaemoproteus or Plasmodium but could not be sequenced and thus were removed from further analyses involving either of these genera. Prevalence of infection varied significantly among study areas (Fig. 2; Table 1). Both Manitoba study sites (Riding Mountain National Park, and Southeast Manitoba) had more overall infections than any other sites, and this trend was driven by the significantly higher prevalence of Parahaemoproteus in Manitoba compared with the other five study sites (Table 1). Both Manitoba study sites and the eastern Ontario study site, Frontenac Arch, had a similar prevalence of Leucocytozoon parasites, but these three sites had greater Leucocytozoon prevalence than the remaining study sites (Table 1). Kentucky, on the other hand, had the fewest infections overall, and tended to have fewer infections of every parasite genus, although this trend was not always statistically significant (Table 1). Both Manitoba sites also had significantly more co-infections (infections of more than one genus of parasite) than nearly all other study sites (Fig. 2; Table 1). The study site that did not have significantly fewer co-infections than Manitoba (Carden) had zero co-infections, but there may be co-infections that are present and undetected due to the smaller number of samples from this site.
Fig. 2.
Prevalence of Plasmodium, Parahaemoproteus, Leucocytozoon infections in golden-winged warblers across five study sites across the golden-winged warbler's range. Different letters represent significant differences (P < 0.1) within the parasite genus among sites. Sample sizes were: Kentucky, n = 38 (2005, 2006, 2007, 2008), Manitoba, n = 100 (2009, 2015), Ontario, n = 71 (2015), WI = Wisconsin (2010), n = 48. Error bars denote 95% confidence intervals.
Table 1.
Comparison of the prevalence of Plasmodium, Parahaemoproteus and Leucocytozoon infections in golden-winged warblers across five study sites across the golden-winged warbler's range
| Genus | Area | Pre- or post-1920 range | Prevalence (%)±s.e. | Comparison with other study areas | s.e. MB | ||||
|---|---|---|---|---|---|---|---|---|---|
| Carden | Frontenac | Kentucky | RMNP | ||||||
| Parahaemoproteus | |||||||||
| Carden, ON (2015) | Post | 8.00 ± 0.7 | |||||||
| Frontenac, ON (2015) | Post | 4.35 ± 0.29 | E = −0.65, P = 0.001 | ||||||
| Kentucky (2005, 2006, 2007, 2008) | Pre | 2.63 ± 0.42 | E = −1.17, P = 0.351 | E = −0.52, P = 0.68 | |||||
| RMNP, MB (2009, 2015) | Post | 34.04 ± 0.99 | E = 1.78, P = 0.03 | E = 2.43, P = 0.002 | E = 2.95, P = 0.005 | ||||
| s.e. MB (2009, 2015) | Post | 31.25 ± 0.97 | E = 1.65, P = 0.04 | E = 2.30, P = 0.003 | E = 2.82, P = 0.008 | E = −0.13, P = 0.77 | |||
| Wisconsin (2010) | Pre | 8.51 ± 0.58 | E = 0.67, P = 0.94 | E = 0.72, P = 0.42 | E = 1.24, P = 0.28 | E = −1.71, P = 0.005 | E = −1.59, P = 0.009 | ||
| Leucocytozoon | |||||||||
| Carden, ON (2015) | Post | 4.00 ± 0.59 | |||||||
| Frontenac, ON (2015) | Post | 21.74 ± 0.59 | E = 1.90, P = 0.08 | ||||||
| Kentucky (2005, 2006, 2007, 2008) | Pre | 0 | E = 1.55, P = 0.36 | E = −3.10, P = 0.037 | |||||
| RMNP (2009, 2015) | Post | 32.65 ± 0.94 | E = 2.52, P = 0.02 | E = 0.62, P = 0.19 | E = 3.69, P = 0.0121 | ||||
| s.e. MB (2009, 2015) | Post | 34.00 ± 0.95 | E = 2.52, P = 0.02 | E = 0.618, P = 0.19 | E = 3.69, P = 0.012 | E < 0.001, P > 0.999 | |||
| Wisconsin (2010) | Pre | 8.33 ± 0.58 | E = 0.78, P = 0.50 | E = −1.53, P = 0.043 | E = 2.05, P = 0.18 | E = −1.74, P = 0.0039 | E = −1.74, P = 0.0039 | ||
| Plasmodium | |||||||||
| Carden, ON (2015) | Post | 24.00 ± 1.12 | |||||||
| Frontenac, ON (2015) | Post | 4.35 ± 0.29 | E = −1.94, P = 0.02 | ||||||
| Kentucky (2005, 2006, 2007, 2008) | Pre | 2.63 ± 0.42 | E = −2.46, P = 0.028 | E = −0.52, P = 0.68 | |||||
| RMNP (2009, 2015) | Post | 4.26 ± 0.43 | E = −1.96, P = 0.023 | E = −0.023, P = 0.98 | E = 0.50, P = 0.69 | ||||
| s.e. MB (2009, 2015) | Post | 6.25 ± 0.50 | E = −1.55, P = 0.04 | E = 0.38, P = 0.68 | E = 0.90, P = 0.43 | E = 0.41, P = 0.67 | |||
| Wisconsin (2010) | Pre | 6.38 ± 0.51 | E = −0.53, P = 0.043 | E = 0.41, P = 0.67 | E = 0.93, P = 0.43 | E = 0.43, P = 0.65 | E = 0.02, P = 0.98 | ||
| Co-infection | |||||||||
| Carden, ON (2015) | Post | 0 | |||||||
| Frontenac, ON (2015) | Post | 0 | E = −0.601, P = 0.769 | ||||||
| Kentucky (2005, 2006, 2007, 2008) | Pre | 0 | E = −0.412, P = 0.84 | E = 0.189, P = 0.93 | |||||
| RMNP (2009, 2015) | Post | 14.58 ± 0.74 | E = 2.22, P = 0.14 | E = 2.82, P = 0.06 | E = 2.63, P = 0.08 | ||||
| s.e. MB (2009, 2015) | Post | 14.28 ± 0.73 | E = 2.19, P = 0.15 | E = 2.78, P = 0.06 | E = 2.61, P = 0.081 | E = −0.24, P = 0.97 | |||
| Wisconsin (2010) | Pre | 0 | E = −0.64, P = 0.75 | E = −0.04, P = 0.98 | E = −0.23, P = 0.91 | E = −2.86, P = 0.05 | E = −2.84, P = 0.06 | ||
RMNP, Riding Mountain National Park in Manitoba, SE MB, southeastern Manitoba; ON, Ontario; E, estimate from the binomial generalized linear model, in this case it is the estimated increase in the log odds of a bird being infected with the specified parasite genus between the compared study areas.
Significant differences are bolded (P < 0.1).
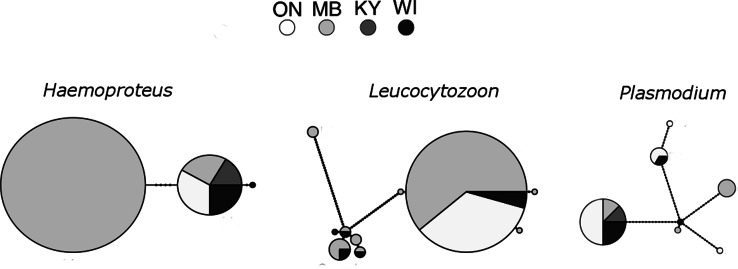
We found 20 unique Haemosporidian lineages, and these displayed some geographic structure. Of these, three belonged to Parahaemoproteus, ten belonged to Plasmodium, and seven belonged to Leucocytozoon. AMOVAs revealed that Plasmodium was not genetically distinct among the study areas (P = 0.214), though FST analysis suggested that the Manitoba and Ontario populations of Plasmodium parasites are the most different from one another (Table 2). Manitoba also had one uncommon parasite lineage, BT7, which was not found in any other population (Fig. 3). Ontario had two unique Plasmodium lineages, one of which is 1.79% divergent from other reported lineages, and may be considered an unrecorded species (Table 3; Ricklefs et al., 2014). The AMOVA for Leucocytozoon displayed a different pattern: the populations were significantly differentiated (P = 0.010), and FST testing suggested that the Manitoba, Ontario, and Wisconsin populations were all significantly different (Table 2). Manitoba and Ontario share one very common lineage that is considerably less common in Wisconsin, and Wisconsin and Manitoba share a complex of similar parasite lineages (Fig. 3). Further, all populations with Leucocytozoon infections had at least one lineage that was unique to that population (Fig. 3). The AMOVA for Parahaemoproteus also revealed that the populations were significantly differentiated (P < 0.001), which was driven by differences between Manitoba and the other sites (Table 2). This is clear from the lineage network, as Manitoba has one unique and very common lineage that is not found in the other study areas (Fig. 3).
Table 2.
FST test results comparing frequency and genetic distance of lineages of Parahaemoproteus, Plasmodium and Leucocytozoon among golden-winged warbler populations
| Manitoba | Ontario | Wisconsin | |
|---|---|---|---|
| Parahaemoproteus | |||
| Ontario | 0.745 (P = 0.010*) | – | 0 (P > 0.999) |
| Wisconsin | 0.754 (P = 0.002*) | 0 (P > 0.999) | 0 (P > 0.999) |
| Kentucky | 0.732 (P = 0.030*) | 0 (P > 0.999) | 0 (P > 0.999) |
| – | |||
| Plasmodium | |||
| Wisconsin | 0 (P = 0.453) | 0 (P = 0.873) | |
| Ontario | 0.147 (P = 0.056*) | – | 0 (P = 0.873) |
| Leucocytozoon | |||
| Wisconsin | 0.184 (P = 0.064*) | 0.791 (P = 0.005*) | – |
| Ontario | 0.092 (P = 0.093*) | – | 0.791 (P = 0.005*) |
Values presented are pairwise FST calculated using the following formula: (average number of base pair differences between populations − average number of base pair differences among populations)/average number of base pair differences among populations. The significance of paired FST values was calculated by permuting lineages among populations, and P indicates the proportion of randomly permuted populations that matched or exceeded the variation found in the actual populations.
Fig. 3.
Haplotype networks displaying frequencies and genetic distance among Plasmodium, Leucocytozoon and Parahaemoproteus parasite lineages in different golden-winged warbler populations. KY = Kentucky, n = 38 (2005, 2006, 2007, 2008), MB = Manitoba, n = 100 (2009, 2015), ON = Ontario, n = 71 (2015), WI = Wisconsin, n = 48 (2010). Each circle is a unique lineage (differs from other lineages by at least one base pair), the size of the circle corresponds to the relatively frequency of the lineage, and the colour of the shape shows which study site it is found in. Labels denote unique lineages (lineages that differ from a central lineage by more than 2 base pairs).
Table 3.
Parasite lineagesa present in golden-winged warblers in Kentucky (2005, 2006), Manitoba (2009, 2010, 2015), Ontario (2015) and Wisconsin (2009, 2010)
| Genus | Lineagea | Study sitesb | Nearest reported lineagec | PNDd (%) | Accession number |
|---|---|---|---|---|---|
| Plasmodium | |||||
| GWWAP01 | MB(1), WI(2), ON(4), KY(1) | GEOTRI09 | 0 | MN114074 | |
| GWWAP02 | WI(1) | RWB01 | 0.34 | MN153504 | |
| GWWAP03 | MB(1) | RWB01 | 0.35 | MN153505 | |
| GWWAP04 | ON(1) | CXRES06, AMMAUR01 | 1.88 | ||
| MN114075 | |||||
| GWWAP05 | MB(3) | BT7 | 0 | MN114076 | |
| GWWAP06 | WI(1), ON(2) | CATUST05, TFUS05, APSPI03 | 0 | MN114077 | |
| GWWAP07 | ON(1) | WW3 | 0 | MN114078 | |
| Parahaemoproteus | |||||
| GWWAH01 | MB(27) | PASILI01 | 0 | MN114079 | |
| GWWAH02 | MB(3), WI(2), ON(4), KY(2) | DENADE01 | 0.42 | MN114080 | |
| GWWAH03 | WI(1) | DENADE01 | 0 | MN153506 | |
| Leucocytozoon | |||||
| GWWAL01 | WI(1) | EMPALN01 | 0.215 | MN114064 | |
| GWWAL02 | MB(1), WI(1) | EMPALN01, COLBF21 | 0 | MN114065 | |
| GWWAL03 | MB(3), WI(1) | DUMCAR03 | 0.216 | MN114066 | |
| GWWAL04 | MB(1), WI(1) | DUMCAR05 | MN114067 | ||
| GWWAL05 | MB(2) | DUMCAR03, DUMCAR02 | 0 | MN114068 | |
| GWWAL06 | MB(2) | DUMCAR01 | 0 | MN114069 | |
| GWWAL07 | MB(1) | COLBF23 | 0 | MN114070 | |
| GWWAL08 | ON(1) | CNEORN01 | 0.215 | MN114071 | |
| GWWAL09 | MB(1) | CNEORN01 | 0.215 | MN114072 | |
| GWWAL10 | MB(13), WI(1), ON(8) | CNEORN01, SPIPAS07, PHEMEL01 | 0 | MN114073 |
Lineages differ from each other by at least one nucleotide difference.
MB, Manitoba; ON, Ontario; WI, Wisconsin; KY, Kentucky.
Bensch et al. (2009).
Pairwise nucleotide distance from nearest reported lineage.
Evidence for breeding vs wintering ground transmission
Of the 14 black-capped chickadees screened, eight were infected with Leucocytozoon, two of those were co-infected with Plasmodium, and one was only infected with Plasmodium. The Plasmodium lineage that was only found in Manitoba, BT7, was also found in black-capped chickadees. Black-capped chickadees in Manitoba were also infected with the Leucocytozoon lineage DUMCAR03, demonstrating that this lineage may be transmitted during the breeding season in Manitoba. No resident black-capped chickadees were infected with Parahaemoproteus parasites.
Based on comparison with the MalAvi database, we ascertained that at least one of our Parahaemoproteus lineages is probably transmitted on the breeding grounds. The common but Manitoba-specific Parahaemoproteus lineage (PASILI01) has been found in hatch-year fox sparrows (Passerella iliaca) that were caught on their breeding grounds in California (Walther et al., 2016); suggesting that this lineage is transmittable in breeding grounds in North America. PASILI01 has also been found in a resident American crow (Corvus brachyrhynchos) in Alaska, and migratory yellow-rumped warblers (Setophaga coronata) in Alaska and New Mexico (Galen et al., unpublished results; Williamson et al., unpublished results). To the best of our knowledge, the uncommon lineage with one match in Wisconsin, DENADE01, has only ever been found in the West Indies before (Ricklefs et al., 2014). The common lineage that was found in all sites, GWWAH02, is not yet reported in the genetic databases (Table 3).
Most of the Leucocytozoon lineages found here are also probably transmitted on the breeding grounds. The lineage COLBF23, which was found in one bird in Manitoba only, has not yet been reported in a host species, but has been found in black flies (Simulium silvestre) in the Rocky Mountains of Colorado (Murdock et al., 2015). Due to the coverage of our sequences, we could not distinguish between DUMCAR03 and DUMCAR01; however, both have been found in resident species in Michigan: the lineage DUMCAR03 has been found in resident tufted titmouse (Baeolophus bicolor) and migrant grey catbird (Dumetella carolinensis), while DUMCAR01 was found resident a resident northern cardinal (Cardinalis cardinalis), and several migratory species (Smith et al., 2017). DUMCAR01 was also found in migratory yellow-rumped warblers in BC and Alberta, Canada (Cozzarolo et al., 2018). DUMCAR05 has only been reported in migratory grey catbird in Michigan (Smith et al., 2017). We were unable to distinguish between COLBF21 and EMPALN01, but both have been found in resident species. EMPALN01 has been found in two resident species in California (Fruend et al., 2016), while COLBF21 has been found in resident northern cardinals in Michigan (Smith et al., 2017). Both have also been found in a number of migratory species in western/southwestern North America (Bensch et al., 2009). For the most common and widespread Leucocytozoon lineage in this study, we were unable to distinguish between CNEORN01, SPIPAS07 and PHEMEL01 due to the coverage of our sequence (Table 3). CNEORN01 is also the most common lineage of the three in the literature, and has been found in black flies in the Rocky Mountains of Colorado (Murdock et al., 2015) resident northern cardinals in Michigan (Smith et al., 2017), and in numerous migratory species in Alaska (Oakgrove et al., 2014; Cozzarolo et al., 2018; Galen unpublished results), California (Walther et al., 2016), and New Mexico (Smith et al., 2017). CNEORN01 is very abundant across the hybrid zone between yellow-rumped warbler subspecies in Alberta and British Columbia (Cozzarolo et al., 2018). SPIPAS07 has only been reported in a migratory chipping sparrow (Spizella passerina) in Michigan (Smith et al., 2017), and PHEMEL01 has only been found in migratory black-headed grosbeak (Pheucticus melanocephalus) in California (Walther et al., 2016).
Our results, in combination with a survey of the literature, strongly support a wide-ranging, continental distribution for Plasmodium parasites. The lineage BT7, the Plasmodium found in local black-capped chickadees, is widely reported in the literature, and appears to be nearly globally distributed (Bensch et al., 2009). BT7 has been found in Alaska, Vermont, and California in the US, and in many regions across Eurasia. Evidence of transmission for this parasite in North America is limited to California (Walther et al., 2016), Alaska (Oakgrove et al., 2014), and now Manitoba. The lineage WW3 has a near-global distribution similar to BT7, with evidence of transmission in breeding and wintering grounds: it has been found in resident northern cardinal, and house finch (Haemorhous mexicanus) in Michigan (Smith et al., 2017), but also multiple resident species in Peru (Fecchio et al., 2019). The lineage GEOTRI09 has only been found in a handful of migratory species across North America (Oakgrove et al., 2014; Sarquis-Adamson and MacDougall-Shackleton, 2016; Smith et al., 2017; Galen et al., unpublished results). Due to differential sequence coverage, we were unable to distinguish between the Plasmodium lineages CATUSTO05, APSP103, and TFUS05. CATUSTO05 may be transmitted on both wintering and summering grounds: it has been found in a number of migratory species in numerous locations across North America (Bensch et al., 2009), but has also been found in resident tufted titmouse in Michigan (Smith et al., 2017), and numerous resident species in South America (González et al., 2015; Fecchio et al., 2019). APSP103 has been found in migratory species in several locations in Canada and the US (Pagenkopp et al., 2008; Sarquis-Adamson and MacDougall-Shackleton, 2016), and has been found in resident thorn-tailed rayadito (aphrastura spinicauda) in Chile (Merino et al., 2008). TFUS05 has only been found in great thrush in Colombia (Mantilla et al., 2013).
Site-scale cover of agriculture was positively associated with the prevalence of Parahaemoproteus (β = 8.22, s.e. = 2.26, P < 0.001) and Leucocytozoon parasites (β = 6.93, s.e. = 3.38, P = 0.0407), but was independent of Plasmodium prevalence (P = 0.717).
Discussion
Our results demonstrate high blood parasite prevalence in some populations of golden-winged warblers. Parasite prevalence generally increases to the north-west across the range of the golden-winged warbler. More than half of the Manitoba golden-winged warblers sampled carried at least one genus of a blood parasite, a frequency ten times higher than the site with the lowest prevalence, Kentucky (see also Vallender et al., 2012). Manitoba also has the highest number of co-infections, which can have synergistic fitness consequences resulting in poor body condition and mortality (Palinauskas et al., 2011). The Manitoba population is of great conservation importance, as it appears to have the lowest rates of hybridization with the blue-winged warbler (Vermivora cyanoptera; Moulton et al., 2018). Hybridization between these sister species is believed to have been a significant cause of precipitous population declines in golden-winged warblers over the past forty years (Gill, 1997; Vallender et al., 2007; Sauer et al., 2011). Unfortunately, the Manitoba population appears to be at a fitness disadvantage in terms of lower survival and reduced reproductive output compared to eastern Canada (Moulton, 2017), and we speculate that high parasite prevalence may contribute to this trend. However, further work is needed to elucidate the effect of these parasites on the Manitoba population, as infection with haemosporidian parasites is not inherently negative, and might even result in positive reproductive outputs (Zylberberg et al., 2015). The extensive variation in parasite prevalence among our sites suggests that populations may face differing pressure from parasites, and this might be of particular conservation concern for the ecologically unique and highly parasitized Manitoba population.
We found a considerable genetic structure to parasite communities within the golden-winged warbler across its range. Golden-winged warblers were infected with a number of population-specific lineages, and two of the three parasite genera (Parahaemoproteus and Leucocytozoon) were significantly genetically differentiated among populations. Parahaemoproteus and Leucocytozoon lineages are more likely to be geographic specialists than Plasmodium lineages, which are typically widespread and are host generalists (Olsson-Pons et al., 2015), consistent with our results.
Golden-winged warbler populations in geographic regions with high cover of agriculture had more infections of Parahaemoproteus and Leucocytozoon. However, there is a strong correlation between latitude and agriculture (r = 0.80); therefore, the effect could be related to either geographic location or disturbance, and we must be cautious with the interpretation of these results. This is especially true in the case of Leucocytozoon, which is known to be more likely to be transmitted at higher latitudes and cooler temperatures (Pérez-Rodríguez et al., 2013; Galen and Witt, 2014), and may actually be constrained by higher temperatures (Fecchio et al., 2019). Our results differ from the findings of some other, particularly tropical studies, that have found an increase in anthropogenic disturbance to be associated with decreased parasite prevalence (see Sehgal, 2010; Loiseau et al., 2012b; Laurance et al., 2013). Variation in responses to landscape change is unsurprising because landscape change results from a wide diversity of human disturbances, and the mechanism of change for parasite communities is likely to change depending on the type of disturbance (Brearley et al., 2013). While contiguous forest in tropical regions may contribute to vector habitat (Bonneaud et al., 2009), agricultural disturbance may increase vector habitat in more temperate regions. Agriculture may increase parasite prevalence if associated irrigation creates wet and open habitat, which is suitable larval habitat for the two species of biting midges that transmit Parahaemoproteus (Kardatzke and Rowley, 1971; Patz et al., 2008). Similarly, though not suitable for all black fly species, irrigation can create larval habitat for Leucocytozoon transmitting black flies (Fredeen and Shemanchuck, 1960; Patz et al., 2008). Though we might expect pesticide use to deter the presence of larvae, some black flies are resistant to non-target commercial pesticides (Montagna et al., 2012).
Our results are not consistent with the Enemy Release hypothesis, because the most recent region to be colonized, Manitoba (Rosenberg et al., 2016), has a high level of parasitism. Our results are more consistent with the Novel Enemies hypothesis: that golden-winged warblers are susceptible to novel parasites as they colonize new areas. We detected an increasing trend in parasite infection from the southeast to the northwest across the golden-winged warbler's range, consistent with its historical range expansion (Confer et al., 2011; Fig. 1). The pattern of Parahaemoproteus and Leucocytozoon lineage distributions is also consistent with the Novel Enemies hypothesis. Golden-winged warblers in our most southeastern study site, Kentucky, had the fewest lineages of parasites, and the one lineage of Parahaemoproteus in Kentucky was also present in every other study area. Consistent with historic and natal dispersal trends from the southeast to the northwest: golden-winged warblers in Ontario and Manitoba share a common Leucocytozoon lineage that is also present but much less common in Wisconsin, and Golden-winged warblers in Wisconsin and Manitoba share a complex of very similar Leucocytozoon parasites that are not found in other study areas. Manitoba also has at least one unique and distinct parasite lineage from each parasite genus, consistent with the Novel Enemies hypothesis, as golden-winged warblers appear to rarely migrate south from Manitoba, which could limit movement of unique parasites (Lopez-Calderon et al., 2019). Further, differences in parasite communities between the two Ontario study sites are also consistent with the Novel Enemies hypothesis. Although the sites in Ontario are close in proximity, McCracken's re-creation (1994) of the golden-winged warbler's expansion into central Ontario indicates that the Frontenac and Carden populations might not be part of the same colonizing wave, but rather originated from separate northward colonization events from either side of Lake Ontario. Consistent with these historical origins, the dominant parasite genus is different between the two sites; Plasmodium is significantly more abundant in the western site and Leucocytozoon is significantly more abundant in the eastern site. Overall, the patterns observed among parasite lineages are correlated with the historical dispersal patterns of golden-winged warblers. However, as with any correlational study, we cannot be certain of the mechanism.
Both Dispersal hypotheses are robust to violations of the assumption that parasites are transmitted on the breeding ground. Migratory connectivity between golden-winged warbler populations is strong (Kramer et al., 2017). If a parasite lineage is transmitted on the wintering ground, the only aspect of the Dispersal hypothesis that changes is that the lineage disperses between wintering ground locations instead of breeding ground locations. In contrast, the Land Cover hypothesis requires the assumption that birds were infected where we analysed the proxies for vector habitat: the breeding grounds. To examine whether parasites may be transmitted on the breeding grounds, we screened parasites of resident black-capped chickadees, and also compared parasite lineages we found in golden-winged warblers with those in the MalAvi database. There are limitations to comparing parasites between different species; black-capped chickadees and golden-winged warblers may have different parasite communities because they are not closely related (Jetz et al., 2012) and use different habitat types (Foote et al., 2010; Confer et al., 2011). Despite these limitations, because both species share some otherwise rare lineages, this and other studies (Bensch et al., 2009) suggest that at least some lineages are probably transmitted on the breeding grounds. Additionally, although further study is needed, recent studies indicate that parasites found on the breeding grounds are likely transmitted there as well (Hellgren et al., 2013; Ricklefs et al., 2016; Pulgarín et al., 2019).
An additional important caveat to this study is that many study areas were sampled in different years, or only 1 year. Although year was not a significant predictor of parasite prevalence (see ‘Methods'), without extensive multi-year sampling in each area, we cannot be sure that we have the definitive parasite composition for each area (Bensch et al., 2007).
Our results suggest that both land-cover characteristics and historical patterns of golden-winged warbler dispersal affect the community composition of blood parasites across the species' northern range. As the golden-winged warbler's range continues to gradually move northwards, as habitat allows (Rosenberg et al., 2016), the Novel Enemies hypothesis suggests that one of the challenges it might increasingly face across its northern extent is exposure to blood parasites to which it is not adapted. The Manitoba population of golden-winged warblers is likely to continue to increase in importance to conservation of the species, and unless the species rapidly evolves to tolerate these parasites, the high prevalence and diversity of parasites in this region may contribute one more threat to an already at-risk population.
Acknowledgements
This work would not have been possible without help from the many scientists that provided data and assistance to this research. Patricia Hartman and David Wesneat collected blood samples for the Kentucky dataset, and Monica Cong collected blood samples for Wisconsin. Rachel Vallender, Steve Van Wilgenberg, Marek Allen and Adam Crosby collected blood samples in Manitoba in 2009. Laurel Moulton assisted with sample collection and laboratory work for the Manitoba samples. Christian Artuso also provided extensive advice in the development of this project. Roger Bull provided assistance, advice, and training in laboratory work. We also thank the technicians and volunteers who helped collect data for this project: Callie Bowman, Sabina Mastrolonardo, Delaney Brooks, Christoph Ng, Marika Olynyk, and Erin Prokopanko. We thank Riding Mountain National Park and the Nature Conservancy of Canada for their generous in-kind support and the Canadian Museum of Nature for providing space in their beautiful and innovative DNA Lab.
Financial support
Financial support was provided by supporting institutions, the University of Manitoba and Bird Studies Canada. Grant funding that supported this research included the National Science and Engineering Council (N.K. 327777-2012, C.E. no grant number); and the Connie Holland Bird Studies Fund (C.E. 20151787).
Conflict of interest
None.
Ethical standards
This work was performed under permit by the University of Manitoba Animal Care Committee Protocol F2015-004 (AC11030). The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182019001240.
click here to view supplementary material
References
- Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology, 403–410. [DOI] [PubMed] [Google Scholar]
- Asghar M, Hasselquist D and Bensch S (2011) Are chronic avian haemosporidian infections costly in wild birds? Journal of Avian Biology 42, 530–537. [Google Scholar]
- Bensch S, Pérez-Tris J, Waldenström J and Olof H (2004) Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution 58, 1617–1621. [DOI] [PubMed] [Google Scholar]
- Bensch S, Waldenström J, Jonzén N, Westerdahl H, Hansson B, Sejberg D and Hasselquist D (2007) Temporal dynamics and diversity of avian malaria parasites in a single host species. Journal of Animal Ecology 76, 112–122. [DOI] [PubMed] [Google Scholar]
- Bensch S, Hellgren O and Pérez-Tris J (2009) Malavi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Molecular Ecology Resources 9, 1353–1358. [DOI] [PubMed] [Google Scholar]
- Bernotiene R and Bartkeviciene G (2013) The relationship between water temperature and the development cycle beginning and duration in three black fly species. Journal of Insect Science 13, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird CE, Karl S, Smouse PE and Toonen RJ (2011) Detecting and measuring genetic differentiation. In Held C, Koenemann S and Schubart C (eds), Crustacean Issues: Phylogeography and Population Genetics in Crustacea. Boca Raton, USA: CRC Press, pp. 31–73. [Google Scholar]
- Bishop AL, Mckenzie HJ, Barchia M and Harris AM (1996) Effect of temperature regimes on the development, survival and emergence of Culicoides brevitarsis Kieffer (Diptera: Ceratopogonidae) in bovine dung. Australian Journal of Entomology 35, 361–368. [Google Scholar]
- Bonneaud C, Sepil I, Milá B, Buermann W, Pollinger J, Sehgal RNM, Valkiūnas G, Lezhova Ta., Saatchi S and Smith TB (2009) The prevalence of avian Plasmodium is higher in undisturbed tropical forests of Cameroon. Journal of Tropical Ecology 25, 439–447. [Google Scholar]
- Brearley G, Rhodes J, Bradley A, Baxter G, Seabrook L, Lunney D, Liu Y and Mcalpine C (2013) Wildlife disease prevalence in human-modified landscapes. Biological Reviews 88, 427–442. [DOI] [PubMed] [Google Scholar]
- Buehler D, Roth A, Vallender R, Will T, Confer JL, Canterbury RA, Swarthout SB, Rosenberg KV and Bulluck LP (2007) Status and conservation priorities of golden-winged warbler (Vermivora chrysoptera) in North America. The Auk 124, 1439–1445. [Google Scholar]
- Burnham KP and Anderson DR (2002) Model Selection and Multimodel Inference, 2nd Edn. New York City, USA: Springer. [Google Scholar]
- Chasar A, Loiseau C, Valkiūnas G, Iezhova T, Smith TB and Sehgal RNM (2009) Prevalence and diversity patterns of avian blood parasites in degraded African rainforest habitats. Molecular Ecology 18, 4121–4133. [DOI] [PubMed] [Google Scholar]
- Confer JL, Hartman P and Roth A (2011) Golden-winged warbler (Vermivora chrysoptera). In Rodewald PG (ed.), The Birds of North America. Ithaca: Cornell Lab of Ornithology. Retrieved from the Birds of North America. Available at https://birdsna.org/Species-Account/bna/species/gowwar. [Google Scholar]
- Córdoba-Lloria S, Serna-Mompeán JP, Giménez-Gras O, Acosta-Aleixandre R and Bueno-Mari R (2017) Notes on black flies of the Júcar River and tributaries in Eastern Spain. The Simuliid Bulletin 48, 8–14. [Google Scholar]
- Cozzarolo C, Christe P, Jenkins T, Toews DPL and Brelsford A (2018) Prevalence and diversity of haemosporidian parasites in the rumped warbler hybrid zone. Ecology and Evolution 8, 9834–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enslow C (2017) Human Disturbance, Host Dispersal, and Hybridization Influence Blood Parasite Communities in a Threatened Songbird Species: The Golden-Winged Warbler (Vermivora Chrysoptera) (thesis). University of Manitoba, Winnipeg, Canada. [Google Scholar]
- ESRI (2011) ArcGIS Desktop: Release 10. Redlands, CA: Environmental Systems Research Institute. [Google Scholar]
- Excoffier L and Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10, 564–567. [DOI] [PubMed] [Google Scholar]
- Fecchio A, Silveira P, Weckstein J and Anciaes M (2019) First record of Leucocytozoon (Haemosporida: Leucocytozoidae) in Amazonia: evidence for rarity in Neotropical lowlands or lack of sampling for this parasite genus? Journal of Parasitology 104, 168–172. [DOI] [PubMed] [Google Scholar]
- Figuerola J, Munoz E, Gutierrez R and Ferrer D (1999) Blood parasites, leucocytes and plumage brightness in the cirl bunting, Emberiza cirlus. Functional Ecology 13, 594–601. [Google Scholar]
- Fitzpatrick BM (2009) Power and sample size for nested analysis of molecular variance. Molecular Ecology 18, 3961–3966. [DOI] [PubMed] [Google Scholar]
- Foote JR, Mennill DJ, Ratcliffe LM and Smith SM (2010) Black-capped chickadee (Poecile atricapillus). In Rodewald PG (ed.), The Birds of North America. Ithaca: Cornell Lab of Ornithology; Retrieved from the Birds of North America. Available at https://birdsna.org/Species-Account/bna/species/bkcchi. [Google Scholar]
- Fredeen FJH and Shemanchuk JA (1960) Black flies (Diptera: Simuliidae) of irrigation systems in Saskatchewan and Alberta. Canadian Journal of Zoology 38, 723–735. [Google Scholar]
- Freeman-Gallant CR, O'Connor KD and Breuer ME (2001) Sexual selection and the geography of Plasmodium infection in Savannah sparrows (Passerculus sandwichensis). Oecologia 127, 517–521. [DOI] [PubMed] [Google Scholar]
- Freund D, Wheeler SS, Townsend AK, Boyce WM, Ernest HB, Cicero C and Sehgal RNM (2016) Genetic sequence data reveals widespread sharing of Leucocytozoon lineages in corvids. Parasitology Research, 1–9. [DOI] [PubMed] [Google Scholar]
- Galen SC and Witt CC (2014) Diverse avian malaria and other haemosporidian parasites in Andean house wrens: Evidence for regional co-diversification by host-switching. Journal of Avian Biology 45, 374–386. [Google Scholar]
- Gill FB (1980) Historical aspects of hybridization between blue-winged and golden-winged warblers. The Auk 97, 1–18. [Google Scholar]
- Gill FB (1997) Local cytonuclear extinction of the golden-winged warbler. Evolution 51, 519–525. [DOI] [PubMed] [Google Scholar]
- González AD, Lotta Ia., García LF, Moncada LI and Matta NE (2015) Avian haemosporidians from Neotropical highlands: Evidence from morphological and molecular data. Parasitology International 64, 48–59. [DOI] [PubMed] [Google Scholar]
- Greiner EC, Bennett GF, White EM and Coombs RF (1975) Distribution of the avian hematozoa of North America. Canadian Journal of Zoology 53, 1762–1787. [DOI] [PubMed] [Google Scholar]
- Guo SW and Thompson EA (1992) Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48, 361–372. [PubMed] [Google Scholar]
- Hellgren O, Waldenström J and Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Parahaemoproteus from avian blood. The Journal of Parasitology 90, 797–802. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Wood M, Hasselquist D, Ottosson U, Stervander M and Bensch S (2013) Circannual variation in blood parasitism in a sub-Saharan migrant passerine bird, the garden warbler. Journal of Evolutionary Biology 26, 1047–1059. [DOI] [PubMed] [Google Scholar]
- Ivanova NV, DeWaard JR and Hebert PDN (2006) An inexpensive, automation-friendly protocol for recovering high-quality DNA. Molecular Ecology Notes 6, 998–1002. [Google Scholar]
- Jetz W, Thomas G, Joy JB, Hartmann K and Mooers A (2012) The global diversity of birds in space and time. Nature 491, 444–448. [DOI] [PubMed] [Google Scholar]
- Kardatzke JT and Rowley WA (1971) Comparison of Culicoides larval habitats and populations in central Iowa. Annals of the Entomological Society of America 64, 215–218. [Google Scholar]
- Keane RM and Crawley MJ (2002) Exotic plant invasions and the enemy release hypothesis. Trends in Ecology and Evolution 17, 164–170. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P and Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16, 111–120. [DOI] [PubMed] [Google Scholar]
- Kimura M, Dhondt AA and Lovette IJ (2006) Phylogeographic structuring of Plasmodium lineages across the North American range of the house finch (Carpodacus mexicanus). The Journal of Parasitology 92, 1043–1049. [DOI] [PubMed] [Google Scholar]
- Kosmidis I (2013) brglm: Bias reduction in binomial-response generalized linear models. Available at http://www.ucl.ac.uk/~ucakiko/software.html.
- Kramer GR, Streby HM, Peterson SM, Lehman JA, Buehler DA, Wood PB, McNeil DJ, Larkin JL and Andersen DE (2017) Nonbreeding isolation and population-specific migration patterns among three populations of golden-winged warblers. The Condor 119, 108–121. [Google Scholar]
- Kramer GR, Andersen DE, Buehler DA, Wood PB, Peterson SM, Lehman JA, Aldinger KR, Bulluck LP, Harding S, Jones JA, Loegering JP and Smalling C (2018) Population trends in Vermivora warblers are linked to strong migratory connectivity. Proceedings of the National Academy of Sciences of the United States of America 115, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachish S, Knowles SCL, Alves R, Wood MJ and Sheldon BC (2011) Fitness effects of endemic malaria infections in a wild bird population: the importance of ecological structure. Journal of Animal Ecology 80, 1196–1206. [DOI] [PubMed] [Google Scholar]
- LaPointe DA, Atkinson CT and Samuel MD (2012) Ecology and conservation biology of avian malaria. Annals of the New York Academy of Sciences 1249, 211–226. [DOI] [PubMed] [Google Scholar]
- Laurance SGW, Jones D, Westcott D, McKeown A, Harrington G and Hilbert DW (2013) Habitat fragmentation and ecological traits influence the prevalence of avian blood parasites in a tropical rainforest landscape. PLoS One 8, e76227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewicki KE, Huyvaert KP, Piaggio AJ, Diller LV and Franklin AB (2015) Effects of barred owl (Strix varia) range expansion on Parahaemoproteus parasite assemblage dynamics and transmission in barred and northern spotted owls (Strix occidentalis caurina). Biological Invasions 17, 1713–1727. [Google Scholar]
- Loiseau C, Harrigan RJ, Cornel AJ, Guers SL, Dodge M, Marzec T, Carlson JS, Seppi B and Sehgal RNM (2012a) First evidence and predictions of Plasmodium transmission in Alaskan bird populations. PLoS One 7, e44729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau C, Harrigan RJ, Robert A, Bowie RCK, Henri A, Smith TB and Sehgal RNM (2012b) Host and habitat specialization of avian malaria in Africa. Molecular Ecology 21, 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Calderón C, Van Wilgenburg SL, Roth AM, Flaspohler DJ and Hobson KA (2019) An evaluation of isotopic (δ2H) methods to provide estimates of avian breeding and natal dispersal. Ecosphere (Washington, D.C) 10, 1–20. [Google Scholar]
- Manitoba Conservation, Manitoba Land Use Initiative (2005) Land use/Cover maps. Available at http://mli2.gov.mb.ca/landuse/index.html.
- Mantilla JS, Gonzáles AD, Moncada L and Matta NE (2013) Description and molecular characterization of Plasmodium (novyella) unalis sp. nov. from the Great Thrush (Turdus fuscater) in highland of Colombia. Parasitology Research 112, 4193–4204. [DOI] [PubMed] [Google Scholar]
- Marzal A, de Lope F, Navarro C and Møller AP (2005) Malarial parasites decrease reproductive success: an experimental study in a passerine bird. Oecologia 142, 541–545. [DOI] [PubMed] [Google Scholar]
- McCracken JD (1994) Golden-winged and blue-winged warblers: their history and future in Ontario. In McNicholl MK and Cranmer-Byng JL (eds), Ornithology in Ontario. Whitby, Ontario: Hawk Owl Publishing, pp. 279–289. [Google Scholar]
- Meixell BW, Arnold TW, Lindberg MS, Smith MM, Runstadler JA and Ramey AM (2016) Detection, prevalence, and transmission of avian hematozoa in waterfowl at the Arctic/sub-Arctic interface: co-infections, viral interactions, and sources of variation. Parasites & Vectors 9, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino S, Moreno J, Vásquez RA, Martínez J, Sánchez-monsálvez I, Estades CF, Ippi S, Sabat P, Rozzi R and Mcgehee S (2008) Haematozoa in forest birds from southern Chile: latitudinal gradients in prevalence and parasite lineage richness. Austral Ecology 33, 329–340. [Google Scholar]
- Møller AP and Szép T (2011) The role of parasites in ecology and evolution of migration and migratory connectivity. Journal of Ornithology 152, S141–S150. [Google Scholar]
- Montagna CM, Gauna LE, Angelo APDD and Anguiano OL (2012) Evolution of insecticide resistance in non-target black flies (Diptera: Simuliidae) from Argentina. Memórias do Instituto Oswaldo Cruz 107, 458–465. [DOI] [PubMed] [Google Scholar]
- Moulton L (2017) Golden-Winged Warbler (Vermivora Chrysoptera) Habitat Selection, Mating Behaviour, and Population Viability in a Fragmented Landscape at the Northern Range Limit (PhD thesis). University of Manitoba, Winnipeg, Canada. [Google Scholar]
- Moulton L and Artuso C (2017) Golden-winged warbler. In Artuso C, Couturier AR, De Smet KD, Koes RF, Lepage D, McCracken J, Mooi R and Taylor P (eds), The Atlas of the Breeding Birds of Manitoba, 2010–2014. Winnipeg, Canada: Bird Studies Canada. [Google Scholar]
- Moulton LL, Vallender R, Artuso C and Koper N (2018) The final frontier: early-stage genetic introgression and hybrid habitat use in the final frontier: early-stage genetic introgression and hybrid habitat use in the northwestern extent of the golden-winged Warbler breeding range. Conservation Genetics 18, 1481–1487. [Google Scholar]
- Murdock CC, Adler PH, Frank J and Perkins SL (2015) Molecular analyses on host-seeking black flies (Diptera: Simuliidae) reveal a diverse assemblage of Leucocytozoon (Apicomplexa: Haemospororida) parasites in an alpine ecosystem. Parasites & Vectors 8, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakgrove KS, Harrigan RJ, Loiseau C, Guers S, Seppi B and Sehgal RNM (2014) Distribution, diversity and drivers of blood-borne parasite co-infections in Alaskan bird populations. International Journal for Parasitology 44, 717–727. [DOI] [PubMed] [Google Scholar]
- Okanga S, Cumming GS, Hockey PAR and Peters JL (2013) Landscape structure influences avian malaria ecology in the Western Cape, South Africa. Landscape Ecology 28, 2019–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson-Pons S, Clark NJ, Ishtiaq F and Clegg SM (2015) Differences in host species relationships and biogeographical influences produce contrasting patterns of prevalence, community composition and genetic structure in two genera of avian malaria parasites in southern Melanesia. Journal of Animal Ecology 84, 985–998. [DOI] [PubMed] [Google Scholar]
- Ontario Ministry of Natural Resources (2008) Southern Ontario Land Resource Information System 1.2. Available at: https://www.javacoeapp.lrc.gov.on.ca/geonetwork/srv/en/main.home?uuid=c42f0216-df61-405f-8b99-0e613e1cfc85.
- Ontario Natural Resources and Forestry (2011) Southern Ontario land resource information system 2.0. Available at https://www.ontario.ca/data/southern-ontario-land-resource-information-system-solris-20.
- Pagenkopp KM, Klicka J, Durrant KL, Garvin JC and Fleischer RC (2008) Geographic variation in malarial parasite lineages in the common yellowthroat (Geothlypis trichas). Conservation Genetics 9, 1577–1588. [Google Scholar]
- Palinauskas V, Valkiūnas G, Bolshakov CV and Bensch S (2011) Plasmodium relictum (lineage SGS1) and Plasmodium ashfordi (lineage GRW2): the effects of the co-infection on experimentally infected passerine birds. Experimental Parasitology 127, 527–533. [DOI] [PubMed] [Google Scholar]
- Patz JA, Olson SH, Uejio CK and Gibbs HK (2008) Disease emergence from global climate and land use change. The Medical Clinics of North America 92, 1473–1491. [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez A, Fernández-González S, de la Hera I and Pérez-Tris J (2013) Finding the appropriate variables to model the distribution of vector-borne parasites with different environmental preferences: climate is not enough. Global Change Biology 19, 3245–3253. [DOI] [PubMed] [Google Scholar]
- Pulgarín PC, Fitzgerald AM, Gómez C, Bayly NJ, Bensch S, Starkloff N, Kirchman JJ, González-prieto AM, Hobson KA, Ungvari-martin J, Skeen H, Isabel M, Carlos C and Cadena D (2019) Migratory birds as vehicles for parasite dispersal? Infection by avian haemosporidians over the year and throughout the range of a long – distance migrant. Journal of Biogeography 46, 83–96. [Google Scholar]
- R Core Team (2015) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available at https://www.R-project.org/. [Google Scholar]
- Ricklefs RE and Fallon SM (2002) Diversification and host switching in avian malaria parasites. Proceedings of the Royal Society B: Biological Sciences 269, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs RE, Outlaw DC, Svensson-Coelho M, Medeiros MCI, Ellis VA and Latta S (2014) Species formation by host shifting in avian malaria parasites. Proceedings of the National Academy of Sciences of the United States of America 111, 14816–14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs RE, Medeiros M, Ellis VA, Svensson-coelho M, Blake JG, Loiselle BA, Soares L, Fecchio A, Outlaw D, Marra PP, Latta SC, Bar R and Jeremoabo D (2016) Avian migration and the distribution of malaria parasites in New World passerine birds. Journal of Biogeography, 1277–1286. [Google Scholar]
- Rosenberg KV, Will T, Buehler DA, Swarthout SB, Thogmartin WE, Bennett RE and Chandler RB (2016) Dynamic distributions and population declines of Golden-winged Warblers. In Streby HM, Andersen DE and Buehler DA (eds), Golden-winged Warbler Ecology, Conservation, and Habitat Management. Boca Raton, FL: CRC Press, pp. 3–28. [Google Scholar]
- Samuel MD, Hobbelen PHF, DeCastro F, Ahumada JA, LaPointe DA, Atkinson CT, Woodworth BL, Hart PJ and Duffy DC (2011) The dynamics, transmission, and population impacts of avian malaria in native Hawaiian birds: a modeling approach. Ecological Applications 21, 2960–2973. [Google Scholar]
- Sarquis-Adamson Y and MacDougall-Shackleton EA (2016) Song sparrows Melospiza melodia have a home-field advantage in defending against sympatric malarial parasites. Royal Society Open Science 3, 160216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JR, Hines JE, Fallon JE, Pardieck KL, Ziolkowski DJ Jr and Link WA (2011) The North American Breeding Bird Survey, Results and Analysis 1966–2010, version 12.07. 2011 USGS Patuxent Wildlife Research Center, Laurel, MD. doi: 10.1128/JB.01736-06. [DOI]
- Sehgal RNM (2010) Deforestation and avian infectious diseases. The Journal of Experimental Biology 213, 955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Gill SA, Baker KM and Vonhof MJ (2017) Prevalence and diversity of avian Haemosporida infecting songbirds in southwest Michigan. Parasitology Research 117, 471–489. [DOI] [PubMed] [Google Scholar]
- Sol D, Jovani R and Torres J (2000) Geographical variation in blood parasites in feral pigeons: the role of vectors. Ecography 23, 307–314. [Google Scholar]
- Species At Risk Act (2002) s. 27. c. 29. Vol 141. No. 26 (2007).
- Szymanski MM and Lovette IJ (2005) High lineage diversity and host sharing of malarial parasites in a local avian assemblage. The Journal of Parasitology 91, 768–774. [DOI] [PubMed] [Google Scholar]
- Takaoka H, Yunus M, Hadi UK, Sigit SH and Miyagi I (2000) Preliminary report of faunistic surveys on black flies (Diptera: Simuliidae) in Sumatra, Indonesia. 28, 157–166.
- US Geological Survey, Multi-Resolution Land Characteristics (MRLC) Consortium (2011) National land cover database 2011. Available at https://www.mrlc.gov/nlcd2011.php.
- Valkiūnas G (2005) Avian Malaria Parasites and Other Haemosporidia. Boca Ratón, USA: CRC Press. [Google Scholar]
- Vallender R, Robertson RJ, Friesen VL and Lovette IJ (2007) Complex hybridization dynamics between golden-winged and blue-winged warblers (Vermivora chrysoptera and Vermivora pinus) revealed by AFLP, microsatellite, intron and mtDNA markers. Molecular Ecology 16, 2017–2029. [DOI] [PubMed] [Google Scholar]
- Vallender R, Bull RD, Moulton LL and Robertson RJ (2012) Blood parasite infection and heterozygosity in pure and genetic-hybrid golden-winged warblers (Vermivora chrysoptera) across Canada. The Auk 129, 716–724. [Google Scholar]
- Walther EL, Carlson JS, Cornel A, Morris BK and Sehgal RNM (2016) First molecular study of prevalence and diversity of avian haemosporidia in a Central California songbird community. Journal of Ornithology 157, 549–564. [Google Scholar]
- White P and Densmore LD (1992) Molecular genetic analysis of populations. In Hoelzel AR (ed.), A Practical Approach. Oxford, UK: Oxford University Press, pp. 29–58. [Google Scholar]
- Zylberberg M, Derryberry EP, Breuner C, Macdougall-shackleton E and Cornelius JM (2015) Haemoproteus infected birds have increased lifetime reproductive success. Parasitology 142, 1033–1043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182019001240.
click here to view supplementary material