Abstract
During three decades, only about 20 new drugs have been developed for malaria, tuberculosis and all neglected tropical diseases (NTDs). This critical situation was reached because NTDs represent only 10% of health research investments; however, they comprise about 90% of the global disease burden. Computational simulations applied in virtual screening (VS) strategies are very efficient tools to identify pharmacologically active compounds or new indications for drugs already administered for other diseases. One of the advantages of this approach is the low time-consuming and low-budget first stage, which filters for testing experimentally a group of candidate compounds with high chances of binding to the target and present trypanocidal activity. In this work, we review the most common VS strategies that have been used for the identification of new drugs with special emphasis on those applied to trypanosomiasis and leishmaniasis. Computational simulations based on the selected protein targets or their ligands are explained, including the method selection criteria, examples of successful VS campaigns applied to NTDs, a list of validated molecular targets for drug development and repositioned drugs for trypanosomatid-caused diseases. Thereby, here we present the state-of-the-art of VS and drug repurposing to conclude pointing out the future perspectives in the field.
Trypanosomatid-caused diseases and treatments
Trypanosomatids are unicellular flagellate organisms, belonging to the clade Trypanosomatida, most of them pathogenic for other organisms including mammals, insects and plants (Adl et al., 2019; Marchese et al., 2018; Menna-Barreto, 2019). Among trypanosomatids, two genera comprise known species pathogenic to humans: Trypanosoma and Leishmania. The first one includes two human-infecting species: Trypanosoma cruzi, causing the American trypanosomiasis or Chagas disease (Chagas, 1909) [https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis)], and Trypanosoma brucei, the etiological agent of the human African trypanosomiasis (HAT) or sleeping sickness (Steverding, 2008) [https://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness)]. The genus Leishmania includes more than 20 species causing a variety of human diseases generically known as leishmaniasis (Maxfield and Crane, 2019) (https://www.who.int/news-room/fact-sheets/detail/leishmaniasis).
Trypanosomatids have a series of peculiarities concerning their cellular organization, control of gene expression and metabolism (Marchese et al., 2018). But, despite these unique characteristics offering a myriad of potential targets for drugs, most of the treatments for trypanosomatid-caused diseases remain unsatisfactory, and even in those cases in which new alternatives have been developed, the emergence of resistant strains is foreseeable (Menna-Barreto, 2019).
Chagas disease affects approximately 8 million people, and an estimated 70 million at risk of contracting the infection (Perez-Molina and Molina, 2018). The disease presents two major phases: acute and chronic. The acute phase happens immediately after infection and is usually asymptomatic. In cases in which clinical symptoms manifest, they are mild and unspecific as presented in Table 1. The acute phase is characterized by a high parasitaemia and the absence of humoral immune response (Bern, 2015; Perez-Molina and Molina, 2018). After the acute phase, which can last for up to 2 months, follows the chronic phase that lasts for the rest of their life. The chronic phase is characterized by the absence of evident parasitaemia and a robust immune humoral response, and presents several clinical forms that can be divided in the indeterminate form, which is asymptomatic and accounts for approximately 70% of the patients; and the symptomatic forms, affecting the remaining 30% of the infected population (Perez-Molina and Molina, 2018; Rassi et al., 2010). The chronic clinical features are mentioned in Table 1. The treatment for Chagas disease consists of only two drugs approved for human use half a century ago: benznidazole (1) and nifurtimox (2). Both drugs are efficient in the acute phase, but frequently fail in the chronic phase when most of the patients are diagnosed (Boscardin et al., 2010; Hall et al., 2011).
Table 1.
Principal features of trypanosomatid-caused diseases
| Disease | Organism | Transmission | Epidemiology | Drugs | Main clinical manifestations |
|---|---|---|---|---|---|
| Chagas disease or American trypanosomiasis | Trypanosoma cruzi | Vectorial (triatomine insect) Mother-to-child transmission Organ transplant and blood transfusion Oral transmission through contaminated food Sexual transmission Laboratory accidents |
Latin America 6–7 million infected people <40 000 new cases per year 70 million people at risk |
Nifurtimox Benznidazole |
Acute phase: fever, inflammation at the inoculation site, increased lymph nodes, muscle pain, headaches Chronic phase: cardiomyopathy (severe arrhythmia, heart muscle failures and embolism) and digestive forms (megaoesophagus, megacolon) |
| Human African trypanosomiasis or sleeping sickness | Trypanosoma brucei | Vectorial (tse-tse fly) Transmission through other blood-sucking insects Mother-to-child transmission Sexual transmission Laboratory accidents |
Sub-Saharan Africa 300 000 infected people <2500 new cases per year 60 million people at risk |
Suramin Pentamidine Melarsoprol Eflornithine/ Nifurtimox Fexinidazole |
First or early stage: fever, headaches, muscle and joint pains, lymphadenopathy Second or brain stage: neurological and psychological symptoms (sleep disorders, ataxia, sensory alterations, hallucinations, personality changes) |
| Leishmaniasis: visceral (VL), cutaneous (CL), mucocutaneous (ML) | Leishmania spp. | Vectorial (sand fly) Mother-to-child transmission Organ transplant and blood transfusion Zooanthroponotic transmission Sexual transmission |
Worldwide, except Australia and Antarctica 12 million infected people 2 million new cases per year 350 million people at risk |
Miltefosine Amphotericin B Paromomycin |
VL: persistent irregular fevers, splenomegaly, pancytopenia, hepatomegaly, hypergammaglobulinemia, weight loss CL: ulcerating lesions ML: destructive lesions of the nasal septum, lips, and palate |
For more information about these diseases see: Chagas disease https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis); human African trypanosomiasis https://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness); leishmaniasis https://www.who.int/news-room/fact-sheets/detail/leishmaniasis.
HAT is considered mostly under control (Bottieau and Clerinx, 2019); in the last two decades, it has been observed a dramatic drop of nearly 85% in the number of reported new cases. However, it still threatens 65 million people living in endemic areas. HAT presents two stages: during the first (or early) stage, the parasites proliferate in the blood and lymphatic system, causing mild and unspecific symptoms, as shown in Table 1. After a variable time, the parasites can cross the brain–blood barrier, reaching the central nervous system, initiating the second (or brain) stage. While the central nervous system infection progresses, neurological and psychological symptoms can be observed (Table 1) (Mogk et al., 2017). If left untreated, sleeping sickness can cause death within several months or several years, depending on which T. brucei subspecies caused the infection (Buscher et al., 2017). The treatment of stage one is mostly based on the administration of suramin (3), the first-line drug, and upon failure pentamidine (4), the second-line treatment. Both are ineffective for stage two since they do not cross the blood–brain barrier. Thus, for the second stage of the disease, melarsoprol (5) has been used since the 1940s. It has the advantage of being useful for both T. brucei sub-species causing HAT; however, it is extremely toxic, and in some cases, it could be fatal. Eflornithine (6; difluoromethylornithine) is less toxic than melarsoprol (5), but is ineffective against T. brucei rhodesiense. More recently, eflornithine (6) was indicated to be used in combination with nifurtimox (2), which made the therapy more efficient (Babokhov et al., 2013). Finally, fexinidazole (7) was approved for being distributed via the World Health Organization (WHO) since 2019 in T. brucei endemic countries to treat HAT first and second stages when caused by the subspecies T. brucei gambiense (which is responsible for 98% of the human reported cases) (Deeks, 2019; Mesu et al., 2018).
Leishmaniasis constitutes a broad spectrum of diseases with different severity, ranging from self-cure skin lesions to visceral damage that can lead to death (Aronson et al., 2017). The disease is endemic of at least 100 countries mostly located in the tropical and sub-tropical belt of the planet and it is estimated that 12 million people are affected. Three main forms of leishmaniasis can be recognized, depending on the Leishmania species involved in the infection: visceral (VL), cutaneous (CL) and mucocutaneous (ML) (Burza et al., 2018). VL can be asymptomatic, however, when symptoms appear they can develop within 2 weeks and several years after the infection. If left untreated, VL can be fatal. CL is the most common form of leishmaniasis, consisting of exposed lesions of the skin or, in a small number of cases, sub-dermal diffuse papules. ML is much more aggressive than CL, usually causing the partial or complete destruction of mucous tissues. CL and ML have serious consequences due to severe disabilities, opportunistic infections and social stigma, producing negative psychological effects. Table 1 shows the main clinical features of the three forms of leishmaniasis. The strategies to treat and manage leishmaniasis must take into account several factors such as parasite species, geographic location and co-infections. Classically, the treatment of VL consists of two pentavalent antimonials: sodium stibogluconate (8; Sb(V)) or meglumine antimoniate (9). Their toxicity and the increasing emergence of resistance led to the search for alternatives. For example in North Bihar, India, where VL caused by L. donovani is endemic, a widespread primary failure to Sb(V) has been reported and its use is not recommended anymore (Croft et al., 2006; Ponte-Sucre et al., 2017). In the last two decades, some drugs were launched to be used as single-treatment or in combination: an oral formulation of miltefosine (10), which constitutes now the first-line treatment in most of the Asian endemic countries (Pinto-Martinez et al., 2018), and later an injectable formulation of paromomycin (11), followed by a liposomal formulation of amphotericin B (12) (Alves et al., 2018; Burza et al., 2018; van Griensven and Diro, 2019). Most of the CL lesions are self-cured in a period between 2 and 18 months in immunocompetent patients. However, accelerating the cure is desirable to reduce the risk of dissemination or progression to ML. The treatments used can be local, such as intralesional injections of sodium stibogluconate (8), physical therapies like cryotherapy or thermotherapy, or topical application of agents such as paromomycin (11). Currently, a combination of locally applied antimonials and cryotherapy are considered the first-line treatment in Asia and African endemic countries (Aronson and Joya, 2019; Burza et al., 2018).
The precise mode of action of the drugs mentioned in this section is not determined, except for eflornithine (6), which functions as an irreversible inhibitor of ornithine decarboxylase (ODC), an enzyme involved in the polyamine biosynthesis (Wilkinson and Kelly, 2009).
Table 1 summarizes the clinical and epidemiological characteristics and the available drugs for the treatment of these trypanosomatid diseases.
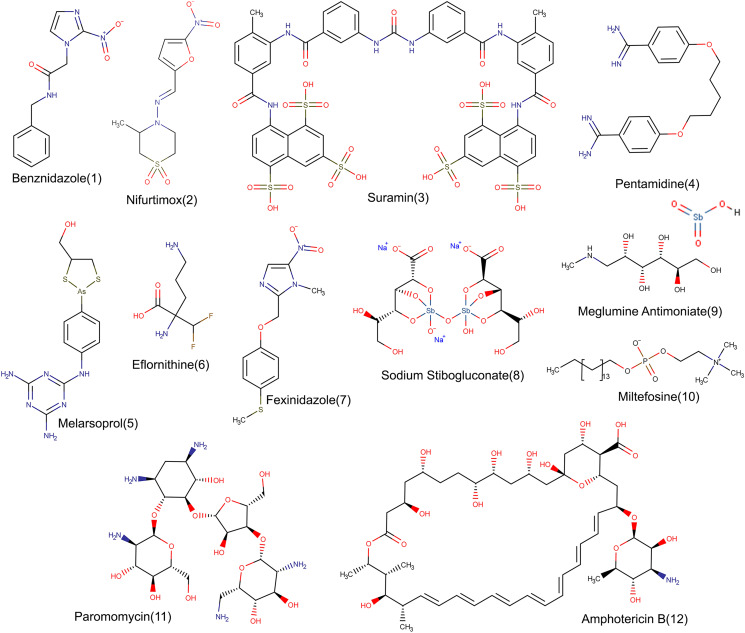
The molecular structures of all the mentioned drugs are presented in Fig. 1.
Fig. 1.
Chemical structures. Detailed structures of approved drugs to treat Chagas disease (1 and 2), human African trypanosomiasis (3–7) and leishmaniasis (8–12).
Molecular targets for trypanosomiasis and leishmaniasis drug development
Traditionally, the way to identify the drug targets relied exclusively on comparative biochemistry and genetics. The completion of the genome projects for human-infecting trypanosomatids is a breakthrough that allows the identification of an increasing number of possible molecular targets, usually enzymes, proteins or biochemical pathways. Strictly, there are three features an ideal target must satisfy: it has to be absent or strikingly different from its homologue in the mammalian hosts, being druggable and essential for the parasite survival (Hughes et al., 2011; Wyatt et al., 2011). The first criterion denotes target selectivity in order to differentially direct to the parasite a given drug. With the availability of trypanosomatid genomes (Berna et al., 2018; Berriman et al., 2005; El-Sayed et al., 2005a,b) and more accurate databases (https://tritrypdb.org/tritrypdb/; https://www.genedb.org/) supporting the computational background with biochemical data, it should be straightforward to verify if a given gene product is absent from the mammalian counterpart, or the degree of divergence they present. But, in practice, a real target does not always meet selectivity, for example, the ODC which is a valid target against African trypanosomiasis. In these cases, selectivity should be provided by improving the affinity of the drug towards the parasite target (Kawasaki and Freire, 2011). The term druggability refers to the capacity of a given target to be affected by a drug; in essence, the target must bind a molecule that modulates its activity (Abi Hussein et al., 2017). This information can be obtained during the preliminary stage of a drug discovery project by accessing accurate computational druggability prediction methods. The Special Programme for Research and Training in Tropical Diseases (TDR) has developed the TDR targets database, which is a very useful tool that facilitates the identification and prioritization of candidate drug targets for the ‘Tritryp’ genomes among other pathogens (Magarinos et al., 2012) (http://tdrtargets.org/). Likewise, more recently, the Target-Pathogen database (Sosa et al., 2018) (http://target.sbg.qb.fcen.uba.ar/patho) was designed and developed as an online resource. This platform has integrated and weighed protein information, such as structural properties including druggability and essentiality, one of the most important steps in the validation of a given target. Nowadays many drug discovery programmes consider the genetic validation a critical point because it reflects the loss of function attributable to therapeutic intervention. This implies that genes are made inoperative by knockout or knock-down procedures which are particularly effective in T. brucei. However, when working with T. cruzi and Leishmania, the situation is more critical because T. cruzi and most species of Leishmania do not possess the RNA interference and the traditional genetic knockouts in many cases are not successful (Burle-Caldas Gde et al., 2015). Also, gene disruption experiments are mainly carried out in the insect stage of the parasite because their easy culture and manipulation (Barrett et al., 1999) and the results not always reflect the biological effect observed in other stages. To avoid target misestimation, the mutants should be also tested for their ability to progress through the life cycle and survive in vivo and in vitro. It is possible that the gene is lethal for the other life cycle stages or generates a conditional lethal phenotype (Barrett et al., 1999). The mentioned limitations have been recently evidenced by Jones et al., who published an overview of the genetic assessments of suitable targets in Leishmania and T. cruzi (Jones et al., 2018). Noteworthy, to date, 65 out of 200 knocked out genes in Leishmania are essential and only 16 out of 36 in T. cruzi (Jones et al., 2018; Osorio-Mendez and Cevallos, 2018); whereas T. brucei has been widely subjected to high-throughput genetic screens covering the whole genome. CRISPR-Cas9 has become one of the most promising methodologies for the genetic validation of trypanosomatid targets (Lander et al., 2016; Soares Medeiros et al., 2017), and it is expected to make further valuable contributions to this field.
An alternative strategy to the genetic validation is the pharmacological validation, but evidence of essentiality is preferred to be supported by both criteria (Field et al., 2017; Gilbert, 2013).
In the evaluation of which targets are better, the fundamentals of metabolic control analysis and metabolic modelling offer new insights into target prioritization. This methodology allows studying the control of cellular metabolic pathways regardless of whether it is a two-step or multiple-step pathway, showing that enzymes with the highest pathway control are the most convenient targets for therapeutic intervention. This idea is supported based on the fact that in any essential pathway, removing an enzyme by genetic manipulations would lead to the same essential phenotype, providing a number of potential drug targets equal or similar to that of the total components (Bakker et al., 2000; Gonzalez-Chavez et al., 2015; Hornberg et al., 2007; Olin-Sandoval et al., 2012). Therefore, this approach emphasizes the point that proving a gene is essential, specific and druggable is no guarantee that it encodes a valid drug target.
Several biochemical pathways that are common to pathogenic trypanosomes and exclusive to them are supposed to be the most promising for drug discovery, for example, mitochondrial metabolic pathways, sterol biosynthesis, the thiol–polyamine metabolism and glycolysis, among others (Alberca et al., 2016; Avilan et al., 2011; Burri and Brun, 2003; Dietrich et al., 2018; Khare et al., 2015; Leroux and Krauth-Siegel, 2016; Lu et al., 2013; Menzies et al., 2018; Morillo et al., 2017; Nowicki et al., 2008; Reigada et al., 2018, 2017; Sharlow et al., 2010a,b; Torrie et al., 2009; Urbina, 2015; Vazquez et al., 2017). In Table 2, some targets are summarized regarding these pathways; some of them are introduced in ‘VS applied to trypanosomatid-caused diseases’ section of this manuscript.
Table 2.
Drug targets in Trypanosoma and Leishmania parasites
| Target | Process | Localization | Organism | Drugs | Reference |
|---|---|---|---|---|---|
| Alternative oxidase | Electron transport chain | Mitochondria | T. brucei | Aromatic hydroxamates | Menzies et al. (2018) |
| Cytochrome b | Electron transport chain | Mitochondria | T. cruzi | GNF7686 | Khare et al. (2015) |
| Lanosterol 14-α-demethylase | Ergosterol biosynthesis | Mitochondria and endoplasmatic reticulum | T. cruzi | Posaconazole Ravuconazole |
Morillo et al. (2017); Urbina (2015) |
| Trypanothione synthase | Thiol metabolism | Cytoplasm |
T. brucei T. cruzi Leishmania spp. |
Revised in the references | Vazquez et al. (2017); Leroux and Krauth-Siegel (2016); Torrie et al. (2009) |
| Trypanothione reductase | Thiol metabolism | Cytoplasm |
T. brucei T. cruzi Leishmania spp. |
Revised in the references | Leroux and Krauth-Siegel (2016); Vazquez et al. (2017); Lu et al. (2013) |
| Ornithine decarboxilase | Polyamine metabolism | Cytoplasm | T. brucei | DFMO | Burri and Brun (2003) |
| TcPAT12 | Polyamine metabolism | Flagellar pocket | T. cruzi | Isotretinoin ANT4 Triclabendazole Sertaconazole Paroxetine Cisapride |
Alberca et al. (2016); Reigada et al. (2018); Reigada et al. (2017); Dietrich et al. (2018) |
| Enolase | Glycolisis | Cytoplasm and cell surface |
T. brucei T. cruzi Leishmania spp. |
Phosphonoacetohydroxamate | Avilan et al. (2011) |
| Piruvate kinase | Glycolisis | Cytoplasm | Leishmania spp. | Furanose sugar amino amides | Nowicki et al. (2008) |
| Phosphofructokinase | Glycolisis | Glycosome |
T. brucei T. cruzi |
ML251 Furanose sugar amino amides |
Brimacombe et al. (2014); Nowicki et al. (2008) |
| Hexokinase | Glycolisis | Glycosome | T. brucei | EbSe Revised in the references |
Lu et al. (2013); Sharlow et al. (2010a,b) |
Finally, in addition to finding a good target, when thinking in a possible therapy, the biological differences between parasite intracellular and extracellular stages inside the host should be considered. Contrary to T. brucei, which is only extracellular, T. cruzi and Leishmania spp. possess intracellular forms, so the in vivo accessibility of a drug is different for each of them. Drugs need to overcome additional barriers to meet its target such as host plasmatic membranes, parasitophorous vacuoles, host metabolism, among others. Drugs must be active in these different environment conditions.
Drug development for trypanosomatid-caused diseases
Along with the history of drug development for trypanosomatid-related diseases, many strategies have been implemented. Through different programmes, an initial chemotherapy arsenal to treat leishmaniasis and trypanosomiasis was introduced and remained unaltered for decades.
Different approaches have been implemented to identify new drug candidates. Classical methods to find and optimize new chemical entities (NCE) have been based mainly on new compounds synthesis (de novo drug discovery) and bio-guided fractionation and isolation of natural products.
The first one has been included in many classical drug discovery pipelines, being a high-cost and usually very long and time-consuming approximation. The second provided NCE with either a known or a new scaffold, which can be structurally complex. To be able to use those compounds as a starting point in a drug discovery programme, it is necessary to develop a complete synthetic route to perform a structure–activity relationship and preclinical studies. Alternatively, the complete biosynthetic pathway has to be elucidated to produce adequate amounts of a natural product, and the heterologous expression of the biosynthetic genes should be optimized (Luo et al., 2015).
On the other hand, short-term approaches have been introduced to speed up the process of candidates' identification. One of those strategies involved drug combinations (Sun et al., 2016) that have been explored for leishmaniasis and trypanosomiasis treatment. Those approaches were implemented looking to increase drug efficacy, shorten the treatments and decrease the administered doses (Alcântara et al., 2018; Nwaka et al., 2009). Additionally, drug combination therapy is a well-established approximation to avoid resistance in pathogenic organisms, being a valuable approach that optimizes the resources and know-how to produce improved therapies with better properties (Walvekar et al., 2019). Tolerability can be also increased, because if the combined drugs can be administered below their individually prescribed dose limits, their side-effects would be significantly reduced. One leading case example of that approach is the nifurtimox (2)–eflornithine (6) combination therapy, which can be safely used as first-line treatment for the second stage of HAT caused by T. brucei gambiense (Kansiime et al., 2018; Priotto et al., 2009).
Pharmaceutical companies have recently recovered their historical role in drug development against neglected tropical diseases (NTDs) (Aerts et al., 2017). Over the last decades, GlaxoSmithKline (GSK), Johnson & Johnson, Merck KGaA, MedPharm, Merck & Co, and Pfizer reassumed the leadership as drugs provision for NTDs. Those companies have donated billions of tablets to treat some NTDs in addition to direct procurement. Beyond those efforts, it is clear that there are not enough investments for NTDs yet from the pharmaceutical industries or participation of non-governmental organizations (NGOs). The WHO is critical to make the medication available for the patients on the endemic regions (Hollingsworth, 2018), working with the public and private sectors, international agencies and NGOs in order to guarantee adequate free of charge medication for millions of people.
Recently, one new approach has been consolidated, the partnership of large pharmaceutical companies with non-profit organizations like the Drugs for Neglected Diseases initiative (DNDi), Wellcome Trust or the Academia. Those partnerships have been actively working on campaigns to characterize new NCE with leishmanicidal and trypanocidal activity. Such efforts included the screening of millions of compounds against TriTryps parasites, in particular, partnerships with GSK and Novartis (Khare et al., 2016). GSK Tres Cantos has also integrated a collaborative research network for more than a decade with the Drug Discovery Unit (DDU, University of Dundee) and Wellcome Trust to discover new candidate drugs for VL and Chagas disease (Wyllie et al., 2019). Thanks to that endeavours, many hit compounds have been identified. In general, the approach involves the screening of drug-like libraries against the etiological agents of these diseases to identify compounds that kill the parasites. This approach provides compounds able to cross the cell membranes and kill the parasite within the parasitophorous vacuole. One logical and important disadvantage is that usually this approach is set to be very stringent, providing few bioactive compounds per campaign. Another obvious disadvantage is that the molecular targets have to be elucidated, having to specifically design strategies to identify them.
There are some successful examples of new structures that have been identified from the phenotypic screening of big libraries on Leishmania parasites. One of them is the identification of GNF6702 by Novartis (Khare et al., 2016) and another is the ‘Leish-Box’ of inhibitors by GSK (Lamotte et al., 2019), just to mention a few.
It is also important to understand an experimental compound's mode of action as this can enable an assessment of the likelihood of resistance mechanisms evolving in the parasite. Strategies of target deconvolution are therefore required to identify the molecular target of a hit compound obtained by phenotypic screening. The usual approach involves a combination of genetic and/or metabolomic approaches or pull-down experiments that afterward must be genetically validated. That is a long and laborious process, even with today's advances such as CRISPR/Cas9 (Beneke et al., 2017; Duncan et al., 2017).
An alternative strategy to the very costly, time-consuming and usually very inefficient phenotypic screening campaigns is the target-based drug discovery approach, which is the most commonly used in the pharmaceutical industry. This strategy has also been applied to drug discovery against trypanosomatid-caused diseases in Academia. In this approach, a validated protein target is selected, requiring a well-developed biochemical or biophysical assay that can be used to identify the inhibitors. Big pharmaceutical companies and some well-equipped academic institutions have performed high-throughput screening (HTS) campaigns looking for new hits. Those hits should eventually go through a hit-to-lead process where they are chemically optimized to improve their properties in terms of potency, selectivity and bioavailability.
Over the last decades, the knowledge on the trypanosomatids biochemistry has allowed the identification of many putative drug targets that can potentially provide the validated hits for drug development. Nevertheless, only a few of them have been extensively explored.
Thiol–polyamine metabolism of trypanosomatids was one of the first examples of enzymes used as target-based drug discovery. The most studied enzyme on that matter is trypanothione reductase. Since the early reports of the activity, substrate specificity and kinetics of T. cruzi trypanothione reductase in the late 80s (Krauth-Siegel et al., 1987), the activity of hundreds of compounds has been reported on the enzyme (Tiwari et al., 2018).
Recently, an HTS campaign to find new inhibitors of T. brucei tryparedoxin peroxidase has been reported (Fueller et al., 2012). On that work, nearly 80 000 compounds were analysed, with only 32 displaying activity. Further studies revealed that the compounds not only targeted the enzyme in vitro but also in the intact parasite, validating the target. Trypanothione synthetase is another enzyme of thiol–polyamine metabolism that has been explored. Benitez et al. have studied the potential of that target by assaying 144 compounds, mostly obtained by chemical synthesis and some natural products (Benitez et al., 2016). Different inhibitors have been found, being paulone derivatives the most promising scaffold, nevertheless, some 5-substituted 3-chlorokenpaullone derivatives were off-target (Orban et al., 2016).
An article reported by Professor Gelb in 2003 highlighted the potential of protein farnesyl and N-myristoyl transferases (NMTs) as piggy-back medicinal chemistry targets for the development of anti-trypanosomatids (Gelb et al., 2003). Those enzymes that produce the co- and post-translational protein modification were studied for drug development in other eukaryotic systems, in particular mainly looking for new anticancer agents. The studies on protein farnesyltransferase as a target of screening libraries against the parasitic enzyme did not produce any interesting compounds to develop new medications.
CYP51 (sterol 14α-demethylase cytochrome P450) has been proposed as a possible target for antikinetoplastids drug discovery. That enzyme is the target of azole drugs in clinical practice. In general, the activity of antifungal drugs is often different on the parasitic orthologues, requiring the optimization of existing structures or introducing NCE to achieve the required selectivity. Many different structures have been prepared and assayed in vitro against parasitic CYP51. Those differences require the optimization of existing structures or the introduction of NCE that were more potent and selective. Between those structures, there are substrate analogues, mostly sterol derivatives, indomethacin amides (Konkle et al., 2009) and imidazoles modified from a collection of vitamin D hydroxylase inhibitors. Interesting examples are imidazolyl benzamides (called VNI) that have been through a hit-to-lead optimization process (Friggeri et al., 2018; Lepesheva et al., 2007) that have been able to cure acute and chronic forms of Chagas disease in mice models (Villalta et al., 2013). Other examples are 4-aminopyridyl derivatives (Calvet et al., 2017; Choi et al., 2013) and the tipifarnib-modified structures (Kraus et al., 2010).
Another enzyme that has been usefully used on target-directed antikinetoplastids drug discovery is the NMT. This enzyme has been genetically and experimentally validated in Leishmania spp. Once its essentiality on the parasite biology was established, in vitro HTS of a diverse subset of the Pfizer corporate collection against LdNMT, Plasmodium falciparum NMT and the two human isoforms (HsNMT) led to the discovery of new and potent inhibitors (Bell et al., 2012). The compounds were subsequently resynthesized and validated leading to a compound 43 that is a potent and neutral NMT inhibitor and a promising candidate for antileishmanial drug development (Hutton et al., 2014).
An initiative led by the Novartis Institute for Tropical Diseases screened 3 million compounds in proliferation assays on L. donovani, T. cruzi and T. brucei. That campaign provided GNF5343, which was later optimized preparing nearly 3000 new analogues that led to GNF6702, a compound 400-fold more active in intra-macrophage L. donovani. Later, the parasite proteasome was identified by different strategies as the target of the lead compound. GNF6702 is shown to be able to eradicate parasites in mouse disease models (Khare et al., 2016). Besides the tremendous work behind that report, there is a remarkable example of wide-spectrum antikinetoplastid drug development.
Despite the extensive work and the profound improvement on the drug discovery and development process over the last decades, there are many gaps in the process and only a few targets have been progressed to preclinical development. The involvement of pharmaceutical companies has improved the process and the budget, but there are still financial and material resources limitations. Consequently, the approaches of drug repurposing and the inclusion of computational resources in the analysis of the ever-growing amount of biochemical and genetic data appear as a logical and convenient approach to optimize the process.
An overview of the computational/virtual screening techniques
Similarly to HTS, a virtual screening (VS) employs computer-generated models to search in libraries of small molecules those with chances of binding a molecular target, commonly, but not restricted to, an enzyme or receptor (Rester, 2008).
Computer-aided drug discovery is hugely advantageous; allowing to test bigger compound libraries at negligible costs. Molecules that are not yet synthesized to expand the chemical space can be also added (Rodriguez et al., 2016) without preparing compounds that most likely will not have the desired biological activity (Gasteiger, 2015; Schneider, 2010).
When using digital means in the search of bioactive molecules, the options and strategies are plentiful (Haga et al., 2016), and the factors to take into account when deciding which ones to employ and how to combine them are addressed below.
The starting point
The first step before planning a VS workflow should always be performing an extensive bibliographical research about the target that one is trying to find drugs for (Gimeno et al., 2019); aspects as, for example, its biological function, availability of techniques to measure its activity, natural ligands, known inhibitors, catalytic mechanism, structure, known homologues and their ligands.
While the results of the literature review will determine what kind of computational tools can be used, every strategy shares the need for a compound library to screen. The confection of the screening library will greatly depend on the specific goals of the VS. There are different small molecule databases available for VS. The ChEMBL (https://www.ebi.ac.uk/chembl/) (Gaulton et al., 2017), PubChem (https://pubchem.ncbi.nlm.nih.gov/) (Kim et al., 2019) and ZINC (https://zinc.docking.org/) (Sterling and Irwin, 2015) are databases with hundreds of millions of compounds and useful search tools. The SWEETLEAD (https://simtk.org/projects/sweetlead) (Novick et al., 2013) and the DrugBank (https://www.drugbank.ca/) (Wishart et al., 2018) databases contain drugs approved for human administration. Also, some compound vendors, such as Enamine (https://enamine.net/) and Asinex (http://www.asinex.com/), offer screening libraries of their products.
As the goal of a VS strategy is finding molecules to test against a molecular target, it is wise to filter out compounds that could give false positives in the binding assays. These compounds, known as Pan-Assay Interference Compounds (PAINs) (Dahlin et al., 2015), can give false results by reacting non-specifically with the target, with several other targets, or interfering with the measurement assays (Baell and Walters, 2014). Some chemical groups are shared by many known PAINs, which make it possible to previously remove any molecule containing the said groups (Baell and Holloway, 2010).
An estimated 50% of the tested drug candidates fail because of inefficiencies in Absorption, Distribution, Metabolism, Excretion and/or Toxicity (ADME/Tox) (Li, 2001). Based on the physical chemical characteristics of known drugs, Lipinski et al. developed the ‘rule of five’ for orally available drugs (H-bond donors ⩽5, H-bond acceptors ⩽10, molecular weight ⩽500 Da, logP ⩽5) (Lipinski et al., 2001). There are computational tools that predict ADME/Tox characteristics, but many of them rely on the Lipinski's rules, excluding administration routes other than oral (Scior et al., 2012); they also have a low predictive performance on more complex properties, e.g. carcinogenesis (Stouch et al., 2003).
In this sense, at the stage of filtering the screening library, one could take into account the pharmacokinetic characteristics of the compounds to be screened, and three scenarios are possible (Oprea, 2002). Some strategies focus on first obtaining high-affinity lead compounds that later would be optimized for good pharmacokinetic properties, modifications to achieve better ADME/Tox could be detrimental to the target binding, leading to a trial–error optimization that consumes time and resources. Another scenario is filtering compounds before the screening, in an attempt to obtain lead compounds with good ADME/Tox properties and later optimize the potency, which can reduce ADME characteristics, but would be a less consuming process towards optimal structures. A third and highly recommended strategy is to simultaneously follow changes that increase affinity and ADME/Tox characteristics (Drews, 1998).
Known ligands of a target can be the starting point in a VS campaign. Also, using experimental data of the molecular structure of the target or a homologue, a receptor-based approximation can be performed (Ghemtio et al., 2012; Table 3).
Table 3.
List of databases of interest for drug virtual screening
| Database | Description | Web-link | Reference |
|---|---|---|---|
| SwissDock | Molecular docking with rigid target | http://www.swissdock.ch/ | Sterling and Irwin (2015) |
| ChEMBL | 1.9 million curated bioactive molecules including when known their activities, molecular targets, tissue absorption, indication, development and approval state, molecular assays, physico-chemical properties and related genomic data | https://www.ebi.ac.uk/chembl/ | Gaulton et al. (2017) |
| PubChem | 102 million compounds with reported bioactivity, safety and toxicity, patents, citations, physico-chemical properties and more. It includes a molecule drawing tool for search | https://pubchem.ncbi.nlm.nih.gov/ | Kim et al. (2019) |
| SWEETLEAD | Chemical structures of >9000 approved medicines, illegal drugs and isolates from traditional medicinal herbs | https://simtk.org/projects/sweetlead | Novick et al. (2013) |
| DrugBank | Contains >13 400 entries including approved drugs, nutraceuticals, experimental and illicit drugs. Additionally, >5000 non-redundant proteins linked to the drug entries | https://www.drugbank.ca/ | Wishart et al. (2018) |
| Enamine | Compound libraries available for purchase in the vendors websites | https://enamine.net/ | - |
| Asinex | http://www.asinex.com/ | - | |
| Maybridge | http://www.maybridge.com/ | - | |
| TDR Targets | Contains information about genes and targets from 21 bacterial and eukaryotic tropical pathogens, phylogeny, >2 million bioactive compounds and the possibility of specifying the search criteria to prioritize drug targets | https://tdrtargets.org | Uran Landaburu et al. (2019) |
| Protein Data Bank | Curated and annotated archive about the experimentally determined 3D shapes of proteins, nucleic acids, and complex assemblies | https://www.rcsb.org/ | Berman et al. (2000) |
| ModBase | Comparative protein structure models calculated by the ModPipe pipeline | https://modbase.compbio.ucsf.edu/ | Pieper et al. (2014) |
Ligand-based virtual screening
Johnson et al. (1990) introduced the concept that similar molecules exhibit similar behaviours, an assumption extended to their biological activity. Based on this principle, if there is knowledge of compounds with the desired effect, finding molecules similar to them is a reasonable starting point in the search of new drugs. However, ‘similarity’ is a tricky concept, to determine if two or more compounds are similar, different characteristics, methods of comparison and metrics that allow such contrasts can be used.
To compare molecules for ligand-based VS, the first step is representing them in numerical terms. To this end, there are different mathematical models to denote different measurable properties of compounds in ways that are usable, these models are called molecular descriptors (Todeschini et al., 2009). Descriptors used in VS can be classified as one-dimensional (1D), two-dimensional (2D) and three-dimensional (3D), depending on the molecules information about what they represent. As not all descriptors correlate with the biological activity of the molecule, the selection of the descriptors and the methods used to compare them is crucial.
Because the ligand binding to its receptor will depend in great extent on the spatial interactions that can occur between them, 3D descriptors are considered a more reliable choice (Danishuddin and Khan, 2016; Mavridis et al., 2007) that, when thoroughly used, enhances the chances of finding structurally diverse candidates (Brown and Jacoby, 2006). However, a molecule can have many 3D configurations, and comparing spatial data is more complex than comparing 2D descriptors, which translates in greater computational costs (Mavridis et al., 2007).
On the other hand, 2D descriptors consume less computational resources, maintaining good performance, but missing key characteristics involved in the interaction (Fradera and Babaoglu, 2017).
Fingerprint similarity methods rely on the abstraction of molecular properties into bit sequences, where the bit value (0 or 1) at each position of the sequence represents the absence or presence of a particular descriptor in a molecule (Banegas-Luna et al., 2018). The sequences can be compared at each position to obtain a metric on how similar are the compounds, given the compared descriptors.
Many comparison algorithms exist, being the Tanimoto coefficient one of the most popular (Bajusz et al., 2015). Regardless of the comparison metric selected, the results can be sorted from more to less similar to the known ligands. At this point comes the thorny choice of where to apply the cut-off after which compounds will be discarded, as there is no universal value for it, so the selectivity/sensitivity trade-off needs to be carefully determined from the retrieval of known actives and inactives (Fradera and Babaoglu, 2017). In addition, because similarity coefficients assign to all the compared bits an equal relevance, compounds similar in the bits important for the biological activity can end down in the list for not sharing enough of the non-important characteristics, and vice versa (Scior et al., 2012).
Machine learning algorithms permit computer-aided drug discovery take a step further, by stop relying on explicit physical representations of what is needed for an expected biological activity, and allow the use of complex pattern recognition algorithms to construct mathematical models that take into account many molecular descriptors at the same time, as well as exploring bigger datasets with low computational costs (Lo et al., 2018). These methods rely on databases of known active and inactive compounds, so the algorithms try to find a set of molecular descriptors that correlate with the desired activity, assigning a level of importance to each of them, and producing a model able to predict the activity of new compounds (Gimeno et al., 2019).
As the algorithm will try and find any patterns, the initial or ‘training’ dataset of active and inactive compounds is extremely important. When the training library is too small or with poor structural diversity, the produced model might be based in chance correlation or be biased towards similar characteristics not determining the biological activity (Ma et al., 2009; Scior et al., 2012), so it is preferred to count with a large and structurally diverse input set. As the amount of inactive compounds will always be greater than the actives (Schierz, 2009), it is important to balance both sets to avoid the production of a model biased towards the correct identification of inactive compounds, but being sub-optimal in the discrimination of true actives. Whenever possible, it is advisable to choose those inactive compounds that are structurally similar to the actives, so the model will have more chances of discriminate between them (Tropsha, 2010).
Ligand-based pharmacophores are ensembles of spatial and electrostatic features shared between a set of known active molecules. These models are used later to search for other candidates containing such features, assuming they are responsible for the interaction with the receptor (Gimeno et al., 2019). In this way, the compounds retrieved can be structurally richer, as the matched features can be contained by a wider range of structures.
Though pharmacophore models can be constructed from one or a few ligands, it is always better to use large sets of known actives (Scior et al., 2012) in order to identify which features seem to be critical to the binding, as well as finding as many important features as possible that may not be shared by every ligand.
Receptor-based virtual screening
Using known ligands of the target macromolecule to find hit compounds is very fast and computationally inexpensive. Nevertheless, for NTDs such as those caused by trypanosomatids, the amount of information available regarding experimentally demonstrated ligands of interesting targets can be scarce or even non-existing. Additionally, ligand-based methods tend to narrow the chemical space by retrieving only molecules similar to the known ligands, leaving out potentially good and structurally diverse hits. Although similar molecules tend to have similar activities, this is not necessarily true, as some of the chemical groups that make the hit different from the known ligands might be detrimental to the ligand–receptor interaction in what is known as activity cliff (Stumpfe and Bajorath, 2012).
Analysing the molecular target allows to discard hits that would be incompatible with the binding site, and allows finding structurally novel hits capable of fitting in and interacting with a given pocket in the receptor. Structural information of some targets can be found in the Protein Data Bank (PDB, https://www.rcsb.org/) (Berman et al., 2000), a database containing experimentally determined protein structures, or in the ModBase (https://modbase.compbio.ucsf.edu/) (Pieper et al., 2014), a database of comparative protein structure models.
A strong drawback of receptor-based techniques is that in many cases the target 3D structure has not been experimentally determined, especially in the case of trypanosomatids. This problem can be circumvented if there are structural data of molecules similar enough to the target to build a homology model.
A general rule of thumb is selecting the protein template with the highest sequence identity with the receptor of interest, particularly on the target pocket, and sequence identities lower than 30% will produce significantly less reliable models (Fiser, 2010). The quality of homology models must be assessed before using them in a receptor-based VS, and while there are many ways of evaluating the quality of a homology model (Bhattacharya et al., 2008; DasGupta et al., 2015; di Luccio and Koehl, 2012; Eramian et al., 2006; Shen and Sali, 2006), and most of the modelling tools include scores for quality assessment, it is important to know their capabilities and limits. Many assessment tools are biased towards the more known structures and may fail with proteins less represented in the databases, as is the case for membrane proteins (Benkert et al., 2011; di Luccio and Koehl, 2012).
Molecular docking is the receptor-based technique most extensively used in VS (Forli et al., 2016; Sousa et al., 2013) to predict if and how a library of ligands would interact with a receptor. It relies on randomly changing the spatial conformation of the ligand and calculating how well the generated poses would interact with the receptor, assigning an interaction score to each. This results in a set of conformations that are scored as the most likely to represent the real binding mode. When a compound library is used, it is possible to rank those molecules according to their biding scores, and obtaining their possible modes of interaction with the receptor.
While molecular docking algorithms give potential ways of a receptor–ligand interaction, it is not definitive proof of the mode of binding, or that there is binding at all. Thus the docked poses should be treated more as hypotheses to test experimentally. In fact, for some receptors and docking algorithm, mode of interaction with known ligands might not be reproduced (Chaput et al., 2016), for that reason it is extremely important, when possible, to validate whether the docking algorithm is capable of reproducing experimental results before using it to predict interactions with new compounds. The two most used methods of validation are re-docking, when the co-crystalized ligand is removed from the protein and docked to test whether the produced pose is the same as in the crystal, and cross-docking, when different co-crystalized ligands are docked with the receptor (Jain, 2009).
One of the most important drawbacks of molecular docking is the treatment of the receptor as a rigid molecule, so compounds that would otherwise bind to a pocket that is different – or not present – in the rigid receptor will be wrongly targeted as non-binders. To manage the flexibility of the receptor (B-Rao et al., 2009), an option would be using all known conformations of the receptor. Some algorithms make it possible to allow some degree of overlapping between the ligand and the receptor, treating some key residues of the target as flexible, and even perform induced fit models (Xu and Lill, 2013); all of them requiring additional computational costs.
Structure-based pharmacophore can also be obtained from the receptor as a set of spatial features capable of interacting with the residues on the binding site. This can be done directly by analysing the electrostatic distribution on the pocket, or by doing molecular docking of small fragments with varying molecular nature to probe the pocket and finding which features are more probable to interact with different parts of the binding site.
A very interesting strategy, although computationally expensive, is to perform molecular dynamic (MD) simulations of the receptor embedded in organic/aqueous mixed solvents containing different molecular features to find which areas of the receptor had more interactions with such solvents (Defelipe et al., 2018). This approach results in a spatial distribution of preferential interactions in the protein surface while taking into account the flexibility of the protein in a span of time.
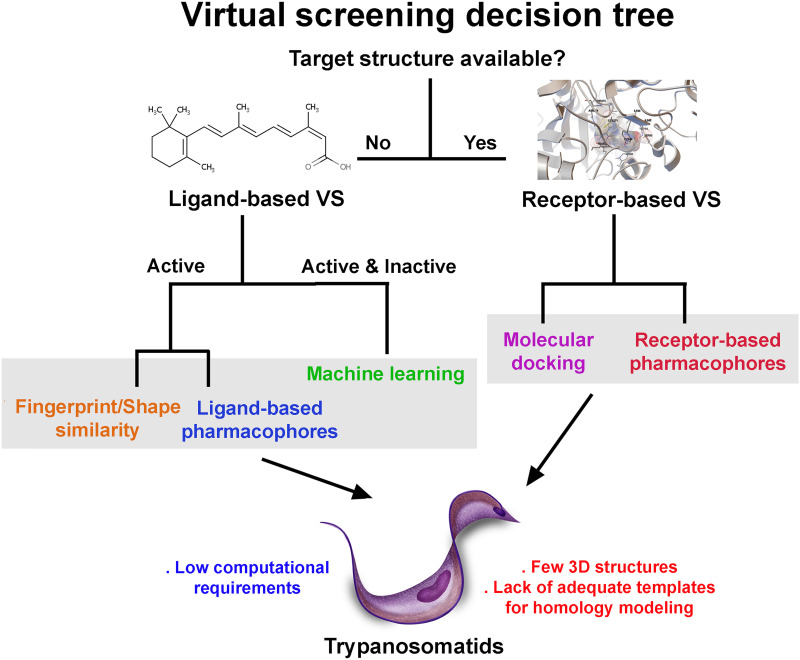
Figure 2 shows a graph on how to choose among the different VS strategies, as well as their advantages and disadvantages.
Fig. 2.
Common virtual screening techniques. Different VS approaches based on the available information about the protein targets and/or ligands. Receptor-based VS requires experimentally determined 3D structures or good-quality homology models that, in the case of trypanosomatid, are scarce. In the case of ligand-based VS, only small molecules (substrates, inhibitors, etc.) that interact with the target protein are needed.
Combined virtual screening strategies
Combinations of different techniques in a single VS workflow are strongly advised, taking advantage of their different strengths while minimizing the downsides they would have when used separately (Talevi et al., 2009). The size of the compound library being used is another factor to take into account at the moment of designing a VS pipeline; as for bigger libraries, it is preferable to start using methods computationally less expensive that allow discarding a huge volume of compounds and data. For smaller libraries or later steps in the workflow, it is plausible to use techniques that employ more computing power but also give more information about the possible mode of interaction that then can be used to bias the VS into finding compounds with such characteristics. As an example, receptor- and ligand-based pharmacophores can be used to adjust docking protocols to prefer the kind of interactions found in the model in what is known as ‘biased’ or ‘guided’ docking (Hu and Lill, 2014), which increases the performance of the docking algorithms.
Computational approaches are of great value in the drug discovery for trypanosomatid-caused diseases, by reducing the testable chemical space to a handful of promising compounds with high chances of having the desired biological activity, and by allowing to better exploit the growing information about the biology of these parasites. The available informatic tools are plentiful, whether the molecular target and its structure are known, or if there is a set of compounds interacting with a specific target in ways that might or might not be known; and combination of diverse tools is always the best choice to draw on their advantages while reducing their short-comes. The use of these approaches is not restricted to the search of active compounds, as the produced models can be harnessed to better understand the chemical characteristics of the ligand–target interaction. Finally, we must always keep in mind that these models have no value until they are experimentally tested, and feedback from the bench is critical for their betterment (Table 4).
Table 4.
Software of interest for computer-aided drug discovery
| Software | Free license | Capabilities | Web-link | Reference |
|---|---|---|---|---|
| SwissDock | Yes | Molecular docking with rigid target | http://www.swissdock.ch/ | Grosdidier et al. (2011) |
| AutoDock 4 | Yes | Molecular docking with rigid target or allowing flexible residues | http://autodock.scripps.edu/ | Morris et al. (2009) |
| AutoDock Vina | Yes | Rigid docking, online server | http://vina.scripps.edu/ | Trott and Olson (2010) |
| ZDOCK | Yes | https://zlab.umassmed.edu/zdock/ | Pierce et al. (2011) | |
| OEDocking | Trial | Molecular docking, flexible fitting, 2D and 3D similarity | https://www.eyesopen.com/oedocking | Kelley et al. (2015) |
| DOCK | Yes | Molecular docking with rigid target | http://dock.compbio.ucsf.edu/ | Allen et al. (2015) |
| GOLD | No | Molecular docking with rigid target, side-chain flexibility, and ensemble docking | https://www.ccdc.cam.ac.uk/solutions/csd-discovery/components/gold/ | Jones et al. (1997) |
| Glide | No | Protein homology modelling | https://www.schrodinger.com/glide | Friesner et al. (2006) |
| FlexX | No | https://www.biosolveit.de/flexx/ | Rarey et al. (1996) | |
| SwissModel | Yes | https://swissmodel.expasy.org/ | Waterhouse et al. (2018) | |
| I-TASSER | Yes | Setting of docking parameters, docking results and molecular visualization | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ | Yang and Zhang (2015) |
| Modeller | Yes | https://salilab.org/modeller/ | Webb and Sali (2016) | |
| AutoDock Tools | Yes | http://autodock.scripps.edu/ | Morris et al. (2009) | |
| Pymol | Yes | Molecular visualization | https://pymol.org/ | Schrödinger (USA) |
| VMD | Yes | Ligand and receptor-based pharmacophores | https://www.ks.uiuc.edu/Research/vmd/ | Humphrey et al. (1996) |
| Ligand Scout | Trial | http://www.inteligand.com/ligandscout/ | Wolber and Langer (2005) | |
| Discovery Studio | Visualizer only | Ligand and receptor-based pharmacophore, docking, ligand design, physico-chemical predictions, molecular graphics | https://www.3dsbiovia.com/ | Dassault Systèmes BIOVIA (2017) |
| Phase | No | Ligand and receptor-based pharmacophores, 3D QSAR | https://www.schrodinger.com/phase/ | Dixon et al. (2006) |
| LiSiCA | Yes | 2D and 3D similarity | http://insilab.org/lisica/ | Lesnik et al. (2015) |
| ShaEP | Yes | 3D small molecule alignment and similarity | http://users.abo.fi/mivainio/shaep/ | Vainio et al. (2009) |
| fPocket | Yes | Protein pocket prediction | http://fpocket.sourceforge.net/ | Schmidtke et al. (2010) |
| Gromacs | Yes | Molecular dynamics | http://www.gromacs.org/ | James Abraham et al. (2015) |
| AMBER suite | No | https://ambermd.org/ | Case et al. (2018) | |
| Dalton | Yes | Calculation of molecular descriptors 1D, 2D, and 3D | https://daltonprogram.org/ | Aidas et al. (2014) |
| PaDEL | Yes | http://www.yapcwsoft.com/dd/padeldescriptor/ | Yap (2011) |
VS applied to trypanosomatid-caused diseases
During the last decade, there was a significant increase in the number of scientific publications about different VS techniques applied to the identification of drug candidates for the treatment of NTDs. Probably one of the reasons for this emergent trend is the power of VS techniques to select active compounds rapidly and with an accessible cost for any laboratory (Bellera et al., 2019).
Many international organizations, such as DNDi (Chatelain and Ioset, 2011), recommend repurposing drugs for the treatment of NTDs in order to reduce the economic cost and the time of implementation of new therapeutic alternatives. In this sense, one of the main approaches for drug repositioning is through the application of computer simulations or VS. These techniques can use libraries of approved drugs to find a molecule with the desired biological activity. Most common approaches usually include a first in silico step based on individual or combined VS campaign followed by in vitro enzymatic or cell viability assays (Kontoyianni, 2017).
To illustrate the capabilities of VS, a few examples applied to drug discovery in NTDs will be detailed below.
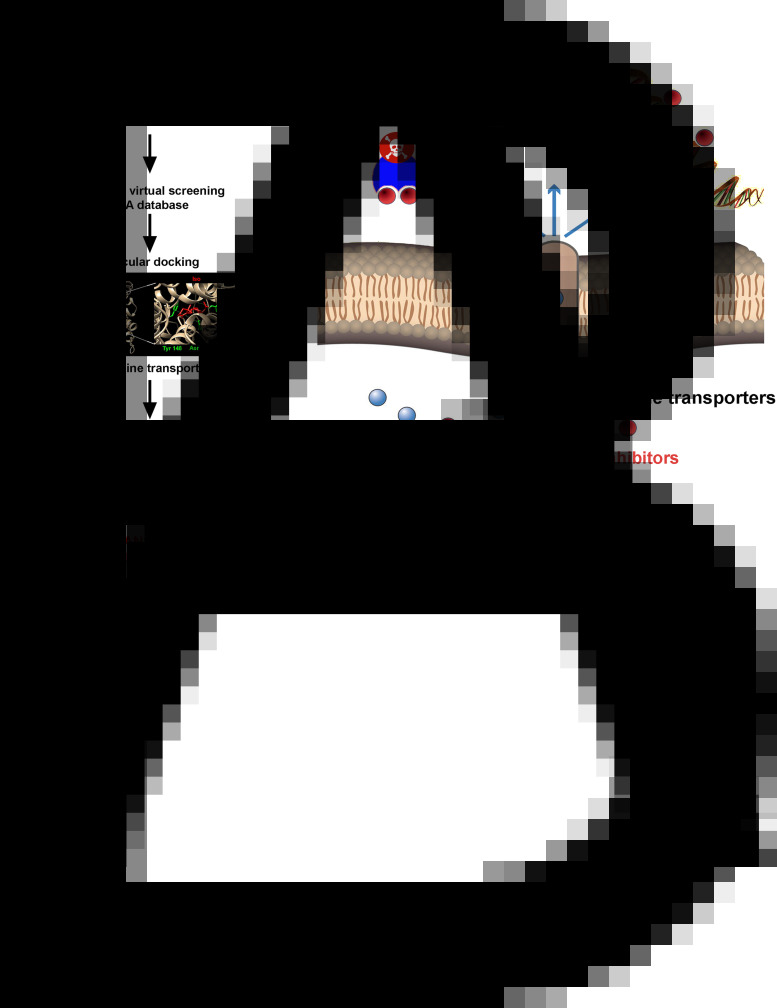
Reigada et al. performed a VS strategy to repurpose drugs to inhibit the T. cruzi polyamine transporter TcPAT12. The authors used the Tanimoto coefficient in LiSiCA v1.0 to search by 2D molecular similarity among 2924 compounds approved by the Food and Drug Administration (FDA) for its use in humans, employing retinol acetate as the reference since this molecule has been reported to decrease the intracellular polyamine concentration in Leishmania. A set of seven retinoids of dermatological use was identified and subsequently used in molecular docking. Among these compounds, isotretinoin, a drug used to treat severe acne, obtained the lowest docking score (−10.78 kcal/mol), which was in the range of the reference molecule (10.02 kcal/mol) and three times higher than the scores obtained for its natural ligands, spermidine and putrescine. Because of this, isotretinoin was tested in vitro, inhibiting the polyamine transport in the parasite and showing a strong trypanocidal effect at nanomolar concentrations (Reigada et al., 2017) (Fig. 3A).
Fig. 3.
Membrane transporters as drug targets. Ligand- and receptor-based VS was applied in this example to identify inhibitors of the T. cruzi polyamine permease (Reigada et al., 2017). Retinol acetate was first reported as a leishmanicidal compound that reduces the intracellular concentration of polyamines (Mukhopadhyay and Madhubala, 1994). Through a similarity VS using a database of FDA-approved drugs and retinol acetate as a reference molecule, a group of candidate drugs was identified. After the second step of receptor-based VS (molecular docking) followed by in vitro assays, it was demonstrated that the retinoid isotretinoin is a polyamine transport inhibitor with a strong anti-T. cruzi activity (A). Some advantages of membrane transporters as drug targets are schematized (B). For example, in many cases transport processes are the only way to obtain essential metabolites (i.e. polyamines in T. cruzi); the presence of extracellular spans in the transporter facilitates the accessibility of the drugs; some inhibitors are incorporated to the cell presenting additional intracellular targets such as enzymes or nucleic acids.
Another approach to identify inhibitors of the same T. cruzi polyamine transporter involved an anthracene–putrescine conjugate (Ant4) that blocks polyamine uptake in cancer cells. Ant4 was also found to inhibit the polyamine transport system in T. cruzi and produced a strong trypanocidal effect. Considering that Ant4 is not currently approved by the FDA, a similarity ligand-based VS using this compound as a reference molecule was applied. Three tricyclic antipsychotic drugs, promazine, chlorpromazine and clomipramine, showed to be effective inhibitors of putrescine uptake, and also revealed a high trypanocidal activity against T. cruzi amastigotes and trypomastigotes with calculated IC50s between 1.3 and 3.8 μm (Reigada et al., 2019)
These are interesting examples of trypanosomatid-caused diseases when little information about the target and its binding molecules is available. In the ligand-based approach, it shows the capabilities of similarity search to find active molecules with high potency starting from a single compound, even when the antecedents are in another organism, a significant advantage in the case of these insufficiently studied organisms, but since similarity search highly depends on the input set, it is worthy to note the small quantity of retrieved compounds. It also highlights a potential problem that should be taken into account; if the molecular target is too similar to a human homologue, it could bind to it as well, the reason why the differences between the parasites and the host are a key aspect to observe in the drug search.
Regarding the receptor-based methods, membrane proteins are more readily accessible for drugs but less structural information about them is available. At the time of publication, there was no crystal structure for a polyamine transporter in Tritryps, and the most related protein deposited in the PDB was an E. coli amino acid transporter (AdiC), with an identity of 30% with TcPAT12 considered the lower limit in the production of a reasonable homology model. Having in mind the previously mentioned bias of the quality assessment towards soluble proteins, the authors had to rely only on a Ramachandran plot to check the produced model was worthy of using in docking assays. Nevertheless, the molecular docking worked on predicting the binding of isotretinoin that was later determined experimentally in the same work. An important thing to have in mind is that docking scores by themselves are not a good indicator of whether a ligand will be a good binder or not, as they do not represent actual binding energies, working only to rank the complementarity of a ligand inside a pocket and hinting to which molecules might be better ligands than the others, the reason why the authors use retinoic acetate, spermidine and putrescine as a reference for what a good score might be for this particular case.
Using the same protein target, Dietrich et al. identified other anti-T. cruzi polyamine transport inhibitor, cisapride, a drug withdrawn for human treatments currently used in veterinary medicine to stimulate the upper gastrointestinal tract. The authors screened the ZINC and DrugBank databases employing similarity search, quantitative structure–activity relationship (QSAR) models and molecular docking-based screening (Dietrich et al., 2018).
For the similarity search, they used six compounds that disrupted the putrescine uptake in T. cruzi. Two different cut-off values were employed, for the DrugBank database, comprising 8261 molecules, those with a Tanimoto coefficient <0.5 were filtered out, while for the ZINC database, due to its greater size (17 900 742 compounds) they set a more stringent cut-off of 0.7, showing how its selection depends entirely on the researchers criteria about the desired quantity and structural diversity of the retrieved molecules. Because of the limitation of this strategy to find few compounds because of the quantity of input molecules, they complement the strategy with a QSAR model designed to find polyamine analogues with trypanocidal activity in micromolar concentrations, whether or not their molecular target was known. Employing both strategies, they find 594 candidates for further filtering by molecular docking.
The authors used the natural ligands and reported inhibitors of the transporter as reference molecules, and as negative controls a set of amino acids that do not bind to it. They compared Autodock 4.2 and Autodock Vina docking software and evaluated the performance of different scoring functions. Additionally, they performed a set of evaluations with rigid receptors, and other sets allowing flexibility on different residues determined by docking or mutagenesis to be involved in the binding of the natural ligands. Although Autodock Vina is reported to have better predictive power than Autodock 4.2 (Gaillard, 2018), in this case, the former ranked the inactive compounds higher than the natural ligands. From all the tested docking conditions, the rigid model with Autodock 4.2 performed the best on discriminating non-binders. The example shows the importance of testing various scoring functions and docking parameters, as their performance is specific to each receptor–ligand system.
By using a set of active and inactive compounds, the researchers could build a Receiving Operating Characteristic (ROC) curve to determine the score cut-off with the better trade-off between specificity and sensibility for the receptor-based filtering. Applying the mentioned model, 203 molecules were classified as possible binders; the top 10% were thoroughly analysed for their physicochemical properties, structural diversity and purchasability, leading to four compounds of which only cisapride inhibited the putrescine uptake in vitro. This illustrates how a richer input dataset can lead to better predictive models capable of processing larger libraries and retrieving active compounds.
Recently, it was demonstrated that crystal violet, a colourant used as an additive in blood banks to prevent transfusion-transmitted Chagas disease, inhibits the T. cruzi proline permease TcAAAP069. Using crystal violet as a query for a drug repurposing ligand-based VS, loratadine, cyproheptadine, olanzapine and clofazimine were identified as structurally related compounds. All these already-approved drugs for clinical use inhibited TcAAAP069 activity with different efficacies, presented trypanocidal action in epimastigotes, trypomastigotes and amastigotes of different T. cruzi strains and also presented a synergistic effect in combination with benznidazole (Saye et al., 2020)
Regarding the above-mentioned examples, some properties of membrane transporters as targets for drug development are outlined in Fig. 3B.
Other approaches using molecular descriptors and QSAR models were applied to find natural products that inhibit the de novo pyrimidine biosynthetic pathway, specifically the enzyme dihydroorotate dehydrogenase (DHODH) from L. major (Chibli et al., 2018). Similarly, inhibitors of the enzyme that reduce trypanothione were identified by linear discriminant analysis using molecular descriptors (Prieto et al., 2006).
For T. brucei, the only fully validated molecular target is the protein ODC (i.e. its disruption is the known target of current clinic treatment for the disease) (Gilbert, 2014), in this sense, great efforts have been made in order to identify other compounds capable of inhibiting its enzymatic activity. In a compelling example (Smithson et al., 2010), the authors started from a commercially available library of compounds and used chemoinformatic tools to filter out potential PAINs, select molecules with good ADME properties and generate clusters containing up to 20 compounds with maximum structural diversity. The generated clusters (with a total of 316 000 compounds) were used for HTS against T. brucei and human ODC. They found a novel chemotype comprising eight tested compounds that were a potent and selective inhibitor of the parasite ODC. Because both active sites have a high identity, the authors found unlikely that these compounds would be binding to the active site considering the high observed selectivity. Therefore, they used informatics tools to identify three other possible binding pockets for these inhibitors, followed by rigid docking simulations with the found active compounds and inactive chemical analogues in the identified pockets as well as the active site. The models predicted that only one of the pockets would bind better to the actives compared with the enzyme active site; also, the dockings in the same pocket yielded better discrimination between actives and inactives. To evaluate the role of the predicted binding residues, the authors analysed the differences in the predicted pocket between the human and parasite, and performed mutagenesis experiments, both analysis further supported their hypothesis. This is a fascinating example of how the feedback loop between computational models and the experimental results lead to a better understanding of the studied molecular systems.
Two promising drug targets for the treatment of HAT are the enzymes pteridine reductase and the N-acetyl-glucosaminyl-phosphatidylinositol deacetylase (GlcNAc-PI de-N-acetylase), involved in the essential pterin metabolism and GPI anchor biosynthesis of membrane proteins, respectively. Different chemical determinants of the T. brucei pteridine reductase activity were identified by pharmacophore mapping and subsequently used to database screening to find potential nanomolar range inhibitors (Dube et al., 2014). A very similar approach was applied to discover GlcNAc-PI de-N-acetylase inhibitors and two approved drugs were repositioned; the antibiotic ethambutol and the vasoconstrictor metaraminol (Rashmi and Swati, 2015).
A combined VS campaign was designed to find specific inhibitors of the L. donovani γ-glutamylcysteine synthetase (Gcs), an enzyme of the trypanothione-based redox system. The receptor-based steps include the homology modelling of the enzyme structure and active site prediction. Using a database of 55 000 commercially available compounds obeying the Lipinski's rules (http://www.maybridge.com/), the authors used molecular docking with three different scoring functions and retrieved five compounds ranked by the three functions better than L-buthionine-S, R-sulfoximine (BSO), a Gcs inhibitor that prolongs the survival in T. brucei mice infections but induces toxicity in the host. The predicted poses were evaluated by MD using GROMACS (James Abraham et al., 2015). These simulations confirmed the stability of the predicted binding modes, allowing the authors better assess the residues important for the binding of these compounds, and to identify other residues in the active site that could be exploited in lead optimization to increase the binding affinity. However, including the docking pose of BSO would be a great addition to the work, as it would work as a positive control of the model and throw some light on what molecular determinants should be retained if an optimization of its toxicity would be carried in the future. The five ligands were successfully validated in vitro, four compounds had better enzymatic inhibition than BSO, dissociation constants comparable to it, and leishmanicidal activity, three of them having negligible toxicity in human cell lines (Agnihotri et al., 2017). These results are a clear example of following in parallel the binding affinity and the ADME/Tox properties, and how information obtained from the predicted models could be of use for further lead optimization.
Another example of combined ligand- and structure-based VS strategy employing similarity VS, molecular docking and MD was applied to find putative T. cruzi enolase (TcENO) inhibitors. The enzyme substrates and two known enolase inhibitors were used as queries for the similarity VS using five different algorithms, resulting in six compounds of medical use (etidronate, pamidronate, fosfomycin, acetohydroxamate, triclofos and aminohydroxybutyrate). Molecular docking simulations and pose re-scoring predicted that etidronate and pamidronate were the best candidates. Finally, using MD calculations, it was proposed that etidronate is the best potential TcENO inhibitor and described the molecular motifs to be taken into account in the repurposing or design of drugs targeting this enzyme active site (Valera-Vera et al., 2020).
A novel approach based on the combination of proteomics and VS was used to identify potential drug targets to treat leishmaniasis. First, by proteome mining, new drug targets essential for the parasite and with low identity to human homologues were detected. One of these proteins related to the N-glycan biosynthesis pathway and a putative inhibitor, miglitol, were predicted in silico and validated in vitro (Chavez-Fumagalli et al., 2019).
An important point that can be remarked from the previous examples of VS strategies is the need of sources of structural variability in the databases screened to increase the chances of finding a compound with the appropriate biological properties. In this sense, the databases of approved drugs used for the drug repositioning have only about 3000 drugs. A widely used alternative are databases of small molecules either of natural or synthetic compounds that have >100 000 structures to find lead compounds for further optimization, always reminding that compounds obtained by VS must be tested in vitro and in vivo, and that following the evolution of potency and ADME/Tox through the drug development is highly recommended. Although the different VS tools can be combined in diverse ways to increase the efficiency in retrieved active compounds, the probability of success with this approach is completely uncertain until biological assays are performed on the protein, the target organism and infection models, results that can in turn be used to the improve the predictive models. For the specific case of trypanosomatid-caused diseases, there are, as yet, no treatments obtained from a VS strategy. Nevertheless, the enrichment in active molecules obtained from computational tools and the growing amount of information about potential targets and compounds binding to them make the discovery and development of chemotherapies against these parasites a more approachable task.
Additional examples of VS techniques applied to trypanosomiasis and leishmaniasis are listed in Supplementary Table S1.
Drug repurposing, an advantageous alternative to new drugs in NTDs
During the period 2016–2018, 130 NCE and 78 drug line extensions, which are products based on a previously approved molecule, were approved and launched to global markets. That is almost 40% of the new treatments in the last years corresponds to new indications, new combinations or new formulations for already marketed drugs (Graul et al., 2017, 2019, 2018). As previously mentioned, finding new indications for approved, withdrawn, abandoned or investigational drugs is called drug repurposing or drug repositioning, and in this section, we will present some advantages of this drug discovery strategy and also will provide examples of repositioned drugs to treat human pathologies, including trypanosomatid-caused diseases.
The classic drug development approach usually takes between 10 and 17 years from target identification to be available in the market. All these years also imply a rough investment of 0.8–2.3 billion dollars (DiMasi et al., 2016), and even then, drugs can fail and never get to the pharmacy. The main reasons for this failure are that the drugs are not as effective in humans as predicted by the preclinical assays, and/or that they are not safe for human administration. Drug repurposing can accelerate the time needed for a drug to reach the market and reduced the financial costs mainly because the preclinical and clinical assays can take advantage of the available safety, toxicity and pharmacokinetics and pharmacodynamics data. This approach can take between 3 and 12 years and diminish the cost around 40% of the traditional development (Ashburn and Thor, 2004; Chong and Sullivan, 2007) (Drug Repurposing and Repositioning: Workshop Summary; https://www.nap.edu/read/18731/chapter/1). The potential repositioned compound can be identified through serendipity or rational approaches, including computational strategies, biological experimental strategies or a combination of both (Xue et al., 2018). One of the most recognized examples of a successful repurposing story involves sildenafil which was first developed as an antihypertensive drug and then repurposed for the treatment of erectile dysfunction and pulmonary arterial hypertension (Ghofrani et al., 2006). Another example is the drug thalidomide that was originally developed for treating morning sickness and was withdrawn from the market because of its teratogenic effects. However, this compound is now used to treat erythema nodosum leprosum and it is also employed in combination with dexamethasone for the treatment of newly diagnosed multiple myeloma (Gupta et al., 2013; Singhal et al., 1999; Zhou et al., 2013). Inspired by these cases and many other success stories of repositioned drugs, several studies are underway to identifying new biological activities for existing drugs (Czech et al., 2019; Ferreira and Andricopulo, 2016; Novac, 2013).
NTDs, like Chagas disease, HAT and leishmaniasis, are usually associated with underdeveloped countries and poverty. Thus, big pharmaceutical companies are not generally interested in the development and production of treatments for these diseases because it is unlikely for them to recover the investment and even less probable to make a profit. In this regard, the drug repurposing approach turns out very appealing since the costs of the drug discovery process are greatly reduced.
Eflornithine (6), a polyamine synthesis inhibitor, constitutes a remarkable case of drug repositioning in trypanosomatid-caused diseases since it was initially evaluated as an antitumor agent, but the clinical studies were discontinued due to adverse effects (Abeloff et al., 1986; Meyskens et al., 1986). However, in the late 1980s, eflornithine (6) was licensed as an orphan drug for treating HAT (Burri and Brun, 2003). Another example involves nifurtimox (2), used for Chagas diseases, which has been combined with eflornithine (6) for first-line treatment of second-stage T. brucei gambiense HAT (Priotto et al., 2009). Another drug tested was fexinidazole (7), which had been in preclinical development in the 1970s–1980s as a broad-spectrum antimicrobial agent (Raether and Seidenath, 1983). The molecule is a DNA synthesis inhibitor rediscovered by the DNDi in 2005 as having an activity against African trypanosomes (Deeks, 2019). The DNDi, in collaboration with Sanofi, have demonstrated that fexinidazole (7) represents the first well-tolerated single-compound oral therapy against first and second stage of HAT due to T. brucei gambiense (Deeks, 2019). The drug is currently undergoing Phase III clinical trials for treating this disease (https://www.dndi.org/diseases-projects/portfolio/fexinidazole/).
Despite no drugs have been successfully repurposed for its use against Chagas disease yet, fexinidazole (7) also represents an important advance for drug discovery in this parasitic disease. Its activity was investigated in vivo on several T. cruzi strains [susceptible, resistant or partially resistant to the current treatment benznidazole (1)] and its efficacy in suppressing parasitaemia and preventing death in infected mice has been demonstrated (Bahia et al., 2012). Another study revealed that fexinidazole (7) is more effective at curing chronic than acute T. cruzi infections in a similar mouse model (Francisco et al., 2016). This drug is currently being evaluated in clinical trials as a treatment for Chagas disease (https://www.dndi.org/diseases-projects/portfolio/fexinidazole-chagas/).
Most of the drugs that are active against leishmaniasis were repurposed from other indications. For instance, amphotericin B (12) was introduced as an antifungal agent obtained from Streptomyces nodosus. In 1997, liposomal amphotericin B (AmBisome) was the first drug approved for the treatment of visceral leishmaniasis (Meyerhoff, 1999). It binds to ergosterol, the predominant sterol in Leishmania (Roberts et al., 2003). Paromomycin (11), an aminoglycoside antibiotic, was isolated in the 1950s from Streptomyces krestomuceticus and it is active against bacteria as well as some protozoa and cestodes (Davidson et al., 2009). The antileishmanial activities of paromomycin (11) were recognized in the 1960s and it is used as an alternative treatment of both visceral and cutaneous leishmaniasis (Jain and Jain, 2013). One mechanism of action of paromomycin (11) involves inhibition of cytoplasmic and mitochondrial protein synthesis (Jhingran et al., 2009). The antileishmanial drug miltefosine (10), an alkylphosphocholine, was originally developed for the treatment of cutaneous cancers but was discontinued for this indication due to its adverse effects (Dorlo et al., 2012). Miltefosine (10) has re-emerged as the only effective oral drug available to treat all of the clinical forms of leishmaniasis; however, it is limited by its relatively high cost and side-effects (Ortega et al., 2017; Sindermann et al., 2004). On the other hand, fexinidazole (7) was also effective in L. donovani-infected mice; however, clinical trials in patients with visceral leishmaniasis have been discontinued due to lack of efficacy (Wyllie et al., 2012) (https://clinicaltrials.gov/ct2/show/NCT01980199).
Many more cases are emerging for developing treatments for neglected diseases through drug repurposing. The computational strategies mentioned previously are becoming an important part of this process. They can be used for identifying potential repositioning candidates systematically and are an excellent complement to experimental techniques (Delavan et al., 2018). These in silico approaches contribute to speed up the process of drug discovery at little extra cost (Ekins et al., 2011). For example, VS methods offer a quick assessment of huge libraries compiling known drugs and reduce the number of compounds that need testing to discover novel treatments (Kontoyianni, 2017). This computer-aided strategy has been signalled as a relevant strategy to aid find new medications for neglected diseases (Ekins et al., 2011; Pollastri and Campbell, 2011; Sardana et al., 2011). Examples of drugs repositioned against trypanosomatid-caused diseases are listed in Table 5.
Table 5.
Repurposed drugs
| Drug/structure | Original indication | Target | Organism | Repurposed/progress | Reference |
|---|---|---|---|---|---|
Isotretinoin 
|
Acute acne | Polyamine permease | T. cruzi | Effectiveness in animal model | Reigada et al. (2017) |
Cisapride 
|
Gastroesophageal reflux | Polyamine permease | T. cruzi | Evaluation in T. cruzi clinically relevant forms | Dietrich et al. (2018) |
Miglitol 
|
Diabetes mellitus type 2 | N-glycan biosynthesis (predicted) |
L. amazonensis L. infantum |
Effectiveness in animal model | Chavez-Fumagalli et al. (2019) |
Eflornithine (6) 
|
Cancer | Polyamine metabolism | T. brucei | Human African trypanosomiasis | Burri and Brun (2003) |
Nifurtimox (2) 
|
Chagas disease | Macromolecules | T. brucei gambiense | Second stage human African trypanosomiasis, in combination with eflornithine | Priotto et al. (2009); Hall et al. (2011) |
Fexinidazole (7) 
|
Broad-spectrum antimicrobial agent | DNA |
T. brucei gambiense T. cruzi |
Phase III clinical trials for African trypanosomiasis Clinical trials for Chagas disease |
Raether and Seidenath (1983); Deeks (2019); Bahia et al. (2012); Francisco et al. (2016) |
Amphotericin B (12) 
|
Antifungal agent | Ergosterol | Leishmania spp. | Visceral leishmaniasis | Meyerhoff (1999); Roberts et al. (2003) |
Paromomycin (11) 
|
Antibiotic (bacteria, protozoa and cestodes) | Cytoplasmic and mitochondrial protein synthesis | Leishmania spp. | Visceral and cutaneous leishmaniasis | Davidson et al. (2009); Jhingran et al. (2009); Jain and Jain (2013) |
Miltefosine (10) 
|
Cutaneous cancer | Lipid biosynthesis | Leishmania spp. | Visceral and cutaneous leishmaniasis | Sindermann et al. (2004); Dorlo et al. (2012); Ortega et al. (2017); Pinto-Martinez et al. (2018) |
Examples of repositioned drugs against trypanosomatid-caused diseases including the actual treatment, protein target and the new drug indication.
Concluding remarks
Since the development of new drugs for neglected diseases is a hard task due to the low investment of resources and the lack of economic interest from most pharmaceutical companies, the use of VS techniques for drug repurposing is a good option. The advantages of this experimental approach are the low time-consuming first stage generating a group of candidate compounds for further testing in vitro and in vivo. In addition, working with drugs already approved for other diseases shortens the subsequent trials and the funds needed for implementing a new therapy against NTDs.
These applications are accessible to any laboratory since a large number of free open source software are available and in most cases can be used with standard personal computers. In addition, drugs used for other pathologies have available information about, for example, their toxicity, pharmacokinetics, pharmacodynamics, bioavailability and half-life.
In addition, in drug repurposing approaches, side-effects are not necessary exclusion factors since the main goal is the development of new therapeutics against deadly diseases and most of the current treatments for NTDs are not safe for the patients.
There are numerous successful examples of drug development, involving VS techniques, for the treatment of different diseases. Some examples are isoniazid (DrugBank ID: DB00951) approved as tuberculostatic, amprenavir (DrugBank ID: DB00701) approved for the treatment of HIV or flurbiprofen (DrugBank ID: DB00712) approved as non-steroidal anti-inflammatory agent with antipyretic and analgesic activity (Batool et al., 2019). However, in the case of trypanosomiasis, these strategies were recently applied to the identification of new drugs and it will still be necessary to wait a few years to evaluate the first results in clinical trials.
Finally, we encourage research groups that work with drug targets to try the VS techniques described in this review. As a very important initial tip, we consider that the most suitable and reliable approach is the use of a combined strategy. However, there are no predetermined schemes to establish the order or the techniques to use, they exclusively depend on each particular case.
Financial support
This work was supported by Consejo Nacional de Investigaciones Científicas y Técnicas, Agencia Nacional de Promoción Científica y Tecnológica (FONCyT PICT 2015–0539, 2017-2096 and 2018-1801). The research leading to these results has, in part, received funding from the UK Research and Innovation via the Global Challenges Research Fund under grant agreement ‘A Global Network for Neglected Tropical Diseases’ grant number MR/P027989/1. CAP, GRL and MRM are members of the career of the scientific investigator; CR and EVV are research fellows from CONICET; and MS is PDRA from the A Global Network for Neglected Tropical Diseases.
Conflict of interest
None.
Ethical standards
Not applicable.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182020000207.
click here to view supplementary material
References
- Abeloff MD, Rosen ST, Luk GD, Baylin SB, Zeltzman M and Sjoerdsma A (1986) Phase II trials of alpha-difluoromethylornithine, an inhibitor of polyamine synthesis, in advanced small cell lung cancer and colon cancer. Cancer Treatment Reports 70, 843–845. [PubMed] [Google Scholar]
- Abi Hussein H, Geneix C, Petitjean M, Borrel A, Flatters D and Camproux AC (2017) Global vision of druggability issues: applications and perspectives. Drug Discovery Today 22, 404–415. [DOI] [PubMed] [Google Scholar]
- Adl SM, Bass D, Lane CE, Lukes J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cardenas P, Cepicka I, Chistyakova L, Del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V and Zhang Q (2019) Revisions to the classification, nomenclature, and diversity of eukaryotes. Journal of Eukaryotic Microbiology 66, 4–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts C, Sunyoto T, Tediosi F and Sicuri E (2017) Are public-private partnerships the solution to tackle neglected tropical diseases? A systematic review of the literature. Health Policy 121, 745–754. [DOI] [PubMed] [Google Scholar]
- Agnihotri P, Mishra AK, Mishra S, Sirohi VK, Sahasrabuddhe AA and Pratap JV (2017) Identification of novel inhibitors of Leishmania donovani gamma-glutamylcysteine synthetase using structure-based virtual screening, docking, molecular dynamics simulation, and in vitro studies. Journal of Chemical Information and Modeling 57, 815–825. [DOI] [PubMed] [Google Scholar]
- Aidas K, Angeli C, Bak KL, Bakken V, Bast R, Boman L, Christiansen O, Cimiraglia R, Coriani S, Dahle P, Dalskov EK, Ekstrom U, Enevoldsen T, Eriksen JJ, Ettenhuber P, Fernandez B, Ferrighi L, Fliegl H, Frediani L, Hald K, Halkier A, Hattig C, Heiberg H, Helgaker T, Hennum AC, Hettema H, Hjertenaes E, Host S, Hoyvik IM, Iozzi MF, Jansik B, Jensen HJ, Jonsson D, Jorgensen P, Kauczor J, Kirpekar S, Kjaergaard T, Klopper W, Knecht S, Kobayashi R, Koch H, Kongsted J, Krapp A, Kristensen K, Ligabue A, Lutnaes OB, Melo JI, Mikkelsen KV, Myhre RH, Neiss C, Nielsen CB, Norman P, Olsen J, Olsen JM, Osted A, Packer MJ, Pawlowski F, Pedersen TB, Provasi PF, Reine S, Rinkevicius Z, Ruden TA, Ruud K, Rybkin VV, Salek P, Samson CC, de Meras AS, Saue T, Sauer SP, Schimmelpfennig B, Sneskov K, Steindal AH, Sylvester-Hvid KO, Taylor PR, Teale AM, Tellgren EI, Tew DP, Thorvaldsen AJ, Thogersen L, Vahtras O, Watson MA, Wilson DJ, Ziolkowski M and Agren H (2014) The Dalton quantum chemistry program system. Wiley Interdisciplinary Reviews. Computational Molecular Science 4, 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberca LN, Sbaraglini ML, Balcazar D, Fraccaroli L, Carrillo C, Medeiros A, Benitez D, Comini M and Talevi A (2016) Discovery of novel polyamine analogs with anti-protozoal activity by computer guided drug repositioning. Journal of Computer-Aided Molecular Design 30, 305–321. [DOI] [PubMed] [Google Scholar]
- Alberca LN, Sbaraglini ML, Morales JF, Dietrich R, Ruiz MD, Pino Martinez AM, Miranda CG, Fraccaroli L, Alba Soto CD, Carrillo C, Palestro PH and Talevi A (2018) Cascade ligand- and structure-based virtual screening to identify new trypanocidal compounds inhibiting putrescine uptake. Frontiers in Cellular and Infection Microbiology 8, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcântara LM, Ferreira TCS, Gadelha FR and Miguel DC (2018) Challenges in drug discovery targeting TriTryp diseases with an emphasis on leishmaniasis. International Journal for Parasitology: Drugs and Drug Resistance 8, 430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PT, Case DA, Kuntz ID and Rizzo RC (2015) DOCK 6: impact of new features and current docking performance. Journal of Computational Chemistry 36, 1132–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves F, Bilbe G, Blesson S, Goyal V, Monnerat S, Mowbray C, Muthoni Ouattara G, Pecoul B, Rijal S, Rode J, Solomos A, Strub-Wourgaft N, Wasunna M, Wells S, Zijlstra EE, Arana B and Alvar J (2018) Recent development of visceral leishmaniasis treatments: successes, pitfalls, and perspectives. Clinical Microbiology Reviews 31, e00048–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson NE and Joya CA (2019) Cutaneous leishmaniasis: updates in diagnosis and management. Infectious Disease Clinics of North America 33, 101–117. [DOI] [PubMed] [Google Scholar]
- Aronson N, Herwaldt BL, Libman M, Pearson R, Lopez-Velez R, Weina P, Carvalho E, Ephros M, Jeronimo S and Magill A (2017) Diagnosis and treatment of leishmaniasis: clinical practice guidelines by the Infectious Diseases Society of America (IDSA) and the American Society of Tropical Medicine and Hygiene (ASTMH). American Journal of Tropical Medicine and Hygiene 96, 24–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn TT and Thor KB (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nature Reviews. Drug Discovery 3, 673–683. [DOI] [PubMed] [Google Scholar]
- Avilan L, Gualdron-Lopez M, Quinones W, Gonzalez-Gonzalez L, Hannaert V, Michels PA and Concepcion JL (2011) Enolase: a key player in the metabolism and a probable virulence factor of trypanosomatid parasites-perspectives for its use as a therapeutic target. Enzyme Research 2011, 932549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B-Rao C, Subramanian J and Sharma SD (2009) Managing protein flexibility in docking and its applications. Drug Discovery Today 14, 394–400. [DOI] [PubMed] [Google Scholar]
- Babokhov P, Sanyaolu AO, Oyibo WA, Fagbenro-Beyioku AF and Iriemenam NC (2013) A current analysis of chemotherapy strategies for the treatment of human African trypanosomiasis. Pathogens and Global Health 107, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell JB and Holloway GA (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of Medicinal Chemistry 53, 2719–2740. [DOI] [PubMed] [Google Scholar]
- Baell J and Walters MA (2014) Chemistry: chemical con artists foil drug discovery. Nature 513, 481–483. [DOI] [PubMed] [Google Scholar]
- Bahia MT, de Andrade IM, Martins TA, do Nascimento AF, Diniz Lde F, Caldas IS, Talvani A, Trunz BB, Torreele E and Ribeiro I (2012) Fexinidazole: a potential new drug candidate for Chagas disease. PLoS Neglected Tropical Diseases 6, e1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajusz D, Racz A and Heberger K (2015) Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? Journal of Cheminformatics 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker BM, Westerhoff HV, Opperdoes FR and Michels PA (2000) Metabolic control analysis of glycolysis in trypanosomes as an approach to improve selectivity and effectiveness of drugs. Molecular and Biochemical Parasitology 106, 1–10. [DOI] [PubMed] [Google Scholar]
- Banegas-Luna AJ, Ceron-Carrasco JP and Perez-Sanchez H (2018) A review of ligand-based virtual screening web tools and screening algorithms in large molecular databases in the age of big data. Future Medicinal Chemistry 10, 2641–2658. [DOI] [PubMed] [Google Scholar]
- Barrett MP, Mottram JC and Coombs GH (1999) Recent advances in identifying and validating drug targets in trypanosomes and leishmanias. Trends in Microbiology 7, 82–88. [DOI] [PubMed] [Google Scholar]
- Batool M, Ahmad B and Choi S (2019) A structure-based drug discovery paradigm. International Journal of Molecular Sciences 20, E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AS, Mills JE, Williams GP, Brannigan JA, Wilkinson AJ, Parkinson T, Leatherbarrow RJ, Tate EW, Holder AA and Smith DF (2012) Selective inhibitors of protozoan protein N-myristoyltransferases as starting points for tropical disease medicinal chemistry programs. PLoS Neglected Tropical Diseases 6, e1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellera CL, Balcazar DE, Alberca L, Labriola CA, Talevi A and Carrillo C (2013) Application of computer-aided drug repurposing in the search of new cruzipain inhibitors: discovery of amiodarone and bromocriptine inhibitory effects. Journal of Chemical Information and Modeling 53, 2402–2408. [DOI] [PubMed] [Google Scholar]
- Bellera CL, Balcazar DE, Alberca L, Labriola CA, Talevi A and Carrillo C (2014) Identification of levothyroxine antichagasic activity through computer-aided drug repurposing. TheScientificWorldJournal 2014, 279618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belllera CL, Sbaraglini ML, Alberca LN, Alice JI and Talevi A (2019) In silico modeling of FDA-approved drugs for discovery of therapies against neglected diseases: a drug repurposing approach. In Roy K. (ed.), In Silico Drug Design: Repurposing Techniques and Methodologies. Massachusetts, USA: Academic Press, pp. 625–648. [Google Scholar]
- Beneke T, Madden R, Makin L, Valli J, Sunter J and Gluenz E (2017) A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. Royal Society Open Science 4, 170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez D, Medeiros A, Fiestas L, Panozzo-Zenere EA, Maiwald F, Prousis KC, Roussaki M, Calogeropoulou T, Detsi A, Jaeger T, Sarlauskas J, Peterlin Masic L, Kunick C, Labadie GR, Flohe L and Comini MA (2016) Identification of novel chemical scaffolds inhibiting trypanothione synthetase from pathogenic trypanosomatids. PLoS Neglected Tropical Diseases 10, e0004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, Biasini M and Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics (Oxford, England) 27, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE (2000) The protein data bank. Nucleic Acids Research 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern C (2015) Chagas’ disease. New England Journal of Medicine 373, 456–466. [DOI] [PubMed] [Google Scholar]
- Berna L, Rodriguez M, Chiribao ML, Parodi-Talice A, Pita S, Rijo G, Alvarez-Valin F and Robello C (2018) Expanding an expanded genome: long-read sequencing of Trypanosoma cruzi. Microbial Genomics 4. doi: 10.1099/mgen.0.000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berneman A, Montout L, Goyard S, Chamond N, Cosson A, d'Archivio S, Gouault N, Uriac P, Blondel A and Minoprio P (2013) Combined approaches for drug design points the way to novel proline racemase inhibitor candidates to fight Chagas’ disease. PLoS ONE 8, e60955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE and El-Sayed NM (2005) The genome of the African trypanosome Trypanosoma brucei. Science (New York, N.Y.) 309, 416–422. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Wunderlich Z, Monleon D, Tejero R and Montelione GT (2008) Assessing model accuracy using the homology modeling automatically software. Proteins 70, 105–118. [DOI] [PubMed] [Google Scholar]
- Boscardin SB, Torrecilhas AC, Manarin R, Revelli S, Rey EG, Tonelli RR and Silber AM (2010) Chagas’ disease: an update on immune mechanisms and therapeutic strategies. Journal of Cellular and Molecular Medicine 14, 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottieau E and Clerinx J (2019) Human African trypanosomiasis: progress and stagnation. Infectious Disease Clinics of North America 33, 61–77. [DOI] [PubMed] [Google Scholar]
- KR Brimacombe, MJ Walsh, Liu L, MG Vásquez-Valdivieso, HP Morgan, McNae I, LA Fothergill-Gilmore, PAM Michels, DS Auld, Simeonov A, MD Walkinshaw, Shen M and MB Boxer (2017) Identification of ML251, a Potent Inhibitor of T. brucei and T. cruzi Phosphofructokinase. ACS Medicinal Chemistry Letters 5, 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N and Jacoby E (2006) On scaffolds and hopping in medicinal chemistry. Mini Reviews in Medicinal Chemistry 6, 1217–1229. [DOI] [PubMed] [Google Scholar]
- Burle-Caldas Gde A, Grazielle-Silva V, Laibida LA, DaRocha WD and Teixeira SM (2015) Expanding the tool box for genetic manipulation of Trypanosoma cruzi. Molecular and Biochemical Parasitology 203, 25–33. [DOI] [PubMed] [Google Scholar]
- Burri C and Brun R (2003) Eflornithine for the treatment of human African trypanosomiasis. Parasitology Research 90, S49–S52. [DOI] [PubMed] [Google Scholar]
- Burza S, Croft SL and Boelaert M (2018) Leishmaniasis. Lancet (London, England) 392, 951–970. [DOI] [PubMed] [Google Scholar]
- Buscher P, Cecchi G, Jamonneau V and Priotto G (2017) Human African trypanosomiasis. Lancet (London, England) 390, 2397–2409. [DOI] [PubMed] [Google Scholar]
- Calvet CM, Choi JY, Thomas D, Suzuki B, Hirata K, Lostracco-Johnson S, de Mesquita LB, Nogueira A, Meuser-Batista M, Silva TA, Siqueira-Neto JL, Roush WR, de Souza Pereira MC, McKerrow JH and Podust LM (2017) 4-aminopyridyl-based Lead compounds targeting CYP51 prevent spontaneous parasite relapse in a chronic model and improve cardiac pathology in an acute model of Trypanosoma cruzi infection. PLoS Neglected Tropical Diseases 11, e0006132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham TE III, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, Gohlke H, Goetz AW, Greene D, Harris R, Homeyer N, Huang Y, Izadi S, Kovalenko A, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Mermelstein DJ, Merz KM, Miao Y, Monard G, Nguyen C, Nguyen H, Omelyan I, Onufriev A, Pan F, Qi R, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Smith J, SalomonFerrer R, Swails J, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xiao L, York DM and Kollman PA (2018) AMBER 2018. San Francisco: University of California. [Google Scholar]
- Chagas C (1909) Nova tripanozomiaze humana: estudos sobre a morfolojia e o ciclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbida do homem. Nova tripanozomiaze humana: estudos sobre a morfolojia e o ciclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbida do homem 1, 1678–8060. [Google Scholar]
- Chaput L, Martinez-Sanz J, Saettel N and Mouawad L (2016) Benchmark of four popular virtual screening programs: construction of the active/decoy dataset remains a major determinant of measured performance. Journal of Cheminformatics 8, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelain E and Ioset JR (2011) Drug discovery and development for neglected diseases: the DNDi model. Drug Design, Development and Therapy 5, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Fumagalli MA, Lage DP, Tavares GSV, Mendonca DVC, Dias DS, Ribeiro PAF, Ludolf F, Costa LE, Coelho VTS and Coelho EAF (2019) In silico Leishmania proteome mining applied to identify drug target potential to be used to treat against visceral and tegumentary leishmaniasis. Journal of Molecular Graphics & Modelling 87, 89–97. [DOI] [PubMed] [Google Scholar]
- Cheleski J, Rocha JR, Pinheiro MP, Wiggers HJ, da Silva AB, Nonato MC and Montanari CA (2010) Novel insights for dihydroorotate dehydrogenase class 1A inhibitors discovery. European Journal of Medicinal Chemistry 45, 5899–5909. [DOI] [PubMed] [Google Scholar]
- Chibli LA, Schmidt TJ, Nonato MC, Calil FA and Da Costa FB (2018) Natural products as inhibitors of Leishmania major dihydroorotate dehydrogenase. European Journal of Medicinal Chemistry 157, 852–866. [DOI] [PubMed] [Google Scholar]
- Choi JY, Calvet CM, Gunatilleke SS, Ruiz C, Cameron MD, McKerrow JH, Podust LM and Roush WR (2013) Rational development of 4-aminopyridyl-based inhibitors targeting Trypanosoma cruzi CYP51 as anti-chagas agents. Journal of Medicinal Chemistry 56, 7651–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong CR and Sullivan DJ Jr (2007) New uses for old drugs. Nature 448, 645–646. [DOI] [PubMed] [Google Scholar]
- Croft SL, Sundar S and Fairlamb AH (2006) Drug resistance in leishmaniasis. Clinical Microbiology Reviews 19, 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech T, Lalani R and Oyewumi MO (2019) Delivery systems as vital tools in drug repurposing. AAPS PharmSciTech 20, 116. [DOI] [PubMed] [Google Scholar]
- Dahlin JL, Nissink JW, Strasser JM, Francis S, Higgins L, Zhou H, Zhang Z and Walters MA (2015) PAINS In the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. Journal of Medicinal Chemistry 58, 2091–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danishuddin M and Khan AU (2016) Descriptors and their selection methods in QSAR analysis: paradigm for drug design. Drug Discovery Today, 21, 1291–1302. [DOI] [PubMed] [Google Scholar]
- DasGupta D, Kaushik R and Jayaram B (2015) From Ramachandran maps to tertiary structures of proteins. Journal of Physical Chemistry B 119, 11136–11145. [DOI] [PubMed] [Google Scholar]
- Dassault Systèmes BIOVIA (2017). Discovery studio, San Diego: Dassault Systèmes, 2017.
- Davidson RN, den Boer M and Ritmeijer K (2009) Paromomycin. Transactions of the Royal Society of Tropical Medicine and Hygiene 103, 653–660. [DOI] [PubMed] [Google Scholar]
- Deeks ED (2019) Fexinidazole: first global approval. Drugs 79, 215–220. [DOI] [PubMed] [Google Scholar]
- Defelipe LA, Arcon JP, Modenutti CP, Marti MA, Turjanski AG and Barril X (2018) Solvents to fragments to drugs: MD applications in drug design. Molecules 23, E3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavan B, Roberts R, Huang R, Bao W, Tong W and Liu Z (2018) Computational drug repositioning for rare diseases in the era of precision medicine. Drug Discovery Today 23, 382–394. [DOI] [PubMed] [Google Scholar]
- Demir O, Labaied M, Merritt C, Stuart K and Amaro RE (2014) Computer-aided discovery of Trypanosoma brucei RNA-editing terminal uridylyl transferase 2 inhibitors. Chemical Biology & Drug Design 84, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza ML, de Oliveira Rezende Junior C, Ferreira RS, Espinoza Chavez RM, Ferreira LLG, Slafer BW, Magalhaes LG, Krogh R, Oliva G, Cruz FC, Dias LC and Andricopulo AD (2019) Discovery of potent, reversible, and competitive cruzain inhibitors with trypanocidal activity: a structure-based drug design approach. Journal of Chemical Information and Modeling 62, 1028–1041. [DOI] [PubMed] [Google Scholar]
- Dietrich RC, Alberca LN, Ruiz MD, Palestro PH, Carrillo C, Talevi A and Gavernet L (2018) Identification of cisapride as new inhibitor of putrescine uptake in Trypanosoma cruzi by combined ligand- and structure-based virtual screening. European Journal of Medicinal Chemistry 149, 22–29. [DOI] [PubMed] [Google Scholar]
- di Luccio E and Koehl P (2012) The H-factor as a novel quality metric for homology modeling. Journal of Clinical Bioinformatics 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMasi JA, Grabowski HG and Hansen RW (2016) Innovation in the pharmaceutical industry: new estimates of R&D costs. Journal of Health Economics 47, 20–33. [DOI] [PubMed] [Google Scholar]
- Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE and Friesner RA (2006) PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. Journal of Computer-Aided Molecular Design 20, 647–671. [DOI] [PubMed] [Google Scholar]
- Dorlo TP, Balasegaram M, Beijnen JH and de Vries PJ (2012) Miltefosine: a review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. Journal of Antimicrobial Chemotherapy 67, 2576–2597. [DOI] [PubMed] [Google Scholar]
- Drews J (1998) Innovation deficit revisited: reflections on the productivity of pharmaceutical R&D. Drugs Discovery Today 3, 491–494. [Google Scholar]
- Dube D, Sharma S, Singh TP and Kaur P (2014) Pharmacophore mapping, In silico screening and molecular docking to identify selective Trypanosoma brucei pteridine reductase inhibitors. Molecular Informatics 33, 124–134. [DOI] [PubMed] [Google Scholar]
- Duncan SM, Jones NG and Mottram JC (2017) Recent advances in Leishmania reverse genetics: manipulating a manipulative parasite. Molecular and Biochemical Parasitology 216, 30–38. [DOI] [PubMed] [Google Scholar]
- Durrant JD, Hall L, Swift RV, Landon M, Schnaufer A and Amaro RE (2010) Novel naphthalene-based inhibitors of Trypanosoma brucei RNA editing ligase 1. PLoS Neglected Tropical Diseases 4, e803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Williams AJ, Krasowski MD and Freundlich JS (2011) In silico repositioning of approved drugs for rare and neglected diseases. Drug Discovery Today 16, 298–310. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD and Andersson B (2005a) The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science (New York, N.Y.) 309, 409–415. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, Ghedin E, Peacock C, Bartholomeu DC, Haas BJ, Tran AN, Wortman JR, Alsmark UC, Angiuoli S, Anupama A, Badger J, Bringaud F, Cadag E, Carlton JM, Cerqueira GC, Creasy T, Delcher AL, Djikeng A, Embley TM, Hauser C, Ivens AC, Kummerfeld SK, Pereira-Leal JB, Nilsson D, Peterson J, Salzberg SL, Shallom J, Silva JC, Sundaram J, Westenberger S, White O, Melville SE, Donelson JE, Andersson B, Stuart KD and Hall N (2005b) Comparative genomics of trypanosomatid parasitic protozoa. Science (New York, N.Y.) 309, 404–409. [DOI] [PubMed] [Google Scholar]
- Eramian D, Shen MY, Devos D, Melo F, Sali A and Marti-Renom MA (2006) A composite score for predicting errors in protein structure models. Protein Science 15, 1653–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LG and Andricopulo AD (2016) Drug repositioning approaches to parasitic diseases: a medicinal chemistry perspective. Drug Discovery Today 21, 1699–1710. [DOI] [PubMed] [Google Scholar]
- Field MC, Horn D, Fairlamb AH, Ferguson MAJ, Gray DW, Read KD, De Rycker M, Torrie LS, Wyatt PG, Wyllie S and Gilbert IH (2017) Anti-trypanosomatid drug discovery: an ongoing challenge and a continuing need. Nature Reviews Microbiology 15, 447. [DOI] [PubMed] [Google Scholar]
- Fiser A (2010) Template-based protein structure modeling. Methods in Molecular Biology 673, 73–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS and Olson AJ (2016) Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nature Protocols 11, 905–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradera X and Babaoglu K (2017) Overview of methods and strategies for conducting virtual small molecule screening. Current Protocols in Chemical Biology 9, 196–212. [DOI] [PubMed] [Google Scholar]
- Francisco AF, Jayawardhana S, Lewis MD, White KL, Shackleford DM, Chen G, Saunders J, Osuna-Cabello M, Read KD, Charman SA, Chatelain E and Kelly JM (2016) Nitroheterocyclic drugs cure experimental Trypanosoma cruzi infections more effectively in the chronic stage than in the acute stage. Scientific Reports 6, 35351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC and Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- Friggeri L, Hargrove TY, Rachakonda G, Blobaum AL, Fisher P, de Oliveira GM, da Silva CF, Soeiro MNC, Nes WD, Lindsley CW, Villalta F, Guengerich FP and Lepesheva GI (2018) Sterol 14alpha-demethylase structure-based optimization of drug candidates for human infections with the protozoan trypanosomatidae. Journal of Medicinal Chemistry 61, 10910–10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fueller F, Jehle B, Putzker K, Lewis JD and Krauth-Siegel RL (2012) High throughput screening against the peroxidase cascade of African trypanosomes identifies antiparasitic compounds that inactivate tryparedoxin. Journal of Biological Chemistry 287, 8792–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard T (2018) Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark. Journal of Chemical Information and Modeling 58, 1697–1706. [DOI] [PubMed] [Google Scholar]
- Gangwar S, Baig MS, Shah P, Biswas S, Batra S, Siddiqi MI and Goyal N (2012) Identification of novel inhibitors of dipeptidylcarboxypeptidase of Leishmania donovani via ligand-based virtual screening and biological evaluation. Chemical Biology & Drug Design 79, 149–156. [DOI] [PubMed] [Google Scholar]
- Gasteiger J (2015) Cheminformatics: computing target complexity. Nature Chemistry 7, 619–620. [DOI] [PubMed] [Google Scholar]
- Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, Davies M, Dedman N, Karlsson A, Magarinos MP, Overington JP, Papadatos G, Smit I and Leach AR (2017) The ChEMBL database in 2017. Nucleic Acids Research 45, D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb MH, Van Voorhis WC, Buckner FS, Yokoyama K, Eastman R, Carpenter EP, Panethymitaki C, Brown KA and Smith DF (2003) Protein farnesyl and N-myristoyl transferases: piggy-back medicinal chemistry targets for the development of antitrypanosomatid and antimalarial therapeutics. Molecular and Biochemical Parasitology 126, 155–163. [DOI] [PubMed] [Google Scholar]
- Ghemtio L, Perez-Nueno VI, Leroux V, Asses Y, Souchet M, Mavridis L, Maigret B and Ritchie DW (2012) Recent trends and applications in 3D virtual screening. Combinatorial Chemistry & High Throughput Screening 15, 749–769. [DOI] [PubMed] [Google Scholar]
- Ghofrani HA, Osterloh IH and Grimminger F (2006) Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nature Reviews. Drug Discovery 5, 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert IH (2013) Drug discovery for neglected diseases: molecular target-based and phenotypic approaches. Journal of Medicinal Chemistry 56, 7719–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert IH (2014) Target-based drug discovery for human African trypanosomiasis: selection of molecular target and chemical matter. Parasitology 141, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno A, Ojeda-Montes MJ, Tomas-Hernandez S, Cereto-Massague A, Beltran-Debon R, Mulero M, Pujadas G and Garcia-Vallve S (2019) The light and dark sides of virtual screening: what Is there to know? International Journal of Molecular Sciences 20, E1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Chavez Z, Olin-Sandoval V, Rodiguez-Zavala JS, Moreno-Sanchez R and Saavedra E (2015) Metabolic control analysis of the Trypanosoma cruzi peroxide detoxification pathway identifies tryparedoxin as a suitable drug target. Biochimica et Biophysica Acta 1850, 263–273. [DOI] [PubMed] [Google Scholar]
- Graul AI, Pina P, Cruces E and Stringer M (2017) The year's new drugs & biologics 2016: part I. Drugs Today (Barc) 53, 27–74. [DOI] [PubMed] [Google Scholar]
- Graul AI, Pina P and Stringer M (2018) The year's new drugs and biologics 2017: part I. Drugs Today (Barc) 54, 35–84. [DOI] [PubMed] [Google Scholar]
- Graul AI, Pina P, Cruces E and Stringer M (2019) The year's new drugs and biologics 2018: part I. Drugs Today (Barc) 55, 35–87. [DOI] [PubMed] [Google Scholar]
- Grosdidier A, Zoete V and Michielin O (2011) Swissdock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Research 39, W270–W277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SC, Sung B, Prasad S, Webb LJ and Aggarwal BB (2013) Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends in Pharmacological Sciences 34, 508–517. [DOI] [PubMed] [Google Scholar]
- Haga JH, Ichikawa K and Date S (2016) Virtual screening techniques and current computational infrastructures. Current Pharmaceutical Design 22, 3576–3584. [DOI] [PubMed] [Google Scholar]
- Hall BS, Bot C and Wilkinson SR (2011) Nifurtimox activation by trypanosomal type I nitroreductases generates cytotoxic nitrile metabolites. Journal of Biological Chemistry 286, 13088–13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigua-Souiai E, Abdelkrim YZ, Bassoumi-Jamoussi I, Zakraoui O, Bouvier G, Essafi-Benkhadir K, Banroques J, Desdouits N, Munier-Lehmann H, Barhoumi M, Tanner NK, Nilges M, Blondel A and Guizani I (2018) Identification of novel leishmanicidal molecules by virtual and biochemical screenings targeting Leishmania eukaryotic translation initiation factor 4A. PLoS Neglected Tropical Diseases 12, e0006160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Mayorga V, Lara-Ramirez EE, Chacon-Vargas KF, Aguirre-Alvarado C, Rodriguez-Paez L, Alcantara-Farfan V, Cordero-Martinez J, Nogueda-Torres B, Reyes-Espinosa F, Bocanegra-Garcia V and Rivera G (2019) Structure-based virtual screening and in vitro evaluation of new Trypanosoma cruzi cruzain inhibitors. International Journal of Molecular Sciences 20, E1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann FC, Lenz M, Jose J, Kaiser M, Brun R and Schmidt TJ (2015) In silico identification and in vitro activity of novel natural inhibitors of Trypanosoma brucei glyceraldehyde-3-phosphate-dehydrogenase. Molecules 20, 16154–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth TD (2018) Counting down the 2020 goals for 9 neglected tropical diseases: what have We learned from quantitative analysis and transmission modeling? Clinical Infectious Diseases 66, S237–S244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberg JJ, Bruggeman FJ, Bakker BM and Westerhoff HV (2007) Metabolic control analysis to identify optimal drug targets. Progress in Drug Research 64, 173–189. [DOI] [PubMed] [Google Scholar]
- Hu B and Lill MA (2014) Pharmdock: a pharmacophore-based docking program. Journal of Cheminformatics 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JP, Rees S, Kalindjian SB and Philpott KL (2011) Principles of early drug discovery. British Journal of Pharmacology 162, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A and Schulten K (1996) VMD: visual molecular dynamics. Journal of Molecular Graphics 14, 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- Hutton JA, Goncalves V, Brannigan JA, Paape D, Wright MH, Waugh TM, Roberts SM, Bell AS, Wilkinson AJ, Smith DF, Leatherbarrow RJ and Tate EW (2014) Structure-based design of potent and selective Leishmania N-myristoyltransferase inhibitors. Journal of Medicinal Chemistry 57, 8664–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AN (2009) Effects of protein conformation in docking: improved pose prediction through protein pocket adaptation. Journal of Computer-Aided Molecular Design 23, 355–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K and Jain NK (2013) Novel therapeutic strategies for treatment of visceral leishmaniasis. Drug Discovery Today 18, 1272–1281. [DOI] [PubMed] [Google Scholar]
- James Abraham TM, Schulz R, Pall S, Smith J, Hess B and Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. [Google Scholar]
- Jhingran A, Chawla B, Saxena S, Barrett MP and Madhubala R (2009) Paromomycin: uptake and resistance in Leishmania donovani. Molecular and Biochemical Parasitology 164, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Maggiora GM and Meeting ACS (1990) Concepts and Applications of Molecular Similarity. New York City, USA: Wiley. [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR and Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. Journal of Molecular Biology 267, 727–748. [DOI] [PubMed] [Google Scholar]
- Jones NG, Catta-Preta CMC, Lima A and Mottram JC (2018) Genetically validated drug targets in Leishmania: current knowledge and future prospects. ACS Infectious Diseases 4, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansiime F, Adibaku S, Wamboga C, Idi F, Kato CD, Yamuah L, Vaillant M, Kioy D, Olliaro P and Matovu E (2018) A multicentre, randomised, non-inferiority clinical trial comparing a nifurtimox-eflornithine combination to standard eflornithine monotherapy for late stage Trypanosoma brucei gambiense human African trypanosomiasis in Uganda. Parasites and Vectors 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashif M, Hira SK, Upadhyaya A, Gupta U, Singh R, Paladhi A, Khan FI, Rub A and Manna PP (2019) In silico studies and evaluation of antiparasitic role of a novel pyruvate phosphate dikinase inhibitor in Leishmania donovani infected macrophages. International Journal of Antimicrobial Agents 53, 508–514. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y and Freire E (2011) Finding a better path to drug selectivity. Drug Discovery Today 16, 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley BP, Brown SP, Warren GL and Muchmore SW (2015) POSIT: flexible shape-guided docking for pose prediction. Journal of Chemical Information and Modeling 55, 1771–1780. [DOI] [PubMed] [Google Scholar]
- Khare P, Gupta AK, Gajula PK, Sunkari KY, Jaiswal AK, Das S, Bajpai P, Chakraborty TK, Dube A and Saxena AK (2012) Identification of novel S-adenosyl-L-homocysteine hydrolase inhibitors through homology-model-based virtual screening, synthesis, and biological evaluation. Journal of Chemical Information and Modeling 52, 777–791. [DOI] [PubMed] [Google Scholar]
- Khare S, Roach SL, Barnes SW, Hoepfner D, Walker JR, Chatterjee AK, Neitz RJ, Arkin MR, McNamara CW, Ballard J, Lai Y, Fu Y, Molteni V, Yeh V, McKerrow JH, Glynne RJ and Supek F (2015) Utilizing chemical genomics to identify cytochrome b as a novel drug target for Chagas disease. PLoS Pathogens 11, e1005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S, Nagle AS, Biggart A, Lai YH, Liang F, Davis LC, Barnes SW, Mathison CJN, Myburgh E, Gao M-y, Gillespie JR, Liu X, Tan JL, Stinson M, Rivera IC, Ballard J, Yeh V, Groessl T, Federe G, Koh HXY, Venable JD, Bursulaya B, Shapiro M, Mishra PK, Spraggon G, Brock A, Mottram JC and Buckner FS (2016) Proteasome inhibition for treatment of leishmaniasis, Chagas disease and sleeping sickness. Nature 537, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J and Bolton EE (2019) Pubchem 2019 update: improved access to chemical data. Nucleic Acids Research 47, D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Keller S, Li M, Smith A, Zheng S, Kaur G, Yang X, Wang B and Docampo R (2010) Chemical validation of phosphodiesterase C as a chemotherapeutic target in Trypanosoma cruzi, the etiological agent of Chagas’ disease. Antimicrobial Agents and Chemotherapy 54, 3738–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkle ME, Hargrove TY, Kleshchenko YY, von Kries JP, Ridenour W, Uddin MJ, Caprioli RM, Marnett LJ, Nes WD, Villalta F, Waterman MR and Lepesheva GI (2009) Indomethacin amides as a novel molecular scaffold for targeting Trypanosoma cruzi sterol 14alpha-demethylase. Journal of Medicinal Chemistry 52, 2846–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyianni M (2017) Docking and virtual screening in drug discovery. Methods in Molecular Biology 1647, 255–266. [DOI] [PubMed] [Google Scholar]
- Kraus JM, Tatipaka HB, McGuffin SA, Chennamaneni NK, Karimi M, Arif J, Verlinde CL, Buckner FS and Gelb MH (2010) Second generation analogues of the cancer drug clinical candidate tipifarnib for anti-Chagas disease drug discovery. Journal of Medicinal Chemistry 53, 3887–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauth-Siegel RL, Enders B, Henderson GB, Fairlamb AH and Schirmer RH (1987) Trypanothione reductase from Trypanosoma cruzi. Purification and characterization of the crystalline enzyme. European Journal of Biochemistry 164, 123–128. [DOI] [PubMed] [Google Scholar]
- Lamotte S, Aulner N, Spath GF and Prina E (2019) Discovery of novel hit compounds with broad activity against visceral and cutaneous Leishmania species by comparative phenotypic screening. Scientific Reports 9, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander N, Chiurillo MA and Docampo R (2016) Genome editing by CRISPR/Cas9: a game change in the genetic manipulation of protists. Journal of Eukaryotic Microbiology 63, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Ramirez EE, Lopez-Cedillo JC, Nogueda-Torres B, Kashif M, Garcia-Perez C, Bocanegra-Garcia V, Agusti R, Uhrig ML and Rivera G (2017) An in vitro and in vivo evaluation of new potential trans-sialidase inhibitors of Trypanosoma cruzi predicted by a computational drug repositioning method. European Journal of Medicinal Chemistry 132, 249–261. [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Ott RD, Hargrove TY, Kleshchenko YY, Schuster I, Nes WD, Hill GC, Villalta F and Waterman MR (2007) Sterol 14alpha-demethylase as a potential target for antitrypanosomal therapy: enzyme inhibition and parasite cell growth. Chemistry & Biology 14, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux AE and Krauth-Siegel RL (2016) Thiol redox biology of trypanosomatids and potential targets for chemotherapy. Molecular and Biochemical Parasitology 206, 67–74. [DOI] [PubMed] [Google Scholar]
- Lesnik S, Stular T, Brus B, Knez D, Gobec S, Janezic D and Konc J (2015) LiSiCA: a software for ligand-based virtual screening and its application for the discovery of butyrylcholinesterase inhibitors. Journal of Chemical Information and Modeling 55, 1521–1528. [DOI] [PubMed] [Google Scholar]
- Li AP (2001) Screening for human ADME/Tox drug properties in drug discovery. Drug Discovery Today 6, 357–366. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW and Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews 46, 3–26. [DOI] [PubMed] [Google Scholar]
- Llanos MA, Sbaraglini ML, Villalba ML, Ruiz MD, Carrillo C, Alba Soto C, Talevi A, Angeli A, Parkkila S, Supuran CT and Gavernet L (2020) A structure-based approach towards the identification of novel antichagasic compounds: Trypanosoma cruzi carbonic anhydrase inhibitors. Journal of Enzyme Inhibition and Medicinal Chemistry 35, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Rensi SE, Torng W and Altman RB (2018) Machine learning in chemoinformatics and drug discovery. Drug Discovery Today 23, 1538–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Vodnala SK, Gustavsson AL, Gustafsson TN, Sjoberg B, Johansson HA, Kumar S, Tjernberg A, Engman L, Rottenberg ME and Holmgren A (2013) Ebsulfur is a benzisothiazolone cytocidal inhibitor targeting the trypanothione reductase of Trypanosoma brucei. Journal of Biological Chemistry 288, 27456–27468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Li BZ, Liu D, Zhang L, Chen Y, Jia B, Zeng BX, Zhao H and Yuan YJ (2015) Engineered biosynthesis of natural products in heterologous hosts. Chemical Society Reviews 44, 5265–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XH, Jia J, Zhu F, Xue Y, Li ZR and Chen YZ (2009) Comparative analysis of machine learning methods in ligand-based virtual screening of large compound libraries. Combinatorial Chemistry & High Throughput Screening 12, 344–357. [DOI] [PubMed] [Google Scholar]
- Magarinos MP, Carmona SJ, Crowther GJ, Ralph SA, Roos DS, Shanmugam D, Van Voorhis WC and Aguero F (2012) TDR Targets: a chemogenomics resource for neglected diseases. Nucleic Acids Research 40, D1118–D1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluf FV, Andricopulo AD, Oliva G and Guido RV (2013) A pharmacophore-based virtual screening approach for the discovery of Trypanosoma cruzi GAPDH inhibitors. Future Medicinal Chemistry 5, 2019–2035. [DOI] [PubMed] [Google Scholar]
- Mansuri R, Kumar A, Rana S, Panthi B, Ansari MY, Das S, Dikhit MR, Sahoo GC and Das P (2017) In vitro evaluation of antileishmanial activity of computationally screened compounds against ascorbate peroxidase to combat amphotericin B drug resistance. Antimicrobial Agents and Chemotherapy 61, e02429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese L, Nascimento JF, Damasceno FS, Bringaud F, Michels PAM and Silber AM (2018) The uptake and metabolism of amino acids, and their unique role in the biology of pathogenic trypanosomatids. Pathogens (Basel, Switzerland) 7, E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavridis L, Hudson BD and Ritchie DW (2007) Toward high throughput 3D virtual screening using spherical harmonic surface representations. Journal of Chemical Information and Modeling 47, 1787–1796. [DOI] [PubMed] [Google Scholar]
- Maxfield L and Crane JS (2019) Leishmaniasis. In StatPearls. Florida, USA: StatPearls Publishing, pp. 1–9. [Google Scholar]
- Meiering S, Inhoff O, Mies J, Vincek A, Garcia G, Kramer B, Dormeyer M and Krauth-Siegel RL (2005) Inhibitors of Trypanosoma cruzi trypanothione reductase revealed by virtual screening and parallel synthesis. Journal of Medicinal Chemistry 48, 4793–4802. [DOI] [PubMed] [Google Scholar]
- Menna-Barreto RFS (2019) Cell death pathways in pathogenic trypanosomatids: lessons of (over)kill. Cell death & disease 10, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies SK, Tulloch LB, Florence GJ and Smith TK (2018) The trypanosome alternative oxidase: a potential drug target? Parasitology 145, 175–183. [DOI] [PubMed] [Google Scholar]
- Mesu V, Kalonji WM, Bardonneau C, Mordt OV, Blesson S, Simon F, Delhomme S, Bernhard S, Kuziena W, Lubaki JF, Vuvu SL, Ngima PN, Mbembo HM, Ilunga M, Bonama AK, Heradi JA, Solomo JLL, Mandula G, Badibabi LK, Dama FR, Lukula PK, Tete DN, Lumbala C, Scherrer B, Strub-Wourgaft N and Tarral A (2018) Oral fexinidazole for late-stage African Trypanosoma brucei gambiense trypanosomiasis: a pivotal multicentre, randomised, non-inferiority trial. Lancet (London, England) 391, 144–154. [DOI] [PubMed] [Google Scholar]
- Meyerhoff A (1999) U.S. Food and Drug Administration approval of AmBisome (liposomal amphotericin B) for treatment of visceral leishmaniasis. Clinical infectious Diseases 28, 42–48, discussion 49–51. [DOI] [PubMed] [Google Scholar]
- Meyskens FL, Kingsley EM, Glattke T, Loescher L and Booth A (1986) A phase II study of alpha-difluoromethylornithine (DFMO) for the treatment of metastatic melanoma. Investigational New Drugs 4, 257–262. [DOI] [PubMed] [Google Scholar]
- Mishra AK, Singh N, Agnihotri P, Mishra S, Singh SP, Kolli BK, Chang KP, Sahasrabuddhe AA, Siddiqi MI and Pratap JV (2017) Discovery of novel inhibitors for Leishmania nucleoside diphosphatase kinase (NDK) based on its structural and functional characterization. Journal of Computer-Aided Molecular Design 31, 547–562. [DOI] [PubMed] [Google Scholar]
- Mogk S, Bosselmann CM, Mudogo CN, Stein J, Wolburg H and Duszenko M (2017) African trypanosomes and brain infection – the unsolved question. Biological Reviews of the Cambridge Philosophical Society 92, 1675–1687. [DOI] [PubMed] [Google Scholar]
- Morillo CA, Waskin H, Sosa-Estani S, Del Carmen Bangher M, Cuneo C, Milesi R, Mallagray M, Apt W, Beloscar J, Gascon J, Molina I, Echeverria LE, Colombo H, Perez-Molina JA, Wyss F, Meeks B, Bonilla LR, Gao P, Wei B, McCarthy M and Yusuf S (2017) Benznidazole and posaconazole in eliminating parasites in asymptomatic T. Cruzi carriers: the STOP-CHAGAS trial. Journal of the American College of Cardiology 69, 939–947. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS and Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. Journal of Computational Chemistry 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpamhanga CP, Spinks D, Tulloch LB, Shanks EJ, Robinson DA, Collie IT, Fairlamb AH, Wyatt PG, Frearson JA, Hunter WN, Gilbert IH and Brenk R (2009) One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment hits discovered by virtual screening. Journal of Medicinal Chemistry 52, 4454–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R and Madhubala R (1994) Effect of antioxidants on the growth and polyamine levels of Leishmania donovani. Biochemical Pharmacology 47, 611–615. [DOI] [PubMed] [Google Scholar]
- Novac N (2013) Challenges and opportunities of drug repositioning. Trends in Pharmacological Sciences 34, 267–272. [DOI] [PubMed] [Google Scholar]
- Novick PA, Ortiz OF, Poelman J, Abdulhay AY and Pande VS (2013) SWEETLEAD: an in silico database of approved drugs, regulated chemicals, and herbal isolates for computer-aided drug discovery. PLoS ONE 8, e79568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki MW, Tulloch LB, Worralll L, McNae IW, Hannaert V, Michels PA, Fothergill-Gilmore LA, Walkinshaw MD and Turner NJ (2008) Design, synthesis and trypanocidal activity of lead compounds based on inhibitors of parasite glycolysis. Bioorganic & Medicinal Chemistry 16, 5050–5061. [DOI] [PubMed] [Google Scholar]
- Nwaka S, Ramirez B, Brun R, Maes L, Douglas F and Ridley R (2009) Advancing drug innovation for neglected diseases – criteria for lead progression. PLoS Neglected Tropical Diseases 3, e440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa R, Watowich SJ, Florez A, Mesa CV, Robledo SM and Muskus C (2016) Drug search for leishmaniasis: a virtual screening approach by grid computing. Journal of Computer-Aided Molecular Design 30, 541–552. [DOI] [PubMed] [Google Scholar]
- Ochoa R, Rocha-Roa C, Marin-Villa M, Robledo SM and Varela MR (2018) Search of allosteric inhibitors and associated proteins of an AKT-like kinase from Trypanosoma cruzi. International Journal of Molecular Sciences 19, E3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa R, Garcia E, Robledo SM and Cardona GW (2019) Virtual and experimental screening of phenylfuranchalcones as potential anti-Leishmania candidates. Journal of Molecular Graphics & Modelling 91, 164–171. [DOI] [PubMed] [Google Scholar]
- Olin-Sandoval V, Gonzalez-Chavez Z, Berzunza-Cruz M, Martinez I, Jasso-Chavez R, Becker I, Espinoza B, Moreno-Sanchez R and Saavedra E (2012) Drug target validation of the trypanothione pathway enzymes through metabolic modelling. FEBS Journal 279, 1811–1833. [DOI] [PubMed] [Google Scholar]
- Oprea TI (2002) Virtual screening in lead discovery: a viewpoint. Molecules 7, 51–62. [Google Scholar]
- Orban OC, Korn RS, Benitez D, Medeiros A, Preu L, Loaec N, Meijer L, Koch O, Comini MA and Kunick C (2016) 5-Substituted 3-chlorokenpaullone derivatives are potent inhibitors of Trypanosoma brucei bloodstream forms. Bioorganic & Medicinal Chemistry 24, 3790–3800. [DOI] [PubMed] [Google Scholar]
- Ortega V, Giorgio S and de Paula E (2017) Liposomal formulations in the pharmacological treatment of leishmaniasis: a review. Journal of Liposome Research 27, 234–248. [DOI] [PubMed] [Google Scholar]
- Osorio-Mendez JF and Cevallos AM (2018) Discovery and genetic validation of chemotherapeutic targets for Chagas’ disease. Frontiers in cellular and infection microbiology 8, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palos I, Lara-Ramirez EE, Lopez-Cedillo JC, Garcia-Perez C, Kashif M, Bocanegra-Garcia V, Nogueda-Torres B and Rivera G (2017) Repositioning FDA drugs as potential cruzain inhibitors from Trypanosoma cruzi: virtual screening, in vitro and in vivo studies. Molecules 22, E1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran S, Saudagar P, Dubey VK and Patra S (2014) Discovery of novel anti-leishmanial agents targeting LdLip3 lipase. Journal of Molecular Graphics & Modelling 49, 68–79. [DOI] [PubMed] [Google Scholar]
- Perez-Molina JA and Molina I (2018) Chagas disease. Lancet (London, England) 391, 82–94. [DOI] [PubMed] [Google Scholar]
- Pieper U, Webb BM, Dong GQ, Schneidman-Duhovny D, Fan H, Kim SJ, Khuri N, Spill YG, Weinkam P, Hammel M, Tainer JA, Nilges M and Sali A (2014) Modbase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Research 42, D336–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BG, Hourai Y and Weng Z (2011) Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE 6, e24657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Martinez AK, Rodriguez-Duran J, Serrano-Martin X, Hernandez-Rodriguez V and Benaim G (2018) Mechanism of action of miltefosine on Leishmania donovani involves the impairment of acidocalcisome function and the activation of the sphingosine-dependent plasma membrane Ca(2+) channel. Antimicrobial Agents and Chemotherapy 62, e01614-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri MP and Campbell RK (2011) Target repurposing for neglected diseases. Future Medicinal Chemistry 3, 1307–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponte-Sucre A, Gamarro F, Dujardin JC, Barrett MP, Lopez-Velez R, Garcia-Hernandez R, Pountain AW, Mwenechanya R and Papadopoulou B (2017) Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS Neglected Tropical Diseases 11, e0006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JJ, Talevi A and Bruno-Blanch LE (2006) Application of linear discriminant analysis in the virtual screening of antichagasic drugs through trypanothione reductase inhibition. Molecular Diversity 10, 361–375. [DOI] [PubMed] [Google Scholar]
- Priotto G, Kasparian S, Mutombo W, Ngouama D, Ghorashian S, Arnold U, Ghabri S, Baudin E, Buard V, Kazadi-Kyanza S, Ilunga M, Mutangala W, Pohlig G, Schmid C, Karunakara U, Torreele E and Kande V (2009) Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet (London, England) 374, 56–64. [DOI] [PubMed] [Google Scholar]
- Prokopczyk IM, Ribeiro JF, Sartori GR, Sesti-Costa R, Silva JS, Freitas RF, Leitao A and Montanari CA (2014) Integration of methods in cheminformatics and biocalorimetry for the design of trypanosomatid enzyme inhibitors. Future Medicinal Chemistry 6, 17–33. [DOI] [PubMed] [Google Scholar]
- Raether W and Seidenath H (1983) The activity of fexinidazole (HOE 239) against experimental infections with Trypanosoma cruzi, trichomonads and Entamoeba histolytica. Annals of Tropical Medicine & Parasitology 77, 13–26. [DOI] [PubMed] [Google Scholar]
- Rarey M, Kramer B, Lengauer T and Klebe G (1996) A fast flexible docking method using an incremental construction algorithm. Journal of Molecular Biology 261, 470–489. [DOI] [PubMed] [Google Scholar]
- Rashmi M and Swati D (2015) In silico drug re-purposing against African sleeping sickness using GlcNAc-PI de-N-acetylase as an experimental target. Computational Biology and Chemistry 59, 87–94. [DOI] [PubMed] [Google Scholar]
- Rassi A Jr, Rassi A and Marin-Neto JA (2010) Chagas disease. Lancet (London, England), 375, 1388–1402. [DOI] [PubMed] [Google Scholar]
- Reigada C, Valera-Vera EA, Saye M, Errasti AE, Avila CC, Miranda MR and Pereira CA (2017) Trypanocidal effect of isotretinoin through the inhibition of polyamine and amino acid transporters in Trypanosoma cruzi. PLoS Neglected Tropical Diseases 11, e0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reigada C, Phanstiel Ot, Miranda MR and Pereira CA (2018) Targeting polyamine transport in Trypanosoma cruzi. European Journal of Medicinal Chemistry 147, 1–6. [DOI] [PubMed] [Google Scholar]
- Reigada C, Saye M, Phanstiel Ot, Valera-Vera E, Miranda MR and Pereira CA (2019) Identification of Trypanosoma cruzi polyamine transport inhibitors by computational drug repurposing. Frontiers in Medicine (Lausanne) 6, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rester U (2008) From virtuality to reality – virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Current Opinion in Drug Discovery & Development 11, 559–568. [PubMed] [Google Scholar]
- Roberts CW, McLeod R, Rice DW, Ginger M, Chance ML and Goad LJ (2003) Fatty acid and sterol metabolism: potential antimicrobial targets in apicomplexan and trypanosomatid parasitic protozoa. Molecular and Biochemical Parasitology 126, 129–142. [DOI] [PubMed] [Google Scholar]
- Rodriguez D, Chakraborty S, Warnick E, Crane S, Gao ZG, O'Connor R, Jacobson KA and Carlsson J (2016) Structure-based screening of uncharted chemical space for atypical adenosine receptor agonists. ACS Chemical Biology 11, 2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KE, Keranen H, Durrant JD, Ratnam J, Doak A, Arkin MR and McCammon JA (2012) Novel cruzain inhibitors for the treatment of Chagas’ disease. Chemical Biology & Drug Design 80, 398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruda GF, Campbell G, Alibu VP, Barrett MP, Brenk R and Gilbert IH (2010) Virtual fragment screening for novel inhibitors of 6-phosphogluconate dehydrogenase. Bioorganic & Medicinal Chemistry 18, 5056–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana D, Zhu C, Zhang M, Gudivada RC, Yang L and Jegga AG (2011) Drug repositioning for orphan diseases. Briefings in Bioinformatics 12, 346–356. [DOI] [PubMed] [Google Scholar]
- Saye M, Gauna L, Valera-Vera E, Reigada C, Miranda MR and Pereira CA (2020) Crystal violet structural analogues identified by in silico drug repositioning present anti-Trypanosoma cruzi activity through inhibition of proline transporter TcAAAP069. PLoS Neglected Tropical Diseases 14, e0007481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierz AC (2009) Virtual screening of bioassay data. Journal of Cheminformatics 1, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke P, Le Guilloux V, Maupetit J and Tuffery P (2010) fpocket: online tools for protein ensemble pocket detection and tracking. Nucleic Acids Research 38, W582–W589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G (2010) Virtual screening: an endless staircase? Nature Reviews. Drug Discovery 9, 273–276. [DOI] [PubMed] [Google Scholar]
- Scior T, Bender A, Tresadern G, Medina-Franco JL, Martinez-Mayorga K, Langer T, Cuanalo-Contreras K and Agrafiotis DK (2012) Recognizing pitfalls in virtual screening: a critical review. Journal of Chemical Information and Modeling 52, 867–881. [DOI] [PubMed] [Google Scholar]
- Sharlow E, Golden JE, Dodson H, Morris M, Hesser M, Lyda T, Leimgruber S, Schroeder CE, Flaherty DP, Weiner WS, Simpson D, Lazo JS, Aube J and Morris JC (2010a) Identification of Inhibitors of Trypanosoma brucei Hexokinases. In Probe Reports from the NIH Molecular Libraries Program. Bethesda, USA: National Center for Biotechnology Information, pp. 1–44. [PubMed] [Google Scholar]
- Sharlow ER, Lyda TA, Dodson HC, Mustata G, Morris MT, Leimgruber SS, Lee KH, Kashiwada Y, Close D, Lazo JS and Morris JC (2010b) A target-based high throughput screen yields Trypanosoma brucei hexokinase small molecule inhibitors with antiparasitic activity. PLoS Neglected Tropical Diseases 4, e659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MY and Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Science 15, 2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindermann H, Croft SL, Engel KR, Bommer W, Eibl HJ, Unger C and Engel J (2004) Miltefosine (Impavido): the first oral treatment against leishmaniasis. Medical Microbiology and Immunology 193, 173–180. [DOI] [PubMed] [Google Scholar]
- Singh J, Srivastava A, Jha P, Sinha KK and Kundu B (2015) L-Asparaginase as a new molecular target against leishmaniasis: insights into the mechanism of action and structure-based inhibitor design. Molecular BioSystems 11, 1887–1896. [DOI] [PubMed] [Google Scholar]
- Singhal S, Mehta J, Desikan R, Ayers D, Roberson P, Eddlemon P, Munshi N, Anaissie E, Wilson C, Dhodapkar M, Zeddis J and Barlogie B (1999) Antitumor activity of thalidomide in refractory multiple myeloma. New England Journal of Medicine 341, 1565–1571. [DOI] [PubMed] [Google Scholar]
- Smithson DC, Lee J, Shelat AA, Phillips MA and Guy RK (2010) Discovery of potent and selective inhibitors of Trypanosoma brucei ornithine decarboxylase. Journal of Biological Chemistry 285, 16771–16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MB, Silva CV, Bastos TM, Guimaraes ET, Figueira CP, Smirlis D and Azevedo WF Jr (2012). Anti-Trypanosoma cruzi activity of nicotinamide. Acta Tropica, 122, 224–229. [DOI] [PubMed] [Google Scholar]
- Soares Medeiros LC, South L, Peng D, Bustamante JM, Wang W, Bunkofske M, Perumal N, Sanchez-Valdez F and Tarleton RL (2017) Rapid, selection-free, high-efficiency genome editing in protozoan parasites using CRISPR-Cas9 ribonucleoproteins. MBio 8, e01788-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa EJ, Burguener G, Lanzarotti E, Defelipe L, Radusky L, Pardo AM, Marti M, Turjanski AG and Fernandez Do Porto D (2018) Target-Pathogen: a structural bioinformatic approach to prioritize drug targets in pathogens. Nucleic Acids Research 46, D413–D418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SF, Ribeiro AJ, Coimbra JT, Neves RP, Martins SA, Moorthy NS, Fernandes PA and Ramos MJ (2013) Protein-ligand docking in the new millennium--a retrospective of 10 years in the field. Current Medicinal Chemistry 20, 2296–2314. [DOI] [PubMed] [Google Scholar]
- Spinks D, Ong HB, Mpamhanga CP, Shanks EJ, Robinson DA, Collie IT, Read KD, Frearson JA, Wyatt PG, Brenk R, Fairlamb AH and Gilbert IH (2011) Design, synthesis and biological evaluation of novel inhibitors of Trypanosoma brucei pteridine reductase 1. ChemMedChem 6, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling T and Irwin JJ (2015) ZINC 15 – ligand discovery for everyone. Journal of Chemical Information and Modeling 55, 2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanovic S, Perdih A, Sencanski M, Glisic S, Duarte M, Tomas AM, Sena FV, Sousa FM, Pereira MM and Solmajer T (2018) In silico discovery of a substituted 6-methoxy-quinalidine with leishmanicidal activity in Leishmania infantum. Molecules 23, E772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanovic S, Sencanski M, Danel M, Menendez C, Belguedj R, Bouraiou A, Nikolic K, Cojean S, Loiseau PM, Glisic S, Baltas M and Garcia-Sosa AT (2019) Synthesis, In silico, and In vitro evaluation of anti-leishmanial activity of oxadiazoles and indolizine containing compounds flagged against anti-targets. Molecules 24, E1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steverding D (2008) The history of African trypanosomiasis. Parasites & Vectors 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouch TR, Kenyon JR, Johnson SR, Chen XQ, Doweyko A and Li Y (2003) In silico ADME/Tox: why models fail. Journal of Computer-Aided Molecular Design 17, 83–92. [DOI] [PubMed] [Google Scholar]
- Stumpfe D and Bajorath J (2012) Exploring activity cliffs in medicinal chemistry. Journal of Medicinal Chemistry 55, 2932–2942. [DOI] [PubMed] [Google Scholar]
- Sun W, Sanderson PE and Zheng W (2016) Drug combination therapy increases successful drug repositioning. Drug Discovery Today 21, 1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevi A, Gavernet L and Bruno-Blanch LE (2009) Combined virtual screening strategies. Current Computer-Aided Drug Design 5, 23–37. [Google Scholar]
- Tiwari N, Tanwar N and Munde M (2018) Molecular insights into trypanothione reductase-inhibitor interaction: a structure-based review. Arch Pharm (Weinheim) 351, e1700373. [DOI] [PubMed] [Google Scholar]
- Todeschini R, Consonni V and Gramatica P (2009) Chemometrics in QSAR. In Brown S, Walczak B and Tauler R (eds), Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, Vol. 4. Amsterdam, Netherlands: Elsevier, pp. 129–172. [Google Scholar]
- Torrie LS, Wyllie S, Spinks D, Oza SL, Thompson S, Harrison JR, Gilbert IH, Wyatt PG, Fairlamb AH and Frearson JA (2009) Chemical validation of trypanothione synthetase: a potential drug target for human trypanosomiasis. Journal of Biological Chemistry 284, 36137–36145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Molecular Informatics 29, 476–488. [DOI] [PubMed] [Google Scholar]
- Trott O and Olson AJ (2010) Autodock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uran Landaburu L, Berenstein AJ, Videla S, Maru P, Shanmugam D, Chernomoretz A and Aguero F. (2019) TDR Targets 6: driving drug discovery for human pathogens through intensive chemogenomic data integration. Nucleic Acids Research 48, gkz999. doi: 10.1093/nar/gkz999 5611677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina JA (2015) Recent clinical trials for the etiological treatment of chronic chagas disease: advances, challenges and perspectives. Journal of Eukaryotic Microbiology 62, 149–156. [DOI] [PubMed] [Google Scholar]
- Vainio MJ, Puranen JS and Johnson MS (2009) ShaEP: molecular overlay based on shape and electrostatic potential. Journal of Chemical Information and Modeling 49, 492–502. [DOI] [PubMed] [Google Scholar]
- Valera-Vera EA, Saye M, Reigada C, Miranda MR and Pereira CA (2020) In silico repositioning of etidronate as a potential inhibitor of the Trypanosoma cruzi enolase. Journal of Molecular Graphics & Modelling 95, 107506. [DOI] [PubMed] [Google Scholar]
- van Griensven J and Diro E (2019) Visceral leishmaniasis: recent advances in diagnostics and treatment regimens. Infectious Disease Clinics of North America 33, 79–99. [DOI] [PubMed] [Google Scholar]
- Vazquez C, Mejia-Tlachi M, Gonzalez-Chavez Z, Silva A, Rodriguez-Zavala JS, Moreno-Sanchez R and Saavedra E (2017) Buthionine sulfoximine is a multitarget inhibitor of trypanothione synthesis in Trypanosoma cruzi. FEBS Letters 591, 3881–3894. [DOI] [PubMed] [Google Scholar]
- Vera V, Saye EA, Reigada M, Damasceno C, Silber FS, Miranda AM, and Pereira MR and A C (2016) Resveratrol inhibits Trypanosoma cruzi arginine kinase and exerts a trypanocidal activity. International Journal of Biological Macromolecules 87, 498–503. [DOI] [PubMed] [Google Scholar]
- Villalta F, Dobish MC, Nde PN, Kleshchenko YY, Hargrove TY, Johnson CA, Waterman MR, Johnston JN and Lepesheva GI (2013) VNI Cures acute and chronic experimental Chagas disease. Journal of Infectious Diseases 208, 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walvekar P, Gannimani R and Govender T (2019) Combination drug therapy via nanocarriers against infectious diseases. European Journal of Pharmaceutical Sciences 127, 121–141. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R and Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B and Sali A (2016) Comparative protein structure modeling using MODELLER. Current Protocols in Protein Science 86, 2 9 1–2 9 37. [DOI] [PubMed] [Google Scholar]
- Wiggers HJ, Rocha JR, Fernandes WB, Sesti-Costa R, Carneiro ZA, Cheleski J, da Silva AB, Juliano L, Cezari MH, Silva JS, McKerrow JH and Montanari CA (2013) Non-peptidic cruzain inhibitors with trypanocidal activity discovered by virtual screening and in vitro assay. PLoS Neglected Tropical Diseases 7, e2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson SR and Kelly JM (2009) Trypanocidal drugs: mechanisms, resistance and new targets. Expert Reviews in Molecular Medicine 11, e31. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C and Wilson M (2018) Drugbank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Research 46, D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolber G and Langer T (2005) Ligandscout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. Journal of Chemical Information and Modeling 45, 160–169. [DOI] [PubMed] [Google Scholar]
- Wyatt PG, Gilbert IH, Read KD and Fairlamb AH (2011) Target validation: linking target and chemical properties to desired product profile. Current Topics in Medicinal Chemistry 11, 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie S, Patterson S, Stojanovski L, Simeons FR, Norval S, Kime R, Read KD and Fairlamb AH (2012) The anti-trypanosome drug fexinidazole shows potential for treating visceral leishmaniasis. Science Translational Medicine 4, 119re111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie S, Brand S, Thomas M, Rycker MD, Chung C-w, Pena I, Shishikura Y, Spinks D, Stojanovski L, Thomas J, Thompson S, Viayna E, Martin J, Gray DW, Miles TJ, Gilbert IH, Read KD, Marco M and Wyatt PG (2019) Preclinical candidate for the treatment of visceral leishmaniasis that acts through proteasome inhibition. Proceedings of the National Academy of Sciences 116, 9318–9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M and Lill MA (2013) Induced fit docking, and the use of QM/MM methods in docking. Drug Discovery Today. Technologies 10, e411–e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Li J, Xie H and Wang Y (2018) Review of drug repositioning approaches and resources. International Journal of Biological Sciences 14, 1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J and Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Research 43, W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wu X, Zhang J, Zhang X, Xu C, Liao S and Tu X (2017) Recognition of hyperacetylated N-terminus of H2AZ by TbBDF2 from Trypanosoma brucei. Biochemical Journal 474, 3817–3830. [DOI] [PubMed] [Google Scholar]
- Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. Journal of Computational Chemistry 32, 1466–1474. [DOI] [PubMed] [Google Scholar]
- Zhou S, Wang F, Hsieh TC, Wu JM and Wu E (2013) Thalidomide-a notorious sedative to a wonder anticancer drug. Current Medicinal Chemistry 20, 4102–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182020000207.
click here to view supplementary material