Abstract
Gastrointestinal helminth infection likely affects the gut microbiome, in turn affecting host health. To investigate the effect of intestinal parasite status on the gut microbiome, parasitic infection surveys were conducted in communities in Nan Province, Thailand. In total, 1047 participants submitted stool samples for intestinal parasite examination, and 391 parasite-positive cases were identified, equating to an infection prevalence of 37.3%. Intestinal protozoan species were less prevalent (4.6%) than helminth species. The most prevalent parasite was the minute intestinal fluke Haplorchis taichui (35.9%). Amplicon sequencing of 16S rRNA was conducted to investigate the gut microbiome profiles of H. taichui-infected participants compared with those of parasite-free participants. Prevotella copri was the dominant bacterial operational taxonomic unit (OTU) in the study population. The relative abundance of three bacterial taxa, Ruminococcus, Roseburia faecis and Veillonella parvula, was significantly increased in the H. taichui-infected group. Parasite-negative group had higher bacterial diversity (α diversity) than the H. taichui-positive group. In addition, a significant difference in bacterial community composition (β diversity) was found between the two groups. The results suggest that H. taichui infection impacts the gut microbiome profile by reducing bacterial diversity and altering bacterial community structure in the gastrointestinal tract.
Key words: Gut microbiome, Haplorchis taichui, helminth, minute intestinal fluke, Thailand
Introduction
Intestinal parasitic infections are a problem worldwide, particularly in underdeveloped, developing and even emerging countries. Soil-transmitted helminths (STHs), e.g. Ascaris lumbricoides, Trichuris trichiura, Strongyloides stercoralis and hookworms, are the most common intestinal parasites worldwide, followed by blood flukes (Schistosoma spp.) and filarial worms (e.g. Wuchereria and Brugia) (Hotez et al., 2008). In addition, food-borne zoonotic helminths have been known to affect populations globally (Pal et al., 2018). These include nematodes, which cause trichinellosis and gnathostomiasis; cestodes, which cause taeniasis, cysticercosis and diphyllobothriasis; and trematodes, which cause fascioliasis, paragonimiasis, opisthorchiasis and minute intestinal fluke (MIF) infection, e.g. haplorchiasis.
Although intestinal parasitic infections are considered low-impact diseases, they are also neglected diseases that can potentially cause problems in some vulnerable groups. For example, chronic STH infection causes malnutrition and impairs physical and cognitive development in children (Pabalan et al., 2017). Immunocompromised patients (e.g. those with HIV or cancer or those who have undergone organ transplantation) may suffer from chronic helminth infections as well as helminth-induced abnormalities in vital organs (Keiser and Nutman, 2004; Muehlenbachs et al., 2015). For example, long-term infection with the liver fluke Opisthorchis viverrini is associated with the development of a high mortality bile duct cancer known as cholangiocarcinoma (Sripa et al., 2007). In terms of global burden, over a quarter of the world's population is estimated to be infected with parasitic helminths (Hotez et al., 2006; Mutapi, 2015), particularly populations living in rural areas and remote villages throughout tropical and subtropical regions, including Thailand.
The highest prevalence of parasitic infection was reported in the north-eastern, southern and northern regions of Thailand (Wongsaroj et al., 2014). Although the national prevalence of helminth infections has decreased in the last 10 years, sporadic infections still occur and remain an important health problem in rural communities. The occurrence of helminth infection varies among regions depending on environmental conditions and social behaviour. High average rainfall, high humidity and optimal temperature were found to be the main factors favouring STHs in the southern provinces of Thailand (Chaiyos et al., 2018; Punsawad et al., 2018). The main parasitic infections in northern and north-eastern parts of Thailand are food- and fish-borne parasitic diseases, e.g. taeniasis, cysticercosis, trichinellosis, opisthorchiasis and haplorchiasis, which is a result of a tradition of raw fish/meat consumption in these regions. Parasitological surveys in Nan Province (bordering Lao PDR) revealed that infection with MIFs, in this case Haplorchis spp., was more common than infection with the liver fluke O. viverrini (Watthanakulpanich et al., 2010; Wijit et al., 2013; Chaisiri et al., 2018). The eggs of these two fish-borne trematodes are very similar and practically impossible to differentiate based on morphology during fecal examination (Tantrawatpan et al., 2014). Laboratory diagnostic techniques (e.g. molecular identification) or worm expulsion experiments for morphological confirmation of the adult stages are required for accurate diagnosis of infection (Chai et al., 2010).
Apart from helminths, the gastrointestinal tract is also home to the gut microbiome, a complex community of microbes (including viruses, fungi, bacteria, archaea and protozoans) that contributes to host homoeostasis. Among those microbes, bacteria are the most abundant, with up to a thousand species present in the gut (Gilbert et al., 2018). In humans, the gut microbiome contributes to several aspects of health and physiological functions, including digestion, the production of nutrients, detoxification, protection against pathogens and regulation of the immune system (Wu and Wu, 2012; Jandhyala et al., 2015). Gut microbiome composition and diversity play a key role in maintaining and balancing host homoeostasis. Disruption of this equilibrium, known as dysbiosis, leading to an imbalance in gut microbial communities (e.g. a decrease in gut microbiome diversity), may result in shifts in gut microbial composition and consequently changes in metabolic activities and physiological functions (Round and Mazmanian, 2009; Kriss et al., 2018).
Both helminths and bacteria are known to independently and synergistically affect host nutrition, immunity and ultimately health (Glendinning et al., 2014). Over the past decade, evidence highlighting the effects of helminth infection on the diversity and composition of the host gut microbiome has grown. Most previous studies on the topic involved experiments in laboratory animals, such as cockroaches, rodents, rabbits, pigs, goats, horses and monkeys (Broadhurst et al., 2012; Wu et al., 2012; Cattadori et al., 2016; Itthitaetrakool et al., 2016; Li et al., 2016; Vicente et al., 2016; Su et al., 2018; Walshe et al., 2019) as well as observational studies in wild animal models, such as fish, birds, rodents and lemurs (Kreisinger et al., 2015; Weldon et al., 2015; Aivelo and Norberg, 2016; Newbold et al., 2017; Fu et al., 2019). However, the number of similar studies in humans remains limited (but see Cantacessi et al., 2014; Lee et al., 2014; Jenkins et al., 2018; Martin et al., 2019).
Here, we conducted a parasitological survey in eight villages of a subdistrict in Nan Province, Thailand. Screening for helminth infection and gut microbiome profiling in relation to intestinal parasite infection were performed by focusing on a MIF, H. taichui, as a dominant parasite in this area. The study was a joint collaboration between local communities, the local public health administration and researchers. The aims of this study were: (1) to determine the prevalence of intestinal parasitic infection among the eight villages of the subdistrict and (2) to analyse the gut microbiome profile response to MIF infection and compare the profiles of infected and non-infected individuals. This study provides up-to-date information on the status of parasitic infections in Nan Province to local communities and the local public health administration for further implementation of campaigns to raise public health awareness and promote good hygiene practices in rural communities. The results of H. taichui-associated gut microbiome analysis will contribute to a deeper understanding of the field. New insights into the effects of MIF infection on the host gut microbiome in areas where parasites are endemic will improve the diagnosis, treatment and control of parasitic infections. In addition, the present study provides the first information on gut microbiome profile in relation to intestinal helminth infection in Thailand.
Materials and methods
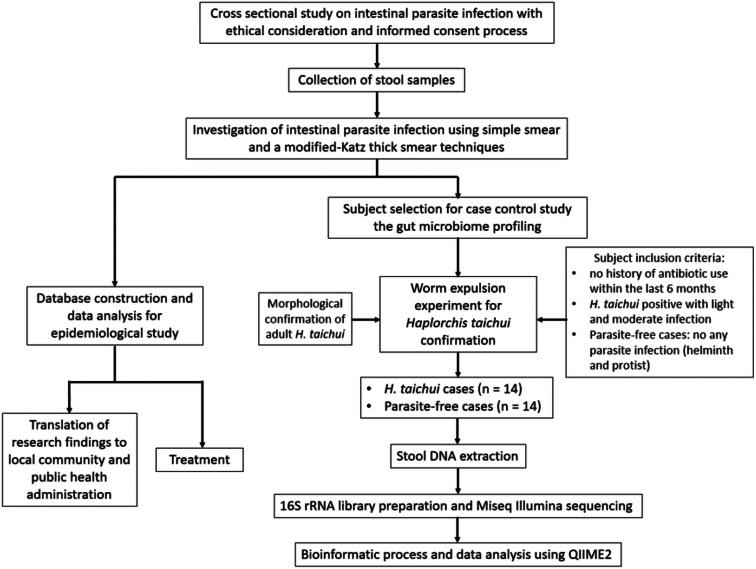
We conducted a cross-sectional study by collecting stool samples from volunteer villagers residing in rural communities in Nan Province, Thailand. Intestinal parasites were microscopically examined using screening and concentration techniques. Groups of helminth (H. taichui-positive) and parasite-free individuals were selected for subsequent profiling of the gut microbiome. An overview of the research workflow is presented in Fig. 1. Procedures concerning human samples, data collection and laboratory investigation were reviewed and approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (document no. MUTM 2018-035-01).
Fig. 1.
Schematic diagram showing research workflow.
Study site
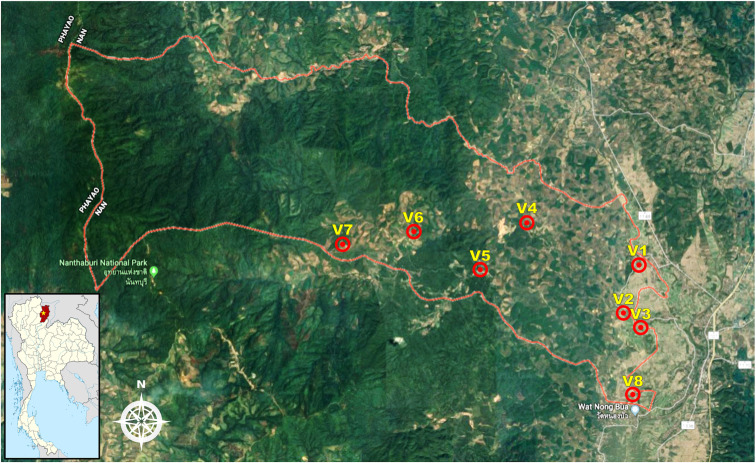
Saen Thong subdistrict (GPS coordinates: latitude 19.1330 and longitude 100.7680), Tha Wang Pha district, Nan Province was selected as the study site. There are eight villages altogether in this subdistrict: Baan Nanoon (Village 1), Baan Nasai (Village 2), Baan Pho (Village 3), Baan Huak (Village 4), Baan Namkrai (Village 5), Baan Huaymuang (Village 6; this village was investigated in a previous study, Chaisiri et al., 2018), Baan Santisuk (Village 7) and Baan Hae (Village 8). The total population size is around 3,840 inhabitants. The majority of the population is employed in the agricultural sector (70%), but some people are government officers (10%) and contractors (15%) and a few have other careers (5%). The landscape of this subdistrict is characterized by flatlands (lowlands) and mountainous areas (uplands). Two small rivers, the Rim and the Yao, run through the area from west to east, sometimes causing flooding in the flatlands, particularly during the wet season. The western part of the subdistrict is within the boundaries of Nantaburi National Park (a conserved forest), whereas the eastern part of the subdistrict is part of the urbanized community of Tha Wang Pha city. On the eastern side, the river Rim runs through the area before flowing into the river Nan in Tha Wang Pha city. Based on these geographical characteristics, the subdistrict can be roughly divided into two landscape types: (1) lowland agriculture in an urbanized setting (Village 1, 2, 3 and 8 in the eastern part of the subdistrict) and (2) upland agriculture in the forest (Village 4, 5, 6 and 7 in the western part of the subdistrict). For details of Saen Thong subdistrict, refer to the map in Fig. 2.
Fig. 2.
Map of Saen Thong subdistrict, Nan Province (Google Maps) showing the landscape of the study area and the distribution of the eight studied villages. Small panel (bottom left) shows Nan Province (red) on a map of Thailand with the location of Saen Thong subdistrict marked with a yellow star.
Stool sample collection
In August 2018, fieldwork was conducted with the help and participation of local communities. The study began with a meeting among investigators, local public health officers, primary health care units, the Subdistrict Administration Organisation, the head of subdistrict, all village chiefs and local health volunteers in order to design the plans for data and sample collection. A number of strategies to encourage the villagers to participate in the research project were created with the help of local public health officers and local health volunteers, such as making participation cost-free, including laboratory examination and treatment. Informed consent/assent and participant information processes were initiated thereafter through the use of community sound amplifiers to inform every household of the purposes and details of the research project.
Stool containers (plastic ziplock bags and spoons) were distributed to households by local health volunteers. Participants submitted stool samples (5–10 g) via local health volunteers, and the samples were subsequently delivered to mobile parasitological laboratory units at the primary health care units. Upon arrival, each sample was divided into two parts: one for parasite examination by experienced parasitologists using modified Katz thick smear and simple smear methods, and one for preservation in liquid nitrogen for gut microbiome profiling and further study.
Intestinal helminth investigation using a modified Katz thick smear and a simple smear for intestinal protozoa
The stool samples were first checked for intestinal parasitic infections using the direct smear technique to screen for helminths and protozoa. Saline wet mounts were prepared by mixing a small quantity of stool material in a drop of saline placed on a clean glass slide. A drop of Lugol's iodine was applied to increase the visibility of protozoan cysts.
The smears were then examined under a microscope. Concurrently, a concentration method, a modified Katz thick smear technique (Katz et al., 1972), was applied to identify and quantify helminth eggs or larvae present in stool samples. Approximately 39.2 mg of stool sample was used for each test. The slides were coated with an equal amount of stool samples, sieved through a wire-net filter and covered with a cellophane strip pre-soaked in Katz's solution (glycerine-malachite green solution). The prepared slides were incubated at room temperature for about 30 min to allow the stool contents to dry and clear, then the slides were microscopically examined. Focusing on MIF infection, the number of parasite egg per gram (EPG) of stool sample was calculated by multiplying the number of eggs per slide by 25.5, the constant factor suggested by the manufacturers of the test kit (Kato-Katz Faecal Examination Kit, Mahidol University, https://www.tm.mahidol.ac.th/helminth/?q=kato). The intensity of MIF infection was classified into three groups according to the criteria set out by Sato et al. (2009): light (1–2000 EPG), moderate (2001–6000 EPG) and heavy infection (>6000 EPG). Two slides were examined for each stool sample. Positive samples were confirmed by two parasitologists (slide readers) and the number of eggs or larvae present in each sample were counted.
The prevalence, mean abundance and mean intensity of intestinal parasite infections were estimated by using Quantitative Parasitology software version 3.0 (Rozsa et al., 2000). The χ2 test was used to determine whether the prevalence of intestinal parasite infection differed between male and female participants. A non-parametric Kruskal–Wallis test with post-hoc multiple pairwise comparisons (Holm–Bonferroni method) and the Mann–Whitney–Wilcoxon test were used to evaluate the effects of village, age group and gender on the level of H. taichui infection (EPG of stool). All statistical analyses were performed using R freeware (R Core Team, 2019).
Worm expulsion experiment for H. taichui confirmation
In total, 36 patients with moderate H. taichui infection participated in the worm expulsion experiment. The patients were informed of their results from the smear experiments and prepared for the subsequent processes. The patients were treated with praziquantel (40 mg kg−1) and saturated magnesium sulphate solution was given as a purgative 2 h later. Whole stool samples from each patient were collected. Stool samples were washed in tap water three times, following a simple sedimentation method, and examined for the presence of parasitic worms. Expelled MIFs were collected in 70% ethanol. Five MIF specimens from each patient were selected for slide preparation and species identification. The worms were washed in normal saline solution, stained in Semichon's acid carmine and decolourized in acid alcohol solution (1% acidulated 70% ethanol). Then, the specimens were subjected to a dehydration process through a graded series of ethanol concentrations (70, 80, 90, 95 and 100%). Finally, the specimens were neutralized in a mixture of 100% ethanol and xylene (1:1 ratio), cleared in xylene and mounted on glass slides with Permount solution.
Preparation of genomic DNA from selected stool samples
There were 28 stool samples altogether (14 H. taichui-positive and 14 parasite-free samples) selected for gut microbiome study (specimen selection criteria in Fig. 1). Genomic DNA was extracted from the frozen stool specimens together with two negative control samples (no DNA template) using the QIAamp® PowerFaecal® DNA Kit (QIAGEN, Thailand). Stool material (around 0.25 g per sample) was homogenized using a bead beating method, and the rest of the procedure was performed according to the manufacturers' instructions.
16S. rDNA library preparation and next-generation sequencing using MiSeq Illumina for gut microbiome investigation
The V3–V4 hypervariable region of the 16S rRNA gene (approx. 460 bp) was amplified with two rounds of nested PCR following the 16S rRNA dual-index nested PCR protocol for MiSeq Illumina sequencing (Caporaso et al., 2011; Illumina, 2013). The first round of PCR was performed using the following primers (Illumina overhang adapter attached): 16SForward (5′)-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-(3′) and 16SReverse (5′)-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-(3′). PCR was conducted in a total reaction volume of 25 μL, containing 8.7 μL of nuclease-free water, 12.8 μL of 2X KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA, USA), 0.5 μL of each primer (1 μm final concentration) and 2.5 μL of DNA template (5 ng μL−1 concentration). The PCR thermal cycler was set to the following programme: initial denaturation at 98 °C for 3 min, followed by 25 cycles of 98 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, then a final extension step at 72 °C for 1 min. After the first round of PCR, the amplicons were cleaned up using a QIAquick PCR Purification Kit (Qiagen, Thailand). A second round of PCR was performed to attach the barcode indices according to the Nextera XT DNA protocol (Illumina, 2013). This step ideally tags DNA products for later bioinformatics processes to track and bin the amplicon sequences to their associated samples. Primers with barcode indices used in this step followed the in-house materials from OMICs Science & Bioinformatics Centre (Chulalongkorn University, Thailand). The PCR components in the second round of PCR were similar to those in the first round. The PCR thermal cycler was set to the following programme: 98 °C for 2 min, followed by eight cycles of 98 °C for 20 s, 60 °C for 20 s and 72 °C for 30 s and a final extension step at 72 °C for 1 min. All indexed amplicons were purified using AMPure XP beads (Beckman Coulter, USA) and separated by agarose gel electrophoresis to observe the amplified PCR amplicon products. All samples were pooled in equimolar amounts (4 nm) and subjected to paired-end sequencing (2 × 250 bp) on an Illumina MiSeq platform (Illumina, USA) according to the standard protocol of the OMICs Sciences & Bioinformatics Centre (Chulalongkorn University, Thailand).
Post-sequencing bioinformatics analyses of gut microbiome and statistical analyses
Analyses of the 16S rRNA microbiome profiles were performed using the Quantitative Insights Into Microbial Ecology 2 platform (QIIME2, https://qiime2.org/, Bolyen et al., 2019). Firstly, individual nucleotide sequences (reads in .fastq format) generated by MiSeq Illumina sequencing were trimmed with Cutadapt to remove the Illumina overhang adapter (Martin, 2011). Sequence data were then transformed to QIIME2 zipped artefact format (.qza). For read quality filtering, the sequences were subjected for demultiplexing and denoising processes (i.e. checking sequence quality, filtering chimeric sequences, assigning reads into operational taxonomic units or ‘OTUs’, sequence alignment for taxonomy assignment and construction of the feature table) using the DADA2 plugin (Callahan et al., 2016). For taxonomic assignment, reads were binned at 99% similarity against SILVA-132-99-nb-classifier (Quast et al., 2013). Finally, the feature table (or OTU table) with the assigned bacterial taxonomy was exported from QIIME2. Singleton OTUs were removed prior to downstream analyses.
To analyse the diversity of the gut microbiomes of participants, α diversity was assessed via calculation of observed OTU richness and Shannon's diversity index using a rarefied dataset (10 000 reads). Statistical testing of the Shannon's diversity index between H. taichui-positive and parasite-free groups was performed using the Mann–Whitney–Wilcoxon test. The α rarefaction plots were also created for comparison of bacterial diversity between groups. To assess differences in bacterial communities, β diversity analysis was conducted using a phylogenetic method (i.e. using weighted and unweighted UniFrac distances). Principal coordinates analysis (PCoA) was performed to visualize clustering patterns in the bacterial compositions of the samples. Non-parametric analysis of similarity (ANOSIM) with 1000 permutations was used to test the statistical significance of the clustering patterns. In addition, the differential abundance of microbiome taxa was analysed with linear discriminant analysis effect size (LEfSe) (Segata et al., 2011) via the algorithm module on the Galaxy platform (http://huttenhower.sph.harvard.edu/galaxy/). LEfSe was used to identify key microbiome taxa OTUs that differentiated in abundance between the H. taichui-positive and parasite-free groups by taking into account statistical significance with biological consistency and effect size estimation.
Results
Demographic information of participants
Altogether, 1047 participants (27.2% of the overall population registered in the subdistrict) signed the research consent/assent form and submitted their stool samples for intestinal parasite investigation. Among them, 495 participants were female (47.3%) and 552 were male (52.7%), so the participant group was balanced in terms of gender ratio. Adults (30–59 years old) were dominant in this cross-sectional study, with 584 participants in this category (55.6% of total participants), followed by the elderly (>60 years old; 381 participants; 36.4% of total participants), children (0–14 years old; 33 participants; 3.2% of total participants) and young adults (15–29 years old; 32 participants; 3.2% of total participants). In 17 cases (1.6%), information on participant age was not recorded. These samples were excluded from the age-related analysis.
Intestinal parasite infections
Intestinal parasite investigation via simple smear and modified Katz techniques revealed 12 species/taxa of intestinal parasites (seven helminths and five protozoans). Seven types of helminth egg and one type of larva were found, including a trematode (Opisthorchis-like eggs, finally identified as Haplorchis taichui), a cestode (Taenia eggs) and five nematodes (eggs of A. lumbricoides, Enterobius vermicularis, hookworms and T. trichiura and S. stercoralis larvae). Cysts of five species of intestinal protozoans were identified, including Blastocystis hominis, Endolimax nana, Giardia duodenalis and Sarcocystis sp.; cysts like those of Entamoeba histolytica (E. histolytica-liked) were also found. Out of 1047 participants, 391 tested positive for intestinal helminths, equating to a 37.3% total prevalence of infection. The most prevalent helminth was the MIF H. taichui (35.9%), followed by Taenia sp. (1.4%), A. lumbricoides (0.8%), S. stercoralis (0.8%), T. trichiura (0.4%), hookworm (0.4%) and E. vermicularis (0.09%). The total prevalence of protozoan infection was low compared to the prevalence of helminth infection: 48 participants (4.6%) tested positive for B. hominis (2.8%), which was the most prevalent protozoa, followed by Sarcocystis sp. (0.9%), E. nana (0.4%) and G. duodenalis (0.4%). Entamoeba histolytica-liked cysts were least prevalent (0.09%). There were no obvious differences in the prevalence and patterns of helminth infection among the eight villages. However, there were some patterns in protozoan species occurrence. Endolimax nana was detected in only four participants residing in the upland villages (Village 4, 5 and 6), while 10 cases of Sarcocystis sp. infection were clustered in the lowland villages (Village 2, 3 and 8). More details of intestinal parasite infection prevalence and patterns are presented in Table 1.
Table 1.
Intestinal parasite infections in villagers residing in eight villages of Saen Thong subdistrict, Tha Wang Pha district, Nan Province, Thailand
| Intestinal parasites | Number of persons infected by intestinal parasite infections (% prevalence), [95% CI] | |||||||
|---|---|---|---|---|---|---|---|---|
| Villages in lowland area | Villages in upland area | |||||||
| Village 1 (n = 191) | Village 2 (n = 160) | Village 3 (n = 120) | Village 8 (n = 93) | Village 4 (n = 185) | Village 5 (n = 93) | Village 6 (n = 153) | Village 7 (n = 52) | |
| Helminth | ||||||||
| Haplorchis taichui | 106 (55.5), [48.1–62.3] | 40 (25.0), [18.7–32.4] | 34 (28.3), [20.7–37.0] | 37 (39.8), [30.1–50.0] | 20 (10.8), [6.9–16.1] | 20 (21.5), [14.0–31.1] | 89 (58.2), [50.0–65.8] | 30 (57.7), [43.8–70.3] |
| Taenia sp. | 2 (1.0), [0.2–3.8] | 3 (2.0), [0.5–5.5] | 3 (2.5), [0.7–7.3] | – | 1 (0.5), [0.03–3.1] | 2 (2.2), [0.4–7.4] | 3 (2.0), [0.5–5.7] | 1 (2.0), [0.1–10.2] |
| Ascaris lumbricoides | – | – | – | 7 (7.5), [35.9–14.9] | – | – | – | 2 (3.8), [0.7–13.2] |
| Trichuris trichiura | 1 (0.5), [0.03–3.0] | – | 1 (0.8), [0.05–4.5] | – | 1 (0.5), [0.03–3.1] | – | – | 1 (1.9), [0.1–10.2] |
| Hookworm | 1 (0.5), [0.03–3.0] | – | – | 1 (1.1), [0.06–5.7] | 1 (0.5), [0.03–3.0] | 1 (1.1), [0.06–5.7] | – | – |
| Enterobius vermicularis | – | – | 1 (0.8), [0.05–4.5] | – | – | – | – | – |
| Strongyloides stercoralis | 2 (1.0), [0.2–3.8] | – | – | – | 4 (2.2), [0.7–5.5] | 2 (2.2), [0.4–7.4] | 1 (0.7), [0.04–3.7] | – |
| Total (N = 1,047) | 106 (55.5), [48.1–62.3] | 43 (26.9), [20.5–34.3] | 35 (29.2), [21.6–37.9] | 38 (40.9), [31.1–51.1] | 25 (13.5), [9.1–19.4] | 24 (25.8), [17.6–35.9] | 90 (58.8), [50.6–66.4] | 30 (57.7), [43.8–70.3] |
| Protozoa | ||||||||
| Blastocystis hominis | 8 (4.2), [1.9–8.0] | 3 (2.0), [0.5–5.5] | 6 (5.0), [2.2–10.7] | – | 10 (5.4), [2.9–9.6] | 1 (1.1), [0.06–5.7] | 1 (0.7), [0.04–3.7] | – |
| Endolimax nana | – | – | – | – | 1 (0.5), [0.03–3.1] | 1 (1.1), [0.06–5.7] | 2 (1.3), [0.2–4.7] | – |
| Entamoeba histolytica cyst-liked | – | – | – | – | – | – | 1 (0.7), [0.04–3.7] | – |
| Giardia duodenalis | 1 (0.5), [0.03–3.0] | – | 1 (0.8), [0.05–4.5] | – | 1 (0.5), [0.03–3.1] | 1 (1.1), [0.06–5.7] | – | – |
| Sacrocystis sp. | – | 3 (2.0), [0.5–5.5] | 5 (4.2), [1.6–9.4] | 2 (2.2), [0.4–7.4] | – | – | – | – |
| Total (N = 1,047) | 9 (4.7), [2.4–8.8] | 6 (3.8), [1.6–8.0] | 12 (10.0), [5.7–16.9] | 2 (2.2), [0.4–7.4] | 12 (6.5), [3.7–11.0] | 3 (3.2), [0.9–8.9] | 4 (2.6), [0.9–6.4] | – |
N = total number of participants, n = number of participants from each village
Infections of H. taichui, the dominant human parasite in the study area
In the worm expulsion experiment with the 36 individuals who tested positive for Opisthorchis-liked eggs, all recovered trematodes appeared to be MIFs, with no Opisthorchis flukes found. We subsampled 75 of the recovered adult flukes for permanent slide preparation (Supplementary Fig. S1). All of these flukes were identified as H. taichui based on clear morphological evidence, i.e. the presence of a ventral sucker armed with around 13–17 fan-shaped spines (mode = 16; n = 35).
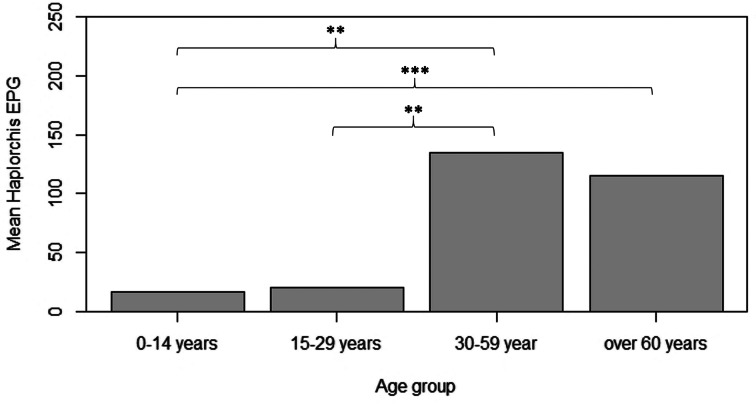
Haplorchis taichui was clearly the dominant species of intestinal parasite in the study area. This parasite was found in all villages, but the highest prevalence of infection was in Village 6 (58.2%), followed by Village 7 (57.7%), Village 1 (55.5%) and Village 8 (39.8%; Table 2). The intensity of H. taichui infection varied from light to moderate, with a few cases of heavy infection. The number of eggs in stool samples ranged from 26 to 7650 EPG of stool. There were no significant differences in the prevalence (χ2 = 0.461, P = 0.49) or EPG intensity (Mann–Whitney–Wilcoxon test = 127 745, P = 0.27) of H. taichui infection between male and female patients. Prevalence was highest in adults (38.2%) and the elderly (47.0%), then gradually decreased in young adults (18.8%) and children (6.1%). The number of H. taichui EPG was significantly higher in older people (adult and elderly groups) than in those in younger age groups (Kruskal–Wallis test = 20.75, d.f. = 3, P < 0.001; Fig. 3).
Table 2.
Prevalence (%), mean intensity (MI), mean abundance (MA) and range (eggs per gram of stool) of Haplorchis taichui infection among groups of different variables (village, gender and age group)
| Category | No. of infection (% prevalence) | MI [95% CI] | MA [95% CI] | Range |
|---|---|---|---|---|
| Village | ||||
| Village 1 (n = 191) | 106 (55.5) | 374.4 [275.8–554.6] | 207.8 [145.9–304.3] | 26–4,896 |
| Village 2 (n = 160) | 40 (25.0) | 141.6 [89.3–300.4] | 35.4 [20.9–72.4] | 26–1,556 |
| Village 3 (n = 120) | 34 (28.3) | 191.4 [120.2–390.9] | 54.2 [31.7–110.8] | 26–1,811 |
| Village 8 (n = 93) | 37 (39.8) | 119.5 [71.9–222.9] | 47.5 [26.7–85.1] | 26–944 |
| Village 4 (n = 185) | 20 (10.8) | 414.7 [144.4–1,237.0] | 44.83 [14.5–148.1] | 26–4,692 |
| Village 5 (n = 93) | 20 (21.5) | 52.7 [34.9–93.5] | 11.3 [6.4–22.1] | 26–281 |
| Village 6 (n = 153) | 89 (58.2) | 570.9 [412.3–880.7] | 332.1 [232.2–512.7] | 26–7,650 |
| Village 7 (n = 52) | 30 (57.7) | 291.8 [164.3–562.9] | 168.4 [89.9–338.0] | 26–2,499 |
| Gender | ||||
| Female (n = 495) | 172 (34.7) | 295.7 [216.1–454.3] | 102.8 [72.9–159.2] | 26–7,650 |
| Male (n = 552) | 204 (36.9) | 364.52 [283.4–470.8] | 134.7 [103.0–178.3 | 26–4,896 |
| Age group | ||||
| 0–14 years old (n = 33) | 2 (6.1) | 268.0 [51.0–485.0] | 16.2 [0–75.0] | 51–485 |
| 15–29 years old (n = 32) | 6 (18.8) | 106.7 [34.3–298.0] | 20.0 [4.8–66.3] | 26–434 |
| 30–59 years old (n = 584) | 223 (38.2) | 378.3 [290.4–513.8] | 144.5 [110.7–196.3] | 26–7,650 |
| >60 years old (n = 381) | 141 (37.0) | 279.5 [210.4–407.7] | 103.4 [74.1–145.3] | 26–4,692 |
Numbers of samples examined (n) are indicated in parentheses.
Fig. 3.
Analysis of difference in mean H. taichui intensity (EPG) among different age groups with multiple pairwise comparison after Kruskal–Wallis with Bonferroni post-hoc test (**P < 0.01 and ***P < 0.001).
Gut microbiome analysis of H. taichui-infected and parasite-free groups
The 16S rRNA (V3–V4) of 28 stool samples (14 H. taichui-positive samples and 14 parasite-free samples, Table 3) was sequenced using the MiSeq Illumina platform. A total of 1 892 476 quality-filtered reads were retained after quality checking and trimming Illumina adaptors and barcodes. These reads were clustered into 3404 OTUs. The mean read frequency per sample was 67 588 (14 716–587 940) and the mean read per OTU was 728 (1–75 418). After OTU filtering (at >10 000 reads/OTU), 60 OTUs were retained and assigned to bacterial taxonomic ranks.
Table 3.
Personal attributes of the 14 H. taichui-positive and 14 parasite-negative participants of the gut microbiome profiling experiment
| Case | Gender | Age (years) | BMI | H. taichui intensity (EPG) | Mean age (years) | Mean BMI |
|---|---|---|---|---|---|---|
| H. taichui-positive | F | 47 | 19.33 | 1148 | 54.28 | 18.18 |
| F | 49 | 15.82 | 1020 | |||
| F | 52 | 20.00 | 281 | |||
| F | 67 | 11.94 | 1403 | |||
| F | 76 | 15.03 | 1275 | |||
| M | 36 | 22.00 | 3774 | |||
| M | 42 | 18.29 | 1913 | |||
| M | 50 | 18.00 | 4896 | |||
| M | 51 | 26.32 | 740 | |||
| M | 53 | 16.77 | 1352 | |||
| M | 53 | 20.86 | 842 | |||
| M | 54 | 18.35 | 1785 | |||
| M | 54 | 19.50 | 408 | |||
| M | 76 | 12.42 | 561 | |||
| Parasite-free | F | 66 | 18.75 | – | 49.21 | 19.01 |
| F | 59 | 14.12 | – | |||
| F | 55 | 20.88 | – | |||
| F | 60 | 17.20 | – | |||
| F | 32 | 26.67 | – | |||
| F | 56 | 17.33 | – | |||
| F | 60 | 21.60 | – | |||
| F | 59 | 15.82 | – | |||
| F | 15 | 15.92 | – | |||
| F | 51 | 20.31 | – | |||
| M | 16 | 25.00 | – | |||
| M | 42 | 19.06 | – | |||
| M | 58 | 16.13 | – | |||
| M | 60 | 17.32 | – |
The 14 patients in the positive group were exclusively infected with H. taichui; no other helminths or protozoa were found during fecal examination.
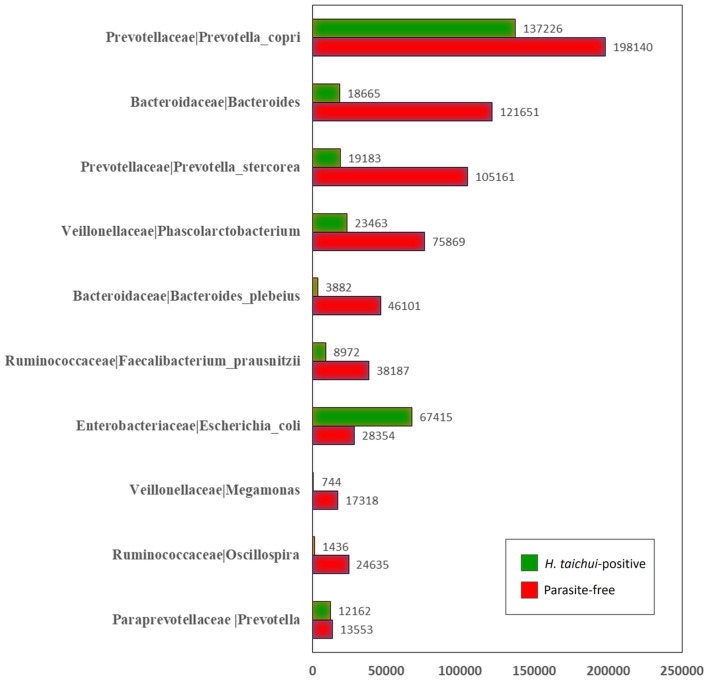
In all samples, the top 10 most abundant bacterial OTUs were as follows: Prevotella copri at 198 140 reads (96%, 27 out of 28 samples); Phascolarctobacterium at 75 869 reads (96%, 27 out of 28 samples); Faecalibacterium prausnitzii at 38 187 reads (96%, 27 out of 28 samples); Escherichia coli at 28 354 reads (96%, 27 out of 28 samples); Oscillospira at 24 635 reads (68%, 19 out of 28 samples); Prevotella stercorea at 105 161 reads (64%, 18 out of 28 samples); Bacteroides plebeius at 46 101 reads (50%, 14 out of 28 samples); Megamonas at 17 318 reads (46%, 13 out of 28 samples); Prevotella at 13 553 reads (43%, 12 out of 28 samples) and Bacteroides at 121 651 reads (29%, 8 out of 28 samples). Most of these OTUs had a higher read abundance in parasite-free samples compared to H. taichui-positive samples; E. coli was the only OTU in this list that was more abundant in H. taichui-positive samples (Fig. 4).
Fig. 4.
The top 10 most abundant bacterial OTUs found in the present case–control study comparing H. taichui-positive (green) and parasite-free groups (red). Number of bacterial OTU reads are given in bar charts.
Bacterial diversity and composition between H. taichui-infected and parasite-free groups
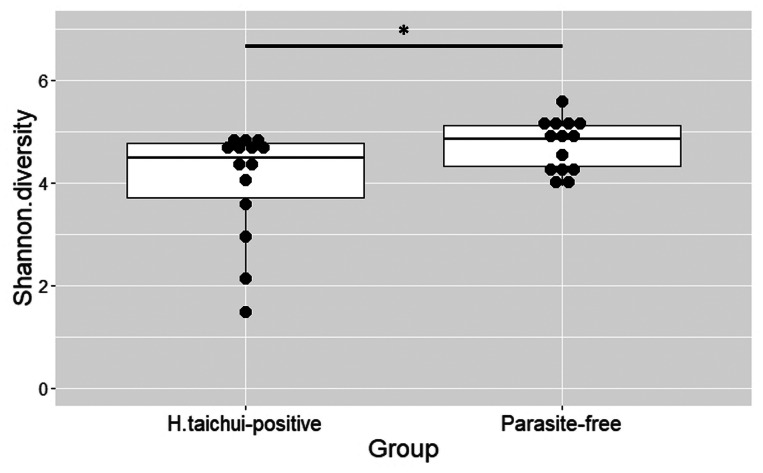
The α diversity of bacterial OTUs between H. taichui-infected and parasite-free groups was determined based on the observed OTU richness and estimation of the diversity index. The Shannon index values were significantly greater in the parasite-free group than in the H. taichui-positive group (Mann–Whitney–Wilcoxon test = 53, P = 0.039; Fig. 5). Rarefaction curves generated for the observed OTUs showed that the H. taichui-positive group had lower bacterial species richness than the parasite-free group (Fig. 6A). The rarefaction curves of the observed OTUs also showed a decreasing trend in bacterial species richness in the samples infected with higher worm EPG (Fig. 6B).
Fig. 5.
Analysis of difference in bacterial α diversity based on the Shannon diversity index between H. taichui-positive and parasite-free groups. *P < 0.05.
Fig. 6.
Analyses of gut microbial diversity and community composition between H. taichui-positive and parasite-free groups. (A) Rarefaction curves comparing bacterial α diversity (based on observed OTUs) between H. taichui-positive (light blue line) and parasite-free groups (dark blue line). (B) Rarefaction curves comparing bacterial α diversity (based on observed OTUs) of H. taichui-positive group varying in intensity (EPG) of infection. Light orange line, parasite-free; light blue line, EPG = 250–1299; dark orange line, EPG = 1300–2500; dark blue line, EPG > 2500. (C) Principal coordinates analysis plots (PCoA) created using unweighted UniFrac metric showing significant differences in clustering patterns (ANOSIM: R = 0.094, P = 0.03) of bacterial communities (β diversity) between H. taichui-positive (red) and parasite-free groups (blue).
The β diversity was analysed by PCoA with weighted and unweighted UniFrac distance matrixes. No significant difference was observed between H. taichui-infected and parasite-free groups using the weighted UniFrac matrix. However, a significant clustering pattern in bacterial composition was observed between the two groups using the unweighted UniFrac method (ANOSIM: R = 0.094, P = 0.03; Fig. 6C).
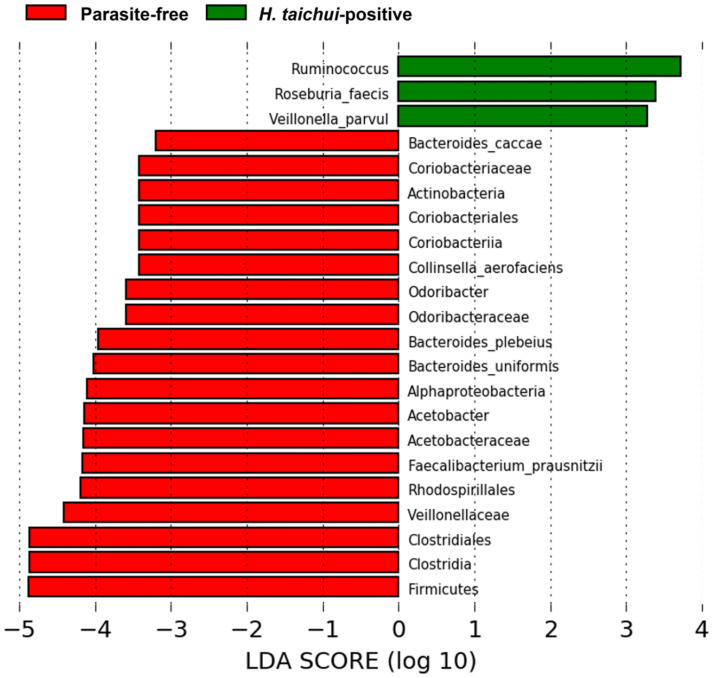
Differential abundance of gut microbial taxa according to LEfSe
Some bacterial taxa were found to be associated with either the H. taichui-infected group or the parasite-free group by linear discriminant analysis. The H. taichui-infected group harboured a higher abundance of certain bacterial taxa, such as Ruminococcus, Roseburia faecis and Veillonella parvula, in comparison to the parasite-free group. The parasite-free group showed an increased abundance of several bacterial taxa, such as Firmicutes, Clostridiales, Clostridia and Veillonellaceae (for more details, Fig. 7).
Fig. 7.
Results of LEfSe analysis showing differential abundance of bacterial OTUs between H. taichui-positive (green) and parasite-free groups (red).
Discussion
A high level of intestinal parasite infection was found in the eight villages of Saen Thong subdistrict, Tha Wang Pha district, Nan Province. The total prevalence of intestinal helminths was 37.3%, while the prevalence of protozoan infection was lower, at 4.6%. The dominant parasite in this region was the MIF H. taichui, with a prevalence of 35.9%. This parasite is known to be one of the causative agents of fish-borne helminthiases in Thailand.
Eggs of MIFs are morphologically similar to those of the liver fluke O. viverrini. Previous surveillance reports from northern Thailand have revealed a prevalence of opisthorchiasis of over 70% based on the detection of O. viverrini-liked eggs in stool samples (Waikagul et al., 2008; Wijit et al., 2013). However, after a previous worm expulsion experiment, only adult stages of Haplorchis spp. were found, suggesting that MIF infection is the dominant trematode infection in the area (Wijit et al., 2013). In the present study, cases of O. viverrini-liked infection were subjected to species confirmation by deworming and worm expulsion methods. All recovered parasites were morphologically identified as H. taichui, suggesting again that the most common fish-borne helminthiasis in northern Thailand, particularly in Nan Province, is caused by MIFs rather than opisthorchiasis.
There was no difference in H. taichui infection between male and female participants, although a higher prevalence and intensity of infection were observed in older people. The lack of differences between genders is probably explained by similar habits of raw fish consumption in males and females. Infection with MIFs is associated with the consumption of particular fish species that act as intermediate hosts for the parasite, including cyprinid fishes found in the local rivers in Nan Province, such as Barbodes schwanenfeldi, Mystacoleucus marginatus, Poropuntius deauratus, P. normani and Systomus orphoides (Boonmekam et al., 2016). Raw fish consumption is very popular in the area as part of the local traditional lifestyle. Together, this information suggests that the consumption of raw fish is an important risk factor for fish-borne helminthiases.
The highest prevalence of H. taichui infections was found in Village 6 (Ban Huay Muang). There is a small river (the Rim river) running along the southern part of this village, where villagers catch local fishes and other aquatic animals for food. Local people mentioned during personal interviews that fishes collected from this river were commonly sold or distributed to neighbours. This could potentially explain the high rate of fish-borne infection in this village. Interestingly, previous research by Chaisiri et al. (2018) reported a high level of fish-borne trematode infection (45.2%) in the same village in 2012. Chaisiri et al. (2018) also showed an immediate reduction in the fish-borne trematode infection level to 4.4% after deworming and a health education programme, which disseminated information about parasitic diseases through participatory activities, including a cooking contest campaign. Surprisingly, the prevalence of the fish-borne trematode observed in the present study in 2018 has increased to a higher level (58.2%) than it was 6 years ago, before the interventions. This suggests that the provision of health education must be improved to be effective in the long term.
A higher prevalence and intensity of H. taichui infection were found in adults and the elderly (ages 30–59 years old and >60 years old, respectively). After finishing their daily work in agricultural fields, villagers usually join in with small parties, preparing traditional dishes with raw fish meat, e.g. ‘lab-pla’ and ‘koi-pla’ (a kind of raw fish salad with chilli, lime juice and fish sauce) and alcoholic beverages. Accordingly, adults are more likely to be exposed to the infective stages of the parasite. In contrast, lower infection rates were found in the younger age groups (0–14 and 15–29 years old), mostly because parents are aware of the parasite and usually do not allow their children to eat the raw fish dishes.
Other intestinal helminth species sporadically found in these villages were STHs, including A. lumbricoides, T. trichiura, S. stercoralis and hookworms. Usually, people acquire STH infections orally or through the skin after exposure to the infective stages in the soil or environment. In previous publications, a lower occurrence of STH infection was reported in the northern provinces of Thailand than in the southern parts of the country (Nithikathkul et al., 2017). Chaiyos et al. (2018) identified a potential climatic factor, i.e. higher mean annual precipitation in southern Thailand, affecting the occurrence and distribution of A. lumbricoides, T. trichiura and S. stercoralis in the southern provinces of Thailand. Rainfall increases humidity, which is known to be an important factor in the survival of the larval stage of STHs (Schule et al., 2014).
In the present study, gut microbiome profiling of H. taichui-positive and parasite-free groups identified 60 bacterial OTUs after quality filtering of sequencing reads. The most abundant OTU was P. copri, a Gram-negative bacterium present as a normal flora of the human gut (Human Microbiome Project Consortium, 2012; Truong et al., 2017). Current research has provided evidence of an association between P. copri and rheumatoid arthritis, an autoimmune disease, with a higher abundance of the bacterium in patients with rheumatoid arthritis than in healthy individuals (Alpizar-Rodriguez et al., 2019). Future research should focus on the status of this autoimmune disease, as we found significant signs of the presence of this bacterium in both H. taichui-infected and non-infected participants.
Escherichia coli was found as an abundant OTU in the H. taichui-positive group (Fig. 4). Although this bacterium is commonly found in the gut of healthy humans and other mammals, some strains can be pathogenic, causing abdominal disorders and dysbiosis in humans. Regarding the association between intestinal parasitic infection and gut microbiota, previous studies have reported similarly a high abundance of E. coli in parasitic infections; for example, Toxoplasma gondii infection induced bacterial dysbiosis and promoted E. coli overgrowth in a mouse model (Wang et al., 2019); greater E. coli O157 shedding was reported in cattle after infection with Fasiola hepatica (Howell et al., 2018) and chronic infection of O. viverrini in a hamster model promoted not only the growth of the carcinogen-inducing bacteria Helicobacter, but also significantly increased the signal of E. coli (Itthitaetrakool et al., 2016).
The abundance of three bacterial taxa, Ruminococcus, R. faecis and V. parvula, was significantly higher in the H. taichui-positive group than in the parasite-free group according to LEfSe. These three bacteria are classified in the phylum Firmicutes and are part of the normal flora in the human gut and oral cavity. A significant increase in Ruminococcus was also associated with infection with other helminths in previous studies, e.g. in primary school children infected with the pinworm E. vermicularis in Taiwan (Yang et al., 2017) and in a population in China infected with the liver fluke Clonorchis sinensis (Xu et al., 2018). Regarding the occurrence of R. faecis, our results contrasted those of a previous study, which showed a decreasing abundance of R. faecis in human subjects experimentally infected with the hookworm Necator americanus (Giacomin et al., 2016). In addition, a decreased abundance of Veillonella was found in individuals infected with E. vermicularis and C. sinensis (Yang et al., 2017; Xu et al., 2018).
We found that H. taichui infection was associated with decreased gut microbial diversity (α diversity) and significantly altered microbial composition (β diversity). The worms may affect host gut microbiota in different ways, for example, by altering gut physiology and permeability, stimulating mucous secretion and producing anti-microbial substances (Zaiss and Harris, 2016). In our case, these activities may cause dysbiosis, leading to a loss of gut microbial diversity and potentially promoting the proliferation of opportunistic pathogens such as E. coli. The reduction in microbial diversity observed in helminth-infected individuals in the present study was in accordance with the findings of a previous study investigating STH infection and gut microbiome in schoolchildren from Ecuador (Cooper et al., 2013). However, several other publications have shown an inverse relationship; increased bacterial α diversity was significantly associated with intestinal helminths in the indigenous people of Malaysia (Lee et al., 2014); Schistosoma haematobium infection in babies and children in Zimbabwe (Kay et al., 2015) and S. stercolaris infection in people from northern Italy (Jenkins et al., 2018) (more details of helminth–microbiome research in Supplementary Table S1). It appears that there are inconsistencies in the results of the analysis of microbiome diversity and composition in certain intestinal helminth infections among different studies. This could be caused by a variety of factors during the course of infection, such as the intensity of infection and the virulence of particular parasite species or strains (Jenkins, 2019).
Limitations and implications of the present study
We were not able to collect clinical data on H. taichui-positive individuals; we did not record, for example, signs of irritable bowel syndrome, which are a potential clinical manifestation of haplorchiasis. Hence, we were not able to draw a final conclusion about how H. taichui infection clinically affects patient health or their associated gut microbiota. Also, the sample size in the present case–control study was relatively small due to the fact that participation in the worm expulsion experiment was voluntary. This is another potential concern, as sample size may affect the estimation of microbiome α diversity or statistical power in downstream analyses. Also, we were unable to control other potential confounding factors, e.g. the diets of individual villagers, which may have affected the results of this study.
Our research provides information on the status of intestinal helminth infection in the communities of Nan Province and consequently may help to increase awareness and improve public health in these communities by preventing intestinal infection, although health education should be improved. For example, the results of this study may be discussed thoroughly with local public health officials and community leaders to foster engagement for improving health education and awareness. The association found between gut microbiome profile and intestinal helminth infection (H. taichui in this case) needs to be researched further to determine the potential synergistic interactions between the gut microbiome, intestinal helminths and health.
Acknowledgement
We would like to express our gratitude to the local administration in Nan Province, i.e. Nan Provincial Public Health Office, Tha Wang Pha District Public Health Office, Tha Wang Pha Hospital and Saen Thong Subdistrict Health Promoting Hospital for supporting and facilitating research in the field. Also, we would like to extend our sincere thanks to the community leaders and the villagers for their involvement in this study. We acknowledge the help from retired expert parasitologists Surapol Sanguankiat and Rungson Praevanit (from the Faculty of Tropical Medicine, Mahidol University) for parasite examination in the field. We thank Professor Dr Saovanee Leelayoova, head of Parasitology Department, Phramongkutklao College of Medicine, Bangkok for her kind reviewing during the preparation of the article.
Data
Sequence data and associated metadata of the 16S rRNA amplicon sequencing have been deposited in the National Centre for Biotechnology Information Sequence Read Archive (SRA) under the SRA accession: PRJNA 626610.
Financial support
This work was supported by the French ANR Project FutureHealthSEA (grant number: ANR-17-CE35-0003-02) ‘Predictive scenarios of health in Southeast Asia: linking land use and climate changes to infectious diseases’. Ajala Prommi is a scholar recipient of the MSc (Tropical Medicine) programme, Faculty of Tropical Medicine, Mahidol University.
Ethical standards
Procedures concerning human samples, data collection and laboratory investigation were reviewed and approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (document no. MUTM 2018-035-01).
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182020000736.
click here to view supplementary material
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this research article.
References
- Aivelo T and Norberg A (2016) Parasite–microbiota interactions potentially affect intestinal communities in wild mammals. Journal of Animal Ecology 87, 438–447. [DOI] [PubMed] [Google Scholar]
- Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, Gabay C, Finckh A and Strowig T (2019) Prevotella copri in individuals at risk for rheumatoid arthritis. Annals of the Rheumatic Diseases 78, 590–593. [DOI] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R and Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonmekam D, Namchote S, Nak-ai W, Glaubrecht M and Krailas D (2016) The prevalence of human intestinal fluke infections, Haplorchis Taichui, in Thiarid snails and cyprinid fish in Bo Kluea District and Pua District, Nan Province, Thailand. Silpakorn University Science and Technology Journal 10, 29–37. [Google Scholar]
- Broadhurst MJ, Ardeshir A, Kanwar B, Mirpuri J, Gundra UM, Leung JM, Wiens KE, Vujkovic-Cvijin I, Kim CC, Yarovinsky F, Lerche NW, McCune JM and Loke P (2012) Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathogens 8, e1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ and Holmes SP (2016) Dada2: high-resolution sample inference from Illumina amplicon data. Nature Methods 13, 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C, Giacomin P, Croese J, Zakrzewski M, Sotillo J, McCann L, Nolan MJ, Mitreva M, Krause L and Loukas A (2014) Impact of experimental hookworm infection on the human gut microbiota. Journal of Infectious Diseases 210, 1431–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N and Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the USA 108(Suppl), 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattadori IM, Sebastian A, Hao H, Katani R, Albert I, Eilertson KE, Kapur V, Pathak A and Mitchel S (2016) Impact of helminth infections and nutritional constraints on the small intestine microbiota. PLoS ONE 11, e0159770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JY, Yong TS, Eom KS, Min DY, Shin EH, Banouvong V, Insisiengmay B, Insisiengmay S, Phommasack B and Rim HJ (2010) Prevalence of the intestinal flukes Haplorchis taichui and H. yokogawai in a mountainous area of Phongsaly Province, Lao PDR. Korean Journal of Parasitology 48, 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisiri K, Jollivet C, Della Rossa P, Sanguankiat S, Wattanakulpanich D, Lajaunie C, Binot A, Tanita M, Rattanapikul S, Sutdan D, Morand S and Ribas A (2018) Parasitic infections in relation to practices and knowledge in a rural village in Northern Thailand with emphasis on fish-borne trematode infection. Epidemiology and Infection 15, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyos J, Suwannatrai K, Thinkhamrop K, Pratumchart K, Sereewong C, Tesana S, Kaewkes S, Sripa B, Wongsaroj T and Suwannatrai AT (2018) Maxent modeling of soil-transmitted helminth infection distributions in Thailand. Parasitology Research 117, 3507–3517. [DOI] [PubMed] [Google Scholar]
- Cooper P, Walker AW, Reyes J, Chico M, Salter SJ, Vaca M and Parkhill J (2013) Patent human infections with the whipworm, Trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS ONE 8, e76573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu PP, Xiong F, Feng WW, Zou H, Wu SG, Li M, Wang GT and Li WX (2019) Effect of intestinal tapeworms on the gut microbiota of the common carp, Cyprinus carpio. Parasites & Vectors 12, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomin P, Zakrzewski M, Jenkins TP, Su X, Al-Hallaf R, Croese J, de Vries S, Grant A, Mitreva M, Loukas A, Krause L and Cantacessi C (2016) Changes in duodenal tissue-associated microbiota following hookworm infection and consecutive gluten challenges in humans with coeliac disease. Scientific Reports 6, 36797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV and Knight R (2018) Current understanding of the human microbiome. Nature Medicine 24, 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glendinning L, Nausch N, Free A, Taylor DW and Mutapi F (2014) The microbiota and helminths: sharing the same niche in the human host. Parasitology 141, 1255–1271. [DOI] [PubMed] [Google Scholar]
- Hotez PJ, Molyneux DH, Fenwick A, Ottesen E, Sachs SE and Sachs JD (2006) Incorporating a rapid-impact package for neglected tropical diseases with programs for HIV/AIDS, tuberculosis, and malaria: a comprehensive pro-poor health policy and strategy for the developing world. PLoS Medicine 3, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ and Jacobson J (2008) Helminth infections: the great neglected tropical diseases. Journal of Clinical Investigation 118, 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AK, Tongue SC, Currie C, Evans J, Williams DJL and McNeilly TN (2018) Co-infection with Fasciola hepatica may increase the risk of Escherichia coli O157 shedding in British cattle destined for the food chain. Preventive Veterinary Medicine 150, 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina (2013) 16S Metagenomic sequencing library preparation guide. Available at https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html (Assessed September 2018).
- Itthitaetrakool U, Pinlaor P, Pinlaor S, Chomvarin C, Dangtakot R, Chaidee A, Wilailuckana C, Sangka A, Lulitanond A and Yongvanit P (2016) Chronic Opisthorchis viverrini infection changes the liver microbiome and promotes Helicobacter Growth. PLoS ONE 11, e0165798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M and Reddy DN (2015) Role of the normal gut microbiota. World Journal of Gastroenterology 21, 8787–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TP (2019) Exploring the impact of gastrointestinal parasitic helminths on the human microbiome using advanced biomolecular and bioinformatics technologies (PhD thesis). University of Cambridge, Cambridge. [Google Scholar]
- Jenkins TP, Formenti F, Castro C, Piubelli C, Perandin F, Buonfrate D, Otranto D, Griffin JL, Krause L, Bisoffi Z and Cantacessi C (2018) A comprehensive analysis of the faecal microbiome and metabolome of Strongyloides stercoralis infected volunteers from a non-endemic area. Scientific Reports 8, 15651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz N, Chaves A and Pellegrino J (1972) A simple device for quantitative stool thick-smear technique in Schistosomiasis mansoni. Revista do Instituto de Medicina Tropical de São Paulo 14, 397–400. [PubMed] [Google Scholar]
- Kay GL, Millard A, Sergeant MJ, Midzi N, Gwisai R, Mduluza T, Ivens A, Nausch N, Mutapi F and Pallen M (2015) Differences in the faecal microbiome in Schistosoma haematobium infected children vs uninfected children. PLoS Neglected Tropical Diseases 9, e0003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser PB and Nutman TB (2004) Strongyloides stercoralis in the immunocompromised population. Clinical Microbiology Reviews 17, 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisinger J, Bastien G, Hauffe HC, Marchesi J and Perkins SE (2015) Interactions between multiple helminths and the gut microbiota in wild rodents. Philosophical Transactions of the Royal Society B 370, 20140295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriss M, Hazleton KZ, Nusbacher NM, Martin CG and Lozupone CA (2018) Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery. Current Opinion in Microbiology 44, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Tang MS, Lim YA, Choy SH, Kurtz ZD, Cox LM, Gundra UM, Cho I, Bonneau R, Blaser MJ, Chua KH and Loke P (2014) Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Neglected Tropical Diseases 8, e2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RW, Li W, Sun J, Yu P, Baldwin RL and Urban JF (2016) The effect of helminth infection on the microbial composition and structure of the caprine abomasal microbiome. Scientific Reports 6, 20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17, 10–12. [Google Scholar]
- Martin I, Kaisar MMM, Wiria AE, Hamid F, Djuardi Y, Sartono E, Rosa BA, Mitreva M, Supali T, Houwing-Duistermaat JJ, Yazdanbakhsh M and Wammes LJ (2019) The effect of gut microbiome composition on human immune responses: an exploration of interference by helminth infections. Frontiers in Genetics 10, 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlenbachs A, Bhatnagar J, Agudelo CA, Hidron A, Eberhard ML, Mathison BA, Frace MA, Ito A, Metcalfe MG, Rollin DC, Visvesvara GS, Pham CD, Jones TL, Greer PW, Vélez Hoyos A, Olson PD, Diazgranados LR and Zaki SR (2015) Malignant transformation of Hymenolepis nana in a human host. New England Journal of Medicine 373, 1845–1852. [DOI] [PubMed] [Google Scholar]
- Mutapi F (2015) The gut microbiome in the helminth infected host. Trends in Parasitology 31, 405–406. [DOI] [PubMed] [Google Scholar]
- Newbold LK, Burthe SJ, Oliver AE, Gweon HS, Barnes CJ, Daunt F and Van der Gast CJ (2017) Helminth burden and ecological factors associated with alterations in wild host gastrointestinal microbiota. ISME Journal 11, 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithikathkul C, Trevanich A, Wongsaroj T, Wongsawad C and Reungsang P (2017) Health informatics model for helminthiasis in Thailand. Journal of Helminthology 91, 528–533. [DOI] [PubMed] [Google Scholar]
- Pabalan N, Singian E, Tabangay L, Jarjanazi H, Boivin MJ and Ezeamama AE (2017) Soil-transmitted helminth infection, loss of education and cognitive impairment in school-aged children: a systematic review and meta-analysis. PLoS Neglected Tropical Diseases 12, e0005523. 10.1371/journal.pntd.0005523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ayele Y, Hadush A, Kundu P and Jadhav VJ (2018) Public health significance of foodborne helminthiasis: a systematic review. Journal of Experimental Food Chemistry 4, 135–140. [Google Scholar]
- Punsawad C, Phasuk N, Bunratsami S, Thongtup K, Viriyavejakul P, Palipoch S, Koomhin P and Nongnaul S (2018) Prevalence of intestinal parasitic infections and associated risk factors for hookworm infections among primary schoolchildren in rural areas of Nakhon Si Thammarat, southern Thailand. BMC Public Health 18, 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J and Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available at http://www.r-project.org/index.html. [Google Scholar]
- Round JL and Mazmanian SK (2009) The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology 9, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozsa L, Reiczigel J and Majoros G (2000) Quantifying parasites in samples of hosts. Journal of Parasitology 86, 228–232. [DOI] [PubMed] [Google Scholar]
- Sato M, Sanguankiat S, Pubampen S, Kusolsuk T, Maipanich W and Waikagul J (2009) Egg laying capacity of Haplorchis taichui (Digenea: Heterophyidae) in humans. Korean Journal of Parasitology 47, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schule SA, Clowes P, Kroidl I, Kowuor DO, Nsojo A, Mangu C, Riess H, Geldmacher C, Laubender RP, Mhina S, Maboko L, Löscher T, Hoelscher M and Saathoff E (2014) Ascaris lumbricoides infection and its relation to environmental factors in the Mbeya region of Tanzania, a cross-sectional, population-based study. PLoS ONE 9, e92032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS and Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biology 12, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripa B, Kaewkes S, Sithithaworn P, Mairiang E, Laha T, Smout M, Pairojkul C, Bhudhisawasdi V, Tesana S, Thinkamrop B, Bethony JM, Loukas A and Brindley PJ (2007) Liver fluke induces cholangiocarcinoma. PLoS Medicine 4, e201. 10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Su L, Li Y, Long SR, Chang J, Zhang W, Walker WA, Xavier RJ, Cherayil BJ and Shi HN (2018) Helminth-induced alterations of the gut microbiota exacerbate bacterial colitis. Mucosal Immunology 11, 144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantrawatpan C, Intapan PM, Thanchomnang T, Sanpool O, Janwan P, Lulitanond V, Sadaow L and Maleewong W (2014) Development of a PCR assay and pyrosequencing for identification of important human fish-borne trematodes and its potential use for detection in faecal specimens. Parasites & Vectors 7, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DT, Tett A, Pasolli E, Huttenhower C and Segata N (2017) Microbial strain-level population structure and genetic diversity from metagenomes. Genome Research 27, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente CS, Ozawa S and Hasegawa K (2016) Composition of the cockroach gut microbiome in the presence of parasitic nematodes. Microbes and Environments 31, 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikagul J, Jongsuksantigul P, Rattanawitoon U, Radomyos P, Kojima S and Takeuchi T (2008) Parasitological monitoring of helminth control program in Northern Thailand. Southeast Asian Journal of Tropical Medicine and Public Health 39, 1008–1014 [PubMed] [Google Scholar]
- Walshe N, Duggan V, Cabrera-Rubio R, Crispie F, Cotter P, Feehan O and Mulcahy G (2019) Removal of adult cyathostomins alters faecal microbiota and promotes an inflammatory phenotype in horses. International Journal for Parasitology 49, 489–500. [DOI] [PubMed] [Google Scholar]
- Wang S, El-Fahmawi A, Christian DA, Fang Q, Radaelli E, Chen L, Sullivan MC, Misic AM, Ellringer JA, Zhu XQ, Winter SE, Hunter CA and Beiting DP (2019) Infection-induced intestinal dysbiosis is mediated by macrophage activation and nitrate production. mBio 10, e00935–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watthanakulpanich D, Waikagul J, Maipanich W, Nuamtanong S, Sanguankiat S, Pubampen S, Praevanit R, Mongkhonmu S and Nawa Y (2010) Haplorchis taichui as a possible etiologic agent of irritable bowel syndrome-like symptoms. Korean Journal of Parasitology 48, 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM and Viney M (2015) The gut microbiota of wild mice. PLoS ONE 10, e0134643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijit A, Morakote N and Klinchid J (2013) High prevalence of haplorchiasis in Nan and Lampang provinces, Thailand, proven by adult worm recovery from suspected opisthorchiasis cases. Korean Journal of Parasitology 51, 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsaroj T, Nithikathkul C, Rojkitikun W, Nakaia W, Royal L and Rammasut P (2014) National survey of helminthiasis in Thailand. Asian Biomedicine 8, 779–783. [Google Scholar]
- Wu HJ and Wu E (2012) The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 3, 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Li RW, Li E, Beshah E, Dawson HD and Urban JF (2012) Worm burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. PLoS ONE 7, e35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Jiang Z, Huang W, Yin J, Ou S, Jiang Y, Meng L, Cao S, Yu A, Cao J and Shen Y (2018) Altered gut microbiota composition in subjects infected with Clonorchis sinensis. Frontiers in Microbiology 9, 2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CA, Liang C, Lin CL, Hsiao CT, Peng CT, Lin HC and Chang JG (2017) Impact of Enterobius vermicularis infection and mebendazole treatment on intestinal microbiota and host immune response. PLoS Neglected Tropical Diseases 11, e0005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss MM and Harris NEL (2016) Interactions between the intestinal microbiome and helminth parasites. Parasite Immunology 38, 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182020000736.
click here to view supplementary material