Abstract
African animal trypanosomiasis (AAT) affects the livestock of 12.3 million Somalis and constrains their development and wellbeing. There is missing data on AAT in the country after the civil war of the 1990s. Therefore, this study has aimed to assess the prevalence of Trypanosoma spp. in 614 blood samples from cattle (n = 202), goats (n = 206) and sheep (n = 206) in Afgoye and Jowhar districts, Somalia using parasitological and molecular methods. Twenty-one out of 614 (3.4%; 95% CI: 2.1–5.2%) and 101/614 (16.4%; 95% CI: 13.6–19.6%) ruminants were positive for Trypanosoma spp. by buffy coat technique (BCT) and internal transcribed spacer 1 (ITS1)-polymerase chain reaction (PCR), respectively. Using ITS1-PCR, the highest prevalence was observed in cattle (23.8%; 95% CI: 18.4–30.1%) followed by goats (17.5%; 95% CI: 12.9–23.3%) and sheep (8.3%; 95% CI: 5.1–12.9%). A total of 74/101 (73.3%; 95% CI: 63.5–81.6%) ruminants were shown coinfection with at least two Trypanosome species. The four T. brucei-positive samples have tested negative for T. b. rhodesiense, by the human-serum-resistance-associated-PCR. Trypanosoma evansi, T. godfreyi, T. vivax, T. brucei, T. simiae and T. congolense were the Trypanosoma species found in this study. This is the first study on the molecular detection of Trypanosoma sp. in ruminants in Somalia. Further investigations and control measures are needed to manage Trypanosomiasis spreading in the country. Studies should also focus on the detection of T. b. rhodesiense in the country.
Key words: AAT, Glossina spp, HAT, Hirshabelle, Somalia, Southwest, Trypanosomiasis
Introduction
African animal trypanosomiasis (AAT) is a parasitic disease caused by protozoans of the genus Trypanosoma (OIE Terrestrial Manual, 2013) and causes economic losses of 4.5 billion US dollars per year (Oluwafemi et al., 2007), as a result of direct (mortality, production losses, costs of prophylactic and curative trypanocidal drugs) and indirect losses due to crop production decay and agricultural workers' involvement (deficiency of animal protein diets) (Harberd, 1988; Angara et al., 2014). In Somalia, the economic impact of AAT has been estimated at 88 million US dollars (Mohamed and Dairri, 1987). Although the National Tsetse and Trypanosomiasis Control Project (NTTCP) has been established in the 1980s (Mohamed and Dairri, 1987), and a tsetse and trypanosomiasis (T & T) control project has been funded by the International Committee of the Red Cross (ICRC) in some villages of Shabelle and Jubba regions (ICRC, 2017), no other control measures or wide coverage area to reduce the losses from trypanosomiasis have been implemented following the collapse of the central government of Somalia in 1991 (Salah, 2016). Effective and sustainable T & T control projects will contribute to improving livestock health that enhances the production and livelihood of dependent families (Swallow, 2000).
Examination of thick and thin peripheral blood or buffy coat films stained with Giemsa stains or fresh wet blood or buffy coat smears has been used for the detection and identification of Trypanosome species (Rosenblatt, 2009; OIE Terrestrial Manual, 2013). However, these direct parasitological techniques lack sensitivity and specificity (Mattioli and Faye, 1996; Picozzi et al., 2002). Therefore, molecular methods provide multi-species-specific detection of trypanosomes in a single polymerase chain reaction (PCR) (Salim et al., 2011) and have been used in epidemiological studies (Desquesnes and Dávila, 2002; Picozzi et al., 2002; Kouadio et al., 2014; Hassan-Kadle et al., 2019). Moreover, the pan-PCR techniques for Trypanosomes would reduce the cost of PCR three to five times (Njiru et al., 2005).
In Somalia, AAT has been reported in different domestic ruminants by standard trypanosome detection methods (STDM) (Macchioni and Abdullatif, 1985; Mohamed and Dairri, 1987; Dirie et al., 1988a, 1988b; Ainanshe et al., 1992). A previous study has found T. congolense (Schoepf et al., 1984; Dirie et al., 1988b) and T. vivax infecting sheep (Dirie et al., 1988b). Both Trypanosome species have also been reported in cattle from southern Somalia (Moggridge, 1936; Dirie et al., 1988a). Additionally, a recent study has confirmed infections with Trypanosoma evansi and Trypanosoma simiae in camels through DNA sequencing (Hassan-Kadle et al., 2019). Moreover, the human African trypanosomiasis (sleeping sickness) is unknown in Somalia, despite the presence of Glossina pallidipes, a known vector of the disease in other areas of East Africa with the practice of transhumance (Harberd, 1988). The widespread infestation of Trypanosome vectors in Somalia is one of the main obstacles to the development of the livestock industry which supports the livelihood of 12.3 million Somali communities living in an area of 640 000 km2 (Bernacca, 1967; UNFPA, 2014). However, there is a lack of data on AAT after the civil war of the 1990s. Hence, the present study aimed to assess the prevalence of Trypanosoma spp. in cattle, goats and sheep from Afgoye and Jowhar districts of Somalia.
Materials and methods
Study area
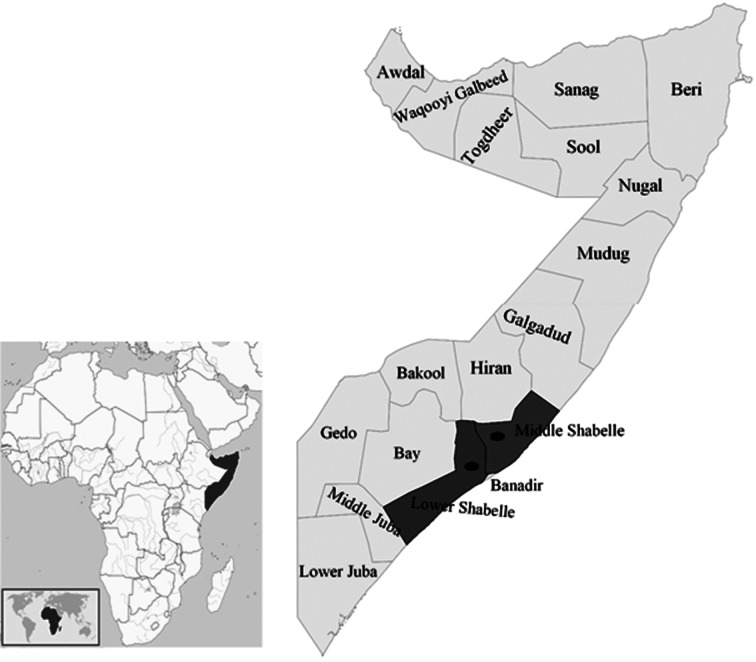
The present study was carried out in Afgoye (2°08′47.67″N 45°07′08.11″E) and Jowhar (2°46′38.72″N 45°30′05.85″E) districts, Somalia (Fig. 1), areas are known to be infested by tsetse and other biting flies (Hursey, 1985; Mohamed and Dairri, 1987; Dirie et al., 1989).
Fig. 1.
Map of Somalia showing the location of blood samples. Sampled regions are highlighted in green (Middle Shabelle) and blue (Lower Shabelle), and the dots indicate the approximate location of the sampled districts (Jowhar and Afgoye). The figure was generated and modified using QGIS software version 2.18.19.
Sampling
From November 2017 to February 2018, which represents the dry season in Somalia, a total of 614 ruminants from Afgoye (n = 304) and Jowhar (n = 310) were evaluated. Blood samples were collected by jugular venipuncture of cattle (n = 202; 183 females and 19 males), sheep (n = 206; 190 females and 16 males) and goats (n = 206; 191 females and 15 males). Five millilitres were placed into EDTA tubes for microscopical detection of Trypanosomes and preparation of blood spots on filter paper (Whatman no.4, Whatman, Springfield Mill, UK) for PCR analysis (Ahmed et al., 2011).
Parasitological diagnosis of Trypanosoma spp.
All blood samples were evaluated for the presence of Trypanosoma spp. by the buffy coat technique (BCT), as previously described (Murray et al., 1977). The BCT was considered positive when Trypanosomes could be visually detected regardless of the species.
DNA extraction and PCR for Trypanosoma spp.
DNA was extracted from all 614 blood spots by Chelex® 100 (Sigma–Aldrich, St. Louis, USA), as previously described (Ahmed et al., 2011). Five different PCR assays were used to detect Trypanosoma spp.: (i) internal transcribed spacer 1 (ITS1)-PCR that amplifies the ITS1 region (Njiru et al., 2005), which is known to vary in size within trypanosome species, except members of Trypanozoon species (T. brucei brucei, T. brucei rhodesiense, T. brucei gambiense, T. evansi, and T. equiperdum), and therefore differentiates Trypanosomes by their ITS sizes, (ii) TBR-PCR specific for T. brucei (Moser et al., 1989), (iii) human-serum-resistance-associated (SRA)-PCR targeting the SRA gene uniquely specific for T. brucei rhodesiense (Radwanska et al., 2002), (iv) RoTat 1.2 PCR which amplifies the Rode Trypanozoon antigenic type (RoTat) 1.2 variable surface glycoprotein (VSG) gene fragment specific for the detection of T. evansi (Urakawa et al., 2001), and (v) TSM-PCR for detection of T. simiae (Masiga et al., 1992).
All samples were initially screened for Trypanosoma spp. by the ITS1-PCR assay (Njiru et al., 2005). Samples positive for Trypanozoon subspecies were further tested for T. brucei using TBR-PCR (Moser et al., 1989) and RoTat 1.2 PCR for the presence of T. evansi (Urakawa et al., 2001). Positive samples on the TBR-PCR were further screened for T. brucei rhodensiense by SRA-PCR (Radwanska et al., 2002). Finally, all T. simiae positive samples on the ITS1-PCR assay were further subjected to the TSM-PCR (Masiga et al., 1992). The PCR primers used in this study are shown in Table 1. A camel sample is known to be positive for T. evansi (Hassan-Kadle et al., 2019) was used as a positive control, in all ITS1, TBR and TSM PCR runs, while brucei rhodesiense DNA (kindly donated by Dr Enock Matovu, Makerere University, Uganda) was used in SRA-PCR assay. Nuclease-free water was used as a negative control in all reactions. The amplified PCR products were analysed by electrophoresis in a 1.5% agarose gel, stained with ethidium bromide and visualized by UV illuminator (UVITEC™ Cambridge, UK).
Table 1.
PCR primers used in the present study
| Taxa | Band size | Target gene | Specific primers | Primer sequence (5′-3′) | Reference |
|---|---|---|---|---|---|
| T. congolense savannah | 700 | ITS1 | ITS1 forward ITS2 reverse |
CCGGAAGTTCACCGATATTG TTGCTGCGTTCTTCAACGAA |
Njiru et al. (2005) |
| T. congolense forest | 710 | ||||
| T. congolense kilifi | 620 | ||||
| T. simiae Tsavo | 370 | ||||
| T. simiae | 400 | ||||
| Trypanozoon | 480 | ||||
| T. vivax | 250 | ||||
| T. godfreyi | 300 | ||||
| T. theileri | – | ||||
| T. evansi | 488 | RoTat1.2 VSG | RoTat 1.2 forward RoTat 1.2 reverse |
GCCACCACGGCGAAAGAC TAATCAGTGTGGTGTGC |
Urakawa et al. (2001) |
| T. simiae | 437 | Sat-DNA | TSM1 forward TSM2 reverse |
CCGGTCAAAAACGCATT AGTCGCCCGGAGTCGAT |
Masiga et al. (1992) |
| T. brucei | 173 | Sat-DNA | TBR1 forward TBR2 reverse |
CGAATGAATATTAAACAATGCGCAGT AGAACCATTTATTAGCTTTGTTGC |
Moser et al. (1989) |
| T. b. rhodesiense | 284 | SRA | SRA forward SRA reverse |
ATAGTGACAAGATGCGTACTCAACGC AATGTGTTCGAGTACTTCGGTCACGCT |
Radwanska et al. (2002) |
Data management and analysis
Either Chi-square or Fisher's exact test was used to assess the association of the individual variables (district and species) with Trypanosoma spp. infection. Odds ratio (OR), 95% confidence intervals (95% CI) and P values were calculated, and results were considered significant when P ⩽ 0.05. Data were compiled and analysed by Statistical Package for Social Sciences (SPSS) version 25 (IBM Corp., Armonk, NY, USA).
Results
Parasitological diagnosis of Trypanosoma spp.
A total of 21/614 (3.4%; 95% CI: 2.1–5.2%) ruminants were positive for Trypanosome species by BCT. Cattle were more likely to be infected by Trypanosoma spp. than sheep (OR: 3.8; χ2 = 6.0, P = 0.01). Ruminants reared in Afgoye district were more likely to be infected to Trypanosoma spp. than those reared in Jowhar district (OR: 3.4, 95% CI: 1.2–9.4, P = 0.01) (Table 2).
Table 2.
Prevalence of Trypanosoma spp. in cattle, goats and sheep from Afgoye and Jowhar districts, Somalia
| Variable | BCT | ITS1-PCR | ||||||
|---|---|---|---|---|---|---|---|---|
| +/n | Prevalence (%) (95% CI) |
P value | OR (95% CI) | +/n | Prevalence (%) (95% CI) |
P value | OR (95% CI) | |
| Afgoye | ||||||||
| Cattle | 10/100 | 10 (4.9–17.6) | 0.089 (χ2 = 2.9) | 2.7 (0.8–8.9) | 35/100 | 35 (25.7–45.2) | <0.001 (χ2 = 18.5) | 4.9 (2.3–10.7) |
| Goat | 2/102 | 1.9 (0.24–6.9) | 0.407 (χ2 = 0.7) | 0.5 (0.1–2.7) | 14/102 | 13.7 (7.7–21.9) | 0.385 (χ2 = 0.8) | 1.5 (0.6–3.5) |
| Sheep | 4/102 | 3.9 (1.1–9.7) | 10/102 | 9.8 (4.8–17.3) | ||||
| Jowhar | ||||||||
| Cattle | 4/102 | 3.9 (1.1–9.7) | 0.041 (χ2 = 4.2) | 0.0 | 13/102 | 12.7 (6.9–20.8) | 0.145 (χ2 = 2.1) | 2 (0.8–5.3) |
| Goat | 1/104 | 0.9 (0.02–5.2) | 0.318 (χ2 = 1.0) | 0.0 | 22/104 | 21.2 (13.8–30.3) | 0.003 (χ2 = 9) | 3.7 (1.5–9.1) |
| Sheep | 0/104 | 0.0 (0.0–3.5) | 7/104 | 6.7 (2.7–13.4) | ||||
| Species | ||||||||
| Cattle | 14/202 | 6.9 (3.8–11.4) | 0.014 (χ2 = 6.0) | 3.8 (1.2–11.6) | 48/202 | 23.8 (18.4–30.1) | <0.001 (χ2 = 18.3) | 3.5 (1.9–6.3) |
| Goat | 3/206 | 1.5 (0.3–4.2) | 0.703 (χ2 = 0.15) | 0.7 (0.2–3.4) | 36/206 | 17.5 (12.9–23.3) | 0.005 (χ2 = 7.8) | 2.4 (1.3–4.3) |
| Sheep | 4/206 | 1.9 (0.5–4.9) | 17/206 | 8.3 (5.1–12.9) | ||||
| District | ||||||||
| Afgoye | 16/304 | 5.3 (3.0–8.4) | 0.013 (χ2 = 6.2) | 3.4 (1.2–9.4) | 59/304 | 19.4 (15.3–24.2) | 0.005 (χ2 = 3.8) | 1.5 (0.9–2.4) |
| Jowhar | 5/310 | 1.6 (0.5–3.7) | 42/310 | 13.6 (10.2–17.8) | ||||
+, number of positive animals; n, number of samples; 95% CI, 95% confidence interval; OR, odds ratio.
Molecular diagnosis of Trypanosoma spp.
Overall, 101/614 (16.4%, 95% CI: 13.6–19.6%) ruminants tested positive for Trypanosoma spp. by the ITS1-PCR assay.
The highest prevalence of Trypanosoma spp. was observed in cattle (48/202; 23.8%) followed by goats (36/206; 17.5%) and sheep (17/206; 8.3%) (Table 2). The ITS1-PCR assay has detected 27/101 (26.7%; 95% CI: 18.4–36.5%) ruminants single infected by Trypanosomes. Additionally, coinfections with different trypanosomes were found in 74/101 (73.3%; 95% CI: 63.5–81.6%) ruminants, with coinfections mainly observed in cattle (35/48; 72.9%) and goats (26/36; 72.2%). The prevalence of Trypanosoma spp. by ITS1-PCR within each ruminant species evaluated are summarized in Table 3.
Table 3.
Single and coinfections with Trypanosoma spp. in cattle, goats and sheep in Somalia
| Single/coinfections | Species | Districts | Total +/ITS1-PCR assay (%) |
|||
|---|---|---|---|---|---|---|
| Cattle +/n (%) |
Goat +/n (%) |
Sheep +/n (%) |
Afgoye +/n (%) |
Jowhar +/n (%) |
||
| Single infection | 13/202 (6.4%) | 10/206 (4.9%) | 4/206 (1.9%) | 9/304 (2.9%) | 18/310 (5.8%) | 27/101 (26.7%) |
| Trypanozoon | 7/202 (3.5%) | 9/206 (4.4%) | 4/206 (1.9%) | 4/304 (1.3%) | 16/310 (5.2%) | 20/101 (19.8%) |
| T. congolense | 3/202 (1.5%) | 0/206 (0%) | 0/206 (0%) | 3/304 (0.9%) | 0/310 (0%) | 3/101 (2.9%) |
| T. godfreyi | 3/202 (1.5%) | 0/206 (0%) | 0/206 (0%) | 2/304 (0.7%) | 1/310 (0.3%) | 3/101 (2.9%) |
| T. simiae | 0/202 (0%) | 1/206 (0.5%) | 0/206 (0%) | 0/304 (0%) | 1/310 (0.3%) | 1/101 (0.9%) |
| Two species | 22/202 (10.9%) | 16/206 (7.8%) | 5/206 (2.4%) | 30/304 (9.9%) | 13/310 (4.2%) | 43/101 (42.6%) |
| T. v & Trypanozoon | 4/202 (1.9%) | 11/206 (5.3%) | 3/206 (1.5%) | 16/304 (5.3%) | 2/310 (0.6%) | 18/101 (17.8%) |
| T. g & Trypanozoon | 8/202 (3.9%) | 1/206 (0.5%) | 2/206 (0.9%) | 6/304 (1.9%) | 5/310 (1.6%) | 11/101 (10.9%) |
| T. s & Trypanozoon | 3/202 (1.5%) | 3/206 (1.5%) | 0/206 (0%) | 3/304 (0.9%) | 3/310 (0.9%) | 6/101 (5.9%) |
| T. c & Trypanozoon | 4/202 (1.9%) | 1/206 (0.5%) | 0/206 (0%) | 3/304 (0.9%) | 2/310 (0.6%) | 5/101 (4.9%) |
| T. v & T. c | 1/202 (0.5%) | 0/206 (0%) | 0/206 (0%) | 0/304 (0%) | 1/310 (0.3%) | 1/101 (0.9%) |
| T. g & T. c | 2/202 (0.9%) | 0/206 (0%) | 0/206 (0%) | 2/304 (0.7%) | 0/310 (0%) | 2/101 (1.9%) |
| Three species | 10/202 (4.9%) | 6/206 (2.9%) | 8/206 (3.9%) | 17/304 (5.6%) | 7/310 (2.3%) | 24/101 (23.8%) |
| T. v, T. g & Trypanozoon | 7/202 (3.5%) | 1/206 (0.5%) | 7/206 (3.4%) | 12/304 (3.9%) | 3/310 (0.9%) | 15/101 (14.9%) |
| T. v, T. s & Trypanozoon | 0/202 (0%) | 4/206 (1.9%) | 0/206 (0%) | 1/304 (0.3%) | 3/310 (0.9%) | 4/101 (3.9%) |
| T. c, T. g & Trypanozoon | 2/202 (0.9%) | 0/206 (0%) | 0/206 (0%) | 2/304 (0.7%) | 0/310 (0%) | 2/101 (1.9%) |
| T. v, T. c & Trypanozoon | 0/202 (0%) | 0/206 (0%) | 1/206 (0.5%) | 1/304 (0.3%) | 0/310 (0%) | 1/101 (0.9%) |
| T. c, T. s & Trypanozoon | 1/202 (0.5) | 0/206 (0%) | 0/206 (0%) | 1/304 (0.3%) | 0/310 (0%) | 1/101 (0.9%) |
| T. g, T. s & Trypanozoon | 0/202 (0%) | 1/206 (0.5%) | 0/206 (0%) | 0/304 (0%) | 1/310 (0.3%) | 1/101 (0.9%) |
| Four species | 3/202 (1.5%) | 4/206 (1.9%) | 0/206 (0%) | 3/304 (0.9%) | 4/310 (1.3%) | 7/101 (6.9%) |
| T. v, T. s, T. g & Trypanozoon | 2/202 (0.9%) | 3/206 (1.5%) | 0/206 (0%) | 2/304 (0.7%) | 3/310 (0.9%) | 5/101 (4.9%) |
| T. v, T. c, T. s & Trypanozoon | 0/202 (0%) | 1/206 (0.5%) | 0/206 (0%) | 1/304 (0.3%) | 0/310 (0%) | 1/101 (0.9%) |
| T. v, T. c, T. g & Trypanozoon | 1/202 (0.5) | 0/206 (0%) | 0/206 (0%) | 0/304 (0%) | 1/310 (0.3%) | 1/101 (0.9%) |
+, number of positive animals; n, number of samples; T.v, T. vivax; T.c, T. congolense; T.g, T. godfreyi; T.s, T. simiae.
Cattle (OR: 3.5, 95% CI: 1.9–6.3, P < 0.001) and goats (OR: 2.4, 95% CI: 1.3–4.3, P = 0.005) were more likely to be infected with Trypanosoma spp. than sheep. Ruminants reared in Afgoye district were likely to be infected by Trypanosoma spp. than those in Jowhar district (OR: 1.5, 95% CI: 0.9–2.4, P = 0.005). The highest prevalence of Trypanosome infection was recorded in cattle and goats from Afgoye and Jowhar districts, respectively (Table 2).
Ninety-one ITS1-PCR Trypanozoon-positive samples were further tested by species-specific PCR to characterize the T. evansi and T. brucei. As a result, four out of 91 (4.4%) ruminants tested positive for T. brucei by the TBR-PCR assay, while the remaining 87/91 (95.6%) ruminants tested positive for T. evansi by the RoTat 1.2 PCR assay. All the four T. brucei-positive samples have tested negative for T. b. rhodesiense by the SRA-PCR assay. Additionally, two PCR assays were used to detect T. simiae in the present study: the ITS1-PCR was able to detect 19/101 (18.8%) ruminants, while 12/19 (63.2%) ruminants tested positive by the TSM-PCR assay.
Discussion
To the author's knowledge, this is the first study to combine BCT and molecular detection of Trypanosome species in ruminants from Somalia. The overall prevalence recorded by ITS1-PCR was significantly higher than the one observed with the BCT technique (P < 0.001, χ2 = 58.3). This was expected due to the known higher sensitivity of molecular methods, as previously reported (Angwech et al., 2015; N'Djetchi et al., 2017).
Herein, overall, 3.4% ruminants were positive to Trypanosoma spp. by using the BCT technique. Previous studies performed in ruminants from Somalia have shown prevalence rates ranging from 4% to 28.6% by STDM with a record of T. congolense, T. vivax and T. brucei (Moggridge, 1936; Schoepf et al., 1984; Dirie et al., 1988a, 1988b; Ainanshe et al., 1992). Considering that T. brucei and T. evansi are morphologically indistinguishable (Gibson, 2003), it is important to consider that previous studies performed in Somalia may have misclassified the Trypanosoma species detected in ruminants.
In the present study, 16.4% ruminants tested positive for Trypanosoma spp. by the ITS1-PCR assay, which is in agreement with a previous study that has reported a prevalence of 17.2% in Tanzania (Simwango et al., 2017). However, our finding is lower than that reported in Sudan (57.7%) (Osman et al., 2016) and Uganda (41%) (Angwech et al., 2015), but it is higher than that reported in Zambia (7.5%) (Laohasinnarong et al., 2015). Differences among the prevalence of Trypanosome species may be explained by the population studied and management, and tsetse seasonal dynamics (rainy vs dry season).
Previous studies have shown that sheep is more frequently infected by Trypanosoma species than goats under natural conditions, suggesting that goats are more refractory to Trypanosome infections than sheep (Masiga et al., 2002; Ng'ayo et al., 2005). In the present study, Trypanosoma spp. infection rates were significantly higher in cattle (23.8%; P < 0.001, χ2 = 18.3) and goats (17.5%; P = 0.005, χ2 = 7.8) than in sheep (8.3%), with goats being 2.4 times more likely to be infected by these protozoa than sheep. This may be attributed by the host susceptibility to Trypanosome infections and the skin coat suitability for fly feeding (Murray et al., 1982; Wilkowsky, 2018).
Trypanosoma evansi (86.1%) followed by T. vivax (45.5%) were the predominant Trypanosoma species detected in ruminants from the studied areas. This may be due to the camel grazing in the area and the presence of other biting flies which may accelerate the mechanical transmission of the parasite (Mossaad et al., 2020). The T. simiae and T. godfreyi were detected in cattle and goats, which may be linked to the presence of warthog (Phacochoerus africanus) and bushpig (Potamochoerus larvatus somaliensis) in the studied districts (data not shown).
Although a previous study has detected T. simiae in sheep from Kenya (Ng'ayo et al., 2005), herein all sheep tested negative for this protozoon. Wild pigs constitute a reservoir for T. simiae and T. godfreyi infections to ruminants (Hamill et al., 2013). To the author's knowledge, this is the first study to detect T. simiae and T. godfreyi in cattle and goats from Somalia.
In the present study, ruminants reared in Afgoye district were 1.5 times more likely to be infected by Trypanosoma spp. than those in Jowhar district (P = 0.005, χ2 = 3.8). Both districts are located along with the Shabelle river, where tsetse and other biting flies are often abundant (Hursey, 1985; Mohamed and Dairri, 1987; Dirie et al., 1989). However, in Jowhar, livestock owners repel flies with smoke from smouldering cattle dung around the animal shelters with cattle being grazed during the night, when flies are inactive, while goats and sheep are grazed during the day. This probably explains why cattle (12.7%) Trypanosome infection rate was lower than goats (21.2%), with goats being 3.7 times more likely to be infected by Trypanosoma spp. than sheep (P = 0.003, χ2 = 9). Conversely, in Afgoye, cattle, goats and sheep are grazed during the day, when flies are active, and no other local strategies for controlling T & T have been implemented. This possibly explains why cattle Trypanosome infection rate (35%) was higher than goats (13.7%) and sheep (9.8%), with cattle being 4.9 times more likely to be infected by Trypanosoma spp. than sheep (P < 0.001, χ2 = 18.5). Thus, this may explain why the overall Trypanosome prevalence rate in Afgoye was higher than in Jowhar district.
Small ruminants constitute a key component of livestock in many regions of East Africa. A previous study has shown that sheep and goats may harbour T. b. rhodesiense, the causative agent of sleeping sickness or human African trypanosomiasis (Ng'ayo et al., 2005). Additionally, previous studies have also implicated that cattle may act as a reservoir of Trypanosomes to humans (Onyango et al., 1966; N'Djetchi et al., 2017). In the present study, all animals tested with the SRA primer set were tested negative for T. b. rhodesiense. Considering that the T. b. rhodesiense vector, Glossina pallidipes, has been reported in Somalia (Mohamed and Dairri, 1987), associated with the fact that this Trypanosome species is present in ecologically similar areas of neighbouring Kenya (Rutto and Karuga, 2009) and Ethiopia (Baker et al., 1970), further studies should focus on the detection of T. b. rhodesiense in the country.
Concluding remarks
The present study has shown that AAT is prevalent in cattle, goats and sheep from Afgoye and Jowhar districts of Somalia, with the identification of Trypanosoma evansi, T. brucei, T. vivax, T. congolense, T. godfreyi and T. simiae. Coinfections with at least two Trypanosome species were also reported in all ruminants in this study. This is the first study on the molecular detection of Trypanosoma spp. in Somali ruminants as well as detection of T. simiae and T. godfreyi in cattle and goats in Somalia. Further large-scale studies and sustainable control programmes against trypanosomes and its vectors are needed in the country. In addition, studies should also focus on the possibility of detecting of T. brucei rhodesiense, HAT causative agent, in the country.
Acknowledgements
The authors would like to thank Hassan Hussein and Abdiwali Mohamud at ARTC lab, Abrar University for the assistance of blood samples collection and kind cooperation of animal owners who participated in this study. We also acknowledge Dr Enock Matovu (Makerere University, Uganda) for the donation of T. b. rhodesiense DNA to be used as a positive control at the Vector and Vector Borne Diseases Institute, Tanzania. The Brazilian National Council of Scientific and Technological Development (CNPq) provided a fellowship of research productivity (PQ) to Dr Rafael Vieira.
Financial support
This study was supported by the Abrar University, Somalia (grant number AURG04012017). The sponsor had no role in the design of the study, collection, analysis and interpretation of data, writing the manuscript and the decision to submit the article for publication.
Conflicts of interest
The authors declare there are no conflicts of interest.
Ethical standards
This study was approved by the ethical committee of Abrar University, Somalia (reference number AU/ARTC/EC/04/2017). All cattle, goats and sheep owners gave consent to sample their animals.
References
- Ahmed AH, MacLeod ET, Hide G, Welburn SC and and Picozzi K (2011) The best practice for preparation of samples from FTA® cards for diagnosis of blood borne infections using African trypanosomes as a model system. Parasites and Vectors 4, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainanshe OA, Jennings FW and Holmes PH (1992) Isolation of drug-resistant strains of Trypanosoma congolense from the Lower Shabelle region of southern Somalia. Tropical Animal Health Production 24, 65–73. [DOI] [PubMed] [Google Scholar]
- Angara T-EE, Ismail AA and Ibrahim AM (2014) An overview on the economic impacts of animal trypanosomiasis. Global Journal for Research Analysis 3, 275–276. [Google Scholar]
- Angwech H, Nyeko JH, Opiyo EA, Okello-Onen J, Opiro R, Echodu R, Malinga GM, Njahira MN and Skilton RA (2015) Heterogeneity in the prevalence and intensity of bovine trypanosomiasis in the districts of Amuru and Nwoya, Northern Uganda. BMC Veterinary Research 11, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JR, McConnell E and Hady J (1970) Human Trypanosomiasis in Ethiopia. Transactions of the Royal Society of Tropical Medicine and Hygiene 64, 523–530. [DOI] [PubMed] [Google Scholar]
- Bernacca JP (1967) Second Report to the Government of Somalia on The Control of the Tsetse Fly. UNDP/FAO, No. TA 2412. TE/AN/SOM.
- Desquesnes M and Dávila AMR (2002) Applications of PCR-based tools for detection and identification of animal trypanosomes: a review and perspectives. Veterinary Parasitology 11, 213–231. [DOI] [PubMed] [Google Scholar]
- Dirie MF, Wallbanks KR, Molyneux DH and Bornstein S (1988a) Haemorrhagic syndrome associated with T. vivax Infections of cattle in Somalia. Acta Tropica 45, 291–292. [PubMed] [Google Scholar]
- Dirie MF, Wardhere MA and Farah MA (1988b) Sheep Trypanosomiasis in Somalia. Tropical Animal Health Production 20, 45–46. [DOI] [PubMed] [Google Scholar]
- Dirie M, Walbanks K, Aden A, Bornstein S and Ibrahim M (1989) Camel trypanosomosis and its vectors in Somalia. Veterinary Parasitology 32, 285–289. [DOI] [PubMed] [Google Scholar]
- Gibson W (2003) Species concepts for trypanosomes: from morphological to molecular definitions? Kinetoplastid Biology and Disease 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill LC, Kaare MT, Welburn SC and Picozzi K (2013) Domestic pigs as potential reservoirs of human and animal trypanosomiasis in Northern Tanzania. Parasites Vectors 6, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd AJ (1988) Studies on the ecology and control of tsetse flies (Diptera: Glossinidae) in Somalia (PhD Thesis). University of London, UK. [Google Scholar]
- Hassan-Kadle A, Ibrahim AM, Nyingilili HS, Yusuf AA, Vieira TSWJ and Vieira RFC (2019) Parasitological, serological and molecular survey of Camel Trypanosomiasis in Somalia. Parasites & Vectors 12, 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hursey BS (1985) Tsetse control. Revue Scientifique et Technique-Office International des Epizooties 14, 287–297. [DOI] [PubMed] [Google Scholar]
- ICRC Somalia (2017) Trapping the tsetse fly to save livestock and income. Retrieved from: http://blogs.icrc.org/somalia/2017/06/19/resilience-trapping-tsetse-fly-save-livestock-income (Accessed 11 April 2018).
- Kouadio IK, Sokouri D, Koffi M, Konaté I, Ahouty B, Koffi A and N'Guetta SP (2014) Molecular characterization and prevalence of Trypanosoma species in cattle from a Northern Livestock area in Côte d'Ivoire. Open Journal of Veterinary Medicine 4, 314–321. [Google Scholar]
- Laohasinnarong D, Goto Y, Asada M, Nakao R, Hayashida K, Kajino K, Kawazu S, Sugimoto C, Inoue N and Namangala B (2015) Studies of trypanosomiasis in the Luangwa valley, north-eastern Zambia. Parasites & Vectors 8, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchioni G and Abdullatif MA (1985) Indagine Epidemiologica Sulla Tripanosomiasi Del Cammello in Somalia. VI Bollettino Scientifico Della Facolta Di Zootecnia e Veterinaria, Universita Nazionale Somala, 175–181.
- Masiga DK, Smyth AJ, Hayes PJ, Bromidge TJ and Gibson WC (1992) Sensitive detection of trypanosomes in tsetse flies by DNA amplification. International Journal of Parasitology 22, 909–918. [DOI] [PubMed] [Google Scholar]
- Masiga DK, Okech G, Irungu P, Ouma J, Wekesa S, Ouma B, Guya SO and Ndung'u JM (2002) Growth and mortality in sheep and goats under high tsetse challenge in Kenya. Tropical Animal Health Production 34, 489–501. [DOI] [PubMed] [Google Scholar]
- Mattioli RC and Faye JAA (1996) Comparative study of the parasitological buffy coat technique and an antigen enzyme immunoassay for trypanosome diagnosis in sequential Trypanosoma congolense infections in N'Dama, Gobra zebu and N'Dama x Gobra crossbred cattle. Acta Tropica 62, 71–81. [DOI] [PubMed] [Google Scholar]
- Moggridge JY (1936) Some observations on the seasonal spread of Glossina pallidipes in Italian Somaliland with notes on G. Brevipalpis and G. Austeni. Bulletin of Entomological Research 27, 449–466. [Google Scholar]
- Mohamed Ahmed MM and Dairri MF (1987) Trypanosome infection rate of Glossina Pallipides During wet and dry seasons in Somalia. Tropical Animal Health Production 19, 11–20. [DOI] [PubMed] [Google Scholar]
- Moser DR, Cook GA, Ochs DE, Bailey CP, McKane MR and Donelson JE (1989) Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology 99, 57–66. [DOI] [PubMed] [Google Scholar]
- Mossaad E, Ismail AA, Ibrahim AM, Musinguzi P, Angara TEE, Xuan X, Inoue N and Suganuma K (2020) Prevalence of different trypanosomes in livestock in Blue Nile and West Kordofan States, Sudan. Acta Tropica 203, 105302. [DOI] [PubMed] [Google Scholar]
- Murray M, Murray PR and Mc Intyre WIM (1977) An improved parasitological technique for diagnosis of African trypanosomiasis. Acta Tropica 27, 384–386. [DOI] [PubMed] [Google Scholar]
- Murray M, Morrison WI and Whitelaw DD (1982) Host Susceptibility to African Trypanosomiasis: Trypanotolerance. Advances in Parasitology 21, 1–68. [DOI] [PubMed] [Google Scholar]
- N'Djetchi MK, Ilboudo H, Koffi M, Kaboré J, Kaboré JW, Kaba D, Courtin F, Coulibaly B, Fauret P, Kouakou L, Ravel S, Deborggraeve S, Solano P, De Meeûs T, Bucheton B and Jamonneau V (2017) The study of trypanosome species circulating in domestic animals in two human African trypanosomiasis foci of Côte d'Ivoire identifies pigs and cattle as potential reservoirs of Trypanosoma brucei gambiense. PLOS Neglected Tropical Diseases 11, e0005993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng'ayo MO, Njiru ZK, Kenya EU, Muluvi GM, Osir EO and Masiga DK (2005) Detection of trypanosomes in small ruminants and pigs in western Kenya: important reservoirs in the epidemiology of sleeping sickness? Kinetoplastid Biology and Disease 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC and Dávila AM (2005) The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitology Research 95, 186–192. [DOI] [PubMed] [Google Scholar]
- OIE Terrestrial Manual (2013) Chapter 2.4.17. Trypanosomosis (tsetse-transmitted). OIE, 1–11.
- Oluwafemi RA, Ilemobade AA and Laseinde EAO (2007) The impact of African animal trypanosomosis and tsetse on the livelihood and well-being of cattle and their owners in the BICOT study area of Nigeria. Scientific Research and Essay 2, 380–383. [Google Scholar]
- Onyango RJ, Van Hoeve K and De Raadt P (1966) The epidemiology of Trypanosoma rhodesiense sleeping sickness in Alego location central Nyanza, Kenya. I. Evidence that cattle may act as a reservoir host of trypanosomes infectious to man. Transactions of the Royal Society of Tropical Medicine and Hygiene 60, 175–182. [DOI] [PubMed] [Google Scholar]
- Osman NM, Njahira M, Skilton R, Ibtisam AG and A/Rahman AH (2016) Molecular detection of some bovine trypanosome isolates from different areas of Sudan. Sudan Journal of Veterinary Research 31, 9–19. [Google Scholar]
- Picozzi K, Tilley A, Fèvre EM, Coleman PG, Magona JW, Odiit M, Eisler MC and Welburn SC (2002) The diagnosis of trypanosome infections: applications of novel technology for reducing disease risk. African Journal of Biotechnology 1, 39–45. [Google Scholar]
- Radwanska M, Chamekh M, Vanhamme L, Claes F, Magez S, Magnus E, De Baetselier P, Büscher P and Pay E (2002) The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. American Journal of Tropical Medicine and Hygiene 67, 684–690. [DOI] [PubMed] [Google Scholar]
- Rosenblatt JE (2009) Laboratory diagnosis of infections due to blood and tissue parasites. Clinical Infectious Diseases 49, 1103–1108. [DOI] [PubMed] [Google Scholar]
- Rutto JJ and Karuga JW (2009) Temporal and spatial epidemiology of sleeping sickness and use of geographical information system (GIS) in Kenya. Journal of Vector Borne Diseases 46, 18–25. [PubMed] [Google Scholar]
- Salah AA (2016) Epidemiological Studies on Camel Trypanosomosis (Surra) and its Control and Economic Impact in Somaliland (PhD Thesis). Murdoch University, CVM, Mudoch, Western Australia, Australia. [Google Scholar]
- Salim B, Bakheit AM, Kamau K, Nakamura I and Sugimoto C (2011) Molecular epidemiology of camel trypanosomiasis based on ITS1 rDNA and RoTat 1.2 VSG gene in the Sudan. Parasites and Vectors 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepf K, Mustafa Mohamed HA and Katende JM (1984) Observations on blood-borne parasites of domestic livestock in the Lower Juba region of Somalia. Tropical Animal Health Production 16, 227–232. [DOI] [PubMed] [Google Scholar]
- Simwango M, Ngonyoka A, Nnko HJ, Salekwa LP, Ole-Neselle M, Kimera SI and Gwakisa PS (2017) Molecular prevalence of trypanosome infections in cattle and tsetse flies in the Maasai Steppe, northern Tanzania. Parasites & Vectors 10, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow BM (2000) Impacts of Trypanosomiasis on African Agriculture. Rome: FAO (PAAT Technical and Scientific Series, no. 2).
- UNFPA (2014) Population Estimation Survey 2014 for the 18 pre-war regions of Somalia. United Nations Population Fund, Somalia Country Office.
- Urakawa T, Verloo D, Moens L, Büscher P and Majiwa PAO (2001) Trypanosoma Evansi: cloning and expression in Spodoptera Fugiperda insect cells of the diagnostic antigen RoTat1.2. Experimental Parasitology 99, 181–189. [DOI] [PubMed] [Google Scholar]
- Wilkowsky SE (2018) Trypanosoma. In Florin-Christensen M and Schnittger L (eds), Parasitic Protozoa of Farm Animals and Pets. Cham: Springer, pp. 271–287. [Google Scholar]