Abstract
Objectives
We aimed to present the state of the art of CT- and MRI-based radiomics in the context of ovarian cancer (OC), with a focus on the methodological quality of these studies and the clinical utility of these proposed radiomics models.
Methods
Original articles investigating radiomics in OC published in PubMed, Embase, Web of Science, and the Cochrane Library between January 1, 2002, and January 6, 2023, were extracted. The methodological quality was evaluated using the radiomics quality score (RQS) and Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2). Pairwise correlation analyses were performed to compare the methodological quality, baseline information, and performance metrics. Additional meta-analyses of studies exploring differential diagnoses and prognostic prediction in patients with OC were performed separately.
Results
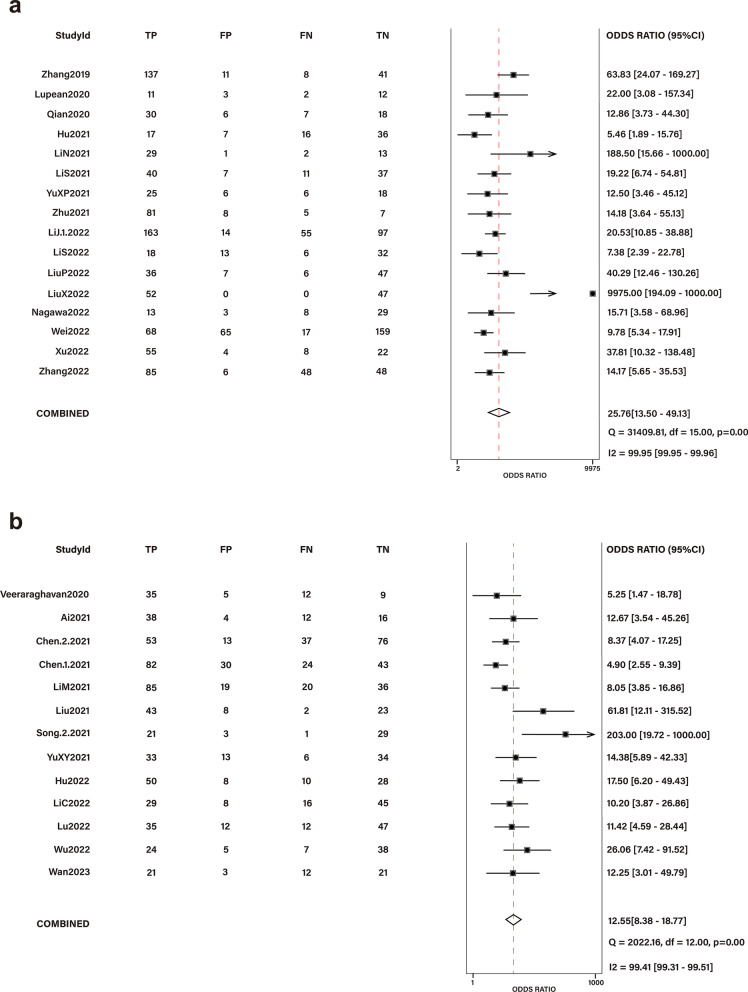
Fifty-seven studies encompassing 11,693 patients were included. The mean RQS was 30.7% (range − 4 to 22); less than 25% of studies had a high risk of bias and applicability concerns in each domain of QUADAS-2. A high RQS was significantly associated with a low QUADAS-2 risk and recent publication year. Significantly higher performance metrics were observed in studies examining differential diagnosis; 16 such studies as well as 13 exploring prognostic prediction were included in a separate meta-analysis, which revealed diagnostic odds ratios of 25.76 (95% confidence interval (CI) 13.50–49.13) and 12.55 (95% CI 8.38–18.77), respectively.
Conclusion
Current evidence suggests that the methodological quality of OC-related radiomics studies is unsatisfactory. Radiomics analysis based on CT and MRI showed promising results in terms of differential diagnosis and prognostic prediction.
Critical relevance statement
Radiomics analysis has potential clinical utility; however, shortcomings persist in existing studies in terms of reproducibility. We suggest that future radiomics studies should be more standardized to better bridge the gap between concepts and clinical applications.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13244-023-01464-z.
Keywords: Ovarian neoplasms, Differential diagnosis, Machine learning, Systematic review
Key points
The methodological quality of current radiomics studies concerning ovarian cancer was unsatisfactory.
Meta-analyses showed high diagnostic odds ratios regarding differential diagnosis and prognostic prediction.
Radiomics analysis in ovarian cancer holds promise for clinical applications.
More standardization should be required for radiomics studies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13244-023-01464-z.
Introduction
Ovarian cancer (OC) is the most lethal gynecological cancer and the fifth-leading cause of cancer-related deaths among women; there were 313,959 newly diagnosed cases and 207,252 deaths worldwide in 2020 [1, 2]. The most recent cancer statistics report indicates that approximately 19,710 new cases of OC will be diagnosed in the USA in 2023, and 13,270 women will die from the disease [2]. The World Health Organization classification of tumors divides OC into dozens of pathological types [3]; furthermore, 70% of patients with OC are diagnosed at an advanced stage, leading to worse outcomes (their 5-year overall survival rates are 20–30% versus 80–95% for those diagnosed at early stages) [4–6]. Treatment involves surgery and chemotherapy and depends on the pathological type and International Federation of Gynaecology and Obstetrics stage [5, 7]. Notwithstanding advances in the diagnosis and treatment of OC, however, mortality rates have not changed appreciably in the last 30 years [2, 8–12]. This is partly due to the difficulty of early detection and the lack of effective therapeutic options for patients in advanced stages.
Computed tomography (CT) and magnetic resonance imaging (MRI) are essential for diagnosing and staging OC [5] and are invaluable for assessing chemotherapy response [13]. However, conventional imaging interpretations rely on the skills of radiologists, and variabilities among reports inevitably exist. Fortunately, medical imaging is advancing toward more standardized, specialized, and quantitative approaches, contributing to greater consistency and communication among radiologists. With this evolution, radiologists are shifting from conventional free-text reporting to structured reporting, enabling more accurate and efficient analysis of the extensive volumes of imaging data [14, 15]. Radiomics is a rapidly emerging field that quantitatively analyzes medical images utilizing artificial intelligence; based on high-throughput mining of quantitative image features, radiomics analysis generates unique markers that may be visually indiscernible yet can support clinical decision-making and increase diagnostic and prognostic accuracy [8, 16, 17].
In recent years, extensive studies using radiomics methods based on CT and MRI have linked quantitative image features to diagnosis, response evaluation, and prognostic prediction in patients with OC [8, 17, 18]. Nevertheless, it has remained difficult to assess the clinical value of radiomics in OC owing to the complexity of the methods and varying study designs. Therefore, we performed this study with two main aims: First, we evaluated the methodological quality of existing studies using the ‘quality of diagnostic accuracy studies-2’ (QUADAS-2) tool as well as the ‘radiomics quality score’ (RQS) [16, 19]. Second, we conducted a meta-analysis to determine the diagnostic performance of radiomics in patients with OC.
Materials and methods
Evidence acquisition protocol and registry
This systematic review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement [20] (Additional file 1: Table S1). A review protocol is available through the International Prospective Register of Systematic Reviews (PROSPERO) (CRD 42022313519).
Literature search and study selection
A structured search was performed independently by two reviewers (M.H. and J.R.) with 5 and 3 years of experience in gynecological imaging interpretation, respectively. Any disagreement was arbitrated by a third reviewer (Y.H.) with 12 years of experience, which was performed in 1.9% of the studies (8/418). The reviewers independently screened the titles, abstracts, and full texts of the extracted articles; uncertainties were discussed and resolved by consensus. Detailed search strategies and selection criteria are described in Additional file 1.
Data extraction and methodological quality assessment
After selecting the relevant studies, the two reviewers developed a data extraction instrument (described in Additional file 1: Table S2). After independently reading the full text of each eligible article, they documented: (1) bibliographical information, (2) baseline study information (including study design, imaging technique parameters, and modeling information), and (3) model performance metrics. The extracted information was recorded using the Excel software (Microsoft Corp., Redmond, WA, USA). The methodological quality of the eligible articles was independently assessed by the two reviewers using the RQS (described in Additional file 1: Table S3) [16] and Quality Assessment of QUADAS-2 tools [19]. Each of the 16 key components identified by the RQS was rated, resulting in a total score ranging from − 8 to 36 points; these were converted to RQS percentages, with − 8 to 0 points defined as 0% and 36 points defined as 100% [16]. The QUADAS-2 tool comprises seven assessment items reflecting four domains: ‘patient selection’, ‘index test’, ‘reference standard’, and ‘flow and timing’. Each item was judged as “low,” “high,” or “unclear” based on responses to signaling questions on the risk of bias and applicability concerns [19]. A summed RQS rating calculated by averaging the scores of the two reviewers was determined for each study. For QUADAS-2 assessment, any disagreement was arbitrated with a third reviewer (H.Y.) to achieve a common appraisal for each item. To facilitate additional analysis, a final risk assessment was performed for each study as follows: studies in which all seven items were rated “low” were defined as “low risk”, studies in which at least one item was rated “high” were defined as “high risk”, and the remainder were defined as “unclear risk”.
Data synthesis and analysis
Statistical analysis was conducted using the SPSS software version 25.0.0.0 (IBM Corp., Armonk, NY, USA), Review Manager (RevMan) version 5.3, and R (version 4.0.5; R Foundation for Statistical Computing) incorporating the ‘tidyverse’ packages. Categorical variables are presented as numbers and percentages, while continuous variables are presented as means and standard deviations or as medians and ranges. The inter-rater agreement for the RQS and QUADAS-2 was determined using Cohen’s kappa [21] and the ratio of agreements [21, 22]. An interclass correlation coefficient (ICC) of ≤ 0.4 was considered poor, 0.4–0.75 moderate, and > 0.75 good. Only training set performance metrics of the proposed radiomics models were recorded, even if validation sets existed, to maintain consistency among studies. Pairwise correlation analyses between methodological quality, baseline information, and performance metrics were conducted. Correlations between numeric variables were evaluated using linear regression analyses, while associations between numeric and categorical variables were assessed using unpaired Student’s t-tests or Mann–Whitney tests. The significance level α was 0.05, and statistical significance was set at a two-tailed p-value of < 0.05.
Meta-analyses were performed using the STATA software version 17.0 (StataCorp LP, College Station, TX, USA) with the ‘midas’ package when a sufficient number of studies attempted to address a similar question, and two-by-two tables could be extracted or reconstructed based on published data (the details are shown in Additional file 1). The sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) with 95% confidence intervals (CIs) were calculated. Summary receiver operating characteristic (SROC) analysis was performed, and the areas under the curve (AUCs) were obtained to describe the diagnostic accuracy. If a particular study involved multiple models, only the radiomics model was selected. Heterogeneity was assessed using Cochrane’s Q test and Higgin’s inconsistency index (I2) test. Any heterogeneity was considered significant if the p-value on Cochran’s Q test was < 0.05, whereupon the random effects model was used. Higgins I2 values of < 25%, 25–50%, and > 50% were associated with low, moderate, and substantial heterogeneity, respectively. In cases of the latter, meta-regression was performed to explore the possible sources of heterogeneity. Deeks’ funnel plots were constructed to illustrate the risk of publication bias.
Results
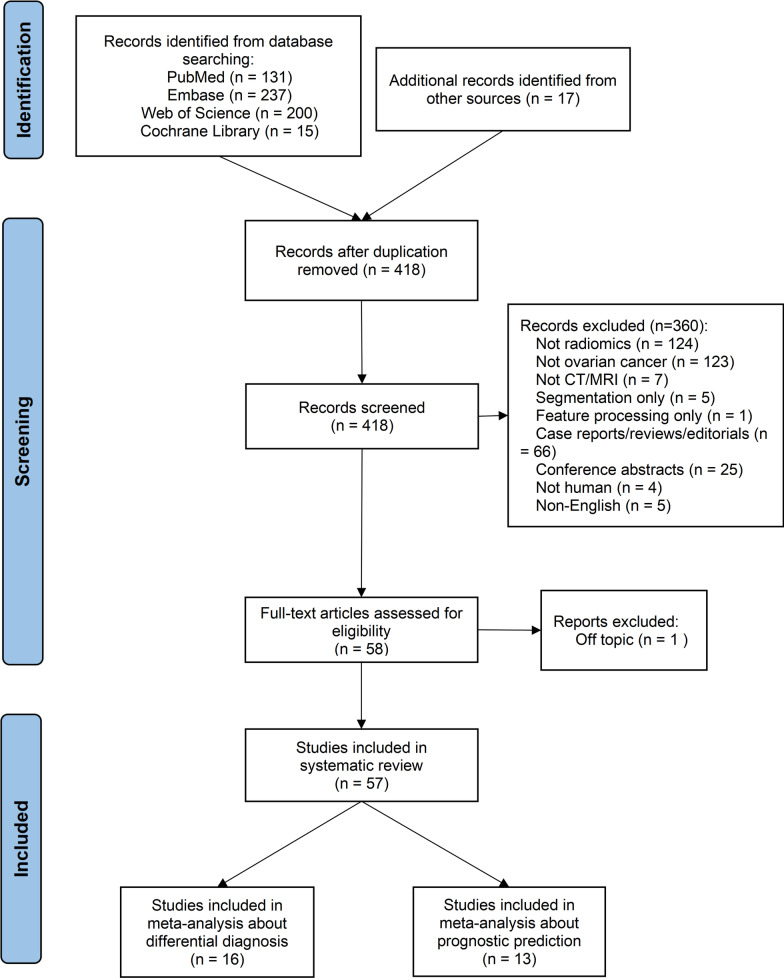
The study selection process is illustrated in Fig. 1; 57 articles were ultimately included in the systematic review [23–79]. We divided these publications based on the studies’ main objectives into three categories: differential diagnosis (24/57, 42.1%), response evaluation (4/57, 7.0%), and prediction of prognosis (28/57, 49.1%). Additionally, one paper (1.8%) described both differential diagnosis and prognostic prediction. We found that assessments of differential diagnosis and prognostic prediction were both commonly performed; thus, 16 articles focusing on the differential diagnosis of OC [24–26, 28-32, 34, 35, 39, 40, 42, 44, 45, 47] and 13 that described studies on prognostic factors [52, 53, 55–57, 64, 66–68, 71–74] were subjected to separate meta-analyses.
Fig. 1.
Flowchart of the study selection process for the systematic review and meta-analysis
Study characteristics
There were between 28 and 1329 patients in each study; their median or mean ages ranged from 38.5 to 75 years. The studies’ baseline information and characteristics are shown in Tables 1 and 2, respectively. Nearly half of the studies were published in imaging journals (28/57, 49.1%); the first authors were mainly specialized in radiology (36/57, 63.2%) and most came from Asia (43/57, 75.4%). Thirty-seven studies used CT and 20 used MRI; most applied the manual segmentation method and three-dimensional analysis. According to the model classification method proposed by the TRIPOD statement [80], a plurality of the studies established developed models validated with random splitting of data (27/57, 47.4%), followed by developed models validated using exactly the same data (10/57, 17.5%) and separate data (9/57, 15.8%). Furthermore, using Zhong et al. method [22], 59.6% of the studies were classified as phase 0 owing to their lack of external validation. Two phase III studies were conducted without sufficient patients or lacked external validation; however, they retained their phase categories owing to their prospective designs. Detailed characteristics of each study’s population and proposed radiomics model are presented in Additional file 1: Tables S4 and S5.
Table 1.
Baseline information of studies included in the systematic review
| StudyID | Research question | #Pat | FIGO stages | Imaging modality | Study design | Outcome | Validation | Reference standard | RQS (%) |
|---|---|---|---|---|---|---|---|---|---|
| Differential diagnosis | |||||||||
| Zheng2022 | Differentiating SBOTs and SMOTs |
D = 125 IV = 31 |
NR | MRI (fs-T2WI, DWI) | R | SBOT or SMOT | Yes (internal) | HP | 30.6 |
| Zhang2022 | Differentiating EOTs and MOTs |
D = 187 IV = 99 |
NR | ceCT (VP) | R | EOT or MOT | Yes (internal) | HP | 38.9 |
| Xu2022 | Differentiating: (1) BEOTs and EOCs; (2) type I and type II EOCs |
D = 89 IV = 114 D = 48 |
30I/82II | MRI (DWI, ADC) | R | BEOTs or EOCs; (2) type I or type II EOCs |
Yes (internal) No |
HP | 38.9 |
| Wei2022 | Differentiating benign and borderline EOTs |
D = 309 IV = 78 EV = 30 |
NR | MRI (T2WI) | R | benign or borderline EOTs | Yes (internal and external) | HP | 38.9 |
| M.Wang2022 | Differentiating HGSC and non-HGSC |
D = 532 IV = 133 |
NR | ceCT | R | HGSC or non-HGSC | Yes (internal) | HP | 34.8 |
| Nagawa2022 | Differentiating OTFGs and OGCTs | D = 53 | 17I/2II/2III | MRI (T2WI) | R | OTFG or OGCT | None | HP | 13.9 |
| LiuX2022 | Differentiating BEOTs and MEOTs |
D = 99 IV = 97 |
NR |
MRI (T2, fs-T2) |
R | BEOT or MEOT | Yes (internal) | HP | 22.2 |
| LiuP2022 | Differentiating benign and malignant ovarian tumors |
D = 96 IV = 39 |
NR | ceCT (AP) | R | Benign or malignant ovarian tumors | Yes (internal) | HP | 29.2 |
| LiS2022 | Differentiating benign and malignant ovarian tumors |
D = 99 IV = 41 |
NR | neCT | R | Benign or malignant ovarian tumors | Yes (internal) | HP | 38.9 |
| LiJ.1.2022 | Differentiating benign and malignant ovarian tumors |
D = 930 IV = 399 |
NR | ceCT (VP) | R | Benign or malignant ovarian tumors | Yes (internal) | HP | 33.3 |
| LiJ.2.2022 | Differentiating type I and type II EOCs |
D = 329 IV = 141 |
237I-II/233III-IV | ceCT (VP) | R | Type I or type II EOCs | Yes (internal) | HP | 38.9 |
| Zhu2021 | Differentiating EOCs and NEOCs | D(IV) = 101 | 28I/14II/57III/2IV | neCT | R | EOC or NEOC | Yes (internal) | HP | 38.9 |
| YuXP2021 | Differentiating SBOTs and SMOTs |
D = 127 IV = 55 |
117I /65II |
ceCT (AP, VP, EP) |
R | SBOT or SMOT | Yes (internal) | HP | 26.4 |
| Ye2021 | Differentiating BEOTs and FIGO stage I/II MEOTs |
D = 62 IV = 26 |
I-II |
MRI (T1, T2, DWI) |
R | BEOT or FIGO stage I/II MEOT | Yes (internal) | HP | 27.8 |
| Song.1.2021 | Differentiating benign, borderline, and malignant ovarian tumors | D + IV = 82 | NR |
MRI (DCE) |
P | Benign, borderline, or malignant ovarian tumors | Yes (internal) | HP | 55.6 |
| Park2021 | Differentiating benign and malignant ovarian tumors | D(IV) = 427 | NR | ceCT | R | Benign or malignant ovarian tumors | Yes (internal) |
Benign: (1) HP, (2) US or MRI, (3) stable after 2 years, or (4) resolved on subsequent imaging without treatment Malignant:HP |
33.3 |
| LiS2021 | Differentiating benign and malignant ovarian tumors |
D = 95 IV = 39 EV = 26 |
NR |
CeCT (VP) |
R | Benign or malignant ovarian tumors | Yes (internal and external) | HP | 41.7 |
| LiN2021 | Differentiating OGCTs and OTCA–FTCA | D = 46 | NR |
MRI (T2) |
R | OGCT or OTCA-FTCA | None | HP | 11.1 |
| Jian2021 | Differentiating type I and type II EOCs |
D = 144 IV = 75 EV = 75 |
NR |
MRI (T1, T2, DWI, ADC) |
R | Type I or type II EOC | Yes (internal and external) | HP | 30.6 |
| Hu2021 | Differentiating POCs and SOCs |
D = 76 IV = 34 |
NR |
ceCT (neCT, AP) |
R | POC or SOC | Yes (internal) | HP | 30.6 |
| An2021 | Differentiating HGSC and non-HGSC |
D = 163 IV = 42 |
44I/25II/106III/30IV |
ceCT (VP) |
R | HGSC or non-HGSC | Yes (internal) | HP | 31.9 |
| Qian2020 | Differentiating type I and type II EOCs | D(IV) = 61 | 26I-II/35III-IV |
MRI (fs-T2, DWI, DCE) |
R | Type I or type II EOC | Yes (internal) | HP | 44.4 |
| Lupean2020 | Differentiating benign and malignant ovarian cysts | D = 28 | NR |
MRI (T2) |
R | Benign or malignant ovarian cysts | None | HP | 5.6 |
| Li2020 | Differentiating BEOTs and MEOTs |
D = 250 IV = 92 EV = 159 |
NR |
MRI (T1, fs-T2, DWI, ADC) |
R | BEOT or MEOT | Yes (internal and external) | HP | 36.1 |
| Zhang2019 | Differentiating (1) benign and malignant ovarian tumors; (2) type I and type II EOC; |
D = 195 IV = 85 |
I-IV |
MRI (T1, T2, fs-T2, DWI) |
R |
(1) Benign or malignant ovarian tumors; (2) type I or type II EOC |
Yes (internal) | HP, immunohistological staining | 30.6 |
| Response evaluation | |||||||||
| Rundo2022 | Predicting response to neoadjuvant chemotherapy in HGSOC |
D = 61 IV = 48 |
77IIIC/32IV | ceCT | R | Non-complete response (CRS1-2) or complete response (CRS3) | Yes (internal) | CRS | 33.3 |
| Zargari2018 | Predicting tumor response to postsurgical chemotherapy in patients with advanced-stage ovarian cancer | D(IV) = 120 | NR |
ceCT (60 s) |
R | Response to postsurgical chemotherapy | Yes (internal) | RECIST 1.1 | 30.6 |
| Danala2017 | Predicting tumor response to chemotherapy in ovarian cancer patients | D = 91 | NR |
ceCT (60 s) |
R | Response to chemotherapy | None | RECIST 1.1 | 8.3 |
| Qiu2016 | Predicting early response of ovarian cancer patients to chemotherapy | D = 30 | NR |
ceCT (60 s, 5 min) |
R | Early response to chemotherapy | None | RECIST 1.1 | 0.0 |
| Prognosis prediction | |||||||||
| Wan2023 | Predicting CCR5 expression level and survival |
D = 57 IV = 89 |
NR | CT | R | CCR5 expression level, survival | Yes (internal) | CCR5 expression data from TCGA, OS | 50.0 |
| Wu2022 | Predicting early recurrence in patients with HGSOC |
D = 74 IV = 36 |
33I-II/77III-IV | ceCT (AP, VP) | R | Cancer recurrence | Yes (internal) | PFS | 33.3 |
| WangT2022 | Predicting EOC prognosis |
D = 130 IV = 56 |
85I/16II/76III/9IV |
MRI (T1, T2, DWI, CE-T1) |
R | Survival | Yes (internal) | DFS | 33.3 |
| Lu2022 | Predicting residual tumor in patients with HGSOC |
D = 106 IV = 22 |
5IIIA/5IIIB/82IIIC/36IV | MRI (T2WI, DWI, ADC) | R | RT status | Yes (internal) | RT status from operative reports | 38.9 |
| LiC2022 | Predicting recurrence in patients with HGSOC |
D = 98 IV = 43 |
19I-II/122III-IV | MRI (fs-T2WI, DWI, T1WI + C) | R | Postoperative recurrence | Yes (internal) | DFS | 33.3 |
| Hu2022 | Predicting OS and DFS in patients with HGSOC |
D = 95 IV = 90 EV = 32 |
32I/43II/121III/8IV | ceCT (VP) | R | Survival | Yes (internal and external) | OS, DFS | 30.6 |
| Hong2022 | Predicting survival in patients with serous ovarian cancer |
D = 80 EV = 39 |
13I/11II/66III/29IV | ceCT (VP) | R | Survival | Yes (external) | OS | 27.8 |
| Gao2022 | Predicting the expression of PD-1 and OS in OC patients |
PDCD1: D + IV = 57 Survival: IV = 89 |
PDCD1: NR Survival: 47II-III/42IV-unknown |
CT | R | PD-1 expression, survival | Yes (internal) | PD-1 expression status from TCGA-OV, OS | 38.9 |
| Fotopoulou2022 | Validating the prognostic value of RPV in patients with HGSOC | V = 547 | 15I/30II/2671III/227IV/8unknown | ceCT (VP) | R | Survival, operability | Yes (external) | PFS, OS, macroscopic tumor clearance | 36.1 |
| Feng2022 | Predicting hypoxia pattern in patient prognostication |
D = 40 IV = 19 |
NR | ceCT (AP) | R | Hypoxia pattern | Yes (internal) | nine-gene panel | 27.8 |
| Boehm2022 | Predicting risk stratification of HGSOC |
D = 298 IV = 40 |
NR | ceCT (VP) | R | Survival | Yes (internal) | OS, PFS | 34.7 |
| Avesani2022 | Predicting BRCA mutation and PFS in patients with HGSOC |
D = 152 EV = 66 |
5I/17II/1471III/45IV/4unknown | ceCT (VP) | R | BRCA mutation, Survival | Yes (external) | BRCA mutation status, PFS | 36.1 |
| YuXY2021 | Predicting peritoneal carcinomatosis in EOC patients before surgery | D = 86 | NR |
MRI (fs-T2, DWI, DCE) |
R | Peritoneal carcinomatosis | None | HP | 19.4 |
| Yi2021 | Predicting platinum resistance for OC treatment |
D = 71 IV = 31 |
8II/80III/14IV |
ceCT (neCT, VP) |
R | Platinum resistance | Yes (internal) | 6-month PFS | 36.1 |
| Song.2.2021 | Predicting peritoneal metastasis in ovarian cancer |
D = 54 IV = 35 |
NR |
MRI (T2, fs-T2, DWI) |
P | Peritoneal metastasis | Yes (internal) | HP | 61.1 |
| Liu2021 | Predicting BRCA gene mutation status in patients with EOC | D = 106 | NR |
ceCT (AP, VP, DP) |
R | BRCA mutation | Yes (internal) | NGS genetic testing | 36.1 |
| LiM2021 | Predicting BRCA gene mutation status in patients with advanced EOC |
95 (D:IV = 7:3) |
32III/63IV |
ceCT (AP, VP, DP) |
R | BRCA mutation | Yes (internal) | NGS genetic testing | 36.1 |
| LiH.1.2021 | Predicting RFS in patients with advanced HGSOC | D(IV) = 117 | III-IV |
MRI (CE-T1, T2) |
R | Survival | Yes (internal) | RFS | 27.8 |
| LiH.2.2021 | Predicting residual disease in patients with advanced HGSOC |
D = 160 IV = 57 |
III-IV |
MRI (CE-T1, T2) |
R | Residual disease | Yes (internal) | R0 resection | 23.6 |
| Chen.1.2021 | Predicting early recurrence in patients with HGSOC |
D = 179 IV = 77 |
55 I-II/201 III-IV |
ceCT (VP) |
R | Early recurrence | Yes (internal) | PFS | 33.3 |
| Chen.2.2021 | Predicting preoperative LN metastasis in patients with HGSOC |
D = 179 IV = 77 |
NR |
ceCT (VP) |
R | Preoperative LN metastasis | Yes (internal) | HP | 41.7 |
| Ai2021 | Predicting metastatic status of OC patients |
D = 70 IV = 31 |
29I/14II/57III/1IV | neCT | R | Preoperative metastasis | Yes (internal) | HP | 22.2 |
| Veeraraghavan2020 | Predicting PFS and platinum resistance in patients with HGSOC |
D(IV) = 40 EV = 35 |
58III/17IV | ceCT | R | Survival, platinum resistance | Yes (internal and external) | PFS | 36.1 |
| Wei2019 | Predicting risk for postoperative advanced HGSOC recurrence |
D = 50 IV = 50 EV = 42 |
112III/30IV |
ceCT (neCT, AP, VP) |
R | Postoperative recurrence | Yes (internal and external) | PFS | 38.9 |
| Meier2019 | Predicting survival and BRCA mutation status in patients with HGSOC | D = 88 | NR |
ceCT (VP) |
R | Survival, BRCA mutation | None | OS, PFS, BRCA mutation status | 0.0 |
| Lu2019 | Predicting prognostic- and molecular-phenotypes of EOC |
D = 136 IV = 77 EV = 70 |
53I-II/223III-IV/18 unknown | ceCT | R | Survival, molecular-phenotypes | Yes (internal and external) | PFS, OS | 40.3 |
| Zhang2019* | Predicting survival among EOC patients |
D = 195 IV = 85 |
I-IV |
MRI (T1, T2, fs-T2, DWI) |
R | Survival | Yes (internal) | DFS | 30.6 |
| Rizzo2018 | Predicting residual tumor at surgery and the risk of PD12 in OC patients | D = 101 | 11II/66III/24IV |
ceCT (VP) |
R | Residual tumor, risk of PD12 | None | RT, PD12 | 8.3 |
| Vargas2017 | Predicting outcomes in patients with HGSOC | D = 38 | 23III/15IV |
ceCT (70 s) |
R | Surgical resection, survival | None | Surgical resection status, molecular analysis, OS | 12.5 |
One study discussed two topics and was described twice, which was marked with “*”
BEOT borderline epithelial ovarian tumor, MEOT malignant epithelial ovarian tumor, EOC epithelial ovarian cancer, NEOC non-epithelial ovarian cancer, SBOT serous borderline ovarian tumor, SMOT serous malignant ovarian tumor, MOT metastatic ovarian tumor, EOT epithelial ovarian tumor, OTFG ovarian granulosa cell tumor, OGCT ovarian granulosa cell tumor, OTCA–FTCA thecoma–fibrothecoma, POC primary ovarian cancer, SOC secondary ovarian cancer, HGSC high-grade serous carcinoma, HGSOC high-grade serous ovarian carcinoma, OC ovarian cancer, RPV Radiomic Prognostic Vector, D development, IV internal validation, EV external validation, V validation, AP arterial phase, VP venous phase, EP equilibrium phase, DP delay phase, R retrospective, P prospective, CRS chemotherapy response score, LN lymph node, PD12 disease progression within 12 months, HP histopathology, DFS disease-free survival, PFS progression-free survival, RFS recurrence-free survival, OS overall survival, RT residual tumor, NRnot reported, NA not applicable
Table 2.
Characteristics of included studies
| Characteristics | No. of studies (%) |
|---|---|
| Journal speciality | |
| Imaging | 28/57 (49.1%) |
| Clinical | 20/57 (35.1%) |
| Generalist | 7/57 (12.3%) |
| Medical physics | 2/57 (3.5%) |
| Specialty of the 1st author* | |
| Radiologist | 36/57 (63.2%) |
| Radiotherapist | 1/57 (1.8%) |
| Clinician | 11/57 (19.3%) |
| Physicist, Computer scientist | 9/57 (15.8%) |
| Geographical origin of the study | |
| Asia | 43/57 (75.4%) |
| Europe | 6/57 (10.5%) |
| North America | 8/57 (14.0%) |
| Imaging modality | |
| CT | 37/57 (64.9%) |
| MRI | 20/57 (35.10%) |
| Study objectives** | |
| Differential diagnosis | 25/57 (43.9%) |
| Response evaluation | 4/57 (7.0%) |
| Prognosis prediction | 29/57 (50.9%) |
| Segmentation method | |
| Manual | 53/57 (93.0%) |
| Semi-automatic | 4/57 (7.0%) |
| Publication of negative results | |
| No | 55/57 (96.5%) |
| Yes | 2/57 (3.5%) |
| Type of ROI | |
| 2D | 11/57 (19.3%) |
| 3D | 43/57 (75.4%) |
| 2D&3D | 3/57 (5.3%) |
| Model type | |
| Type 1a: developed model validated with exactly the same data | 10/57 (17.5%) |
| Type 1b: developed model validated with resampling data | 6/57 (10.5%) |
| Type 2a: developed model validated with randomly splitting data | 27/57 (47.4%) |
| Type 2b: developed model validated with non-randomly splitting data | 4/57 (7.0%) |
| Type 3: developed model validated with separate data | 9/57 (15.8%) |
| Type 4: validation only | 1/57 (1.8%) |
| Phase classification*** | |
| Discovery science: experimental | 10/57 (17.5%) |
| Phase 0: < 100 patients; retrospective; internal validation Or > 100 patients; retrospective; internal validation | 34/57 (59.6%) |
| Phase I: < 100 patients; retrospective; external validation | 2/57 (3.5%) |
| Phase II: > 100 patients; retrospective; external validation | 9/57 (15.8%) |
| Phase III: > 100 patients; prospective; external validation | 2/57 (3.5%) |
| Phase IV: real world | 0/57 |
*Thirteen studies claimed joint first author, the author whose name was written first was assessed
**One study discussed two topics
***Two studies were classified as phase III due to prospective design, although they were conducted with less than 100 patients and without external validation
Methodological quality assessment
The 57 studies had a mean RQS of 12 (interquartile range 10–14), with RQS values ranging from − 4 to 22. The mean percentage RQS was 30.7%, with a maximum of 61.1%. The average ICC for the RQS was 0.80 (95% confidence interval (CI) 0.69–0.91), i.e., a ‘good’ rating. The average rating and inter-rater agreement per RQS component are shown in Table 3. In most studies, the lack of reproducibility and repeatability analysis of imaging features (e.g., phantom study and imaging at multiple time points), insufficient cost-effectiveness analyses, and inadequate access to the data led to a low RQS. Additionally, biological correlation and cutoff analyses were seldom performed in the aftermath, and the rated validation score (2 [2]) was suboptimal. Discrimination statistics to assess model performance were available for all studies; 21 of them also reported calibration statistics. Fifty-five studies (96.5%) were retrospective analyses, while only two (3.5%) were based on prospectively acquired data. The RQS assessments by each reviewer (M.H. and J.R.) are shown in Additional file 1: Table S6.
Table 3.
Average rating and inter-rater agreement per component of RQS
| No. | RQS item | Range | Average | Median (range) | Inter-rater agreement | |
|---|---|---|---|---|---|---|
| ICC | 95%CI | |||||
| 1 | Image protocol quality | 0–2 | 0.89 | 1 (0–2) | 0.90 | 0.76–1.00 |
| 2 | Multiple segmentations | 0–1 | 0.63 | 1 (0–1) | 1.00 | 1.00–1.00 |
| 3 | Phantom study on all scanners | 0–1 | 0.05 | 0 (0–1) | 1.00 | 1.00–1.00 |
| 4 | Imaging at multiple time points | 0–1 | 0 | 0 (0–0) | 1.00 | 1.00–1.00 |
| 5 | Feature reduction or adjustment for multiple testing | − 3 to 3 | 2.79 | 3 (− 3 to 3) | 1.00 | 1.00–1.00 |
| 6 | Multivariable analysis with non-radiomics features | 0–1 | 0.72 | 1 (0–1) | 0.91 | 0.80–1.00 |
| 7 | Detect and discuss biological correlate | 0–1 | 0.23 | 0 (0–1) | 0.90 | 0.77–1.00 |
| 8 | Cutoff analyses | 0–1 | 0.13 | 0 (0–1) | 0.77 | 0.52–1.00 |
| 9 | Discrimination statistics | 0–2 | 1.42 | 0 (0.5–2) | 0.72 | 0.54–0.90 |
| 10 | Calibration statistics | 0–2 | 0.42 | 0 (0–2) | 1.00 | 1.00–1.00 |
| 11 | Prospective study registered in a trial database | 0–7 | 0.25 | 0 (0–7) | 1.00 | 1.00–1.00 |
| 12 | Validation | − 5 to 5 | 1.39 | 2 (− 5 to 5) | 0.94 | 0.85–1.00 |
| 13 | Comparison to 'gold standard' | 0–2 | 1.26 | 2 (0–2) | 0.93 | 0.82–1.00 |
| 14 | Potential clinical utility | 0–2 | 0.70 | 0 (0–2) | 1.00 | 1.00–1.00 |
| 15 | Cost-effectiveness analysis | 0–1 | 0 | 0 (0–0) | 1.00 | 1.00–1.00 |
| 16 | Open science and data | 0–4 | 0.15 | 0 (0 ≥ 2) | 0.92 | 0.78–1.00 |
| Total points: − 8 to 0 = 0%, 36 = 100% | 0–100% | 11.04 = 30.7% | 12 (− 4 to 22) | 0.80 | 0.69–0.91 | |
CI confidence interval, ICC interclass correlation coefficient, RQS Radiomics Quality Score
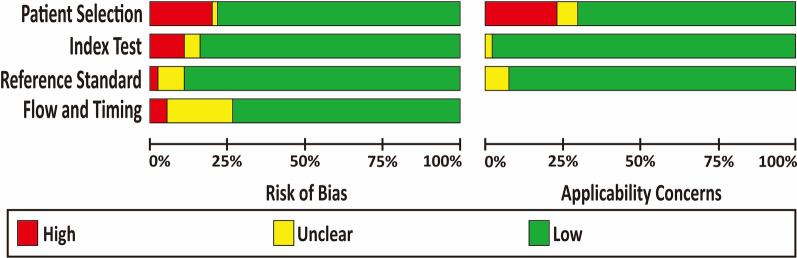
The summarized and individual QUADAS-2 results are presented in Fig. 2 and Additional file 1: Table S7. A high risk of publication bias (19.3%) and applicability concerns (22.8%) were observed in terms of patient selection, mainly because of inappropriate exclusion. Twelve studies (21.1%) were regarded as having an unclear risk of bias in flow and timing, as they did not provide sufficient information regarding the interval between index tests and reference standards. Complete agreement between the two reviewers concerning the seven QUADAS-2 items ranged from 80.7 to 96.5%.
Fig. 2.
Quality assessment of included studies by QUADAS-2 tool. The authors’ judgments for each domain of each included study were reviewed. The proportion of included studies that indicated low, unclear, high risk, and applicability concerns were shown in green, yellow and gray, respectively
Pairwise correlation analysis of methodological quality, baseline information, and performance metrics
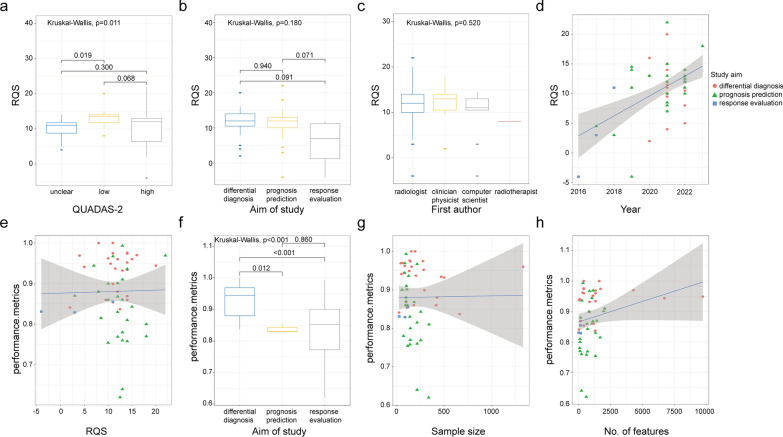
The pairwise correlation analysis results are presented in Fig. 3. One article that discussed both differential diagnosis and prognostic prediction was analyzed twice as two separate studies. The RQS values were significantly different between studies with different QUADAS-2 risk assessments (p = 0.011). Importantly, studies deemed low risk had a higher mean RQS than those with high or unclear risk; the difference between low-risk and unclear-risk studies was significant (p = 0.002). The study aim (p = 0.180) and specialty of the first author (p = 0.520) did not influence the RQS rating. Studies published more recently received significantly higher scores (adjusted R2 = 0.264, p < 0.001).
Fig. 3.
Correlations between radiomics quality score (RQS), performance metrics and baseline information. a–d Correlation between RQS and QUADAS-2, study aim, first author, and publication year. The vertical dashed line corresponds to the year of publication of the RQS. e–h Correlation between performance metrics and RQS, study aim, number of patients, and number of features. Each point corresponds to a study. The regression line and its 95% confidence interval are shown in gray with adjusted R2 and p-value
The performance metrics were represented by the AUC in 49 studies (86.0%) and by the c-index in three (5.3%); they were missing in the remaining five (8.8%). Two of the studies explored two radiomics models with different aims; hence, their performance metrics were documented separately. The best radiomics model used in each study had an AUC or c-index value between 0.620 and 1.000. Moreover, the performance of the radiomics model was closely related to the study aim (p < 0.001); performance was significantly better in studies exploring differential diagnosis than in those evaluating response (p = 0.012) or predicting prognosis (p < 0.001). No significant correlation was found between performance metrics and RQS (adjusted R2 = − 0.019, p = 0.892), sample size (adjusted R2 = − 0.019, p = 0.938), or number of features extracted (adjusted R2 = 0.046, p = 0.068).
Meta-analysis
Sixteen studies that focused on differential diagnosis and 13 that investigated prognostic prediction were subjected to a meta-analysis. For studies of differential diagnosis, the pooled diagnostic odds ratio (DOR) was 25.76 (95% CI 13.50–49.13) (Fig. 4a); the pooled sensitivity, specificity, positive likelihood ratio (PLR), and negative likelihood ratio (NLR) were 0.84 (95% CI 0.76–0.89), 0.83 (95% CI 0.77–0.88), 5.00 (95% CI 3.58–6.97), and 0.19 (95% CI 0.13–0.30), respectively (Additional file 1: Figs. S1a and Fig. S2a). The SROC curve suggested good diagnostic performance with an AUC of 0.90 (95% CI 0.87–0.92) (Additional file 1: Fig. S3a). For studies analyzing prognostic prediction, the pooled DOR, sensitivity, specificity, PLR, and NLR were 12.55 (95% CI 8.38–18.77), 0.78 (95% CI 0.71–0.83), 0.78 (95% CI 0.72–0.82), 3.59 (95% CI 2.80–4.59), and 0.29 (95% CI 0.22–0.37), respectively (Fig. 4b, Additional file 1: Figs. S1b, and S2b). The AUC calculated from the SROC curve was 0.85 (95% CI 0.81–0.88), indicating good performance of the prognostic prediction models (Additional file 1: Fig. S3b). Per Deeks’ funnel plots, the likelihood of publication bias was low for differential diagnosis studies (p = 0.760) but high for prognostic prediction studies (p = 0.040) (Additional file 1: Fig. S4).
Fig. 4.
Forrest plot of the effect size calculated as diagnostic odds ratio for studies discussing the diagnostic accuracy of radiomics models in the a differential diagnosis; b prognosis prediction of ovarian tumors. The numbers are pooled estimates with 95% CIs in parentheses; horizontal lines indicate 95% CIs. TP, FP, FN, and TN were defined according to the original articles' descriptions
Cochrane’s Q test (p < 0.001 and p < 0.001) and Higgin’s I2 test (I2 = 94% and I2 = 91%) indicated high heterogeneity; therefore, a meta-regression analysis was conducted to identify its source (Table 4 and Additional file 1: Fig. S5). The number of patients was significantly associated with heterogeneity in specificity (p < 0.001) for differential diagnosis studies and contributed to heterogeneity in sensitivity (p < 0.001), specificity (p < 0.001), and the AUC (p = 0.04) in prognostic prediction studies. Additionally, the imaging method (CT vs. MRI) influenced heterogeneity in both sensitivity (p < 0.001 for differential diagnosis and p < 0.01 for prognostic prediction studies) and specificity (p < 0.01 for both study types). The heterogeneity of sensitivity and specificity in the differential diagnosis subgroup was associated with the type of region of interest (p = 0.04 for sensitivity and p < 0.01 for specificity). In the prognostic prediction subgroup, however, the region of interest type influenced AUC (p = 0.03), sensitivity (p < 0.001), and specificity (p < 0.01). Meanwhile, the type of features only contributed to heterogeneity in specificity (p < 0.01 for both differential diagnosis and prognostic prediction).
Table 4.
The results of meta-regression analysis of studies about differential diagnosis and prognosis prediction of OC
| Covariates | Subgroup | No. of studies | Sensitivity [95%CI] | PSEN | Specificity [95%CI] | PSPE | P |
|---|---|---|---|---|---|---|---|
| Differential diagnosis | |||||||
| Number of patients | < 100 | 11 | 0.83 [0.75–0.92] | 0.15 | 0.86 [0.80–0.91] | < 0.001 | 0.42 |
| ≥ 100 | 5 | 0.84 [0.74–0.95] | 0.78 [0.69–0.88] | ||||
| Imaging modality | MRI | 8 | 0.89 [0.82–0.95] | < 0.001 | 0.85 [0.78–0.93] | < 0.01 | 0.17 |
| CT | 8 | 0.78 [0.68–0.88] | 0.81 [0.73–0.89] | ||||
| Type of ROI | 2D | 7 | 0.85 [0.76–0.95] | 0.04 | 0.84 [0.75–0.92] | < 0.01 | 0.92 |
| 3D | 9 | 0.83 [0.74–0.92] | 0.83 [0.76–0.90] | ||||
| Type of features | Low-order | 12 | 0.83 [0.75–0.91] | 0.26 | 0.84 [0.78–0.90] | < 0.01 | 0.77 |
| High-order | 4 | 0.86 [0.75–0.97] | 0.81 [0.71–0.91] | ||||
| Prognosis prediction | |||||||
| Number of patients | < 100 | 9 | 0.79 [0.73–0.85] | < 0.001 | 0.82 [0.76–0.87] | < 0.001 | 0.04 |
| ≥ 100 | 4 | 0.73 [0.64–0.82] | 0.73 [0.64–0.82] | ||||
| Imaging modality | MRI | 5 | 0.79 [0.71–0.88] | < 0.01 | 0.79 [0.70–0.87] | < 0.01 | 0.85 |
| CT | 8 | 0.76 [0.69–0.84] | 0.78 [0.71–0.85] | ||||
| Type of ROI | 2D | 4 | 0.84 [0.77–0.92] | < 0.001 | 0.82 [0.74–0.91] | < 0.01 | 0.03 |
| 3D | 9 | 0.74 [0.68–0.80] | 0.76 [0.69–0.83] | ||||
| Type of features | Low-order | 6 | 0.74 [0.65–0.82] | 0.09 | 0.80 [0.70–0.85] | < 0.01 | 0.46 |
| High-order | 7 | 0.81 [0.74–0.88] | 0.77 [0.70–0.85] | ||||
Discussion
Our systematic review found that the methodological quality of CT and MRI radiomics for patients with OC was relatively low, while our meta-analysis revealed that radiomics has promising potential in discriminating between OC subtypes and predicting patient prognosis.
Several narrative reviews described the prospects of applying radiomics in OC [8, 17, 81, 82]. Rizzo et al.’s systematic review [83] included six studies that lacked RQS ratings and meta-analyses, whereas that by Ponsiglione et al. [18] used the RQS to evaluate the methodological quality of studies involving CT-, MRI-, ultrasonography-, or positron emission tomography-based radiomics in ovarian disorders published as of November 2021. The field of radiomics is in a period of rapid growth; 42.1% of the studies we analyzed were published in 2022 or 2023. Aside from the RQS, QUADAS-2 was applied to assess potential publication bias and applicability concerns. While previously published reviews usually describe only qualitative analyses, we performed quantitative evaluation of the effect of radiomics models by conducting a meta-analysis; our mean RQS (30.7%) was acceptable when compared to those published previously (5.6–36.1%) [18, 22, 84-87]. However, the overall scientific quality was still unsatisfactory, given that the assessment values were considerably below 100%. The main reasons for the low RQS ratings, as in previous analyses [22, 86, 88], included low levels of feature robustness, insufficient model assessment, and lack of concern for clinical applications.
The primary challenge for feature robustness was the high variability in radiomics features; few of the publications we analyzed described phantom studies [74, 77, 78], multiple time-point imaging, or automatic segmentation [49–51]. Regarding model assessment, radiomics studies usually included discrimination statistics; however, cutoff analyses and calibration statistics were often neglected, thus complicating the risk evaluation of overly optimistic results and accuracy of model predictions. Classification indexes including sensitivity and specificity, which are critical for quantitative analysis, were lacking in some studies. Our results suggest the need for performing additional technical validation before considering radiomics models for clinical applications.
Although most studies involved internal validation (which is indispensable for the clinical translation and broad application of radiomics models), the absence of external validation at several centers undermined the credibility and generalizability of the models. Very few prospective studies [37, 66], which are considered to have a high level of evidence, were performed. Furthermore, analysis of cost-effectiveness was absent from all our included studies, and the lack of decision curve analysis in approximately two-thirds of them also obscured the applicability of the models to clinical settings. As such, technical and clinical validations remain critical unmet requirements for incorporating radiomics analysis into clinical applications.
The importance of open science is emphasized in the RQS ratings to ensure the transparency and reproducibility of research findings, but such access was lacking in our studies. As such, we suggest that researchers should at least include the computed numerical values of any investigated features in their publications.
In terms of correlations between the RQS and original variables, there was a significant relationship between RQS and publication year. More modern studies presumably have a larger sample size, collect data from multiple centers, use a prospective design, and/or apply more strict inclusion and exclusion criteria, thereby contributing to a higher RQS. Additionally, a higher RQS was significantly correlated with low-level risk according to QUADAS-2, which may support using the former to improve research quality. We also found that studies exploring differential diagnosis had significantly higher performance metrics, whereas studies of response evaluation or prognostic prediction may be more affected by non-relevant factors, such as lifestyle and other medications, given that they have longer follow-up periods. Contrary to our expectation, we did not find a significant relationship between performance metrics and the number of features extracted, which could be explained by the heterogeneity in feature selection and modeling.
The results of our meta-analysis were promising; the pooled AUCs reached 0.90 for differential diagnosis and 0.85 for prognostic prediction, indicating a relatively high accuracy in categorizing different pathological subtypes and predicting the prognoses of patients with OC. According to Cochrane’s Q and Higgin’s I2 tests, studies included in the meta-analysis had high levels of heterogeneity, which a meta-regression analysis attributed to the influence of patient number, imaging modality, region of interest, and feature type.
Our study had some limitations. First, most included articles did not report the numbers of true/false positives or true/false negatives, and our calculation of these data from the available information might have introduced some errors. Second, studies predicting chemotherapy response were not included in the meta-analysis owing to insufficient data. Third, studies involved in the meta-analysis showed high heterogeneity; although we found significant correlations between such heterogeneity and certain factors, the latter may not have sufficiently explained the former. Fourth, the prognostic prediction studies included in our meta-analysis had a significant risk of publication bias, likely because we only included English-language articles and also because half of the studies comprised small sample sizes; reluctance to publish negative data may also have been a factor.
In conclusion, radiomics analysis showed promise in terms of overcoming some current obstacles in determining differential diagnosis, chemotherapy response, and prognosis in patients with OC. Pairwise correlation analysis revealed a significant relationship between RQS and QUADAS-2 result or publication year, as well as between performance metrics and study aims. Additionally, our meta-analysis demonstrated the suitability of radiomics analysis for discriminating between various subtypes of OC and identifying prognostic factors through quantitative analysis. Given the generally low RQS ratings of all the included studies, the methodological quality of radiomics studies involving OC is lower than desirable; hence, more high-level evidence is required to develop effective radiomics models.
Supplementary Information
Additional file 1. Electronic Supplementary Materials.
Abbreviations
- AUC
Area under the curve
- CI
Confidence interval
- DOR
Diagnostic odds ratio
- ICC
Interclass correlation coefficient
- NLR
Negative likelihood ratio
- OC
Ovarian cancer
- PLR
Positive likelihood ratio
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-analysis
- PROSPERO
International Prospective Register of Systematic Reviews
- QUADAS-2
Quality Assessment of Diagnostic Accuracy Studies 2
- RQS
Radiomics quality score
- SROC
Summary receiver operating characteristic
Author contributions
ZYJ, HDX, YL and YLH contributed to conceptualization; HDX, YLH, MLH and JR were involved in protocol finalization; JR and XYL were involved in technical contributions; JR, XYL and MLH contributed to article selection; YLH was involved in article consensus; MLH, JR and YLH contributed to manuscript writing; MLH, JR and XYL were involved in visualization; HDX, YL and YLH contributed to writing—review and editing; YLH, ZYJ and HDX were involved in supervision. All authors read and approved the final manuscript.
Funding
This study has received funding by grants from Natural Science Foundation of China (Grant No. 81901829), National High Level Hospital Clinical Research Funding (Grant No. 2022-PUMCH-A-004) and Natural Science Foundation of China (Grant No. 82271886).
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its Additional files].
Declarations
Ethics approval and consent to participate
Institutional Review Board approval was not required because our data are extracted from existing manuscripts.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Meng-Lin Huang and Jing Ren contributed equally to this work.
Contributor Information
Yong-Lan He, Email: heyonglan@pumch.cn.
Yuan Li, Email: liyuan10833@pumch.cn.
Hua-Dan Xue, Email: bjdanna95@hotmail.com.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 3.WHO classification of Tumours Editorial Board (2020) WHO classification of tumours. In: Female genital tumours, 5th ed. IARC Press, Lyon
- 4.Park HK, Ruterbusch JJ, Cote ML. Recent trends in ovarian cancer incidence and relative survival in the United States by race/ethnicity and histologic subtypes. Cancer Epidemiol Biomark Prev. 2017;26:1511–1518. doi: 10.1158/1055-9965.EPI-17-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network (2023) Ovarian cancer (Version 1.2023). Available via https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf. Accessed 3 Jan 2023
- 6.Peres LC, Cushing-Haugen KL, Köbel M, et al. Invasive epithelial ovarian cancer survival by histotype and disease stage. J Natl Cancer Inst. 2019;111:60–68. doi: 10.1093/jnci/djy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaona-Luviano P, Medina-Gaona LA, Magaña-Pérez K. Epidemiology of ovarian cancer. Chin Clin Oncol. 2020;9:47–47. doi: 10.21037/cco-20-34. [DOI] [PubMed] [Google Scholar]
- 8.Nougaret S, McCague C, Tibermacine H, Vargas HA, Rizzo S, Sala E. Radiomics and radiogenomics in ovarian cancer: a literature review. Abdom Radiol (NY) 2021;46:2308–2322. doi: 10.1007/s00261-020-02820-z. [DOI] [PubMed] [Google Scholar]
- 9.Lee JM, Minasian L, Kohn EC. New strategies in ovarian cancer treatment. Cancer. 2019;125:4623–4629. doi: 10.1002/cncr.32544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara K, Hasegawa K, Nagao S. Landscape of systemic therapy for ovarian cancer in 2019: primary therapy. Cancer. 2019;125:4582–4586. doi: 10.1002/cncr.32475. [DOI] [PubMed] [Google Scholar]
- 11.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 12.Arend R, Martinez A, Szul T, Birrer MJ. Biomarkers in ovarian cancer: To be or not to be. Cancer. 2019;125:4563–4572. doi: 10.1002/cncr.32595. [DOI] [PubMed] [Google Scholar]
- 13.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 14.Faggioni L, Coppola F, Ferrari R, Neri E, Regge D. Usage of structured reporting in radiological practice: results from an Italian online survey. Eur Radiol. 2017;27:1934–1943. doi: 10.1007/s00330-016-4553-6. [DOI] [PubMed] [Google Scholar]
- 15.Granata V, Faggioni L, Grassi R, et al. Structured reporting of computed tomography in the staging of colon cancer: a Delphi consensus proposal. Radiol Med. 2022;127:21–29. doi: 10.1007/s11547-021-01418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 17.Arezzo F, Loizzi V, La Forgia D, et al. Radiomics analysis in ovarian cancer: a narrative review. Appl Sci Basel. 2021;11:7833. doi: 10.3390/app11177833. [DOI] [Google Scholar]
- 18.Ponsiglione A, Stanzione A, Spadarella G, et al. Ovarian imaging radiomics quality score assessment: an EuSoMII radiomics auditing group initiative. Eur Radiol. 2022 doi: 10.1007/s00330-022-09180-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 20.McInnes MDF, Moher D, Thombs BD, et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies. JAMA. 2018;319:388. doi: 10.1001/jama.2017.19163. [DOI] [PubMed] [Google Scholar]
- 21.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 22.Zhong J, Hu Y, Si L, et al. A systematic review of radiomics in osteosarcoma: utilizing radiomics quality score as a tool promoting clinical translation. Eur Radiol. 2021;31:1526–1535. doi: 10.1007/s00330-020-07221-w. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Y, Wang H, Li Q, Sun H, Guo L. Discriminating between benign and malignant solid ovarian tumors based on clinical and radiomic features of MRI. Acad Radiol. 2022 doi: 10.1016/j.acra.2022.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Zhang A, Hu Q, Ma Z, Song J, Chen T. Application of enhanced computed tomography-based radiomics nomogram analysis to differentiate metastatic ovarian tumors from epithelial ovarian tumors. J Xray Sci Technol. 2022;30:1185–1199. doi: 10.3233/XST-221244. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Luo HJ, Ren J, Guo LM, Niu J, Song X. Diffusion-weighted imaging-based radiomics in epithelial ovarian tumors: assessment of histologic subtype. Front Oncol. 2022;12:978123. doi: 10.3389/fonc.2022.978123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei M, Zhang Y, Bai G, et al. T2-weighted MRI-based radiomics for discriminating between benign and borderline epithelial ovarian tumors: a multicenter study. Insights Imaginging. 2022;13:130. doi: 10.1186/s13244-022-01264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, Perucho JAU, Hu Y, et al. Computed tomographic radiomics in differentiating histologic subtypes of epithelial ovarian carcinoma. JAMA Netw Open. 2022;5:e2245141. doi: 10.1001/jamanetworkopen.2022.45141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagawa K, Kishigami T, Yokoyama F, et al. Diagnostic utility of a conventional MRI-based analysis and texture analysis for discriminating between ovarian thecoma-fibroma groups and ovarian granulosa cell tumors. J Ovarian Res. 2022;15:65. doi: 10.1186/s13048-022-00989-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Wang T, Zhang G, et al. Two-dimensional and three-dimensional T2 weighted imaging-based radiomic signatures for the preoperative discrimination of ovarian borderline tumors and malignant tumors. J Ovarian Res. 2022;15:22–22. doi: 10.1186/s13048-022-00943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P, Liang X, Liao S, Lu Z. Pattern classification for ovarian tumors by integration of radiomics and deep learning features. Curr Med Imaging. 2022;18:1486–1502. doi: 10.2174/1573405618666220516122145. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Liu J, Xiong Y, et al. Application values of 2D and 3D radiomics models based on CT plain scan in differentiating benign from malignant ovarian tumors. Biomed Res Int. 2022;2022:5952296. doi: 10.1155/2022/5952296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Zhang T, Ma J, Zhang N, Zhang Z, Ye Z. Machine-learning-based contrast-enhanced computed tomography radiomic analysis for categorization of ovarian tumors. Front Oncol. 2022;12:934735. doi: 10.3389/fonc.2022.934735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Li X, Ma J, Wang F, Cui S, Ye Z. Computed tomography-based radiomics machine learning classifiers to differentiate type I and type II epithelial ovarian cancers. Eur Radiol. 2022 doi: 10.1007/s00330-022-09318-w. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H, Ai Y, Zhang J, et al. Preoperative nomogram for differentiation of histological subtypes in ovarian cancer based on computer tomography radiomics. Front Oncol. 2021;11:642892. doi: 10.3389/fonc.2021.642892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X-p, Wang L, Yu H-y, et al. MDCT-based radiomics features for the differentiation of serous borderline ovarian tumors and serous malignant ovarian tumors. Cancer Manag Res. 2021;13:329–336. doi: 10.2147/CMAR.S284220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye R, Weng S, Li Y, et al. Texture analysis of three-dimensional MRI images may differentiate borderline and malignant epithelial ovarian tumors. Korean J Radiol. 2021;22:106–117. doi: 10.3348/kjr.2020.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song X-l, Ren J-L, Zhao D, Wang L, Ren H, Niu J. Radiomics derived from dynamic contrast-enhanced MRI pharmacokinetic protocol features: the value of precision diagnosis ovarian neoplasms. Eur Radiol. 2021;31:368–378. doi: 10.1007/s00330-020-07112-0. [DOI] [PubMed] [Google Scholar]
- 38.Park H, Qin L, Guerra P, Bay CP, Shinagare AB. Decoding incidental ovarian lesions: use of texture analysis and machine learning for characterization and detection of malignancy. Abdom Radiol (NY) 2021;46:2376–2383. doi: 10.1007/s00261-020-02668-3. [DOI] [PubMed] [Google Scholar]
- 39.Li S, Liu J, Xiong Y, et al. A radiomics approach for automated diagnosis of ovarian neoplasm malignancy in computed tomography. Sci Rep. 2021;11:8730. doi: 10.1038/s41598-021-87775-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li NY, Shi B, Chen YL, et al. The value of MRI findings combined with texture analysis in the differential diagnosis of primary ovarian granulosa cell tumors and ovarian thecoma-fibrothecoma. Front Oncol. 2021;11:758036. doi: 10.3389/fonc.2021.758036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jian J, Li Y, Pickhardt PJ, et al. MR image-based radiomics to differentiate type Iota and type Iota Iota epithelial ovarian cancers. Eur Radiol. 2021;31:403–410. doi: 10.1007/s00330-020-07091-2. [DOI] [PubMed] [Google Scholar]
- 42.Hu Y, Weng Q, Xia H, et al. A radiomic nomogram based on arterial phase of CT for differential diagnosis of ovarian cancer. Abdom Radiol (NY) 2021;46:2384–2392. doi: 10.1007/s00261-021-03120-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An H, Wang Y, Wong EMF, et al. CT texture analysis in histological classification of epithelial ovarian carcinoma. Eur Radiol. 2021;31:5050–5058. doi: 10.1007/s00330-020-07565-3. [DOI] [PubMed] [Google Scholar]
- 44.Qian L, Ren J, Liu A, et al. MR imaging of epithelial ovarian cancer: a combined model to predict histologic subtypes. Eur Radiol. 2020;30:5815–5825. doi: 10.1007/s00330-020-06993-5. [DOI] [PubMed] [Google Scholar]
- 45.Lupean R-A, Sefan P-A, Feier DS, et al. Radiomic analysis of MRI images is instrumental to the stratification of ovarian cysts. J Personal Med. 2020;10:127. doi: 10.3390/jpm10030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ya Li, Jian J, Pickhardt PJ, et al. MRI-based machine learning for differentiating borderline from malignant epithelial ovarian tumors: a multicenter study. J Magn Reson Imag. 2020;52:897–904. doi: 10.1002/jmri.27084. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Mao Y, Chen X, et al. Magnetic resonance imaging radiomics in categorizing ovarian masses and predicting clinical outcome: a preliminary study. Eur Radiol. 2019;29:3358–3371. doi: 10.1007/s00330-019-06124-9. [DOI] [PubMed] [Google Scholar]
- 48.Rundo L, Beer L, Escudero Sanchez L, et al. Clinically interpretable radiomics-based prediction of histopathologic response to neoadjuvant chemotherapy in high-grade serous ovarian carcinoma. Front Oncol. 2022;12:2423. doi: 10.3389/fonc.2022.868265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zargari A, Du Y, Heidari M, et al. Prediction of chemotherapy response in ovarian cancer patients using a new clustered quantitative image marker. Phys Med Biol. 2018;63:155020. doi: 10.1088/1361-6560/aad3ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danala G, Thai T, Gunderson CC, et al. Applying quantitative CT image feature analysis to predict response of ovarian cancer patients to chemotherapy. Acad Radiol. 2017;24:1233–1239. doi: 10.1016/j.acra.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu Y, Tan M, McMeekin S, et al. Early prediction of clinical benefit of treating ovarian cancer using quantitative CT image feature analysis. Acta Radiol. 2016;57:1149–1155. doi: 10.1177/0284185115620947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan S, Zhou T, Che R, et al. CT-based machine learning radiomics predicts CCR5 expression level and survival in ovarian cancer. J Ovarian Res. 2023;16:1. doi: 10.1186/s13048-022-01089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y, Jiang W, Fu L, Ren M, Ai H, Wang X. Intra- and peritumoral radiomics for predicting early recurrence in patients with high-grade serous ovarian cancer. Abdom Radiol (NY) 2022 doi: 10.1007/s00261-022-03717-9. [DOI] [PubMed] [Google Scholar]
- 54.Wang T, Wang H, Wang Y, et al. MR-based radiomics-clinical nomogram in epithelial ovarian tumor prognosis prediction: tumor body texture analysis across various acquisition protocols. J Ovarian Res. 2022;15:6. doi: 10.1186/s13048-021-00941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J, Cai S, Wang F, et al. Development of a prediction model for gross residual in high-grade serous ovarian cancer by combining preoperative assessments of abdominal and pelvic metastases and multiparametric MRI. Acad Radiol. 2022 doi: 10.1016/j.acra.2022.12.019. [DOI] [PubMed] [Google Scholar]
- 56.Li C, Wang H, Chen Y, et al. A nomogram combining MRI multisequence radiomics and clinical factors for predicting recurrence of high-grade serous ovarian carcinoma. J Oncol. 2022;2022:1716268. doi: 10.1155/2022/1716268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu J, Wang Z, Zuo R, et al. Development of survival predictors for high-grade serous ovarian cancer based on stable radiomic features from computed tomography images. iScience. 2022;25:104628. doi: 10.1016/j.isci.2022.104628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong Y, Liu Z, Lin D, et al. Development of a radiomic-clinical nomogram for prediction of survival in patients with serous ovarian cancer. Clin Radiol. 2022;77:352–359. doi: 10.1016/j.crad.2022.01.038. [DOI] [PubMed] [Google Scholar]
- 59.Gao L, Jiang W, Yue Q, et al. Radiomic model to predict the expression of PD-1 and overall survival of patients with ovarian cancer. Int Immunopharmacol. 2022;113:109335. doi: 10.1016/j.intimp.2022.109335. [DOI] [PubMed] [Google Scholar]
- 60.Fotopoulou C, Rockall A, Lu H, et al. Validation analysis of the novel imaging-based prognostic radiomic signature in patients undergoing primary surgery for advanced high-grade serous ovarian cancer (HGSOC) Br J Cancer. 2022;126:1047–1054. doi: 10.1038/s41416-021-01662-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng S, Xia T, Ge Y, et al. Computed tomography imaging-based radiogenomics analysis reveals hypoxia patterns and immunological characteristics in ovarian cancer. Front Immunol. 2022;13:868067. doi: 10.3389/fimmu.2022.868067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boehm KM, Aherne EA, Ellenson L, et al. Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat Cancer. 2022;3:723–733. doi: 10.1038/s43018-022-00388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Avesani G, Tran HE, Cammarata G, et al. CT-based radiomics and deep learning for BRCA mutation and progression-free survival prediction in ovarian cancer using a multicentric dataset. Cancers. 2022;14:2739. doi: 10.3390/cancers14112739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu XY, Ren J, Jia Y, et al. Multiparameter MRI radiomics model predicts preoperative peritoneal carcinomatosis in ovarian cancer. Front Oncol. 2021;11:765652. doi: 10.3389/fonc.2021.765652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi X, Liu Y, Zhou B, et al. Incorporating SULF1 polymorphisms in a pretreatment CT-based radiomic model for predicting platinum resistance in ovarian cancer treatment. Biomed Pharmacother. 2021;133:111013. doi: 10.1016/j.biopha.2020.111013. [DOI] [PubMed] [Google Scholar]
- 66.Song X-L, Ren J-L, Yao T-Y, Zhao D, Niu J. Radiomics based on multisequence magnetic resonance imaging for the preoperative prediction of peritoneal metastasis in ovarian cancer. Eur Radiol. 2021;31:8438–8446. doi: 10.1007/s00330-021-08004-7. [DOI] [PubMed] [Google Scholar]
- 67.Liu M, Ge Y, Li M, Wei W. Prediction of BRCA gene mutation status in epithelial ovarian cancer by radiomics models based on 2D and 3D CT images. BMC Med Imaging. 2021 doi: 10.1186/s12880-021-00711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li MR, Liu MZ, Ge YQ, Zhou Y, Wei W. Assistance by routine CT features combined With 3D texture analysis in the diagnosis of BRCA gene mutation status in advanced epithelial ovarian cancer. Front Oncol. 2021;11:696780. doi: 10.3389/fonc.2021.696780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li HM, Gong J, Li RM, et al. Development of MRI-based radiomics model to predict the risk of recurrence in patients with advanced high-grade serous ovarian carcinoma. AJR Am J Roentgenol. 2021;217:664–675. doi: 10.2214/AJR.20.23195. [DOI] [PubMed] [Google Scholar]
- 70.Li H, Zhang R, Li R, et al. Noninvasive prediction of residual disease for advanced high-grade serous ovarian carcinoma by MRI-based radiomic-clinical nomogram. Eur Radiol. 2021;31:7855–7864. doi: 10.1007/s00330-021-07902-0. [DOI] [PubMed] [Google Scholar]
- 71.Chen H-z, Wang X-r, Zhao F-m, et al. A CT-based radiomics nomogram for predicting early recurrence in patients with high-grade serous ovarian cancer. Eur J Radiol. 2021;145:110018. doi: 10.1016/j.ejrad.2021.110018. [DOI] [PubMed] [Google Scholar]
- 72.Chen H-z, Wang X-r, Zhao F-m, et al. The development and validation of a CT-based radiomics nomogram to preoperatively predict lymph node metastasis in high-grade serous ovarian cancer. Front Oncol. 2021;11:3362. doi: 10.3389/fonc.2021.711648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ai Y, Zhang J, Jin J, Zhang J, Zhu H, Jin X. Preoperative prediction of metastasis for ovarian cancer based on computed tomography radiomics features and clinical factors. Front Oncol. 2021;11:610742. doi: 10.3389/fonc.2021.610742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Veeraraghavan H, Vargas HA, Alejandro-Jimenez S, et al. Integrated multi-tumor radio-genomic marker of outcomes in patients with high serous ovarian carcinoma. Cancers. 2020;12:3403. doi: 10.3390/cancers12113403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei W, Liu Z, Rong Y, et al. A computed tomography-based radiomic prognostic marker of advanced high-grade serous ovarian cancer recurrence: a multicenter study. Front Oncol. 2019;9:255. doi: 10.3389/fonc.2019.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meier A, Veeraraghavan H, Nougaret S, et al. Association between CT-texture-derived tumor heterogeneity, outcomes, and BRCA mutation status in patients with high-grade serous ovarian cancer. Abdom Radiol (NY) 2019;44:2040–2047. doi: 10.1007/s00261-018-1840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu H, Arshad M, Thornton A, et al. A mathematical-descriptor of tumor-mesoscopic-structure from computed-tomography images annotates prognostic- and molecular-phenotypes of epithelial ovarian cancer. Nat Commun. 2019;10:764. doi: 10.1038/s41467-019-08718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rizzo S, Botta F, Raimondi S, et al. Radiomics of high-grade serous ovarian cancer: association between quantitative CT features, residual tumour and disease progression within 12 months. Eur Radiol. 2018;28:4849–4859. doi: 10.1007/s00330-018-5389-z. [DOI] [PubMed] [Google Scholar]
- 79.Vargas HA, Veeraraghavan H, Micco M, et al. A novel representation of inter-site tumour heterogeneity from pre-treatment computed tomography textures classifies ovarian cancers by clinical outcome. Eur Radiol. 2017;27:3991–4001. doi: 10.1007/s00330-017-4779-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162:55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 81.Martin-Gonzalez P, Crispin-Ortuzar M, Rundo L, et al. Integrative radiogenomics for virtual biopsy and treatment monitoring in ovarian cancer. Insights Imaging. 2020 doi: 10.1186/s13244-020-00895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nougaret S, Tardieu M, Vargas HA, et al. Ovarian cancer: an update on imaging in the era of radiomics. Diagn Interv Imaging. 2019;100:647–655. doi: 10.1016/j.diii.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 83.Rizzo S, Manganaro L, Dolciami M, Gasparri ML, Papadia A, Del Grande F (2021) Computed Tomography Based Radiomics as a Predictor of Survival in Ovarian Cancer Patients: A Systematic Review. Cancers 13. [DOI] [PMC free article] [PubMed]
- 84.Staal FCR, Aalbersberg EA, Van Der Velden D, et al. GEP-NET radiomics: a systematic review and radiomics quality score assessment. Eur Radiol. 2022 doi: 10.1007/s00330-022-08996-w. [DOI] [PubMed] [Google Scholar]
- 85.Li Y, Liu Y, Liang Y, et al. Radiomics can differentiate high-grade glioma from brain metastasis: a systematic review and meta-analysis. Eur Radiol. 2022 doi: 10.1007/s00330-022-08828-x. [DOI] [PubMed] [Google Scholar]
- 86.Gao Y, Cheng S, Zhu L, et al. A systematic review of prognosis predictive role of radiomics in pancreatic cancer: heterogeneity markers or statistical tricks? Eur Radiol. 2022 doi: 10.1007/s00330-022-08922-0. [DOI] [PubMed] [Google Scholar]
- 87.Brancato V, Cerrone M, Lavitrano M, Salvatore M, Cavaliere C. A systematic review of the current status and quality of radiomics for glioma differential diagnosis. Cancers. 2022;14:2731. doi: 10.3390/cancers14112731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ursprung S, Beer L, Bruining A, et al. Radiomics of computed tomography and magnetic resonance imaging in renal cell carcinoma: a systematic review and meta-analysis. Eur Radiol. 2020;30:3558–3566. doi: 10.1007/s00330-020-06666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Electronic Supplementary Materials.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its Additional files].