Abstract
Viruses adopt strategies to efficiently utilize their compact genome. Members of the family Paramyxoviridae, exhibit a cotranscriptional RNA editing mechanism wherein polymerase stuttering generates accessory proteins from Phosphoprotein (P) gene. Newcastle disease virus (NDV), an avian paramyxovirus, expresses two accessory proteins, V and W, by RNA editing. While P and V proteins are well studied, very little is known about W protein. Recent studies confirmed W protein expression in NDV and the unique subcellular localization of W proteins of virulent and avirulent NDV. We characterized the W protein of NDV strain Komarov, a moderately virulent vaccine strain. W mRNA expression ranged between 7 and 9% of total P gene transcripts similar to virulent NDV. However, W protein expression, detectable by 6 h, peaked at 24 h and dropped by 48 h post infection in DF1 cells indicating a kinetically regulated expression by the virus. The W protein localized in the nucleus and by mutations, a strong nuclear localization signal was identified in the C-terminal region of W protein. The viral growth kinetics study suggested neither supplementation of W protein nor subcellular localization pattern of the supplemented W protein influenced viral replication in vitro similar to that noticed in avirulent NDV. A cytoplasmic mutant of W protein localized in cytoplasm unlike specific mitochondrial colocalization as recorded in velogenic NDV strain SG10 indicating a possible role of W protein in determining the viral pathogenicity. This study describes for the first time, the distinct features of W protein of moderately virulent NDV.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13337-023-00813-2.
Keywords: Newcastle disease virus, W protein, Ivermectin, Mesogenic NDV strain, Strain Komarov
Introduction
Newcastle disease (ND) is a highly contagious and an economically important poultry viral disease known to affect various domestic and wild avian species across the globe [3]. ND is caused by Newcastle disease virus (NDV) that belongs to the genus Orthoavulavirus in the family Paramyxoviridae. NDV is the prototype virus of subfamily Avulavirinae and is the most well characterized amongst the avian paramyxoviruses. NDV strains are broadly grouped into three pathotypes based on the severity of disease in the chickens: lentogenic, mesogenic and velogenic [23]. The lentogenic NDV strains are avirulent causing sub-clinical infections with minimal respiratory signs, mesogenic NDV strains cause moderate respiratory and enteric symptoms and velogenic NDV strains lead to severe disease and high mortality in naive chickens. The velogenic NDV strains are further classified as neurotropic velogenic and viscerotropic velogenic which have affinity for nervous tissues and visceral organs, respectively [26]. The mesogenic NDV strain Komarov referred in our present study was derived from a virulent isolate by serial passages in ducklings by A. Komarov in Palestine [8]. NDV strain Komarov is used as vaccine in India and in many African countries and is less pathogenic than other mesogenic strains such as NDV strain Mukteswar [11, 36].
NDV has a non-segmented, negative sense, single stranded RNA genome coding for six structural proteins: nucleocapsid protein (NP), phosphoprotein (P), matrix (M), fusion (F), hemagglutinin–neuraminidase (HN), and the large polymerase (L) protein [9]. Similar to other paramyxoviruses, the P gene undergoes cotranscriptional RNA editing through polymerase slippage during transcription leading to non-template additions of single or two guanosine (G) residues at a conserved edit site (3′-UUUUUCC-5′). A + 1 frameshift in the ORF created by a single G nucleotide insertion leads to V mRNA with a frequency of 25 to 35% and + 2 frameshift in the ORF by addition of two G nucleotides results in W mRNA with a frequency of 2 to 8.5% and the unedited mRNA (60–70%) codes for P protein [19, 34, 42]. All the three P gene products share common N-terminal region, but their C-terminal regions vary in length and amino acid composition [34]. Their unique C terminal sequences probably contribute to their specific functions.
Among the three P gene-derived products, P protein plays an indispensable role as a non-catalytic subunit of the viral RNA polymerase. It also helps in packaging of the nascent RNA genome along with the nucleocapsid protein [12]. Studies on V protein of NDV and other paramyxoviruses have revealed that V protein is multifunctional, targets STAT1 degradation, interferes with MDA5, is an interferon antagonist [5, 13, 24, 25] inhibits apoptosis [7, 38] assist in viral replication [6, 13], plays important roles in tissue tropism, virulence [1, 13, 19] and host range determination [24]. On the other hand, there is very limited information on the W protein of paramyxoviruses. The W protein of Nipah virus (NiV) localizes in the nucleus [31], and has been shown to reduce minigenome replication [33], to evade host immunity through interaction with STAT1 and STAT2 and by inhibiting TLR 3 pathway [2, 30, 32]. Furthermore, W protein of Nipah virus impacts viral pathogenesis and determines the disease course [29]. The expression of W protein of NDV lentogenic and velogenic strains, their subcellular localization and incorporation of W protein into the virion have been recently reported [14, 35, 42, 43]. W protein of NDV lentogenic strains Clone30, LaSota localized in nucleus and that of velogenic strain SG10 localized in cytoplasm specifically in mitochondria [14, 42, 43]. W protein is one of the only two viral proteins of NDV to enter the nucleus probably with roles in viral replication, budding and pathogenesis etc. Yang et al. showed that varying lengths of W protein of NDV strains exhibited different cellular compartmentalization [42]. While there was no effect of W protein on viral replication in case of avirulent NDV strain Clone 30 [14], the deletion of W protein in virulent NDV strain SG10 affected NDV replication both in vitro and in vivo [42, 43]. Mutated nuclear localizing W protein of strain SG10 induced lesser IFNβ production which in turn enhanced the viral replication, virulence and pathogenicity when compared with NDV expressing the cytoplasm localizing W protein [43]. Till date, there is no information on the W protein of mesogenic NDV strains, we present here a detailed characterization of W protein of mesogenic NDV strain Komarov.
Materials and methods
Virus and cells
Newcastle disease virus strain Komarov, a mesogenic pathotype of NDV, used in this study was obtained from VBRI, AP. African green monkey kidney (Vero) cell line, chicken embryo fibroblasts (DF-1) cell line and baby hamster kidney (BHK-21) cell line were used in transfection and infection experiments. The cells were cultured and maintained in high glucose Dulbeco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin–Streptomycin solution (growth medium) at 37 °C in a 5% CO2 incubator.
Next generation sequencing of P gene transcripts of NDV strain Komarov
DF-1 cells infected with NDV strain Komarov were collected at 6, 12 and 24 h post infection (hpi) for total RNA isolation. The mRNAs were selectively reverse transcribed using oligo (dT) primer. PCR was performed on this cDNA with primers complementary to sequences on either side of the P gene RNA editing site (Supplementary Fig. 1a). The amplicons generated were gel cleaned (Supplementary Fig. 1b) and subjected to next generation sequencing with Illumina MiSeq platform with 150 bp paired-end read output. Sequence reads with additional G nucleotides in the edit region were identified and their percentage of total sequence reads was estimated to conclude the editing frequencies.
Western blotting analysis
The total cell protein lysates were extracted from the NDV infected DF1 cells at 6, 12, 24 and 48 hpi in RIPA buffer. Briefly, the cells were lysed in RIPA buffer (supplemented with 1 × protease cocktail inhibitor) by sonication and the supernatant with the whole cell protein lysate was collected after centrifugation. The total protein concentration in the supernatant was determined by Bradford assay (Sigma). The extracted proteins were separated with 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences). The membranes were blocked with 5% BSA and 0.1% Tween 20 in phosphate buffered saline (PBST) for 2 h at room temperature, and then incubated overnight at 4 °C with primary antibody. The following primary antibodies were used in our experiments: W-Ab (peptide antibody against the region 153–167 aa of W protein of NDV strain Komarov, raised in rabbit) for detection of the W protein; anti-Newcastle Disease virus antibody (Anti-NDV ab), a polyclonal antibody against NDV raised in chicken (Abcam—Cat. No. ab34402), for the detection of NDV proteins; and horseradish peroxidase (HRP)—conjugated anti-β-actin mAb (ThermoFisher Scientific—Catalog # MA5-15739-HRP) to detect the β-actin as the loading control. The membranes were washed three times with PBST, they were incubated with corresponding HRP-conjugated anti-rabbit antibody or CF™ 660C conjugated anti-chicken antibody (in dark) for 1 h at room temperature. HRP was detected with SuperSignal™ West Pico Chemiluminescent Substrate and the CF™ 660C was detected at far red range.
Bioinformatics
The amino acid sequences derived from P gene transcripts (P, V and W mRNA) of NDV strain Komarov were aligned using clustal alignment software [18]. The subcellular localization of W protein was analysed using LocTree3, an online Protein subcellular localization prediction system which applies machine learning (profile kernel SVM) to predict the native sub-cellular localization and improves over it through the addition of homology-based inference [10]. The nuclear localization signal (NLS) in W protein sequence of NDV strain Komarov was predicted by using online tool, cNLS mapper with a cut-off score of 5.0 which predicts the NLS specific to the importin αβ pathway [16]. The online tool, NLStradamus which is based on Hidden Markov Model (HMM) was used to analyse the results generated by the cNLS mapper [20]. The MODELLER program was used to produce the homology model of W protein of NDV strain Komarov [39]. LocNES, a Support Vector Machine (SVM) predictor that locates classical nuclear export signals (NESs) in CRM1 cargoes was used to predict the presence of NES in the W protein [40].
Plasmid construction
The ORFs of P, V and W were amplified from NDV strain Komarov (GenBank accession no. KT445901.1) infected DF1 cells and cloned into pCMV-HA (Clontech) to generate HA tagged P, V and W proteins. Additionally, plasmids coding for HA tagged mutated W ORFs were generated by substituting the basic amino acids in the proposed NLS region with Alanine (Table 1). A non-NLS mutant of W, where the basic amino acid (Arginine outside the NLS region) at position 176 was mutated to alanine, was also generated. Furthermore, wild type W ORF sequence and mutant W ORF that predominantly localized in the cytoplasm (Wµ162-163-165-166) were cloned into pEYFP-N1 (Clonetech) to express EYFP-W fusion proteins.
Table 1.
Bipartite NLS of wild type W protein of NDV strain Komarov as predicted by NLStradamus and the mutant of W proteins that were generated by introducing mutations
| W-Wild type | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ132 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ133 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ138 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ141 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ142 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ162 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ163 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ165 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ166 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ132-133 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ141-142 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ162-163 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ165-166 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ132-133-134 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ141-142-162-163 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
| Wµ162-163-165-166 | 132 KKGAHGRAPKRGTTNVRLNSREVNPAAETVRKDRRTK 168 |
*The bold and underlined amino acids represent the amino acids that were replaced by alanine
Transfection and localization studies
Vero, DF1 and BHK21 cells were transfected with pCMV-HA plasmids encoding P, V, W and mutated W proteins individually using lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Briefly, the plasmid DNA and the lipofectamine were diluted in Opti-MEM media and they were mixed and incubated for 5 min at room temperature (RT) to form plasmid DNA-lipid complex and the DNA-lipid complex was added to the cells. The localization of P, V, W and mutated W proteins were observed after immunostaining the transfected cells using anti-HA-FITC antibody (Sigma) using confocal microscopy Leica Micro Systems (SP8) at different time points post transfection. The fluorescence intensities observed in the nucleus (Fn) and the cytoplasm (Fc) of the transfected cells were calculated using Image-J software to arrive at Fn/Fc ratio. Further to explore the possibility of mitochondrial localization of cytoplasmic mutant W, we stained both wild type and cytoplasmic mutant W plasmids transfected cells with MitoTracker® Deep Red FM and imaged the cells using confocal microscopy Leica Micro Systems (SP8).
Ivermectin treatment
Vero cells were transfected with pCMV-HA encoding wild type W protein using lipofectamine 2000 as described earlier. The transfected cells were treated with 25 µM of ivermectin (sigma) at 24 h post transfection for 1 h. Transfected but untreated cells served as control. The cells were fixed with 4% paraformaldehyde and stained with anti-HA-FITC antibody as described earlier. The cells were observed using fluorescence microscope (ZEISS) for studying the effect of ivermectin on the localization of W proteins. The fluorescence intensities observed in the nucleus (Fn) and the cytoplasm (Fc) of the transfected cells were calculated using Image-J software to arrive at Fn/Fc ratio.
Viral growth kinetics by plaque assay
DF-1 cells were transfected with EYFP tagged wild type W (W − Wt) and cytoplasmic mutant W (Wµ162-163-165-166) protein encoding plasmids using polyethylenimine (PEI). The DF-1 cells over expressing the wild type W protein and cytoplasmic mutant W protein (Supplementary Fig. 3) were infected with NDV strain Komarov at a multiplicity of infection (MOI) of 0.5. Plaque assay was performed in Vero cells as described below with the cell supernatants collected at 3, 6, 12, 24 and 48 hpi to compare the viral growth kinetics upon W protein supplementation.
Plaque assay
The supernatants collected from infected cells were serially diluted and inoculated into Vero cells in 24-well plates for virus adsorption for 1 h. The cells were washed with PBS thrice and overlaid with DMEM containing 0.8% methyl cellulose supplemented with 1% penicillin and streptomycin. The cells were incubated at 37 °C in a 5% CO2 incubator for 6 days. Later, the overlay medium was removed, the cells were fixed in 100% ice cold methanol for 20 min at 4 °C and cells were stained for plaques with 1% crystal violet for 30 min. The crystal violet solution was removed and the plates were rinsed briefly under running tap water and dried. Plaques were counted in the virus dilution at which minimum number of countable plaques were observed. The virus titre was expressed as log PFU/mL.
Results
Relative levels of P, V and W mRNA in NDV strain Komarov infected DF1 cells
The amplicon sequencing analysis of P, V and W mRNA from NDV strain Komarov infected DF1 cells at 6, 12 and 24 hpi indicated that amplicons with + 2Gs (W mRNA) ranged between 7 and 9%. The unedited (P mRNA) increased from 65% at 6 hpi to 69% at 12 hpi and 76% at 24 hpi while the +1G (V mRNA) amplicons decreased from 26% at 6 hpi to 23% at 12 hpi and 16% at 24 hpi (Fig. 1).
Fig. 1.
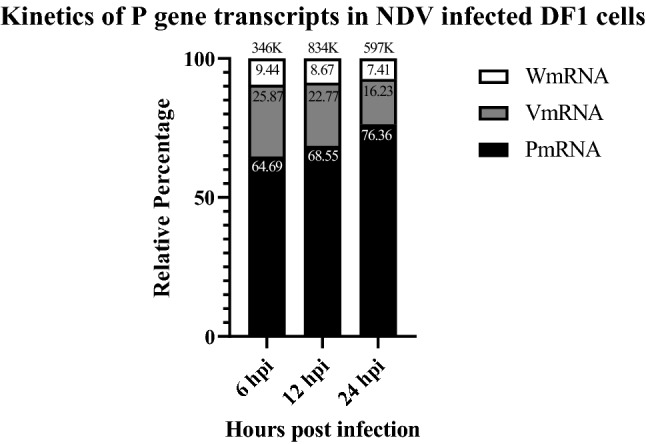
Kinetics of P gene transcripts in NDV strain Komarov infected DF1 cells. The amplicon sequencing analysis showed differential expression of P gene transcripts at different time points post infection. While the levels of P and V transcripts varied at different time points post infection, the levels of W transcripts remained almost constant. The numbers at the top of each bar denote the total number of reads obtained during amplicon sequencing
Detection of W protein expression and its kinetics in the NDV strain Komarov infected DF1 cells
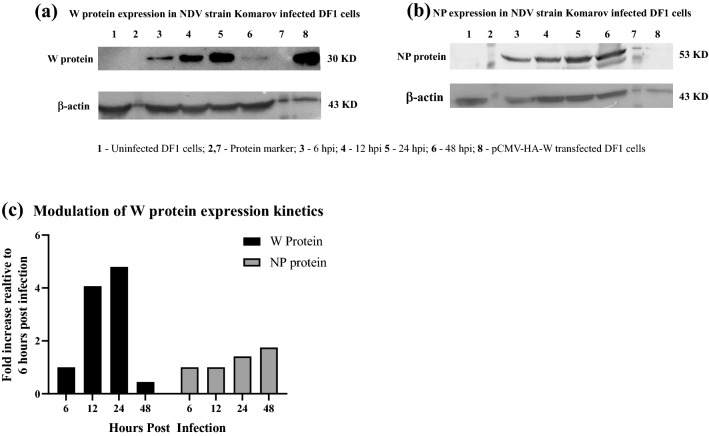
The expression kinetics of viral proteins, W and NP, was studied by western blot of total protein obtained from NDV strain Komarov infected DF1 cells. In contrast to the increase in the expression of NP over time, the W protein expression detected at 6 hpi, increased at 24 hpi but decreased at 48 hpi (Fig. 2). Fold change in the expression levels of W protein as determined by ImageJ analysis of the western blot showed that W protein expression increased atleast 4 folds at 24 hpi since its initial detection at 6 hpi. However, W protein level reduced to less than half from the early stages of infection (6hpi) at 48 hpi (Fig. 2c).
Fig. 2.
W protein expression in NDV strain Komarov infected DF1 cells: a Immunoblotting of W protein in the infected DF1 cells at different time points post infection showed increasing levels of W protein up to 24 h post infection which reduced at 48 h post infection. b The levels of NP of NDV showed increasing trend till 48 h post infection. c Bar graph representing the fold change in the levels of W protein and NP relative to 6 h post infection as determined by ImageJ
Bioinformatic analysis of W protein of NDV strain Komarov
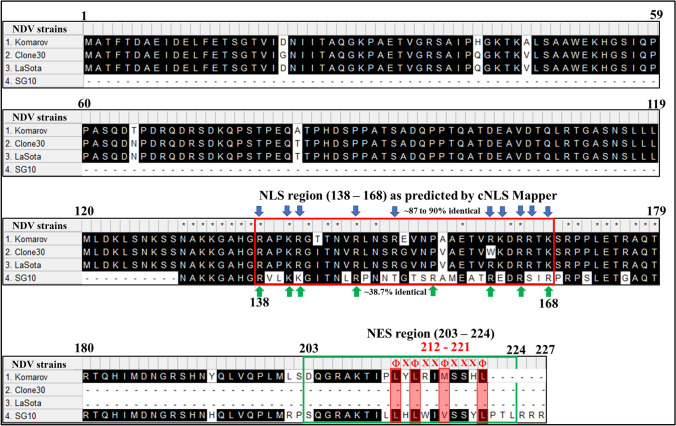
LocTree3, an online tool for predicting protein subcellular localization, suggested that the W protein of NDV strain Komarov could be localized in the nucleus of eukaryotic cells with a 93% accuracy and a prediction score of 60 (Supplementary Fig. 2a). Analysis of W protein of NDV strain Komarov with online tool cNLS Mapper, showed the presence of a bipartite NLS specific to importin αβ pathway in the unique C-terminal region of W protein (Supplementary Fig. 2b). The results of cNLS mapper were confirmed by using online tool NLStradamus (Supplementary Fig. 2c). Computer program, Modeller, was used to develop the homology model of W protein of NDV strain Komarov which illustrated the exposed basic amino acids of the NLS region suggesting their amenability for recognition by importins (Supplementary Fig. 2d). The predicted NLS sequence of W protein of NDV strain Komarov was in consensus (87–90% amino acid identity) with the previously reported NLS of W proteins of NDV lentogenic strains Clone 30 and LaSota (Fig. 3). However, the NLS of W protein of NDV strain Komarov showed only 38.7% amino acid identity with that of W protein of virulent NDV strain SG10 (Fig. 3). The nuclear export signal (NES) in the W protein of NDV strain Komarov predicted by online tool LocNES showed a putative leucine rich region at the C-terminal region in consensus with a class 1a-R NES (Fig. 3). Comparison of C-terminal region of W protein of NDV mesogenic strain Komarov with that of the virulent strain SG10 showed highly dissimilar amino acids in the NLS region (138–168 aa) but the amino acids in the NES region (203–224 aa) were conserved (Fig. 3).
Fig. 3.
Multiple sequence alignment of W proteins of NDV strains Komarov, Clone 30, LaSota and SG10: C-terminal region of NDV strains Komarov, LaSota and Clone 30 showed high percentage of identical amino acids (87–90) in the predicted NLS region between 138 and 168 aa (basic amino acids in the NLS region are represented by blue arrows) whereas the C-terminal region of NDV strain Komarov and SG10 had lower percentage of amino acids identity (38.7) in the predicted NLS region (basic amino acids in the NLS region are represented by green arrows). NES region (203–224 aa) in W protein of NDV strains Komarov and SG10 as predicted by LocNES showed higher percentage of amino acids identity
Subcellular localization patterns exhibited by P gene products of NDV strain Komarov
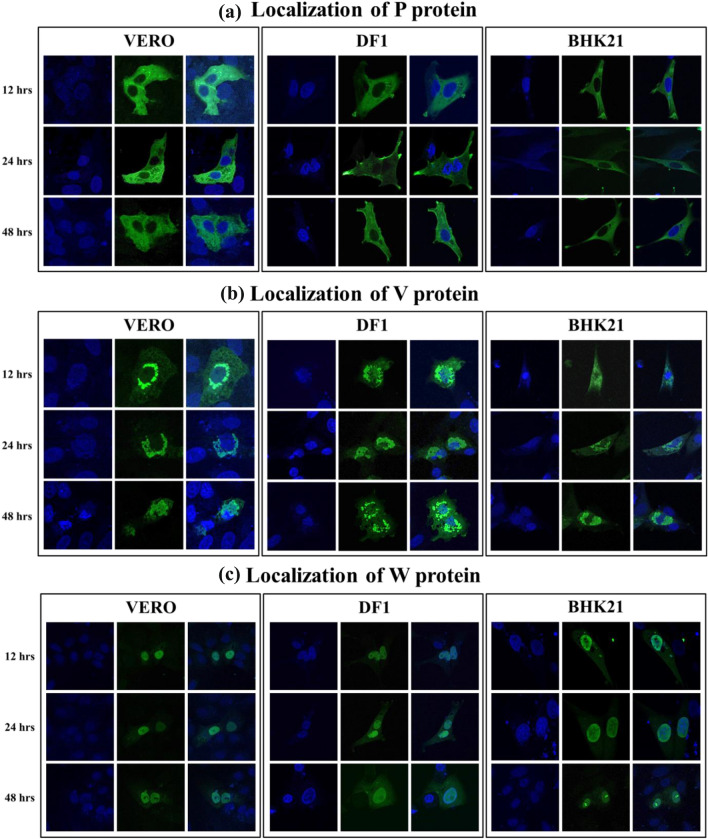
We compared the subcellular localization patterns of P gene products in Vero, DF1 and BHK21 cell lines at 12, 24 and 48 hpi. We observed that P protein was cytoplasmic, V protein aggregated in perinuclear region and W protein was mostly accumulated inside the nucleus (Fig. 4). This distinct pattern of subcellular localization of P, V and W proteins was conserved in all cell lines studied irrespective of the time post transfection.
Fig. 4.
Subcellular localization of P, V and W proteins. Different cell lines (Vero, DF-1 and BHK-21) were transfected with pCMV-HA-P, pCMV-HA-V and pCMV-HA-W plasmids and were subjected to immunofluorescence analysis at different time points (12, 24 and 48 h) post transfection to study the subcellular localization of P gene products. a The P protein of NDV strain Komarov localized predominantly in the cytoplasm, b the V protein of NDV strain Komarov localized in perinuclear region and c the W protein of NDV strain Komarov localized predominantly in the nucleus irrespective of the cell type or the time post transfection
Importin α/β-mediated nuclear transport of W protein of NDV strain Komarov
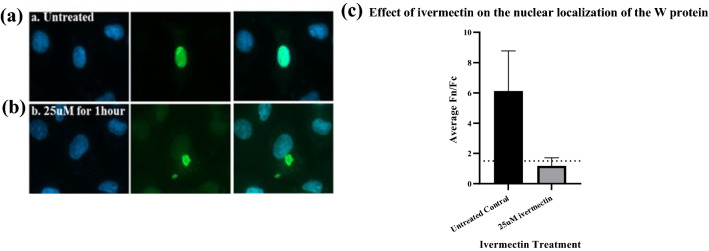
Ivermectin, a recently recognized antiviral agent and a broad-spectrum anti-parasite drug, was found to disrupt the nuclear import of W protein in the transfected cells (Fig. 5) confirming that nuclear transport of W protein of NDV strain Komarov is mediated through importin α/β1 pathway.
Fig. 5.
Effect of Ivermectin on the Nuclear localization of W protein: a-b Vero cells transfected with pCMV-HA-W were treated with ivermectin (25 µM) 24 h post transfection for 1 h and were subjected to immunofluorescence analysis to study the effect of ivermectin on the localization of the W protein. c The ratio of relative nuclear to cytoplasmic fluorescence intensities in the ivermectin treated and untreated cells revealed that the ivermectin treatment limited the accumulation of the W protein in the nucleus
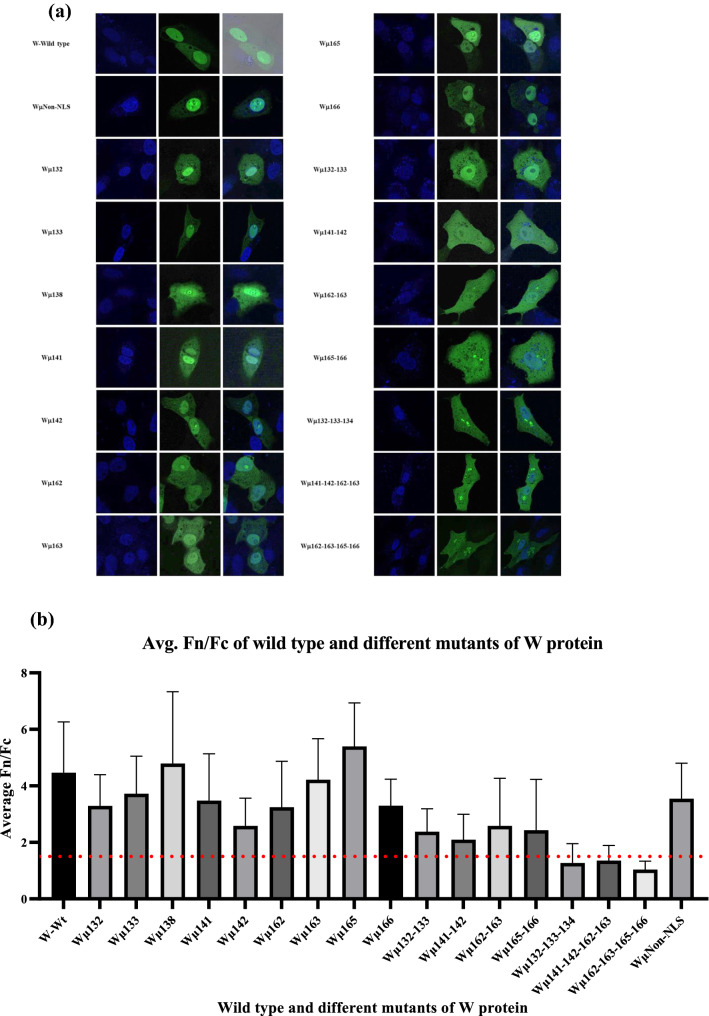
Key NLS sequence of the W protein
We generated 17 mutants (16 mutants with alanine mutations of basic amino acids within the predicted NLS region and 1 mutation of arginine at position 176 to alanine outside the NLS region) and studied their localization pattern in Vero cells. The ratio of nuclear to cytoplasmic fluorescence intensities of the expressed proteins were calculated for the W protein mutants at 24 h post transfection (Fig. 6b). It was observed that mutations of at least 2 basic amino acids within the NLS region of the W protein abrogated the nuclear import of W protein (Fig. 6). While mutations of 3 or 4 basic amino acids in the NLS region resulted in higher accumulation of W protein within the cytoplasm. A quadruplet mutant (Wµ162-163-165-166) showed maximum retention of W protein in cytoplasm (Fig. 6) and hereby this mutant is referred as cytoplasmic W mutant.
Fig. 6.
Subcellular localization different mutants of W protein. a Vero cells were transfected with pCMV-HA vector encoding different NLS mutants of W protein and were subjected to immunofluorescence analysis to study the effect of mutations on the localization of W protein. b The ratio of relative nuclear to cytoplasmic fluorescence intensities of the wild type and different mutants of W proteins were calculated and it is observed that transfection with the quadruplet mutant Wµ162-163-165-166 retained most of the W protein in cytoplasm
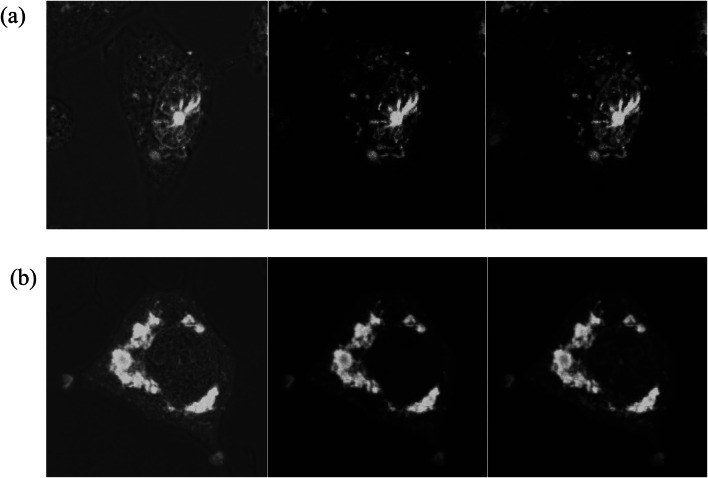
Cytoplasmic W mutant does not localize with mitochondria
Unlike the previously described W protein of virulent NDV strain SG10, the cytoplasmic W mutant of NDV strain Komarov did not localize with mitochondria (Fig. 7) showing a distinct difference in localization pattern of W proteins of moderately virulent and virulent NDV strains.
Fig. 7.
Cytoplasm-localized W protein does not colocalize with mitochondria: DF1 cells transfected with wild type W (Fig. 7a) and cytoplasmic quadruplet mutant Wµ162-163-165-166 (Fig. 7b) stained with MitoTracker® Deep Red FM showed that the cytoplasmic quadruplet mutant W protein does not colocalize with the mitochondria. The localization of W protein can be seen as green fluorescence while the red fluorescence represents the mitochondria
Effect of W protein overexpression on viral replication
The growth kinetics of NDV strain Komarov in DF1 cells that over expressed either the wild type W protein or the cytoplasmic mutant W protein (Wµ162-163-165-166) did not show any difference when compared with control (DF1 cells infected with NDV strain Komarov) suggesting that neither the overexpression of W protein nor the localization pattern of W protein influenced the viral replication in vitro (Fig. 8).
Fig. 8.
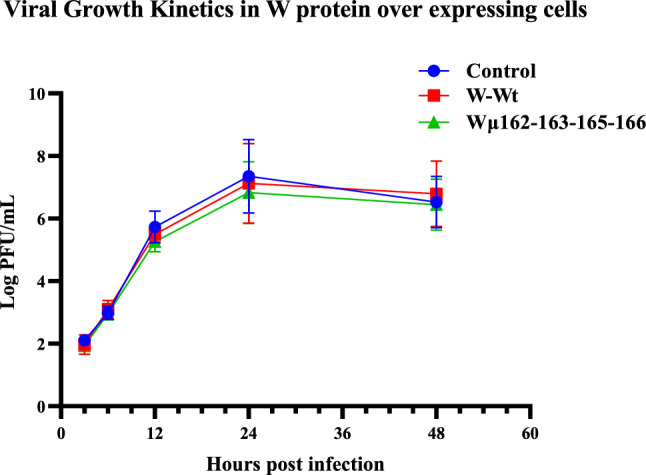
Growth kinetics of NDV in cells over expressing wild type W protein and cytoplasmic mutant W protein: DF1 cells over expressing Wild type W protein (W-EYFP) and cytoplasmic mutant W protein (Wµ-EYFP) were infected with NDV strain Komarov to study the effect of W protein overexpression on viral growth. The viral titres at different time points were measured by plaque assay in Vero cells and the data showed that neither the W protein overexpression nor its localization pattern impacted the viral replication
Discussion
Steward et al. [34] detected W mRNA of NDV while the W protein expression in infected cells and the presence of W protein in purified virions were confirmed by Karsunke et al. in 2019 [14]. Among paramyxoviruses, the W protein of Nipah virus (NiV) has been extensively studied but only limited information is available regarding the W protein of NDV and other avian paramyxoviruses [14, 28, 35, 42, 43]. The W protein of NiV localizes in nucleus and plays anti-interferon role by inhibiting JAK-STAT and TLR3 pathways [25, 31–33]. The W protein of NiV was shown to modulate immune response and determine the disease course [29].
There are more than 1000 NDV strains across the globe with a wide spectrum of pathogenicity and virulence. These strains are broadly classified as avirulent (lentogenic), moderately virulent (mesogenic) and highly virulent (velogenic) strains based on their pathogenicity in the host. While majorly the viral glycoproteins, F and HN proteins, are implicated in the virulence of NDV, recent studies have shown the role of V protein in determination of virulence and host range [5, 7, 13, 24, 25]. It is interesting to study the role of the accessory viral proteins that are expressed in very low quantities by the virus and only during the early stages of infection. The P:V:W mRNA ratio of Nipah Virus (NiV) was identified as 1:1:1 [17], however, the proportions of P, V, and W mRNAs in avirulent NDV strains Ulster 2C and Clone 30 were reported as 68:29:2 [19, 34]. Recent studies have shown that the percentages of these mRNAs vary in different strains of NDV and also depend on the stage of the infection [27, 42]. Our results on the kinetics of P gene transcripts in mesogenic NDV strain Komarov showed a time dependent RNA editing, the ratios of P:V:W observed at 6 hpi and 24 hpi were P64.7%:V25.9%:W9.44% and P76.36%:V16.23%:W7.41%, respectively. Higher percentage of edited transcripts was observed at 6 hpi than at 24 hpi, suggesting that the editing frequency was higher during early stages of infection. This kinetic modulation of RNA editing could be favouring viral replication and or immune evasion which are both required during the early stages of virus infection. Also, we observed higher V transcripts compared to W transcripts as previously noted for velogenic NDV strains [4]. Though the levels of W mRNA were relatively constant until 24 hpi, the level of W protein expression in the NDV infected cells reached peak at 24 hpi suggesting the stability of W mRNA. The decline in W protein expression by 48 hpi indicated that W protein’s role may be restricted to early stages of viral infection and that W protein is probably dispensable at later stages of virus life cycle.
The bioinformatic analyses of the W protein sequence of mesogenic NDV strain Komarov (221 amino acids) predicted nuclear localization, detected bipartite NLS and leucine rich NES in the C-terminal region. Comparison of the NLS sequences of NDV strains showed that the W protein of NDV strain Komarov showed higher similarity with avirulent strains than with virulent NDV strain. The C-terminal region of the W protein of strain Komarov carried the predicted NES (212LYLRIMSSHL221) at the 3’ end of the protein which was in consensus with the predicted NES of the W protein of virulent NDV strain SG10 however lacked anchoring sequences at the 3’ end. The W protein of NDV strain Komarov localized in the nucleus despite the predicted NES unlike the previously reported NDV strain SG10 (Yang et al. [43]). Furthermore, NES score of the cytoplasmic mutant W protein of NDV strain Komarov (where the amino acids in the C-terminal NLS region of the W protein were mutated) showed higher NES score suggesting that the presence of NLS in the C-terminal region of W protein masked the function of the predicted NES. Based on previous data from Yang et al. [42], it appears that strain SG10 has an active NLS in N-terminal region and a strong NES in C-terminal region thus enabling nuclear cytoplasmic shuttling of the W protein. The nuclear transport of W protein of NDV strain Komarov was mediated by importin α/β pathway as reported earlier in other NDV strains [14, 43] but did not show nuclear cytoplasmic shuttling. Furthermore, irrespective of the cells transfected or the time of transfection, the P gene products of strain Komarov showed distinct localization pattern.
Viral proteins localizing in nucleus or showing nuclear cytoplasmic shuttling are often associated with functions such as regulating viral replication, assembly, virion formation, in subverting host immunity and or leading to persistent infection [15, 21, 22, 37, 41]. The role of W protein in viral replication couldn’t be established in avirulent strain Clone 30 as neither the virus lacking W expression nor the disruption of the NLS had any effect on viral replication in vitro [14]. In virulent strain SG10, it was found that the C-terminally deleted W proteins has slightly lower replication ability that the parental strains and supplementation of appropriate doses of W protein during infection enhanced the viral replication but not at higher doses [42]. Further, the nuclear localizing mutant of W protein of strain SG10 showed slightly higher virulence and growth kinetics both in vitro and in vivo compared to the mitochondria localizing W protein [43]. The authors concluded that this effect was either due to the reduction of interferon beta levels in the cells infected with virus expressing nuclear localizing W protein or stimulation of interferon beta in the cells infected with virus expressing mitochondrial localizing W protein. However, in our study, overexpression of neither the nuclear localizing W protein nor the mutant W protein that predominantly localized in cytoplasm, had any effect on virus replication. It is possible that in moderately virulent NDV strains and in those NDV strains where W protein is either localized in nucleus or in cytoplasm but not in mitochondria, the W protein plays no role in viral replication. Also, the absence of nuclear cytoplasmic shuttling of W protein of strain Komarov points to possibilities for unique functions of W protein in different NDV strains. This study attempts to fill the knowledge gap with respect to W protein of the three NDV pathotypes. With the variations observed in the W protein among the three pathotypes of NDV, it will be interesting to study further if the diverse C-terminal regions of W proteins could be attributing to the wide spectrum of pathogenicity and virulence exhibited by the NDV strains.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank their lab members, the National Institute of Animal Biotechnology and Department of Biotechnology, Government of India for supporting the study. The first author is currently pursuing his doctoral work and acknowledges his fellowship (JRF and SRF) received for his PhD work from Council of Scientific and Industrial Research (CSIR).
Funding
This work was funded by the National Institute of Animal Biotechnology core grant (#C0007), the Department of Biotechnology, Government of India (#BT/PR8740/AGR/36/772/2013) and CSIR, Government of India, File No. 09/1150(0001)/2016-EMR-1.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This study required no ethical clearance.
Consent to participate
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alamares JG, Elankumaran S, Samal SK, Iorio RM. The interferon antagonistic activities of the V proteins from two strains of Newcastle disease virus correlate with their known virulence properties. Virus Res. 2010;147:153–157. doi: 10.1016/j.virusres.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audsley MD, Jans DA, Moseley GW. Nucleocytoplasmic trafficking of Nipah virus W protein involves multiple discrete interactions with the nuclear import and export machinery. Biochem Biophys Res Commun. 2016;479:429–433. doi: 10.1016/j.bbrc.2016.09.043. [DOI] [PubMed] [Google Scholar]
- 3.Aziz Ul R, Munir M, Shabbir MZ. Comparative evolutionary and phylogenomic analysis of Avian avulaviruses 1–20. Mol Phylogenet Evol. 2018;127:931–951. doi: 10.1016/j.ympev.2018.06.040. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Jia Y, Ren S, et al. Identification of newcastle disease virus P-gene editing using next-generation sequencing. J Vet Med Sci. 2020;82:1231–1235. doi: 10.1292/jvms.18-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Childs KS, Andrejeva J, Randall RE, Goodbourn S. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J Virol. 2009;83:1465–1473. doi: 10.1128/JVI.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu Z, Ma J, Wang C, et al. Newcastle disease virus V protein promotes viral replication in HeLa cells through the activation of MEK/ERK signaling. Viruses. 2018;10:489. doi: 10.3390/v10090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu Z, Wang C, Tang Q, et al. Newcastle disease virus V protein inhibits cell apoptosis and promotes viral replication by targeting CacyBP/SIP. Front Cell Infect Microbiol. 2018;8:304. doi: 10.3389/fcimb.2018.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czegledi A, Wehmann E, Lomniczi B. On the origins and relationships of Newcastle disease virus vaccine strains Hertfordshire and Mukteswar, and virulent strain Herts'33. Avian Pathol. 2003;32:271–276. doi: 10.1080/0307945031000097868. [DOI] [PubMed] [Google Scholar]
- 9.Ganar K, Das M, Sinha S, Kumar S. Newcastle disease virus: current status and our understanding. Virus Res. 2014;184:71–81. doi: 10.1016/j.virusres.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg T, Hecht M, Hamp T (2014) LocTree3 prediction of localization. Nucleic acids research 42(Web Server issue): W350-W355 [DOI] [PMC free article] [PubMed]
- 11.Grimes SE. A basic laboratory manual for the small-scale production and testing of 1–2 Newcastle disease vaccine. FAO Regional Office for Asia and the Pacific (RAP) 2002.
- 12.Hamaguchi M, Yoshida T, Nishikawa K, Naruse H, Nagai Y. Transcriptive complex of Newcastle disease virus I Both L and P proteins are required to constitute an active complex. Virology. 1983;128:105–117. doi: 10.1016/0042-6822(83)90322-7. [DOI] [PubMed] [Google Scholar]
- 13.Huang Z, Krishnamurthy S, Panda A, Samal SK. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J Virol. 2003;77:8676–8685. doi: 10.1128/JVI.77.16.8676-8685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karsunke J, Heiden S, Murr M, et al. W protein expression by Newcastle disease virus. Virus Res. 2019;263:207–216. doi: 10.1016/j.virusres.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Kim YH, Han ME, Oh SO. The molecular mechanism for nuclear transport and its application. Anat Cell Biol. 2017;50:77–85. doi: 10.5115/acb.2017.50.2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulkarni S, Volchkova V, Basler CF, et al. Nipah virus edits its P gene at high frequency to express the V and W proteins. J Virol. 2009;83:3982–3987. doi: 10.1128/JVI.02599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madeira F, Park YM, Lee J, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mebatsion T, Verstegen S, De Vaan LT, Römer-Oberdörfer A, Schrier CC. A recombinant Newcastle disease virus with low-level V protein expression is immunogenic and lacks pathogenicity for chicken embryos. J Virol. 2001;75:420–428. doi: 10.1128/JVI.75.1.420-428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov model for nuclear localization signal prediction. BMC Bioinform. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishio M, Tsurudome M, Ito M, Kawano M, Komada H, Ito Y. Characterization of sendai virus persistently infected L929 cells and Sendai virus pi strain: recombinant Sendai viruses having Mpi protein shows lower cytotoxicity and are incapable of establishing persistent infection. Virology. 2003;314:110–124. doi: 10.1016/S0042-6822(03)00404-5. [DOI] [PubMed] [Google Scholar]
- 22.Nishio M, Nagata A, Tsurudome M, Ito M, Kawano M, Komada H, Ito Y. Recombinant sendai viruses with L1618V mutation in their L polymerase protein establish persistent infection, but not temperature sensitivity. Virology. 2004;329:289–301. doi: 10.1016/j.virol.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 23.OIE (2019) Infection with Newcastle disease virus. OIE—Terrestrial Animal Health Code 2019
- 24.Park MS, García-Sastre A, Cros JF, et al. Newcastle disease virus V protein is a determinant of host range restriction. J Virol. 2003;77:9522–9532. doi: 10.1128/JVI.77.17.9522-9532.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park MS, Shaw ML, Jorge Muñoz-Jordan J, et al. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol. 2003;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pringle CR. Virus taxonomy–San Diego. Arch Virol. 1998;143(7):1449–1459. doi: 10.1007/s007050050389. [DOI] [PubMed] [Google Scholar]
- 27.Qiu X, Fu Q, Meng C, et al. Kinetic analysis of RNA editing of Newcastle disease virus P gene in the early period of infection. Acta Virol. 2016;60:71–77. doi: 10.4149/av_2016_01_71. [DOI] [PubMed] [Google Scholar]
- 28.Rao PL, Gandham RK, Subbiah M. Molecular evolution and genetic variations of V and W proteins derived by RNA editing in Avian Paramyxoviruses. Sci Rep. 2020;10:9532. doi: 10.1038/s41598-020-66252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satterfield BA, Cross RW, Fenton KA, et al. The immunomodulating V and W proteins of Nipah virus determine disease course. Nat Commun. 2015;6:7483. doi: 10.1038/ncomms8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satterfield BA, Geisbert TW, Mire CE. Inhibition of the host antiviral response by Nipah virus: current understanding and future perspectives. Futur Virol. 2016;11:331–344. doi: 10.2217/fvl-2016-0027. [DOI] [Google Scholar]
- 31.Shaw ML, García-Sastre A, Palese P, Basler CF. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J Virol. 2004;78:5633–5641. doi: 10.1128/JVI.78.11.5633-5641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw ML, Cardenas WB, Zamarin D, et al. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J Virol. 2005;79:6078–6088. doi: 10.1128/JVI.79.10.6078-6088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleeman K, Bankamp B, Hummel KB. The C, V and W proteins of Nipah virus inhibit minigenome replication. J Gen Virol. 2008;89:1300–1308. doi: 10.1099/vir.0.83582-0. [DOI] [PubMed] [Google Scholar]
- 34.Steward M, Vipond IB, Millar NS, Emmerson PT. RNA editing in Newcastle disease virus. J Gen Virol. 1993;74:2539–2547. doi: 10.1099/0022-1317-74-12-2539. [DOI] [PubMed] [Google Scholar]
- 35.Vaidyanathan SP, Gawai S, Subbiah M. Elucidation of the role of non-structural viral protein (W) of Newcastle disease virus. Int J Infect Dis. 2016;45:337–338. doi: 10.1016/j.ijid.2016.02.730. [DOI] [Google Scholar]
- 36.Visnuvinayagam S, Thangavel K, Lalitha N, Malmarugan S, Sukumar K. Assessment of the pathogenicity of cell-culture-adapted Newcastle disease virus strain Komarov. Braz J Microbiol. 2015;46:861–865. doi: 10.1590/S1517-838246320140051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Harmon A, Jin J, et al. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles. J Virol. 2010;84:4673–4681. doi: 10.1128/JVI.02306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, Chu Z, Liu Z, et al. Newcastle disease virus V protein inhibits apoptosis in DF-1 cells by downregulating TXNL1. Vet Res. 2018;49:102. doi: 10.1186/s13567-018-0599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinform 2016;54:5.6.1–37 [DOI] [PMC free article] [PubMed]
- 40.Xu D, Marquis K, Pei J, et al. LocNES: a computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics. 2015;31:1357–1365. doi: 10.1093/bioinformatics/btu826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu C, Sun L, Liu W, Duan Z. Latent membrane protein 1 of Epstein-Barr virus promotes RIG-I degradation mediated by proteasome pathway. Front Immunol. 2018;9:1446. doi: 10.3389/fimmu.2018.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Bu Y, Zhao J, et al. Appropriate amount of W protein of avian avulavirus 1 benefits viral replication and W shows strain-dependent subcellular localization. Virology. 2019;538:71–85. doi: 10.1016/j.virol.2019.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Xue J, Teng Q, et al. Mechanisms and consequences of Newcastle disease virus W protein subcellular localization in the nucleus or mitochondria. J Virol. 2021;95:e02087–e2120. doi: 10.1128/JVI.02087-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.