SUMMARY
Biomolecular condensation underlies the biogenesis of an expanding array of membraneless assemblies, including stress granules (SGs) which form under a variety of cellular stresses. Advances have been made in understanding the molecular grammar of a few scaffold proteins that make up these phases, but how the partitioning of hundreds of SG proteins is regulated remains largely unresolved. While investigating the rules that govern the condensation of ataxin-2, a SG protein implicated in neurodegenerative disease, we unexpectedly identified a short 14aa sequence that acts as a condensation switch and is conserved across the eukaryote lineage. We identify poly(A)-binding proteins as unconventional RNA-dependent chaperones that control this regulatory switch. Our results uncover a hierarchy of cis and trans interactions that fine-tune ataxin-2 condensation and reveal an unexpected molecular function for ancient poly(A)-binding proteins as regulators of biomolecular condensate proteins. These findings may inspire approaches to therapeutically target aberrant phases in disease.
Keywords: Protein phase separation, short linear motif, polyQ, protein aggregation, PABPC, stress granules, ATXN2, amyotrophic lateral sclerosis, spinocerebellar ataxia, microtubule-binding protein
eTOC BLURB

A growing list of proteins has been found to moonlight as chaperones. Boeynaems et al. show that poly(A)-binding proteins bind a short linear motif in the stress granule and disease protein ataxin-2, hereby regulating its condensation in homeostasis and stress. This function is dependent on RNA binding and evolutionary conserved.
INTRODUCTION
Stress granules (SGs) are cytoplasmic ribonucleoprotein assemblies that form when cells are exposed to stress (e.g., heat, chemical, osmotic stress)1. SGs were initially thought to function in translational regulation, but emerging evidence indicates that they may serve a protective role in the cell by preventing the irreversible aggregation of proteins and even mRNA2–5. Hundreds of proteins and thousands of mRNAs have been found to localize to SGs6–10. Many of these SG-localized proteins harbor “sticky” and aggregation-prone domains11–13, and the ability of these proteins to interact with RNA might help protect them from aggregation3,14. Given their aggregation-prone nature, several SG proteins have been implicated in human protein aggregation diseases. For instance, missense mutations in RNA-binding proteins TDP-43, FUS, EWSR1, TAF15, and other heterogeneous nuclear ribonucleoproteins (hnRNPs) are linked to hereditary forms of degenerative diseases amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and myopathies15,16. Additionally, loss-of-function mutations have been found in the proteins that regulate SG clearance17,18, indicating that—while SGs might be initially protective—their misregulation may underlie eventual pathological aggregation15,16.
SGs and other so-called biomolecular condensates form via phase separation19,20. The set of rules dictating this behavior remains unresolved, but for RNA-centric condensates—including SGs—a picture emerges where condensation is driven by a complex interplay between specific and non-specific interactions over a range of interaction strengths and length scales. Recent work has uncovered a hierarchy of such interactions between oligomerization, RNA-binding, and disordered domains, and elucidated how these rules govern the phase separation of the key SG scaffold protein G3BP14,21,22. Additionally, the disordered prion-like domains of several ALS-related RNA-binding proteins are involved in their liquid-like behavior23–25. Whether these same simple rules apply to other SG proteins remains unknown. Defining these principles will be crucial for devising novel ways to therapeutically modulate phase separation and aggregation of these disease-linked proteins.
Ataxin-2 (ATXN2) is another SG protein that has been implicated in two neurodegenerative diseases—spinocerebellar ataxia-2 (SCA2) and ALS. ATXN2 harbors a polyglutamine (polyQ) repeat. Long expansions (>34 repeats) of this polyQ repeat cause SCA226–28, whereas intermediate-length expansions (27–33 repeats) are associated with ALS29,30. Notably, lowering levels of ATXN2 is sufficient to mitigate neurodegeneration and extend survival in multiple model systems29,31,32. Mechanistically, ATXN2 localizes to SGs and promotes TDP-43 mislocalization31. The protein has also been implicated in dendritogenesis and memory formation in flies, potentially by regulating mRNA stability and transport33,34. These latter functions are dependent on the ability of ATXN2 to condense into RNA granules via its intrinsically disordered regions (IDRs). Lastly, ATXN2 has been implicated in the regulation of circadian rhythm in flies35,36 and mice37, and perturbed sleeping patterns have been observed in both SCA and ALS cases38,39. Thus, defining the mechanisms connecting ATXN2 to condensate form and function will provide insight into this important disease gene and could suggest additional ways to safely target its function therapeutically40.
Here, we set out to define the molecular determinants regulating ATXN2’s phase separation behavior. ATXN2 is a 140 kDa intrinsically disordered RNA-binding protein, but contrary to our expectations, we found that its SG targeting is exclusively mediated by a short linear motif (SLiM) that confers interaction with cytoplasmic poly(A)-binding proteins (PABPCs). We provide evidence that PABPC acts as an ATXN2 chaperone by preventing its spontaneous condensation and driving its mixing into stress granules in times of stress. The identification of PABPCs as an RNA-dependent chaperone provides an unexpected novel function for this ancient class of RNA-binding proteins. These findings have direct implications for our understanding of the complex regulatory networks governing protein condensation, and point to potential therapeutic approaches to target and modulate this behavior in human disease.
RESULTS
The PAM2 motif acts as a switch that regulates ATXN2 condensation in time and space
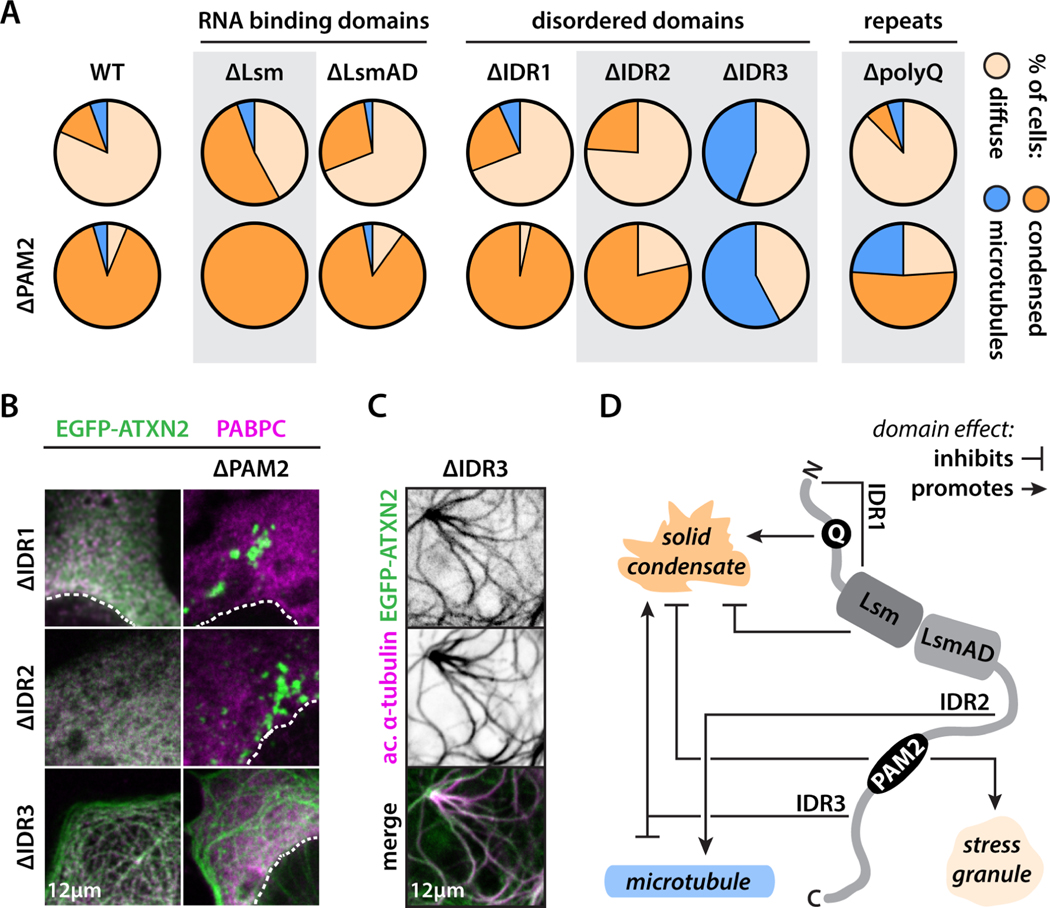
While hundreds of proteins condense into SGs upon cellular stress, the functional contributions of each protein to condensation remain largely unknown (Fig. 1A). Some of these proteins have been shown to be essential for SG formation41,42, so-called scaffold proteins, whereas others are called client proteins and partition into condensates but are not required for their formation. To test if ATXN2 functions as a SG scaffold or client, we generated an ATXN2 knock-out HeLa cell line. These cells were still able to form SGs upon treatment with arsenite, indicating that ATXN2 is not a required scaffold but acts as a client protein (Fig. 1B-C), consistent with previous observations from knock-down studies31,41,43. Notably, stress granules in ATXN2 knock-out cells were smaller and more numerous, suggesting that partitioning of the protein does modulate stress granules (Fig. S1A). To define the molecular features that drive ATXN2 to partition into SGs, we performed systematic domain deletions and evaluated their effect on SG partitioning in U2OS cells (Fig. 1D). ATXN2 consists of two RNA-binding domains (Lsm and LsmAD) and three intrinsically disordered domains (IDR1, 2 and 3). A polyQ repeat (residues 166–189) is embedded in IDR1, while a PAM2 short linear motif (SLiM) (residues 909–923) separates IDR2 and IDR3. The PAM2 motif mediates ATXN2’s interaction with cytoplasmic poly(A)-binding protein (PABPC)44–46. Although RNA-binding and disordered domains have been extensively implicated in protein phase separation23–25, these domains were dispensable for SG targeting and mixing of ATXN2 into the condensed phase (measured as heterogeneity, see STAR Methods) (Fig. 1E–F). However, deleting the PAM2 SLiM, a fourteen amino acid sequence that makes up only ~1% of the ATXN2 sequence, prevented proper mixing of ATXN2 into SGs. Instead, ΔPAM2 ATXN2 formed small condensates that dotted the surface of SGs (Fig. 1E–F). Because ΔPAM2 ATXN2 demixed from SGs, we tested if its condensation was stress-dependent. Expressing this mutant in non-stressed cells resulted in spontaneous condensation under standard conditions (Fig. 1G, Fig. S1B). This behavior was not dependent on the position of the EGFP tag (Fig. S1B). Since high levels of wildtype ATXN2 expression can also lead to aggregation, we confirmed that the ΔPAM2 ATXN2 protein was expressed at comparable levels to the WT protein (Fig. 1H). We further show—via two independent methods—that this spontaneous condensation of ΔPAM2 ATXN2 happens within its physiological window of expression (Fig. S1C–D). Fluorescence recovery after photobleaching (FRAP) experiments indicate that both spontaneous wildtype and mutant condensates have solid-like properties (Fig. S1E). Thus, our findings indicate that the PAM2 motif acts as a switch for the spatiotemporal regulation of ATXN2’s phase separation behavior: PAM2 is required for (1) proper targeting to and mixing of ATXN2 in SGs under times of stress, and (2) preventing its spontaneous condensation into solid-like gels under normal conditions.
Figure 1: The PAM2 switch regulates ATXN2’s behavior in stress and non-stress conditions.
(A) Stress granules form upon cellular stress. (B) ATXN2 is dispensable for arsenite-induced stress granule assembly. Average percentage of cells with stress granules is shown. n = 3 experiments with a total of [255–285] cells. Dashed lines highlight nuclei. EGFP is shown in inverse gray scale (see also Fig. S1A). (C) Phase separation of scaffold proteins and RNA drives stress granule assembly, with subsequent recruitment of non-essential client proteins, such as ATXN2. (D) Domain structure and disorder prediction of ATXN2. Metapredict98 score: 1 = disordered, 0 = folded. (E) Deletion of PAM2 prevents homogeneous partitioning of ATXN2 in stress granules. (F) Quantification of the heterogeneity of ATXN2 deletion mutant distribution within the stress granule compartment (see STAR methods). One-way ANOVA. n = 30 stress granules. (G) PAM2 deletion drives spontaneous condensation of ATXN2 into small granules under non-stress conditions (see also Fig. S1B). (H) Wildtype ATXN2 can spontaneously condense upon overexpression. ΔPAM2 ATXN2 condensation is not an overexpression artefact (see also Fig. S2). Scatterplots show cells with diffuse or condensed ATXN2 localization. Average cytoplasmic EGFP intensity. Cells combined from 3 experiments. Mann-Whitney. * p-value < 0.05, **** p-value < 0.0001. Panel (B) shows HeLa cells. All other panels show U2OS cells. Every picture shows endogenous PABPC staining. EGFP-ATXN2 (mutants) are expressed from a plasmid.
The PAM2 switch is conserved across eukaryotes
ATXN2 is a pan-eukaryote protein. While the Lsm RNA-binding domain is common in Archaea and Bacteria, we could not find any examples of ATXN2 orthologs in these clades. Since IDRs are notoriously fast-evolving sequences, we wondered whether acquiring the PAM switch was a more recent or ancient event. The PAM2 motif is found across all eukaryote lineages44–46 and we observed that it is always centered between IDR2 and IDR3 (Fig. S2A). Exceptions exist though—both budding yeast and C. elegans lost the PAM2 sequence, but flanking motifs now serve as the PABPC interaction site, indicating functional conservation of a SLiM-based interaction between these proteins. In contrast, the polyQ repeat can be found at different locations in different lineages or is absent46. Given this striking conservation of the PAM2 motif, we wondered whether its role as a regulatory switch would also be conserved (Fig. 2A). We tested this in two protists, where we identified and synthesized their respective ATXN2 orthologs and generated ΔPAM2 mutants. First, Capsaspora owczarzaki is a close unicellular relative of animals and a model for the study of the origin of multicellularity47. Wildtype CoATXN2 (CAOG_07908) spontaneously formed several small granules, whereas PAM2 deletion resulted in the formation of one single large granule (Fig. 2B, Fig. S2B–D). Second, Trypanosoma brucei, the parasite causing sleeping sickness, is a member of the basal eukaryote lineage Discoba. Wildtype TbATXN2 (Tb927.8.4540) localized diffusely to the cytoplasm and PAM2 deletion resulted in the appearance of spontaneous condensates (Fig. 2C, Fig. S2E–F). To extend this analysis to other multicellular eukaryotes, we performed the same experiment in the model plant Arabidopsis thaliana. We generated stable transgenic lines carrying GFP-tagged wildtype and ΔPAM2 AtATXN2 (CID4) (Fig. 2D). By assaying cotyledon (embryonic leaf) and root tissue in Arabidopsis seedlings, we found that the PAM2 motif prevents spontaneous condensation in vivo. To evaluate whether the PAM2 motif was also required for SG targeting, we transiently co-infected tobacco leaves with AtATXN2-GFP and PAB2-RFP, which is the Arabidopsis ortholog of PABPC (Fig. 2E). Identical to our observations in human cells, wildtype AtATXN2 partitioned into heat shock-induced PABPC-positive SGs, whereas ΔPAM2 condensates did not mix with SGs. Together, these data indicate that the PAM2 switch modulates ATXN2 condensation across the eukaryote lineage, providing evidence for the biological importance of this ancient regulatory element.
Figure 2: The PAM2 switch is functionally conserved across eukaryotes.
(A) Eukaryote tree highlighting different clades and model organisms. Archaea are the outgroup. Black stars indicate organisms compared in this figure. (B) PAM2 deletion results in the formation of a single large CoATXN2 granule upon expression in C. owczarzaki, opposed to multiple small wildtype granules. N denotes nucleus (see also Fig. S3B–D). (C) TbATXN2 localizes diffusely in trypanosomes, whereas the ΔPAM2 mutant spontaneously condenses (see also Fig. S3E). N denotes nucleus, K denotes kinetoplast. (D) AtATXN2 localizes diffusely to the cytoplasm in vivo, whereas ΔPAM2 AtATXN2 spontaneously condenses. Data are shown for the cotyledon (embryonic leaf) and root of 3-day old A. thaliana seedling. V denotes vacuole, C denotes cytoplasm. Representative images from three biological replicates from three independent transgenic lines. (E) Expression of wildtype and ΔPAM2 AtATXN2 in tobacco leaves recapitulates phenotypes from A. thaliana seedlings. PAB2 (AtPABPC) localizes diffusely to the cytoplasm, and targets stress granules upon heat shock (30 min @ 37°C). Cytoplasm outlined in white. Wildtype AtATXN2 partitions into PAB2 stress granules (white condensates), whereas ΔPAM2 AtATXN2 remains demixed (magenta and green condensates).
Complex interactions between ATXN2 domains tune its biophysical behavior
Our initial set of domain deletion mutants identified ATXN2’s PAM2 motif as the key regulator of its phase separation behavior. Next, we sought to define how this regulatory switch interacts with other ATXN2 domains (Fig. 3A, Fig S3). Deletion of the main RNA-binding domain LSM potentiated the spontaneous condensation of both the single (ΔLSM) and double (ΔPAM2-ΔLSM) mutants (Fig. 3A). This observation is in line with recent work on other prion-like proteins showing that RNA binding can prevent their spontaneous condensation3,14,48,49. Conversely, deletion of the polyQ repeat—an established driver of aggregation50—decreased condensation of ΔPAM2 ATXN2. Interestingly, a significant population of ΔpolyQ ΔPAM2 ATXN2 cells exhibited localization of ATXN2 to microtubules (Fig. 3A). We next tested the impact of each of the three IDRs on ATXN2’s behavior. Deleting IDR1 (ΔIDR1) did not have a strong impact on ATXN2 condensation, a result that is surprising given this IDR contains the polyQ repeat, therefore suggesting a role for the flanking regions. In contrast, ΔIDR2 and ΔIDR3 had opposing effects on ATXN2’s behavior (Fig. 3A–C). Deletion of IDR2 modestly reduced condensation of the ΔPAM2 mutant, but more importantly, abolished any microtubule localization of both the single and double mutant. Lastly, deletion of IDR3 completely prevented spontaneous condensation of ATXN2, and conferred microtubule localization in about half of the cells regardless of the presence of the PAM2 motif.
Figure 3: ATXN2 IDRs modulate condensation and localization.
(A) Differential contribution of ATXN2 domains to its phase behavior. Grey boxes highlight domain deletions that strongly affect ATXN2 behavior. Cells combined from 3 experiments. n = [96–155] cells (see also Fig. S4). (B) Example pictures of IDR deletion and IDR-PAM2 double deletion mutants. (C) Example picture showing microtubule localization (acetylated tubulin antibody) of the IDR3 deletion mutant. (D) Scheme highlighting the complex interactions between different domains on ATXN2 behavior. U2OS cells. Endogenous PABPC and tubulin staining. EGFP-ATXN2 (mutants) expressed from a plasmid.
Since IDR2 and IDR3 had opposing effects on promoting condensation versus microtubule targeting, we wondered whether the precise localization of the PAM2 switch in between these IDRs (Fig. S2A) was of functional importance. To test this, we generated add-back mutants where we reintroduced the PAM2 motif to the N- or C-terminus of a ΔPAM2 mutant. Even though both of these mutants were able to bind PABPC, they only partially rescued the spontaneous condensation phenotype (Fig. S3C), suggesting that the precise interplay of PABPC and the ATXN2 IDRs is important for its proper regulation (Fig. 3D).
A balance in IDR-IDR interactions controls ATXN2’s behavior
The surprising observation of microtubule targeting seen for some of our IDR mutants encouraged us to investigate how this behavior—and its control—is encoded in sequence. We performed a bioinformatics analysis using MAPAnalyzer, a prediction software trained on experimentally verified microtubule-binding proteins51. Unexpectedly, ATXN2 is predicted to be a high confidence microtubule-binding protein. More specifically, this prediction seems to be driven by an accumulation of basic short linear motifs in the ATXN2 IDR2 domain (Fig. 4A, see also Discussion). Indeed, as we have seen above, deleting IDR2 completely prevents microtubule localization of ATXN2 (Fig. 3A). This suggests that IDR3 effectively quenches the microtubule-binding activity of IDR2. A PLAAC analysis52 predicts that IDR3 is a classic prion-like domain (Fig. 4A). Prion-like domains can drive the condensation of a wide array of proteins across the tree of life53,54. This is consistent with our earlier findings that ΔIDR3 mutants are completely unable to spontaneously condense, even when missing the PAM2 motif (Fig. 3A). IDRs are often poorly conserved at the sequence level55, but they do tend to display conservation in amino acid composition56. By comparing ATXN2 IDRs spanning 1.7 billion years of eukaryote evolution, we observed that IDR2 and IDR3 have conserved differences in their composition (Fig. 4B); IDR2 is enriched in basic amino acids whereas IDR3 is enriched in aromatic residues (Fig. 4C–D, Fig. S4A). We and others have previously shown that a combination of basic and aromatic IDRs can strongly promote condensation of SG proteins through the formation of cation-pi and pi-pi interactions54,57,58. Coarse-grained simulations of the ATXN2 IDR2–3 region indeed suggest that such interactions can be at play (Fig. 4E). Given its strong aromatic and hydrophobic character, IDR3 is predicted to be prone to homotypic condensation—in line with our observations in cells. IDR2 on the other hand is predicted to be in a more expanded conformation due to its charged nature, but can directly engage with IDR3 through cation-pi and pi-pi interactions involving its arginine residues. To test whether these interactions occur in vitro we purified recombinant IDR2 and IDR3. Both were able to condense individually (Fig. 4F). Consistent with the predictions, IDR3 condensates were more solid-like (i.e., less dynamic, and more heterogeneous and irregular) than those formed by IDR2 (Fig. S4B–C). When combined, IDR2 would partition in (Fig. S4D) and wet IDR3 condensates (Fig. 4F, Fig. S4D). These experiments provide direct evidence that IDR2 can engage IDR3, at least in trans. We aimed to test these interactions in cis, but due to its length and extreme aggregation-propensity we were unable to purify a recombinant IDR2-IDR3 construct. Nonetheless, our test tube experiments and simulations suggest that these interactions are possible, and we and others have previously used similar trans assays to deduce pathologically relevant cis IDR-IDR interactions of FUS54,57.
Figure 4: IDR3 is a quencher of IDR2-mediated microtubule binding.
(A) MAPanalyzer predicts IDR2 to be a microtubule binding domain. PLAAC predicts IDR3 and the polyQ repeat to be prion-like domains. Pie charts same as in Fig. 3. (B) Evolutionary comparison of IDR2 and IDR3 amino acid composition across the eukaryote lineage. Yellow dot highlights last common eukaryote ancestor. (C Specific classes of amino acids are differentially enriched/depleted in IDR2 and IDR3. Only mammalian species are shown. Aromatic and basic residues are highlighted red and blue shades respectively. (D) For all tested eukaryote species, the differential IDR enrichment of basic versus aromatic residues is conserved. Dashed line connects IDR2 and IDR3 of the same species. (E) All-atom simulation of IDR2-IDR3 indicate a more compact IDR3 conformation engaged in interactions with a more expanded IDR2. (F) IDR2 condensates wet IDR3 condensates (see also Fig. S4). (G) Increasing the relative charge of IDR2 or decreasing the aromatic character of IDR3 promotes microtubule binding. n = [108–157] cells (see also Fig. S4). Arrowheads highlight interaction of ATXN2 with microtubules. (H) Scheme highlighting the balance in IDR2 and IDR3 interactions that regulate ATXN2 behavior. Shown are electrostatic, cation-pi, pi-pi, and hydrophobic interactions. We highlight how IDR2 and IDR3 mutants perturb these and promote microtubule binding. U2OS cells. Endogenous tubulin staining. EGFP-ATXN2 (mutants) expressed from a plasmid.
Our results suggest the balance of inter- and intra-IDR interactions tune IDR2’s microtubule binding activity, with IDR3 functioning to quench this interaction. To directly test this in cells, we generated three sets of mutants (Fig. 4G). (1) To test the hypothesis that tyrosine residues in IDR3 engage arginine residues in IDR2, precluding their microtubule-binding capacity, we replaced all the tyrosine stickers in IDR3 with serine. In line with our expectation, almost half of the cells show strong microtubule binding for this ATXN2 mutant. (2) We also know that IDR2 possesses many acidic residues (Fig. 4C) which may attenuate its microtubule binding activity via electrostatic repulsion with the negatively-charged tubulin tails. To test this, we replace glutamic acid residues with glycine, which as expected, led to enhance IDR2:microtubule interaction. (3) Scrambling the IDR2 and IDR3 sequence—while keeping the PAM2 motif intact—similarly disrupts the balance of interactions, increasing microtubule-binding propensity of this mutant (Fig. S3D). Combined, the bioinformatics, simulations, in vitro condensation assays and expression of specific IDR mutants in cells support a simple model where the precise balance of interactions within and between IDR2 and IDR3 is required to prevent ATXN2 from erroneously binding to microtubules (see also Discussion).
Thus, both a precise balance of IDR-IDR interactions (see IDR2 and IDR3 mutants) and the interaction with PABPC and RNA (see ΔPAM2 and ΔLsm mutants) are required to prevent condensation and microtubule binding of ATXN2. These data point to the complex hierarchy of regulatory interactions in cis and trans needed to keep such long sticky proteins from misbehaving.
PABPC acts like a holdase
We have provided evidence that ATXN2 requires PABPC binding to its C-terminal IDR, comprised of IDR2 and IDR3, to remain soluble. This suggests the possibility that PABPC effectively functions as a holdase. Holdases are chaperones that bind to unfolded or disordered proteins to prevent their condensation59. To directly test the hypothesis that PABPC could have holdase-like properties, we performed experiments using orthogonal designer condensates. We recently developed the PopTag system—a modular platform for generating synthetic and orthogonal condensates in human cells60. This condensation module can be functionalized with a variety of fusion proteins. Using protein-protein interaction domains (recruitment domain, recD), we can recruit specific clients to these synthetic condensates (Fig. 5A). The ATXN2-PABPC interaction is mediated by the PAM2 motif on ATXN2 and the MLLE domain in PABPC44–46. Whereas standard PopTag (GFP-Pop) condensates exclude ATXN2, a PopTag construct fused to the MLLE domain drives the recruitment of ATXN2 into MLLE-PopTag condensates (Fig. 5B). Likewise, PopTag condensates exclude PABPC, but fusing the PAM2 motif to the PopTag construct results in the recruitment of PABPC to these condensates. Importantly, the PAM2-PopTag was no longer able to coalesce into large condensates (Fig. 5C), but instead formed numerous small granules—reminiscent of the size differences we observed for wildtype and ΔPAM2 CoATXN2 condensates in C. owczarzaki (Fig. 2B). We confirmed that MLLE-PopTag, but not PAM2-PopTag, condensates recruit PAM2-containing proteins (ATXN2 and NFX1, Fig. 5D-E), and that only the PAM2-PopTag condensates recruit PABPC (Fig. 5F), highlighting the specificity of our system. These two observations support the physiological role of the PAM2 switch in regulating ATXN2 condensation in time and space (Fig. 5G) and provide evidence that PABPC has holdase-like activity. First, ATXN2 is recruited into MLLE-containing condensates via its PAM2 motif, and these can be physiological PABPC-positive SGs (Fig. 1E) or synthetic MLLE-PopTag condensates (Fig. 5B). Second, PABPC binding counteracts the condensation of PAM2-containing proteins, like ATXN2 (Fig. 1G) but also PAM2-functionalized PopTag condensates (Fig. 5C).
Figure 5: PABPC acts as a holdase and promotes wetting of immiscible condensates.
(A) Scheme illustrating the design and use of the PopTag system for the recruitment (via a recruitment domain, recD) of clients to synthetic condensates. (B) Functionalizing PopTag condensates with the PABPC-derived MLLE domain drives ATXN2 partitioning into PopTag condensates. (C) Fusing GFP-PopTag to the ATXN2-derived PAM2 motif recruits PABPC and prevents formation of large PopTag condensates. (D) Only MLLE-PopTag fusions recruit ATXN2. (E) Only MLLE-PopTag fusions recruit the PAM2-containing protein NFX1. (F) Only PAM2-PopTag fusions recruit PABPC and prevent coalescence of small PopTag granules into larger condensates. (G) Scheme highlighting the effect of functionalizing synthetic PopTag condensates with the PAM2 motif or MLLE domain. (H) G3BP1 is not recruited to PopTag condensates under non-stress conditions. (I-J) Arsenite stress or G3BP1 overexpression drive stress granule formation. Small PAM2-PopTag granules coalesce into larger condensates that mix with G3BP1-positive stress granules. Other PopTag condensates do not mix with stress granules. (K) Scheme highlighting how the amphiphilic nature of PABPC drives the wetting of PAM2-containing condensates and stress granules. RNA is not shown in the schemes for clarity. U2OS cells. Endogenous PABPC, G3BP1, and ATXN2 staining. PopTag (mutants), mCherry-G3BP1, and NFX1-FLAG expressed from a plasmid.
PABPC promotes condensate wetting
We next used our synthetic system to investigate the mechanism by which PABPC regulates protein phase separation. PABPC is a key translation initiation factor that oligomerizes on poly(A)-tails and recruits an array of effector proteins involved in translational regulation61. Under times of stress, ribosomes will dissociate from these mRNA granules, which causes the mRNA to condense into SGs. An emerging view is that scaffold proteins drive the early stages of stress granule formation, while late recruitment of clients builds on the initial assemblies4,6,62. One example of these kinds of scaffold proteins is the essential SG protein G3BP1 and its paralog G3BP241,42. Under standard conditions, G3BP1 does not interact with any of our PopTag-derived condensates (Fig. 5H). However, when we induced G3BP1 phase separation via cellular stress or overexpression, we observe that although G3BP1 SGs do not interact with PopTag and MLLE-PopTag condensates, they do intermix with the PAM2-PopTag granules (Fig. 5I–J). PABPC is amphiphilic, as it has both affinity for PAM2-PopTag granules (via its MLLE domain) and SGs (via its RRMs). By acting as a “bridge” PABPC promotes extensive wetting of two normally incompatible phases (Fig. 5K). This observation is consistent with a recent network-based framework that explains the wetting/mixing behaviors of SGs and P-bodies22. By increasing the number of “bridges” between the stress granule and P-body interaction networks, one can drive mixing of both phases. Here, we find the reverse to be true as well. By deleting the ΔPAM2 motif, ATXN2 effectively loses a critical bridge to the SG network, thus driving its demixing into separate granules that dot the surface of SGs (Fig. 1E,G)—similar to how P-bodies normally interact with SGs. Hence, in the context of network theory we can describe PABPC as a bridge that brings protein networks together. When we view the same observation in light of different phases instead of different protein interaction networks, we can propose that PABPC acts in a manner reminiscent of emulsifiers—also called wetting agents—since it (1) has affinity for two normally immiscible phases (i.e., PAM2-PopTag and SGs) and (2) drives their mixing/wetting.
RNA-binding is required for PABPC holdase activity
Using both natural and synthetic condensate systems, we have provided evidence that PABPC has holdase-like activity. Yet, what remains unclear is how it exerts this effect. Because there are multiple compensatory paralogs63 and PABPC’s function in translational control is essential for normal cell physiology64, we needed to generate a designer holdase system to circumvent any potentially confounding loss-of-function effects when introducing mutant PABPCs.
As we have shown above, PABPC recognizes its clients via the PAM2-MLLE interaction (Fig. 6A). Given our experiments are done with the endogenous PABPC, we next sought to create a synthetic, orthogonal PABPC where the MLLE domain was replaced by an alternative protein interaction domain. To do this, we replaced the MLLE domain with an engineered HA-binding nanobody (a Frankenbody, F-body)65, enabling us to place F-body-binding HA-tags on putative PABPC partners to create artificial heterodimeric interactions (Fig. 6B). Whereas PAM2-PopTag cannot coalesce into large condensates, HA-PopTag can (Fig. 6C). Having established this result, we next asked if heterodimeric interactions automatically suppress the formation of large condensates (as was the case for PABPC and PAM2-PopTag). An mCherry-tagged F-body is strongly recruited to HA-PopTag condensates (Fig. 6D), but importantly the mCherry-F-body does not prevent the formation of large condensates (Fig. 6G, Fig. S5A–C). In contrast, F-PABPC, which is full-length PABPC with its MLLE domain replaced by the F-body (Fig. 6E), completely prevents HA-PopTag coalescence (Fig. 6D,G). These results clearly show that the non-MLLE regions of PABPC are sufficient to ‘emulsify’ condensates upon recruitment, providing us with a platform to disentangle the molecular basis for this activity.
Figure 6: RNA binding is required for PABPC holdase-like activity.
(A) Scheme highlighting PABPC domain structure. (B) Examples of a natural and synthetic SLiM-based interaction pair. (C) PAM2-PopTag fails to condense into large condensates due to PABPC interaction. HA-PopTag condensates do not interact with endogenous PABPC. This allows us to interrogate sequence requirements of holdase activity using designer holdases. (D) mCherry is diffusely localized throughout the cytoplasm and Ha-PopTag condensates. F-body strongly partitions into HA-PopTag condensates. F-PABPC binds to HA-PopTag granules and prevents their coalescence into large condensates, whereas this was not the case for the RNA-binding deficient F-PABPC* mutant. (E) Scheme highlighting domain architecture of designer condensates and RNA-binding mutants. (F) Holdase activity is dependent on the involved RNA-binding domains. (G) Quantification of activity of designer holdases. Cells combined from three experiments (see also Fig. S5). One-way ANOVA. (H) Replacing the PAM2 motif with an HA-tag drives the spontaneous condensation of ATXN2. (I) PAM2>HA ATXN2 fails to properly partition into PAPC stress granules. (J-K) F-PABPC but not F-PABPC* rescues spontaneous condensation (J) and stress granule demixing (K) of PAM2>HA ATXN2. Mann-Whitney. *** p-value < 0.001, **** p-value < 0.0001. U2OS cells. Endogenous PABPC1 and G3BP1. PopTag (mutants), mCherry and mCherry-holdases, and EGFP-ATXN (mutants) expressed from a plasmid.
To test the importance of RNA binding to F-PABPC’s activity in this assay, we engineered an RNA-binding deficient mutant by substituting key aromatic residues in the RRMs for leucines66, called F-PABPC*. This mutant is unable to efficiently prevent HA-PopTag condensation, indicating that RNA binding is important for PABPC’s holdase activity (Fig. 6D,G). To test whether poly(A) binding is specifically required, or whether RNA binding in general can confer this activity, we engineered holdase versions where we substituted the four PABPC RRM domains for other tetravalent RNA binding domains (hnRNPI, MBNL1 and FXR1) (Fig. 6E). Synthetic holdases carrying the hnRNPI or MBNL1 RNA-binding domains are active but the FXR1 chimera is not (Fig. 6F–G). These data suggest that specific RNA recruitment could be sufficient for driving the observed effects. To test this, we fused these three RNA binding domains directly to the PopTag. Identically to our observations using the designer holdases, hnRPI-PopTag and MBLN1-PopTag form small granules, compared to the large PopTag and FXR1-PopTag ones. Upon further examination, we found that the FXR1-PopTag was not able to effectively recruit mRNA, whereas hnRNPI- and MBLN1-PopTag were (Fig. S5D–E). This experiment shows that the efficient recruitment of RNA—directly or indirectly—is sufficient to counteract condensation in this system. Thus, PABPC holdase activity depends solely on the recruitment of mRNA species or complete mRNA particles to its target protein, thereby emulsifying it in the cytoplasm.
The results of our PopTag system provide evidence that RNA binding is required for PABPC’s holdase activity. To test if these findings also applied to ATXN2, we first created a mutant where we replaced the PAM2 motif with an HA-tag. As with the ΔPAM2 mutant, this mutant spontaneously condenses under standard conditions and is unable to properly mix into SGs, suggesting its emulsification into the SG phase via PABPC is impaired (Fig. 6H–I). Co-expressing our engineered holdases revealed that RNA-binding by F-PABPC is also required to prevent spontaneous condensation and to drive proper SG mixing of the HA-tag mutant ATXN2 (Fig. 6J–K). Thus, our data indicate that PABPC is a holdase regulating ATXN2 protein phase separation, and that this function is licensed by RNA binding.
DISCUSSION
SGs have been extensively studied as model biomolecular condensates1,19,20,67–69. However, an understanding of the complex molecular interactions that govern their behavior in health and disease is still incomplete. How hundreds of different proteins specifically partition into these assemblies—and their functional implications—remains largely unknown. Since several SG proteins have both RNA-binding and sticky disordered domains, an initial hypothesis was that the combination of these domains was sufficient to drive partitioning of many proteins into these condensates. Here, we focused on ATXN2 as a case study of a disordered RNA-binding protein, and surprisingly found that its SG targeting is exclusively mediated by a small fourteen amino acid SLiM that engages PABPC. SLIMs are abundant in the proteome70 and have been previously implicated in the regulation of SG proteins, as sites of post-translational modifications71, or as localization sequences for nucleocytoplasmic transport72. Another example is the FGDF motif that mediates the interaction between USP10 and G3BPs. Interestingly, this FGDF motif has evolved in several viruses as a way to block SG assembly73. We now report another example of a SLiM that can specifically drive SG partitioning. Moreover, two recent studies demonstrated that several viruses evolved PAM2 motifs that are important for viral replication74,75. For example, Respiratory Syncytial Virus specifically recruits PABPCs to its viral factory74. Exactly how this SLiM-mediated recruitment of PABPC aids in viral replication—and whether its newfound function described in this study is involved—remains to be determined.
Karyopherins mediate nuclear import of client proteins by binding nuclear localization signals, a class of SLIMs. In addition to their role in nuclear transport, karyopherins have been recently shown to counteract (aberrant) protein condensation58,76,77. Like karyopherins, our work suggests that PABPC also moonlights as an unconventional holdase. Holdases are chaperones that maintain disordered or misfolded clients in a specific conformation, preventing their condensation59. Via a synthetic biology approach making use of designer condensates and engineered holdases, we provide evidence that PABPC acts as a general holdase—regulating the condensation of natural or synthetic PAM2 proteins. This activity is dependent on RNA binding. Moreover, we find that this chaperone-client interaction between PABPC and ATXN2 is functionally conserved across eukaryotes, highlighting the biological importance of this ancient regulatory switch.
PABPC seems to prevent ATXN2 condensation by regulating its IDRs. The PAM2 motif has a conserved location between IDR2 and IDR3, and placing it on either N- or C-terminus was not able to fully prevent aberrant condensation. Since RNA binding is essential for PABPC activity as a holdase, a likely model is that the recruitment of RNA to the ATXN2 IDR enables IDR2 to engage with RNA and/or other mRNA granule components. When these interactions are not at play—in a ΔPAM2 mutant or when bound by and RNA-binding deficient holdase—the ATXN2 IDRs drive condensation into aberrant granules. The prion-like IDR3 is essential for this condensation. If this IDR is the problem, why would evolution not just get rid of it? Compellingly, we find that IDR3 serves as an essential quencher of IDR2. Perturbing this quencher activity causes the mislocalization of ATXN2 to the microtubule lattice—indistinguishable from the behavior of classic microtubule-associated proteins. Interestingly, all eukaryote ATXN2 homologs have this exact same domain structure. While speculative, it is very likely that ATXN2 has evolved in a eukaryote ancestor when an LSM domain gained a long C-terminal IDR. Given their conservation, all three components of this IDR—two mutually sticky IDRs connected by a chaperone recruitment motif—likely must all have been there from ATXN2’s birth, locking them into a shared fate as each of them is required to keep the ensemble from misbehaving.
Besides ATXN2, many SG and other RNA-binding proteins contain IDRs that are differentially enriched in basic or aromatic residues, which can engage one another via pi-pi and cation-pi interactions54. Since it is exactly the interplay between such domains that seems to drive the aggregation propensity and neurotoxicity of these proteins, one question remains as to why this combination of extremely sticky domains is so prevalent? Our ATXN2 dissection suggests that more generally these domains may in part exist together to quench—and therefore regulate—one another. Several positive charged proteins have microtubule-binding activity. While we do not know whether the behavior we observed in this study could be physiological in certain instances, it is worth noting that ATXN2 has been implicated in the regulation of the microtubule cytoskeleton in both Drosophila78 and Caenorhabditis79,80, with one study showing its enrichment directly on the mitotic spindle79. Potentially circumstantial, but ATXN2 and other SG proteins are commonly found in mass spectrometry datasets of microtubule-binding proteins (Fig. S6). Recently, a study found convincing evidence for the spontaneous interaction of SGs with microtubule filaments in cells81, further showing that the involved proteins—which are enriched in positively charged IDRs13—may directly engage with acidic tubulin tails besides RNA. These observations warrant a further exploration of how prion-like domains may broadly act as quenchers of basic IDRs—regulating their affinities for specific interaction partners. In this way, basic disordered peptides produced by abnormal translation of expanded repeats, have now been identified in four independent degenerative conditions82–87. These peptides show extreme toxicity in an array of disease models and can broadly dysregulate cellular metabolism88. Notably, such basic peptides potently interact with both prion-like proteins13 and the microtubule cytoskeleton89. These “biological accidents” leading to the uncontrolled expression of an unquenched basic IDR show how important the correct regulation of these cationic sequences is to normal cell physiology90.
Our data indicates that the precise balance between cis (i.e., IDR2-IDR3) and trans (i.e., Lsm-RNA and PAM2-MLLE) interactions tunes ATXN2’s behavior—a finding further validated by similar observations in an independent Drosophila study91. However, the dependence of this system on the client-holdase interaction may form its Achilles’ heel in disease: polyQ expansions drive irreversible ATXN2 aggregation in patients and disease models92. Despite chaperoning the C-terminal IDR against spontaneous condensation, PABPC is seemingly unable to counteract aggregation caused by the N-terminal polyQ domain. Pathology data from murine models has shown that PABPC colocalizes with ATXN2 inclusions93,94. Since PABPC serves an important role in translational regulation, its sequestration into these irreversible aggregates could potentially affect neuronal health via its loss of function. Such a mechanism would explain previous results from a Drosophila ATXN2 model that required the PAM2-mediated interaction with PABPC for neurotoxicity95. This finding illuminates a new aspect of SCA2 and ALS pathogenesis, and could inspire novel opportunities for therapeutic intervention (e.g., designer chaperones).
In conclusion, by using a multidisciplinary approach involving evolutionary analyses combined with synthetic biology, we uncover a conserved and unexpected function for an ancient family of RNA-binding proteins. Additionally, our results highlight how a complex hierarchy of cis and trans interactions–with a central role for SLiMs—mediates the precise behavior of SG proteins, and provide a starting point for further evaluation of how these principles may be hijacked in infectious and neurodegenerative disease.
LIMITATIONS OF THIS STUDY
In this study we find that in cells recruitment of PABPC via the PAM2 motif to both natural and synthetic condensates results in changes that are in line with holdase activity of this protein. Unfortunately, due to the extreme length and aggregation-propensity of ATXN2, preventing us from recombinantly purifying the protein, we were unable to test this activity in an in vitro reconstituted system. Future work should be aimed at replicating these findings in such a test tube setting to provide direct evidence of the anti-condensation effect of RNA-bound PABPC. Additionally, such an approach would allow us to rule out the potential binding of other factors to the ATXN2 PAM2 motif and flanking regions96. We note though that our observations on the counteracting effect of the RNA recruitment to the PopTag condensate surface, are in line with another recent study showing similar observations97.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Aaron D. Gitler (agitler@stanford.edu).
Materials availability
All unique materials generated in this study will be made available on request from the Lead Contact upon completion of a Materials Transfer Agreement.
Data and code availability
The published article includes all datasets generated or analyzed during this study.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Human cell lines
U2OS (ATCC, HTB-96) and HeLa cells (ATCC, CCL-2) were grown at 37°C in a humidified atmosphere with 5% CO2 for 24h in Dulbecco’s Modified Eagle’s Medium (DMEM), high glucose, GlutaMAX + 10% Fetal Bovine Serum (FBS) and pen/strep (Thermo Fisher Scientific).
Capsapora owczarzaki
C. owczarzaki was grown axenically in cell culture at 23°C in ATCC medium 1034 (modified PYNFH medium).
Trypanosoma brucei
T. brucei PCF 29–13 (T7RNAP NEO TETR HYG) co-expressing T7 RNA polymerase and Tet repressor was grown in SDM-79 medium as previously described99, supplemented with hemin (7.5 μg/ml) and 10% heat-inactivated fetal bovine serum and at 27°C in the presence of G418 (15 μg/ml) and hygromycin (50 μg/ml) to maintain the integrated genes for T7 RNA polymerase and tetracycline repressor, respectively.
Plant growth conditions
Nicotiana benthamiana (tobacco) plants and Arabidopsis thaliana plants used for seed propagation were grown in soil (PRO-MIX® HP Mycorrhizae) inside growth chambers held at 22 °C with a 16/8 hour photoperiod (130 μmol.m−2.s−1). Arabidopsis thaliana seedlings used for microscopy were grown on Murashige and Skoog (MS) medium (0.5X Murashige and Skoog basal salt mixture (PhytoTechnologies Laboratories) at a pH of 5.7 supplemented with 0.8 % agar (Difco) and 1 % sucrose (Sigma-Aldrich)) inside growth cabinets (Percival) held at 22 ˚C with a 16/8 hour photoperiod (130 μmol.m−2.s−1). Their seeds were first sterilized in 70 % ethanol by vortexing for 5 minutes followed by replacement of the solution with 100 % ethanol. Seeds were then immediately placed on pre-sterilized filter papers (Grade 410, VWR) and left to dry in a laminar flow hood. They were then sown on square petri dishes (120 × 120 wide x 15 mm high (VWR)) containing 40 mL of MS medium. Plates were then sealed with micropore surgical tape (3M) and covered in aluminum foil before being placed at 4˚C for three days to break seed dormancy.
METHOD DETAILS
Plasmid construction
Sequences of all mutants, orthologs, and holdases are shown in Table S1.
Constructs for human expression were generated through custom synthesis and subcloned into a pcDNA3.1 backbone by Genscript (Piscataway, USA). The G3BP1-mCherry construct was a kind gift of Dr. Nancy Kedersha and Dr. Paul Anderson (Harvard Medical School, USA).
Constructs for CoATXN2 (CAOG_07908) were synthesized by Genscript and subcloned into the pONSY vector (Addgene, 111873) using Gibson assembly 100. Prior to assembly, the backbone vector and the insert were prepared as following: the pONSY vector was linearized with EcoRV-HF restriction enzyme (New England BioLabs); the cDNA coding for ATXN2-GFP or dPAM-ATXN2-GFP was amplified using Phusion High-Fidelity DNA Polymerase (New England BioLabs) with the forward and reverse primers (see below). TOP10 E. coli cells were transformed with the product of the assembly reaction and positive clones were confirmed by Sanger sequencing.
Forward primer CoATXN2: CGGGACTAGTGATATCATGAGCAAGGGCGAGGAG
Reverse primer CoATXN2: GCAAACACAAAATTCAAACGGGCCCTGCCTT
Constructs for TbATXN2 (Tb927.8.4540) were synthesized by Genscript and subcloned into the T. brucei expression vector pLew100v5_bsd to generate pLew100 (GFP-TbATXN2) and pLew100(GFP-dPAM-TbATXN2) 99. Prior to subcloning, the backbone vector and the insert were prepared as following: the pLew100v5_bsd vector was digested with XbaI and BamHI restriction enzymes (New England BioLabs) and gel-purified; the cDNA coding for GFP-TbATXN2 or GFP-dPAM-TbATXN2 was amplified using Q5 High-Fidelity DNA Polymerase (New England BioLabs) with forward and reverse primers (see below, digested with XbaI and BamHI, and purified. The enzyme-cut pLew100v5_bsd and GFP-TbATXN2 or GFP-dPAM-TbATXN2 were ligated and transformed into DH5α E. coli cells, and the positive clones were confirmed by Sanger sequencing at Genewiz (Research Triangle Park, NC).
Forward primer TbATXN2: 5’GCTCTAGATAAGGCACCATGAGCAAGGGCGAGGAGCTG-3’ (XbaI site)
Reverse primer TbATXN2: 5’CGGGATCCCTATTTCCCAACTCGTTTCTTCGGCC-3’ (BamHI site)
Constructs for AtATXN2 (AT3G14010) and ΔPAM2 AtATXN2 were synthesized by GenScript Biotech Corporation (Piscataway, NJ) with flanking Gateway attB sites. They were then BP recombined using the Gateway system (Thermo Fisher Scientific) into pDONR221, and then subcloned using LR recombination (Thermo Fisher Scientific) into pGWB606 (https://shimane-u.org/nakagawa/pgwb-tables/4.htm) 101 to generate p35S:GFP-AtATXN2 and p35S:GFP-ΔPAM2AtATXN2. To generate p35S:RFP-PAB2, a pENTR223 vector (GC104970) containing PAB2’s cDNA (AT4G34110) was first obtained by the Arabidopsis Biological Resource Center (ABRC). Since it contained the same resistance, spectinomycin, as the destination vector pGWB661 (https://shimane-u.org/nakagawa/pgwb-tables/4.htm) 101, it was first subcloned into pDEST15 (Thermo Fisher Scientific) using LR recombination (Thermo Fisher Scientific) and then subcloned into another entry vector, pDONR221, (Thermo Fisher Scientific) using BP recombination (Thermo Fisher Scientific). Finally, the PAB2 cDNA sequence was then subcloned into pGWB661 to generate p35S:RFP-PAB2.
Recombinant protein purification
The coding sequences of ATXN2 IDR2 (aa 477–909) and ATXN2 IDR3 (aa 1025–1312) were amplified from human cDNA. ATXN2 IDR2 was cloned into pRSFDuet vector with an N-terminal MBP tag and C-terminal 8×His tag. ATXN2 IDR3 was cloned into pHIS-parallel vector with an N-terminal 6×His tag. BL21(DE3) cells were transformed with the plasmids and cultured in a 5 ml LB culture medium with ampicillin selection overnight at 37 °C in a shaker at 220 rpm. The culture was added to 1L of LB media and grown at 37 °C till the OD600 reached 0.6.
Transformed cells expressing ATXN2 IDR2 were induced with final 500 μM IPTG at 16 °C for 16 hours, 220 rpm. Cells were harvested at 4°C for 20 mins. The pellet was resuspended in lysis buffer (25 mM Tris-HCl pH 7.5, 500 mM NaCl, 5% glycerol and protease inhibitors) and then sonicated for 10 min (10 s on / 10 s off, 180W) using a cell ultrasonic crusher. After centrifugation at 20,000g for 40 minutes, the supernatant was collected and incubated with Ni-NTA Resin (GenScript) for 30 minutes at 4 °C. The Ni-NTA Resin was washed with wash buffer (25 mM Tris-HCl pH 7.5, 500 mM NaCl, 5% glycerol and 20 mM imidazole) and eluted with elution buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% glycerol, and 300 mM imidazole). The proteins were further loaded onto a MBPTrap HP column (Cytiva) and eluted with the MBP elution buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% glycerol and 10 mM maltose). MBP tags were then cleaved by Tobacco Etch Virus (TEV) protease using a protease-to-target protein ratio (w/w) of 1:50 at 4 °C overnight. The cleaved MBP tag was then removed by another step of Ni-NTA purification. The ATXN2 IDR2–8×His proteins were elution with the buffer containing 25 mM Tris-HCl pH 7.5, 150 mM NaCl, and concentrated to 2–3 mM.
Cells expressing ATXN2 IDR3 were induced with 500 μM IPTG at 37 °C for 4 hours, 220 rpm, and harvested by centrifuging at 4,000 rpm for 20 minutes, at 4°C. The pellet was resuspended in denaturing lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 6 M Guanidine-HCl, 0.1 mM PMSF, 20 mM β-Mercaptoethanol and 1× protease inhibitors) and then sonicated for 20 minutes (10 s on / 10 s off, 180 W) with a cell ultrasonic crusher. After centrifugation at 20,000 g for 40 minutes, the supernatant was collected and loaded onto Ni-NTA Resin for 30 minutes at 4 °C. The Ni-NTA Resin was washed with denaturing wash buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 6 M Guanidine-HCl, 0.1 mM PMSF, 20 mM β-Mercaptoethanol and 20 mM imidazole) and eluted with denaturing elution buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 6 M Guanidine-HCl, 0.1 mM PMSF, 20 mM β-Mercaptoethanol and 300 mM imidazole). The purified proteins were further concentrated to 2–3 mM. The purity of all proteins was confirmed by SDS-PAGE.
In vitro phase separation assays
The purified proteins were conjugated with fluorescent dyes by the addition of 200 ng/ml Alexa Fluor 488/568-C5-maleimide (Thermo Fisher Scientific, A10254) and incubated in dark for 1 hour at 4 °C. Spin desalting columns (Cytiva, 28918006PD) were used to remove free dyes. After equilibration, protein samples were loaded onto the desalting column and centrifuge at 2,000 g for 1 minute at 4°C. Proteins flowing through the column were collected for tube tests.
Phase-separated condensates of the purified proteins were formed by a dilution of the purified protein into a dilution buffer containing 25 mM Tris-HCl pH 7.5, 150 mM NaCl to reach the final protein concentration of 50 μM. For mixing experiment, the two IDR proteins were simultaneously added into the dilution buffer, each with a final protein concentration of 50 μM. The phase-separated droplet solution was loaded into a glass bottom 384-well plate (Cellvis) and incubated for 30 min at room temperature. Images of the condensates were taken using Olympus IXplore Spin Spinning Disk Microscope.
FRAP measurements in vitro
In vitro FRAP experiments were carried out with Nikon A1 HD25 confocal microscope. ATXN2 IDR proteins were bleached with a 488/561 nm laser pulse. For FRAP recordings, images were recorded for 10 s prior to bleaching to establish baseline fluorescence. Recovery from photobleaching was recorded for 10 minutes. For FRAP data analysis, NIS-Elements AR 5.4 software was used to calibrate the fluorescence intensity of the bleached part. Statistical analyses were carried out using Excel, GraphPad Prism.
Human cell culture, treatments and microscopy
U2OS and HeLa cells were grown at 37 °C in a humidified atmosphere with 5 % CO2 for 24 h in Dulbecco’s Modified Eagle’s Medium (DMEM), high glucose, GlutaMAX + 10 % Fetal Bovine Serum (FBS) and pen/strep (Thermo Fisher Scientific). Cells were transiently transfected using Lipofectamine 3000 (Thermo Fisher Scientific) according to manufacturer’s instructions. Cells grown on cover slips were fixed for 24 h after transfection in 4 % formaldehyde in PBS. Slides were mounted using ProLong Gold antifade reagent (Life Technologies). Confocal images were obtained using a Zeiss LSM 710 confocal microscope. Images were processed and analyzed using Fiji102. To induce stress granule formation, cells were treated for 1 hour with 250 μM of sodium arsenite (Sigma-Aldrich).
Antibodies: Anti-PABP antibody (Abcam ab21060), anti-G3BP1 (Abcam ab181150), anti-FLAG (Sigma-Aldrich F7425 and F1807), anti-acetylated tubulin (Abcam ab24610).
FRAP measurements in human cells
U2OS cells were cultured in glass bottom dishes (Ibidi) and transfected with GFP-ATXN2 constructs as described above. After 24 h, GFP-ATXN2 condensates were bleached and fluorescence recovery after bleaching was monitored using ZEN software on a Zeiss LSM 710 confocal microscope with incubation chamber at 37 °C and 5 % CO2. Data were analyzed as described previously 103. In brief, raw data were background subtracted and normalized using Excel, and subsequently plotted using GraphPad Prism 8.4.1 software. To allow for proper comparison between conditions, we only quantified highly expressing wildtype and ΔPAM2 cells, which both develop larger condensates.
ATXN2 KO generation in human cells
An ATXN2-targeting sgRNA sequence (GATGGCATGGAGCCCCGATCC) was cloned into a lentiviral backbone containing mCherry and puromycin resistance cassette. We then transfected this construct into HEK293T cells (from ATCC) at 70–80% confluency in 6-well plates. The resulting supernatant, collected 48 hours later using a syringe through a 0.45um filter (EMD Millipore; SLHP033RS), was used to infect low-passage HeLa-cas9 cells (HeLa cells expressing cas9) for generation of the knockout line. HeLa-Cas9 cells were cultured in DMEM containing high glucose, 10% FBS, and pen/strep in a 10cm plate. Virus titering was performed such that MOI of the sgRNA sequence-containing construct was <40% (as determined by % of cells that were mCherry-positive). The media was changed 24 hours after infection to get rid of lentivirus-containing media. One week after infection, mCherry-positive cells (cells that incorporated a sgRNA) were single-cell sorted into a 96-well plate. Clones were grown up then Sanger sequenced at the ATXN2 locus to determine successful knockout. One of the confirmed clonal knockout lines was used for this study.
Human cells Western blots
Ice-cold RIPA buffer (Sigma-Aldrich R0278) containing protease inhibitor cocktail (Thermo Fisher 78429) and phosphatase inhibitor (Thermo Fisher 78426) was placed on cells for lysis. After 1–2 min, the lysates were moved to Protein LoBind tubes (Eppendorf 02243108), vortexed, and placed on ice. The lysates were vortexed two more times after 10 min intervals then pelleted at maximum speed on a table-top centrifuge for 15 minutes at 4°C. After moving the supernatant to new Protein LoBind tubes, protein concentrations were determined using bicinchoninic acid (Invitrogen 23225) assays. Samples were denatured at 70°C in LDS sample buffer (Invitrogen NP0008) containing 2.5% 2-mercaptoethanol (Sigma-Aldrich) for 10 min. Samples were run on 4–12% Bis–Tris gels (Thermo Fisher) using gel electrophoresis, then wet-transferred (Bio-Rad Mini Trans-Blot Electrophoretic Cell 170–3930) onto 0.45 μm nitrocellulose membranes (Bio-Rad 162–0115) at 100V for 90 min. Odyssey Blocking Buffer (LI-COR 927–40010) was applied to membranes for one hour then replaced with Odyssey Blocking Buffer containing antibodies against ataxin-2 (1:1000, ProteinTech 21776–1-AP) and β-actin (1:2000, Thermo Fisher Scientific MA1–744) and placed on a shaker overnight at room temperature. After rinsing three times in PBS-Tween (0.1%) for 10 min each, membranes were incubated in Odyssey Blocking Buffer containing HRP-conjugated anti-rabbit IgG (H + L) (1:2000, Life Technologies 31462) or anti-mouse IgG (H + L) (1:2000, Fisher 62–6520) secondary antibodies for one hour. After rinsing the blots three additional times in PBS-Tween (0.1%), the membranes were developed using ECL Prime kit (Invitrogen) and imaged using ChemiDoc XRS + System and Image Lab software (Bio-Rad Laboratories).
Capsaspora cell transfection and microscopy
Confluent Capsaspora cells were co-transfected with the pONSY-H2B-mCherry and pONSY-ATXN2-GFP or pONSY-dPAM2-ATXN2-GFP plasmids as described elsewhere104. For life imaging, cells were seeded in a μ-Slide 4-well glass-bottom dish (Ibidi). Pictures were taken 2 days after transfection on a Zeiss Axio Observer Z.1 epifluorescence inverted microscope equipped with LED illumination and a Axioscan 503 mono camera. A 100x immersion oil objective was used. All pictures were taken at the same laser intensity and exposure settings.
Trypanosoma cell transfection, immunoblotting and microscopy
The plasmid pLew100 (GFP-TbATXN2) or pLew100(GFP-dPAM-TbATXN2) was NotI-linearized and transfected into mid-log phase T. brucei PCF, as described previously99. The stable transformants were obtained in SDM-79 medium supplemented with 15% FBS plus the appropriate antibiotic (15 μg/ml G418, 50 μg/ml hygromycin, and 10 μg/ml blasticidin). Expression of GFP-TbATXN2 or GFP-dPAM-TbATXN2 was induced with 1 μg/ml fresh tetracycline and confirmed by immunoblotting and microscopy, as described below.
The blots were incubated with rabbit antibodies against GFP (1:2,500) or mouse antibodies against tubulin (1:10,000) for 1 h. After five washings with PBS-T, the blots were incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (H+L) antibody at a dilution of 1:15,000 for 1 h. After washing five times with PBS-T, the immunoblots were visualized using Pierce ECL western blotting substrate according to the manufacturer’s instructions.
The tetracycline-induced trypanosomes were fixed with paraformaldehyde, adhered to poly-L-lysine-coated coverslips, permeabilized with Triton X-100, and blocked with BSA, as described previously 99. After blocking, trypanosomes were stained in 3% BSA/PBS with rabbit polyclonal antibody against GFP (1:250) for 1 h. After thoroughly washing with PBS containing 3% BSA, cells were incubated with Alexa 488-conjugated goat anti-rabbit antibody at 1:1,000 for 1 h. The cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) before mounting with Gold ProLong Gold antifade reagent (Molecular Probes). Differential interference contrast (DIC) and fluorescent optical images were captured using an Olympus IX-71 inverted fluorescence microscope with a Photometrix CoolSnapHQ charge-coupled device camera driven by DeltaVision software (Applied Precision, Seattle, WA). Images were deconvolved for 15 cycles using SoftwoRx deconvolution software.
Transgenic Arabidopsis thaliana lines
Transgenic plants were generated using Agrobacterium tumefaciens-mediated (GV3101 strain) transformation 105 of Col-0 with the constructs described in the Plant plasmid construction section. Transgenic seedlings (T1) were selected with Basta and left to self to generate T2 seeds. These were then selected on MS medium supplemented with Basta to select only those that carried one T-DNA construct as determined by the Mendelian segregation ratio (3:1) of the Basta-resistance trait. Basta-resistant seedlings from the selected lines were transferred to soil and left to self. Only lines with 100% Basta-resistant progeny (T3), indicating their homozygosity, were used for microscopy experiments
Tobacco transient assays
Agrobacterium lines (GV3101 strain) carrying p35S:GFP-AtATXN2, p35S:GFP-ΔPAM2AtATXN2 and p35S:RFP-PAB2, were grown overnight at 28 ˚C in LB broth (Fisher BioReagents) containing 25 mg/L rifampicin (Fisher BioReagents), 50 mg/mL gentamicin (GoldBio) and 50 mg/L spectinomycin (GoldBio). Cultures were washed four times with infiltration buffer (10 mM MgCl2 (omniPur, EMD), 10 mM MES (pH 5.6) (J. T. Baker), and 100 uM acetosyringone (Sigma-Aldrich)) and diluted to reach an OD600 of 0.8. An equal amount of cultures carrying p35S:RFP-PAB2 and p35S:GFP-AtATXN2 or p35S:GFP-ΔPAM2AtATXN2 were then pre-mixed. The mixtures were then infiltrated into 4th or 5th leaves from 6-week-old tobacco plants using Monoject 1mL Tuberculin Syringes (Covidien). For each pair of constructs, four individual tobacco plants were infiltrated. Three days after infiltration, small leaf squares (approximately 0.5 cm side length) were cut from the infiltrated regions and mounted between two cover slips in water. They were then either directly imaged (no stress condition) or placed in a 37 °C incubator for 30 min to induce SGs (heat shock) before imaging.
Plant microscopy
Arabidopsis thaliana seedlings and tobacco leaves were imaged at room temperature on a LEICA TCS SP8 laser scanning confocal microscope in resonant scanning mode using the Leica Application Suite X software. Seedlings or tobacco samples were mounted in water and then imaged using the HC PL APO CS2 63X/1.20 water objective. GFP and RFP fluorescence signals were detected by exciting with a white light laser at 488 nm and 555 nm, respectively, and by collecting emission from 500–550 nm and 565–615 nm, respectively, on a HyD SMD hybrid detector (Leica) with a lifetime gate filter of 1–6 ns to reduce background autofluorescence. Z-stacks (Arabidopsis) were collected with a bidirectional 64-line averaging while single-frame images (tobacco) were collected with a bidirectional 1024-line averaging. For colocalization experiments, samples were imaged sequentially between each line to ensure that the colocalization signals were not due to bleed-throughs. Images are representative of at least three biological replicates for each construct (tobacco) and of at least three independently generated transgenic lines (Arabidopsis).
Disorder prediction
Intrinsic disorder in ATXN2 was predicted using metapredict V2 (https://metapredict.net/)98.
Coarse-grained simulations
Coarse-grained molecular dynamics (MD) simulations were performed using the LAMMPS simulation engine with Mpipi model using the default parameters106. Simulations were run in the NVT ensemble. Mpipi is a one-bead-per-residue coarse-grained force field developed specifically for working with intrinsically disordered proteins. Non-bonded interactions are driven by a short-range Lennard-Jones-like potential (Wang-Frankel potential), while long-range electrostatics are driven by a Coulombic potential attenuated by a Debye-Hückel screening term. Bonded interactions are encoded via a simple harmonic potential. All parameters and simulation settings were run as described by Joseph et al.106.
Simulations were run on ATXN2476−1313, an IDR of 837 amino acids in length. Thirty independent simulations were run for an aggregate simulation time of 2.4 μ in a 60 nm3 simulation box. The ensemble average radius of gyration for this sequence is 84.6 Å ± 1.3. Given the underlying forcefield’s granularity and the IDR’s size, we interpret our simulations as a qualitative assessment of ensemble-average intramolecular chemical interaction. Simulations were analyzed using SOURSOP107.
The scaling map shown in Fig. 4E were calculated by fitting the internal scaling profile of the full IDR2-IDR3 simulated ensemble against a polymer scaling model, leading to ensemble-average scaling parameters of R0 = 6.68 and ν = 0.518. Using this model, all intra-residue distances were normalized against the expected distances obtained if the chain behaved as a homopolymer using the get_polymer_scaled_distance_map() function in SOURSOP using mode signed-fractional-change. Code for all simulation analyses along with the full trajectory are provided at https://github.com/holehouselab/supportingdata/tree/master/2023/boeynames_2023_ATXN2.
Evolutionary analysis
The cladogram depicted in Figure 3 and Figure S2 was generated using phyloT v2 (https://phylot.biobyte.de/) and iTOL v6 (https://itol.embl.de/)108. species list for cladograms for amino acid composition and PAM2 (from top to bottom): Homo sapiens, Mus musculus, Bos taurus, Gallus gallus, Falco peregrinus, Alligator mississippiensis, Anolis carolinensis, Xenopus tropicalis, Latimeria chalumnae, Danio rerio, Apis mellifera, Drosophila melanogaster, Ustilago maydis, Aspergillus niger, Arabidopsis thaliana, Oryza sativa, Chlamydomonas reinhardtii, Naegleria gruberi, Toxoplasma gondii. Amino acid percentages were calculated via ProtParam (https://web.expasy.org/protparam/)109 and plotted in heatmaps via Morpheus (https://software.broadinstitute.org/morpheus/).
QUANTIFICATION AND STATISTICAL ANALYSIS
All data was analyzed using GraphPad Prism 8.4.1 and Excel. Statistical tests, p values, number of samples, replicates, and experiments are indicated in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-PABP antibody | Abcam | ab21060 |
| Anti-G3BP1 | Abcam | ab181150 |
| Anti-acetylated tubulin | Abcam | ab24610 |
| Anti-FLAG | Sigma-Aldrich | F7425 |
| Anti-FLAG | Sigma-Aldrich | F1807 |
| Anti-ATXN2 | ProteinTech | 21776–1-AP |
| Anti-beta actin | Thermo-Fischer | MA1–744 |
| Anti-GFP | Molecular Probes | A-6455 |
| Anti-alpha tubulin | Sigma-Aldrich | T6199 |
| Bacterial and virus strains | ||
| TOP10 E. coli cells | Thermo-Fischer | C404010 |
| Stbl3 E. coli cells | Thermo-Fischer | C737303 |
| BL21(DE3) E. coli cells | Thermo-Fischer | EC0114 |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Sodium arsenite | Sigma-Aldrich | S7400 |
| Recombinant IDR2 | This study | N/A |
| Recombinant IDR3 | This study | N/A |
| Critical commercial assays | ||
| Lipofectamine 3000 | Thermo-Fischer | L3000001 |
| Deposited data | ||
| Experimental models: Cell lines | ||
| U2OS | ATCC | HTB-96 |
| HeLa | ATCC | CRM-CCL-2 |
| Experimental models: Organisms/strains | ||
| Capsaspora owczarzaki | ATCC | 50974 |
| Trypanosoma brucei | Docampo lab | PCF 29–13 |
| Arabidopsis thaliana | Rhee lab | Col-0 |
| Nicotiana benthamiana | Rhee lab | N/A |
| Oligonucleotides | ||
| Custom oligonucleotides | IDT | N/A |
| Recombinant DNA | ||
| Custom constructs | Genscript | N/A |
| Software and algorithms | ||
| FIJI | Shindelin et al. (2012) | https://imagej.net/software/fiji/ |
| Metapredict v2.2 | Emenecker et al. (2021) | https://metapredict.net/# |
| GraphPad Prism 8.4.1 | GraphPad | https://www.graphpad.com/features |
| ProtParam | Wilkins et al. (1999) | https://web.expasy.org/protparam/ |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| LAMMPS | Thompson et al. (2022) | https://www.lammps.org/#gsc.tab=0 |
| Mpipi | Joseph et al. (2021) | https://doi.org/10.6084/m9.figshare.1677281270 |
| Simulations for this study | This study | https://www.doi.org/10.5281/zenodo.7927158 |
| MAPanalyzer | Zhou et al. (2015) | http://systbio.cau.edu.cn/mappred/ |
| PLAAC | Alberti et al. (2009) | http://plaac.wi.mit.edu/ |
| Other | ||
HIGHLIGHTS.
A conserved short linear motif acts as an ataxin-2 condensation switch
Poly(A)-binding protein binds this motif and prevents ataxin-2 condensation
This chaperone activity is dependent on RNA binding
ACKNOWLEDGEMENTS:
We thank all members of the Gitler lab as well as the Carnegie-Stanford Intrinsically Disordered Protein Scientific Interest Group (IDPSIG) for helpful discussion and suggestions. We thank the Stanford Neuroscience Microscopy Service for use of the core facility. We thank Dr. Seung Y. Rhee for resources and feedback, G. Materassi-Shultz for plant growth facilities management, and the Carnegie Advanced Imaging facility.
FUNDING:
Work in the A.D.G. lab is supported by NIH (grant R35NS097263). A.D.G. is a Chan Zuckerberg Biohub Investigator. Work in the S.B. lab is supported by CPRIT (RR220094) and NSF (WALII, DBI grant # 2213983). S.B. acknowledges an EMBO Long Term Fellowship. Y.D. was supported by the Stanford Graduate Fellowship in Science and Engineering, Carnegie Institution for Science, and Brigitte Berthelemot. G.K. is supported by a fellowship from the Knight-Hennessy Scholars Program at Stanford University. The Stanford Neuroscience Microscopy Service is supported by NIH (grant NS069375). Work in the I.R-T. lab was supported by grant (BFU2017-90114-P) from Ministerio de Economía y Competitividad (MINECO), Agencia Estatal de Investigación (AEI), and Fondo Europeo de Desarrollo Regional (FEDER) to I.R.-T. Work in the R.D. lab was supported by NIH (grant AI140421). Work in the Y.L. lab is supported by National Natural Science Foundation of China (Grant # 32170684). A.S.H. is supported by the Human Frontier Science Program (RGP0015/2022) and NSF (WALII, DBI grant # 2213983). D.G. is supported by an NSF Graduate Research Fellowship (DGE-2139839).
Footnotes
DECLARATION OF INTERESTS:
A.D.G is a scientific founder of Maze Therapeutics. A.S.H. is a scientific consultant for Dewpoint Therapeutics and on the Scientific Advisory Board for Prose Foods. All other authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wolozin B, and Ivanov P. (2019). Stress granules and neurodegeneration. Nat Rev Neurosci 20, 649–666. 10.1038/s41583-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGurk L, Gomes E, Guo L, Mojsilovic-Petrovic J, Tran V, Kalb RG, Shorter J, and Bonini NM (2018). Poly(ADP-Ribose) Prevents Pathological Phase Separation of TDP-43 by Promoting Liquid Demixing and Stress Granule Localization. Mol Cell 71, 703–717 e709. 10.1016/j.molcel.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG, Copley KE, Hurtle B, Portz B, Pyles NJ, et al. (2019). RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron 102, 321–338 e328. 10.1016/j.neuron.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, et al. (2020). RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361 e317. 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. (2015). Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 162, 1286–1298. 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R. (2016). ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–498. 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, and Parker R. (2017). The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol Cell 68, 808–820 e805. 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. (2018). Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604 e513. 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namkoong S, Ho A, Woo YM, Kwak H, and Lee JH (2018). Systematic Characterization of Stress-Induced RNA Granulation. Mol Cell 70, 175–187 e178. 10.1016/j.molcel.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marmor-Kollet H, Siany A, Kedersha N, Knafo N, Rivkin N, Danino YM, Moens TG, Olender T, Sheban D, Cohen N, et al. (2020). Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. Mol Cell 80, 876–891 e876. 10.1016/j.molcel.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King OD, Gitler AD, and Shorter J. (2012). The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462, 61–80. 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuechler ER, Budzynska PM, Bernardini JP, Gsponer J, and Mayor T. (2020). Distinct Features of Stress Granule Proteins Predict Localization in Membraneless Organelles. J Mol Biol 432, 2349–2368. 10.1016/j.jmb.2020.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, et al. (2017). Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol Cell 65, 1044–1055 e1045. 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann TM, et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramaswami M, Taylor JP, and Parker R. (2013). Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–736. 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li YR, King OD, Shorter J, and Gitler AD (2013). Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201, 361–372. 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monahan Z, Shewmaker F, and Pandey UB (2016). Stress granules at the intersection of autophagy and ALS. Brain Res 1649, 189–200. 10.1016/j.brainres.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchan JR, Kolaitis RM, Taylor JP, and Parker R. (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, et al. (2018). Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol 28, 420–435. 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357. 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 21.Han X, Yu D, Gu R, Jia Y, Wang Q, Jaganathan A, Yang X, Yu M, Babault N, Zhao C, et al. (2020). Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat Struct Mol Biol 27, 333–341. 10.1038/s41594-020-0394-8. [DOI] [PubMed] [Google Scholar]
- 22.Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. (2020). Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324 e328. 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 24.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y, Protter DS, Rosen MK, and Parker R. (2015). Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60, 208–219. 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pulst SM, Nechiporuk A, Nechiporuk T, Gispert S, Chen XN, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, et al. (1996). Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet 14, 269–276. 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 27.Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, et al. (1996). Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet 14, 277–284. 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 28.Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier JM, Weber C, Mandel JL, Cancel G, Abbas N, et al. (1996). Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet 14, 285–291. 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 29.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. (2010). Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075. 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass JD, Dewan R, Ding J, Gibbs JR, Dalgard C, Keagle PJ, Shankaracharya, Garcia-Redondo A, Traynor BJ, Chia R, and Landers JE (2022). ATXN2 intermediate expansions in amyotrophic lateral sclerosis. Brain 145, 2671–2676. 10.1093/brain/awac167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, et al. (2017). Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371. 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scoles DR, Meera P, Schneider MD, Paul S, Dansithong W, Figueroa KP, Hung G, Rigo F, Bennett CF, Otis TS, and Pulst SM (2017). Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544, 362–366. 10.1038/nature22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh A, Hulsmeier J, Kandi AR, Pothapragada SS, Hillebrand J, Petrauskas A, Agrawal K, Rt K, Thiagarajan D, Jayaprakashappa D, et al. (2021). Antagonistic roles for Ataxin-2 structured and disordered domains in RNP condensation. Elife 10. 10.7554/eLife.60326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakthavachalu B, Huelsmeier J, Sudhakaran IP, Hillebrand J, Singh A, Petrauskas A, Thiagarajan D, Sankaranarayanan M, Mizoue L, Anderson EN, et al. (2018). RNP-Granule Assembly via Ataxin-2 Disordered Domains Is Required for Long-Term Memory and Neurodegeneration. Neuron 98, 754–766 e754. 10.1016/j.neuron.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 35.Lim C, and Allada R. (2013). ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science 340, 875–879. 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Ling J, Yuan C, Dubruille R, and Emery P. (2013). A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science 340, 879–882. 10.1126/science.1234746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeffer M, Gispert S, Auburger G, Wicht H, and Korf HW (2017). Impact of Ataxin-2 knock out on circadian locomotor behavior and PER immunoreaction in the SCN of mice. Chronobiol Int 34, 129–137. 10.1080/07420528.2016.1245666. [DOI] [PubMed] [Google Scholar]
- 38.Huebra L, Coelho FM, Filho FMR, Barsottini OG, and Pedroso JL (2019). Sleep Disorders in Hereditary Ataxias. Curr Neurol Neurosci Rep 19, 59. 10.1007/s11910-019-0968-1. [DOI] [PubMed] [Google Scholar]
- 39.Boentert M. (2020). Sleep and Sleep Disruption in Amyotrophic Lateral Sclerosis. Curr Neurol Neurosci Rep 20, 25. 10.1007/s11910-020-01047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auburger G, Sen NE, Meierhofer D, Basak AN, and Gitler AD (2017). Efficient Prevention of Neurodegenerative Diseases by Depletion of Starvation Response Factor Ataxin-2. Trends Neurosci 40, 507–516. 10.1016/j.tins.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. (2020). G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 181, 325–345 e328. 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, and Anderson P. (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212, 845–860. 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lastres-Becker I, Nonis D, Eich F, Klinkenberg M, Gorospe M, Kotter P, Klein FA, Kedersha N, and Auburger G. (2016). Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation. Biochim Biophys Acta 1862, 1558–1569. 10.1016/j.bbadis.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozlov G, Menade M, Rosenauer A, Nguyen L, and Gehring K. (2010). Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J Mol Biol 397, 397–407. 10.1016/j.jmb.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 45.Xie J, Kozlov G, and Gehring K. (2014). The “tale” of poly(A) binding protein: the MLLE domain and PAM2-containing proteins. Biochim Biophys Acta 1839, 1062–1068. 10.1016/j.bbagrm.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Jimenez-Lopez D, and Guzman P. (2014). Insights into the evolution and domain structure of Ataxin-2 proteins across eukaryotes. BMC Res Notes 7, 453. 10.1186/1756-0500-7-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrer-Bonet M, and Ruiz-Trillo I. (2017). Capsaspora owczarzaki. Curr Biol 27, R829–R830. 10.1016/j.cub.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt HB, and Rohatgi R. (2016). In Vivo Formation of Vacuolated Multi-phase Compartments Lacking Membranes. Cell Rep 16, 1228–1236. 10.1016/j.celrep.2016.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H, Lu S, Gasior K, Singh D, Vazquez-Sanchez S, Tapia O, Toprani D, Beccari MS, Yates JR 3rd, Da Cruz S, et al. (2021). HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science 371. 10.1126/science.abb4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lieberman AP, Shakkottai VG, and Albin RL (2019). Polyglutamine Repeats in Neurodegenerative Diseases. Annu Rev Pathol 14, 1–27. 10.1146/annurev-pathmechdis-012418-012857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Yang S, Mao T, and Zhang Z. (2015). MAPanalyzer: a novel online tool for analyzing microtubule-associated proteins. Database (Oxford) 2015. 10.1093/database/bav108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alberti S, Halfmann R, King O, Kapila A, and Lindquist S. (2009). A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158. 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newby GA, and Lindquist S. (2013). Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol 23, 251–259. 10.1016/j.tcb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018). A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699 e616. 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown CJ, Johnson AK, and Daughdrill GW (2010). Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol 27, 609–621. 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zarin T, Strome B, Nguyen Ba AN, Alberti S, Forman-Kay JD, and Moses AM (2019). Proteome-wide signatures of function in highly diverged intrinsically disordered regions. Elife 8. 10.7554/eLife.46883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bogaert E, Boeynaems S, Kato M, Guo L, Caulfield TR, Steyaert J, Scheveneels W, Wilmans N, Haeck W, Hersmus N, et al. (2018). Molecular Dissection of FUS Points at Synergistic Effect of Low-Complexity Domains in Toxicity. Cell Rep 24, 529–537 e524. 10.1016/j.celrep.2018.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Strohl F, et al. (2018). FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 173, 720–734 e715. 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall D. (2020). On the nature of the optimal form of the holdase-type chaperone stress response. FEBS Lett 594, 43–66. 10.1002/1873-3468.13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lasker K, Boeynaems S, Lam V, Stainton E, Jacquemyn M, Daelemans D, Villa E, Holehouse AS, Gitler AD, and Shapiro L. (2021). A modular platform for engineering function of natural and synthetic biomolecular condensates. BioRxiv. 10.1101/2021.02.03.429226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goss DJ, and Kleiman FE (2013). Poly(A) binding proteins: are they all created equal? Wiley Interdiscip Rev RNA 4, 167–179. 10.1002/wrna.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wheeler JR, Matheny T, Jain S, Abrisch R, and Parker R. (2016). Distinct stages in stress granule assembly and disassembly. Elife 5. 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie J, Wei Y, Chen Y, and Gehring K. (2021). Loss of PABPC1 is compensated by elevated PABPC4 and correlates with transcriptome changes. BioRxiv. 10.1101/2021.02.07.430165. [DOI] [Google Scholar]
- 64.Gorgoni B, Richardson WA, Burgess HM, Anderson RC, Wilkie GS, Gautier P, Martins JP, Brook M, Sheets MD, and Gray NK (2011). Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc Natl Acad Sci U S A 108, 7844–7849. 10.1073/pnas.1017664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao N, Kamijo K, Fox PD, Oda H, Morisaki T, Sato Y, Kimura H, and Stasevich TJ (2019). A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nat Commun 10, 2947. 10.1038/s41467-019-10846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buratti E, and Baralle FE (2001). Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276, 36337–36343. 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 67.Alberti S, Mateju D, Mediani L, and Carra S. (2017). Granulostasis: Protein Quality Control of RNP Granules. Front Mol Neurosci 10, 84. 10.3389/fnmol.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang P, Fan B, Yang P, Temirov J, Messing J, Kim HJ, and Taylor JP (2019). Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife 8. 10.7554/eLife.39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Protter DSW, and Parker R. (2016). Principles and Properties of Stress Granules. Trends Cell Biol 26, 668–679. 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tompa P, Davey NE, Gibson TJ, and Babu MM (2014). A million peptide motifs for the molecular biologist. Mol Cell 55, 161–169. 10.1016/j.molcel.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 71.Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng W, et al. (2017). Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J 36, 2951–2967. 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dormann D, Madl T, Valori CF, Bentmann E, Tahirovic S, Abou-Ajram C, Kremmer E, Ansorge O, Mackenzie IR, Neumann M, and Haass C. (2012). Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J 31, 4258–4275. 10.1038/emboj.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Panas MD, Schulte T, Thaa B, Sandalova T, Kedersha N, Achour A, and McInerney GM (2015). Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog 11, e1004659. 10.1371/journal.ppat.1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouillier C, Cosentino G, Leger T, Rincheval V, Richard CA, Desquesnes A, Sitterlin D, Blouquit-Laye S, Eleouet JF, Gault E, and Rameix-Welti MA (2019). The Interactome analysis of the Respiratory Syncytial Virus protein M2–1 suggests a new role in viral mRNA metabolism post-transcription. Sci Rep 9, 15258. 10.1038/s41598-019-51746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mihalic F, Simonetti L, Giudice G, Sander MR, Lindqvist R, Akprioro Peters MP, Benz C, Kassa E, Badgujar D, Inturi R, et al. (2022). Large-scale phage-based screening reveals extensive pan-viral mimicry of host short linear motifs. BioRxiv. 10.1101/2022.06.19.496705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp MD, Simons M, Niessing D, Madl T, and Dormann D. (2018). Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 173, 706–719 e713. 10.1016/j.cell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 77.Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O’Donovan K, Fare CM, Diaz Z, Singh N, et al. (2018). Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 173, 677–692 e620. 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.del Castillo U, Norkett R, Serpinskaya A, and Gelfand VI (2021). Ataxin-2 is essential for neurodevelopment in Drosophila and is a major regulator of the cytoskeleton. BioRxiv. 10.1101/2021.01.07.425768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gnazzo MM, Uhlemann EE, Villarreal AR, Shirayama M, Dominguez EG, and Skop AR (2016). The RNA-binding protein ATX-2 regulates cytokinesis through PAR-5 and ZEN-4. Mol Biol Cell 27, 3052–3064. 10.1091/mbc.E16-04-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stubenvoll MD, Medley JC, Irwin M, and Song MH (2016). ATX-2, the C. elegans Ortholog of Human Ataxin-2, Regulates Centrosome Size and Microtubule Dynamics. PLoS Genet 12, e1006370. 10.1371/journal.pgen.1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boddeker TJ, Rosowski KA, Berchtold D, Emmanouilidis L, Han Y, Allain FHT, Style RW, Pelkmans L, and Dufresne ER (2022). Non-specific adhesive forces between filaments and membraneless organelles. Nat Phys 18, 571–578. 10.1038/s41567-022-01537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, et al. (2014). C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194. 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zu T, Cleary JD, Liu Y, Banez-Coronel M, Bubenik JL, Ayhan F, Ashizawa T, Xia G, Clark HB, Yachnis AT, et al. (2017). RAN Translation Regulated by Muscleblind Proteins in Myotonic Dystrophy Type 2. Neuron 95, 1292–1305 e1295. 10.1016/j.neuron.2017.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Todd TW, McEachin ZT, Chew J, Burch AR, Jansen-West K, Tong J, Yue M, Song Y, Castanedes-Casey M, Kurti A, et al. (2020). Hexanucleotide Repeat Expansions in c9FTD/ALS and SCA36 Confer Selective Patterns of Neurodegeneration In Vivo. Cell Rep 31, 107616. 10.1016/j.celrep.2020.107616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, et al. (2013). Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646. 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. (2013). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 87.Reyes CJ, Asano K, Todd PK, Klein C, and Rakovic A. (2022). Repeat-Associated Non-AUG Translation of AGAGGG Repeats that Cause X-Linked Dystonia-Parkinsonism. Mov Disord 37, 2284–2289. 10.1002/mds.29183. [DOI] [PubMed] [Google Scholar]
- 88.Odeh HM, and Shorter J. (2020). Arginine-rich dipeptide-repeat proteins as phase disruptors in C9-ALS/FTD. Emerg Top Life Sci 4, 293–305. 10.1042/ETLS20190167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fumagalli L, Young FL, Boeynaems S, De Decker M, Mehta AR, Swijsen A, Fazal R, Guo W, Moisse M, Beckers J, et al. (2021). C9orf72-derived arginine-containing dipeptide repeats associate with axonal transport machinery and impede microtubule-based motility. Sci Adv 7. 10.1126/sciadv.abg3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boeynaems S, Ma XR, Yeong V, Ginell GM, Chen JH, Blum JA, Nakayama L, Sanyal A, Briner A, Van Haver D, et al. (2023). Aberrant phase separation is a common killing strategy of bioactive peptides in biology and human disease. BioRxiv. [Google Scholar]
- 91.Petrauskas A, Fortunati DL, Singh A, Kandi AR, Pothapragada SS, Agrawal K, Huelsmeier J, Hillebrand J, Brown G, Chaturvedi D, et al. (2022). Structured and disordered regions of Ataxin-2 contribute differently to the specificity and efficiency of mRNP granule formation. BioRxiv. 10.1101/2022.02.15.480566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alves-Cruzeiro JM, Mendonca L, Pereira de Almeida L, and Nobrega C. (2016). Motor Dysfunctions and Neuropathology in Mouse Models of Spinocerebellar Ataxia Type 2: A Comprehensive Review. Front Neurosci 10, 572. 10.3389/fnins.2016.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Damrath E, Heck MV, Gispert S, Azizov M, Nowock J, Seifried C, Rub U, Walter M, and Auburger G. (2012). ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet 8, e1002920. 10.1371/journal.pgen.1002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sen NE, Canet-Pons J, Halbach MV, Arsovic A, Pilatus U, Chae WH, Kaya ZE, Seidel K, Rollmann E, Mittelbronn M, et al. (2019). Generation of an Atxn2-CAG100 knock-in mouse reveals N-acetylaspartate production deficit due to early Nat8l dysregulation. Neurobiol Dis 132, 104559. 10.1016/j.nbd.2019.104559. [DOI] [PubMed] [Google Scholar]
- 95.Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, Lee VM, Finkbeiner S, Gitler AD, and Bonini NM (2014). Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet 46, 152–160. 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Inagaki H, Hosoda N, Tsuiji H, and Hoshino SI (2020). Direct evidence that Ataxin-2 is a translational activator mediating cytoplasmic polyadenylation. J Biol Chem 295, 15810–15825. 10.1074/jbc.RA120.013835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cochard A, Garcia-Jove Navarro M, Piroska L, Kashida S, Kress M, Weil D, and Gueroui Z. (2022). RNA at the surface of phase-separated condensates impacts their size and number. Biophys J 121, 1675–1690. 10.1016/j.bpj.2022.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Emenecker RJ, Griffith D, and Holehouse AS (2021). Metapredict: a fast, accurate, and easy-to-use predictor of consensus disorder and structure. Biophys J 120, 4312–4319. 10.1016/j.bpj.2021.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang G, Vercesi AE, and Docampo R. (2013). Essential regulation of cell bioenergetics in Trypanosoma brucei by the mitochondrial calcium uniporter. Nat Commun 4, 2865. 10.1038/ncomms3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 101.Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71, 2095–2100. 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 102.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boeynaems S, De Decker M, Tompa P, and Van Den Bosch L. (2017). Arginine-rich Peptides Can Actively Mediate Liquid-liquid Phase Separation. Bio-protocol 7. 0.21769/BioProtoc.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Parra-Acero H, Ros-Rocher N, Perez-Posada A, Kozyczkowska A, Sanchez-Pons N, Nakata A, Suga H, Najle SR, and Ruiz-Trillo I. (2018). Transfection of Capsaspora owczarzaki, a close unicellular relative of animals. Development 145. 10.1242/dev.162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clough SJ (2005). Floral dip: agrobacterium-mediated germ line transformation. Methods Mol Biol 286, 91–102. 10.1385/1-59259-827-7:091. [DOI] [PubMed] [Google Scholar]
- 106.Joseph JA, Reinhardt A, Aguirre A, Chew PY, Russell KO, Espinosa JR, Garaizar A, and Collepardo-Guevara R. (2021). Physics-driven coarse-grained model for biomolecular phase separation with near-quantitative accuracy. Nat Comput Sci 1, 732–743. 10.1038/s43588-021-00155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lalmansingh JM, Keeley AT, Ruff KM, Pappu RV, and Holehouse AS (2023). SOURSOP: A Python package for the analysis of simulations of intrinsically disordered proteins. bioRxiv. 10.1101/2023.02.16.528879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Letunic I, and Bork P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, and Hochstrasser DF (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112, 531–552. 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 110.Burki F, Roger AJ, Brown MW, and Simpson AGB (2020). The New Tree of Eukaryotes. Trends Ecol Evol 35, 43–55. 10.1016/j.tree.2019.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.