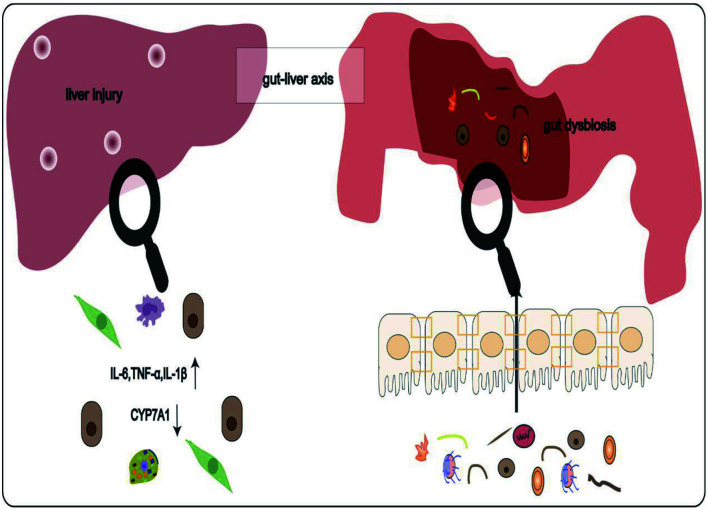
Abstract
Primary biliary cholangitis (PBC) is a complex cholestatic liver disease with an unresolved etiology. The gut microbiota is composed of a dynamic community of bacteria, archaea, fungi, and viruses that have a key role in physiological processes related to nutrition, immunity, and host defense responses. A number of recent studies found that the composition of the gut microbiota of PBC patients was significantly altered, and reported that gut dysbiosis might arise during PBC development because of the close interactions of the liver and the gut. In light of the growing interest in this topic, the focus of this review is to characterize PBC gut microbiota alterations, the correlation between PBC pathology and the gut microbiota, and prospective therapies that target the altered gut microbiota, such as probiotics and fecal microbiota transplantation.
Keywords: Primary biliary cholangitis, Gut microbiota, Lipopolysaccharides, Bile acids, Probiotics, Fecal microbiota transplantation
Graphical abstract
Introduction
Primary biliary cholangitis (PBC) is a cholestatic liver disease characterized by chronic nonsuppurative destructive lymphocytic cholangitis, specific antimitochondrial antibodies (AMAs), and a female predominance.1 It is found in most regions of the world, and both its incidence and prevalence are rising globally.2 Even though the PBC condition usually progresses slowly, it adversely impacts patient overall well being and gives rise to liver fibrosis and eventually cirrhosis.3 To date, the pathophysiology of PBC is still poorly understood. Genetic predisposition, immune dysregulation, and environmental risk factors are thought to act together in PBC development.4 Numerous studies5–13 focusing on the gut-liver axis have reported that gut microbiota dysbiosis was linked to the pathophysiology of PBC.
The gut, which accounts for the majority of the human microbiome, is the least understood of the four host tissues, the gut, oral cavity, vagina, and skin.14 The gut microbiota consists of a dynamic collection of bacteria, archaea, fungi, and viruses that are active in numerous host physiological functions, such as digestion, metabolism, resistance to pathogenic infections, and the maturation of immunity.15 Hence, even slight alterations in the gut microbiota might negatively impact organismal health at different levels. Not surprisingly, gut dysbiosis has been cited as a major contributor to the pathological process of autoimmune liver diseases, such as primary sclerosing cholangitis (PSC) and PBC.16
Currently, the first-line medication for PBC treatment is ursodeoxycholic acid (UDCA). However, approximately 40% of the patients respond poorly.1 An increasing number of studies5–13,16 have established that the pathogenesis of PBC depends heavily on the gut microbiota, making the gut microbiota a promising novel PBC diagnostic and therapeutic target. Therefore, more emphasis should be placed on further exploring the specific influence of gut microbiota on PBC, as well as its relevant pathogenetic mechanisms and potential therapies. We searched the MEDLINE literature database through PubMed for the purpose of review by entering keywords, including “gut microbiota,” “microbiota,” “gut,” “PBC,” “primary biliary cholangitis,” and “cholestatic liver disease”.
Gut-liver axis
The gut-liver axis was initially characterized in 198717 and was first reported to be implicated in liver pathogenesis in 1998.18 The portal vein and the bile acid enterohepatic circulation establish bidirectional communication of the gut-liver axis, in which the liver directly receives gut-derived products.14,19 In healthy conditions, the liver’s innate immune system is robust, allowing it to maintain immune tolerance against gut-derived antigens.20 However, in the context of gut dysbiosis or gut leakiness, pathogenic bacteria or metabolites that translocate to the liver induce corresponding immune responses that subsequently lead to liver damage.21 As the gut-liver axis has come to light as a major contributor to the occurrence and development of hepatobiliary disorders,22 a better understanding of it will help shed light on the relevant processes by which gut dysbiosis impacts PBC.
Association between gut microbiota and PBC
PBC has a significant association with the gut microbiota, as patients with recurrent urinary tract infections (UTIs) are more likely to develop PBC.23,24 Animal models of chronic bacterial exposure were shown to induce autoantibody production and PBC-like histological features.25 In addition, female dnTGFβRII mice, a transgenic murine model of PBC, had more severe phenotypes than male mice, and they could be reversed by oral antibiotic therapy.26 The aforementioned data suggest a strong link between the etiology of PBC and gut microbiota.
Furthermore, several studies5–13 have demonstrated that, compared with healthy controls, bacterial diversity declined and the overall gut microbiota composition was significantly changed in PBC patients and animal models (Table 1).5,7-13 Overall, the abundance of beneficial bacteria was decreased in PBC, while the abundance of opportunistic pathogens increased. At the phylum level, Firmicutes and Proteobacteria were over-represented in PBC, whereas Bacteroidetes were significantly decreased.5,11,13 At the genus level, compared with healthy controls, patients with PBC had an increased abundance of Sphingomonas, Haemophilus, Veillonella, Clostridium, Lactobacillus, Streptococcus, Bifidobacterium, Pseudomonas, Curvibacter, Klebsiella, Enterobacteriaceae,g, Carnobacterium, Megasphaera micronuciformis, Anaeroglobus geminatus, γ-Proteobacteria, Spirochaetaceae, Actinobacillus pleuropneumoniae, Paraprevotella clara, Methylobacterium, Acinetobacter, and Clostridiaceae and a decreased abundance of Leptotrichia, Bacteroides, Atopobium, Sutterella, Bulleidia, Oscillospira, Faecalibacterium, Acidobacteria, Lachnobacterium, Bacteroides eggerthii, Ruminococcus bromii, Morganella, Lautropia, Mogibacterium, Eikenella, Paludibacter, and F16,g.7–9,11–13 Remarkably, PBC patients who were stratified by their total bilirubin and albumin levels in the serum showed no meaningful difference in bacterial diversity and the relative abundance of the associated PBC taxa.11 Yet, the taxonomic analysis of fecal microbes from PBC patients with or without advanced fibrosis revealed a reduced alpha diversity, an increase in Weissella, and a unique microbiota composition in PBC patients with advanced fibrosis compared with patients without advanced fibrosis.10 In addition to the taxonomic analysis of fecal and mucosal samples from PBC patients, it is important to note that patients with a history of recurrent UTIs are more likely to develop PBC.23 Further, Escherichia coli was the most common pathogen isolated from patients with UTIs.24 It was also a key factor resulting from the production of AMAs,27 highlighting its role in the pathogenesis of PBC.
Table 1. Change in gut microbiota associated with PBC.
| Participants | Sample origin | Comparison | Change of gut microbiota |
Method | Ref | |
|---|---|---|---|---|---|---|
| Increased | Decreased | |||||
| dnTGFβRII mice; WT mice | Feces | PBC vs. Healthy | Firmicutes; Lachnospiraceae; Bacteroidaceae | Bacteroidetes; S24-7; Ruminococcaceae; Rikenellaceae; Porphyromonadaceae | 16SrRNA; 454 Genome Sequencer FLX Titanium platform | Ma et al.5 |
| PBC patients (n=42); Healthy controls (n=30) | Feces | PBC vs. Healthy | γ-Proteobacteria; Spirochaetaceae; Enterobacteriaceae; Veillonella; Neisseriaceae; Enterobacter asburiae; Streptococcus; Enterobacterasburiae; Actinobacillus pleuropneumoniae; Anaeroglobus geminatus; Haemophilus parainfluenzae; Megasphaera micronuciformis; Paraprevotella Clara; Klebsiella; | Acidobacteria; Lachnobacterium sp.; Bacteroides eggerthii; Ruminococcus bromii | 16SrRNA; Illumina MiSeq | Lv et al.13 |
| PBC patients (n=79); Healthy controls (n=114) | Feces | PBC vs. Healthy | f-Enterobacteriaceae; Prevotella; Sneathia; Veillonella; Fusobacterium; Haemophilus; Streptococcus; Pseudomonas; f-Clostridiaceae; Citrobacter; Lactobacillus; Salmonella; Clostridium; Klebsiella; | Bacteroides; f-Mogibacteriaceae; Blautia; f-Christensenellaceae; Butyricimonas; Akkermansia; Odoribacter; Dialister; f-S24-7; f-Rikenellaceae; Oscillospira; Faecalibacterium; Sutterella | 16SrRNA; Illumina MiSeq | Chen et al.12 |
| PBC patients (n=60); Healthy controls (n=80) | Feces | PBC vs. Healthy | Fusobacteria; Proteobacteria spp; Haemophilus; Veillonella; Klebsiella; Clostridium; Lactobacillus; Streptococcus; Pseudomonas; Enterobacteriaceae,g; | Bacteroidetes spp; Sutterella; Oscillospira; Faecalibacterium | 16SrRNA; Illumina MiSeq | Tang et al.11 |
| PBC patients (n=23) | Feces | PBC patients with advanced fibrosis (15) vs. with non-advanced fibrosis (8) | Weissella | 16SrRNA; Illumina MiSeq | Lammert et al.10 | |
| PBC patients(n=34); Healthy controls (n=21) | Ileal mucosa | PBC vs. Healthy | Sphingomonas; Pseudomonas; Methylobacterium; Carnobacterium; Acinetobacter; Curvibacter; Clostridiaceae; | Leptotrichia; Morganella; Eikenella; Lautropia; Bulleidia; Atopobium; Paludibacter; Mogibacterium; an unknown genus belonging to the class TM7_3; F16,g (an unknown genus of the family F16) | 16SrRNA; Illumina MiSeq | Kitahata et al.9 |
| PBC patients (n=76); Healthy controls (n=23) | Feces | PBC vs. Healthy | Streptococcus; Lactobacillus; Bifidobacterium; Enterococcus; | Lachnospiraceae; Ruminococcaceae | 16SrRNA; Illumina MiSeq | Furukawa et al.8 |
| PBC patients (n=39); Healthy controls (n=15) | Feces; Saliva | PBC vs. Healthy | Lactobacillales in feces; Eubacterium and Veillonella in saliva; | Clostridium subcluster XIVa in feces; Fusobacterium in saliva; | 16SrRNA; Terminal restriction fragment length polymorphism | Abe et al.7 |
Comparison of condition A vs. condition B: ↑signifies an increase in condition A relative to condition B; ↓signifies a decrease in condition A relative to condition B. PBC, primary biliary cholangitis; WT, wild type.
Some specific microbiota were reported as probably associated with PBC. For example, increased bacterial invasion of epithelial was ascribed to the elevated abundance of two genera, Enterobacter and Klebsiella in family Enterobacteriaceae.11 Chronic nonsuppurative destructive cholangitis was linked to Sphingomonadaceae abundance in PBC.9 Furthermore, some altered microbiota were positively linked to liver function and serum cytokines in PBC patients,11,13 such as Veillonella with interleukin (IL)-18 and IL16, Megasphaera micronuciformis and Enterobacter asburiae with IL18, Klebsiella with IL2A and total bilirubin, and Eggerthella sp. with direct bilirubin. Such correlations indicate that the microbes may be associated with disease states. In addition, some microbiota may act indirectly in PBC patients by modulating metabolites, such as indoleacrylate, which is produced by gut bacteria through tryptophan degradation and can trigger endothelial dysfunction and leukocyte activation.13 Indoleacrylate, enriched in the urine and feces of PBC patients, has been found to be positively correlated with enrichment of the genera Neisseriaceae and Klebsiella in PBC,13 suggesting that the two genera may be related to indoleacrylate-related signaling pathways and act together on the development of PBC. However, it still needs to be explicitly proven in future studies using the correct methods.
In summary, patients with PBC have decreased bacterial diversity and significantly different gut microbiota composition. Of note, some specific alterations of gut microbiota have a close association with the emergence of PBC.
Potential mechanisms that affect the gut microbiota in PBC
Production of metabolites
The gut microbiota, also known as “the new vital metabolic organ”, is essential for human well-being.28 Therefore it is unavoidable that any perturbation of the gut microbiota may adversely affect the body. For instance, accumulation of microbial products in the liver, such as lipopolysaccharide (LPS), leads to tissue damage by triggering inappropriate inflammatory immune responses, and impaired microbial bile-acid metabolism can result in cholestasis and liver disorder. The potential role of these two metabolites, LPS and bile acids, in the pathogenesis of PBC is summarized below.
LPS
LPS, the primary element of the Gram-negative bacterial outer membrane, consists of lipid A, core polysaccharide, and O-antigen, wherein lipid A is linked to the virulence of Gram-negative bacterial infections.29 LPS affects the immune system by activating Toll-like receptor 4 (TLR4),30 a pattern recognition receptor that confers resistance against pathogens infections and bridges innate and adaptive immunity.31,32 Low doses of LPS build a nonspecific antibacterial and antiviral defense, whereas high doses can induce inappropriate or excessive immune activation that leads to cellular injury and multiple organ damage.29 The liver is able to directly receive gut-derived products, such as pathogens, LPS, and nutrients, via the portal vein because to its anatomical location. Under healthy conditions, the liver exhibits no indication of immune response because of low hepatic TLR4 expression and negative TLR4 signaling regulation.33 Increasing evidence indicates that the LPS-TLR4 pathway is implicated in several diseases, including PBC, by activating inappropriate immune responses (Fig. 1).34–36
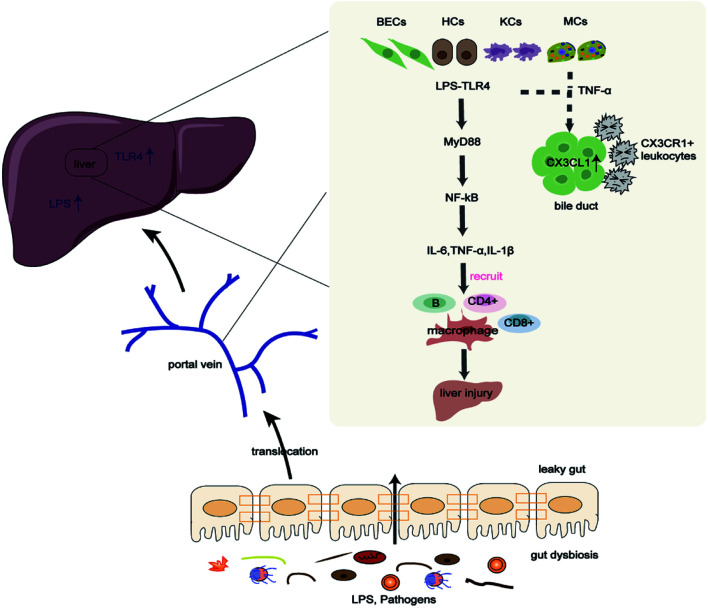
Fig. 1. Gut microbiota contributes to the pathogenesis of PBC through the LPS/TLR4 signaling pathway.
Gut dysbiosis and increased gut permeability promote pathogens and metabolite translocation to the liver, contributing to liver injury through the LPS/TLR4 signaling pathway. In addition, sensitized monocytes and TNF-α stimulate BECs to upregulate CX3CL1, causing cholangitis by interacting with CX3CR1+ leukocytes. BECs, bile duct epithelial cells; HCs, hepatocytes; IL, interleukin; KCs, Kupffer cells; LPS, lipopolysaccharide; MCs, monocytes; MyD88, myeloid differentiation factor 88; NF- κB, nuclear factor kappa-B; TLR4, Toll-like receptor 4; TNF-α, tumor necrosis factor-alpha.
Compared with healthy controls, serum LPS levels were shown to be elevated in PBC patients, and were positively linked to serum alkaline phosphatase, total bilirubin, and γ-glutamyltransferase levels.36 Increased LPS and TLR4 have also been reported in injured biliary epithelial cells (BECs), hepatocytes, and infiltrating inflammatory cells of patients with PBC.34,36,37 Increased levels of LPS activated TLR4, contributing to liver injury by promoting the recruitment of myeloid differentiation factor 88 (MyD88), the activation of the pro-inflammatory nuclear factor kappa-B (NF- κB) pathway, and the release of pro-inflammatory factors IL1β, IL6, and tumor necrosis factor-alpha (TNF-α), which facilitate recruitment of effector lymphocytes CD4+T and CD8+T into the portal tracts of PBC patients.36,38–40 Beyond that, peripheral blood mononuclear cells (PBMCs) in PBC patients are also highly sensitive to LPS. Compared with healthy controls, PBMCs isolated from PBC patients produced higher levels of IL1β, IL6, IL8, and TNF-α after LPS stimulation.36,41 Further, sensitized monocytes and TNF-α stimulated BECs to upregulate the chemokine-adhesion molecule CX3CL1. Upregulation of CX3CL1 in damaged bile ducts attracted CD4+T and CD8+T cells and interacted with CX3CR1+ leukocytes to cause cholangitis.42 In summary, the interaction between LPS and TLR4 and the upregulation of CX3CL1 in the liver promote inflammatory immune responses, causing the breakdown of local immune tolerance and liver injury.
Bile acids
Bile acids, the primary functional elements of bile, are synthesized in the liver from cholesterol via classical and unconventional pathways.43 After meals, bile acids are released into the small intestine to facilitate digestion and absorption of dietary lipids.44 Approximately 95% of the bile acids are actively reabsorbed and delivered back to the liver by portal circulation.45 The remaining 5% of bile acids are modified by various bacterial transformations, with a portion passively reabsorbed into the liver and the rest lost in the feces.46 Some bile acids are harmful to the organism. For instance, hydrophobic bile acids are considered to be hepatotoxic, leading to irreversible hepatocyte death by inducing mitochondrial oxidative stress and ER stress, with increased mitochondrial permeability.43,47 Conversely, bile acids have well-known antimicrobial properties and subsequently affect the gut microbial composition.46,48 The loss of secondary bile acids is related to susceptibility to pathogens infections, which was reversed by restoring the secondary bile acid pool.49 Taken together, the evidence of mutual regulation between bile acids and gut microbes is crucial for bile acid metabolism and microbiota composition, and perturbation may impact the pathophysiological processes of the host.
Evidence has shown that the serum and fecal bile acids profiles in PBC patients without UDCA treatment are significantly different from those of controls.12 Particularly, PBC patients without UDCA treatment showed an increase in the ratio of conjugated/unconjugated bile acids and a decrease in the ratio of secondary/primary bile acids, indicating abnormal microbial bile-acid metabolism (Fig. 2). Importantly, variation in bile acids pool of PBC patients was linked to gut dysbiosis. For instance, the level of secondary bile acids was negatively associated with genera that were enriched in PBC (Veillonella, Klebsiella), and positively associated with genera enriched in controls (Bacillus spp, Oscillatory spp).12 It is widely accepted that conversion of bile acids remaining in the intestine is primarily dependent on the activity of microbial enzymes.50,51 For example, bile salt hydrolases (BSHs), which are present in all major bacteria in the human intestine, including Bacteroides, lactobacilli, bifidobacteria, and Clostridium, deconjugate bile acids to unconjugated forms for further modification, which is the first step of bile acid metabolism in the intestine.52 Therefore, alterations of bile acids are likely the result of changes in the gut microbiota composition in PBC and subsequently dysfunction of the responsible microbial enzymes leads to lack of bile acid conversion. Conversely, because of the antimicrobial properties of bile acids, it is plausible that alterations of bile acids in PBC may profoundly affect microbial composition. Indeed, cholestyramine treatment modulated endogenous bile acids in PBC patients and had benefits associated with changes in gut microbiota and their metabolites.53
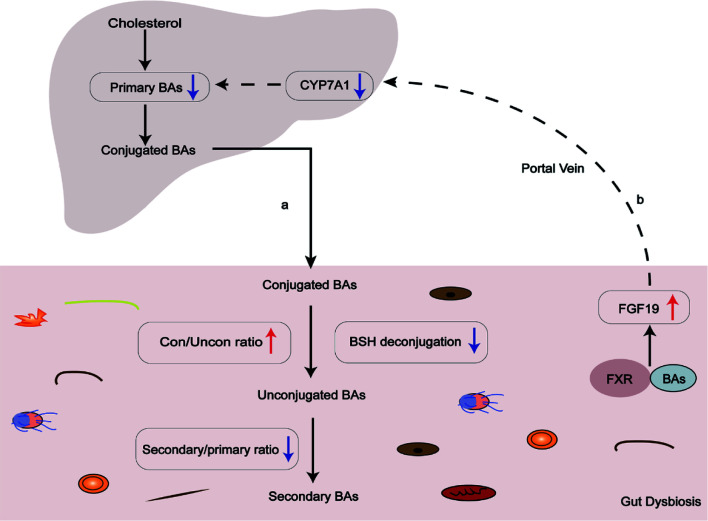
Fig. 2. Bile acid-microbiota interactions are impaired in PBC patients.
(A) Gut dysbiosis and decreased BSH deconjugation result in impaired microbial bile acid metabolism, particularly an increase in the ratio of conjugated/unconjugated bile acids and a decrease in the ratio of secondary/primary bile acids. (B) Serum FGF19 is elevated in patients with PBC, further inhibiting CYP7A1 transcription and bile acid synthesis. BAs, bile acids; BSH, bile salt hydrolase; CYP7A1, cholesterol 7-alpha-hydroxylase; FGF19, fibroblast growth factor 19; FXR, farnesoid X receptor.
In addition to the abnormal microbial bile acid metabolism described above, the gut microbiota also suppress bile acid synthesis by regulating the bile acid receptor farnesoid X receptor (FXR). FXR is a bile acid sensor that acts to regulate bile acid synthesis and homeostasis together with its downstream effector, fibroblast growth factor (FGF) 15/19.54 A recent study showed that in conventionally raised mice, bile acid synthesis was inhibited because of the alleviation of ileum FXR inhibition.55 Bile acid levels were dramatically lowered in the gallbladder compared with germ-free (GF) mice due to the presence of gut microbiota, especially the level of taurine-conjugated β-muricholic acid (TβMCA), which is an FXR antagonist. Decreased levels of TβMCA activated FXR and increased the downstream expression of FGF15, leading to the suppression of cholesterol 7-alpha-hydroxylase (CYP7A1)-mediated bile acid synthesis.55 Another study reported that FXR antagonists cannot be metabolized without gut bacteria.56 Along line, the gut microbiota might modulate bile acid biosynthesis by altering the bile acid composition and by regulating the FXR-FGF15/19 negative feedback system. Furthermore, patients with PBC had increased serum FGF19 levels, suggesting that the bile acid pool composition was changed, which was consistent with a microbial bile acid metabolism disorder in PBC.12 In summary, the gut microbiota and bile acids modulate each other. Any perturbation of the gut microbiota and bile acids may profoundly impact the organismal pathophysiology process. Significant alterations in gut microbiota composition and impaired microbial bile acids metabolism in PBC patients may account for disease consequences, such as cholestasis.
Others mechanisms
Molecular mimicry
PBC is an autoimmune liver disease with highly specific AMAs that primarily target the mitochondrial PDC-E2 complex, but what triggers this autoimmune mechanism is unclear.57,58 The involvement of molecular mimicry in the pathogenesis of PBC has been proposed, in which host cells recognize exogenous bacterial or viral epitopes and subsequently trigger AMA production and effector T-cell responses against their own proteins.59,60 Streptococcus intermedius or Novosphingobium aromaticivorans exposure led to specific antibody production and liver injury resembling PBC in mice.25,61 Furthermore, AMAs in the serum of PBC patients also crossed-react to bacterial proteins of Escherichia coli and Novosphingobium aromaticivorans.59,62,63 Both showed the close cross-reactivity between the microbes and antibodies in PBC. Accordingly, a PBC hypothesis is based on the “leaky gut concept”, in which the gut microbiota induces anti-PDC-E2 responses and biliary damage through the translocation of bacterial molecules and byproducts from the gut. Details of the molecular mimicry of microorganisms in PBC, including E. coli, mycoplasma, lactobacilli, H. pylori, and Novosphingobium aromaticivorans, have been extensively reviewed elsewhere, and some plausible mechanisms have been proposed.64–68
Regulation of gut permeability
Studies have revealed that gut permeability was increased in PBC patients,69,70 but the cause of this increase remains poorly understood. It is well established that the intestinal commensal bacteria act as an innate immune barrier to resist pathogen infections together with mucins, antimicrobial peptides, and immunoglobulins.22,71 Intestinal commensal bacteria, such as Ruminococcaceae and Eubacterium, affect gut barrier function by producing butyrate that can stimulate tight junctions and mucus production.72 Also, microbes themselves impact the intestinal barrier function by controlling the production of tight junction proteins.73 In that way, dysbiosis of the gut microbiota may disrupt tight junction and thereby increase gut permeability. As described above, Ruminococcaceae abundance was reduced in PBC patients, which might account for the increased gut permeability.8,13 Furthermore, secreted immunoglobulin A (IgA), which contributes to gut microbiome composition and is essential for maintaining intestinal homeostasis,74 was abnormal in the intestinal epithelium of PBC patients.75 In addition, the presence of gut microbes can enhance gut permeability during cholestasis.76 With compromised gut barrier function, gut-derived antigens translocate from the gut to mesenteric lymph nodes, extraintestinal organs, and the systematic circulation.34,36,77 As previously stated, translocated products may trigger inflammatory immune responses through the LPS/TLR4 pathway in the liver of PBC patients or induce autoantibodies production by molecular mimicry, thereby leading to exacerbating liver damage.
PBC and intestinal mucosal immune activation
Intestinal mucosal immunity is essential for resisting pathogen infections and maintaining intestinal homeostasis. Its dysregulation has been implicated in autoimmune diseases.78–80 The gut microbiota is required for intestinal mucosal immunity maturation,81 and lack of gut microbiota in GF mice leads to immune system deficiency.82–84 It is thus no surprise that gut dysbiosis can influence the intestinal mucosal immune balance. For example, gut dysbiosis increases inflammatory bowel disease (IBD) inflammation by inducing abnormal immune responses that are improved by active bacterial products.85–88 Prebiotic treatment decreased the levels of the proinflammatory factors in the mesenteric lymph nodes in a mice model.89 In addition, IL17-producing T helper (Th17) cells are associated with inflammatory disorders and are enriched in the liver and the gut.90 In the gut, Th17 cells are maintained by commensal bacteria that induce the secretion of a factor, serum amyloid A, needed for Th17-specific IL17A expression,91 which promotes inflammation via leucocyte recruitment.92 Indeed, Th17 cells have been linked to PBC.93 Given the significance of gut microbiota in intestinal mucosal immunity, it seems plausible to hypothesize that gut dysbiosis and imbalanced mucosal immunity contribute to the progression of PBC, warranting the emergence of new evidence.
Therapy
Knowing the importance of gut microbiota dysbiosis in PBC progression, treatments aimed at restoring the gut microbiota, such as the administration of probiotics, fecal microbiota transplantation (FMT), and the intervention of mechanistic pathways might benefit PBC patients. In addition, existing PBC medications, such as UDCA, treat the disease by both regulating bile acid metabolism and adjusting gut microbiota composition.
Probiotics
Probiotics are live, beneficial microorganisms that can help with a variety of diseases by promoting the balance of gut microbiota, improving metabolic profiles, and repairing intestinal barrier dysfunction.94–96 For example, the administration of the probiotic VSL#3 to mice, and BSH-active Lactobacillus reuteri NCIMB 30242 in humans, was shown to improve microbial bile-acid metabolism by promoting the enrichment of BSH-retaining species, mainly Lactobacillus and Bifidobacteria, in the intestinal microbiota.97,98 Furthermore, daily consumption of probiotic rich foods, such as yogurt, had positive outcomes in patients with nonalcoholic fatty liver disease (NAFLD).99 Along those lines, it is reasonable that probiotics treatment is beneficial for patients with PBC despite the lack of published evidence and warrants further study.
FMT
FMT is emerging as a way to restore healthy microbiota by transferring a fecal suspension from a healthy donor to the digestive system of a patient.100 In addition to treating recurrent Clostridium difficile infections, FMT has been found to be beneficial for autoimmune diseases, such as PSC.101–103 FMT restores gut microbiota diversity and improves biochemical indicators in patients with PSC, but needs to be validated in large study cohorts. Although FMT treatment has not been studied in PBC patients, it is intriguing to attempt that in future studies.
Intervention pathway in pathogenesis
As defective microbial bile acid metabolism and aberrant immune responses have been found in PBC, the focus is on reestablishing disrupted microbial bile acid metabolism, inhibiting abnormal immune responses, or preserving immune tolerance.1 Drugs other than UDCA and obeticholic acid (OCA) for treating PBC are still in experimental or clinical stages. Currently, drugs targeting bile acids metabolism, including FXR agonists, apical sodium-dependent bile acid transporter (ASBT) inhibitors, and peroxisome proliferator-activated receptor (PPAR) agonists, delay disease progression by decreasing cytotoxicity and inflammation.104 Furthermore, it is necessary to repair the damaged intestinal barrier to avoid inappropriate intestinal immune responses. Dietary therapies and anti-TNF-α drugs proposed for restoring gut permeability in IBD105 might also be evaluated in PBC in the future. Immunosuppressive therapy, long-term glucocorticoid treatment, and classical immunosuppressants have failed to provide significant benefits to PBC patients.106 The development of B-cell, T-cell, and cytokine/chemokine-targeted treatments is accelerating. In addition, the use of nanomedicines to optimize treatment of autoimmune disease is under development, and has the potential to overcome many of the drawbacks of current immunosuppressive therapies.107 The specific mechanisms of these drugs have been described in detail elsewhere.104
UDCA
The current first-line treatment for PBC patients is UDCA, which improves liver function and graft-free survival by several mechanisms, including enhancing the hydrophilicity of bile acid pools, immune modulation, anti-inflammatory properties, and antifibrotic properties.108,109 Remarkably, UDCA might also adjust gut microbiota composition. Studies have shown that PBC patients treated with UDCA for 6 months had a partial restoration of gut dysbiosis, with a reduced abundance of Haemophilus spp, Streptococcus spp, and Pseudomonas spp that were enriched in PBC patients without UDCA treatment. UDCA also increased Bacteroidetes spp, Oscillospira spp and Sutterella spp, which were enriched in healthy controls.11 In addition, bacteria Bilophila spp, which metabolizes taurine,110 was elevated in patients after UDCA treatment, resulting in a reduction in taurine-conjugated bile acids and the conjugated/unconjugated ratio, which was associated with a decrease in liver enzyme levels.11 Furthermore, UDCA increased the abundance of intestinal Bacteroides that express BSH, which is crucial for microbial bile acid metabolism, in pregnant intrahepatic cholestasis patients.111 The evidence suggests UDCA treatment could potentially ameliorate PBC by modifying the gut microbiota composition, but, details of the mechanism of action of UDCA on the gut microbiota of the host are poorly understood and should be explored in future studies.
Conclusions
PBC is a cholestatic liver disease with an obscure etiology. Mounting evidence points to the significance of gut-liver crosstalk in PBC progression. Overall, there exist significant differences between PBC patients and healthy individuals in gut microbiota composition and microbial metabolite levels. Gut dysbiosis promotes hepatobiliary injury in PBC patients by several mechanisms, including influencing the intestinal mucosal immune balance, increasing gut permeability that in turn promotes bacterial translocation, inducing abnormal immune activation through the LPS/TLR4 signaling pathway and molecular mimicry mechanisms, and suppressing the microbial metabolism of bile acids. For patients who do not respond effectively to UDCA/OCA treatment, novel treatments such as probiotics, FMT, and some pharmacological agents targeting gut microbiota-associated pathways represent new avenues for improving PBC by restoring gut microbiota composition and modulating immune responses. Furthermore, it should be highlighted that numerous additional variables, such as nutrition, geographical location, and medications, affect the metataxonomic analysis of human gut microbes. Thus, we should also focus on inter-individual variation.
However, there are several limitations in existing microbiome research that should be addressed in the future. Firstly, the number of studies is too small to infer any meaningful association between microbiota composition and disease severity or to control for confounding factors, such as diet, drugs, and environment. Secondly, most feces samples do not completely reflect the profiles of mucosal communities. Finally, fungi and viruses should be considered even though they only make up a modest fraction of the gut microbiota. A better overall understanding of gut dysbiosis in PBC patients will help us have a better appreciation of the pathological mechanisms of PBC.
Acknowledgments
The authors thank Yixin Zhu for help in polishing the paper’s language and grammar.
Abbreviations
- AMA
anti-mitochondrial antibody
- ASBT
apical sodium-dependent bile acid transporter
- BEC
bile duct epithelial cell
- BSH
bile salt hydrolase
- CYP7A1
cholesterol 7-alpha-hydroxylase
- FXR
farnesoid X receptor
- FGF
fibroblast growth factor
- FMT
fecal microbiota transplantation
- GF
germ-free
- IL
interleukin
- IgA
immunoglobulin A
- IBD
inflammatory bowel disease
- LPS
lipopolysaccharide
- MyD88
myeloid differentiation factor 88
- NF-κB
nuclear factor kappa-B
- NAFLD
nonalcoholic fatty liver disease
- OCA
obeticholic acid
- PBC
primary biliary cholangitis
- PSC
primary sclerosing cholangitis
- PBMC
peripheral blood mononuclear cell
- PPAR
peroxisome proliferator-activated receptor
- TLR4
Toll-like receptor 4
- TNF-α
tumor necrosis factor-alpha
- TβMCA
taurine-conjugated β-muricholic acid
- Th
T helper
- UDCA
ursodeoxycholic acid
- UTI
urinary tract infection
References
- 1.Gulamhusein AF, Hirschfield GM. Primary biliary cholangitis: pathogenesis and therapeutic opportunities. Nat Rev Gastroenterol Hepatol. 2020;17(2):93–110. doi: 10.1038/s41575-019-0226-7. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths L, Dyson JK, Jones DE. The new epidemiology of primary biliary cirrhosis. Semin Liver Dis. 2014;34(3):318–328. doi: 10.1055/s-0034-1383730. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan MM, Gershwin ME. Primary biliary cirrhosis. N Engl J Med. 2005;353(12):1261–1273. doi: 10.1056/NEJMra043898. [DOI] [PubMed] [Google Scholar]
- 4.Mattner J. Impact of Microbes on the Pathogenesis of Primary Biliary Cirrhosis (PBC) and Primary Sclerosing Cholangitis (PSC) Int J Mol Sci. 2016;17(11):1864. doi: 10.3390/ijms17111864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma HD, Zhao ZB, Ma WT, Liu QZ, Gao CY, Li L, et al. Gut microbiota translocation promotes autoimmune cholangitis. J Autoimmun. 2018;95:47–57. doi: 10.1016/j.jaut.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Ji F, Guo J, Shi D, Fang D, Li L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci Rep. 2016;6:34055. doi: 10.1038/srep34055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abe K, Takahashi A, Fujita M, Imaizumi H, Hayashi M, Okai K, et al. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS One. 2018;13(7):e0198757. doi: 10.1371/journal.pone.0198757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furukawa M, Moriya K, Nakayama J, Inoue T, Momoda R, Kawaratani H, et al. Gut dysbiosis associated with clinical prognosis of patients with primary biliary cholangitis. Hepatol Res. 2020;50(7):840–852. doi: 10.1111/hepr.13509. [DOI] [PubMed] [Google Scholar]
- 9.Kitahata S, Yamamoto Y, Yoshida O, Tokumoto Y, Kawamura T, Furukawa S, et al. Ileal mucosa-associated microbiota overgrowth associated with pathogenesis of primary biliary cholangitis. Sci Rep. 2021;11(1):19705. doi: 10.1038/s41598-021-99314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lammert C, Shin A, Xu H, Hemmerich C, O’Connell TM, Chalasani N. Short-chain fatty acid and fecal microbiota profiles are linked to fibrosis in primary biliary cholangitis. FEMS Microbiol Lett. 2021;368(6):fnab038. doi: 10.1093/femsle/fnab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67(3):534–541. doi: 10.1136/gutjnl-2016-313332. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Wei Y, Xiong A, Li Y, Guan H, Wang Q, et al. Comprehensive Analysis of Serum and Fecal Bile Acid Profiles and Interaction with Gut Microbiota in Primary Biliary Cholangitis. Clin Rev Allergy Immunol. 2020;58(1):25–38. doi: 10.1007/s12016-019-08731-2. [DOI] [PubMed] [Google Scholar]
- 13.Lv LX, Fang DQ, Shi D, Chen DY, Yan R, Zhu YX, et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol. 2016;18(7):2272–2286. doi: 10.1111/1462-2920.13401. [DOI] [PubMed] [Google Scholar]
- 14.Kho ZY, Lal SK. The Human Gut Microbiome - A Potential Controller of Wellness and Disease. Front Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 2019;76(3):473–493. doi: 10.1007/s00018-018-2943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kummen M, Hov JR. The gut microbial influence on cholestatic liver disease. Liver Int. 2019;39(7):1186–1196. doi: 10.1111/liv.14153. [DOI] [PubMed] [Google Scholar]
- 17.Volta U, Bonazzi C, Bianchi FB, Baldoni AM, Zoli M, Pisi E. IgA antibodies to dietary antigens in liver cirrhosis. Ric Clin Lab. 1987;17(3):235–242. doi: 10.1007/BF02912537. [DOI] [PubMed] [Google Scholar]
- 18.Pereira SP, Rhodes JM, Campbell BJ, Kumar D, Bain IM, Murphy GM, et al. Biliary lactoferrin concentrations are increased in active inflammatory bowel disease: a factor in the pathogenesis of primary sclerosing cholangitis? Clin Sci (Lond) 1998;95(5):637–644. doi: 10.1042/cs0950637. [DOI] [PubMed] [Google Scholar]
- 19.Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol. 2020;72(3):558–577. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Gao B, Jeong WI, Tian Z. Liver: An organ with predominant innate immunity. Hepatology. 2008;47(2):729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 21.Ma HD, Wang YH, Chang C, Gershwin ME, Lian ZX. The intestinal microbiota and microenvironment in liver. Autoimmun Rev. 2015;14(3):183–191. doi: 10.1016/j.autrev.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15(7):397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corpechot C, Chretien Y, Chazouilleres O, Poupon R. Demographic, lifestyle, medical and familial factors associated with primary biliary cirrhosis. J Hepatol. 2010;53(1):162–169. doi: 10.1016/j.jhep.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Burroughs AK, Rosenstein IJ, Epstein O, Hamilton-Miller JM, Brumfitt W, Sherlock S. Bacteriuria and primary biliary cirrhosis. Gut. 1984;25(2):133–137. doi: 10.1136/gut.25.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haruta I, Kikuchi K, Hashimoto E, Nakamura M, Miyakawa H, Hirota K, et al. Long-term bacterial exposure can trigger nonsuppurative destructive cholangitis associated with multifocal epithelial inflammation. Lab Invest. 2010;90(4):577–588. doi: 10.1038/labinvest.2010.40. [DOI] [PubMed] [Google Scholar]
- 26.Huang MX, Yang SY, Luo PY, Long J, Liu QZ, Wang J, et al. Gut microbiota contributes to sexual dimorphism in murine autoimmune cholangitis. J Leukoc Biol. 2021;110(6):1121–1130. doi: 10.1002/JLB.3MA0321-037R. [DOI] [PubMed] [Google Scholar]
- 27.Floreani A, Leung PS, Gershwin ME. Environmental Basis of Autoimmunity. Clin Rev Allergy Immunol. 2016;50(3):287–300. doi: 10.1007/s12016-015-8493-8. [DOI] [PubMed] [Google Scholar]
- 28.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21(29):8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Quinn PJ. Lipopolysaccharide: Biosynthetic pathway and structure modification. Prog Lipid Res. 2010;49(2):97–107. doi: 10.1016/j.plipres.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Ciesielska A, Matyjek M, Kwiatkowska K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol Life Sci. 2021;78(4):1233–1261. doi: 10.1007/s00018-020-03656-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 33.Pimentel-Nunes P, Soares JB, Roncon-Albuquerque R, Jr, Dinis-Ribeiro M, Leite-Moreira AF. Toll-like receptors as therapeutic targets in gastrointestinal diseases. Expert Opin Ther Targets. 2010;14(4):347–368. doi: 10.1517/14728221003642027. [DOI] [PubMed] [Google Scholar]
- 34.Sasatomi K, Noguchi K, Sakisaka S, Sata M, Tanikawa K. Abnormal accumulation of endotoxin in biliary epithelial cells in primary biliary cirrhosis and primary sclerosing cholangitis. J Hepatol. 1998;29(3):409–416. doi: 10.1016/s0168-8278(98)80058-5. [DOI] [PubMed] [Google Scholar]
- 35.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42(2):145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Zhao J, Zhao S, Zhou G, Liang L, Guo X, Mao P, et al. Altered biliary epithelial cell and monocyte responses to lipopolysaccharide as a TLR ligand in patients with primary biliary cirrhosis. Scand J Gastroenterol. 2011;46(4):485–494. doi: 10.3109/00365521.2010.539624. [DOI] [PubMed] [Google Scholar]
- 37.Wang AP, Migita K, Ito M, Takii Y, Daikoku M, Yokoyama T, et al. Hepatic expression of toll-like receptor 4 in primary biliary cirrhosis. J Autoimmun. 2005;25(1):85–91. doi: 10.1016/j.jaut.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Honda Y, Yamagiwa S, Matsuda Y, Takamura M, Ichida T, Aoyagi Y. Altered expression of TLR homolog RP105 on monocytes hypersensitive to LPS in patients with primary biliary cirrhosis. J Hepatol. 2007;47(3):404–411. doi: 10.1016/j.jhep.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 39.Yokoyama T, Komori A, Nakamura M, Takii Y, Kamihira T, Shimoda S, et al. Human intrahepatic biliary epithelial cells function in innate immunity by producing IL-6 and IL-8 via the TLR4-NF-kappaB and -MAPK signaling pathways. Liver Int. 2006;26(4):467–476. doi: 10.1111/j.1478-3231.2006.01254.x. [DOI] [PubMed] [Google Scholar]
- 40.Harada K, Isse K, Nakanuma Y. Interferon gamma accelerates NF-kappaB activation of biliary epithelial cells induced by Toll-like receptor and ligand interaction. J Clin Pathol. 2006;59(2):184–190. doi: 10.1136/jcp.2004.023507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao TK, Lian ZX, Selmi C, Ichiki Y, Ashwood P, Ansari AA, et al. Altered monocyte responses to defined TLR ligands in patients with primary biliary cirrhosis. Hepatology. 2005;42(4):802–808. doi: 10.1002/hep.20859. [DOI] [PubMed] [Google Scholar]
- 42.Shimoda S, Harada K, Niiro H, Taketomi A, Maehara Y, Tsuneyama K, et al. CX3CL1 (fractalkine): a signpost for biliary inflammation in primary biliary cirrhosis. Hepatology. 2010;51(2):567–575. doi: 10.1002/hep.23318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Tang R, Leung PSC, Gershwin ME, Ma X. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun Rev. 2017;16(9):885–896. doi: 10.1016/j.autrev.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Ticho AL, Malhotra P, Dudeja PK, Gill RK, Alrefai WA. Intestinal Absorption of Bile Acids in Health and Disease. Compr Physiol. 2019;10(1):21–56. doi: 10.1002/cphy.c190007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50(10):1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24(1):41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 47.Liu HX, Keane R, Sheng L, Wan YJ. Implications of microbiota and bile acid in liver injury and regeneration. J Hepatol. 2015;63(6):1502–1510. doi: 10.1016/j.jhep.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hofmann AF, Eckmann L. How bile acids confer gut mucosal protection against bacteria. Proc Natl Acad Sci U S A. 2006;103(12):4333–4334. doi: 10.1073/pnas.0600780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306(4):G310–319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joyce SA, Gahan CG. Bile Acid Modifications at the Microbe-Host Interface: Potential for Nutraceutical and Pharmaceutical Interventions in Host Health. Annu Rev Food Sci Technol. 2016;7:313–333. doi: 10.1146/annurev-food-041715-033159. [DOI] [PubMed] [Google Scholar]
- 52.Jones H, Alpini G, Francis H. Bile acid signaling and biliary functions. Acta Pharm Sin B. 2015;5(2):123–128. doi: 10.1016/j.apsb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Zhang J, Chen Y, Wang Q, Yan L, Wang R, et al. Alterations in microbiota and their metabolites are associated with beneficial effects of bile acid sequestrant on icteric primary biliary Cholangitis. Gut Microbes. 2021;13(1):1946366. doi: 10.1080/19490976.2021.1946366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kliewer SA, Mangelsdorf DJ. Bile Acids as Hormones: The FXR-FGF15/19 Pathway. Dig Dis. 2015;33(3):327–331. doi: 10.1159/000371670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, et al. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48(12):2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Invernizzi P, Lleo A, Podda M. Interpreting serological tests in diagnosing autoimmune liver diseases. Semin Liver Dis. 2007;27(2):161–172. doi: 10.1055/s-2007-979469. [DOI] [PubMed] [Google Scholar]
- 58.Lleo A, Selmi C, Invernizzi P, Podda M, Coppel RL, Mackay IR, et al. Apotopes and the biliary specificity of primary biliary cirrhosis. Hepatology. 2009;49(3):871–879. doi: 10.1002/hep.22736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bogdanos DP, Baum H, Grasso A, Okamoto M, Butler P, Ma Y, et al. Microbial mimics are major targets of crossreactivity with human pyruvate dehydrogenase in primary biliary cirrhosis. J Hepatol. 2004;40(1):31–39. doi: 10.1016/s0168-8278(03)00501-4. [DOI] [PubMed] [Google Scholar]
- 60.Bogdanos DP, Choudhuri K, Vergani D. Molecular mimicry and autoimmune liver disease: virtuous intentions, malign consequences. Liver. 2001;21(4):225–232. doi: 10.1034/j.1600-0676.2001.021004225.x. [DOI] [PubMed] [Google Scholar]
- 61.Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O, et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe. 2008;3(5):304–315. doi: 10.1016/j.chom.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selmi C, Balkwill DL, Invernizzi P, Ansari AA, Coppel RL, Podda M, et al. Patients with primary biliary cirrhosis react against a ubiquitous xenobiotic-metabolizing bacterium. Hepatology. 2003;38(5):1250–1257. doi: 10.1053/jhep.2003.50446. [DOI] [PubMed] [Google Scholar]
- 63.Fussey SP, Ali ST, Guest JR, James OF, Bassendine MF, Yeaman SJ. Reactivity of primary biliary cirrhosis sera with Escherichia coli dihydrolipoamide acetyltransferase (E2p): characterization of the main immunogenic region. Proc Natl Acad Sci U S A. 1990;87(10):3987–3991. doi: 10.1073/pnas.87.10.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olafsson S, Gudjonsson H, Selmi C, Amano K, Invernizzi P, Podda M, et al. Antimitochondrial antibodies and reactivity to N. aromaticivorans proteins in Icelandic patients with primary biliary cirrhosis and their relatives. Am J Gastroenterol. 2004;99(11):2143–2146. doi: 10.1111/j.1572-0241.2004.40397.x. [DOI] [PubMed] [Google Scholar]
- 65.Vilagut L, Parés A, Viñas O, Vila J, Jiménez de Anta MT, Rodés J. Antibodies to mycobacterial 65-kD heat shock protein cross-react with the main mitochondrial antigens in patients with primary biliary cirrhosis. Eur J Clin Invest. 1997;27(8):667–672. doi: 10.1046/j.1365-2362.1997.1690724.x. [DOI] [PubMed] [Google Scholar]
- 66.Bogdanos DP, Baum H, Okamoto M, Montalto P, Sharma UC, Rigopoulou EI, et al. Primary biliary cirrhosis is characterized by IgG3 antibodies cross-reactive with the major mitochondrial autoepitope and its Lactobacillus mimic. Hepatology. 2005;42(2):458–465. doi: 10.1002/hep.20788. [DOI] [PubMed] [Google Scholar]
- 67.Bogdanos DP, Baum H, Gunsar F, Arioli D, Polymeros D, Ma Y, et al. Extensive homology between the major immunodominant mitochondrial antigen in primary biliary cirrhosis and Helicobacter pylori does not lead to immunological cross-reactivity. Scand J Gastroenterol. 2004;39(10):981–987. doi: 10.1080/00365520410003236. [DOI] [PubMed] [Google Scholar]
- 68.Hopf U, Möller B, Stemerowicz R, Lobeck H, Rodloff A, Freudenberg M, et al. Relation between Escherichia coli R(rough)-forms in gut, lipid A in liver, and primary biliary cirrhosis. Lancet. 1989;2(8677):1419–1422. doi: 10.1016/s0140-6736(89)92034-5. [DOI] [PubMed] [Google Scholar]
- 69.Feld JJ, Meddings J, Heathcote EJ. Abnormal intestinal permeability in primary biliary cirrhosis. Dig Dis Sci. 2006;51(9):1607–1613. doi: 10.1007/s10620-006-9544-z. [DOI] [PubMed] [Google Scholar]
- 70.Di Leo V, Venturi C, Baragiotta A, Martines D, Floreani A. Gastroduodenal and intestinal permeability in primary biliary cirrhosis. Eur J Gastroenterol Hepatol. 2003;15(9):967–973. doi: 10.1097/00042737-200309000-00005. [DOI] [PubMed] [Google Scholar]
- 71.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 72.Ohira H, Tsutsui W, Fujioka Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J Atheroscler Thromb. 2017;24(7):660–672. doi: 10.5551/jat.RV17006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allam-Ndoul B, Castonguay-Paradis S, Veilleux A. Gut Microbiota and Intestinal Trans-Epithelial Permeability. Int J Mol Sci. 2020;21(17):6402. doi: 10.3390/ijms21176402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inamine T, Schnabl B. Immunoglobulin A and liver diseases. J Gastroenterol. 2018;53(6):691–700. doi: 10.1007/s00535-017-1400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Floreani A, Baragiotta A, Pizzuti D, Martines D, Cecchetto A, Chiarelli S. Mucosal IgA defect in primary biliary cirrhosis. Am J Gastroenterol. 2002;97(2):508–510. doi: 10.1111/j.1572-0241.2002.05521.x. [DOI] [PubMed] [Google Scholar]
- 76.Isaacs-Ten A, Echeandia M, Moreno-Gonzalez M, Brion A, Goldson A, Philo M, et al. Intestinal Microbiome-Macrophage Crosstalk Contributes to Cholestatic Liver Disease by Promoting Intestinal Permeability in Mice. Hepatology. 2020;72(6):2090–2108. doi: 10.1002/hep.31228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fukui H. Gut-liver axis in liver cirrhosis: How to manage leaky gut and endotoxemia. World J Hepatol. 2015;7(3):425–442. doi: 10.4254/wjh.v7.i3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li B, Selmi C, Tang R, Gershwin ME, Ma X. The microbiome and autoimmunity: a paradigm from the gut-liver axis. Cell Mol Immunol. 2018;15(6):595–609. doi: 10.1038/cmi.2018.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014;35(11):507–517. doi: 10.1016/j.it.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 81.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4(3):269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 83.Ostman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. Eur J Immunol. 2006;36(9):2336–2346. doi: 10.1002/eji.200535244. [DOI] [PubMed] [Google Scholar]
- 84.Crabbé PA, Bazin H, Eyssen H, Heremans JF. The normal microbial flora as a major stimulus for proliferation of plasma cells synthesizing IgA in the gut. The germ-free intestinal tract. Int Arch Allergy Appl Immunol. 1968;34(4):362–375. doi: 10.1159/000230130. [DOI] [PubMed] [Google Scholar]
- 85.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52(2):237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44(11):4136–4141. doi: 10.1128/jcm.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carasi P, Racedo SM, Jacquot C, Romanin DE, Serradell MA, Urdaci MC. Impact of kefir derived Lactobacillus kefiri on the mucosal immune response and gut microbiota. J Immunol Res. 2015;2015:361604. doi: 10.1155/2015/361604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475(7357):514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, et al. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell. 2015;163(2):381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29(6):947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harada K, Shimoda S, Sato Y, Isse K, Ikeda H, Nakanuma Y. Periductal interleukin-17 production in association with biliary innate immunity contributes to the pathogenesis of cholangiopathy in primary biliary cirrhosis. Clin Exp Immunol. 2009;157(2):261–270. doi: 10.1111/j.1365-2249.2009.03947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang CJ, Lin TL, Tsai YL, Wu TR, Lai WF, Lu CC, et al. Next generation probiotics in disease amelioration. J Food Drug Anal. 2019;27(3):615–622. doi: 10.1016/j.jfda.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 96.Li C, Niu Z, Zou M, Liu S, Wang M, Gu X, et al. Probiotics, prebiotics, and synbiotics regulate the intestinal microbiota differentially and restore the relative abundance of specific gut microorganisms. J Dairy Sci. 2020;103(7):5816–5829. doi: 10.3168/jds.2019-18003. [DOI] [PubMed] [Google Scholar]
- 97.Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 2014;7(1):12–18. doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 98.Martoni CJ, Labbé A, Ganopolsky JG, Prakash S, Jones ML. Changes in bile acids, FGF-19 and sterol absorption in response to bile salt hydrolase active L. reuteri NCIMB 30242. Gut Microbes. 2015;6(1):57–65. doi: 10.1080/19490976.2015.1005474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ebrahimi-Mousavi S, Alavian SM, Sohrabpour AA, Dashti F, Djafarian K, Esmaillzadeh A. The effect of daily consumption of probiotic yogurt on liver enzymes, steatosis and fibrosis in patients with nonalcoholic fatty liver disease (NAFLD): study protocol for a randomized clinical trial. BMC Gastroenterol. 2022;22(1):102. doi: 10.1186/s12876-022-02176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gupta A, Khanna S. Fecal Microbiota Transplantation. Jama. 2017;318(1):102. doi: 10.1001/jama.2017.6466. [DOI] [PubMed] [Google Scholar]
- 101.Hvas CL, Dahl Jørgensen SM, Jørgensen SP, Storgaard M, Lemming L, Hansen MM, et al. Fecal Microbiota Transplantation Is Superior to Fidaxomicin for Treatment of Recurrent Clostridium difficile Infection. Gastroenterology. 2019;156(5):1324–1332.e1323. doi: 10.1053/j.gastro.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 102.Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. 2018;24(12):1804–1808. doi: 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Allegretti JR, Kassam Z, Carrellas M, Mullish BH, Marchesi JR, Pechlivanis A, et al. Fecal Microbiota Transplantation in Patients With Primary Sclerosing Cholangitis: A Pilot Clinical Trial. Am J Gastroenterol. 2019;114(7):1071–1079. doi: 10.14309/ajg.0000000000000115. [DOI] [PubMed] [Google Scholar]
- 104.Gao L, Wang L, Woo E, He X, Yang G, Bowlus C, et al. Clinical Management of Primary Biliary Cholangitis-Strategies and Evolving Trends. Clin Rev Allergy Immunol. 2020;59(2):175–194. doi: 10.1007/s12016-019-08772-7. [DOI] [PubMed] [Google Scholar]
- 105.Michielan A, D’Incà R. Intestinal Permeability in Inflammatory Bowel Disease: Pathogenesis, Clinical Evaluation, and Therapy of Leaky Gut. Mediators Inflamm. 2015;2015:628157. doi: 10.1155/2015/628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carbone M, Invernizzi P. Treatment of PBC-A step forward. Liver Int. 2017;37(4):503–505. doi: 10.1111/liv.13353. [DOI] [PubMed] [Google Scholar]
- 107.Gharagozloo M, Majewski S, Foldvari M. Therapeutic applications of nanomedicine in autoimmune diseases: from immunosuppression to tolerance induction. Nanomedicine. 2015;11(4):1003–1018. doi: 10.1016/j.nano.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 108.Ridlon JM, Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B. 2015;5(2):99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee JY, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res. 2013;54(11):3062–3069. doi: 10.1194/jlr.M039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Laue H, Denger K, Cook AM. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl Environ Microbiol. 1997;63(5):2016–2021. doi: 10.1128/aem.63.5.2016-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ovadia C, Perdones-Montero A, Fan HM, Mullish BH, McDonald JAK, Papacleovoulou G, et al. Ursodeoxycholic acid enriches intestinal bile salt hydrolase-expressing Bacteroidetes in cholestatic pregnancy. Sci Rep. 2020;10(1):3895. doi: 10.1038/s41598-020-60821-w. [DOI] [PMC free article] [PubMed] [Google Scholar]