Abstract
Aberrant alternative splicing is one of the important causes of cancer. Polypyrimidine tract binding protein 1 (PTBP1) has been found to be involved in splicing regulation in a variety of tumors. Here, we observed significant up-regulation of PTBP1 in primary hepatocellular carcinoma (HCC) tissues. High levels of PTBP1 expression were associated with poor prognosis and increased metastatic potential in HCC. In vitro studies demonstrated that elevated PTBP1 promoted both migration and invasion by HCC cells. In contrast, knockdown of PTBP1 significantly inhibited the migration and invasion of HCC cells in vitro. Further, up-regulation of PTBP1 markedly accumulated the expression of oncogenic isoform of NUMB, NUMB-PRRL. We observed two isoforms of NUMB, NUMB-PRRL and NUMB-PRRS exhibit opposite function in HCC cells, which partially explain PTBP1 plays the tumor promoting roles in a NUMB splicing-dependent manner. In summary, our study indicates that PTBP1 may serve as an oncogene in HCC patients by regulating the alternative splicing of NUMB exon 9 and could potentially serve as a prognostic indicator.
Keywords: Hepatocellular carcinoma, Alternative splicing, PTBP1, NUMB
1. Introduction
Hepatocellular carcinoma (HCC), one of the most common liver tumors, is expected to affect up to 1 million people globally by 2025 [1], with Chinese patients accounting for about 53%. The worldwide 5-year survival rate for HCC patients is less than 20%, owing to delayed diagnosis and high recurrence/metastasis rates during treatments. It has been found that the interaction of multiple biological processes, such as chronic hepatitis B virus (HBV) infection, tumor microenvironment, intercellular signaling pathways, and cellular metabolic system, could accelerate the progress of HCC [2,3]. At present, several studies focus on the application of nanobiotechnology in the precise theranostics of HCC, but the selection of therapy targets still deserves further exploration [4,5].
Alternative splicing is one of the critical mechanisms of eukaryotic gene expressions, which contributes to proteomic diversity [6]. And 95% of human genes have been spliced during transcription. Alternative splicing usually functions during post-transcriptional processing. During alternative splicing, a single sequence of precursor mRNA (pre-mRNA) is processed into multiple mature mRNAs, because of the different sequences of UTR regions or coding regions [7]. Polypyrimidine tract binding protein 1 (PTBP1), also known as hnRNP I, is a member of the non-homogeneous nuclear ribonucleoproteins (hnRNPs) family that can recognize pyrimidine-enriched regions in pre-mRNAs. As an RNA binding protein, PTBP1 was capable of directly bind the pre-mRNAs or through ligands, causing the bound RNA to become circular, wrapping the variable exons and ultimately affecting mRNA splicing [8,9].
Increasing evidences suggested that PTBP1 is associated with tumor malignant progression. PTBP1 was participated in the metastasis of tumors by affecting alternative splicing of the related genes. For example, overexpression of PTBP1 in ovarian cancer cells could promote cell metastasis and colony formation due to the fact that PTBP1 could promote the exon 6B skipping of CDC42 [10]. It has been found that PTBP1 was correlated with poor prognosis in the patients with advanced colorectal cancer [11]. Additionally, knockdown of PTBP1 regulated the alternative splicing processing of several tumor metastasis-related genes (such as CTTN, PKM2, and EGFR), ultimately inhibiting the metastasis of colorectal cancer cells [12]. However, there are fewer studies on PTBP1 in HCC cells and whether there is regulation of alternative splicing.
Our study revealed an upregulation of PTBP1 expression in HCC tissues. PTBP1 facilitated the proliferation, migration, and invasion of HCC cells. Moreover, we observed that PTBP1 modulated the expressions of pro-oncogenic NUMB isoforms that respectively promote HCC cell proliferation and migration.
2. Materials and methods
2.1. Cell lines, RNA interference and plasmids
We obtained the human normal liver cell line L-02 and five HCC cell lines (Huh7, SMMC7721, HepG2, PLC/PRF/5 and HCCLM3) from the China Center for Type Culture Collection (CCTCC) in Wuhan City. These cells were cultured in our laboratory under standard conditions of 37 °C with 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM, Gibco, Australia) supplemented with 10% fetal bovine serum (FBS, Gibco, Australia), as well as penicillin and streptomycin. The siRNAs targeting different isoforms of NUMB and the control siRNAs were synthesized by Ribobio (Guangzhou, China), and transfected using the RiboFECTTM CP transfection kit (Ribobio, China) according to the manufacturer's instructions. To stably knock down the expression of PTBP1, the sequences of short hairpin RNAs (shRNAs) targeting PTBP1 were cloned into the pGreenPuro vector. Target sequences were used as follows: shPTBP1-1: GGACGGCATTGTCCCAGATAT; shPTBP1-2: GCGTGAAGATCCTGTTCAATA. The intact sequences of PTBP1 were synthesized by Biomed Co. (Beijing, China) and cloned into the pLVX-Puro vector (Clontech, USA) for overexpression of PTBP1. These vectors were then subject to lentiviral packaging to construct HCC cell lines with stable overexpression or knockdown of PTBP1.
2.2. Cells proliferation, migration and invasion assays
The CCK-8 kit (CK04, Dojindo, Japan) was used to measure the proliferation of HCC cells (9 × 103 cells per well), following the manufacturer's instructions. A total of 5 × 104 cells (200 μL) were seeded on the top chamber of each insert (353097, BD Biosciences, USA) or matrigel invasion chamber (354480, BD Biosciences, USA). The lower chambers contained DMEM supplemented with 20% FBS. Cells adhering to the lower side of the inserts were stained with 0.1% crystal violet solution and counted using an IX71 inverted microscope (Olympus, Japan) after incubation at 37 °C for 24–48 h.
2.3. RNA extraction and semi-quantitative PCR assays
Total RNAs were extracted from cultured cells using RNApure tissue&cell kit (CW0584S, CWBIO, China) and cDNAs were synthesized from 500 ng of total RNAs using the PrimeScript RT reagent kit (RR037A, Takara, Japan). The semi-quantitative PCR assays for NUMB and its spliced isoforms were conducted using KOD-Plus-Neo reagent (KOD401, TOYOBO, Japan) with gene-specific primers: NUMB forward 5′-GCATCAGCTCCCTGTGCTCAC-3′ and reverse 5′-GGTCGGCCTCAGAGGGAGTAC-3′; GAPDH forward 5′-CGGAGTCAACGGATTTGGTCGT-3′ and reverse 5′- TCTCAGCCTTGACGGTGCCA-3’. The quantification of PCR products was performed using the Image J software (USA) after agarose gel electrophoresis.
2.4. Immunoblotting assays
For protein analysis, cells were lysed in RIPA buffer (CW2333S, CWBIO, China) with EDTA-free protease inhibitor cocktail (1:100; 04693132001, Roche, Switzerland). Total proteins (10–20 μg) were electrophoresed on SDS-polyacrylamide gels and transferred to nitrocellulose filter membrane for incubation with primary and secondary antibodies. The immunoreactive bands were detected using the SuperSignal™ West Pico chemiluminescent substrate kit (34577, Thermo, USA) and immunoblotting detection system (BioRad, USA). The following primary antibodies were used: anti-GAPDH (1:1,000; ab9485, Abcam, USA) and anti-PTBP1 (1:1,000; 32–4800, Thermo, USA).
2.5. Statistics and reproducibility
The experiments, including Western blotting, were repeated at least twice with consistent results. Data is presented as means ± standard deviation (s.d.). Student's t-test was used to compare quantitative data between two groups assuming equal variances. If the measured values did not meet normality and homogeneity assumptions, log-transformation was applied before t-tests. Fisher's exact test was used to analyze contingency tables based on sample sizes. Disease-free survival (DFS) was calculated from tumor resection until the first HCC recurrence, death, or last follow-up. Overall survival (OS) was defined as the time between the surgery and death or last follow-up. Patients lost to follow-up or who died from causes unrelated to HCC were considered censored events. Univariate and multivariate Cox regression analyses were used to assess the impact of variables on survival rates. Kaplan-Meier method was applied to calculate survival curves, and log-rank test was conducted for significance determined (P < 0.05). Statistical analysis was performed using R software (version 3.5.2; www.rproject.org) or GraphPad Prism (version 8), unless otherwise specified.
3. Result
3.1. Overexpression of PTBP1 in HCC predicts poor outcomes of HCC patients
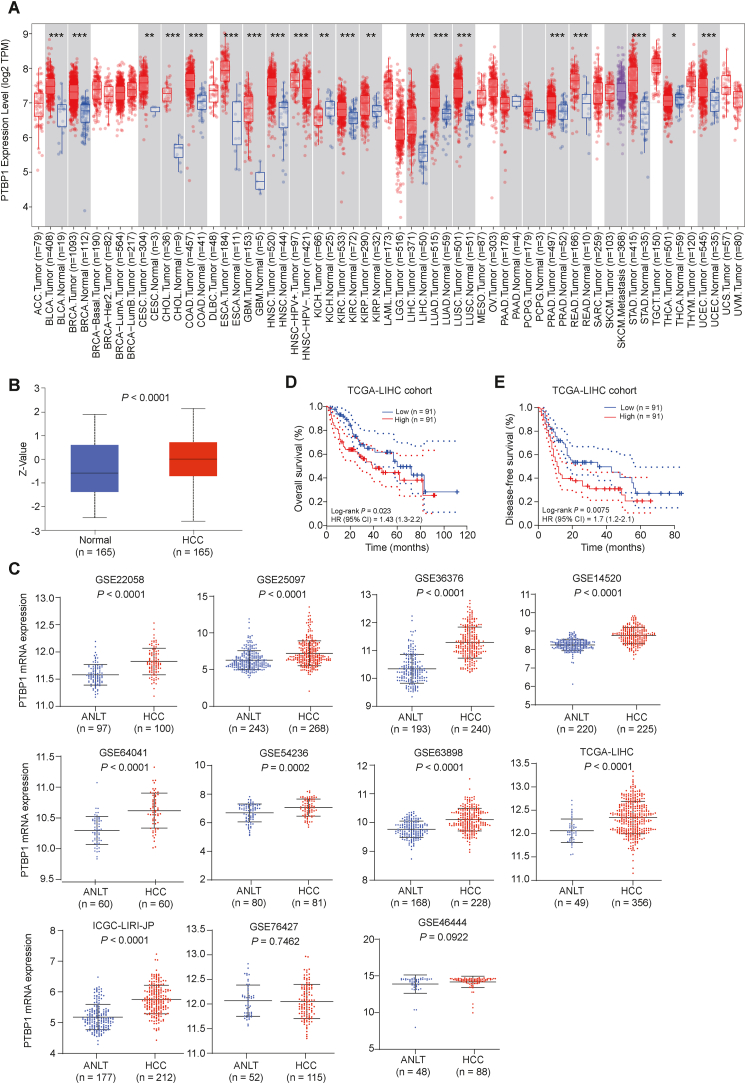
Previous studies have found that PTBP1 usually plays the role of oncogene in tumors [[13], [14], [15]]. In this study, we were aimed to elucidate the molecular mechanisms by which PTBP1 promotes HCC progression. First, we utilized the TIMER2 database [16] to analyze the mRNA expression status of PTBP1 across various cancer types of TCGA. As shown in Fig. 1A, PTBP1 was expressed at apparent higher levels in such kinds of solid tumor tissues than in the corresponding normal tissues, including in HCC. Then, we profiled the pattern of PTBP1 protein expression in the TCGA-LIHC cohort through the UALCAN website [17] (http://ualcan.path.uab.edu/index.html). Consistently, the protein expression level of PTBP1 was significantly elevated in HCC tissues (P < 0.0001; Fig. 1B). We next evaluated PTBP1 transcription levels in multiple HCC studies from HCCDB database [18] (http://lifeome.net/database/hccdb/home.html; including 11 datasets: GSE22058, GSE25097, GSE36376, GSE14520, GSE64041, GSE54236, GSE63898, TCGA-LIHC, ICGC-LIRI-JP, GSE76427 and GSE46444). An analysis of 11 HCC cohorts revealed that PTBP1 mRNA expression significantly increased in HCC compared to adjacent non-carcinoma tissues in 9 HCC cohorts (Fig. 1C). We also examined the correlation between PTBP1 expression and patient prognosis in the TCGA-LIHC cohort. The study showed that high PTBP1 expression was associated with worse overall survival (Log-rank P = 0.023, HR = 1.43; Fig. 1D) and progression-free survival in HCC patients compared to those with low PTBP1 expression (Log-rank P = 0.0075, HR = 1.7; Fig. 1E). These findings suggested that increased PTBP1 expression may serve as an oncogene in HCC and is linked to poor prognosis.
Fig. 1.
Abnormal expression of PTBP1 in HCC. (A) mRNA expression levels of PTBP1 in 33 solid tumor samples (red) and the corresponding normal tissues (blue). (B) The protein expression level of PTBP1 in the TCGA-LIHC cohort identified by the UALCAN website. (C) Up-regulation of PTBP1 in 11 independent cohorts of HCC patients (including TCGA-LIHC, ICGC-LIRI-JP, GSE22058, GSE25097, GSE36376, GSE14520, GSE64041, GSE54236, GSE63898, GSE76427 and GSE46444). (D¸ E) Kaplan-Meier analyses of overall survival (D) and disease-free survival (E) in HCCs according to the mRNA expression status of PTBP1. Data was obtained from the TCGA HCC dataset. HR, hazard ratio. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. PTBP1 enhances the abilities of proliferation and migration of HCC cells
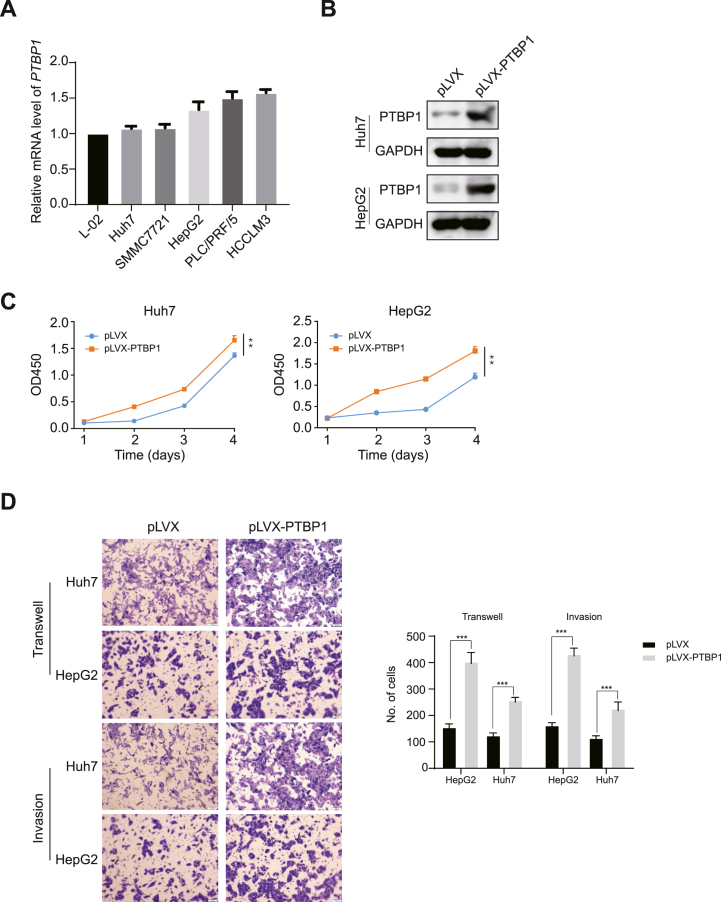
To better understand PTBP1's biology in HCC, we examined its expression in five HCC cell lines and a control liver cell line. Endogenous PTBP1 mRNA levels were lower in L-02, Huh7, SMMC7721, and HepG2 compared to other HCC cell lines (Fig. 2A). We then created stable PTBP1-expressing Huh7 and HepG2 cells (Fig. 2B) and found that it promoted their proliferation using CCK-8 assays (Fig. 2C). Previous studies had shown that HCC cell's unlimited proliferation and strong invasive and metastatic abilities are the main causes of high malignancy degree and worse overall survival [19]. As expected, PTBP1 overexpression accelerated HCC cell migration and invasion (Fig. 2D).
Fig. 2.
Over-expression of PTBP1 is required for HCC cell survival. (A) The mRNA levels of PTBP1 were determined in multiple human hepatocyte cell lines utilizing real-time quantitative PCR (RT-qPCR) assays, including one immortalized hepatocyte cell line (L-02) and five HCC cell lines (Huh7, SMMC7721, HepG2, PLC/PRF/5 and HCCLM3). (B) Construction of Huh7 and HepG2 cells stably expressing exogenous PTBP1. (C) CCK-8 assays revealed that up-redulated of PTBP1 has pro-growth effect on Huh7 and HepG2 cells. (D) Transwell assays revealed that overexpression of PTBP1 promotes migration and invasion of Huh7 and HepG2 cells. All quantification data are mean ± SD from three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 (Student's t-test). n.s., not significant.
3.3. Depletion of PTBP1 impaired the proliferation and migration of HCC cells
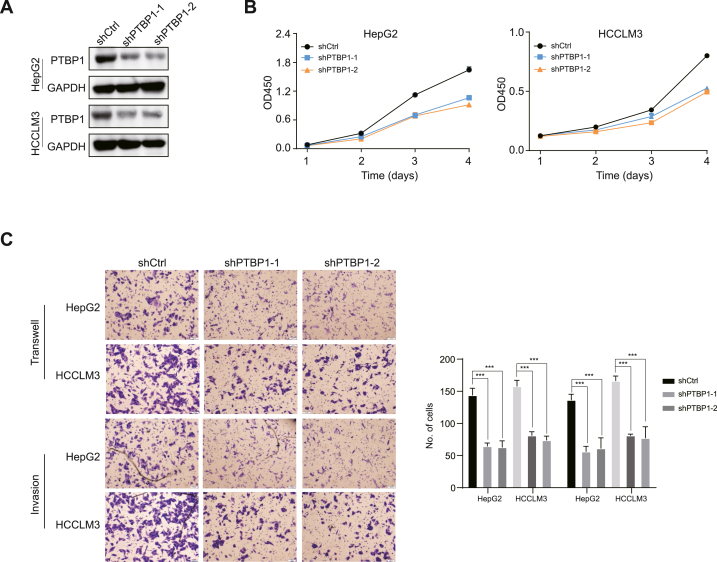
We further utilized shRNAs targeting PTBP1 to stably weaken the endogenous expression of PTBP1 in HepG2 and HCCLM3 cell lines (Fig. 3A). In contrast, the proliferation rates of HepG2 and HCCLM3 with knockdown of PTBP1 were significantly slower than that of the control group of corresponding cells (Fig. 3B). Knockdown of PTBP1 significantly reduced the migration and invasion of HCC cells (Fig. 3C), indicating its vital role in regulating their tumorigenicity through proliferation and migration.
Fig. 3.
Knockdown of PTBP1 damages HCC cell survival. (A) Stable knockdown of PTBP1 by two independent shRNAs. (B) CCK-8 assays revealed that down-regulated of PTBP1 has a suppressive effect on HepG2 and HCCLM3 cells. (C) Transwell assays revealed that knockdown of PTBP1 decreases migration and invasion of HepG2 and HCCLM3 cells. All quantification data are mean ± SD from three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 (Student's t-test). n.s., not significant.
3.4. PTBP1 regulated the expressions of pro-oncogenic isoforms of NUMB
As an oncogene, PTBP1 can promote tumorigenesis by playing the role of a splicing factor that regulates target genes, such as NUMB [20]. PTBP1, by regulating the splicing of NUMB exon 9, allows NUMB to produce both PRRL (proline-rich domain long) and PRRS (proline-rich domain short) isoforms. In particular, NUMB-PRRL expression is upregulated in a variety of tumor tissues and promotes tumor progression, while NUMB-PRRS can inhibit AKT signaling pathway activation, thereby suppressing cancer cell proliferation and survival [[21], [22], [23], [24]]. To confirm whether PTBP1 regulates the splicing of NUMB in HCC, we examined the effect of PTBP1 expression on NUMB-PRRL and NUMB-PRRS by the semi-quantitative PCR assay. As shown in Fig. 4A, knockdown of PTBP1 significantly promoted the expression of the NUMB-PRRS, while suppressed the expression of the NUMB-PRRL. The opposite regulatory effect was observed in the assay which PTBP1 was overexpressed (Fig. 4B). This suggested that PTBP1 is involved in regulating NUMB alternative splicing in HCC cells.
Fig. 4.
NUMB isoforms exert different tumor biological functions. (A) Semi-quantitative PCR assays reveals that knockdown of PTBP1 promoted the expression of the NUMB-PRRS and suppressed the expression of the NUMB-PRRL in HepG2 and HCCLM3 cells. (B) Semi-quantitative PCR assays reveals that up-regulated of PTBP1 promoted the expression of the NUMB-PRRL and suppressed the expression of the NUMB-PRRS in HepG2 and Huh7 cells. (C) Instant knockdown of NUMB-PRRL and NUMB-PRRS by two independent siRNAs respectively. (D) CCK-8 assays revealed that down-regulated of NUMB isoforms have an opposite effect on HepG2 and Huh7 cells. (E) Transwell assays revealed that knockdown of NUMB isoforms decrease or increase migration and invasion of HepG2 and Huh7 cells. All quantification data are mean ± SD from three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 (Student's t-test). n.s., not significant.
Further, in order to evaluate the tumorigenicity of NUMB-PRRL and NUMB-PRRS in HCC cells, we utilizing siRNAs respectively targeting two NUMB isoforms in HepG2 and HCCLM3 cell lines (Fig. 4C). The CCK-8 assays revealed that knockdown of NUMB-PRRS promoted HepG2 and HCCLM3 cell proliferation. In contrast, the proliferation rates of HepG2 and HCCLM3 with knockdown of NUMB-PRRL was significantly slower than that of the control group of corresponding cells (Fig. 4D). The results of Transwell and Invasion assays showed that knockdown of NUMB-PRRL significantly reduced the number of migrating and invading HCC cells, while knockdown of NUMB-PRRS increased the number of cells (Fig. 4E). Therefore, these results indicated that PTBP1 regulates the splicing of NUMB isoforms which benefit to the generation of NUMB-PRRL. Furthermore, NUMB-PRRL promotes HCC progression, meanwhile NUMB-PRRS inhibits HCC progression.
3.5. Splicing of NUMB isoforms was required for PTBP1s tumor-promoting function
We then investigated whether the oncogenic isoform of NUMB is necessary for PTBP1 to promote HCC cell proliferation, migration and invasion. To do so, we conducted a series of in vitro assays to determine if PTBP1 plays a role in tumor promotion through NUMB splicing. As expected, knockdown of NUMB-PRRL abolished PTBP1-induced promotion of cells proliferation, migration and invasion (Fig. 5A and B), while knockdown of NUMB-PRRS reversed the inhibitory effects caused by PTBP1 depletion on those malignant phenotypes (Fig. 5C and D). Furthermore, we confirmed in three HCC cohorts (GSE25097, GSE54263 and TCGA-LIHC) that NUMB exon9 expression is positively correlated with PTBP1 expression (Fig. 5E). These results suggest that PTBP1 promotes oncogenesis by facilitating the isoform switch from NUMB-PRRS to NUMB-PRRL.
Fig. 5.
PTBP1 exerts its tumor-promoting role through splicing of the NUMB isoforms. (A) siNUMB-PRRL treatment abolishes the pro-proliferative effects of PTBP1 overexpression on HepG2 and Huh7 cells. (B) siNUMB-PRRL treatment eliminates the pro-migratory effects of PTBP1 overexpression on HepG2 and Huh7 cells. (C) siNUMB-PRRS treatment offsets the pro-proliferative effects of PTBP1 depletion on HepG2 and HCCLM3 cells. (D) siNUMB-PRRS treatment eliminates the pro-migratory effects of PTBP1 overexpression on HepG2 and HCCLM3 cells. All quantification data are mean ± SD from three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 (Student's t-test). n. s., not significant. (E) Pearson correlation analyses of the mRNA levels of PTBP1 and the exon 9 of NUMB in GSE25097, GSE54236 and the TCGA-LIHC cohort. TPM, transcripts per million.
4. Discussion
In this study, we demonstrated that the splicing factor PTBP1 in HCC acts as an oncogene by causing abnormal isoform switching of NUMB. We observed PTBP1 was increased in primary HCC tissues from the TCGA-LIHC cohort and higher levels of it were associated with worse OS and DFS rates in with HCC patients, indicating its potential as a valuable prognostic biomarker for further investigation. Additionally, our findings showed that overexpression of PTBP1 significantly enhances HCC proliferation and metastasis in vitro. Our findings also demonstrated that PTBP1 overexpression promotes HCC metastasis by upregulating NUMB-PRRL and downregulating NUMB-PRRS expression.
Previous reports demonstrated that PTBP1 is often up-regulated in multi-types of cancer cells and reinforces the tumorigenicity of these cancer cells including colorectal cancer and breast cancer [[25], [26], [27]]. PTBP1 can serve as splicing factor to bind pre-mRNAs, such as PKM, regulating the expressions of their splicing isoforms [28,29]. However, less research has carried on in HCC. In this study, we found that PTBP1 is increased in HCC and predicts poor prognosis for patients with this disease, highlighting its clinical relevance. Additionally, PTBP1 promoted the malignant progression of HCC cells and regulates NUMB isoform splicing.
NUMB is a membrane-associated protein, which encodes several mRNA transcripts via alternative splicing [30]. To our best knowledge, PTBP1 is responsible for the inclusion of NUMB exon 9 and the activation of the MAPK/ERK pathway is beneficial for NUMB exon 9 inclusion in cancer cells [31]. In our research, we preliminarily elucidated the regulation effect of PTBP1 on NUMB exon 9 splicing, which further enriched the research on the splicing of NUMB. We further revealed that two transcripts of NUMB perform different tumorigenic functions in HCC. This could be due to exon 9 of NUMB, corresponding to a 49-amino-acid region, is located within the proline-rich region (PRR) of the protein [31]. Alternative splicing is achieved by regulating the inclusion or exclusion of exons, which are then translated into protein sequences that perform different functions [32]. For example, alternative splicing of FGFR2 generates two transcripts, Ⅲb and Ⅲc, which promote or inhibit the EMT process, respectively [33]. In addition, alternative splicing of Bcl-x exon 2 produces two isoforms with opposing effects on cell survival: the anti-apoptotic long isoform (Bcl-xL) and the pro-apoptotic short isoform (Bcl-xS) [34]. However, further evidence is needed to clarify NUMB's mechanism in HCC, which could aid in developing a novel treatment strategy for this malignancy.
5. Conclusions
In conclusion, the research revealed that PTBP1 functions as an oncogene by promoting NUMB exon 9 inclusion and contributing to HCC development. High levels of PTBP1 expression indicate poor prognosis in HCC patients. The inclusion of NUMB exon 9 triggers malignant proliferation and metastasis in HCC cells (Fig. 6). However, further research is important to understand the mechanism behind NUMB splicing in HCC, which could be a potential therapeutic target for HCCs with NUMB exon 9 inclusion.
Fig. 6.
A schematic model for the function and mechanisms of NUMB splicing in HCC progression. Overexpression of PTBP1 in HCC cells leads to inclusion of the exon 9 in NUMB. Upregulation of NUMB-PRRL increases HCC cells proliferation and metastasis, leading to progress of HCC. However, NUMB-PRRS without exon 9 inhibits the inordinate proliferation and metastasis of cells, resulting to maintaining normal cell growth.
Declarations
Author contribution statement
Yongsheng Duo: Conceived and designed the experiments.
Zhentao He: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Qianhua Ni, Xichun Li, Mingyu Zhao and Qingguo Mo: Contributed reagents, materials, analysis tools or data.
Data availability statement
Data included in article/supp. material/referenced in article. Conflict of Interests.
Additional information
No additional information is available for this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17387.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bray F., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Llovet J.M., et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2016;2 doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q., et al. Anti-rheumatic drug-induced hepatitis B virus reactivation and preventive strategies for hepatocellular carcinoma. Pharmacol. Res. 2022;178 doi: 10.1016/j.phrs.2022.106181. [DOI] [PubMed] [Google Scholar]
- 4.Mahmoud K., et al. Lipid based nanoparticles as a novel treatment modality for hepatocellular carcinoma: a comprehensive review on targeting and recent advances. J. Nanobiotechnol. 2022;20(1):109. doi: 10.1186/s12951-022-01309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu M., et al. Emerging nanobiotechnology for precise theranostics of hepatocellular carcinoma. J. Nanobiotechnol. 2022;20(1):427. doi: 10.1186/s12951-022-01615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J., et al. Second-generation antipsychotics induce cardiotoxicity by disrupting spliceosome signaling: implications from proteomic and transcriptomic analyses. Pharmacol. Res. 2021;170 doi: 10.1016/j.phrs.2021.105714. [DOI] [PubMed] [Google Scholar]
- 7.Bates D.O., et al. Pharmacology of modulators of alternative splicing. Pharmacol. Rev. 2017;69(1):63–79. doi: 10.1124/pr.115.011239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romanelli M.G., Diani E., Lievens P.M. New insights into functional roles of the polypyrimidine tract-binding protein. Int. J. Mol. Sci. 2013;14(11):22906–22932. doi: 10.3390/ijms141122906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng S., et al. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012;15(3):381–388. doi: 10.1038/nn.3026. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X., Yuan C., Yang J. Regulation and functional significance of CDC42 alternative splicing in ovarian cancer. Oncotarget. 2015;6(30):29651–29663. doi: 10.18632/oncotarget.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi H., et al. Significance of polypyrimidine tract-binding protein 1 expression in colorectal cancer. Mol. Cancer Therapeut. 2015;14(7):1705–1716. doi: 10.1158/1535-7163.MCT-14-0142. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z.N., et al. High expression of PTBP1 promote invasion of colorectal cancer by alternative splicing of cortactin. Oncotarget. 2017;8(22):36185–36202. doi: 10.18632/oncotarget.15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo J., Jia J., Jia R. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci. Rep. 2015;5 doi: 10.1038/srep14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang J., et al. Polypyrimidine Tract-Binding Protein 1 promotes proliferation, migration and invasion in clear-cell renal cell carcinoma by regulating alternative splicing of PKM. Am. J. Canc. Res. 2017;7(2):245–259. [PMC free article] [PubMed] [Google Scholar]
- 15.Jo Y.K., et al. Polypyrimidine tract-binding protein 1-mediated down-regulation of ATG10 facilitates metastasis of colorectal cancer cells. Cancer Lett. 2017;385:21–27. doi: 10.1016/j.canlet.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Li T., et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrashekar D.S., et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lian Q., et al. HCCDB: a database of hepatocellular carcinoma expression atlas. Dev. Reprod. Biol. 2018;16(4):269–275. doi: 10.1016/j.gpb.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J.D., et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollander D., et al. A network-based analysis of colon cancer splicing changes reveals a tumorigenesis-favoring regulatory pathway emanating from ELK1. Genome Res. 2016;26(4):541–553. doi: 10.1101/gr.193169.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L., et al. ATP11A promotes EMT by regulating Numb PRR(L) in pancreatic cancer cells. PeerJ. 2022;10 doi: 10.7717/peerj.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y.J., et al. Alternative splicing of VEGFA, APP and NUMB genes in colorectal cancer. World J. Gastroenterol. 2015;21(21):6550–6560. doi: 10.3748/wjg.v21.i21.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng K.L., et al. Alternative splicing of NUMB, APP and VEGFA as the features of pancreatic ductal carcinoma. Int. J. Clin. Exp. Pathol. 2015;8(6):6181–6191. [PMC free article] [PubMed] [Google Scholar]
- 24.Muto J., et al. RNA-binding protein Musashi1 modulates glioma cell growth through the post-transcriptional regulation of Notch and PI3 kinase/Akt signaling pathways. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen L., et al. Skipping of exon 10 in Axl pre-mRNA regulated by PTBP1 mediates invasion and metastasis process of liver cancer cells. Theranostics. 2020;10(13):5719–5735. doi: 10.7150/thno.42010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J., et al. Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics. 2021;11(15):7507–7526. doi: 10.7150/thno.59546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., et al. PTBP1 promotes the growth of breast cancer cells through the PTEN/Akt pathway and autophagy. J. Cell. Physiol. 2018;233(11):8930–8939. doi: 10.1002/jcp.26823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mochizuki Y., et al. Alternative microexon splicing by RBFOX2 and PTBP1 is associated with metastasis in colorectal cancer. Int. J. Cancer. 2021;149(10):1787–1800. doi: 10.1002/ijc.33758. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H., et al. Metabolic and proliferative state of vascular adventitial fibroblasts in pulmonary hypertension is regulated through a MicroRNA-124/PTBP1 (polypyrimidine tract binding protein 1)/pyruvate kinase muscle Axis. Circulation. 2017;136(25):2468–2485. doi: 10.1161/CIRCULATIONAHA.117.028069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dho S.E., et al. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 1999;274(46):33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 31.Rajendran D., et al. Regulation of Numb isoform expression by activated ERK signaling. Oncogene. 2016;35(39):5202–5213. doi: 10.1038/onc.2016.69. [DOI] [PubMed] [Google Scholar]
- 32.Ule J., Blencowe B.J. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol. Cell. 2019;76(2):329–345. doi: 10.1016/j.molcel.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 33.Ranieri D., et al. The aberrant expression in epithelial cells of the mesenchymal isoform of FGFR2 controls the negative crosstalk between EMT and autophagy. J. Cell Mol. Med. 2021;25(8):4166–4172. doi: 10.1111/jcmm.16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens M., Oltean S. Modulation of the apoptosis gene bcl-x function through alternative splicing. Front. Genet. 2019;10:804. doi: 10.3389/fgene.2019.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article. Conflict of Interests.