Abstract
Macrophages are efficient reservoirs for viruses that enable the viruses to survive over a longer period of infection. Alphaviruses such as chikungunya virus (CHIKV) are known to persist in macrophages even after the acute febrile phase. The viral particles replicate in macrophages at a very low level over extended period of time and are localized in tissues that are often less accessible by treatment. Comprehensive experimental studies are thus needed to characterize the CHIKV-induced modulation of host genes in these myeloid lineage cells and in one such pursuit, we obtained global transcriptomes of a human macrophage cell line infected with CHIKV, over its early and late timepoints of infection. We analyzed the pathways, especially immune related, perturbed over these timepoints and observed several host factors to be differentially expressed in infected macrophages in a time-dependent manner. We postulate that these pathways may play crucial roles in the persistence of CHIKV in macrophages.
Keywords: CHIKV, Macrophages, Transcriptome, Metabolic pathways
1. Introduction
Macrophages are the key players of the immune system in combating viral infection by modulating physiological and pathological changes in a host [1,2]. The phagocytic activity of macrophages is critical for host defense against infection as well as sustaining cellular activities such as the removal of apoptotic cells and repairing of the extracellular matrix [3]. Upon viral infection, macrophages respond with an enhanced expression of inflammatory cytokines (-pro and -anti) which leads to an activated antiviral immune response [4]. Several studies report that viruses persist in macrophages for extended periods that may have a direct bearing on disease progression [[5], [6], [7], [8], [9]].
Chikungunya virus (CHIKV) is an arthropod-borne alphavirus virus belonging to the Togaviridae family [10]. The mosquito-transmitted virus is a major public health concern capable of causing huge outbreaks and impacting the disability-adjusted life years (DALYs) of the affected population [11]. The RNA virus has a single-stranded positive-sense RNA genome containing two open reading frames that encode structural and non-structural proteins [12,13]. The structural proteins are mainly involved in the formation of viral particles while non-structural proteins form the replication machinery and with interactions with several host factors, generate viral particles in the infected host cell [14].
During CHIKV infection, it has been reported that the virus replicates in synovial macrophages leading to viral persistence in synovial tissues for a prolonged period of time that may result in the chronic phase of infection lasting for several years [5,15]. As a consequence, these synovial macrophages secrete various inflammatory molecules such as cytokines, chemokines, growth factors, and matrix metalloproteinases (MMPs) that are involved in prolonged inflammation of the joint. Reports have established that the plasma of CHIKV patient contains increased immune molecules such as type I IFN, interleukin 1 beta (IL-1β), interleukin 6 (IL-6), monocyte chemoattractant protein (MCP1/CCL2), and tumor necrosis alpha (TNFα) that directly correlated with high viral load and severity of disease [[16], [17], [18]]. These molecules also play an important role in cellular infiltration leading to the development of chronic arthralgia [19].
The advent of next-generation sequencing (NGS) revolutionized the analysis of global gene profiles enabling deriving snapshots of the expression pattern of transcripts in tissues/cells in real-time, thereby making it convenient to infer and postulate the probable underlying mechanisms during a specific condition in a comprehensive manner. In this study, we performed a global transcriptional analysis of a human macrophage cell line, THP-1, in response to CHIKV infection over a period of time. We segregated the infection timepoints into two phases – namely early phase of infection that comprised of 8hrs and 24hrs post-infection (hpi); and the late phase of infection, comprising 36hpi and 42hpi. Upon generating the global transcriptome of these timepoints of infection, we performed different types of analyses to identify the regulated cellular host factors and putative pathways that are involved during CHIKV pathogenesis in human macrophages. In the first analysis, we performed a paired analysis of the individual timepoints against the uninfected sample to arrive at the timepoint-specific perturbations in the THP-1 transcriptome upon CHIKV infection. Further, we performed a combined analysis of all the timepoints against the uninfected to arrive at those transcripts that were uniquely modulated at that specific timepoint and also those transcripts that were modulated at all timepoints at varied levels. Pathway analysis of all these different conditions helped in understanding the molecular fluctuations in the macrophages over time owing to CHIKV infection.
2. Results
2.1. Infection kinetics of CHIKV in THP-1 derived human macrophages
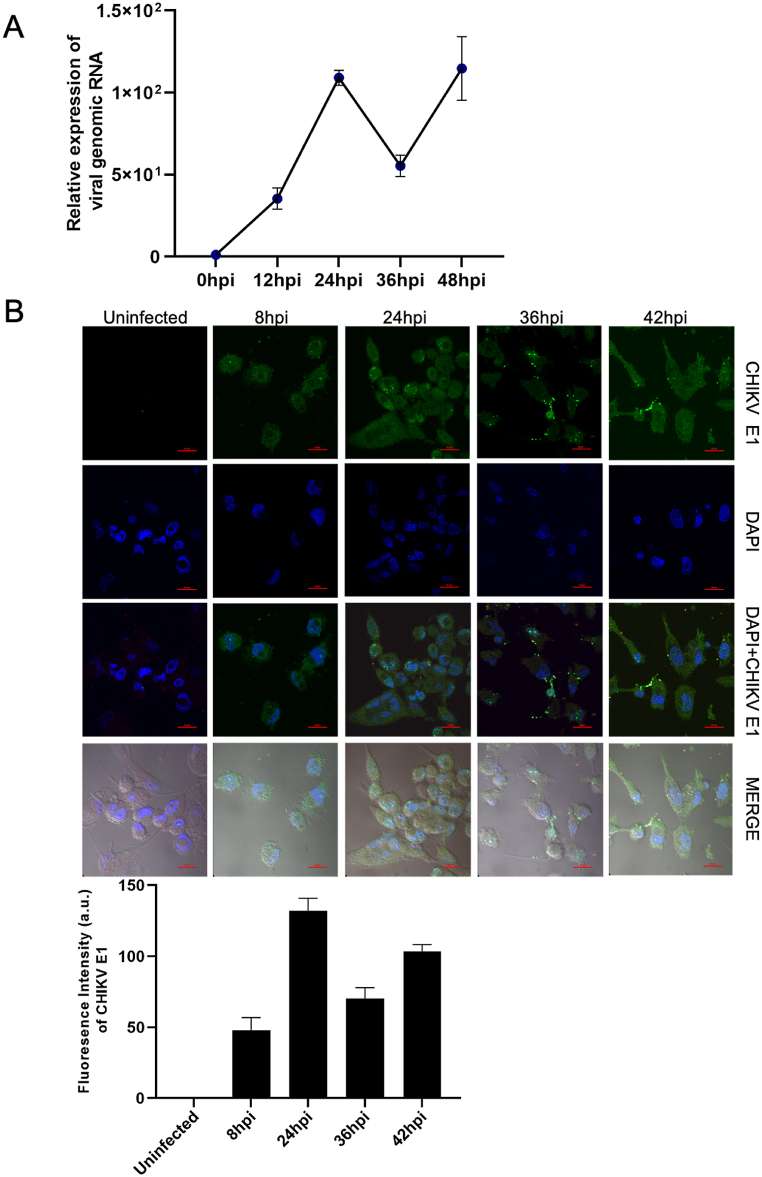
CHIKV infects a wide range of human cells including fibroblasts, epithelial, muscle cells, and macrophages [20,21]. Several studies have established that CHIKV persists in macrophages in meagre numbers and the subsequent clearance or persistence of the virus had a direct bearing on disease resolution or disease progression [5,22,23]. To confirm this, we evaluated CHIKV infection kinetics in THP-1 derived macrophages over a period of time starting from 12hpi to 48hpi using qPCR. We observed that at 12hpi, the virus had established infection in the cells and the viral titer peaked at 24hpi after which it significantly declined at 36hpi. At 48hpi, the virus titer peaked again and was coupled with significant host cell death (Fig. 1a). Since we were interested in evaluating the progression of infection in host cells upon virus infection, we performed confocal imaging of the infected cells using CHIKV E1 antibodies. Based on the results obtained by qPCR, we decided on 8hpi, 24hpi, 36hpi, and 42hpi timepoints to capture early timepoints of infection (ie, 8hpi instead of 12hpi) and to ensure integrity of cells at later timepoints (i.e., 42hpi instead of 48hpi). We detected infection to be just established at 8hpi and the 24hpi and 36hpi timepoints followed a pattern similar to qPCR results. At 42hpi, the viral titers increased more than 36hpi and the host cell integrity was retained (Fig. 1b). We hypothesized that distinct pathways were regulated in the macrophages over time since the establishment of CHIKV infection, and identifying these pathways may provide important insights towards understanding CHIKD pathogenesis in the macrophages. As a first step in this pursuit, we sought to map the global transcriptome of macrophages upon CHIKV infection (MOI1) based on regulated genes and their pathways in a pair-wise analysis over all the timepoints tested. We profiled the transcriptomes of CHIKV-infected macrophages at the following timepoints – 8hpi, 24hpi, 36hpi, and 42hpi. Uninfected macrophages cells served as control. Based on CHIKV infection kinetics data, we chose two early timepoints, namely 8hpi, i.e., the timepoint of initial stages of viral infection, and 24hpi, coinciding with the peak of infection, and two later timepoints of infection, namely, 36hpi and 42hpi and performed global transcriptome profiling of CHIKV infected macrophages.
Fig. 1.
CHIKV kinetics of infected THP-1 derived macrophages a. Relative expression of viral genomic RNA of CHIKV at different timepoints i.e., 0hpi, 12hpi, 24hpi, 36hpi and 48hpi in infected THP-1 derived macrophages. b. THP-1 cells were infected with CHIKV (MOI 1) at different timepoints i.e., 8hpi, 24hpi, 36hpi, and 42hpi. Infected cells were taken at different timepoints and fixed it. Further, cells were stained with an anti-CHIKV E1 (Green) antibody. DNA was visualized with DAPI (blue) (Fig. 1b, upper panel). Graphical representation of the fluorescence intensity of CHIKV E1 upon CHIKV infection at different timepoints (Fig. 1b, lower panel). Quantifications are measured in at least 6 different images per sample using NIS image processing software. The experiments were repeated at least thrice, and each experiment included at least three treatments. Data are expressed as mean ± SD. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. Transcriptome data analysis of CHIKV infected THP-1 derived human macrophages
Reads post-paired-end RNA sequencing was pre-processed for mapping by assessing quality using FastQC and adapters were removed using Cutadapt. After adapter removal, 8.269202 × 106 reads were identified from libraries of control samples while 6.758941 × 106 reads were obtained from 8hpi library samples. In 36hpi and 42hpi libraries, 8.749197 × 106 reads and 1.1395790 × 107 reads were acquired respectively. The quality trimmed reads were mapped against the GRCh38. p13 human genome using STAR with default parameters. Mapping of reads from each library to the human genome showed 88.8% mapping in the control library while 84.3%, 88.01%, 89.33%, and 89.38% reads of 8hpi, 24hpi, 36hpi, and 42hpi libraries respectively. A total of 26,000–40,000 genes were reported in the human database out of which 15034 genes in control, 15143 genes in 8hpi, 15638 genes in 24hpi, 16042 genes in 36hpi and 16518 genes in 42hpi libraries were identified (Table 1).
Table 1.
Summary of RNAseq data: reads, mapping percent and number of genes identified in each library.
| Sample Name | Number of reads | Number of reads after trimming | Mapping percentage | Number of genes |
|---|---|---|---|---|
| Uninfected_1 | 3.497256 × 106 | 3.493146 × 106 | 89.26% | 15642 |
| Uninfected _2 | 2.295436 × 106 | 2.292945 × 106 | 88.01% | 14456 |
| Uninfected _3 | 2.476510 × 106 | 2.473815 × 106 | 89.23% | 15003 |
| Infection_8hrs_1 | 1.292474 × 106 | 1.291886 × 106 | 87.51% | 14159 |
| Infection_8hrs_2 | 2.271973 × 106 | 2.165559 × 106 | 82.26% | 15315 |
| Infection_8hrs_3 | 3.194494 × 106 | 3.031140 × 106 | 83.20% | 15956 |
| Infection_24hrs_1 | 4.285212 × 106 | 4.283470 × 106 | 84.74% | 17451 |
| Infection_24hrs_2 | 1.747034 × 106 | 1.745137 × 106 | 89.82% | 14257 |
| Infection_24hrs_3 | 2.716861 × 106 | 2.714146 × 106 | 89.48% | 15205 |
| Infection_36hrs_1 | 3.581904 × 106 | 3.979116 × 106 | 89.96% | 17374 |
| Infection_36hrs_2 | 2.584487 × 106 | 2.581532 × 106 | 89.70% | 15185 |
| Infection_36hrs_3 | 3.062558 × 106 | 3.059011 × 106 | 88.33% | 15567 |
| Infection_42hrs_1 | 3.345994 × 106 | 3.342521 × 106 | 88.85% | 15685 |
| Infection_42hrs_2 | 5.712274 × 106 | 5.710146 × 106 | 90.03% | 18877 |
| Infection_42hrs_3 | 2.337522 × 106 | 2.334867 × 106 | 89.26% | 14991 |
Post mapping, intra library and inter library normalization between samples was performed. After normalization, pair-wise comparison of each of the infection timepoints against the uninfected as well as comparison of all infection timepoints against uninfected were made and the differentially expressed genes were identified using the edgeR package (Supplementary Fig. 1b). Concerning pair wise analysis of the individual timepoints against the uninfected samples, a total of 2136 genes were found to be regulated of which 1715 were found to be regulated only at 8hpi. In case of 24hpi, a total of 658 were found to be regulated of which 190 were uniquely regulated only at 24hpi. Similarly, at 36hpi, a total of 384 genes were modulated of which 54 genes were found to be uniquely regulated at 36hpi. In case of 42hpi, a total of 446 genes were found to be regulated of which 105 was unique to this hpi. An in-depth analysis of these regulated genes and their pathways is described below.
2.3. Genes regulated at 8hrs post-infection
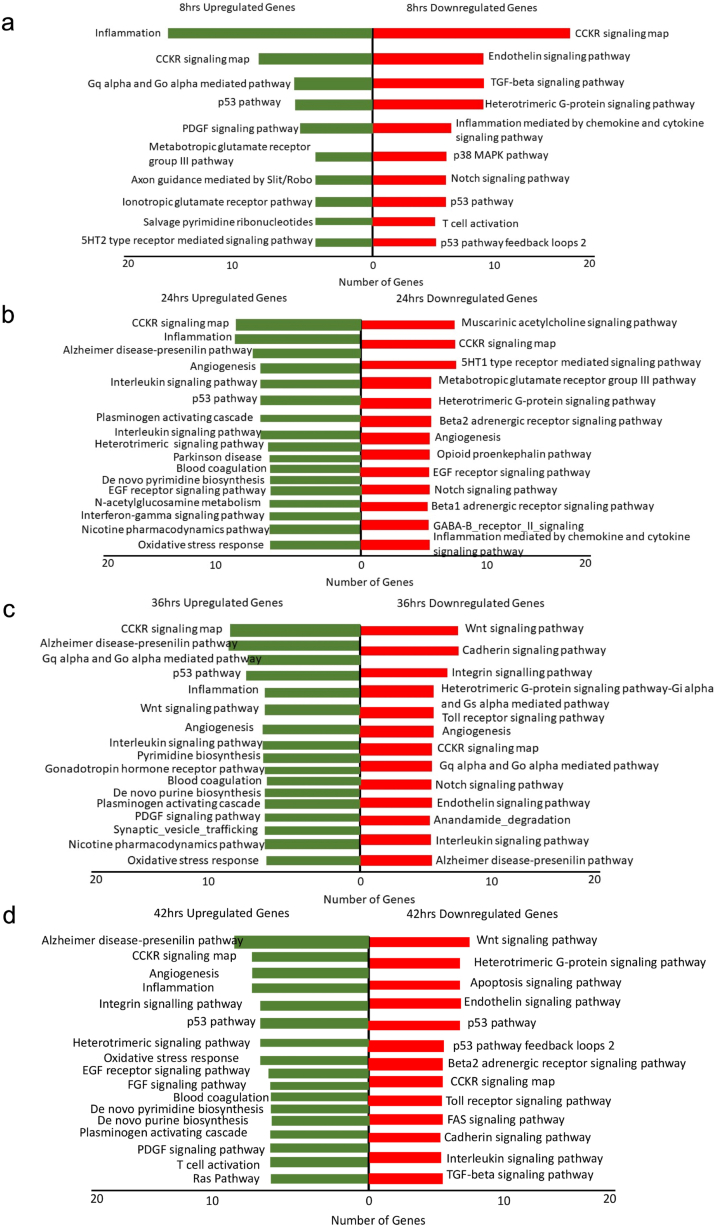
At 8hpi, the number of infected cells was observed to be less as per the confocal results. However, regulation of the global transcriptome was found to be quite significant at this timepoint, with a total of 2136 genes found to be significantly regulated when compared with uninfected sample. Of the regulated genes, 707 genes were significantly upregulated while 1429 genes were significantly downregulated (Supplementary File 1). Pathway analysis of differentially expressed genes (DEGs) revealed that inflammation, CCKR signaling pathway, p53 signaling pathway, G-alpha mediated pathway, salvage nucleotide pathway, endothelin signaling pathway, TGF-beta signaling pathway, p38 MAPK pathway, notch signaling pathway, and T-cell activation pathway were significantly regulated. (Fig. 2a). Further, to identify the cellular component, molecular mechanism, and/or biological process of DEGs at 8hpi, we performed a Gene Ontology (GO) enrichment analysis on the DEG list. The GO analysis of upregulated genes at 8hpi revealed that genes belong to different functional classes mainly biological regulation, cell communication, and metabolic processes. Molecular functions of these upregulated genes showed functions like protein binding, ion binding, nucleic acid binding, hydrolase, and transferase activity. The GO analysis of downregulated genes at 8hpi explained that biological regulation, metabolic processes, cell communication, and response to stimuli were the functional classes of downregulated genes. While the molecular functions of these genes were transporter activity, enzymatic activity, and protein/nucleic acid binding activity (Supplementary Fig. 2a). Noteworthy, in spite of low infection that was observed at this timepoint, the host immune system and metabolic pathways were getting regulated at same timepoint. A bird's eye view of DEGs at 8hpi suggested that various immune related genes such as CCL2, IFIH1, AIM2, IFIT2, IFIT1, CXCL5, TNFSF10, CXCL6, DDX58, TNFSF13B, CXCL8, CXCL1, CCL1, CCL8, PPBP, CD1B, IL11, CCL28, SERPING1, CD80, SLAMF6, and EXO1 were upregulated. Immune genes that were downregulated at 8hpi included HLA-E, TNFRSF14, CD74, RIPK2, CXCR4, ECSIT, IRF3, HLA-DRB1, SPP1, IL-4R, LAT2, HLA-B, HLA-A, CD300A, TNF, TNFRSF1A, PRG2, CLEC4A, TNFSF9, EBI3, C4B, OSM, CD6, LIF and TLR-9. Furthermore, we examined those genes that were found to be regulated only at 8hpi and identified a total of 1715 genes unique to 8hpi (Supplementary Fig. 1b); pathway analysis of these significantly regulated unique genes using KOBAS web server revealed that a total of three significantly regulated pathways (with corrected p-value ≤0.05), namely, metabolic pathways, Cysteine and methionine metabolism, and Other types of O-glycan biosynthesis (Supplementary File 2). Taken together, it is evident that global transcriptional changes in macrophages upon CHIKV infection gets kickstarted early on in infection with important genes of the host immune and metabolic pathways aligning to combat the infection.
Fig. 2.
Pathway analysis of significantly upregulated and downregulated genes at different timepoints a. 8hpi b. 24hpi c. 36hpi d. 42hpi.
Genes regulated at 24hrs post-infection: In case of 24hpi, a total of 658 genes were found to be regulated of which 320 genes were significantly upregulated and 338 genes were significantly downregulated (Supplementary File 1). Pathway analysis of DEGs at 24hpi demonstrated that CCKR signaling pathway, inflammatory pathway, interleukin signaling pathway, p53 pathway, oxidative stress, GABA-B receptor signaling pathway, chemokine signaling pathway, and EGF receptor signaling pathway were mainly modulated (Fig. 2b). Further, GO analysis of both upregulated and downregulated genes at 24hpi showed genes share some common functional classes namely, biological regulation, metabolic process, cell communication, and development process. The molecular function of DEG revealed its functional property like protein binding, nucleic acid binding, enzymatic activity, and transporter activity (Supplementary Fig. 2b). Immune-related genes namely CCL3, CCL2, CCL8, CXCL1, IFIT1, CXCL8, PPBP, CXCL5, CCL7, CXCL7, IFITM1, CXCL6, CXCL2, IL11, SELPLG, ITGA2, CCL1, CCL22, CSF3 were found to be upregulated while CD300A, CLEC4A, and, TLR9 were found to be downregulated at 24hpi. Of interest were the upregulation of the interferon inducible genes such as CXCL1, IFIT1, IFITM1, IFITM3, IFNβ1, and IFIT2 at this timepoint.
Analysis of those genes regulated only at 24hpi revealed that 190 genes were uniquely regulated only at this timepoint. Pathway analysis of these unique genes of 24hpi unveiled that the chemokine signaling pathway was found to be regulated. The genes that were found to be involved in chemokine signaling pathway were CXCL2(MIP-2), G Protein Subunit Alpha I1, CCL3(MIP-1α), CXCL3(MIP-2b), CCL22, and Protein Kinase cAMP-Activated Catalytic Subunit Beta (Supplementary File 2). In conclusion, with respect to 24hpi, it appears that a more focused defense is being embarked by the host system, as evidenced by distinct immune molecules, against the increasing viral titer.
Genes regulated at 36hrs post-infection: In 36hpi samples compared with uninfected samples, 384 genes were found to be regulated in which 230 genes were upregulated and 154 genes were downregulated (Supplementary File 1). Pathway analysis of regulated genes at 36hpi unveiled that some of the pathways namely, CCKR signaling pathway, p53 pathway, inflammation, interleukin signaling pathway, pyrimidine biosynthesis, wnt signaling pathway, cadherin signaling pathway, integrin signaling pathway, Notch signaling pathway, toll receptor signaling pathway, and oxidative stress pathway were found to be regulated (Fig. 2c). The GO analysis of DEGs at 36hpi revealed that genes belonged to functional classes mainly biological process, metabolic process, cellular communication and response to stress and stimulus while molecular functions of DEGs showed its function mainly in protein binding, nucleic acid binding, and ion binding (Supplementary Fig. 2c). However, immune related genes that are upregulated at 36hpi included PPBP, CXCL5, CSF1, CXCL8, IL11, CCL2, CCL7, FCGR2B, CR1, and CD1A. Out of totally regulated 230 genes, a total of 54 genes uniquely regulated only at this time point. Due to a smaller number of unique genes at this timepoint, we didn't extract significant pathways that might be related to infection (Supplementary File 2).
Genes regulated at 42hrs post-infection: In 42hpi group, a total of 446 genes were regulated; 273 genes were upregulated and 173 genes were downregulated at 42hpi (Supplementary File 1). Pathway analysis of DEGs at 42hpi showed that some of the pathways namely, CCKR signaling pathway, p53 pathway, inflammation, FGF signaling pathway, de novo nucleotide biosynthesis, integrin signaling pathway, wnt signaling pathway, apoptosis, toll-like receptor pathway, fas signaling pathway, interleukin signaling pathway, TGF signaling pathway, and oxidative stress pathway were found to be regulated (Fig. 2d). Further, GO analysis of DEGs at 42hpi revealed its functional classes namely, biological processes, metabolic process, cell communication and response to stress and stimulus. Annotation of DEGs based on molecular function showed that these genes were involved in protein binding, ion binding, hydrolase activity, transferase activity, and transporter activity (Supplementary Fig. 2d). At 42hpi, immune-related genes namely ECSIT, TP53, and IL15, HLA-DPA1 were found to significantly downregulated while FCGR2B, IL11, CCL2, CD1A, CXCL5, PPBP, CSF1, CRLF2, and CCL3L1 genes were upregulated.
Further, of the total regulated 446 genes, 105 genes regulated only at this time point. The pathway analysis of regulated unique genes showed two pathways were found to be significant and these include the cAMP signaling pathway and aldosterone synthesis. The genes that found to be impacted in these pathways are natriuretic peptide receptor 1 (NPR1), ATPase plasma membrane Ca2+ transporting 4 (ATP2B4), ATPase Na+/K+ transporting subunit alpha 2 (ATP1A2), calcium/calmodulin dependent protein kinase II beta (CAMK2B), and endothelin 1 (EDN1) (Supplementary File 2).
2.4. Commonly regulated genes in all timepoints
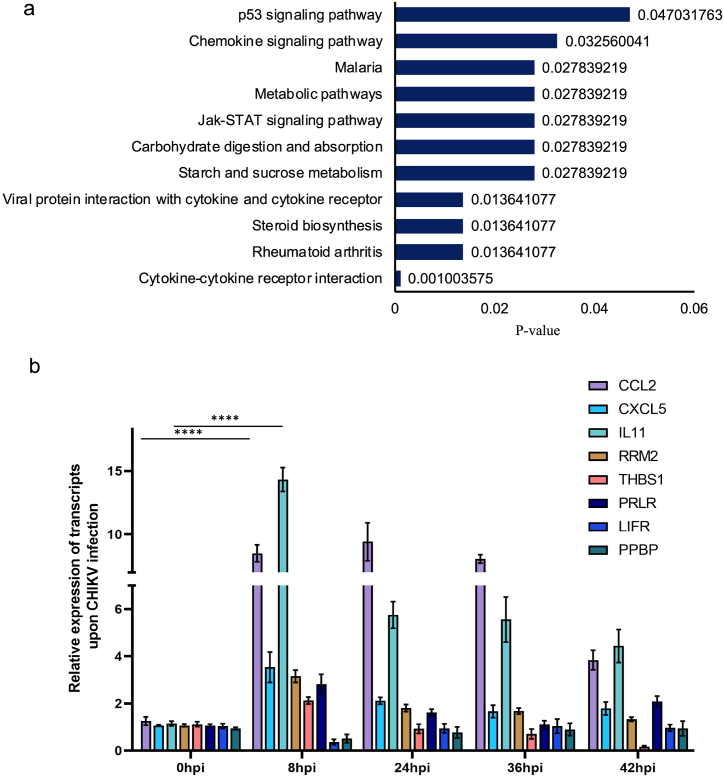
Further comparisons of all the genes in all different infection groups divulged 122 genes, common in all groups, to be significantly regulated (p-value ≤0.05) with a log fold change of log2(FC) ≥ ± 1.5 (Supplementary File 3). These significantly regulated common genes were subjected to analysis using KOBAS web server for pathway analysis (Supplementary File 3). Pathway analysis of regulated common genes from all infected datasets revealed that a total of eleven pathways to be significantly regulated upon infection (with corrected p-value ≤0.05) (Fig. 3a). Out of these, six pathways are mainly involved in cellular signaling cascade i.e., p53 signaling pathway, chemokine signaling pathway, JAK-STAT signaling pathway, rheumatoid arthritis, cytokine-cytokine receptor interaction and viral protein interaction with cytokine and cytokine receptors. Out of 122 commonly regulated genes, only 8 genes were impacted on these pathways namely, Pro-Platelet Basic Protein (PPBP), LIF Receptor Subunit Alpha (LIFR), Interleukin11 (IL11), C–C Motif Chemokine Ligand 2 (CCL2), Prolactin receptor (PRLR), Ribonucleotide Reductase Regulatory Subunit M2 (RRM2), C-X-C Motif Chemokine Ligand 5 (CXCL5), Thrombospondin 1 (THBS1) to be regulated in all the 4 datasets when compared against the uninfected sample. Several of these genes showed multifold differences, such as in case of PPBP (3-5-fold), CCL2 (2-6-fold), and CXCL5 (3-6-fold).
Fig. 3.
CHIKV induces major changes in the transcriptome of infected macrophages a. Regulated pathways of total of 122 significantly regulated common genes of all timepoints using KOBAS web tool. b. qRT-PCR analysis of expression profiles of selected genes regulated in significant pathways of CHIKV-infected macrophage cell relatives to uninfected controls. Relative expression was determined using the 2−ΔΔCT method and RNA levels were normalized by GAPDH. Data are presented as the mean ± standard deviation (SD) (****< 0.05).
To validate the differential expression of significantly regulated common targets obtained by RNA-seq analysis upon CHIKV infection, we performed quantitative RT-PCR of selected targets namely, PPBP, LIFR, IL11, CCL2, PRLR, RRM2, CXCL5, and THBS1. Maximum upregulation of most of the molecules was observed at 8hpi at varying levels. Out of eight targets, CCL2, and IL11 were found to be most significantly upregulated at all timepoints investigated. CCL2 showed a 7-fold upregulation at 12hpi, whereas IL11 showed a similar upregulation of almost 7-8-fold at 8hpi. CXCL5 was upregulated almost 2-fold at 8hpi. Factors PRLR, RRM2, and THBS1 showed a maximum upregulation of 1.5-fold at 8hpi after which the expression was reduced. PPBP and LIFR did not reveal any change in their expression pattern during the timepoints investigated (Fig. 3b). RT-PCR results of these molecules show consistency with respective RNA-seq data analysis. Although some differences were observed between these two types of analyses due to intrinsic differences between the techniques, the qRT-PCR results displayed the same relative regulation of commonly regulated genes as the RNA-seq data.
3. Discussion
Global changes to transcriptome and proteome of susceptible and model host cell systems have been previously described [21,[24], [25], [26]]. Amongst the susceptible cells, macrophages hold a critical role in viral infection owing to the defense they mount against the virus and in case of CHIKV, they are also the site where the virus may persist thereby resulting in a prolonged manifestation of arthritic symptoms [5]. A recent study described global transcriptional modulations owing to CHIKV infection in U937 macrophages at a single timepoint and identified potential pathways that could be targeted for therapeutic interventions [27]. Our study describes the first detailed RNA-Seq analysis of different timepoints of THP-1-derived CHIKV-infected macrophages. To understand the molecular changes in these infected macrophages over time, we generated and analyzed the transcriptome data of different timepoints in two different ways: firstly, we performed a pairwise analysis of the transcriptome at the individual timepoint to arrive at the repertoire of genes that were regulated at that time of CHIKV infection. This helped in understanding gene regulation at a specific timepoint in a nutshell. Furthermore, we analyzed all the datasets together to arrive at those unique genes modulated at a specific infection timepoint as well as to arrive at those common genes that are regulated at all infection timepoints. KEGG pathway analysis of significantly up-regulated and down-regulated unique genes at 8hpi revealed metabolic pathways to be significantly regulated with p-value ≤0.05. Interestingly, serum metabolome study of CHIKV-infected patients also showed perturbation in metabolic pathways mainly glycine, serine, threonine, and pyrimidine metabolisms [28]. Several studies have previously elucidated that metabolic processes are modulated upon viral infection not only at the transcript level but also at the proteomic level. For instance, proteomic profiling of CHIKV-infected mosquito cells showed the regulation of host factors that are involved in cysteine and methionine metabolism [29]. Similarly, another study of Ross River virus (RRV) and Semliki Forest virus (SFV) showed that both viruses trigger the activation of AKT which leads to an increase in cellular metabolism for the synthesis of new biomass [30]. The perturbation in the metabolic pathway at 8hpi could be attributed to the initiation of the translational shut off that is well described in alphavirus infections in the host [31]. In addition, the biological processes of DEGs at 8hpi reveal that these genes are involved in cell communication, metabolic process, and cellular process. Considering the low viral titer at this timepoint and it is known that cell communication is one of the most important roles in cell to cell spread of virus, regulation of these pathways is expected for the successful establishment of infection in the host [32]. Viruses are dependent on host for viral genome replication, propagation, and reproduction and use the host resources for their own survival and rapid multiplication by upregulating the host metabolic process [28,33].
At the time of the peak of viral infection in macrophages, namely 24hpi, chemokine signaling pathway was found to be regulated at 24hpi. Several studies report the robust production of chemokines and cytokines during CHIKV infection is associated with regulation of inflammatory responses [33,34]. Activation of chemokine signaling pathway suggests probable macrophage polarization upon infection since the onset of infection can have a direct bearing on disease progression into the joints. In our study, some genes that are regulated in chemokine signaling pathway are CXCL2(MIP-2), CCL3(MIP-1α), CXCL3(MIP-2b), and CCL22. These molecules are mainly synthesized from macrophages upon CHIKV infection and act as chemoattractant. The upregulation of MIP-1α and MIP-2 was found in our 24hpi dataset. Similarly in a study on mononuclear dengue-infected cells, MIP-1α production was significantly increased at early stage of infection [35]. This indicates that MIP-1α and MIP-2 molecules might be playing a role in early recruitment of different subsets of immune cells to generate a robust inflammatory response. In CHIKV-infected human astrocytoma cells, the upregulated expression of CXCL3 factor was found and its expression was partially dependent on the induction of Early growth response 1(EGR1). EGR1 is a well-known transcriptional factor that is involved in inflammatory response and cell death. In EGR1(−/−) astrocytoma cells with CHIKV infection, a significant downregulation of CXCL3 was found when compared to wild type-infected cells [36]. In our data, 1.6-fold significant upregulation of CXCL3 was found at 24hpi. Similarly, in human primary fibroblasts, CXCL3 was significantly upregulated at 24hrs post CHIKV infection which may be associated with the activation of the inflammatory response [37]. Another factor, CCL22 (MDC) is primarily generated by macrophages in response to pathogenic products and plays an important role in the recruitment of Th-2 lymphocytes at inflammatory sites of infection [38].
At the later timepoints, especially at 36hpi, where the infection was at its nadir amongst the timepoints evaluated, we observed that there were very few regulated genes suggesting that the host cells were mounting a robust defense against the virus and almost reaching a pre-infection state. Probably, a more detailed analysis between the 24hpi and 36hpi involving tightly rendered timepoints for scrutiny might throw light as to how the host is at its fighting best. This could be elucidated to an extent at the next timepoint, namely 42hpi, where two important pathways were modulated mainly, cAMP signaling pathway and aldosterone synthesis pathway. In studies on Japanese encephalitis virus (JEV) infected mice brains, calcium/calmodulin-dependent protein kinase type II (CAMPK2) was found to be downregulated which ultimately led to virus induced neuropathogenesis [39]. In contrast, CAMPK2 was upregulated in late stage of CHIKV infection. Another study of HIV-1 infection explained that infection is associated with increased levels of cAMP and hyperactivation activation of PKA [40]. Interestingly, cAMP is a key inducer of anti-inflammatory responses [41]. We hypothesize that activation of cAMP signaling pathway might in turn helps the macrophages to cope the viral infection by activating anti-inflammatory responses.
Several pathways are modulated in all different infection timepoints mainly p53 signaling pathway, JAK-STAT pathway, and chemokine signaling pathway. Regulation of these pathways during viral infection has been well documented in earlier reports. For instance, one study showed that p53 signaling pathway upregulated upon viral infection in mouse embryonic fibroblast cells [42]. Similarly, transcriptome profiling of CHIKV-infected human MDMs revealed activation of JAK-STAT pathway and induced robust proinflammatory and anti-inflammatory responses [43]. The study however inspected the transcriptomic changes only at one timepoint, i.e., 24hpi, due to which no information could be inferred regarding infection progression. Owing to longer timepoint analyses in our current study, we provide a more comprehensive insight as to the host response starting from the early stages of infection, especially those pertaining to host defense and adaptive immunity and those factors that persist during the course of infection. Molecules such as PPBP, CCL2, and CXCL5 are nodal in cellular infiltration at the site of infection, as reported in previous studies [44,45]. The significant fold change in the expression of these molecules in the present study provides credence to their role in disease progression in CHIKV infection. Some of the factors such as CCL2, CCL5, and CXCL5 are known atherogenic factors and are found to be upregulated in both rheumatoid arthritis patients and CHIKV-infected patients [[46], [47], [48]]. In the present study, the expression of CCL2 was found to be significantly upregulated in all timepoints compared with control; an observation corroborating with published information from clinical studies [34,49]. Another factor, CXCL5 was found to be significantly upregulated at an early timepoint i.e., 8hpi, and further its expression level decreases. Likewise, another study unveiled that decrease in CXCL5 level was associated with chronicity of chikungunya and works as a potential biomarker in chikungunya disease severity [50]. The expression level of IL11 was found to be significantly upregulated in all timepoints. Similar results were observed in CHIKV-infected mouse astrocytes and were found to be involved in inflammatory process [51].
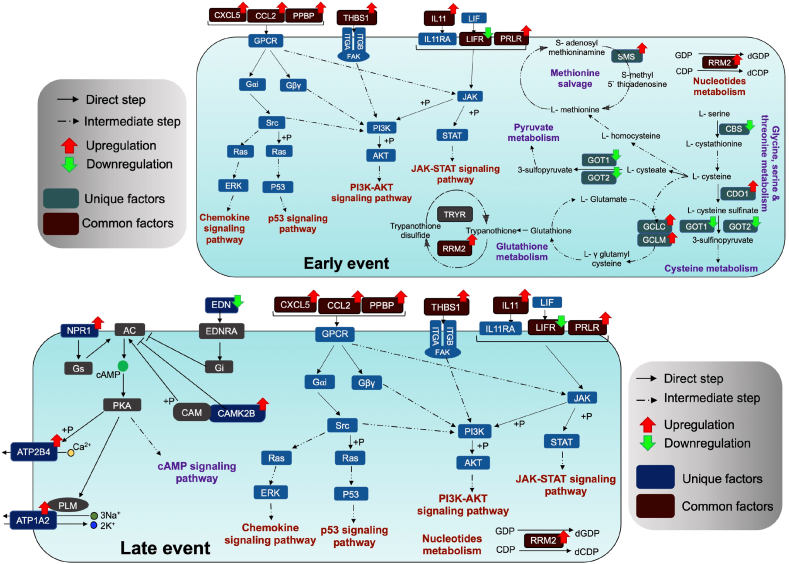
Analyses of the DEGs regulated at the four timepoints yielded important insight concerning the regulated pathways involved in macrophage physiology and cell immunity specifically in the early timepoint, namely, 8hpi, and in the later timepoint, 42hpi. These reports corroborate our present findings of the pathways regulated, thereby prompting us to develop a probable model of CHIKV in macrophages depicting the pathways that could be involved during the early and late phase of infection in the macrophages and the cellular processes that might be triggered early on in infection and how they alter in the due course of CHIKV infection (Fig. 4).
Fig. 4.
Pathways regulated during early and late stages of CHIKV infection.
3.1. Limitations of the study
THP-1 cells are a human monocytic cell line of a homogenous population of cells, thereby providing robust data with respect to overall modulations that occur upon infection; however, this may not reflect a real time scenario of the modulations that would occur upon infection in primary macrophages and this can be perceived as a limitation of the present study. Including macrophages from PBMCs as a group for transcriptomic analysis would definitely provide additional information with respect to CHIKV infection in macrophages per se, but was beyond the scope of the present study. Further, the inherent low efficiency of infection and transfection in the THP-1 cells prevented us from planning further validation experiments to characterize the perturbed molecules. Similarly, the poor CHIKV infection in the THP-1 cells did not seem to elicit much IFN or inflammatory responses that is reported to occur in natural infections. And these aspects could not be addressed through specific experiments owing to the low transfection efficiency in manipulating the THP-1 cells. Similarly, for the same reason mentioned above, specific questions pertaining to the role of viral proteins/genes in modulating macrophages could not be addressed. However, in a bid to overcome these limitations, the present study obtained the global picture of infection at multiple timepoints and then by performing multiple analyses, attempted to understand the role of the pathways modulated during early and late CHIKV infection in THP-1 cells.
4. Conclusion
In conclusion, our study provides insights into the pathways regulated in macrophages early and in the later stages of CHIKV infection. Our study is the first to show the global transcriptional changes that occur in THP-1 derived macrophage cells over such an extended period of time enabling consolidating and substantiating several of the isolated evidence of CHIKV infection in the macrophages in a nutshell. Since the macrophages serve as key reservoirs of the virus, the virus must initiate its roadmap to survival early on in infection. The modulation of metabolic pathways and chemokine signaling as observed in our study during the early stages of infection might be of the schemes employed by the virus in this pursuit. It is noteworthy that other pathways such as JAK-STAT pathway and chemokine signaling pathway are upregulated even during the later stages of infection emphasizing the continued battle between the virus and the host in disease progression and resolution. Studying the impact of these pathways upon challenging higher doses of virus can provide clues on how a more effective defense mechanism might get kicked in. Activation of the cAMP signaling pathway triggers the antiviral response in the later stages of infection as observed in our study might be one of the strategies employed by the macrophages to combat CHIKV infection.
5. Materials and methods
Cell Culture: THP-1 monocytes were grown in RPMI media supplemented with 10% FBS, l-glutamine, penicillin, and streptomycin at 37 °C with 5% Co2 in Co2 incubator. Further, monocytes were activated using PMA at a working concentration of 20 ng/ml in RPMI media. Post 48hrs treatment with PMA, the adherent macrophages were washed with complete media and used for infection experiments.
Virus Propagation and Infection: Chikungunya virus (accession no. JF950631.1) was used in this study [52]. The virus was initially propagated in C6/36 cells grown in DMEM media supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% l-glutamine. The virus was harvested at 96hpi and used in infection experiments in THP-1 cells after quantifying it using plaque assay [53]. THP-1 cells were seeded in 6-well plate followed by PMA activation. Post 48hrs activation, the differentiated macrophages were infected with CHIKV (MOI1) diluted in sera free media. 2 h post-infection, media was changed by RPMI media supplemented with 2% FBS, 1% penicillin/streptomycin, and 1% l-glutamine. Cells were harvested at different timepoints in trizol for RNA isolation.
Sample processing:CHIKV infected cells at different timepoints were collected separately in Trizol reagent. RNA was isolated using standard trizol RNA isolation method. The quality and quantity checks of extracted RNA samples were done using NanoDrop 2000/2000c Spectrophotometers (Thermo Fisher Scientific) and Bioanalyzer/TapeStation 4200 (Agilent Technologies). The final concentration was assessed on Qubit 3.0 Fluorometer (Thermo Fisher Scientific).
Transcriptome library preparation: 1 μg RNA of each sample (RIN≥7) was taken for transcriptome library preparation using the Truseq Stranded mRNA kit (Illumina) which generates mRNA-focused sequencing libraries from total RNA, as per the manufacturer's protocol. The quality and quantity analysis of libraries was done on Bioanalyzer/TapeStation 4200 (Agilent Technologies) and Qubit 3.0 fluorometer (Thermo Fisher Scientific). A graphical representation of work flow followed for high-throughput RNA seq sample processing and data analysis was shown in Supplementary Fig. 1a.
Transcriptome Library Sequencing:The mRNA libraries for sequencing were prepared according to the MiSeq System User Guide (Illumina, San Diego, CA, USA). Briefly, the pooled sample library was diluted to 4 nM, denatured with 0.2 N fresh NaOH, diluted to 12.5 pM by addition of Illumina HT1 buffer, and then supplemented with 1% spike-in of 12.5 pM PhiX (Illumina) as an in-lane positive control for alignment calculations and quantification efficiency. The total volume of 600 μl of the combined library was loaded on a 150-cycle (2 × 75 paired ends) Miseq v3 reagent cartridge (Illumina, San Diego, CA, USA) and sequenced on MiSeq instrument (Illumina, San Diego, CA, USA). The quality of the sequence run was monitored by Sequencing Analysis Viewer (SAV) (Illumina).
Transcriptome data analysis: We assessed Phred scores with FastQC, to evaluate RNA-seq data accuracy. Reads with average minimum quality scores corresponding to 99% base call accuracy at every nucleotide position (Phred score> 20) were retained. Cutadapt, a python extension module was used for adapter removal. We used GRCh38. p 13 GTF file for transcript/gene annotations. Post alignment and annotation, the resulting read counts were processed to obtain the differentially expressed genes. Differential gene expression analysis was performed with EdgeR. To define a gene as differentially expressed, the EdgeR output was filtered to retain genes that have multiple testing corrected p-value of <0.05 and a log2 fold threshold of ± 1.5. A gene having positive log2 fold values was considered as upregulated whereas negative log2 fold indicated a gene was downregulated upon infection.
Gene ontology (GO) and Pathway analysis: For functional annotation analysis of the whole gene set, KOBAS web server was used to identify genes expressed differently between control and infected samples. Genes with log2 fold threshold of ± 1.5 and p-value of <0.05 was considered as significantly regulated. Also, pathway analysis of significantly upregulated and downregulated genes of different timepoints were done by pathfindR script using default parameters. GO analysis was done using WEB-based GEne SeT Analysis Toolkit (Webgestalt tool) to categorize functional classification, molecular and biological process of genes.
Transcript Expression profiling using qRT-PCR: For transcripts expression profiling, THP-1 infected cells of different timepoints were used to isolate total RNA using trizol method. The concentration of isolated RNA was measured by NanoDrop 2000 spectrophotometer (Thermo Scientific). The quality of RNA was checked by calculating the ratio absorbance at 260 and 280 nm. qRT-PCR assay was performed using QuantiTect SYBR Green RT-PCR Kit (Qiagen) as per the manufacturer's instruction. The initial concentration of RNA was set at 300 ng and used in qRT-PCR using specific forward and reverse primers (Supplementary Table 1). All reactions were performed in triplicate using PikoReal 96 Real-Time PCR system (Thermo Scientific). Expression levels of transcripts were compared with those in control after normalization against the housekeeping gene, GAPDH.
Confocal Imaging: THP-1 cells were seeded on glass coverslips in 6-well plates with 100 ng/ml PMA for macrophage activations. Post-activation, cells were infected with CHIKV (MOI 1) and processed at different infection timepoints for confocal study. Firstly, cells were fixed with 4% paraformaldehyde in PBS for 15min and then permeabilized for 30min in 100% chilled methanol at 4 °C. After washing with PBS, cells were incubated with blocking buffer (1%BSA and 0.3% Triton X-100 in PBS) for 45min. Post-blocking, cells were incubated in CHIKV E1 mice antibody (in-house generated, 1:50) primary antibody diluted in 1% BSA and 0.5% Triton X-100 in PBS for 1hrs. Further, Alexa Fluor 591/488-conjugated secondary antibodies (Invitrogen) were added for 1hrs. Cells were washed three times with PBS and stained with DAPI (Invitrogen) to visualize nuclei. Images of the different infected cells were taken using a Nikon confocal microscope.
Measurement of fluorescence intensity: Images for each infected conditions were acquired from five different fields across the slide using bright-field and standard filters. The mean intensity was obtained for cells greater than 20 μm in size from a 16-bit image. The mean fluorescence intensity data were expressed as a mean (±SD) percentage of fixed, PBS-treated cells. The fluorescence value was calculated using NIS Element Software.
Statistical analysis: Standard error of the mean was taken to represent the data. Each experiment was conducted at least thrice as a biological replicate. The error bars were generated using experimental replicates. Statistical analysis of experimental data was performed using GraphPad Prism (version9) using Student's t-test. Analysis of variance (ANOVA) was used for data from different treatments. For multiple comparisons, Tukey's test or Dunnett's test was performed. Significant values were P < 0.05 represented with an asterisk. In KOBAS, Statistical analysis was performed using Hypergeometric test/Fisher's exact test as statistical method, and Benjamin and Hochberg method (1995) was used for FDR correction.
Funding
This work was supported by financial grant from the Department of Biotechnology, Government of India (BT/PR14725/AGR/36/672/2010) to SS.
Author contribution statement
Priyanshu Srivastava: Designed and performed the experiments; Analyzed and interpreted the data; Wrote the paper; review the paper.
Sakshi Chaudhary: Analyzed and interpreted the data
Surbhi Malhotra: Performed the experiments
Binuja Varma: Contributed reagents, materials, analysis tools or data.
Sujatha Sunil: Conceived and designed the experiments; Wrote and review the paper.
Data availability statement
Raw data has been to submitted in Sequence 547 Read Archive (SRA), National Center for Biotechnology Information (NCBI and) database.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
P.S., was supported through fellowships from University Grants Commission, Government of India.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17158.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Clatch R.J., et al. Monocytes/macrophages isolated from the mouse central nervous system contain infectious Theiler's murine encephalomyelitis virus (TMEV) Virology. 1990;176(1):244–254. doi: 10.1016/0042-6822(90)90249-q. [DOI] [PubMed] [Google Scholar]
- 2.Nagra R.M., Wong P.K., Wiley C.A. Expression of major histocompatibility complex antigens and serum neutralizing antibody in murine retroviral encephalitis. J. Neuropathol. Exp. Neurol. 1993;52(2):163–173. doi: 10.1097/00005072-199303000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Okabe Y., Medzhitov R. Tissue biology perspective on macrophages. Nat. Immunol. 2016;17(1):9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 4.Sintiprungrat K., et al. Alterations in cellular proteome and secretome upon differentiation from monocyte to macrophage by treatment with phorbol myristate acetate: insights into biological processes. J. Proteonomics. 2010;73(3):602–618. doi: 10.1016/j.jprot.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Labadie K., et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J. Clin. Invest. 2010;120(3):894–906. doi: 10.1172/JCI40104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbas W., et al. Eradication of HIV-1 from the macrophage reservoir: an uncertain goal? Viruses. 2015;7(4):1578–1598. doi: 10.3390/v7041578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abreu C.M., et al. Infectious virus persists in CD4(+) T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J. Virol. 2019;93(15) doi: 10.1128/JVI.00065-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perrone L.A., et al. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4(8) doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linn M.L., Aaskov J.G., Suhrbier A. Antibody-dependent enhancement and persistence in macrophages of an arbovirus associated with arthritis. J. Gen. Virol. 1996;77(Pt 3):407–411. doi: 10.1099/0022-1317-77-3-407. [DOI] [PubMed] [Google Scholar]
- 10.Morrison C.R., Plante K.S., Heise M.T. Chikungunya virus: current perspectives on a reemerging virus. Microbiol. Spectr. 2016;4(3) doi: 10.1128/microbiolspec.EI10-0017-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dileep Mavalankar P.S. Lancet Infect Diseases.; 2007. Parvathy Raman, Chikungunya Epidemic in India: a Major Public-Health Disaster. [DOI] [PubMed] [Google Scholar]
- 12.Ahola T., Merits A. Chikungunya Virus. Springer; 2016. Functions of chikungunya virus nonstructural proteins; pp. 75–98. [Google Scholar]
- 13.Metz S.W., Pijlman G.P. Chikungunya Virus. Springer; 2016. Function of Chikungunya virus structural proteins; pp. 63–74. [Google Scholar]
- 14.Lum F.M., Ng L.F. Cellular and molecular mechanisms of chikungunya pathogenesis. Antivir. Res. 2015;120:165–174. doi: 10.1016/j.antiviral.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Jaffar-Bandjee M.C., et al. Chikungunya virus takes centre stage in virally induced arthritis: possible cellular and molecular mechanisms to pathogenesis. Microb. Infect. 2009;11(14–15):1206–1218. doi: 10.1016/j.micinf.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Chow A., et al. Persistent arthralgia induced by Chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J. Infect. Dis. 2011;203(2):149–157. doi: 10.1093/infdis/jiq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy V., et al. Correlation of plasma viral loads and presence of Chikungunya IgM antibodies with cytokine/chemokine levels during acute Chikungunya virus infection. J. Med. Virol. 2014;86(8):1393–1401. doi: 10.1002/jmv.23875. [DOI] [PubMed] [Google Scholar]
- 18.Poo Y.S., et al. Multiple immune factors are involved in controlling acute and chronic chikungunya virus infection. PLoS Neglected Trop. Dis. 2014;8(12):e3354. doi: 10.1371/journal.pntd.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szekanecz Z., Koch A.E. Macrophages and their products in rheumatoid arthritis. Curr. Opin. Rheumatol. 2007;19(3):289–295. doi: 10.1097/BOR.0b013e32805e87ae. [DOI] [PubMed] [Google Scholar]
- 20.Her Z., et al. Active infection of human blood monocytes by Chikungunya virus triggers an innate immune response. J. Immunol. 2010;184(10):5903–5913. doi: 10.4049/jimmunol.0904181. [DOI] [PubMed] [Google Scholar]
- 21.Hussain K.M., et al. Establishment of a novel primary human skeletal myoblast cellular model for chikungunya virus infection and pathogenesis. Sci. Rep. 2016;6 doi: 10.1038/srep21406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava P., et al. Disease resolution in chikungunya-what decides the outcome? Front. Immunol. 2020;11:695. doi: 10.3389/fimmu.2020.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S., et al. Mouse macrophage innate immune response to Chikungunya virus infection. Virol. J. 2012;9:313. doi: 10.1186/1743-422X-9-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubey S.K., et al. An E3 ubiquitin ligase scaffolding protein is proviral during chikungunya virus infection in Aedes aegypti. Microbiol. Spectr. 2022;10(3) doi: 10.1128/spectrum.00595-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jungfleisch J., et al. CHIKV infection reprograms codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. Nat. Commun. 2022;13(1):4725. doi: 10.1038/s41467-022-31835-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pott F., et al. Single-cell analysis of arthritogenic alphavirus-infected human synovial fibroblasts links low abundance of viral RNA to induction of innate immunity and arthralgia-associated gene expression. Emerg Microbes Infect. 2021;10(1):2151–2168. doi: 10.1080/22221751.2021.2000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray M., et al. Chikungunya virus time course infection of human macrophages reveals intracellular signaling pathways relevant to repurposed therapeutics. PeerJ. 2022;10 doi: 10.7717/peerj.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jatin Shrinet J.S.S., RajniGaind Neel, Bhavesh Sarovar, Sunil Sujatha. Serum metabolomics analysis of patients with chikungunya and dengue mono/co-infections reveals distinct metabolite signatures in the three disease conditions. Sci. Rep. 2016;6 doi: 10.1038/srep36833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee R.C., Chu J.J. Proteomics profiling of chikungunya-infected Aedes albopictus C6/36 cells reveal important mosquito cell factors in virus replication. PLoS Neglected Trop. Dis. 2015;9(3) doi: 10.1371/journal.pntd.0003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzon M., et al. Alphavirus-induced hyperactivation of PI3K/AKT directs pro-viral metabolic changes. PLoS Pathog. 2018;14(1) doi: 10.1371/journal.ppat.1006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fros J.J., Pijlman G.P. Alphavirus infection: host cell shut-off and inhibition of antiviral responses. Viruses. 2016;8(6) doi: 10.3390/v8060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 2008;6(11):815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 33.Chirathaworn C., Chansaenroj J., Poovorawan Y. Cytokines and chemokines in chikungunya virus infection: protection or induction of pathology. Pathogens. 2020;9(6) doi: 10.3390/pathogens9060415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venugopalan A., Ghorpade R.P., Chopra A. Cytokines in acute chikungunya. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0111305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sierra B., et al. MCP-1 and MIP-1alpha expression in a model resembling early immune response to dengue. Cytokine. 2010;52(3):175–183. doi: 10.1016/j.cyto.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Lehman C.W., et al. EGR1 upregulation during encephalitic viral infections contributes to inflammation and cell death. Viruses. 2022;14(6) doi: 10.3390/v14061210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekchariyawat P., et al. Inflammasome signaling pathways exert antiviral effect against Chikungunya virus in human dermal fibroblasts. Infect. Genet. Evol. 2015;32:401–408. doi: 10.1016/j.meegid.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita U., Kuroda E. 2002. Regulation Of Macrophage-Derived Chemokine (MDC/CCL22) Production. Crit. Rev. Immunol. 22(2) [PubMed] [Google Scholar]
- 39.Saha S., Datta K., Rangarajan P. Characterization of mouse neuronal Ca2+/calmodulin kinase II inhibitor alpha. Brain Res. 2007;1148:38–42. doi: 10.1016/j.brainres.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann B., et al. Human immunodeficiency virus proteins induce the inhibitory cAMP/protein kinase A pathway in normal lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1993;90(14):6676–6680. doi: 10.1073/pnas.90.14.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tavares L.P., et al. Blame the signaling: role of cAMP for the resolution of inflammation. Pharmacol. Res. 2020;159 doi: 10.1016/j.phrs.2020.105030. [DOI] [PubMed] [Google Scholar]
- 42.Takaoka A., et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 2003;424(6948):516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 43.Valdes-Lopez J.F., Fernandez G.J., Urcuqui-Inchima S. Interleukin 27 as an inducer of antiviral response against chikungunya virus infection in human macrophages. Cell. Immunol. 2021;367 doi: 10.1016/j.cellimm.2021.104411. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz Silva M., et al. Mechanism and role of MCP-1 upregulation upon chikungunya virus infection in human peripheral blood mononuclear cells. Sci. Rep. 2016;6 doi: 10.1038/srep32288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos I., et al. CXCL5-mediated recruitment of neutrophils into the peritoneal cavity of Gdf15-deficient mice protects against abdominal sepsis. Proc. Natl. Acad. Sci. U. S. A. 2020;117(22):12281–12287. doi: 10.1073/pnas.1918508117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adamopoulos I.E., Pflanz S. The emerging role of Interleukin 27 in inflammatory arthritis and bone destruction. Cytokine Growth Factor Rev. 2013;24(2):115–121. doi: 10.1016/j.cytogfr.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burska A., Boissinot M., Ponchel F. Cytokines as biomarkers in rheumatoid arthritis. Mediat. Inflamm. 2014;2014 doi: 10.1155/2014/545493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yap H.Y., et al. Pathogenic role of immune cells in rheumatoid arthritis: implications in clinical treatment and biomarker development. Cells. 2018;7(10) doi: 10.3390/cells7100161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rulli N.E., et al. Protection from arthritis and myositis in a mouse model of acute chikungunya virus disease by bindarit, an inhibitor of monocyte chemotactic protein-1 synthesis. J. Infect. Dis. 2011;204(7):1026–1030. doi: 10.1093/infdis/jir470. [DOI] [PubMed] [Google Scholar]
- 50.Ng L.F., et al. IL-1beta, IL-6, and RANTES as biomarkers of Chikungunya severity. PLoS One. 2009;4(1):e4261. doi: 10.1371/journal.pone.0004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das T., et al. Multifaceted innate immune responses engaged by astrocytes, microglia and resident dendritic cells against Chikungunya neuroinfection. J. Gen. Virol. 2015;96(Pt 2):294–310. doi: 10.1099/vir.0.071175-0. [DOI] [PubMed] [Google Scholar]
- 52.Shrinet J., et al. Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol. J. 2012;9:100. doi: 10.1186/1743-422X-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tripathi P.K., et al. Evaluation of novobiocin and telmisartan for anti-CHIKV activity. Virology. 2020;548:250–260. doi: 10.1016/j.virol.2020.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data has been to submitted in Sequence 547 Read Archive (SRA), National Center for Biotechnology Information (NCBI and) database.