Abstract
The calmodulin-activated α-kinase, eukaryotic elongation factor 2 kinase (eEF-2K), serves as a master regulator of translational elongation by specifically phosphorylating and reducing the ribosome affinity of the guanosine triphosphatase, eukaryotic elongation factor 2 (eEF-2). Given its critical role in a fundamental cellular process, dysregulation of eEF-2K has been implicated in several human diseases, including those of the cardiovascular system, chronic neuropathies, and many cancers, making it a critical pharmacological target. In the absence of high-resolution structural information, high-throughput screening efforts have yielded small-molecule candidates that show promise as eEF-2K antagonists. Principal among these is the ATP-competitive pyrido-pyrimidinedione inhibitor, A-484954, which shows high specificity toward eEF-2K relative to a panel of “typical” protein kinases. A-484954 has been shown to have some degree of efficacy in animal models of several disease states. It has also been widely deployed as a reagent in eEF-2K-specific biochemical and cell-biological studies. However, given the absence of structural information, the precise mechanism of the A-484954-mediated inhibition of eEF-2K has remained obscure. Leveraging our identification of the calmodulin-activatable catalytic core of eEF-2K, and our recent determination of its long-elusive structure, here we present the structural basis for its specific inhibition by A-484954. This structure, which represents the first for an inhibitor-bound catalytic domain of a member of the α-kinase family, enables rationalization of the existing structure–activity relationship data for A-484954 variants and lays the groundwork for further optimization of this scaffold to attain enhanced specificity/potency against eEF-2K.
Keywords: protein translation, atypical kinase, alpha-kinase, calmodulin-dependent kinase, ATP-competitive kinase inhibitor, eukaryotic elongation factor 2 kinase
Eukaryotic elongation factor 2 kinase (eEF-2K) is an atypical serine/threonine kinase that is activated by calmodulin (CaM) (1, 2, 3, 4) and is one of six members of the α-kinase family encoded in the human genome (5, 6). eEF-2K plays a critical role in regulating the elongation stage of translation by phosphorylating its only currently validated cellular target, the ribosome-associated guanosine triphosphatase (GTPase), eukaryotic elongation factor 2 (eEF-2) (7) that facilitates translocation of the nascent chain from the ribosomal aminoacyl site (A-site) to the peptidyl site (P-site) to accommodate an incoming aminoacyl-tRNA during elongation. The eEF-2K-mediated phosphorylation of eEF-2 on a specific threonine residue (Thr-56) compromises its ability to engage the ribosome and leads to a global downregulation of elongation (7, 8, 9) with differential effects on the translation of specific mRNA messages (10). Dysregulation of eEF-2K has been implicated in a variety of human disorders, including those of the cardiovascular system (11) and chronic neuropathies such as Alzheimer’s (12) and Parkinson’s (13) diseases. Aberrant eEF-2K activity has also been linked to the progression of many forms of cancer, including medulloblastoma (14), neuroblastoma (15), and breast cancer (16). Given its functional correlation with diverse disease states, eEF-2K has been recognized as a potential therapeutic target for a range of cardiovascular diseases (17), neurological conditions (18) and several cancers (19).
For several years, there have been consistent efforts to discover specific modulators of eEF-2K function (20, 21, 22). Among these, the pyrido-pyrimidinedione inhibitor, A-484954 (Fig. 1A), discovered through high-throughput screening, is the best-established eEF-2K inhibitor (20) that has been widely deployed in a variety of functional and in vivo studies. Although the potency of this inhibitor (20, 21) is modest in pharmacological terms, it has shown some efficacy in animal models of Alzheimer’s disease (23), Dravet syndrome (24) and hypertension (25). Although individually these studies should be interpreted with caution since off-target effects are possible, they collectively suggest that the A-484954 scaffold may lend to further optimization for more efficiently targeting eEF-2K.
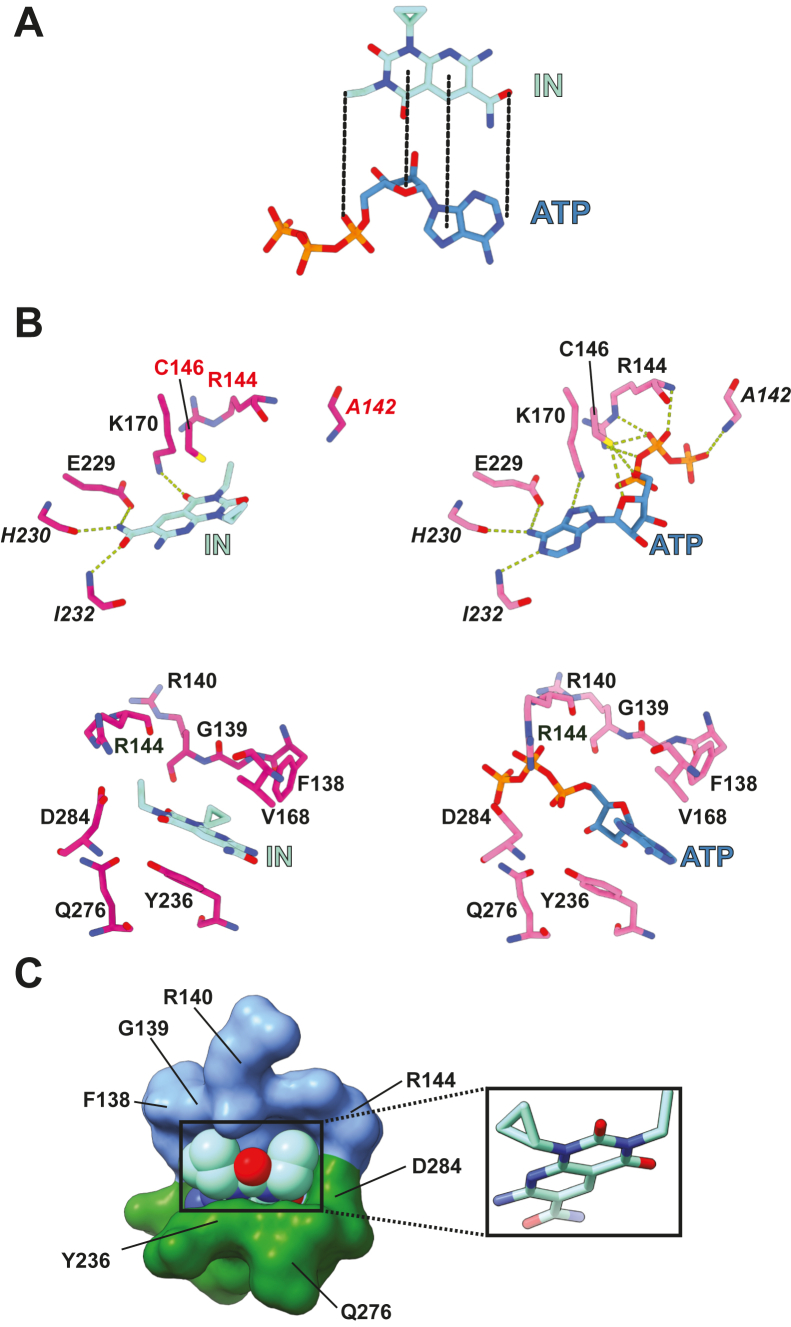
Figure 1.
The inhibitor-bound CaM•peEF-2KTRcomplex.A, chemical structure of the eEF-2K inhibitor, A-484954 (IN). B, schematic representation of the eEF-2KTR construct, showing the calmodulin-targeting motif (CTM, blue), the α-kinase domain (KD, lime), and the C-terminal domain (CTD, orange). The eEF-2KTR construct is missing (indicated by the dashed lines) the first 70 N-terminal residues (S70 is restored as the remnant of an affinity tag), and a portion of the regulatory loop (R-loop, 359–489) but includes the sites of primary (T348) and secondary (S500) activating autophosphorylation therein. C, the structure of the CaM•peEF-2KTR complex bound to ADP and IN (INB complex) is shown on the left panel. Also shown for comparison in the same orientation, on the right panel, is the NB structure (bound to ADP and ATP; PDB: 8FNY). The various domains have been colored as in (B). Only the C-lobe of calmodulin (colored purple) within the INB and NB complexes could be modeled. The phosphorylated T348 residue (pT348) is also indicated.
The rational design of small molecules antagonists of eEF-2K activity, or its interactions with CaM, a critical step in activation (26), had thus far been hindered by the absence of a three-dimensional structure of this enzyme or a functional construct thereof. To elucidate the mechanism of the CaM-mediated activation of eEF-2K and provide a structural framework for rational drug design, we engineered a minimal CaM-activatable construct representing the functional core of eEF-2K (eEF-2KTR, Fig. 1B) (27). We subsequently determined the crystal structure of the complex of eEF-2KTR phosphorylated at the primary activating site, T348, with CaM (CaM•peEF-2KTR) (28). As a necessary next step in designing and optimizing highly potent and selective ATP-competitive eEF-2K inhibitors, here we present the structural basis for its inhibition by A-484954. We expect that our structure, the first of an inhibitor-bound α-kinase catalytic domain, will lay the groundwork for future efforts in the rational design of antagonists targeting the active site of eEF-2K.
Results and discussion
To establish the mode of nucleotide recognition by eEF-2K, we had previously determined the structure of the CaM•peEF-2KTR complex bound to ATP and ADP (NB, for nucleotide-bound). In the NB structure, ATP was bound at the kinase active site, while ADP was unexpectedly engaged to a basic pocket (BP) at the interface between the C-lobe of CaM (CaMC) and the N-lobe of the kinase domain (KD) (Fig. 1C) (29). Using conditions similar to those used for the NB complex, we crystallized and solved the structure of the CaM•peEF-2KTR complex bound to A-484954 and ADP (INB for inhibitor-bound; Fig. 1C).
In the INB structure (see Table S1 for details about data collection, structure refinement, and statistics), ADP is bound at the BP in a conformation identical to that seen in the NB complex (29). The inhibitor, A-484954, replaces ATP at the kinase active site (compare the left and right panels of Fig. 1C). The functional implications of the engagement of ADP at the BP and the resultant enhancement in the CaM-mediated activation of eEF-2K were discussed at length previously (29) and are therefore not revisited here. In the NB structure, A-484954 is stabilized at the kinase active site (Fig. S1) through numerous interactions reflective of its sub-μM IC50 (20, 21) in an alignment such that its pyridine ring is in an equivalent spatial position to the imidazole ring of ATP (Fig. 2A). The sidechain of K170, which engages the adenine N7 of ATP in the NB complex, now forms a hydrogen bond with the 4-oxo moiety of the dioxopyrimidine ring of the inhibitor. The carboxamide moiety is oriented through several hydrogen bonds involving the H230 and I232 backbones and the E229 sidechain (Fig. 2B, top left). These residues are also involved in the recognition of ATP in the NB complex, with the adenine N6 forming hydrogen bonds with the sidechain and mainchain of E229 and H230, respectively, while N1 engages the I232 mainchain (Fig. 2B, top right). The Y236 ring takes part in π-π stacking interactions with the dioxopyrimidine ring of the inhibitor (Fig. 2B, bottom left), substituting a corresponding stacking with the adenine ring of ATP in the NB complex, albeit with a slightly altered geometry (Fig. 2B, bottom right). This interaction is supplemented by a donor-π interaction (30) with C146 (Fig. 2B, top left) and hydrophobic contacts with V168 (Fig. 2B, bottom left). In contrast, the ethyl and cyclopropyl moieties of the inhibitor show few specific, directional contacts with the kinase. However, several van der Waals contacts of the cyclopropyl ring with F138, G139, and R140, and the ethyl group with R144, Q276, and D284, are seen. Y236 contacts both the cyclopropyl and the ethyl moieties of the inhibitor. These non-specific interactions appear to be important since significant modifications of the ethyl or the cyclopropyl groups significantly weaken binding (21). We suspect these contacts are important to stabilize a relatively close-packed structure at the entrance to the kinase active site (Fig. 2C). Consistent with the structure presented here, the replacement of the ethyl moiety by smaller entities (a hydrogen or a methyl group), that would result in reduced packing, lead to ∼15-fold increases in the IC50. While replacement with a slightly larger propyl moiety is tolerated (∼2-fold increase in the IC50), introducing a significantly bulkier group, for example, CH2-Ph-NH2, that would require more substantial conformational rearrangements to prevent steric clashes, results in a >50-fold increase in IC50. The importance of the hydrogen bonds involving the carboxamide moiety, discussed above, is also evident since even minor changes, for example, the replacement of the oxygen with sulfur, result in a >50-fold increase in IC50 (21).
Figure 2.
The inhibitor occupies the active site of peEF-2KTR.A, comparison of the spatial positioning of A-484954 (in the INB complex) with respect to ATP (in the NB complex; PDB: 8FNY) at the peEF-2KTR active site. The dashed vertical lines indicate equivalent spatial positions. The pyridine ring of the inhibitor occupies the same spatial position as the imidazole ring of ATP. B, hydrogen-bonding patterns (top) and other key contacts (bottom) are shown for A-484954 (left) and ATP (right) in the INB and NB complexes, respectively. Residues that form hydrogen bonds (green dashed lines) using only their backbone atoms are indicated in italicized font. Hydrogen bonds involving A142, R144, and C146 with ATP seen in the NB complex are missing in the INB complex. These residues are labeled in red font on the top left panel. Note that the NB structure has two molecules in the asymmetric unit. The two protomers have subtle differences in the orientations of the triphosphate moieties of ATP (given the absence of essential Mg2+ ions in the structure). This leads to small differences in the corresponding hydrogen bonding networks (29). The ATP molecule bound to chain A is shown here. Hydrogen bonds were calculated using UCSF ChimeraX (49) by relaxing constraints by 0.5 Å and 20°. The π-stacking interactions of Y236 (bottom panel) are somewhat different between A-484954 and ATP. The inter-centroid distance and interplanar angle between Y236 and the dioxopyrimidine ring of the inhibitor are 4.0 Å and 14.2º, respectively; the corresponding values with the imidazole ring of ATP are 4.9 Å and 27.3º. C, van der Waals contacts of A-484954 bound at the peEF-2KTR active site. The local surface encompassing key kinase residues that contact the cyclopropyl and the ethyl moieties of A-484954 is shown. Elements corresponding to the N- and C-lobes of the kinase domain are colored blue and green, respectively. The inset illustrates the orientation of the inhibitor in the kinase active site in this view.
We have previously noted a closure of the phosphate-binding P-loop relative to the N/D-loop in peEF-2KTR upon comparing the NB structure with that in the absence of nucleotides (nucleotide-free, NF) (29). A comparison of the NB and INB structures suggests a further closure of the P-loop upon replacing ATP with A-484954 at the kinase active site (Fig. 3A). It is notable that F138, G139, R140, and R144, mentioned above, are all part of the P-loop on the N-lobe of the kinase, while Y236, Q276, and D284 are housed on the C-lobe (Fig. 2C). Therefore, it appears that the ethyl and cyclopropyl groups on A-484954 serve to bridge the N- and C-lobes of the kinase domain and stabilize a closed state. This enhanced degree of closure appears to be a common feature in nucleotide-bound α-kinases KDs (29, 31).
Figure 3.
Conformationalvariabilityin the inhibitor-bound complex.A, Comparison of the relative conformations of the P- and N/D-loops in the nucleotide-free (NF; light pink; PDB: 7SHQ), nucleotide bound (NB; dark pink; PDB: 8FNY), and inhibitor-bound (INB; magenta) complexes. The ATP (blue) and A-484954 (IN, cyan) molecules are shown in stick representation. B, structure of the INB2 complex, peEF-2KTR, is shown in magenta for conformation A, and green for conformation B; CaM is in orange (only CaMC is resolved in the complex). The right panel shows an expansion of the variable region in ball and stick representation. S500, which is only visible in conformation A, is circled. The termini of the segments represented for the two conformations are labeled. The features corresponding to conformation A are seen in the NF and NB complexes, while those corresponding to conformation B are seen in the INB complex.
In addition to the relatively modest differences at/near the active site between the NB and INB structures, substantial differences are noted at a distance more than 35 Å away from the catalytic center. In the NF and NB structures, the A501-N510 segment at the N-terminal end of the C-terminal domain folds into a helix (α1’; secondary structural elements as defined by Piserchio et al. (28)). This element is absent in the INB structure, in which the entire segment extending to K516 could not be modeled due to weak/missing density. In contrast to the NF and NB structures, the N-terminus of the subsequent helix (α2′) is lengthened by a single turn in the INB structure. We also obtained a second inhibitor-bound structure (INB2, see Table S2 for data collection, structure refinement, and statistics) that shows density suggesting partial occupancies of both these conformations, that is, the presence of α1′ and a longer α2’ (Figs. 3B and S2). This indicates significant conformational heterogeneity around a region proximal to S500, a site of activating autophosphorylation (32) also targeted by PKA (33). Perhaps disorder in this region, a feature likely to be enhanced by an intact R-loop (absent in eEF-2KTR; see Fig. 1B), is necessary to facilitate phosphorylation on S500. It is also possible that this conformational variability is an artifact of the R-loop truncation used to generate eEF-2KTR. Notably, while the CaM-driven autophosphorylation at the primary activating site, T348, is rapid, we have been unable to detect autophosphorylation on S500 in eEF-2KTR even after an hour of incubation with ATP and Mg2+. The overall orientations of the inhibitor bound at the eEF-2KTR active site are indistinguishable between the INB and INB2 structures (Fig. S3).
A-484954 has been suggested to be highly specific for eEF-2K and inactive “against a wide panel of serine/threonine and tyrosine kinases” (20). To understand the origin of this selectivity, we compared the structure of eEF-2KTR with PKA and the kinases included in the screen to determine residues that are spatially equivalent to those (C146, V168, K170, E229, and Y236; see Table S3 for a representative subset) whose sidechains appear to be involved in A-484954 recognition. Of these residues, only the “catalytic lysine” K170 seems common within this group. In conventional kinases (K72 in PKA; Table S3), this residue forms a critical salt bridge with a conserved glutamate (E91 in PKA) on the αC helix to maintain an “αC in” conformation and correctly position the α− and β−phosphates of ATP in the active state (34). The positions equivalent to C146 (V57 in PKA) and V168 (A70) comprise key hydrophobic components of the so-called catalytic spine (C-spine) in conventional kinases (35, 36). The spatial position of E229 corresponds to that of the so-called gatekeeper (37, 38), generally a bulky hydrophobic residue in most conventional kinases. Finally, the position corresponding to Y236 is never aromatic in the conventional kinases used in the panel (20). Based on this simple analysis, it appears unlikely that the critical interactions that stabilize A-484954 within the eEF-2K active site can be recapitulated in the active sites of conventional protein kinases, reflective of its previously-determined selectivity towards eEF-2K (20).
In contrast to the conventional kinases, eEF-2K residues that engage A-484954 show a high degree of conservation among human α-kinases, with K170 and E229 being invariant in all α-kinases (39). The other critical positions are also quite similar in human α-kinases. V168 is a valine in ALPK1 and ALPK2, and isoleucine in ALPK3, TRPM6 and TRPM7. Y236 is a phenylalanine in ALPK1, ALPK2, TRPM6, and TRPM7, except in ALPK3, where it is a leucine, potentially substituting the π-π stacking seen in eEF-2K by a methyl-π interaction. Based on these observations, whether A-484594 would provide sufficient specificity towards eEF-2K over the other five human α-kinases is unclear. However, the position corresponding to C146, an alanine in the other five human α-kinases, is distinct in eEF-2K (6). This residue could perhaps provide a means to enhance specificity and potency through reversible covalent targeting. For example, the cysteine Sγ atom is less than 5 Å away from both the ethyl and the cyclopropane moieties of A-484954. A previous study employing carbonitrile-based inhibitors in eEF-2K indicated that the C146 sidechain could be covalently targeted (40). However, careful thought is required for designing and placing such a modifier on the A-484954 scaffold since the donor-π interaction involving C146, mentioned earlier, could potentially affect its reactivity. Nevertheless, we expect that the present structure, the first of an inhibitor-bound eEF-2K, and indeed of any α-kinase bound to an inhibitor, will provide a framework for the rational design of ATP-competitive antagonists targeting this critical regulator of translational elongation.
Experimental procedures
Crystallization
CaM and peEF-2KTR were expressed and purified, and their 1:1 complex was generated as described previously (28). For crystallization, the CaM•peEF-2KTR complex prepared in buffer containing 20.0 mM Tris (pH 7.5), 100.0 mM NaCl, 3.0 mM CaCl2, 1.0 mM TCEP was concentrated to 10.3 mg/ml using a spin column (Amicon Ultra, Millipore) with a 3.0 kDa cutoff. The ligand stock solution comprising 44.4 mM A-484954 (TargetMol, Catalog number: TQ0137) dissolved in 2H-DMSO was added immediately before plating to the protein solution at concentrations of 0.3 mM (∼0.7% 2H-DMSO) or 1.5 mM (∼3.1% 2H-DMSO), together with 0.3 mM ADP and 1.5 mM MgCl2. Under these conditions, the presence of ADP in the buffer was essential for the success of the co-crystallization experiments. Optimal crystallization conditions were established by simultaneous application of PEG-3350 (Hampton Research, 12 steps from 16% to 22%) and pH gradients (8 steps from 5.9 to 7.0, or from 6.5 to 8.5) in the presence of 100.0 mM Bis-Tris Propane and 200.0 mM NaF (Hampton Research) using Art-Robbins low profile, 3-well Intelli-Plate 96 plates. The crystallization cocktails were prepared using an SPT Labtech Dragonfly liquid handling robot and mixed with the protein stock in ratios of 2:1, 1:1, and 1:2 (0.2 μl total volume) using an SPT Labtech Mosquito. The structures presented in this study were derived from crystals obtained from a batch (two parts) containing 0.3 mM inhibitor, ∼0.7% 2H-DMSO, mixed with a cocktail containing 16.5% PEG-3350, pH 6.2 (one part); and a batch (two parts) containing 1.5 mM inhibitor, 3.1% 2H-DMSO, mixed with 16.5% PEG-3350, pH 6.5 (one part). These crystals were looped directly from the Intelli-Plate wells, cryoprotected with LV CryoOil (Mitegen), and stored in liquid N2 before data collection.
Data collection and structure determination
Diffraction data were acquired on the FMX beamline at NSLS-II and processed using the autoPROC toolbox (41). Phases were obtained by molecular replacement and our initial structure of the CaM•peEF-2KTR complex (PDB: 7SHQ) (28) using Phaser after removing the N-lobe of CaM, and refined using phenix.refine (42, 43). The ligands, ADP and A-484954, were prepared using eLBOW (44) and fitted using phenix.ligandfit (45, 46). The metal ions were assigned using the CheckMyBlob server (47). The first dataset, truncated with a spherical cutoff (2.009 Å), was initially refined at 2.327 Å. The resolution limit was extended to 2.120 Å using PAIRREF (48) following prescribed guidelines, with a visual inspection of the 2FO-FC map in the region around the bound A-484954. The second dataset (corresponding to INB2) was refined with data integrated using an ellipsoidal shell, as previously described (29).
Data availability
Structure factors and atomic coordinates have been deposited in the Protein Data Bank with PDB IDs 8GM4 and 8GM5.
Supporting information
This article contains supporting information (20, 35, 37, 49, 50).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
A. P. investigation; A. P. and E. A. I investigation and formal analysis; K. N. D. and R. G. conceptualization, supervision, and resources; R. G. writing – original draft; A. P., E. A. I., K. N. D. and R. G. writing – review and editing.
Funding and additional information
This work is supported by NIH award R01 GM123252 (to K. N. D. and R. G.) and a CPRIT award RP210088 (to K. N. D.). Use of the NYX beamline (19-ID) at the National Synchrotron Light Source II (NSLS-II) is supported by the member institutions of the New York Structural Biology Center. NSLS-II is a United States Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract DE-SC0012704. Dr Kevin Battaile (New York Structural Biology Center) and Dr Dale Kreitler (NSLS-II) are thanked for their support during data collection and analysis.
Reviewed by members of the JBC Editorial Board. Edited by Wolfgang Peti
Contributor Information
Kevin N. Dalby, Email: dalby@austin.utexas.edu.
Ranajeet Ghose, Email: rghose@ccny.cuny.edu.
Supporting information
References
- 1.Nairn A.C., Bhagat B., Palfrey H.C. Identification of calmodulin-dependent protein kinase III and its major Mr 100,000 substrate in mammalian tissues. Proc. Natl. Acad. Sci. U. S. A. 1985;82:7939–7943. doi: 10.1073/pnas.82.23.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nairn A.C., Palfrey H.C. Identification of the major Mr 100,000 substrate for calmodulin-dependent protein kinase III in mammalian cells as elongation factor-2. J. Biol. Chem. 1987;262:17299–17303. [PubMed] [Google Scholar]
- 3.Ryazanov A.G. Ca2+/calmodulin-dependent phosphorylation of elongation factor 2. FEBS Lett. 1987;214:331–334. doi: 10.1016/0014-5793(87)80081-9. [DOI] [PubMed] [Google Scholar]
- 4.Ryazanov A.G., Natapov P.G., Shestakova E.A., Severin F.F., Spirin A.S. Phosphorylation of the elongation factor 2: the fifth Ca2+/calmodulin-dependent system of protein phosphorylation. Biochimie. 1988;70:619–626. doi: 10.1016/0300-9084(88)90245-3. [DOI] [PubMed] [Google Scholar]
- 5.Drennan D., Ryazanov A.G. Alpha-kinases: analysis of the family and comparison with conventional protein kinases. Prog. Biophys. Mol. Biol. 2004;85:1–32. doi: 10.1016/S0079-6107(03)00060-9. [DOI] [PubMed] [Google Scholar]
- 6.Middelbeek J., Clark K., Venselaar H., Huynen M.A., van Leeuwen F.N. The alpha-kinase family: an exceptional branch on the protein kinase tree. Cell. Mol. Life Sci. 2010;67:875–890. doi: 10.1007/s00018-009-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryazanov A.G., Shestakova E.A., Natapov P.G. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988;334:170–173. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- 8.Ryazanov A.G., Davydova E.K. Mechanism of elongation factor 2 (EF-2) inactivation upon phosphorylation. Phosphorylated EF-2 is unable to catalyze translocation. FEBS Lett. 1989;251:187–190. doi: 10.1016/0014-5793(89)81452-8. [DOI] [PubMed] [Google Scholar]
- 9.Carlberg U., Nilsson A., Nygard O. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 1990;191:639–645. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- 10.Sossin W.S., Costa-Mattioli M. Translational control in the brain in health and disease. CSH Persp. Biol. 2019;11:a032912. doi: 10.1101/cshperspect.a032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Usui T., Okada M., Hara Y., Yamawaki H. Eukaryotic elongation factor 2 kinase regulates the development of hypertension through oxidative stress-dependent vascular inflammation. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H756–768. doi: 10.1152/ajpheart.00373.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W., Zhou X., Ryazanov A.G., Ma T. Suppression of the kinase for elongation factor 2 alleviates mGluR-LTD impairments in a mouse model of Alzheimer's disease. Neurobiol. Aging. 2021;98:225–230. doi: 10.1016/j.neurobiolaging.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jan A., Jansonius B., Delaidelli A., Bhanshali F., An Y.A., Ferreira N., et al. Activity of translation regulator eukaryotic elongation factor-2 kinase is increased in Parkinson disease brain and its inhibition reduces alpha synuclein toxicity. Acta Neuropathol. Commun. 2018;6:54. doi: 10.1186/s40478-018-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leprivier G., Remke M., Rotblat B., Dubuc A., Mateo A.R., Kool M., et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaidelli A., Negri G.L., Jan A., Jansonius B., El-Naggar A., Lim J.K.M., et al. MYCN amplified neuroblastoma requires the mRNA translation regulator eEF2 kinase to adapt to nutrient deprivation. Cell Death Differ. 2017;24:1564–1576. doi: 10.1038/cdd.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meric-Bernstam F., Chen H., Akcakanat A., Do K.A., Lluch A., Hennessy B.T., et al. Aberrations in translational regulation are associated with poor prognosis in hormone receptor-positive breast cancer. Breast Cancer Res. 2012;14:R138. doi: 10.1186/bcr3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P., Riazy M., Gold M., Tsai S.H., McNagny K., Proud C., et al. Impairing eukaryotic elongation factor 2 kinase activity decreases atherosclerotic plaque formation. Can. J. Cardiol. 2014;30:1684–1688. doi: 10.1016/j.cjca.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beretta S., Gritti L., Verpelli C., Sala C. Eukaryotic elongation factor 2 kinase a pharmacological target to regulate protein translation dysfunction in neurological diseases. Neuroscience. 2020;445:42–49. doi: 10.1016/j.neuroscience.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Temme L., Asquith C.R.M. eEF2K: an atypical kinase target for cancer. Nat. Rev. Drug Discov. 2021;20:577. doi: 10.1038/d41573-021-00124-5. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z., Gopalakrishnan S.M., Bui M.H., Soni N.B., Warrior U., Johnson E.F., et al. 1-Benzyl-3-cetyl-2-methylimidazolium iodide (NH125) induces phosphorylation of eukaryotic elongation factor-2 (eEF2): a cautionary note on the anticancer mechanism of an eEF2 kinase inhibitor. J. Biol. Chem. 2011;286:43951–43958. doi: 10.1074/jbc.M111.301291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edupuganti R., Wang Q., Tavares C.D., Chitjian C.A., Bachman J.L., Ren P., et al. Synthesis and biological evaluation of pyrido[2,3-d]pyrimidine-2,4-dione derivatives as eEF-2K inhibitors. Bioorg. Med. Chem. 2014;22:4910–4916. doi: 10.1016/j.bmc.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupt K.A., Jia Z. eEF2K Inhibitor design: the progression of exemplary structure-based drug design. Molecules. 2023;28:1095. doi: 10.3390/molecules28031095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasica N.P., Zhou X., Yang Q., Wang X., Yang W., Zimmermann H.R., et al. Antagonists targeting eEF2 kinase rescue multiple aspects of pathophysiology in Alzheimer's disease model mice. J. Neurochem. 2022;160:524–539. doi: 10.1111/jnc.15562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beretta S., Gritti L., Ponzoni L., Scalmani P., Mantegazza M., Sala M., et al. Rescuing epileptic and behavioral alterations in a Dravet syndrome mouse model by inhibiting eukaryotic elongation factor 2 kinase (eEF2K) Mol. Autism. 2022;13:1. doi: 10.1186/s13229-021-00484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodama T., Okada M., Yamawaki H. Eukaryotic elongation factor 2 kinase inhibitor, A484954 lowered blood pressure in spontaneously hypertensive rats via inducing vasorelaxation. J. Pharmacol. Sci. 2020;144:165–171. doi: 10.1016/j.jphs.2020.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Tavares C.D., Ferguson S.B., Giles D.H., Wang Q., Wellmann R.M., O'Brien J.P., et al. The molecular mechanism of eukaryotic elongation factor 2 kinase activation. J. Biol. Chem. 2014;289:23901–23916. doi: 10.1074/jbc.M114.577148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Will N., Lee K., Hajredini F., Giles D.H., Abzalimov R.R., Clarkson M., et al. Structural dynamics of the activation of elongation factor 2 kinase by Ca2+-calmodulin. J. Mol. Biol. 2018;430:2802–2821. doi: 10.1016/j.jmb.2018.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piserchio A., Isiorho E.A., Long K., Bohanon A.L., Kumar E.A., Will N., et al. Structural basis for the calmodulin-mediated activation of eukaryotic elongation factor 2 kinase. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piserchio A., Long K.J., Browning L.S., Bohanon A.L., Isiorho E.A., Dalby K.N., et al. ADP enhances the allosteric activation of eukaryotic elongation factor 2 kinase by calmodulin. Proc. Natl. Acad. Sci. U. S. A. 2023;120 doi: 10.1073/pnas.2300902120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes C.R., Sinha S.K., Ganguly H.K., Bai S., Yap G.P., Patel S., et al. Insights into thiol-aromatic interactions: A stereoelectronic basis for S-H/π interactions. J. Am. Chem. Soc. 2017;139:1842–1855. doi: 10.1021/jacs.6b08415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Q., Yang Y., van Staalduinen L., Crawley S.W., Liu L., Brennan S., et al. Structure of the Dictyostelium myosin-II heavy chain kinase A (MHCK-A) alpha-kinase domain apoenzyme reveals a novel autoinhibited conformation. Sci. Rep. 2016;6 doi: 10.1038/srep26634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavares C.D., O'Brien J.P., Abramczyk O., Devkota A.K., Shores K.S., Ferguson S.B., et al. Calcium/calmodulin stimulates the autophosphorylation of elongation factor 2 kinase on Thr-348 and Ser-500 to regulate its activity and calcium dependence. Biochemistry. 2012;51:2232–2245. doi: 10.1021/bi201788e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redpath N.T., Proud C.G. Cyclic AMP-dependent protein kinase phosphorylates rabbit reticulocyte elongation factor-2 kinase and induces calcium-independent activity. Biochem. J. 1993;293:31–34. doi: 10.1042/bj2930031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modi V., Dunbrack R.L., Jr. Defining a new nomenclature for the structures of active and inactive kinases. Proc. Natl. Acad. Sci. U. S. A. 2019;116:6818–6827. doi: 10.1073/pnas.1814279116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornev A.P., Haste N.M., Taylor S.S., Eyck L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornev A.P., Taylor S.S. Defining the conserved internal architecture of a protein kinase. Biochim. Biophys. Acta. 2010;1804:440–444. doi: 10.1016/j.bbapap.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y., Shah K., Yang F., Witucki L., Shokat K.M. A molecular gate which controls unnatural ATP analogue recognition by the tyrosine kinase v-Src. Bioorg. Med. Chem. 1998;6:1219–1226. doi: 10.1016/s0968-0896(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 38.Taylor S.S., Kornev A.P. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye Q., Crawley S.W., Yang Y., Cote G.P., Jia Z. Crystal structure of the alpha-kinase domain of Dictyostelium myosin heavy chain kinase A. Sci. Signal. 2010;3:ra17. doi: 10.1126/scisignal.2000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devkota A.K., Edupuganti R., Yan C., Shi Y., Jose J., Wang Q., et al. Reversible covalent inhibition of eEF-2K by carbonitriles. ChemBioChem. 2014;15:2435–2442. doi: 10.1002/cbic.201402321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwart P.H., Afonine P.V., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., et al. Automated structure solution with the PHENIX suite. Meth. Mol. Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 44.Moriarty N.W., Grosse-Kunstleve R.W., Adams P.D. Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terwilliger T.C., Adams P.D., Moriarty N.W., Cohn J.D. Ligand identification using electron-density map correlations. Acta Crystallogr. D Biol. Crystallogr. 2007;63:101–107. doi: 10.1107/S0907444906046233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terwilliger T.C., Klei H., Adams P.D., Moriarty N.W., Cohn J.D. Automated ligand fitting by core-fragment fitting and extension into density. Acta Crystallogr. D Biol. Crystallogr. 2006;62:915–922. doi: 10.1107/S0907444906017161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brzezinski D., Porebski P.J., Kowiel M., Macnar J.M., Minor W. Recognizing and validating ligands with CheckMyBlob. Nucl. Acids Res. 2021;49:W86–W92. doi: 10.1093/nar/gkab296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maly M., Diederichs K., Dohnalek J., Kolenko P. Paired refinement under the control of PAIREF. IUCrJ. 2020;7:681–692. doi: 10.1107/S2052252520005916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liebschner D., Afonine P.V., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., et al. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 2017;73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure factors and atomic coordinates have been deposited in the Protein Data Bank with PDB IDs 8GM4 and 8GM5.