Abstract
Pkc53E is the second conventional protein kinase C (PKC) gene expressed in Drosophila photoreceptors; it encodes at least six transcripts generating four distinct protein isoforms including Pkc53E-B whose mRNA is preferentially expressed in photoreceptors. By characterizing transgenic lines expressing Pkc53E-B-GFP, we show Pkc53E-B is localized in the cytosol and rhabdomeres of photoreceptors, and the rhabdomeric localization appears dependent on the diurnal rhythm. A loss of function of pkc53E-B leads to light-dependent retinal degeneration. Interestingly, the knockdown of pkc53E also impacted the actin cytoskeleton of rhabdomeres in a light-independent manner. Here the Actin-GFP reporter is mislocalized and accumulated at the base of the rhabdomere, suggesting that Pkc53E regulates depolymerization of the actin microfilament. We explored the light-dependent regulation of Pkc53E and demonstrated that activation of Pkc53 E can be independent of the phospholipase C PLCβ4/NorpA as degeneration of norpAP24 photoreceptors was enhanced by a reduced Pkc53E activity. We further show that the activation of Pkc53E may involve the activation of Plc21C by Gqα. Taken together, Pkc53E-B appears to exert both constitutive and light-regulated activity to promote the maintenance of photoreceptors possibly by regulating the actin cytoskeleton.
Keywords: eye-PKC, actin cytoskeleton, Drosophila, retinal degeneration, PLCβ, PLC21C
Conventional protein kinase C (cPKC) requires both diacylglycerol (DAG) and Ca2+ for its activity (1, 2) and is one of the major regulatory proteins following the activation of phospholipase C (PLC) that includes PLCβ and PLCγ (3). PLCγ is one of the key effectors of the growth factor receptors, and cPKC has been shown critical for processes associated with morphological changes leading to the growth and differentiation of cells (4). In contrast, in PLCβ-mediated signaling events such as the visual signaling that takes place in Drosophila photoreceptors, in which rhodopsin couples to the heterotrimeric Gq protein leading to the activation of PLCβ4 (NorpA) (5, 6), the role of cPKC has not been fully understood. cPKC has been linked to the regulation of the actin cytoskeleton (7, 8). Indeed, several PKC substrates have been identified and characterized (8). However, the mechanisms by which cPKC regulates cell morphology or the cytoskeleton remain to be explored.
There are two cPKCs expressed in Drosophila photoreceptors, eye-PKC (9) and Pkc53E (10), both of which share more than 70% sequence homology. Eye-PKC is critical for the negative regulation of the visual response and is localized in the rhabdomere (11), the visual organelle in which the visual signaling takes place. Moreover, eye-PKC is constitutively associated with a multimeric signaling complex organized by the scaffolding protein INAD (inactivation-no-afterpotential D) (12, 13). Eye-PKC has been shown to phosphorylate both INAD and TRP (transient receptor potential) in vitro and in vivo (14, 15), both of which are integral parts of the signaling complex. In contrast, the role of Pkc53E in photoreceptors has not been investigated.
Studies have linked cPKCs to the regulation of the actin cytoskeleton (8). The actin cytoskeleton consists of a network of actin microfilaments, which can be found in the cell cortex, the stress fiber, and various extensions including filopodia, lamellipodia, and microvilli. The actin cytoskeleton is critical for maintaining cell shape and it also regulates diverse processes including cytokinesis, chemotaxis, and endocytosis (16).
Here we performed molecular characterization of the pkc53E locus and demonstrate that the B-isoform is expressed only in photoreceptors. We show that GFP-tagged Pkc53E-B is present in the cytoplasm and the rhabdomere. We characterized a loss of function allele of pkc53E that lacks the transcript for the B isoform and show the mutant undergoes light-dependent retinal degeneration. Further investigations using Actin-GFP reporter demonstrate that a reduction in the pkc53E activity leads to defects in the actin cytoskeleton of rhabdomeres. To address the light-dependent regulation of Pkc53E, we first examined the contribution of PLCβ4 (NorpA) that mediates the visual response (17). Unexpectedly, the knockdown of pkc53E greatly exacerbated the retinal degeneration of norpA mutants, indicating a NorpA-independent regulation of Pkc53E. To explore the alternate pathways leading to the activation of Pkc53E, we show that Plc21C (18) may be involved. Moreover, activation of Plc21C may require Gqα leading to the generation of DAG thereby activating PKC when PLCβ4 is absent.
Results
Molecular characterization of the pkc53E locus
The pkc53E (CG6622) gene is the second cPKC gene in Drosophila (9), which is located on the second chromosome about 25 kb 3′ of the previously characterized eye-PKC gene (inaC). While eye-PKC has been shown involved in the regulation of visual signaling (11), the role of Pkc53E in photoreceptors has not been investigated.
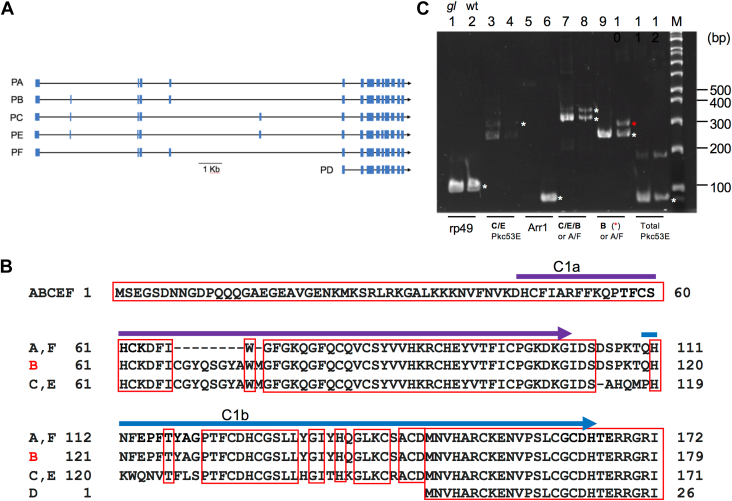
Based on the genome annotation at FlyBase, pkc53E generates six transcripts (A-F) leading to the translation of four distinct polypeptides with different N-terminal sequences (Fig. 1, A and B). These Pkc53 E isoforms include PB (679 amino acids, aa), PC/PE (678 aa), PA/PF (670 aa), and PD (525 aa) with variations in the C1 domains that are known to bind DAG (Fig. 1B). Specifically, all isoforms contain two C1 domains (C1a and C1b) except PD which lacks 145 aa at the N-terminus. These isoforms are likely to have different affinities toward DAG due to differences in the respective C1 domains (Fig. 1B).
Figure 1.
Molecular characterization reveals that pkc53-B is preferentially expressed in photoreceptors.A, a graphic map depicting the coding exons (filled boxes) of six alternatively spliced transcripts, A to F, in the pkc53E locus. B, the alignment of the N-terminal sequences from the six Pkc53E isoforms. All isoforms have two distinct C1 domains (arrows) of about 50 aa (C1a, aa 45–110, and C1b, aa 120–173, in the B isoform), except the D isoform. Identical amino acids in all isoforms are boxed in red. C, identification of the photoreceptor-specific isoform by RT/PCR. Shown are PCR products analyzed by polyacrylamide gel (8%). Even-numbered lanes represent products from wild-type and odd-numbered lanes, glass mutants (gl). Rp49 was served as a positive control whereas arrestin 1 (Arr1), a positive control for wild-type but a negative control for gl. DNA fragments corresponding to the predicted PCR products are marked with asterisks (∗) next to wild-type lanes. The B isoform of pkc53E (red ∗) appears highly expressed in photoreceptors as its expression is drastically reduced in the gl mutant (lane 9). Specific amplification of genes is indicated below. DNA size standards are shown on the right.
We investigated whether any pkc53E isoform is expressed in photoreceptors by comparing the expression between wild-type and glass (gl) mutants that lack photoreceptors (19). We first demonstrated that mRNA for the PB/PC/PE group is present in wild-type (Fig. 1C, lane 8) but greatly reduced in heads of gl mutants (lane 7), indicating that the PB/PC/PE group is preferentially expressed in photoreceptors. To explore further, we employed isoform-specific primers and observed that the PB isoform was highly expressed in wild-type (Fig. 1C, lane 10) but not gl heads (lane 9). In contrast, mRNA for the PC/PE group was present in both wild-type and gl heads (Fig. 1C, lanes 3 and 4). Taken together, our results support the notion that pkc53E-B which encodes a polypeptide of 679 aa is preferentially expressed in photoreceptors. Pkc53E-B shares 57% sequence identities with eye-PKC.
Loss of function in pkc53E-B leads to light-dependent retinal degeneration
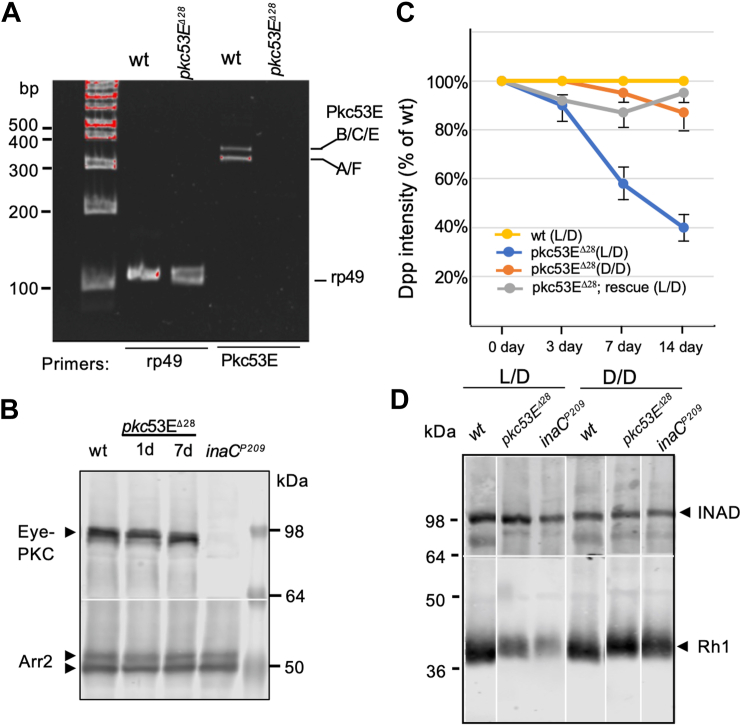
We obtained a mutant allele, pkc53EΔ28, which was generated by imprecise excision of the P-element in P{EPgy2}Pkc53EEY14093 (FBrf0241761). This P-element is located about 2 kb at the 5′ of all isoforms (Fig. S1) except pkc53E-D. We analyzed the genomic DNA from the mutant to uncover the extent of the deletion within the pkc53E locus; we show that pkc53EΔ28 lacks the 5′ sequence including the first three exons leading to the deletion of the first 76 aa for Pkc53E-B (Figs 1 and S1). In contrast, the 3′ sequence including the fourth exon and beyond remains intact. Taken together, pkc53EΔ28 is devoid of the promoter and some of the N-terminal coding sequence, which would greatly affect the transcription of all pkc53E transcripts except the short form, pkc53E-D. Consistently, pkc53EΔ28 displays a drastically reduced level of the major pkc53E transcripts including that coding for the B isoform, supporting that pkc53EΔ28 is a loss of function allele of pkc53E-B (Fig. 2A). To ensure that the excision of the P-element did not affect the expression of the adjacent eye-PKC gene, we analyzed the eye-PKC content by Western blotting and showed it is not affected in pkc53EΔ28 (Fig. 2B).
Figure 2.
Loss of function in pkc53E leads to light-dependent retinal degeneration.A, a greatly reduced expression of pkc53E in the null allele, pkc53EΔ28. Shown are the RT/PCR results comparing the expression between the wild-type and the mutant. The pkc53E transcripts including A to C, E, and F isoforms are drastically reduced in the mutant (6.2 ± 1.7%, n = 3). DNA size markers are indicated on the left. B, pkc53EΔ28 does not affect the expression of eye-PKC. Shown is a Western blot consisting of the merged images of the same blot probing with either anti-eye-PKC or anti-Arr2 (loading control) antibodies. Protein molecular weight standards are indicated on the right. C, the light-dependent retinal degeneration in pkc53EΔ28 and the rescue by the pkc53E-B-GFP transgene. Shown are the time courses depicting the intensity of dpp in pkc53EΔ28 and pkc53EΔ28; rescue, under either 12 h L/D (L/D) or constant-dark (D/D) conditions. D, the light-dependent retinal degeneration in pkc53EΔ28 or inaCP209 as determined by a reduction of Rh1. Shown is a Western blot consisting of the merged images from the same blot probing with either anti-Rh1 or anti-INAD (loading control) antibodies in extracts from 7-day-old wild-type, inaCP209, and pkc53EΔ28 raised in either L/D or D/D conditions. Selected lanes with representative results from the same Western blot, as indicated, were chosen and assembled.
To uncover the function of Pkc53E in photoreceptors, we explored whether pkc53EΔ28 undergoes light-dependent retinal degeneration similar to inaCP209 which lacks eye-PKC. Experimentally, we monitored the reduction of deep pseudopupil (dpp) in the eye, which is commonly employed to detect retinal degeneration. Dpp reflects the optical superposition of rhodopsin epifluorescence in the rhabdomere (20). Indeed, the intensity of dpp in pkc53EΔ28 is progressively reduced under 12 h L/D (L/D) but not significantly in the constant dark (D/D) conditions (Fig. 2C). We also analyzed the total Rh1 content in the mutants by Western blotting (Fig. 2D); we show that 7-day-old pkc53EΔ28 (L/D) contained a reduced Rh1 content (51.2 ± 3.2%), similar to inaCP209 (56.4 ± 1.5%), further supporting that pkc53EΔ28 undergoes the light-dependent retinal degeneration.
Rescue of pkc53EΔ28via transgenic expression of GFP tagged pkc53E-B
To investigate whether the light-dependent degeneration defect in pkc53EΔ28 is caused by the loss of the Pkc53E-B isoform specifically, we performed a rescue experiment by transgenically expressing in R1-6 photoreceptors a modified pkc53E-B containing an enhanced GFP tag. We show that the transgene prevented the light-dependent reduction of dpp (Fig. 2C) and the Rh1 level (Fig. S2) in pkc53EΔ28 under 12 h L/D conditions. These findings further support that a lack of Pkc53E-B is responsible for the light-dependent retinal degeneration in pkc53EΔ28 mutants.
Subcellular localization of Pkc53E-B via GFP tagged pkc53E-B
We investigated the subcellular distribution of Pkc53E-B for insights into localization and functions in vivo. It is known that cPKC present in the cytosol becomes tethered to the plasma membrane by associating with DAG following the activation of PLC. Subsequently, activated cPKC may translocate close to its substrates by interacting with protein scaffolds or adaptor proteins such as receptors for activated C Kinase (21). For example, a scaffolding protein INAD anchors eye-PKC to a multi-protein signaling complex in the rhabdomere of photoreceptors (12, 13).
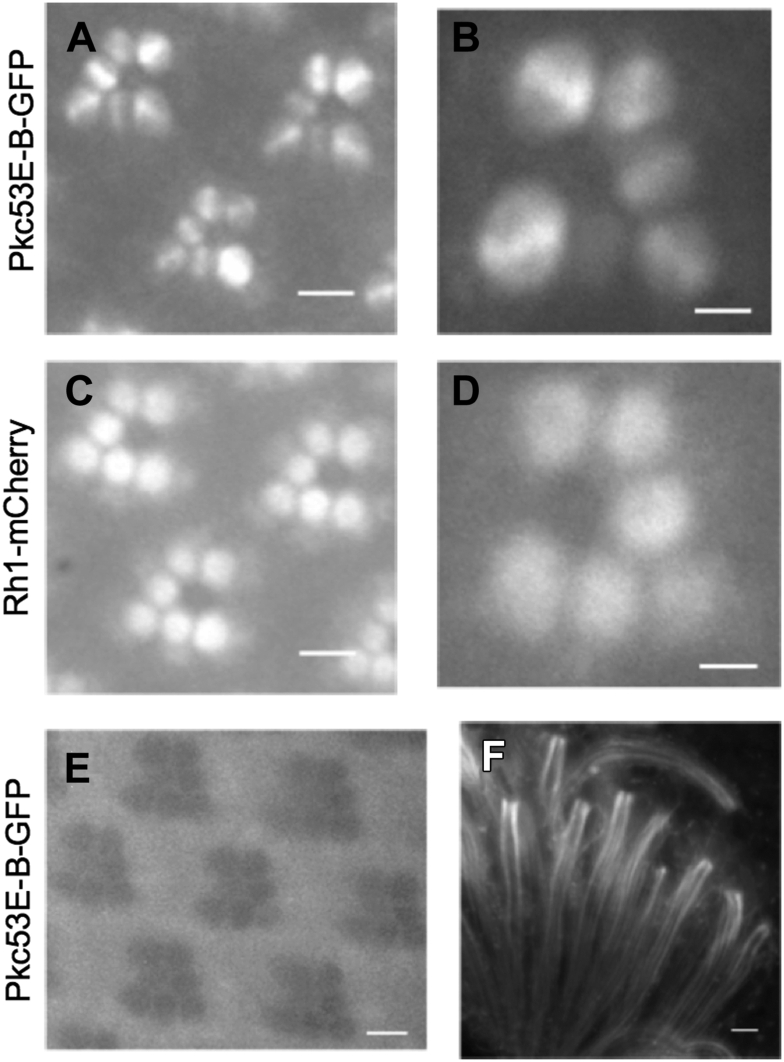
To investigate the subcellular distribution of the GFP-tagged Pkc53E-B we employed water-immersion fluorescence microscopy in live retinas. We show that Pkc53E-B could be observed in both rhabdomeres and the cytosol of photoreceptors (Fig. 3). When Pkc53-B was detected in the cytosol, additional blue light stimulation (1300 lux) for 10 min failed to traffic the kinase to the rhabdomere, indicating that membrane recruitment appears not solely regulated by DAG and Ca2+ following light stimulation. Furthermore, the distribution and intensity of Pkc53E-GFP in the rhabdomere are not significantly altered when shifting flies to the dark for 1 h (not shown), suggesting that Pkc53E-B is not readily released from the rhabdomere membrane when the visual signaling has been terminated. In contrast, the intensity of the rhabdomere localized Pkc53E-B was reduced upon continued blue light stimulation for 5 min (not shown), suggesting that persistent light stimulation promotes the translocation out of rhabdomeres possibly due to desensitization or downregulation of the kinase. Together, the rhabdomere localization of the kinase is not acutely regulated by DAG but possibly by binding to adaptor proteins. Interestingly, Pkc53E-B is not uniformly distributed but concentrated along the horizontal axis of rhabdomeres (Fig. 3, A and B), which is consistent with its association with adaptor proteins in the rhabdomere. This unique localization is different from that of Rh1-mCherry which appears uniformly distributed (Fig. 3, C and D).
Figure 3.
Subcellular localization of GFP tagged Pkc53E-B in photoreceptors. The distribution of GFP-tagged Pkc53E-B was analyzed in live retinas (A and B). Pkc53E-B is detected in the rhabdomere, but also in the cytosol. In the rhabdomere, Pkc53E is not uniformly distributed but appears enriched along the axis of photoreceptors (B). In contrast, Rh1-mCherry is uniformly present in the rhabdomere (C and D). When raised in either constant light or constant-dark conditions, Pkc53E-B is not detected in the rhabdomere but appears distributed and/or sequestered in the cytoplasm (E). In the absence of GFP signals, the R1-R7 rhabdomeres appear as clusters of dark circles under the blue light illumination. In dissociated photoreceptors, Pkc53E can be observed in both rhabdomeres and the cytoplasm (F). Scale bars, 5 μm (A, C, and E); 2 μm (B and D); 20 μm (F).
We further explored the role of light that influences subcellular localization. Importantly, Pkc53E-B was found only in the cytosol when flies were raised in either constant dark or light conditions (Fig. 3E), suggesting that alternating light and dark conditions similar to the diurnal cycle are required for the rhabdomere localization. For example, the diurnal cycle may influence the abundance of the Pkc53E interacting proteins in the rhabdomere. Thus, we propose that adaptor proteins are integral for the localization of Pkc53E-B in rhabdomeres. The identity of the Pkc53E-B interacting proteins and their regulation of the kinase remain to be explored.
Characterization of retinal degeneration in pkc53E mutants and knockdowns in either eye-PKC or pkc53E
We examined the light-dependent degeneration of the pkc53E knockdown caused by RNA-mediated interference (RNAi) (22, 23) and compared it to that of the null mutant and eye-PKC knockdown. Specifically, we employed the GMR driver (24) to direct the expression of double-strand RNA using the UAS/GAL4 binary system (25). Subsequently, we examined the retinas of live flies using Arrestin 2-GFP (Arr2-GFP) (26) or Actin-GFP (27, 28). This use of two GFP reporters allows us to explore specifically whether Rh1 or the actin cytoskeleton of the rhabdomere may be affected (Fig. S3).
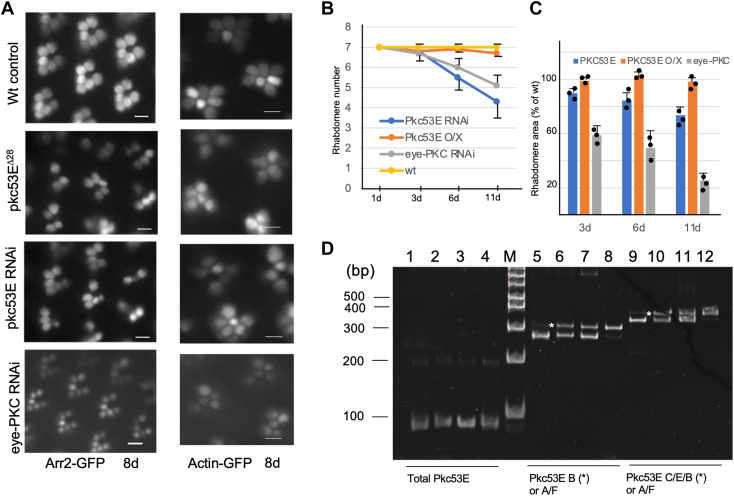
Using Arr2-GFP we show pkc53 EΔ28 displays an age-dependent retinal degeneration that is characterized by distorted ommatidia clusters with missing rhabdomeres (Fig. 4A, left panel), which were also observed in the pkc53E knockdown. For example, we detected the loss of about one or two rhabdomeres with 4.8 ± 0.7 (n = 3) remaining in most ommatidia clusters in 8-day-old flies. In contrast, the knockdown of eye-PKC led to more severe degeneration with reduced GFP intensity but less impact on the number, orientation, and arrangement of rhabdomeres within the cluster (Fig. 4A). The difference in retinal morphology between eye-PKC RNAi and pkc53E RNAi may reflect distinct underlying mechanisms leading to degeneration.
Figure 4.
Knockdown of pkc53E or eye-PKC leads to distinct degeneration phenotype.A, retinal degeneration caused by pkc53EΔ28 or knockdown of either pkc53E or eye-PKC using Arr2-GFP (left) or Actin-GFP (right) as the reporter. Shown are representative retinal morphology of 8-day-old flies. Scale bars on the left panel, 5 μm, and on the right panel, 2 μm. B, comparison of the age-dependent loss of rhabdomeres in various pkc mutants (pkc53E RNAi, pkc53E overexpression, eye-PKC RNAi) using Actin-GFP as the reporter. Each time point represents the mean of three flies (mean ± S.E.M, n = 3). C, the age-dependent changes of rhabdomere area in pkc mutants. Shown are mean ± S.E.M (n = 3) from three independent experiments. D, changes of the pkc53E-B mRNA expression following photoreceptor-targeted RNAi or overexpression by RT/PCR. Three sets of oligonucleotide primers (below) were used. Shown are PCR products analyzed using 8% polyacrylamide gel. The mRNA level for pkc53E-B (∗) is drastically reduced to 5.1 ± 4.1 (n = 3) when compared to the wild-type control of 45.3 ± 8.5 (n = 3, lanes 5). In contrast, overexpression of pkc53E-B leads to an increase of about two-fold (203.5 ± 17.2%; lanes 8). The first-strand cDNA templates derived from pkc53E RNAi (lanes 1, 5, and 9), wild-type (2, 6, and 10), flies expressing GMR-GAL4 driver alone (3, 7, and 11), and pkc53E overexpressing flies (4, 8, and 12) were used. The PCR products corresponding to or enriched with the B-isoform are marked with asterisks next to the wild-type lanes.
Conventional PKC has been implicated in regulating the actin cytoskeleton. Therefore, we examined how the actin cytoskeleton of rhabdomeres might be affected using Actin-GFP. We show that both pkc53EΔ28 and pkc53E RNAi resulted in reduced or loss of actin cytoskeleton, which is accompanied by the abnormal arrangement of ommatidia clusters (Fig. 4A, right). Moreover, in pkc53E RNAi, the number of rhabdomeres was reduced from seven to about five in 10-day-old retinas (Fig. 4B), a reduction similarly observed using Arr2-GFP. In contrast, eye-PKC RNAi initially affected the rhabdomere diameter, and later the rhabdomere number (Fig. 4, B and C). The age-dependent reduction of the rhabdomere number (Fig. 4B) or the area (Fig. 4C) in either pkc53E or eye-PKC knockdown was compared. We also investigated the functional consequence of overexpressing pkc53E-B in the wild-type background. However, overexpression did not have a significant impact on the retinal morphology (Fig. 4, B and C).
Knockdown and overexpression of pkc53E were confirmed by RT/PCR, which shows transcripts corresponding to pkc53E-B were greatly reduced to about 11.3% of wild-type [from 45.3 ± 8.5 (n = 3) to 5.1 ± 4.1 (n = 3)] (Lanes 5, 9, Fig. 4D). In contrast, overexpression increased the mRNA about two-fold (203.5 ±17.2%, n = 3) (Lanes 8, 12, Fig. 4D). The knockdown of eye-PKC was analyzed by Western blotting that showed the eye-PKC level was decreased to 4.9 ± 3.4% (n = 4) when compared to wild-type (not shown).
Taken together, our findings support the notion that a reduced Pkc53E-B activity mostly affects the integrity of the actin cytoskeleton. We propose that Pkc53E-B is required for the maintenance of rhabdomeres.
Abnormal distribution of Actin-GFP in pkc53E RNAi photoreceptors when raised in the dark
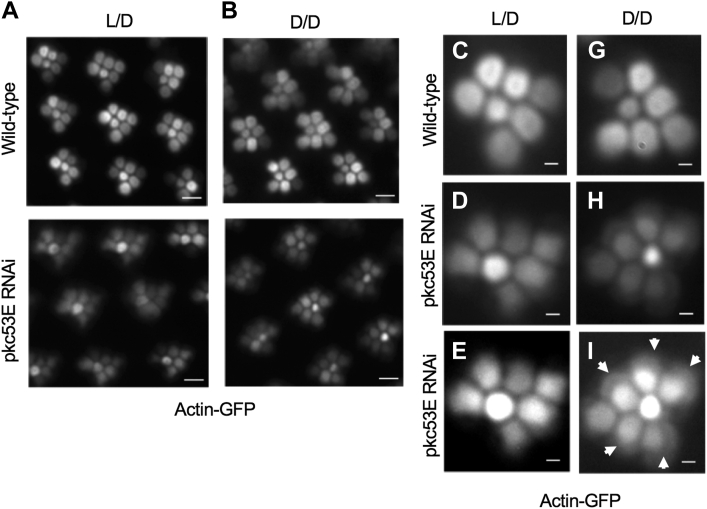
To modulate visual sensitivity, many invertebrate photoreceptors exhibit light-dependent turnover of the visual organelle such as rhabdomere (29). We investigated how Pkc53E is involved in the light-dependent remodeling of the actin cytoskeleton in the rhabdomere. We compared the distribution of Actin-GFP in photoreceptors of pkc53E knockdown to that of wild-type, both of which were subjected to either constant dark or 12 h L/D conditions for 5 days (Fig. 5). Significantly, we show that in either condition pkc53E-RNAi retinas contained smaller rhabdomeres (Fig. 5, A and B, bottom panel; Fig. 5, C, D, G, and H) indicating that Pkc53E is involved in both the light-dependent and light-independent regulation of the actin cytoskeleton. Furthermore, Actin-GFP appeared to accumulate in the cytoplasm including the base of the rhabdomere when pkc53E-RNAi flies were raised in constant dark condition (Fig. 5I, arrows), compared to those in 12 h L/D (Fig. 5E).
Figure 5.
The light-independent regulation of the actin cytoskeleton by Pkc53E. A reduced Pkc53E activity impacts the actin cytoskeleton of the rhabdomere in flies raised in either 12 h L/D (A) or constant dark (B) conditions. Shown are the retinal morphology of wild-type and knockdown mutant marked with Actin-GFP that is enriched in the actin cytoskeleton of rhabdomeres. Scale bars, 5 μm. When examining closely at individual rhabdomere level, Actin-GFP was also present in the cytosol from pkc53E knockdown flies raised in D/D (H), when compared with those in L/D (D). Wild-type controls are shown in (C) (L/D) and (G) (D/D). Shown below (E and I) are enhanced images marked with arrows to indicate the cytosolic accumulation of Actin-GFP. Scale bars in (C–I), 2 μm.
Based on the findings, we speculate that Pkc53E exerts at least two different functions in photoreceptors. Particularly, Pkc53E appears critical for the light-independent remodeling of the actin cytoskeleton to ensure the maintenance of the rhabdomere.
Knockdown of pkc53E or eye-PKC enhanced the degeneration of norpAP24 photoreceptors
We explored the light-dependent regulation of Pkc53E and investigated whether Pkc53E is operating downstream of PLCβ4/NorpA that mediates the visual response. It is important to note that norpA flies lacking PLCβ4 undergo light-dependent retinal degeneration (26, 30), which is characterized by the age-dependent reduction in rhabdomeres (Fig. 6A), a phenotype different from that of pkc53E RNAi (Figs. 4 and 6A).
Figure 6.
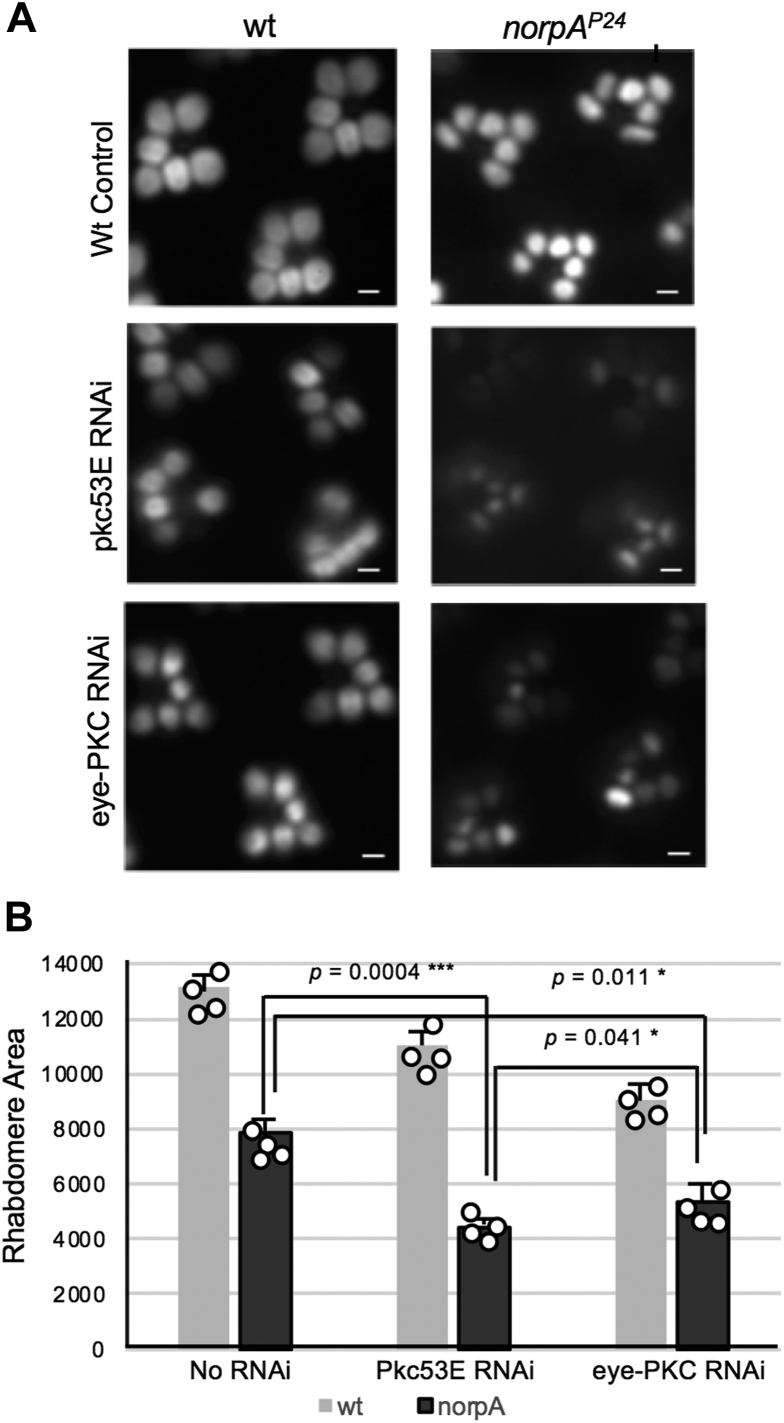
Genetic interactions between norpAP24and pkc53E or eye-PKC.A, knockdown of either pkc53E or eye-PKC enhanced the degeneration of norpAP24 photoreceptors. Shown is the retinal morphology of 7-day-old flies in either wild-type (left) or norpAP24 (right) genetic background. norpAP24 displays light-dependent retinal degeneration leading to reduced rhabdomeres (top right). Similarly, pkc53E RNAi (middle) and eye-PKC RNAi (bottom) also result in degeneration. The reduction of rhabdomere size in norpAP24 is further decreased when the activity of either Pkc53E (middle right) or eye-PKC (bottom right) is decreased. Scale bars, 2 μm. B, comparison of rhabdomere areas in wild-type, single and double mutants using Arr2-GFP as the reporter. Shown is a histogram depicting rhabdomere areas (in arbitrary units, n = 4) that show enhancement of degeneration by knockdown of either pkc53E or eye-PKC. Significant differences between norpAP24 and norpAP24; pkc53E RNAi (p = 0.0004) or norpAP24 and norpAP24; eye-PKC RNAi (p = 0.011) were determined by a two-tailed Student’s t test.
We generated and characterized double mutants between norpAP24 and pkc53E RNAi. We speculate that double mutants (norpAP24; pkc53E RNAi) would lead to a phenotype similar to that of norpAP24 if Pkc53E is acting downstream of PLCβ4 solely. Unexpectedly, we observed more severely degenerated retinas in the double mutant when compared to norpAP24 or pkc53E RNAi, indicating that knockdown of pkc53E enhances the retinal degeneration of norpAP24 or vice versa (Fig. 6, A and B). Similarly, the knockdown of eye-PKC also accelerated the degeneration of norpAP24 photoreceptors (Fig. 6, A and B).
Our findings suggested that every single mutant utilizes distinct pathways, which may act synergistically leading to a more severe phenotype in the double mutant. Indeed, it has been proposed that the degeneration of norpA is due to enhanced metarhodopsin-mediated internalization (31) while perturbation of the actin cytoskeleton is likely involved in the degeneration of pkc53E RNAi (Fig. 4). Thus, both Pkc53E and NorpA appear to work independent of each other. We conclude that Pkc53E remains active in norpAP24 mutants missing PLCβ4, suggesting the contribution of either another PLCβ or an alternate pathway leading to the synthesis of DAG, such as the activation of phospholipase D (PLD) (26, 30) to activate Pkc53E.
plc21C RNAi but not pld RNAi enhanced the degeneration of norpAP24 photoreceptors
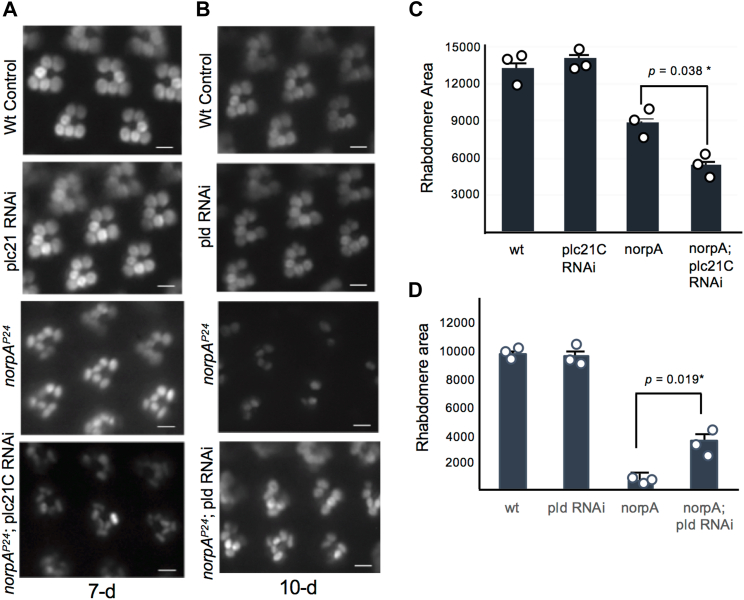
We first investigated the contribution of phospholipase C at 21C (Plc21C) (18), which was reported to participate in the light-dependent regulation of the circadian clock (32). Plc21C also functions in olfaction (33). We speculate if Plc21C is critical for promoting the activation of Pkc53E, the knockdown of plc21C would similarly enhance the degeneration of norpAP24 photoreceptors. Indeed, this was observed as shown in 7-day-old flies (Fig. 7, A and C). However, plc21C RNAi alone did not significantly modify the retinal morphology (Fig. 7, A and C), indicating that Plc21C is not required or exerts minimal contribution in the wild-type genetic background.
Figure 7.
Knockdown of plc21 C enhances the degeneration of norpAP24photoreceptors.A, genetic interactions between norpAP24 and plc21C RNAi. plc21C RNAi does not affect the retinal morphology in the wild-type background but exacerbates the retinal degeneration of norpAP24 (bottom panel). Shown are the retinas of 7-day-old flies using Arr2-GFP as the reporter. Scale bars, 5 μm. B, genetic interactions between norpAP24 and pld RNAi. pld RNAi alone does not modify retinal morphology but delays the degeneration of norpAP24 photoreceptors (bottom). Shown are the retinas of 10-day-old flies. Scale bars, 5 μm. C, comparison of rhabdomere areas in a histogram to show the enhancement of norpAP24 degeneration by plc21C RNAi (in arbitrary units, n = 3). D, comparison of rhabdomere areas in a histogram to support the delay of norpAP24 degeneration by pld RNAi (in arbitrary units, n = 3). Significant differences between norpAP24 and norpAP24; plc21C RNAi (p = 0.038) or norpAP24 and norpAP24; pld RNAi (p = 0.019) were determined by a two-tailed Student’s t test.
We also investigated whether PLD played a role to activate two cPKCs. PLD is known to catalyze the hydrolysis of phosphatidylcholine to release phosphatidic acid (34), which can be dephosphorylated by Laza to generate DAG (35, 36). It was reported that overexpression of pld led to retinal degeneration but rescued degeneration of norpA photoreceptors (37, 38). In contrast, a loss of PLD did not affect retinal morphology under 12 h L/D condition (37). We speculate if PLD is critical for promoting the activation of Pkc53E in the norpAP24 background, degeneration of norpAP24 photoreceptors would be enhanced by the knockdown of pld.
We characterized 10-day-old pld RNAi; norpAP24 and observed the double mutant displayed a better retinal morphology than that of norpAP24 alone, strongly suggesting that activation of PLD is not critical for the Pkc53E activity. Indeed, the knockdown of pld appeared to alleviate the degeneration of norpAP24 photoreceptors (Fig. 7, B and D), suggesting that the PLD activity may lessen the metarhodopsin-mediated retinal degeneration. However, the knockdown of pld alone did not significantly modify retinal morphology (Fig. 7, B and D).
We validated each knockdown by RT/qPCR analyses. We show mRNA levels for pld and plc21C in the fly head were reduced by 55 ± 14% (n = 3), and 41 ± 11% (n = 3), respectively, following RNAi. As both pld and plc21 are widely expressed in the fly head, the residual mRNA levels may be contributed by the expression in non-photoreceptors.
Taken together, our findings strongly suggest that Plc21C but not PLD is critical for the generation of DAG, thereby activating Pkc53E and eye-PKC in norpAP24 photoreceptors when PLCβ4 is absent.
Gqα RNAi leads to retinal degeneration that is not modified by pkc53E RNAi
We explored the mechanism by which Plc21C becomes activated in norpAP24 photoreceptors. Similar to PLCβ4, Plc21C may be activated by the heterotrimeric Gq protein (33, 39). Indeed, Gqα, the α-subunit of Gq, was shown to couple to Plc21C in norpAP24 (32). We speculate if Gqα is directly involved in activating Plc21C leading to the activation of Pkc53E, the knockdown of Pkc53E would not modify the degeneration in Gqα RNAi.
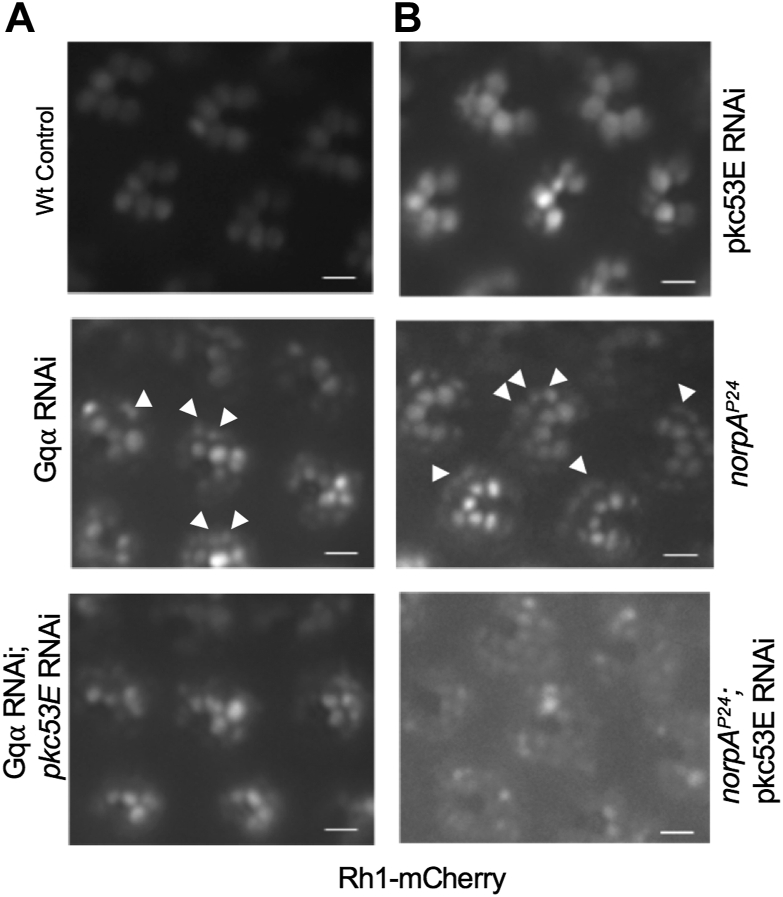
To uncover the relationship between Gqα and pkc53E, we employed a transgene expressing a modified Rh1 with a mCherry tag (40). In the wild-type background, Rh1-mCherry was mostly localized in the rhabdomere of R1-6 photoreceptors (Fig. 8A, top). In contrast, the knockdown of Gqα led to retinal degeneration, which is characterized by a reduction of Rh1-mCherry in the rhabdomere and the accumulation in vesicles within the cytoplasm of photoreceptors, as shown in 5-day-old retinas (Fig. 8A, middle). This phenotype with internalized Rh1-mCherry was similarly observed in norpAP24 photoreceptors (Fig. 8B, middle), consistent with the notion that a reduction of either PLCβ4/NorpA or Gqα activity that greatly diminishes the Ca2+ influx appears to impact the recycling of the activated rhodopsin (31, 41).
Figure 8.
Genetic interactions between pkc53E and Gqα using Rh1-mCherry as the reporter.A, knockdown of pkc53E does not modify retinal degeneration caused by Gqα RNAi. In wild-type photoreceptors, Rh1-mCherry is mostly localized in the rhabdomere (top). Gqα RNAi leads to retinal degeneration that is accompanied by the accumulation of internalized Rh1 rhodopsin (arrows, middle panel). Retinal degeneration of Gqα RNAi is not affected by the knockdown of pkc53E (bottom). B, knockdown of pkc53E enhances the degeneration in norpAP24 photoreceptors. Retinal degeneration in norpAP24 also results in the accumulation of internalized Rh1-mCherry (arrows, middle panel) similar to that of Gqα RNAi. In contrast, pkc53E RNAi does not affect the subcellular distribution of Rh1-mCherry (top) but enhances the retinal degeneration of norpAP24 (bottom panel). Shown are the retinas of 6-day-old flies. Scale bars, 5 μm.
Importantly, we show that double knockdown (Gqα RNAi; pkc53E RNAi) and Gqα knockdown resulted in a similar phenotype (Fig. 8A, bottom), indicating that knockdown of pkc53E did not modify retinal degeneration of Gqα knockdown. This finding supports that Gqα may be required for the activation of Pkc53E in norpAP24 photoreceptors. Similarly, eye-PKC RNAi did not enhance the retinal degeneration caused by Gqα RNAi (not shown). Therefore, activation of both eye-PKC and Pkc53E in norpAP24 appears to involve Gqα. Taken together, we propose that Gqα may couple to Plc21C leading to the activation of Pkc53E in norpA photoreceptors. Knockdown of the Gq subunit was confirmed by Western blotting using polyclonal antibodies (data not shown).
Discussion
Pkc53E-B participates in distinct regulatory mechanisms in photoreceptors
In Drosophila photoreceptors, light initiates a cascade of biochemical events leading to the breakdown of PIP2 by PLCβ to generate IP3 and DAG, which subsequently activates eye-PKC and probably Pkc53E. While eye-PKC plays a major role in the negative regulation of the visual response, the function of Pkc53E remains elusive. Here we report the identification and molecular characterization of a photoreceptor-specific isoform, Pkc53E-B, and show a loss of function of pkc53E-B leads to the light-dependent retinal degeneration and light-independent disruption of the rhabdomere actin cytoskeleton. The morphological characteristics of degeneration revealed by two GFP reporters in pkc53E mutants are different from those of inaC missing eye-PKC, supporting the notion that Pkc53E-B exerts a distinct role different from that of eye-PKC in photoreceptors.
Pkc53E-B is critical for the light-dependent and light-independent maintenance of the actin cytoskeleton
We explored the contribution of Pkc53E-B in regulating the actin cytoskeleton of photoreceptors. The actin cytoskeleton provides mechanical support for the rhabdomere membrane. It is important to note that the actin cytoskeleton is dynamic as it undergoes constant remodeling by depolymerization at the pointed end and polymerization at the barbed end (42). We show that a reduced or loss of pkc53E-B activity negatively impacted the actin cytoskeleton of rhabdomeres independent of the light condition. Furthermore, we observed the accumulation of Actin-GFP in the cytosol including at the base of rhabdomeres, a compartment that is critically involved in membrane cytoskeletal reorganization (43, 44). This phenotype becomes more pronounced in flies that were raised in the constant dark condition, suggesting that Pkc53E-B is involved in the light-independent actin depolymerization.
Pkc53E-B also appears to participate in the light-dependent maintenance of the actin cytoskeleton of rhabdomeres and a loss of Pkc53E leads to retinal degeneration. Pkc53E-B may coordinate the remodeling of the actin cytoskeleton with the light-stimulated turnover of the rhabdomere membrane proteins possibly by regulating proteins that serve as a link between the membrane and the cytoskeleton (44). Mis-regulation of the membrane-cytoskeleton interaction is likely to enhance the turnover of rhabdomere proteins leading to a reduced Rh1 content. As mentioned before, Pkc53E appears to modulate the light-independent remodeling of the actin cytoskeleton by fine-tuning the microfilament depolymerization. Therefore, in the absence of light stimulation, actin becomes accumulated at the base of rhabdomeres of pkc53E mutants. However, accumulation of Actin-GFP is not observed in the mutant under 12 h L/D condition, which could be due to the contribution of additional mechanisms and/or the potential involvement of eye-PKC. How does Pkc53E regulate the turnover of actin filaments? We speculate that Pkc53E may regulate actin-binding proteins to perturb the dynamics of actin polymerization and depolymerization (42).
Subcellular distribution and trafficking of Pkc53E-B in photoreceptors
It is well established that cPKC is recruited to the membrane by binding to DAG following PLC activation. In photoreceptors, Pkc53E-B can be found in both cytosol and rhabdomeres. Translocation between these two compartments appears not acutely affected by the activation of visual signaling as it occurs with much slower kinetics when compared to the light-dependent trafficking of two visual arrestins. For example, Arr2 is rapidly translocated to the membrane with a time constant in seconds upon binding to photoactivated rhodopsin (26, 45). Similarly, translocation of arrestin 1 (Arr1) occurs within minutes of light stimulation (46) upon binding to activated rhodopsin which is also phosphorylated by rhodopsin kinase.
The slow kinetics of rhabdomere translocation by Pkc53E-B strongly suggest that the abundance and/or competence of the Pkc53E-B adaptor/substrate proteins may be subjected to complex regulation. The identity of the Pkc53E-B interacting proteins remains to be investigated. We propose that rhabdomere localized kinase is required for the light-dependent maintenance of photoreceptors critical for averting retinal degeneration. In contrast, cytosolic Pkc53E-B may play a role in the light-independent maintenance of the actin cytoskeleton. Thus, the subcellular localization of Pkc53E-B in photoreceptors supports both the light-dependent and the light-independent activities during the diurnal cycle.
An alternate pathway involved in the activation of two cPKCs when PLCβ4 is missing
We explored the light-dependent regulation of Pkc53E and observed a reduced Pkc53E activity accelerates retinal degeneration of norpAP24 mutants that lack PLCβ4, the major PLCβ involved in the visual signaling. To explore further how Pkc53E can be activated in norpAP24, we show that the knockdown of Plc21C also exacerbated the degeneration of norpAP24, suggesting that Plc21C may be substituted for PLCβ4 for activating Pkc53E. Our findings support the notion that both PLCβ4 and Plc21C are critical for activating Pkc53E (and eye-PKC). Moreover, Plc21C is required when PLCβ4 is absent.
To explore the upstream regulator of Plc21C, we investigated the role of Gq. We show knockdown of Gqα results in retinal degeneration leading to the accumulation of internalized Rh1-mCherry, similar to norpAP24 mutants. Importantly, retinal degeneration of Gqα RNAi is not modified by pkc53E RNAi, strongly suggesting that Gq is required for the Pkc53E activation. Taken together, we propose that activation of Pkc53E may involve Gqα to turn on Plc21C in norpAP24. It has been shown that a loss of PLCβ4 leads to unregulated Gqα, as NorpA appears to serve as GTPase activating protein for inactivating Gqα (47). Consequently, the unregulated Gq activity may promote its coupling to Plc21C. Together, our results support the notion that the light-dependent activation of Pkc53E involves either PLCβ4 and/or Plc21C, both of which are activated by Gq. In contrast, the light-independent activity of Pkc53E to modulate actin turnover might be due to the basal activity of the kinase.
In visual signaling, activation of rhodopsin leads to an increase of cytosolic Ca2+ and transient activation of cPKC. Both Ca2+ and cPKC are critically involved in the stability and maintenance of the actin cytoskeleton. Indeed, when the canonical pathway to turn on cPKC is missing, an alternate pathway that utilizes Plc21C is deployed. We propose that regulation of the actin cytoskeleton is a fundamental event that fine-tunes the PLCβ-mediated signaling response.
Experimental procedures
Fluorescence microscopy of live photoreceptors
Adult flies were anesthetized by CO2 and immobilized in clay with compound eyes facing upward in a 50 mm Petri dish for imaging. The compound eye was examined using an upright Olympus AX70 microscope equipped with a 40X (LUMPlan 40X) or 100X (LUMPlan 100X) water immersion lens for revealing multiple ommatidia of live flies. Image acquisition was performed at 400X or 1000X for rhabdomeres/ommatidia using IPLab image acquisition software (BioVision Technologies) and the Retiga camera from QImaging. Exposure time was made constant throughout each experiment based on the brightest signal in the control group. Multiple flies (n ≥ 3) of each genotype were analyzed. For investigating the subcellular distribution of pkc53E-B-GFP, immobilized flies of the white-eyed genetic background were dark-adapted for 30 min before imaging.
Fly handling for microscopy
Newly eclosed adult flies were collected and aged under 12 h light/12 h dark (12 h L/D; L, 600 lux) condition unless noted otherwise. For imaging, flies were sorted and manipulated under a dissecting microscope with a light source of 600 lux for less than 1 min. During imaging, compound eyes were subjected to the blue light (1300 lux) or green light (4500 lux) from the fluorescent microscope, and images were taken immediately for Actin-GFP, Arr2-GFP, Pkc53E-B-GFP or Rh1-mCherry.
Fluorescent image analysis
All image manipulation was performed under the guideline of Rossner and Yamada (48). Fluorescent images included in the Figures are similar in appearance to the raw images. Experimentally, we collected newly eclosed flies of the desirable genotype, placed them in a vial, and aged them at 25 °C for various amounts of time. Live flies were analyzed for GFP or mCherry-marked rhabdomeres by water-immersion fluorescence microscopy. Retinal morphology was scored based on either the rhabdomere area or the number of rhabdomeres present in an ommatidium (unit eye) similar to that described by Cerny et al. (49). Briefly, retinas from three flies from each compound eye eight ommatidia were selected and counted to obtain the average of the rhabdomere number. To score rhabdomere areas. Four ommatidia clusters were selected in which the total rhabdomere area of each cluster was measured and averaged via ImageJ. Wild-type flies of the same age were used as controls. To score retinal degeneration, we compared the intensity of dpp (20) that is quantified by Image J.
Quantitative Western blotting
A single fly head was dissected and total proteins were extracted with 15 μl of 2X Laemmli sample buffer by sonication. Proteins were size-fractionated by SDS/PAGE (10–12%) and transferred onto a nitrocellulose filter. Filters were incubated with desired primary antibodies followed by the fluorophore-conjugated secondary antibodies (IRDye 680LT Goat anti-Rabbit IgG, or IRDye 800CW Goat anti-mouse IgG, LI-COR). The fluorophore signal was detected by the Odyssey Infrared Imaging System (LI-COR) and analyzed by Image Studio 5.2.
Individual protein content was normalized by using INAD (50) as the loading control. We carried out three to five analyses using one fly head for each analysis. Polyclonal antibodies against the α-subunit of Gq were generated in rabbits using a bacterial fusion protein corresponding to 1 to 200 aa of Gqα. The monoclonal antibody for Rh1 (4C5) was obtained from the Developmental Studies Hybridoma Bank (University of Iowa).
Recombinant DNA and molecular biology
A full-length Rh1 cDNA lacking the 3′ stop codon was generated by PCR with the engineered SacI (5′) and EcoRI (3′) restriction enzyme sites. The mCherry cDNA sequence with the flanking EcoRI (5′) and XhoI (3′) restriction enzyme sites was generated by PCR using pUAST-mCherry [a gift from Dr Amy Kiger (UCSD)] as the template. The mCherry nucleotide sequence was inserted in-frame into the 3′ of the Rh1 cDNA and the resulting Rh1-mCherry chimera DNA was subcloned into YC4 for the expression under the control of the Drosophila Rh1 promoter (26). A full-length cDNA of pkc53E-B (GH03188) was obtained from Drosophila Genomic Resource Center (Indiana University, Bloomington, IN). The nucleotide sequence of an enhanced GFP was inserted at the 3′ of the pkc53E cDNA after the removal of the 3′ stop codon by PCR. The recombinant pkc53E-B-GFP cDNA was subcloned intoYC4 and injected into yw embryos.
Reverse transcription-PCR
Total RNA from 20 fly heads was extracted by a modified method of Chirqwin et al. (51) and dissolved in 20 μl water. Five μl of total RNA were used for the first-strand cDNA synthesis via Superscript III (Invitrogen) primed with random hexamers. Quantitative PCR in triplicate was performed via CFX96 Real-Time System (BIO-RAD) using iQTM SYBR Green Supermix (BIO-RAD). All expression values were normalized to RpL32 (rp49). To compare isoform-specific transcripts in various genetic backgrounds, RT/PCR products were analyzed by polyacrylamide gel (8%) and relative band intensity was quantified using Image Lab (Bio-Rad). Most of the primer sequences used were selected from the FlyPrimerBank (52) and listed below: rp49 (113 nt), AGCATACAGGCCCAAGATCG (5′), TGTTGTCGATACCCTTGGGC (3′); pkc53E (for total pkc53E, 90 nt), AGACTCGCACCATTAAGGCTT (5′), GGATGCGTCGATCCTTGTCTT (3′); pkc53E (for distinguishing between C/E/B isoforms, 356/359 nt and A/F isoforms, 332 nt), CACGTTCTGCTCCCACTGCA (5′), GCTCCGTGTGATCGCATCC (3′); pkc53E (for F isoform, 262 nt or B isoform, 289 nt), AGCCCTCAAGAAGAAGAACGT (5′), TCCTGCGTATGTGAATGGCTC (3′); pkc53E for C/E isoforms (282 nt) AGCCCTCAAGAAGAAGAACGT (5′), AGGAAGGTGACATTCTGCCA (3′); GTCGGAGAAACTGGGCAAG (5′), GAAACCGCAGAATGATGGTCC (3′); arr1 (90 nt), CATGAACAGGCGTGATTTTGTAG (5′), TTCTGGCGCACGTACTCATC (3′); Plc21C (159 nt), GAGAAGACAGTGACGGTATGC (5′), CAGGAACATAATCGCCGAGC (3′); Pld (132 nt), GATGAGACCCTCGCTTTTCCT (5′), GACTACACTGTTGTTTTCCTCGT(3′).
Drosophila stocks
Drosophila lines including mutants for eye-PKC (inaCP209, #42241) and Pkc53E (pkc53EΔ28, #80988) were obtained from Bloomington Drosophila Stock Center (BDSC) (NIH P40OD018537). Transgenic flies for GMR-GAL4 (stock #1104), RNAi lines for the following genes including inaC (#36776), pkc53E (#55864, #27491), Gqα (#63987), plc21C (#31269, #33719), and pld (#32839) were also obtained from BDSC. UAS-driven overexpressing lines including Pkc53E-B (#80989), and Actin-GFP (#9253) were from BDSC. Fly cultures were maintained in the standard cornmeal medium at 25 °C. Standard crosses were used to introduce suitable genetic backgrounds.
Statistical analysis
One-way ANOVA and two-tailed Student's t test were employed for statistical analysis.
Data availability
Data are available within the article or its Supplementary materials.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
B.-H. S. funding acquisition; B.-H. S., D. F., and W. S. investigation; B.-H. S. and D. F. formal analysis; B.-H. S. writing; W. S. methodology; D. F. supervision.
Funding and additional information
This work was supported by NIH grants [R01, to B.-H. S]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Enrique De La Cruz
Supporting information
References
- 1.Lipp P., Reither G. Protein kinase C: the "masters" of calcium and lipid. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newton A.C. Protein kinase C: perfectly balanced. Crit. Rev. Biochem. Mol. Biol. 2018;53:208–230. doi: 10.1080/10409238.2018.1442408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebecchi M.J., Pentyala S.N. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol. Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y.R., Choi J.H., Chang J.S., Kwon H.M., Jang H.J., Ryu S.H., et al. Diverse cellular and physiological roles of phospholipase C-gamma1. Adv. Biol. Regul. 2012;52:138–151. doi: 10.1016/j.advenzreg.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Hardie R.C., Juusola M. Phototransduction in drosophila. Curr. Opin. Neurobiol. 2015;34:37–45. doi: 10.1016/j.conb.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Montell C. Drosophila visual transduction. Trends Neurosci. 2012;35:356–363. doi: 10.1016/j.tins.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keenan C., Kelleher D. Protein kinase C and the cytoskeleton. Cell. Signal. 1998;10:225–232. doi: 10.1016/s0898-6568(97)00121-6. [DOI] [PubMed] [Google Scholar]
- 8.Larsson C. Protein kinase C and the regulation of the actin cytoskeleton. Cell. Signal. 2006;18:276–284. doi: 10.1016/j.cellsig.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Schaeffer E., Smith D., Mardon G., Quinn W., Zuker C. Isolation and characterization of two new drosophila protein kinase C genes, including one specifically expressed in photoreceptor cells. Cell. 1989;57:403–412. doi: 10.1016/0092-8674(89)90915-x. [DOI] [PubMed] [Google Scholar]
- 10.Rosenthal A., Rhee L., Yadegari R., Paro R., Ullrich A., Goeddel D.V. Structure and nucleotide sequence of a Drosophila melanogaster protein kinase C gene. EMBO J. 1987;6:433–441. doi: 10.1002/j.1460-2075.1987.tb04773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith D.P., Ranganathan R., Hardy R.W., Marx J., Tsuchida T., Zuker C.S. Photoreceptor deactivation and retinal degeneration mediated by a photoreceptor-specific protein kinase C. Science. 1991;254:1478–1484. doi: 10.1126/science.1962207. [DOI] [PubMed] [Google Scholar]
- 12.Adamski F.M., Zhu M.Y., Bahiraei F., Shieh B.H. Interaction of eye protein kinase C and INAD in Drosophila. Localization of binding domains and electrophysiological characterization of a loss of association in transgenic flies. J. Biol. Chem. 1998;273:17713–17719. doi: 10.1074/jbc.273.28.17713. [DOI] [PubMed] [Google Scholar]
- 13.Tsunoda S., Zuker C.S. The organization of INAD-signaling complexes by a multivalent PDZ domain protein in Drosophila photoreceptor cells ensures sensitivity and speed of signaling. Cell Calcium. 1999;26:165–171. doi: 10.1054/ceca.1999.0070. [DOI] [PubMed] [Google Scholar]
- 14.Peng L., Popescu D.C., Wang N., Shieh B.H. Anchoring TRP to the INAD macromolecular complex requires the last 14 residues in its carboxyl terminus. J. Neurochem. 2008;104:1526–1535. doi: 10.1111/j.1471-4159.2007.05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popescu D.C., Ham A.J., Shieh B.H. Scaffolding protein INAD regulates deactivation of vision by promoting phosphorylation of transient receptor potential by eye protein kinase C in Drosophila. J. Neurosci. 2006;26:8570–8577. doi: 10.1523/JNEUROSCI.1478-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svitkina T. The actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloomquist B.T., Shortridge R.D., Schneuwly S., Perdew M., Montell C., Steller H., et al. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- 18.Shortridge R.D., Yoon J., Lending C.R., Bloomquist B.T., Perdew M.H., Pak W.L. A Drosophila phospholipase C gene that is expressed in the central nervous system. J. Biol. Chem. 1991;266:12474–12480. [PubMed] [Google Scholar]
- 19.Moses K., Ellis M.C., Rubin G.M. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- 20.Franceschini N. Springer-Verlag, Berlin Heidelberg; New York, NY: 1972. Pupil and Pseudopupil in the Compound Eye of Drosophila. [Google Scholar]
- 21.Schechtman D., Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal N., Dasaradhi P.V., Mohmmed A., Malhotra P., Bhatnagar R.K., Mukherjee S.K. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrimon N., Ni J.Q., Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 25.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 26.Kristaponyte I., Hong Y., Lu H., Shieh B.H. Role of rhodopsin and arrestin phosphorylation in retinal degeneration of Drosophila. J. Neurosci. 2012;32:10758–10766. doi: 10.1523/JNEUROSCI.0565-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roper K., Mao Y., Brown N.H. Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. J. Cell Sci. 2005;118:3937–3948. doi: 10.1242/jcs.02517. [DOI] [PubMed] [Google Scholar]
- 28.Shieh B.H., Nuzum L., Kristaponyte I. Exploring excitotoxicity and regulation of a constitutively active TRP Ca(2+) channel in Drosophila. Fly (Austin) 2021;15:8–27. doi: 10.1080/19336934.2020.1851586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams D.S. Rhabdom size and photoreceptor membrane turnover in a muscoid fly. Cell Tissue Res. 1982;226:629–639. doi: 10.1007/BF00214790. [DOI] [PubMed] [Google Scholar]
- 30.Chinchore Y., Mitra A., Dolph P.J. Accumulation of rhodopsin in late endosomes triggers photoreceptor cell degeneration. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alloway P.G., Howard L., Dolph P.J. The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron. 2000;28:129–138. doi: 10.1016/s0896-6273(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 32.Ogueta M., Hardie R.C., Stanewsky R. Non-canonical phototransduction mediates synchronization of the Drosophila melanogaster circadian clock and retinal light responses. Curr. Biol. 2018;28:1725–1735.e3. doi: 10.1016/j.cub.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kain P., Chakraborty T.S., Sundaram S., Siddiqi O., Rodrigues V., Hasan G. Reduced odor responses from antennal neurons of G(q)alpha, phospholipase Cbeta, and rdgA mutants in Drosophila support a role for a phospholipid intermediate in insect olfactory transduction. J. Neurosci. 2008;28:4745–4755. doi: 10.1523/JNEUROSCI.5306-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frohman M.A. The phospholipase D superfamily as therapeutic targets. Trends Pharmacol. Sci. 2015;36:137–144. doi: 10.1016/j.tips.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Murillas I., Pettitt T., Macdonald E., Okkenhaug H., Georgiev P., Trivedi D., et al. Lazaro encodes a lipid phosphate phosphohydrolase that regulates phosphatidylinositol turnover during Drosophila phototransduction. Neuron. 2006;49:533–546. doi: 10.1016/j.neuron.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Kwon Y., Montell C. Dependence on the lazaro phosphatidic acid phosphatase for the maximum light response. Curr. Biol. 2006;16:723–729. doi: 10.1016/j.cub.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 37.LaLonde M.M., Janssens H., Rosenbaum E., Choi S.Y., Gergen J.P., Colley N.J., et al. Regulation of phototransduction responsiveness and retinal degeneration by a phospholipase D-generated signaling lipid. J. Cell Biol. 2005;169:471–479. doi: 10.1083/jcb.200502122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thakur R., Panda A., Coessens E., Raj N., Yadav S., Balakrishnan S., et al. Phospholipase D activity couples plasma membrane endocytosis with retromer dependent recycling. Elife. 2016;5 doi: 10.7554/eLife.18515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott K., Becker A., Sun Y., Hardy R., Zuker C. Gq alpha protein function in vivo: genetic dissection of its role in photoreceptor cell physiology. Neuron. 1995;15:919–927. doi: 10.1016/0896-6273(95)90182-5. [DOI] [PubMed] [Google Scholar]
- 40.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 41.Kiselev A., Socolich M., Vinos J., Hardy R.W., Zuker C.S., Ranganathan R. A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron. 2000;28:139–152. doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 42.Pollard T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karagiosis S.A., Ready D.F. Moesin contributes an essential structural role in Drosophila photoreceptor morphogenesis. Development. 2004;131:725–732. doi: 10.1242/dev.00976. [DOI] [PubMed] [Google Scholar]
- 44.Neisch A.L., Fehon R.G. Ezrin, radixin and moesin: key regulators of membrane-cortex interactions and signaling. Curr. Opin. Cell Biol. 2011;23:377–382. doi: 10.1016/j.ceb.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satoh A.K., Xia H., Yan L., Liu C.H., Hardie R.C., Ready D.F. Arrestin translocation is stoichiometric to rhodopsin isomerization and accelerated by phototransduction in Drosophila photoreceptors. Neuron. 2010;67:997–1008. doi: 10.1016/j.neuron.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shieh B.H., Kristaponyte I., Hong Y. Distinct roles of arrestin 1 protein in photoreceptors during Drosophila development. J. Biol. Chem. 2014;289:18526–18534. doi: 10.1074/jbc.M114.571224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cook B., Bar-Yaacov M., Cohen Ben-Ami H., Goldstein R.E., Paroush Z., Selinger Z., et al. Phospholipase C and termination of G-protein-mediated signalling in vivo. Nat. Cell Biol. 2000;2:296–301. doi: 10.1038/35010571. [DOI] [PubMed] [Google Scholar]
- 48.Rossner M., Yamada K.M. What's in a picture? the temptation of image manipulation. J. Cell Biol. 2004;166:11–15. doi: 10.1083/jcb.200406019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerny A.C., Oberacker T., Pfannstiel J., Weigold S., Will C., Huber A. Mutation of light-dependent phosphorylation sites of the Drosophila transient receptor potential-like (TRPL) ion channel affects its subcellular localization and stability. J. Biol. Chem. 2013;288:15600–15613. doi: 10.1074/jbc.M112.426981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shieh B.H., Niemeyer B. A novel protein encoded by the InaD gene regulates recovery of visual transduction in Drosophila. Neuron. 1995;14:201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- 51.Chirgwin J.M., Przybyla A.E., MacDonald R.J., Rutter W.J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 52.Hu Y., Sopko R., Foos M., Kelley C., Flockhart I., Ammeux N., et al. FlyPrimerBank: an online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 (Bethesda) 2013;3:1607–1616. doi: 10.1534/g3.113.007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available within the article or its Supplementary materials.