Abstract
Noncovalent adsorption of biopolymers on the surface of gold nanoparticles (AuNPs) forms a corona phase that drastically diversify AuNP functions. However, mechanical stabilities of corona phase are still obscure, hindering the application of biopolymer-coated AuNPs. Here, using optical tweezers we have observed for the first time that DNA corona phase adsorbed on a 5 nm AuNP via two (dA)21 strands in proximity can withstand an average desorption force of 40 pN, which is higher than the stall force of DNA/RNA polymerases. This suggests a new role for AuNPs to modulate replications or transcriptions after binding to prevalent poly(dA) segments in eukaryotic genomes. We have also revealed that with increasing AuNP size (1.8 – 10 nm), DNA corona becomes harder to remove, likely due to the larger surfaces and flatter facets on bigger AuNPs. These findings provide guidance to design AuNP corona that can withstand harsh environment for biological and materials applications.
Graphical Abstract
Introduction
With tuneable physical and chemical properties, gold nanoparticles (AuNPs) have demonstrated versatile applications across many fields.1–3 In particular, owing to their excellent biocompatibility, versatile bifunctionalities, and easy derivatization with bioactive molecules, AuNPs have been widely employed in the biomedical applications.1,4 Functionalization of AuNPs plays an essential role to modulate activities of AuNPs. A diverse set of molecules such as proteins,5,6 nucleic acids (DNA or RNA),7–9 and organic molecules10,11 can be conjugated to the AuNP surface, drastically expanding the applications of AuNPs.
Among different functionalization strategies, anchoring DNA molecules onto AuNPs represents a particularly attractive approach due to the biocompatibility, functional diversity, and bottom-up programmability in DNA.7,9,12 However, since both DNA and AuNPs are negatively charged, challenge exists to overcome their charge repulsion while maintaining the stability of AuNP colloids.7,12 Salt-aging and pH-based DNA loading are among the first techniques to functionalize thiolated DNA on gold nanospheres.7,13–15 These processes are tedious to perform and require chemical modification of DNA with a sulfhydryl group. Recently, it has been found that natural poly-deoxyadenosine (poly(dA)) can adsorb to AuNP surfaces to form a DNA corona phase via noncovalent Au-N interactions16–20 using either salt-aging17 or low-pH assisted21 procedures. This strategy has been further streamlined using a simple freeze and thaw procedure to displace original AuNP coating ligands with poly(dA) within 15 mins.22
When AuNPs are used in biological environment, it becomes important to maintain a stable corona surface for bioavailability while preserving desired activities of AuNPs. Research has found that thermodynamic stability of coated ligands on AuNPs depends on the nanoparticle size, salt concentration, and buffer pH.23–27 Several studies in the past have revealed mechanical stabilities of covalent Au-S interactions.28,29 Dependent on the sulfhydryl group in various molecules,30,31 rupture forces up to 1.5 nN have been detected, which may come from the rupture events of Au-Au atoms, instead of Au-S bonds30.
However, the mechanical stability of noncovalently adsorbed corona phase, such as the poly(dA) coating on AuNPs, is yet to be revealed. Due to rather convenient procedures, these noncovalent adsorptions have been recently exploited to prepare multi-domain AuNP containing nanodevices with hierarchical structures and functionalities.32,33 Given that many applications of AuNPs occur in hydrogels and polymers which may experience high mechanical stresses such as swelling and shrinking, as well as centrifugation and shear flows, it becomes urgent to reveal mechanical properties of the corona phase on the AuNPs. In this work, we have studied the mechanical stability of noncovalent Au-N based corona adsorption for the first time using optical tweezers. Given the high sensitivity of optical tweezers in the mechanical force measurement, and its superiority over other force spectroscopy methods to minimize surface effect,34 we used this method to study the mechanical stability of DNA corona phase on AuNPs. Based on the widely used poly(dA)-AuNP conjugates, we have found that two (dA)21 strands in close proximity show much improved adsorption to AuNPs than one (dA)21 strand or strands with shorter poly(dA) segments.
We have observed desorption force of >20 pN (average 37 – 43 pN) is needed to break the noncovalent interaction between the DNA and AuNPs. This mechanical stability is lower than that of covalent Au-S bonds, which is in the range of several hundreds to >1000 pN.30,31 However, it is higher than that to maintain DNA secondary structures such as duplexes and tetraplexes (<20 pN35,36) or that to stall motor proteins such as DNA or RNA polymerases37–39. As DNA duplexes and tetraplexes routinely exist inside cells40, this suggests that AuNP coated with poly(dA) corona phase is strong enough to withstand the cellular environment from a mechanical perspective. Since poly(dA) is ubiquitous in genomes,41–43 a new function of AuNP may exist to modulate replication or transcription processes via binding to the poly(dA) segments.
Results and discussion
Single-molecule setup to study the poly(dA) corona on gold nanospheres
To investigate the noncovalent binding of poly(dA) DNA to AuNP, we designed a DNA construct consisting of a hairpin with two (dA)21 internal loops in the stem (shown in red in Figure 1A). In the microfluidic chamber in optical tweezers instrument (see SI section S9), the hairpin was tethered between two optically trapped beads using 1558 bp and 2391 bp dsDNA handles. The tethering was achieved by biotin/streptavidin and digoxigenin/anti-digoxigenin affinity interactions (detailed synthesis of the construct can be found in Figures S1 & S2). Tension in the DNA construct was developed when the two optically trapped beads were moved apart by steering one of the trapping lasers. When the tension was sufficiently high, the hairpin was unfolded, generating distinct unfolding features in the force-extension (FX) curve as shown in Figure 1B. The internal loops were ripped apart before the unfolding of the terminal hairpin since the former experienced the tension before the latter did due to their relative locations with respect to the direction of the applied force. Upon relaxing the stretched DNA construct to ~12.9 pN, the unfolded terminal hairpin refolded faster than the rest of the structure due to its small size and close location of the two complementary hairpin stems. The region below the terminal hairpin didn’t show immediate refolding during relaxing because of a long non-complementary region made of (dA)21 in internal loop. However, upon providing sufficient time (~15 s) at zero force, the complementary region below the poly(dA) internal loop refolded completely and the rupture feature was observed again in the next FX curve. The forces at which the internal loops and the terminal loop were unfolded were centered at 12.9 pN (Figure 1 C&D), which is close to the unzipping force of duplex DNA.35,44 In the change-in-contour length (ΔL) histogram (Figure 1 C&E), two distinct populations represent the unfolding’s of the terminal hairpin (ΔL = 15.4 nm) and the internal (ΔL = 36.5 nm) loops, respectively. While the unfolding to release the internal loops shows the ΔL (36.5 nm (observed) vs 39.6 nm (expected)) close to that expected (see Figure S7), the terminal hairpin gives smaller values (15.4 nm (observed) vs 22.3 nm (expected)), which may be due to the breathing of the duplex DNA at the end of hairpin stems35,45.
Figure 1.
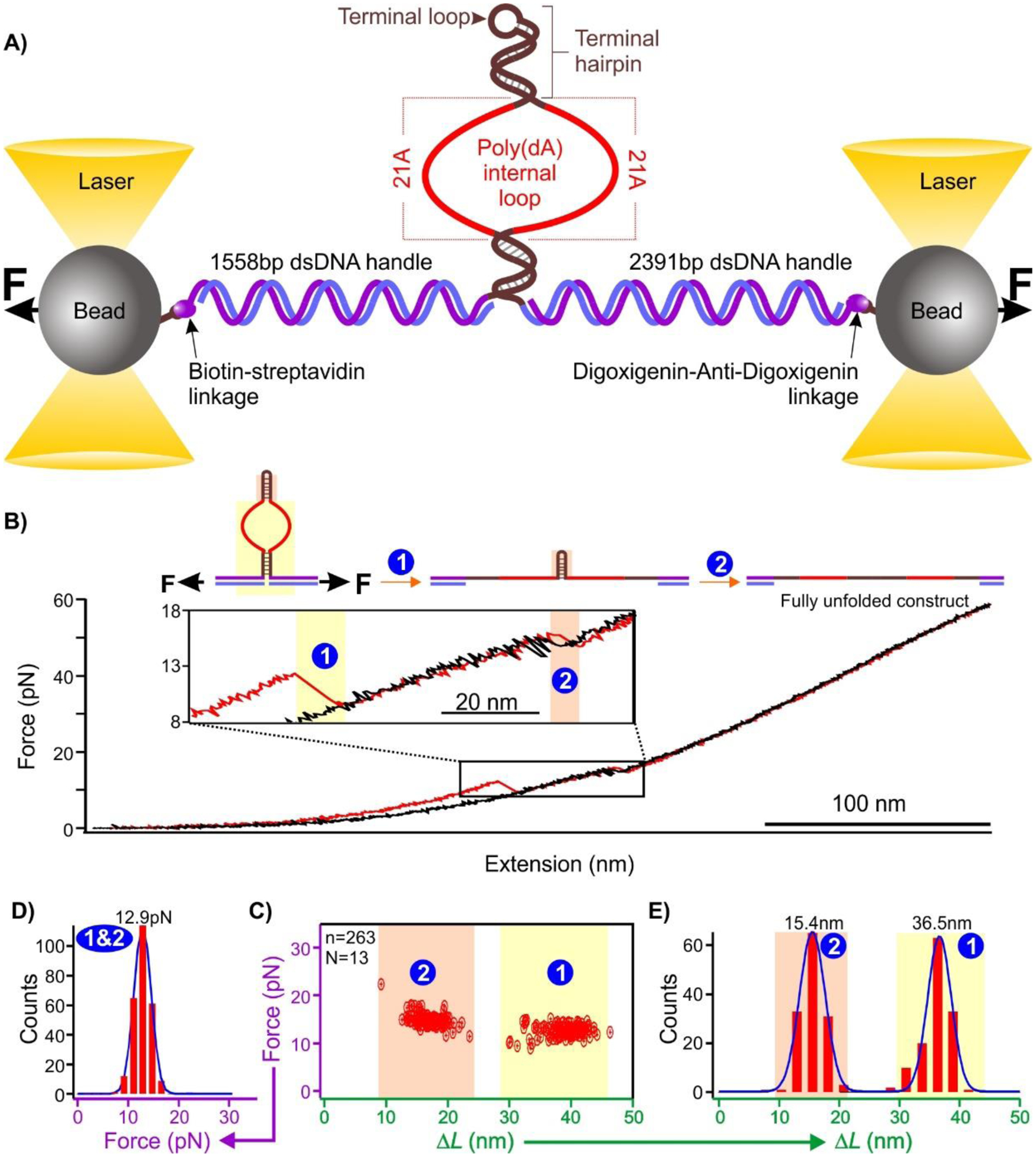
A) Schematic of the single-molecule platform to investigate the binding of poly(dA) to gold nanospheres. See Table S1 for DNA sequences. B) A typical force extension curve when a DNA construct is stretched (red) and relaxed (black) in the optical tweezers instrument. C) Scatter plot of force vs change-in-contour length (ΔL). D) Unfolding force and E) change-in-contour-length (ΔL) histograms of the unfolding features shown in C). Solid curves are Gaussian fittings. N represents the number of molecules from which the FX curves were collected, and n represents the total number of unfolding features measured.
Adsorption of DNA corona to AuNPs via the poly(dA) internal loops
Gold nanospheres are known to noncovalently bind to the poly(dA) via Au-N interactions.19,22,46 To evaluate this binding, we prepared a AuNP bound hairpin-based DNA construct. To prepare it, in brief, we incubated the DNA construct with 5 nm gold nanospheres at −80 °C for 15 mins in a 10:1 mole ratio before thawing at room temperature.22 This freeze-thaw procedure enabled the binding of AuNP to the (dA)21 containing construct (see SI sections 7 & 8, and Figure S4). The force ramp experiment was performed after the construct was tethered between the two trapped beads in optical tweezers instrument. During mechanical unfolding of the AuNP bound DNA construct, a new rupture feature at a force higher than that to unfold terminal hairpin (14.9 pN, average force from Figure 1C) or internal loops (12.7 pN, average force from Figure 1C) was observed (state “3” in Figure 2 A - C, average 37.0 pN). Close inspection on the ΔL histogram (state “3” in Figure 2D) revealed an average unfolding population of 7.0 nm of this feature, which is much smaller than either the terminal hairpin (15.4 nm) or the internal loop (36.5 nm). This observation plus the fact that a higher force is required for its unfolding suggests this feature is associated with the AuNP binding to the DNA construct.
Figure 2.
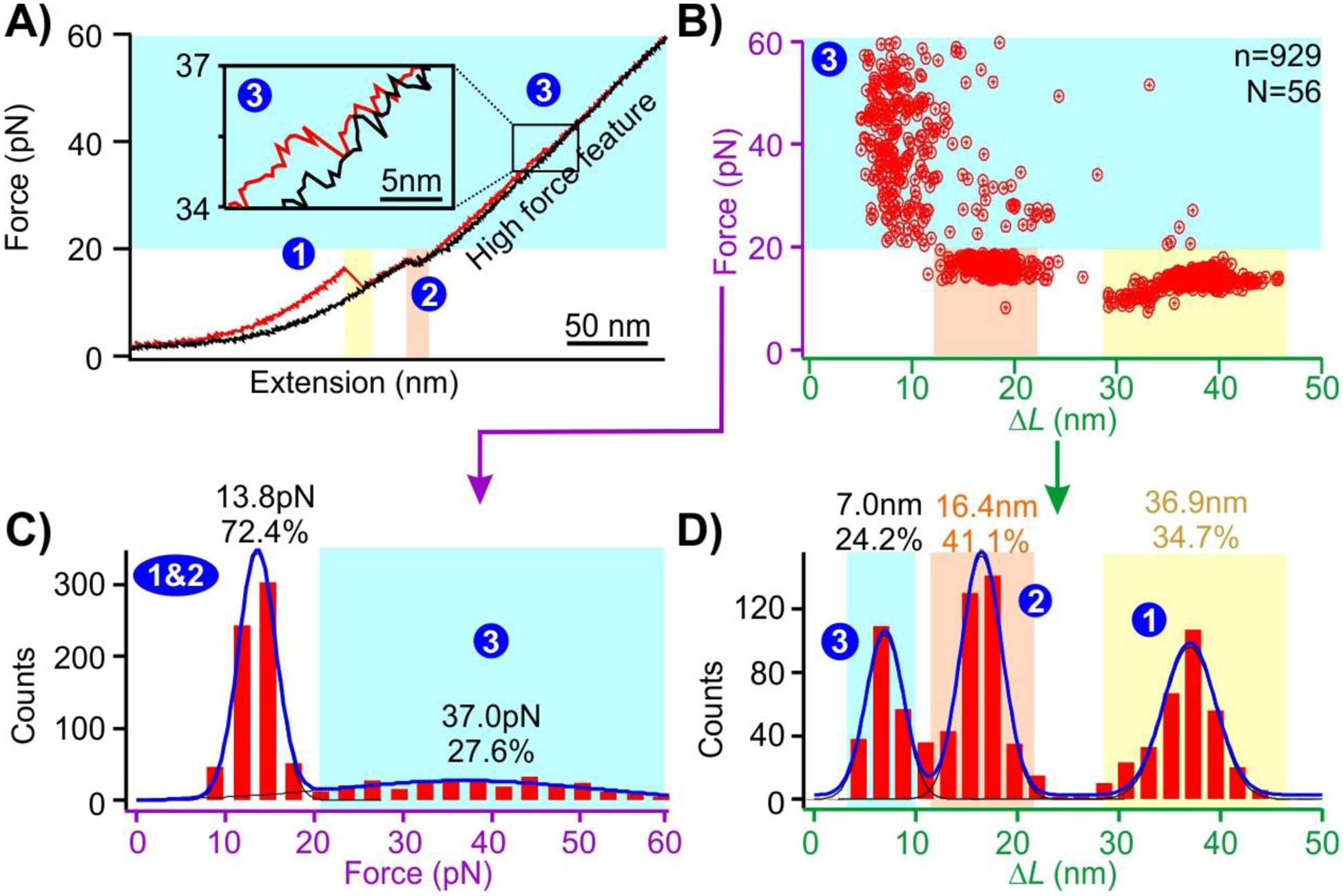
A) A typical force extension curve when the DNA construct (shown in Figure 1) is stretched (red) and relaxed (black) in the presence of a 5 nm AuNP. A new unfolding feature (marked as ‘3’) is observed in the presence of nanogold. B) Plot of the unfolding force vs ΔL of the unfolding events. C) and D) Unfolding force and ΔL histograms of the features in the presence of the 5 nm AuNP, respectively. Solid curves are Gaussian fittings. N represents the number of molecules from which the FX curves were collected, and n represents the total number of unfolding features measured.
To confirm that the AuNP is bound to the poly(dA) via the internal loops, we performed a control experiment in which both internal loops contain random nucleotide sequences (see SI Table S1 for sequences, and Figure S3 for predicted lowest energy structures of the hairpins). As shown in Figure 3A, the low-ΔL, high-force population (state “3”) is significantly reduced (p<0.05) from 25% (state “3” in Figure 2) to 7.5%. In another experiment, we placed (dA)21 only in one strand of the internal loop while keeping random sequence in the other loop strand. As shown in Figure 3B, the population of interest (state “3”, 8.7% of entire population) was again significantly smaller (p<0.05) than the two (dA)21 internal loops. When we reduced (dA)21 to (dA)10 (Figure 3C) in each internal loop, we found the new species reduced to 8.0% of the total population. Finally, when only one internal loop contains (dA)10 whereas the other loop has a random sequence (Figure 3D), the new species further reduced to 7.4% of the whole population. These experiments indicated that (dA)21 in both internal loops are most efficient to bind to the 5 nm AuNP. It is conceivable that closer locations of the two poly(dA) strands assist with each other to accommodate their better bindings to the AuNP surface.
Figure 3.
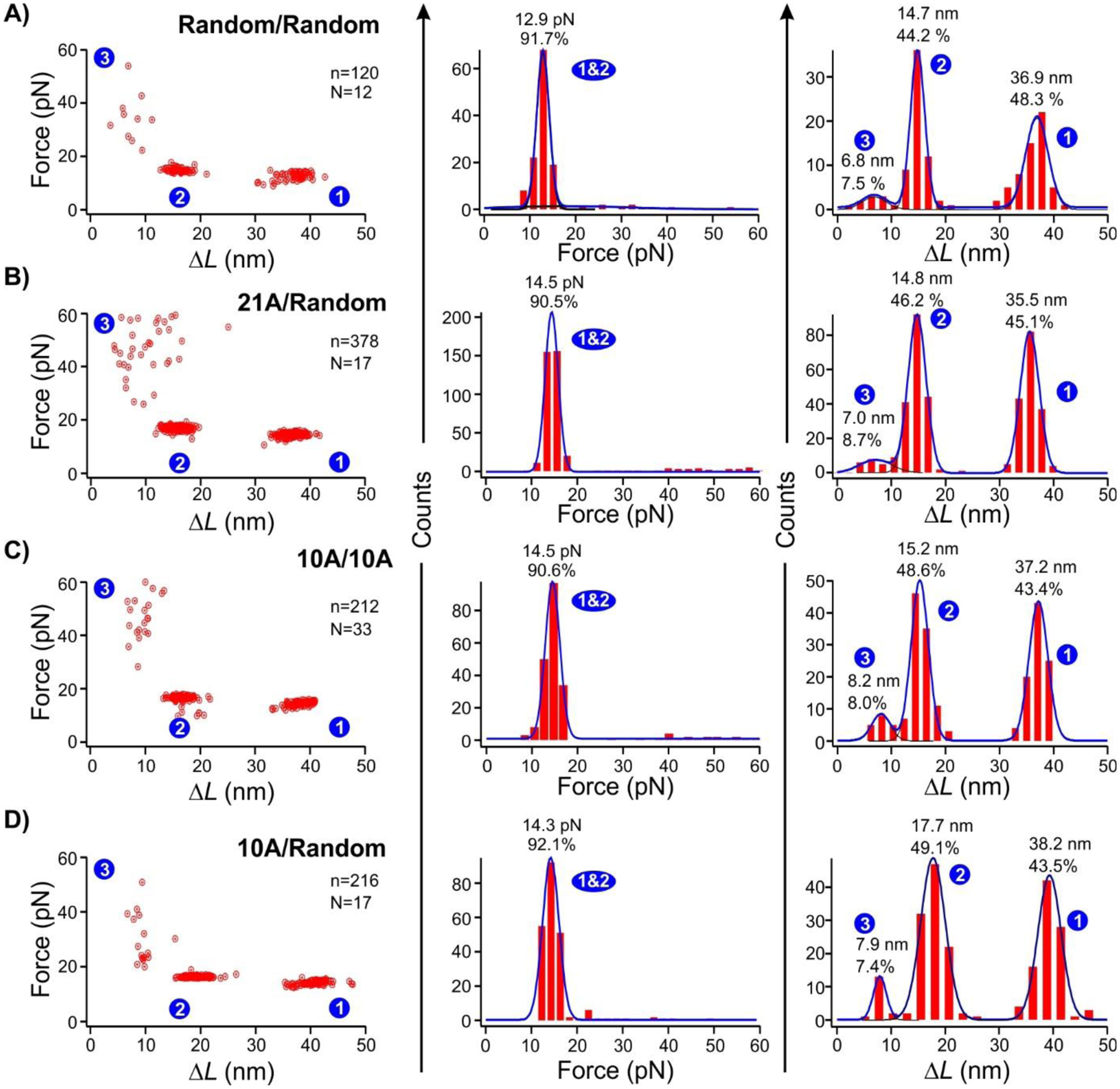
Force vs ΔL plots (left), unfolding force histograms (middle), and ΔL histograms (right) of the unfolding features when the two DNA strands in the internal loops have different poly(dA) sequences. (A) Two random sequences. (B) One (dA)21 and one random sequence. (C) Two (dA)10 sequences. (D) One (dA)10 and one random sequence. Solid curves are Gaussian fittings. N represents the number of molecules from which the FX curves were collected, and n represents the total number of unfolding features measured. All experiments were performed in presence of 5 nm AuNP.
The fact that removal of (dA)21 or reduction in the length of the (dA) segments results in much reduced AuNP binding population while neither rupture force nor ΔL varies with the length of the poly(dA) suggest only one AuNP is bound to (dA)21 in both internal loops. AFM images by others with similar poly(dA) and AuNP interactions also proved binding of one AuNP by poly(dA) segment.32 Finally, the observation of only one rupture event with the high-force and low-ΔL feature (state “3” in Figures 2&3) in each force-extension curve confirmed the binding of single AuNP for each DNA construct used here.
Binding model between gold nanospheres and poly(dA)
Based on the observations from these control experiments (Figure 3), it is clear that poly(dA) on both strands of the internal loop facilitates the binding of one Au nanosphere. Given that the rupture event corresponding to the unfolding of the AuNP-DNA conjugate (state “3” in Figure 2) occurs at the later stage of the unfolding sequence where the force is high (Figures 2&3), we reasoned that the terminal hairpin (unfolding force: 13.8 pN, see Figure 2C) must unfold before the unbinding of the AuNP-DNA conjugate. This can only occur when one (dA)21 strand has a looser binding than the other stand in the internal loops (Figure 4). Thus, the unfolding starts with the ripping of the loosely adsorbed (dA)21 strand from the AuNP, generating a large ΔL feature (step 1, Figure 4) with the size and force similar to those without AuNP (compare the state “1” in Figure 1B – D and Figure 2). After the step 1, the force experienced by the AuNP-bound DNA construct is exerted on the terminal hairpin (Figure 4). This leads to the step 2 in which the unfolding of the terminal hairpin occurs (see state “2” in Figures 2 and 4). Completion of this step results in the force exerted on the remaining (dA)21 loop adsorbed to the AuNP. The small ΔL observed during step 3 (7.0 nm, the state “3” in Figure 2B&D) is either due to the stretching of some looped-out (dA)21 region in the DNA-AuNP conjugate or the change in the apparent contour length of the AuNP-bound (dA)21 when the construct experiences higher force (from the winding conformation of the poly(dA) adsorbed to the AuNP surface to the straight conformation, see step 3 in Figure 4).
Figure 4.
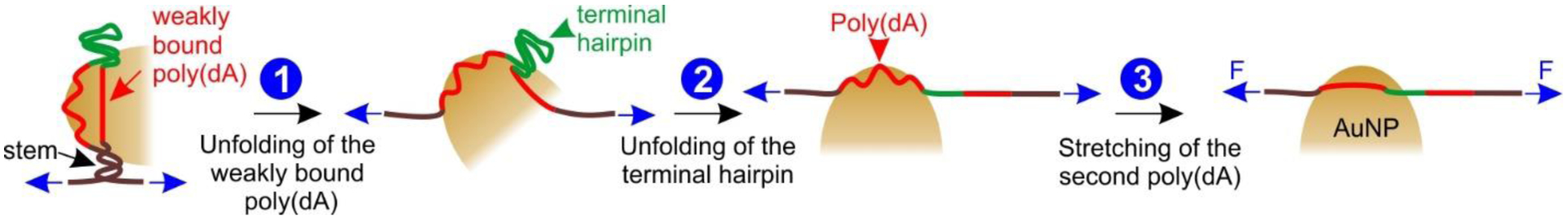
Mechanical desorption pathway of the DNA adsorbed on the AuNP surface. The DNA adsorption is achieved via the two (dA)21 internal loops (red). Blue arrows indicate external forces.
We argue that after step 3, the AuNP is still associated with the poly(dA) since steps 1–3 are repeatedly observed in consecutive F-X curves, indicating rebinding of the AuNP to the (dA)21 strands at reduced force. The remaining adsorption is likely due to the Au-N bonds contributed from a limited number of deoxyadenosines without resulting in any looped-out poly(dA) segments. Such an adsorption is not expected to experience any stretching force in optical tweezers.
Mechanical binding strength of the (dA)21 to the AuNPs with different sizes
To study the size effect of the AuNP on the (dA)21 binding (state “3” in Figures 1–3 and step 3 in Figure 4), we repeated force-ramping experiments using the hairpin construct with the two (dA)21 internal loops in the presence of nanogold particles with different sizes. As shown in Figure 5 (state “3”), the unfolding force of the characteristic AuNP-(dA)21 conjugates gradually shifted towards higher force with the increasing nanogold size. With the 1.8 nm nanogold (Figure 5A), the binding populations can be well fit by three force regimes centered at 27.9 pN (21.5%), 40.8 pN (12.6%), and 52.3 pN (5.9%) (average 36.9 pN). We attribute this distribution to the three possible binding modes between (dA)21 and AuNP (Figure S8). From the unfolding ΔL of 6.4 nm (Figure 5, state “3”), we estimated ~16 deoxyadenosines were contained in the sticking-out loop (Figure S8), leaving five deoxyadenosines to serve as additional anchoring points (a total of 7) on the AuNP surface. The three unfolding force populations in Figure 5A can therefore be attributed to the one, two, or three anchoring deoxyadenosines located on each side of the sticking-out loop, which corresponded the 27.9 pN, 40.8 pN, and 52.3 pN unfolding forces, respectively. Due to the small size of the 1.8 nm AuNP, however, only a limited number of deoxyadenosines can adsorb on AuNP surface. Therefore, the more the anchoring deoxyadenosines, the higher the unfolding force, but the lower the populations as the random poly(dA) adsorption process may not be able to accommodate all 7 anchoring deoxyadenosines for the 1.8 nm AuNP. It is likely that in the most stable binding mode (52.3 pN), the sticking-out poly(dA) loop straddles between two AuNP facets to fully accommodate the 3 anchoring deoxyadenosines on each side (Figure S8).
Figure 5.
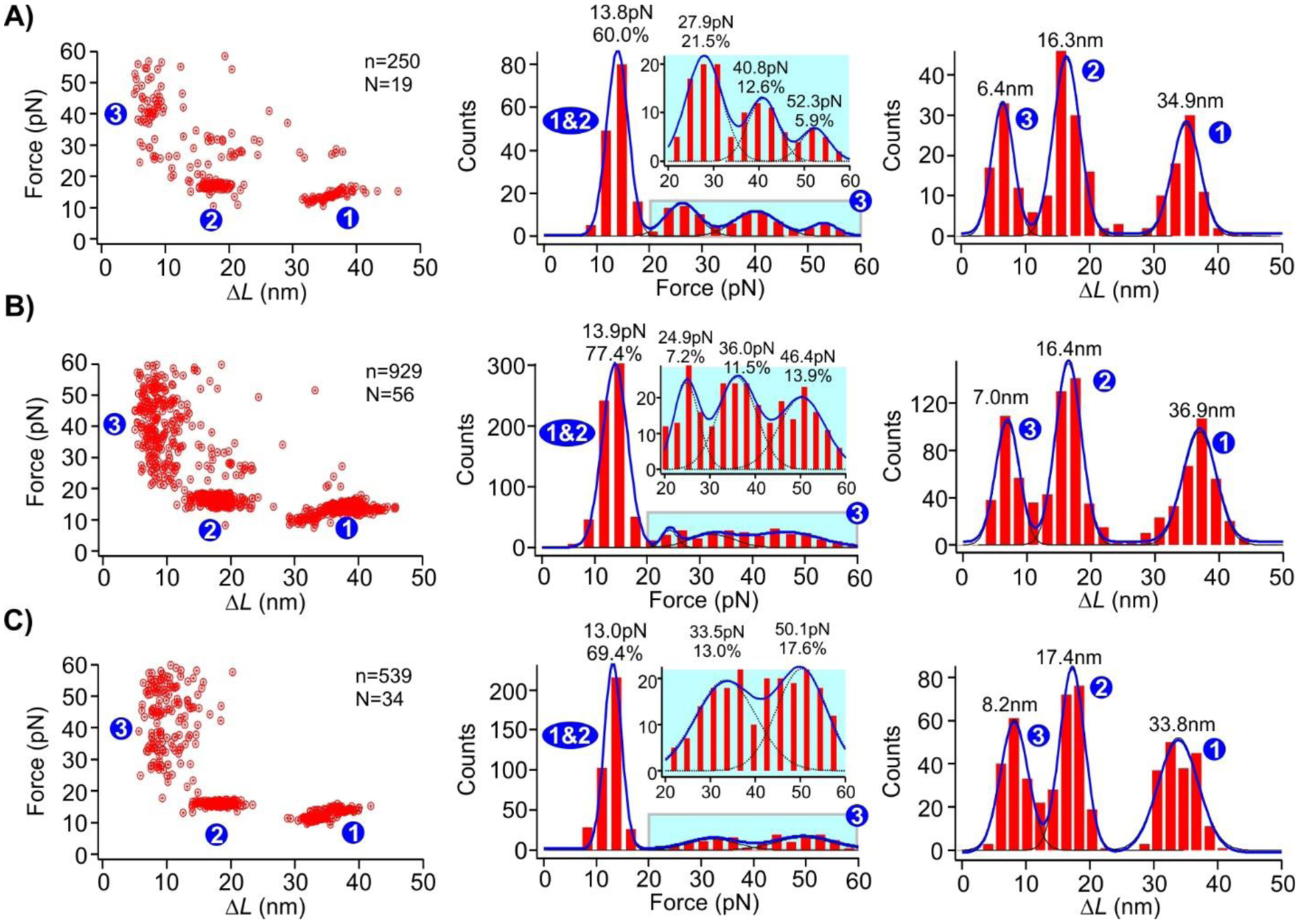
Force vs ΔL plot (left), unfolding force histograms (middle), and ΔL (right) histograms of the (dA)21 containing DNA constructs in the presence of AuNPs with diameters of A) 1.8 nm, B) 5.0 nm, and C) 10.0 nm. Dotted curves in the blown-up insets in the unfolding force histograms represent the gaussian fits for individual populations. Solid curves are Gaussian fittings for overall populations. N represents the number of molecules from which the FX curves were collected, and n represents the total number of unfolding features measured.
With the 5.0 nm AuNP (Figure 5B), the three unfolding species (24.9, 36.0, and 46.4 pN, average 39.5 pN), which respectively correspond to the 1, 2, and 3 anchoring deoxyadenosines on each side of the sticking-out loop (a total of 6 anchoring deoxyadenosines, see Figure S8), increase their populations (7.2%, 11.5%, and 13.9%, respectively). We attribute this to the bigger surface area of the 5 nm nanogold than the 1.8 nm AuNP, which makes it easier for all the anchoring deoxyadenosines to bind to the same AuNP facet (Figure S8), thereby increasing the mechanical stabilities of the adsorbed DNA as well as respective populations. Finally, the DNA adsorbed on the 10.0 nm AuNP showed only two populations (33.5 pN (13.0%) and 50.1 pN (17.6%), average 42.6 pN, Figure 5C). This is expected as a maximum of four anchoring deoxyadenosines is allowed in this case (see Figure S8 for calculation). This trend can be explained as larger facets in the 10 nm nanogold have increased opportunity to accommodate one (the 33.5 pN population) or two (the 50.1 pN species) anchoring deoxyadenosines at each side of the sticking-out loop on the same facet (Figure S8). Since the 33.5 pN population contains only one deoxyadenosine anchored at one side of the sticking-out loop, its force is lower than those of the 1.8 nm (40.8 pN) and 5.0 nm AuNP (36.0 pN), both of which have at least two anchoring deoxyadenosines at each side of the sticking-out loop.
We found that average mechanical stability of (dA)21 strands adsorbed on these three AuNPs is ~40 pN. This value is significantly higher than the stall force of DNA/RNA polymerases.37–39 Therefore, similar to the G-quadruplex that may serve as a mechanical block47,48 to DNA replication or RNA transcription processes, the AuNP-poly(dA) may also serve similar functions given there are plenty of poly(dA) segments in genomes of many species41–43. This potential function further expands the applicability of AuNP in biomedicines. This approach of determining the mechanical stability of DNA corona phase on nanoparticles can also be extended to other materials such as carbon nanotubes which are of growing interest in recent years.49 However, the use of optical tweezers also comes with the limitation that it cannot measure very high forces unlike AFM.34
Conclusions
In summary, we have found the mechanical stability of the DNA corona phase on gold nanospheres increased with the size of AuNP. The average desorption force of the DNA corona is 36.9 – 42.6 pN, which is lower than the covalent S-Au attachment, but higher than those to maintain DNA duplex or tetraplex structures inside cells, suggesting that the poly(dA) adsorption on AuNPs can withstand intracellular environment from mechanical perspective. The fact that the desorption force of poly(dA) from AuNP is higher than the stall force of motor proteins brings the possibility of using AuNP as a new agent to modulate various processes catalysed by motor proteins, which include replication and transcriptions. Given numerous applications of AuNPs inside cells as well as in materials explorations, our findings here provide convincing support that noncovalent DNA corona adsorption can serve as a mechanically competent approach to tailor functions of gold or other nanospheres.
Supplementary Material
Acknowledgements
HM thanks NSF (CBET1904921) and NIH (R01 CA236350) for the grant support.
Footnotes
The supporting information is available free of charge in the ACS publications website.
Materials, List of oligos, Preparation of dsDNA handles for the synthesis of single-molecule hairpin construct, Synthesis of single-molecule hairpin construct to study mechanical stability of poly(dA) and AuNPs, (dA)21 hairpin structure, Predicted structures of the DNA construct studied, Binding of gold nanospheres to the poly(dA) containing hairpin-internal-loop construct, UV/vis spectra of AuNP and poly(dA) mixture before and after freeze-thaw process, Three-channel microfluidic chamber, Mechanochemical study of poly(dA) and AuNP binding in optical tweezers, Calculation of the theoretical change-in-contour length (ΔL) of the construct during mechanical unfolding, Possible binding modes of the (dA)21 to the AuNPs with different sizes.
Conflicts of interest
The authors declare no conflicts of interest.
References
- (1).Giljohann DA; Seferos DS; Daniel WL; Massich MD; Patel PC; Mirkin CA Gold nanoparticles for biology and medicine Angewandte Chemie (International ed. in English) 2010, 49, 3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yeh Y-C; Creran B; Rotello VM Gold nanoparticles: preparation, properties, and applications in bionanotechnology Nanoscale 2012, 4, 1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dreaden EC; Alkilany AM; Huang X; Murphy CJ; El-Sayed MA The golden age: gold nanoparticles for biomedicine Chemical Society Reviews 2012, 41, 2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Tiwari PM; Vig K; Dennis VA; Singh SR Functionalized Gold Nanoparticles and Their Biomedical Applications Nanomaterials (Basel, Switzerland) 2011, 1, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Guo Y; Wang Z; Qu W; Shao H; Jiang X Colorimetric detection of mercury, lead and copper ions simultaneously using protein-functionalized gold nanoparticles Biosensors and Bioelectronics 2011, 26, 4064. [DOI] [PubMed] [Google Scholar]
- (6).Abad JM; Mertens SFL; Pita M; Fernández VM; Schiffrin DJ Functionalization of Thioctic Acid-Capped Gold Nanoparticles for Specific Immobilization of Histidine-Tagged Proteins Journal of the American Chemical Society 2005, 127, 5689. [DOI] [PubMed] [Google Scholar]
- (7).Zhang X; Servos MR; Liu J Instantaneous and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA Using a pH-Assisted and Surfactant-Free Route Journal of the American Chemical Society 2012, 134, 7266. [DOI] [PubMed] [Google Scholar]
- (8).Delong RK; Reynolds CM; Malcolm Y; Schaeffer A; Severs T; Wanekaya A Functionalized gold nanoparticles for the binding, stabilization, and delivery of therapeutic DNA, RNA, and other biological macromolecules Nanotechnology, science and applications 2010, 3, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ A DNA-based method for rationally assembling nanoparticles into macroscopic materials Nature 1996, 382, 607. [DOI] [PubMed] [Google Scholar]
- (10).Thomas KG; Kamat PV Chromophore-Functionalized Gold Nanoparticles Accounts of Chemical Research 2003, 36, 888. [DOI] [PubMed] [Google Scholar]
- (11).Gibson JD; Khanal BP; Zubarev ER Paclitaxel-Functionalized Gold Nanoparticles Journal of the American Chemical Society 2007, 129, 11653. [DOI] [PubMed] [Google Scholar]
- (12).Liu B; Liu J Methods for preparing DNA-functionalized gold nanoparticles, a key reagent of bioanalytical chemistry Analytical Methods 2017, 9, 2633. [Google Scholar]
- (13).Hurst SJ; Lytton-Jean AKR; Mirkin CA Maximizing DNA loading on a range of gold nanoparticle sizes Analytical chemistry 2006, 78, 8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Storhoff JJ; Elghanian R; Mucic RC; Mirkin CA; Letsinger RL One-Pot Colorimetric Differentiation of Polynucleotides with Single Base Imperfections Using Gold Nanoparticle Probes Journal of the American Chemical Society 1998, 120, 1959. [Google Scholar]
- (15).Elghanian R; Storhoff James J; Mucic Robert C; Letsinger Robert L; Mirkin Chad A Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles Science 1997, 277, 1078. [DOI] [PubMed] [Google Scholar]
- (16).Li H; Rothberg L Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles Proceedings of the National Academy of Sciences 2004, 101, 14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Pei H; Li F; Wan Y; Wei M; Liu H; Su Y; Chen N; Huang Q; Fan C Designed Diblock Oligonucleotide for the Synthesis of Spatially Isolated and Highly Hybridizable Functionalization of DNA–Gold Nanoparticle Nanoconjugates Journal of the American Chemical Society 2012, 134, 11876. [DOI] [PubMed] [Google Scholar]
- (18).Opdahl A; Petrovykh Dmitri Y; Kimura-Suda H; Tarlov Michael J; Whitman Lloyd J Independent control of grafting density and conformation of single-stranded DNA brushes Proceedings of the National Academy of Sciences 2007, 104, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhu D; Song P; Shen J; Su S; Chao J; Aldalbahi A; Zhou Z; Song S; Fan C; Zuo X; Tian Y; Wang L; Pei H PolyA-Mediated DNA Assembly on Gold Nanoparticles for Thermodynamically Favorable and Rapid Hybridization Analysis Analytical chemistry 2016, 88, 4949. [DOI] [PubMed] [Google Scholar]
- (20).Koo KM; Sina AAI; Carrascosa LG; Shiddiky MJA; Trau M DNA–bare gold affinity interactions: mechanism and applications in biosensing Analytical Methods 2015, 7, 7042. [Google Scholar]
- (21).Zhang X; Liu B; Dave N; Servos MR; Liu J Instantaneous Attachment of an Ultrahigh Density of Nonthiolated DNA to Gold Nanoparticles and Its Applications Langmuir 2012, 28, 17053. [DOI] [PubMed] [Google Scholar]
- (22).Hu M; Yuan C; Tian T; Wang X; Sun J; Xiong E; Zhou X Single-Step, Salt-Aging-Free, and Thiol-Free Freezing Construction of AuNP-Based Bioprobes for Advancing CRISPR-Based Diagnostics Journal of the American Chemical Society 2020, 142, 7506. [DOI] [PubMed] [Google Scholar]
- (23).Li F; Zhang H; Dever B; Li X-F; Le XC Thermal Stability of DNA Functionalized Gold Nanoparticles Bioconjugate Chemistry 2013, 24, 1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhou J; Ralston J; Sedev R; Beattie DA Functionalized gold nanoparticles: Synthesis, structure and colloid stability Journal of Colloid and Interface Science 2009, 331, 251. [DOI] [PubMed] [Google Scholar]
- (25).Kang H; Buchman JT; Rodriguez RS; Ring HL; He J; Bantz KC; Haynes CL Stabilization of Silver and Gold Nanoparticles: Preservation and Improvement of Plasmonic Functionalities Chemical Reviews 2019, 119, 664. [DOI] [PubMed] [Google Scholar]
- (26).Gao J; Huang X; Liu H; Zan F; Ren J Colloidal Stability of Gold Nanoparticles Modified with Thiol Compounds: Bioconjugation and Application in Cancer Cell Imaging Langmuir 2012, 28, 4464. [DOI] [PubMed] [Google Scholar]
- (27).Mei BC; Oh E; Susumu K; Farrell D; Mountziaris TJ; Mattoussi H Effects of Ligand Coordination Number and Surface Curvature on the Stability of Gold Nanoparticles in Aqueous Solutions Langmuir 2009, 25, 10604. [DOI] [PubMed] [Google Scholar]
- (28).Grandbois M; Beyer M; Rief M; Clausen-Schaumann H; Gaub Hermann E How Strong Is a Covalent Bond? Science 1999, 283, 1727. [DOI] [PubMed] [Google Scholar]
- (29).Huang; Chen F; Bennett PA; Tao Single Molecule Junctions Formed via Au−Thiol Contact: Stability and Breakdown Mechanism Journal of the American Chemical Society 2007, 129, 13225. [DOI] [PubMed] [Google Scholar]
- (30).Xue Y; Li X; Li H; Zhang W Quantifying thiol–gold interactions towards the efficient strength control Nature Communications 2014, 5, 4348. [DOI] [PubMed] [Google Scholar]
- (31).Wei W; Sun Y; Zhu M; Liu X; Sun P; Wang F; Gui Q; Meng W; Cao Y; Zhao J Structural Insights and the Surprisingly Low Mechanical Stability of the Au–S Bond in the Gold-Specific Protein GolB Journal of the American Chemical Society 2015, 137, 15358. [DOI] [PubMed] [Google Scholar]
- (32).Chen X; Wang Y; Dai X; Ding L; Chen J; Yao G; Liu X; Luo S; Shi J; Wang L; Nechushtai R; Pikarsky E; Willner I; Fan C; Li J Single-Stranded DNA-Encoded Gold Nanoparticle Clusters as Programmable Enzyme Equivalents Journal of the American Chemical Society 2022, 144, 6311. [DOI] [PubMed] [Google Scholar]
- (33).Wang S-T; Zhang H; Xuan S; Nykypanchuk D; Zhang Y; Freychet G; Ocko BM; Zuckermann RN; Todorova N; Gang O Compact Peptoid Molecular Brushes for Nanoparticle Stabilization Journal of the American Chemical Society 2022, 144, 8138. [DOI] [PubMed] [Google Scholar]
- (34).Neuman KC; Nagy A Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy Nat Meth 2008, 5, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Woodside Michael T; Behnke-Parks William M; Larizadeh K; Travers K; Herschlag D; Block Steven M Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins Proceedings of the National Academy of Sciences 2006, 103, 6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Koirala D; Dhakal S; Ashbridge B; Sannohe Y; Rodriguez R; Sugiyama H; Balasubramanian S; Mao H A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands Nature Chemistry 2011, 3, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Galburt EA; Grill SW; Wiedmann A; Lubkowska L; Choy J; Nogales E; Kashlev M; Bustamante C Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner Nature 2007, 446, 820. [DOI] [PubMed] [Google Scholar]
- (38).Wang Michelle D; Schnitzer Mark J; Yin H; Landick R; Gelles J; Block Steven M Force and Velocity Measured for Single Molecules of RNA Polymerase Science 1998, 282, 902. [DOI] [PubMed] [Google Scholar]
- (39).Maier B; Bensimon D; Croquette V Replication by a single DNA polymerase of a stretched single-stranded DNA Proceedings of the National Academy of Sciences 2000, 97, 12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Huppert JL; Balasubramanian S Prevalence of quadruplexes in the human genome Nucleic Acids Research 2005, 33, 2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lustig AJ; Petes TD Long poly(A) tracts in the human genome are associated with the Alu family of repeated elements Journal of Molecular Biology 1984, 180, 753. [DOI] [PubMed] [Google Scholar]
- (42).Shenkin A; Burdon RH Deoxyadenylate-rich and deoxyguanylate-rich regions in mammalian DNA Journal of Molecular Biology 1974, 85, 19. [DOI] [PubMed] [Google Scholar]
- (43).Tubbs A; Sridharan S; van Wietmarschen N; Maman Y; Callen E; Stanlie A; Wu W; Wu X; Day A; Wong N; Yin M; Canela A; Fu H; Redon C; Pruitt SC; Jaszczyszyn Y; Aladjem MI; Aplan PD; Hyrien O; Nussenzweig A Dual Roles of Poly(dA:dT) Tracts in Replication Initiation and Fork Collapse Cell 2018, 174, 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Mandal S; Zhang X; Pandey S; Mao H Single-Molecule Topochemical Analyses for Large-Scale Multiplexing Tasks Analytical Chemistry 2019, 91, 13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).von Hippel PH; Johnson NP; Marcus AH Fifty years of DNA “Breathing”: Reflections on old and new approaches Biopolymers 2013, 99, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Yin J; Wang J; Yang X; Wu T; Wang H; Zhou X Poly(adenine)-mediated DNA-functionalized gold nanoparticles for sensitive detection of mercury ions in aqueous media RSC Advances 2019, 9, 18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Yu Z; Schonhoft JD; Dhakal S; Bajracharya R; Hegde R; Basu S; Mao H ILPR G-Quadruplexes Formed in Seconds Demonstrate High Mechanical Stabilities Journal of the American Chemical Society 2009, 131, 1876. [DOI] [PubMed] [Google Scholar]
- (48).Mandal S; Kawamoto Y; Yue Z; Hashiya K; Cui Y; Bando T; Pandey S; Hoque ME; Hossain MA; Sugiyama H; Mao H Submolecular dissection reveals strong and specific binding of polyamide–pyridostatin conjugates to human telomere interface Nucleic Acids Research 2019, 47, 3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Cheung W; Pontoriero F; Taratula O; Chen AM; He H DNA and carbon nanotubes as medicine Advanced Drug Delivery Reviews 2010, 62, 633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


