Abstract
Background
It remains unknown why ~30% of patients with psychotic disorders fail to respond to treatment. Previous genomic investigations of treatment-resistant psychosis have been inconclusive, but some evidence suggests a possible link between rare disease-associated copy number variants (CNVs) and worse clinical outcomes in schizophrenia. Here, we identified schizophrenia-associated CNVs in patients with treatment-resistant psychotic symptoms and then compared the prevalence of these CNVs to previously published schizophrenia cases not selected for treatment resistance.
Methods
CNVs were identified using chromosomal microarray (CMA) and whole exome sequencing (WES) in 509 patients with treatment-resistant psychosis (a lack of clinical response to ≥3 adequate antipsychotic medication trials over at least 5 years of psychiatric hospitalization). Prevalence of schizophrenia-associated CNVs in this sample was compared to that in a previously published large schizophrenia cohort study.
Results
Integrating CMA and WES data, we identified 47 cases (9.2%) with at least one CNV of known or possible neuropsychiatric risk. 4.7% (n = 24) carried a known neurodevelopmental risk CNV. The prevalence of well-replicated schizophrenia-associated CNVs was 4.1%, with duplications of the 16p11.2 and 15q11.2-q13.1 regions, and deletions of the 22q11.2 chromosomal region as the most frequent CNVs. Pairwise loci-based analysis identified duplications of 15q11.2-q13.1 to be independently associated with treatment resistance.
Conclusions
These findings suggest that CNVs may uniquely impact clinical phenotypes beyond increasing risk for schizophrenia and may potentially serve as biological entry points for studying treatment resistance. Further investigation will be necessary to elucidate the spectrum of phenotypic characteristics observed in adult psychiatric patients with disease-associated CNVs.
Keywords: treatment-resistant psychosis, copy number variants, ultra-TRS, 16p11.2 duplication, 15q11.2-q13.1 duplication, schizophrenia, neurodevelopmental disorders, rare variants
Introduction
Approximately one-third of individuals diagnosed with a psychotic disorder experience treatment-resistant psychotic symptoms (TRS; commonly defined as “treatment-resistant schizophrenia”).1,2 Here we consider treatment resistance across the spectrum of psychotic disorders. These patients have chronic severe psychotic symptoms and marked impairments in social-community functioning.3 They experience higher rates of suicide and greater cognitive deficits than treatment-responsive patients.4,5 Apart from the patients’ suffering, direct healthcare costs for TRS in the United States are 3-11x higher than for the population of patients with schizophrenia as a whole, adding an estimated direct care cost of $32 billion annually to the national mental healthcare budget.6
Numerous studies have attempted to identify clinical predictors of TRS, with limited success.7 Recent studies have focused on potential genetic risk factors for treatment resistance, including the role of common risk alleles8–12 and genome-wide burden of copy number variants (CNVs).8,11 These metrics contribute to the overall genetic liability for schizophrenia13,14 and may have unique contributions with regard to differences in treatment response, but so far do not provide predictive power and thus clinical utility for treating TRS.11,12
Apart from common variants that individually contribute minimal disease-risk, other sources of genetic risk are rare, highly penetrant, and/or deleterious variants.15 Recent studies of TRS have indicated an increased burden of rare, damaging variants in mutation-intolerant genes,16 and in gene sets implicated in Mendelian diseases or specific biochemical pathways, such as neurotransmission and gene targets of antipsychotic drugs (APDs).17–19 Focusing on highly penetrant variants can also identify rare recurrent CNVs that may impact loci associated with clinical phenotypes (ie, disease-associated CNVs). Disease-associated CNVs have large phenotypic effects and increase risk for neurodevelopmental and psychiatric outcomes by several-fold,20,21 but cumulatively effect only a small percentage (~2%) of patients with schizophrenia.15,20,22
Legge et al.10 found no apparent excess in the prevalence of disease-associated CNVs, or of large CNVs (>500 kb or >1 Mb) in patients having TRS compared to general schizophrenia cohorts. However, their study, and others,9,11,17 used surrogate measures such as clozapine prescriptions or antipsychotic polypharmacy to define TRS without measures of clinical outcomes. While these studies make excellent use of large deidentified datasets, a major limitation is the inability to clinically confirm treatment resistance in participants, adding potential heterogeneity to the analysis. Another study,23 in which TRS was determined by examining clinical improvement in social/occupational functioning, found a significantly higher rate of treatment resistance (OR = 2.79) among CNV carriers compared to non-carriers.
We sought to: (1) identify rare CNVs with potential clinical relevance in a sample of 509 patients with clinically confirmed TRS, and (2) compare the prevalence of schizophrenia-associated CNVs (SCZ CNVs) in this sample with a previously published schizophrenia cohort not selected for treatment resistance. If specific CNVs increase the risk for TRS, a thorough investigation may reveal clues to the biological mechanisms underlying treatment resistance. Understanding the impact of CNVs across the lifespan, including aspects of treatment response, will also become increasingly important as more children with genetic diagnoses enter adult psychiatric services.
Methods
Subject Recruitment and Screening
Participants were recruited between April 2015 and August 2019 from 5 Pennsylvania state hospitals (PASHs) and their affiliated long-term structured residence (LTSR) facilities. LTSRs are smaller, locked facilities with 24-hour staffing and medication management allowing for “deinstitutionalization” of patients back into the community, but do not necessarily indicate clinical improvement. The study was approved by the Institutional Review Boards at the Drexel University College of Medicine and the University of North Carolina at Chapel Hill, the Chief Medical Officer of the Commonwealth of Pennsylvania, Department of Public Welfare, and individual recruitment sites. Research was carried out in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants (or legal guardian with participant assent).
Subject selection was based on information derived from direct patient interviews and medical records. Inclusion criteria were: (1) ability and willingness to give informed consent (or written consent of a legal guardian with subject assent); (2) age ≥18 years; (3) a current diagnosis of schizophrenia, schizoaffective disorder, bipolar I disorder with psychotic features, major depressive disorder with psychotic features, or psychotic disorder NOS as documented in medical records; (4) continuous psychiatric hospitalization for ≥5 years; and (5) lack of clinical improvement despite ≥3 APD trials of adequate dose and duration. Exclusion criteria were: (1) a psychotic disorder associated with substance dependence; (2) a medical condition known to cause psychosis; (3) any instance of sustained treatment response. No study participants were prisoners or on forensic units at the time of enrollment. A senior psychiatrist not associated with the study (T.S., see acknowledgments) and one of the senior authors (R.C.J.) reviewed cases to confirm diagnostic inclusion/exclusion criteria (see supplementary material for additional details).
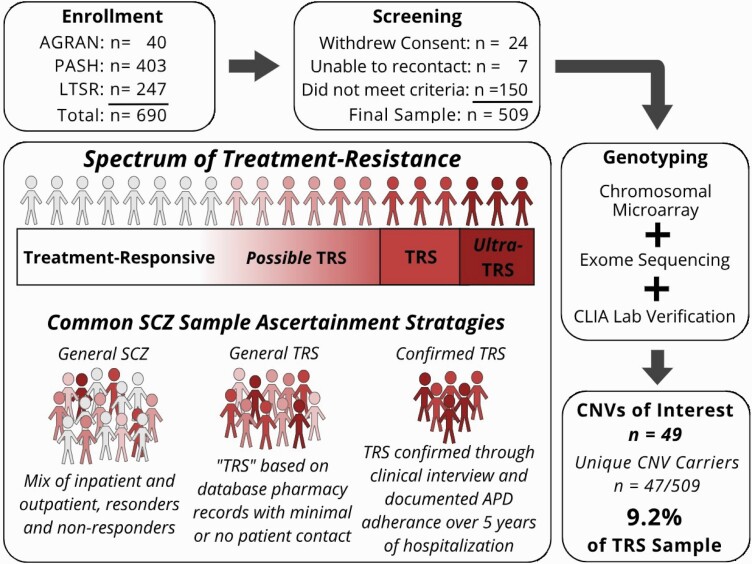
Structured interviews were conducted with subjects willing and able to participate (see table 1). The Ammons Quick Test24 was used to estimate intelligence, and the Positive and Negative Syndrome Scale (PANSS)25 was administered to document the presence and severity of current psychotic symptoms. Demographic and phenotypic data were extracted from medical records and entered into a secure electronic data capture platform (REDCap).26,27Figure 1 provides an overview of subject selection. From 690 initial participants, 509 were included in the final sample.
Table 1.
Sociodemographic and Clinical Characteristics of the TRS Sample and CNV Carriers
| Characteristic | Entire Sample (n = 509) | CNV Carriers (n = 47) | ||||
|---|---|---|---|---|---|---|
| N | % | N | % | |||
| Sex | ||||||
| Male | 334 | 65.6 | 32 | 68.1 | ||
| Female | 175 | 34.4 | 15 | 31.9 | ||
| Ancestry | ||||||
| Hispanic | 11 | 2.2 | 1 | 2.1 | ||
| White | 381 | 74.9 | 42 | 89.4 | ||
| Black | 98 | 19.3 | 4 | 8.5 | ||
| Native American | 1 | 0.2 | — | — | ||
| Asian/Pacific Islander | 6 | 1.2 | — | — | ||
| Mixed ancestry | 12 | 2.4 | — | — | ||
| Primary chart DSM-IV diagnosisa | ||||||
| Schizophrenia (all subtypes) | 238 | 46.8 | 23 | 48.9 | ||
| Schizophrenia, disorganized | 14 | — | 1 | — | ||
| Schizophrenia, paranoid | 119 | — | 12 | — | ||
| Schizophrenia, undifferentiated | 82 | — | 6 | — | ||
| Schizophrenia, unspecified | 23 | — | 4 | — | ||
| Psychotic disorder NOS | 14 | 2.7 | 3 | 6.4 | ||
| Schizoaffective disorder | 236 | 46.3 | 17 | 36.2 | ||
| Bipolar disorder I with psychosis | 15 | 2.9 | 3 | 6.4 | ||
| Major depression with psychosis | 6 | 1.2 | 1 | 2.1 | ||
| Clozapine exposure | 259 | 50.9 | 25 | 53.2 | ||
| Adequate clozapine trial | 175 | 34.3 | 14 | 29.8 | ||
| N | Mean | SD | N | Mean | SD | |
|---|---|---|---|---|---|---|
| Age (years)b | 509 | 53.6 | 13.2 | 47 | 54.5 | 13.1 |
| Age at illness onset (years)c | 479 | 21.2 | 8.7 | 45 | 22.6 | 11.7 |
| Duration of illness (years)a | 479 | 32.4 | 12.6 | 45 | 31.7 | 12.8 |
| Duration of current hospitalization (years) | 509 | 13.4 | 10.1 | 47 | 15.8 | 11.7 |
| Total number of hospitalizationsa | 509 | 8.1 | 8.0 | 47 | 8.4 | 9.0 |
| Average GAF scorea, d | 472 | 31.4 | 5.3 | 46 | 30.8 | 3.4 |
| Most current GAF | 484 | 32.3 | 6.3 | 45 | 31.1 | 5.4 |
| Modified PANSS scores | ||||||
| Positive symptom score | 363 | 19.1 | 5.7 | 33 | 21.0 | 4.8 |
| Negative symptom score | 363 | 17.0 | 6.7 | 33 | 16.2 | 5.5 |
| Ammons quick test | 341 | 86.5 | 15.5 | 27 | 88.1 | 14.4 |
Note: CNV, copy number variant; PANSS, positive and negative syndrome scale.
aBased initially on medical records;
bage at time of blood collection;
cage at onset of psychotic symptoms (hallucinations or delusions);
dGlobal Assessment of Functioning Scale—scores range from 1 (severe impairment) to 100 (extremely high functioning).
Fig. 1.
Spectrum of treatment resistance and study workflow. Ultra-TRS describes those who failed to respond to clozapine treatment; AGRAN, samples taken from a previous study of agranulocytosis in patients with TRS28. Note: PASH, Pennsylvania State Hospital; LTSR, long term structured residence; CLIA, clinical laboratory improvement amendment; SCZ, schizophrenia.
Genomic Analysis
DNA was extracted from a blood sample and genome-wide SNP genotypes obtained using Illumina Infinium Global Screening Array (v2.0+MD+Psych) per standard protocols (Life & Brain GmbH, Bonn, Germany). CNVs were called from array intensity data using PennCNV,29 QuantiSNP,30 and iPattern,31 as packaged by EnsembleCNV.32 CNV calling was performed for chromosomes 1–22 and chromosome X. Individual samples were removed (n = 17) due to outliers in quality control (QC) metrics (standard deviations [SD] of log R ratio [LRR], SD of B allele frequency [BAF], wave factor in LRR, BAF drift, and/or the number of CNVs detected). Additional QC steps were then followed to remove low confidence CNV calls spanning <10 probes, confidence score <10 for PennCNV, Log Bayes Factor <10 for QuantiSNP, score <1 or complex CNVs for iPattern, or with >50% reciprocal overlap with large genomic gaps (eg, centromeres) or regions subject to rearrangement in white blood cells. We annealed adjoining CNVs that appeared to be artificially split by the CNV calling algorithm by recursively joining CNVs if the called region is ≥75% of the entire region to be joined and then removed CNVs with <100 kb length. CNVs detected by two of the three calling algorithms were intersected by requiring the same subject, same CNV type, and >50% reciprocal overlap. We excluded CNVs detected by only one algorithm and merged the remaining concordant CNVs using outer probe boundaries. This improved specificity (detected by ≥2 algorithms) as well as sensitivity (combining all concordant calls among three complementary algorithms). We imposed a 0.01 frequency threshold by removing CNVs with >50% of its length spanning a region in >1% of cases, thus retaining only large rare CNVs. A subset of CNV calls were confirmed using a clinical-grade Agilent comparative genome hybridization array in a CLIA-certified lab (Allele Diagnostics, Spokane, WA).
Exome sequencing data were available for 478 subjects and used as a second approach for CNV identification, and as the primary source for CNV calling in cases excluded from SNP-based CNV analysis. Exome sequencing was performed using Agilent SureSelectXT Clinical Research Exome capture and was sequenced using paired-end 150 bp reads on Illumina NovaSeq (GeneByGene, Houston, Texas, USA). Sequences were aligned to hg19, variants called using the GATK pipeline33 and annotated using Variant Effect Predictor.34 CNVs were called using XHMM35 and CN-Learn.36 For the training input to CN-Learn, CLIA-confirmed CNVs from the current study (n = 21) along with other high confidence CNVs called from the Global Screening Array (n = 10) were used (recommended minimum number of training CNVs for CN-Learn is 31). See supplementary material for CNV calling workflow.
Called CNVs were cross-referenced with a list of CNVs associated with neurodevelopmental disorders (NDD CNVs; see supplementary table S2) based on large case-control studies, including 12 SCZ CNVs.15,37,38 Large (>1 Mb) CNVs that do not overlap with known NDD CNV loci are also considered to have potential clinical value due to their size, and are reported here.39 Three large CNVs (>1 Mb) were considered incidental findings and not included in the reported prevalence as they are unlikely to be relevant to psychiatric disorders (a mosaic copy loss of the Y chromosome, a 17p12 deletion associated with Hereditary Neuropathy, and a 16p11.2-p11.1 duplication in a region non-sensitive to CNV [hg19 chr16:34,197,143-35,257,261; https://dosage.clinicalgenome.org/). Our methods integrated array data (CMA), exome sequencing data (WES), and independent CLIA lab verification as outlined above. The SCZ CNVs identified from exome sequencing data and “missed” in our array-based CNV call set were 4 (of the 6) 16p11.2 duplications (see table 2). The raw signal plots for all six 16p11.2 duplications are shown in supplementary figure S2.
Table 2.
CNVs Associated With Neurodevelopmental/Psychosis Phenotypes
| SCZ CNVs | Canonical CNV Coordinates (Size in kb) | Cases (N) | Case CNV Coordinates (Size in kb) | Source | Prevalence (%) | P valuea | ||
|---|---|---|---|---|---|---|---|---|
| TRS (n = 509) | SCZ15 (n = 21094) | Uncorrected | Adjusted for FDR | |||||
| 3q29 Del | chr3:195.72–197.35 (1635) | 1 | chr3:195.8–197.1 (1302) | CMA, CLIA | 0.20 | 0.076 | .324 | .714 |
| 7q11.23 Dup | chr7:72.74–74.14 (1398) | 1 | chr7:72.74–74.14 (1395) | CMA | 0.20 | 0.062 | .276 | .714 |
| 15q11.2 Del | chr15:22.81–23.09 (298) | 3 | Cases 1–3: chr15:22.82–23.09 (263) | CMA, CLIA | 0.59 | 0.45 | .491 | .900 |
| 15q11.2-13.1 Dup | chr15:22.81–28.39 (5585) | 4 | Case 1: chr15:23.72–28.51 (4796) | CMA, CLIA | 0.78 | 0.071 | .000787 | .00866* |
| Cases 2–3: chr15:22.82–28.51 (5691) | CMA, CLIA | |||||||
| Case 4: chr15:22.82–29.57 (6750) | CMA, CLIA | |||||||
| 16p11.2 (distal) Del | chr16:28.82–29.05 (224) | 1 | chr16:28.49–29.05 (558) | CMA, CLIA | 0.20 | 0.052 | .242 | .714 |
| 16p11.2 (proximal) Dup | chr16:29.65–30.20 (550) | 6 | Cases 1–4: chr16:29.66–30.19 (535) | WES, CLIA | 1.18 | 0.30 | .661 | 1.00 |
| Cases 5–6: chr16:29.66–30.19 (535) | CMA, CLIA | |||||||
| 16p13.11 Dup | chr16:15.51–16.29 (782) | 1 | chr16:15.12–16.29 (1162) | CLIA | 0.20 | 0.40b | ||
| 22q11.21 Del | chr22:19.04–21.47 (2429) | 4 | Cases 1–4: chr22:18.92–21.46 (2541) | CMA, CLIA | 0.78 | 0.28 | .0529 | .291 |
| NDD CNVs | ||||||||
| 1q21.1 (proximal TAR region) Dup | chr1:145.39–145.81 (413) | 1 | chr1:145.4–145.8 (412) | WES | ||||
| 15q13.3 Dup | chr15:30.94–32.52 (1,572) | 1 | chr15:31.16–32.44 (1280) | CMA, CLIA | ||||
| 16p12.2 Del | chr16:21.95–22.43 (482) | 1 | chr16:21.95–22.43 (478) | CMA, CLIA | ||||
| Total case count (% of TRS sample, n = 509) | 24 (4.7%) | 4.13 | 1.95c | |||||
Note: SCZ, schizophrenia; kb, kilobase pair; TRS, treatment resistant psychotic symptoms; FDR, false discovery rate; Del, deletion; CMA, chromosomal microarray; CLIA, Clinical Laboratory Improvement Amendment (These CNVs were clinically verified in an independent CLIA certified lab); Dup, duplication; WES, whole exome sequencing.
CNVs overlapping >50% with the canonical coordinates of a known neurodevelopmental/psychiatric CNV loci. All coordinates aligned to hg19. For CNVs called from multiple sources (CMA, WES, and CLIA lab), the shown coordinates are those provided in the CLIA certified lab report. *Statistically significant associations with adjusted P-value < .05.
aStatistical tests are based on the rate of CNVs identified using CMA in this TRS sample (N = 492 with CMA data after QC). Four of the 16p11.2 duplications identified from WES were not included. These rates are different than what is shown in the prevalence column, which considers CNVs called from all sources, and from our entire sample (N = 509).
bThis prevalence reflects both duplications and deletions of 16p13.11 region and thus this CNV is not included in loci-based association tests.
cTotal prevalence of SCZ CNVs counting only those present in this TRS sample. For statistical analysis of cumulative SCZ CNV rate, all 12 SCZ CNV loci shown in supplementary table 2 are counted, including both deletions and duplication of 16p13.11 region, and the final prevalence in SCZ was 2.18% (460/21094; Marshall et al.15).
Statistical Analysis
To test whether SCZ CNVs, in aggregate, increase risk for TRS, we compared the prevalence of SCZ CNVs in this sample to previously published data from a large schizophrenia sample (21 094 subjects not selected for treatment resistance, in which CNVs were called using CMA)15 using a chi-squared test of independence. The combined prevalence of SCZ CNVs was determined across genotyping methods (CMA + WES + CLIA; n = 509 cases genotyped using one or more methods), as well as for CNVs identified using only CMA (n = 492 cases after QC). These same data were used for loci-based comparisons of SCZ CNVs using 2-sided Fisher exact tests, with P-values adjusted to correct for multiple testing using Benjamini–Hochberg false discovery rate (BHFDR; adjusted P-values <.05 were considered statistically significant). To control against sensitivity differences from our combined use of genotyping technologies, only CNVs identified using CMA were included in loci-based tests. All SCZ CNVs for which data were available in Marshall et al.15 (CNV probe-level results) were included in loci-based analyses (n = 11; the 16p13.11 duplication was excluded from this analysis because Marshall et al.15 provided only the combined prevalence of deletions and duplications in this region). All statistical analyses were performed using R Statistical Software (v3.6.3)40 and the epitools R package (v0.5-10.1).41
Results
Demographic data are displayed in table 1. Of 509 participants, 47 (9.2%) carried at least one CNV with potential relevance to their clinical presentation, which were grouped as follows: (1) 24 patients (4.7% of the sample) carried one of the neurodevelopmental risk CNVs (NDD CNVs) when they overlapped by more than 50% with loci previously associated with neurodevelopmental and/or psychotic disorders (see supplementary table 2); (2) 11 patients carried large CNVs (>1 Mb) which did not overlap with NDD/SCZ CNV loci, but nonetheless may impact clinical presentation39; and (3) 12 cases carried variants of uncertain significance (VUS), including CNVs partially overlapping (<50% coverage) with known NDD CNV loci and duplications of regions where only deletions are associated with disease risk.
We examined demographic differences between CNV carriers and non-carriers across all variables listed in table 1; Positive symptom scores as measured by PANSS were significantly different between groups t(361) = 1.996; uncorrected (P = .046), with CNV carriers (M = 20.97; SD = 4.825) having higher average scores than non-carriers (M = 18.91; SD = 5.729).
Known Risk for Neurodevelopmental/Psychotic Phenotypes
Of 24 cases with NDD CNVs, 21 cases were carriers of CNVs previously implicated in schizophrenia (see table 2). The combined prevalence of SCZ CNVs identified (integrating CMA + WES + CLIA) in this TRS sample (4.1%) represents an approximately 2-fold statistically significant increase (χ2 [1, N = 21 603] = 7.77, P = .005) compared to a previously published schizophrenia cohort (2.2%).15 However, when limited to only CNVs identified through CMA (n = 16 out of 492 TRS cases with CMA data), this difference was no longer statistically significant (χ2 [1, N = 21 586] = 2.0861, P = .148).
We then examined differences in the prevalence of each SCZ CNV against that observed in Marshall et al.15 (see table 2). There was a statistically significant differences in prevalence rate of duplications at 15q11.2-q13.1 reflecting a greater than 11-fold increase in the prevalence of this CNV (n = 4 out of 492 TRS cases with CMA data, 0.81%; corrected P = .0087; OR = 11.5; 95% CI = 3.81–34.8). There was an approximately 4-fold increase in the rate of 16p11.2 duplications when considering both CMA and WES data (n = 6 TRS cases; OR = 3.98, 95% CI = 1.72–9.24, see table 2); however, there is no difference in the rate of this CNV considering only cases identified using CMA (n = 2; corrected P = 1.00).
Additional Copy Number Variant Findings
As shown in table 3, 11 participants (2.2% of the total sample) carried large CNVs (>1 Mb) in regions not previously associated with NDD/SCZ phenotypes in case-control studies (one case carried two large CNVs). For most of these loci, case reports describe neurodevelopmental phenotypes in individuals with overlapping CNVs (see supplementary table S3). Many of these loci involve genes highly expressed in the brain or previously implicated in the central nervous system or neurodevelopmental disorders (table 3).
Table 3.
Large (>1 Mb) CNVs Not Implicated in Neurodevelopmental Phenotypes in Case-Control Studies
| CNV | Cases (N) | Genomic Coordinates (Size) | Gene Count (Brain Expressed Genes)a | Source | CLIA Lab Annotationb |
|---|---|---|---|---|---|
| 2p12-p11.2 Del | 1 | chr2:75.99–84.39 (8.4 Mb) | 7 (CTNNA2, LRRTM1, LRRTM4) | CMA | — |
| 4q28.3 Dup | 1 | chr4:131.71–133.77 (2.1 Mb) | — | CMA | — |
| 4q33 Dup | 1 | chr4:171.04–172.04 (1.0 Mb) | — | CMA | — |
| 5p14.3 Del | 1 | chr5:20.26–23.03 (2.8 Mb) | 2 (CDH12, CDH18) | CMA | — |
| 5q23.2 Del | 1 | chr5:125.14–126.33 (1.2 Mb) | 6 (ALDH7A1, GRAMD3, LMNB1, MARCHF3) | CMA, CLIA | Unclear clinical significance |
| 6q22.31 Dup | 1 | chr6:117.99–120.65 (2.7 Mb) | 8 (CEP85L, FAM184A, MAN1A1, MCM9, NUS1, SLC35F1) | CMA, WES | — |
| 7q21.13 Dup | 2c | chr7:88.17–90.17 (2.0 Mb) | 9 (CDK14, CFAP69, FAM237B, STEAP2, ZNF804B) | CMA | — |
| 9q33.1 Dup | 1 | chr9:119.92–122.03 (2.1 Mb) | 3 (ASTN2, BRINP1, TLR4) | CMA | — |
| 13q33.1-q34 Dup | 1 | chr13:104.45–115.11 (10.7 Mb) | 44 (ARHGEF7, ATP11A, CARS2, CFAP97D2, CHAMP1, COL4A1, COL4A2, DCUN1D2, EFNB2, GAS6, GRK1, GRTP1, LIG4, MCF2L, MYO16, NALF1, NAXD, RASA3, SOX1, TEX29, TMCO3, TUBGCP3) | CMA, WES | — |
| 17q25.3 Dup | 1 | chr17:76.41–78.94 (2.5 Mb) | 26 (CANT1, CCDC40, CEP295NL, DNAH17, EIF4A3, ENDOV, GAA, NPTX1, RBFOX3, RNF213, RPTOR, SGSH, SLC26A11, TBC1D16) | WES | — |
| 18p11.31-p11.23 Dup | 1 | chr18:6.84–8.05 (1.2 Mb) | 4 (LAMA1, PTPRM) | CMA, WES | — |
| Total case count (% of TRS sample, n = 509) | 11 (2.2%) |
Note: Mb, megabase
Genomic coordinates aligned to hg19.
aTotal count of protein coding genes in CNV region from NCBI RefSeq Database,42 displaying genes with brain tissue or cell type expression enrichment from proteinatlas.org43 and/or involvement in CNS or developmentally related disease (bolded) based on OMIM and ClinVar data.
bInterpretation of the clinical significance of CNVs provided by Allele Diagnostics based on review of available literature at the time of CNV call.
cOne carrier of the 7q21.13 duplications is also the carrier of the 2p12p11.2 deletion.
Finally, 12 participants (2.4%) carried one or more VUS CNVs which do not fit the above categories but nevertheless may influence phenotypic expression in carriers (see table 4). Seven carried duplications in regions where deletions are known to be associated with NND phenotypes, including 2q37, 15q11.2, 16p12.2, and distal 22q11.2. Six carried CNVs which overlapped with known NDD CNVs by less than 50%, including deletions of 1q21.1 and 22q11.2, as well as deletions and duplications of 15q13.3. These regions have known susceptibility to structural rearrangements that vary in size and breakpoint location due to the presence of several genomic low-copy repeats, with some data supporting the phenotypic effects of smaller or atypical CNVs.44–47 One case was a carrier of both 15q13.3 and 2q37 duplications.
Table 4.
Variants of Uncertain Significance With Potential Relevance to Clinical Phenotypes
| CNV | Cases (N) | Genomic Coordinates (Size) | Source | CLIA Lab Annotationa | CNV Groupb |
|---|---|---|---|---|---|
| 15q11.2 Dup | 4 | chr15:22.78–23.23 (449 kb) | CMA, WES | — | 1 |
| 16p12.2 Dup | 1 | chr16:21.96–22.59 (623 kb) | CMA, WES | — | 1 |
| 2q37 (HDAC4) Two copy gain | 1 | chr2:239.98–240.26 (270 kb) | CMA, CLIA | Unclear clinical significance | 1,2 |
| 15q13.3 (CHRNA7) Del | 1 | chr15:32.03–32.44 (409 kb) | CMA, CLIA | Pathogenic | 2 |
| 15q13.3 (CHRNA7) Dup | 3c | chr15:32.03–32.44 (409 kb) | CMA, CLIA | Population variant | 2 |
| 1q21.1 (proximal TAR region) Del | 1 | chr1:145.64–145.80 (163 kb) | CMA | — | 2 |
| 22q11.2 (distal) Dup | 1 | chr22:22.13–22.58 (454 kb) | CMA | — | 2 |
| 22q11.2 Del | 1 | chr22:21.06–21.41 (358 kb) | CMA, WES | — | 2 |
| Total case count (% of TRS sample, n = 509) | 12 (2.4%) |
Note: Del, deletion; Dup, duplication; WES, whole exome sequencing; CMA, chromosomal microarray.
Genomic coordinates aligned to hg19.
aInterpretation of the clinical significance of CNVs provided by Allele Diagnostics based on review of available literature at the time of CNV call.
bCNV groups are: (1) those that have an opposite copy number change of, or (2) partially overlap (<50% coverage) with known neurodevelopmental CNV loci (supplementary table S3).
cOf the three 15q13.3 duplication carriers, Case 1 is also the carrier of the 2q37 (HDAC4) duplication and is the mother of Case 2, while Case 3 is also the carrier of the 17p12 deletion (incidental finding, see “Methods”). CLIA, clinical laboratory improvement amendment (these CNVs were clinically verified in an independent CLIA-certified lab).
Discussion
Treatment-resistant psychosis is a complex and severe form of neuropsychiatric disorder in need of novel therapeutic insights. Several clinical variables (ie, age of onset, social dysfunction) may have potential for predicting treatment response,10 but do not directly lead to novel mechanistic insights into TRS. We identified a potentially increased prevalence of schizophrenia-associated CNVs in our TRS sample compared to previously published findings of schizophrenia patients not selected for treatment resistance. This is consistent with findings from a previous schizophrenia cohort23 that indicated an increased rate of treatment resistance in patients carrying disease-associated CNVs vs non-carriers. A non-ambiguous understanding of the possible link between rare-disease-associated CNVs and TRS will require a comprehensive comparison of CNV carriers and non-carriers using the same technology, choice of CNV calling platforms, and QC methods. Such an effort would require international collaboration, considerable resources, and time well beyond those available to this investigation. However, initial smaller investigative steps can begin to add clarity.
If the increased prevalence rate of SCZ CNVs identified in this sample is replicated in future studies, an important consideration will be whether it is: (1) the result of specific CNVs affecting downstream biological pathways with direct relevance to treatment resistance etiopathology (ie, dopamine system functioning, APD pharmacology, or other neurotransmitter systems such as glutamate or GABA signaling)48; or alternatively, (2) a secondary result of the more generalized effects of CNVs on reduced social/cognitive functioning indirectly impacting treatment outcomes. This is akin to separate models (summarized by Mulle et al.)49 that view CNVs as having either specific risk for specific diagnostic outcomes, or generalized risk for neuropsychiatric disorders more broadly.
Generalized vs Specific Risk
Any generalized effect that disease-associated CNVs exert on TRS risk may be mediated first through early neurocognitive impact. There is compelling evidence for neurodevelopmental factors being implicated in TRS. Treatment resistance has been genetically correlated with traits related to IQ and cognition in terms of having overlap in polygenic risk alleles.12 Studies have found lower premorbid IQ, younger age at onset, and poorer premorbid social adjustment are associated with TRS.10,11,50 Similar clinical features are also predictive of which individuals with schizophrenia carry clinically relevant CNVs. Specifically, a high prevalence of disease-associated CNVs are found in schizophrenia patients with comorbid intellectual disability (24%; IQ < 70), and/or childhood-onset psychosis (11.9%; onset <13 years of age).11,51,52
Kushima et al.23 provide additional evidence that neurodevelopmental burden in CNV carriers may be related to APD response. Among patients with schizophrenia carrying disease-associated CNVs, treatment resistance was seen more than twice as often in patients with co-occurring congenital and/or developmental phenotypes (48% had TRS) vs those without developmental phenotypes (21.4% had TRS). Importantly, no CNVs have been identified which increase the risk for psychosis alone. All SCZ CNVs also increase risk for other neurodevelopmental disorders (eg, developmental delay, intellectual disability, autism spectrum disorder), and often appear at higher rates in neurodevelopmental cohorts than in schizophrenia cohorts. In combination with our findings, these data suggest that SCZ CNVs may offer unique opportunities to study processes and shared mechanisms among neurodevelopmental burden, psychosis, and APD response.
Conversely, there is an important aspect of specificity in the contribution of CNVs to neuropsychiatric outcomes, especially psychotic phenotypes. It is likely that the distinct effects that CNVs have on neurobiology underly such genotype-specific outcomes.53,54 From the CNVs we screened for (supplementary table S2), 88% (21/24) of the NDD CNVs identified in our TRS sample affected one of the 12 schizophrenia-risk CNVs (table 2).
15q11.2-q13.1 Duplications
Duplications at the 15q11.2-q13.1 region are associated with early clinical findings such as global developmental delays, neurological disturbances, and autism spectrum disorder among others.55–61 However, the longitudinal effects of this variant have remained largely unexplored in clinically affected adults, particularly regarding APD response. To the best of our knowledge, no reports exist of APD treatment response in adult 15q11.2-q13.1 duplications carriers. One study62 indicated a relative inefficacy of benzodiazepines in the treatment of seizures in 15q11.2-q13.1 duplication carriers, which was hypothesized to be related to disruption in GABAergic signaling (several GABAβ3 receptor genes are located in the 15q11.2-13.1 region). Disruption in GABAergic inhibitory signaling has previously been implicated in psychosis and APD response, and may be one convergent mechanism of these CNV sites for TRS.63,64 Importantly, the 15q11.2-q13.1 region contains several dozen genes, and therefore mechanisms of action related to APD response may be complex and multigenic. Yet, our findings suggest that 15q11.2-q13.1 duplications may provide opportunities to model treatment resistance in experimental systems, with the aim of uncovering bio-mechanisms or potential drug targets.
Clinical Implications
The potential role of specific CNVs in treatment resistance is also relevant to discussions surrounding the use of genetic testing in psychiatry. Genome-wide screening technologies are routinely used to identify genomic etiologies in pediatric settings and the benefits of testing in these populations are well established.65,66 It remains an open question as to whether testing is similarly justified for psychiatric conditions.67 As we have previously argued,68,69 there is a shortage of detailed clinical data needed to investigate the impact that large-effect CNVs have on health comorbidities and treatment outcomes in adult psychiatric patients. This shortage has made it impossible to assess areas in which specific genetic etiologies may provide guidance to treatment management and understanding of individual health risks.70
The 22q11.2 deletion syndrome (22q11DS) stands out among the group of SCZ CNVs, in that there are thorough investigations to characterize the associated phenotype in both pediatric and adult carriers. Thus, clinical care guidelines have been established,71 and a diagnosis of 22q11DS even has implications for APD prescribing practices.72 These resources will become increasingly important in adult psychiatric practice as children diagnosed with 22q11DS (and other CNV syndromes) enter adulthood and seek psychiatric care. We did not identify a significant increase of 22q11DS in our TRS cohort, which is consistent with the prevailing idea that psychosis associated with 22q11DS responds to APD treatment at rates similar to idiopathic schizophrenia.73,74 Whether other SCZ CNVs (such as duplications of 15q11.2-q13.1 and 16p11.2) influence treatment-response, and through which biological pathways, should be the focus of future research efforts.
Strengths and Limitations
There remains pervasive heterogeneity in the definitions of TRS in research. Since large, well-defined TRS cohorts are difficult to acquire, many studies base subject selection on surrogate measures of TRS, such as filled clozapine prescriptions or the use of polypharmacy, without direct patient contact to confirm the TRS diagnoses or rule out instances of pseudo-TRS (ie, medication noncompliance, substance abuse). Each subject in our TRS cohort resided in the protected environment of the PASH system for at least 5 years with continuous clinical care, medication management, adequate sustenance, and minimal access to substances of abuse. State hospital policies also separate psychiatric patients from those with primarily developmental and intellectual disabilities. The latter are transferred into more appropriate treatment programs, which minimized the risk of oversampling patients with intellectual disability (which is associated with high rates of NDD CNVs).
Our approach to CNV detection (combining research-grade microarrays, exome sequencing, and clinical CLIA lab confirmation) allowed us to maximize sensitivity while minimizing false positives. However, this combined approach may have captured CNVs that microarray alone would have missed, which must be considered when making comparisons of our findings to studies that used differing methods. For example, of the six 16p11.2 duplications identified in our TRS sample, only 2 were called from SNP-based microarray through exome data, whereas all 6 were called from analysis of exome sequencing data, visual inspection of SNP data, and confirmed by a CLIA-certified lab. Larger case-control studies are necessary to confirm and further investigate the association of specific CNVs with TRS; however, this will require significant collaboration. Furthermore, there is a need for a greater depth of phenotypic data regarding adult psychiatric patients with disease-associated CNVs to translate genomic research into clinical improvement.
Conclusions
In this carefully selected sample of patients with TRS, we identified an increased prevalence of CNVs at specific loci previously implicated in schizophrenia. These data support previous indications that disease-associated CNVs may be more common in treatment-resistant patients and identifies the 15q11.2-q13.1 genomic region for further investigation into treatment resistance. There is a need to carefully examine the global clinical outcomes, in psychiatry and other medical domains, across the lifespan of carriers of clinically relevant CNVs.
Supplementary Material
Acknowledgments
The authors thank Dale Adair, MD, Chief Medical Officer, Office of Mental Health and Substance Abuse Services, Commonwealth of Pennsylvania, for his encouragement and support for this project. The authors also thank Takahiro Soda, MD, PhD, Department of Psychiatry, University of Florida, Gainesville Florida for reviewing the PASH cases in terms of inclusion/exclusion criteria. At the time of his assistance Dr Soda held a position in the Department of Psychiatry, University of North Carolina (Chapel Hill). Syed Sikandar Shah, MD participated in preliminary hospital chart reviews. Marice Davis, Kelly Bingaman, and Katina Geiger provided phlebotomy services. We thank the NIMH and Rutgers University Cell and DNA Repository for support in sample collection and processing. PFS reports potentially competing financial interests from H. Lundbeck A/S (advisory committee), the H. Lundbeck Foundation (grant recipient), and RBNC Therapeutics (advisory committee). Some of the sample recruitment and genomic assays were supported by H. Lundbeck A/S. AEF, JN, and MD are employed by H. Lundbeck A/S.
Contributor Information
Martilias Farrell, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Tyler E Dietterich, Translational Neuroscience, LLC, Conshohocken, PA, USA.
Matthew K Harner, Translational Neuroscience, LLC, Conshohocken, PA, USA.
Lisa M Bruno, Translational Neuroscience, LLC, Conshohocken, PA, USA.
Dawn M Filmyer, Translational Neuroscience, LLC, Conshohocken, PA, USA.
Rita A Shaughnessy, Translational Neuroscience, LLC, Conshohocken, PA, USA.
Maya L Lichtenstein, Department of Neurology, Geisinger Health System, Wilkes Barre, PA, USA.
Allison M Britt, School of Nursing, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Tamara F Biondi, Office of the Vice Chancellor for Research, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
James J Crowley, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Department of Psychiatry, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden.
Gabriel Lázaro-Muñoz, Center for Bioethics, Harvard Medical School, Boston, MA, USA; Department of Psychiatry, Massachusetts General Hospital, Boston, MA, USA.
Annika E Forsingdal, Department of Bioinformatics, H. Lundbeck A/S, Valby, Denmark.
Jacob Nielsen, Division of Neuroscience, H. Lundbeck A/S, Valby, Denmark.
Michael Didriksen, Division of Neuroscience, H. Lundbeck A/S, Valby, Denmark.
Jonathan S Berg, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Jia Wen, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Jin Szatkiewicz, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Rose Mary Xavier, School of Nursing, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Patrick F Sullivan, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Department of Psychiatry, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden.
Richard C Josiassen, Translational Neuroscience, LLC, Conshohocken, PA, USA.
Funding
This work was supported by the National Institute of Mental Health (K01 MH108894, R01MH110427 and K99HG008689); H. Lundbeck A/S; the Vernik Family Trust; and the Samuel and Paul Lofgren Family Trust.
Data Availability
Data are available upon request from corresponding author R.C.J. and will also be available from national repositories with the NIMH study number of 150.
References
- 1. Lally J, Gaughran F, Timms P, Curran SR.. Treatment-resistant schizophrenia: current insights on the pharmacogenomics of antipsychotics. Pharmgenomics Pers Med. 2016;9:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Howes OD, McCutcheon R, Agid O, et al. Treatment-resistant schizophrenia: treatment response and resistance in psychosis (TRRIP) Working Group Consensus guidelines on diagnosis and terminology. Am J Psychiatry. 2017;174(3):216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iasevoli F, Giordano S, Balletta R, et al. Treatment resistant schizophrenia is associated with the worst community functioning among severely-ill highly-disabling psychiatric conditions and is the most relevant predictor of poorer achievements in functional milestones. Prog Neuropsychopharmacol Biol Psychiatry. 2016;65:34–48. [DOI] [PubMed] [Google Scholar]
- 4. de Bartolomeis A, Balletta R, Giordano S, Buonaguro EF, Latte G, Iasevoli F.. Differential cognitive performances between schizophrenic responders and non-responders to antipsychotics: correlation with course of the illness, psychopathology, attitude to the treatment and antipsychotics doses. Psychiatry Res. 2013;210(2):387–395. [DOI] [PubMed] [Google Scholar]
- 5. Hor K, Taylor M.. Suicide and schizophrenia: a systematic review of rates and risk factors. J Psychopharmacol. 2010;24(4 Suppl):81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kennedy JL, Altar CA, Taylor DL, Degtiar I, Hornberger JC.. The social and economic burden of treatment-resistant schizophrenia: a systematic literature review. Int Clin Psychopharmacol. 2014;29(2):63–76. [DOI] [PubMed] [Google Scholar]
- 7. Smart SE, Kepinska AP, Murray RM, MacCabe JH.. Predictors of treatment resistant schizophrenia: a systematic review of prospective observational studies. Psychol Med. 2021;51(1):44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martin AK, Mowry B.. Increased rare duplication burden genomewide in patients with treatment-resistant schizophrenia. Psychol Med. 2016;46(3):469–476. [DOI] [PubMed] [Google Scholar]
- 9. Wimberley T, Gasse C, Meier SM, Agerbo E, MacCabe JH, Horsdal HT.. Polygenic risk score for schizophrenia and treatment-resistant schizophrenia. Schizophr Bull. 2017;43(5):1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Legge SE, Dennison CA, Pardinas AF, et al. Clinical indicators of treatment-resistant psychosis. Br J Psychiatry. 2020;216(5):259–266. [DOI] [PubMed] [Google Scholar]
- 11. Kowalec K, Lu Y, Sariaslan A, et al. Increased schizophrenia family history burden and reduced premorbid IQ in treatment-resistant schizophrenia: a Swedish National Register and Genomic Study. Mol Psychiatry. 2021;26:4487–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pardinas AF, Smart SE, Willcocks IR, et al. ; Genetics Workstream of the Schizophrenia Treatment Resistance and Therapeutic Advances (STRATA) Consortium and the Schizophrenia Working Group of the Psychiatric Genomics Consortium (PGC). Interaction testing and polygenic risk scoring to estimate the association of common genetic variants with treatment resistance in schizophrenia. JAMA Psychiatry. 2022;79(3):260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Szatkiewicz JP, O’Dushlaine C, Chen G, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19(7):762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marshall CR, Howrigan DP, Merico D, et al. ; Psychosis Endophenotypes International Consortium. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49(1):27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zoghbi AW, Dhindsa RS, Goldberg TE, et al. High-impact rare genetic variants in severe schizophrenia. Proc Natl Acad Sci USA. 2021;118(51):e2112560118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruderfer DM, Charney AW, Readhead B, et al. Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. Lancet Psychiatry. 2016;3(4):350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Q, Man Wu H, Yue W, et al. ; Chinese Antipsychotics Pharmacogenomics Consortium. Effect of damaging rare mutations in synapse-related gene sets on response to short-term antipsychotic medication in Chinese patients with schizophrenia: a randomized clinical trial. JAMA Psychiatry. 2018;75(12):1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sriretnakumar V, Harripaul R, Vincent JB, Kennedy JL, So J.. Enrichment of pathogenic variants in genes associated with inborn errors of metabolism in psychiatric populations. Am J Med Genet B Neuropsychiatr Genet. 2019;180(1):46–54. [DOI] [PubMed] [Google Scholar]
- 20. Kirov G, Rees E, Walters JT, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry. 2014;75(5):378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torres F, Barbosa M, Maciel P.. Recurrent copy number variations as risk factors for neurodevelopmental disorders: critical overview and analysis of clinical implications. J Med Genet. 2016;53(2):73–90. [DOI] [PubMed] [Google Scholar]
- 22. Rees E, Walters JT, Georgieva L, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014;204(2):108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kushima I, Aleksic B, Nakatochi M, et al. High-resolution copy number variation analysis of schizophrenia in Japan. Mol Psychiatry. 2017;22(3):430–440. [DOI] [PubMed] [Google Scholar]
- 24. Ammons RB, Ammons CH.. Quick Test.: Psychological Test Specialists; 1962. [Google Scholar]
- 25. Kay SR, Fiszbein A, Opler LA.. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13(2):261–276. [DOI] [PubMed] [Google Scholar]
- 26. Harris PA, Taylor R, Minor BL, et al. ; REDCap Consortium. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG.. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goldstein JI, Jarskog LF, Hilliard C, et al. Clozapine-induced agranulocytosis is associated with rare HLA-DQB1 and HLA-B alleles. Nat Commun. 2014;5:4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang K, Li M, Hadley D, et al. An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Colella S, Yau C, Taylor JM, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35(6):2013–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pinto D, Darvishi K, Shi X, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29(6):512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Z, Cheng H, Hong X, et al. EnsembleCNV: an ensemble machine learning algorithm to identify and genotype copy number variation using SNP array data. Nucleic Acids Res. 2019;47(7):e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11 10 11–11 10 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91(4):597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pounraja VK, Jayakar G, Jensen M, Kelkar N, Girirajan S.. A machine-learning approach for accurate detection of copy number variants from exome sequencing. Genome Res. 2019;29(7):1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rees E, Kendall K, Pardinas AF, et al. Analysis of intellectual disability copy number variants for association with schizophrenia. JAMA Psychiatry. 2016;73(9):963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin J, Tammimies K, Karlsson R, et al. Copy number variation and neuropsychiatric problems in females and males in the general population. Am J Med Genet B Neuropsychiatr Genet. 2019;180(6):341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kirov G, Grozeva D, Norton N, et al. ; International Schizophrenia Consortium. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18(8):1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. R: A language and environment for statistical computing [computer program]. Version: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 41. epitools: Epidemiology Tools [computer program]. Version; 2020. [Google Scholar]
- 42. O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sjostedt E, Zhong W, Fagerberg L, et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367(6482). [DOI] [PubMed] [Google Scholar]
- 44. Budisteanu M, Papuc SM, Streata I, et al. The phenotypic spectrum of 15q13.3 region duplications: report of 5 patients. Genes (Basel). 2021;12(7):1025-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gillentine MA, Berry LN, Goin-Kochel RP, et al. The cognitive and behavioral phenotypes of individuals with CHRNA7 duplications. J Autism Dev Disord. 2017;47(3):549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pang H, Yu X, Kim YM, et al. Disorders associated with diverse, recurrent deletions and duplications at 1q21.1. Front Genet. 2020;11:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vervoort L, Demaerel W, Rengifo LY, et al. ; International 22q11.2 Brain. Atypical chromosome 22q11.2 deletions are complex rearrangements and have different mechanistic origins. Hum Mol Genet. 2019;28(22):3724–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wada M, Noda Y, Iwata Y, et al. Dopaminergic dysfunction and excitatory/inhibitory imbalance in treatment-resistant schizophrenia and novel neuromodulatory treatment. Mol Psychiatry. 2022;27:2950–2967. [DOI] [PubMed] [Google Scholar]
- 49. Mulle JG, Sullivan PF, Hjerling-Leffler J.. Editorial overview: Rare CNV disorders and neuropsychiatric phenotypes: opportunities, challenges, solutions. Curr Opin Genet Dev. 2021;68:iii–iix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chan SKW, Chan HYV, Honer WG, et al. Predictors of treatment-resistant and clozapine-resistant schizophrenia: a 12-year follow-up study of first-episode schizophrenia-spectrum disorders. Schizophr Bull. 2021;47(2):485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ahn K, Gotay N, Andersen TM, et al. High rate of disease-related copy number variations in childhood onset schizophrenia. Mol Psychiatry. 2014;19(5):568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Foley C, Heron EA, Harold D, et al. Identifying schizophrenia patients who carry pathogenic genetic copy number variants using standard clinical assessment: retrospective cohort study. Br J Psychiatry. 2020;216(5):275–279. [DOI] [PubMed] [Google Scholar]
- 53. Modenato C, Kumar K, Moreau C, et al. ; 16p11.2 European Consortium. Effects of eight neuropsychiatric copy number variants on human brain structure. Transl Psychiatry. 2021;11(1):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Seidlitz J, Nadig A, Liu S, et al. Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nat Commun. 2020;11(1):3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Urraca N, Cleary J, Brewer V, et al. The interstitial duplication 15q11.2-q13 syndrome includes autism, mild facial anomalies and a characteristic EEG signature. Autism Res 2013;6(4):268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wolpert CM, Menold MM, Bass MP, et al. Three probands with autistic disorder and isodicentric chromosome 15. Am J Med Genet. 2000;96(3):365–372. [DOI] [PubMed] [Google Scholar]
- 57. Shaaya EA, Pollack SF, Boronat S, Davis-Cooper S, Zella GC, Thibert RL.. Gastrointestinal problems in 15q duplication syndrome. Eur J Med Genet. 2015;58(3):191–193. [DOI] [PubMed] [Google Scholar]
- 58. Bernier R, Hudac CM, Chen Q, et al. ; Simons VIP consortium. Developmental trajectories for young children with 16p11.2 copy number variation. Am J Med Genet B Neuropsychiatr Genet. 2017;174(4):367–380. [DOI] [PubMed] [Google Scholar]
- 59. Hudac CM, Bove J, Barber S, et al. Evaluating heterogeneity in ASD symptomatology, cognitive ability, and adaptive functioning among 16p11.2 CNV carriers. Autism Res. 2020;13(8):1300–1310. [DOI] [PubMed] [Google Scholar]
- 60. Kim SH, Green-Snyder L, Lord C, et al. Language characterization in 16p11.2 deletion and duplication syndromes. Am J Med Genet B Neuropsychiatr Genet. 2020;183(6): 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Steinman KJ, Spence SJ, Ramocki MB, et al. ; Simons VIP Consortium. 16p11.2 deletion and duplication: characterizing neurologic phenotypes in a large clinically ascertained cohort. Am J Med Genet A. 2016;170(11):2943–2955. [DOI] [PubMed] [Google Scholar]
- 62. Conant KD, Finucane B, Cleary N, et al. A survey of seizures and current treatments in 15q duplication syndrome. Epilepsia. 2014;55(3):396–402. [DOI] [PubMed] [Google Scholar]
- 63. Frankle WG, Cho RY, Prasad KM, et al. In vivo measurement of GABA transmission in healthy subjects and schizophrenia patients. Am J Psychiatry. 2015;172(11): 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kaster TS, de Jesus D, Radhu N, et al. Clozapine potentiation of GABA mediated cortical inhibition in treatment resistant schizophrenia. Schizophr Res. 2015;165(2-3):157–162. [DOI] [PubMed] [Google Scholar]
- 65. Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5): 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Srivastava S, Love-Nichols JA, Dies KA, et al. ; NDD Exome Scoping Review Work Group. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med. 2019;21(11): 2413–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Morris E, O’Donovan M, Virani A, Austin J.. An ethical analysis of divergent clinical approaches to the application of genetic testing for autism and schizophrenia. Hum Genet. 2022;141(5):1069–1085. [DOI] [PubMed] [Google Scholar]
- 68. Farrell M, Lichtenstein M, Harner MK, et al. Treatment-resistant psychotic symptoms and the 15q11.2 BP1-BP2 (Burnside-Butler) deletion syndrome: case report and review of the literature. Transl Psychiatry. 2020;10(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Harner MK, Lichtenstein M, Farrell M, et al. Treatment-resistant psychotic symptoms and early-onset dementia: a case report of the 3q29 deletion syndrome. Schizophr Res. 2020;224:195–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sullivan PF, Owen MJ.. Increasing the clinical psychiatric knowledge base about pathogenic copy number variation. Am J Psychiatry. 2020;177(3):204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fung WL, Butcher NJ, Costain G, et al. Practical guidelines for managing adults with 22q11.2 deletion syndrome. Genet Med. 2015;17(8):599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mosheva M, Korotkin L, Gur RE, Weizman A, Gothelf D.. Effectiveness and side effects of psychopharmacotherapy in individuals with 22q11.2 deletion syndrome with comorbid psychiatric disorders: a systematic review. Eur Child Adolesc Psychiatry. 2020;29(8):1035–1048. [DOI] [PubMed] [Google Scholar]
- 73. Dori N, Green T, Weizman A, Gothelf D.. The effectiveness and safety of antipsychotic and antidepressant medications in individuals with 22q11.2 deletion syndrome. J Child Adolesc Psychopharmacol. 2017;27(1):83–90. [DOI] [PubMed] [Google Scholar]
- 74. Butcher NJ, Fung WL, Fitzpatrick L, et al. Response to clozapine in a clinically identifiable subtype of schizophrenia. Br J Psychiatry. 2015;206(6):484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request from corresponding author R.C.J. and will also be available from national repositories with the NIMH study number of 150.