Abstract
Radiomics is an emerging approach to analyze clinical images with the purpose of revealing quantitative features that are unvisible to the naked eye. Radiomic features can be further combined with clinical data and genomic information to formulate prediction models using machine learning algorithms or manual statistical analysis. While radiomics has been classically applied to tumor analysis, there is promising research in its application to spine surgery, including spinal deformity, oncology, and osteoporosis detection. This article reviews the fundamental principles of radiomic analysis, the current literature relating to the spine, and the limitations of this approach.
Keywords: artificial intelligence, radiomics, spine, machine learning
Introduction
Radiomics is a technique in which diagnostic images can be processed and analyzed to extract quantitative and ideally reproducible information that is not perceptible to the human eye.1,2 The foundational principle is that qualitative and quantitative information found within clinical images reflect underlying pathophysiology, for example, genetic mutations within tumors.3,4 Although radiomics was primarily rooted in basic science research, it has recently gained the interest of clinical researchers as the clinical potentials are being increasingly recognized.5 For instance, a clinical radiographer can utilize a radiomics approach to classify whether a lesion on a computed tomography (CT) image could be malignant or benign.
In the era of personalized medicine, radiomics presents an exciting potential opportunity, as machine learning (ML) algorithms can be subsequently applied to predict outcomes such as survival or adverse effects. This enables radiomics to be a potentially powerful clinical tool to assist clinicians in decision-making and forming clinical decision support systems.6 The novel approach to convert clinical images into a large quantifiable data source for clinical decision algorithms contrasts with the traditional view that clinical images are intended purely for visual interpretation.
While a great proportion of radiomics research has stemmed from oncology, research is growing in the spine domain.7 This article summarizes the multistep process involved in radiomics, how radiomics can be applied to spine surgery, and the potential for future development.
What is Radiomics?
The term “radiomics” was first coined in 2012 by Phillippe Lambin when describing the automated extraction of large quantities of image features from radiographical images.8 In 2014, radiomics was applied to the field of oncology, where CT images of lungs were analyzed to predict outcomes in lung cancer. Since that time, there has been rapid growth in the range of its applications.7 Radiomics can be utilized across the breadth of imaging modalities, including radiography, magnetic resonance imaging (MRI), CT, and positron emission tomography.9
The radiomics process is illustrated in Figure 1.10 When an image is acquired, the region of interest (ROI) must be identified for image segmentation.11 This is the area of the clinical image (by location of pixels/voxels), which contains the prognostic value and where radiomic features will be extracted.10 This is a complex step in radiomics because the segmentation process varies across studies. Segmentation can be a manual process, but developments in deep learning algorithms have enabled automatic processing, which can be advantageous as it limits inter- and intraobserver variations.12 For larger datasets, automated processes are often needed because manual techniques are more time-consuming and less reproducible.
Figure 1.
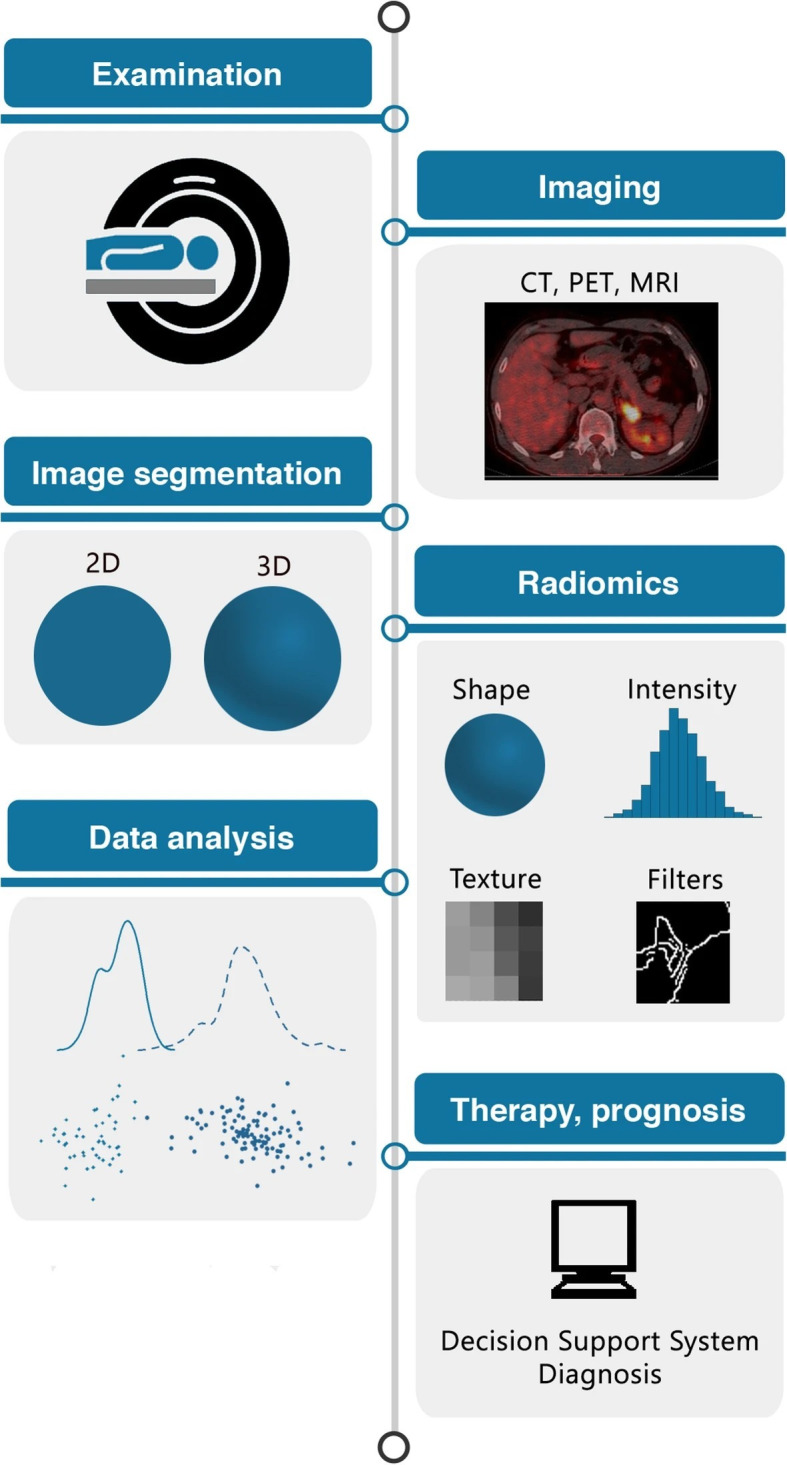
The radiomics process and sequential steps, including image acquisition, segmentation of region of interest, extraction of radiomic features, and predictive modeling and subsequent validation.10 2D, 2-dimensional; 3D, 3-dimensional; CT, computed tomography; MRI, magnetic resonance imaging; PET, positron emission tomography. Source: Reprinted from van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging—”how-to” guide and critical reflection. Insights Imaging. 2020;11:91. https://doi.org/10.1186/s13244-020-00887-2.10
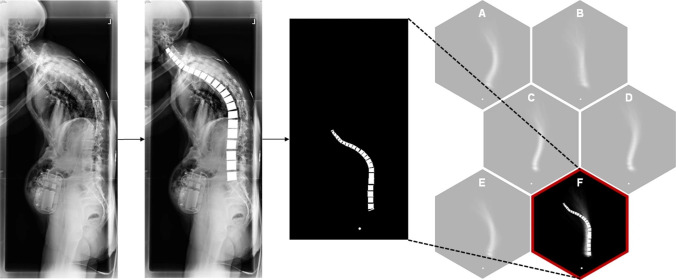
The second step is image processing to standardize the images for subsequent feature extraction. Durand et al demonstrated how data preprocessing is an important step in the artificial intelligence clustering of adult spinal deformity morphology using lateral long-cassette spinal radiographs.13 The femoral heads and distance from the femoral head to the sacral endplate were standardized to a set location to ensure consistent feature extraction of the spine shape and size (Figure 2). Image processing allowed for subsequent vertebral landmark identification and mapping of the spinal morphology to a cluster with similar patients. It is important to note the importance of designing preprocessing with the algorithm’s ultimate outcome in mind—this approach was appropriate specifically given the goal of clustering overall spine “shape” with an emphasis on thoracolumbar deformity.
Figure 2.
The importance of image processing. Durand et al used an artificial intelligence approach to cluster patients with adult spinal deformity according to their spine morphology. Reprinted with permission from Durand et al. Artificial intelligence clustering of adult spinal deformity sagittal plane morphology predicts surgical characteristics, alignment, and outcomes. European Spine Journal. 2021.13
The purpose of image segmentation and processing is to enable an accurate extraction of radiomic features from clinical images obtained. These are numerous but can include intensity, size, texture, shape, and location or relationship to adjacent tissues.14 There are 2 overall types of features that can be extracted using radiomic approaches: “semantic” and “agnostic” features.5 Semantic features are common language terms used to describe an ROI—for example, size, shape, and vascularity. However, agnostic features are mathematically calculated quantitative signifiers that may reflect relationships between spatially unrelated image features.
The cross-sectional area (CSA) of the psoas muscle is a good example of a radiomic feature that is identifiable on MRI imaging and may have utility in predicting outcomes of spine surgery. Prior studies investigated how the CSA of the paraspinal muscles and psoas muscle relates to lean body mass, muscle strength, and short-term outcomes of minimally invasive thoracolumbar interbody fusion.15–17 Banno et al found that the CSA of the multifidus muscle was lower in patients with adult spinal deformity and may correlate with severity of sagittal deformity.18 Additionally, another study found that the CSA of the multifidus muscle was reduced by lumbar disc herniation.19 Through a radiomic approach, future studies can improve upon the predictive value of the CSA in spine surgery outcomes.
With such complexity of radiomic features, feature selection is an essential step in the process. If all possible features were included in a model, there would be overfitting of the model, which limits its wider applicability.9 Therefore, reducing the number of features to build into algorithmic models is critical for generating generalizable results. Although there is no definitive rule for defining feature selection, certain principles are helpful to consider. If a feature has a high inter- or intraobserver variability, depending on how the image was segmented, this is unlikely to be useful in building a model.20
Radiomic modeling, for prognostic analysis and clinical prediction making, involves 3 key elements: feature selection, modeling methodology, and validation.9 The most comprehensive models should include sources of data beyond radiomic features, which may include genetic information, patient-reported outcome measures, health-related quality of life measures, patient health data, and other details relating to the patient’s clinical course. By combining data from radiomic analysis with clinical and molecular data, there is a greater ability to form more accurate clinical predictions. Multiple ML models should then be applied to the data to identify the most effective algorithm at creating a clinical decision support system, which is stable and clinically relevant. Finally, models should be validated to assess their performance, ideally with both internal and external prospective validation, to demonstrate generalizability on independent datasets.
When considering the strength of radiomic analysis, “repeatability” and “reproducibility” are 2 key concepts.21 Repeatability refers to radiomic features that remain consistent when imaged multiple times in the same subject using the same image acquisition method. Reproducibility refers to radiomic features that remain constant when different equipment (such as CT scanners), different software, and different parameters are applied.
Radiomics in Spine Surgery
In spinal pathologies, ML algorithms have been previously applied to identify subtle abnormalities on plain radiographs. For example, ossification of the posterior longitudinal ligament can have elusive findings on plain radiographs, but it is a critical pathology to identify due to the possibility of adverse outcomes both with nonoperative treatment as well as choosing the type of operative intervention. An ML model applied to plain radiographs was able to detect ossification of the posterior longitudinal ligament with 90% accuracy, outperforming the accuracy of spine surgeons (75%).22 Radiomic modeling is another approach used to identify pathology from clinical images of the spine and can be combined with ML algorithms.
Detecting Pathology
Radiomics has been previously applied in the orthopedic domain to identify pathology, such as osteoarthritis. Hirvasniemi et al developed a model to predict knee osteoarthritis using MRI-based radiomic features from tibial bone.23 Xue et al then furthered this by analyzing radiomic features in tibial and femoral subchondral bone to predict osteoarthritis.24 Osteoporosis is an important condition that spine surgeons consider when planning potential surgical intervention and preoperative optimization. Although this condition is traditionally diagnosed using dual-energy x-ray absorptiometry, most spine patients have their pathology evaluated by MRI. He et al applied a radiomics model to 109 patients who underwent both dual-energy x-ray absorptiometry and MRI with 396 features extracted.25 The area under the curve (AUC) was calculated to evaluate the discriminative ability of these models. The AUC for the optimal classification of normal vs osteopenia was 0.810, normal vs osteoporosis was 0.797, and osteopenia vs osteoporosis was 0.769. This suggests good discriminative ability between these bone densities and the potential utility in the diagnosis of osteoporosis without the need for additional testing.
Spinal Oncology
Radiomics has been frequently applied in oncologic research.26,27 In the orthopedic literature, Gitto et al have utilized radiomics and ML to classify cartilaginous bone tumors and differentiate between atypical cartilaginous tumors and grade II chondrosarcoma of long bones.28,29 Naturally, this has given interest in the role of radiomics specifically in spinal oncology. One of the challenges in this field is how MRI can differentiate between lesion types. This is an important clinical question because the spine is the most common site of bone metastasis, and management largely depends on the characteristics of the lesion.30 Chianca et al performed a radiomics study using 146 patients who underwent MRI for a single vertebral lesion.31 They found that a radiomics and ML approach was helpful in differentiating metastatic lesions from both malignant and benign primary bone tumors. A recent study by Gitto et al performed a similar investigation to differentiate malignant spinal bone tumors using diffusion and T2-weighted MRI. A total of 1702 radiomic features were extracted in this study, with 76.4% of these considered stable to variations in the ROI.32 The ML algorithm achieved 76% accuracy in differentiating between benign and malignant lesions when tested using the extracted radiomic features with a sensitivity of 78%, specificity of 68%, and AUC of 0.78.
Treatment Prediction
Once a patient is diagnosed with a condition, it is also helpful to predict how their treatment will alter their long-term prognosis. Patients with spinal metastasis may undergo stereotactic body radiation therapy to alleviate pain and control tumor progression. However, an undesirable sequela of this therapy is radiation-induced vertebral compression fracture (VCF). Gui et al were able to build an ML algorithm that could predict the risk of VCF after stereotactic body radiation therapy using the radiomic features from pretreatment imaging.33 In addition to extracting radiomic features from CT images and T1-weighted MRIs, clinical features, such as patient demographics and treatment characteristics, were also incorporated into the algorithm. The best performing model was able to predict VCF at 1 year, with a sensitivity of 84.4% and specificity of 80.0%.
Radiomic approaches may also be used to predict survival of patients with tumors, especially if used in conjunction with clinical data points. For example, Wang et al formulated a nomogram to predict 1- and 2-year survival for a wild-type glioblastoma using the radiomic signature and clinical risk factors.34 This tool may benefit clinicians, as patients with a shorter predicted life expectancy can potentially be spared invasive and painful procedures. Interestingly, radiomic feature extraction does not always improve the predictive power of models. Sanli et al performed a study on 250 patients who were treated for spinal metastasis with a Prognostic Index calculated for each patient; this was termed a radscore (radiomics score) and a clinscore (clinical score).35 The clinscore model showed good model accuracy, whereas the radiomics model added little predictive information (AUC 0.731 vs 0.623).
Limitations
Despite the growing evidence in radiomics and its application to a variety of clinical questions, there are several limitations to consider. Many of the limitations stem from the complex subprocesses involved in radiomics from image acquisition, feature extraction, and modeling. Each of these steps is affected by a wide range of nonstandardized decisions and parameter selections.9
Technical Limitations
To quantify radiomic features, several technical factors are involved and are subject to variability. To perform automatic segmentation in ROI, texture is a commonly utilized feature property for image classification. Galavis et al evaluated 50 textual features from positron emission tomography CT images of 20 patients with solid tumors. These features were classified according to their variability, which occurs due to different reconstruction parameters, acquisition modes, matrix sizes, and iteration numbers.36 Forty of the features (80%) were found to have a variation greater than 30%, including contrast, coarseness, and busyness.
The scanner used to acquire images for radiomic analysis can also pose a limitation. Mackin et al found that different CT scanners resulted in variability in the values of radiomics features extracted and concluded that the interscanner differences should be considered for future radiomic studies.37
Reproducibility and Overfitting
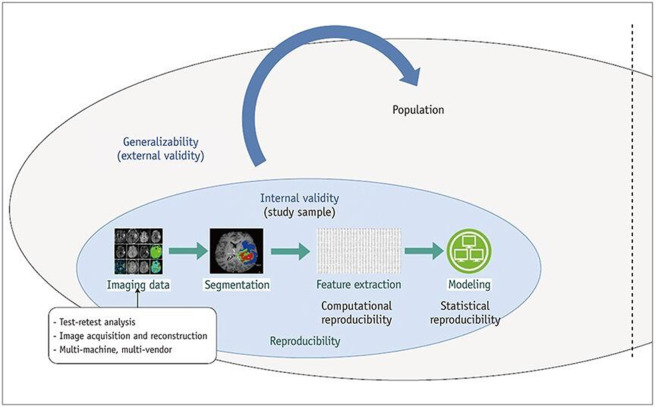
In radiomics, reproducibility depends on image acquisition, segmentation, and feature extraction.38 Reproducibility contributes to the internal validity of the study where the relationship between predicting variables and outcomes is explained without additional “noise.” Generalizability refers to the external validity when the model is applied across different population groups (Figure 3). Radiomics studies are typically retrospective and involve small patient datasets. Although these studies are important for proof of concept, the number of radiomic features extracted is much greater than the number of patients, and, therefore, this can lead to false positive results and feature selection bias.39 To confirm the predictive importance of models built upon radiomic features, a prospective external validation dataset should be utilized.9,40
Figure 3.
Diagram illustrating how reproducibility relates to internal validity and the subsequent generalizability across the population. Source: Reprinted from Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and generalizability in radiomics modeling: possible strategies in radiologic and statistical perspectives. Korean J Radiol. 2019;20(7):1124-1137. https://doi.org/10.3348/kjr.2018.0070.38
Overfitting is an associated concept and is a common problem in ML models. It occurs when an algorithm is trained too exactly with a dataset, thereby limiting its generalizability.41 Radiomic approaches result in “high dimensionality,” which refers to the production of a very large number of features implemented in the subsequent modeling and can risk overfitting.42 To mitigate overfitting, the number of radiomic features can be minimized using a dimensionality reduction algorithm. Dimensionality reduction removes the noise in the dataset and, therefore, keeps the most important features for the model to learn and apply to other datasets, not just the one the model is trained with.43 Other strategies to mitigate overfitting include using an expanded dataset and using data resampling techniques, such as bootstrapping aggregating.42,44 Cross-validation, such as K-fold cross-validation, is an additional method in which the complete dataset is split into parts to evaluate how the algorithms perform on unseen data.44
Class Imbalances
Class balance is another challenge where radiomic approaches are combined with ML algorithms. Class imbalance occurs when the number of examples available in 1 class is far less than in other classes; the class with a large number of samples is the “majority class,” and the one with few samples is the “minority class.” This is a problem because most ML algorithms assume an equal distribution of data between classes, and, therefore, a class imbalance will cause bias toward the majority class.
Class imbalance can be addressed through data handling techniques (eg, synthetic minority oversampling technique). For example, Kha et al built a model where radiomic features of low-grade gliomas with known genetic mutations were extracted from MRIs to predict a specific gene mutation of the tumor.45 An oversampling data technique was applied to address the class imbalance because a greater proportion of the patients did not have the gene mutation of interest. Therefore, the model would be less specific and biased toward identifying patients without the gene mutation.
Future Directions
Radiomics will no doubt play a role in the development of useful clinical tools for clinicians in the future. However, for radiomics to integrate within standard clinical use, several developments must occur. Radiomic studies are typically based on retrospectively collected data, which have limitations and serve primarily as proofs of concept.10 The retrospective nature of these radiomic studies results in the lack of standardization for imaging protocols and the introduction of unmeasured confounding variables.10 To confirm the utility of radiomics, validation studies using prospective datasets are necessary.
Furthermore, future radiomic studies should have coherent evaluation criteria and reporting guidelines. The transparent reporting of a multivariable prediction model for individual prognosis or diagnosis statement is a good example of recommendation guidelines for prediction models.46 Lambin et al outlined a radiomics quality score (RQS) to help reviewers, editors, and readers to evaluate whether the investigators are compliant with best practice methodology.9 The RQS includes 16 criteria, including image protocol quality to allow reproducibility/replicability, imaging at multiple time points, and reporting specific metrics. Each criterion, if achieved in the study, is assigned variable points to give a score out of possible 36 points. Notably, the highest point awarded is 7 points if a study is prospective and registered in a trial database for validation of the radiomics signature. Points can also be deducted if the study lacks a particular requirement. For example, 5 points are deducted if validation is missing; however, if validation is based on 3+ databases from multiple institutes, then 5 points are added. Therefore, the RQS clearly accentuates the principles that are required for future radiomic studies to have clinical utility—prospective datasets and appropriate validation. Koçak et al also provided a checklist of important criteria which should be transparently reported in radiomic studies, incorporating artificial intelligence analysis.42
To achieve the goal of acquiring a validated high-quality dataset, data sharing across intuitions should be employed. The power of prediction models created with radiomics and ML algorithms depends largely on the size and quality of the data.5 Although the image acquisition quality is important, the covariates collected must also be considered. Overall survival is a commonly reported outcome in the oncologic radiomics literature, although this may include all-cause mortality rather than the exact disease studied. More precise measures, such as disease-free survival, would be more clinically applicable but require laborious medical record reviews. Such a process could be simplified with a multicenter approach to collect more patient samples. One of the existing challenges is that imaging data must be shared in a Health Insurance Portability and Accountability Act–compliant manner. Deidentifying data, such as in the Cancer Imaging Archive, is one solution; or processing the data through a multisite institutional review board is another potential solution.
The ambition of radiomics is to enable clinicians to be increasingly informed in their diagnostic process and effectively counsel patients regarding their individualized risks and prognosis. Future radiomic studies may involve the integration of other data. Radiogenomics is one such approach where quantitative information extracted from radiomic analysis of clinical images is combined with individual genomic phenotype to construct prediction models.47 Imaging data are seldom used to predict clinical outcomes in isolation but are rather interpreted in the context of history, physical examination findings, laboratory data, etc. It is likely unreasonable to expect that, ultimately, the utility of radiomics would differ significantly from this integrated approach.
Conclusion
In the growing era of personalized medicine, spinal radiomics offers clinicians a promising decision-making tool.48 However, further work must be done to ensure these models are validated appropriately on high-quality datasets before being introduced within the clinical environment. As in many ML applications, future work in spinal radiomics will be required to reduce the opacity of these algorithms, facilitating both clinical utilization and the generation of novel clinical information.
References
- 1. Mayerhoefer ME, Materka A, Langs G, et al. Introduction to radiomics. J Nucl Med. 2020;61(4):488–495. 10.2967/jnumed.118.222893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wagner MW, Namdar K, Biswas A, Monah S, Khalvati F, Ertl-Wagner BB. Radiomics, machine learning, and artificial intelligence-what the neuroradiologist needs to know. Neuroradiology. 2021;63(12):1957–1967. 10.1007/s00234-021-02813-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2(1):36. 10.1186/s41747-018-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lo Gullo R, Daimiel I, Morris EA, Pinker K. Combining molecular and imaging metrics in cancer: radiogenomics. Insights Imaging. 2020;11(1):1. 10.1186/s13244-019-0795-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arimura H, Soufi M, Kamezawa H, Ninomiya K, Yamada M. Radiomics with artificial intelligence for precision medicine in radiation therapy. J Radiat Res. 2019;60(1):150–157. 10.1093/jrr/rry077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ding H, Wu C, Liao N, et al. Radiomics in oncology: a 10-year bibliometric analysis. Front Oncol. 2021;11:689802. 10.3389/fonc.2021.689802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 10. van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-'' how-to'' guide and critical reflection. Insights Imaging. 2020;11(1):91. 10.1186/s13244-020-00887-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Genomics and Radiomics: Tools to See the Unseen to Personalize Radiation Therapy. https://appliedradiationoncology.com/articles/genomics-and-radiomics-tools-to-see-the-unseen-to-personalize-radiation-therapy. 1 June 2022.
- 12. Hill BG, Krogue JD, Jevsevar DS, Schilling PL. Deep learning and imaging for the orthopaedic surgeon: how machines ''read'' radiographs. J Bone Joint Surg Am. 2022;104(18):1675–1686. 10.2106/JBJS.21.01387 [DOI] [PubMed] [Google Scholar]
- 13. Durand WM, Lafage R, Hamilton DK, et al. Artificial intelligence clustering of adult spinal deformity sagittal plane morphology predicts surgical characteristics, alignment, and outcomes. Eur Spine J. 2021;30(8):2157–2166. 10.1007/s00586-021-06799-z [DOI] [PubMed] [Google Scholar]
- 14. Kumar V, Gu Y, Basu S, et al. Qin radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–1248. 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Honkanen T, Mäntysaari M, Leino T, et al. Cross-Sectional area of the paraspinal muscles and its association with muscle strength among fighter pilots: a 5-year follow-up. BMC Musculoskelet Disord. 2019;20(1):170. 10.1186/s12891-019-2551-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morrell GR, Ikizler TA, Chen X, et al. Psoas muscle cross-sectional area as a measure of whole-body lean muscle mass in maintenance hemodialysis patients. J Ren Nutr. 2016;26(4):258–264. 10.1053/j.jrn.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Urakawa H, Sato K, Vaishnav AS, et al. 220. the cross-sectional area of psoas muscle impacts functional outcomes of MI-TLIF for lumbar degenerative diseases. The Spine Journal. 2020;20(9):S109. 10.1016/j.spinee.2020.05.631 [DOI] [Google Scholar]
- 18. Banno T, Yamato Y, Hasegawa T, et al. Assessment of the cross-sectional areas of the psoas major and multifidus muscles in patients with adult spinal deformity: a case-control study. Clin Spine Surg. 2017;30(7):E968–E973. 10.1097/BSD.0000000000000480 [DOI] [PubMed] [Google Scholar]
- 19. Kim WH, Lee SH, Lee DY. Changes in the cross-sectional area of multifidus and psoas in unilateral sciatica caused by lumbar disc herniation. J Korean Neurosurg Soc. 2011;50(3):201–204. 10.3340/jkns.2011.50.3.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haarburger C, Müller-Franzes G, Weninger L, Kuhl C, Truhn D, Merhof D. Radiomics feature reproducibility under inter-rater variability in segmentations of CT images. Sci Rep. 2020;10(1):12688. 10.1038/s41598-020-69534-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jha AK, Mithun S, Jaiswar V, et al. Repeatability and reproducibility study of radiomic features on a phantom and human cohort. Sci Rep. 2021;11(1):2055. 10.1038/s41598-021-81526-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ogawa T, Yoshii T, Oyama J, et al. Detecting ossification of the posterior longitudinal ligament on plain radiographs using a deep convolutional neural network: a pilot study. Spine J. 2022;22(6):934–940. 10.1016/j.spinee.2022.01.004 [DOI] [PubMed] [Google Scholar]
- 23. Hirvasniemi J, Klein S, Bierma-Zeinstra S, Vernooij MW, Schiphof D, Oei EHG.. A machine learning approach to distinguish between knees without and with osteoarthritis using MRI-based radiomic features from tibial bone. Eur Radiol. 2021;31(11):8513-8521. 10.1007/s00330-021-07951-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xue Z, Wang L, Sun Q, et al. Radiomics analysis using MR imaging of subchondral bone for identification of knee osteoarthritis. J Orthop Surg Res. 2022;17(1):414. 10.1186/s13018-022-03314-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He L, Liu Z, Liu C, et al. Radiomics based on lumbar spine magnetic resonance imaging to detect osteoporosis. Acad Radiol. 2021;28(6):e165–e171. 10.1016/j.acra.2020.03.046 [DOI] [PubMed] [Google Scholar]
- 26. Chen B, Zhang R, Gan Y, Yang L, Li W. Development and clinical application of radiomics in lung cancer. Radiat Oncol. 2017;12(1):154. 10.1186/s13014-017-0885-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aerts H, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gitto S, Cuocolo R, van Langevelde K, et al. Mri radiomics-based machine learning classification of atypical cartilaginous tumour and grade II chondrosarcoma of long bones. EBioMedicine. 2022;75:103757. 10.1016/j.ebiom.2021.103757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gitto S, Cuocolo R, Albano D, et al. Mri radiomics-based machine-learning classification of bone chondrosarcoma. Eur J Radiol. 2020;128:109043. 10.1016/j.ejrad.2020.109043 [DOI] [PubMed] [Google Scholar]
- 30. Kakhki VRD, Anvari K, Sadeghi R, Mahmoudian AS, Torabian-Kakhki M. Pattern and distribution of bone metastases in common malignant tumors. Nucl Med Rev Cent East Eur. 2013;16(2):66–69. 10.5603/NMR.2013.0037 [DOI] [PubMed] [Google Scholar]
- 31. Chianca V, Cuocolo R, Gitto S, et al. Radiomic machine learning classifiers in spine bone tumors: a multi-software, multi-scanner study. Eur J Radiol. 2021;137:109586. 10.1016/j.ejrad.2021.109586 [DOI] [PubMed] [Google Scholar]
- 32. Gitto S, Bologna M, Corino VDA, et al. Diffusion-weighted MRI radiomics of spine bone tumors: feature stability and machine learning-based classification performance. Radiol Med. 2022;127(5):518–525. 10.1007/s11547-022-01468-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gui C, Chen X, Sheikh K, et al. Radiomic modeling to predict risk of vertebral compression fracture after stereotactic body radiation therapy for spinal metastases. J Neurosurg Spine. 2021;1:1–9. 10.3171/2021.3.SPINE201534 [DOI] [PubMed] [Google Scholar]
- 34. Wang S, Xiao F, Sun W, et al. Radiomics analysis based on magnetic resonance imaging for preoperative overall survival prediction in isocitrate dehydrogenase wild-type glioblastoma. Front Neurosci. 2021;15:791776. 10.3389/fnins.2021.791776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sanli I, Osong B, Dekker A, et al. Radiomics biopsy signature for predicting survival in patients with spinal bone metastases (SBMs). Clin Transl Radiat Oncol. 2022;33:57–65. 10.1016/j.ctro.2021.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncologica. 2010;49(7):1012–1016. 10.3109/0284186X.2010.498437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mackin D, Fave X, Zhang L, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015;50(11):757–765. 10.1097/RLI.0000000000000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and generalizability in radiomics modeling: possible strategies in radiologic and statistical perspectives. Korean J Radiol. 2019;20(7):1124–1137. 10.3348/kjr.2018.0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150–R166. 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cui Y, Song J, Pollom E, et al. Quantitative analysis of (18) F-fluorodeoxyglucose positron emission tomography identifies novel prognostic imaging biomarkers in locally advanced pancreatic cancer patients treated eith stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys. 2016;96(1):102–109. 10.1016/j.ijrobp.2016.04.034 [DOI] [PubMed] [Google Scholar]
- 41. Alsoof D, McDonald CL, Kuris EO, Daniels AH. Machine learning for the orthopaedic surgeon: uses and limitations. J Bone Joint Surg Am. 2022;104(17):1586–1594. 10.2106/JBJS.21.01305 [DOI] [PubMed] [Google Scholar]
- 42. Koçak B, Durmaz EŞ, Ateş E, Kılıçkesmez Ö. Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol. 2019;25(6):485–495. 10.5152/dir.2019.19321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ubaldi L, Valenti V, Borgese RF, et al. Strategies to develop radiomics and machine learning models for lung cancer stage and histology prediction using small data samples. Phys Med. 2021;90:13–22. 10.1016/j.ejmp.2021.08.015 [DOI] [PubMed] [Google Scholar]
- 44. Overfitting in Machine Learning: What It Is and How to Prevent It. EliteDataScience. Published September 7, 2017. https://elitedatascience.com/overfitting-in-machine-learning. 3 June 2022.
- 45. Kha QH, Le VH, Hung TNK, Le NQK. Development and validation of an efficient MRI radiomics signature for improving the predictive performance of 1p/19q co-deletion in lower-grade gliomas. Cancers (Basel). 2021;13(21):5398. 10.3390/cancers13215398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD). Ann Intern Med. 2015;162(10):735–736. 10.7326/L15-5093-2 [DOI] [PubMed] [Google Scholar]
- 47. Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol. 2015;12(8):862–866. 10.1016/j.jacr.2015.04.019 [DOI] [PubMed] [Google Scholar]
- 48. Miles K. Radiomics for personalised medicine: the long road ahead. Br J Cancer. 2020;122(7):929–930. 10.1038/s41416-019-0699-8 [DOI] [PMC free article] [PubMed] [Google Scholar]