Significance
Lipid nanoparticles (LNPs) are the most successful RNA delivery carriers to date and are used in FDA-approved products like the COVID-19 mRNA vaccine. Expanding LNP-based therapies hinges on efficient delivery to a variety of tissues and cells, a process often hindered by endosomal entrapment. It is well known that depending on lipid molecular properties, LNPs assemble into different nanostructures, but how these impart endosomal escape remains unknown. We demonstrate that combining lipid composition with nanostructure synergistically impacts the ability of LNPs to escape endosomes. LNP–RNA complexes prescribed with bicontinuous cubic and inverse hexagonal internal structures facilitate the topological transition of LNP–endosome fusion-pore formation. We show that nanostructure is a potent handle to engineer highly efficient LNPs for RNA delivery.
Keywords: lipids, cubosomes, RNA delivery, membrane-fusion, endosomal escape
Abstract
RNA therapeutics have the potential to resolve a myriad of genetic diseases. Lipid nanoparticles (LNPs) are among the most successful RNA delivery systems. Expanding their use for the treatment of more genetic diseases hinges on our ability to continuously evolve the design of LNPs with high potency, cellular-specific targeting, and low side effects. Overcoming the difficulty of releasing cargo from endocytosed LNPs remains a significant hurdle. Here, we investigate the fundamental properties of nonviral RNA nanoparticles pertaining to the activation of topological transformations of endosomal membranes and RNA translocation into the cytosol. We show that, beyond composition, LNP fusogenicity can be prescribed by designing LNP nanostructures that lower the energetic cost of fusion and fusion–pore formation with a target membrane. The inclusion of structurally active lipids leads to enhanced LNP endosomal fusion, fast evasion of endosomal entrapment, and efficacious RNA delivery. For example, conserving the lipid make-up, RNA–LNPs having cuboplex nanostructures are significantly more efficacious at endosomal escape than traditional lipoplex constructs.
RNA therapeutics has gained wide attention due to its power to regulate the expression of disease-related genes that drive a myriad of human diseases, including cancer (1), neurodegeneration (2), and metabolic disorders (3), among others (4–8). Delivery vectors play an important role in the development of such therapeutics in preventing RNA degradation and successfully deliver RNA to target cells. Compared to viral vectors, nonviral systems have the advantage of being less immunogenic and easier to manufacture (9). Among the different materials used for nonviral delivery, lipid nanoparticles (LNPs) have been particularly successful, as many LNP–RNA formulations are clinically available (10, 11) or have advanced to clinical trials. The first Food and Drug Administration (FDA)–approved small interfering RNA (siRNA) treatment, patisiran (12), utilized LNPs for delivery. The effectiveness of LNPs has been further manifested by the development of lipid–mRNA (messenger RNA) vaccines for coronavirus disease 2019 (COVID-19) (13).
Despite the astonishing progress LNP delivery achieved, many barriers are still present in realizing the full potential of LNP–RNA systems (14). While most LNP systems are efficiently taken up by the cell via endocytosis, they often remain trapped in endosomal compartments and degrade through the endosome–lysosome acidification pathway (15–18). One of the main bottlenecks of LNP-based RNA delivery is poor endosomal escape (19, 20). Viral delivery systems have membrane proteins that undergo a conformation change allowing them to easily fuse with the plasma or endosomal membranes to release their payload. Previous studies have shown that for LNP mediated siRNA delivery, less than 2% of the siRNA can successfully escape the endosome and reach the cytosol (21). With low cytosolic delivery efficacy, much research has been done in an attempt to improve endosomal escape and transfection efficiency of LNPs by modifying lipid compositions (22–24). Sahay et al. have shown that LNPs containing cholesterol demonstrate improved delivery efficiencies, potentially caused by enhanced endosomal membrane fusion (25). Other ways of optimizing lipid compositions have also shown success (26), such as the inclusion of ionizable lipids (27–31). Ionizable lipids are able to stay neutral under physiological pH but become cationic under the acidic environment of endosomes, being able to electrostatically bind the endosomal membrane which may allow LNPs to destabilize it and facilitate escape (10). Siegwart’s team has also shown that the lipid composition of LNPs can be used to target the particles to specific organs (32, 33).
However, compared to the extensive research done exploring different lipid formulations, the nanostructure of LNPs or the specific packing of lipids and nucleic acids into the LNP has not been fully investigated. In this paper, we establish structural activity as an LNP design principle. Due to their amphiphilic nature, lipid molecules can form a variety of self-assembled structures in aqueous environments (Fig. 1B) that are conserved when encapsulating nucleic acids. This includes lamellar vesicles (L), inverse hexagonal (HII), and bicontinuous cubic phases (QII) (34–44). The optimal mechanism to boost LNPs endosomal escape is to promote LNP–endosomal membrane fusion, a process that should take place in less than 30 s (45) and be independent of endosome acidification or the debated “proton sponge” effect (46, 47). The design of “fusogenic” LNPs has been exclusively attributed to tuning the molecular packing parameter of lipid molecules such that their spontaneous membrane curvature (C0) is negative (48, 49). However, the elastic energy cost of membrane fusion or, more importantly, the development of the required fusion pore is controlled by a topological transformation that should mostly depend on the Gaussian moduli of membranes . The membrane elastic energy can be represented by the Helfrich equation (50, 51), , where κ is the bending modulus, J is the total/extrinsic curvature which equals the sum of the principle curvatures C1 + C2, and C0 is the spontaneous curvature. The second term comprises the Gaussian curvature K = C1C2 and the Gaussian modulus . Taking into account the Gauss–Bonnet theorem (52) and the Helfrich framework, one can calculate that the elastic energy cost of the topological transformation from two nested membrane vesicles () to two fused vesicles () to be (53) (Fig. 1A). It is immediately clear that modulating the Gaussian modulus and Gaussian curvature K will significantly impact membrane fusion events and the formation of fusion pores through which RNA cargo can be delivered into the cytosol (Fig. 1C). There are three main categories of reported LNP–nucleic acid nanostructures: L, HII, and QII. In the lamellar L phase, nucleic acids are sandwiched between lipid bilayers and are often referred to as “lipoplexes” (54). In HII, nucleic acids are inserted in water tubes decorated by lipids, and in QII, cuboplexes, siRNA has been shown to locate in the water nanochannel domains (39–41). Positively curved vesicular lamellar phases (L) have but cubosomes (55) or cuboplexes [cubosomes loaded with RNA (39, 40)], which are made of bicontinuous cubic phases, have intrinsically negative Gaussian curvature K and positive , and the activation energy for fusion with endosomal membranes and fusion pore formation should be the lowest. Siegel (56–61) first suggested that bicontinuous cubic phases should be highly fusogenic, and we conjecture that QII LNPs have a greater potential in fusing with the endosomal membrane and successfully release their cargo (Fig. 1C). In addition, the bicontinuous nature of membranes can endow LNPs with gene/drug release properties sustained over longer periods of time which has been demonstrated for polymer-based systems by Scott’s team (62–65).
Fig. 1.
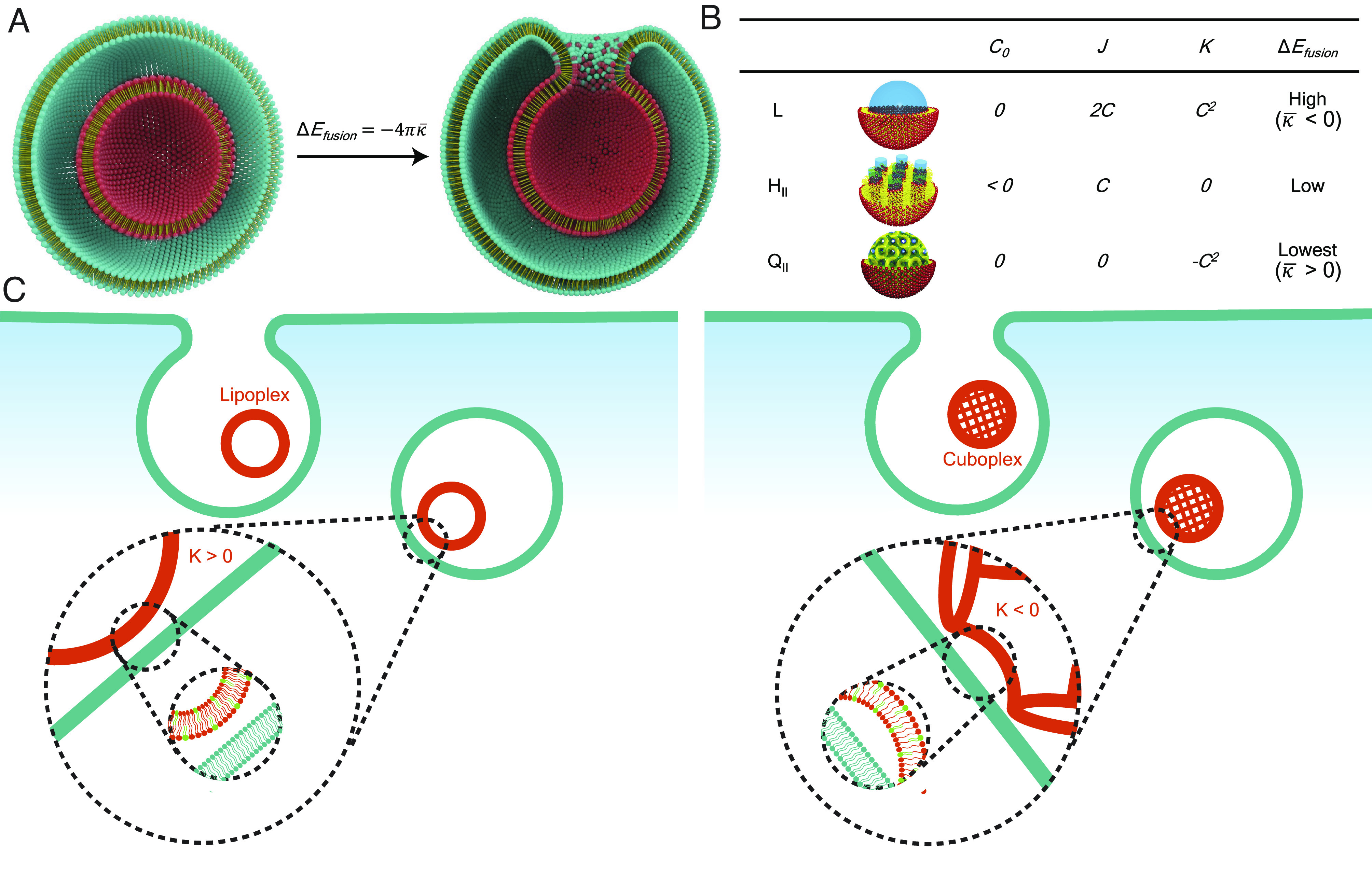
Schematic representation of the cell entry of LNP–RNA complexes. (A) LNPs (red particle) enter the cells via endocytosis and are entrapped in the endosome (teal vesicle). Illustration credits: Alex D. Jerez, Imaging Technology Group at the Beckman Institute University of Illinois, Urbana-Champaign, IL. LNP–endosome membrane fusion involves a topological transformation of activation elastic energy . (B) LNPs of different nanostructures (L, HII, and QII) have distinct spontaneous curvature C0, total curvature J, and Gaussian curvature K properties. (C) The escape of LNPs from endosomal entrapment will be enhanced by lowering the energetic barrier for fusion and fusion pore formation with the endosomal membrane for which QII LNPs should have preferential properties.
In this paper, we show that, in addition to lipid molecular properties and composition, the structure of LNP–RNA complexes is a powerful design handle to boost the extent of endosomal-membrane fusion and concomitant efficacy of RNA cytoplasmic delivery.
Results
Formation of LNP–RNA Complexes of Precise Nanostructures.
In our study, we judiciously select specific neutral lipid molecular systems and compositions to control the nanostructure of LNP–RNA complexes. Glycerol monooleate (GMO) is a neutral lipid approved by the FDA for in vivo use and is utilized as an adjuvant (66, 67). GMO is well known to be stabilize into a variety of bicontinuous cubic phases (68) that can be formulated into cubosome LNPs (QII) encapsulating siRNA which we termed cuboplexes (39, 40). The cuboplex LNP formulation comprised GMO, a cationic lipid 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) just enough to electrostatically bind RNA without raising toxicity as well as 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)]-2000 (DOPE-PEG) to optimize the colloidal stability of the LNPs (69–71). Modulating the amount of neutral lipid GMO in the ternary GMO, DOTAP, and DOPE-PEG mixture results in precise tuning of different bicontinuous cubic structures of distinct space groups (Ia3d— gyroid, Im3m—primitive, and Pn3m—diamond) as well as the formation of hexagonal phases (HII) (34, 39). To form the traditional lamellar phase (L), GMO contents are low (< 25 mol%) (34), or the neutral lipid was switched to a phosphatidylcholine, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). LNP–RNA complexes are formed with an optimal (39) charge ratio of ρ=3, where charge ratio represents the number of positive charges from lipid molecules (nDOTAP) over the number of negative charges from RNA (nNA). When nucleic acid is added to the lipid systems, the nanostructures are mostly conserved, but there is a preference to adopt HII phases as the 1D straight cylindrical water channels can accommodate long-chain nucleic acids (72). Fig. 2 shows structural information of dilute LNP–RNA complexes (siRNA and mRNA) at different amounts of neutral GMO obtained by synchrotron small-angle X-ray scattering (SAXS). LNP–siRNA complexes with a GMO/DOTAP/DOPE-PEG (molar ratio 85/14/1) composition show four distinct diffraction peaks that indicate the presence of a bicontinuous cubic gyroid nanostructure (QII) coexisting with a 2D inverse hexagonal phase (HII) (Fig. 2A). The first two peaks show reciprocal lattice vectors q/(2π/a) = Ghkl/(2π/a) = (h2 + k2 + l2)1/2 = , , which corresponds to the {211}, {220} planes of a QII gyroid phase. The first, third, and fourth peaks show reciprocal lattice vectors q/() = Ghk / (4) = (h2 + hk + k2)1/2 = , , and , corresponding to {10}, {11}, and {20} of the HII phase. The lattice spacings are aQII = 127 Å and aHII = 60 Å. LNP–mRNA complexes with the same lipid composition have a similar scattering profile and display HII / QII phase coexistence with lattice spacings aQII = 124 Å and aHII = 51 Å. Due to the electrostatic interaction with negatively charged nucleic acids and the cationic lipid headgroups, nucleic acids are preferentially located in the water channels (40). We previously imaged using cryo-EM gold-tagged siRNA colocalized with the water domains of cuboplexes (40). Using the unit cell size of the cubic phase (a = 127 Å), the diameter of the water channels can be estimated (34, 40) to be 33 Å which is just enough to tightly pack RNA (diameter of 22 Å with a hydration layer). This short-range electrostatic interaction will be dominant, and RNA is not expected to unbind and diffuse out of the cuboplex during organic solvent removal (by dialysis or evaporation). At intermediate GMO contents (Fig. 2B), LNP–RNA complexes mostly adopt the HII phase in coexistence with the L phase. The lattice spacings of the complexes are similar, with LNP–siRNA aHII = 63 Å and aL = 59 Å, and LNP–mRNA aHII = 103 Å and aL = 91 Å. At low GMO content (below 25 mol%), LNP–RNA complexes adopt the L phase (SI Appendix, Fig. S1). Switching GMO for a phosphatidylcholine neutral lipid always results in a L phase shown in Fig. 2C for DOPC/DOTAP/DOPE-PEG (molar ratio 85/14/1). The complexes show similar lattice parameters, with LNP–siRNA aL = 332 Å and LNP–mRNA aQII = 334 Å.
Fig. 2.
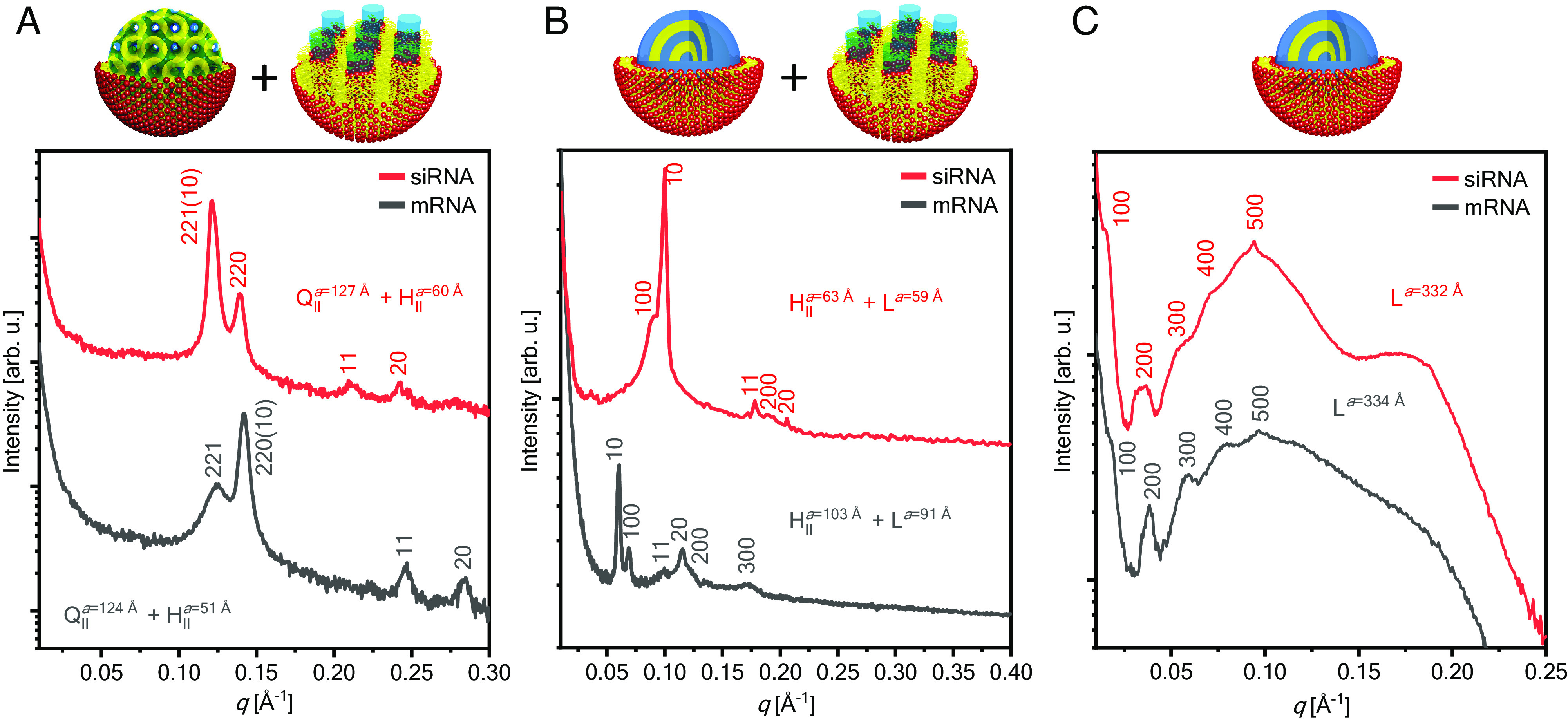
Synchrotron SAXS data of LNP–siRNA and LNP–mRNA complexes under dilute conditions show bicontinuous cubic (QII), 2D reversed hexagonal (HII) and lamellar phases (L). (A) QII, HII LNP–RNA complexes were formed with a lipid composition of GMO/DOTAP/DOPE-PEG at molar ratio 85/14/1 and mixed with siRNA or mRNA at ρ = 3. (B) HII, L LNP–RNA complexes were formed with a lipid composition of GMO/DOPC/DOTAP/DOPE-PEG at molar ratio 50/35/14/1 and mixed with siRNA at ρ = 3. (C) L LNP–RNA complexes were formed with a lipid composition of DOPC/DOTAP/DOPE-PEG at molar ratio 85/14/1 and mixed with siRNA or mRNA at ρ = 3.
The structural diversity of LNP–RNA complexes is not only shown by SAXS but also manifested by cryogenic electron microscopy (cryo-EM). With the ability to image LNPs in their near-native hydrated state, cryo-EM has become an important technique to evaluate the structure of LNPs (73). Fig. 3 shows representative cryo-EM images of LNP–RNA complexes prepared by microfluidic methods with different nanostructures. Fig. 3A shows the lamellar structure of LNP–mRNA lipoplexes. In this “onion-like” structure, lipid domains (represented in yellow) intercalate with mRNA located in the water-layer domains (represented in blue). This is readily observable by fast Fourier transform (FFT, Inset) image analysis revealing periodic layers of electron density. Fig. 3B shows an LNP–siRNA complex of the HII phase. In this nanostructure, siRNA aligns in the hexagonally packed water tubes that are decorated with lipids. The FFT image analysis clearly demonstrates the hexagonal symmetry of this LNP–siRNA complex. A hybrid LNP–mRNA complex of both QII and HII is shown in Fig. 3C. In this case, FFT image analysis of different regions of the particle reveals that this is a hybrid LNP system where both QII and HII are present. This is significant because SAXS data (Fig. 2A) indicated the coexistence of these phases both for siRNA and mRNA systems. With SAXS being an average method, it was possible that both pure QII and pure HII LNPs coexisted in a test tube. The cryo-EM data indicate that, importantly, coexistence is present at a single–LNP level. Fig. 3D shows an LNP–siRNA cuboplex (QII) and respective FFT analysis where siRNA inserts in the water nanochannels. (Adapted from previous studies of Leal’s team) (40). We also seen that while the preparation method does not significantly impact the nanostructure of the LNPs (SI Appendix, Fig. S2), top–down film hydration yields larger and more polydisperse systems compared to bottom–up nanoprecipitation in microfluidics (dialysis).
Fig. 3.
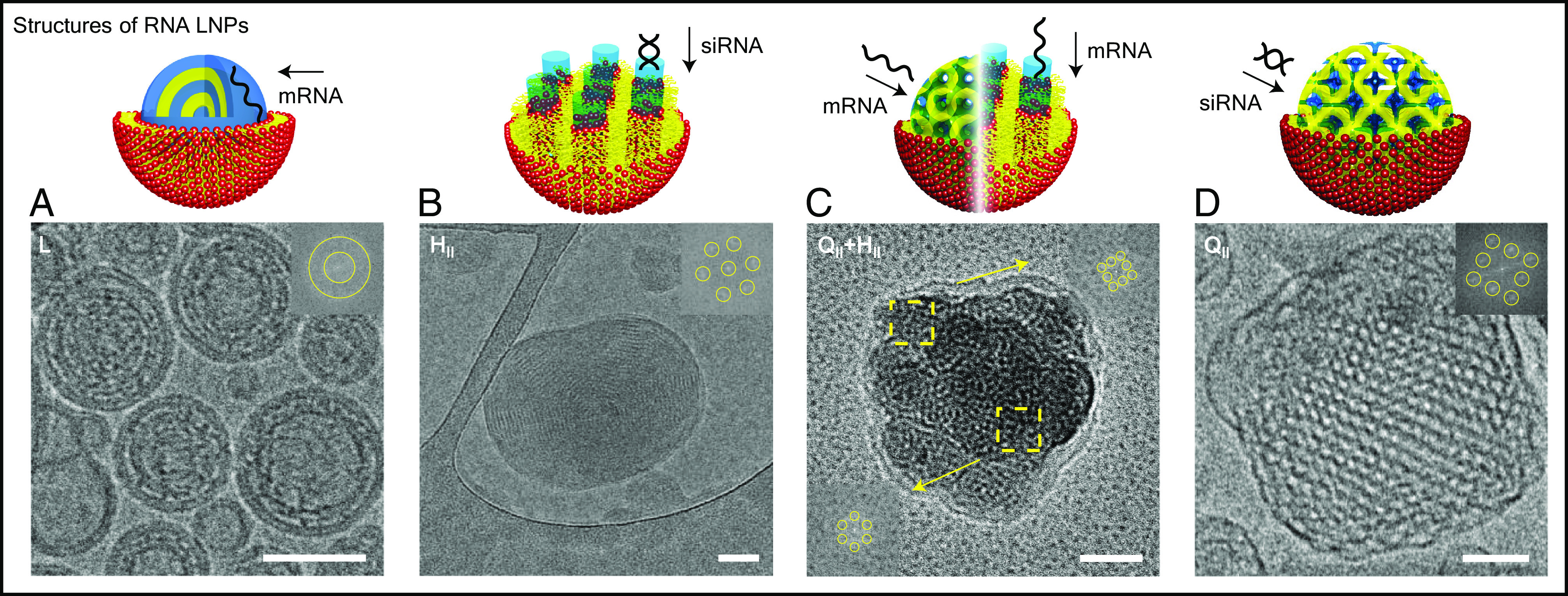
Representative cryo-EM images of LNP–RNA complexes with insets representing the fast Fourier transforms (FFT). (Scale bar, 50 nm.) (A) Cryo-EM micrograph of lamellar phase LNP–RNA complexes. These complexes were formed with a lipid composition of DOPC/DOTAP/DOPE-PEG at molar ratio 85/14/1 and with mRNA at ρ = 6. (B) Cryo-EM micrograph of a hexagonal phase LNP–RNA complex. This complex has a lipid composition of GMO/DOTAP/DOPE-PEG at molar ratio 85/14/1 and with siRNA at ρ = 3. (C) Cryo-EM micrograph of an LNP–RNA complex with coexisting hexagonal and bicontinuous cubic phases. The complex was formed with a lipid composition of GMO/DOTAP/DOPE-PEG at molar ratio 85/14/1 and with mRNA at ρ = 6. (D) Cryo-EM micrograph of an LNP–RNA complex with the bicontinuous cubic phase. Adapted with permission from ref. 40. Copyright 2018 American Chemical Society.
The modulation of neutral lipid amount and identity yields rich structural diversity of LNP–RNA complexes without disrupting their stability. LNP–RNA complexes remain colloidally stable regardless of the nanostructure having size distributions between 150 and 250 nm (SI Appendix, Table S1).
Fusion of LNP–RNA Complexes with Endosomes Is Regulated by LNP Nanostructure.
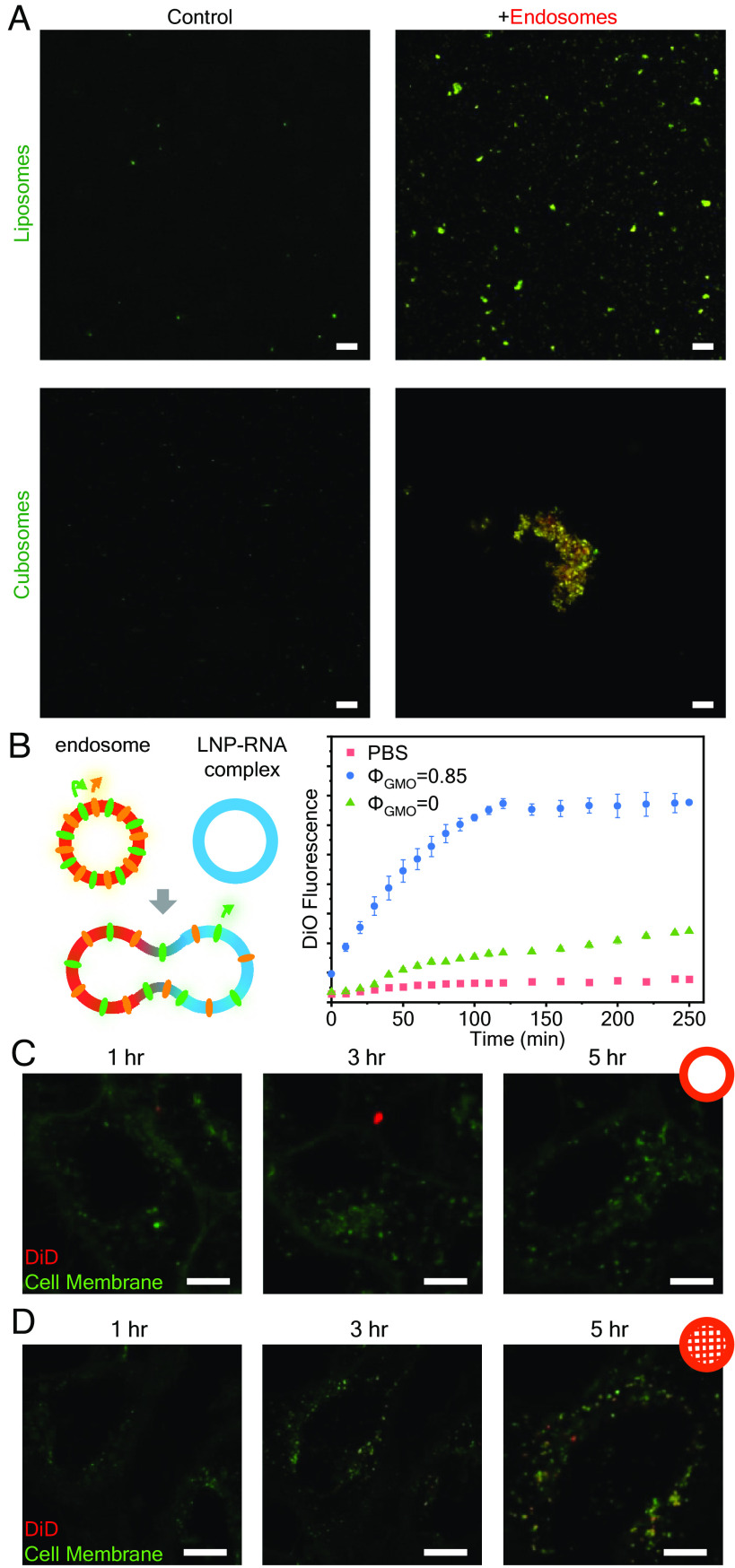
The interaction between endosomes and LNP–RNA complexes of different nanostructures in vitro was explored using confocal laser scanning microscopy (CLSM), fluorescence resonance energy transfer (FRET) assays, and live-cell self-quenching CLSM experiments (Fig. 4). Fig. 4A shows CLSM images of isolated endosomes incubated with pure LNPs (no RNA) having the L (liposome) and QII (cubosomes) nanostructures. Endosomes were isolated by standard spin–column techniques from epithelial human cervix cancer (HeLa) cells and fluorescently labeled by incubating in 20 μM Nile Red. The LNPs were tagged with 0.1% 1,1′-dioctadecyl-3,3,3′,3′- tetramethylindodicarbocyanine (DiD). LNPs do not aggregate significantly on their own, but when incubated with endosomes, LNP–endosome fusion and aggregation are readily visible by colocalization of DiD and Nile Red fluorescence in both systems but is significantly more pronounced for QII LNPs.
Fig. 4.
Cubic and hexagonal LNPs fuse more readily with endosomes compared to lamellar LNPs. (A) Visualization of LNP–endosome interaction with CLSM after 6 h incubation. (Scale bar, 20 μm.) (B) FRET assay to evaluate membrane fusion between endosomes and LNP–siRNA complexes of different structures. The fusion extent is indicated by DiO fluorescence recovery (n = 3, data presented as mean ± SD). (C and D) Live cell imaging of HeLa cells treated with lipoplexes (C) and cuboplexes (D) labeled with 1% self-quenching dye DiD. Recovery of DiD signal (red) implies LNP–endosomal fusion. (Scale bar, 10 μm.)
After observing that QII LNPs bind with isolated endosomes more favorably, we conducted an FRET assay (74) to evaluate the fusion process extent between endosomes and RNA–loaded LNPs of different nanostructures. Isolated endosomes were colabeled with 1,1′-dioctadecyl-3,3,3′,3′- tetramethylindocarbocyanine perchlorate (DiI) and 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO), with DiO acting as a donor fluorophore and DiI acting as an acceptor fluorophore. The proximity of the dye molecules enables FRET to occur, leading to enhanced acceptor (DiI) fluorescence. If endosomes fuse with another membrane, the dye molecules diffuse, increasing the distance between each other and FRET occurrences diminish. Reduced FRET allows donor (DiO) fluorescence to recover. Thus, the extent of membrane fusion can be evaluated by the increase in DiO fluorescence. To see how LNP structure influences LNP–endosome membrane fusion, we prepared LNP–siRNA complexes with the same DOTAP molar fractions (ΦDOTAP = 0.14) but different GMO molar fractions. At ΦGMO = 0.85, RNA–loaded LNPs display the QII structure (in coexistence with HII) termed cuboplexes (39, 40) and at ΦGMO = 0 the lamellar L phase (Fig. 2) termed lipoplexes. The intensity of DiO fluorescence as a function of time (Fig. 4B) is significantly higher for LNPs comprising ΦGMO = 0.85 (blue data points) compared to ΦGMO = 0 (green data points). This is consistent with a considerably higher extent of fusion of endosomes with “nonlamellar” LNPs (QII/HII) compared to regular “lipoplex” LNPs. Additional data are shown in SI Appendix, Fig. S3.
The fact that QII/HII RNA–loaded LNPs fuse with isolated endosomes side-by-side on a glass slide is encouraging for their application as fusogenic LNPs for RNA delivery, but this is not representative of the fusion process in live cells where LNPs have to break out of an enclosing endosomal membrane. To evaluate how LNPs of different nanostructures fuse with the entrapping endosomal membranes in live cells, we conducted a fluorescence self-quenching experiment in live HeLa cells (Fig. 4 C and D). We employed two types of siRNA-loaded LNPs: cuboplexes (QII/HII) and lipoplexes (L) labeled with high molar percentages (1%) of a self-quenching dye, DiD (75, 76). When the membranes of the LNP–siRNA complexes fuse with endosomes, DiD molecules will diffuse across the membrane, reducing the self-quenching of DiD and increasing its fluorescence. Fig. 4 C and D displays the obtained DiD fluorescence recovery at three time points (1, 3, and 5 h) after incubation with L (Fig. 4C, lipoplexes) and QII/HII (Fig. 4D, cuboplexes) siRNA-loaded LNPs. As time progresses, an increasing DiD signal (red fluorescence signal) is observed in cells treated with cuboplexes, indicating that indeed LNPs successfully fused with the endosomal membrane that internalized them. Comparatively at the same time point, cells treated with lipoplexes show little DiD fluorescence recovery, implying less effective endosomal membrane fusion. These results show that the nanostructure of LNP–RNA complexes plays a critical role in their ability to fuse and break out of endosomal compartments.
RNA Delivery of LNP–RNA Complexes of Different Nanostructures.
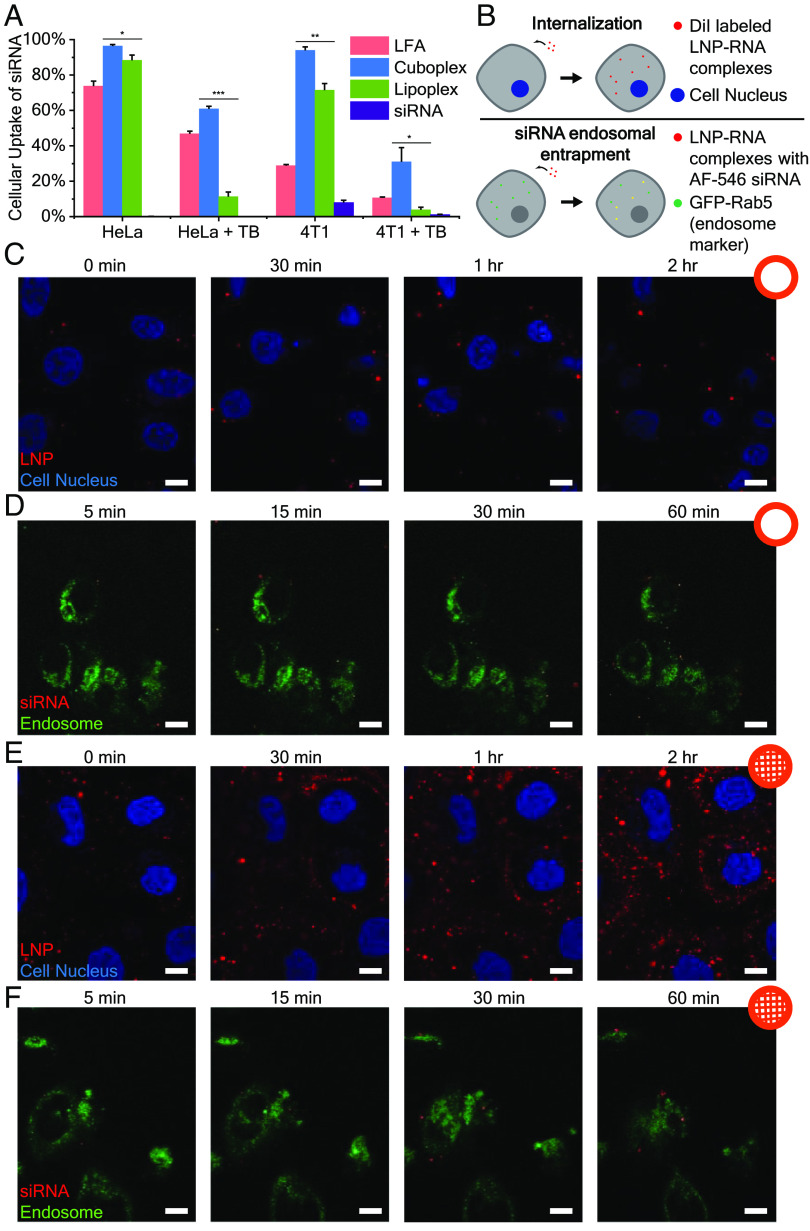
We next investigated whether the ability for QII/HII RNA-loaded LNPs (cuboplexes) to efficiently fuse with endosomal membranes translates to better delivery of cargo and more efficient endosomal escape. Uptake of siRNA in HeLa cells and murine breast cancer (4T1) cells was quantified by flow cytometry (Fig. 5A). To verify that the siRNA signal is not from siRNA outside the cell membrane or from dead cells, trypan blue (TB) was added to quench the fluorescence of siRNA outside the cellular membrane and that of membrane-compromised cells (77, 78). The results show that HeLa cells treated with cuboplexes have significantly more cytoplasmic siRNA compared to cells treated with lipoplexes, and the siRNA uptake is comparable to that of Lipofectamine (LFA), a commercially available siRNA transfection agent. HeLa cells are well known to be relatively easy to transfect; however, in more resilient cell lines like 4T1 cells (79), we observe that cuboplexes continue to outperform lipoplexes in siRNA delivery capabilities.
Fig. 5.
Cuboplexes deliver siRNA more effectively compared to lipoplexes. (A) Uptake of siRNA in HeLa cells and 4T1 cells quantified by flow cytometry. Trypan blue (TB) was added to quench the fluorescence of siRNA outside the cellular membrane and that of membrane-compromised cells (n = 3, data presented as mean ± SD). Significance between groups was calculated with two-sided Student’s t-test and indicated as *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001. (B) Schematic representation of live cell imaging experiments evaluating the internalization and siRNA endosomal entrapment of LNP–RNA complexes. (C and D) HeLa cells were treated with DiO-labeled LNP–siRNA complexes (red) and imaged every 5 min. Less lipoplexes (C) enter the cells compared to cuboplexes (D). (Scale bar, 10 μm.) (E and F) Endosome-labeled cells were treated with cuboplexes (E) and lipoplexes (F) containing AF546-siRNA and imaged every 5 min. Less siRNA (red) are entrapped in the endosome in cuboplex-treated cells compared to those treated with lipoplexes. (Scale bar, 10 μm.)
To test the hypothesis that cuboplexes deliver more RNA to cells compared to lipoplexes because they more effectively fuse and break out of endosomal entrapment, we utilized live cell imaging to compare the cell internalization as well as endosomal entrapment of these two different LNP–RNA complexes (Fig. 5B). DiO-labeled LNP–siRNA complexes (shown as red) were added to HeLa cells stained with Hoescht nuclei dye (blue) and imaged as a function of time (0, 30, 60, and 120 min). The higher amount of red signal around the nuclei would correspond to a higher extent of nanoparticle internalization (Fig. 5 C and E). Lipoplexes show lower levels of internalization by HeLa cells compared to cuboplexes. To investigate the endosomal entrapment of the different LNP–siRNA complexes, early endosomes of HeLa cells were labeled with a fusion construct of Rab5a-GFP (shown as green). The cells were treated with LNP–siRNA complexes prepared with siRNA fluorescently labeled with Alexa Fluor 546 (AF546), shown as red, and monitored over time (5, 15, 30, and 60 min). When siRNA resides entrapped in the endosome, the fluorescent signal of siRNA and endosome would colocalize, appearing as yellow. Less colocalization, or yellow, would be an indication of less endosomal entrapment and more occurrences of successful endosomal escape. In Fig. 5 D and F, compared to lipoplexes, cuboplexes showed less colocalization with the endosomes, indicating that less cuboplexes were entrapped.
Combined, lipoplexes show low levels of internalization and high amounts of siRNA–endosome colocalization (Fig. 5 C and D) throughout the investigated timeline of up to 2 h (internalization) and 1 h (entrapment). This indicates that cuboplexes have superior ability not only to get internalized by cells at faster rates but also to efficiently evade endosomal entrapment and translocate RNA into the cytosol.
We have previously shown that cuboplexes are more efficient at transfecting siRNA into cells compared to lipoplexes leading to better specific gene-knockdown performance (39–41). When replacing siRNA for mRNA, LNPs retain their structural identity (Fig. 2) as well as their shape and size (SI Appendix, Table S1). To show that cuboplexes containing siRNA silence genes more effectively than lipoplexes with siRNA, we performed an siRNA knockdown experiment with cuboplexes, lipoplexes, and LNPs formed using the state-of-the-art ionizable lipid formulations (IL-LNPs). The LNPs used for these transfection studies were prepared by nanoprecipitation in microfluidics, which is a well-established LNP preparation method. Dialysis was utilized to remove the solvent to minimize disruption to the LNPs. Other studies have shown that dialysis does have an impact on the structure of IL-LNPs (30, 31). Specifically, changes in pH during dialysis drive IL-LNPs to have a more solid core, and the presence of ethanol disrupts the stability. However, in our case, LNPs use cationic lipid DOTAP, and there is no pH difference present during the dialysis process. Therefore, a structural change seen in IL-LNPs from dialysis is not observed. HeLa cells stably expressing green fluorescent protein (GFP) were treated with LNPs containing siRNA targeting GFP (siGFP) or scrambled siRNA (siNeg). LNP–siNeg samples were included as a negative control to test the effects of nonspecific gene knockdown. The level of GFP expression was evaluated by its fluorescence using flow cytometry. As shown in Fig. 6A, cuboplexes successfully reduced GFP expression, while lipoplexes showed only minimal GFP silencing. IL-LNPs showed the strongest knockdown capabilities of all. This is consistent with our understanding of how ILs operate such that ILs become cationic inside the endosome electrostatically binding the endosomal membrane and facilitating fusion (10). To test how general the effect is with respect or other RNA-editing approaches, we employed cuboplex LNPs to deliver firefly luciferase–expressing mRNA (mFluc) to cells. It is clear that cuboplexes lead to more efficient activation of luciferase expression (Fig. 6B) compared to parent lipoplex mRNA–LNPs. Both siRNA-mediated silencing and mRNA-mediated expression reveal that LNPs that are inherently more fusogenic (ILs due to electrostatic attraction and cuboplexes due to effects) are more efficacious. These data highlight the importance of designing not only the lipid identity and composition but also the overall nanostructure of the LNP.
Fig. 6.
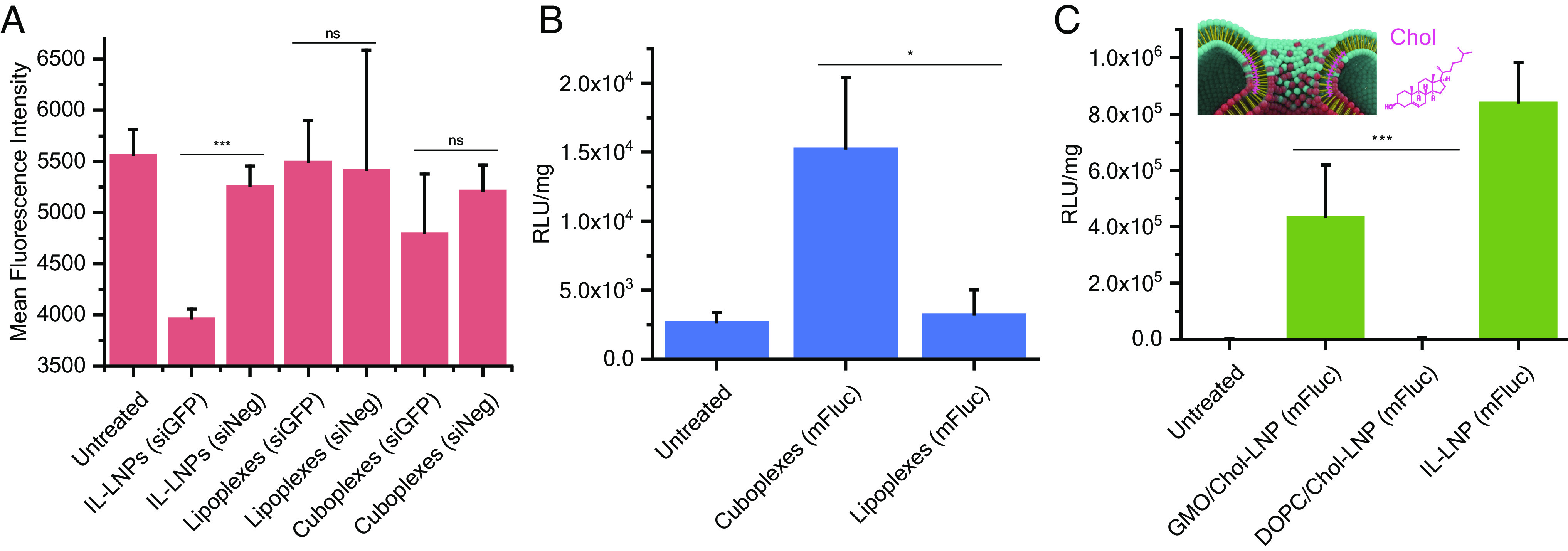
Cuboplexes show higher transfection efficiency of siRNA and mRNA compared to lipoplexes. (A) siRNA knockdown of GFP in HeLa–GFP cells with IL-LNPs, cuboplexes, and lipoplexes. GFP expression is determined by fluorescence intensity measured by flow cytometry (n = 3, data presented as mean ± SD). (B) Luciferase activity of HeLa cells transfected with firefly luciferase mRNA with mRNA alone, cuboplexes, and lipoplexes (n = 5, data presented as mean ± SD). (C) Luciferase activity of HeLa cells transfected with firefly luciferase mRNA using IL-LNPs and LNP–mRNA complexes with composition of GMO/DOPC:Chol:DOTAP:DOPE-PEG 55:30:14:1 mol% (n = 5, data presented as mean ± SD). Significance between groups was calculated with two-sided Student’s t-test and indicated as *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001.
To get further insight into the importance of LNP fusogenicity for efficient RNA delivery, we evaluated the approach of Sahay’s lab that has demonstrated the beneficial effects of cholesterol on endosomal escape (24, 25, 80). We designed a GMO–LNP (with coexisting QII/HII phases) containing 30 mol % of cholesterol (GMO/DOPC:Chol:DOTAP:DOPE-PEG 55:30:14:1), which matches the composition utilized in state-of-the-art IL-LNPs. Previous studies have shown that cholesterol helps stabilize high-curvature membrane bud necks (81, 82), and they should have a stabilizing effect also on membrane fusion pores. As schematically depicted in the inset of Fig. 6C, the presence of fusion pores that are formed after merging two apposing membranes is fundamental to the endosomal escape process. It is expected that cholesterol from LNPs has the ability to stabilize high-curvature regions of the membrane fusion pores aiding in successful endosomal escape. The transfection data show (Fig. 6C) that for cells treated with complexes containing GMO and Chol (GMO/Chol-LNPs), a significant, more amount of luciferase is expressed compared to cells treated with complexes with DOPC and Chol (DOPC/Chol-LNPs). This is important because it reveals that cholesterol alone is not enough to elicit fusion. However, after the onset of membrane attachment enhanced by electrostatic effects in the case of ILs and in the case of GMO, cholesterol accumulation in the membrane pore would reduce the energetic cost of the topological transformation from endosomes encapsulating LNPs to a single endosome-LNP fused object. A lot more fusion pores are created when the cells are treated with GMO/Chol-LNPs or IL/Chol-LNPs, and membrane fusion plays an important role in the endosomal escape of LNPs. It is noteworthy that a plasma membrane integrity assay confirmed that all LNP–RNA complexes used in this study have negligible cytotoxicity (SI Appendix, Fig. S4). The possibility of replacing DOTAP with IL was also explored, and the immediate finding is that replacing cationic DOTAP with SM-102 ionizable lipids does not result in a well-ordered cuboplex (or any interesting nanostructure), and as expected, their performance in mRNA delivery and expression is quite poor compared to a cuboplex (SI Appendix, Fig. S5). These results suggest that a different ionizable lipid structure (not commercially available) would be needed to construct ionizable cuboplexes.
Discussion
In this study, we show that fusogenicity is a fundamental LNP property for efficacious RNA delivery and that LNP nanostructure identity is an additional handle to enhance it. Compared to lamellar structured LNP–RNA complexes, cubic and inverse hexagonal structured LNP–RNA complexes are able to fuse with endosomal membranes easier, hence leading to a higher extent of endosomal escape. These findings provide valuable insight in how nanostructures can affect nanoparticle–cell interactions, but also highlight the potential in utilizing structurally active lipids and nanoparticle structure as an additional handle to controlling the efficacy of drug delivery systems.
Endosomal escape has been a major hurdle in the development of successful RNA therapeutics. Our studies were able to show that by controlling the nanostructure of LNP–RNA complexes, we can influence the efficiency of endosomal escape without any aid from proteins, endosome acidification, or lipid ionization. Specifically, complexes with periodic bicontinuous cubic membrane interiors are able to promote membrane fusion between LNPs and endosomal membranes. When cholesterol is included, the process of membrane-fusion pore formation is facilitated. Cuboplexes have an intrinsic ability to lower the elastic cost of inducing membrane fusion followed by the topological transformation from an endosome with a nested LNP to a fused LNP–endosome state having a pore through which RNA can be transported into the cytosol. This enhancement in endosomal escape for cuboplexes led to better RNA delivery compared to its lipoplex counterparts. These results show that in addition to a judicious choice of lipid composition, LNP nanostructure is a critical factor controlling fusogenicity. In other words, the inclusion of structurally active lipids is a potential design handle of nonviral delivery systems, and more research is required to fully unravel how nanoparticles of various structures interact with cells. Our paper highlights the importance of quantifying and predicting LNP fusogenicity. We show that LNP fusion ability cannot simply be justified by the effect of “inverted conical” lipids (negative spontaneous C0< 0) as bicontinuous cubic structures (C0 = 0) are highly fusogenic. Rather than evaluating the ratio of bicontinuous cubic to hexagonal phases, we propose a more general description in that the Gaussian modulus, or rather the ratio between the Gaussian and the Bending moduli (), could be a better quantifier of LNP fusogenicity as it accounts for contributions of topology (generation of Gaussian curvature, e.g., during the formation of a fusion pore), elasticity, and C0. However, unlike C0 and κ that are experimentally accessible, is hard to measure as it requires a controllable and reversible change of membrane topology. Nevertheless, SAXS methods have been successfully employed (59, 60, 83, 84) to quantify the Gaussian to bending moduli ratios of the monolayers interfacing with the water channels in hexagonal and bicontinuous cubic phases. For a ternary GMO/DOPC/DOPE (58:38:4 mol%) mixture, SAXS measurements yield (59) comparing to pure DOPE where (85) that is less fusogenic. In addition, as the importance of LNP nanostructure becomes more appreciated, further research needs to be done to fully explore how different preparation and purification methods might affect the nanostructure of LNPs beyond size and encapsulation efficiency (86).
Materials and Methods
Materials.
Glycerol monooleate (GMO) was purchased from Sigma-Aldrich (MO, USA). 8-[(2-hydroxyethyl)[6-oxo-6-(undecyloxy)hexyl]amino]-octanoic acid, 1-octylnonyl ester (SM-102) was purchased from Cayman Chemical Company (MI, USA). 1,2-dioleoyl-sn-glycero-3- phosphocholine (DOPC), 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)- 2000] (DOPE-PEG), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2000), and cholesterol (Chol) were purchased from Avanti Polar Lipids (AL, USA). siRNA targeting GFP and scrambled siRNA were purchased from ThermoFisher Scientific (IL, USA). Alexa Fluor 546 labeled siRNA (AF546-siRNA) was purchased from Qiagen (MD, USA). mRNA targeting firefly luciferase was purchased from TriLink Biotechnologies (CA, USA).
Preparation of LNPs and LNP–RNA Complexes.
Lipid chloroform solutions were mixed at desired volumetric ratios. Chloroform was removed by treating the solution with a stream of nitrogen and then placing it under vacuum for at least 8 h. DOPE-PEG was dried in a separate vial. The dried lipid film was hydrated with sterile Milli-Q water and incubated overnight at 37 °C. LNPs were obtained by sonicating the suspension with a cup horn system (Fisher Scientific) for 6 min at 100 % amplitude. The temperature was maintained at below 4 °C during sonication by a water chiller (Qsonica). The LNPs were transferred to the vial with dried DOPE-PEG and incubated for 1 h at 60 °C for postpegylation.
LNP–RNA complexes were formed by mixing LNPs with siRNA or mRNA at a charge ratio ρ (nDOTAP/nNA) of 3.
For transfection experiments, LNP–RNA complexes were formed by NanoAssemblr Ignite (Precision NanoSystems). Ionizable lipid (IL) LNPs were formed with a formulation of SM-102:DSPC:Chol:DMG-PEG2000 at molar ratios 50:10:38.5:1.5. Lipid chloroform solutions were mixed at desired volumetric ratios, and the solvent was removed as described above. The lipids were then dissolved in ethanol at a concentration of 10 mM. The total flow rate was maintained at 12 mL per min for all formulations. For cuboplexes, lipoplexes, GMO/Chol LNPs, and DOPC/Chol LNPs, 4:1 ratio of aqueous to ethanol phase was used, while a 3:1 ratio was used for formulating IL-LNPs. All formulations were made at a charge ratio ρ (nDOTAP or IL/nNA) of 6. Ethanol was removed by dialysis using Slide-A-LyzerTM cassettes with a molecular weight cutoff of 3.5k. The encapsulation efficiency was measured using QuantiFluor®RNA System (Promega).
Small-Angle X-Ray Scattering.
LNP–RNA complexes (total lipid concentration 20 mM) were prepared and transferred to quartz capillaries (Hilgenberg Glas, Germany). Synchrotron SAXS was performed at beamline 12-ID-B of the Advanced Photon Source at Argonne National Laboratory. The average photon energy was 14 keV, and the data were radially averaged upon acquisition on a Pilatus 2M detector.
Cryogenic Electron Microscopy.
To prepare samples for cryo-EM imaging, lacey carbon–coated 300 mesh copper grids (Electron Microscopy Sciences) were glow discharged at 15 mA for 30 s with the PELCO easiGLOWTM glow discharge system (Ted Pella). Then, 4 μL of sample was applied to the grids and incubated for 10 min. The grids were blotted with filter paper, and another 4 μL of sample was applied. The grids were then blotted for 2.5 s and plunge frozen in liquid ethane using Vitrobot Mark IV, under 4 °C and 100% humidity. The grids were kept in liquid nitrogen until imaging. Cryo-EM images were collected with Glacios Cryo-TEM (ThermoFisher) at 200 kV with a Falcon 4 direct electron detector. Images were taken at −2 μm defocus to improve contrast. Fast Fourier transforms (FFTs) of images were obtained by ImageJ software.
Cell Culture.
HeLa cells (ATCC), 4T1 cells (ATCC), HeLa–GFP cells (Cell BioLabs), and HeLa-Luc cells (Signosis) were cultured in full cell media consisting of Dulbecco’s modified Eagle’s medium (Corning), 10% fetal bovine serum (Gibco), and 1% penicillin–streptomycin (Gibco) at 37 °C with 5% of carbon dioxide.
Endosome Isolation and Characterization.
Endosomes were isolated with the Trident endosome isolation kit (GeneTex). Particle concentration and size were measured by nanoparticle tracking analysis with NanoSight NS300 (Malvern Panalytical).
Membrane Fusion Studies.
For confocal microscopy analysis, DiD (Biotum)-labeled LNPs were prepared by including 0.1% DiD in the lipid mixture. Endosomes were labeled by incubating with 20 μM NileRed for 1 h at 37 °C. Free dye was removed from endosomes by centrifuging for 30 min at 10,000×g and discarding the supernatant. The endosomes were resuspended in PBS and incubated with DiD-labeled LNPs for 6 h at 37 °C. The mixture was imaged on an LSM 800 (Carl Zeiss) confocal microscope.
For the FRET assay, endosomes were incubated with 20 μM DiO and 20 μM DiI for 1 h at 37 °C for colabeling. Free dye was removed, and endosomes were resuspended in PBS. Endosomes and lipid–siRNA complexes were incubated at 37 °C and measured for DiO fluorescence every 5 min. Fluorescence was detected with the Synergy Neo 2 microplate reader (Biotek).
Live Cell Imaging of the Uptake of LNP–RNA Complexes.
Labeled LNP–siRNA complexes were prepared by including 0.1% DiI in the lipid mixture. HeLa-Luc were cultured on coverslip bottom dishes (ibidi) and stained with Hoescht dye. Complexes were added to the cells and imaged immediately on an LSM 800 (Carl Zeiss) confocal microscope: 405-nm laser was used for the Hoescht dye channel, and 488-nm laser was used for DiO and DiI. Images were acquired every 5 min for 3 h.
Cells were seeded onto poly-lysine-coated glass-bottom dishes at 1,000 cells/cm2 density, and CellLightTM Early Endosomes-GFP reagent (ThermoFisher Scientific) was added after cell adherence. After 16 h of incubation, complexes with Alexa Fluor 546 labeled siRNA were added to the cells with a final siRNA concentration of 33 nM. The dishes were observed immediately under confocal microscopy. Images were acquired every 5 min for 4 h.
Flow Cytometry.
HeLa cells (for siRNA uptake experiments) or HeLa–GFP cells (for siRNA transfection experiments) were seeded onto 12-well plates at 1,000 cells/cm2 density prior to transfection. The following day, LNP–siRNA complexes were added to the cell media in which the cells were cultured in with a final siRNA concentration of 33 nM, and then incubated for 24 h. After incubation, the cells were trypsinized and washed with PBS. Cells were later suspended in FACS buffer (98% PBS, 2% FBS) and analyzed by flow cytometry (BD LSRFortessa X-20). For siRNA uptake experiments, the fluorescence of AF-546 was measured, and after the sample data were acquired, 0.4% trypan blue was added to the samples at a 1:10 ratio and analyzed by flow cytometry. For siRNA transfection experiments, the fluorescence of GFP was measured.
Luciferase Assay.
Cells were seeded onto 96-well plates at 1,000 cells/cm2 density prior to transfection. The following day, LNP–mRNA complexes were added to the cell media in which the cells are cultured in with 0.1 μg mRNA per well. The cells were incubated for 24 h, and the luciferase activity was evaluated by a luciferase assay (Promega). Luminescence was measured with the Synergy Neo 2 microplate reader (Biotek) and normalized by the cell protein mass of each well. Cell protein mass was quantified by the BCA assay (ThermoFisher Scientific).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the NIH under Grant Nos. 1DP2EB024377-01 and R01GM143723-01A1. This research used resources of the Advanced Photon Source, beamline 12-ID-B, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This research was in part conducted in the Materials Research Laboratory in University of Illinois Urbana-Champaign. We would like to acknowledge Professor Markus Deserno at the Department of Physics at Carnegie Mellon University for fruitful discussion regarding the Gaussian modulus and membrane fusion.
Author contributions
L.Z. and C.L. designed research; L.Z., S.R.B., and Z.T. performed research; L.Z. and C.L. analyzed data; and L.Z. and C.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. Previously published data were used for this work (40).
Supporting Information
References
- 1.Hou J., et al. , YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 18, 163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cochran J. N., et al. , Non-coding and Loss-of-Function Coding Variants in TET2 are Associated with Multiple Neurodegenerative Diseases. Am. J. Hum. Genet. 106, 632–645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matharu N., et al. , CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363, eaau0629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni J. A., et al. , The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 16, 630–643 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Feng R., Patil S., Zhao X., Miao Z., Qian A., RNA Therapeutics - Research and Clinical Advancements. Front. Mol. Biosci. 8, 710738. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines – a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berryer M. H., et al. , Mutations in SYNGAP1 Cause Intellectual Disability, Autism, and a Specific Form of Epilepsy by Inducing Haploinsufficiency. Hum. Mutat. 34, 385–394 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Li J., Parker B., Martyn C., Natarajan C., Guo J., The PMP22 Gene and Its Related Diseases. Mol. Neurobiol. 47, 673–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S.-D., Huang L., Non-viral is superior to viral gene delivery. J. Control. Release. 123, 181–183 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Hou X., Zaks T., Langer R., Dong Y., Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rietwyk S., Peer D., Next-Generation Lipids in RNA Interference Therapeutics. ACS Nano 11, 7572–7586 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Wood H., FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat. Rev. Neurol. 14, 570 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Baden L. R., et al. , Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolgin E., Better lipids to power next generation of mRNA vaccines. Science 376, 680–681 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Patel S., et al. , Brief update on endocytosis of nanomedicines. Adv. Drug Deliv. Rev. 144, 90–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degors I. M. S., Wang C., Rehman Z. U., Zuhorn I. S., Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors. Acc. Chem. Res., acs.accounts.9b00177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ur Rehman Z., Hoekstra D., Zuhorn I. S., Mechanism of Polyplex- and Lipoplex-Mediated Delivery of Nucleic Acids: Real-Time Visualization of Transient Membrane Destabilization without Endosomal Lysis. ACS Nano 7, 3767–3777 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Lamb C. A., Dooley H. C., Tooze S. A., Endocytosis and autophagy: Shared machinery for degradation. Bioessays 35, 34–45 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Wittrup A., et al. , Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 33, 870–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahay G., et al. , Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 31, 653–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilleron J., et al. , Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31, 638–646 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Semple S. C., et al. , Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 28, 172–176 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Kauffman K. J., et al. , Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 15, 7300–7306 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Patel S., et al. , Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA. Nano Lett. 17, 5711–5718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel S., et al. , Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 11, 983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paunovska K., Loughrey D., Dahlman J. E., Drug delivery systems for RNA therapeutics (Nat. Rev, Genet, 2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu B., et al. , Thermostable ionizable lipid-like nanoparticle (iLAND) for RNAi treatment of hyperlipidemia. Sci. Adv. 8, eabm1418 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabnis S., et al. , A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 26, 1509–1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S., et al. , Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 20, 701–710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanez Arteta M., et al. , Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. PNAS 115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulkarni J. A., et al. , On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano 12, 4787–4795 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Dilliard S. A., Cheng Q., Siegwart D. J., On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 118, e2109256118. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Q., et al. , Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung S. S. W., Leal C., The stabilization of primitive bicontinuous cubic phases with tunable swelling over a wide composition range. Soft Matter 15, 1269–1277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H., Song Z., Leal C., Super-swelled lyotropic single crystals. PNAS 114, 10834–10839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang M., Kim H., Leal C., Self-organization of nucleic acids in lipid constructs. Curr. Opin. Colloid Interface Sci 26, 58–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rueben J., Jayaraman A., Mahanthappa M. K., Leal C., Near-Infrared-Triggered Reversible Transformations of Gold Nanorod-Laden Lipid Assemblies: Implications for Cellular Delivery. ACS Appl. Nano Mater. 5, 710–717 (2022). [Google Scholar]
- 38.Porras-Gomez M., Leal C., Lipid-based liquid crystalline films and solutions for the delivery of cargo to cells. Liq. Cryst. Rev. 7, 167–182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H., Leal C., Cuboplexes: Topologically Active siRNA Delivery. ACS Nano 9, 10214–10226 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Kim H., Sung J., Chang Y., Alfeche A., Leal C., Microfluidics Synthesis of Gene Silencing Cubosomes. ACS Nano 12, 9196–9205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leal C., Bouxsein N. F., Ewert K. K., Safinya C. R., Highly Efficient Gene Silencing Activity of siRNA Embedded in a Nanostructured Gyroid Cubic Lipid Matrix. J. Am. Chem. Soc. 132, 16841–16847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan C.-L., et al. , Optimizing cationic and neutral lipids for efficient gene delivery at high serum content: Lipoplexes resistant to high serum. J. Gene Med. 16, 84–96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirlekar R., Jain S., Patel H., Garse M., Kadam V., Hexosomes: A novel drug delivery system. Curr. Drug Deliv. 7, 28–35 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Leal C., Ewert K. K., Shirazi R. S., Bouxsein N. F., Safinya C. R., Nanogyroids Incorporating Multivalent Lipids: Enhanced Membrane Charge Density and Pore Forming Ability for Gene Silencing. Langmuir 27, 7691–7697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porras-Gómez M., Shoaib T., Steer D., Espinosa-Marzal R. M., Leal C., Pathological cardiolipin-promoted membrane hemifusion stiffens pulmonary surfactant membranes. Biophys. J. 121, 886–896 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vermeulen L. M., De Smedt S. C., Remaut K., Braeckmans K., The proton sponge hypothesis: Fable or fact? Euro. J. Pharm. Biopharm. 129, 184–190 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Benjaminsen R. V., Mattebjerg M. A., Henriksen J. R., Moghimi S. M., Andresen T. L., The Possible "Proton Sponge " Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal pH. Mol. Ther. 21, 149–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen F., Melikyan G., The Energetics of Membrane Fusion from Binding, through Hemifusion, Pore Formation, and Pore Enlargement. J. Membrane. Biol. 199, 1–14 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Zuhorn I. S., et al. , Nonbilayer phase of lipoplex–membrane mixture determines endosomal escape of genetic cargo and transfection efficiency. Mol. Ther. 11, 801–810 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Helfrich W., Elastic Properties of Lipid Bilayers: Theory and Possible Experiments. Z. Für Naturforschung C 28, 693–703 (1973). [DOI] [PubMed] [Google Scholar]
- 51.S. Germain, Recherches sur la théorie des surfaces élastiques (V. Courcier, 1821).
- 52.Hyde S., Blum Z., Landh T., Lidin S., Ninham B. W., The Language of Shape (Elsevier Science B.V, Amsterdam, 1997). [Google Scholar]
- 53.Bassereau P., et al. , The 2018 biomembrane curvature and remodeling roadmap. J. Phys. D: Appl. Phys. 51, 343001. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rädler J. O., Koltover I., Salditt T., Safinya C. R., Structure of DNA-Cationic Liposome Complexes: DNA Intercalation in Multilamellar Membranes in Distinct Interhelical Packing Regimes. Science 275, 810–814 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Barriga H. M. G., Holme M. N., Stevens M. M., Cubosomes: The Next Generation of Smart Lipid Nanoparticles? Angew. Chem. Int. Ed. 58, 2958–2978 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegel D. P., Inverted micellar intermediates and the transitions between lamellar, cubic, and inverted hexagonal lipid phases. ii. implications for membrane-membrane interactions and membrane fusion. Biophys. J. 49, 1171–1183 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siegel D. P., Epand R. M., The mechanism of lamellar-to-inverted hexagonal phase transitions in phosphatidylethanolamine: Implications for membrane fusion mechanisms. Biophys. J. 73, 3089–3111 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siegel D. P., Inverted micellar intermediates and the transitions between lamellar, cubic, and inverted hexagonal amphiphile phases. iii. isotropic and inverted cubic state formation via intermediates in transitions between lα and hII phases. Chem. Phy. Lipids 42, 279–301 (1986). [DOI] [PubMed] [Google Scholar]
- 59.Siegel D. P., Determining the Ratio of the Gaussian Curvature and Bending Elastic Moduli of Phospholipids from QII Phase Unit Cell Dimensions. Biophys. J. 91, 608–618 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siegel D., Kozlov M., The Gaussian Curvature Elastic Modulus of N-Monomethylated Dioleoylphosphatidylethanolamine: Relevance to Membrane Fusion and Lipid Phase Behavior. Biophys. J. 87, 366–374 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellens H., et al. , Membrane fusion and inverted phases. Biochemistry 28, 3692–3703 (1989). [DOI] [PubMed] [Google Scholar]
- 62.Allen S. D., Bobbala S., Karabin N. B., Scott E. A., On the advancement of polymeric bicontinuous nanospheres toward biomedical applications. Nanoscale Horiz. 4, 258–272 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Allen S. D., Bobbala S., Karabin N. B., Modak M., Scott E. A., Benchmarking Bicontinuous Nanospheres against Polymersomes for in Vivo Biodistribution and Dual Intracellular Delivery of Lipophilic and Water-Soluble Payloads. ACS Appl. Mater. Interfaces 10, 33857–33866 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Bobbala S., Allen S. D., Scott E. A., Flash nanoprecipitation permits versatile assembly and loading of polymeric bicontinuous cubic nanospheres. Nanoscale 10, 5078–5088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Modak M., et al. , Magnetic Nanostructure-Loaded Bicontinuous Nanospheres Support Multicargo Intracellular Delivery and Oxidation-Responsive Morphological Transitions. ACS Appl. Mater. Interfaces 12, 55584–55595 (2020). [DOI] [PubMed] [Google Scholar]
- 66.S. Food, D. Administration, Cfr-code of federal regulations title 21. (2017).
- 67.Upadhyay M. S., Pathak K., Glyceryl monooleate-coated bioadhesive hollow microspheres of riboflavin for improved gastroretentivity: Optimization and pharmacokinetics. Drug Deliv. and Transl. Res. 3, 209–223 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Larsson K., Two cubic phases in monoolein - water system. Nature 304, 664–664 (1983). [Google Scholar]
- 69.Mare R., et al. , Post-insertion parameters of PEG-derivatives in phosphocholine-liposomes. Int. J. Pharm. 552, 414–421 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Oliveira A. C., et al. , Stealth monoolein-based nanocarriers for delivery of siRNA to cancer cells. Acta Biomater. 25, 216–229 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Eliasen R., Andresen T. L., Larsen J. B., PEG-Lipid Post Insertion into Drug Delivery Liposomes Quantified at the Single Liposome Level. Adv. Mater. Interfaces 6, 1801807 (2019). [Google Scholar]
- 72.Leal C., et al. , Stacking of short DNA induces the gyroid cubic-to-inverted hexagonal phase transition in lipid–DNA complexes. Soft Matter 9, 795–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tonggu L., Wang L., Cryo-EM sample preparation method for extremely low concentration liposomes. Ultramicroscopy 208, 112849. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao X., et al. , Rapid Detection of Exosomal MicroRNAs Using Virus-Mimicking Fusogenic Vesicles. Angew. Chem. Int. Ed. 58, 8719–8723 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Joo K.-I., Wang P., Visualization of targeted transduction by engineered lentiviral vectors. Gene. Ther. 15, 1384–1396 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lakadamyali M., Rust M. J., Babcock H. P., Zhuang X., Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 100, 9280–9285 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wonder E., et al. , Competition of charge-mediated and specific binding by peptide-tagged cationic liposome–DNA nanoparticles in vitro and in vivo. Biomaterials 166, 52–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ewert K. K., et al. , Synthesis of linear and cyclic peptide-PEG–lipids for stabilization and targeting of cationic liposome–DNA complexes. Bioorg. Med. Chem. Lett. 26, 1618–1623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neuhaus B., et al. , Nanoparticles as transfection agents: A comprehensive study with ten different cell lines. RSC Adv. 6, 18102–18112 (2016). [Google Scholar]
- 80.Eygeris Y., Gupta M., Kim J., Sahay G., Chemistry of Lipid Nanoparticles for RNA Delivery. Acc. Chem. Res. 55, 2–12 (2022). [DOI] [PubMed] [Google Scholar]
- 81.Ryu Y.-S., et al. , Reconstituting ring-rafts in bud-mimicking topography of model membranes. Nat. Commun. 5, 4507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang J., Xue R., Ong W.-Y., Chen P., Roles of Cholesterol in Vesicle Fusion and Motion. Biophys. J. 97, 1371–1380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Templer R. H., Khoo B. J., Seddon J. M., Gaussian Curvature Modulus of an Amphiphilic Monolayer. Langmuir 14, 7427–7434 (1998). [Google Scholar]
- 84.Hu M., Briguglio J. J., Deserno M., Determining the Gaussian Curvature Modulus of Lipid Membranes in Simulations. Biophys. J. 102, 1403–1410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Siegel D. P., The Gaussian Curvature Elastic Energy of Intermediates in Membrane Fusion. Biophys. J. 95, 5200–5215 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jung H. N., Lee S.-Y., Lee S., Youn H., Im H.-J., Lipid nanoparticles for delivery of RNA therapeutics: Current status and the role of in vivo imaging. Theranostics 12, 7509–7531 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix. Previously published data were used for this work (40).