Significance
Bacteroides is a prominent genus of the human gut microbiota, which produces large amounts of OMVs (outer membrane vesicles) carrying specific protein cargo. Here, we show that sorting of proteins into OMVs depends on a negatively charged “lipoprotein export signal” (LES). We employed OM (outer membrane) and OMV markers fused to fluorescent proteins to show OMV biogenesis via time-lapse fluorescence microscopy. Additionally, we performed comparative proteomic analyses showing that Bt (Bacteroides thetaiotaomicron) actively alters the OMV content to optimize the utilization of polysaccharides. We conclude that OMV production is highly regulated in Bt. Despite the recognized roles of OMVs in Bacteroides, little is known about their biogenesis. Future studies will focus on further elucidating this undercharacterized process.
Keywords: Bacteroides, OMV, vesicles
Abstract
Extracellular vesicles are produced in all three domains of life, and their biogenesis has common ancient origins in eukaryotes and archaea. Although bacterial vesicles were discovered several decades ago and multiple roles have been attributed to them, no mechanism has been established for vesicles biogenesis in bacteria. For this reason, there is a significant level of skepticism about the biological relevance of bacterial vesicles. Bacteroides thetaiotaomicron (Bt), a prominent member of the human intestinal microbiota, produces significant amounts of outer membrane vesicles (OMVs) which have been proposed to play key physiological roles. Here, we employed a dual marker system, consisting of outer membrane- and OMV-specific markers fused to fluorescent proteins to visualize OMV biogenesis by time-lapse microscopy. Furthermore, we performed comparative proteomic analyses to show that, in Bt, the OMV cargo is adapted for the optimal utilization of different polysaccharides. We also show that a negatively charged N-terminal motif acts as a signal for protein sorting into OMVs irrespective of the nutrient availability. Our results demonstrate that OMV production is the result of a highly regulated process in Bt.
The human gastrointestinal tract harbors one of the densest microbial communities known in nature. A healthy adult human gut microbiota is primarily colonized by two bacterial phyla: Firmicutes and Bacteroidota (1, 2). Bacteroides spp. are prominent members of the human intestinal microbiota, representing nearly one-third of the total composition in industrialized countries (3). It has been shown that Bacteroides spp. promote gut homeostasis by shaping host immunity (4–6), and preventing pathogen colonization (7, 8). Moreover, Bacteroides is well known as a glycan-degrading specialist that can metabolize a wide array of polysaccharides, derived from host glycans and dietary fibers (9–12). This metabolic flexibility resides in a series of gene clusters termed polysaccharide utilization loci (PULs), colocalized and coregulated loci in response to specific glycans that enable the degradation and uptake of specific carbon sources (13). Several studies have shown that Bacteroides spp. produce outer membrane vesicles (OMVs) that participate in glycan utilization and the modulation of host immune responses in the gut (6, 14, 15), among other functions.
In gram-negative bacteria, OMVs are generated by the blebbing of the outer membrane (OM). OMVs are composed mainly of phospholipids, lipopolysaccharide, or lipooligosaccharide, as well as periplasmic and membrane proteins (16). Numerous bacterial processes have been proposed to be mediated by OMVs, including envelope stress responses, delivery of virulence factors, modulation of host immune responses, and digestion of extracellular nutrients (16). Despite being discovered more than 50 y ago (17, 18), the bacterial vesicle field has struggled to progress due to three crucial flaws: 1) There is no method to visually distinguish between genuine OMVs from lysis byproducts that occur during normal cell growth, 2) no reliable ways to quantify OMV production under physiologically relevant conditions are available, and 3) no general mechanism for OMV biogenesis has been established. For these reasons, part of the microbiology community remains skeptical about the biological significance of the OMVs in gram-negative bacteria (19).
Bacteroides spp. produce a large amount of OMVs. Previous mass spectrometry (MS) analyses have demonstrated that in Bacteroides thetaiotaomicron (Bt) and Bacteroides fragilis (Bf ), a subset of proteins is selectively targeted to OMVs, while others are retained in the OM and not directed to vesicles (20, 21). Furthermore, many OMV-localized proteins were shown to be lipoproteins that share common functionalities (i.e., glycosylhydrolases and proteases), and contain a negatively charged “lipoprotein export signal (LES)” motif that was shown to be necessary for surface localization and possibly to efficiently targeting them to OMVs (21, 22). In addition, it has been shown that OMVs from Bacteroides spp. retain glycolytic activity and can be used to supplement the growth of bacteria on carbon sources they normally could not utilize (23, 24). To the best of our knowledge, these features are exclusively found in Bacteroides-derived vesicles. In this work, we employed time-lapse fluorescence microscopy to visualize OMV biogenesis and performed comparative proteomic analyses to demonstrate that Bt actively alters the content of their OMVs to gain a fitness advantage in different growth conditions. Our results show that the production of OMVs in Bt is the result of a highly regulated process.
Results
Vesicle Cargo Proteins Localize at Defined Foci on the Bacterial Membrane.
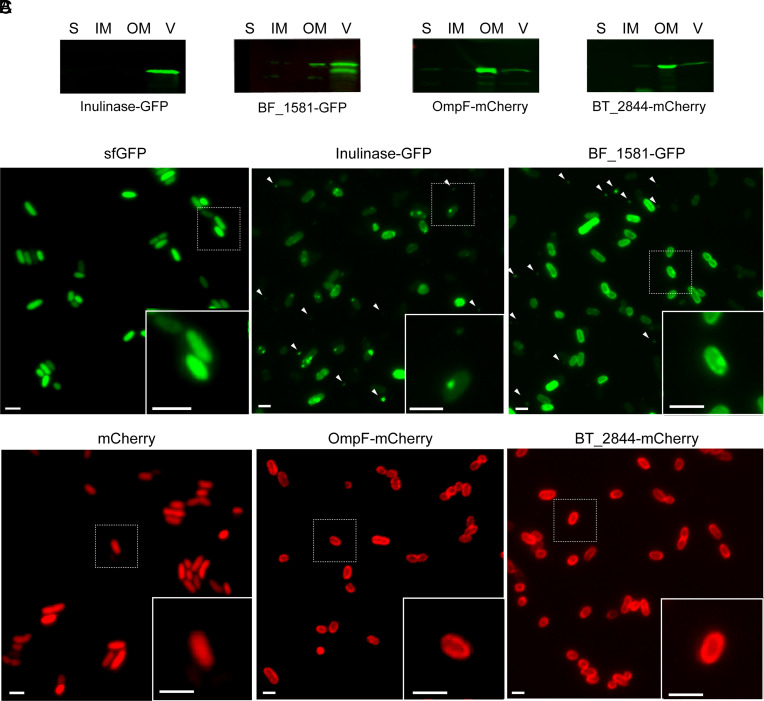
Due to the unique properties, we hypothesized that Bacteroides OMV production is the result of an orchestrated cellular process. To test this hypothesis, we exploited the differential sorting of proteins into OMVs to visualize their biogenesis by fluorescence microscopy. First, we identified proteins that were either enriched in OMVs or retained in the OM and fused them to the fluorophores Superfolder Green Fluorescent Protein (sfGFP) or mCherry. As OMV markers we chose three different lipoproteins previously reported to be preferentially packed in vesicles: BACOVA_04502 (inulinase) from Bacteroides ovatus (Bo) (20, 23, 25), BF_1581 from Bf (20), and BT_3698 (SusG) from Bt (21). As OM markers we chose the integral OM protein BT_0418 (OmpF) and the lipoprotein BT_2844 (21). The chimeric versions of all candidates were cloned under constitutive promoters into the pNBU2 integrative vector in Bt (SI Appendix, Table S1 and Materials and Methods). Western blot analysis of subcellular fractions demonstrated that the fusion of fluorescent proteins did not alter the partitioning between OM and OMVs of each construct (Fig. 1A and SI Appendix, Fig. S1).
Fig. 1.
OMV and OM chimeric markers show a differential distribution in Bt. (A) Western blot of 10 μg of protein from soluble fraction (S), inner membrane (IM), outer membrane (OM), and OMV (V) fractions of Bt expressing Inulinase-GFP, BF_1581-GFP, OmpF-mCherry, or BT_2844-mCherry. Anti-His and anti-mCherry antibodies were employed to identify GFP and mCherry chimeric markers, respectively. (B) Representative widefield fluorescence microscopy images of OMV chimeric markers Inulinase-GFP and BF_1581-GFP. White arrows indicate the presence of OMVs. (C) Representative widefield fluorescence microscopy images of OM chimeric markers OmpF-mCherry and BT_2844-mCherry. Cytosolic expression of sfGFP and mCherry are used as reference (Left). (Scale bar: 2 μm.)
Bt is an anaerobe, however, both mCherry and sfGFP require oxygen for fluorophore maturation. It has been shown that this limitation can be overcome by exposing bacteria to nanaerobic concentrations of oxygen prior to their visualization by fluorescence microscopy (25). Although in these conditions we were able to visualize GFP, fluorescence of chimeric proteins containing mCherry was not achieved. Moreover, fusion to other red fluorescent proteins able to mature in nanaerobic conditions, like mKate2, did not display fluorescence, and the fusions were not identified by Western blot. However, proper fluorophore maturation and fluorescence were achieved after exposure to aerobic conditions (Materials and Methods). Although atmospheric concentrations of oxygen are not physiological for Bacteroides spp., neither cell viability nor the distribution of our chimeric markers was affected after 4 h of exposure to air (SI Appendix, Fig. S2). In all cases, fluorescent chimeric proteins were detected at the cell surface. We frequently observed the OMV markers localized at defined foci (Fig. 1 B, Inset). This phenotype was not always present, and the number of foci was variable. On the contrary, OM markers invariably exhibited a more homogeneous distribution along the cell borders (Fig. 1 C, Inset). Interestingly, round-shaped extracellular structures of a size compatible with vesicles were observed in Bt expressing OMV chimeric markers inulinase, BT_1581, or SusG fused to sfGFP (Fig. 1B and SI Appendix, Fig. S3). These structures were not detected in the cells expressing the OM chimeric markers OmpF and BT_2844 fused to mCherry (Fig. 1C). These results suggest that OMV-enriched chimeric fluorescence proteins can be employed to visualize OMVs, and we hypothesize that the punctate distribution observed for the OMV markers is related to the recruitment of vesicle-targeted proteins to specific foci prior to vesicle formation.
Visualizing OMV Formation.
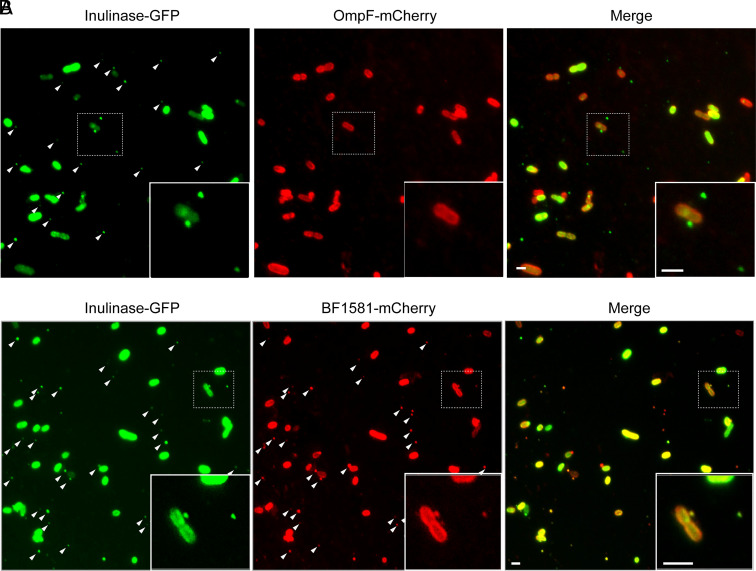
We subsequently analyzed whether the coexpression of an OMV marker along with an OM marker enables the distinction between OMVs and cells or lysis byproducts by fluorescence microscopy. We reasoned that lysis byproducts would be generated by the nonspecific breakage of cellular membranes and, due to this, should contain OM marker or both OM and OMV markers. On the contrary, genuine OMVs are not expected to contain OM markers and, instead, should only be detected via the proper OMV markers. Western blot analysis confirmed that coexpression of the different chimeric proteins did not affect their original OMV vs. OM distribution (SI Appendix, Fig. S4). Bt cells coexpressing Inulinase-GFP and OmpF-mCherry were detected in both, green and red fluorescence channels. Remarkably, extracellular structures were exclusively detected in the green channel (Fig. 2A, white arrows). When analyzing late stationary cultures coexpressing OM and OMV markers, we were able to distinguish cell debris from bonafide vesicles (SI Appendix, Fig. S5) Furthermore, when we simultaneously expressed two different OMV markers, Inulinase-GFP and BF_1581-mCherry, both colocalized in the extracellular structures (Fig. 2B, white arrows).
Fig. 2.
Coexpression of OMV and OM markers do not show colocalization in vesicles. Representative widefield fluorescent microscopy images of Bt coexpressing (A) Inulinase-GFP and OmpF-mCherry, or (B) Inulinase-GFP and Bf_1581-mCherry. (Scale bar: 2 μm.)
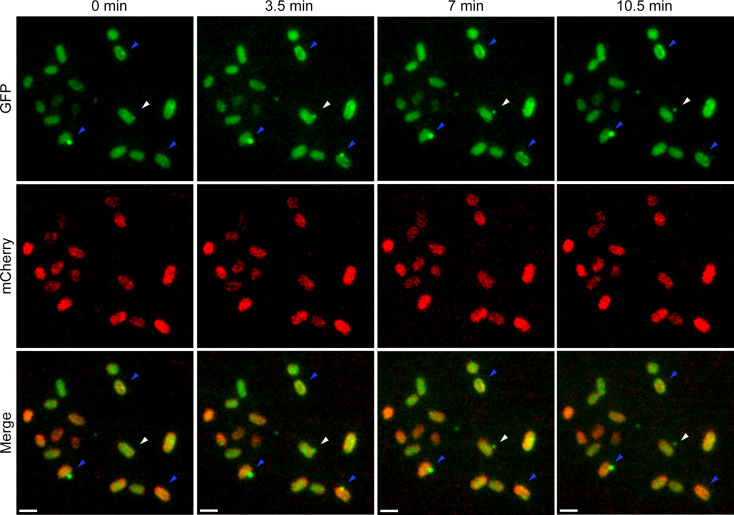
To capture OMV biogenesis in live cells, we utilized time-lapse widefield fluorescent microscopy of Bt coexpressing Inulinase-GFP and OmpF-mCherry. Here, we observed that the OM marker invariably remained in the cell, while the OMV markers were found to be released from the cell (Fig. 3, white arrows and Movie S1). It was also observed that some OMVs were not released by the bacteria (Fig. 3, blue arrows and Movie S1), evidencing that OMV cargo proteins have dynamic distributions over time. These results show vesicle biogenesis dynamics in live bacteria. Altogether, the observations support the existence of OMV protein-sorting mechanisms as a part of an orchestrated process in Bacteroides spp.
Fig. 3.
Time-lapse of OMV formation. Bt coexpressing Inulinase-GFP and OmpF-mCherry were grown overnight in anaerobic conditions in liquid minimal media supplemented with glucose. Cultures were then incubated at 37 °C in the presence of oxygen for 4 h for fluorophore maturation. Cultures were diluted in prewarmed minimal media supplemented with glucose and loaded onto 1% agarose pads for image acquisition. Images were acquired in a 37 °C prewarmed microscope every 3.5 min. (Scale bar: 2 μm.) (Movie S1).
Proteomic Analysis of Membrane and OMV Fractions.
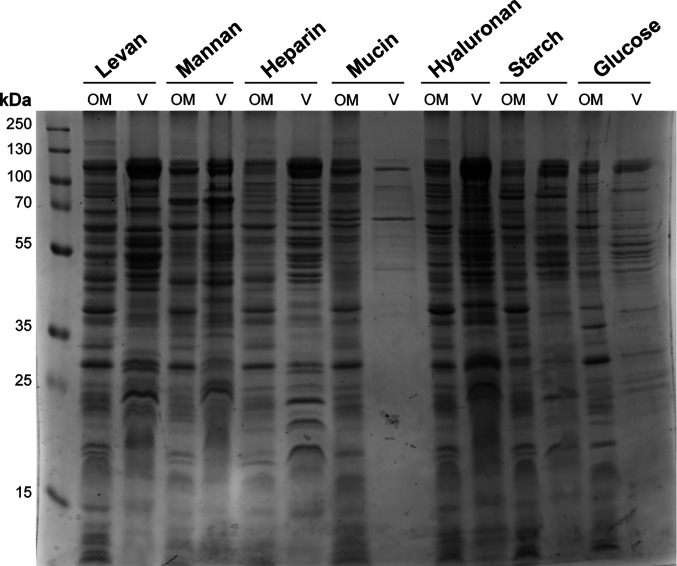
Bacteroides is highly specialized in the degradation of host- and diet-derived glycans (9–12). We have previously shown that OMVs are equipped with numerous glycosyl hydrolases and other degradative enzymes required for the digestion of nutrients (20, 21). We hypothesized that Bt can modulate the protein cargo of their OMVs to effectively utilize diverse array of glycans. To test this, we cultured Bt in minimal media supplemented with different polysaccharides or glycosaminoglycans: levan, starch, mannan, hyaluronan, heparin, or mucin; glucose was employed as a reference condition (SI Appendix, Fig. S6). Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of OM and OMV fractions from Bt grown under the different conditions revealed significant variations in protein profiling (Fig. 4). Surprisingly, the amounts of protein detected in OMV fractions from cell culture in mucin and glucose were significantly lower than those in the other conditions.
Fig. 4.
Bt modulates OMV cargo in different nutrient conditions. Coomassie blue staining after SDS-PAGE of Bt OM and OMV (V) fractions obtained after growth in minimal media supplemented with levan, mannan, heparin, mucin, hyaluronan, starch, or glucose. Fractions were normalized by optical density (OD) at 600 nm (Materials and Methods).
To further analyze their composition, we performed comparative proteomic analyses of OMVs and OMs from Bt cultured in all these nutrients. Principal component analysis (PCA) of OMV and OM proteomics showed that the biological replicates cluster together (SI Appendix, Fig. S7). This clustering was also observed by Pearson correlation analysis, with correlation coefficients of above 0.85 in most of the cases (SI Appendix, Fig. S8), revealing highly similar biological replicates. When comparing OMV proteomic datasets, we observed higher correlations between samples obtained after growth in levan and starch, as well as between heparin and hyaluronan, indicating that the proteome is more similar between these groups (SI Appendix, Fig. S8). Instead, OMVs obtained under growth in mucin were the most divergent ones, displaying the lowest correlation coefficients when compared to all the conditions (SI Appendix, Fig. S8). This is also reflected in the PCA, where the mucin proteome clusters separately (SI Appendix, Fig. S7). Remarkably, this divergent behavior was not observed in the OM comparative proteomic analysis, where OM proteome from Bt grown in mucin is found to be more similar to the OM proteome from Bt grown in hyaluronan and heparin (SI Appendix, Figs. S7 and S8). This suggests that the changes in the content of the OMVs do not necessarily correspond with the changes in the OM, likely as a result of a differential protein sorting into vesicles depending on the glycan available.
Lipoproteins Containing the Negatively Charged LES Signal Are Specifically Packed in OMVs of Bacteria Grown in All Conditions.
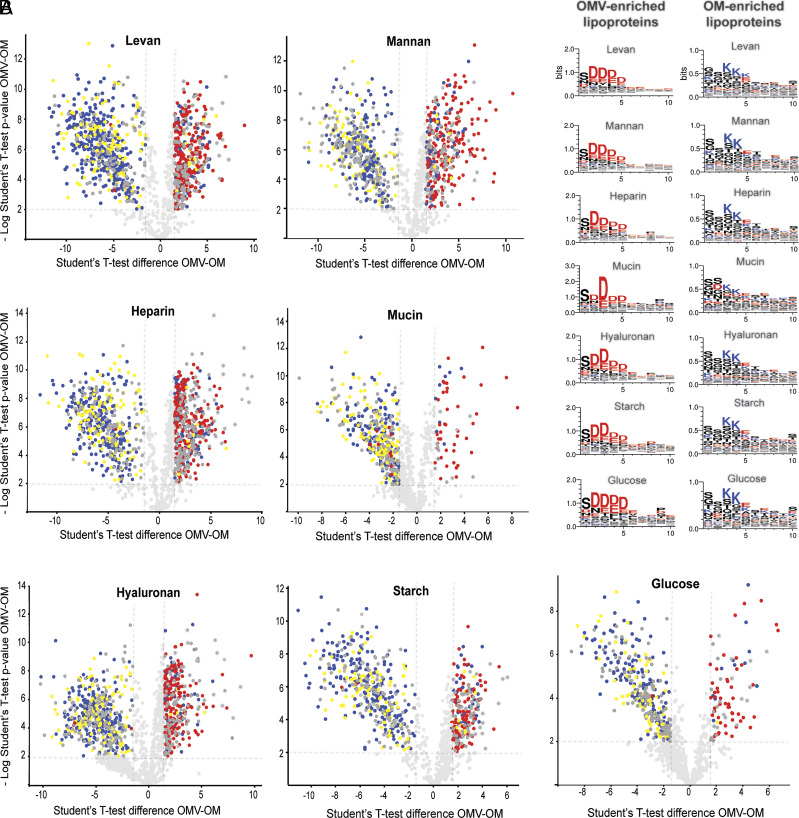
We previously observed that, when Bt was cultured in rich media, most OMVs-enriched lipoproteins contain the N-terminal motif S(D/E)3 (21). Moreover, mutations in this motif in the lipoprotein SusG abrogates its sorting into OMVs (21). A similar motif, K-(D/E)2 or Q-A-(D/E)2, named the LES motif, was proposed by Lauber and colleagues to be required for lipoproteins surface exposure in the OM in the oral pathogen Capnocytophaga canimorsus, another member of the Bacteroidota phylum (22). Through our comparative proteomics experiments we tested whether the LES-based partition occurred in the presence of seven different carbohydrate nutrients. Volcano plots presented in Fig. 5A indicate that in all conditions, a great majority of the proteins preferentially sorted into OMVs contain the LES motif (labeled in red in Fig. 5 A and B and Datasets S1–S7). On the contrary, integral components of the membrane were mostly retained in the OM fraction (labeled in blue). Additionally, we found that lipoproteins lacking the LES motif (labeled in yellow in Fig. 5A) are mostly retained in the OM (Fig. 5A), irrespective of the glycan present in the media. These experiments strongly support a model in which the LES motif acts as a conserved signal for lipoprotein sorting into OMVs.
Fig. 5.
OMV-enriched lipoproteins expressed in different culture conditions harbor a conserved N-terminal LES motif. (A) Volcano plot representations of OM and OMV-enriched proteins. Integral membrane proteins are represented in blue, lipoproteins with LES motifs are indicated in red, lipoproteins lacking the LES motif are depicted in yellow, and soluble proteins are indicated in dark gray. (B) Lipoprotein amino acid residues next to the cysteine required for acylation were aligned. Left panels show prevalent amino acid residues in OMV-enriched lipoproteins (cut-off: OMV/OM fold change >2) and Right panels show prevalent amino acid residues in OM-enriched lipoproteins (cut-off: OMV/OM fold change <−2). Negatively charged amino acid residues are colored in red and positively charged in blue. The consensus sequences were generated using WebLogo (https://weblogo.berkeley.edu/logo.cgi).
OMV Cargo Is Tailored for the Digestion of Specific Polysaccharides.
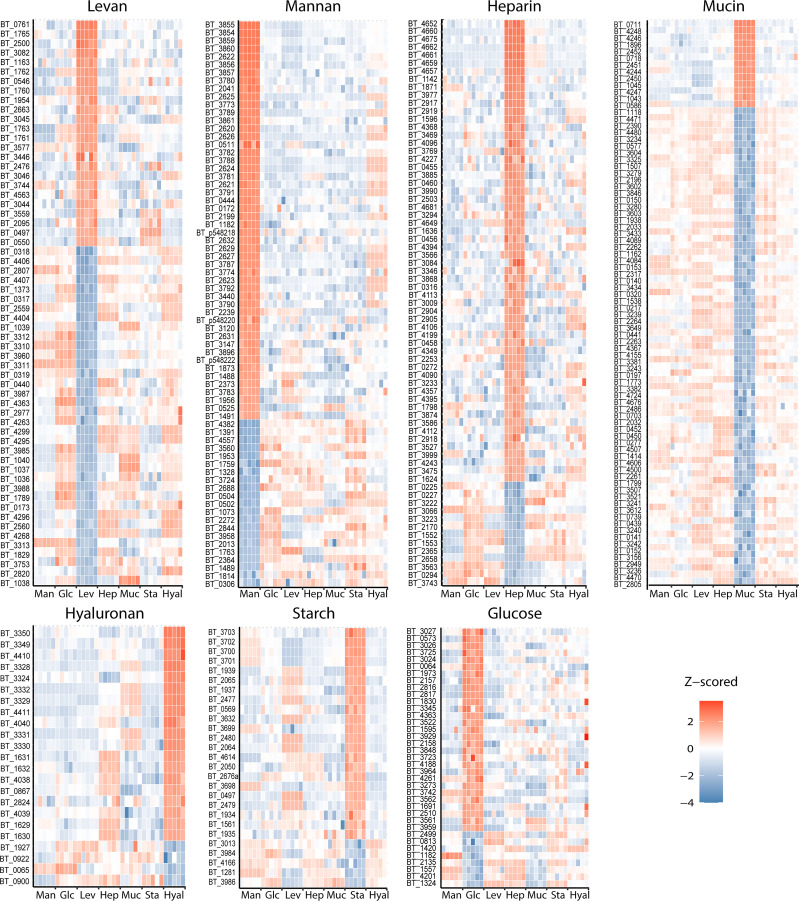
Comparative proteomic analysis between OMVs from Bt grown in the different conditions determined that specific subsets of proteins are preferentially sorted into vesicles depending on the glycan present in the media (Fig. 6, red hits and Dataset S8). Proteins that are underrepresented in the OMVs for each growth condition are also illustrated (Fig. 6, blue hits). Notably, although the OMV-enriched proteins vary for each glycan, most of these proteins are functionally and, in many cases, genetically related. Many of the proteins are glycosyl hydrolases encoded within PULs, which contain proteins involved in the binding, uptake, and degradation of specific polysaccharides that are coregulated in response to specific nutrients to enable glycan utilization. For example, proteins belonging to the heparin utilization PUL 85, such as BT_4652, BT_4675, and BT_4662 were particularly enriched in OMVs produced by cells cultured in the presence of heparin but not in the presence of the other glycans. Similarly, proteins encoded in PUL 57, proposed to be involved in hyaluronan and chondroitin sulphate degradation, were enriched in OMVs produced by bacteria grown in hyaluronan (Dataset S8). Most of the induced PULs identified in our proteomic analysis correlate with findings from previous transcriptomic studies performed with Bt in vivo and in vitro (9). Additionally, we identified that components of unassigned PULs were specifically induced and enriched in OMVs in response to specific nutrients. For example, protein components of PULs 41 and 42 are specially induced and targeted to OMVs in the presence of heparin (Dataset S8).
Fig. 6.
OMV cargo is tailored for the digestion of specific polysaccharides. Heat maps of protein levels (Z-score) showing most enriched (red) and excluded (blue) proteins found in OMVs from Bt grown in minimal media supplemented with levan (Lev), mannan (Man), heparin (Hep), mucin (Muc), hyaluronan (Hyal), or glucose (Glc) as carbon sources. Five biological replicates were performed per growth condition.
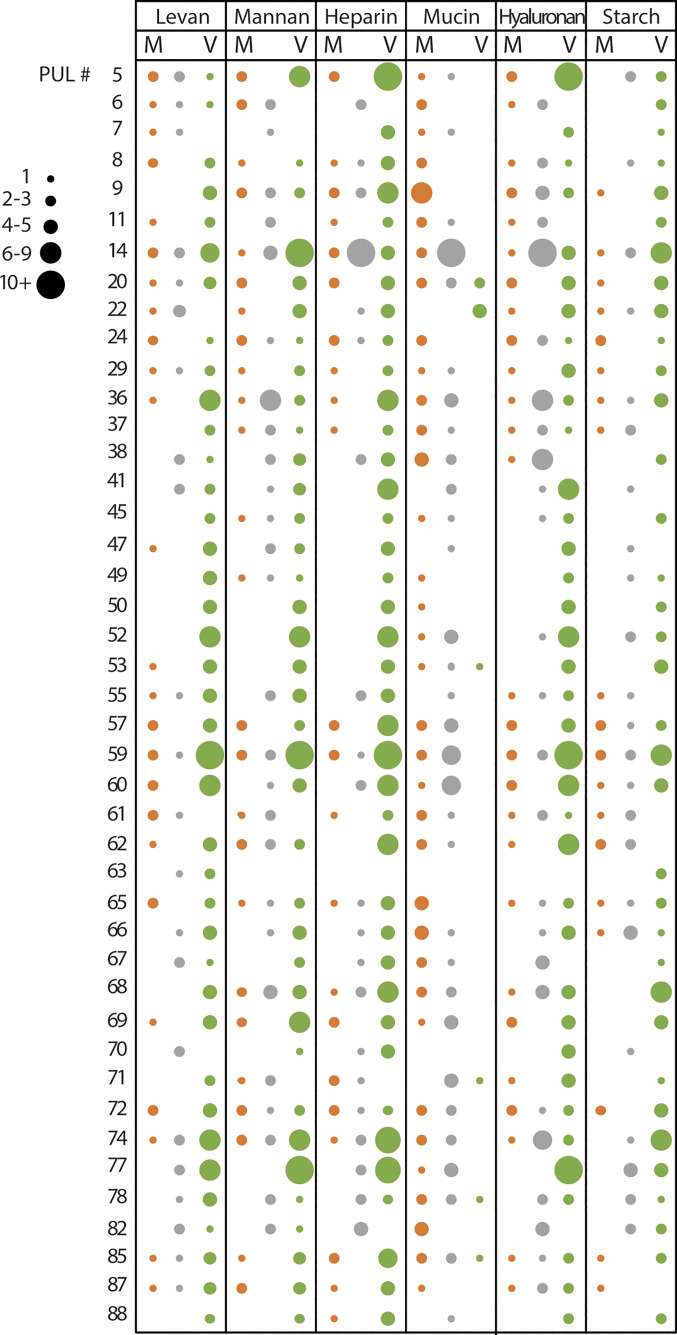
Surprisingly, despite mucin being the most complex carbon source employed in these experiments, mucin-derived OMVs displayed a reduced number of proteins targeted to vesicles (Fig. 6). However, the comparative OM proteome dataset indicates that the PULs previously reported to be required for mucin degradation are indeed expressed (Dataset S9). In addition, OM fractions from bacteria grown in mucin displayed a higher number of LES-containing lipoproteins in comparison to all other growth conditions (Dataset S4). This suggests that proteins that would usually be targeted to the OMVs are retained in the presence of mucin. Fig. 7 illustrates the behavior of PUL components in response to different nutrients. PUL-derived proteins were enriched in the OMV fraction (green circles) for all the conditions except for mucin. In the presence of mucin, PUL-derived proteins were either retained in the OM (orange circles) or equally distributed in both fractions (gray circles). The complete PUL dataset analysis is presented in SI Appendix, Fig. S7.
Fig. 7.
PUL-encoded proteins show OMV enrichment except for Bt grown in mucin. PUL-encoded proteins were identified and classified as OM-enriched (OMV/OM fold change <−1, M column, colored in orange), OMV-enriched (OMV/OM fold change >1, V column, colored in green), or unclassified (OMV/OM fold change between −1 and 1, colored in gray). Circle size represents the number of identified proteins for each PUL. Main representative PULs are shown (complete PUL dataset in SI Appendix, Fig. S9).
Finally, we analyzed of the distribution of all predicted hydrolases between OMV and OM fractions, irrespective of their presence in PULs. Results revealed that about 60 to 80% of the total predicted hydrolases are enriched in OMV fractions in all the conditions, except for mucin, that exhibits only ~20% of the total predicted hydrolases enriched in OMVs (SI Appendix, Fig. S10). Together, these results demonstrate that Bt can customize the OMVs protein cargo to optimize glycan utilization and suggest that vesicles are heavily involved in utilization of complex polysaccharides but not in breakdown of mucin.
OMVs Liberate Carbohydrate Breakdown Products Supporting Growth of Other Species.
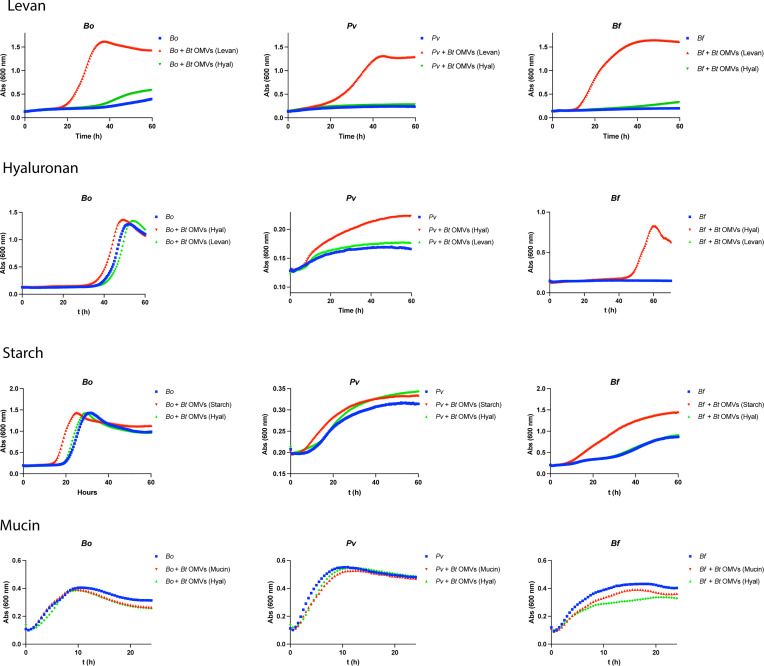
Bacteroides vesicles have been referred as “public goods” due to their ability to degrade extracellular polysaccharides whose breakdown products can be utilized by surrounding bacteria (23). To functionally validate our proteomic findings, we analyzed the ability of OMVs to enhance the growth of other Bacteroides spp. in minimal media supplemented with different polysaccharides. We found that OMVs with cargo optimized for degradation of levan support the growth of poor levan utilizing bacteria, such as Phocaeicola vulgatus [Pv, reclassified from Bacteroides vulgatus (26)], Bo, and Bf, in the presence of levan. However, as predicted from our MS data, OMVs optimized for degradation of hyaluronan did not support growth of these bacteria in the presence of levan (Fig. 8). We also demonstrated the reciprocal, OMVs produced by Bt in hyaluronan support the growth of Pv, Bo, and Bf in the same carbon source, but OMVs from Bt grown in levan do not. Similar results were obtained for other sugars (Fig. 8). As suggested by our MS data, OMVs of Bt grown in mucin are not well equipped to degrade mucin, and consistently, these did not improve growth of Pv, Bo, or Bf in the presence of mucin. Our results are consistent with the proposed roles of OMVs as public goods in the human gut (23).
Fig. 8.
Tailored OMVs from Bt grown in a specific glycan enhance growth of Bacteroides spp. under the same culture condition. Growth curves of Bo, Pv, and Bf in minimal media with levan, hyaluronan, starch, or mucin. Cultures were not supplemented (blue lines), supplemented with 1 μg/mL of Bt OMVs obtained after growth in the same glycan (red lines), or supplemented with 1 μg/mL of Bt OMVs obtained after growth in a different glycan (green lines).
Discussion
In all three domains of life, cells secrete a large variety of membrane-bound extracellular vesicles (EV) that transport numerous molecules and mediate intercellular communication in both physiological and pathological conditions (27). In eukaryotes and archaea, EV biogenesis is known to rely on endosomal sorting complex required for transport machinery (28–30), which consists of a collection of highly conserved proteins that drive membrane budding and scission. However, despite the fast-growing literature related to vesicle production in bacteria, researchers have been unable to answer fundamental questions regarding how vesicle biogenesis occurs in bacteria, and how this process is regulated. In most gram-negative bacterial species, OMVs and OMs share very similar composition. In some cases, OMVs are reported to carry cytoplasmic components, such as ribosomal proteins, DNA, and RNA, that, according to the current knowledge of bacterial physiology, should not be packed into OMVs if vesicles are formed by blebbing of the OM. However, these elements can be associated with vesicles if they result from bacterial lysis. Indeed, some reports suggest that vesicles are formed by “explosive bacterial lysis” (31), and most of the proposed mechanisms for OMVs biogenesis involve destabilization of the OM (32). In consequence, the scientific community remains partially skeptical about the physiological relevance of OMVs. Based on our proteomic data, we have previously proposed the existence of a cargo selection process for Bacteroides vesicles (20, 21). This unique characteristic of Bacteroides-derived OMVs prompted us to label bacteria and vesicles with different markers, allowing us to visualize vesicle biogenesis by time-lapse microscopy. Our microscopy analysis together with our exhaustive comparative proteomic analysis unequivocally demonstrate that OMVs are not the result of lysis but a highly orchestrated process in Bacteroides. We also show that based on a specific signal, the LES motif, OMV protein cargo is tailored to specifically degrade diet- and host-derived polysaccharides. It remains to be demonstrated whether similar dedicated OMV protein sorting mechanisms are present in other gram-negative bacterial species.
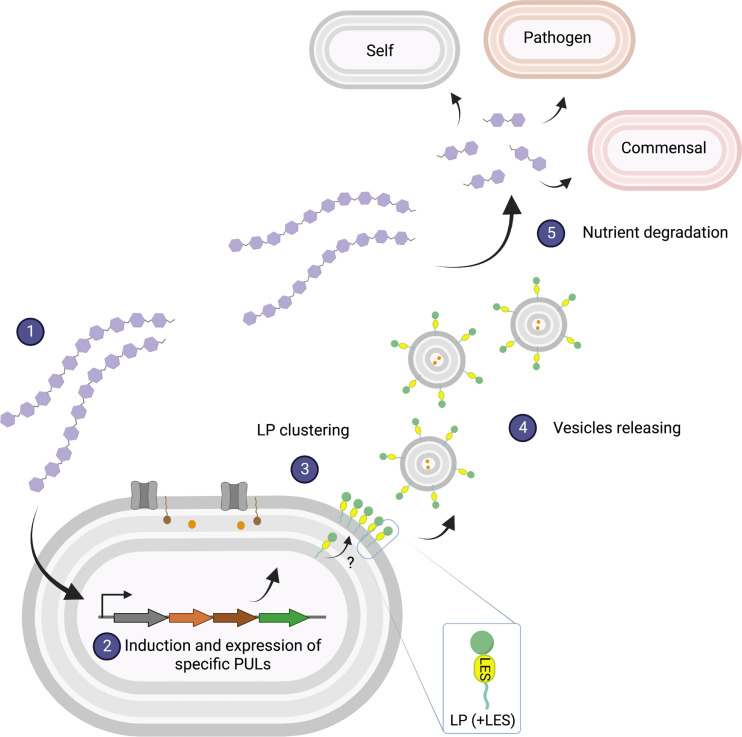
Our observations led us to propose a model for vesicle production in Bacteroides (Fig. 9). As it has been shown in previous studies, in the presence of a given polysaccharide, Bacteroides induces the expression of specific PULs required for binding, uptake, and degradation of that glycan. Glycosyl hydrolases encoded in these PULs containing the LES signal are translocated across the OM, exposing them on the bacterial surface, where they cluster at defined foci in the OM. OMVs are subsequently generated by a still unknown machinery and subsequently released to the media. Production of OMVs enables the degradation of distal polysaccharides, and the resulting breakdown products can then be utilized by kin bacteria, other members of the microbiota, and even potential pathogens. This model does not exclude additional roles for OMVs in the context of interbacterial or host–bacteria interactions.
Fig. 9.
Model of OMV biogenesis. In the presence of a specific glycan (1), Bacteroides induces specific PULs (2). Glycosyl-hydrolases containing the LES signal are transported to the OM and exposed to the cell surface. OMV-targeted lipoproteins cluster at defined foci in the OM (3), where they are formed (4) by an unknown mechanism. OMVs diffuse and hydrolyze glycans that can be imported by the OMV-producing bacteria, as well as other commensals and pathogens (5). Created with Biorender.
In our microscopy experiments, we observed that OMV markers frequently localized at defined foci that were not detected in cells expressing OM markers (Fig. 1). We propose that these foci represent regions of the OM where OMV proteins are recruited before the vesicles are released. It was also observed that bacterial cells do not always release the vesicles from these foci, which dynamically moved over time (Fig. 3 and Movie S1), in line with previously reported dynamic behavior for SusG (33).
Multiple comparative proteomic analyses revealed the existence of an underlying regulatory mechanism for OMV protein packing in Bt. We observed that, for each nutrient, a different subset of enzymes, several of them encoded in PULs, were packed into OMVs. In general, our proteomics data are in agreement with previous transcriptomic analyses that identified the PULs induced for the utilization of these glycans in vitro and in vivo (9). However, we were able to demonstrate that an increase in the expression of PULs does not necessarily correlate with changes in the protein content of the OMVs. For each glycan analyzed in this work, most of the carbohydrate-induced enzymes containing the LES motif were targeted to OMVs, suggesting that OMVs play a role in their utilization. On the contrary, numerous proteins containing the LES motif were induced but retained in the OM in the presence of mucin. We hypothesize that OMV biogenesis machinery is not induced in the presence of mucin. Supporting these hypotheses, we observed significantly lower protein packed into OMVs when cultured in mucin as the sole carbon source (Figs. 4 and 5). Previous experiments performed in Bt in which mucin glycans were included together with other carbon sources show that degradation of mucin is not preferred in the presence of alternative substrates, even when Bt has been previously acclimated to growth in pure mucin glycans (34). Altogether, these observations indicate that Bt has evolved to degrade mucus but as a last resource. It is tempting to speculate that Bt utilizes mucin “selfishly” rather than providing breakdown products for other bacteria at the mucus layer. The fact that they pack less proteins into OMVs probably also represents an evolutionary response to avoid exacerbated degradation of the host intestinal mucus layer (35).
Due to their size, OMVs diffuse beyond the bacterial surface, allowing the breakdown of nutrients that are out of reach for the producing bacteria. This is beneficial for the producing bacteria, but other microorganisms can also profit from OMV hydrolytic activities. Supporting this notion, Bt grown in starch displays a shorter lag phase when supplemented with physiological amounts of Bt OMVs produced in the same polysaccharide (SI Appendix, Fig. S11). In addition, OMVs can also enhance the growth of other species (Fig. 8). An interesting example of cooperative feeding based on OMVs occurs between Bo and Pv. Bo secretes inulinases through OMVs, which supports the growth of Pv, unable to grow on inulin (23, 24). Interestingly, it has been shown that the secreted and surface-exposed inulinases of Bo are not required for its growth in inulin and, in fact, represent a fitness cost for the bacterium. However, this commensal relationship is reciprocal, as Bo benefits from Pv through an unknown mechanism. Also, pathogen–commensal interactions mediated via OMVs have been observed. The gastrointestinal human pathogen Campylobacter jejuni shows enhanced growth in mucin when cocultured with Pv due to the secretion of fucosidases via OMVs (36). Altogether, these examples reflect the coevolution of commensal and pathogen species within the microbiota environment and highlight the diverse roles OMVs.
Our understanding of the physiological role of Bacteroides OMVs is still in its infancy. The use of a dual marker system that allows the distinction between vesicles from cellular debris is essential to identify the molecular machinery responsible for OMV biogenesis and to understand which are the signals that trigger vesicle production in vitro and in vivo.
Materials and Methods
Strains, oligonucleotides, and plasmids are described in SI Appendix, Table S1. Bacteroides strains were routinely grown in an anaerobic chamber (Coy Laboratories) using an atmosphere of 10% H2, 5% CO2, and 85% N2. Bacteria were cultured in brain heart infusion medium supplemented with 5 µg/mL Hemin and 1 µg/mL vitamin K3. When applicable, antibiotics were used as follows: 100 µg/mL ampicillin, 200 µg/mL gentamicin, and 25 µg/mL erythromycin. When required, Bacteroides was grown in minimal medium (MM) containing 100 mM KH2PO4 (pH 7.2), 15 mM NaCl, 8.5 mM (NH4)2SO4, 4 mM L-cysteine, 1.9 mM hematin/200 mM L-histidine (prepared together as a 1,000× solution), 100 mM MgCl2, 1.4 mM FeSO4.7H2O, 50 mM CaCl2, 1 µg/mL vitamin K3, and 5 ng/mL vitamin B12. Carbohydrates used to supplement MM include glucose (G7528, Sigma), levan (P-Levan, Neogen corp.), mannan (M7504, Sigma) starch (S2004, Sigma), hyaluronan (FH76335, Biosynth Carbosynth), heparin (YH09354, Biosynth Carbosynth), and mucin (M2378, Sigma). All carbon sources were added to MM in a final concentration of 0.5% (w/v). A full description of methods is available in the SI Appendix, Materials and Methods. All data are available in the main text or the SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Live OMV formation in Bt co-expressing Inulinase-GFP and OmpF-mCherry.
Acknowledgments
We thank all members of the Feldman lab for critically reading the manuscript. We also thank Wandy Beatty for assistance with fluorescence microscopy experiments. This work was supported by NIH grants R21AI151873 and R21AI168719.
Author contributions
M.F.F. designed research; M.G.S., E.J.P., and N.E.S. performed research; N.E.S. and M.F.F. contributed new reagents/analytic tools; M.G.S. and N.E.S. analyzed data; and M.G.S., E.J.P., and M.F.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The mass spectrometry proteomics data have been deposited in the Proteome Xchange Consortium (37) via the PRIDE partner repository with the data set identifier PXD036181 (38) PXD036275 (39) and PXD036272 (40). All data are available in the main text or the SI Appendix.
Supporting Information
References
- 1.Huttenhower C., et al. , Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yatsunenko T., et al. , Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wexler A. G., Goodman A. L., An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2, 17026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stappenbeck T. S., L. v. Hooper, J. I. Gordon, Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U.S.A. 99, 15451–15455 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown E. M., et al. , Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe 25, 668–680.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazmanian S. K., Liu C. H., Tzianabos A. O., Kasper D. L., An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Pacheco A. R., et al. , Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng D., Liwinski T., Elinav E., Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martens E. C., Chiang H. C., Gordon J. I., Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 4, 447–457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens E. C., et al. , Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9, e1001221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luis A. S., et al. , Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat. Microbiol. 3, 210–219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ndeh D., et al. , Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus. Nat. Commun. 11, 646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjursell M. K., Martens E. C., Gordon J. I., Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, bacteroides thetaiotaomicron, to the suckling period. J. Biol. Chem. 281, 36269–36279 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Shen Y., et al. , Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stentz R., et al. , Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β-lactam antibiotics. J. Antimicrob. Chemother. 70, 701–709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sartorio M. G., Pardue E. J., Feldman M. F., Haurat M. F., Bacterial outer membrane vesicles: From discovery to applications. Annu. Rev. Microbiol. 75, 609–630 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop D. G., Work E., An extracellular glycolipid produced by Escherichia coli grown under lysine-limiting conditions. Biochem. J. 96, 567–576 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knox K. W., Vesk M., Work E., Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 92, 1206–1217 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coelho C., Casadevall A., Answers to naysayers regarding microbial extracellular vesicles. Biochem. Soc. Trans. 47, 1005–1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elhenawy W., Debelyy M. O., Feldman M. F., Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5, e00909-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valguarnera E., Scott N. E., Azimzadeh P., Feldman M. F., Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in bacteroides. mSphere 3, e00559-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauber F., Cornelis G. R., Renzi F., Identification of a new lipoprotein export signal in gram-negative bacteria. mBio 7, 10.1128/mbio.01232-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rakoff-Nahoum S., Coyne M. J., Comstock L. E., An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 24, 40–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum S., Foster K. R., Comstock L. E., The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Bayona L., et al. , Nanaerobic growth enables direct visualization of dynamic cellular processes in human gut symbionts. Proc. Natl. Acad. Sci. U.S.A. 117, 24484–24493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.García-López M., et al. , Analysis of 1,000 type-strain genomes improves taxonomic classification of bacteroidetes. Front. Microbiol. 10, 2083 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gill S., Catchpole R., Forterre P., Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 43, 273–303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Niel G., D’Angelo G., Raposo G., Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Schöneberg J., Lee I.-H., Iwasa J. H., Hurley J. H., Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., et al. , Archaeal extracellular vesicles are produced in an ESCRT-dependent manner and promote gene transfer and nutrient cycling in extreme environments. ISME J. 15, 2892–2905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turnbull L., et al. , Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7, 11220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwechheimer C., Kuehn M. J., Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karunatilaka K. S., Cameron E. A., Martens E. C., Koropatkin N. M., Biteen J. S., Superresolution imaging captures carbohydrate utilization dynamics in human gut symbionts. mBio 5, e02172-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pudlo N. A., et al. , Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6, e01282-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desai M. S., et al. , A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garber J. M., et al. , The gastrointestinal pathogen Campylobacter jejuni metabolizes sugars with potential help from commensal Bacteroides vulgatus. Commun. Biol. 3, 2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deutsch, E. W., et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res 51, D1539–D1548 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sartorio M. G., Scott N. E., Feldman M. F., Comparison of OMV to membrane proteomes across different media (OMV vs membrane experiments). PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD036181 Deposited 25 August 2022.
- 39.Sartorio M. G., Scott N. E., Feldman M. F., Comparison of OMV to membrane proteomes across different media (OMV). PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD036275. Deposited 25 August 2022.
- 40.Sartorio M. G., Scott N. E., Feldman M. F., Comparison of OMV to membrane proteomes across different media (membrane experiments). PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD036272. Deposited 25 August 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Live OMV formation in Bt co-expressing Inulinase-GFP and OmpF-mCherry.
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the Proteome Xchange Consortium (37) via the PRIDE partner repository with the data set identifier PXD036181 (38) PXD036275 (39) and PXD036272 (40). All data are available in the main text or the SI Appendix.