Abstract
Background
“Artificial intelligence” and “big data” increasingly take the step from just being interesting concepts to being relevant or even part of our lives. This general statement holds also true for transfusion medicine. Besides all advancements in transfusion medicine, there is not yet an established red blood cell quality measure, which is generally applied.
Summary
We highlight the usefulness of big data in transfusion medicine. Furthermore, we emphasize in the example of quality control of red blood cell units the application of artificial intelligence.
Key Messages
A variety of concepts making use of big data and artificial intelligence are readily available but still await to be implemented into any clinical routine. For the quality control of red blood cell units, clinical validation is still required.
Keywords: Red blood cell quality, Storage lesion, Erysense, Personalized transfusion
Introduction
Blood donations, the processing of blood products (plasma, platelet units, and red blood cell [RBC] units), and their use to treat patients are success stories at least since the discovery of the ABO blood group system by Landsteiner [1]. The technology and the knowledge developed ever since improved almost all aspects of transfusion medicine. Today, more than 100 million RBC units are transfused each year worldwide [2, 3] and the use of big data in these transfusion data has the potential to further enhance the safety, efficiency, and effectiveness of transfusion medicine [4]. One way to conceptualize the application of big data to transfusion medicine is to combine various types of health data, including electronic health records [5, 6], electronic medical records [7], personal health records [8], laboratory information systems [9], medical practice management [10] software, and hemovigilance data [11]. This combination creates a large database that healthcare professionals can use to identify patterns and trends, leading to improved practices in blood product usage, inventory management, and more, ultimately, improving patient outcomes [4, 12]. By including hemovigilance data in the discussion of big data in transfusion medicine, it highlights the importance of monitoring and ensuring the safety and quality of blood products, which is a critical aspect of transfusion medicine. Blood unit safety in terms of infection risks is very low in high-income countries. For example, in Germany, it was estimated to be as low as 1 in 10.9 million for hepatitis C virus, 1 in 4.3 million for human immunodeficiency virus (HIV)-1, and 1 in 360,000 for hepatitis B virus [13]. The general infection risk is higher (up to 2%) in low-income countries [14]. In particular, the measures implemented after the HIV crisis in the 1980s [15] made blood products very safe in terms of contamination. In addition to the risk of infection, the storage conditions of blood units are another important aspect to consider. In many countries, blood units can be stored for up to 42 days under controlled conditions before transfusion. However, studies have shown that after the first 14 days of storage [16], RBCs can accumulate biochemical and morphological lesions that can irreversibly alter their biological functions [17]. These lesions, known as RBC storage lesions, have been widely discussed in the literature, but it remains unclear which parameters are useful in assessing the quality of blood units for transfusion. Such parameters could be crucial to establish if RBCs should be transfused and in which number. Moreover, possible donor-dependent RBC quality is discussed [18], opening room to single-donor blood quality tests and shelf-life dependence to improve RBC transfusion decision-making and outcomes. This is of particular importance since there is a shortage of blood products all over the world, e.g., [19]. Although quality control per se does not increase the total amount of blood products, a need which is even increased with the COVID-19 pandemic [20, 21], transfusion product management concepts, and putative increased shelf life (at least for a fraction of the donors) may ensure the most effective application for each blood donation (including a best donor-patient match). Artificial intelligence (AI) has emerged as a promising method to access and analyze such parameters providing valuable insights into single RBC quality and function [22]. Moreover, AI can establish reliable quality databases for blood products, enabling efficient management of the manufacturing and clinical application of blood [23]. Computer simulation and algorithmic predictions will facilitate this process, aided by advanced robotics and information technology to standardize production processes. The incorporation of AI as a tool for quality control analysis in big data represents a new and promising approach to single-cell quality control in transfusion medicine, as illustrated in Figure 1. Its ultimate goal is personalized transfusion therapies [24] while maintaining high-quality standards. Here, we review the current situation and perspective for the application of big data and AI analysis in transfusion medicine in general and for RBC quality control in particular.
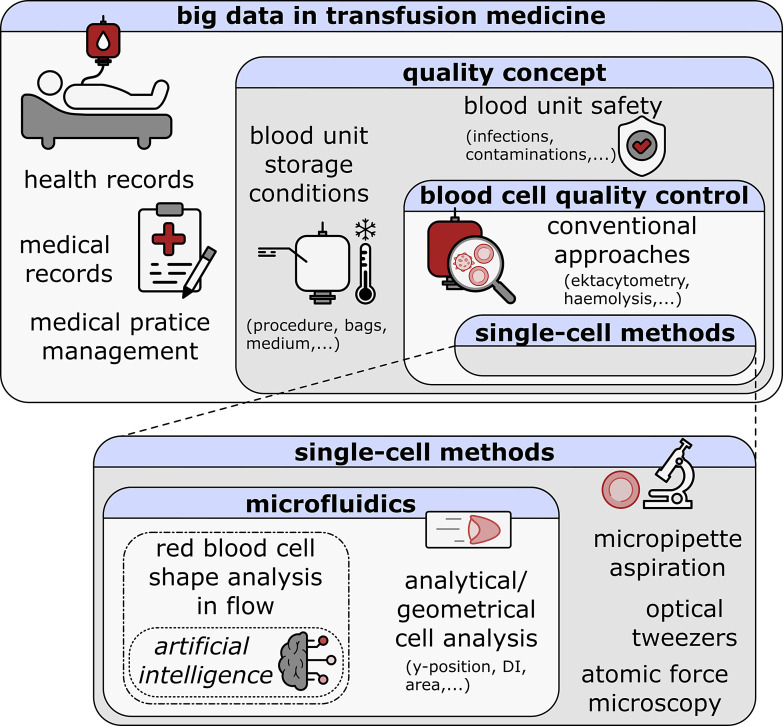
Fig. 1.
Schematic overview of big data in transfusion medicine. The Matryoshka-structured rectangles represent the concept of this review.
Big Data in Transfusion Medicine
The concept of big data, although popular, is still not completely clear. However, it is guided by its four essential features: data attributes, technological requirements, analysis, and limitations [25]. Taken together, big data can be defined as a dataset that increases exponentially in variety, volume, and velocity [26] and that requires specific technology and analytical methods for storage, processing, and extracting information [27].
In the healthcare system, the use and management of electronic health records have expanded the scope for big data and its approaches since the early 2000s. In transfusion medicine, big data has been extensively used at various levels [4], such as in the REDS-III program [28], which utilizes big data techniques and infrastructure to establish a comprehensive research database that connects information from blood donors, their donations, the components derived from these donations, and the recipients of these components. The interconnected database allows for numerous analyses that characterize blood component utilization patterns in diverse settings, inform the design of future clinical trials, and potentially determine the effects of blood donors/donations on the clinical outcomes of recipients.
REDS-III also accesses the SCANDAT [29] database, a large linked electronic donor-recipient database consisting of blood donation records and transfusion activities dating back to the 1960s–1980s in Sweden and Denmark. The upgrade of SCANDAT to SCANDAT2 [30] aims to facilitate systematic surveillance programs for blood safety by enabling complete tracking of subjects regarding cancer, hospital care, and causes of death up to 2012. SCANDAT2 can be integrated with big data through various methods, including linking it with other large-scale health databases like national cancer registries to examine the long-term health outcomes of blood donors and transfusion recipients. Additionally, big data analysis techniques like machine learning algorithms can be utilized to identify patterns in the vast amount of data contained in SCANDAT2, which may indicate transfusion-transmitted infections or adverse health outcomes related to blood transfusions.
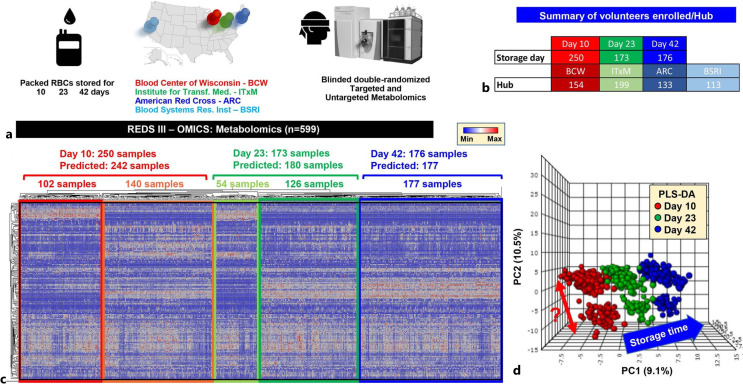
Large databases offer additional insights beyond the influence of donor factors on recipient outcomes, as demonstrated by various studies [31–33], e.g., importance of standardized procedures in blood processing and storage [32]. Using data and samples collected as part of the REDS-III program, the study [32] investigated how the metabolism of stored RBCs is impacted by the heterogeneity of blood processing and storage additives at different centers, emphasizing the crucial role of standardization in blood processing and storage to minimize the variability of blood products across different centers. The relative abundance of metabolites (energy metabolism, amino acids, and nucleotides) in RBCs stored for varying durations (1, 7, 14, 21, 28, and 42 days) from three different centers is shown in Figure 2. The figure highlights the importance of standardizing blood processing and storage procedures to reduce heterogeneity between centers. And it indicates that the heterogeneity of blood processing and storage additives at different centers significantly impacted RBC metabolism, and this impact was as significant as the effect of storage time.
Fig. 2.
Metabolomics analyses of REDS-III samples [32]. a An overview of the study design. b A breakdown of the number of patients enrolled in this arm of the study at four different blood centers (BCW; ITxM, Pittsburgh; ARC at Yale; University of California at San Francisco and BSRI). c The heat map shows the results from unsupervised hierarchical clustering of 599 samples based on metabolic phenotypes. A total of 242, 180, and 177 samples were predicted to belong to storage day 10, 23, and 42 categories, with only 8 samples misclassified as storage day 23 belonging to the day 10 and day 42 groups. d Partial least square discriminant analysis in dataset done after unblinding showing sample clustering based on storage time across principal component 1 (explaining 9.1% of the total variance). Of note, a clear subcluster was observed across PC2 (10.5% of the total variance. Color code: red – day 10 samples; green – day 23 samples; blue – day 42 samples. This figure is reproduced from D’Alessandro et al. [32]. BCW, Blood Center of Wisconsin; ITxM, Institute for Transfusion Medicine; ARC, American Red Cross; BSRI, Blood Systems Research Institute.
Other studies utilize extensive data on each participant, including demographic information, medical history, and blood donation history. For instance, the INTERVAL [34] study investigated whether varying the frequency of whole blood donation affected the safety and efficiency of blood collection, while the PROTON [35] study utilized a large dataset for cost-effectiveness analyses of blood safety interventions. The PROTON study in the Netherlands created the first national dataset of blood product recipients allowing the analysis of the cost-effectiveness of blood safety interventions such as nucleic acid amplification testing, leucocyte depletion, and pathogen reduction. The dataset can be used for economic evaluations and cost-effectiveness analyses to assess the effects of blood safety measures on different age groups, sex, and diagnoses of recipients.
Additional information can also be integrated into such databases, such as the effects of nonmatching patient and donor sex after transfusion [36, 37] and the implementation of various blood preservation methods [38] that may contribute to post-transfusion complications. Moreover, the impact of a blood transfusion monitoring program [39] on financial and allogeneic blood product costs, as well as the optimization of current tools in managing blood orders for specific surgical procedures [40], can be examined to conserve financial and allogeneic resources without adverse effects on patient health. When integrated in a large-scale health database, machine learning algorithms can be applied to identify patterns in the data that may lead researchers to a deeper understanding of the factors that influence blood safety and identify potential areas for improvement in blood screening and transfusion practices [30].
RBC Unit Quality
Investigating RBC quality is an example where new technology is used to generate a large amount of (annotated) imaging data, too large to be analyzed by manual inspection (a definition of big data). Therefore, AI can be used for data analysis, and in the following, we will describe (and review) the entire process in detail.
We need to discriminate between RBC unit quality and safety. The safety of RBC units was improved considerably by screening for HIV, hepatitis B, hepatitis C, syphilis, and (in some countries) malaria [14, 41]. In contrast, until now there is no clinically established graded measure for RBC quality. In some instances, the approaches to improve RBC safety may even compromise RBC quality [42].
An easy-to-measure and straightforward-to-judge parameter is the extracellular hemoglobin indicating the number of hemolyzed RBCs. However, this is an “all or nothing” response just indicating the lysed cells without any chance to judge the remaining cells for putative damage.
There are changes in RBC properties occurring during the time course of the RBC storage, which are usually referred to as “storage lesions” [43, 44]. Additionally, there are indications that the risk for post-transfusion complications increases with storage time [45, 46].
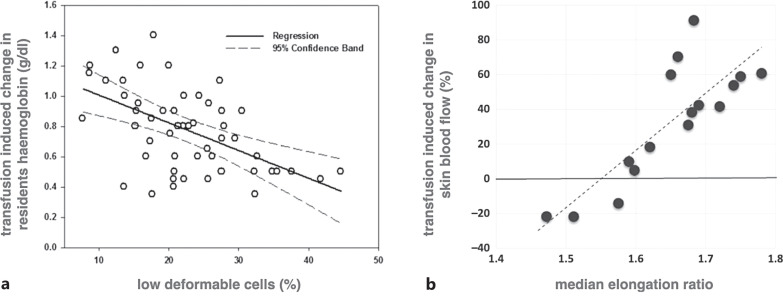
Furthermore, there are indications that the efficiency of the transfusion is indeed related to RBC properties. The seminal work of Barshtein and colleagues showed correlations between single RBC properties (for assays to measure such properties, see below) and parameters, which are influenced by transfusion efficiency [47, 48]. To this end and to judge these results, we extracted and reprinted some of these correlations in Figure 3.
Fig. 3.
Correlation between single RBC properties and parameters related to transfusion efficiency. a shows the change in transfusion-induced hemoglobin plotted against the percentage of low deformable cells for a total of 51 transfusions of 24 beta-thalassemia major patients. The hemoglobin concentration was determined immediately prior to and 10 min after completion of the transfusion. The correlation coefficient is R = −0.48, and the significance level is p = 0.0006. This panel is reproduced from Barshtein et al. [48]. b shows the change of transfusion-induced skin blood flow vs. median RBC elongation ratio. The skin blood flow was determined with a Laser Doppler Imager (Aimago EAsyLDI, Switzerland) on the patient’s wrist immediately prior to and 10 min after completion of the transfusion. The correlation coefficient is R = 0.82, and the significance level is p = 0.00009. This panel is reproduced from Barshtein et al. [47].
Toward Single-Cell Assays for RBC Quality Control
The Implementation of Single-Cell Techniques in General
In the current millennium, single-cell assays have been established as gold standards. This includes microscopic life cell imaging [49], patch clamp [50], flow cytometry but also biochemical methods such as single-cell PCR [51]. This trend in principle also applies to human RBCs and RBC analysis in research and diagnostics [52, 53]. Prominent examples are the application of automated patch clamp in identifying channelopathies [54] or the use of single-cell Ca2+ imaging to decipher general principles in disease mechanisms [55]. However, all these methods require extensive (pre)processing and/or contain time-consuming procedures. Therefore, methods to be applied in clinical routine would benefit from label-free methods and decent throughput.
RBC Characteristics: From Shape to Deformability
To assess the quality of RBCs, traditional methods often characterize the RBC morphology. This ranges from high-resolution scanning electron microscopy [56] to the judgment of blood smears. The latter can be exploited in 3D fluorescence microscopy in stasis [57] and flow [58]. As a result, many RBC-related diseases have derived their names from the shape of the RBCs [59] ranging from sickle cell disease [60] to neuroacanthocytosis syndrome [61]. Furthermore, it is known from rheological and ektacytometric cell population measurements that a crucial parameter (also influenced by RBC storage time) is RBC deformability. In such ensemble average methods, it was shown that the overall RBC deformability reduces as storage progresses [62–65]. However, these traditional bulk flow techniques require a large sample volume of up to a few milliliters. Moreover, these methods do not consider the heterogeneity within the sample population or cannot detect fractions of abnormal RBCs in a sample that contains primarily normal cells. Therefore, single-cell techniques, such as micropipette aspiration, optical tweezers, atomic force microscopy/spectroscopy, and microfluidics, have emerged to assess RBC deformability as a biomarker for numerous pathologies and to evaluate storage lesions [66, 67]. While measuring RBC properties, employing traditional micropipette aspiration, optical tweezers, and atomic force microscopy is tedious, highly skill-dependent, and often low throughput [68, 69].
Microfluidic Approaches to Investigate RBCs
Microfluidic devices provide straightforward single-cell measurements. Thus, microfluidic measurements can be performed with higher throughput, which is paramount for point-of-care applications [52]. Significant progress has been achieved in measuring RBC deformability in microfluidics using various sophisticated designs and mechanisms, including wedging in tapered constrictions, transition, and deformation through constricted channels, and microcapillary flow (see, e.g., [67, 70, 71]). Such microfluidic techniques are often coupled with high-speed imaging and automated image analysis, enabling to test large numbers of cells.
Different parameters have been utilized to assess storage-induced changes in RBC deformability. One property is the clogging of narrow constrictions in microfluidic chips [72–74] or the perfusion in capillary networks of microfluidic chips [75, 76]. As the RBC storage progresses, the degradation of RBC deformability increases the number of cells that are not able to transit through tapered constrictions, smaller than the RBC diameter [77]. Consequently, the perfusion rate in artificial microcapillary networks declines while the number of plugging events increases during storage [65].
Microfluidic Assays to Measure Deformability
In straight microcapillaries, RBC deformability is often evaluated by the elongation of single cells in the flow field, which is described by the deformability index. The deformability index is defined as the ratio between the length L and the diameter D of the deformed RBC, as shown in Figure 4a, for cells in a microfluidic capillary with a height of 8 μm and width of 11 μm. It is calculated for each cell based on the dimensions of its projection in the image plane. Therefore, measuring the deformability index requires optical measurements with sub-micrometer accuracy using high-speed imaging in combination with optical microscopy. By data on storage-associated changes in deformability obtained by ektacytometry [62, 64], a decrease in the deformability index during storage has been reported using microfluidic approaches [78]. In contrast, other microfluidic studies did not observe a significant difference in the change of the deformability index, measured in human-capillary-like microchannels during storage [79, 80]. Remarkably, significant changes in the time constants for cell relaxation and cell circularity during storage have been reported [80]. Cell elongation in microchannels and, thus, the deformability index depend on the exact cell velocity [81], which generally depends on the applied pressure drop, cell size, channel geometry, and used medium for cell suspension. Hence, assessing RBC quality based on geometrical cell parameters alone, such as the deformability index, is prone to experimental and post-processing errors. Therefore, alternative parameters, such as storage-induced morphological changes, have been investigated. The number of pathological RBC morphologies increases during storage [22, 77, 82, 83].
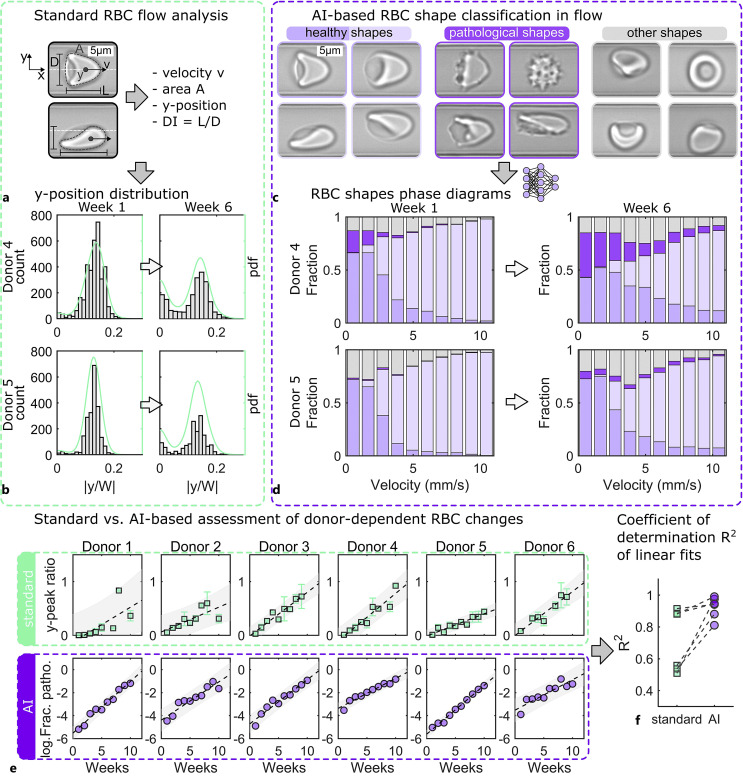
Fig. 4.
Standard and artificial intelligence (AI)-based analysis of single-cell microfluidic flow behavior of stored RBCs. a Representative images of a croissant-shaped RBC (top) and a slipper-shaped RBC (bottom) in a microfluidic capillary with a height of H = 8 µm and width of W = 11 µm recorded by the Erysense technology. Based on these images, standard flow analysis can determine the RBC velocity, projection area, y-position, and deformability index DI. b Histogram and probability density functions (pdf) of the normalized RBC y-position for two donors at week one and week six after storage. c Characteristic healthy, pathological, and other RBC shapes during capillary flow. d AI-based assessment of RBCs during flow. RBC shape phase diagrams, i.e., the fraction of the RBC shape classes shown in c as a function of the RBC velocity for the same donors and points in time as for b. e Comparison of the standard (top) and AI-based (bottom) evaluation of donor-dependent changes in the RBC microcapillary flow behavior. Dashed black lines correspond to linear fits. Gray areas indicate an estimate of a 95% prediction interval. In the standard approach, the y-peak ratio describes the ratio between centered and off-centered RBCs concerning the channel center in the y-direction based on probability density functions (pdfs) at velocities between 7 mm/s and 10 mm/s. For the AI approach, the y-axis of e shows the logarithm of the fraction of pathological RBC shapes based on the shape phase diagrams of d. f Comparison of the coefficient of determination for the linear fits shown in e. Dashed lines connect data points for the same donor analyzed with the standard and the AI approach. This figure was reproduced from data presented by Recktenwald et al. [22].
AI to Judge RBCs
To determine the RBC morphology quickly and unbiasedly, machine learning approaches are used nowadays, similar to other aspects in hematology and transfusion medicine [24, 84, 85]. In the context of RBCs, artificial neural networks and deep learning-based techniques have been used to assess cell phenotypes both in stasis [57, 82, 86–88] and during deformation [71, 83, 89–91]. Kim et al. [82] employed a generative adversarial network to evaluate RBC phenotypes based on phase images obtained by digital holographic microscopy at rest. During storage, they observed a transition from discocytes to echinocytes and, finally, sphero-echinocytes [82]. Recently, Lamoureux et al. [87] proposed a technique to assess the deformability of stored RBCs from brightfield microscopy images. Therefore, cells were first sorted in a microfluidic device and subsequently used to train a convolutional neural network (CNN) to classify cell-based image features related to cell deformability. The CNN correctly predicted the deformability of individual RBCs with 81 ± 11% accuracy averaged across ten donors [87]. However, approaches to assess storage-induced changes in RBCs at rest usually require the sample to be dropped or fixed on a glass slide, which often renders the techniques unsuitable for high throughput. Additionally, blood in vivo is under permanent flow, while blood in stasis is not compatible with life. Therefore, recent AI-based approaches aimed at assessing RBC morphologies in microfluidic devices, mimicking in vivo flow conditions. Recently, imaging flow cytometry was combined with a deep-learning-based morphological assessment to provide an objective evaluation of stored RBC quality [83, 91]. In these studies, images of single RBCs flowing through a cytometer were recorded. Subsequently, a CNN was trained to categorize cells into six different morphological classes. This fully supervised model was able to provide a fast and automated morphological assessment with accuracy values comparable to the accuracy between different human experts. However, it was noted that the classification of RBC shapes into discrete bins, by either human experts or AI, could be an inadequate description of a continuous biological process [91]. Therefore, Doan et al. [91] proposed a weakly supervised network that learned the morphological properties of RBCs independently from visual categories previously defined by experts. This network revealed a chronological progression of RBC morphological changes that better predicted RBC quality than the previously employed shape classification system [91].
RBC Quality Measurements in Microcapillaries Using AI
Despite these advances in RBC shape classification under flow using imaging flow cytometers, the dimensions of the flow channel’s cross-section in such instruments are generally larger than the RBC size to achieve high throughput. However, this results in a large degree of freedom regarding the cell orientation, rotation, and resulting shapes. To overcome these challenges, microfluidic channels with cross sections on the order of 10 μm were previously used to achieve stable RBC shapes that primarily depend on the cell velocity or channel shear rate [58, 92]. In such microcapillaries, healthy RBCs predominantly deform into centered croissant-like shapes (shown in Fig. 4a, top) at low velocities and off-centered slipper-like shapes (shown in Fig. 4a, bottom) at high velocities. Combining these microfluidic measurements with AI-based shape assessment provides new opportunities in RBC flow characterization. Recent advantages have been reported for fast and unbiased classification of healthy RBCs [89], for study changes during infection (e.g., COVID-19) [93], and for detecting donor-dependent RBC aging curves [22].
To highlight the practicability of artificial neural networks and deep-learning-based techniques, we reanalyzed the data presented in [22]. We further compare the possible advantages of this AI-based evaluation with the standard approach reported in [22] that relies on geometric or analytic cell parameters. Figure 4 illustrates the results using standard flow analysis versus AI-based shape classification techniques, evaluating RBC changes of six donors over 10 weeks. In the standard approach, multiple parameters can be extracted from the captured images shown in Figure 4a to assess changes in the RBC capillary flow behavior. These include the cell velocity, which depends on the applied pressure drop in the microfluidic chip, the projection area of the cell, the equilibrium lateral position of the RBC across the channel width, and the deformability index DI, which is calculated based on the geometrical size of the RBC. One standard parameter that has been shown to capture dynamical RBC transitions in capillary flow is the equilibrium cell position across the channel width. Figure 4b showcases the evolution of the normalized cell position distribution for two donors and 2 weeks at cell velocities between 8 mm/s and 10 mm/s. The off-centered peak position is characteristic of the occurrence of slipper-shaped RBCs at such velocities, while other cell shapes preferentially flow at the channel center. Consequently, the ratio between the central and off-centered peak positions was previously used as an analytical parameter in [22] to assess changes in the flow behavior of stored RBCs. However, it was shown that the microcapillary flow behavior is crucially governed by RBC morphology [94]. Hence, RBCs that flow at a similar equilibrium y-position yet exhibit different shapes can have different effects on hemorheology. Therefore, characterizing the cell shape in flow could potentially improve the assessment of RBC quality and flow behavior. Such an automated RBC shape classification, using a CNN, is used as a comparison to the standard analytical approach. The CNN can discriminate between various RBC shapes in flow, including specific cell shapes that are characteristic of a specific pathology, e.g., sickle cells or acanthocytes. To assess storage-induced RBC changes, cells are classified as either healthy or pathological shapes shown in Figure 4c. The so-called shape phase diagram, i.e., the fraction of RBC shapes as a function of the velocity, is one straightforward parameter that can be used in the AI-based approach to depicting the shape dynamics of RBCs in capillary flow [95]. Figure 4d shows representative shape phase diagrams for the same data as in Figure 4b. To assess donor-dependent RBC in vitro aging and predict metadata and prospective changes in a stored sample, data reduction is crucial. Consequently, Figure 4e shows one parameter derived from the standard method (top) and one obtained from the AI-based approach (bottom). In the standard approach, the above-described y-peak ratio based on the probability density functions in Figure 4b is shown in Figure 4e. This quality overall increases over time for the different donors yet exhibits pronounced scattering, indicated by the broad prediction intervals. For the AI approach, the fraction of pathological RBC shapes based on the shape phase diagrams is employed to evaluate donor-dependent RBC changes (Fig. 4e, bottom). Comparing both the standard and the AI approaches, as shown in Figure 4f, we observe an improved estimation of time-dependent change for the AI method, expressed by the R-squared values from the linear regression models in Figure 4e. Given the continuous advancement in the field of microfluidics and AI, new opportunities for assessing RBC quality arise. Simple, rapid, and unbiased approaches that rather require a single instrument, such as the Erysense recently reported [22], instead of a sophisticated cascade of elaborated equipment, will be critical for the future of RBC research and quality control.
Future Activities for RBC Quality Control in Transfusion Medicine
We can measure the integrity of individual cells. However, we do need proof of correlation between the parameters we measure in vitro and the behavior of the RBCs in vivo. To this end, transfused RBCs need to be labeled before transfusion. Historically, 51Cr or other radioactive labels have been used, which are nowadays ethically no longer justifiable. However, with biotin labeling an alternative labeling technology with various advantages is readily available [96].
If the relation between in vitro parameters and in vivo RBC survival could be proven, it has the potential to revolutionize the practice of transfusion medicine: the composition of RBC units can be investigated before transfusion or in intervals over the entire storage period. Similar to those depicted in Figure 4, storage lesions could be quantified and RBC units be selected according to the intended treatment (e.g., acute vs. chronic transfusion need). The judgment of RBC quality over time, which is donor-dependent (compare Fig. 4), may also influence the storage time of particular units, which should be defined by the RBC quality and may, in particular cases, be beyond the current administrative regulations. Future developments based on AI may even predict storage quality over time improving the process even further.
Finally, similar to the complementary blood group determination and bedside tests today, the flow properties of RBCs of particular units could be pretested in the plasma of the patients, and such best match transfusions are realized without knowing (or determining) all RBC properties. This holds especially since not all transfusion-related properties of RBCs are currently known as the recent discovery of the ion channel Piezo1 being determinant of the Er blood group proofed [97, 98]. This RBC – plasma matching on minimal sample volumes, could be a stairway to personalized transfusion medicine.
Further Applications for AI in Transfusion Medicine
Although intensively discussed, the microfluidic-driven, image-based judgment of RBC quality is not the only possible application of AI in transfusion medicine (e.g., [24, 99, 100]). However, so far, all these approaches have in common that they are only used at the academic level or in clinical trials without routine application in clinical laboratories. To achieve the latter, great progress needs to be made in teaching the concepts of AI not only to the next generation of scientists but to the decision-makers today [101].
In addition to aiding decision-making, AI has been researched for other applications in certain studies such as [102, 103]. Sibinga [102] has demonstrated the potential of AI in the prediction of donor behavior and preferences, allowing blood banks (1) to more effectively recruit and retain donors, (2) to match donor blood with recipient blood more efficiently and accurately, (3) to predict blood demand and ensure that the appropriate blood components are available for transfusion, and (4) to predict whether a patient will require a blood transfusion during a surgical procedure, allowing for proactive management of blood product. On the other hand, Meier and Tschoellitsch [103] have explored the use of AI in patient blood management, including its potential in the blood manufacturing processes.
Conflict of Interest Statement
M.L. and S.Q. are employees of Cysmic GmbH, the manufacturer of Erysense, used for data generation presented in this study. In addition, S.R., G.S., S.Q., and L.K. are shareholders of Cysmic GmbH. H.E. and C.W. have no conflicts of interest to declare.
Funding Sources
This work was supported by the European Framework Horizon 2020 under grant agreement number 860436 (EVIDENCE), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, Grant No. WA 1336/13) and the Saarland University through the program “Anschubfinanzierung”. Furthermore, we acknowledge support by the DFG and Saarland University within the funding programme Open Access Publishing.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding Statement
This work was supported by the European Framework Horizon 2020 under grant agreement number 860436 (EVIDENCE), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, Grant No. WA 1336/13) and the Saarland University through the program “Anschubfinanzierung”. Furthermore, we acknowledge support by the DFG and Saarland University within the funding programme Open Access Publishing.
References
- 1. Landsteiner K. [Agglutination phenomena of normal human blood]. Wien Klin Wochenschr. 1901;113(20-21):768–9. [PubMed] [Google Scholar]
- 2. Bruun-Rasmussen P, Kragh Andersen P, Banasik K, Brunak S, Johansson PI. Intervening on the storage time of RBC units and its effects on adverse recipient outcomes using real-world data. Blood. 2022;139(25):3647–54. 10.1182/blood.2022015892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruun-Rasmussen P, Andersen PK, Banasik K, Brunak S, Johansson PI. Estimating the effect of donor sex on red blood cell transfused patient mortality: a retrospective cohort study using a targeted learning and emulated trials-based approach. Eclinicalmedicine. 2022;51:101628. 10.1016/j.eclinm.2022.101628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pendry K. The use of big data in transfusion medicine. Transfus Med. 2015;25(3):129–37. 10.1111/tme.12223. [DOI] [PubMed] [Google Scholar]
- 5. Reisman M. EHRs: the challenge of making electronic data useable and interoperable. P T. 2017;42(9):572–5. [PMC free article] [PubMed] [Google Scholar]
- 6. Murphy GF. Electronic health records: changing the vision. Philadelphia: W. B. Saunders Company; 1999. [Google Scholar]
- 7. Janett RS, Yeracaris PP. Electronic medical records in the American health system: challenges and lessons learned. Cien Saude Colet. 2020;25(4):1293–304. 10.1590/1413-81232020254.28922019. [DOI] [PubMed] [Google Scholar]
- 8. Roehrs A, da Costa CA, Righi RD, de Oliveira KSF. Personal health records: a systematic literature review. J Med Internet Res. 2017;19(1):e13. 10.2196/jmir.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henricks WH. Laboratory information systems. Surg Pathol Clin. 2015;8(2):101–8. 10.1016/j.path.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 10. Majchrowski B. Medical software’s increasing impact on healthcare and technology management. Biomed Instrum Technol. 2010;44(1):70–4. 10.2345/0899-8205-44.1.70. [DOI] [PubMed] [Google Scholar]
- 11. Vuk T, Politis C, Laspina S, Lozano M, Haddad A, de Angelis V, et al. Thirty years of hemovigilance: achievements and future perspectives. Transfus Clin Biol. 2023;30(1):166–72. 10.1016/j.tracli.2022.09.070. [DOI] [PubMed] [Google Scholar]
- 12. Ning S, Li N, Barty R, Arnold D, Heddle NM. Database-driven research and big data analytic approaches in transfusion medicine. Transfusion. 2022;62(7):1427–34. 10.1111/trf.16939. [DOI] [PubMed] [Google Scholar]
- 13. Hourfar MK, Jork C, Schottstedt V, Weber-Schehl M, Brixner V, Busch MP, et al. Experience of German Red Cross blood donor services with nucleic acid testing: results of screening more than 30 million blood donations for human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus. Transfusion. 2008;48(8):1558–66. 10.1111/j.1537-2995.2008.01718.x. [DOI] [PubMed] [Google Scholar]
- 14.WHO. Blood safety and availability. [cited 2022 Dec 7]. Available from: https://www.who.int/news-room/fact-sheets/detail/blood-safety-and-availability.
- 15. Starr D. Blood: an epic history of medicine and commerce. New York: Knopf Doubleday Publishing Group; 1998. [Google Scholar]
- 16. D’Alessandro A, Kriebardis AG, Rinalducci S, Antonelou MH, Hansen KC, Papassideri IS, et al. An update on red blood cell storage lesions, as gleaned through biochemistry and omics technologies. Transfusion. 2015;55(1):205–19. 10.1111/trf.12804. [DOI] [PubMed] [Google Scholar]
- 17. D’Alessandro A, D’Amici GM, Vaglio S, Zolla L. Time-course investigation of SAGM-stored leukocyte-filtered red bood cell concentrates: from metabolism to proteomics. Haematologica. 2012;97(1):107–15. 10.3324/haematol.2011.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mykhailova O, Olafson C, Turner TR, DʼAlessandro A, Acker JP. Donor-dependent aging of young and old red blood cell subpopulations: metabolic and functional heterogeneity. Transfusion. 2020;60(11):2633–46. 10.1111/trf.16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eichler H, Feyer AK, Weitmann K, Hoffmann W, Henseler O, Opitz A, et al. Population-based analysis of the impact of demographics on the current and future blood supply in the saarland. Transfus Med Hemoth. 2021;48(3):175–82. 10.1159/000512645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nieto-Calvache AJ, Quintero-Santacruz M, Macia-Mejía C, López-Girón MC, Vergara-Galliadi LM, Ariza F. Dangerous shortage of blood banks as an indirect effect of SARS-CoV-2: an obstetrics perspective. Int J Gynaecol Obstet. 2020;151(3):424–30. 10.1002/ijgo.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barnes LS, Stanley J, Bloch EM, Pagano MB, Ipe TS, Eichbaum Q, et al. Status of hospital-based blood transfusion services in low-income and middle-income countries: a cross-sectional international survey. Bmj Open. 2022;12(2):e055017. 10.1136/bmjopen-2021-055017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Recktenwald SM, Lopes MGM, Peter S, Hof S, Simionato G, Peikert K, et al. Erysense, a lab-on-a-chip-based point-of-care device to evaluate red blood cell flow properties with multiple clinical applications. Front Physiol. 2022;13:884690. 10.3389/fphys.2022.884690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Futur Healthc J. 2019;6(2):94–8. 10.7861/futurehosp.6-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smit Sibinga CT. Artificial intelligence in transfusion medicine and its impact on the quality concept. Transfus Apher Sci. 2020;59(6):103021. 10.1016/j.transci.2020.103021. [DOI] [PubMed] [Google Scholar]
- 25. De Mauro A, Greco M, Grimaldi M. A formal definition of big data based on its essential features. Libr Rev. 2016;65(3):122–35. 10.1108/lr-06-2015-0061. [DOI] [Google Scholar]
- 26. Laney D. 3D data management: controlling data volume, velocity, and variety. Stamford: META Group Inc.; 2001. [Google Scholar]
- 27. Dash S, Shakyawar SK, Sharma M, Kaushik S. Big data in healthcare: management, analysis and future prospects. J Big Data. 2019;6(1):54. 10.1186/s40537-019-0217-0. [DOI] [Google Scholar]
- 28. Kleinman S, Busch MP, Murphy EL, Shan H, Ness P, Glynn SA, et al. The national heart, lung, and blood institute recipient epidemiology and donor evaluation study (REDS-III): a research program striving to improve blood donor and transfusion recipient outcomes. Transfusion. 2014;54(3 Pt 2):942–55. 10.1111/trf.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edgren G, Hjalgrim H, Tran TN, Rostgaard K, Shanwell A, Titlestad K, et al. A population-based binational register for monitoring long-term outcome and possible disease concordance among blood donors and recipients. Vox Sang. 2006;91(4):316–23. 10.1111/j.1423-0410.2006.00827.x. [DOI] [PubMed] [Google Scholar]
- 30. Edgren G, Rostgaard K, Vasan SK, Wikman A, Norda R, Pedersen OB, et al. The new Scandinavian Donations and Transfusions database (SCANDAT2): a blood safety resource with added versatility. Transfusion. 2015;55(7):1600–6. 10.1111/trf.12986. [DOI] [PubMed] [Google Scholar]
- 31. Lanteri MC, Kanias T, Keating S, Stone M, Guo Y, Page GP, et al. Intradonor reproducibility and changes in hemolytic variables during red blood cell storage: results of recall phase of the REDS-III RBC-Omics study. Transfusion. 2019;59(1):79–88. 10.1111/trf.14987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. D’Alessandro A, Culp-Hill R, Reisz JA, Anderson M, Fu X, Nemkov T, et al. Heterogeneity of blood processing and storage additives in different centers impacts stored red blood cell metabolism as much as storage time: lessons from REDS-III: omics. Transfusion. 2019;59(1):89–100. 10.1111/trf.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stone M, Keating SM, Kanias T, Lanteri MC, Lebedeva M, Sinchar D, et al. Piloting and implementation of quality assessment and quality control procedures in RBC-Omics: a large multi-center study of red blood cell hemolysis during storage. Transfusion. 2019;59(1):57–66. 10.1111/trf.15099. [DOI] [PubMed] [Google Scholar]
- 34. Di Angelantonio E, Thompson SG, Kaptoge S, Moore C, Walker M, Armitage J, et al. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45 000 donors. Lancet. 2017;390(10110):2360–71. 10.1016/S0140-6736(17)31928-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Borkent-Raven BA, Janssen MP, van der Poel CL, Schaasberg WP, Bonsel GJ, van Hout BA. The PROTON study: profiles of blood product transfusion recipients in The Netherlands. Vox Sang. 2010;99(1):54–64. 10.1111/j.1423-0410.2010.01312.x. [DOI] [PubMed] [Google Scholar]
- 36. Heddle NM, Cook RJ, Liu Y, Zeller M, Barty R, Acker JP, et al. The association between blood donor sex and age and transfusion recipient mortality: an exploratory analysis. Transfusion. 2019;59(2):482–91. 10.1111/trf.15011. [DOI] [PubMed] [Google Scholar]
- 37. Holzmann MJ, Sartipy U, Olsson ML, Dickman P, Edgren G. Sex-discordant blood transfusions and survival after cardiac surgery: a nationwide cohort study. Circulation. 2016;134(21):1692–4. 10.1161/CIRCULATIONAHA.116.025087. [DOI] [PubMed] [Google Scholar]
- 38. Roubinian NH, Escobar GJ, Liu V, Swain BE, Gardner MN, Kipnis P, et al. Trends in red blood cell transfusion and 30-day mortality among hospitalized patients. Transfusion. 2014;54(10 Pt 2):2678–86. 10.1111/trf.12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaserer A, Rössler J, Braun J, Farokhzad F, Pape HC, Dutkowski P, et al. Impact of a Patient Blood Management monitoring and feedback programme on allogeneic blood transfusions and related costs. Anaesthesia. 2019;74(12):1534–41. 10.1111/anae.14816. [DOI] [PubMed] [Google Scholar]
- 40. Frank SM, Rothschild JA, Masear CG, Rivers RJ, Merritt WT, Savage WJ, et al. Optimizing preoperative blood ordering with data acquired from an anesthesia information management system. Anesthesiology. 2013;118(6):1286–97. 10.1097/ALN.0b013e3182923da0. [DOI] [PubMed] [Google Scholar]
- 41. Association for the Advancement of Blood and Biotherapies . Standards for blood banks and blood transfusion services. 33rd ed. Bethesda: Association for the Advancement of Blood and Biotherapies; 2022. [DOI] [PubMed] [Google Scholar]
- 42. Kaestner L, Juzeniene A, Moan J. Erythrocytes-the “house elves” of photodynamic therapy. Photochem Photobiol Sci. 2004;3(11–12):981–9. 10.1039/b403127j. [DOI] [PubMed] [Google Scholar]
- 43. Flatt JF, Bawazir WM, Bruce LJ. The involvement of cation leaks in the storage lesion of red blood cells. Front Physiol. 2014;5:214. 10.3389/fphys.2014.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Melzak KA, Spouge JL, Boecker C, Kirschhöfer F, Brenner-Weiss G, Bieback K. Hemolysis pathways during storage of erythrocytes and inter-donor variability in erythrocyte morphology. Transfus Med Hemoth. 2021;48(1):39–47. 10.1159/000508711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aung HH, Tung JP, Dean MM, Flower RL, Pecheniuk NM. Procoagulant role of microparticles in routine storage of packed red blood cells: potential risk for prothrombotic post-transfusion complications. Pathology. 2017;49(1):62–9. 10.1016/j.pathol.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 46. Barshtein G, Manny N, Yedgar S. Circulatory risk in the transfusion of red blood cells with impaired flow properties induced by storage. Transfus Med Rev. 2011 Jan;25(1):24–35. 10.1016/j.tmrv.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 47. Barshtein G, Pries AR, Goldschmidt N, Zukerman A, Orbach A, Zelig O, et al. Deformability of transfused red blood cells is a potent determinant of transfusion-induced change in recipient’s blood flow. Microcirculation. 2016;23(7):479–86. 10.1111/micc.12296. [DOI] [PubMed] [Google Scholar]
- 48. Barshtein G, Goldschmidt N, Pries AR, Zelig O, Arbell D, Yedgar S. Deformability of transfused red blood cells is a potent effector of transfusion-induced hemoglobin increment: a study with β-thalassemia major patients. Am J Hematol. 2017;92(9):E559–60. 10.1002/ajh.24821. [DOI] [PubMed] [Google Scholar]
- 49. Kaestner L, Scholz A, Tian Q, Ruppenthal S, Tabellion W, Wiesen K, et al. Genetically encoded Ca2+ indicators in cardiac myocytes. Circ Res. 2014 May;114(10):1623–39. 10.1161/CIRCRESAHA.114.303475. [DOI] [PubMed] [Google Scholar]
- 50. Thomas SL, Egee S, Lapaix F, Kaestner L, Staines HM, Ellory JC. Malaria parasite Plasmodium gallinaceum up-regulates host red blood cell channels. FEBS Lett. 2001 Jun;500(1–2):45–51. 10.1016/s0014-5793(01)02579-0. [DOI] [PubMed] [Google Scholar]
- 51. Teles J, Enver T, Pina C. Single-cell PCR profiling of gene expression in hematopoiesis. Methods Mol Biol. 2014;1185:21–42. 10.1007/978-1-4939-1133-2_3. [DOI] [PubMed] [Google Scholar]
- 52. Kaestner L, Bianchi P. Trends in the development of diagnostic tools for red blood cell-related diseases and anemias. Front Physiol. 2020;11:387. 10.3389/fphys.2020.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hertz L, Flormann D, Birnbaumer L, Wagner C, Laschke MW, Kaestner L. Evidence of in vivo exogen protein uptake by red blood cells: a putative therapeutic concept. Blood Adv. 2023;7(6):1033–9. 10.1182/bloodadvances.2022008404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rotordam MG, Fermo E, Becker N, Barcellini W, Brüggemann A, Fertig N, et al. A novel gain-of-function mutation of Piezo1 is functionally affirmed in red blood cells by high-throughput patch clamp. Haematologica. 2019;104(5):e179–83. 10.3324/haematol.2018.201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hertz L, Huisjes R, Llaudet-Planas E, Petkova-Kirova P, Makhro A, Danielczok JG, et al. Is increased intracellular calcium in red blood cells a common component in the molecular mechanism causing anemia? Front Physiol. 2017;8:673. 10.3389/fphys.2017.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bessis M. Corpuscles, atlas of red blood cell shapes. Berlin: Springer; 1974. [Google Scholar]
- 57. Simionato G, Hinkelmann K, Chachanidze R, Bianchi P, Fermo E, van Wijk R, et al. Red blood cell phenotyping from 3D confocal images using artificial neural networks. Plos Comput Biol. 2021;17(5):e1008934. 10.1371/journal.pcbi.1008934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Quint S, Christ AF, Guckenberger A, Himbert S, Kaestner L, Gekle S, et al. 3D tomography of cells in micro-channels. Appl Phys Lett. 2017 Sep;111(10):103701. 10.1063/1.4986392. [DOI] [Google Scholar]
- 59. Simionato G, van Wijk R, Quint S, Wagner C, Bianchi P, Kaestner L. Rare anemias: are their names just smoke and mirrors? Front Physiol. 2021;12, 690604. 10.3389/fphys.2021.690604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bessis M, Delpech G. Sickle cell shape and structure: images and concepts (1840-1980). Blood Cell. 1982;8(2):359–435. [PubMed] [Google Scholar]
- 61. Darras A, Peikert K, Rabe A, Yaya F, Simionato G, John T, et al. Acanthocyte sedimentation rate as a diagnostic biomarker for neuroacanthocytosis syndromes: experimental evidence and physical justificationfication. Cells. 2021;10(4):788. 10.3390/cells10040788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Henkelman S, Dijkstra-Tiekstra MJ, de Wildt-Eggen J, Graaff R, Rakhorst G, van Oeveren W. Is red blood cell rheology preserved during routine blood bank storage? Transfusion. 2010;50(4):941–8. 10.1111/j.1537-2995.2009.02521.x. [DOI] [PubMed] [Google Scholar]
- 63. Frank SM, Abazyan B, Ono M, Hogue CW, Cohen DB, Berkowitz DE, et al. Decreased erythrocyte deformability after transfusion and the effects of erythrocyte storage duration. Anesth Analg. 2013;116(5):975–81. 10.1213/ANE.0b013e31828843e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bennett-Guerrero E, Veldman TH, Doctor A, Telen MJ, Ortel TL, Reid TS, et al. Evolution of adverse changes in stored RBCs. Proc Natl Acad Sci. 2007;104(43):17063–8. 10.1073/pnas.0708160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Piety NZ, Stutz J, Yilmaz N, Xia H, Yoshida T, Shevkoplyas SS. Microfluidic capillary networks are more sensitive than ektacytometry to the decline of red blood cell deformability induced by storage. Sci Rep. 2021;11(1):604. 10.1038/s41598-020-79710-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tomaiuolo G. Biomechanical properties of red blood cells in health and disease towards microfluidics. Biomicrofluidics. 2014;8(5):051501. 10.1063/1.4895755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Matthews K, Lamoureux ES, Myrand-Lapierre ME, Duffy SP, Ma H. Technologies for measuring red blood cell deformability. Lab Chip. 2022;22(7):1254–74. 10.1039/d1lc01058a. [DOI] [PubMed] [Google Scholar]
- 68. Steffen P, Jung A, Nguyen DB, Müller T, Bernhardt I, Kaestner L, et al. Stimulation of human red blood cells leads to Ca2+-mediated intercellular adhesion. Cell Calcium. 2011 May;50(1):54–61. 10.1016/j.ceca.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 69. Kaestner L, Steffen P, Nguyen DB, Wang J, Wagner-Britz L, Jung A, et al. Lysophosphatidic acid induced red blood cell aggregation in vitro. Bioelectrochemistry. 2012;87:89–95. 10.1016/j.bioelechem.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 70. Danielczok JG, Terriac E, Hertz L, Petkova-Kirova P, Lautenschläger F, Laschke MW, et al. Red blood cell passage of small capillaries is associated with transient Ca2+-mediated adaptations. Front Physiol. 2017;8:979. 10.3389/fphys.2017.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rizzuto V, Mencattini A, Álvarez-González B, Di Giuseppe D, Martinelli E, Beneitez-Pastor D, et al. Combining microfluidics with machine learning algorithms for RBC classification in rare hereditary hemolytic anemia. Sci Rep. 2021;11(1):13553. 10.1038/s41598-021-92747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Myrand-Lapierre ME, Deng X, Ang RR, Matthews K, Santoso AT, Ma H. Multiplexed fluidic plunger mechanism for the measurement of red blood cell deformability. Lab Chip. 2015;15(1):159–67. 10.1039/c4lc01100g. [DOI] [PubMed] [Google Scholar]
- 73. Man Y, Kucukal E, An R, Watson QD, Bosch J, Zimmerman PA, et al. Microfluidic assessment of red blood cell mediated microvascular occlusion. Lab Chip. 2020;20(12):2086–99. 10.1039/d0lc00112k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Islamzada E, Matthews K, Guo Q, Santoso AT, Duffy SP, Scott MD, et al. Deformability based sorting of stored red blood cells reveals donor-dependent aging curves. Lab Chip. 2020;20(2):226–35. 10.1039/c9lc01058k. [DOI] [PubMed] [Google Scholar]
- 75. Burns JM, Yang X, Forouzan O, Sosa JM, Shevkoplyas SS. Artificial microvascular network: a new tool for measuring rheologic properties of stored red blood cells. Transfusion. 2012;52(5):1010–23. 10.1111/j.1537-2995.2011.03418.x. [DOI] [PubMed] [Google Scholar]
- 76. Robidoux J, Laforce-Lavoie A, Charette SJ, Shevkoplyas SS, Yoshida T, Lewin A, et al. Development of a flow standard to enable highly reproducible measurements of deformability of stored red blood cells in a microfluidic device. Transfusion. 2020;60(5):1032–41. 10.1111/trf.15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Matthews K, Myrand-Lapierre ME, Ang RR, Duffy SP, Scott MD, Ma H. Microfluidic deformability analysis of the red cell storage lesion. J Biomech. 2015;48(15):4065–72. 10.1016/j.jbiomech.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 78. Wang Y, You G, Chen P, Li J, Chen G, Wang B, et al. The mechanical properties of stored red blood cells measured by a convenient microfluidic approach combining with mathematic model. Biomicrofluidics. 2016;10(2):024104. 10.1063/1.4943861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cluitmans JCA, Chokkalingam V, Janssen AM, Brock R, Huck WTS, Bosman GJ. Alterations in red blood cell deformability during storage: a microfluidic approach. BioMed Res Int. 2014;2014:764268. 10.1155/2014/764268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zheng Y, Chen J, Cui T, Shehata N, Wang C, Sun Y. Characterization of red blood cell deformability change during blood storage. Lab Chip. 2014;14(3):577–83. 10.1039/c3lc51151k. [DOI] [PubMed] [Google Scholar]
- 81. Xu Z, Zheng Y, Wang X, Shehata N, Wang C, Sun Y. Stiffness increase of red blood cells during storage. Microsyst Nanoeng. 2018;4(1):17103. 10.1038/micronano.2017.103. [DOI] [Google Scholar]
- 82. Kim E, Park S, Hwang S, Moon I, Javidi B. Deep learning-based phenotypic assessment of red cell storage lesions for safe transfusions. IEEE J Biomed Health Inform. 2021;95(3):1318–28. [DOI] [PubMed] [Google Scholar]
- 83. Pinto RN, Sebastian JA, Parsons MJ, Chang TC, Turner TR, Acker JP, et al. Label-free analysis of red blood cell storage lesions using imaging flow cytometry. Cytometry. 2019;95(9):976–84. 10.1002/cyto.a.23846. [DOI] [PubMed] [Google Scholar]
- 84. Kaestner L. Artificial intelligence meets hematology. Transfus Apher Sci. 2020;59(6):102986. 10.1016/j.transci.2020.102986. [DOI] [PubMed] [Google Scholar]
- 85. Shouval R, Fein JA, Savani B, Mohty M, Nagler A. Machine learning and artificial intelligence in haematology. Br J Haematol. 2021;192(2):239–50. 10.1111/bjh.16915. [DOI] [PubMed] [Google Scholar]
- 86. Sebastian JA, Kolios MC, Acker JP. Emerging use of machine learning and advanced technologies to assess red cell quality. Transfus Apher Sci. 2020;59(6):103020. 10.1016/j.transci.2020.103020. [DOI] [PubMed] [Google Scholar]
- 87. Lamoureux ES, Islamzada E, Wiens MVJ, Matthews K, Duffy SP, Ma H. Assessing red blood cell deformability from microscopy images using deep learning. Lab Chip. 2022;22(1):26–39. 10.1039/d1lc01006a. [DOI] [PubMed] [Google Scholar]
- 88. Routt AH, Yang N, Piety NZ, Lu M, Shevkoplyas SS. Deep ensemble learning enables highly accurate classification of stored red blood cell morphology. Sci Rep. 2023;13(1):3152. 10.1038/s41598-023-30214-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kihm A, Kaestner L, Wagner C, Quint S. Classification of red blood cell shapes in flow using outlier tolerant machine learning. Plos Comput Biol. 2018 Jun;14(6):e1006278. 10.1371/journal.pcbi.1006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang Y, Sang J, Ao R, Ma Y, Fu B. Numerical simulation of deformed red blood cell by utilizing neural network approach and finite element analysis. Comput Method Biomec. 2020;23(15):1190–200. 10.1080/10255842.2020.1791836. [DOI] [PubMed] [Google Scholar]
- 91. Doan M, Sebastian JA, Caicedo JC, Siegert S, Roch A, Turner TR, et al. Objective assessment of stored blood quality by deep learning. Proc Natl Acad Sci. 2020;117(35):21381–90. 10.1073/pnas.2001227117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Recktenwald SM, Graessel K, Maurer FM, John T, Gekle S, Wagner C. Red blood cell shape transitions and dynamics in time-dependent capillary flows. Biophys J. 2022;121(1):23–36. 10.1016/j.bpj.2021.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Recktenwald SM, Simionato G, Lopes MG, Gamboni F, Dzieciatkowska M, Meybohm P, et al. Cross-talk between red blood cells and plasma influences blood flow and omics phenotypes in severe COVID-19. Elife. 2022;11:e81316. 10.7554/eLife.81316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lanotte L, Mauer J, Mendez S, Fedosov DA, Fromental JM, Claveria V, et al. Red cells’ dynamic morphologies govern blood shear thinning under microcirculatory flow conditions. Proc Natl Acad Sci U S A. 2016 Nov;113(47):13289–94. 10.1073/pnas.1608074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Guckenberger A, Kihm A, John T, Wagner C, Gekle S. Numerical–experimental observation of shape bistability of red blood cells flowing in a microchannel. Soft Matter. 2018;14(11):2032–43. 10.1039/c7sm02272g. [DOI] [PubMed] [Google Scholar]
- 96. Mock DM, Nalbant D, Kyosseva SV, Schmidt RL, An G, Matthews NI, et al. Development, validation, and potential applications of biotinylated red blood cells for posttransfusion kinetics and other physiological studies: evidenced-based analysis and recommendations. Transfusion. 2018;58(8):2068–81. 10.1111/trf.14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Crew VK, Tilley LA, Satchwell TJ, AlSubhi SA, Jones B, Spring FA, et al. Missense mutations in PIEZO1, encoding the Piezo1 mechanosensor protein, define the Er red blood cell antigens. Blood. 2023;141(2):135–46. 10.1182/blood.2022016504. [DOI] [PubMed] [Google Scholar]
- 98. Bianchi P, Makhro A, Egee S, Kaestner L. Blood group meets ion channel: an unexpected marriage. Blood. 2023;141(2):online-only. [Google Scholar]
- 99. Smit Sibinga CT. Artificial intelligence and the future of transfusion medicine. Neurosci Chron. 2021;2(2). 10.46439/Neuroscience.2.011. [DOI] [Google Scholar]
- 100. Zhu S, Zhou L, Feng Y, Zhu J, Shu Q, Li H. Understanding the risk factors for adverse events during exchange transfusion in neonatal hyperbilirubinemia using explainable artificial intelligence. BMC Pediatr. 2022;22(1):567. 10.1186/s12887-022-03615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kaestner L. Artificial intelligence: training the trainer. Br J Haematol. 2022;198(5):805–6. 10.1111/bjh.18358. [DOI] [PubMed] [Google Scholar]
- 102. Sibinga CTS. Digital Health. 2022:107–20. [Google Scholar]
- 103. Meier JM, Tschoellitsch T. Artificial intelligence and machine learning in patient blood management: a scoping review. Anesth Analg. 2022;135(3):524–31. 10.1213/ANE.0000000000006047. [DOI] [PubMed] [Google Scholar]