Abstract
Pleiotropy occurs when a genetic variant influences more than one trait. This is a key property of the genomic architecture of psychiatric disorders and has been observed for rare and common genomic variants. It is reasonable to hypothesize that the microscale genetic overlap (pleiotropy) across psychiatric conditions and cognitive traits may lead to similar overlaps at the macroscale brain level such as large-scale brain functional networks.
We took advantage of brain connectivity, measured by resting-state functional MRI to measure the effects of pleiotropy on large-scale brain networks, a putative step from genes to behaviour. We processed nine resting-state functional MRI datasets including 32 726 individuals and computed connectome-wide profiles of seven neuropsychiatric copy-number-variants, five polygenic scores, neuroticism and fluid intelligence as well as four idiopathic psychiatric conditions.
Nine out of 19 pairs of conditions and traits showed significant functional connectivity correlations (rFunctional connectivity), which could be explained by previously published levels of genomic (rGenetic) and transcriptomic (rTranscriptomic) correlations with moderate to high concordance: rGenetic—rFunctional connectivity = 0.71 [0.40–0.87] and rTranscriptomic—rFunctional connectivity = 0.83 [0.52; 0.94]. Extending this analysis to functional connectivity profiles associated with rare and common genetic risk showed that 30 out of 136 pairs of connectivity profiles were correlated above chance. These similarities between genetic risks and psychiatric disorders at the connectivity level were mainly driven by the overconnectivity of the thalamus and the somatomotor networks. Our findings suggest a substantial genetic component for shared connectivity profiles across conditions and traits, opening avenues to delineate general mechanisms—amenable to intervention—across psychiatric conditions and genetic risks.
Keywords: pleiotropy, psychiatry, functional connectivity, autism spectrum disorder, copy-number variant
Moreau et al. provide evidence suggesting a substantial genetic component for shared brain connectivity dimensions across psychiatric conditions and cognitive traits. The results open up new avenues to help reshape psychiatric nosology and delineate general mechanisms—amenable to intervention—across disorders.
Introduction
Genetic pleiotropy, a key feature of psychiatric conditions, refers to the situation in which a genetic variant or gene has effects on more than one phenotype.1 Genetic correlation (rG), a measure of the average effect of pleiotropy across genomic loci, has been computed using common variants (i.e. single nucleotide polymorphisms, SNPs) based on genome-wide association study (GWAS) summary statistics.2 SNP-based rG are moderate to high between schizophrenia (SZ), bipolar disorder (BIP) and major depressive disorder, and lower between these three conditions and autism spectrum disorder (ASD).3–5 Moderate to mild genetic correlations are also observed between these psychiatric conditions and cognitive abilities or personality traits such as neuroticism and fluid intelligence.6,7 Similar levels of correlations between pairs of these same psychiatric conditions have been shown at the brain transcriptomic level (rT).8
Although rG has only been computed for common variants, pleiotropy has also been reported for rare variants such as copy-number variants (CNVs),9,10 which are often associated with a broad range of psychiatric diagnoses and cognitive traits.
It is reasonable to assume that overlap at the microscopic scale (i.e. genetic and transcriptomic) between conditions and traits may lead to similar overlaps at the macroscopic scale, such as large-scale functional networks. The latter can be inferred using resting-state functional MRI (rs-fMRI). This imaging technique measures spontaneous, low-frequency temporal synchronization of the activity in different brain regions during rest.11,12 An overlap between functional connectivity (FC) profiles of eight psychiatric disorders has been previously reported as driven by the default mode, salience and frontoparietal networks.13 A complementary dimensional reduction approach has identified a latent dimension mainly involving the somatosensory-subcortical networks spanning four psychiatric diagnoses.14
FC similarity has also been investigated between two rare CNVs (i.e. 16p11.2 and 22q11.2 deletion) that both confer large risks for ASD, SZ and cognitive deficits. Connectivity profiles of the thalamus, somatomotor, posterior insula and cingulate showed similarities between these two CNVs, as well as groups of individuals with either idiopathic ASD or SZ. Beyond these two genomic loci, nothing is known about the effects of rare high-risk variants on brain FC. Furthermore, little is known about the FC effects of common variants increasing risks for psychiatric conditions (i.e. polygenic scores, PGS).15
Knowledge gaps
The relationship between the level of pleiotropy at the genetic (SNP-based) and large-scale functional brain connectivity network is unknown.
Pleiotropy observed for rare genomic variants associated with psychiatric disorder has not been investigated at the level of functional brain connectivity.
Our overarching aim was to investigate the relationship between pleiotropy at the genetic and functional connectivity levels
Specifically, we aimed to: (i) investigate the concordance between previously established genetic correlations and FC correlations between conditions and traits; and (ii) identify brain networks driving FC correlations observed between rare and common genetic risks, psychiatric conditions and traits.
To this end, we used the same pipeline to analyse rs-fMRI data in n = 32 726 individuals from four genetics-first clinical cohorts (e.g. recruited because they carry a high-risk genetic variant), four case-control idiopathic psychiatric datasets [ASD, SZ, attention-deficit/hyperactivity disorders (ADHD), BIP] and one unselected population. We performed 19 connectome-wide association studies (CWAS) for seven CNVs, five PGS, four idiopathic psychiatric conditions and one non-brain related disease (inflammatory bowel disease, IBD), fluid intelligence and neuroticism. We included 279 CNV carriers, 1022 individuals with either autism, SZ, BIP or ADHD and 31 425 controls.
Materials and methods
Selecting CNVs, conditions and traits
We analysed all of the available rs-fMRI data for neuropsychiatric CNVs with at least n = 20 carriers to allow for the detection of large effect sizes (Cohen’s d > 0.8) previously reported for CNVs. As a result, selected CNVs are those most frequently identified in the clinic: 22q11.2, 1q21.1, 15q11.2, 16p11.2. Fluid intelligence and neuroticism were selected because (i) CNVs that increase risk for ASD and/or SZ decrease cognitive ability10,16; and (ii) both traits show the highest genetic correlation, among commonly measured traits, with ASD4 as well as with SZ.6,7 IBD was selected as a non-psychiatric control condition with a sample size similar to those available for the psychiatric conditions included in the study.
Cohorts
Our analysis included 32 726 individuals from nine datasets (Table 1). Each study of the corresponding dataset was approved by the research ethics review boards of the respective institutions. This project was approved by the research ethics review board at the Centre Hospitalier Universitaire Sainte Justine.
Table 1.
Data demographics
| Genetic variants conditions traits | Status | n total/n clin | Age | Sex (F/M) | Cohorts | IQ loss | OR ASD | OR SZ |
|---|---|---|---|---|---|---|---|---|
| Previously published | ||||||||
|
1q21.1
1: 146.53–147.39 7 genes (CHD1L) |
DEL | 25/15 | 44.4 (19) | 12/13 | UKBB-MRG- Cardiff- SFARI |
15 | 3.2 | 6.4 |
| DUP | 19/6 | 50.9 (19) | 13/6 | 25 | 5.3 | 2.9 | ||
|
22q11.2
22: 19.04–21.47 49 genes (TBX1) |
DEL | 43/43 | 16.9 (7) | 19/24 | UCLA | 28.8 | 32.3 | 23 |
| DUP | 22/12 | 39.4 (23) | 12/10 | UCLA-UKBB Cardiff-MRG |
8.3 | 2 | 0.2 | |
|
16p11.2
16: 29.65–30.20 27 genes (KCTD13) |
DEL | 32/28 | 21.7 (20) | 13/19 | SFARI - MRG -UKBB |
26 | 14.3 | 1.1 |
| DUP | 35/29 | 34.1 (19) | 14/21 | 11 | 10.5 | 11.7 | ||
|
15q11.2
15: 22.81–23.09 4 genes (CYFIP1) |
DEL | 103/0 | 64.3 (7) | 55/48 | UKBB | 3 | 1.3 | 1.9 |
| Idiopathic psychiatric conditions | SZ | 283 | 33.9 (9.2) | 73/210 | Montreal-SZ, CNP | - | - | - |
| BIP | 44 | 35 (9) | 20/24 | CNP | - | - | - | |
| ASD | 472 | 14.9 (6) | 0/472 | ABIDE1, ABIDE2 | - | - | - | |
| ADHD | 223 | 14.8 (9.5) | 66/157 | ADHD-200 CNP | - | - | - | |
| Non-psychiatric condition | IBD | 287 | 64.7(7.5) | 144/143 | UKBB | |||
| Polygenic scores | ASD | 29 460 | 64.2 (7.5) | 15 840/13 620 | UKBB | - | 2.7 | - |
| SZ | - | - | 3.5 | |||||
| BIP | - | - | - | |||||
| MDD | ||||||||
| Cross-D | - | - | - | |||||
| Traits | FI | 27 522 | 64 (7.5) | 14 777/12 745 | - | - | - | |
| NT | 24 025 | 64 (7.5) | 12 723/11 302 | - | - | - | ||
| Controls | UKBB | 30 185 | 64.1 (7.5) | 16 260/13 925 | UKBB | - | - | - |
| SFARI | 84 | 26.7 (15) | 35/49 | SFARI | - | - | - | |
| MRG | 39 | 34 (16) | 25/14 | MRG | - | - | - | |
| Cardiff | 8 | 39.8 (4) | 4/4 | Cardiff | ||||
| UCLA | 43 | 13 (4.6) | 22/21 | UCLA | - | - | - | |
| Psychiatric cohorts | 1066 | 20 (11) | 244/822 | - | - | - | - | |
CNV carriers, individuals with idiopathic psychiatric conditions and controls after MRI quality control. Chr = chromosome number, and coordinates are presented in Megabases (Mb, Hg19). The number of genes encompassed in each CNV is detailed below the genomic coordinates, followed by a well-known gene to help identify the CNV. n = total/clin: total number of participants/number of participants clinically ascertained. Age (in years, mean ± standard deviation). All sites scanned controls and sensitivity analyses were performed to investigate the potential bias introduced by differences in scanning site, age and sex. IQ loss = mean decrease in IQ points associated with each CNV.27,62 Odd-ratios (OR) for the enrichment of CNVs in ASD and SZ were previously published.62–71 ORs for the enrichment of CNVs in ADHD were not available. Detailed information relative to diagnosis, IQ, and motion, are available in Supplementary Tables 2–4. DEL = deletion; DUP = duplication; F = female; M = male; MDD = major depression disorder; CrossD = cross-disorder; CNP = Consortium for Neuropsychiatric Phenomics; IQ = intelligence quotient.
Clinical genetic datasets
We used four ‘genetics-first’ CNV datasets, which were recruited on the basis of the presence of a CNV associated with risk of neurodevelopmental and psychiatric disorders, regardless of symptomatology (detailed in the Supplementary material). These included the Simons Variation in Individuals Project (SVIP for 16p11.2 and 1q21.1 CNVs),17 the University of California, Los Angeles 22q11.2 CNV project, the University of Cardiff and the Montreal Rare Genomic Disorder (MRG) datasets.
Unselected population
CNVs associated with neurodevelopmental and psychiatric disorders were also identified in the UK Biobank dataset18 (Supplementary material).
Idiopathic psychiatric conditions cohorts
We used the ABIDE1,19 ABIDE2,20 ADHD-200,21 the Consortium for Neuropsychiatric Phenomics (CNP)22 and an aggregate dataset of 10 SZ studies23,24; collectively, these datasets include individuals with idiopathic ASD, ADHD, SZ and BIP, as well as their respective controls (Supplementary material).
CNV calling and polygenic scores computation
CNVs were identified in the UK Biobank using PennCNV25 and QuantiSNP26 following previously published methods27 (Supplementary material).
We computed five PGS for individuals of European ancestry in the UK Biobank using PRS-CS, a polygenic prediction via Bayesian regression and continuous shrinkage priors28 (Table 1, Supplementary material and Supplementary Table 1).
Resting-state functional MRI preprocessing
All datasets were preprocessed using the same parameters of Neuroimaging Analysis Kit.29 Preprocessed data were visually controlled for quality of the coregistration, head motion and related artefacts (Supplementary material).
Computing connectomes
We segmented the brain into 64 functional regions defined by the multi-resolution MIST brain parcellation30 to compute connectomes—defined by 2080 connections between 64 regions, which are grouped into 12 functional networks30: https://simexp.github.io/multiscale_dashboard/index.html. The MIST atlas was chosen as it has the advantage of including the cerebellum, which seems to play a critical role in neurodevelopmental disorders and psychiatric conditions.31–34 MIST parcellation also performs on a par with or superior to other templates (such as AAL or Power) on several prediction benchmarks—in particular, those regarding ASD and SZ prediction.35–37
Statistical analyses were performed using scikit-learn38 and stats39 libraries.
Connectome-wide association studies
We performed 19 CWAS by either:
contrasting cases and respective controls for seven CNVs associated with neurodevelopmental and psychiatric disorders (Table 1), and four idiopathic psychiatric disorder cohorts (ASD, SZ, BIP and ADHD). Controls refer to (a) individuals without a CNV for analyses to investigate the effect of CNVs; or (b) individuals without a psychiatric diagnosis in analyses to investigate the effects of psychiatric conditions.
or by investigating the linear effects of five continuous PGS: ASD, BIP, SZ, cross-disorder and major depressive disorder, as well as two continuous traits provided by UK Biobank: neuroticism and fluid intelligence.
FC was z-scored on the basis of the variance of the pooled controls used for each CWAS. They were conducted by linear regression, in which z-scored FC values were the dependent variables and genetic or diagnostic status or traits were the explanatory variables. PGS and traits were normalized within the UKBB sample.
Models were adjusted for sex, scanning site, head motion, age and global signal (defined as the mean of all 2080 Fisher’s Z-values). FC profiles were defined as the 2080 β values of 2080 connections.
This linear regression was applied for each of the 2080 functional connections. Since all raw connectomes were normalized on the variance of the controls, regression estimates (beta) can be interpreted as z-scores. We corrected for multiple testing using false discovery rate (FDR) (q < 0.05) as well as a permutation procedure. We corrected for the number of tests (2080) using the Benjamini–Hochberg correction for FDR at a threshold of q < 0.05.40,41 We also computed an empirical P-value (‘pval effect’) by conducting a permutation test, shuffling the genetic or clinical status labels of the individuals included in each CWAS (5000 permutations). We estimated the empirical P-value by calculating the frequency of obtaining an effect size equal to or greater than the original observation.42 Effect size of genetic risk, conditions and traits on connectivity was defined as the top decile of the 2080 absolute β values.
Concordance between functional, genetic and transcriptomic correlations
We computed correlations of whole-brain connectome profiles across pairs of conditions and traits (Pearson correlation) using the 2080 beta values of each CWAS.
We obtained genetic correlation (rG) values across pairs of conditions and traits [neuroticism,7 intelligence,6 cross-disorders (eight psychiatric conditions)3,43] from previously published GWAS. We also used previously published8 correlation values of transcriptomic profiles between six pairs of conditions.
We performed concordance analyses between correlation at the genetic (rG) and FC (rFC) levels, as well as the transcriptomic (rT) and FC (rFC), levels using DescTools R package to extract Lin’s concordance correlation coefficient (CCC).39,44 The bias correction factor quantifies how far the best fit line deviates from 45°.
Atlas of functional connectivity correlations across genetic risk, traits and conditions
We computed Pearson correlations between the 17 out of 19 whole-brain FC profiles with significantly altered connections (FDR corrected). For the significance of correlations between FC profiles, we generated a null distribution of 10 000 correlation values for each pair of conditions and traits. These 10 000 null correlations were computed using null FC profiles. The latter were obtained by conducting 5000 CWAS after shuffling the clinical status or trait values. To obtain a P-value, the correlation value was compared to the null distribution. We corrected for the number of correlations (n = 136) using the Benjamini–Hochberg correction for FDR at a threshold of q < 0.05.41
Principal component analysis
To identify the FC networks driving the correlations, we conducted a principal component analysis (PCA) on the 17 scaled FC profiles using the prcomp function from stats R package. Functional connections with 5% top loadings for principal components 1 and 2 (PC1, PC2) were represented on chord diagrams using the circlize R package (code available on GitHub). We also reported—per network—the average of absolute loadings of each connection, divided by the number of regions encompassed in each network (Supplementary Fig. 1).
Data and materials availability
Data from UK Biobank was downloaded under the application 40980, and can be accessed via their standard data access procedure (see http://www.ukbiobank.ac.uk/register-apply). UK Biobank CNVs were called using the pipeline developed in Jacquemont Laboratory, and described in https://github.com/labjacquemont/MIND-GENESPARALLELCNV. The final CNV calls are available from UK Biobank returned datasets (return ID: 3104, https://biobank.ndph.ox.ac.uk/ukb/dset.cgi? id=3104).
ABIDE1, COBRE, ADHD200, CNP, 16p11.2 SVIP data are publicly available: http://fcon_1000.projects.nitrc.org/indi/abide/abide_I.html, http://schizconnect.org/queries/new, http://fcon_1000.projects.nitrc.org/indi/adhd200/, https://www.openfmri.org/dataset/ds000030/ and https://www.sfari.org/funded-project/simons-variation-in-individuals-project-simons-vip/. The 22q11.2 UCLA raw data are currently available by request from the PI (C.E.B.). Raw imaging data for the MRG disorder family dataset are going to be available on the LORIS platform in 2023. The Cardiff raw data are not publicly available yet: contact the PI for further information (D.E.J.L.).
All processed connectomes are available through a request to the corresponding authors.
Code for all analyses and visualizations, beta values and P-values for the 19 FC profiles are available online through the GitHub platform with Jupyter notebook at https://github.com/claramoreau9/NeuropsychiatricCNVs_Connectivity.
Results
Pleiotropy: similarities between genetic and functional connectivity correlations across psychiatric conditions and traits
To investigate overlap and pleiotropy at the connectivity level, we first computed seven brain-wide FC profiles across four psychiatric conditions, fluid intelligence, neuroticism and one control non-brain related condition (IBD). Patients diagnosed with idiopathic SZ, BIP and ASD, but not ADHD nor IBD, showed altered FC compared to controls (significance required both FDR and permutation test; Table 2).
Table 2.
CWAS summary
| Genetic variants/conditions/traits | Status | Connections | Beta values | Top-decile β values | P-value effect | ||
|---|---|---|---|---|---|---|---|
| pos | neg | min | max | ||||
| 1q21.1 | DEL | 1 | 11 | −1.07 | 0.62 | 0.44 | 0.002 |
| DUP | 4 | 0 | −0.62 | 0.84 | 0.48 | 0.002 | |
| 22q11.2 | DEL | 4 | 13 | −1.48 | 1 | 0.65 | <2 × 10−4 |
| DUP | 0 | 2 | −0.78 | 0.69 | 0.43 | 0.04 | |
| 16p11.2 | DEL | 124 | 149 | −0.98 | 1.67 | 0.57 | <2 × 10−4 |
| DUP | 4 | 3 | −1.04 | 0.55 | 0.38 | 0.002 | |
| 15q11.2 | DEL | 1 | 0 | −0.29 | 0.36 | 0.2 | 0.01 |
| Idiopathic psychiatric conditions | SZ | 221 | 258 | −0.41 | 0.51 | 0.30 | <2 × 10−4 |
| BIP | 33 | 24 | −0.66 | 0.65 | 0.43 | <2 × 10−4 | |
| ASD | 51 | 55 | −0.26 | 0.36 | 0.16 | <2 × 10−4 | |
| ADHD | 0 | 0 | −0.22 | 0.22 | 0.15 | <2 × 10−4 | |
| Non-psychiatric condition | IBD | 0 | 0 | −0.16 | 0.16 | 0.11 | ns |
| Polygenic scores | Autism | 3 | 1 | −0.02 | 0.02 | 0.01 | 0.04 |
| SZ | 93 | 115 | −0.02 | 0.04 | 0.02 | <2 × 10−4 | |
| BIP | 16 | 2 | −0.02 | 0.03 | 0.01 | 0.002 | |
| MDD | 6 | 21 | −0.02 | 0.03 | 0.01 | 0.003 | |
| Cross-Disorder | 23 | 22 | −0.02 | 0.03 | 0.01 | <2 × 10−4 | |
| Traits | Fluid intelligence | 311 | 281 | −0.04 | 0.04 | 0.02 | <2 × 10−4 |
| Neuroticism | 208 | 208 | −0.03 | 0.04 | 0.02 | <2 × 10−4 | |
The number of significantly altered connections (FDR corrected) for each CWAS (n = 19). min-max = minimum-maximum of z-scored beta values; top decile = top decile of beta values; Connection pos = number of positive connections surviving FDR; Connection neg = number of negative connections surviving. DEL = deletion; DUP = duplication; MDD = major depression disorder; Cross Dis = cross-disorder.
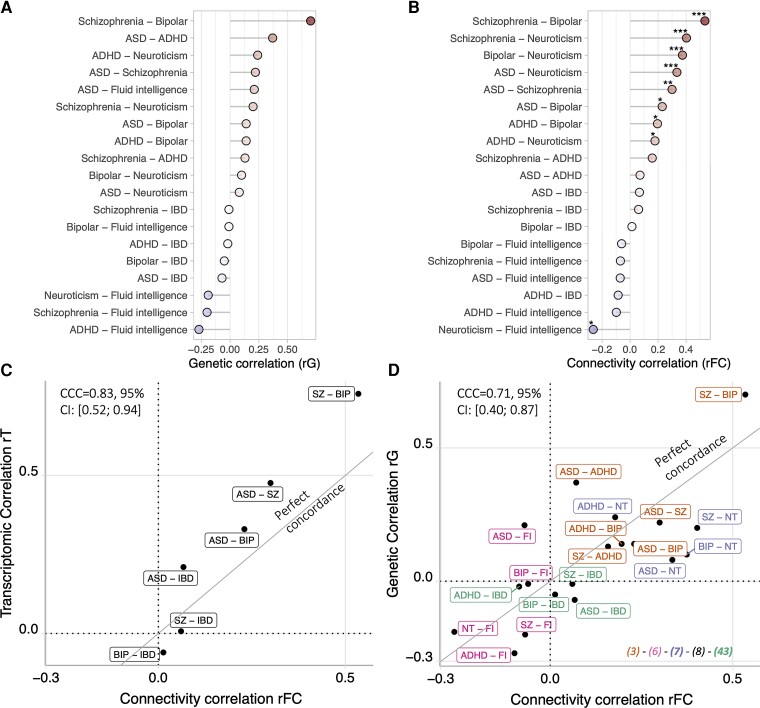
To quantify FC overlap between conditions and traits, we performed correlations between FC profiles (rFC) across 19 pairs of conditions and traits. Nine out of the 19 pairs showed correlation above chance (permutations and FDR) (Fig. 1B). The control trait (IBD) did not correlate with any of the psychiatric conditions or traits.
Figure 1.
Concordance across genetic, transcriptomic and connectomic correlations. (A) Previously published genetic correlations between pairs of conditions and traits.3,6,7,43 (B) FC correlations between pairs of conditions and traits. Correlation values are available in Supplementary Table 5. Stars represent significant correlations (*P < 0.05, **P < 0.005, ***q FDR). (C) Concordance between FC correlation across pairs of conditions and previously published transcriptomic correlation.8 (D) Concordance between FC correlations across pairs of conditions and traits (cognitive ability and neuroticism) and previously published genetic correlation. x- and y-axes: r-values of correlations. The brain FC correlations (rFC) represent the correlation between the FC profiles of a pair of conditions traits. The diagonal represents a perfect concordance. Colours indicate papers that computed rG: green,3 brown,43 purple,7 pink6 and black.8 CCC = Lin’s concordance correlation coefficient; CI = confidence interval; MDD = major depressive disorder; NT = neuroticism; Del = deletion; Dup = duplication; fluid intel = fluid intelligence; IQ = intelligence quotient.
We then asked whether the level of FC correlation (rFC) could be explained by previously published levels of genetic or transcriptomic correlations (rG and rT) between the same pairs of conditions and traits (Fig. 1A and B).
We first observed a high concordance between rT and rFC across six pairs of conditions and traits [Lin’s CCC,45 CCC = 0.83, 95%CI: (0.52; 0.94), without any bias correction factor = 0.85; Fig. 1C].
We also showed a significant concordance between rG and rFC across 19 pairs of conditions and traits [CCC = 0.71, 95%CI: (0.40; 0.87) without any bias (bias correction factor = 0.99; Fig. 1D]. In other words, FC similarity between conditions and traits was neither systematically higher nor lower than rG. All concordance remained significant even after removing the SZ-BIP pair, which showed the strongest correlations at the genetic and functional levels.
A landscape of functional connectivity correlation across genetic risk, psychiatric conditions and traits
We asked whether pleiotropy previously published for rare CNVs and PGS (i.e. a CNV confers risk for several psychiatric conditions)3,34 was also observed at the level of brain FC. We therefore calculated the correlation for FC profiles associated with genomic risk, psychiatric conditions and traits. We first computed brain-wide FC profiles associated with seven CNVs and five PGS (Table 2). All seven CNVs and PGS altered from five to 208 connections that survived FDR q < 0.05 and permutation analyses; Table 2). Of note, an alternative PGS-SZ computed using an older and smaller GWAS was associated with a much lower number of connections. Nevertheless, FC profiles of the old46 and new GWAS47 were correlated (r = 0.89).
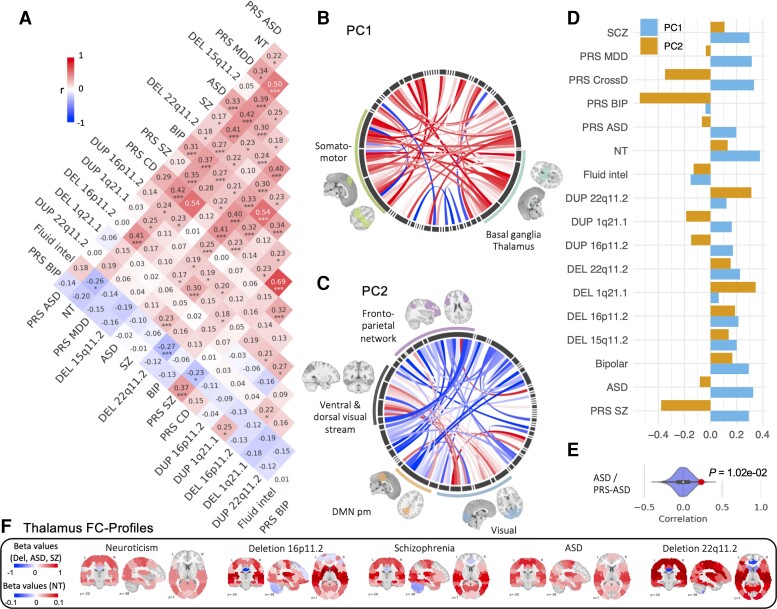
We computed correlations between the FC profiles of CNVs, PGS, conditions and traits. This analysis was limited to the 17 whole-brain FC profiles with significantly altered connections (Table 2) and showed that 30 out of 136 pairs of FC profiles have correlations above what is expected by chance (10 000 permutations and FDR; Fig. 2). FC correlations (rFC) between genetic risks, conditions and traits ranged from weak to moderate, similar to those observed for rG (Fig. 1).
Figure 2.
Atlas of FC relationships across psychiatric conditions, genetic risks and traits. (A) Pearson correlation between 17 FC profiles (2080 beta values from CWAS). Stars represent significant correlations (*P < 0.05, **P < 0.005, ***q FDR). (B–D) PCA conducted on the 17 FC profiles: (B and C) Loadings of functional connections on PC1 (B) and PC2 (C) (overconnectivity in red, underconnectivity in blue). Each chord diagram shows the top 5% of connections’ loadings. All 64 seed regions are represented in the black inner circle. Seed regions are grouped into functional networks. The width of the seed region in the black inner circle corresponds to the contribution of regions to the PC. Dimension 1 was dominated by overconnectivity of the thalamus, basal ganglia and the somatomotor network. Dimension 2 was dominated by altered connectivity between the visual network and the posterior-medial default mode network. (D) Loadings of conditions and traits on PC1 (blue) and PC2 (orange) explaining, respectively, 24 and 10% of the connectome-wide variance across FC profiles. (E) Density plots show examples of null distributions of correlations used to determine significance. FC profiles of ASD and PGS ASD have the lowest correlation that survives FDR. (F) Brain maps represent thalamic FC profiles (64 beta values for each connection between the thalamus and all other functional regions). Red shows overconnectivity and blue underconnectivity. The colour scale represents the beta value (z-score). MDD = major depressive disorder; CD = Cross-disorder; NT = Neuroticism; fluid intel = fluid intelligence; Del = deletion; Dup = duplication; DMN pm = posteromedial default mode network.
Thalamo-sensorimotor alterations are shared across CNVs, PGS and idiopathic conditions
We sought to investigate whether specific functional networks underlied the FC correlations observed previously. We performed a PCA across the 17 FC profiles. The two first dimensions explained 24 and 10% of the variance, respectively, of the FC profiles. Dimension 1 was dominated by increased connectivity between the thalamus and the ventrolateral-, dorsolateral- and medial-somatomotor, as well as the lateral default mode and auditory networks. Dimension 2 was characterized by decreased connectivity between the posterior cingulate, the precuneus and the visual networks (Fig. 2C). Beyond these dominant networks, both latent dimensions were distributed broadly across all 12 networks (Supplementary Fig. 1).
Neuroticism and psychiatric conditions showed higher loadings on dimension 1 than CNVs (Fig. 2D). As a sensitivity analysis, we performed a second PCA on CNVs separately, demonstrating that similar networks and connections were contributing to the main dimension (r = 0.70 between PC1 of CNV + PGS + conditions + traits, and PC1 of CNVs only). The regional FC profiles of the thalamus (Fig. 2F and Supplementary Fig. 2) and dorsolateral motor network (Supplementary Fig. 3) showed, as expected, much higher similarities among genetic risk, conditions and traits (16 and 45 out of 136 correlations survived FDR respectively) compared to whole-brain correlations.
Discussion
Main findings
Our study provided the first systematic analysis of FC across genetic risk, psychiatric conditions and traits. Results demonstrated a stable level of similarities between conditions and traits from genetics, to transcriptomics to brain connectomics. We posit that FC overlap measured by rFC reflects pleiotropy at the level of functional networks. FC profiles associated with rare psychiatric CNVs, psychiatric PGS, psychiatric conditions and traits shared mild to moderate signatures. Although multivariate analyses showed that this shared FC dimension was dominated by overconnectivity of the thalamus and somatomotor networks as well as the underconnectivity of the visual network, similarities were distributed across all networks.
Shared functional connectivity profiles across conditions and traits parallel genetic and transcriptomic overlap
Stable concordance of pleiotropy from genes to connectivity suggests that a major component of FC-profile correlations (rFC) reflects genetically based biological processes, consistent with the previously reported SNP-based heritability of the interindividual differences in brain functional networks.48,49 Previous studies have shown that similarity in cortical thickness or surface between psychiatric conditions were associated with SNP-based genetic similarity (rG) between the same conditions albeit with lower levels of concordance.50,51
This suggests that genetic pleiotropy is reflected across multiple MRI modalities with seemingly similar levels of concordance. All of the well-studied rare variants (i.e. CNVs) have been associated with more than one condition (i.e. ASD, SZ and ADHD) but genetic correlations used in this study were only based on SNPs. It is unknown if rG may be higher or lower once rare variants are included.2
Genetic risks converge on the thalamus and somatomotor network
Overlap between genetic risk, psychiatric conditions and neuroticism was driven by shared overconnectivity of the thalamus/basal ganglia and the somatomotor networks. The implication of the somatomotor and basal ganglia/thalamus network across genetic risk and psychiatric conditions is in line with previous transdiagnostic and single-condition neuroimaging studies.14,52 These functional hubs may be highly sensitive to a broad range of genetic risks for neuropsychiatric conditions. This is consistent with the fact that (i) most if not all rare CNVs, and rare deleterious variants in general, that increase the risk for psychiatric conditions are also associated with delayed gross motor milestones10,53 and development coordination disorders54; and (ii) delay in motor milestones has been demonstrated in individuals with SZ55 and ASD.56 Of note, functional and structural measures of the thalamus, basal ganglia57 and unimodal regions (i.e. somatomotor) show less interindividual variability and higher heritability compared to heteromodal regions.49 Fluid intelligence showed the opposite thalamic pattern. This is in line with (i) negative genetic correlation between cognitive ability and most psychiatric conditions; and (ii) prior fMRI studies demonstrating that thalamocortical pathways are engaged in memory, attention and mental representations.58,59
Clinical translation
Sensory-motor alterations are important dimensions that may underlie some of the pleiotropic effects of genomic risk for psychiatric conditions (Fig. 3). This is in line with the fact that gross and fine motor skills are widely impaired in patients who are referred to autism and neurodevelopmental disorder clinics.56 Furthermore, motor impairments are greater in ASD patients with rare genetic mutations.53 Also, studies demonstrate that soft motor neurological signs in SZ are present in neuroleptic naive patients, and are associated with the severity and persistence of psychopathological symptoms and with poor social functioning.60,61 However, motor abnormalities of severe mental disorders have been neglected both in clinical practice and research. These results represent additional evidence in favour of including motor symptoms in the dimensional assessments of psychiatric conditions.
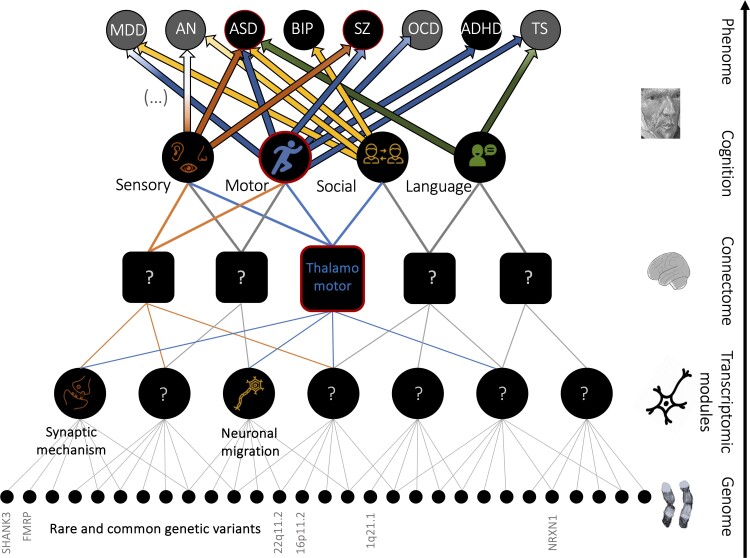
Figure 3.
Schematic diagram summarizing some of the main results and their interpretations. They are integrated into a broader bottom-up perspective representing mechanistic convergence from genes to diagnoses. Rare genetic variants (bottom level) converge on a limited set of transcriptomic modules. The latter may converge on brain alterations (e.g. thalamo-somatomotor overconnectivity, middle-level). Brain alterations may underly differences in cognitive and clinical dimensions altered across several diagnoses (e.g. ASD and SZ, top-level). We showed convergence on sensory-motor FC networks and a pleiotropic effect of sensory-motor dimensions across psychiatric diagnoses.
While psychiatric disorders continue to be defined by their symptoms, course and age of onset, it is reasonable to expect that future efforts to build nosological classifications will be influenced by the increasingly refined characterization of overlaps between conditions at the genetic, transcriptomic and large-scale brain network levels.1
Limitations
FC correlations performed at the whole-brain level are dependent on the sample size used to determine the FC profiles for each genetic risk, condition and trait. Larger samples will probably improve our correlation estimates. This is especially true for conditions such as ADHD, which have been associated with very small effect sizes and will probably require larger samples to identify robust rs-fMRI differences. The same issue applies to genetic correlations that are dependent on the sample size used in the GWAS. As an example, two FC profiles associated with two PGS-SZ computed on the basis of two GWAS of different sample sizes were correlated (r = 0.89) but the number of significant connections was lower for the profile associated with the older SZ-GWAS (computed with 23 585 participants with SZ) compare to the new one (computed with 69 369 subjects with SZ). However, our sensitivity analysis showed that the levels of rFC were not confounded by sample size.
This multisite study including clinically and non-clinically ascertained cohorts may have introduced biases. Confounding factors include sex bias, age differences and medication status, which may have influenced some of the results. However, carefully conducted sensitivity analyses, matching case and control groups for sex, site, age, motion and excluding individuals with medications (in idiopathic psychiatric cohorts) provided similar results (see Supplementary material).
Finally, and because our dataset spans a broad age range, and some CNVs affect total brain volume, we showed in sensitivity analyses that covarying for brain volume did not influence some of the results (Supplementary material and Supplementary Fig. 4).
Conclusion
The level of brain architecture similarities across genetic risks, conditions and traits is consistent with the level of genetic pleiotropy measured across the same conditions and traits. We therefore posit that research on psychiatric conditions will benefit from a neuroimaging genomic multiscale approach. Results highlight the critical contribution of the thalamus and the somatomotor networks across genetic risks and psychiatric conditions suggesting that more attention should be directed towards motor symptoms and mechanisms in psychiatric conditions. Such strategies open promising avenues to help reshape psychiatric nosology as well delineate general mechanisms—amenable to intervention—across conditions and genetic risks.
Supplementary Material
Contributor Information
Clara A Moreau, Human Genetics and Cognitive Functions, Institut Pasteur, UMR3571 CNRS, Université Paris Cité, Paris, France; Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada; Centre de Recherche de l’Institut Universitaire de Gériatrie de Montréal, UdeM, Montreal, QC H3W 1W5, Canada.
Kuldeep Kumar, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Annabelle Harvey, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada; Centre de Recherche de l’Institut Universitaire de Gériatrie de Montréal, UdeM, Montreal, QC H3W 1W5, Canada.
Guillaume Huguet, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Sebastian G W Urchs, Centre de Recherche de l’Institut Universitaire de Gériatrie de Montréal, UdeM, Montreal, QC H3W 1W5, Canada; Montreal Neurological Institute, McGill University, Montreal, QC H3A 2B4, Canada.
Laura M Schultz, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Hanad Sharmarke, Centre de Recherche de l’Institut Universitaire de Gériatrie de Montréal, UdeM, Montreal, QC H3W 1W5, Canada.
Khadije Jizi, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Charles-Olivier Martin, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Nadine Younis, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Petra Tamer, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Jean-Louis Martineau, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Pierre Orban, Centre de Recherche de l’Institut Universitaire en Santé Mentale de Montréal, UdeM, Montréal, QC H1N 3V2, Canada; Département de Psychiatrie et d’Addictologie, Université de Montréal, Pavillon Roger-Gaudry, C.P. 6128, Succursale Centre-ville, Montréal, QC H3C 3J7, Canada.
Ana Isabel Silva, Neuroscience and Mental Health Research Institute, Cardiff University, Cardiff, UK; MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, UK; School for Mental Health and Neuroscience, Maastricht University, Maastricht, The Netherlands.
Jeremy Hall, Neuroscience and Mental Health Research Institute, Cardiff University, Cardiff, UK; MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, UK.
Marianne B M van den Bree, Neuroscience and Mental Health Research Institute, Cardiff University, Cardiff, UK; MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, UK.
Michael J Owen, Neuroscience and Mental Health Research Institute, Cardiff University, Cardiff, UK; MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, UK.
David E J Linden, MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, UK; School for Mental Health and Neuroscience, Maastricht University, Maastricht, The Netherlands.
Sarah Lippé, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Carrie E Bearden, Integrative Center for Neurogenetics, Semel Institute for Neuroscience and Human Behavior, Los Angeles, CA 90095, USA; Department of Psychiatry, University of California, Los Angeles, Los Angeles, CA 90095, USA; Department of Biobehavioral Sciences and Psychology, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Laura Almasy, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Genetics, University of Pennsylvania, Philadelphia, PA, USA; Lifespan Brain Institute, Children's Hospital of Philadelphia, Philadelphia, PA, USA.
David C Glahn, Department of Psychiatry, Harvard Medical School, Cambridge, MA 02115, USA; Boston Children’s Hospital, Tommy Fuss Center for Neuropsychiatric Disease Research, 300 Longwood Avenue, Boston, MA 02115, USA.
Paul M Thompson, Imaging Genetics Center, Stevens Institute for Neuroimaging and Informatics, Keck USC School of Medicine, Marina del Rey, CA, USA.
Thomas Bourgeron, Human Genetics and Cognitive Functions, Institut Pasteur, UMR3571 CNRS, Université Paris Cité, Paris, France.
Pierre Bellec, Centre de Recherche de l’Institut Universitaire de Gériatrie de Montréal, UdeM, Montreal, QC H3W 1W5, Canada.
Sebastien Jacquemont, Sainte Justine Research Center, University of Montréal, Montréal, QC H3T 1C5, Canada.
Funding
This research was supported by Compute Canada (ID 3037 and gsf-624), the Brain Canada Multi-Investigator Research Initiative (MIRI), Canada First Research Excellence Fund, IVADO, Canada First Research Excellence Fund (Healthy Brain Healthy Lives) (S.J.). S.J. is a recipient of a Canada Research Chair in neurodevelopmental disorders, and a chair from the Jeanne et Jean Louis Levesque Foundation. This work was supported by a grant from the 'Fondation Brain Canada' Multi-Investigator initiative (S.J.) and a grant from The Canadian Institutes of Health Research (CIHR 400528, S.J.). The Cardiff CNV cohort was supported by the Wellcome Trust Strategic Award ‘DEFINE’ and the National Centre for Mental Health with funds from Health and Care Research Wales (code 100202/Z/12/Z). The CHUV cohort was supported by the Swiss National Science Foundation (Maillard Anne, Project, PMPDP3 171331). Data from the UCLA cohort provided by C.E.B. (participants with 22q11.2 deletions or duplications and controls) was supported through grants from the NIH (U54EB020403), NIMH (R01MH085953, R01MH100900, 1U01MH119736, R21MH116473) and the Simons Foundation (SFARI Explorer Award). Finally, data from another study were obtained through the OpenFMRI project (http://openfmri.org) from the Consortium for Neuropsychiatric Phenomics (CNP), which was supported by National Institutes of Health Roadmap for Medical Research grants UL1-DE019580, RL1MH083268, RL1MH083269, RL1DA024853, RL1MH083270, RL1LM009833, PL1MH083271 and PL1NS062410. P.B. is a fellow (‘Chercheur boursier Junior 2’) of the ‘Fonds de recherche du Québec—Santé’, Data preprocessing and analyses were supported in part by the Courtois foundation (P.B.). This work was supported by Simons Foundation grant nos. SFARI219193 and SFARI274424. We thank all of the families at the participating SVIP (VIP) sites, as well as the Simons VIP Consortium. We appreciate obtaining access to imaging and phenotypic data on SFARI Base. Approved researchers can obtain the Simons VIP population dataset described in this study by applying at https://base.sfari.org. We are grateful to all families who participated in the 16p11.2 European Consortium. P.M.T. was funded in part by the National Institutes of Health grants R01MH116147, P41EB015922, R01MH111671 and U01 AG068057. P.T. received the Canadian Institute of Health Research (CIHR) Scholarship.
Competing interests
P.M.T. received partial research grant support from Biogen, Inc., for research unrelated to this study. M.J.O., J.H. and M.V.B. have a research grant from Takeda Pharmaceuticals outside the scope of the present work. J.H. is a founding director of the company Meomics (unrelated to this work). The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Lee PH, Feng YCA, Smoller JW. Pleiotropy and cross-disorder genetics among psychiatric disorders. Biol Psychiatry. 2021;89:20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee PH, Anttila V, Won H, et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–82.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grove J, Ripke S, Als TD, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pardiñas AF, Holmans P, Pocklington AJ, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Savage JE, Jansen PR, Stringer S, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagel M, Jansen PR, Stringer S, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–7. [DOI] [PubMed] [Google Scholar]
- 8. Gandal MJ, Haney JR, Parikshak NN, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chawner SJRA, Owen MJ, Holmans P, et al. Genotype-phenotype associations in children with copy number variants associated with high neuropsychiatric risk in the UK (IMAGINE-ID): A case-control cohort study. Lancet Psychiatry. 2019;6:493–505. [DOI] [PubMed] [Google Scholar]
- 10. Douard E, Zeribi A, Schramm C, et al. Effect sizes of deletions and duplications on autism risk across the genome. Am J Psychiatry. 2021;178:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–41. [DOI] [PubMed] [Google Scholar]
- 12. van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: A review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20:519–34. [DOI] [PubMed] [Google Scholar]
- 13. Sha Z, Wager TD, Mechelli A, He Y. Common dysfunction of large-scale neurocognitive networks across psychiatric disorders. Biol Psychiatry. 2019;85:375–88. [DOI] [PubMed] [Google Scholar]
- 14. Kebets V, Holmes AJ, Orban C, et al. Somatosensory-motor dysconnectivity spans multiple transdiagnostic dimensions of psychopathology. Biol Psychiatry. 2019;86:779–91. [DOI] [PubMed] [Google Scholar]
- 15. Cao H, Zhou H, Cannon TD. Functional connectome-wide associations of schizophrenia polygenic risk. Mol Psychiatry. 2021;26:2553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stefansson H, Meyer-Lindenberg A, Steinberg S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–6. [DOI] [PubMed] [Google Scholar]
- 17. Simons VIP Consortium . Simons Variation in Individuals Project (Simons VIP): A genetics-first approach to studying autism spectrum and related neurodevelopmental disorders. Neuron. 2012;73:1063–7. [DOI] [PubMed] [Google Scholar]
- 18. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Di Martino A, Yan CG, Li Q, et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 2014;19:659–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Di Martino A, O’Connor D, Chen B, et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci Data. 2017;4:170010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. ADHD-200 Consortium . The ADHD-200 consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Front Syst Neurosci. 2012;6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poldrack RA, Congdon E, Triplett W, et al. A phenome-wide examination of neural and cognitive function. Sci Data. 2016;3:160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Orban P, Desseilles M, Mendrek A, Bourque J, Bellec P, Stip E. Altered brain connectivity in patients with schizophrenia is consistent across cognitive contexts. J Psychiatry Neurosci. 2017;42:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moreau CA, Urchs SGW, Kuldeep K, et al. Mutations associated with neuropsychiatric conditions delineate functional brain connectivity dimensions contributing to autism and schizophrenia. Nat Commun. 2020;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang K, Li M, Hadley D, et al. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Colella S, Yau C, Taylor JM, et al. QuantiSNP: An objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huguet G, Schramm C, Douard E, et al. Measuring and estimating the effect sizes of copy number variants on general intelligence in community-based samples. JAMA Psychiatry. Published online 21 March 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ge T, Chen CY, Ni Y, Feng YCA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bellec P, Carbonell FM, Perlbarg V, et al. A neuroimaging analysis kit for MATLAB and Octave. In: Proceedings of the 17th International Conference on Functional Mapping of the Human Brain. 2011:2735–46.
- 30. Urchs S, Armoza J, Benhajali Y, St-Aubin J, Orban P, Bellec P. MIST: A multi-resolution parcellation of functional brain networks. MNI Open Res. 2017;1:3. [Google Scholar]
- 31. Fu Z, Sui J, Turner JA, et al. Dynamic functional network reconfiguration underlying the pathophysiology of schizophrenia and autism spectrum disorder. Hum Brain Mapp. 2021;42:80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laidi C, Boisgontier J, Chakravarty MM, et al. Cerebellar anatomical alterations and attention to eyes in autism. Sci Rep. 2017;7:12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guell X, Anteraper SA, Ghosh SS, Gabrieli JDE, Schmahmann JD. Neurodevelopmental and psychiatric symptoms in patients with a cyst compressing the cerebellum: An ongoing enigma. Cerebellum. 2020;19:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreau CA, Raznahan A, Bellec P, Chakravarty M, Thompson PM, Jacquemont S. Dissecting autism and schizophrenia through neuroimaging genomics. Brain. 2021;144:1943–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dadi K, Rahim M, Abraham A, et al. Benchmarking functional connectome-based predictive models for resting-state fMRI. Neuroimage. 2019;192:115–34. [DOI] [PubMed] [Google Scholar]
- 36. Dadi K, Varoquaux G, Machlouzarides-Shalit A, et al. Fine-grain atlases of functional modes for fMRI analysis. Neuroimage. 2020;221:117126. [DOI] [PubMed] [Google Scholar]
- 37. Mellema CJ, Nguyen KP, Treacher A, Montillo A. Reproducible neuroimaging features for diagnosis of autism spectrum disorder with machine learning. Sci Rep. 2022;12:3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pedregosa F, Varoquaux G, Gramfort A. Scikit-learn: machine learning in python. J Mach Learn Res. Published online 2011. http://www.jmlr.org/papers/v12/pedregosa11a [Google Scholar]
- 39. Team RC . R: A Language and Environment for Statistical Computing. Version 3.5. 2, R Foundation for Statistical Computing, Vienna, Austria, 2018.
- 40. Bellec P, Benhajali Y, Carbonell F, et al. Impact of the resolution of brain parcels on connectome-wide association studies in fMRI. Neuroimage. Published online 1 August 2015. [DOI] [PubMed] [Google Scholar]
- 41. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 42. Phipson B, Smyth GK. Permutation P-values should never be zero: Calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol. 2010;9:Article39. [DOI] [PubMed] [Google Scholar]
- 43. Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee SH, Ripke S, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Signorell A. Tools for Descriptive Statistics [R package DescTools version 0.99.44]. Accessed 1 April 2022. https://CRAN.R-project.org/package=DescTools
- 45. Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–68. [PubMed] [Google Scholar]
- 46. Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Electronic address: douglas.ruderfer@vanderbilt.edu, Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 2018;173:1705–15.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. The Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S, Walters JTR, O’Donovan MC. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv. [Preprint]https://doi.org/2020.09.12.20192922 [Google Scholar]
- 48. Elliott LT, Sharp K, Alfaro-Almagro F, et al. Genome-wide association studies of brain imaging phenotypes in UK biobank. Nature. 2018;562:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anderson KM, Ge T, Kong R, et al. Heritability of individualized cortical network topography. Proc Natl Acad Sci USA. 2021;118:e2016271118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Writing Committee for the Attention-Deficit/Hyperactivity Disorder, Autism Spectrum Disorder, Bipolar Disorder , et al. Virtual histology of cortical thickness and shared neurobiology in 6 psychiatric disorders. JAMA Psychiatry. 2021;78:47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Radonjić NV, Hess JL, Rovira P, et al. Structural brain imaging studies offer clues about the effects of the shared genetic etiology among neuropsychiatric disorders. Mol Psychiatry. 2021;26:2101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Van Dam NT, O’Connor D, Marcelle ET, et al. Data-driven phenotypic categorization for neurobiological analyses: beyond DSM-5 labels. Biol Psychiatry. 2017;81:484–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bishop SL, Farmer C, Bal V, et al. Identification of developmental and behavioral markers associated with genetic abnormalities in autism spectrum disorder. Am J Psychiatry. 2017;174:576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cunningham AC, Fung W, Massey TH, et al. Movement disorder phenotypes in children with 22q11.2 deletion syndrome. Mov Disord. 2020;35:1272–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sørensen HJ, Mortensen EL, Schiffman J, Reinisch JM, Maeda J, Mednick SA. Early developmental milestones and risk of schizophrenia: A 45-year follow-up of the Copenhagen perinatal cohort. Schizophr Res. 2010;118:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bishop SL, Thurm A, Farmer C, Lord C. Autism spectrum disorder, intellectual disability, and delayed walking. Pediatrics. 2016;137:e20152959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roshchupkin GV, Gutman BA, Vernooij MW, et al. Heritability of the shape of subcortical brain structures in the general population. Nat Commun. 2016;7:13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hwang K, Bertolero MA, Liu WB, D’Esposito M. The human thalamus is an integrative hub for functional brain networks. J Neurosci. 2017;37:5594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolff M, Vann SD. The cognitive thalamus as a gateway to mental representations. J Neurosci. 2019;39:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jahn T, Hubmann W, Karr M, et al. Motoric neurological soft signs and psychopathological symptoms in schizophrenic psychoses. Psychiatry Res. 2006;142:191–9. [DOI] [PubMed] [Google Scholar]
- 61. Peralta V, Cuesta MJ. Motor abnormalities: From neurodevelopmental to neurodegenerative through “functional” (neuro)psychiatric disorders. Schizophr Bull. 2017;43:956–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huguet G, Schramm C, Douard E, et al. Genome-wide analysis of gene dosage in 24,092 individuals estimates that 10,000 genes modulate cognitive ability. Mol Psychiatry. 2021;26:2663–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Davies RW, Fiksinski AM, Breetvelt EJ, et al. Using common genetic variation to examine phenotypic expression and risk prediction in 22q11.2 deletion syndrome. Nat Med. 2020;26:1912–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kendall KM, Rees E, Escott-Price V, et al. Cognitive performance among carriers of pathogenic copy number variants: Analysis of 152,000 UK biobank subjects. Biol Psychiatry. 2017;82:103–10. [DOI] [PubMed] [Google Scholar]
- 65. Kirov G, Rees E, Walters JTR, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry. 2014;75:378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Malhotra D, Sebat J. CNVs: Harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Marshall CR, Howrigan DP, Merico D, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rees E, Kirov G, Sanders A, et al. Evidence that duplications of 22q11.2 protect against schizophrenia. Mol Psychiatry. 2014;19:37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sanders SJ, He X, Willsey AJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bernier R, Steinman KJ, Reilly B, et al. Clinical phenotype of the recurrent 1q21.1 copy-number variant. Genet Med. 2016;18:341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. D’Angelo D, Lebon S, Chen Q, et al. Defining the effect of the 16p11.2 duplication on cognition, behavior, and medical comorbidities. JAMA Psychiatry. 2016;73:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.