Abstract
Bridging integrator 1 (BIN1) is the second most prevalent genetic risk factor identified by genome-wide association studies (GWAS) for late-onset Alzheimer’s disease. BIN1 encodes an adaptor protein that regulates membrane dynamics in the context of endocytosis and neurotransmitter vesicle release. In vitro evidence suggests that BIN1 can directly bind to tau in the cytosol. In addition, BIN1’s function limits extracellular tau seed uptake by endocytosis and subsequent propagation as well as influences tau release through exosomes. However, the in vivo roles of BIN1 in tau pathogenesis and tauopathy-mediated neurodegeneration remain uncharacterized.
We generated conditional knockout mice with a selective loss of Bin1 expression in the forebrain excitatory neurons and oligodendrocytes in P301S human tau transgenic background (line PS19). PS19 mice develop age-dependent tau neuropathology and motor deficits and are commonly used to study Alzheimer’s disease tau pathophysiology. The severity of motor deficits and neuropathology was compared between experimental and control mice that differ with respect to forebrain BIN1 expression. BIN1’s involvement in tau pathology and neuroinflammation was quantified by biochemical methods and immunostaining. Transcriptome changes were profiled by RNA-sequencing analysis to gain molecular insights.
The loss of forebrain BIN1 expression in PS19 mice exacerbated tau pathology in the somatosensory cortex, thalamus, spinal cord and sciatic nerve, accelerated disease progression and caused early death. Intriguingly, the loss of BIN1 also mitigated tau neuropathology in select regions, including the hippocampus, entorhinal/piriform cortex, and amygdala, thus attenuating hippocampal synapse loss, neuronal death, neuroinflammation and brain atrophy. At the molecular level, the loss of forebrain BIN1 elicited complex neuronal and non-neuronal transcriptomic changes, including altered neuroinflammatory gene expression, concomitant with an impaired microglial transition towards the disease-associated microglial phenotype. These results provide crucial new information on in vivo BIN1 function in the context of tau pathogenesis.
We conclude that forebrain neuronal BIN1 expression promotes hippocampal tau pathogenesis and neuroinflammation. Our findings highlight an exciting region specificity in neuronal BIN1 regulation of tau pathogenesis and reveal cell-autonomous and non-cell-autonomous mechanisms involved in BIN1 modulation of tau neuropathology.
Keywords: BIN1, Alzheimer’s disease, tau pathology, ApoE, GWAS risk factor, neuroinflammation
Ponnusamy et al. show that BIN1 modulates tau pathology in a region-specific manner in the PS19 tauopathy model. Loss of BIN1 curbs brain atrophy, preserves hippocampal synapses, decreases neuroinflammation and results in complex gene expression changes underlying cell-autonomous and non-cell-autonomous responses.
Introduction
Genome-wide association studies (GWAS) have identified the Bridging integrator 1 (BIN1) gene within the second most common susceptibility locus for late-onset Alzheimer’s disease (LOAD).1 Alternate splicing of BIN1 generates more than 15 tissue- and cell-type-specific and ubiquitous isoforms that participate in multiple cellular functions, including endocytosis, membrane remodelling, actin cytoskeleton regulation, DNA repair and apoptosis.2,3 In humans and mice, the loss, downregulation or aberrant alternate splicing of BIN1 in peripheral tissue has been linked to centronuclear myopathy, myotonic dystrophy, ventricular cardiomyopathy, ventricular arrhythmia and cancer progression.2 Most LOAD-associated BIN1 single-nucleotide polymorphisms and an indel lie several kilobases upstream of the BIN1 coding region and have been proposed to increase the risk for Alzheimer’s disease by altering cellular BIN1 expression.4–6
In the brain, BIN1 is expressed in neurons, oligodendrocytes, microglia and at lower levels in astrocytes.3,7,8 In neurons, BIN1 localizes to pre- and post-synaptic sites.3,9–11 Our recent characterization of hippocampal CA1 excitatory synapses by slice electrophysiology revealed BIN1’s indispensable role in synaptic transmission by regulating neurotransmitter vesicle dynamics at the presynaptic terminal.12 In agreement, neuron-specific BIN1 expression has been correlated with excitability in cultured primary neurons.13,14 Thus, neuronal BIN1 has a fundamental function in synaptic physiology. Relatively little is known about BIN1’s function in oligodendrocytes. Recently, we reported that microglial BIN1 plays an essential role in neuroinflammation by regulating the activation of proinflammatory gene expression.8
Interestingly, the BIN1 insertion risk allele s59335482 was associated with higher post-mortem tau pathology but not Aβ when compared to non-carrier Alzheimer’s disease patients.4 Furthermore, among older individuals without dementia, carriers of the BIN1 rs744373 risk allele were found to have similar amyloid pathology but increased tau pathology and significantly impaired memory performance.15 Independent groups have reported a significant decrease in neuronal BIN1 expression in the brains of patients with LOAD.3,16–20 Neuronal loss in BIN1 neuronal conditional knockout (cKO) mice was observed in the context of tauopathy, but how BIN1 loss modified tau pathogenesis was not characterized.13 A more recent study reported the identification of BIN1 rs6733839-T as a risk allele associated with Lewy body dementia.21
It is widely accepted that tau pathology propagates via cell-to-cell spread across neuronal circuits.22,23 BIN1’s role in endocytosis and synaptic activity could profoundly influence tau pathology propagation. However, there is inconsistency in vitro data regarding BIN1’s role in tau pathology. In one study, the loss of BIN1 in cultured neurons was found to facilitate neuron-to-neuron tau pathology propagation by promoting aggregate endocytosis.24 However, another study found that BIN1 knockdown increases phosphorylated tau (p-tau) levels in synapses but reduces basal and stimulated tau release in cultured neurons.25 Existing evidence suggests at least three different modes by which BIN1 might impact tau pathology. First, cytosolic BIN1 limits extracellular pathogenic tau seed uptake and neuron-to-neuron propagation in primary cultures.24 Second, the BIN1 SH3 domain interacts with the proline-rich motif in tau in the cytosol, and tau phosphorylation at Thr231 weakens this interaction. This interaction was speculated to influence Alzheimer’s disease risk by an unknown mechanism.4,26 Third, microglial BIN1 was found to influence the release of tau in extracellular vesicles.27
To explain how BIN1 function relates to disease risk for Alzheimer’s disease, it is imperative to understand BIN1’s role in tau pathogenesis and disease progression better by using appropriate animal models. Here, we generated mice lacking BIN1 expression in forebrain neurons and oligodendrocytes (Bin1-cKO) in the P301S tau transgenic background (line PS19).28 Our results show that BIN1 influences tau pathogenesis in a region-specific manner. Whereas the loss of BIN1 exacerbates tau inclusions in the somatosensory cortex and spinal cord, it attenuates pathology in the hippocampus, entorhinal/piriform cortex and amygdala. Notably, BIN1 loss preserves hippocampal synapses, decreases neuroinflammation and results in complex gene expression changes that underlie cell-autonomous and non-cell-autonomous responses. Collectively, these in vivo results show that BIN1 promotes region-specific tauopathy and neuroinflammation, which is highly relevant to Alzheimer’s disease pathophysiology.
Materials and methods
Ethics statement
All experimental procedures related to animal care and treatment conformed to the Institutional Animal Care and Use Committee policies at the University of South Florida, Tampa.
Animal breeding and husbandry
PS19 transgenic (B6; C3-Tg(Prnp-MAPT*P301S)PS19Vle/J; JAX stock #008169) and Emx1-IRES-Cre (B6.129S2-Emx1tm1(cre)Krj/J; JAX stock #005628) mice were purchased from The Jackson Laboratory. Bin1fl/fl mice were a generous gift from Dr George C. Prendergast (Lankenau Institute for Medical Research). All mice were maintained in C57BL/6J background. PS19 transgenic mice, which express P301S mutant human tau under the control of the mouse prion protein promoter, develop tau pathology in an age-dependent manner.28Emx1-IRES-Cre driver line, which has an IRES-Cre cassette inserted into the exon encoding the 3′ untranslated region of the mouse Emx1 gene, mediates deletion of the floxed alleles in forebrain neurons and glia.29Bin1fl/fl strain was crossed to either PS19 line or Emx1-IRES-Cre mice to generate progenies of PS19:Bin1fl/fl and Bin1fl/fl:Emx-IRES-Cre/Emx-IRES-Cre, respectively. These progenies were crossed to each other to confirm the generation of experimental mice PS19:Bin1fl/fl:Emx-IRES-Cre (referred to as PS19:Bin1-cKO). In parallel, the PS19 line was crossed to the Emx1-IRES-Cre driver line to generate PS19:Emx1-IRES-Cre control mice, henceforth named PS19:Cre. All mice were maintained in the C57BL/6 background and aged 8–9 months to allow age-dependent tau pathogenesis before being euthanized. Aged male PS19 mice develop more robust tau pathology than female mice28; therefore, the results from males and females are indicated in figure legends. All mice were housed a 22 ± 2°C under a 12 h light/dark cycle with ad libitum access to food and water.
Survival analysis
All mice were carefully monitored throughout the study. As soon as the mice developed mild paraparesis, they were provided with HydroGel and DietGel Recovery (ClearH2O) and monitored daily to ensure that paresis of the hindlimbs does not impair their ability to reach for food or drink. Mice were euthanized if more than 15% weight loss occurred. Kaplan–Meier survival curve was used to estimate the median survival time, and Mantel–Cox log-rank test was used to compare the survival curves of the groups.
Behavioural analysis
All behavioural analyses were carried out on animals between 8 and 9 months of age. The number of animals used per genotype for individual tests is indicated in the figure legends. The hindlimb clasping test was performed using a standard procedure.30 The animals scored between 0 (no impairment) and 3 (severe impairment). Mice were examined for deficits in motor coordination and balance on the rotarod using fixed speeds of 5, 10, 15 or 20 rpm for a maximum of 60 s per trial. Each mouse was given three trials per speed, with a 60 s inter-trial interval and at least 5 min rest between the different speeds. The latency to fall was recorded, and the average of the three trials was calculated.31
Tissue preparation
After behavioural tests were completed, the mice were deeply anaesthetized with isoflurane and immediately transcardially perfused with chilled phosphate-buffered saline (10 mM, pH 7.4). The brains were removed and carefully cut into two hemispheres. The right hemibrains were immediately dissected into hippocampus-cortex, midbrain-hindbrain and cerebellum, snap-frozen, and kept at −80°C until further use for biochemical analyses. The left hemibrains were post-fixed in phosphate-buffered saline containing 4% paraformaldehyde and embedded in paraffin. Serial 5-μm thin sections were cut from each paraffin block. The sections were deparaffinized in xylene and rehydrated through a series of increasingly dilute ethanol solutions before use in immunostaining.
Immunofluorescence staining
Multiplex immunostaining was performed on intelliPATH FLX automated slide-staining system (Biocare Medical), using the manufacturer’s reagents except as noted. For epitope retrieval, the sections were incubated with Reveal Decloaker solution at 95°C for 30 min in Decloaking Chamber NxGen. After cooling to room temperature, the slides were rinsed in Tris-buffered saline (TBS) and loaded into the automated staining system. After a wash in TBS auto wash solution, non-specific binding sites were blocked for 10 min using Background Punisher. Da Vinci Green antibody diluent was used to dilute primary and secondary antibodies. Brain sections were sequentially incubated with primary antibodies and Alexa Fluor-conjugated secondary antibodies (Supplementary Table 1). For BIN1 staining, we used a rabbit mAb (Abcam) or a mouse mAb 19H3 generated against full-length BIN1 in this study. The mAb 19H3 reacts with an epitope within the SH3 domain of BIN1 and does not cross-react with the BIN1 orthologue, Amphiphysin 1. Nuclei were stained using Hoechst 33 342 before mounting.
Immunohistochemistry
Tissue sections were treated with antigen retrieval buffer (DAKO) in a steamer for 20 min for epitope retrieval, followed by a 5 min incubation with 3% H2O2, and permeabilized with TBS-T (TBS containing 0.025% Triton × 100). Sections were blocked with 10% normal rabbit serum TBS-T for 1 h or with a MOM kit (Vector Laboratories) and then incubated with the indicated primary antibodies (Supplementary Table 1) for 1 h at room temperature in a humidity chamber followed by species-specific secondary antibodies. The antigen-antibody binding was detected with Envision+ kit and DAB+ chromogen (DAKO). Nuclei were counterstained using haematoxylin.
Imaging acquisition and analysis
Immunofluorescence-stained sections were imaged on an automated Nikon Ti2 microscope fitted with a Yokogawa spinning disc field scanning confocal system. Z-stacks of images were acquired using a 4×, 20×, 60× or 100× objective and deconvolved in NIS-Elements software (Nikon). Z-stacks were then converted to 2D projections by maximum intensity using Fiji software. Immunohistochemical slides were scanned using Aperio ImageScope Scan Whole Slide Scanner (Leica), and 7–9 sections per mouse were analysed by Aperio ImageScope analysis or Fiji software for quantification.32
Pathology quantification and heat map generation
For quantification of regional BIN1 expression, immunofluorescence images were imported into QuPATH software33 to annotate brain regions according to the anatomical pattern in Allen Mouse Brain Atlas (http://mouse.brain-map.org/). Then, the relative integrated densities were quantified from individual regions of interest using Fiji. A custom-designed Microsoft Excel template,34 was used to create an expression heat map representing the mean integrated densities of different brain regions. For regional p-tau pathology quantification, MC1+ or AT180+ stained areas in brain regions were measured. The percentage stained area values were used to calculate the log2 fold-changes in respective regions between PS19:Cre and PS19:Bin1-cKO mice and generate the heatmaps representing differential staining.
Nissl staining and volumetric analysis of brain sections
Five-μm thin paraffin tissue sections were selected at 120-μm intervals and stained with Cresyl Violet to visualize Nissl substance in neurons and evaluate brain atrophy.35 Briefly, deparaffinated and rehydrated sections were first rinsed in water and then stained with 0.1% Cresyl Violet for 10 min at room temperature. The slides were scanned using Aperio ImageScope whole slide scanner with a 40× objective. The volumes were quantified by tracing the lateral ventricle, cerebrum, hippocampus, and entorhinal/piriform cortex/amygdala using QuPATH33 or Fiji.32
Gallyas silver staining
Gallyas silver staining of neurofibrillary tangles was performed essentially as described previously.36 Sections were imaged using a 20× objective on a Zeiss AxioScan.Z1 slide scanner.
Electrochemiluminescence quantification of total tau and p-tau levels
Forebrain tissue (cortex and hippocampus) was sequentially extracted into RAB- and RIPA-soluble and insoluble (FA-soluble) fractions as described previously.37 The RAB and RIPA buffers were supplemented with 0.02 M NaF, 1 mM Na3VO4, 1× PhosSTOP phosphatase inhibitor cocktail and 1× complete protease inhibitor cocktail (Roche), and all steps were performed on ice. Total tau and p-tau (pThr231) in each fraction were quantified using the Meso Scale Discovery (MSD) pThr231/total tau kit on a MesoQuickPlex SQ 120 system using the manufacturer’s software (MSD). Aliquots of RAB and RIPA fractions were diluted 1:400 with 10% MSD blocker A in 1× Tris wash buffer, whereas aliquots of formic acid-soluble fractions were neutralized 1:20 in the neutralization buffer (1 M Tris base, 0.5 M NaH2PO4, pH 11) before performing the MSD assay. All data were normalized to total protein concentrations of the samples and reported as nanograms or units per milligram of total protein for tau and p-tau, respectively.
Immunoblot analysis
Aliquots of RAB and RIPA fractions were diluted in 3 × Laemmli buffer, boiled for 5 min, and resolved on 4–20% Criterion TG precast gels (Bio-Rad). Membranes with transferred proteins were incubated with primary antibodies listed in Supplementary Table 1. The blots were developed with infrared dye-conjugated IR680 anti-rabbit or IR-800 anti-mouse secondary antibodies and visualized with Li-COR Odyssey infrared imaging system (LiCOR), and densitometric analysis was performed using ImageJ.38
RNA-sequencing (RNA-seq) and real-time quantitative polymerase chain reaction (qPCR)
Total RNA was isolated from forebrain tissue using the Qiagen RNeasy mini kit. The RNA concentration and integrity were assessed using a 2100 Bioanalyzer (Agilent Technologies). Samples with an RNA Integrity Number >9.5 were used for RNA-seq and real-time quantitative PCR (qPCR). The RNA-seq library’s quality control analysis and quantification were performed using Agilent Technologies 2100 Bioanalyzer High Sensitivity DNA Chip. Paired-ended sequencing was performed on Illumina’s NovaSeq 6000 sequencing system by LC Sciences.
The primer pairs used for real-time qPCR were chosen from the PrimerBank database39 and are listed in Supplementary Table 2. A high-capacity cDNA reverse transcription kit (Applied Biosystems) was used for reverse transcription. Real-time qPCR was performed using a Power SYBR Green Master Mix on a QuantStudio3 (Applied Biosystems). The PCR cycle parameters were as follows: 95°C for 3 min, then 40 cycles of 95°C for 15 and 60°C for 30 s. The amplification procedure was performed with two repetitions for each sample. The 2−ΔΔCt method was used to calculate the relative abundance of transcripts.
Bioinformatics analysis of RNA-seq data
Sequence adapters and low-quality reads were removed using Cutadapt. Quality control checks on raw sequence data were performed with FastQC. Sequencing reads were then mapped to the mouse reference genome (release-96) using the HISAT program. The mapped reads of each sample were assembled using StringTie. Then, all transcriptomes were merged to reconstruct a comprehensive transcriptome using Perl scripts and GffCompare. After the final transcriptome was generated, R package DESeq2 was used to estimate the expression levels of all transcripts. The differentially expressed mRNAs were selected with statistical significance (P < 0.05).
Principal component analysis was performed to identify the variability and repeatability of samples. A volcano plot was used to visualize the overall distribution of differentially expressed genes (DEGs). Gene ontology (GO) analysis of DEGs was performed using the R Package cluster Profiler (v.3.18.1). The significance level for GO terms and the KEGG pathway was set with P < 0.05. String APP (v.1.6.0) in Cytoscape (v.3.8.2) was used to depict a network of enriched pathways. Gene set enrichment analysis (GSEA, v.4.2.3)40 was performed using the RNA-seq data (Supplementary Table 3) against the gene sets described in Molecular Signatures Database v.7.5.1 C2 category, which covers curated gene sets from online pathway databases, publications in PubMed and knowledge of domain experts.
Quantification and statistical analysis
GraphPad Prism (v.9.0.1) was used for all data analyses. Statistical analyses were carried out by using two-tailed unpaired t-test pairwise comparisons. All values are represented as mean ± SEM. Data were considered statistically significant at P values < 0.05.
Data availability
In accordance with the Journal’s policy, data will be made available upon reasonable request.
Results
Worsened motor deficits and reduced survival in PS19:Bin1-cKO mice
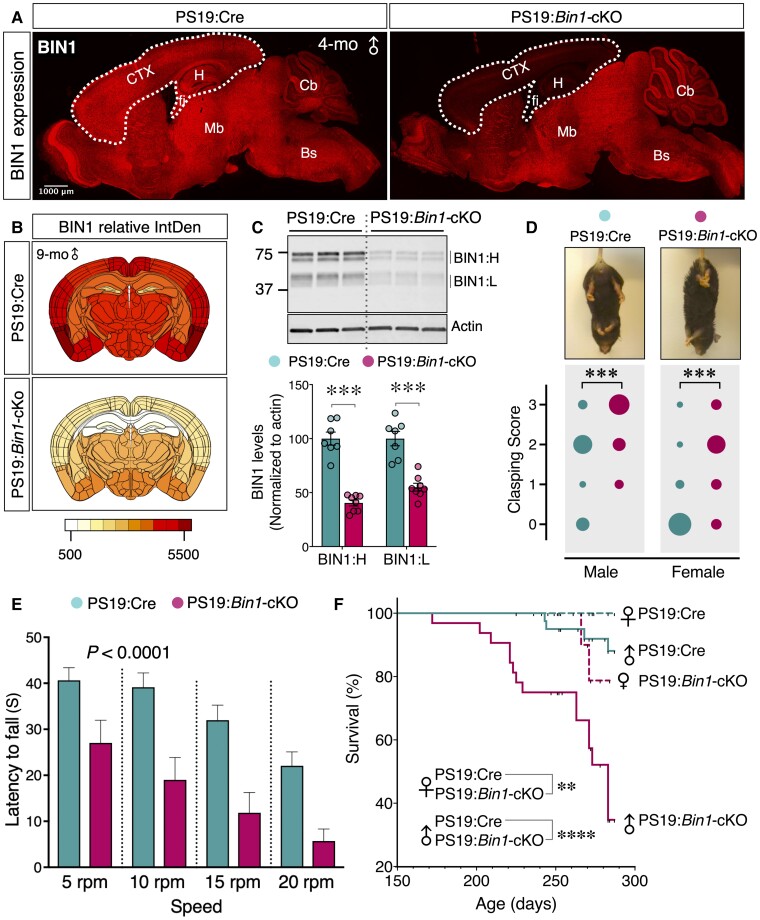
To investigate the in vivo function of BIN1 on tauopathy, we crossed Bin1-cKO mice with the P301S tau transgenic line PS19 (Supplementary Fig. 1A). In the resulting PS19:Bin1-cKO mice, there is a striking loss of BIN1 expression in the forebrain (Fig. 1A and B), consistent with reported mouse BIN1 expression and the pattern of Emx-Cre expression in the forebrain excitatory neurons and oligodendrocytes.3,29 Immunoblot analysis confirmed a significant reduction in the levels of the neuron-specific BIN1:H and ubiquitously expressed BIN1:L isoforms in the cortex and hippocampus of PS19:Bin1-cKO mice as compared with PS19:Emx1-IRES-Cre control mice (PS19:Cre) (P < 0.001) (Fig. 1C). We performed immunostaining to determine whether inhibitory neurons and glial cells account for the residual low-level BIN1 expression in forebrains of cKO mice. Indeed, there was a clear overlap between BIN1 and GAD65/67 in PS19:Bin1-cKO mice (Supplementary Fig. 1B–D). Whereas most of the BIN1 signal overlapped with GAD65/67 staining in some cortical layers (layer V), others (layer IV) showed a partial overlap (Supplementary Fig. 1C). Thus, it is likely that in addition to cortical inhibitory neurons, thalamocortical projections and glial cells contribute to BIN1 expression in the forebrain (Supplementary Fig. 1E). Further analysis showed that BIN1 expression overlapped with the microglial marker IBA1 (Ionized calcium binding adaptor molecule 1) but not with the astrocyte marker GFAP (glial fibrillary acidic protein) (Supplementary Fig. 1F). Moreover, as expected, BIN1 expression was ablated in oligodendrocytes in PS19:Bin1-cKO mice, as evidenced by staining with the oligodendrocyte markers ASPA or TPPP (Supplementary Fig. 1G). Together, these results confirm that most forebrain BIN1 expression is ablated in the forebrain excitatory neurons and oligodendrocytes in PS19:Bin1-cKO mice.
Figure 1.
Accelerated disease progression and reduced survival in PS19:Bin1-cKO mice. (A) Immunofluorescence analysis shows a significant decrease of BIN1 expression (rabbit mAb) in the cortex (CTX) and hippocampus (H) of PS19:Bin1-cKO 4-month-old male mice as compared with PS19:Cre controls (sagittal section). Note that BIN1 expression in the midbrain (Mb), brainstem (Bs) and cerebellum (Cb) remains unaffected. (B) Heatmaps of BIN1 expression. Coronal sections from 9-month-old male PS19:Cre (n = 7) and PS19:Bin1-cKO (n = 7) mice were immunostained using BIN1 mAb 19H3. The relative integrated densities (IntDen) were quantified and colour-coded onto an anatomical map of the mouse brain. The heat map uses a yellow-red colour scale (low-high) representing the IntDen. (C) Representative immunoblots for BIN1 levels in the forebrain (cortex and hippocampus) of PS19:Cre (n = 7) and PS19:Bin1-cKO (n = 8) mice. The levels of BIN1 were normalized to actin, and the means ± SEMs were plotted. BIN1:H, neuronal BIN1 isoforms; BIN1:L, ubiquitous BIN1 isoforms. (D) Hindlimb clasping analysis in PS19:Bin1-cKO mice compared with PS19:Cre mice. The bubble chart displays the hindlimb clasping scores in a cohort of PS19:Cre (n = 32, 15 males and 17 females) and PS19:Bin1-cKO (n = 14, 8 males and 6 females). The variable size bubbles represent the fraction of mice for each score. Bin1-cKO mice exhibit worsened hindlimb clasping behaviour at 9 months of age. Two-way ANOVA P < 0.001. (E) Mice were examined for deficits in motor coordination and balance by the fixed rotarod test at 9 months. The latency to fall is significantly decreased in PS19:Bin1-cKO mice compared to PS19:Cre by Sidak’s multiple comparisons following two-way ANOVA (P < 0.0001). (F) Kaplan–Meier survival curves. Animal survival was monitored throughout the study. The plot demonstrates the reduced survival in PS19:Bin1-cKO mice. Males: PS19:Cre, 88.11% (n = 41); PS19:Bin1-cKO, 34.76% (n = 32). Females: PS19:Cre, 100% (n = 40); PS19:Bin1-cKO, 78.75% (n = 21). The significance was determined using Mantel–Cox log-rank test. **P < 0.01; ***P < 0.001; ****P < 0.0001.
PS19 mice develop tau neuropathology in the brainstem and spinal cord in addition to the neocortex, hippocampus, amygdala and other CNS regions. Consequently, PS19 mice exhibit motor deficits concomitant with pathogenesis in an age-dependent manner, with the males showing more robust deficits.28 At 8–9 months of age, both male and female PS19:Bin1-cKO mice displayed a higher hindlimb clasping reflex score when compared to PS19:Cre controls [F(1,42) = 13.36, P < 0.001; Fig. 1D], consistent with earlier symptomatic disease manifestation in the absence of forebrain BIN1 expression. Moreover, PS19:Bin1-cKO mice exhibited significantly greater deficits in motor coordination and balance as assessed by the fixed rotarod test [F(1,43) = 32.00, P < 0.0001, Fig. 1E]. By 9 months of age, most PS19:Bin1-cKO mice had progressed to significant paralysis and kyphosis. Whereas only 12% of male PS19:Cre mice died before reaching 9 months of age, 65% of male and 21% of female PS19:Bin1-cKO mice died by 9 months of age, demonstrating reduced survival of PS19:Bin1-cKO mice (P < 0.0001; Fig. 1F). Thus, the loss of BIN1 expression in forebrain excitatory neurons and mature oligodendrocytes of PS19 mice exacerbates motor deficits, resulting in earlier death than PS19:Cre control mice.
Elevation of insoluble and hyperphosphorylated tau in PS19:Bin1-cKO mice
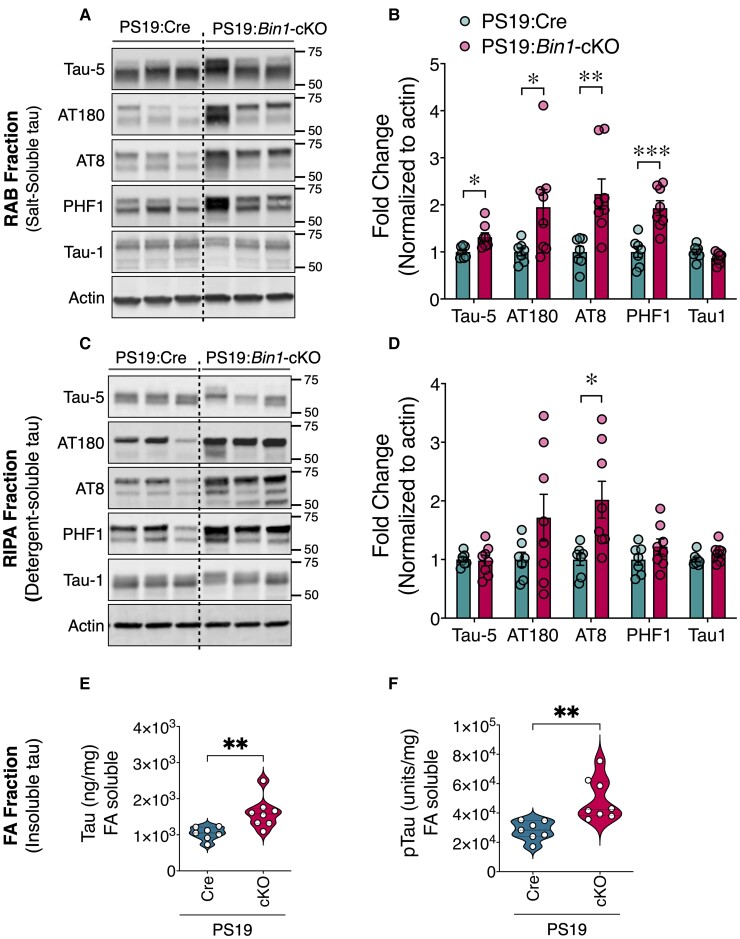
PS19 mice show an age-related increase in tau aggregation, rendering up to 10% of total tau insoluble by 6 months.28 To investigate the effects of forebrain neuronal and oligodendrocyte BIN1 loss on tau pathogenesis, we performed immunoblots on proteins extracted from the cortex and hippocampus of 9-month-old PS19:Bin1-cKO mice and PS19:Cre controls. Because of the significant sex differences in the PS19 line, we separately analysed males (Fig. 2) and females (not shown). High-salt RAB and RIPA buffer extracts, which contained soluble tau species, were probed using a set of tau monoclonal antibodies (mAb) on immunoblots to quantify the levels of total tau (Tau-5), tau phosphorylated at pThr231 (AT180), pSer202/Thr205 (AT8), pSer396/Ser404 (PHF1) and non-p-tau (Tau1). There was a significant increase in total and p-tau in RAB homogenates of PS19:Bin1-cKO mice (Fig. 2A). The levels of all three p-tau forms examined were increased by ∼2-fold in PS19:Bin1-cKO compared with PS19:Cre mice (Fig. 2B). The levels of pSer202/Thr205 tau were also significantly elevated in RIPA fractions (Fig. 2C–D). In contrast, probing with the Tau1 antibody, which recognizes the dephosphorylated state of tau at residues Ser195/Ser198/Ser199/Ser202, showed no differences between PS19:Bin1-cKO mice and controls in either fraction (Fig. 2A–D). Insoluble tau aggregates were extracted in 70% formic acid and quantified using the MSD electrochemiluminescence assay. The levels of total and pThr231 tau in the formic acid-soluble fractions were elevated by 54 and 76%, respectively, in PS19:Bin1-cKO mice relative to PS19:Cre controls (P < 0.01; Fig. 2E and F). Overall, these results show a marked increase in phosphorylated and insoluble tau in forebrain extracts of PS19:Bin1-cKO mice.
Figure 2.
Elevation of insoluble and hyperphosphorylated tau in PS19:Bin1-cKO mice. (A and B) RAB and (C and D) RIPA fractions of forebrain (cortex and hippocampus) were immunoblotted with antibodies against total tau (Tau-5), tau phosphorylated at Thr231s (AT180), Ser202/Thr205 (AT8), Ser396/Ser404 (PHF1) or tau dephosphorylated at Ser195/Ser198/Ser199/Ser202 (Tau1) in 9-month-old male PS19:Cre (n = 7) and PS19:Bin1-cKO (n = 8) mice. Densitometric analyses are expressed as the ratio of total tau, AT180, AT8 and PHF1 to internal control in RAB and RIPA fractions and Tau1 to internal control in both fractions. (E and F) RIPA-insoluble aggregates of 9-month-old mice PS19:Cre (n = 7) and PS19:Bin1-cKO (n = 8) mice were extracted in formic acid (FA) and analysed by MSD assay to quantify total tau (E) and p-tau (Thr231) species (F). All data are shown as mean ± SEM using an unpaired, two-tailed Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
BIN1 differentially regulates tau inclusions in PS19:Bin1-cKO mice
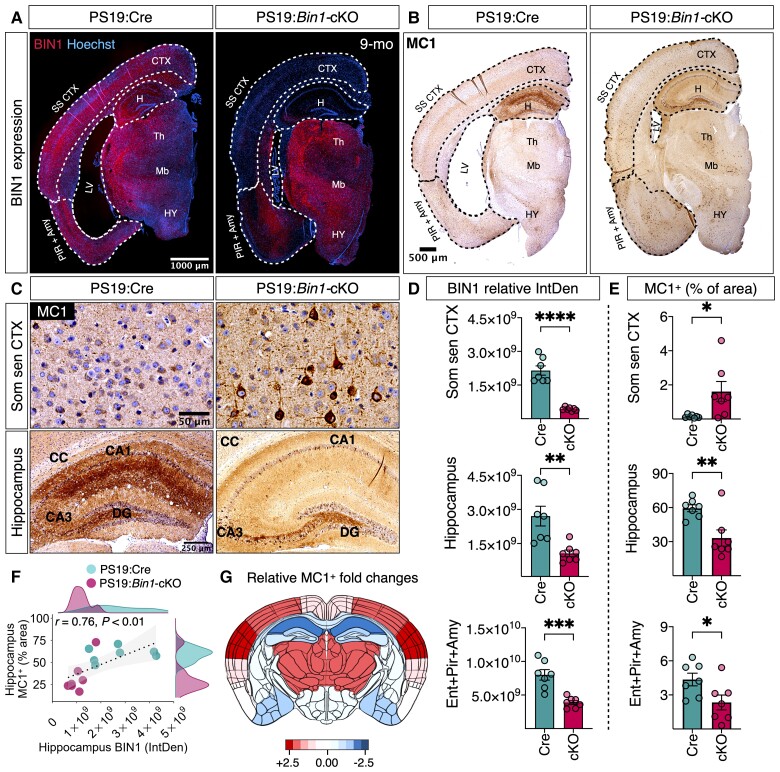
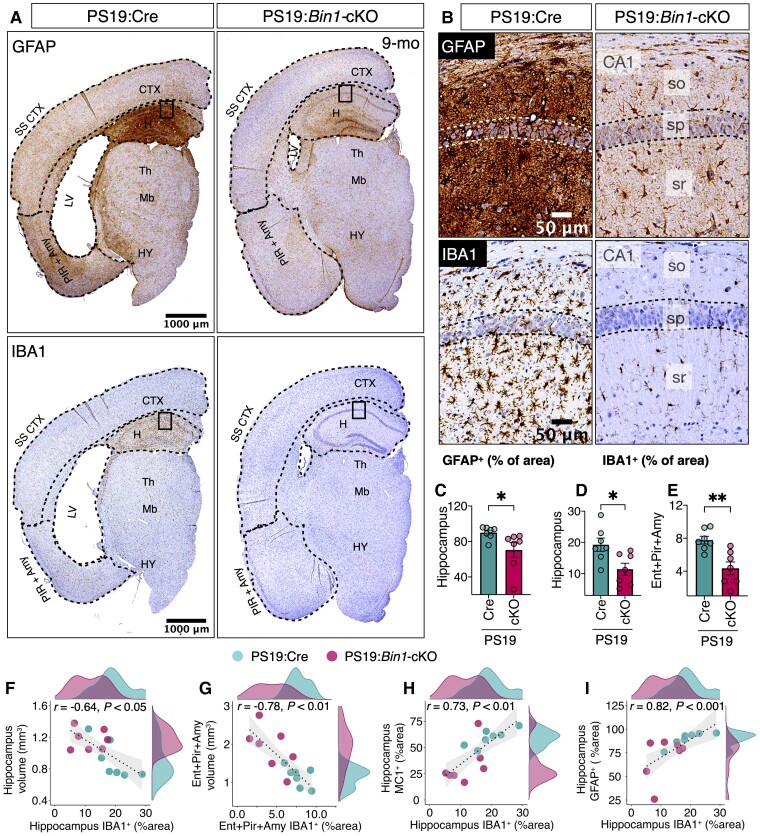
Next, we characterized tau pathology by immunohistological staining in 9-month-old PS19:Cre and PS19:Bin1-cKO mice using mAb MC1 (recognizes tau pathological conformation) and mAb AT8 (recognizes p-tau Ser202/Thr205). The results revealed pronounced differences in MC1 (Fig. 3) and AT8 (Supplementary Fig. 2A and B) immunoreactivity between PS19:Cre and PS19:Bin1-cKO mice. Notably, pathological tau immunoreactivity was higher in the somatosensory cortex but lower in the hippocampus and entorhinal/piriform cortex/amygdala of PS19:Bin1-cKO compared with PS19:Cre (Fig. 3B and C). This regional difference was surprising since BIN1 expression was decreased in the somatosensory cortex (P < 0.0001), hippocampus (P < 0.01) and entorhinal/piriform cortex/amygdala (P < 0.001) of PS19:Bin1-cKO as compared with PS19:Cre mice (Fig. 3A and D). The quantification of pathological tau accumulation in the somatosensory cortex revealed a significantly increased MC1+ area (P < 0.05) and the number of MC1+ cells/mm2 (P < 0.05, not shown) in PS19:Bin1-cKO mice (Fig. 3B, C and E). In contrast, the MC1+ area in the hippocampus (P < 0.01) and entorhinal/piriform cortex/amygdala (P < 0.05) were significantly less in PS19:Bin1-cKO than PS19:Cre mice (Fig. 3C and E). The relative fold-change in the MC1+ area between PS19:Cre and PS19:Bin1-cKO mice in different regions of coronal brain sections is depicted as a heat map in Fig. 3G. Whereas MC1 staining of mossy fibres and the dentate gyrus granule cell layer predominated in PS19:Bin1-cKO, the MC1 staining in PS19:Cre control mice was intense throughout the hippocampus, including the CA1 pyramidal cells, the dentate granule cells and the molecular layer but more diffuse in mossy fibres (Fig. 3C). Similar results were observed by p-tau immunostaining with mAb AT8 (Supplementary Fig. 2A and B), despite uniform hippocampal tau expression in PS19:Cre and PS19:Bin1-cKO mice as observed by Tau-5 staining (Supplementary Fig. 2D).
Figure 3.
Loss of BIN1 differentially regulates tau inclusions in the somatosensory cortex and hippocampus in PS19:Bin1-cKO mice. (A) Immunofluorescence analysis for BIN1 expression in hemibrains of PS19:Cre and PS19:Bin1-cKO 9-month-old male mice. Note that BIN1 expression is dramatically reduced in the cortex and hippocampus. (B) Immunostaining of 9-month-old mice with p-tau mAb MC1 (a confirmation-dependent tau epitope) antibody shows an overall increase of p-tau staining in the cortex despite the attenuation of brain atrophy in PS19:Bin1-cKO mice. (C) Representative MC1+-positive confirmation-dependent tau staining images show differential tau burden in the somatosensory cortex (Som sen CTX) and hippocampus. (D) Quantification of BIN1 expression in the somatosensory cortex and hippocampus. (E) Quantification of MC1+ staining (% area) in the hippocampus, dentate gyrus and somatosensory cortex, and MC1+ cells/mm2 in the somatosensory cortex. (F) Spearman’s correlation analysis shows a positive relationship between hippocampus MC1+ (% area) staining and hippocampal BIN1 expression. PS19:Cre (n = 7) and PS19:Bin1-cKO (n = 7). (G) The heat map shows the relative differences between PS19:Cre and PS19:Bin1-cKO in the extent of MC1+ tau pathology across the brain regions. The MC1+ immunoreactivity (% area covered) was quantified in different brain regions from 9-month-old male PS19:Cre (n = 7) and PS19:Bin1-cKO (n = 7) mice, and the relative regional log2 fold-changes were calculated and colour-coded onto an anatomical map of mouse brain coronal section.
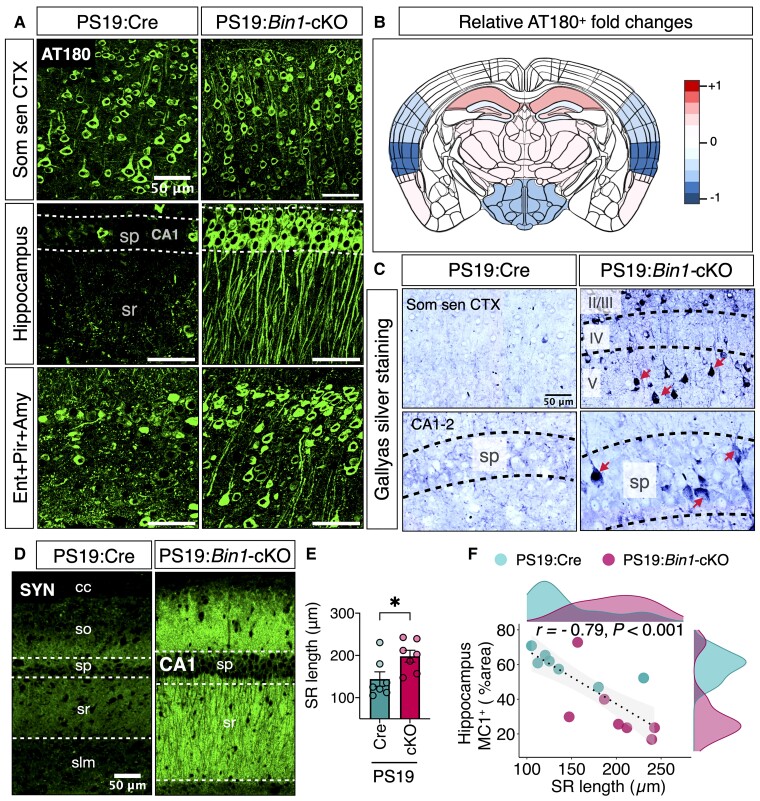
Antibodies MC1 and AT8 detect pathological tau conformation within paired-helical filaments and late-type tau hyperphosphorylation, respectively.41,42 We also performed immunostaining using mAb AT180, which detects p-tau Thr231, one of the earliest phosphorylation of tau occurring at the pre-tangle stage.42 The results show increased levels of AT180+ p-tau in the hippocampus and entorhinal/piriform cortex/amygdala but lower levels in the somatosensory cortex of PS19:Bin1-cKO as compared with PS19:Cre mice (Fig. 4A). A heat map representation of the relative differences in AT180 immunoreactivity (Fig. 4B) reveals regional differences in AT180 levels occur despite significant decreases in BIN1 staining in the cortex and hippocampus (Fig. 1B). Together, these data indicate that the loss of forebrain neuronal BIN1 expression in PS19:Bin1-cKO mice results in significantly lower late-stage characteristic tau pathology in the hippocampus and entorhinal/piriform cortex/amygdala regions.
Figure 4.
Loss of BIN1 differentially regulates early phosphorylation of tau in PS19:Bin1-cKO mice. (A) Nine-month-old PS19:Cre (n = 7) and PS19:Bin1-cKO (n = 7) mouse brains were immunostained with mAb AT180 antibody (p-tau Thr231 epitope). Representative images show differential AT180+ p-tau staining in the somatosensory cortex (Som sen CTX), hippocampus and entorhinal/piriform/amygdala (Ent + Pir + Amy). (B) The heat map shows the relative differences (log2 fold-changes) between PS19:Cre and PS19:Bin1-cKO in the extent of AT180+ tau across the brain regions. (C) Photomicrographs show Gallyas silver-stained tangle-like inclusions (red arrows) in coronal sections of the somatosensory cortex (Som sen CTX) and the hippocampus (CA1-2) from the 9-month-old male PS19:Cre and PS19:Bin1-cKO mice. *P < 0.05; **P < 0.01; ****P < 0.0001. (D) Immunofluorescence staining of synaptophysin (SYN) within the hippocampus CA1 region of 9-month-old PS19:Cre and PS19:Bin1-cKO mice. (E) Quantification of stratum radiatum (SR) length in deconvolved confocal images (5 µm-step size) of brain sections immunostained for synaptophysin (PS19:Cre, n = 7 and PS19:Bin1-cKO, n = 7). (F) Spearman’s correlation coefficient analysis shows a negative correlation between hippocampal tau pathology and SR length. Data are shown as mean ± SEM using an unpaired, two-tailed Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
We performed Spearman’s correlation coefficient analysis to ascertain whether the regional differences observed in tau pathology between the somatosensory cortex and hippocampus are independent of animal-to-animal variations. The results showed a statistically significant and positive correlation between hippocampal MC1 staining (% area) and hippocampal BIN1 expression (integrated staining density) (r = 0.76, P < 0.01) (Fig. 3F). However, no significant correlation in MC1 staining was observed between the hippocampus and somatosensory cortex (r = −0.084, P = 0.78) (Supplementary Fig. 2C). We also noted that cortical layers II–III and V–VI show a high density of MC1+ neurons as compared with layer I and IV in PS19:Bin1-cKO mice, despite uniform tau expression as observed by Tau-5 staining (Supplementary Fig. 2D–F). Layer IV had minimal p-tau staining, which is interesting because BIN1 expression was relatively high in this specific layer in PS19:Bin1-cKO mice. These results suggest that BIN1 expression differentially regulates tau pathology in particular brain regions and that BIN1 promotes hippocampal tau pathology.
To determine whether the p-tau immunostaining observed in PS19:Cre and PS19:Bin1-cKO mice represents neurofibrillary tangles (NFTs), we performed Gallyas silver staining. Results show that Gallyas silver staining was more evident in the somatosensory cortex of PS19:Bin1-cKO mice (Fig. 4C), which paralleled the increase in MC1+ and AT8+ tau staining. Intriguingly, despite the significantly lower hippocampal MC1 and AT8 staining, silver-positive NFTs were more prevalent in the hippocampus of PS19:Bin1-cKO than PS19:Cre mice (Fig. 4C). This finding indicates that the loss of BIN1 in the hippocampus might be associated with p-tau conformational changes that lead to the formation of argentophilic inclusions, despite an overall reduction of MC1+ and AT8+ pathological tau in the hippocampus. Together, these results suggest that the loss of BIN1 has a complex effect on the progression of tau pathology in the mouse brain.
Relative preservation of hippocampal neurons in PS19:Bin1-cKO mice
Older PS19 mice develop neurodegeneration and brain atrophy, which manifest as enlarged lateral ventricles and decreased hippocampal volume.28,35 Therefore, we examined whether the loss of BIN1 expression in the PS19:Bin1-cKO model impacts neurodegeneration by performing a detailed volumetric analysis of mice at 9 months of age. The results showed that the lateral ventricular enlargement was more significant in PS19:Cre than in PS19:Bin1-cKO mice (Supplementary Fig. 3A). We observed significantly smaller lateral ventricle (P < 0.05) and relative preservation of the hippocampus (P < 0.05), entorhinal/piriform cortex/amygdala (P < 0.01), dentate gyrus (P < 0.05) and CA3 (P < 0.001) in PS19:Bin1-cKO as compared with PS19:Cre mice (Supplementary Fig. 3A and C, and data not shown). In addition, we also found that the thickness of the CA1 pyramidal cell layer (P = 0.0002), the CA3 pyramidal cell layer (P < 0.0001) and the dentate gyrus granule cell layer (P = 0.0001), were significantly larger in PS19:Bin1-cKO mice as compared with PS19:Cre mice (Supplementary Fig. 3D). Spearman’s correlation analysis revealed that the lateral ventricle volume positively correlated with hippocampal MC1+ tau pathology (r = 0.77, P < 0.01), and negatively correlated with hippocampal volume (r = −0.61, P < 0.01) and entorhinal/piriform cortex/amygdala volume (r = −0.9, P < 0.0001) (Supplementary Fig. 3C). Further, the hippocampal volume positively correlated with the thickness of the CA1 pyramidal cell layer (r = 0.67, P < 0.05), the CA3 pyramidal cell layer (r = 0.57, P < 0.05) and the dentate gyrus granule cell layer (r = 0.64, P < 0.05) (Supplementary Fig. 3D). These volumetric analyses indicated significant preservation of cortical and hippocampal volumes in the PS19:Bin1-cKO mouse brain.
Next, we sought to ascertain whether the differences observed in tau pathology and neurodegeneration are reflected as differences in synapse density between PS19:Cre and PS1:Bin1-cKO mice. Previous studies have reported an age-dependent synapse loss in the hippocampus of PS19 mice.28,43 Consistent with more severe hippocampal tau pathology in PS19:Cre mice, synaptophysin immunoreactivity was significantly lower in the hippocampus of PS19:Cre mice as compared with PS19:Bin1-cKO mice (Fig. 4B). Similarly, Amphiphysin 1 (a BIN1 orthologue that localizes to presynaptic terminals)44 staining intensity was also higher in PS19:Bin1-cKO mice (Supplementary Fig. 4). These findings indicate the relative preservation of synapses in PS19:Bin1-cKO mice. Furthermore, quantification of synaptophysin-stained sections revealed that the length of the stratum radiatum is significantly higher (P < 0.05) in PS19:Bin1-cKO (198.1 ± 14.08 µm) mice in comparison to that of PS19:Cre (144.1 ± 17.09 µm) (Fig. 4E). Next, we performed Spearman’s correlation analysis of stratum radiatum length, ventricle volume and hippocampal MC1+ tau pathology in the experimental mice to determine whether variations within the groups unduly influence these parameters. The results showed that the stratum radiatum length had a significant negative correlation with the hippocampal MC1+ tau pathology (r = −0.79, P < 0.001, Fig. 4F) and the ventricle volume (r = −0.97, P < 0.0001; Supplementary Fig. 3B). Taken together, the reduction in lateral ventricular atrophy, the larger hippocampal volume, and the relative synapse preservation in PS19:Bin1-cKO mice suggests that the loss of excitatory neuronal BIN1 attenuates hippocampal tau neuropathology.
Loss of excitatory neuronal BIN1 in PS19 mice attenuates glial activation
Neuroinflammation is a hallmark feature of Alzheimer’s disease pathogenesis, and PS19 mice develop age-dependent gliosis concomitant with tau pathogenesis.28 We investigated whether the loss of BIN1 expression and the concomitant attenuation of tau pathogenesis in Bin1-cKO affects astrocyte and microglial activation characteristics of PS19 mice. The immunoreactivity of the astrocyte marker GFAP was significantly less in the hippocampus of 9-month-old PS19-Bin1-cKO mice relative to that of PS19:Cre (P < 0.05; Fig. 5A–C). Moreover, immunostaining analysis using the microglial marker IBA1 revealed significantly reduced microgliosis in the hippocampus (P < 0.05) and in the entorhinal/piriform cortex/amygdala (P < 0.01) of PS19:Bin1-cKO mice as compared with PS19:Cre mice (Fig. 5A–B and D–E). The degree of IBA1 staining in the hippocampus and the entorhinal/piriform cortex/amygdala of each animal showed a negative correlation with the respective region volume (r = −0.64, P < 0.05; and r = −0.78, P < 0.01; Fig. 5F–G). Further, Spearman’s correlation analysis revealed that IBA1 levels in the hippocampus positively correlated (r = 0.73, P < 0.01) with p-tau pathology (% MC1+ area) and the extent of GFAP immunostaining (r = 0.82, P < 0.001) (Fig. 5H–I). These data indicate an overall attenuation of both astrogliosis and microgliosis concomitant with diminished tau pathogenesis in the hippocampus of PS19:Bin1-cKO mice.
Figure 5.
Significant attenuation of neuroinflammation in the hippocampus of PS19:Bin1-cKO mice. (A) Representative images of GFAP and IBA1 immunostaining in hemibrains from 9-month-old mice. (B) Higher magnification of the hippocampus (boxed areas of images in A). (C–E) Quantification of GFAP or Iba1 expression in the hippocampus and Entorhinal/piriform/amygdala revealed significant decreases in PS19:Bin1-cKO (n = 7) mice compared to PS19:Cre (n = 7). (F–I) Spearman’s correlation coefficient analysis shows a negative correlation between IBA1 expression and the volumes of the hippocampus or Entorhinal/piriform/amygdala. Moreover, there is a positive correlation between hippocampal IBA1 expression and tau pathology or GFAP expression. PS19:Cre (n = 7) and PS19:Bin1-cKO (n = 7) mice. *P < 0.05; **P < 0.01.
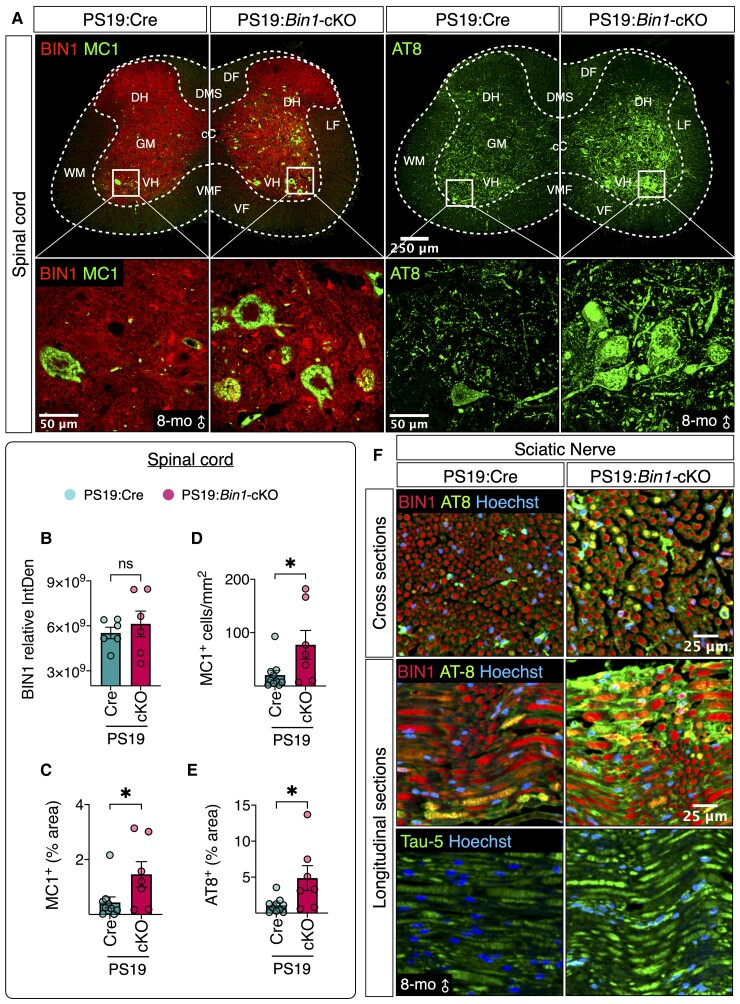
Exacerbated tau pathology in the spinal cord and sciatic nerve of PS19:Bin1-cKO mice
How do we reconcile the preservation of the hippocampus and weaker neuroinflammation with lower survival observed in PS19:Bin1-cKO mice? Early death due to spinal cord tau pathology and consequent muscle atrophy has been described in independent P301S tau transgenic lines.28,45,46 This is relevant to human disease because spinal cord tau pathology has been documented in individuals with Alzheimer’s disease.47 Therefore, we analysed BIN1 expression and tau pathology in the spinal cord and sciatic nerve of PS19:Cre and PS19:Bin1-cKO mice at 8 months of age. On the basis of the previous characterization of Emx-Cre expression,48,49 we expected Cre-mediated recombination of Bin1 alleles would occur in the forebrain corticospinal neurons that project to the spinal cord, but local BIN1 expression in the spinal cord would not change. In agreement, the results showed that BIN1 staining intensities in the spinal cord grey matter and sciatic nerve in PS19:Cre and PS19:Bin1-cKO were comparable (Fig. 6A–B, F and Supplementary Fig. 11). However, immunostaining with MC1 and AT8 antibodies revealed a dramatic increase of p-tau in the spinal cord, specifically in the ventral horn of PS19:Bin1-cKO mice (Fig. 6A). Quantification revealed that PS19:Bin1-cKO mice had elevated levels of MC1+ area (P < 0.05), number of MC1+ cells/mm2 (P < 0.05) and AT8+ area (P < 0.05) as compared with PS19:Cre mice (Fig. 6C–E). Similarly, we observed increased AT8 and Tau-5 immunoreactivities and signs of axonal degeneration in the sciatic nerve of PS19:Bin1-cKO mice relative to PS19:Cre, despite comparable axonal BIN1 staining (Fig. 6F). These results indicate that the loss of BIN1 in forebrain excitatory neurons and oligodendrocytes exacerbate tau pathology in the spinal cord and sciatic nerve of PS19:Bin1-cKO, which probably contributes to severe motor impairments, a hunched-back posture, feeding difficulties and early death (Fig. 1).
Figure 6.
Elevation of tau pathology in the spinal cord of PS19:Bin1-cKO mice. (A) Representative images of BIN1, conformation-dependent pathogenic P-tau (mAb MC1) and pSer202/Thr205 tau (mAb AT8) immunostaining in lumbar spinal cord sections from 8-month-old mice. The boxed region in the ventral horn grey matter is magnified in the bottom row. (B–E) The bar graphs show the quantification of BIN1 expression, the number of MC1+ cells/mm2 and MC1 or AT8 immunoreactivity (% area). (F) Representative images of BIN1, AT8 and Tau-5 immunostaining of sciatic nerve from 8-month-old mice. *P < 0.05. ns = non-significant.
Loss of BIN1 in PS19 mice elicits complex transcriptome changes, including altered neuroinflammation-related gene expression
We analysed forebrain transcriptomes by RNA-seq to gain unbiased insights into the molecular changes associated with the loss of BIN1 expression in forebrain excitatory neurons and oligodendrocytes. We used a false discovery rate (FDR) <0.01 as the cut-off for the differential gene expression analysis. The overall transcriptomic profile of PS19:Cre versus PS19:Bin1-cKO revealed 484 DEGs, of which 248 were upregulated and 236 were downregulated (Supplementary Fig. 4A and Supplementary Table 3). Principal component analysis based on all RNA-seq read counts demonstrated that PS19:Cre and PS19:Bin1-cKO were distinctly clustered (Supplementary Fig. 5A), verifying acceptable reproducibility within the genotypes.
To explore potential biological mechanisms, we performed a GSEA of the 484 DEGs and found a significant over-representation of six molecular functions (MF) and five cellular component terms while using FDR < 0.05 as a cut-off (Supplementary Fig. 5B and Supplementary Table 4). Interestingly, gene sets related to MF included: actin binding, actin filament binding and calmodulin binding. Of these, 16 DEGs that specifically belong to the actin binding and actin filament binding category were analysed in the transcript levels by real-time qPCR. Aside from Bin1, the levels of 6 DEGs, Cnga2, Parvg, Tnnc1, Ldb3, Myl4 and Eps8l1 were confirmed to be significantly different between PS19:Cre and PS19:Bin1-cKO mice (Supplementary Fig. 5C). A protein-protein interaction network of these DEGs, constructed using the STRING database, revealed five stronger edges (connections): Myl4, Tpm2 and Tnnc1 connected to each other, Myoz2 and Ldb3, and Ceacam1 and Ceacam2 (Supplementary Fig. 5D). A group of genes from the Takusan family (gene names indicated as Gmxxxx) was differentially expressed (mostly upregulated) in PS19:Bin1-cKO mice (Supplementary Fig. 6A). Interestingly, when referenced to the list of 136 mouse genes associated with ageing and/or longevity from the GenAge database,50 four transcripts, Brca1, Cisd2, Ercc2 and Sod3, were downregulated in PS19:Bin1-cKO mice (Supplementary Fig. 6B).
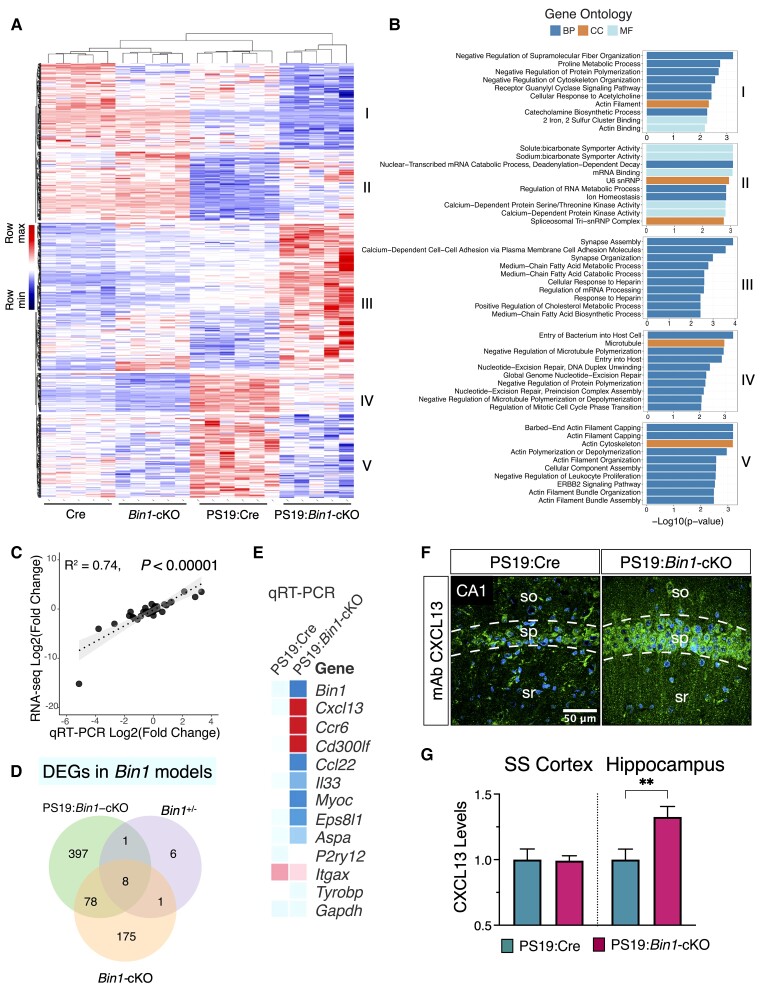
To capture changes associated explicitly with tau pathogenesis, we further generated RNA-seq data from the forebrain tissue of Emx-Cre (Cre) and Bin1-cKO mice (no tau pathology, Supplementary Table 5). We compiled a list of 928 DEGs identified from Cre versus Bin1-cKO mice or PS19:Cre versus PS19:Bin1-cKO mice RNA-seq analyses (cut-off of P < 0.01 and FDR < 0.05; Supplementary Table 6). GO analysis (g:Profiler; FDR < 0.05) revealed the functional profiling of these genes that significant over-representation of 10 MF, 14 MP and 24 corpus callosum terms (Supplementary Fig. 6C and Supplementary Table 7), and many MFs were annotated to ‘actin binding’. We further explored the MF of these DEGs by hierarchical clustering and identified five clusters of genes that showed distinct patterns of regulation by tau pathology and BIN1 expression (Fig. 7A and B, Supplementary Fig. 6D, and Supplementary Table 6 and 8). Cluster I contained genes positively regulated by BIN1 (i.e. downregulated by BIN1 deficiency) in the context of tau pathology, which are involved in the negative regulation of supramolecular fibre organization (Stmn1, Twf2, Cryab, Coro1a and Kank4), proline metabolic process (Dao, Pycr1) and negative regulation of protein polymerization (Stmn1, Twf2 and Kank4). Cluster II genes were generally expressed at lower levels in tau pathology, were negatively regulated by BIN1 and their function included sodium:bicarbonate symporter activity (Slc4a7, Slc4a10), nuclear-transcribed mRNA catabolic process (Ddx6, Lsm7, Lsm6 and Rc3h2) and mRNA binding (Ddx6, Rbfox1, Rbfox2, Ythdc1, Serbp1, Rc3h2, Ptbp2 and Cpeb4). Cluster III genes were negatively regulated by BIN1 in the context of tau pathology and included genes involved in synapse assembly, calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules (including protocadherin family; Supplemental Fig. 7B) and genes functioning in synapse organization (including Lrfn5, Pcdhb2, Flrt3, F2r, Pcdhb6, Pcdhb5, Pcdhb3 and Pcdhb13). Cluster IV and Cluster V genes were upregulated by tau pathology and positively regulated by BIN1. Cluster IV gene function included entry of bacterium into host cell genes (including Cbl and Cbll1) and negative regulation of microtubule polymerization (including Tbcd and Tubb4a). Cluster V genes are involved in actin filament capping, actin cytoskeleton and actin filament organization (Svil, Capza1, VIll, Myo1e, Pstpip1, Plekhh2, Fam107a, Pls3, Shroom3 and Myoz2). The accuracy of the RNA-seq data was validated by real-time qPCR of 42 DEGs (chosen based on low, medium and high expression levels). The RNA-seq results were found to correlate significantly with the real-time qPCR results over a wide range (R2 = 0.74, P < 0.00001; Fig. 7C), confirming the reliability of the RNA-seq data.
Figure 7.
Transcriptomic profiling of Bin1-cKO mice identifies CNS cell-type-specific DEGs. (A and B) Heat map of the upregulated or downregulated DEGs from a combined analysis of the RNA-seq data from Bin1-cKO with- and without tau pathology mice. Hierarchical clustering identified five clusters of genes in this dataset of 928 DEGs (Padj value <0.05). (C) Validation of the RNA-seq by the real-time qPCR revealed a tight correlation (R2 = 0.74, P < 0.0001) between the two methods for 42 selected DEGs. (D) Venn diagram of DEGs (Padj <0.05) identified in each cohort. The 16 DEGs in Bin1+/− mice were identified in a previous study.51 In Bin1-cKO mice (no tau pathology), 262 genes were identified, whereas in PS19:Bin1-cKO mice, 484 genes were identified. Eight common DEGs were found in all three datasets. 397 DEGs were unique to Bin1-cKO mice in the context of tau pathology. (E) qRT-qPCR results of immune response DEGs are represented as a heat map. (F) Immunostaining shows a higher neuronal expression of CXCL13 in the hippocampus of PS19:Bin1-cKO mice. (G) Quantification of CXCL13 immunostaining in the hippocampus and Som sen CTX revealed significant differences in the hippocampus (PS19:Cre, n = 7 and PS19:Bin1-cKO, n = 7). **P < 0.01.
We also compared our RNA datasets described previously to DEGs identified in a previous characterization of Bin1+/− mice.51 This analysis identified eight shared DEGs in all three independent datasets, which indicated that their altered expression, regulated in a similar pattern, exclusively corresponded to the loss of Bin1 allele(s) (Fig. 7D). Of these eight shared DEGs, six belong to the protocadherin family (Supplementary Fig. 7A). Moreover, 11 protocadherin family members were identified as DEGs in the comparison between PS19:Cre and PS19:Bin1-cKO; among this group, nine were upregulated in PS19:Bin1-cKO mice (Supplementary Fig. 7B). A combined analysis of all three datasets identified 397 DEGs unique to Bin1-cKO mice in the context of tau pathogenesis (Fig. 7D). When these 397 genes were referenced to a published CNS cell- type-specific RNA-seq dataset,52 22% of DEGs were annotated to neurons and oligodendrocytes, 22% to microglia, astrocytes or endothelial cells and 55% of DEGs to multiple cell types (Supplementary Fig. 7C and D), revealing a complex non-cell-autonomous response to Emx-Cre-mediated Bin1 ablation in PS19 mice.
Further, ShinyGO enrichment analysis of the 928 unique DEGs suggested that these genes function in regulating the immune system process or immune response (Supplementary Fig. 7E and Supplementary Table 9). We also performed a GSEA, v.4.2.3)40 against curated gene sets in Molecular Signatures Database v.7.5.1. The results showed significant downregulation of several pathways but no significant upregulation of any pathway (Supplementary Table 10). Notably, genes in the Inflammatory Response Pathway were found to be negatively enriched (FDR = 0.002), including the core enrichment of 12 genes, including Cd40, Il5, Il2rg, Cd28, Ifng, Il4r and Il2ra (Supplementary Fig. 8). Despite the marked reduction in gliosis observed in PS19:Bin1-cKO mice (Fig. 5), three genes implicated in inflammation, Cxcl13, Ccr6 and Cd300lf, were upregulated in PS19:Bin1-cKO in comparison to PS19:Cre mice. In addition, several downregulated transcripts were annotated to gliosis function, including Ccl22, Il33, Aspa, Myoc and Trf. Up- or downregulation of these transcripts was verified by real-time qRCR (Fig. 7E). Finally, we performed immunostaining to ascertain the cellular expression of CXCL13, a chemokine that functions as a major B cell attractant in the periphery and is implicated in neuroinflammation in the CNS.53,54 CXCL13 expression was mainly observed in cortical and hippocampal neurons in PS19 mice (Fig. 7F). Neuronal CXCL13 immunoreactivity was undetectable in age-matched Emx-Cre mice, suggesting that expression of this cytokine is induced in PS19 mouse brain (data not shown). Notably, CXCL13 immunoreactivity was significantly higher in the hippocampus of PS19:Bin1-cKO mice than PS19:Cre mice (Fig. 7F and G). In contrast, CXCL13 staining intensity in the somatosensory cortex was comparable between the genotypes (Fig. 7G). Together, these results elucidate non-cell-autonomous functions of BIN1 in the brain and highlight a brain region-specific difference in neuronal CXCL13 expression in mice lacking BIN1 expression in the context of tau pathology.
Loss of forebrain BIN1 reduces hippocampal ApoE expression and impedes homeostatic to disease-associated microglial transition
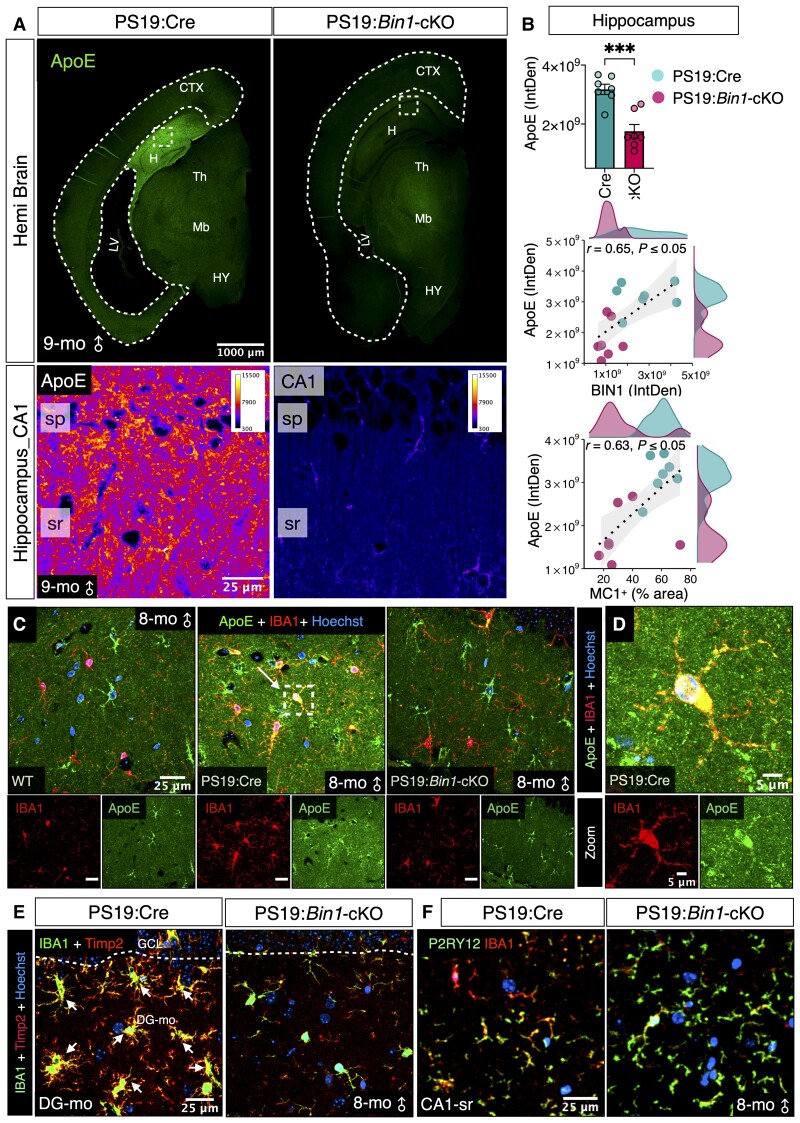
APOE is among Alzheimer’s disease-associated genes that profoundly affect tau pathogenesis and tau-mediated neurodegeneration.55 We asked whether the loss of BIN1 in PS19:Bin1-cKO mice affected ApoE expression. Immunofluorescence analysis revealed high levels of ApoE expression in 9-month-old PS19:Cre mice. In contrast, ApoE expression was significantly lower in the hippocampus of PS19:Bin1-cKO mice (P < 0.001; Fig. 8A and B). Spearman’s correlation analysis revealed that hippocampal ApoE expression positively correlated with hippocampal BIN1 expression (r = 0.65, P < 0.05) and hippocampal MC1+ tau pathology (r = 0.63, P < 0.05) (Fig. 8B). Because ApoE expression was too intense in 9-month-old animals to discern cellular expression, we examined in PS19:Cre mice at 8 months of age at the onset of overt gliosis. At this stage, ApoE was expressed in astrocytes of both PS19:Cre and PS19:Bin1-cKO mice, as expected (Fig. 8C and Supplementary Fig. 9A). In addition, we observed a notable overlap between ApoE staining and IBA1+ microglia in PS19:Cre mice, consistent with the finding that ApoE is upregulated in microglia in mouse models tauopathy and amyloidosis.56–58 In contrast, there was rarely any microglial ApoE expression in PS19:Bin1-cKO mice (Fig. 8C–D). Furthermore, all BIN1+ microglia in PS19:Cre mice were CD68+, consistent with the microglial activation in animals with tauopathy (Supplementary Fig. 9B). Interestingly, there was a significant decrease in the number of CD68+ activated microglia in the hippocampus and the entorhinal/piriform cortex/amygdala of PS19:Bin1-cKO mice as compared with PS19:Cre animals (P < 0.01) (Supplementary Fig. 9C and D). To extend these observations, we analysed the expression of TIMP2, another marker of disease-associated microglia.58 IBA1 + microglia in the hippocampus of PS19:Cre mice showed robust TIMP2 immunostaining, but the staining was absent in microglia of PS19:Bin1-cKO mice (Fig. 8E). In contrast, the expression of P2RY12, a homeostatic microglial marker, was higher in PS19:Bin1-cKO mice than PS19:Cre mice (Fig. 8F and Supplementary Fig. 9E). These results reveal a crucial non-cell-autonomous role of neuronal/oligodendrocyte BIN1 in tau pathology, which mediates microglial transition towards a disease-associated microglial phenotype.
Figure 8.
Bin1-cKO downregulates hippocampal ApoE expression in PS19:Bin1-cKO mice. (A) Immunofluorescence analysis of ApoE expression analysis in the brains of PS19:Cre and PS19:Bin1-cKO mice. Higher magnification of the boxed region in the hippocampus is shown as pseudocolour images in the same intensity scale. (B) Quantification of ApoE expression in the hippocampus (top). Spearman’s correlation coefficient analysis (bottom) shows a positive correlation between the hippocampal ApoE expression and BIN1 expression or tau pathology. PS19:Bin1-cKO (n = 7) and PS19:Cre (n = 7) mice. (C) Tau pathology-dependent ApoE expression in IBA1+ microglia. Brain sections from 8-month-old WT, PS19:Cre and PS19:Bin1-cKO mice were co-immunostained with antibodies against ApoE and IBA1. Nuclei were stained with Hoechst. (D) Higher magnification of the boxed region from PS19:Cre shows ApoE expression in microglia. (E) Immunostaining of Timp2, a DAM marker, together with IBA1 in 8-month-old PS19:Cre and PS19:Bin1-cKO mice. (F) Immunostaining of P2RY12, a marker for homeostatic microglia, and IBA1 in 8-month-old PS19:Cre and PS19:Bin1-cKO mice.
Discussion
Our study demonstrates that BIN1 function in the forebrain modulates tau pathophysiology in a region-specific manner through cell-autonomous and non-cell-autonomous pathways. The loss of BIN1 in excitatory neurons and oligodendrocytes limited hippocampal tau pathogenesis, gliosis and neuroinflammation. Still, it promoted an overall increase of pathogenic tau levels in some cortical regions and the spinal cord. Consequently, hippocampal synapses were preserved, and general brain atrophy characteristic of the PS19 line was curbed in PS19:Bin1-cKO mice. Furthermore, neurons in select brain regions were protected from degeneration, indicated by little or no lateral ventricular enlargement and relatively larger volumes of the hippocampus, piriform and entorhinal cortex regions and increased cell layer thickness in hippocampal subregions. Forebrain BIN1 loss in PS19 mice elicited a complex neuronal and glial response, including significantly reduced expression of ApoE and other disease-associated microglial phenotype genes. These results provide novel insights into in vivo BIN1 function in the context of tau pathogenesis.
Neuronal BIN1 isoform expression is reduced in the brains of LOAD patients.3,16–20 However, the loss of neuronal BIN1 expression had no effect on Aβ production and a 50% reduction of BIN1 expression did not alter cerebral Aβ deposition.59 The details linking BIN1 to tau pathology are only beginning to emerge. The BIN1 SH3 domain can interact with the proline-rich region of tau,26 but whether this interaction has any influence on tau pathogenesis has not been directly tested. In vitro studies from cultured cells predicted that BIN1 expression will limit neuron-to-neuron pathology propagation of tau seeds24 and reduce synaptic p-tau levels while promoting tau release.25 Furthermore, loss of BIN1 in neurons was also found to impede non-p-tau release.25 Surprisingly, the loss of neuronal and oligodendrocyte BIN1 expression correlated with an increase of p-tau Thr231, but lower levels of pSer202/Thr205 and MC1+ pathogenic tau in the hippocampus concomitant with attenuated neurodegeneration. Thus, in contrast to findings from in vitro models, the in vivo results described here reveal that forebrain BIN1 function is required for the development of tau pathology and neuroinflammation in the hippocampus. Nevertheless, BIN1’s influence on tauopathy has some regional specificity as there was an increase in tau pathology in the somatosensory cortex and the spinal cord in the same cKO animals (Fig. 3). These intriguing finding in a commonly used experimental tauopathy mouse model is reminiscent of hippocampal-sparing Alzheimer’s disease subtype described by post-mortem analysis60,61 and recently categorized as medial temporal lobe sparing S2 spatiotemporal tau deposition trajectory observed in about one-fifth of individuals with Alzheimer’s disease.62
Our observations that the loss of BIN1 attenuates hippocampal tauopathy and neuroinflammation are analogous to finding reported on tau pathology modulation by the loss of expression of two Alzheimer’s disease-associated genes, Apoe and Trem2.35,55 PS19 mice lacking ApoE expression developed milder tau pathology; they were protected from neurodegeneration, neuroinflammation and brain atrophy, whereas the expression of human ApoE worsened tau pathology and other outcomes.55 Similarly, PS19 mice lacking TREM2 expression had attenuated brain atrophy and neuroinflammation, despite no discernible effect on pathogenic tau accumulation by the loss of TREM2 function.35 The results reported here show that, like ApoE and TREM2, BIN1 facilitates tau-mediated hippocampal neurodegeneration, astrocyte activation and microglial response. Exacerbated tau-mediated neurodegeneration observed in human ApoE4 knock-in mice was moderated by the ablation of APOE4 alleles in astrocytes, implicating astrocyte-derived ApoE as a key player in tau pathophysiology.63 In contrast, TREM2 is expressed predominantly in microglia. Its function is vital for microglial response to tau pathology, which underlies neurodegeneration in the PS19 model.35 An important distinction between a comparison of BIN1, ApoE and TREM2 function in tau pathology is that we ablated Bin1 alleles in the forebrain excitatory neurons and oligodendrocytes without affecting BIN1 expression in microglia and other cell types. Whereas hippocampal ApoE immunoreactivity dramatically increased between 8 and 9 months of age in PS19:Cre concomitant with tau accumulation, ApoE expression in PS19:Bin1-cKO mice remained at lower levels (Fig. 8). Thus, the attenuation of brain atrophy and gliosis observed in the PS19:Bin1-cKO hippocampus could be secondary to reduced pathogenic tau accumulation. Interestingly, while we observed a significant increase of Bin1 expression in PS19:Cre mice when compared to Cre controls, the levels were comparable between Bin1-cKO and PS19:Bin1-cKO mice (Supplementary Fig. 10A). BIN1 immunoreactivity was also higher in most brain regions of PS19:Cre mice compared with Cre controls (Supplementary Fig. 10B). Nevertheless, our results show an absence of compensatory increase of microglial BIN1 protein levels in PS19:cKO mice compared with PS19:Cre controls (Supplementary Fig. 1C–F).
We found that abrogating BIN1 expression in forebrain excitatory neurons and oligodendrocytes in the context of tau pathogenesis affected the expression of nearly 400 genes, including a significant number of genes uniquely expressed in microglia, astrocytes, endothelial cells and other multiple cell types. Accordingly, significant enrichment in KEGG pathways and GO analysis of biological processes spanning multiple cell types, from actin binding and cytoskeleton localization, cellular entry, immune system process, insulin receptor signalling and synapse organization, assembly and function, was observed. Notable among the DEGs identified in PS19:Bin1-cKO mice are Cxcl13 and Cd300lf, two genes that regulate neuroinflammation. The chemokine CXCL13 is constitutively expressed in the periphery within lymphoid organs. Its function as a B and T lymphocyte attractant through its receptor CXCR5 has been implicated in numerous inflammatory diseases. Infiltrating myeloid and lymphoid cells and microglia account for CXCL13 expression in the CNS,54 with a few exceptions. Neuronal CXCL13 expression gets induced in patients with intractable epilepsy and a rodent model of temporal lobe epilepsy,64 in mutant SOD1 mice exhibiting amyotrophic lateral sclerosis pathology65 and in mice following experimental ligation-induced nerve injury.66,67 Upregulation of CXCL13 in spinal cord motor neurons in fast-progressing amyotrophic lateral sclerosis mice attenuated inflammation and prevented neuromuscular denervation.65 In PS19:Bin1-cKO mice, higher levels of CXCL13 in hippocampal neurons correlate with lower pathogenic tau accumulation, reduced gliosis and overall neuroprotection (Fig. 7F and G). CD300f is a tyrosine-based inhibition motif-containing Ig-like family immunoreceptor involved in immune response regulation whose expression in microglia attenuates proinflammatory cytokine production, thus impeding neuroinflammation and autoimmune demyelination.68,69 Moreover, CD300f was found to be neuroprotective following acute brain injury.70 Despite an overall reduction in microglial activation, we observe a significant upregulation of Cd300lf in PS19:Bin1-cKO mice (Fig. 7E). This finding may have important implications for widespread neuroinflammation activation in the PS19:Bin1-cKO brain because CD300lf expression has been negatively correlated with hippocampal microglial numbers and microglial metabolic fitness.71 These findings suggest the loss of neuronal BIN1 expression influences microglial signalling in the context of tau pathogenesis.
Apart from pathology attenuation in the hippocampus, our study revealed forebrain BIN1 loss exacerbated tau pathology in the somatosensory cortex and spinal cord of PS19:Bin1-cKO mice. Consequently, PS19:Bin1-cKO mice manifested more significant motor deficits than PS19:Cre controls, probably resulting from severe tau pathology-induced muscle atrophy. Since BIN1 expression in spinal cord grey matter and sciatic nerves were relatively unchanged, the likely explanation for higher levels of pathogenic tau accumulation in the spinal cord is the exacerbated propagation of pathology in axonal projections from corticospinal neurons.48,49 Greater deficits in motor coordination and balance translated to accelerated disease progression and lower survival in symptomatic PS19:Bin1-cKO mice. They frequently displayed a hunched posture and were too weak to eat or drink. A comparison with our RNA-seq data to the GenAge database50 revealed decreases in the expression of Brca1, Cisd2, Ercc2 and Sod3, genes that have been associated with premature death. Deletion of Brca1,72Cisd2,73Ercc274 and alteration of Sod375 all lead to a shorter life span through cell senescence, mitochondrial breakdown, DNA repair and innate immune response, respectively. We hypothesize that the loss of BIN1 and subsequent downregulation of these anti-longevity genes in the forebrain tissue of PS19:Bin1-cKO mice might have contributed to a lower survival rate in PS19:Bin1-cKO mice. Thus, these results indicate that the loss of BIN1 expression in forebrain excitatory neurons and oligodendrocytes causes accelerated disease progression and pathology in the spinal cord leading to an earlier death.
GO analysis on the overall PS19:Cre versus PS19:Bin1-cKO transcriptome profile comparison highlighted several genes with functions annotated to the actin cytoskeleton, actin binding, actin filament organization and bundle assembly (Supplementary Fig. 4B and C). Cytoskeletal microfilament actin polymerization/depolymerization is critical for microglial migration, phenotypic transition and phagocytosis.76 Dystrophic and reactive microglia are found in the proximity of both senile plaques and neurofibrillary tangles in the brains of patients with Alzheimer’s disease. Homeostatic microglia have a modest capacity to phagocytose extracellular tau aggregates but could be overwhelmed if the tau load exceeds their phagocytic capacity. Tau aggregates also cause microglial activation, leading to morphology alteration as they lose their homeostatic function and transition towards disease-associated phenotype.77,78 Microglia play a significant role in tau pathology propagation and mediate ApoE-dependent neurodegeneration, as demonstrated by microglial depletion studies.79,80 Moreover, microglial engulfment of synapses underlies tau-induced synapse loss and ensuing neurodegeneration.81 The conservation of the microglial homeostatic phenotype (higher P2RY12 and lower ApoE and TIMP2 expression; Fig. 8E) in PS19:Bin1-cKO correlates well with the significant preservation of hippocampal synapses and attenuation of neurodegeneration in comparison with PS19:Cre controls. Thus, in addition to highlighting region specificity, our investigation highlights cell-autonomous and non-cell-autonomous regulations involved in BIN1 modulation of tau neuropathology in vivo.
Supplementary Material
Acknowledgements
We thank Drs David Borchelt (University of Florida), Laura Blair (University of South Florida) and an anonymous reviewer for their insightful suggestions on tau pathology characterization. We thank Tristan Williams and Paramita Chakrabarty for the discussion on generating IHC heatmaps. We acknowledge Ha-Na Shim and Jeffrey Lenz for their assistance in mouse colony maintenance and histology. We are grateful to the late Dr Peter Davies for MC1 and PHF1 antibodies. We thank Drs Chao Wang and David Holtzman for ApoE antibody and helpful discussions.
Contributor Information
Moorthi Ponnusamy, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Shuai Wang, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Melike Yuksel, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Mitchell T Hansen, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Danielle M Blazier, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Joseph D McMillan, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Xiaolin Zhang, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Eric B Dammer, Department of Biochemistry, Emory University, Atlanta, GA 30322, USA.
Lisa Collier, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Gopal Thinakaran, Byrd Alzheimer’s Center and Research Institute, University of South Florida, Tampa, FL 33613, USA; Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA.
Funding
This work was supported by National Institues of Health grants AG019070, AG056061, AG057290 and AG077610 from the National Institute on Aging.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Karch CM, Goate AM. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prokic I, Cowling BS, Laporte J. Amphiphysin 2 (BIN1) in physiology and diseases. J Mol Med (Berl). 2014;92:453–463. [DOI] [PubMed] [Google Scholar]
- 3. De Rossi P, Buggia-Prévot V, Clayton BL, et al. . Predominant expression of Alzheimer's disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts. Mol Neurodegener. 2016;11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chapuis J, Hansmannel F, Gistelinck M, et al. . Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18:1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martiskainen H, Viswanathan J, Nykänen NP, et al. . Transcriptomics and mechanistic elucidation of Alzheimer's disease risk genes in the brain and in vitro models. Neurobiol Aging. 2015;36:1221.e15–28. [DOI] [PubMed] [Google Scholar]
- 6. Nott A, Holtman IR, Coufal NG, et al. . Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science. 2019:366:1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adams SL, Tilton K, Kozubek JA, Seshadri S, Delalle I. Subcellular changes in bridging integrator 1 protein expression in the cerebral cortex during the progression of Alzheimer disease pathology. J Neuropathol Exp Neurol. 2016;75:779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sudwarts A, Ramesha S, Gao T, et al. . BIN1 is a key regulator of proinflammatory and neurodegeneration-related activation in microglia. Mol Neurodegener. 2022;17:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Rossi P, Buggia-Prevot V, Andrew RJ, et al. . BIN1 localization is distinct from tau tangles in Alzheimer's disease. Matters (Zur). Published online 12 January 2017. 10.19185/matters.201611000018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schürmann B, Bermingham DP, Kopeikina KJ, et al. . A novel role for the late-onset Alzheimer's disease (LOAD)-associated protein Bin1 in regulating postsynaptic trafficking and glutamatergic signaling. Mol Psychiatry. 2020;25:2000–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J Biol Chem. 1997;272:16700–6. [DOI] [PubMed] [Google Scholar]
- 12. De Rossi P, Nomura T, Andrew RJ, et al. . Neuronal BIN1 regulates presynaptic neurotransmitter release and memory consolidation. Cell Rep. 2020;30:3520–3535.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McAvoy KM, Rajamohamed Sait H, Marsh G, et al. . Cell-autonomous and non-cell autonomous effects of neuronal BIN1 loss in vivo. PLoS ONE. 2019;14:e0220125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voskobiynyk Y, Roth JR, Cochran JN, et al. . Alzheimer's disease risk gene BIN1 induces tau-dependent network hyperexcitability. eLife. 2020;9:e57354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franzmeier N, Neitzel J, Rubinski A, et al. . Functional brain architecture is associated with the rate of tau accumulation in Alzheimer's disease. Nat Commun. 2020;11:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glennon EB, Whitehouse IJ, Miners JS, et al. . BIN1 Is decreased in sporadic but not familial Alzheimer's disease or in aging. PLoS ONE. 2013;8:e78806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holler CJ, Davis PR, Beckett TL, et al. . Bridging integrator 1 (BIN1) protein expression increases in the Alzheimer's disease brain and correlates with neurofibrillary tangle pathology. J Alzheimers Dis. 2014;42:1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu L, Chibnik LB, Srivastava GP, et al. . Association of brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol. 2015;72:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKenzie AT, Moyon S, Wang M, et al. . Multiscale network modeling of oligodendrocytes reveals molecular components of myelin dysregulation in Alzheimer's disease. Mol Neurodegener. 2017;12:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marques-Coelho D, Iohan LDCC, Melo de Farias AR, et al. . Differential transcript usage unravels gene expression alterations in Alzheimer's disease human brains. NPJ Aging Mech Dis. 2021;7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chia R, Sabir MS, Bandres-Ciga S, et al. . Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. 2021;53:294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11:155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dujardin S, Hyman BT. Tau prion-like propagation: State of the art and current challenges. Adv Exp Med Biol. 2019; 1184:305–325. [DOI] [PubMed] [Google Scholar]
- 24. Calafate S, Flavin W, Verstreken P, Moechars D. Loss of Bin1 promotes the propagation of tau pathology. Cell Rep. 2016;17:931–940. [DOI] [PubMed] [Google Scholar]
- 25. Glennon EB, Lau DH, Gabriele RMC, et al. . Bridging integrator-1 protein loss in Alzheimer's disease promotes synaptic tau accumulation and disrupts tau release. Brain Commun. 2020; 2(1): fcaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sottejeau Y, Bretteville A, Cantrelle FX, et al. . Tau phosphorylation regulates the interaction between BIN1's SH3 domain and tau's proline-rich domain. Acta Neuropathol Commun. 2015;3:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crotti A, Sait HR, McAvoy KM, et al. . BIN1 favors the spreading of tau via extracellular vesicles. Sci Rep. 2019;9:9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yoshiyama Y, Higuchi M, Zhang B, et al. . Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. [DOI] [PubMed] [Google Scholar]
- 29. Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guyenet SJ, Furrer SA, Damian VM, Baughan TD, La Spada AR, Garden GA. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J Vis Exp. 2010;39:1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carter RJ, Morton J, Dunnett SB. Motor coordination and balance in rodents. Curr Protoc Neurosci. 2001. Chapter 8:Unit 8.12. [DOI] [PubMed] [Google Scholar]
- 32. Schindelin J, Arganda-Carreras I, Frise E, et al. . Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bankhead P, Loughrey MB, Fernández JA, et al. . Qupath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Williams T, Sorrentino Z, Weinrich M, Giasson BI, Chakrabarty P. Differential cross-seeding properties of tau and α-synuclein in mouse models of tauopathy and synucleinopathy. Brain Commun. 2020;2:fcaa090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leyns CEG, Ulrich JD, Finn MB, et al. . TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc Natl Acad Sci U S A. 2017;114:11524–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Criado-Marrero M, Gebru NT, Blazier DM, et al. . Hsp90 co-chaperones, FKBP52 and Aha1, promote tau pathogenesis in aged wild-type mice. Acta Neuropathol Commun. 2021;9:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ising C, Gallardo G, Leyns CEG, et al. . Correction: AAV-mediated expression of anti-tau scFvs decreases tau accumulation in a mouse model of tauopathy. J Exp Med. 2017;214:2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spandidos A, Wang X, Wang H, Seed B. Primerbank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jicha GA, Bowser R, Kazam IG, Davies P. Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J Neurosci Res. 1997;48:128–132. [DOI] [PubMed] [Google Scholar]
- 42. Augustinack JC, Schneider A, Mandelkow EM, Hyman BT. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer's disease. Acta Neuropathol. 2002;103:26–35. [DOI] [PubMed] [Google Scholar]
- 43. Crescenzi R, DeBrosse C, Nanga RP, et al. . Longitudinal imaging reveals subhippocampal dynamics in glutamate levels associated with histopathologic events in a mouse model of tauopathy and healthy mice. Hippocampus. 2017;27:285–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lichte B, Veh RW, Meyer HE, Kilimann MW. Amphiphysin, a novel protein associated with synaptic vesicles. Embo j. 1992;11:2521–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allen B, Ingram E, Takao M, et al. . Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J Neurosci. 2002;22:9340–9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leroy K, Bretteville A, Schindowski K, et al. . Early axonopathy preceding neurofibrillary tangles in mutant tau transgenic mice. Am J Pathol. 2007;171:976–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dugger BN, Hidalgo JA, Chiarolanza G, et al. . The distribution of phosphorylated tau in spinal cords of Alzheimer's disease and non-demented individuals. J Alzheimers Dis. 2013;34:529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bareyre FM, Kerschensteiner M, Misgeld T, Sanes JR. Transgenic labeling of the corticospinal tract for monitoring axonal responses to spinal cord injury. Nat Med. 2005;11:1355–1360. [DOI] [PubMed] [Google Scholar]
- 49. Ni Y, Nawabi H, Liu X, et al. . Characterization of long descending premotor propriospinal neurons in the spinal cord. J Neurosci. 2014;34:9404–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tacutu R, Thornton D, Johnson E, et al. . Human ageing genomic resources: New and updated databases. Nucleic Acids Res. 2017;46:D1083–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pandey RS, Graham L, Uyar A, Preuss C, Howell GR, Carter GW. Genetic perturbations of disease risk genes in mice capture transcriptomic signatures of late-onset Alzheimer’s disease. Mol Neurodegener. 2019; 14:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Chen K, Sloan SA, et al. . An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Legler DF, Loetscher M, Roos RS, Clark-Lewis I, Baggiolini M, Moser B. B cell-attracting chemokine 1, a human CXC chemokine expressed in lymphoid tissues, selectively attracts B lymphocytes via BLR1/CXCR5. J Exp Med. 1998;187:655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Irani DN. Regulated production of CXCL13 within the central nervous system. J Clin Cell Immunol. 2016;7:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi Y, Yamada K, Liddelow SA, et al. . Apoe4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017:549:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Friedman BA, Srinivasan K, Ayalon G, et al. . Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer's disease not evident in mouse models. Cell Rep. 2018;22:832–847. [DOI] [PubMed] [Google Scholar]
- 57. Rexach JE, Polioudakis D, Yin A, et al. . Tau pathology drives dementia risk-associated gene networks toward chronic inflammatory states and immunosuppression. Cell Rep. 2020;33:108398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Keren-Shaul H, Spinrad A, Weiner A, et al. . A unique microglia type associated with restricting development of Alzheimer's disease. Cell. 2017;169:1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- 59. Andrew RJ, De Rossi P, Nguyen P, et al. . Reduction of the expression of the late-onset Alzheimer's disease (AD) risk-factor BIN1 does not affect amyloid pathology in an AD mouse model. J Biol Chem. 2019;294:4477–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Whitwell JL, Dickson DW, Murray ME, et al. . Neuroimaging correlates of pathologically defined subtypes of Alzheimer's disease: A case-control study. Lancet Neurol. 2012;11:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW. Neuropathologically defined subtypes of Alzheimer's disease with distinct clinical characteristics: A retrospective study. Lancet Neurol. 2011;10:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vogel JW, Young AL, Oxtoby NP, et al. . Four distinct trajectories of tau deposition identified in Alzheimer's disease. Nat Med. 2021;27:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang C, Xiong M, Gratuze M, et al. . Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia. Neuron. 2021;109:1657–1674.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li R, Ma L, Huang H, et al. . Altered expression of CXCL13 and CXCR5 in intractable temporal lobe epilepsy patients and pilocarpine-induced epileptic rats. Neurochem Res. 2017;42:526–540. [DOI] [PubMed] [Google Scholar]
- 65. Trolese MC, Mariani A, Terao M, et al. . CXCL13/CXCR5 signalling is pivotal to preserve motor neurons in amyotrophic lateral sclerosis. EBioMedicine. 2020;62:103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jiang BC, Cao DL, Zhang X, et al. . CXCL13 drives spinal astrocyte activation and neuropathic pain via CXCR5. J Clin Invest. 2016;126:745–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Q, Cao DL, Zhang ZJ, Jiang BC, Gao YJ. Chemokine CXCL13 mediates orofacial neuropathic pain via CXCR5/ERK pathway in the trigeminal ganglion of mice. J Neuroinflammation. 2016;13:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Keswani T, Roland J, Herbert F, et al. . Expression of CD300lf by microglia contributes to resistance to cerebral malaria by impeding the neuroinflammation. Genes Immun. 2020;21:45–62. [DOI] [PubMed] [Google Scholar]
- 69. Xi H, Katschke KJ Jr., Helmy KY, et al. . Negative regulation of autoimmune demyelination by the inhibitory receptor CLM-1. J Exp Med. 2010;207:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Peluffo H, Alí-Ruiz D, Ejarque-Ortíz A, et al. . Overexpression of the immunoreceptor CD300f has a neuroprotective role in a model of acute brain injury. Brain Pathol. 2012;22:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lago N, Kaufmann FN, Negro-Demontel ML, et al. . CD300f immunoreceptor is associated with major depressive disorder and decreased microglial metabolic fitness. Proc Natl Acad Sci U S A. 2020;117:6651–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cao L, Li W, Kim S, Brodie SG, Deng C-X. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen Y-F, Kao C-H, Chen Y-T, et al. . Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23:1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. De Boer J. Premature aging in mice deficient in DNA repair and transcription. Science. 2002:296:1276–1279. [DOI] [PubMed] [Google Scholar]
- 75. Kwon MJ, Lee KY, Lee HW, Kim JH, Kim TY. SOD3 variant, R213G, altered SOD3 function, leading to ROS-mediated inflammation and damage in multiple organs of premature aging mice. Antioxid Redox Signal. 2015;23:985–999. [DOI] [PubMed] [Google Scholar]
- 76. Das R, Chinnathambi S. Actin-mediated microglial chemotaxis via G-protein coupled purinergic receptor in Alzheimer's disease. Neuroscience. 2020;448:325–336. [DOI] [PubMed] [Google Scholar]
- 77. Hopp SC, Lin Y, Oakley D, et al. . The role of microglia in processing and spreading of bioactive tau seeds in Alzheimer's disease. J Neuroinflammation. 2018;15:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Perea JR, Llorens-Martín M, Ávila J, Bolós M. The role of microglia in the spread of tau: relevance for tauopathies. Front Cell Neurosci. 2018;12:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Asai H, Ikezu S, Tsunoda S, et al. . Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci. 2015;18:1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shi Y, Manis M, Long J, et al. . Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model. J Exp Med. 2019;216:2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dejanovic B, Huntley MA, De Mazière A, et al. . Changes in the synaptic proteome in tauopathy and rescue of tau-induced synapse loss by C1q antibodies. Neuron. 2018;100:1322–1336.e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In accordance with the Journal’s policy, data will be made available upon reasonable request.