Abstract
The corpus callosum is a bundle of axon fibres that connects the two hemispheres of the brain. Neurodevelopmental disorders that feature dysgenesis of the corpus callosum as a core phenotype offer a valuable window into pathology derived from abnormal axon development. Here, we describe a cohort of eight patients with a neurodevelopmental disorder characterized by a range of deficits including corpus callosum abnormalities, developmental delay, intellectual disability, epilepsy and autistic features. Each patient harboured a distinct de novo variant in MYCBP2, a gene encoding an atypical really interesting new gene (RING) ubiquitin ligase and signalling hub with evolutionarily conserved functions in axon development. We used CRISPR/Cas9 gene editing to introduce disease-associated variants into conserved residues in the Caenorhabditis elegans MYCBP2 orthologue, RPM-1, and evaluated functional outcomes in vivo. Consistent with variable phenotypes in patients with MYCBP2 variants, C. elegans carrying the corresponding human mutations in rpm-1 displayed axonal and behavioural abnormalities including altered habituation. Furthermore, abnormal axonal accumulation of the autophagy marker LGG-1/LC3 occurred in variants that affect RPM-1 ubiquitin ligase activity. Functional genetic outcomes from anatomical, cell biological and behavioural readouts indicate that MYCBP2 variants are likely to result in loss of function. Collectively, our results from multiple human patients and CRISPR gene editing with an in vivo animal model support a direct link between MYCBP2 and a human neurodevelopmental spectrum disorder that we term, MYCBP2-related developmental delay with corpus callosum defects (MDCD).
Keywords: corpus callosum, neurodevelopmental disorder, MYCBP2, epilepsy, habituation, Phr1
AlAbdi et al. describe a cohort of eight patients with a neurodevelopmental disorder characterized by corpus callosum abnormalities, developmental delay, intellectual disability, epilepsy, and autistic features, and show that the disorder is caused by loss-of-function variants in MYCBP2.
Introduction
The corpus callosum is the largest body of commissural axons in the brain, often referred to as callosal axons, which connects the two cerebral hemispheres.1 It is composed of (anteriorly to posteriorly) a rostrum, genu, rostral body, midbody, isthmus and splenium. Each portion of the callosum is enriched with projecting axons from distinct cortical regions. Development of the human corpus callosum starts with pioneer axons that cross the midline at gestational age (GA) 11–12 weeks in a rostrocaudal direction, which is followed by further axon growth. The corpus callosum becomes a recognizable structure by GA 20 weeks. It reaches a maximum thickness coinciding with axonal exuberance at GA 30 weeks before it undergoes refinement resulting in 21% reduction in thickness. Development of corpus callosum continues postnatally with myelination continuing until 9 years of age.1 The corpus callosum is functionally important for cognition and facilitates information transfer, including sensory and motor information, between brain hemispheres.2–6
Dysgenesis of the corpus callosum is a broad term that encompasses a wide range of developmental defects ranging from complete agenesis to enlarged axon tracts.7–9 Dysgenesis is a challenging developmental defect to study because it arises from alterations in a wide range of processes, such as axonal outgrowth and guidance as well as abnormal migration of neuronal (and glial) progenitors. In addition, the clinical consequences of dysgenesis are difficult to predict. For example, up to 80% of patients with isolated agenesis of corpus callosum have no discernible neurological phenotype.10 Nonetheless, dysgenesis is a pathological characteristic in a large number of neurodevelopmental disorders that are highly heterogeneous clinically and genetically.11–14 Interestingly, numerous variants in a key autophagy component, EPG5, are known to cause Vici syndrome, a developmental disorder that features agenesis of the corpus callosum as an anatomical hallmark.15
One molecule previously shown to affect corpus callosum development in rodents is MYCBP2, an atypical really interesting new gene (RING) family ubiquitin ligase and signalling hub.16–18 Human MYCBP2 (also called PAM) has a single conserved orthologue in rodents (Phr1), zebrafish (Phr/Esrom), Drosophila (Highwire) and Caenorhabditis elegans (RPM-1).16 MYCBP2 has prominent roles in nervous system development including important functions in axon development from C. elegans through mice.19–24 In mice, Phr1 deficiency results in impaired axon development in the brain including partial agenesis of corpus callosum, and complete agenesis of the anterior commissure and internal capsule.25 Impaired axon extension of motor neurons and overgrowth of peripheral sensory axons also occurs in Phr1 mutant mice.21 In C. elegans, impairing RPM-1 function results in prominent defects in axon termination resulting in overgrowth of sensory and motor axons.19,20 One recently identified signalling mechanism that RPM-1 utilizes to regulate axon termination is ubiquitination and inhibition of the autophagy initiating kinase UNC-51/ULK.26 As a consequence, RPM-1 restrains subcellular autophagosome formation at axon termination sites in the C. elegans nervous system. Findings with human cell-based studies indicate that MYCBP2 is sufficient to ubiquitinate ULK1 in a proteasome-dependent manner.
While little is known about how Phr1 affects behaviour in mammals, studies using invertebrate models have begun to address this question. In Drosophila, Highwire regulates aversive learning.27 Behavioural studies in C. elegans have shown that RPM-1 is required for habituation to repeated sensory stimulation.28,29 Habituation represents a simple conserved form of learning that occurs from C. elegans through vertebrates.30–33
Despite more than 20 years of research on MYCBP2 in several model organisms and occasional identification of MYCBP2 variants of uncertain significance in large-scale human sequencing projects,34–36 two key questions remain unanswered: (i) Do variants in human MYCBP2 cause neurological phenotypes? and (ii) Does altered MYCBP2 function result in abnormal axon development in humans?
Here, we describe a neurodevelopmental disorder based on a cohort of eight patients, each harbouring a distinct de novo variant in MYCBP2. In depth clinical evaluation indicates that patients display variable corpus callosum defects consistent with dysgenesis, and a broad spectrum of neurobehavioural deficits including developmental delay, intellectual disability, epilepsy, and autistic features. To evaluate the functional impact of MYCBP2 variants, we used CRISPR/Cas9 gene editing to introduce the corresponding variants from human patients into rpm-1 in C. elegans. Our results using this whole-animal model demonstrate that these variants result in abnormal axon development, increased axonal autophagosome formation, and abnormal behavioural habituation. Thus, both human genetic results and outcomes from gene editing with an in vivo model organism suggest that genetic changes in MYCBP2 are the likely mechanism for this new clinical entity.
Materials and methods
Human subjects
Patients were identified clinically by the respective clinical groups who were connected through GeneMatcher.37 Informed consent was obtained from all participating families as part of an institutional review board (IRB)-approved research protocol (KFSHRC REC# 2080 006) or from the respective IRB-approved protocols from each institution. Clinical and radiological data were collected and compared.
Sequencing and variant identification
The technical details of exome sequencing and analysis pipeline of human disease variants have been described elsewhere.38 Variants filtered through this pipeline for each patient are summarized in Supplementary Table 3. The de novo nature of the candidate variants was based on trio-Whole Exome Sequencing or Sanger sequencing that showed absence of MYCBP2 variants in paternal and maternal blood-derived DNA despite normal Mendelian inheritance of other variants confirming maternity and paternity. Further details of RNA-seq analysis were described previously.39 To investigate the possible effect of nonsense-mediated decay (NMD), we conducted a cycloheximide-induced suppression of NMD experiment. Equal numbers of fibroblast cells derived from a skin biopsy taken from patient 21DG0819 and foreskin fibroblast isolated from healthy control individual were seeded in a six-well plate. After the cells adhered overnight, they were serum-starved for 24 h and then two wells were treated with 200 μg/ml cycloheximide for 4 h and another two wells for 8 h. Two wells were left untreated. Cells were harvested and RNA extraction and RT-PCR amplification of MYCBP2 was performed. RT-PCR amplicons were Sanger sequenced to assess the level of the mutant allele by comparing untreated with cycloheximide treated samples.
C. elegans strains
C. elegans N2 isolate was used for all experiments. Animals were maintained using standard procedures. The following integrated transgenes were used: jsIs973 [Pmec-7 mRFP] III, bggSi14 [Pmec-7mCherry::GFP::LGG-1] II and muIs32 [Pmec-7GFP] II. The following CRISPR alleles were used: bgg6 [RPM-1::GFP CRISPR], bgg6 bgg96 [RPM-1::GFP R3645C CRISPR], bgg6 bgg97 [RPM-1::GFP L2544G CRISPR], bgg6 bgg100 [RPM-1::GFP L3070Q CRISPR], bgg6 bgg107 [RPM-1::GFP C3674Y (null) CRISPR], bgg6 bgg115 [RPM-1::GFP K2278stop CRISPR].
C. elegans CRISPR/Cas9 gene editing
MYCBP2 variants were CRISPR edited into the corresponding residues of rpm-1 in C. elegans by direct injection of Cas9 ribonucleoprotein complexes and a rol-6 co-injection marker. Injection mixes contained tracrRNA (IDT), crRNA (IDT), repair template [ssODN (Ultramers DNA oligo, IDT)] and Cas9 protein made in-house. Ribonucleoprotein complexes were heated at 37°C for 15 min prior to injection. All gene edits were confirmed by sequencing and subsequently outcrossed at least twice to wild-type animals (Supplementary Fig. 5). All injection conditions for CRISPR gene editing are shown in Supplementary Table 4. All crRNA and repair template sequences are shown in Supplementary Table 5.
C. elegans CRISPR-based native biochemistry
Mixed-stage CRISPR engineered strains were grown in liquid culture (S complete media and HB101 E. coli) for 3 days at room temperature. Worms were harvested by low-speed centrifugation, and bacteria/debris removed by 30% sucrose flotation centrifugation. Animals were subsequently washed three times with 0.1 M NaCl solution and clean, packed animals were frozen as pellets in liquid nitrogen. Frozen worm pellets (3 g) were ground to submicron particles under liquid nitrogen cooling using a cryomill (Retsch) in the presence of EDTA-free protease inhibitor tablets (Roche). Grindates were lysed in four times volume lysis buffer [50 mM Tris HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 0.1% NP-40, 1 mM DTT, 1 mM PMSF, 1× Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific)]. Whole animal lysates were rotated for 5 min at 4°C and high speed centrifuged (20 000g) for 15 min to remove debris.
Owing to low amounts of endogenous RPM-1 in the C. elegans nervous system, RPM-1::GFP CRISPR levels were evaluated by immunoprecipitation performed at non-saturating levels (Supplementary Fig. 6). Whole animal lysates (1 mg total protein) were incubated with 1.2 μg anti-GFP (3E6, Invitrogen) antibodies for 30 min. Protein-antibody complexes were precipitated with 20 μl of protein-G agarose (Roche) for 1 h at 4°C and washed three times in lysis buffer. Samples were boiled in LDS sample buffer (NuPAGE™, Invitrogen) and 50% was run on 3–8% Tris-acetate gels. Gels were wet transferred onto PVDF membranes (Millipore) overnight at 4°C. Immunoblotting was performed with anti-GFP (1:1000 dilution mouse monoclonal, Roche) antibodies. Immunoblots were visualized using horseradish peroxidase-conjugated secondary antibodies (1:20 000 dilution; GE Healthcare Life Sciences, Thermo Fisher Scientific), ECL (1:3 diluted in TBS Supersignal West Femto, Thermo Fisher Scientific) and X-ray film.
C. elegans axon termination and autophagy analysis
All images were acquired using confocal microscopy. For image acquisition, young adult animals grown at 23°C were anaesthetized (3% (v/v) 1-phenoxy-2-propanol or 10 µM of levamisole in M9 buffer) on a 3% agarose pad on glass slides. Images were acquired using a Zeiss LSM 710 laser scanning confocal microscope (40× objective).
Axon termination in anterior lateral microtubule (ALM) neurons was evaluated using the transgene jsIs973 (Pmec-7::mRFP). Failed termination was defined as ALM axons presenting a short or long hook that extended beyond the normal termination point observed in wild-type animals. For quantitation, young adult animals were anesthetized (10 µM levamisole in M9 buffer) on a 2% agar pad on glass slides and visualized with a Leica DM5000 B (CTR5000) epifluorescent microscope (40× oil-immersion objective). All strains were outcrossed at least twice with wild-type animals prior to scoring.
For autophagy analysis, young adult animals carrying the autophagosome marker, bggSi14 (Pmec-7mCherry::GFP::LGG-1/LC3), and a morphology marker specific for the mechanosensory neurons, muIs32 (Pmec-7GFP), were imaged as stated above. mCherry::LGG-1 signal was used to evaluate axonal autophagy while cell-specific GFP (muIs32) was used to ensure accurate identification of ALM neurons and their axon termination sites. For quantitation of LGG-1/LC3 levels, images were processed in Fiji/ImageJ software. Z-projections of ALM axon tips were generated for each genotype, and images were blinded for analysis using custom software. A region-of-interest (ROI) of 6 µm2 was drawn around the axon tip and ROI average pixel intensity was measured. For all CRISPR edited strains, only axons presenting failed termination that could be confidently identified were imaged and analysed.
C. elegans behavioural assays
Aged-synchronized animals (60–100) were grown until adulthood (72–80 h post laying) at 23°C on a fine layer of E. coli (OP50). Animal behaviour was recorded using a multi-worm tracker for 550 s, during which 45 plate taps were delivered every 10 s, after an initial 100 s of baseline recording. Response to tap is measured by the fraction of animals that reversed their locomotion within 2 s of the tap (reversal probability). Initial response was defined as the reversal probability of animals at the first tap. To evaluate habituation, we performed two types of behavioural analysis. First, we examined response to repeated tap as defined by the overall reversal probability from first to last tap based on an exponential best-fit curve. Second, we analysed habituation level, which is defined as the asymptote of the best-fit curve at the final stimulus. Further technical details were previously described.28 All strains were outcrossed four times prior to data acquisition for behavioural assays.
Structural analysis of MYCBP2 residues involved in MDCD
To predict the structure of the putative MYCBP2 zinc finger domain and MYCBP2 helical segment, the MYCBP2 sequence between amino acid residues 3233–3324 and 3542–4200 were used, respectively. AlphaFold predictions were run with ColabFold:AlphaFold2 using MMseqs2, a publicly available GitHub notebook.40 Five different AlphaFold predictions for each run were aligned and analysed in PyMOL (https://pymol.org/2/) and ChimeraX.41 The predicted models with the highest predicted local distance difference test (plDDT) scores are shown. The zinc ions were manually placed in the centre of the zinc fingers. Crystal structure (PDB ID: 6T7F) was used for the analysis involving MYCBP2 catalytic core. MacPyMOL (The PyMOL Molecular Graphics System, Version 1.7.6.6 Enhanced for Mac OS X Schrödinger, LLC), UCSF ChimeraX (version 1.4.dev202203170703) and Adobe Illustrator CS4 (Version 14.0.0) were used to generate figures.
Statistical analysis
GraphPad Prism software was used for all statistical analysis.
Quantitation of immunoblots
Densitometry was performed using Fiji/ImageJ software. Mean RPM-1::GFP CRISPR band intensity was determined for three replicates run simultaneously from two independent experiments. Dots in graphs represent individual replicates. Significance was tested using unpaired Student’s t-test with Bonferroni correction and defined as P < 0.05.
Axon termination analysis
Significance was tested using unpaired Student’s t-test and defined as P < 0.05. Data were derived from six to eight counts (20–30 animals/count) obtained from three or more independent experiments for each genotype.
Initial mechanical stimulation response
Significance was tested using unpaired Student’s t-test and defined as P < 0.05. Data were derived from 29–30 individual replicates (one replicate is defined as 60–100 worms on a single plate) from eight independent experiments for all genotypes, except for RPM-1::GFP R2669stop CRISPR which was derived from 16 replicates from four independent experiments.
Habituation analysis
For repeated mechanical stimulation response, significance was tested using two-way ANOVA and defined as P < 0.05. Comparisons were made based on data from 29–30 replicates (60–100 animals per replicate) for each genotype obtained from eight independent experiments, except for RPM-1::GFP R2669stop CRISPR which was based on 16 replicates from four independent experiments. In Fig. 4C, we present the exponential fit curve that was generated using means of individual responses at each time point from 29–30 replicates. However, we note that raw data were used for two-way ANOVA comparisons. In Fig. 4D, habituation level was the asymptote generated from the exponential fit curve. Significance was tested using unpaired Student’s t-test and defined as P < 0.05.
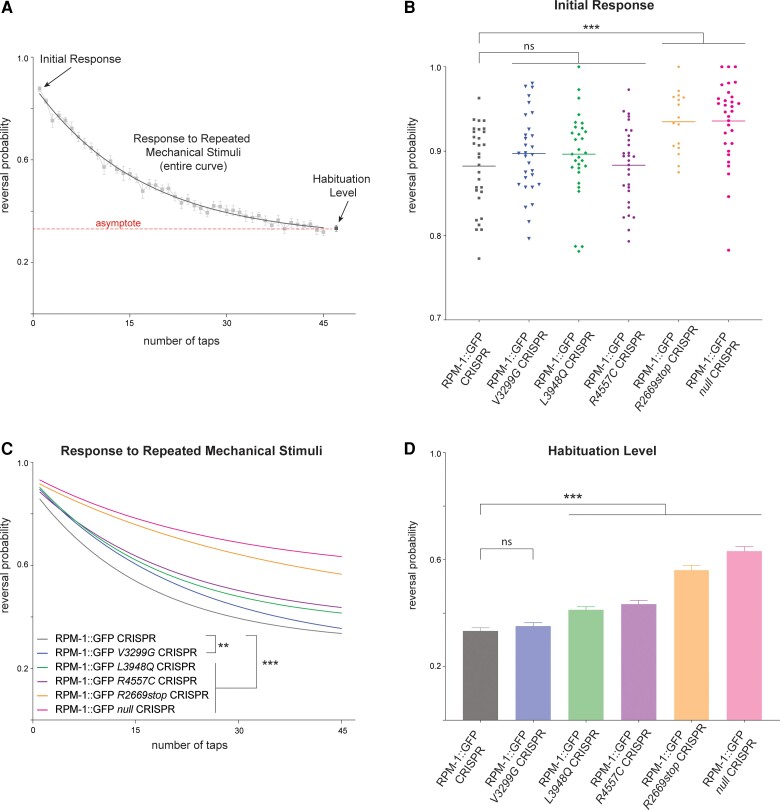
Figure 4.
C. elegans behavioural responses are altered by MDCD-related variants. (A) Multiple behavioural responses to mechanical stimulation were evaluated using computationally automated assays in C. elegans. Shown is raw control data (boxes) for RPM-1::GFP CRISPR animals and analysis points for initial response, response to repeated mechanical stimulation, and habituation level. Exponential fit curve across assays (solid line) determines habituation in two ways, response to repeated mechanical stimulation (overall curve) and habituation level (value of fit curve asymptote, dashed line). (B) Quantitation of initial response for indicated genotypes. RPM-1::GFP CRISPR animals carrying further CRISPR edits for MDCD-related missense variants have normal initial responses to mechanical stimulation. RPM-1::GFP R2669stop and null CRISPR animals show higher sensitivity to initial stimulation. (C) Quantitation showing all MDCD-related variants display abnormalities in response to repeated mechanical stimulation. See Supplementary Fig. 7 for raw data plots for all genotypes. (D) Quantitation of habituation levels for indicated genotypes. Habituation is impaired in RPM-1::GFP CRISPR animals carrying L3948Q, R4557C, and R2669stop MDCD-related variants. For C and D, averages are shown from 29–30 replicates (60–100 animals per replicate) for each genotype obtained from eight independent experiments, except for R2669stop where averages represent 16 replicates from four experiments. Error bars represent SEM. For B, data-points represent each replicate. For B and D, significance was determined using Student’s t-test. For C, significance was determined using two-way ANOVA. **P < 0.01; ***P < 0.001; ns = not significant.
Quantitation of autophagy levels
Significance was tested using unpaired Student’s t-test with Bonferroni correction and defined as P < 0.05. Data were derived from 10 or more animals imaged from three or more independent experiments for each genotype. Individual dots in graphs represent outcomes from one axon from an individual animal.
Data availability
Data collected and analysed for this manuscript are available upon request.
Results
Patients with MYCBP2 variants have neurobehavioural phenotypes and axon defects
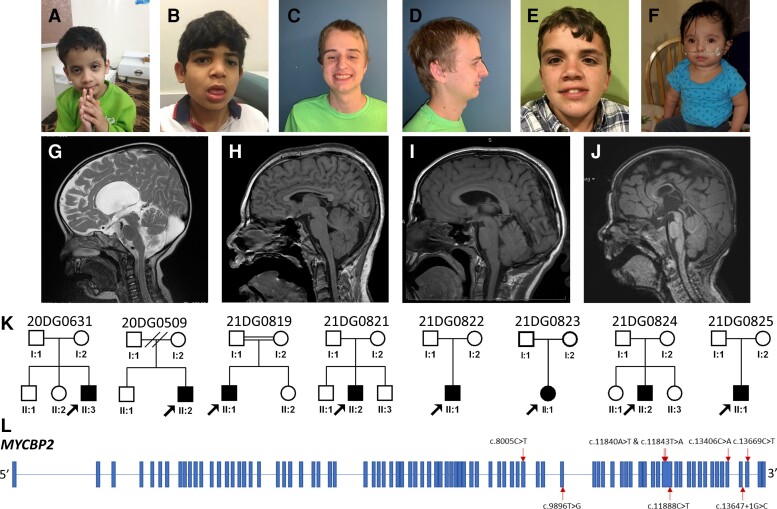
As part of the clinical workup of patients with neurodevelopmental phenotypes, eight independently identified patients underwent exome sequencing (Fig. 1 and Table 1). Patients were found to have variable facial dysmorphisms (Fig. 1A–F). Corpus callosum defects were present in four of the five patients who underwent brain imaging and ranged from mild thinning to complete agenesis (Fig. 1G–J). Table 1 summarizes clinical findings for all eight patients (see Supplementary Table 1 and Supplementary material for further details). Neurodevelopmental deficits observed in the patients included developmental delay (7/8 patients), intellectual disability (4/8), autistic features (5/8) and epilepsy (3/8). Accompanying comorbidities were uncommon. We observed two patients with bilateral hearing impairment. One case each was found for gut malrotation and feeding difficulty, unilateral renal agenesis, hypopituitarism and hypothyroidism, and large palpebral fissures.
Figure 1.
Overview of patients with MYCBP2 variants identified in this study. (A–G) Clinical pictures highlighting the variable facial dysmorphisms in patients (see Supplementary Table 1 for further details): (A) 20DG0631, (B) 21DG0819, (C and D) 21DG0821, (E) 20DG0509, and (F) 21DG0825. (G–K) Sagittal MRI images highlighting the range of corpus callosum defects in the study cohort: (G) 20DG0631, with agenesis of corpus callosum, (H and I) 21DG0819 and 20DG0509, respectively, with mild thinning of the corpus callosum, (J) 21DG0822 with dysgenesis of the corpus callosum. (K) Pedigrees for all the patients participating in this study. (L) Schematic of MYCBP2 with variants annotated.
Table 1.
Summary of the clinical data and the variants for each case
| Case | 21DG0819 | 21DG0821 | 20DG0509 | 21DG0822 | 20DG0631 | 21DG0823 | 21DG0824 | 21DG0825 |
|---|---|---|---|---|---|---|---|---|
| Variant | ||||||||
| cDNA | NM_015057.4:c.8005C>T; p.(R2669*) | NM_015057.4:c.9896T>G; p.(V3299G) | NM_015057.4:c.11840A>T;p.(D3947V) | NM_015057.4:c.11843T>A;p.(L3948Q) | NM_015057.4:c.11888C>T; p.(T3963I) | NM_015057.4:c.13647+1 G>C | NM_015057.4:c.13669C>T; p.(R4557C) | NM_015057.4:c.13406C>A;p.(T4469K) |
| Phenotype/clinic | ||||||||
| Sex | Male | Male | Male | Male | Male | Female | Male | Male |
| Age, years | 12 | 20 | 16 | 3 | 6 | 4 | 6 | 2.5 |
| Intellectual disability | Severe | No | Yes | Not formally tested | Yes | No | Moderate | N/A |
| Autism | Yes | Yes | Yes | None | Mild autistic features | None | Yes | N/A |
| Epilepsy/seizure | Seizures | None | None | Abnormal EEG | None | None | Seizures | None |
| Developmental delay | Yes | Yes | Yes | Yes | Yes | None | Yes | Yes |
| Gross-Motor | Early gross motor skills normal | No | Delayed | Motor delay | Yes | Her motor development is behind given absent leg | Yes | Yes |
| Fine-Motor | Unknown | Yes | Delayed | Motor delay | Yes | None | Yes | None |
| Speech | Currently non-verbal | Yes | Difficult to understand speech | Speech delay | Yes | None | Mostly non-verbal | Three deliberate words |
| Corpus callosum | Mild thinning | N/A | Thinning of the corpus callosum | Dysgenesis of the corpus callosum | Agenesis of corpus callosum | N/A | Unremarkable | N/A |
| Optic nerve | No abnormalities noted | Unremarkable | Unknown | None | hypoplasia of right optic nerve extending to the optic chiasma | N/A | Unremarkable | N/A |
| Optic chiasm | No abnormalities noted | Unknown | Unknown | None | Yes | N/A | Unremarkable | N/A |
| Fundus examination | No retinal involvement | No retinal involvement | Normal | Fundus oculi sharply confined | Cone dystrophy | No ophthalmology exam | Unremarkable | Retinopathy of prematurity |
| Impaired vision | Strabismus | Unknown | Left amblyopia and congenital L posterior capsule cataract | Esotropia (right), strabismus, astigmatism, and hyperopia. | Yes | No ophthalmology exam | Mild myopia and strabismus | Strabismus |
| Facial dysmorphism | Yes | Slightly large ears | Yes | Yes | Yes | Yes | Does not appear dysmorphic | Does not appear dysmorphic |
| Hearing loss (type) | Hearing believed to be normal | None | Congenital bilateral mild sensorineural hearing loss | Bilateral hearing impairment | No | None | None | None |
| Other findings | N/A | Celiac disease | Two posterior hair whorls, widows peak | N/A | Hypopituitarism and hypothyroidism | None observed | None | Mild hypospadias and bilateral inguinal hernia |
In each of the eight patients, exome sequencing and subsequent variant analysis independently identified de novo variants in MYCBP2 (NM_015057.4), making it a likely candidate (Fig. 1K and L, Table 1 and Supplementary Table 1). Collaborative evaluation of patients and MYCBP2 variants was enabled through GeneMatcher.37 The variants included six missense, one nonsense, and one splicing variant. The candidacy of these variants is further supported by consistent in silico analysis of the variants (Supplementary Table 2) and the observation that MYCBP2 is constrained against loss of function (pLI = 1) and missense variants (Z = 6.05).42
RNA sequencing (RNA-seq) of one patient, 21DG0819 [NM_015057.4:c.8005C>T; p.(R2669*)], showed decreased overall MYCBP2 expression and allele-specific expression (ASE) at this locus (Supplementary Fig. 1A and B). This suggests the occurrence of NMD resulting from an early stop codon. To confirm this finding, we performed RT-PCR on patient-derived fibroblasts at baseline and after treatment with cycloheximide. As shown in Supplementary Fig. 1C, the near complete absence of the mutant allele was reversed after cycloheximide treatment. Of note, this patient also had candidate variants in two other genes. One de novo heterozygous variant in G3BP1 (NM_005754.2:c.351+1G>T) creates an in-frame 24 amino acid deletion, and he also has a homozygous frameshift variant in PPP2CB (NM_001009552.1: c.352delC; p.H118ThrfsTer41). Neither of these genes is known to be associated with any disease in humans so their contribution to the patient phenotype (if any) remains unknown. Furthermore, the pLI score of PPP2CB is 0.34 and homozygous knockout mice are viable and fertile suggesting loss-of-function variants in PPP2CB may be tolerated.43
In sum, our patient characterization reveals a new neurodevelopmental disorder which we propose to name MYCBP2-related developmental delay with corpus callosum defects (MDCD). It features a broad spectrum of neurodevelopmental deficits, chief among them being developmental delay and corpus callosum abnormalities. Our genetic analysis reveals that de novo variants in MYCBP2 are associated with MDCD and may be causal.
MDCD-related variants do not affect protein localization or stability in vivo
To test the functional effects of MYCBP2 variants identified in our patient cohort, we turned to C. elegans as an in vivo genetic model system. The value of C. elegans as our model of choice is illustrated on several levels. C. elegans has been used to functionally evaluate pathological variants associated with neurodevelopmental disorders.44–46 CRISPR/Cas9 gene editing has expanded how we use C. elegans to study the molecular genetic basis of neurodevelopmental disorders.32,47–49 Finally, C. elegans has a single well-conserved MYCBP2 orthologue, RPM-1, that is an important regulator of axon development.26,50–52 Thus, C. elegans is well-suited for testing the functional effects of MYCBP2 variants in vivo.
To facilitate our studies in C. elegans, we first needed to determine which human MYCBP2 variants affect conserved domains and residues in C. elegans RPM-1. To do so, we mapped the six missense variants and one nonsense variant onto both MYCBP2 and RPM-1 protein architecture (Fig. 2A and Supplementary Figs 2 and 3). The splicing variant was not considered because MYCBP2 splice junctions and intronic sequence are not well conserved between humans and C. elegans. Our analysis revealed that several MYCBP2 variants occur in conserved protein domains including the RING and tandem cysteine (TC) domains required for ubiquitin ligase activity, and a conserved domain (CD) of unknown function. One variant (R2669stop) would generate at minimum a C-terminal truncation that removes several domains including the RING and TC domains, or could result in NMD similar to what was observed for the human MYCBP2 variant. Interestingly, we identified three MYCBP2 missense variants (V3299G, L3948Q, and R4557C) that affect conserved residues (L2544G, L3070Q and R3645C) in C. elegans RPM-1 (Fig. 2A and Supplementary Figs 2 and 3). Based on structure predictions by AlphaFold in conjunction with a crystal structure for the C-terminal region of MYCBP2,53 these three residues are located in a putative zinc finger domain, an α-helical segment preceding the C-terminal catalytic core and the TC domain, respectively (Supplementary Fig. 4). V3299G is expected to compromise the proper fold of the zinc finger domain, while L3948Q would affect the helical packing of the α-helical region. Being situated right next to the catalytic cysteine C4558, the R4557C variant would most likely impair the sequential transfer of ubiquitin by either competing with C4558 for conjugating the modifier or hindering the catalytic cascade. Based on conservation and structural predictions, these variants were prioritized for further evaluation using C. elegans. For clarity of presentation, we will use the human MYCBP2 variant nomenclature going forward.
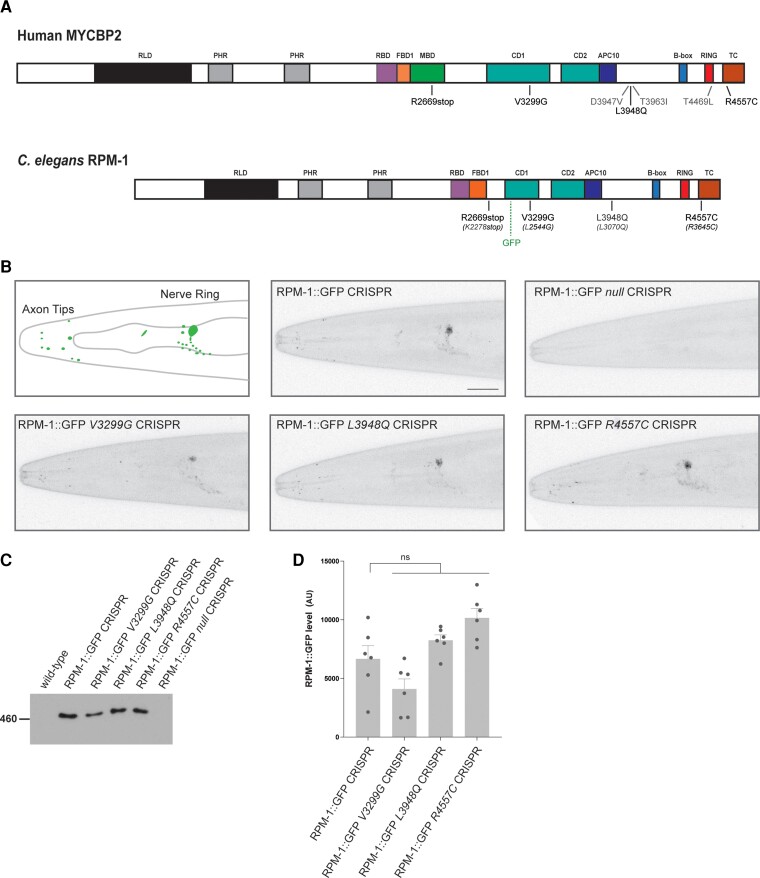
Figure 2.
CRISPR editing MDCD-related variants into rpm-1 does not impair protein localization or stability in C. elegans. (A) Schematic of human MYCBP2 and its C. elegans orthologue RPM-1. Shown are protein sequence changes caused by MYCBP2 missense and nonsense variants from MDCD patients. Also noted is the location of the GFP inserted using CRISPR/Cas9 gene engineering in C. elegans and annotated protein domains. RCC1 like domain (RLD), Pam/Highwire/RPM-1 (PHR) domain, RAE-1 binding domain (RBD), FSN-1 binding domain (FBD), conserved domain of unknown function (CD), anaphase-promoting complex subunit 10 like domain (APC10). (B) CRISPR engineering was used to fuse GFP with endogenous RPM-1. CRISPR editing then introduced conserved MDCD-related variants (V3299G, L3948Q and R4557C) or a known null mutation (C4587Y) into RPM-1::GFP CRISPR animals. Shown is a schematic of RPM-1::GFP CRISPR localization at axon tips and the nerve ring in the head of C. elegans (left). Representative confocal images are shown for all genotypes. Note localization and levels of RPM-1::GFP are normal in CRISPR edited animals carrying MDCD-related variants (V3299G, L3948Q and R4557C). (C) Immunoprecipitation of RPM-1::GFP from whole C. elegans lysates for indicated genotypes. Representative of three independent experiments. (D) Quantitation of RPM-1::GFP for indicated genotypes. Means are shown for six replicates from three independent experiments for each genotype. Error bars represent SEM. Significance determined using Student’s t-test with Bonferroni correction. ns = not significant. Scale bar = 20 μm.
To test MYCBP2 missense variants for potential effects on protein trafficking and function, we CRISPR/Cas9 edited similar mutations into the corresponding, conserved residues of RPM-1 in C. elegans. To evaluate protein localization, expression and stability, genome edits were performed in a C. elegans strain where we CRISPR engineered GFP onto endogenous RPM-1 (RPM-1::GFP CRISPR). Previous studies showed that this GFP insertion does not affect RPM-1 function or localization.20,26 As a result, we were able to generate animals in which endogenous RPM-1::GFP harbours mutations corresponding to the human MYCBP2 variants V3299G, L3948Q and R4557C. All three CRISPR edits were confirmed by sequencing (Supplementary Fig. 5A–C). As a control, we generated RPM-1::GFP C3674Y animals mimicking a known null mutation (RPM-1::GFP null CRISPR).54 Consistent with prior findings,20,26 RPM-1::GFP CRISPR localized to discrete puncta at axon tips located in the head and in a large bundle of axons that form the nerve ring of C. elegans (Fig. 2B). As expected of a protein null allele, we did not observe any GFP fluorescence in RPM-1::GFP null animals (Fig. 2B). RPM-1::GFP V3299G, L3948Q and R4557C CRISPR strains all displayed RPM-1::GFP puncta at axon tips in the head and in the nerve ring (Fig. 2B). This localization pattern was similar to what was observed in RPM-1::GFP CRISPR animals (Fig. 2B).
To expand these studies, we turned to a native biochemical approach to evaluate the effect of the MDCD-related variants on endogenous RPM-1 stability. RPM-1::GFP CRISPR was detected by anti-GFP immunoprecipitation (IP) from whole animal lysates (Fig. 2C and Supplementary Fig. 6). Wild-type animals and RPM-1::GFP null CRISPR animals were used as negative controls. Similar to results from imaging experiments, we were able to detect RPM-1::GFP harbouring patient-related missense variants using IP (Fig. 2C). Quantitation indicated that the RPM-1::GFP signal in missense MDCD-related variants was not significantly different compared to levels in control RPM-1::GFP CRISPR animals (Fig. 2D).
Taken as a whole, these results indicate that CRISPR editing MYCBP2 patient-related variants into endogenous RPM-1 in C. elegans does not affect localization to axonal compartments (Fig. 2B) or global protein stability across the nervous system (Fig. 2C and D). As a result, it is likely that MYCBP2 protein trafficking and stability are normal in patients harbouring MYCBP2 V3299G, L3948Q and R4557C missense variants.
CRISPR edited MDCD-related variants impair axon development in C. elegans
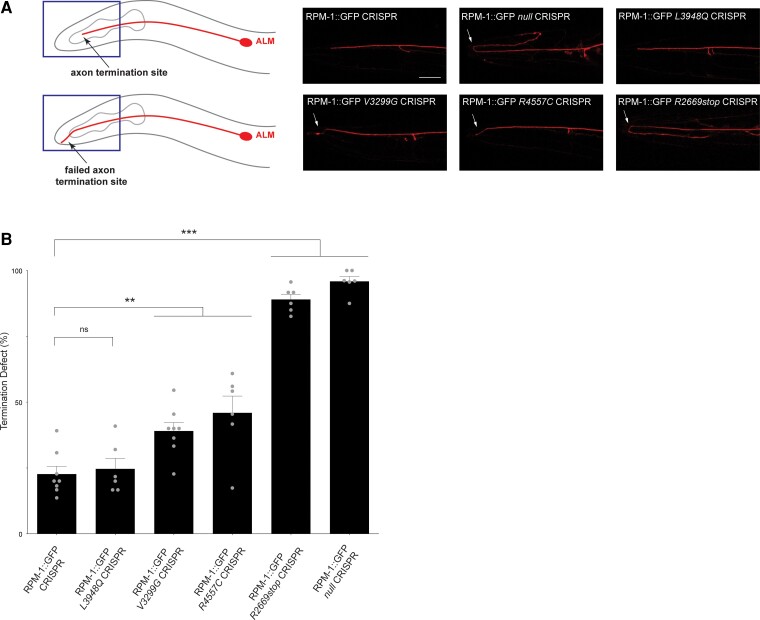
Since MYCBP2 patient-related variants do not affect protein localization and stability, we next tested their functional effects. To do so, we initially evaluated axon development in C. elegans carrying these MDCD-related variants. One of the major defects observed in the mechanosensory neurons of rpm-1 loss-of-function mutants is failure to terminate axon growth. Previous developmental studies showed that this results from impaired growth cone collapse in rpm-1 loss-of-function (lf) mutants.55
We examined axon termination in the ALM mechanosensory neurons using a transgene that expresses RFP specifically in these neurons (Fig. 3A). C. elegans has two ALM neurons on either side of the body and each has a single axon that terminates growth at a precise anatomical location near the nose (Fig. 3A). Similar to prior observations with traditional rpm-1 (lf) mutants,19,55 we observed severe axon termination defects in RPM-1::GFP null CRISPR mutants (Fig. 3A). Quantitation indicated that this phenotype occurs at high frequency (Fig. 3B). The first MDCD-related variant we examined, RPM-1::GFP L3948Q CRISPR, did not display axon termination defects compared to RPM-1::GFP CRISPR controls (Fig. 3A and B). However, we did observe axon termination defects, albeit less severe than in RPM-1::GFP null, in animals carrying two MDCD-related variants, RPM-1::GFP V3299G and R4557C CRISPR (Fig. 3A). Quantitation indicated that these defects were significant compared to RPM-1::GFP controls, but less frequent than defects in RPM-1::GFP null mutants (Fig. 3B).
Figure 3.
MDCD-related variants affect in vivo axon development in C. elegans. (A) Top: Axon termination site (arrow) for ALM mechanosensory neuron of C. elegans. Bottom: Failed termination (arrow) observed in MDCD-related variants. Shown are representative images of ALM axons for indicated genotypes visualized using transgenic, cell-specific RFP. Axon termination defects (arrow) are observed in RPM-1::GFP null CRISPR animals and in RPM-1::GFP CRISPR animals harbouring MDCD-related variants (V3299G, R4557C and R2669stop). (B) Quantitation of axon termination defects for indicated genotypes. Means are shown from six to eight counts (20–30 animals per count) for each genotype, and error bars represent SEM. Significance determined using Student’s t-test. **P < 0.01; ***P < 0.001; ns = not significant. Scale bar = 20 μm.
An early stop codon in MYCBP2, R2669*, was also identified in one patient (Fig. 2A and Table 1). Although our results indicate that NMD occurs with this MYCBP2 variant (Supplementary Fig. 1), we wanted to independently test the functional effects of a similar mutation using RPM-1 in C. elegans as a readout. Because R2669* occurs in the MYC binding domain (MBD) which is not present in C. elegans RPM-1, we generated a CRISPR edited animal with a premature stop codon that occurs at an approximately similar location in rpm-1 (Fig. 2A and Supplementary Figs 3 and 5). RPM-1::GFP R2669stop CRISPR mutants displayed severe axon termination defects (Fig. 3A), which occurred at high penetrance (Fig. 3B).
These results indicate that two MDCD-related variants, V3299G and R4557C, result in partial loss of function and impair axon development. Results with the R2669stop variant suggest it is likely to be a strong loss-of-function allele. Thus, functional genetic outcomes indicate that multiple MDCD-related variants alter axon development in a model organism in vivo and result in variable loss-of-function.
MDCD-related variants cause behavioural abnormalities in C. elegans
Next, we tested how MDCD-related variants influence whole animal behaviour using C. elegans. We used an automated behavioural assay to monitor animal responses to mechanosensory input and assessed multiple behavioural readouts (Fig. 4A): (i) response to the first mechanical stimulation, which is a measure of initial sensitivity; and (ii) habituation to repeated mechanosensory input, which is a simple form of learning. This was analysed in two ways, overall response to repeated mechanical stimulation and habituation level.
We began by evaluating the initial response to mechanical stimulation in CRISPR edited animals carrying MDCD-related variants. As a positive control, we evaluated RPM-1::GFP null mutants, which showed a significant increase in initial response compared to RPM-1::GFP CRISPR control animals (Fig. 4B; see Supplementary Fig. 7 for all raw data). None of the MDCD-related missense variants resulted in altered initial sensitivity to mechanical stimulation (Fig. 4B). In contrast, all missense variants displayed significant defects in their response to repeated mechanical stimulation compared to the control RPM-1::GFP CRISPR strain (Fig. 4C). Finally, we calculated the habituation level for CRISPR strains with missense variants (Fig. 4D). As expected, RPM-1::GFP null animals showed habituation defects in which animals are still sensitive following repeated mechanical stimulation (Fig. 4D). Both RPM-1::GFP L3948Q and R4557C CRISPR mutants also showed significant defects in habituation level (Fig. 4D). In contrast, habituation level was not altered in RPM-1::GFP V3299G CRISPR animals (Fig. 4D). Similar to RPM-1::GFP null CRISPR animals, RPM-1::GFP R2669stop CRISPR mutants displayed significant abnormalities in initial response to mechanical stimulation (Fig. 4B) and habituation (Fig. 4C and D).
Outcomes from our behavioural experiments support several conclusions. First, all MDCD-related variants we tested showed impaired habituation. Second, V3299G, L3948Q and R4557C variants showed partial loss-of-function phenotypes while the R2669stop variant is likely to be a strong loss-of-function allele. These results from multiple behavioural readouts provide further evidence indicating that MDCD-related variants impair gene function.
Increased axonal autophagy in MDCD-related variants
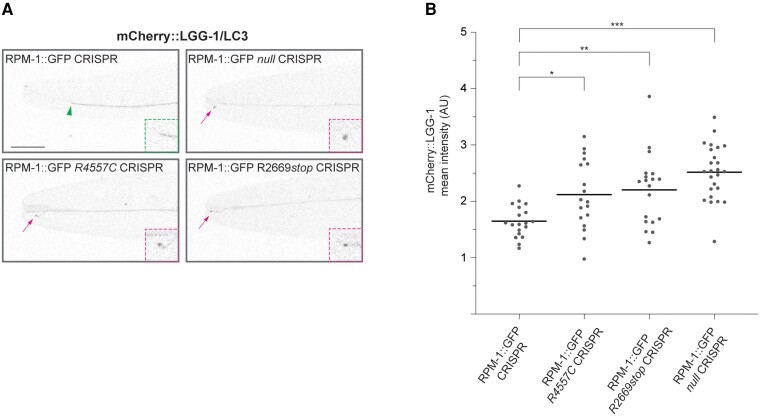
Previous studies showed that RPM-1 ubiquitin ligase activity inhibits initiation of autophagy and restricts autophagosome formation broadly across the nervous system of C. elegans.26 Thus, we tested if the MDCD-related variants that are likely to affect ubiquitin ligase activity, R4557C and R2669stop, result in abnormal axonal autophagosome formation in ALM mechanosensory neurons.
To evaluate autophagy in ALM neurons, we used a single copy transgene that expresses the autophagosome marker mCherry::LGG-1/LC3 specifically in these neurons. Consistent with prior findings,26 we observed abnormal accumulation of mCherry::LGG-1 puncta at the tip of axons that displayed failed termination in RPM-1::GFP null CRISPR mutants (Fig. 5A). Quantitation indicated this was a significant increase compared to RPM-1::GFP CRISPR control animals (Fig. 5B).
Figure 5.
MDCD-related variants result in increased axonal accumulation of the autophagosome marker LGG-1/LC3. (A) Representative images showing mCherry::LGG-1/LC3 expressed in ALM mechanosensory neurons for indicated genotypes. Arrowhead indicates wild-type axon termination site. Arrows indicate impaired axon termination sites with mCherry::LGG-1 accumulation. Insets: Enlarged images of axon termination sites. (B) Quantitation of mCherry::LGG-1 at axon termination sites for indicated genotypes. Means are shown from 18 or more animals for each genotype. Significance determined using Student’s t-test with Bonferroni correction. *P < 0.05, ** P < 0.01; *** P < 0.001. Scale bar = 20 μm.
Similarly, we observed concentration of mCherry::LGG-1 puncta at the ALM axon tip in animals presenting a failed termination phenotype in RPM-1::GFP R4557C CRISPR and RPM-1::GFP R2669stop CRISPR mutants (Fig. 5A). Quantitative analysis demonstrated a significant increase in axonal LGG-1/LC3 in RPM-1::GFP R4557C CRISPR and RPM-1::GFP R2669stop CRISPR mutants compared to RPM-1::GFP CRISPR control animals (Fig. 5B). These findings suggest that two MDCD-related variants, R4557C and R2669stop, lead to impaired function and increased autophagosome formation in subcellular axonal compartments.
Discussion
MYCBP2 functions as both a ubiquitin ligase and signalling hub that regulates several events in nervous system development.16 Studies in mouse, zebrafish, Drosophila and C. elegans have shown that the respective orthologues Phr1, Phr/Esrom, Highwire and RPM-1 play conserved functions in axon and synapse development.19,21–25,54,56,57 In Phr1 loss of function mice, death occurs shortly after birth due to abnormal diaphragm innervation and impaired neuromuscular junction formation.25 With regard to axon development, one principal defect observed in C. elegans rpm-1 mutants is impaired termination of axon growth.19,50 In mammals, the cellular context is important for how Phr1 influences axon development. Phr1 mutant mice have brains with altered axon guidance and outgrowth including corpus callosum agenesis.25 In contrast, mammalian sensory neurons in the dorsal root ganglia display phenotypes consistent with failed axon termination in the periphery and in the spinal cord.21,58 While the signals that mediate axon termination are not completely understood, it is clear that MYCBP2 plays an important, conserved role in this stage of axon development.
As axons elongate, proper axon guidance is required to reach synaptic partners. Vertebrate models of MYCBP2/Phr1 deficiency have notable axon guidance and extension problems. Indeed, similar to patients harbouring MYCBP2 variants (Fig. 1), Phr1 mutant mice display corpus callosum defects.25Phr1 mutant mice show further signs of abnormal axon guidance including loss of the anterior commissure, internal capsule, and thalamocortical projections.25 Likewise, studies in zebrafish identified deficits in axon connections between brain hemispheres in Phr/Esrom mutants.59 To date, we still know relatively little about how Phr1 regulates axon guidance and extension in the mammalian brain. Nonetheless, it is valuable to consider some mechanisms that could be involved in generating corpus callosum defects in MDCD patients. Phr1 ubiquitin ligase activity can inhibit the TSC complex and influence mTOR signalling,60,61 which has been suggested to contribute to corpus callosum defects in Phr1 mutant mice.62 Studies with mammalian olfactory neurons and C. elegans suggest that Phr1 and rpm-1 interact genetically with the axon guidance cue Robo2/sax-3.63,64 Finally, Phr1 and RPM-1 also affect cytoskeletal organization, which influences the morphology, dynamics and collapse of axonal growth cones in mammals, zebrafish and C. elegans.21,55,65
Our findings here indicate that multiple patients with heterozygous de novo variants in MYCBP2 have a neurodevelopmental disorder. The majority of patients have corpus callosum defects suggesting this is a pathological hallmark of the disorder. Previous human genetic studies have begun to reveal roles for axon guidance cues, such as the DCC Netrin receptor and Semaphorins, in neurodevelopmental disorders.66,67 For example, patients with variants in DCC display corpus callosum defects,68,69 similar to our cohort with MYCBP2 variants (Fig. 1). Our findings expand upon these previous discoveries by showing that human MYCBP2, an intracellular signalling molecule that regulates axon development, is involved in corpus callosum development in humans and a novel neurodevelopmental disorder.
Anatomical defects observed in the corpus callosum are likely to contribute to developmental delay, intellectual disability, epilepsy, and autistic features in patients with MYCBP2 variants. However, as noted above, Phr1 mutant mice also have defects in several other major axon tracts of the brain.25 Additionally, MYCBP2 is an important regulator of synapse formation and maintenance in motor and sensory neurons.25,54,56,57,70 Thus, it is possible that abnormal axon development outside the corpus callosum, as well as altered synaptic connectivity or stability might further contribute to cognitive impairment and development delay in MDCD patients.
In our cohort of patients, we argue that the observed disorder and anatomical defects in corpus callosum are caused by haploinsufficiency involving partial or strong loss-of-function variants in MYCBP2. This is based on several lines of reasoning: (i) we independently identified multiple patients (eight) with de novo variants in MYCBP2 who display an overlapping spectrum of neurodevelopmental phenotypes and corpus callosum defects (Fig. 1 and Table 1); (ii) MYCBP2 is a highly constrained gene in the human population with very high scores on intolerance to haploinsufficiency (pLI of 1.0) and constrained against missense variation (Z score 6.05); (iii) prior findings from multiple rodent models demonstrated that impairing MYCBP2/Phr1 function results in corpus callosum defects25,62; (iv) We used CRISPR/Cas9 gene editing in C. elegans to demonstrate that several MDCD-related variants impair gene function resulting in abnormal axon development (Fig. 3) and abnormalities in behavioural habituation (Fig. 4). Our results indicate that deleterious MDCD-related variants impair function resulting in partial (V3299G, L3948Q and R4557C) and strong (R2669*) loss of function. Interestingly, patients with the most severe MDCD clinical presentation carry variants that yield the most severe phenotypes or display phenotypes across the widest range of assays in the in vivo model organism C. elegans. Although three mutations, D3947V, T3963I, and T4469L are not conserved in C. elegans, two of them (D3947V and T3963I) are found in close proximity to L3948 at an α-helical segment (Supplementary Fig. 4). The third variant, T4469L, is located in the middle of the RING domain (Supplementary Fig. 4). It is conceivable that these MDCD variants might negatively impact the structure and function of MYCBP2 similar to the three missense variants we characterized functionally in C. elegans.
Previous studies in C. elegans showed that RPM-1/MYCBP2 ubiquitin ligase activity restricts autophagy in the nervous system by ubiquitinating and degrading the autophagy initiating kinase UNC-51/ULK.26 In this way, RPM-1 restrains autophagosome formation in subcellular axonal compartments. Our results indicate that two MDCD-related variants result in increased accumulation of LGG-1/LC3 at sites of failed axon termination in C. elegans (Fig. 5). Thus, a cell biological readout of autophagy further indicates that MDCD-related variants result in loss of function. Our findings and these prior studies suggest that altered corpus callosum development, developmental delay and neurobehavioural abnormalities observed in patients with MDCD could result from increased neuronal autophagy. Importantly, our observations contribute to a growing body of evidence indicating that mutations that alter autophagy result in developmental disorders that affect the nervous system.71,72 For example, prior studies have shown that mutations in EPG5, a component of the autophagy machinery, cause Vici syndrome.15,73 Patients with Vici syndrome display developmental delay and corpus callosum agenesis similar to our MDCD cohort. Genetic variants in the core autophagy genes ATG5 and ATG7 also result in neurodevelopmental disorders that feature developmental delay, ataxia and intellectual disability.74,75 In the case of ATG7 variants, corpus callosum thinning was observed in several patients.75 Our findings now suggest that MDCD could be added to a growing number of congenital disorders of autophagy.
In summary, our findings with multiple human patients and an in vivo genetic model, C. elegans, suggest that de novo deleterious variants in MYCBP2 cause MDCD. Moving forward, further studies on the molecular genetic basis of MDCD using animal models and effects of MDCD-related variants on MYCBP2 signalling and biochemistry remain important. Moreover, human genetic studies aimed at identifying further MYCBP2 variants and their clinical outcomes will be necessary to fully define this new neurodevelopmental disorder.
Supplementary Material
Acknowledgements
First and foremost, we would like to thank the patients and their families for their participation in this study.
Contributor Information
Lama AlAbdi, Department of Zoology, College of Science, King Saud University, Riyadh 11362, Saudi Arabia; Department of Translational Genomics, Center for Genomic Medicine, King Faisal Specialist Hospital and Research Center, Riyadh 11564, Saudi Arabia.
Muriel Desbois, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA 98101, USA.
Domniţa-Valeria Rusnac, Department of Pharmacology, University of Washington School of Medicine, Seattle, WA 98195, USA; Howard Hughes Medical Institute, Department of Pharmacology, University of Washington, Seattle, WA 98195, USA.
Raashda A Sulaiman, Department of Medical Genetics, King Faisal Specialist Hospital and Research Center, Riyadh 11564, Saudi Arabia.
Jill A Rosenfeld, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Seema Lalani, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
David R Murdock, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Lindsay C Burrage, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Undiagnosed Diseases Network, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Ping Yee Billie Au, Department of Medical Genetics, Alberta Children’s Hospital Research Institute, Cumming School of Medicine, University of Calgary, Calgary, AB T2N 4N1, Canada.
Shelley Towner, Pediatric Genetics, University of Virginia, Charlottesville, VA 22903, USA.
William G Wilson, Pediatric Genetics, University of Virginia, Charlottesville, VA 22903, USA.
Lawrence Wong, Department of Genetics, Northern California Kaiser Permanente, Oakland, CA 94611, USA.
Theresa Brunet, Institute of Human Genetics, Klinikum rechts der Isar, School of Medicine, Technical University of Munich, 81675 Munich, Germany; Institute of Neurogenomics (ING), Helmholtz Zentrum München, German Research Center for Environmental Health, 85764 Neuherberg, Germany.
Gertrud Strobl-Wildemann, Department of Human Genetics, MVZ Humangenetik Ulm, 89073 Ulm, Germany.
Jennifer E Burton, Department of Genetics, University of Illinois College of Medicine at Peoria, Peoria, IL 61605, USA.
George Hoganson, Department of Genetics, University of Illinois College of Medicine at Peoria, Peoria, IL 61605, USA.
Kirsty McWalter, Genedx, Inc., 207 Perry Parkway, Gaithersburg, MD 20877, USA.
Amber Begtrup, Genedx, Inc., 207 Perry Parkway, Gaithersburg, MD 20877, USA.
Yuri A Zarate, Section of Genetics and Metabolism, Department of Pediatrics, University of Arkansas for Medical Sciences, Little Rock, AR 72202, USA.
Elyse L Christensen, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA 98101, USA.
Karla J Opperman, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA 98101, USA.
Andrew C Giles, Division of Medical Sciences, University of Northern British Columbia, Prince George, BC V2N 4Z9, Canada.
Rana Helaby, Department of Translational Genomics, Center for Genomic Medicine, King Faisal Specialist Hospital and Research Center, Riyadh 11564, Saudi Arabia.
Artur Kania, Institut de recherches cliniques de Montréal (IRCM), Montréal, QC H2W 1R7, Canada; Integrated Program in Neuroscience, McGill University, Montréal, QC H3A 2B4, Canada; Division of Experimental Medicine, McGill University, Montréal, QC H3A 2B2, Canada; Department of Anatomy and Cell Biology, McGill University, Montréal, QC H3A 0C7, Canada.
Ning Zheng, Department of Pharmacology, University of Washington School of Medicine, Seattle, WA 98195, USA; Howard Hughes Medical Institute, Department of Pharmacology, University of Washington, Seattle, WA 98195, USA.
Brock Grill, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA 98101, USA; Department of Pharmacology, University of Washington School of Medicine, Seattle, WA 98195, USA; Department of Pediatrics, University of Washington School of Medicine, Seattle, WA 98101, USA.
Fowzan S Alkuraya, Department of Translational Genomics, Center for Genomic Medicine, King Faisal Specialist Hospital and Research Center, Riyadh 11564, Saudi Arabia.
Funding
B.G. was supported by National Institutes of Health grant R01 NS072129. A.K. was supported by operating and project grants from the Canadian Institutes of Health Research (PJT-162225, MOP-77556, PJT-153053, and PJT-159839). N.Z. is a Howard Hughes Medical Institute Investigator. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number(s) U01HG007709. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would also like to thank Researchers Supporting Project number (RSP-2021/181), King Saud University, Riyadh, Saudi Arabia for providing funding for L.A. The project described was supported in part by the Clinical Translational Core at Baylor College of Medicine which is supported by the IDDRC grant number P50HD103555 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the National Institutes of Health.
Competing interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories. K.M. and A.B. are employees of GeneDx, Inc. N. Z. is a co-founder and Scientific Advisory Board Member of Coho Therapeutics Inc. and SEED Therapeutics Inc. The remaining authors declare no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. De Leon Reyes NS, Bragg-Gonzalo L, Nieto M. Development and plasticity of the corpus callosum. Development. 2020;147:dev189738. [DOI] [PubMed] [Google Scholar]
- 2. Seymour SE, Reuter-Lorenz PA, Gazzaniga MS. The disconnection syndrome. Basic findings reaffirmed. Brain. 1994;117(1):105–115. [DOI] [PubMed] [Google Scholar]
- 3. Gazzaniga MS. Forty-five years of split-brain research and still going strong. Nat Rev Neurosci. 2005;6:653–659. [DOI] [PubMed] [Google Scholar]
- 4. Hinkley LB, Marco EJ, Findlay AM, et al. . The role of corpus callosum development in functional connectivity and cognitive processing. PLoS One. 2012;7:e39804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sauerwein HC, Lassonde MC, Cardu B, Geoffroy G. Interhemispheric integration of sensory and motor functions in agenesis of the corpus callosum. Neuropsychologia. 1981;19:445–454. [DOI] [PubMed] [Google Scholar]
- 6. Jeeves MA, Silver PH, Jacobson I. Bimanual co-ordination in callosal agenesis and partial commissurotomy. Neuropsychologia. 1988;26:833–850. [DOI] [PubMed] [Google Scholar]
- 7. Edwards TJ, Sherr EH, Barkovich AJ, Richards LJ. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain. 2014; 137:1579–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paul LK, Brown WS, Adolphs R, et al. . Agenesis of the corpus callosum: Genetic, developmental and functional aspects of connectivity. Nat Rev Neurosci. 2007;8:287–299. [DOI] [PubMed] [Google Scholar]
- 9. Kendall BE. Dysgenesis of the corpus callosum. Neuroradiology. 1983;25:239–256. [DOI] [PubMed] [Google Scholar]
- 10. Bayram AK, Kutuk MS, Doganay S, et al. . An analysis of 109 fetuses with prenatal diagnosis of complete agenesis of corpus callosum. Neurol Sci. 2020;41:1521–1529. [DOI] [PubMed] [Google Scholar]
- 11. Heide S, Spentchian M, Valence S, et al. . Prenatal exome sequencing in 65 fetuses with abnormality of the corpus callosum: Contribution to further diagnostic delineation. Genet Med. 2020;22:1887–1891. [DOI] [PubMed] [Google Scholar]
- 12. Palmer EE, Mowat D. Agenesis of the corpus callosum: A clinical approach to diagnosis. Am J Med Genet C Semin Med Genet. 2014;166C:184–197. [DOI] [PubMed] [Google Scholar]
- 13. Doherty D, Chudley AE, Coghlan G, et al. . GPSM2 mutations cause the brain malformations and hearing loss in Chudley-McCullough syndrome. Am J Hum Genet. 2012;90:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Accogli A, Calabretta S, St-Onge J, et al. . De Novo pathogenic variants in N-cadherin cause a syndromic neurodevelopmental disorder with corpus collosum, axon, cardiac, ocular, and genital defects. Am J Hum Genet. 2019;105:854–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cullup T, Kho AL, Dionisi-Vici C, et al. . Recessive mutations in EPG5 cause vici syndrome, a multisystem disorder with defective autophagy. Nat Genet. 2013;45:83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grill B, Murphey RK, Borgen MA. The PHR proteins: Intracellular signaling hubs in neuronal development and axon degeneration. Neural Dev. 2016;11:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pao K-C, Wood NT, Knebel A, et al. . Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature. 2018;556:381–385. [DOI] [PubMed] [Google Scholar]
- 18. Desbois M, Crawley O, Evans PR, et al. . PAM Forms an atypical SCF ubiquitin ligase complex that ubiquitinates and degrades NMNAT2. J Biol Chem. 2018;293:13897–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schaefer AM, Hadwiger GD, Nonet ML. . rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. [DOI] [PubMed] [Google Scholar]
- 20. Opperman KJ, Grill B. RPM-1 is localized to distinct subcellular compartments and regulates axon length in GABAergic motor neurons. Neural Dev. 2014;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewcock JW, Genoud N, Lettieri K, Pfaff SL. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 2007;56:604–620. [DOI] [PubMed] [Google Scholar]
- 22. D’Souza J, Hendricks M, Le Guyader S, et al. . Formation of the retinotectal projection requires Esrom, an ortholog of PAM (protein associated with Myc). Development. 2005;132:247–256. [DOI] [PubMed] [Google Scholar]
- 23. Shin JE, DiAntonio A. Highwire regulates guidance of sister axons in the Drosophila mushroom body. J Neurosci. 2011;31:17689–17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borgen M, Rowland K, Boerner J, Lloyd B, Khan A, Murphey R. Axon termination, pruning, and synaptogenesis in the giant fiber system of Drosophila melanogaster is promoted by highwire. Genetics. 2017;205:1229–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bloom AJ, Miller BR, Sanes JR, DiAntonio A. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 2007;21:2593–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crawley O, Opperman KJ, Desbois M, et al. . Autophagy is inhibited by ubiquitin ligase activity in the nervous system. Nat Commun. 2019;10:5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang C, Zheng X, Zhao H, et al. . A permissive role of mushroom body alpha/beta core neurons in long-term memory consolidation in Drosophila. Curr Biol. 2012;22:1981–1989. [DOI] [PubMed] [Google Scholar]
- 28. Giles AC, Opperman KJ, Rankin CH, Grill B. Developmental function of the PHR protein RPM-1 is required for learning in Caenorhabditis elegans. G3 (Bethesda). 2015;5:2745–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crawley O, Giles AC, Desbois M, Kashyap S, Birnbaum R, Grill B. A MIG-15/JNK-1 MAP kinase cascade opposes RPM-1 signaling in synapse formation and learning. PLoS Genet. 2017;13:e1007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bozorgmehr T, Ardiel EL, McEwan AH, Rankin CH. Mechanisms of plasticity in a caenorhabditis elegans mechanosensory circuit. Front Physiol. 2013;4:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leussis MP, Bolivar VJ. Habituation in rodents: A review of behavior, neurobiology, and genetics. Neurosci Biobehav Rev. 2006;30:1045–1064. [DOI] [PubMed] [Google Scholar]
- 32. McDiarmid TA, Belmadani M, Liang J, et al. . Systematic phenomics analysis of autism-associated genes reveals parallel networks underlying reversible impairments in habituation. Proc Natl Acad Sci U S A. 2020;117:656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wolman MA, Jain RA, Marsden KC, et al. . A genome-wide screen identifies PAPP-AA-mediated IGFR signaling as a novel regulator of habituation learning. Neuron. 2015;85:1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neale BM, Kou Y, Liu L, et al. . Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kosmicki JA, Samocha KE, Howrigan DP, et al. . Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bertoli-Avella AM, Kandaswamy KK, Khan S, et al. . Combining exome/genome sequencing with data repository analysis reveals novel gene-disease associations for a wide range of genetic disorders. Genet Med. 2021;23:1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sobreira N, Schiettecatte F, Valle D, Hamosh A. Genematcher: A matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Monies D, Abouelhoda M, AlSayed M, et al. . The landscape of genetic diseases in Saudi Arabia based on the first 1000 diagnostic panels and exomes. Hum Genet. 2017;136:921–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murdock DR, Dai H, Burrage LC, et al. . Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. J Clin Invest. 2021;131:e141500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. Colabfold: Making protein folding accessible to all. Nat Methods. 2022;19:679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goddard TD, Huang CC, Meng EC, et al. . UCSF Chimerax: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karczewski KJ, Francioli LC, Tiao G, et al. . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gu P, Qi X, Zhou Y, Wang Y, Gao X. Generation of Ppp2Ca and Ppp2Cb conditional null alleles in mouse. Genesis. 2012;50:429–436. [DOI] [PubMed] [Google Scholar]
- 44. Tong X-J, Hu Z, Liu Y, Anderson D, Kaplan JM. A network of autism linked genes stabilizes two pools of synaptic GABA(A) receptors. eLife. 2015;4:e09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Opperman KJ, Mulcahy B, Giles AC, et al. . The HECT family ubiquitin ligase EEL-1 regulates neuronal function and development. Cell Rep. 2017;19:822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu B, Mak JCH, Morris AP, et al. . Functional analysis of epilepsy-associated variants in STXBP1/Munc18-1 using humanized Caenorhabditis elegans. Epilepsia. 2020;61:810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wong WR, Brugman KI, Maher S, et al. . Autism-associated missense genetic variants impact locomotion and neurodevelopment in Caenorhabditis elegans. Hum Mol Genet. 2019;28:2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang D, Dao M, Muntean BS, Giles AC, Martemyanov KA, Grill B. Genetic modeling of GNAO1 disorder delineates mechanisms of Galphao dysfunction. Hum Mol Genet. 2022;31:510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Di Rocco M, Galosi S, Lanza E, et al. . Caenorhabditis elegans provides an efficient drug screening platform for GNAO1-related disorders and highlights the potential role of caffeine in controlling dyskinesia. Hum Mol Genet. 2022;31:929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Desbois M, Opperman KJ, Amezquita J, Gaglio G, Crawley O, Grill B. Ubiquitin ligase activity inhibits Cdk5 to control axon termination. PLoS Genet. 2022;18:e1010152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baker ST, Opperman KJ, Tulgren ED, Turgeon SM, Bienvenut W, Grill B. RPM-1 Uses both ubiquitin ligase and phosphatase-based mechanisms to regulate DLK-1 during neuronal development. PLoS Genet. 2014;10:e1004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tulgren ED, Turgeon SM, Opperman KJ, Grill B. The nesprin family member ANC-1 regulates synapse formation and axon termination by functioning in a pathway with RPM-1 and beta-catenin. PLoS Genet. 2014;10:e1004481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mabbitt PD, Loreto A, Dery MA, et al. . Structural basis for RING-Cys-Relay E3 ligase activity and its role in axon integrity. Nat Chem Biol. 2020;16:1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. [DOI] [PubMed] [Google Scholar]
- 55. Borgen MA, Wang D, Grill B. RPM-1 regulates axon termination by affecting growth cone collapse and microtubule stability. Development. 2017;144:4658–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, Goodman CS. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. [DOI] [PubMed] [Google Scholar]
- 57. Borgen MA, Giles AC, Wang D, Grill B. Synapse maintenance is impacted by ATAT-2 tubulin acetyltransferase activity and the RPM-1 signaling hub. eLife. 2019;8:e44040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Holland S, Coste O, Zhang DD, Pierre SC, Geisslinger G, Scholich K. The ubiquitin ligase MYCBP2 regulates transient receptor potential vanilloid receptor 1 (TRPV1) internalization through inhibition of p38 MAPK signaling. J Biol Chem. 2011;286:3671–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hendricks M, Mathuru AS, Wang H, Silander O, Kee MZ, Jesuthasan S. Disruption of Esrom and Ryk identifies the roof plate boundary as an intermediate target for commissure formation. Mol Cell Neurosci. 2008;37:271–283. [DOI] [PubMed] [Google Scholar]
- 60. Murthy V, Han S, Beauchamp RL, et al. . Pam and its ortholog highwire interact with and may negatively regulate the TSC1.TSC2 complex. J Biol Chem. 2004;279:1351–1358. [DOI] [PubMed] [Google Scholar]
- 61. Han S, Witt RM, Santos TM, Polizzano C, Sabatini BL, Ramesh V. Pam (protein associated with Myc) functions as an E3 ubiquitin ligase and regulates TSC/mTOR signaling. Cell Signal. 2008;20:1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Han S, Kim S, Bahl S, et al. . The E3 ubiquitin ligase, protein associated with Myc (Pam) regulates mammalian/mechanistic target of rapamycin complex 1 (mTORC1) signaling in vivo through N- and C-terminal domains. J Biol Chem. 2012;287:30063–30072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. James G, Key B, Beverdam A. The E3 ubiquitin ligase Mycbp2 genetically interacts with Robo2 to modulate axon guidance in the mouse olfactory system. Brain Struct Funct. 2013;219:861–874. [DOI] [PubMed] [Google Scholar]
- 64. Li H, Kulkarni G, Wadsworth WG. RPM-1, a Caenorhabditis elegans protein that functions in presynaptic differentiation, negatively regulates axon outgrowth by controlling SAX-3/robo and UNC-5/UNC5 activity. J Neurosci. 2008;28:3595–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bremer J, Marsden KC, Miller A, Granato M. The ubiquitin ligase PHR promotes directional regrowth of spinal zebrafish axons. Commun Biol. 2019;2:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Battum EY, Brignani S, Pasterkamp RJ. Axon guidance proteins in neurological disorders. Lancet Neurol. 2015;14:532–546. [DOI] [PubMed] [Google Scholar]
- 67. Engle EC. Human genetic disorders of axon guidance. Cold Spring Harb Perspect Biol. 2010;2:a001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jamuar SS, Schmitz-Abe K, D’Gama AM, et al. . Biallelic mutations in human DCC cause developmental split-brain syndrome. Nat Genet. 2017;49:606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Srour M, Riviere JB, Pham JM, et al. . Mutations in DCC cause congenital mirror movements. Science. 2010;328:592. [DOI] [PubMed] [Google Scholar]
- 70. Burgess RW, Peterson KA, Johnson MJ, Roix JJ, Welsh IC, O’Brien TP. Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol Cell Biol. 2004;24:1096–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ebrahimi-Fakhari D, Saffari A, Wahlster L, et al. . Congenital disorders of autophagy: An emerging novel class of inborn errors of neuro-metabolism. Brain. 2016;139(Pt ):317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Deneubourg C, Ramm M, Smith LJ, et al. . The spectrum of neurodevelopmental, neuromuscular and neurodegenerative disorders due to defective autophagy. Autophagy. 2022;18:496–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Byrne S, Jansen L, U-King-Im J-M, et al. . EPG5-related Vici syndrome: A paradigm of neurodevelopmental disorders with defective autophagy. Brain. 2016;139:765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim M, Sandford E, Gatica D, et al. . Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. eLife. 2016;5:e12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Collier JJ, Guissart C, Olahova M, et al. . Developmental consequences of defective ATG7-mediated autophagy in humans. New Engl J Med. 2021;384:2406–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data collected and analysed for this manuscript are available upon request.